Abstract
The genus Primula is extremely diverse in the east Himalaya-Hengduan Mountains (HHM) in China as a result of rapid radiation. In order to overcome the difficulty of morphological classification of this genus, we surveyed three plastid regions (rbcL, matK, and trnH-psbA) and two nuclear markers (ITS and ITS2) from 227 accessions representing 66 Primula species across 18 sections, to assess their discriminatory power as barcodes. We found that ITS alone or combined with plastid regions showed the best discrimination across different infrageneric ranks and at species level. We suggest rbcL + matK + ITS as the first choice at present to barcode Primula plants. Although the present barcoding combination performed poorly in many closely related species of Primula, it still provided many new insights into current Primula taxonomy, such as the underlying presence of cryptic species, and several potential improper taxonomic treatments. DNA barcoding is one useful technique in the integrative taxonomy of the genus Primula, but it still requires further efforts to improve its effectiveness in some taxonomically challenging groups.
Introduction
There is a critical need for rigorously delineated species for many theoretical studies and practical applications [1]. However, using traditional morphology-based taxonomy is difficult to discover morphologically cryptic taxa [2]. Species that are the product of rapid radiations within single genera can represent suites of morphologically similar taxa that are difficult to distinguish both in the field and the herbarium [3]. DNA barcoding is a valuable addition to the taxonomic tool box. After 10 years development of DNA barcoding, it has been found that large genera with rapid evolutionary radiations still pose a significant challenge for a universal barcoding system [4,5,6]. In order to understand better the overall discriminatory power of the plant barcoding loci, future work should focus on groups that experienced rapid evolutionary radiations, for example, the closely related species within a single genus.
Primula L. is an extraordinarily species-rich group within the east Himalaya-Hengduan Mountains (HHM) in China. The genus consists of about 500 species with over 300 of these found in China, and most of them (approximately 200 species) are restricted to populations in Southwest China, and are mainly confined to the HHM [7]. The HHM and its adjacent regions have been considered to represent the modern diversification centre of the genus [8]. The exceptionally high Primula species and/or lineage diversity in China occurred no more than 10 Mya [9], and may have been triggered by the extensive uplifts of the Qinghai-Tibet Plateau (QTP) since the early Miocene and strengthened by topographical complexity of the QTP and climate oscillations during the Quaternary [10]. Like other large plant groups co-occurring on the QTP (such as Pedicularis, Rhododendron, Gentiana and Saussurea), Primula is a taxonomically challenging group because: 1) many key diagnostic features are tiny and empirical, and cannot be determined correctly by non-specialists, these features include the shape of calyx and bracts [7]; 2) many dwarf species (such as Primula section Minutissimae) are too small in size to separate; and 3) frequent hybridization or introgression can confuse the Primula species boundaries. Primula species, even distantly related ones, can be hybridized readily in greenhouse conditions [11] and in the wild, as reported recently [12–14]. In addition, new Primula species in the HHM and adjacent area have been described a number of times in recent years [15–20]. This suggests that the species diversity of Primula is still underestimated. Although monographs describing Primula do exist [7,11,21,22], the use of keys for the genus requires a high level of specialized expertise. A more efficient approach to facilitate delimitating Primula species and discovering cryptic species or lineages in the genus is urgently required. Despite the promise of DNA barcoding, only a few studies have used it in plant groups that have a high diversity in the HHM or in neighboring regions [23–27].
Although the limited ability of DNA barcoding to discriminate species in large genera is well known, the following questions are still unclear: 1) to what extent could DNA barcoding discriminate infrageneric levels (i.e., subgenus, section, and series) within large genera? 2) Could DNA barcodes discriminate between certain closely related species pairs? 3) In rapidly evolved genera, could DNA barcoding reveal cryptic species? As a typical rapidly evolved plant taxon in the HHM, the genus Primula provides a good opportunity to answer these questions. In the current study, we sampled 66 species representing 18 sections of Primula in China; these contained many closely related groups. The discriminatory ability of three common plastid barcoding candidates (rbcL, matK, and trnH-psbA) and nuclear regions (ITS and ITS2) were evaluated.
Materials and Methods
Ethics statement
All samples employed in this study are not endangered nor protected in the sampled area, and none of the sampled locations are privately owned or protected by any law. No specific permits were required for the described field studies.
Taxon sampling, DNA extraction and sequencing
During this study we examined a total of 227 accessions of 66 Primula species from 18 of the 24 sections of the genus in China recognized by Hu [21]. We used Omphalogramma delavayi Franch. as an outgroup [28,29]. In order to explore the pattern of genetic variability in morphological species, more than two individuals of each species were collected. Taking account of the effect of geographical sampling scale on DNA barcoding [30], more individuals (> 10) were sampled from widespread species, such as P. secundiflora Franch. and P. Poissonii Franch., across their ranges to allow for their intraspecific variability.
To test the effectiveness of DNA barcoding in more closely related groups, section Proliferae was exhaustively sampled in this study. There are approximately 23 species in this section [11]. In China, nineteen species have been described [7,8,21], and a new record species, P. burmanica Balf. f. et Ward, in the section was recently discovered on the south side of Ailao Mountain in Simao (Szemao) region, China (Yan et al., unpublished data). We collected 84 accessions representing all species of the section in China except P. stenodonta Balf. f. ex W. W. Smith et Fletcher. In addition, we selected several of the most closely related species groups in the genus, such as P. chungensis Balf. f. et Ward vs. P. cockburinana Hemsl., P. ovalifolia Franch. vs. P. tardiflora C. M. Hu, P. prattii Hemsl. vs. P. pulchella Franch., P. fasciculata Balf. f. et Ward vs. P. munroi ssp. yagongensis (Petitm) W. W. Smith et Forr., and the P. poissonii complex. Collection details, voucher numbers, taxonomy, and GenBank accession numbers are listed in S1 Table.
Genomic DNA was extracted from silica gel-dried leaf material following a modified version of the cetyltrimethyl ammonium bromide (CTAB) protocol of Doyle & Doyle [31]. Five candidate DNA barcodes, containing two coding plastid genes (rbcL and matK), one intergenic plastid spacer (trnH-psbA), the nuclear ribosomal internal transcribed spacer (ITS, including ITS1, 5.8s and ITS2) and the internal transcribed spacer2 (ITS2), were evaluated in this study. RbcL was amplified using the primer combination (rbcLa_f and 724R) as suggested by Fay et al. [32] and Kress & Erickson [33], respectively. The amplification of matK was achieved using the primer pair 3F-KIM and XF ([34]; Kim unpublished data). For trnH-psbA, the primers trnH05 and psbA3 were used [35,36]. ITS was amplified with the primers proposed by White et al.[37]. PCR amplification and sequencing conditions followed Yan et al. [24]. ITS2 was retrieved from the ITS data in this study.
Data analyses
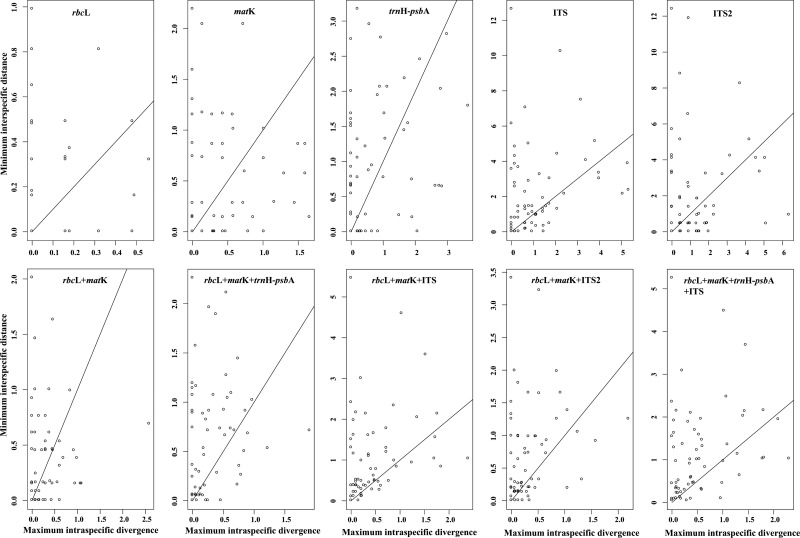
Sequences for each marker were aligned with Muscle 3.8 [38] and then manually adjusted using Se-Al 2.0a11 [39]. We focused on evaluating five single markers and their combinations (rbcL + matK, rbcL + matK + trnH-psbA, rbcL + matK + ITS, rbcL + matK + ITS2, rbcL + matK + trnH-psbA + ITS, and rbcL + matK + trnH-psbA + ITS2). For the pair-wise genetic distance (PWG-distance) method, the genetic pairwise distance was determined by MEGA6 using the Kimura two-parameter distance model (K2P) with pairwise deletion of missing sites [40]. Three parameters (average intraspecific distance, average theta (ө), and coalescent depth) were calculated for all markers. In order to evaluate the ‘local’ barcoding gap for each species [41,42], we plotted the maximum intraspecific divergences against the smallest interspecific distances for each species [41,43].
To test whether accurate species assignments can be made among the samples using a single marker or combinations of markers, we used another two distance-based methods the ‘best match’ (BM) and ‘best close match’ (BCM) using the TaxonDNA/Species Identifier 1.7.7-dev3 program [44]. BM assigns the query to the species with the smallest distance sequence, whereas BCM only identifies the query when the closest sequence is within a distance threshold. The threshold value is determined by using the distance less than 95% of all intraspecific distances, which was calculated by the pairwise summary function [44].
For the tree-building method, we calculated the proportion of monophyletic groups using a Neighbor-Joining (NJ) tree. The test was performed using PAUP* v4b10 with the K2P model [45]. If all individuals of a species cluster together with a bootstrap value above 70%, then the species was considered as having been successfully identified.
Results
Sequence characteristics and genetic divergence
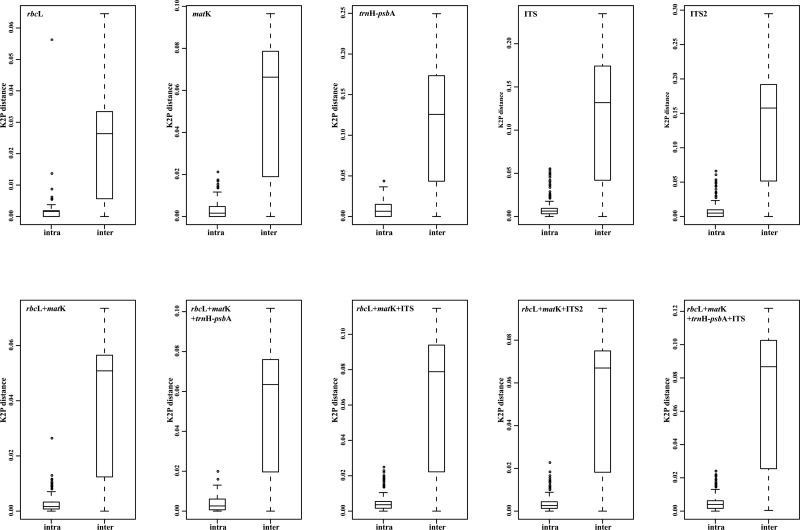
All plastid markers (rbcL, matK, trnH-psbA) were successfully amplified across all individuals, but amplification of ITS failed in two Primula species (P. virginis Lévl. and P. duclouxii Petitm.) and one accession of P. gemmifera Batal. (GXJ253, voucher: Hao940) in this study (S1 Table). The characteristics of the five DNA markers are presented in Table 1. Overall, the aligned length of the five markers ranged from 241 bp (ITS2) to 857 bp (trnH-psbA). The proportion of variable sites were the lowest for rbcL and highest for ITS2. RbcL exhibited the lowest intra-specific and/or inter-specific divergence as well, whilst trnH-psbA showed the highest intra-specific divergence (0.87%), followed by ITS2 (0.80%). However, the greatest interspecific distance was found in ITS2 (12.73%), followed by trnH-psbA (11.69%). The box-and-whisker plots (Fig 1) indicate the distance distribution of inter- and intra-specific distances for all single markers.
Table 1. Summary of genetic variability and sequence characteristics of the candidate barcodes and their main combinations in this study.
| rbcL | matK | trnH-psbA | ITS | ITS2 | R + M | R + M + T | R + M + I | R + M + I2 | R + M + T + I | |
|---|---|---|---|---|---|---|---|---|---|---|
| Aligned length (bp) | 614 | 718 | 857 | 680 | 241 | 1333 | 2191 | 2015 | 1575 | 2872 |
| Average intra-distance | 0.14% | 0.33% | 0.87% | 0.75% | 0.80% | 0.24% | 0.36% | 0.41% | 0.32% | 0.47% |
| Average inter-distance | 2.14% | 5.12% | 11.69% | 11.10% | 12.73% | 3.71% | 5.06% | 6.04% | 4.92% | 6.70% |
| Average theta (ө) | 0.17% | 0.24% | 0.25% | 0.48% | 0.58% | 0.21% | 0.21% | 0.29% | 0.26% | 0.29% |
| Coalescent Depth | 5.57% | 2.04% | 4.15% | 5.30% | 6.36% | 2.58% | 1.91% | 2.39% | 2.19% | 2.31% |
| Proportion of variable sites | 15.79% | 33.43% | 47.37% | 50.88% | 51.19% | 25.36% | 32.63% | 32.90% | 29.21% | 37.05% |
| Proportion of parsimony sites | 12.38% | 27.72% | 32.56% | 43.09% | 48.13% | 20.63% | 25.33% | 27.84% | 24.57% | 29.18% |
| Rate of PCR and sequencing success | 100% | 100% | 100% | 97.80% | 97.80% | N/a | N/a | N/a | N/a | N/a |
R, rbcL; M, matK; T, trnH-psbA; I, ITS; I2, ITS2.
Fig 1. Comparisons of the distribution ranges of inter- and intraspecific distances using boxplots.
The mean intra and inter-specific genetic divergence for the main combinations varied in the ranges 0.24% to 0.47% and 3.71% to 6.70%, respectively (Table 1). The combination of rbcL + matK + trnH-psbA + ITS exhibited the highest mean intra- and inter-specific distance, followed by rbcL + matK + ITS. The core barcode rbcL + matK exhibited the smallest intra-and inter-specific genetic difference (Table 1).
Discrimination success of candidate barcodes
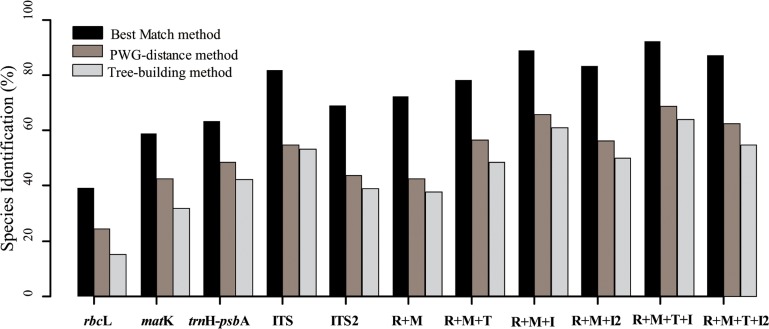
The local barcoding gap, with an interspecific distance larger than the intraspecific distance for a species, directly reveals the species discrimination ability of barcodes. The proportion of the local barcoding gap varied between the regions tested (Figs 2 and 3, S2 Table). ITS showed the best discriminatory power (54.69%) among the five single candidate barcodes, followed by trnH-psbA (48.40%). In contrast, rbcL provided the lowest discrimination rate (24.24%). Of all the combinations tested, the proportion of the barcoding gap of the core barcode combination (rbcL + matK) was the lowest (42.42%) (Fig 3, S2 Table), while rbcL + matK + trnH-psbA + ITS exhibited the highest local barcoding gap (68.75%) followed by rbcL + matK + ITS and rbcL + trnH-psbA + ITS (65.63%). TrnH-psbA and ITS2 individually and/or combined with other plastid markers did not perform well enough to discriminate Primula species in this study (Fig 3, S2 Table). For example, rbcL + matK + trnH-psbA and rbcL + matK + ITS2 could identify 36 Primula species (56.65%), while rbcL + matK + ITS performed better and identified 65.63% of the Primula species.
Fig 2. Relationships between inter- and intraspecific distance indicating the local gaps for species.
Fig 3. Species discrimination rates of several main barcodes in Primula. R, rbcL; M, matK; T, trnH-psbA; I, ITS; I2, ITS2.
Compared with the PWG-distance method, the BM and BCM analyses all showed better discrimination success. BCM always had a lower identification rate than BM analysis (S2 Table). Based on the BM model, ITS performed best among the five single DNA regions, and successfully assigned 81.98% sequences to the correct species (Fig 3). The identification rate of the two-locus combinations ranged from 71.36% to 89.63%. Among them, the core barcode combination rbcL + matK correctly identified 72.24% of specimens, which was only slightly better than rbcL + trnH-psbA (71.36%). For three-locus combinations, matK + trnH-psbA + ITS, rbcL + trnH-psbA + ITS, and rbcL + matK + ITS provided similar discrimination rates (90.99%, 90.54%, and 89.18%), followed by rbcL + matK + trnH-psbA (78.41%). In addition, combinations with ITS2 always produced a lower identification rate compared to combinations with ITS (Fig 3, S2 Table).
The tree-building method provided a similar result to the distance-based method. In this analysis, we found that ITS was the best of all single markers, successfully identifying 53.13% of species. Of the combinations, rbcL + matK showed the poorest discriminatory power (37.88%), while rbcL + matK + trnH-psbA + ITS was the best one with a 64.06% discrimination rate, followed by rbcL + matK + ITS and matK + trnH-psbA + ITS (60.94%) (Fig 3, S2 Table).
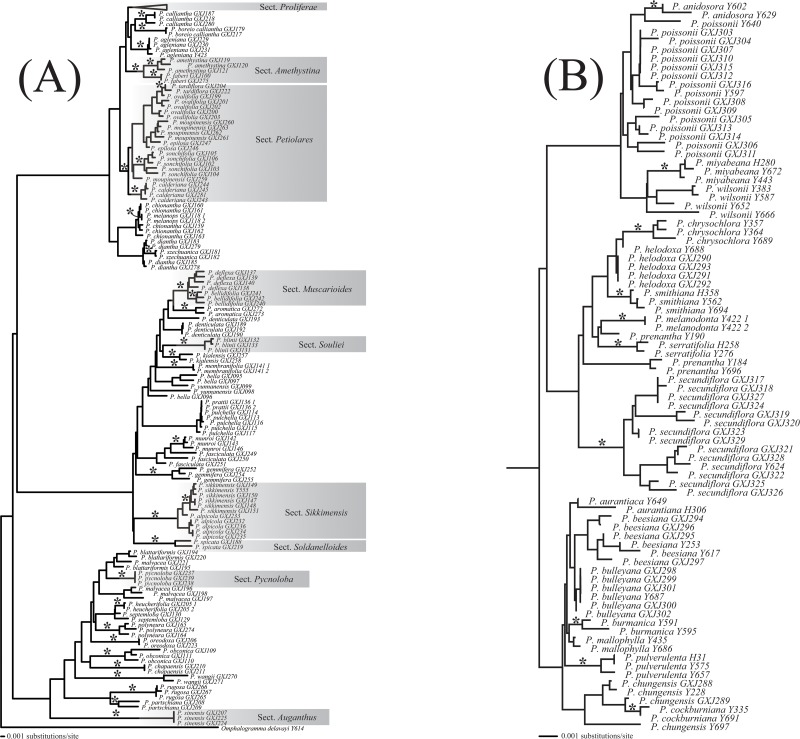
When we considered the previously recognized infrageneric taxa (the twenty four sections, [21]), rbcL and trnH-psbA each only identified four sections (S1 Fig). The discrimination rate of ITS was the best among all single barcodes, distinguishing eight sections (section Pycnoloba, section Auganthus, section Souliei, section Soldanelloides, section Sikkimensis, section Amethystina, section Muscarioides, and section Petiolares) (S1 Fig). Among the main combinations, the core barcode (rbcL + matK) only successfully identified five sections (section Pycnoloba, section Auganthus, section Souliei, section Soldanelloides, and section Sikkimensis), followed by the combinations rbcL + matK + trnH-psbA + ITS2, rbcL + matK + trnH-psbA + ITS and rbcL + matK + trnH-psbA, which all identified the same eight same sections as ITS. In contrast, rbcL + matK + ITS was the best combination, and was able to discriminate nine sections (including section Proliferae) (Fig 4). Our sampling represented four subgenera (subgenus Auriculastrum, subgenus Auganthus, subgenus Carolinella and subgenus Aleuritia) according to the revised classification of Primula [11], nevertheless, the majority of DNA barcodes singly or jointly could only separate out subgenus Auriculastrum correctly.
Fig 4. Neighbor-joining tree based on the combination rbcL + matK + ITS with the K2P distance model. (A) The whole tree of Primula except section Proliferae. (B) The tree of section Proliferae.
Asterisks along branches indicate monophyletic species with bootstrap values above 70%. Accessions are suffixed by sample ID. Monophyletic sections are highlighted with grey shading.
Discrimination ability of DNA barcoding in closely related groups
Section Proliferae is an example containing closely related taxa that is suitable for testing the discriminatory performance of DNA barcoding. Using the tree-building method, the core barcode (rbcL + matK) could only correctly identify P. smithiana Craib with a relatively high bootstrap value (i.e. over 70%), whereas ITS alone could distinguish five species (S1 Fig). rbcL + matK + ITS was the most efficient and precise combination in this study, as stated above, but it only discriminated 10 species correctly (52.63%) in this section (Fig 4). Section Proliferae contained three taxonomically challenging groups (or species complexes). Although the three groups could be easily distinguished by rbcL + matK + ITS, the species within each group were difficult to discriminate using the current barcodes singly and/or in combination. For example, the P. poissonii complex is resolved as monophyletic with high support by rbcL + matK + ITS, however, only two narrowly distributed species (P. anisodora Balf. f. et. Forr. and P. miyabeana Ito et Kawakami) could be readily distinguished (Fig 4).
Compared with section Proliferae, the discrimination performance of DNA barcoding in other Primula species was much better (64.44%, based on the tree-building result of rbcL + matK + ITS). However, we found that a failure often occurred in the most closely related species groups (Table 2), such as P. chungensis vs. P. cockburinana, P. ovalifolia vs. P. tardiflora, P. prattii vs. P. pulchella, and P. fasciculatavs. P. munroi ssp. yagongensis. In addition, some species showed extremely high intraspecific divergence (>1%); these included P. moupingensis Franch., P. bella Franch., P. fasciculata, P. malvacea Franch. and P. yunnanensis Franch. (Table 2). Most of the species with extremely high intraspecific divergence cannot be correctly distinguished by any of the three methods, which probably indicates their status should be further examined.
Table 2. Summary of the candidate barcode rbcL + matK + ITS divergence pattern for unidentified species.
| Taxon | The nearest relative | Mean intraspecific divergence (%) | Maximum intraspecific distance (%) | Minimum interspecific distance (%) |
|---|---|---|---|---|
| P. prattii | P pulchella | 0 | 0 | 0 |
| P. pulchella | P. prattii | 0.1 | 0.15 | 0 |
| P. burmanica | P. mallophylla | 0.19 | 0.21 | 0.16 |
| P. chungensis | P. cockburniana | 0.17 | 0.21 | 0.11 |
| P. bulleyana | P. aurantiaca | 0.32 | 0.21 | 0.11 |
| P. aurantiaca | P. bulleyana | 0.13 | 0.22 | 0.11 |
| P. chrysochlora | P. helodoxa | 0.13 | 0.26 | 0.26 |
| P. poissonii | P. anidosora | 0.12 | 0.38 | 0.38 |
| P. chionantha | P. melanops | 0.18 | 0.41 | 0.1 |
| P. beesiana | P. bulleyana | 0.02 | 0.43 | 0.21 |
| P. wilsonii | P. miyabeana | 0.25 | 0.48 | 0.37 |
| P. septemloba | P. heucherifolia | 0.32 | 0.56 | 0.46 |
| P. prenantha | P. helodoxa | 0.21 | 0.58 | 0.26 |
| P. ovalifolia | P. tardiflora | 0.42 | 0.61 | 0.36 |
| P. alpicola | P. sikkimensis | 0.26 | 0.77 | 0.48 |
| P. blattariformis | P. malvacea | 0.6 | 0.93 | 0.83 |
| P. moupinensis | P. epilosa | 0.54 | 1.24 | 0.61 |
| P. denticulata | P. kialensis | 0.48 | 1.6 | 1.3 |
| P. bella | P. yunnanensis | 0.9 | 1.7 | 1.03 |
| P. fasciculata | P. munroi | 0.99 | 1.72 | 1.55 |
| P. malvacea | P. blattariformis | 0.85 | 1.84 | 0.83 |
| P. yunnanensis | P. bella | 1.82 | 2.39 | 1.03 |
Discussion
The resolution of the tested DNA markers in Primula
In this study, all the three plastid regions tested individually showed a relatively low discriminatory efficacy ranging from 15.16% to 31.82% (based on monophyletic analysis) in Primula species (S2 Table). The core barcode rbcL + matK also provided low discrimination at a rate of 37.88% (S2 Table). One of the most promising supplementary plastid barcodes, trnH-psbA, varied in size from 154 bp (P. poissonii) to 523 bp (P. polynuera Franch.), so there were a large number of gaps in the alignment matrix. Based on tree-building analysis, trnH-psbA identified 42.19% of species; this was the best among the plastid regions but lower than the nuclear markers (ITS) (S2 Table). The combination of trnH-psbA with rbcL or matK did not result in higher resolution (S2 Table), which demonstrated that trnH-psbA is not a preferred barcode in Primula.
The strong identification ability of ITS has been verified based on a comprehensive study [46], even in some complex plant groups, such as Panassia [25], Ficus [47], Lysimachia [27], and Sisyrinchium [48]. In this study, ITS exhibited the highest discriminatory power among all five markers, and any combinations with ITS were able to discriminate more species than combinations without ITS (Fig 3, S2 Table). Of the three-locus combinations, rbcL + matK + ITS and matK + trnH-psbA + ITS all distinguished 60.94% of monophyletic species, which was the best discrimination performance (Appendix S2). Therefore, as suggested by Yan et al. [24], rbcL + matK + ITS should be the first choice to barcode Primula plants. Compared with primer problems associated with ITS, ITS2 has conserved regions for designing universal primers, and can be readily amplified in various groups [49]. However, ITS2 itself or combined with plastid markers did not produce better results than ITS and/or corresponding combinations (S2 Table). We suggest that ITS2 may be an ideal supplementary barcode when ITS amplification fails.
Discrimination performance on section rank in Primula
DNA barcoding should be able to help identify some groups within large genera, thus reducing the time required for morphological studies to produce definitive species lists. Although it is well known that DNA barcoding has difficulties in resolving closely related species, it is not clear whether such barcoding could identify samples correctly to section level within large genera. There are more than 200 Primula species concentrated in the HHM region in China [11]. Primula has always been divided into subgroups, usually with the rank of section [22,50]. In a well-accepted infrageneric system, Smith and Fletcher divided the genus into a total of 31 sections [22]. Twenty-four sections of the Chinese Primula were adopted by Hu [21].
In this study, DNA barcoding performed well for distinguishing sections, and could resolve nine of the current 18 sections [21]. However, of the resolved sections, three (namely section Auganthus, section Souliei, and section Soldanelloides) together with the monotypic section Pycnoloba were each represented by one species in the current study. Considering the fact that the phylogeny of many sections and their close relatives, such as section Soldanelloides, Minutissimae, and Souliei, still lack detailed studies [28,29], the discrimination rate of DNA barcoding would probably drop further if we expanded the number of members in these sections. These results demonstrated that DNA barcoding is useful in some sections of Primula. In addition to barcoding discriminatory ability, the infragenetric classification system will also influence the results. A reliable and well-recognized infrageneric rank in a large genus is a prerequisite for applying DNA barcoding. Therefore, a number of new sectional delimitations will be necessary in the genus Primula [11,28].
Resolving ability of DNA barcoding in section Proliferae
The Primula section Proliferae is a well-delimited and natural group characterized by numerous whorls of flowers resembling candelabra [11]. It is mainly concentrated in the HHM [7]. In China, 19 species have been described and, with the exception of P. miyabeana (endemic to Taiwan), they are narrowly distributed in southwest China [7]. This section contains several taxonomically challenging groups, such as the P. poissonii complex, which consists of P. anisodora, P. wilsonii Dunn and P. poissonii, and the P. beesiana group with P. beesiana Forr., P. bulleyana Forr., P. burmanica and P. pulverulenta Duthie. This complex section provides a good example to test the discriminatory ability of candidate barcodes in closely related species, especially those formed through rapid evolutionary radiation.
Although the discriminatory power of DNA barcoding is limited in section Proliferae (discrimination rate of 52.63%), in the current study it confirmed the monophyly of section Proliferae (tree-building method) (Fig 4), and divided the section into three clades with high support (over 85%), which agree well with the study based on their morphology [51]. It is convenient for us to assign unknown Primula specimens to a rough position in the section. This could help to narrow the scope of identification. Within each complex or clade, DNA barcoding could still provide some clues for identification and taxonomic treatment. For example, P. poissonii and P. anisodora have the closest relationship and they were confirmed by the current barcodes (Fig 4), but only P. anisodora exhibited monophyly. DNA barcoding could also help to solve several classification disputes in this section. For example, barcoding supported treating P. wilsonii and P. anisodora, P. burmarica and P. beesiana as separate species [7,11,21,51] (Fig 4). Therefore, even for very closely related species, DNA barcoding may still provide help to some extent, and narrow the identification range.
It is well known that using the universal DNA barcode (two core barcodes and two alternative barcodes, trn H -psbA, ITS) it is almost impossible to separate very closely related species formed through rapid radiation. Therefore, species-specific barcodes need to be developed for difficult taxa [6]. These markers may be based on other rapidly evolved molecular markers such as low or single copy nuclear genes (e.g. waxy and leafy) [52] or even using high-throughput sequencing methods (such as RAD and GBS).
Biological implications of DNA barcoding in Primula
Traditional taxonomy mainly depends on morphological diagnosis, and it should be corroborated by other sources of data, such as geographical, ecological, reproductive and DNA sequence information [53]. However, constructing a robust taxonomy for recently diverged plant taxa is more difficult, because they often show little difference in their morphological and genetic profiles. In addition, many other aspects could also cause the failure of DNA barcoding, such as imperfect taxonomy, interspecific hybridization, paralogy, and incomplete lineage sorting [42,52,54,55]. For many such taxa, DNA barcoding provides an opportunity to solve some of the taxonomic problems through discovering the underlying biological issues.
By surveying the non-monophyletic taxa at species level and examining genetic distance (Fig 4, Table 2), we filtered out barcoding failures in several species probably caused by incomplete lineage sorting. For example, narrowly distributed P. tardiflora, P. prattii, and P. cockburiana each experienced peripheral isolated speciation from their widely distributed relatives (putative parents) (P. ovalifolia, P. pulchella, and P. chungensis) [55]. The barcoding results were partially supported by a complementary phylogeographic study [56]. It is a question for taxonomy to reflect on these incomplete speciation processes by synonymizing the nested and parent species or elevating lineages in the paraphyletic lineage to species status [55]. In the context, we prefer to treat the nested and parent species as one species because of their similarity in morphology [7,21], but of course additional research is necessary.
Imperfect taxonomy in several plant and animal taxa has been detected by DNA barcoding (e.g. [27,42,57–59]), providing significant support for the taxonomic value of the technique. P. bella examined in this study is an excellent example of over-lumping in traditional taxonomy, as the species appeared polyphyletic and exhibited unexpectedly large intraspecific divergence (Table 2, Fig 4). Given the variable morphological characters (such as shape of bracts and the stem length), there are classification disputes about the delimitation of P. bella [7,11,21,60]. DNA barcoding supported the suggestion that the anomalous individual P. bella GXJ096 (voucher: Hao & Yan 956) should be raised to species status (P. cyclostegia Hand.-Mazz.) on the basis of its genetic profile, although additional work is essential to validate this as a robust species. A similar situation is also probably the case for P. denticulata Smith.
Discovering the potential presence of cryptic species and/or lineages is an important application of DNA barcoding, and this remains within the domain of taxonomy [53]. The taxonomic usefulness of DNA barcoding has been validated in a wide range of animals (see, for example [61–67]), but there are few studies of large plant groups that have recently experienced evolutionary radiation. It is plausible that the frequent occurrence of cryptic species in Chinese Primula represents adaptation to the variable habitats on the HHM and rapid radiation evolution in a relatively short time [7,9,11,21]. By iteratively reexamining peculiar specimens detected by DNA barcoding (such as P. yunnanensis GXJ099, P. fasciculata GXJ249, and P. moupinensis GXJ259) (Table 2, Fig 4), several tiny morphological or geographical divergences may be identified in these taxa, which indicate the possibility of cryptic species; however, further taxonomic scrutiny is required.
Another great challenge for barcoding plant species is linked to hybridization events [23,52,54,68]. Natural or artificial hybrids in Primula have been reported recently [11–14], and these may cause a failure in barcoding Primula species. In the current study, underlying hybridization might occur in P. anisodora and its most close relative P. poissonii. They were found in the same populations, and a putative hybrid (P. poissonii Y640) was also discovered (S2 Fig). Additional research is needed to resolve the biological situation (e.g. [69,70]).
Conclusion
Primula species examined in the present study are difficult to distinguish using the core barcode (rbcL + matK). Another plastid marker, trnH-psbA, varied in size and exhibited lower discrimination compared to ITS, suggesting that it is not a suitable barcode for studies of Primula. In contrast, ITS showed the best discriminatory ability of all the single markers tested, discriminating 65.63% and 60.94% of species (according to the PWG-distance method and tree-building method) when combined with rbcL + matK, which performed best among all three-locus combinations. We propose that rbcL + matK + ITS should be treated as the first local barcode in the genus Primula at present, although its discrimination rates with respect to infrageneric rank and separating closely related Primula species are limited.
Despite the limited discrimination for closely related pairs, DNA barcoding provided many new insights into the current Primula taxonomy, such as detecting potential cryptic species, and revealing several probably improper taxonomic treatments. Obviously, it is difficult to resolve all closely related groups based on the current limited and relatively conserved molecular markers, especially in taxa such as Primula, which have experienced recent rapid radiation. Other more rapidly evolved molecular markers should be incorporated into future DNA barcoding projects, for example low or single copy nuclear genes, nuclear SNPs, nuclear SSRs [23,52], and the complete chloroplast genome [71–74]. As proposed by Twyford [6], we are building a robust phylogeny framework for the Primula section Proliferae using RAD (restriction-site-associated DNA, [75]), and expect to resolve the true evolutionary relationships; these may be necessary to develop robust species-specific barcodes in the future [6]. Overall, DNA barcoding is a useful technique for the integrative taxonomy of the genus, but it still requires further work to improve its value for studying taxonomically challenging groups.
Supporting Information
Asterisks along branches indicate monophyletic species with bootstrap values above 70%. Accessions are suffixed by sample ID. Monophyletic sections are highlighted with grey shading.
(PDF)
(DOCX)
(XLSX)
(DOCX)
Acknowledgments
We are grateful to Drs. Lian-Ming Gao, Zhi-Kun Wu, Xun Gong, Yu-Ming Shui, Zhong-Lai Luo, Yuan Xu, Bing-Qiang Xu, and Xin Wu for their help in collecting plant material. We also thank Dr. Juan Liu and Ms. Yu-Ying Zhou for help in data analysis and laboratory work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was financially supported by the National Natural Science Foundation of China (grant nos. 31270009, 31170205) [http://www.nsfc.gov.cn/].
References
- 1. Dayrat B (2005) Towards integrative taxonomy. Biological Journal of the Linnean Society 85: 407–415. [Google Scholar]
- 2. Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elliott TL, Davies TJ (2014) Challenges to barcoding an entire flora. Molecular Ecology Resources 14: 883–891. 10.1111/1755-0998.12277 [DOI] [PubMed] [Google Scholar]
- 4. Kekkonen M, Hebert PDN (2014) DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Molecular Ecology Resources 14: 706–715. 10.1111/1755-0998.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller SE (2007) DNA barcoding and the renaissance of taxonomy. Proceedings of the National Academy of Sciences of the United States of America 104: 4775–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Twyford AD (2014) Testing evolutionary hypotheses for DNA barcoding failure in willows. Molecular Ecology 23: 4674–4676. 10.1111/mec.12892 [DOI] [PubMed] [Google Scholar]
- 7. Hu CM, Kelso S (1996) Primulaceae; Wu ZY, Raven PH, editors. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. [Google Scholar]
- 8. Hu CM (1994) On the geographical distribution of the Primulaceae. Journal of Tropical and Subtropical Botany 2: 1–14. [Google Scholar]
- 9. de Vos JM, Hughes CE, Schneeweiss GM, Moore BR, Conti E (2014) Heterostyly accelerates diversification via reduced extinction in primroses. Proceedings of the Royal Society B: Biological Sciences 281: 20140075 10.1098/rspb.2014.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H (2014) Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Frontiers in Genetics 5: 4 10.3389/fgene.2014.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richards J (2003) Primula. Portland, OR, USA: Timber Press. [Google Scholar]
- 12. Cianchi R, Arduino P, Mosco MC, Bullini L (2013) Evidence of hybrid speciation in the North American primroses Primula suffrutescens, P. parryi, P. rusbyi and P. angustifolia (Primulaceae). Plant Biosystems-An International Journal Dealing with All Aspects of Plant Biology: 10.1080/11263504.2013.826745 [DOI] [Google Scholar]
- 13. Ma Y, Xie W, Tian X, Sun W, Wu Z, Milne R (2014) Unidirectional hybridization and reproductive barriers between two heterostylous primrose species in north-west Yunnan, China. Annals of Botany: 113: 763–775. 10.1093/aob/mct312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu XF, Li Y, Wu GL, Fang ZD, Li QJ, Liu JQ (2009) Molecular and morphological evidence for natural hybridization between Primula secundiflora Franchet and P. poissonii Franchet (Primulaceae). Acta Biologica Cracoviensia Series Botanica 51: 29–36. [Google Scholar]
- 15. Gong X, Fang RC (2003) Primula calyptrata, a new species in section Carolinella (Primulaceae) from Yunnan, China. Novon 13: 193–195. 12727512 [Google Scholar]
- 16. Hu CM, Hao G (2011) New and noteworthy species of Primula (Primulaceae) from China. Edinburgh Journal of Botany 68: 297–300. [Google Scholar]
- 17. Hu CM (1990) A new species of Primula from Thailand with critical notes on the section Carolinella. Nordic Journal of Botany 10: 399–401. [Google Scholar]
- 18. Hu CM, Geng YY (2003) Two new species of Primula (Primulaceae) from China. Novon: 196–199. [Google Scholar]
- 19. Li R, Hu CM (2009) Primula lihengiana (Primulaceae), a new species from Yunnan, China. Annales Botanici Fennici 46: 130–132. [Google Scholar]
- 20. Wu X, Xu Y, Hu CM, Hao G (2013) Primula mianyangensis (Primulaceae), a new species from Sichuan, China. Phytotaxa 131: 49–52. [Google Scholar]
- 21. Hu CM (1990) Primulaceae; Chen FW, Hu CM, editors. Beijing: Science Press. [Google Scholar]
- 22.Smith WW, Fletcher HR (1977) The genus Primula: A facsimile reprint of 22 papers published in various journals, reprinted with original pagination as well as new continuous pagination. Plant Monograph Reprints II.: Copies: BR, FAS, G, M, NY.
- 23. Feng JJ, Jiang DC, Shang HY, Dong M, Wang GN, He XY, et al. (2013) Barcoding Poplars (Populus L.) from Western China. PLoS ONE 8 (8): e71710 10.1371/journal.pone.0071710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan HF, Hao G, Hu CM, Ge XJ (2011) DNA barcoding in closely related species: A case study of Primula L. sect. Proliferae Pax (Primulaceae) in China. Journal of Systematics and Evolution 49: 225–236. [Google Scholar]
- 25. Yang JB, Wang YP, Moller M, Gao LM, Wu D (2012) Applying plant DNA barcodes to identify species of Parnassia (Parnassiaceae). Molecular Ecology Resources 12: 267–275. 10.1111/j.1755-0998.2011.03095.x [DOI] [PubMed] [Google Scholar]
- 26. Yu WB, Huang PH, Ree RH, Liu ML, Li DZ, Wang H (2011) DNA barcoding of Pedicularis L.(Orobanchaceae): Evaluating four universal barcode loci in a large and hemiparasitic genus. Journal of Systematics and Evolution 49: 425–437. [Google Scholar]
- 27. Zhang CY, Wang FY, Yan HF, Hao G, Hu CM, Ge XJ (2012) Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Molecular Ecology Resources 12: 98–108. 10.1111/j.1755-0998.2011.03076.x [DOI] [PubMed] [Google Scholar]
- 28. Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DMS, Conti E (2001) Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. International Journal of Plant Sciences 162: 1381–1400. [Google Scholar]
- 29. Yan HF, He CH, Peng CI, Hu CM, Hao G (2010) Circumscription of Primula subgenus Auganthus (Primulaceae) based on chloroplast DNA sequences. Journal of Systematics and Evolution 48: 123–132. [Google Scholar]
- 30. Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, et al. (2012) The effect of geographical scale of sampling on DNA barcoding. Systematic Biology 61: 851–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- 32. Fay MF, Swensen SM, Chase MW (1997) Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae). Kew Bulletin: 111–120. [Google Scholar]
- 33. Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2 (6): e508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ford CS, Ayres KL, Toomey N, Haider N, Stahl JV, Kelly LJ, et al. (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Botanical Journal of the Linnean Society 159: 1–11. [Google Scholar]
- 35. Sang T, Crawford DJ, Stuessy TF (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120–1136. [PubMed] [Google Scholar]
- 36. Tate JA, Simpson BB (2003) Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Systematic Botany 28: 723–737. [Google Scholar]
- 37. White TJ, Bruns T, Lee S, Taylor J (1990) Amplication and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis M, Gelfand D, Sninsky J, White TJ, editors. PCR protocols: A guide to methods and applications. San Diego: Academic Press; pp. 315–322. [Google Scholar]
- 38. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambaut A (2002) Se-al version2.0a11. http://treebioedacuk/software/seal/.
- 40. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Collins RA, Cruickshank RH (2013) The seven deadly sins of DNA barcoding. Molecular Ecology Resources 13: 969–975. 10.1111/1755-0998.12046 [DOI] [PubMed] [Google Scholar]
- 42. Meyer CP, Paulay G (2005) DNA barcoding: Error rates based on comprehensive sampling. PLoS Biology 3: 2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robinson EA, Blagoev GA, Hebert PDN, Adamowicz SJ (2009) Prospects for using DNA barcoding to identify spiders in species-rich genera. Zookeys: 27–46. [Google Scholar]
- 44. Meier R, Shiyang K, Vaidya G, Ng PKL (2006) DNA barcoding and taxonomy in diptera: A tale of high intraspecific variability and low identification success. Systematic Biology 55: 715–728. [DOI] [PubMed] [Google Scholar]
- 45. Swofford DL (2003) PAUP*: Phylogenetic analysis using parsimony (* and other methods), version 4.0b10. Sunderland: Sinauer. [Google Scholar]
- 46. Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences of the United States of America 108: 19641–19646. 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li HQ, Chen JY, Wang S, Xiong SZ (2012) Evaluation of six candidate DNA barcoding loci in Ficus (Moraceae) of China. Molecular Ecology Resources 12: 783–790. 10.1111/j.1755-0998.2012.03147.x [DOI] [PubMed] [Google Scholar]
- 48. Alves TLS, Chauveau O, Eggers L, de Souza-Chies TT (2014) Species discrimination in Sisyrinchium (Iridaceae): assessment of DNA barcodes in a taxonomically challenging genus. Molecular Ecology Resources 14: 324–335. 10.1111/1755-0998.12182 [DOI] [PubMed] [Google Scholar]
- 49. Yao H, Song JY, Liu C, Luo K, Han JP, Li Y, et al. (2010) Use of ITS2 region as the universal dna barcode for plants and animals. PLoS ONE 5(10): e13102 10.1371/journal.pone.0013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trift I, Kallersjo M, Anderberg AA (2002) The monophyly of Primula (Primulaceae) evaluated by analysis of sequences from the chloroplast gene rbcL. Systematic Botany 27: 396–407. [Google Scholar]
- 51. Smith WW, Fletcher HR (1941) The Genus Primula: Section Candelabra, Balf. Fil; 1941. Taylor & Francis. pp. 122–181. [Google Scholar]
- 52. Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6(5): e19254 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DeSalle R, Egan MG, Siddall M (2005) The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philosophical Transactions of the Royal Society B: Biological Sciences 360: 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, et al. (2009) Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Molecular Ecology Resources 9: 130–139. 10.1111/j.1755-0998.2009.02652.x [DOI] [PubMed] [Google Scholar]
- 55. Funk DJ, Omland KE (2003) Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology Evolution and Systematics 34: 397–423. [Google Scholar]
- 56. Xie XF, Yan HF, Wang FY, Ge XJ, Hu CM, Hao G (2012) Chloroplast DNA phylogeography of Primula ovalifolia in central and adjacent southwestern China: Past gradual expansion and geographical isolation. Journal of Systematics and Evolution 50: 284–294. [Google Scholar]
- 57. Edwards D, Horn A, Taylor D, Savolain V, Hawkins JA (2008) DNA barcoding of a large genus, Aspalathus L. (Fabaceae). Taxon 57: 1317–1327. [Google Scholar]
- 58. Huang JH, Zhang AB, Mao SL, Huang Y (2013) DNA Barcoding and species boundary delimitation of selected species of Chinese Acridoidea (Orthoptera: Caelifera). PLoS ONE 8(12): e82400 10.1371/journal.pone.0082400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pettengill JB, Neel MC (2010) An evaluation of candidate plant dna barcodes and assignment methods in diagnosing 29 species in the Genus Agalinis (Orobanchaceae). American Journal of Botany 97: 1391–1406. 10.3732/ajb.0900176 [DOI] [PubMed] [Google Scholar]
- 60. Smith WW, Fletcher HR (1942) The Genus Primula: Section Minutissimae; 1942. Taylor & Francis. pp. 227–266. [Google Scholar]
- 61. Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN (2008) DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences of the United States of America 105: 6350–6355. 10.1073/pnas.0712181105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clare EL, Lim BK, Engstrom MD, Eger JL, Hebert PDN (2007) DNA barcoding of Neotropical bats: species identification and discovery within Guyana. Molecular Ecology Notes 7: 184–190. [Google Scholar]
- 63. Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . Proceedings of the National Academy of Sciences of the United States of America 101: 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Janzen DH, Hallwachs W, Burns JM, Hajibabaei M, Bertrand C, Hebert PDN (2011) Reading the complex skipper butterfly fauna of one tropical place. PLoS ONE 6(8): e19874 10.1371/journal.pone.0019874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Janzen DH, Hallwachs W, Harvey DJ, Darrow K, Rougerie R, Hajibabaei M, et al. (2012) What happens to the traditional taxonomy when a well-known tropical saturniid moth fauna is DNA barcoded? Invertebrate Systematics 26: 478–505. [Google Scholar]
- 66. Puckridge M, Andreakis N, Appleyard SA, Ward RD (2013) Cryptic diversity in flathead fishes (Scorpaeniformes: Platycephalidae) across the Indo-West Pacific uncovered by DNA barcoding. Molecular Ecology Resources 13: 32–42. 10.1111/1755-0998.12022 [DOI] [PubMed] [Google Scholar]
- 67. Saitoh T, Sugita N, Someya S, Iwami Y, Kobayashi S, Kamigaichi H, et al. (2014) DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago. Molecular Ecology Resources 15: 177–186.68. 10.1111/1755-0998.12282 [DOI] [PubMed] [Google Scholar]
- 68. Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, Burgess KS, et al. (2014) Understanding the spectacular failure of DNA barcoding in willows (Salix): Does this result from a trans-specific selective sweep? Molecular Ecology 23: 4737–4756. 10.1111/mec.12837 [DOI] [PubMed] [Google Scholar]
- 69. Liu BB, Abbott RJ, Lu ZQ, Tian B, Liu JQ (2014) Diploid hybrid origin of Ostryopsis intermedia (Betulaceae) in the Qinghai-Tibet Plateau triggered by Quaternary climate change. Molecular Ecology 23: 3013–3027. 10.1111/mec.12783 [DOI] [PubMed] [Google Scholar]
- 70. Sun YS, Abbott RJ, Li LL, Li L, Zou JB, Liu JQ (2014) Evolutionary history of Purple cone spruce (Picea purpurea) in the Qinghai-Tibet Plateau: homoploid hybrid origin and Pleistocene expansion. Molecular Ecology 23: 343–359. [DOI] [PubMed] [Google Scholar]
- 71. Kane NC, Cronk Q (2008) Botany without borders: barcoding in focus. Molecular Ecology 17: 5175–5176. 10.1111/j.1365-294X.2008.03972.x [DOI] [PubMed] [Google Scholar]
- 72. Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL (2014) Plant DNA barcoding: from gene to genome. Biological Reviews 90: 157–166. 10.1111/brv.12104 [DOI] [PubMed] [Google Scholar]
- 73. Parks M, Cronn R, Liston A (2009) Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7: 84 10.1186/1741-7007-7-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang JB, Tang M, Li HT, Zhang ZR, Li DZ (2013) Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evolutionary Biology 13: 84 10.1186/1471-2148-13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Research 17: 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Asterisks along branches indicate monophyletic species with bootstrap values above 70%. Accessions are suffixed by sample ID. Monophyletic sections are highlighted with grey shading.
(PDF)
(DOCX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.