Disulfide bonds involving cysteine 367 in K14 play a crucial role in the assembly, dynamics, and organization of K14-containing filaments in epidermal keratinocytes.
Abstract
We recently reported that a trans-dimer, homotypic disulfide bond involving Cys367 in keratin 14 (K14) occurs in an atomic-resolution structure of the interacting K5/K14 2B domains and in keratinocyte cell lines. Here we show that a sizable fraction of the K14 and K5 protein pools participates in interkeratin disulfide bonding in primary cultures of mouse skin keratinocytes. By comparing the properties of wild-type K14 with a completely cysteine-free variant thereof, we found that K14-dependent disulfide bonding limited filament elongation during polymerization in vitro but was necessary for the genesis of a perinuclear-concentrated network of keratin filaments, normal keratin cycling, and the sessile behavior of the nucleus and whole cell in keratinocytes studied by live imaging. Many of these phenotypes were rescued when analyzing a K14 variant harboring a single Cys residue at position 367. These findings establish disulfide bonding as a novel and important mechanism regulating the assembly, intracellular organization, and dynamics of K14-containing intermediate filaments in skin keratinocytes.
Introduction
Fibrous cytoskeletal polymers, of which there are three major types in most multicellular eukaryotes, perform a wide array of important if not essential functions in vivo. The pleiotropic roles of F-actin, microtubules, and intermediate filaments (IFs) are defined not only by their intrinsic properties but also by the manner with which they are organized and regulated relative to the cytoarchitectural features of the cells harboring them (Alberts et al., 2007). Dissecting the mechanisms responsible for the intracellular organization of cytoskeletal fibers, and its context-specific regulation, is key to furthering our understanding of how cells behave under virtually any circumstance. Relative to F-actin and microtubules, there is a general paucity of knowledge about the mechanisms responsible for the assembly, organization, and interphase dynamics of IFs in vivo.
Keratin IF proteins are encoded by a large family of conserved genes, numbering ∼54 in the human genome (Schweizer et al., 2006). As the most abundant structural proteins in the cytoplasm of most epithelial cells (e.g., ∼17–27% of total cellular proteins in newborn mouse keratinocytes; Feng et al., 2013), keratin IFs provide crucial structural and mechanical support to protect epithelial cells and tissues from mechanical and nonmechanical stresses alike (Fuchs and Cleveland, 1998; Coulombe and Omary, 2002; Herrmann et al., 2007). Interference with this structural and mechanical support role results in cell and tissue fragility and underlies many keratin-associated diseases, e.g., epidermolysis bullosa simplex (Omary et al., 2004; Coulombe et al., 2009; Seltmann et al., 2013). Unfortunately, there is no effective treatment for disorders rooted in defective IFs (McLean and Moore, 2011; Coulombe and Lee, 2012). A better understanding of the mechanisms regulating the assembly, organization, and dynamics of keratin IFs in vivo may well foster the development of strategies for the effective treatment of these disorders.
Studies published some time ago showed that epidermal keratins (e.g., K1 and K10) form disulfide-bonded polymers in vitro and in vivo (Sun and Green, 1978; Steinert and Parry, 1993). Intermolecular disulfide bonding was postulated, early on, to be important for the stabilization of keratin IFs (Giroud and Leblond, 1951) and formation of a cornified envelope at the final stage of terminal differentiation in the stratum corneum (Matoltsy and Matoltsy, 1970), but has been virtually ignored since then. Recently, our laboratory has reported that a trans-dimer, homotypic disulfide bond involving cysteine 367 (C367) in K14 occurs in a 3.0-Å crystal of the interacting 2B domains from human K5/K14 and in mouse epidermal keratinocytes in vivo, and we have implicated this bond in the genesis of a perinuclear network of keratin IFs able to impact the size and shape of the nucleus (Lee et al., 2012). To further explore the role of disulfide bonding, we devised a cysteine-free variant in K14 (K14CF) and characterized its properties toward the assembly and organization of keratin IFs in vitro and in transfected Krt14-null keratinocytes in primary culture, including live imaging studies. Reintroducing C367 back into the K14CF backbone reveals the central role of this residue toward the assembly, organization, and dynamics of K14-containing filaments in epidermal keratinocytes. The data we report point to a defining role for interkeratin disulfide bonding during keratin IF assembly, organization, and dynamics in skin keratinocytes.
Results
Disulfide bonding in the K5/K14 keratin pairing
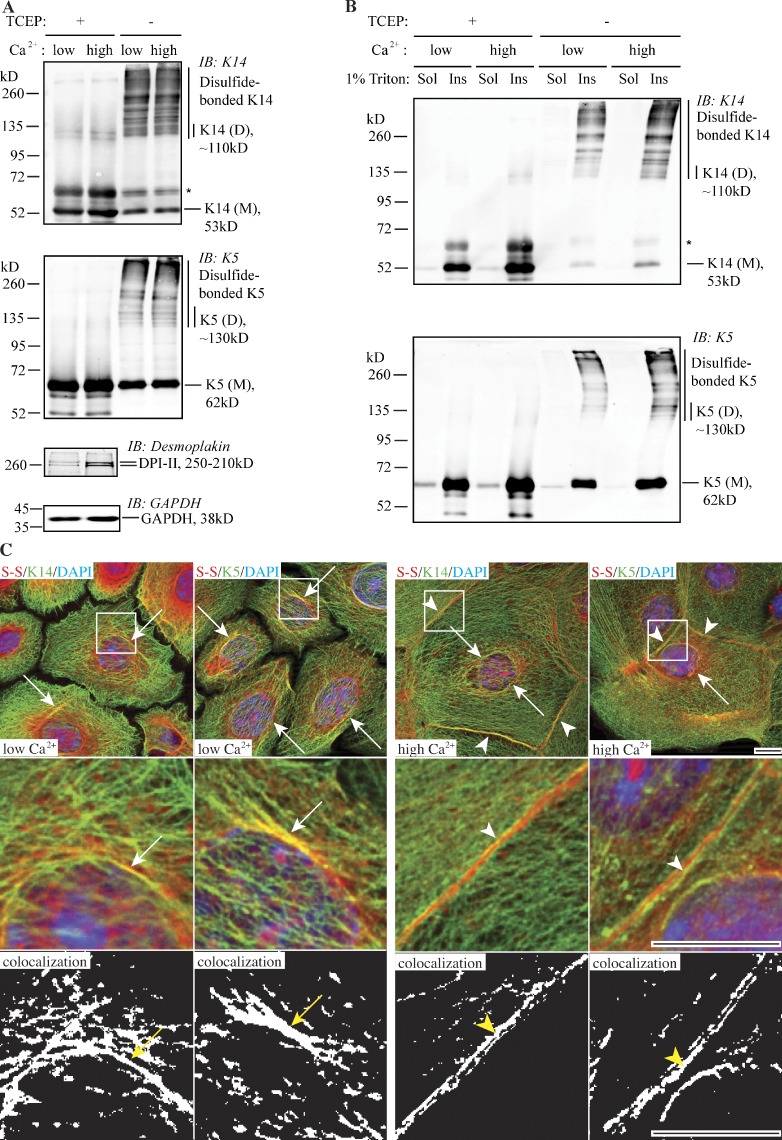
We recently described the occurrence of disulfide-bonded, homodimeric forms of the epidermal type I keratins K14 and K10 in mouse epidermis in situ, and how the elevated calcium concentration in the culture medium increases the yield of disulfide-linked K14 dimers in the mouse 308 skin keratinocyte cell line (Lee et al., 2012). Here, we examined disulfide linkages among keratins expressed in primary cultures of skin keratinocytes isolated from C57BL/6 newborn mice, with a focus on the K5-K14 pairing. We found that a sizable fraction of the K5 and K14 protein pools are engaged in intermolecular disulfide bonding in such cells, under both basal and calcium-promoted differentiation conditions (Fig. 1 A). Under the specific basal culture conditions we used, disulfide-bonded K5 and K14 in primary mouse keratinocytes consist of a series of high-molecular-weight species in addition to the dimers previously described for the 308 mouse keratinocyte line (Lee et al., 2012), which indicates that interkeratin disulfide bonding can be a quantitatively important event in normal mouse skin keratinocytes (Fig. 1 A). The total amount of disulfide-bonded K14 is modestly elevated upon calcium switch (Fig. S1 C), which is consistent with previous observations made in 308 cells (Lee et al., 2012). Irrespective of calcium concentration, the bulk of these disulfide-linked keratin species are Triton X-100–insoluble (Fig. 1 B), suggesting that they are part of the polymerized pool of keratin IFs.
Figure 1.
Prevalence of K5 and K14 disulfide bonding in wild-type newborn mouse skin keratinocytes in primary culture. (A) Immunoblotting analysis shows the occurrence of multiple disulfide-bonded species immunoreactive for K5 and K14 epitopes. M, keratin monomer; D, keratin dimer species. The asterisk denotes cross-reactivity of anti-K14 antibody with K5. (B) Analysis of the solubility of disulfide-bonded K5- and K14-containing species using detergent extraction and Western immunoblotting analysis. Sol, Triton-soluble fraction; Ins, Triton-insoluble fraction. (C) Immunofluorescence-based imaging for keratin (green channel) and disulfides (red channel) in mouse keratinocytes under low and high calcium culture conditions. The middle and bottom rows show magnified views of the boxed areas in the top row. The bottom row shows the spatial overlap between K5/K14-specific signals and the global signal for disulfide bonds. S-S, disulfide bonds. Arrows depict colocalization between K5/K14 and S-S in the perinuclear region. Arrowheads indicate colocalization at the cell–cell interface. Bar, 10 µm.
We next analyzed the localization patterns of keratin-associated disulfide bonds in mouse keratinocytes in primary culture. In cell preparations fixed with methanol (reflecting insoluble material), part of the signal for disulfide bonding manifests as fibrous elements in the cytoplasm and accumulated in the perinuclear region (Fig. S1, A and B). When performing dual staining, the respective signals for K5/K14 and disulfide bonds colocalize in the cytoplasm and perinuclear region under low-calcium culture conditions. Upon calcium switch, cells display a distinct colocalization pattern for keratins and disulfides, particularly at the cell–cell interfaces where stable desmosome cell–cell contacts are now present (Fig. 1 C). We observed no significant difference in perinuclear colocalization in mouse keratinocytes in primary culture in low versus high calcium conditions. This is different from what we previously reported for 308 mouse skin keratinocytes (Lee et al., 2012), and is likely due to the intrinsic difference between these two types of cells and the specific culture conditions used. These findings suggest that the formation and/or maintenance of a perinuclear-concentrated network is a significant occurrence in normal mouse epidermal keratinocytes in primary culture, under a range of culture conditions.
Formation of disulfide bonds limits elongation of K5/K14 filaments in vitro
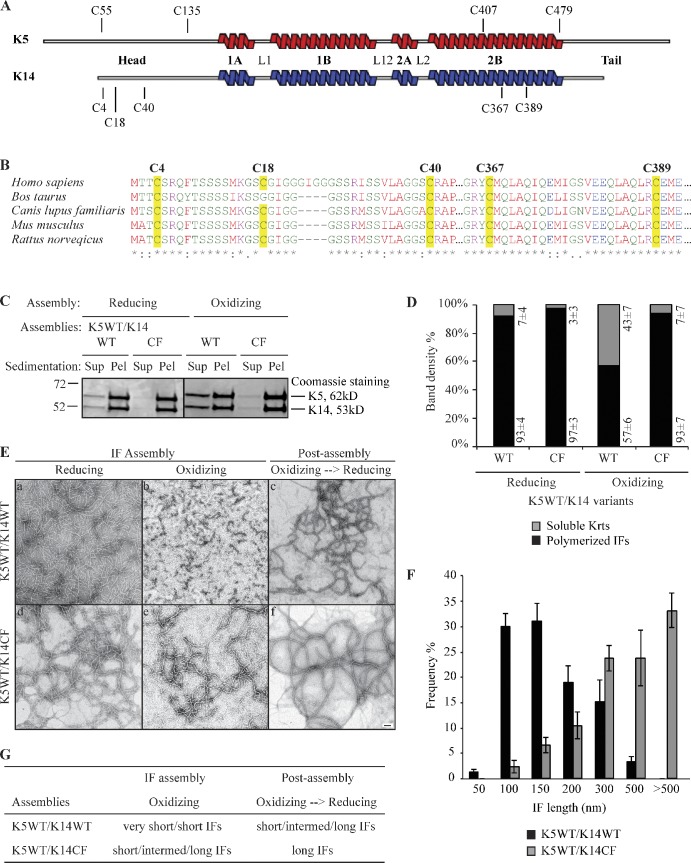
Human K14 protein contains five cysteine residues: C4, C18, and C40 in its N-terminal head domain, and C367 and C389 in its central rod domain, all of which are conserved across mammals (Fig. 2, A and B). Residue C367, but not C389, was found to mediate a trans-dimer homotypic disulfide bond in a crystal of interacting K5/K14 2B rod domains (Lee et al., 2012). We devised a cysteine-free variant of K14, designated K14CF, in which all five cysteine residues are substituted with alanines, to examine the impact of eliminating all K14-dependent disulfide bonding for filament assembly and network formation.
Figure 2.
Limitation of filament elongation via the formation of K14-dependent intermolecular disulfide bonds in vitro. (A) Location of cysteine residues in human K5 and K14 proteins. (B) Alignment of K14 sequence from several mammals: Homo sapiens (NP_000517), Bos Taurus (NP_001160047), Canis lupus familiaris (NP_001240670), Mus musculus (NP_058654), and Rattus norvegicus (NP_001008751). Cysteine residues are highlighted in yellow. (C) High-speed sedimentation of keratin assemblies (150,000 g, 30 min) followed by reducing SDS-PAGE analysis of supernatant (Sup) and pellet (Pel) fractions. K14WT and K14CF have different assembly efficiencies when coassembled with K5WT under oxidizing buffer conditions. The black line indicates that intervening lanes have been spliced out. (D) Quantitation of filament assembly efficiency by densitometry-based analysis of supernatant (S) and pellet (P) fractions as reported in C. Data are presented as the mean ± SD from three independent experiments. (E) Negative staining and transmission electron microscopy showing the structural features of K14WT- and K14CF-containing filaments in an oxidized environment, and upon exposure to a reducing environment at the post-assembly stage. Bar, 100 nm. (F) Measurements of filament length when assembled under oxidizing buffer conditions. Data were obtained from three independent experiments. At least 100 filaments were measured per experiment. Error bars represent SD. (G) Summary of in vitro assembly data for K5/K14WT and K5/K14CF under different buffer conditions. “Very short IFs” are <150 nm, “short IFs” are 200–500 nm, “intermed IFs” are 500–1,000 nm, and “long IFs” are >1 µm.
In vitro, K14CF polymerizes with high efficiency (∼98%) and forms normal 10-nm filaments when coassembled with wild-type K5 under standard polymerization buffer conditions (which include a reducing agent; Aebi et al., 1983). This is confirmed by comparing K5/K14CF to K5/K14WT assemblies using either negative staining/electron microscopy or high-speed sedimentation assays (Fig. 2, C–E). Along with the outcome of transfection assays in cultured Krt14−/− keratinocytes (see the next section), these findings suggest that replacing Cys with Ala at amino acid positions 4, 18, 40, 367, and 389 in K14 does not significantly impair its assembly competency, owing to a biochemical/structural defect.
Performing the in vitro assembly reactions in the absence of a reducing agent provides an oxidation-prone environment that allows for the formation of inter- and/or intramolecular disulfide bonds, and yielded different results. Under such conditions, K14WT-containing assemblies display a series of disulfide-bonded species that, as shown by Western blotting, show retarded mobility and involve both K14 and K5. By comparison, K14CF-containing assemblies exhibit only K5-mediated disulfide bonds (Fig. S2), establishing that cysteines in K5 also contribute to formation of intermolecular disulfide bonding in vitro. In addition, the K5/K14WT pairing polymerizes with significantly lower efficiency (∼35% reduction in the high speed sedimentation assay) and yields short filaments (∼100–150 nm in length, as measured on electron micrographs), whereas the K5/K14CF pairing maintains high assembly efficiency and produces longer filaments (mostly >300 nm; Fig. 2, D and F). These findings are consistent with early observations by Steinert et al. (1976), and indicate that formation of intermolecular disulfide bonds limits the elongation of (wild-type) K5/K14 filaments in vitro. Disulfide bonding involving K5 (which features four Cys residues; Fig. 2 A) likely accounts for the subnormal length of K14CF-containing filaments under oxidizing buffer conditions (Fig. 2 E). Of note, the length of K5/K14WT-containing filaments normalizes when final assemblies are directly transferred to the standard reducing buffer environment at the post-assembly stage (Fig. 2, E and G). These observations are significant, as they suggest that an oxidation-prone environment might regulate the elongation of K5/K14 filaments in epidermal keratinocytes in vivo.
Loss of K14-bound cysteines and/or disulfide bonding interferes with the formation and/or maintenance of perinuclear-concentrated IF networks in keratinocytes
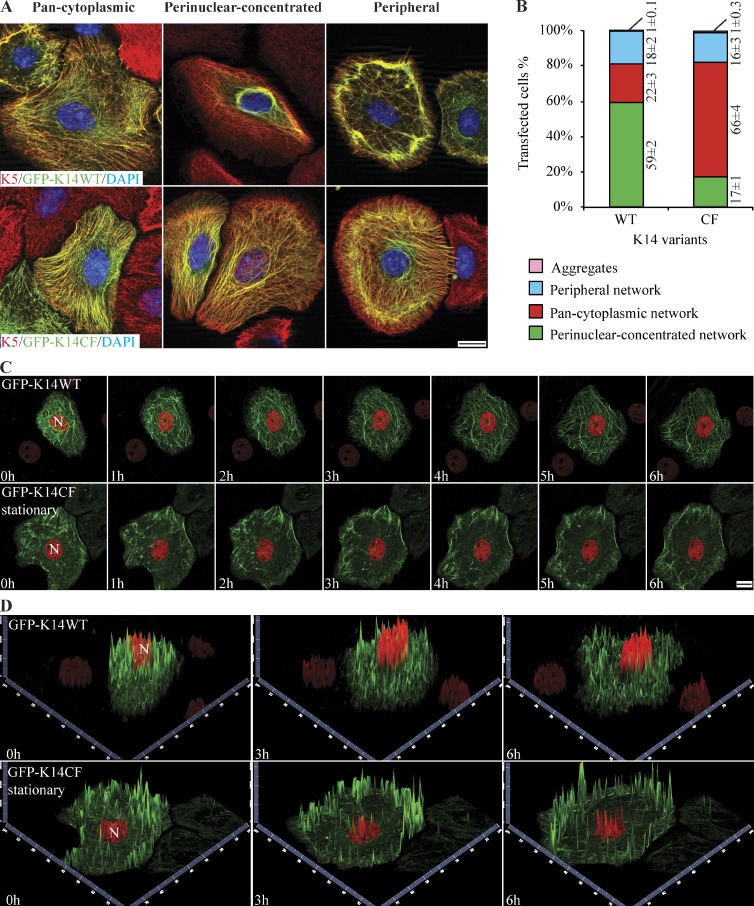
In Lee et al. (2012), we proposed that a homotypic, K14 Cys367-mediated trans-dimer disulfide bond contributes to the formation of a perinuclear-concentrated network of keratin IFs. We next tested whether the complete loss of K14-bound cysteines and/or disulfide bonding interferes with the intracellular organization of keratin IFs in keratinocytes by conducting transfection assays in Krt14−/− mouse keratinocytes in primary culture, which are devoid of K14 but harbor K5/K17 and K5/K15 preexisting IF networks (Chan et al., 1994; Lloyd et al., 1995). Expression of GFP-K14WT in Krt14−/− keratinocytes reveals three modes of keratin network organization in fixed preparations subjected to fluorescence imaging: “pan-cytoplasmic” (i.e., keratin filaments appear evenly distributed throughout the cytoplasm), “perinuclear-concentrated” (most filaments are concentrated in the perinuclear region), and “peripheral” IF networks (filaments are concentrated at the cell periphery; Fig. 3 A; see Fig. S3 for quantitative definition of these categories). The predominant mode of IF organization for GFP-K14WT is perinuclear-concentrated, occurring in 59% of transfected cells, whereas the pan-cytoplasmic mode of IF organization occurs in 22% of cells (Fig. 3 B). By comparison, GFP-K14CF–containing filaments exhibit perinuclear-concentrated IF networks in only 17% of transfected Krt14−/− keratinocytes, whereas a high proportion of them (66%) show pan-cytoplasmic networks (Fig. 3, A and B). These findings suggest that loss of K14-bound cysteines and/or disulfide bonding has no obvious impact on 10-nm filament formation but interferes with the formation and/or maintenance of perinuclear-concentrated IF networks in mouse skin keratinocytes in primary culture. Importantly, the observation of an incomplete loss of perinuclear-concentrated networks in K14CF-expressing cells may reflect contributions from other type I keratins, e.g., K16 and K17, as they both contain several cysteines, including one that corresponds to K14’s C367, along with the fact that K17 is up-regulated in Krt14−/− mouse keratinocytes (Troy and Turksen, 1999; unpublished data). K15, another type I keratin expressed under these conditions, likely does not play a role in this incompletely penetrant phenotype because it is missing a cysteine residue at the position corresponding to K14’s C367 (Lee et al., 2012).
Figure 3.
Formation and/or maintenance of perinuclear keratin networks are compromised by loss of K14-bound cysteines and/or disulfide bonding. (A) Immunofluorescence imaging reveals three distinct modes of organization for K14-containing filament networks in primary culture of Krt14−/− mouse keratinocytes transfected with either GFP-K14WT or GFP-K14CF: “pan-cytoplasmic,” “perinuclear-concentrated,” and “peripheral” IF networks. Bar, 10 µm. (B) Frequency of each type of IF network as illustrated in A. Data are presented as mean ± SD from three independent experiments. (C) Still images from live movie recordings comparing keratinocytes expressing GFP-K14WT and GFP-K14CF (green channel). H2B-mCherry (red channel) was cotransfected to visualize nuclei. The images shown correspond to seven time points, from 0 to 6 h, in movies recorded using laser scanning confocal microscopy (see Videos 1 and 2). N, nucleus. Bar, 10 µm. (D) 2.5-dimensional views of z-stacked images at time 0, 3, and 6 h show GFP-K14WT– or GFP-K14CF-containing networks in the cytoplasm (green channel) and H2B-mCherry–labeled nucleus (red channel) in transfected keratinocytes.
We surmised that the three distinct modes of keratin filament network organization observed in fixed cell preparations (Fig. 3 A) could represent distinct stages along a temporal continuum rather than terminal or stable phenotypes. Consistent with the former, R.E. Leube and colleagues have shown that keratin IFs undergo a spatially and temporally patterned cycle in cultured epithelial cells: very short filaments are first assembled at the cell periphery, then elongate as they move in as part of a continuous flow toward to the cell center, where they become bundled and form a perinuclear-concentrated network (e.g., Windoffer and Leube, 2004, 2011; Kölsch et al., 2010). By imaging live cells using laser-scanning confocal microscopy, we analyzed IF dynamics in Krt14−/− mouse keratinocytes expressing either GFP-K14WT or GFP-K14CF (Fig. 3, C and D; and Videos 1 and 2). A total of seven z stacks (0.43-µm-thick each, totaling 3 µm in thickness) were acquired for each position at every time point. In mouse keratinocytes, GFP-K14WT behaves in a manner consistent with the keratin cycle described by Leube and colleagues (Windoffer et al., 2011): short filaments appear to form at the cell periphery, then elongate as they move directionally and accumulate in the perinuclear region (Fig. 3 C and Video 1). Even though it is expressed in amounts comparable to its wild-type counterpart (Fig. S4 B), GFP-K14CF appears to carry out only half of the cycle: short filaments do form in the cell periphery and initiate progress toward the cell center, but the process becomes disrupted at the midpoint of the cell such that few long filaments make it stably to the perinuclear region (Fig. 3 C and Video 2). Instead, discrete “small dots” containing GFP-K14CF or “empty space” is what prevails near the nucleus. We generated 2.5-dimensional views of z stack images (Fig. 3 D), and these confirm that GFP-K14WT–containing filaments readily form perinuclear-concentrated networks that tightly wrap around H2B-mCherry–labeled nuclei, whereas the GFP-K14CF–containing filaments are concentrated at the cell periphery and in the mid-cytoplasm, but conspicuously lack in the perinuclear region of the cell. Furthermore, we observed that GFP-K14CF–expressing cells tend to exhibit markedly enhanced movements of the nucleus and the whole cell (>60% of cells; Fig. S5, A, B, D, and F; and Video 3) relative to GFP-K14WT–expressing cells (<15% of cells; Fig. S5, C and F). Together, these data suggest that K14-bound cysteines and/or disulfide bonding play an important role in the keratin cycle whereby they regulate the progress toward, and/or the stability of, the perinuclear network of keratin IFs. Further, in addition to impacting nuclear dimensions as reported (Lee et al., 2012), live imaging studies revealed that loss of the perinuclear keratin network is associated with enhanced movement of the nucleus and the whole cell.
Transfection of a K14 variant harboring a single cysteine residue at position 367 partially rescues the keratin cycle and perinuclear IF organization
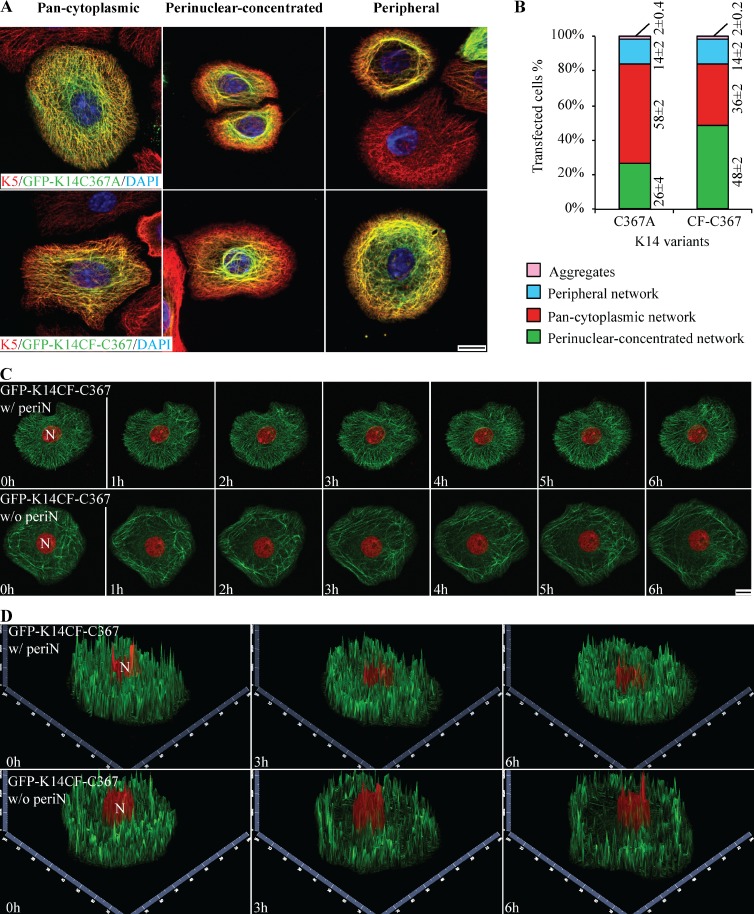
We reported that replacing Cys with Ala at position 367 (K14C367A variant) impairs K14’s ability to impact the size and the shape of the nucleus upon transfection in a tumor-derived keratinocyte cell line (Lee et al., 2012). We engineered the same variant in the GFP vector backbone (GFP-K14C367A), and engineered a new, complementary K14 variant by putting a cysteine back at position 367 in the K14CF backbone (K14CF-C367 variant; Fig. S4 A). We then examined the impact of expressing these two variants in Krt14−/− mouse keratinocytes in primary culture. GFP-K14C367A–expressing keratinocytes show a significant decrease in the frequency of perinuclear IF networks (seen in only 26% of cells; Fig. 4, A and B), a finding that is consistent with Lee et al. (2012). However, 48% of cells expressing the GFP-K14CF-C367 variant display a perinuclear-concentrated network (Fig. 4, A and B). This represents a significant increase relative to cells expressing the parental GFP-K14CF backbone (see Fig. 3 B).
Figure 4.
Partial rescue of perinuclear-concentrated networks in Krt14−/− keratinocytes transfected with K14CF-C367. (A) Immunofluorescence-based imaging shows three modes of keratin filament network organization resulting from overexpression of GFP-tagged K14C367A and K14CF-C367 in Krt14−/− mouse keratinocytes in primary culture. Bar, 10 µm. (B) Population frequency of each type of IF network as illustrated in A. Data are presented as mean ± SD from three independent experiments. (C) Still fluorescence images from video recordings of live mouse keratinocytes expressing GFP-K14CF-C367 (green channel). H2B-mCherry (red channel) was cotransfected to visualize nuclei. Seven time points from 0 to 6 h are shown (see Videos 4 and 5). Maximum intensity projections identify two different populations of mouse keratinocytes expressing the GFP-K14CF-C367: with (w/periN) or without (w/o periN) perinuclear-concentrated networks (45% vs. 55%, respectively). Bar, 10 µm. (D) 2.5-dimensional views of z-stacked images corresponding to the two-dimensional images at time 0, 3, and 6 h in C.
Next, GFP-K14CF-C367–expressing Krt14−/− mouse keratinocytes were subjected to live cell imaging. Two distinct cell populations are observed (Fig. 4, C and D): 45% of transfected cells show a normal keratin cycle whereby keratin filaments elongate and continuously move toward the nucleus, thereby forming a perinuclear-concentrated network of filaments (Video 4), whereas 55% of cells display an impaired keratin filament cycle and, ultimately, fail to form a perinuclear network even though filament elongation initially appears to be normal (Video 5). Quantitation of fluorescence intensity suggests that differences in expression levels do not account for these two behaviors (Fig. S4 C). It seems likely, then, that the difference reflects a partial involvement of other cysteines in K14, or in K16 and/or K17. Altogether these findings indicate that C367 in K14 participates in, but does not completely suffice for, the establishment of a normal filament cycle along with normal perinuclear keratin filament network organization in skin keratinocytes. Besides, we observed that keratinocytes expressing GFP-K14CF-C367 show a marked normalization of the very abnormal cell motility behavior of Krt14−/− keratinocytes (Fig. S5, E and F), which suggests that C367 in K14 plays an important role in this regard.
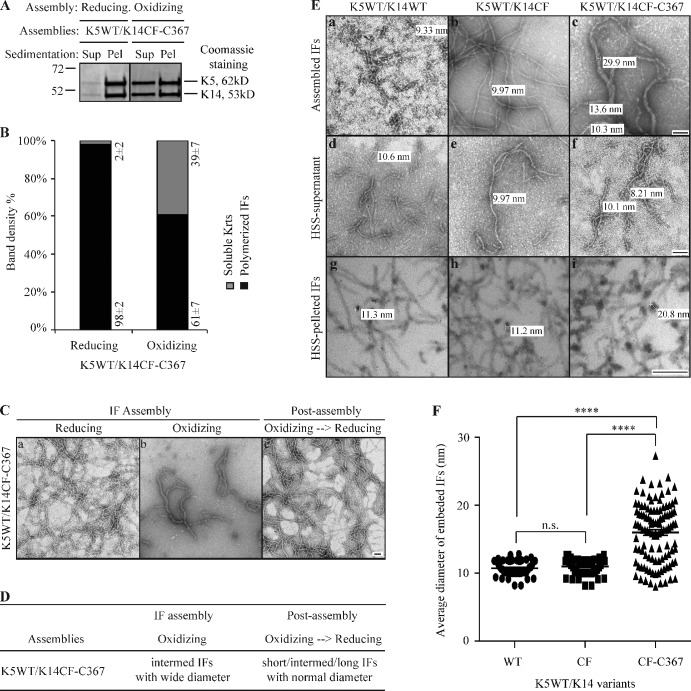
Formation of K14CF-C367–mediated interdisulfide bonds yields abnormal IFs with a large and variable diameter in vitro
Residue Cys367 in K14 is conserved in several type I keratins, including K10, K16, and K17, and is located in the stutter region, a four-residue interruption in the long-range heptad repeats that is conserved in the 2B portion of the central rod domain in all IF proteins (Lee et al., 2012). Position 367 in K14, or the corresponding position in other type I keratins, has not been found to be mutated in the setting of genodermatoses (Szeverenyi et al., 2008). To assess whether formation of disulfide bonds via this specific residue’s side chain impacts keratin IF structure, we performed in vitro assembly assays with purified proteins under oxidizing buffer conditions (see Fig. 2). Relative to K5/K14WT, K5/K14CF-C367 assemblies are very abnormal (Fig. 5, A–C). Specifically, filaments are intermediate-to-long in length and feature a variable, often wider-than-normal diameter (Fig. 5 C), correlating with the formation of intermolecular disulfide bonds mediated by C367 (Fig. S2). Importantly, the various anomalies exhibited by K5/K14CF-C367 filaments are rescued when disulfide bonds are reduced at the post-assembly stage (Fig. 5, C and D).
Figure 5.
Formation of K14CF-C367–mediated disulfide bonds yields abnormal IFs with wide diameters in vitro. (A) High-speed sedimentation assay (150,000 g, 30 min) followed by SDS-PAGE analysis of supernatant (Sup) and pellet (Pel) fractions. K14CF-C367 was coassembled with K5WT under both reducing and oxidizing buffer conditions. (B) Quantitation of filament assembly efficiency using densitometry-based analysis of the corresponding fractions in A. Data are presented as mean ± SD from three independent experiments. (C) Negative staining and transmission electron microscopy reveals the abnormal morphology of K14CF-C367–containing filaments under oxidizing buffer conditions, which can be restored upon exposure to a reducing environment at the post-assembly stage. Bar, 100 nm. (D) Summary of in vitro assembly data for K5/K14CF-C367 under various buffer conditions. Short IFs, 200–500 nm; intermed IFs, 500–1,000 nm; long IFs, >1 µm; normal diameter, 8–12 nm; wide diameter, >13 nm. (E) Morphology of WT, cysteine-free (CF), and CF-C367 of K14-containing filaments assembled under oxidizing buffer conditions. The top row shows examples of negatively stained intact assemblies upon completion of the reaction, while the middle and bottom row are examples, respectively, of negatively stained supernatant fractions after a high-speed sedimentation assay and cross sections of epoxy resin-embedded pellet fractions. HSS, high-speed sedimentation. Bar, 100 nm. (F) Measurements of diameters of cross-sectioned filaments corresponding to HSS-pelleted IFs (bottom row in E). More than 50 cross sectioned IFs were measured for K14WT and K14CF, and >100 cross sectioned IFs were measured for K14CF-C367. P < 0.0001 (****).
Further analysis of IFs reconstituted in vitro from the K5/K14WT, K5/K14CF, and K5/K14CF-C367 pairings highlights interesting differences. Thus, in spite of differences in their length, K14WT- and K14CF-containing filaments show a normal diameter (∼10–11 nm) whether the filaments are directly visualized by negative staining or first subjected to pelleting, epoxy resin-embedding, and sectioning before transmission electron microscopy (Fig. 5 E, top and bottom rows). In striking contrast, the diameter of K14CF-C367–containing filaments is variable and can be threefold wider (Fig. 5 E, top and bottom rows). Consistent with the current understanding of IF assembly and structure (Herrmann et al., 2007; see Discussion), three main possibilities might lead to a wider diameter of IFs: (1) two or more filaments intertwining along one another, (2) loss of control of subunit number per filament cross section, and (3) loss of “compaction” (filament narrowing) at a late stage of assembly. To help discriminate among these three possibilities, we resolved the soluble and insoluble fractions via high-speed sedimentation, and observed that all three samples (including K5/K14CF-C367) contain short filaments with a normal diameter in the supernatant fraction (see middle row in Fig. 5 E). Analysis of pellet fractions by epoxy-embedding, thin-sectioning, and electron microscopy with a focus on filaments cross-sectioned at right angles showed that K5/K14WT and K5/K14CF filaments have a normal diameter (8–12 nm), whereas the K5/K14CF-C367 exhibit a variable diameter ranging from 8 to 27 nm (bottom row in Fig. 5 E; and Fig. 5 F) but do not appear to be composites of multiple 10-nm filaments. The latter implies that, when present as the sole cysteine residue in K14, C367 interferes with the control of subunit number per filament and/or with compaction-maturation during assembly in vitro.
Discussion
The notion that disulfide bonding may be important for keratins and play a functional role at the final stage of terminal differentiation in epidermis is more than 60 years old (Giroud and Leblond, 1951). Yet, very few studies (e.g., Steinert et al., 1976; Sun and Green, 1978) have examined this phenomenon in any detail. Our interest in this issue originates in the discovery of a homotypic, trans-dimer disulfide bond mediated by K14’s C367 in the crystal structure of a heterocomplex of the L2-2B domains of K5/K14 and in mouse epidermal keratinocytes in vivo (Lee et al., 2012). The findings we report here (Table 1) significantly extend previous work by P. Steinert and colleagues in showing that the formation of disulfide bonding limits the axial growth (elongation) of keratin filaments during in vitro assembly (Steinert et al., 1976). We demonstrate, for the first time, that interkeratin disulfide bonding is also required for normal keratin cycling and formation and/or maintenance of a perinuclear network of keratin IFs, the latter being a defining feature in keratinocytes of the epidermis and related surface epithelia. Our findings also implicate a specific cysteine residue in K14, C367, in these capabilities, thus significantly extending the work of Lee et al. (2012). These observations introduce a completely new and exciting element in our quest for understanding the determinants of keratin IF assembly, organization, and dynamics in epithelial cells.
Table 1.
Summary of the assembly properties of K14WT and the K14CF and K14CF-C367 variants
| Settings | Features | K14 variant analyzed | ||
| K14WT | K14CF | K14CF-C367 | ||
| In vitroa (purified proteins) | ||||
| Reducing conditions | IF length | Long | Long | Long |
| IF width | Normal | Normal | Normal | |
| Assembly efficiency | High | High | High | |
| Oxidizing conditions | IF length | Very short | Intermediate | Intermediate |
| IF width | Normal | Normal | Aberrant | |
| Assembly efficiency | Reduced | High | Reduced | |
| Oxidizing→reducing | IF length | Long | Long | Long |
| IF width | Normal | Normal | Normal | |
| Assembly efficiency | High | High | High | |
| Krt14−/− newborn skin keratinocytes in primary cultureb | Frequency of perinuclear-concentrated networks | High | Low | Significantly restored (i.e. wild type–like) |
| Frequency of pan-cytoplasmic networks | Low | High | Significantly restored | |
| Keratin cycling | Normal | Impaired | 45%: Significantly restored55%: Impaired | |
| Movement of nucleus and whole cell | Limited | Exaggerated | Significantly restored | |
(1) Formation of disulfide bonds limits the elongation of filaments; this effect is fully reversible. (2) When present as the only cysteine residue in K14, C367 interferes with elongation and causes a dramatic loss of filament width control.
(1) K14-bound cysteines and/or disulfide bonds are required for the centripetal keratin cycle to occur in live cells. (2) When present as the only cysteine residue in K14, C367 significantly rescues the keratin cycle, even though it exhibits aberrant properties in vitro.
The prevailing view regarding the pathway to IF assembly is based on in vitro polymerization assays and recognizes three main stages: (1) formation of “unit length filaments” (ULFs), which are short (50–60 nm for vimentin and 50–120 nm for keratins K8/K18) but exhibit the full complement of subunits per filament cross section; (2) annealing of ULFs to yield long filaments (elongation); and (3) compaction of immature filaments across their lateral axis to reduce their diameter from ∼16 nm to ∼10 nm (Herrmann et al., 1999, 2002). This model was borne out of studies focused on vimentin, a type III IF protein that can readily homopolymerize into mature 10-nm filaments, and has since then been extended to many other types of IF proteins (Herrmann et al., 2002). The extent to which this pathway is physiologically relevant and applies to keratin proteins, which polymerize very rapidly (Lichtenstern et al., 2012) under atypically low ionic strength buffer conditions in vitro (Steinert et al., 1976; Aebi et al., 1983), remains unclear. Meanwhile, Steinert’s data on K1-K10 (Steinert and Parry, 1993), and ours on K5-K14, clearly implicate that, when allowed to occur, disulfide bonding interferes with the elongation of keratin filaments during in vitro assembly. The short filament species (100–150 nm range) that form under oxidizing conditions in vitro can be viewed, at least conceptually, as ULF-like. Our data show that the interference with filament elongation is fully reversible for wild-type K14, and significantly mitigated when using a cysteine-free form of K14 in vitro. Our findings also show that formation of normal-looking IFs requires cysteines when K14 is expressed in vivo, and that disulfide bonds form primarily between, rather than within, keratin molecules in keratinocytes. Clearly, therefore, the formation (and dynamic regulation) of disulfide bonds regulates both the assembly and organization of keratin filaments in vitro and in vivo. Beyond its conceptual value, this newly uncovered property may provide a path to isolating and characterizing relevant keratin assembly precursors from cellular sources.
Assembly of the cornified envelope at a late stage of terminal differentiation in epidermis sets a precedent regarding biochemical circumstances allowing for the formation of disulfide linkages in the cytoplasm of keratinocytes (Ma and Sun, 1986), an environment that is generally considered as strongly reducing (Krebs 1967). In this regard, residue C367 in K14 is conserved in the corresponding position of K10, the major type I keratin in differentiating epidermis (Lee et al., 2012). K10 can be retrieved as a disulfide-bonded homodimer from skin tissue, and as part of larger oligomers along with its partner K1 in vivo (Sun and Green, 1978; Lee et al., 2012). Another physiologically prominent example of interkeratin disulfides occurs in hard epithelia such as hair and nail, which express a distinct set of highly cysteine-rich keratin proteins (Fraser et al., 2003). In hair trichocytes, keratin subunits “slip” at a specific stage of maturation so as to align and bring the side chains of cysteines closer to one another, promoting the formation of intrafilamentous disulfide bonds that are believed to increase the resilience of IFs and the entire cell and tissue during terminal differentiation (Wang et al., 2000; Fraser and Parry, 2007, 2012, 2014). Based on the literature, both enzymatic and nonenzymatic mechanisms could be involved in catalyzing interkeratin disulfide bonds in the cytoplasm of epidermal or hair keratinocytes. A family of sulfhydryl oxidases has been described as being involved in catalyzing the formation of disulfide bonds in keratinocytes (Hashimoto et al., 2001). Otherwise, changes in redox potential, and/or oxidative stress, have been shown to promote terminal differentiation in epidermis (see Thiele et al., 2001; Hamanaka et al., 2013; Kennedy et al., 2013). Though the cytoplasm is generally considered as a reducing environment, genuine differences in the redox potential do occur across the cytoplasmic space, and are seemingly organized in a gradient: the cell periphery and lysosomes are oxidizing, the general cytoplasm is reducing, and the perinuclear region and mitochondria are oxidizing as well (Clarke et al., 2006). An intriguing possibility that takes into account Leube’s keratin cycle (Windoffer et al., 2011) is that interkeratin disulfide bonding may initially occur at the cell periphery to limit filament elongation and thereby pace the process of 10-nm filament assembly, in order to then be modulated (i.e., decreased or reconfigured) to allow for filament elongation, and later on promoted again in the perinuclear space of skin keratinocytes (see Lee et al., 2012). Differences among keratin proteins, e.g., in terms of the number and location of cysteine residues, as well as differences in redox gradients or enzyme activities among various types of cells, are likely to impact this hypothetical scenario. Of course, mechanisms other than disulfide bonding are most certainly at play in a variety of settings to promote the formation and/or maintenance of a dense perinuclear network of IFs.
In addition to defining the mechanisms involved, two important issues that need attention in the short term are the physiological relevance of interkeratin disulfide linkages involving K14, and the general relevance of K14’s case for other keratins and IF proteins. The issue of functional relevance can be best addressed by generating knock-in alleles, with an obvious first possibility being a C367A substitution in the Krt14 locus, in the germ line of mice, and ascertain the consequences for epidermal structure and function. Interestingly, Werner et al. (2004) have shown that a hot spot mutation, Arg125-Cys, which maps to the amino-terminal part of K14’s rod domain (Coulombe et al., 1991) and is responsible for ∼40% of EBS cases (Coulombe et al., 2009), disrupts the keratin cycle at an early stage in live transfected epithelial cells. Omary and his colleagues showed that mutant G62C, which introduces a Cys residue in an otherwise cysteine-free K8 and is associated with acute and chronic human liver diseases (Omary et al., 2009; Strnad et al., 2010), decreases keratin solubility and impairs K8/K18 filament reorganization upon oxidative stress in transfected cells (Ku et al., 2001, 2005). Though de novo cysteine introduction occurs in only ∼5% of disease-associated keratin missense mutations (Strnad et al., 2011), such observations, along with ours, raise the prospect that aberrant disulfide bonding could be at play in select cases of keratinopathies. For the second issue, our data underscore the participation of cysteines other than C367 in K14-dependent disulfide bonds (Lee et al. 2012; this study) as well as the existence of K5- and K10-dependent bonds in skin keratinocytes. As virtually all IF proteins (i.e., in addition to the so-called sulfur-containing hard keratins) feature cysteine residues, it appears likely that disulfide bonding will emerge as a significant mechanism regulating the assembly, organization, and function of IFs in vivo.
Materials and methods
Plasmids
For in vitro studies, plasmids pT7HMT-K14WT and pT7HMT-K5WT were created by subcloning the relevant cDNAs (1,419 bp for K14WT; 1,773 bp for K5WT) into a pT7HMT modified vector (which features a T7 promoter) using NcoI–BamHI and NdeI–BamHI restriction sites, respectively. For expression in mammalian cells, plasmids pBK-CMV His-GFP-K14WT and pBK-CMV His-GFP-K14C367A were generated by subcloning the cDNAs for K14WT and K14C367A fused to a His-GFP tag at the N terminus into a pBK-CMV vector backbone using SalI–NotI restriction sites (Lee et al., 2012). The cysteine-free variant of K14 (K14CF) was generated by replacing all five cysteines in K14WT with alanines using a QuikChange lightning multi site-directed mutagenesis kit (Agilent Technologies). Otherwise, cysteine point mutations were introduced into the K14WT cDNA via a QuikChange lightning site-directed mutagenesis kit (Agilent Technologies). For live cell imaging, plasmid pShuttle-CMV-H2B-mCherry was created by subcloning the H2B-mCherry cDNA (1,098 bp) from plasmid PGK-H2BmCherry (Addgene) into a pShuttle-CMV backbone (a gift from B. Vogelstein, Johns Hopkins University School of Medicine, Baltimore, MD) via KpnI–HindIII restriction sites.
Antibodies and other reagents
The following antibodies were used: anti-K14 (rabbit, 1:1,000 for immunofluorescence, 1:5,000 for immunoblotting) and anti-K5 (rabbit, 1:1,000 for immunofluorescence, 1:3,000 for immuno-blotting) were from Covance, anti-desmoplakin (a rabbit polyclonal antibody generated against a C-terminal fragment conserved in desmoplakin I and II; 1:2,000; Angst et al., 1990) was a gift from K. Green (Northwestern University Feinberg School of Medicine, Chicago, IL), anti-GAPDH (rabbit, 1:1,000) and anti-protein disulfide isomerase (PDI; rabbit, 1:1,000) were from Cell Signaling Technology, IRDye 800CW goat anti–rabbit IgG (1:20,000) was from LI-COR Biosciences, and Alexa Fluor 594 goat anti–rabbit IgG (1:1,000) and Alexa Fluor 488 goat anti–rabbit IgG (1:1,000) were from Life Technologies. Alexa Fluor 594 C5-maleimide and DAPI were from Life Technologies, tris(2-carboxyethyl) phosphine (TCEP) was from Thermo Fisher Scientific, and N-ethylmaleimide (NEM) was from Sigma-Aldrich. DMEM/calcium free/low glucose and DMEM/calcium free/low glucose/no phenyl red were used in preparation of flavin adenine dinucleotide (FAD) medium, and FAD imaging medium was from US Biologicals. Restriction endonuclease products were from New England Biolabs, Inc. All other chemicals were from Sigma-Aldrich unless noted otherwise.
Keratin purification, assembly of keratin filaments in vitro, and polymerization efficiency
pT7HMT-K5WT, pT7HMT-K14WT, pT7HMT-K14CF, and pT7HMT-K14CF-C367 were transformed into Escherichia coli BL21(DE3) to produce the corresponding recombinant human keratin proteins as inclusion bodies. Keratin proteins were purified using HiTrapQ and MonoQ columns (GE Healthcare) as described previously (Lee and Coulombe, 2009). Inclusion bodies were dissolved in urea “buffer A” (50 mM Tris-HCl, 6.5 M urea, 1 mM EGTA, 1 mM PMSF, and 2 mM dithiothreitol, pH 8.5), and then applied onto a HiTrapQ column followed by a MonoQ column. Keratin proteins were eluted using a linear gradient elution of urea-guanidine hydrochloride “buffer B” (50 mM Tris-HCl, 6.5 M urea, 0.5 M guanidine hydrochloride, 1 mM EGTA, 1 mM PMSF, and 2 mM dithiothreitol, pH 8.5). Heterotypic complexes containing K5 and K14 in an equimolar ratio were prepared and purified using the MonoQ column (Lee and Coulombe, 2009). The concentration of purified keratin complexes was adjusted to 0.15 mg/ml for filament assembly experiments. Keratin mixtures were assembled via a serial dialysis using the following three buffers: (1) 9 M urea and 25 mM Tris-HCl, pH 7.5, for 4 h at room temperature; (2) 2 M urea and 5 mM Tris-HCl, pH 7.5, for 1 h at room temperature; and (3) 5 mM Tris-HCl, pH 7.5, overnight at 4°C. The presence or absence of a reducing reagent β-mercaptoethanol was used to provide a reducing or oxidizing environment during keratin assembly. In some experiments, keratin filaments assembled under oxidizing conditions were subjected to reducing conditions at the post-assembly stage. The physical state of the keratin polymers was analyzed using a high-speed sedimentation assay (150,000 g for 30 min; Ma et al., 2001), followed by SDS-PAGE of the pellet and supernatant fractions and densitometry using ImageJ (Schneider et al., 2012). Data are presented as mean ± SD from three independent experiments.
Transmission electron microscopy
Ultrastructure of keratin assemblies was examined by negative staining (1% uranyl acetate) and transmission electron microscopy using either 7600 or HU-12A instruments (Hitachi). Measurement of filament length was performed on recorded micrographs (80,000× nominal magnification) using the AmtV602 and ImageJ freeware programs. Data are presented as mean ± SD from three independent experiments. To access keratin filaments in cross section, polymerized keratins were pelleted by high-speed sedimentation and fixed in 2% glutaraldehyde, 0.2% tannic acid, 0.1 M cacodylate buffer, pH 7.2, overnight at 4°C, post-fixed in 2% osmium tetroxide/0.1 M cacodylate buffer for 30 min on ice, en bloc stained with 2% uranyl acetate for 1 h at room temperature, embedded in 1% agar, dehydrated in graded ethanol, and epoxy resin-embedded (Electron Microscopy Sciences). Samples were reembedded in “OO” beem capsules and polymerized for 24 h at 60°C. Thin (60–80 nm) sections were counterstained with uranyl acetate and lead citrate, and visualized using the Hitachi 7600 microscope. Measurement of filament diameter was performed on micrographs (80,000× nominal magnification) using AmtV602 and ImageJ software. Only isolated dense dots showing no branching were included when measuring the diameter of cross-sectioned filaments. Data are presented as mean ± SD from three independent experiments.
Wild-type mouse keratinocytes in primary culture and calcium switch experiments
All studies involving mice were reviewed and approved by the Johns Hopkins’ Institutional Animal Care and Use Committee. Keratinocytes from 1- or 2-d-old C57BL/6 newborn mouse skin were isolated as described previously (Bernot et al., 2004). The epidermis was recovered by treating surgically removed mouse trunk skin using 0.25% trypsin/0.02 mM EDTA for 16–20 h at 4°C. Keratinocytes were isolated from epidermis using a Lymphoprep (Stemcell Technologies)-based density gradient centrifugation (1,600 rpm for 20 min at 4°C), and cultured in Epidermal Keratinocyte Medium CnT57 (low calcium, 0.07 mM; CELLnTEC) until reaching 90% confluence. Calcium switch experiments were performed by switching to Keratinocyte Basal Medium 2 (KBM-2, Lonza) supplemented with 1.8 mM calcium and 10% FBS (Atlanta Biologicals; Hennings et al., 1980; Poumay and Pittelkow, 1995). Keratinocytes were harvested 24 h after the calcium switch.
Preparation of cell lysates, protein gel electrophoresis, and immunoblotting analysis
To prepare whole cell lysates, cells were lysed in cold urea lysis buffer (pH 7.0, 6.5 M urea; 50 mM Tris-HCl; 150 mM sodium chloride; 5 mM EDTA; 0.1% Triton X-100; 50 µM NEM; 1 mM PMSF; 1 µg/ml each of chymostatin, leupeptin, and pepstatin; 10 µg/ml each of aprotinin and benzamidine; 2 µg/ml antipain; and 50 mM sodium fluoride). To examine solubility of disulfide-bonded keratin proteins, keratinocytes were first lysed in cold Triton X-100 lysis buffer (similar to urea lysis buffer except no urea and including 1 mM sodium orthovanadate), and pelleted via centrifugation (14,000 rpm for 30 min at 4°C) to separate Triton-soluble versus insoluble fractions. Detergent-insoluble proteins were then dissolved in the same volume of urea lysis buffer.
For immunoblotting, protein concentration in cell lysates was determined using the Bradford assay (Bio-Rad Laboratories) with bovine serum albumin as a standard. Samples for SDS-PAGE were prepared in lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) in the presence of 20 mM TCEP and incubated at room temperature for 1 h to reduce disulfide bonds. Untreated lysates were prepared directed in LDS sample buffer. Equal amounts of cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membrane (0.45 µm; Bio-Rad Laboratories, Inc.). Quantitative infrared immunoblotting analysis (LI-COR Biosciences) was performed using primary antibodies, followed by secondary antibodies conjugated to infrared fluorescent dye IRDye800 (Feng et al., 2013).
Krt14−/− mouse keratinocytes in primary culture and transfection assays
Freshly isolated keratinocytes from 1 or 2-d-old Krt14−/− newborn mouse skin (Lloyd et al., 1995) were transfected by pBK-CMV His-GFP-K14WT or cysteine variants using a P1 primary cell 4D-Nucleofection X kit (Lonza). To study keratin IF organization, transfected keratinocytes were plated on collagen-coated coverslips (VWR), cultured in CnT57 medium for 72 h, fixed using cold methanol, and processed for immunofluorescence staining. To analyze keratin dynamics, Krt14−/− keratinocytes were cotransfected by pShuttle-CMV-H2BmCherry along with the GFP-tagged K14 variants (1:3 molar ratio), and plated on collagen-coated glass-bottom dishes (35 mm; MatTek Corporation), cultured in FAD medium (consisting of DMEM/Ham’s F12; [3.5:1.1 vol/vol] and low calcium [0.05mM Ca2+]; Reichelt and Haase, 2010) for 48 h, and processed for live cell imaging experiments.
Immunofluorescence microscopy and image processing
Cytoskeleton-bound cysteine bonds were stained as described previously (Lee et al., 2012). In brief, methanol-fixed keratinocytes were incubated in 5 mM NEM/PBS, pH 7.0, for 2 h at room temperature to block all of free cysteine sulfhydryl groups, incubated in 10 mM TCEP/PBS, pH 7.0, for 1 h at room temperature to reduce disulfide bonds, and incubated in 2.5 µM maleimide conjugated to AF594/PBS, pH 7.0, overnight at 4°C to stain thiol groups that were engaged in disulfide bonding at the time of harvesting. Immunofluorescence staining was then performed in a dark setting.
For indirect immunofluorescence, fixed cell samples were blocked in 10% normal goat serum/0.1% Triton X-100/PBS for 1 h at room temperature, incubated in primary antibody solution for 1 h, washed in PBS, incubated in the secondary antibody solution, counterstained with DAPI, and mounted in FluorSave Reagent mounting medium (EMD Millipore). Fluorescence images were acquired as single focal planes using a fluorescence microscope (Axio Observer.Z1) equipped with an ApoTome II attachment and an AxioCam MRm camera (all from Carl Zeiss). Focal planes were selected by focusing on the center of the nuclei. Pictures were taken using a 63× Plan-Apochromat oil immersion objective lens, with the system controlled using AxioVision 4.8.2 software (Carl Zeiss). The colocalization module in ImageJ freeware was used to evaluate colocalization between the signals for keratin and disulfides (threshold channels 1 and 2 [range 0–255] = 125) and generate colocalization channels (8-bit grayscale).
Three distinct modes of keratin filament network organization were distinguished when analyzing transfected mouse keratinocytes (Fig. S3). Using ImageJ, we measured the relative intensity of the fluorescence signal for keratin filaments across the space extending from the outer edge of the cell to the nuclear envelope to typify the organization of its keratin IF network. To do so, an arbitrary line was drawn on a 16-bit grayscale image from the cell outer membrane to the nuclear membrane (aligned through the center of the nucleus) using the Freehand command. The distance from the cell periphery to the nuclear envelope was arbitrarily set as 1 to facilitate comparisons between cells having slightly different sizes and shapes due to heterogeneity of keratinocytes isolated from newborn mouse skin (Dotto, 1998). A graph was built up using relative fluorescence intensity (FI%, ranging from 0 to 100) as the y axis and the corresponding relative distance (RD, ranging from 0 for the outer cell membrane to 1 for the nuclear envelope) as the x axis (see Fig. S3 for definition criteria). 50 images for each type of transfectants were randomly selected and subjected to this quantitation to ascertain consistency within the dataset, which was obtained from a blindly performed qualitative assessment.
Live cell imaging and image processing
Mouse keratinocytes were washed in PBS and switched to FAD imaging medium 48 h after transfection. Images were recorded using a single-point laser scanning confocal microscope (LSM780-FCS; Carl Zeiss). Pictures were acquired using the 40× Plan-Neofluar oil immersion objective lens together with the Zen software (Carl Zeiss). Cells in a closed culture dish were kept in an environmental chamber at 37°C with 5% CO2 and 25°C humidity. Excitation was set at 488 nm for GFP and 561 nm for mCherry. Imaging parameters were selected based on previous live cell imaging studies of keratin dynamics (Windoffer and Leube, 2004). Recording intervals were 5 min, color depth was 12 bit, and image resolution was 1,024 × 1,024 pixels. Seven focal planes (optical slice thickness was 0.43 µm per stack), focusing on the center of the cell, were recorded for each time point, and video recordings lasted 6 h. Maximum intensity projections were generated by compressing 3D stacks of time series using Zen software, and regions of interest were selected from projected images using the AxioVision 4.8.2 software (both from Carl Zeiss), and used for preparing figures and converting images into QuickTime movies (Apple). Reconstruction of 2.5-dimentional images from z stacks was done using Zen software. Additionally, maximum intensity projections were subjected to analyses of movement of nuclei. Nuclei were automatically selected and labeled, and their corresponding positions and speeds were tracked using Imaris software (Bitplane).
Definitions
In the context of our live cell imaging studies, we refer to a “normal” keratin cycle when short keratin filaments form at the cell periphery, then elongate as they move toward the cell center as part of a continuous and steady flow, and appear to accumulate in the cell center as they form a perinuclear-concentrated network. Alternatively, we refer to an “impaired” keratin cycle when keratin filament nucleation or elongation at the cell periphery is disrupted or there is a failure to form or maintain a perinuclear-concentrated network during keratin cycling.
Statistical analysis
Statistical analyses were performed using Excel 2011 (Microsoft) and Prism version 5 (GraphPad Software, Inc.). The t test was used when comparing two conditions or samples (see Figs. S1 C and S4, B and C). One-way analysis of variance (ANOVA) was used when comparing three conditions or samples (see Figs. 5 F and S5 F). A P-value of <0.05 was considered significant.
Online supplemental material
Fig. S1 reports on global disulfide bond staining in primary cultures of newborn mouse skin keratinocytes. Fig. S2 reports on the formation of intermolecular disulfide bonds involving K14WT, K14CF-C367, and K5WT in vitro. Fig. S3 illustrates the criteria used to assign keratin filament network organization to one of three categories (pan-cytoplasmic, perinuclear-concentrated, and peripheral networks). Fig. S4 shows fluorescence intensity comparison of GFP-K14WT–, GFP-K14CF–, and GFP-K14CF-C367–expressing keratinocytes during movie recordings. Fig. S5 reports on the enhanced movements of the nucleus and the whole cell in GFP-K14CF–expressing keratinocytes. Videos 1–5 show GFP-tagged K14WT (Video 1)-, K14CF (Videos 2 and 3)-, and K14CF-C367 (Videos 4 and 5)–transfected skin keratinocytes derived from Krt14−/− mice and in primary culture. Online supplement material is available at http://www.jcb.org/cgi/content/full/jcb.201408079/DC1.
Supplementary Material
Acknowledgments
We are grateful to members of the Coulombe laboratory for support, to Jill Hakim and Janet Folmer for technical assistance, to the School of Medicine Microscope Facility for live cell imaging experiments (National Institutes of Health [NIH] grant S10 OD016374), and to Valeria Culotta, Michael Matunis, Andrew Ewald, Ryan Hobbs, and Carole Parent for advice.
This research was funded by grant AR042047 from National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH (to P.A. Coulombe).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FAD
- flavin adenine dinucleotide
- IF
- intermediate filament
- NEM
- N-ethylmaleimide
References
- Aebi U., Fowler W.E., Rew P., and Sun T.T.. 1983. The fibrillar substructure of keratin filaments unraveled. J. Cell Biol. 97:1131–1143 10.1083/jcb.97.4.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., and Walter P.. 2007. The cytoskeleton. Molecular Biology of the Cell. Fifth edition Garland Science, New York: 965–1010. [Google Scholar]
- Angst B.D., Nilles L.A., and Green K.J.. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257. [DOI] [PubMed] [Google Scholar]
- Bernot K.M., Coulombe P.A., and Wong P.. 2004. Skin: an ideal model system to study keratin genes and proteins. Methods Cell Biol. 78:453–487 10.1016/S0091-679X(04)78016-4 [DOI] [PubMed] [Google Scholar]
- Chan Y., Anton-Lamprecht I., Yu Q.C., Jäckel A., Zabel B., Ernst J.P., and Fuchs E.. 1994. A human keratin 14 “knockout”: the absence of K14 leads to severe epidermolysis bullosa simplex and a function for an intermediate filament protein. Genes Dev. 8:2574–2587 10.1101/gad.8.21.2574 [DOI] [PubMed] [Google Scholar]
- Clarke S.J., Hollmann C.A., Zhang Z., Suffern D., Bradforth S.E., Dimitrijevic N.M., Minarik W.G., and Nadeau J.L.. 2006. Photophysics of dopamine-modified quantum dots and effects on biological systems. Nat. Mater. 5:409–417 10.1038/nmat1631 [DOI] [PubMed] [Google Scholar]
- Coulombe P.A., and Lee C.H.. 2012. Defining keratin protein function in skin epithelia: epidermolysis bullosa simplex and its aftermath. J. Invest. Dermatol. 132:763–775 10.1038/jid.2011.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P.A., and Omary M.B.. 2002. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 14:110–122 10.1016/S0955-0674(01)00301-5 [DOI] [PubMed] [Google Scholar]
- Coulombe P.A., Hutton M.E., Letai A., Hebert A., Paller A.S., and Fuchs E.. 1991. Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell. 66:1301–1311 10.1016/0092-8674(91)90051-Y [DOI] [PubMed] [Google Scholar]
- Coulombe P.A., Kerns M.L., and Fuchs E.. 2009. Epidermolysis bullosa simplex: a paradigm for disorders of tissue fragility. J. Clin. Invest. 119:1784–1793 10.1172/JCI38177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto P.1998. The keratinocyte growth-differentiation switch. Front. Biosci. 3:d502–d508. [DOI] [PubMed] [Google Scholar]
- Feng X., Zhang H., Margolick J.B., and Coulombe P.A.. 2013. Keratin intracellular concentration revisited: implications for keratin function in surface epithelia. J. Invest. Dermatol. 133:850–853 10.1038/jid.2012.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser R.D., and Parry D.A.. 2007. Structural changes in the trichocyte intermediate filaments accompanying the transition from the reduced to the oxidized form. J. Struct. Biol. 159:36–45 10.1016/j.jsb.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Fraser R.D.B., and Parry D.A.. 2012. The role of disulfide bond formation in the structural transition observed in the intermediate filaments of developing hair. J. Struct. Biol. 180:117–124 10.1016/j.jsb.2012.05.020 [DOI] [PubMed] [Google Scholar]
- Fraser R.D., and Parry D.A.. 2014. Keratin intermediate filaments: differences in the sequences of the Type I and Type II chains explain the origin of the stability of an enzyme-resistant four-chain fragment. J. Struct. Biol. 185:317–326 10.1016/j.jsb.2013.12.012 [DOI] [PubMed] [Google Scholar]
- Fraser R.D., Steinert P.M., and Parry D.A.. 2003. Structural changes in trichocyte keratin intermediate filaments during keratinization. J. Struct. Biol. 142:266–271 10.1016/S1047-8477(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Fuchs E., and Cleveland D.W.. 1998. A structural scaffolding of intermediate filaments in health and disease. Science. 279:514–519 10.1126/science.279.5350.514 [DOI] [PubMed] [Google Scholar]
- Giroud A., and Leblond C.P.. 1951. The keratinization of epidermis and its derivatives, especially the hair, as shown by x-ray diffraction and histochemical studies. Ann. N. Y. Acad. Sci. 53:613–626 10.1111/j.1749-6632.1951.tb31963.x [DOI] [PubMed] [Google Scholar]
- Hamanaka R.B., Glasauer A., Hoover P., Yang S., Blatt H., Mullen A.R., Getsios S., Gottardi C.J., DeBerardinis R.J., Lavker R.M., and Chandel N.S.. 2013. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci. Signal. 6:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Suga Y., Matsuba S., Mizoguchi M., Takamori K., Seitz J., and Ogawa H.. 2001. Inquiry into the role of skin sulfhydryl oxidase in epidermal disulfide bond formation: implications of the localization and regulation of skin SOx as revealed by TPA, retinoic acid, and UVB radiation. J. Invest. Dermatol. 117:752–754 10.1046/j.0022-202x.2001.01488.x [DOI] [PubMed] [Google Scholar]
- Hennings H., Michael D., Cheng C., Steinert P., Holbrook K., and Yuspa S.H.. 1980. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 19:245–254 10.1016/0092-8674(80)90406-7 [DOI] [PubMed] [Google Scholar]
- Herrmann H., Häner M., Brettel M., Ku N.O., and Aebi U.. 1999. Characterization of distinct early assembly units of different intermediate filament proteins. J. Mol. Biol. 286:1403–1420 10.1006/jmbi.1999.2528 [DOI] [PubMed] [Google Scholar]
- Herrmann H., Wedig T., Porter R.M., Lane E.B., and Aebi U.. 2002. Characterization of early assembly intermediates of recombinant human keratins. J. Struct. Biol. 137:82–96 10.1006/jsbi.2002.4466 [DOI] [PubMed] [Google Scholar]
- Herrmann H., Bär H., Kreplak L., Strelkov S.V., and Aebi U.. 2007. Intermediate filaments: from cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 8:562–573 10.1038/nrm2197 [DOI] [PubMed] [Google Scholar]
- Kennedy L.H., Sutter C.H., Leon Carrion S., Tran Q.T., Bodreddigari S., Kensicki E., Mohney R.P., and Sutter T.R.. 2013. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated production of reactive oxygen species is an essential step in the mechanism of action to accelerate human keratinocyte differentiation. Toxicol. Sci. 132:235–249 10.1093/toxsci/kfs325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch A., Windoffer R., Würflinger T., Aach T., and Leube R.E.. 2010. The keratin-filament cycle of assembly and disassembly. J. Cell Sci. 123:2266–2272 10.1242/jcs.068080 [DOI] [PubMed] [Google Scholar]
- Krebs H.A.1967. The redox state of nicotinamide adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Adv. Enzyme Regul. 5:409–434 10.1016/0065-2571(67)90029-5 [DOI] [PubMed] [Google Scholar]
- Ku N.O., Gish R., Wright T.L., and Omary M.B.. 2001. Keratin 8 mutations in patients with cryptogenic liver disease. N. Engl. J. Med. 344:1580–1587 10.1056/NEJM200105243442103 [DOI] [PubMed] [Google Scholar]
- Ku N.O., Lim J.K., Krams S.M., Esquivel C.O., Keeffe E.B., Wright T.L., Parry D.A.D., and Omary M.B.. 2005. Keratins as susceptibility genes for end-stage liver disease. Gastroenterology. 129:885–893 10.1053/j.gastro.2005.06.065 [DOI] [PubMed] [Google Scholar]
- Lee C.H., and Coulombe P.A.. 2009. Self-organization of keratin intermediate filaments into cross-linked networks. J. Cell Biol. 186:409–421 10.1083/jcb.200810196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., Kim M.S., Chung B.M., Leahy D.J., and Coulombe P.A.. 2012. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat. Struct. Mol. Biol. 19:707–715 10.1038/nsmb.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstern T., Mücke N., Aebi U., Mauermann M., and Herrmann H.. 2012. Complex formation and kinetics of filament assembly exhibited by the simple epithelial keratins K8 and K18. J. Struct. Biol. 177:54–62 10.1016/j.jsb.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Yu Q.C., Cheng J., Turksen K., Degenstein L., Hutton E., and Fuchs E.. 1995. The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J. Cell Biol. 129:1329–1344 10.1083/jcb.129.5.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A.S., and Sun T.T.. 1986. Differentiation-dependent changes in the solubility of a 195-kD protein in human epidermal keratinocytes. J. Cell Biol. 103:41–48 10.1083/jcb.103.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Yamada S., Wirtz D., and Coulombe P.A.. 2001. A ‘hot-spot’ mutation alters the mechanical properties of keratin filament networks. Nat. Cell Biol. 3:503–506 10.1038/35074576 [DOI] [PubMed] [Google Scholar]
- Matoltsy A.G., and Matoltsy M.N.. 1970. The chemical nature of keratohyalin granules of the epidermis. J. Cell Biol. 47:593–603 10.1083/jcb.47.3.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean W.H., and Moore C.B.. 2011. Keratin disorders: from gene to therapy. Hum. Mol. Genet. 20(R2):R189–R197 10.1093/hmg/ddr379 [DOI] [PubMed] [Google Scholar]
- Omary M.B., Coulombe P.A., and McLean W.H.. 2004. Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 351:2087–2100 10.1056/NEJMra040319 [DOI] [PubMed] [Google Scholar]
- Omary M.B., Ku N.O., Strnad P., and Hanada S.. 2009. Toward unraveling the complexity of simple epithelial keratins in human disease. J. Clin. Invest. 119:1794–1805 10.1172/JCI37762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poumay Y., and Pittelkow M.R.. 1995. Cell density and culture factors regulate keratinocyte commitment to differentiation and expression of suprabasal K1/K10 keratins. J. Invest. Dermatol. 104:271–276 10.1111/1523-1747.ep12612810 [DOI] [PubMed] [Google Scholar]
- Reichelt J., and Haase I.. 2010. Establishment of spontaneously immortalized keratinocyte lines from wild-type and mutant mice. Methods Mol. Biol. 585:59–69 10.1007/978-1-60761-380-0_5 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer J., Bowden P.E., Coulombe P.A., Langbein L., Lane E.B., Magin T.M., Maltais L., Omary M.B., Parry D.A., Rogers M.A., and Wright M.W.. 2006. New consensus nomenclature for mammalian keratins. J. Cell Biol. 174:169–174 10.1083/jcb.200603161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann K., Fritsch A.W., Käs J.A., and Magin T.M.. 2013. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc. Natl. Acad. Sci. USA. 110:18507–18512 10.1073/pnas.1310493110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert P.M., and Parry D.A.. 1993. The conserved H1 domain of the type II keratin 1 chain plays an essential role in the alignment of nearest neighbor molecules in mouse and human keratin 1/keratin 10 intermediate filaments at the two- to four-molecule level of structure. J. Biol. Chem. 268:2878–2887. [PubMed] [Google Scholar]
- Steinert P.M., Idler W.W., and Zimmerman S.B.. 1976. Self-assembly of bovine epidermal keratin filaments in vitro. J. Mol. Biol. 108:547–567 10.1016/S0022-2836(76)80136-2 [DOI] [PubMed] [Google Scholar]
- Strnad P., Zhou Q., Hanada S., Lazzeroni L.C., Zhong B.H., So P., Davern T.J., Lee W.M., and Omary M.B.. Acute Liver Failure Study Group. 2010. Keratin variants predispose to acute liver failure and adverse outcome: race and ethnic associations. Gastroenterology. 139:828–835: e1–e3 10.1053/j.gastro.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Usachov V., Debes C., Gräter F., Parry D.A.D., and Omary M.B.. 2011. Unique amino acid signatures that are evolutionarily conserved distinguish simple-type, epidermal and hair keratins. J. Cell Sci. 124:4221–4232 10.1242/jcs.089516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.T., and Green H.. 1978. Keratin filaments of cultured human epidermal cells. Formation of intermolecular disulfide bonds during terminal differentiation. J. Biol. Chem. 253:2053–2060. [PubMed] [Google Scholar]
- Szeverenyi I., Cassidy A.J., Chung C.W., Lee B.T., Common J.E., Ogg S.C., Chen H., Sim S.Y., Goh W.L., Ng K.W., et al. . 2008. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 29:351–360 10.1002/humu.20652 [DOI] [PubMed] [Google Scholar]
- Thiele J.J., Schroeter C., Hsieh S.N., Podda M., and Packer L.. 2001. The antioxidant network of the stratum corneum. Curr. Probl. Dermatol. 29:26–42 10.1159/000060651 [DOI] [PubMed] [Google Scholar]
- Troy T.C., and Turksen K.. 1999. In vitro characteristics of early epidermal progenitors isolated from keratin 14 (K14)-deficient mice: insights into the role of keratin 17 in mouse keratinocytes. J. Cell. Physiol. 180:409–421 [DOI] [PubMed] [Google Scholar]
- Wang H., Parry D.A., Jones L.N., Idler W.W., Marekov L.N., and Steinert P.M.. 2000. In vitro assembly and structure of trichocyte keratin intermediate filaments: a novel role for stabilization by disulfide bonding. J. Cell Biol. 151:1459–1468 10.1083/jcb.151.7.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner N.S., Windoffer R., Strnad P., Grund C., Leube R.E., and Magin T.M.. 2004. Epidermolysis bullosa simplex-type mutations alter the dynamics of the keratin cytoskeleton and reveal a contribution of actin to the transport of keratin subunits. Mol. Biol. Cell. 15:990–1002 10.1091/mbc.E03-09-0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R., and Leube R.E.. 2004. Imaging of keratin dynamics during the cell cycle and in response to phosphatase inhibition. Methods Cell Biol. 78:321–352 10.1016/S0091-679X(04)78012-7 [DOI] [PubMed] [Google Scholar]
- Windoffer R., Beil M., Magin T.M., and Leube R.E.. 2011. Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J. Cell Biol. 194:669–678 10.1083/jcb.201008095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.