The function of Sac2/INPP5F in the endocytic pathway and its activity as a 4-phosphatase suggest that Sac2/INPP5F and OCRL may cooperate in the sequential dephosphorylation of PI(4,5)P2 in a partnership that mimics that of the two phosphatase modules of synaptojanin.
Abstract
The recruitment of inositol phosphatases to endocytic membranes mediates dephosphorylation of PI(4,5)P2, a phosphoinositide concentrated in the plasma membrane, and prevents its accumulation on endosomes. The importance of the conversion of PI(4,5)P2 to PtdIns during endocytosis is demonstrated by the presence of both a 5-phosphatase and a 4-phosphatase (Sac domain) module in the synaptojanins, endocytic PI(4,5)P2 phosphatases conserved from yeast to humans and the only PI(4,5)P2 phosphatases in yeast. OCRL, another 5-phosphatase that couples endocytosis to PI(4,5)P2 dephosphorylation, lacks a Sac domain. Here we show that Sac2/INPP5F is a PI4P phosphatase that colocalizes with OCRL on endocytic membranes, including vesicles formed by clathrin-mediated endocytosis, macropinosomes, and Rab5 endosomes. An OCRL–Sac2/INPP5F interaction could be demonstrated by coimmunoprecipitation and was potentiated by Rab5, whose activity is required to recruit Sac2/INPP5F to endosomes. Sac2/INPP5F and OCRL may cooperate in the sequential dephosphorylation of PI(4,5)P2 at the 5 and 4 position of inositol in a partnership that mimics that of the two phosphatase modules of synaptojanin.
Introduction
Phosphoinositides (PIs), the seven metabolites resulting from the phosphorylation of phosphatidylinositol at the 3, 4, and 5 position of the inositol ring, are key regulatory phospholipids localized on the cytosolic leaflets of cellular membranes. Via interactions with proteins mediated by their differentially phosphorylated headgroups, they control various aspects of cell physiology including, but not limited to, signaling, membrane traffic and interactions, cytoskeleton dynamics, and lipid homeostasis (De Matteis and Godi, 2004; Di Paolo and De Camilli, 2006; Balla, 2013). Not surprisingly, in view of these pleiotropic effects, connections between malfunctions of PI metabolism and human disease are becoming progressively evident (McCrea et al., 2008; Ooms et al., 2009; Staiano et al., 2014).
The seven PIs are differentially localized on cellular membranes and help define their distinct properties (De Matteis and Godi 2004; Idevall-Hagren and De Camilli, 2014). This implies the occurrence of mechanisms that coordinate membrane transport from one compartment to another with a change in the PI signature. One of the best characterized examples of this conversion is the metabolism of PIs that occurs in the endocytic pathway. Although PI(4,5)P2 is primarily concentrated in the plasma membrane, PI3P is the predominant PI in early endosomes (Simonsen et al., 2001; Di Paolo and De Camilli, 2006). Thus, endocytosis, which starts with the PI(4,5)P2-dependent recruitment of endocytic factors to the plasma membrane, is closely coupled to PI(4,5)P2 dephosphorylation (Cremona et al., 1999; Stefan et al., 2002; Milosevic et al., 2011; Nández et al., 2014; Posor et al., 2014), whereas arrival of the endocytic membrane is signaled by the acquisition of PI3P (Patki et al., 1997; Simonsen et al., 2001; Zoncu et al., 2009).
The first enzyme to be implicated in the coupling between endocytosis and PI(4,5)P2 dephosphorylation was synaptojanin 1, a PI phosphatase that is expressed at very high levels in neurons, where it participates in the endocytosis of synaptic vesicles (McPherson et al., 1996; Cremona et al., 1999). Synaptojanin 1 and its close homologue synaptojanin 2 (Nemoto et al., 1997) dephosphorylate PI(4,5)P2 via two phosphatase modules arranged in tandem: an N-terminal Sac1 domain (henceforth referred to as Sac domain) and a central inositol 5-phosphatase domain (McPherson et al., 1996; Pirruccello and De Camilli, 2012). The Sac domain of synaptojanin primarily has inositol 4-phosphatase activity (Guo et al., 1999; Nemoto et al., 2000) and derives its name from the Sac1 protein, an ER-localized PI4P phosphatase that plays a critical role in the regulation of PI4P homeostasis (Guo et al., 1999; Foti et al., 2001; Hsu and Mao, 2013). As the Sac domain of synaptojanin acts on PI4P and not on PI(4,5)P2, it may function in coordination with the 5-phosphatase module to convert PI(4,5)P2 to PtdIns.
Another 5-phosphatase implicated in PI(4,5)P2 dephosphorylation during endocytosis is OCRL, an enzyme whose loss of function results in Lowe syndrome and Dent’s disease (Attree et al., 1992; Hoopes et al., 2005; Pirruccello and De Camilli, 2012; Mehta et al., 2014). OCRL, which is broadly distributed on organelles of the endocytic pathway, is thought to dephosphorylate PI(4,5)P2 upon endocytosis and then to prevent its ectopic accumulation on downstream stations of the endocytic pathway (Hyvola et al., 2006; Erdmann et al., 2007; Mao et al., 2009; Vicinanza et al., 2011; Mehta et al., 2014; Nández et al., 2014). While synaptojanin 1 has a major role at synapses (Cremona et al., 1999; Harris et al., 2000; Verstreken et al., 2003), OCRL appears to be the dominant endocytic 5-phosphatase in nonneuronal cells (Zhang et al., 1998; Nández et al., 2014). A close homologue of OCRL, INPP5B, is also expressed by mammalian genomes and cooperates with OCRL in PI(4,5)P2/PI(3,4,5)P3 dephosphorylation on endocytic membranes (Shin et al., 2005; Pirruccello and De Camilli, 2012). Neither OCRL nor INPP5B, however, contain a Sac domain. Thus, it remains possible that they may act in cooperation with a separate Sac domain–containing protein with 4-phosphatase activity.
Mammalian genomes encode five Sac domain–containing proteins: the ER protein Sac1, the two synaptojanins, Sac2 (also referred to as INPP5F), and Fig4 (also referred to as Sac3; Hughes et al., 2000; Hsu and Mao, 2013). However, Fig4 was shown to be part of a PI(3,5)P2 metabolizing complex on late endosomes (Gary et al., 2002), and Sac2/INPP5F, whose subcellular localization has not been characterized, was reported to function as a 5-phosphatase for PI(4,5)P2 and to a lower extent for PI(3,4,5)P3 (Minagawa et al., 2001).
The goal of this study was to gain new insight into the properties of Sac2/INPP5F, whose gene was recently identified as a risk locus in Parkinson’s disease (Nalls et al., 2014). Our results implicate this enzyme in the endocytic pathway at sites that closely overlap with sites of action of OCRL. They further demonstrate that its Sac domain functions predominantly as a 4-phosphatase, and not as a 5-phosphatase, which is consistent with its very strong similarity to the Sac domain of the Sac1 protein and to the catalytically active Sac domains of the synaptojanins. We suggest that Sac2/INPP5F may function in close cooperation with other inositol phosphatases including OCRL and INPP5B and that the partnership of Sac2/INPP5F with the OCRL or INPP5B pair may represent a functional homologue of synaptojanin. Note that both the NCBI (www.ncbi.nlm.nih.gov) and Uniprot database (www.uniprot.org) currently also assign the name INPP5F to OCRL. To avoid such confusion, and in view of our demonstration that Sac2/INPP5F is a 4-phosphatase, we will use henceforth only the name Sac2.
Results
Localization of Sac2 on early endosomes
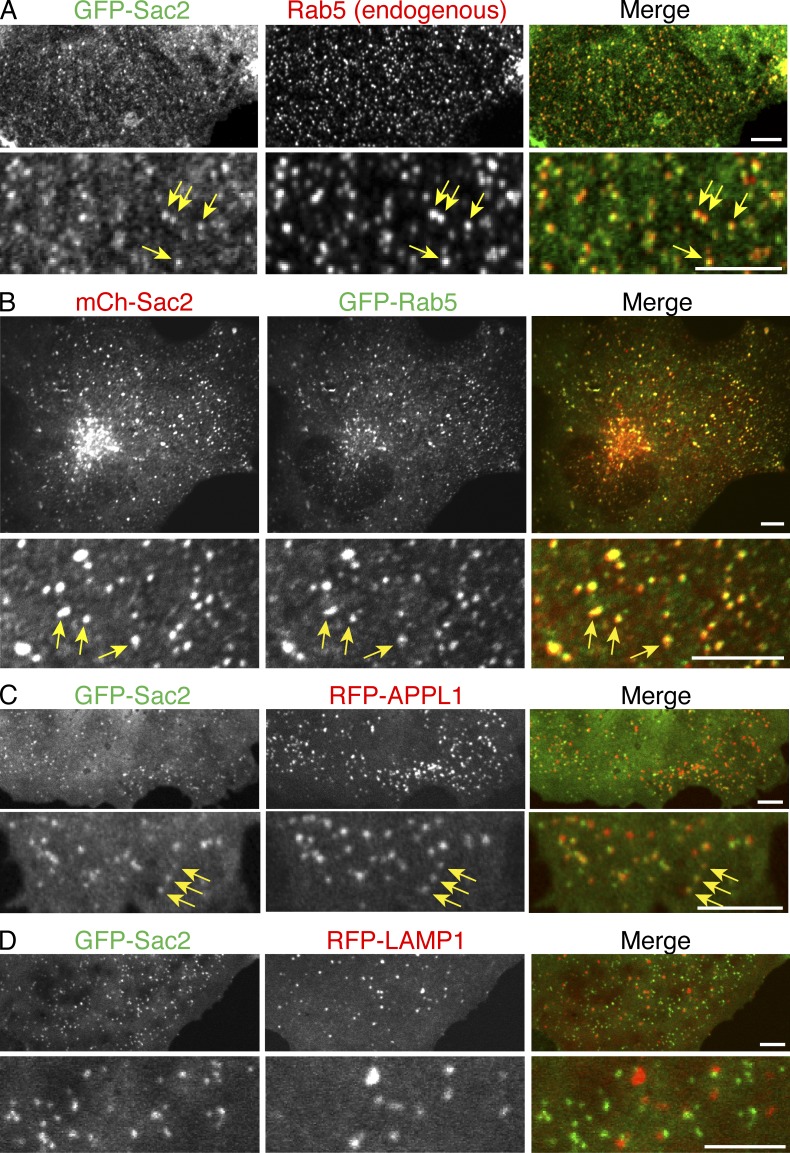
The subcellular targeting of Sac2 was examined by expressing GFP-tagged Sac2 in COS7 cells and observing cells with low/moderate levels of expression (antibodies directed against Sac2 did not yield detectable immunofluorescence signal for the endogenous protein in all of several cell lines tested). GFP-Sac2 had a punctate distribution (Fig. 1 A). The great majority of these puncta colocalized with the early endosomal marker Rab5 (Zerial and McBride, 2001), as detected by immunofluorescence (Fig. 1 A) and by coexpression with GFP-Rab5 (Fig. 1 B). Quantification analysis demonstrates that 76.7% of GFP-Sac2 or 80.0% of mCh-Sac2 colocalized with endogenous Rab5 or GFP-Rab5, respectively. The most peripheral fraction of Sac2 puncta also colocalized with APPL1, a Rab5 effector that marks a very early endosomal station upstream of PI3P-positive endosomes (Miaczynska et al., 2004; Zoncu et al., 2009; 54.2% of GFP-Sac2 colocalized with RFP-APPL1; Fig. 1 C). Sac2, however, did not show an obvious colocalization with the lysosomal maker protein LAMP1 (3.51% of GFP-Sac2 colocalized with RFP-LAMP1; Fig. 1 D). This indicates that Sac2 is localized on organelles of the endocytic pathway, but only, or primarily, on early stations of this pathway.
Figure 1.
Localization of Sac2 on endosomes. (A and B) GFP- or mCherry (mCh)-Sac2 is present on Rab5-positive endosomes, as shown by colocalization with endogenous Rab5 immunoreactivity (A) or with GFP-Rab5 (B). (C and D) GFP-Sac2 also colocalizes with the majority of APPL1 (RFP-APPL1)-positive endosomes (C), but does not colocalize with a lysosomal marker protein, RFP-LAMP1 (D). In A, B, and C, examples of colocalizations are indicated by arrows. Images were taken by spinning-disc confocal microscopy. Bars, 5 µm.
Overlap with the localizations of OCRL on early endocytic stations
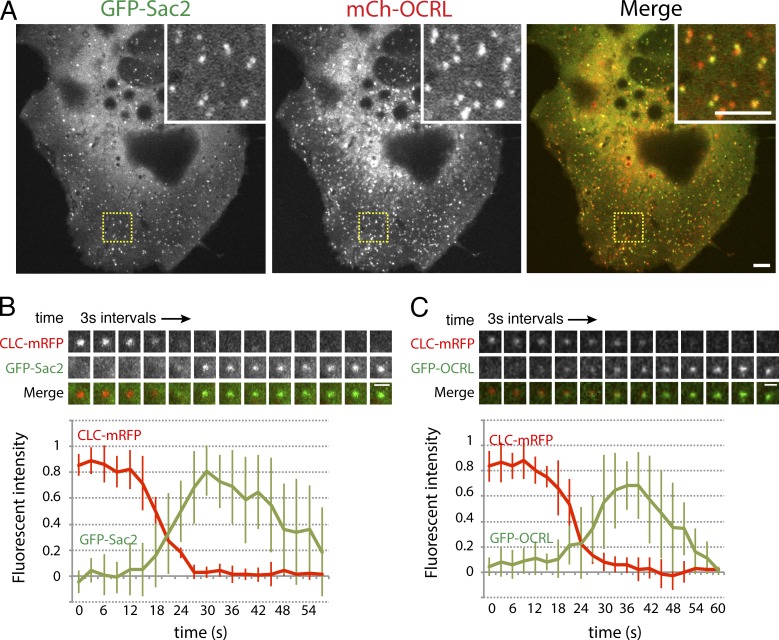
A localization throughout the Rab5-positive compartment including APPL1-positive endosomes had been previously reported for the inositol 5-phosphatase OCRL and its close homologue INPP5B, both of which are Rab5 effectors (Shin et al., 2005; Hyvola et al., 2006; Erdmann et al., 2007; Vicinanza et al., 2011). In fact, many of the puncta positive for Sac2 were also positive for OCRL (Fig. 2 A; 69.8% colocalization with OCRL). In the case of OCRL, a localization at the earliest stages of endocytosis—both clathrin dependent and clathrin independent—had also been demonstrated (Erdmann et al., 2007; Coon et al., 2009; Swan et al., 2010; Bohdanowicz et al., 2012; Nández et al., 2014). More specifically, OCRL is recruited to very late-stage clathrin-coated pits, at the transition stage between clathrin-coated pits and vesicles (Erdmann et al., 2007; Mao et al., 2009; Nández et al., 2014), with the same recruitment signature at GAK (Taylor et al., 2012), a cofactor of the clathrin uncoating ATPase (Lee et al., 2006). OCRL is also recruited to macropinosomes as they separate from the plasma membrane (Coon et al., 2009; Swan et al., 2010). Thus, we explored whether Sac2 is also present at sites of clathrin-mediated endocytosis and on macropinosomes.
Figure 2.
Colocalization of Sac2 with OCRL on early endosomes and at the last stage of clathrin-mediated endocytosis. Live spinning-disc confocal microscopy. (A) Colocalization of GFP-Sac2 and mCherry-OCRL. The majority of the fluorescent puncta positive for both proteins are expected to be early endosomes, given the close colocalization of both proteins with Rab5 (see Fig. 1 A and Hyvola et al., 2006; Erdmann et al., 2007). Insets show enlarged views of the regions boxed in yellow. (B and C) Time course of CLC-mRFP and either GFP-Sac2 (B) or GFP-OCRL (C) fluorescence intensities at very late stage clathrin-coated pits demonstrating that Sac2 and OCRL appear when clathrin is disappearing, and that Sac2 and OCRL display similar recruitment patterns. The disappearance of the Sac2 and OCRL fluorescence reflects the newly formed vesicle moving out of the focal plane. Bars: (A) 5 µm; (B and C) 1 µm.
To assess the localization of Sac2 at sites of clathrin-mediated endocytosis, time-lapse spinning-disc confocal microscopy was performed on COS7 cells expressing GFP-Sac2 and mRFP-tagged clathrin light chain (CLC-mRFP). Analysis of blinking clathrin spots on the ventral surface of the cells, i.e., endocytic clathrin-coated pits, revealed that Sac2 appeared at sites of clathrin-positive puncta as the clathrin signal had nearly disappeared and remained associated with them as they moved to a deeper position in the cell out of the focal plane (Video 1 and Fig. 2 B). The time-course of this recruitment was very similar to that of OCRL, which also remains associated with the newly formed vesicles (Fig. 2 C; Erdmann et al., 2007; Nández et al., 2014). Although in the case of OCRL the recruitment to sites of clathrin-mediated endocytosis is mediated at least in part by its clathrin boxes (Mao et al., 2009), the mechanism underlying the recruitment of Sac2 remains to be determined, as mutations of a potential clathrin-binding site in its C-terminal region (1047LLELE1051, a so-called clathrin box; Lafer, 2002) did not abolish recruitment.
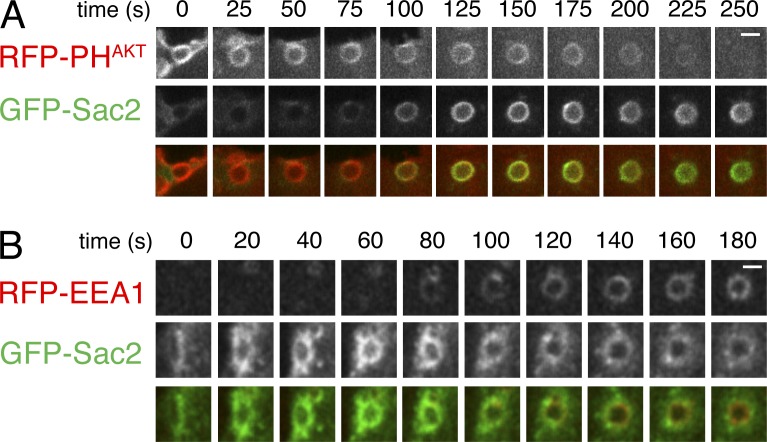
To determine the potential presence of Sac2 on macropinosomes, live spinning-disk confocal microscopy was performed on COS7 cells expressing constitutively active H-RasV12G. This H-Ras mutant induces an abundant formation of macropinosomes, at least in part through the stimulation of PI(3,4,5)P3 production (Porat-Shliom et al., 2008). This assay revealed the recruitment of GFP-Sac2 to macropinosomes when PI(3,4,5)P3 had started to decrease, as detected by the PI(3,4,5)P3 reporter RFP-PHAKT(Fig. 3 A). GFP-Sac2 stayed on these macropinosomes as they became positive for EEA1, a marker of PI3P-positive endosomes (Fig. 3 B).
Figure 3.
Recruitment of Sac2 to macropinosomes. Mouse fibroblasts expressing GFP-Sac2 and either RFP-PHAKT (A) or RFP-EEA1 (B) were also cotransfected with H-RasV12G to induce the formation of macropinosomes. The gallery of confocal images shows that Sac2 is recruited to these vesicles when the AKT signal, which primarily reflects PI(3,4,5)P3, is disappearing, and remains associated with the vesicles as they mature to EEA1-positive endosomes. Bars, 1 µm.
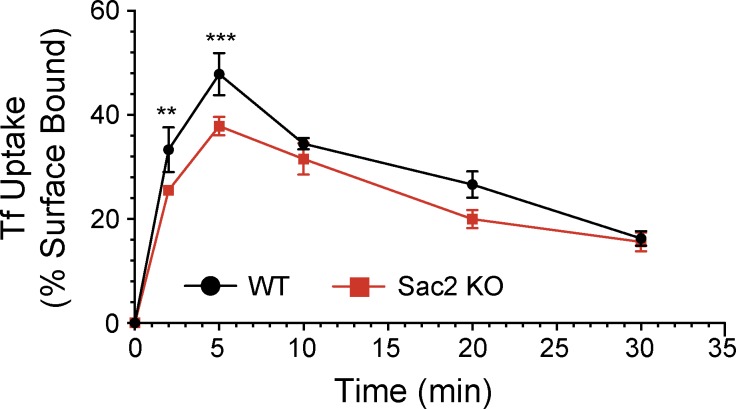
In view of these localizations of Sac2, we explored whether its absence would result in a disruption of the endocytic system. To this aim, we generated mouse embryonic fibroblasts (MEFs) from previously described Sac2/INPP5F knockout (KO) mice (Zhu et al., 2009). In agreement with the lack of major pathological phenotypes at the organismal levels in Sac2 KO mice, no major defects were observed in the endosomal system of Sac2 KO MEFs, which suggests a functional overlap with other PI 4-phosphatases. The steady-state levels of PIs in Sac2 KO MEFs, as measured by HPLC of metabolically labeled cells, demonstrated only a very minor (∼5%, and at the limit of significance) increase in PI4P (PI4P levels expressed as the percentage of total detectable PIs in wild-type (WT) control and KO MEFs were 0.058 ± 0.0010 and 0.061 ± 0.0012 [P < 0.375], respectively). Levels of other PIs were not different from controls (unpublished data). Analysis of the localization of several endocytic markers in KO MEFs (clathrin, AP2, OCRL, Rab5, APPL1, EEA1, and LAMP1) also did not reveal major abnormalities relative to control WT MEFs (Fig. S1). Sac2 KO MEFs, however, showed a defect in the internalization of transferrin, an assay used to monitor clathrin-meditated endocytosis (Fig. 4). This defect, which is reminiscent of the defect in clathrin-mediated endocytosis observed in cells that lack OCRL (Nández et al., 2014), provides genetic evidence for a functional involvement of Sac2 in the endocytic pathway.
Figure 4.
Defect in the internalization of transferrin in Sac2 KO MEFs. Internalization of biotinylated transferrin over time was analyzed biochemically in WT and Sac2 KO MEFs using an ELISA-based assay (see Materials and methods for details). Data were expressed as mean ± SD (error bars) of three independent experiments (Student’s t test, **, P < 0.01; ***, P < 0.001).
Rab5 is a major regulator of Sac2 localization
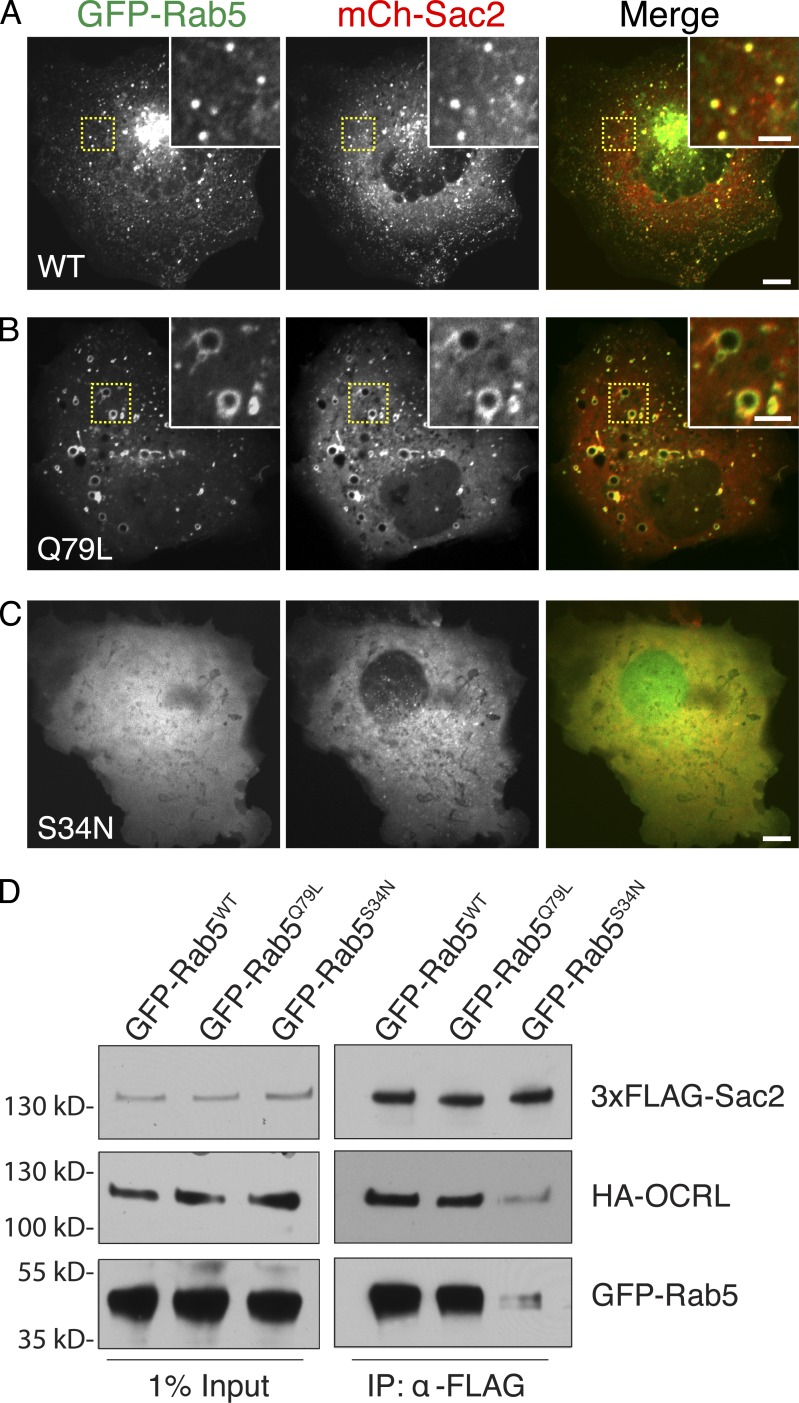
The major colocalization of Sac2 with Rab5 raised the possibility that, as in the case of OCRL (Hyvola et al., 2006; McCrea et al., 2008) and INPP5B (Shin et al., 2005), this small GTPase may have a major role in the localization of Sac2. To explore this possibility, the distribution of mCherry-Sac2 was assessed in COS7 cells expressing GFP-tagged WT Rab5 (Rab5WT), constitutively active Rab5 (Rab5Q79L), and dominant-negative mutant Rab5 (Rab5S34N). mCherry-Sac2 closely colocalized with Rab5WT and Rab5Q79L on endosomes, including the large endosomes typically induced by Rab5Q79L-expressing cells (Fig. 5, A and B). In contrast, the bright spots of mCherry-Sac2 were no longer visible in a cell expressing Rab5S34N, which also had a diffuse distribution (Fig. 5 C), even though endosomes positive for an endosomal marker (RFP-2xFyveEEA1) were still present in these cells (Fig. S2). We conclude that Sac2, like OCRL, is a Rab5 effector, although it may not bind Rab5 directly and although Rab5 may not be the only determinant of its localization. Importantly, Rab5 not only recruits both OCRL and Sac2 to the same membranes, but also enhances formation of a complex comprising the two proteins, which can be demonstrated by anti-Sac2 immunoprecipitation from COS7 cells coexpressing HA-tagged OCRL, Flag-tagged Sac2, and GFP-Rab5 constructs. A pool of HA-tagged OCRL was recovered along with GFP-Rab5 constructs in anti-Flag (i.e., Flag-Sac2 enriched) immunoprecipitates obtained from cells expressing GFP-Rab5Q79L or GFP-Rab5WT (Fig. 5 D). In contrast, recovery of HA-OCRL and Rab5 was drastically reduced when using cells expressing Rab5S34N, i.e., dominant-negative Rab5 (Fig. 5 D). As Rab5 cannot bind two effectors simultaneously, this finding speaks against a direct interaction between Rab5 and Sac2.
Figure 5.
Sac2 functions downstream of Rab5 and is part of a complex also comprising OCRL. (A–C) COS7 cells were cotransfected with mCherry (mCh)-Sac2 together with GFP-Rab5WT, GFP-Rab5Q79L (constitutively active; B), or GFP-Rab5S34N (dominant negative; C), as indicated, and imaged by confocal microscopy. mCh-Sac2 colocalizes with WT and constitutively active Rab5. In contrast, it has a predominant cytosolic localization in cells expressing dominant-negative Rab5. Inset panels show enlarged views of the regions boxed in yellow. Bars: (full-size images) 5 µm; (insets) 2 µm for inset image. (D) COS7 cells were cotransfected with 3×Flag-Sac2, HA-OCRL, and GFP-tagged Rab5 constructs (Rab5WT, GFP-Rab5Q79L, or GFP-Rab5S34N). Cell lysates were immunoprecipitated with anti-Flag antibody to enrich for Sac2. Immunoblotting of the starting lysates (left) and of the immunoprecipitates (right) for the Flag, HA, and GFP epitopes confirms enrichment of Sac2 and shows robust coprecipitation of Rab5 and OCRL only from cells expressing GFP-Rab5WT or GFP-Rab5Q79L. Recovery of OCRL and Rab5 was dramatically reduced from cells expressing Rab5S34N.
Sac2 is an inositol 4-phosphatase
OCRL is a member of the inositol 5-phosphatase family, whereas Sac2 is a member of the Sac family of inositol phosphatases. The Sac phosphatase domains of Sac1 and of the synaptojanins act primarily on PI4P (Guo et al., 1999; Nemoto et al., 2000). However, based on the enzymatic characterization of GST-(human) Sac2 purified from Sf9 insect cells, Sac2 was reported to act as a 5-phosphatase (Minagawa et al., 2001), i.e., with a substrate specificity similar to OCRL, which dephosphorylates PI(4,5)P2, and to a lower extent also PI(3,4,5)P3, at the 5 position (Zhang et al., 1998). The similar localization of two structurally distinct enzymes with the same catalytic activity, Sac2 and OCRL, seemed puzzling. In synaptojanin, the Sac domain and the 5-phosphatase domain are arranged in tandem as part of the same polypeptide. We considered the possibility that the presence of Sac2 in the proximity of OCRL may reflect a similar functional partnership of two enzymes with distinct activities, but in this instance encoded by two separate genes. Thus, we revisited the catalytic function of Sac2.
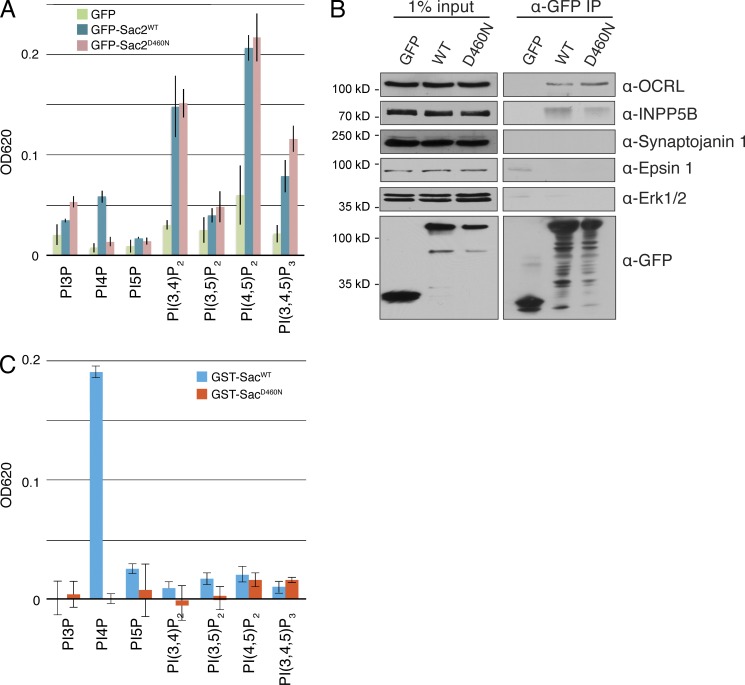
Inspection of Sac2 reveals no other catalytic modules besides its Sac domain, which is very similar to other Sac domain–containing phosphatases (Hsu and Mao, 2013). Although the central region of Sac2 contains another conserved module, the so-called “hSac2 domain” (http://pfam.xfam.org/family/hSac2), structural predictions of this domain suggest a pleckstrin homology (PH) domain (Fig. S3). Initial experiments to assess substrate specificity of the phosphatase activity of Sac2 were performed using anti-GFP immunoprecipitates from HEK cells expressing GFP-tagged WT full-length Sac2 or either GFP alone or GFP-Sac2D460N as controls. GFP-Sac2D460N, which like GFP-Sac2WT has an endosomal localization (Fig. S4), harbors a mutation in the Sac domain expected to abolish its catalytic activity, based on studies of other Sac phosphatase domains (Nemoto et al., 2000; Manford et al., 2010). A malachite-based in vitro phosphatase assay to test activity against the seven PIs revealed that immunoprecipitates generated from GFP-Sac2WT–expressing cells had the highest activity against PI(4,5)P2, PI(3,4)P2, and PI(3,4,5)P3 (Fig. 6 A). Lower activities were detected against other PIs, including an activity against PI4P. However, only the activity directed against PI4P was abolished in immunoprecipitates from cells expressing GFP-Sac2D460N. The other activities were unchanged (Fig. 6 A).
Figure 6.
Sac2 is an inositol 4-phosphatase. (A) GFP, GFP-Sac2, and GFP-Sac2D460N were transiently overexpressed in Expi293 HEK cells and semi-purified by immunoprecipitation using anti-GFP antibody. The presence of inositol-phosphatase activity in the immunoprecipitated material was assessed with a malachite-based assay. Data shown represent mean ± SD (error bars). (B) Lysates of COS-7 cells expressing GFP, GFP-Sac2WT, or GFP-Sac2D460N were subjected to anti-GFP immunoprecipitation, and immunoprecipitates were processed by immunoblotting to assess the enrichment of GFP and GFP fusion proteins (bottom), of inositol phosphatases (OCRL, INPP5B, and synaptojanin 1), and of control proteins (epsin 1 and Erk1/2). Note the enrichment of OCRL and INPP5B in anti-GFP immunoprecipates from COS7 cells expressing GFP-Sac2WT or GFP-Sac2D460N. (C) GST-SacWT and GST-SacD460N were expressed in bacteria, purified on glutathione Sepharose beads, and analyzed for inositol phosphatase activity against the seven PIs via the malachite assay. Data shown represent mean ± SD (error bars).
This result strongly suggested that the activities observed both with the WT and with the mutant Sac2 may have reflected coprecipitation of other inositol phosphatases with robust activities present in HEK cells. One such enzyme could be OCRL, in view of the coprecipitation of this PI(4,5)P2 and PI(3,4,5)P3 5-phosphatase with Sac2 shown in Fig. 5 D. Accordingly, when the same material used for the malachite-based assay was examined by Western blotting for the presence of OCRL, an OCRL immunoreactive band was specifically detected in extracts of cells expressing either GFP-Sac2WT or Sac2D460N (Fig. S5 A). Similar results were obtained using anti-GFP immunoprecipitates generated from COS7 cells transiently expressing GFP, GFP-Sac2WT, or GFP-Sac2D460N (Fig. 6 B). Further Western blot analysis of these immunoprecipitates for additional inositol phosphatases revealed the presence not only of OCRL, but also of the 5-phosphatase INPP5B. These samples did not coprecipitate synaptojanin 1, which is not a Rab5 effector (Fig. 6 B), nor other cytosolic control proteins, such as epsin 1 (an endocytic clathrin adaptor) and the MAPK Erk1/2 (a soluble kinase). In spite of the detection of PI(3,4)P2 phosphatase activity in the malachite-based phosphatase assay (Fig. 6 A), we did not detect PI(3,4)P2 phosphatases with available antibodies in the immunoprecipitates. Thus, we used anti-GFP immunoprecipitates (i.e., immunoprecipitates enriched in GFP, GFP-Sac2WT, or GFP-Sac2D460N, respectively) as bait to affinity-purify material from COS7 cell lysates. This assay confirmed the selective association of OCRL and INPP5B, but not of synaptojanin 1 (and control proteins) with Sac2 (Fig. S5 B). It also demonstrated an enrichment of INPP4A, a Rab5 effector and a 4-phosphatase that dephosphorylates PI(3,4)P2, (Shin et al., 2005) in GFP-Sac2WT or GFP-Sac2D460N affinity-purified materials (Fig. S5 B). These results demonstrate the occurrence of complexes in which Sac2 is associated with other “endocytic” phosphatases including OCRL, and suggest that the phosphatase activities against PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2 observed in the in vitro phosphatase assay (Fig. 6 A) were likely accounted for by such inositol phosphatases.
To more directly assess the substrate specificity of the Sac domain of Sac2, we generated this domain and its D460N mutant form as GST fusion proteins in bacteria. When tested in the malachite-based phosphatase assay for activity against the seven PIs, robust activity was only observed when PI4P was the substrate. This activity was abolished by the D460N mutation (Fig. 6 C). We conclude that the Sac domain of Sac2 functions primarily as a PI4P 4-phosphatase.
Discussion
This study demonstrates that the inositol phosphatase Sac2 is localized on early endocytic membranes and thus likely participates, along with other phosphatases (Cremona et al., 1999; Stefan et al., 2002; Shin et al., 2005; Erdmann et al., 2007; Vicinanza et al., 2011; Nández et al., 2014; Posor et al., 2014), in the modification of the PI signature of plasma membrane patches that undergo internalization.
Our results also shed light on the discrepancy between the reported 5-phosphatase activity of the Sac domain of Sac2 (Minagawa et al., 2001; Hsu and Mao, 2013) and the close similarity of this domain to the Sac domains of Sac1 and of the synaptojanins, which strongly prefer PI4P as a substrate (Guo et al., 1999; Nemoto et al., 2000; Manford et al., 2010; Hsu and Mao, 2013). Our results show that the purified recombinant Sac domain of Sac2 has a strong preference for PI4P and that this activity is abolished by a mutation expected to disrupt its catalytic activity (Nemoto et al., 2000; Manford et al., 2010), the D to N mutation at position 460 in human and mouse Sac2 (D394 in yeast Sac1). They also strongly suggest that other phosphatase activities detected in immunoprecipitates from mammalian cells expressing full-length Sac2WT—primarily activities against PI(4,5)P2, PI(3,4)P2, and PI(3,4,5)P3—are due to coprecipitating phosphatases, as they are also detected when immunoprecipitates are generated from the catalytically inactive GFP-Sac2D460N–expressing cells. OCRL, which as we show here is present in anti-Sac2 immunoprecipitates along with its homologue INPP5B, may account for the activity on PI(4,5)P2 and PI(3,4,5)P3, its two preferred substrates (Zhang et al., 1998). INPP4A may account for the PI(3,4)P2 phosphatase activity. Interestingly, this enzyme, like OCRL and INPP5B, is also a Rab5 effector (Shin et al., 2005), which explains its presence in endocytic complexes.
In the previous study reporting 5-phosphatase activity of human Sac2, the analysis of such activity was performed on GST-Sac2 expressed in, and affinity-purified from, metazoan cells (insect cells; Minagawa et al., 2001). Such activity was reported (based on unpublished data) to be abolished by an asparagine-to-alanine mutation at amino acid position 460 of the human Sac2 used for that study (possibly an oversight, as position 460 is represented by an aspartate in both human and mouse Sac2/INPP5F). This finding in principle argues against the possibility that a protein with robust 5-phosphatase activity and endogenously expressed in insect cells could have copurified with GST-Sac2 and account for the 5-phosphatase activity in the affinity-purified material. This is in contrast to our findings that the presence of 5-phosphatase activity in anti-Sac2 immunoprecipitates was unaffected by mutation of aspartate 460 to asparagine. However, when we mutated aspartate 460 to alanine, the resulting mutant Sac2 was cytosolic and did not have the characteristic punctate localization in COS7 cells, which suggests misfolding (Fig. S4). Misfolding of Sac2 could have affected its interaction with 5-phosphatases in the study of Minagawa et al. (2001).
The 4-phosphatase activity of Sac2 and a partial, but quite strong, colocalization with OCRL points to the interesting possibility that Sac2 and OCRL/INPP5B may be functionally linked as in synaptojanin, where the Sac domain and the 5-phosphatase domain are arranged in tandem within the same protein (McPherson et al., 1996). The partnership of a 4-phosphatase and a 5-phosphatase domain may facilitate the sequential dephosphorylation of PI(4,5)P2 to PI4P and then to PtdIns, which is required for the conversion of the PI(4,5)P2 signature of the plasma membrane to the PI3P signature of early endosomes (Simonsen et al., 2001; Zoncu et al., 2009). Our present findings complement our recent report that OCRL has an important role in clathrin uncoating in nonneuronal cells (Nández et al., 2014), as they demonstrate a new similarity with synaptojanin, which has a major role in clathrin uncoating at synapses (McPherson et al., 1996; Cremona et al., 1999). Given the strong colocalization of both Sac2 and OCRL with Rab5, these two enzymes may also have a role in preventing PI(4,5)P2 and PI4P accumulation on endosomal membranes, where both PI 4-kinases (Balla et al., 2002; Salazar et al., 2005; Minogue et al., 2006) and PI4P 5-kinases (Sun et al., 2013) have been localized.
The next important step will be to elucidate the role of the enzymatic activity of Sac2 in cell physiology. KO mice for Sac2 have no obvious major phenotypes under basal conditions (Zhu et al., 2009). The one phenotype reported, in the context of studies specifically focused on heart function, was an augmented hypertrophy and reactivation of the fetal gene program in response to stress, but the underlying mechanisms remained unclear (Trivedi et al., 2007; Zhu et al., 2009). A survey of the distribution of several endosomal markers (Fig. S1) and the localization of PI4P by genetically encoded probes in Sac2 KO MEFs did not reveal major abnormalities in the endosomal system, which suggests a possible compensation by synaptojanin. However, we detected a defect of clathrin-mediated endocytosis in MEFs derived from Sac2 KO mice, thus confirming a role of Sac2 in the endocytic pathway.
It is possible that the function of Sac2 may be compensated by the function of the Sac domains of synaptojanin, another protein that functions in the endocytic pathway. We also note that in the case of synaptojanin the physiological function of Sac domains remains elusive. In mouse cells (Mani et al., 2007) and in zebrafish (George et al., 2014), rescue experiments of synaptojanin KO phenotypes have demonstrated a much more critical role of the 5-phosphatase domain than of the Sac domain. Accordingly, although absence of synaptojanin 1 results in perinatal lethality in mice (Cremona et al., 1999), a homozygous mutation of its Sac domain resulting in loss of catalytic activity is compatible with life in humans (Krebs et al., 2013; Quadri et al., 2013). However, patients with such mutation develop early onset progressive Parkinsonism with generalized seizures, indicating a subtle role that becomes manifest with postnatal life (Krebs et al., 2013; Quadri et al., 2013). In this context, it is of special interest that a recent genome-wide association study has identified the gene encoding Sac2, a protein highly expressed in brain, as a risk locus in Parkinson’s disease (Nalls et al., 2014).
Materials and methods
Plasmids, purified proteins, and antibodies
Mouse Sac2 cDNA was amplified by PCR from the cDNA (Genbank accession no. BC125437) obtained from GE Healthcare, and cloned into the pEGFP-C2 vector (Takara Bio Inc.) driven by CMV promoter or the p3×FLAG-CMV-10 vector (Sigma-Aldrich) to construct GFP-Sac2 and 3×Flag-Sac2, respectively. GST-SacWT was obtained by cloning the appropriate PCR fragment (amino acids 1–591) into the pGEX6 vector (tac promoter). GFP-Sac2D460N and GST-SacD460N were generated by site-directed mutagenesis. Sources of other plasmids (and their backbone vector shown in parenthesis) are as follows. Note that all plasmids are driven by a CMV promoter: CLC-mRFP (pcDNA3; Zoncu et al., 2007); RFP-EEA1 (pEGFP; provided by S. Corvera, University of Massachusetts Medical School, Worcester, MA); GFP-OCRL (pEGFP; Erdmann et al., 2007); mCherry-OCRL (pmCherry; Nández et al., 2014); HA-OCRL (pcDNA3; our laboratory); GFP-tagged Rab5 including WT, Q79L, and S34N mutants (pEGFP; provided by M. Zerial, Max Plank Institute, Munich, Germany); RFP-LAMP1 (pDs-Red; provided by W. Mothes, Yale University, New Haven, CT); and RFP-APPL1 (pcDNA3; Zoncu et al., 2009). Sources of antibodies were as follows: rabbit anti-EEA1 (Thermo Fisher Scientific), mouse anti-LAMP1 (clone H4A3; Developmental Studies Hybridoma Bank), mouse anti-Flag M2 mAb and anti-OCRL (Sigma-Aldrich), rat anti-HA mAb (clone 3F10; Roche), mouse anti-GFP monoclonal antibody (Takara Bio Inc.), rabbit anti-CLC and rabbit anti-INPP5B (EMD Millipore), rabbit anti-Rab5 (clone C8B1) and rabbit anti-Erk1/2 (Cell Signaling Technology), rabbit anti–α-Adaptin (raised against full-length recombinant protein), rabbit anti–Synaptojanin 1 (raised against recombinant protein containing proline-rich region), rabbit anti-INNP4A (raised against full-length recombinant protein), rabbit anti–epsin 1 (raised against recombinant protein containing DPW-NPW motif), and rabbit anti-APPL1 (raised against peptide: SQSEESDLGEGGKKRESEA; our laboratory).
GST-SacWT and GST-SacD460N recombinant proteins were purified from Escherichia coli BL21 using glutathione Sepharose beads (GE Healthcare).
Mice and MEFs
Sac2/INPP5F KO mice, established from gene-trap embryonic stem cells (ES clone no. XL0571; International Gene Trap Consortium) in which Sac2/inpp5f locus is disrupted by insertional mutagenesis, were a gift from J.A. Epstein (University of Pennsylvania, Philadelphia, PA; Zhu et al., 2009). WT and KO MEFs were established from newborn pups obtained from intercrossing of heterozygous mice, as described previously (Nakatsu et al., 2012). In brief, newborn pups were minced, trypsinized, and cultured using DMEM supplemented with 10% FBS to obtain primary MEFs.
Immunoprecipitation
Expi293 HEK cells (Thermo Fisher Scientific) grown in suspension, or COS7 cells, expressing GFP, GFP-Sac2WT, or GFP-Sac2D460N were lysed with lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40) supplemented with protease inhibitor cocktail (Roche) and centrifuged for 30 min at 16,000 g. The supernatant of Expi293 HEK cells was incubated at 4°C with Chromotek GFP-trap agarose (Allele Biotech) or anti-Flag M2 mAb conjugated to agarose (Sigma-Aldrich) for lysates from COS7 cells. The agarose beads were washed with lysis buffer and bead-attached material was used for the in vitro phosphatase assay or for protein analysis by Western blotting. To assess binding of phosphatases to Sac2 enriched by immunoprecipitation, immunoprecipitates (bead bound material) generated from COS7 cells were incubated for 3 h at 4°C with COS7 lysate and then subjected to multiple washes with lysis buffer. SDS-PAGE and Western blotting were performed using standard procedures.
In vitro phosphatase assay
The in vitro phosphatase malachite green-based assay was performed as described previously (Nakatsu et al., 2010). In brief, purified proteins were incubated with water-soluble PI lipids (diC8-acyl chain; Echelon) for 30 min at 37°C. Free phosphate released was detected by the malachite green solution, and development of the color was measured at 620 nm wavelength using a plate reader. Data were expressed as the mean ± SD of three independent experiments.
Microscopy
Image acquisition was performed with an UltraView VoX system (PerkinElmer) on an inverted microscope (Ti-E Eclipse; Nikon) equipped with a spinning-disc confocal scanner (CSU-X1; Yokogawa Electric Corporation), a camera (C9100-50; Hamamatsu Photonics), 60 or 100× oil objective lenses (1.4 NA, CFI Plan Apochromat VC), and perfect focus controlled by Volocity software (PerkinElmer). All images were acquired with live cells (either snapshots or videos) with the incubation chamber maintained at 37°C.
Image analysis
Image analysis was performed using Volocity software (PerkinElmer), MetaMorph software (Molecular Devices), and Fiji software (http://fiji.sc/wiki/index.php/Fiji). For the quantification of clathrin-coated pit fluorescence, the fluorescence signal at each time point was normalized to the peak fluorescent intensity. More than 10 clathrin-coated pits were analyzed for each sample. Data are expressed as mean ± SD.
Transferrin uptake
Uptake of biotinylated transferrin was performed as described previously (Yarar et al., 2005), with slight modifications. In brief, WT and Sac2 KO MEFs were starved for 1.5 h, then chilled on ice for 30 min and finally incubated with biotinylated transferrin (10 µg/ml) in ice-cold DMEM on ice for 45 min. After washing with cold PBS, cells were incubated with prewarmed culture media (DMEM; Life Technologies) at 37°C for the indicated times. Internalization was stopped by placing the cells on ice and washing them three times with cold PBS. Cells were then incubated on ice with avidin (0.05 mg/ml) for 1 h followed by incubation with biocytin (0.05 mg/ml) for 15 min. Cells were then washed three times with PBS and lysed (1% Triton X-100, 0.1% SDS, 0.2% BSA, 50 mM NaCl, and 1 mM Tris, pH 7.4). Cell lysates (1 µg) were then added to ELISA plates coated with anti–human transferrin antibody (Abcam) and assayed for detectable biotinylated transferrin using chromogen-conjugated streptavidin as indicated in the manufacturer’s protocol. Internalized biotinylated transferrin was expressed as the percentage of total surface bound at 4°C, which was not incubated with avidin or biocytin.
HPLC analysis for PI measurement
Control and Sac2 KO MEFs were metabolically labeled with [3H]myo-inositol for 3 d. Cells were lysed in 4.5% perchloric acid, and the pellet was rinsed three times with 0.1 M EDTA, followed by deacylation with a mixture of methylamine/water/n-butanol/methanol (36:8:9:47) for 45 min at 50°C and dried in a SpeedVac. The residue was extracted with a mixture of n-butanol/petroleum ether/ethyl formate (20:40:1) and water. The aqueous phase was resolved using anion-exchange HPLC with an ammonium phosphate gradient (LC-20AT UFLC [Shimadzu] equipped with an HPLC column [Partisil 5 SAX 4.6 × 125 mm; Whatman]) and identified using a radiometric detector (β-RAM 4B; LabLogic). The identity of each peak was determined by comparison to known compounds. Data are expressed as mean ± SD of three independent experiments.
Online supplemental material
Fig. S1 shows the localization of various endocytic proteins in WT and Sac2 KO MEFs. Fig. S2 shows the presence of PI3P-positive endosomes in control COS7 cells and in COS7 cells expressing GFP-tagged Rab5-S34N. Fig. S3 shows the result of a bioinformatics analysis of the domain structure of Sac2 predicting that the hSac2 domain has a PH domain fold. Fig. S4 shows the punctate endosomal localization of GFP-Sac2D460N and the diffuse cytosolic localization of GFP-Sac2D460A. Fig. S5 shows the biochemical interaction of inositol phosphatases including OCRL with Sac2. Video 1 shows GFP-Sac2 dynamics at the late-stage clathrin-coated pits. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201409064/DC1.
Supplementary Material
Acknowledgments
We are thankful to Lijuan Liu and Louise Lucast for outstanding technical assistance. Generous gifts of mice and key reagents are acknowledged in the Materials and methods section.
This work was supported in part by National Institutes of Health grants DK082700, R37NS036251, DK45735, and DA018343 (to P. De Camilli).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CLC
- clathrin light chain
- KO
- knockout
- PH
- pleckstrin homology
- PI
- phosphoinositide
- WT
- wild type
References
- Attree O., Olivos I.M., Okabe I., Bailey L.C., Nelson D.L., Lewis R.A., McInnes R.R., and Nussbaum R.L.. 1992. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 358:239–242 10.1038/358239a0 [DOI] [PubMed] [Google Scholar]
- Balla T.2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla A., Tuymetova G., Barshishat M., Geiszt M., and Balla T.. 2002. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 277:20041–20050 10.1074/jbc.M111807200 [DOI] [PubMed] [Google Scholar]
- Bohdanowicz M., Balkin D.M., De Camilli P., and Grinstein S.. 2012. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol. Biol. Cell. 23:176–187 10.1091/mbc.E11-06-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon B.G., Mukherjee D., Hanna C.B., Riese D.J. II, Lowe M., and Aguilar R.C.. 2009. Lowe syndrome patient fibroblasts display Ocrl1-specific cell migration defects that cannot be rescued by the homologous Inpp5b phosphatase. Hum. Mol. Genet. 18:4478–4491 10.1093/hmg/ddp407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona O., Di Paolo G., Wenk M.R., Lüthi A., Kim W.T., Takei K., Daniell L., Nemoto Y., Shears S.B., Flavell R.A., et al. 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 99:179–188 10.1016/S0092-8674(00)81649-9 [DOI] [PubMed] [Google Scholar]
- De Matteis M.A., and Godi A.. 2004. PI-loting membrane traffic. Nat. Cell Biol. 6:487–492 10.1038/ncb0604-487 [DOI] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Erdmann K.S., Mao Y., McCrea H.J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., and De Camilli P.. 2007. A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell. 13:377–390 10.1016/j.devcel.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M., Audhya A., and Emr S.D.. 2001. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell. 12:2396–2411 10.1091/mbc.12.8.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary J.D., Sato T.K., Stefan C.J., Bonangelino C.J., Weisman L.S., and Emr S.D.. 2002. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell. 13:1238–1251 10.1091/mbc.01-10-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A.A., Hayden S., Holzhausen L.C., Ma E.Y., Suzuki S.C., and Brockerhoff S.E.. 2014. Synaptojanin 1 is required for endolysosomal trafficking of synaptic proteins in cone photoreceptor inner segments. PLoS ONE. 9:e84394 10.1371/journal.pone.0084394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Stolz L.E., Lemrow S.M., and York J.D.. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274:12990–12995 10.1074/jbc.274.19.12990 [DOI] [PubMed] [Google Scholar]
- Harris T.W., Hartwieg E., Horvitz H.R., and Jorgensen E.M.. 2000. Mutations in synaptojanin disrupt synaptic vesicle recycling. J. Cell Biol. 150:589–600 10.1083/jcb.150.3.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes R.R. Jr, Shrimpton A.E., Knohl S.J., Hueber P., Hoppe B., Matyus J., Simckes A., Tasic V., Toenshoff B., Suchy S.F., et al. 2005. Dent Disease with mutations in OCRL1. Am. J. Hum. Genet. 76:260–267 10.1086/427887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., and Mao Y.. 2013. The Sac domain-containing phosphoinositide phosphatases: structure, function, and disease. Front Biol (Beijing). 8:395–407 10.1007/s11515-013-1258-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes W.E., Cooke F.T., and Parker P.J.. 2000. Sac phosphatase domain proteins. Biochem. J. 350:337–352 10.1042/0264-6021:3500337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvola N., Diao A., McKenzie E., Skippen A., Cockcroft S., and Lowe M.. 2006. Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J. 25:3750–3761 10.1038/sj.emboj.7601274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idevall-Hagren O., and De Camilli P.. 2014. Detection and manipulation of phosphoinositides. Biochim. Biophys. Acta. 10.1016/j.bbalip.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C.E., Karkheiran S., Powell J.C., Cao M., Makarov V., Darvish H., Di Paolo G., Walker R.H., Shahidi G.A., Buxbaum J.D., et al. 2013. The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum. Mutat. 34:1200–1207 10.1002/humu.22372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer E.M.2002. Clathrin-protein interactions. Traffic. 3:513–520 10.1034/j.1600-0854.2002.30801.x [DOI] [PubMed] [Google Scholar]
- Lee D.-W., Wu X., Eisenberg E., and Greene L.E.. 2006. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell Sci. 119:3502–3512 10.1242/jcs.03092 [DOI] [PubMed] [Google Scholar]
- Manford A., Xia T., Saxena A.K., Stefan C., Hu F., Emr S.D., and Mao Y.. 2010. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 29:1489–1498 10.1038/emboj.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani M., Lee S.Y., Lucast L., Cremona O., Di Paolo G., De Camilli P., and Ryan T.A.. 2007. The dual phosphatase activity of synaptojanin1 is required for both efficient synaptic vesicle endocytosis and reavailability at nerve terminals. Neuron. 56:1004–1018 10.1016/j.neuron.2007.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Balkin D.M., Zoncu R., Erdmann K.S., Tomasini L., Hu F., Jin M.M., Hodsdon M.E., and De Camilli P.. 2009. A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 28:1831–1842 10.1038/emboj.2009.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea H.J., Paradise S., Tomasini L., Addis M., Melis M.A., De Matteis M.A., and De Camilli P.. 2008. All known patient mutations in the ASH-RhoGAP domains of OCRL affect targeting and APPL1 binding. Biochem. Biophys. Res. Commun. 369:493–499 10.1016/j.bbrc.2008.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson P.S., Garcia E.P., Slepnev V.I., David C., Zhang X., Grabs D., Sossin W.S., Bauerfeind R., Nemoto Y., and De Camilli P.. 1996. A presynaptic inositol-5-phosphatase. Nature. 379:353–357 10.1038/379353a0 [DOI] [PubMed] [Google Scholar]
- Mehta Z.B., Pietka G., and Lowe M.. 2014. The cellular and physiological functions of the Lowe syndrome protein OCRL1. Traffic. 15:471–487 10.1111/tra.12160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler-Joseph S., Habermann B., Wilm M., Parton R.G., and Zerial M.. 2004. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 116:445–456 10.1016/S0092-8674(04)00117-5 [DOI] [PubMed] [Google Scholar]
- Milosevic I., Giovedi S., Lou X., Raimondi A., Collesi C., Shen H., Paradise S., O’Toole E., Ferguson S., Cremona O., and De Camilli P.. 2011. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 72:587–601 10.1016/j.neuron.2011.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa T., Ijuin T., Mochizuki Y., and Takenawa T.. 2001. Identification and characterization of a sac domain-containing phosphoinositide 5-phosphatase. J. Biol. Chem. 276:22011–22015 10.1074/jbc.M101579200 [DOI] [PubMed] [Google Scholar]
- Minogue S., Waugh M.G., De Matteis M.A., Stephens D.J., Berditchevski F., and Hsuan J.J.. 2006. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J. Cell Sci. 119:571–581 10.1242/jcs.02752 [DOI] [PubMed] [Google Scholar]
- Nakatsu F., Perera R.M., Lucast L., Zoncu R., Domin J., Gertler F.B., Toomre D., and De Camilli P.. 2010. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 190:307–315 10.1083/jcb.201005018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Baskin J.M., Chung J., Tanner L.B., Shui G., Lee S.Y., Pirruccello M., Hao M., Ingolia N.T., Wenk M.R., and De Camilli P.. 2012. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199:1003–1016 10.1083/jcb.201206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., et al. 2014. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46:989–993 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nández R., Balkin D.M., Messa M., Liang L., Paradise S., Czapla H., Hein M.Y., Duncan J.S., Mann M., and De Camilli P.. 2014. A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. eLife. 3:e02975 10.7554/eLife.02975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y., Arribas M., Haffner C., and DeCamilli P.. 1997. Synaptojanin 2, a novel synaptojanin isoform with a distinct targeting domain and expression pattern. J. Biol. Chem. 272:30817–30821 10.1074/jbc.272.49.30817 [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Kearns B.G., Wenk M.R., Chen H., Mori K., Alb J.G. Jr, De Camilli P., and Bankaitis V.A.. 2000. Functional characterization of a mammalian Sac1 and mutants exhibiting substrate-specific defects in phosphoinositide phosphatase activity. J. Biol. Chem. 275:34293–34305 10.1074/jbc.M003923200 [DOI] [PubMed] [Google Scholar]
- Ooms L.M., Horan K.A., Rahman P., Seaton G., Gurung R., Kethesparan D.S., and Mitchell C.A.. 2009. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419:29–49 10.1042/BJ20081673 [DOI] [PubMed] [Google Scholar]
- Patki V., Virbasius J., Lane W.S., Toh B.H., Shpetner H.S., and Corvera S.. 1997. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA. 94:7326–7330 10.1073/pnas.94.14.7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirruccello M., and De Camilli P.. 2012. Inositol 5-phosphatases: insights from the Lowe syndrome protein OCRL. Trends Biochem. Sci. 37:134–143 10.1016/j.tibs.2012.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat-Shliom N., Kloog Y., and Donaldson J.G.. 2008. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol. Biol. Cell. 19:765–775 10.1091/mbc.E07-08-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y., Eichhorn-Grünig M., and Haucke V.. 2014. Phosphoinositides in endocytosis. Biochim. Biophys. Acta. 10.1016/j.bbalip.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Quadri M., Fang M., Picillo M., Olgiati S., Breedveld G.J., Graafland J., Wu B., Xu F., Erro R., Amboni M., et al. 2013. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum. Mutat. 34:1208–1215 10.1002/humu.22373 [DOI] [PubMed] [Google Scholar]
- Salazar G., Craige B., Wainer B.H., Guo J., De Camilli P., and Faundez V.. 2005. Phosphatidylinositol-4-kinase type II α is a component of adaptor protein-3-derived vesicles. Mol. Biol. Cell. 16:3692–3704 10.1091/mbc.E05-01-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.-W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M.R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., et al. 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170:607–618 10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Wurmser A.E., Emr S.D., and Stenmark H.. 2001. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13:485–492 10.1016/S0955-0674(00)00240-4 [DOI] [PubMed] [Google Scholar]
- Staiano L., De Leo M.G., Persico M., and De Matteis M.A.. 2014. Mendelian disorders of PI metabolizing enzymes. Biochim. Biophys. Acta. 10.1016/j.bbalip.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Audhya A., and Emr S.D.. 2002. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 13:542–557 10.1091/mbc.01-10-0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Hedman A.C., Tan X., Schill N.J., and Anderson R.A.. 2013. Endosomal type Iγ PIP 5-kinase controls EGF receptor lysosomal sorting. Dev. Cell. 25:144–155 10.1016/j.devcel.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan L.E., Tomasini L., Pirruccello M., Lunardi J., and De Camilli P.. 2010. Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc. Natl. Acad. Sci. USA. 107:3511–3516 10.1073/pnas.0914658107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.J., Lampe M., and Merrifield C.J.. 2012. A feedback loop between dynamin and actin recruitment during clathrin-mediated endocytosis. PLoS Biol. 10:e1001302 10.1371/journal.pbio.1001302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi C.M., Luo Y., Yin Z., Zhang M., Zhu W., Wang T., Floss T., Goettlicher M., Noppinger P.R., Wurst W., et al. 2007. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat. Med. 13:324–331 10.1038/nm1552 [DOI] [PubMed] [Google Scholar]
- Verstreken P., Koh T.-W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., and Bellen H.J.. 2003. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 40:733–748 10.1016/S0896-6273(03)00644-5 [DOI] [PubMed] [Google Scholar]
- Vicinanza M., Di Campli A., Polishchuk E., Santoro M., Di Tullio G., Godi A., Levtchenko E., De Leo M.G., Polishchuk R., Sandoval L., et al. 2011. OCRL controls trafficking through early endosomes via PtdIns4,5P2-dependent regulation of endosomal actin. EMBO J. 30:4970–4985 10.1038/emboj.2011.354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D., Waterman-Storer C.M., and Schmid S.L.. 2005. A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell. 16:964–975 10.1091/mbc.E04-09-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., and McBride H.. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107–117 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hartz P.A., Philip E., Racusen L.C., and Majerus P.W.. 1998. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 273:1574–1582 10.1074/jbc.273.3.1574 [DOI] [PubMed] [Google Scholar]
- Zhu W., Trivedi C.M., Zhou D., Yuan L., Lu M.M., and Epstein J.A.. 2009. Inpp5f is a polyphosphoinositide phosphatase that regulates cardiac hypertrophic responsiveness. Circ. Res. 105:1240–1247 10.1161/CIRCRESAHA.109.208785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M., Sebastian R., Nakatsu F., Chen H., Balla T., Ayala G., Toomre D., and De Camilli P.V.. 2007. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 104:3793–3798 10.1073/pnas.0611733104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Perera R.M., Balkin D.M., Pirruccello M., Toomre D., and De Camilli P.. 2009. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 136:1110–1121 10.1016/j.cell.2009.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.