TMEM231, a functional component of the MKS complex at the ciliary transition zone, is mutated in orofaciodigital syndrome type 3 and Meckel syndrome.
Abstract
The Meckel syndrome (MKS) complex functions at the transition zone, located between the basal body and axoneme, to regulate the localization of ciliary membrane proteins. We investigated the role of Tmem231, a two-pass transmembrane protein, in MKS complex formation and function. Consistent with a role in transition zone function, mutation of mouse Tmem231 disrupts the localization of proteins including Arl13b and Inpp5e to cilia, resulting in phenotypes characteristic of MKS such as polydactyly and kidney cysts. Tmem231 and B9d1 are essential for each other and other complex components such as Mks1 to localize to the transition zone. As in mouse, the Caenorhabditis elegans orthologue of Tmem231 localizes to and controls transition zone formation and function, suggesting an evolutionarily conserved role for Tmem231. We identified TMEM231 mutations in orofaciodigital syndrome type 3 (OFD3) and MKS patients that compromise transition zone function. Thus, Tmem231 is critical for organizing the MKS complex and controlling ciliary composition, defects in which cause OFD3 and MKS.
Introduction
Primary cilia project from the surfaces of many cell types and communicate intercellular signals during development and adult tissue homeostasis (Goetz et al., 2009). Consistent with roles in cell–cell communication, membrane-associated proteins involved in signal transduction, such as Polycystic kidney disease 2 (Pkd2) and Smoothened (Smo), localize to cilia (Yoder et al., 2002; Corbit et al., 2005). Smo is a critical component of Hedgehog signaling that, in vertebrates, patterns the anteroposterior axis of the limb buds (Hooper and Scott, 2005). Pkd2 restrains the growth of the kidney epithelium (Wu et al., 1998). Consequently, disruption of either ciliogenesis or the control of ciliary protein composition disturbs vertebrate Hedgehog signaling and cell proliferation, resulting in polydactyly and kidney cysts (Huangfu and Anderson, 2005; Garcia-Gonzalo et al., 2011).
To enter or exit the cilium, proteins pass through the transition zone (TZ), located just distal to the basal body. Several protein complexes have been identified that localize to the TZ, including the Meckel syndrome (MKS) and Nephronophthisis (NPHP) complexes (Garcia-Gonzalo et al., 2011; Sang et al., 2011; Williams et al., 2011; Chih et al., 2012). In mammals, defects in TZ function can disrupt ciliary membrane composition, including the mislocalization of transmembrane signaling proteins (Hu et al., 2010; Garcia-Gonzalo et al., 2011; Chih et al., 2012). In support of a conserved role for the TZ, orthologues of the genes encoding components of the MKS and NPHP complexes are present in other ciliated organisms, including Caenorhabditis elegans and Chamydomonas reinhardtii (Craige et al., 2010; Williams et al., 2011; Reiter et al., 2012).
Inherited defects in ciliary function underlie a diverse set of diseases called ciliopathies. Mutations in TZ complex genes cause a subset of ciliopathies, including NPHP, characterized by corticomedullary kidney cysts; Joubert syndrome, marked by cerebellar vermis aplasia; and MKS, distinguished by cystic kidneys, hepatic ductal plate malformations, polydactyly, and encephalocele (Smith et al., 2006; Arts et al., 2007; Baala et al., 2007; Gorden et al., 2008; Mougou-Zerelli et al., 2009; Otto et al., 2009; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Hildebrandt et al., 2011; Huang et al., 2011; Sang et al., 2011; Shaheen et al., 2011; Chih et al., 2012; Davis and Katsanis, 2012; Srour et al., 2012; Shaheen et al., 2013; Tuz et al., 2013). For example, B9D1 and TMEM231 mutations are associated with MKS (Hopp et al., 2011; Shaheen et al., 2013) as well as Joubert syndrome (Srour et al., 2012; Romani et al., 2014).
B9D1 encodes one of three B9 domain–containing proteins in mammals, all of which are components of the MKS complex and are involved in MKS (Weatherbee et al., 2009; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Hopp et al., 2011; Chih et al., 2012). Mouse embryos lacking B9d1 function display MKS-like phenotypes (Dowdle et al., 2011). The genomes of diverse ciliated organisms encode B9 domain–containing proteins, suggesting that this protein family has an evolutionarily conserved role in ciliary function (Bialas et al., 2009; Dowdle et al., 2011). We detected Tmem231, a 36-kD two-pass transmembrane protein, as a potential B9d1 interactor (Dowdle et al., 2011). Tmem231 mutant mouse embryos display ciliopathy hallmarks including polydactyly, microphthalmia, and dorsalization of the neural tube (Chih et al., 2012).
Here, we demonstrate that B9d1 and Tmem231 are required to localize a subset of MKS complex components to the TZ and to maintain ciliary protein composition in diverse tissues and cilia types in vivo and in cultured cells. We show that the role of Tmem231 in controlling TZ and ciliary membrane protein composition are conserved in C. elegans. Mouse embryos of mixed background and homozygous for a loss-of-function Tmem231 mutation exhibit MKS-like phenotypes including polycystic kidneys, polydactyly, and hepatic ductal plate malformations. Additionally, we identify eight novel human TMEM231 mutations that cause MKS and orofaciodigital syndrome type 3 (OFD3). The missense mutations identified in our ciliopathy cohorts compromise the ability of the TZ to control ciliary membrane-associated protein composition.
Results
Tmem231 is a component of the MKS complex
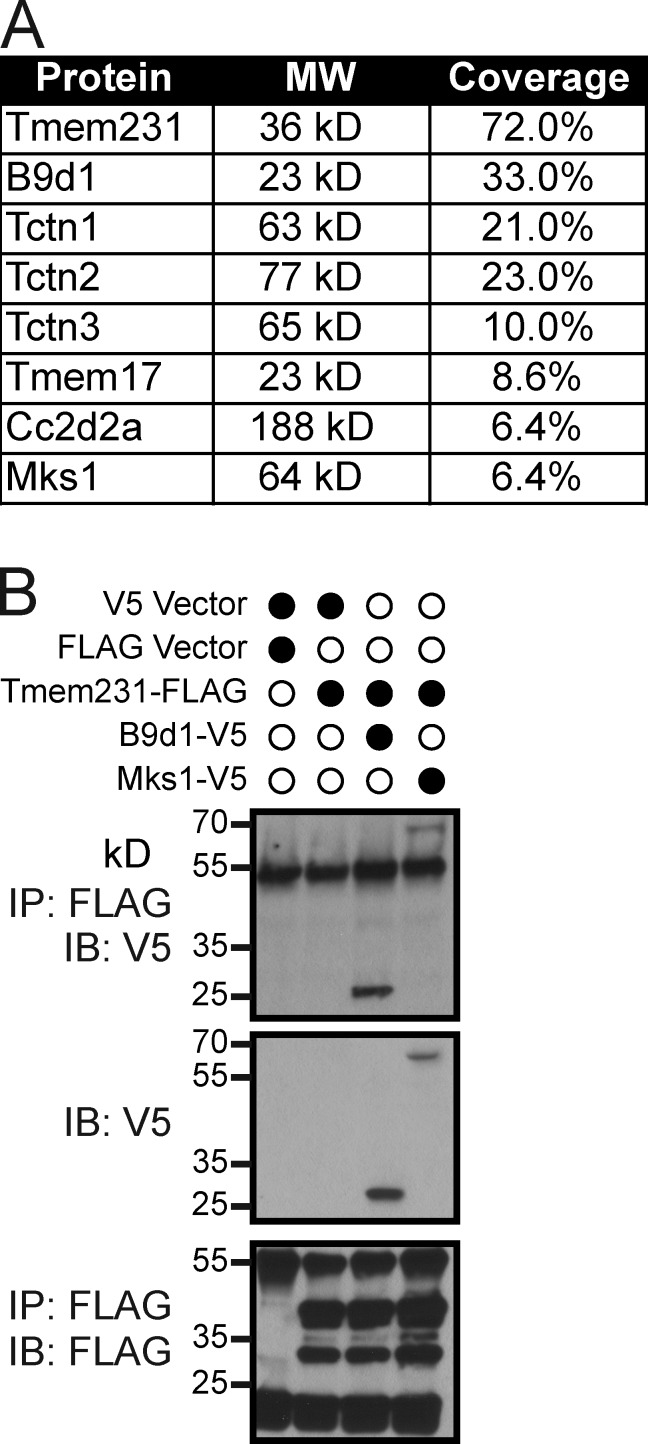
Tmem231 was identified as an interaction partner of B9d1 (Dowdle et al., 2011; Chih et al., 2012). To determine if Tmem231 interacts with only B9d1 or additional constituents of the TZ, we fused Tmem231 to a localization and affinity purification (LAP) tag and subjected copurifying proteins to mass spectrometric identification. This experiment confirmed the interaction between B9d1 and Tmem231, and revealed that Tmem231 interacts with multiple other components of the MKS complex, including Mks1, Tctn1, Tctn2, Tctn3, Cc2d2a (Mks6), and Tmem17 (Fig. 1 A; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Sang et al., 2011; Chih et al., 2012). We validated interactions by coimmunoprecipitating epitope-tagged versions of Tmem231, B9d1, and Mks1 (Fig. 1 B). These data reveal that Tmem231 is a biochemical component of the MKS complex.
Figure 1.
Tmem231 is a component of the MKS complex. (A) Percentage of coverage of known MKS complex proteins identified by mass spectrometry as copurifying with Tmem231. (B) Immunoprecipitation of FLAG-tagged Tmem231 coimmunoprecipitates B9d1 and Mks1, as shown in the third and fourth lanes in the top blot.
Tmem231 mutant mice exhibit ciliopathy phenotypes including kidney cysts, hepatic ductal plate malformations, and polydactyly
Some mouse mutants affecting MKS complex components, such as Mks1, display hallmarks of MKS including kidney cysts, polydactyly, and malformation of the hepatic ductal plate (Weatherbee et al., 2009). Based on our observation that Mks1 and Tmem231 are interacting partners (Fig. 1), we assessed whether loss of Tmem231 has similar effects on embryonic development using mutant mice in which the Tmem231 transcript is truncated at the second exon by an insertion cassette (Chih et al., 2012). On a C57BL/6 background, homozygous Tmem231 mutant embryos die at embryonic day (E) 15.5 and exhibit abrogated Hedgehog signaling, microphthalmia, and polydactyly (Chih et al., 2012). To analyze later stages of development, we generated a homozygous Tmem231 mutation on a C57BL/6-CD1 mixed background; these Tmem231−/− embryos survive until birth (unpublished data).
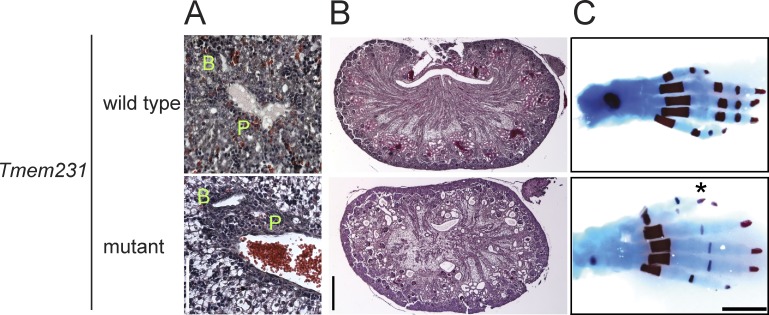
Human MKS fetuses and mouse B9d1−/− embryos display hepatic ductal plate malformations, where the portal vein fails to separate from the bile duct and the portal mesenchyme does not remodel (Salonen, 1984; Weatherbee et al., 2009; Dowdle et al., 2011). Similarly, Tmem231−/− embryos display hepatic ductal plate malformations at E15.5 (Fig. 2 A). Diverse ciliopathies manifest kidney cysts and polydactyly. At E18.5, Tmem231−/− embryos exhibit kidney cysts, predominantly at the corticomedullary border and hindlimb preaxial polydactyly (Fig. 2, B and C). Thus, loss of mouse Tmem231 function causes a spectrum of phenotypes similar to those observed in mice with mutations in some other MKS complex components and human MKS–affected individuals (Salonen, 1984; Weatherbee et al., 2009; Dowdle et al., 2011).
Figure 2.
Tmem231 mutant mice display MKS-like phenotypes. (A) Hematoxylin and eosin–stained sections of portal triads of E15.5 livers of wild-type and Tmem231−/− embryos. In Tmem231−/− embryos, the portal vein (P) and the bile duct (B) are surrounded by increased mesenchyme. Bar, 100 µm. (B) Hematoxylin and eosin–stained sections of E18.5 kidneys show cyst formation in Tmem231−/− embryos. (C) Postnatal day 0 hindlimb skeletons stained with alcian blue and alizarin red reveal an extra digit (*) in Tmem231−/− embryos. Bars, 1 mm.
Tmem231 and B9d1 control ciliary membrane composition
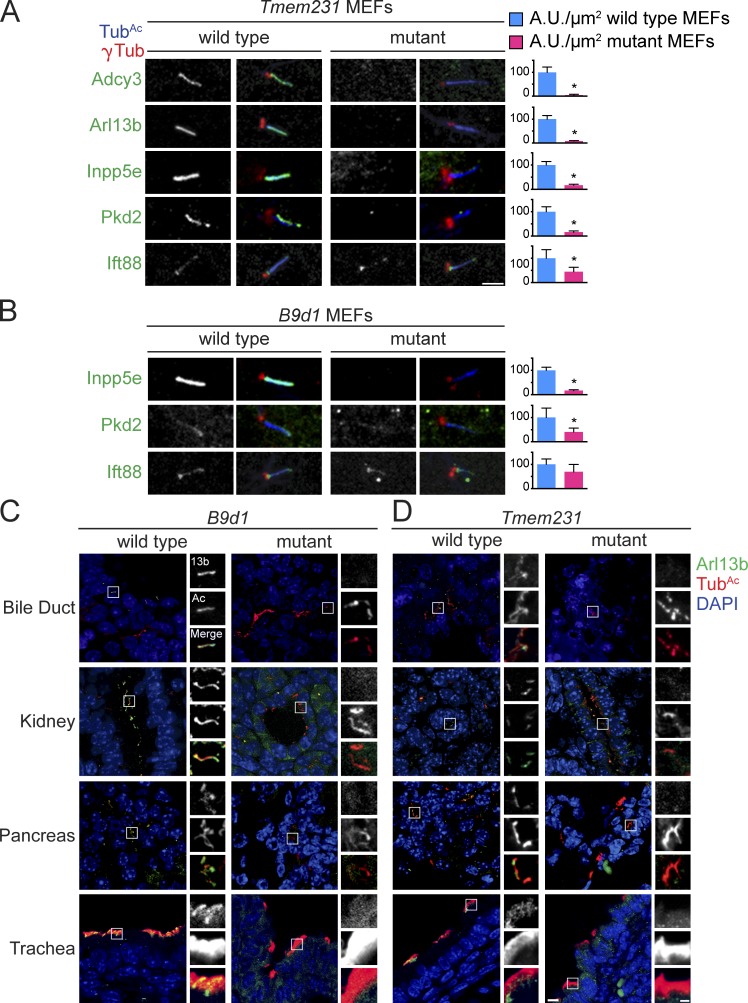
Other MKS components participate in developmental signaling, at least in part, by controlling the membrane composition of cilia. For example, Tctn1 and B9d1 are essential for Smo, Adenylyl cyclase 3 (Adcy3), and Arl13b to localize to cilia (Dowdle et al., 2011; Garcia-Gonzalo et al., 2011). Smo, Adcy3, and Arl13b are all associated with the membrane: Smo and Adcy3 are transmembrane proteins and Arl13b associates with the ciliary membrane through its palmitoyl moiety (Cevik et al., 2010). Given that Tmem231 is a biochemical component of the MKS complex, we tested whether Tmem231 participates with Tctn1 and B9d1 in MKS complex function. Like Tctn1−/− or B9d1−/− mouse embryonic fibroblasts (MEFs), Tmem231−/− MEFs fail to localize Adcy3 and Arl13b to cilia (Fig. 3 A and Fig. S1, A and C), indicating that Tmem231 is an essential component of the MKS complex required to control the composition of the ciliary membrane.
Figure 3.
Tmem231 and B9d1 regulate ciliary membrane protein localization in vitro and in vivo. Tmem231 wild-type and mutant MEFs (A) and B9d1 wild-type and mutant MEFs (B) stained for γ-tubulin (γTub, red), acetylated tubulin (TubAc, blue), and either Adcy3, Arl13b, Inpp5e, Pkd2, or Ift88 (green). Tmem231 and B9d1 are required for ciliary membrane protein localization, but not Ift88, a nonmembrane-associated ciliary protein. Bar, 2.5 µm. Quantitation of at least 10 cilia per condition are shown next to each figure. *, P < 0.05, as measured by Student’s t test with Welch’s correction, throughout. Error bars represent the 95% confidence interval, also throughout. B9d1 (C) and Tmem231 (D) wild-type and mutant tissues stained for TubAc (green), Arl13b (red), and DAPI (blue). Insets are a magnification of the indicated portion. Bars: (full field) 10 µm; (inset) 2.5 µm.
Arl13b is required for the ciliary localization of Inpp5e, a membrane-associated protein implicated in Joubert syndrome (Humbert et al., 2012). As the MKS complex was essential for the ciliary localization of Arl13b, we examined whether Inpp5e also fails to localize to cilia in B9d1−/− and Tmem231−/− MEFs and found that both B9d1 and Tmem231 are necessary for the ciliary localization of Inpp5e (Fig. 3, A and B; and Fig. S1, A and C). Additionally, B9d1 and Tmem231 are required for Pkd2 to localize to cilia (Fig. 3, A and B; and Fig. S1, A and C). In contrast, localization of intraflagellar transport protein Ift88 along the axoneme is similar between wild-type and Tmem231−/− or B9d1−/− MEFs: although there is a decrease in the fluorescence intensity of Ift88, the percentage of Ift88-positive cilia is unaffected by disruption of the TZ complex (Fig. 3, A and B; and Fig. S1, A and C). Furthermore, loss of Tmem231 or B9d1 in MEFs results in longer cilia but in a decrease in the proportion of cells possessing cilia (Fig. S1, D and E). Thus, Tmem231 and B9d1 are essential for controlling the composition of the ciliary membrane, but are not essential for ciliogenesis.
It is not clear if the function of the MKS complex and the TZ in MEFs is similar to their function in other cell types in vivo. Therefore, to determine if Tmem231 and B9d1 control ciliary composition in vivo, we analyzed ciliary membrane protein composition across diverse tissues in wild-type, Tmem231−/−, or B9d1−/− embryos. In wild-type embryos, Arl13b localizes to cilia in the pancreas, bile duct, trachea, and kidney (Fig. 3, C and D). In the absence of either Tmem231 or B9d1, Arl13b no longer localizes to cilia in these tissues (Fig. 3, C and D). Together with our in vitro data, these results demonstrate that the MKS complex is essential for regulating the membrane composition of varied types of cilia in vivo.
Tmem231 and B9d1 are required for assembly of the MKS complex at the TZ
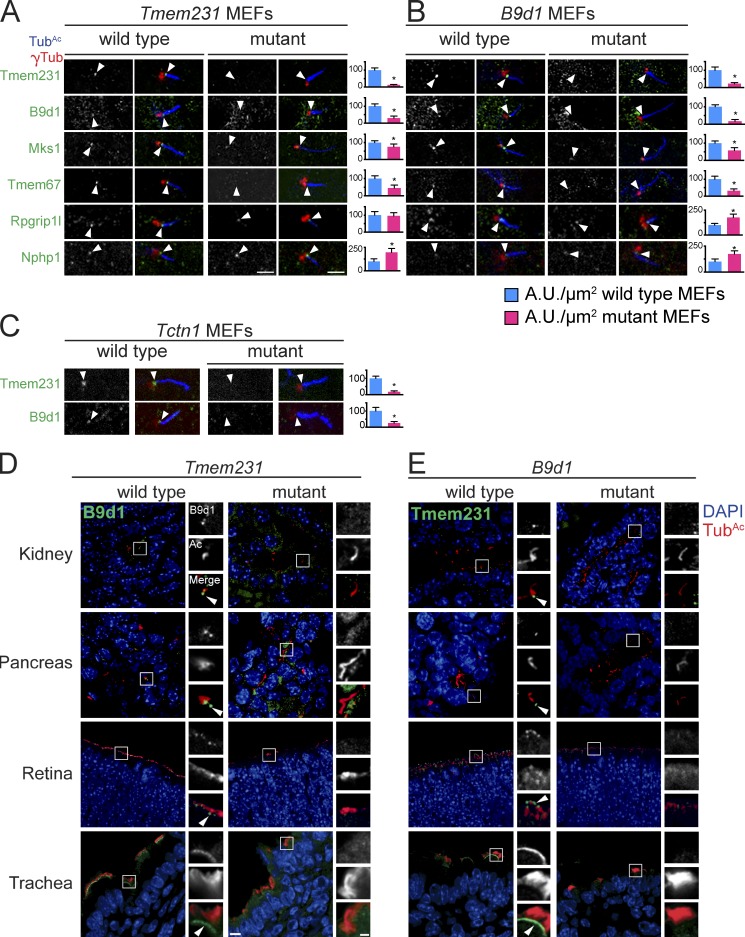
At least 20 proteins localize to the TZ, a subdomain at the base of cilia critical for controlling ciliary composition and function (Garcia-Gonzalo and Reiter, 2012; Reiter et al., 2012). To understand how Tmem231 and B9d1 control ciliary composition, we investigated whether Tmem231 and B9d1 are required to localize other MKS complex components to the TZ. In wild-type MEFs, Tmem231 and B9d1 are present at the TZ (Fig. 4, A–C; and Fig. S2, A–C). In Tmem231−/− MEFs, B9d1 fails to localize to the TZ (Fig. 4 A and Fig. S2, A and C). Similarly, in B9d1−/− MEFs, Tmem231 no longer concentrates at the TZ (Fig. 4 B and Fig. S2, B and C). Thus, B9d1 and Tmem231 are reciprocally required for their localization to the TZ. Moreover, two additional MKS complex components, Mks1 and Tmem67 (Mks3), are lost from the TZ in the absence of either B9d1 or Tmem231 (Fig. 4, A and B; and Fig. S2, A–C). In contrast, neither Rpgrip1l nor Nphp1 require Tmem231 or B9d1 to localize to the TZ (Fig. 4, A and B; and Fig. S2, A–C). Instead, loss of either Tmem231 or B9d1 results in increased accumulation of Nphp1 at the TZ (Fig. 4, A and B). These data indicate that B9d1 and Tmem231 are both indispensible for, and perform similar roles in, the assembly of the MKS complex.
Figure 4.
Tmem231, B9d1, and Tctn1 are required for TZ MKS complex formation. Ciliary TZ of Tmem231 (A), B9d1 (B), and Tctn1 (C) wild-type and mutant MEFs, stained for γ-tubulin (red), TubAc (blue), and either Tmem231, B9d1, Mks1, Tmem67, Rpgrip1l, or Nphp1 (green). Tmem231 and B9d1 are necessary for Mks1 and Tmem67 but not Rpgrip1L or Nphp1 localization. Tctn1 is required for both Tmem231 and B9d1 localization. TZs are denoted by white arrowheads. Bars, 2.5 µm. Quantitation of TZ fluorescence intensity from at least 10 cilia are shown next to each set of images. Error bars represent the 95% confidence interval. *, P < 0.05, as measured by Student’s t test with Welch’s correction. (D) Tmem231 wild-type and mutant bile duct, kidney, pancreas, retina, and trachea stained for B9d1 (green), TubAc (red), and DAPI (blue). (E) B9d1 wild-type and mutant bile duct, kidney, pancreas, retina, and trachea stained for Tmem231 (green), TubAc (red), and DAPI (blue). Bars: (full field) 10 µm; (inset) 2.5 µm.
Mammalian B9d1 was identified as an MKS complex component via LAP-tandem affinity purification and mass spectrometry analysis of Mks1 and Tctn1 (Garcia-Gonzalo et al., 2011; Sang et al., 2011). Like B9d1 and Tmem231, Tctn1 is necessary to localize Mks1 and Tmem67 to TZs, suggesting that Tctn1 may function with B9d1 and Tmem231 to construct the TZ (Garcia-Gonzalo et al., 2011). To test this hypothesis, we analyzed B9d1 and Tmem231 localization in wild-type and Tctn1−/− MEFs. In the absence of Tctn1, B9d1 and Tmem231 are no longer at the TZ (Fig. 4 C and Fig. S2 C), demonstrating that Tctn1 participates with B9d1 and Tmem231 in TZ organization.
Cilia perform different roles in different mammalian cell types, raising the possibility that their TZs may have different compositions. Moreover, loss of certain MKS complex components, including B9d1 or Tctn1, compromises ciliogenesis in some but not all tissues, which indicates that TZ components can have tissue-specific functions (Garcia-Gonzalo et al., 2011). We tested whether these differential requirements for ciliogenesis reflect tissue-specific differences in TZ composition and found that Tmem231 and B9d1 localize to the TZ of cilia in the kidney, bile duct, pancreas, trachea, and retina of mice (Fig. 4, D and E), suggesting that TZ composition is similar in many ciliated cell types.
Similar to our observations in MEFs, B9d1 fails to localize to the TZ in all Tmem231 mutant tissues examined (Fig. 4 D). Reciprocally, in all B9d1 mutant tissues examined, Tmem231 is absent from the TZ (Fig. 4 E). These data reveal that the requirements for MKS complex assembly are similar in diverse ciliated mammalian tissues.
TMEM-231 control of TZ and ciliary composition is conserved in C. elegans
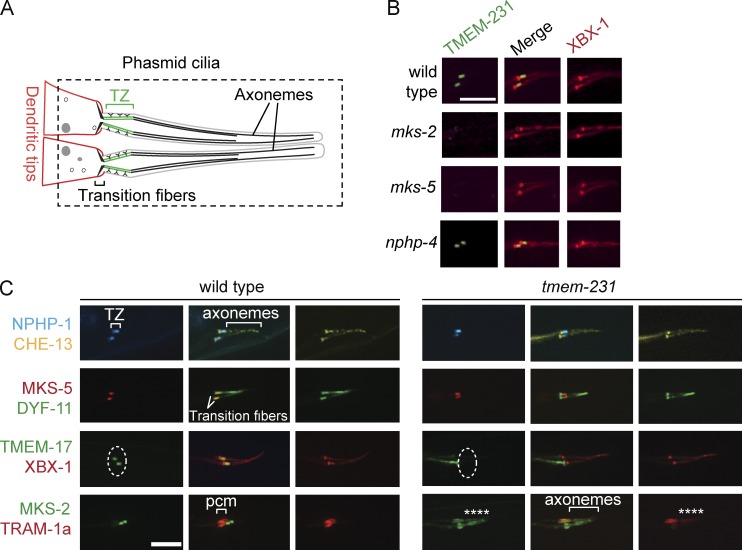
Given that the localization and function of Tmem231 in TZ assembly and control of ciliary membrane protein composition is conserved among multiple cell types in mouse, we explored whether the localization and function of Tmem231 is conserved in an evolutionarily distant organism. To examine the localization of the C. elegans orthologue of Tmem231, TMEM-231 (also called T26A8.2), we created a transgenic line that expresses a fusion of GFP and TMEM-231. This translational fusion, under the control of the tmem-231 promoter, is expressed exclusively in ciliated sensory neurons, similar to other TZ proteins (Fig. 5, A and B; Bialas et al., 2009; Williams et al., 2011). Also consistent with a role in the TZ, TMEM-231 localizes specifically to this ciliary region, just distal to the basal body (Fig. 5, A and B; basal body and axoneme marked with the IFT protein XBX-1).
Figure 5.
C. elegans TMEM-231 functionally interacts with other TZ proteins and is required for ciliary gating. (A) Schematic of C. elegans phasmid ciliary structure. The region delineated by the dashed line is depicted in subsequent panels. (B) TMEM-231::GFP (green) is enriched at the TZ of wild-type animals, but is mislocalized in the mks-2 and mks-5 mutants, but not in the nphp-4 mutants. The Dynein 2 component XBX-1::tdTomato (red) marks the basal body transition fibers and axonemes. (C) NPHP-1::CFP (blue) localizes to the TZ of wild-type and tmem-231 mutant animals. Similarly, MKS-5::tdTomato (red) localizes to the TZ of wild-type and tmem-231 mutant animals. CHE-13::YFP (yellow), the C. elegans orthologue of Ift57, and DYF-11::GFP (green), the orthologue of Ift54, mark the transition fibers and axonemes. In contrast, TMEM-17::GFP (green) fails to localize to the TZ (dotted line) in tmem-231 mutants. MKS-2::GFP (green) fails to localize to the TZ in tmem-231 mutants and is instead within the more distal cilia (asterisks). TRAM-1a::tdTomato (red) localizes to the periciliary membrane (pcm) in wild-type animals. However, in tmem-231 mutants, TRAM-1a::tdTomato enters cilia, indicating defects in ciliary gating (asterisks). The dashed ovals delineate the region of the transition zone. Bars, 5 µm.
We tested whether, as in mammals, TMEM-231 localization to the TZ requires other TZ proteins. TMEM-231 localization to the TZ depends on MKS-2 (the orthologue of mammalian Mks2/Tmem216; Huang et al., 2011) and MKS-5 (the orthologue of mammalian Nphp8/Rpgrip1l), but not NPHP-4 (the orthologue of mammalian Nphp4, Fig. 5 B). Reciprocally, NPHP-1 and MKS-5 localize normally to the TZ in nematodes lacking TMEM-231, but TMEM-17 and MKS-2 no longer concentrate at the TZ in mutants lacking exons 2 and 3 of tmem-231 (Fig. 5 C). The mammalian orthologue of TMEM-17 was identified as a biochemical interactor of Tmem231 (Fig. 1) and B9d1 and is required for B9d1 to localize to the TZ in MEFs (Chih et al., 2012). These data suggest that TMEM-17 is a functional component of the MKS complex in both mammals and nematodes.
To ascertain whether TMEM-231 also plays a role in restricting the entry of nonciliary proteins into the cilium, we examined the localization of the periciliary membrane component TRAM-1a, the orthologue of mammalian translocation-associated membrane protein 1. TRAM-1a is excluded from cilia in wild-type animals (Fig. 5 C, bottom). Loss of TMEM-231 allows TRAM-1a to inappropriately leak into cilia (Fig. 5 C). In accordance with observations of its mammalian orthologue, these experiments demonstrate that C. elegans TMEM-231 is a TZ protein that interacts functionally with other TZ proteins to control the composition of the TZ and ciliary membrane.
Mutation of TMEM231 is associated with OFD3
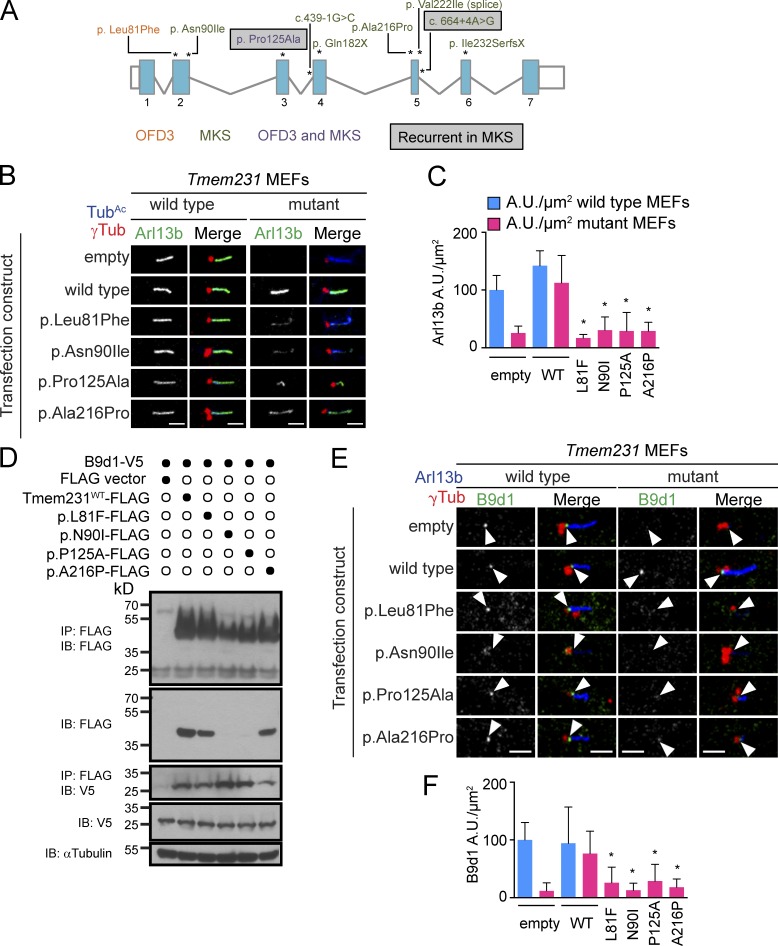
To further assess whether mutations in TMEM231 contribute to human ciliopathies, we performed targeted exon and splice site sequencing of TMEM231 in 1,056 individuals with NPHP-related ciliopathies, applying a recently developed high-throughput mutation analysis approach (Halbritter et al., 2012). We identified compound heterozygous mutations of TMEM231 (A3472-21/-24: c.241C>T, p.Leu81Phe/c.373C>G, p.Pro125Ala; Table S1, Fig. 6 A, and Fig. S4 A) in two affected siblings with OFD3 (Sugarman et al., 1971; Smith and Gardner-Medwin, 1993). Both alleles were predicted in silico to be disease causing (PolyPhen2-HumVar: 0.976 and 0.998) and affect residues conserved among chordates (Fig. S3). Pro125 is even conserved beyond chordates, including among several ciliated protists. TMEM231 orthologues are not found in unciliated organisms, further suggesting that this protein has conserved roles in ciliary biology. The variants were either absent or exceedingly rare in publicly available single nucleotide polymorphism databases (1000 Genomes, Exome Variant Server: c.241C>T, p.Leu81Phe: TT = 0/TC = 1/CC = 6155) and segregated with the OFD3 phenotype within the family (Fig. S4 A).
Figure 6.
TMEM231 mutations are associated with OFD3 and MKS. (A) Schematic of the TMEM231 locus and mutations identified in OFD3- and MKS-affected individuals. Mutation locations are indicated with asterisks and mutation names correspond to GenBank accession no. NM_001077418. Blue boxes, exons; white boxes, untranslated regions; gray lines, introns; black numbers exons. (B) Arl13b localization in Tmem231−/− MEFs is rescued by transient transfection of wild-type Flag-tagged Tmem231. The mutant forms of Tmem231 associated with OFD3 and MKS partially restore Arl13b localization to cilia. Bars, 2.5 µm. (C) Quantitation of the normalized fluorescence intensity of Arl13b at wild-type and Tmem231−/− cilia (at least 10 cilia per condition) transfected with the indicated expression constructs. The data shown are from a single representative experiment out of three repeats. (D) Coimmunoprecipitation of FLAG-tagged Tmem231 constructs and V5-tagged B9d1. Despite two mutant forms of Tmem231 being expressed at lower levels (lanes 3–7, second blot), all four mutant proteins were able to immunoprecipitate B9d1 (third blot, third through seventh lanes). The data are shown are from a single representative experiment out of two repeats. (E) Transfection of wild-type Flag-tagged Tmem231 restores B9d1 localization to Tmem231−/− MEFs. Ciliopathy-associated point mutations restore B9d1 localization to a lesser extent. Arrowheads show the TZ. Bars, 2.5 µm. (F) Quantitation of the normalized fluorescence intensity of B9d1 at wild-type and Tmem231−/− cilia (at least 10 cilia per condition) transfected with the indicated expression constructs. The data shown are from a single representative experiment from two repeats. Error bars represent the 95% confidence interval. *, P < 0.05, as measured by Student’s t test with Welch’s correction.
Although overlapping phenotypically with Joubert syndrome and OFD6, OFD3 is thought to be a clinically distinct autosomal recessive disorder characterized by “metronome” eye movements, lingual hamartomas, and postaxial polydactyly in addition to cerebellar vermis hypoplasia and moderate intellectual disability, all of which were present in both affected siblings (Baraitser, 1986). The younger sister also exhibited a large Dandy-Walker malformation with cystic dilation of the fourth ventricle. Notably, both affected individuals presented with normal renal morphology and function at birth, but developed end-stage renal failure at 13 and 24 years old (Table S1).
Recurrent TMEM231 mutations are associated with MKS
To further investigate the mutational contribution of TMEM231 in MKS, we conducted whole exome sequencing (MKS-1163, MKS-855, and MKS-43), targeted ciliary gene exome sequencing (MKS-947/MKS-948 and GEF1200207), or bidirectional Sanger sequencing (n = 95) of samples from individuals with MKS or MKS-like clinical features. We detected variants inherited under a recessive model of disease and segregating with the disease phenotype in nine pedigrees (Table S1, Fig. 6 A, and Fig. S4). The MKS-associated variants were not present in the exomes of 6,503 control individuals (4,300 European American and 2,203 African American individuals; Exome Variant Server).
Four families of northern European descent harbored the same c.373C>G, p.Pro125Ala change as the OFD3 family (Table S1 and Fig. S4, B–E). In one nonconsanguineous pedigree (NTDMGS 80399), p.Pro125Ala was present in homozygosity (Fig. S4 B). Three other families each had TMEM231 variants in trans with p.Pro125Ala. One pedigree (MKS-692) also had a heterozygous nonsense mutation (c.544C>T, p.Gln182X); a French proband (MKS-79) harbored a heterozygous c.270C>T, p.Asn90Ile mutation; and another family (siblings MKS-947 and MKS-948) had a heterozygous c.664+4A>G intronic change (Fig. 6 A and Fig. S4, C–E). All probands with p.Pro125Ala displayed postaxial polydactyly on at least three limbs, hydrocephalus (MKS-947 and MKS-948), Dandy-Walker malformation with either hydrocephalus (NTDMGS 80399) or cerebellar vermis agenesis (MKS-79 and MKS-692), renal cysts, and hepatic portal fibrosis (Table S1). Like the residues affected in OFD3, Asn90 is a highly conserved residue possessed by chordates including Ciona intestinalis (Fig. S3).
The intronic c.664+4A>G variant was not unique to the MKS family with two affected individuals (MKS-947/MKS-948), as three additional MKS pedigrees carried this putative splice-altering variant (Table S1 and Fig. S4, F–H). Two consanguineous families of Turkish and Libyan descent harbored this change in homozygosity (MKS-1163 and MKS-855, respectively). Further, we detected a maternally inherited heterozygous c.664+4A>G variant in a fourth family of Turkish/European origin (MKS-374), with a paternally inherited heterozygous frameshift mutation (p.Ile232SerfsX). To test the functional consequences of the c.644+4A>G change, we assessed TMEM231 transcripts present in cultured primary skin fibroblasts from MKS-374. Direct sequencing of RT-PCR products using primers spanning exons 4–6 revealed the abolishment of the exon 5 splice donor site, utilization of a novel splice site in intron 5–6, and the retention of 47 bp of intronic sequence to produce a truncated message encoding p.Val222fsX21 (Fig. S4 K).
We identified two additional MKS pedigrees with TMEM231 mutations that segregated appropriately under a recessive model (Table S1, Fig. 6 A, and Fig. S4, I and J): a consanguineous Tunisian family with a homozygous c.664G>A, p.Val222Ile mutation, reported previously to disrupt splicing of exons 5–6 (Shaheen et al., 2013), and a family with novel compound heterozygous splice acceptor (c.439-1G>C) and missense (c.646G>C; p.Ala216Pro) mutations (GEF1200207). In contrast to the affected individuals with p.Pro125Ala mutations who had cerebellar malformations and/or hydrocephalus, the other MKS individuals displayed severe neural tube defects including anencephaly (MKS-374) or encephalocele (MKS-1163, MKS-855, MKS43, and GEF1200207; Table S1).
TMEM231 disease-associated alleles are hypomorphic
To test the effect of the disease-associated nonsynonymous mutations on the ability of Tmem231 to function with the TZ complex, we created a cell-based assay to assess the membrane protein–associated composition of cilia. In the absence of Tmem231, Arl13b fails to localize to cilia (Fig. 6, B and C; and Fig. S5 A). Expression of amino terminally Flag-tagged wild-type Tmem231 in Tmem231−/− MEFs restores Arl13b localization to cilia, as assessed by the fluorescence intensity of Arl13b at cilia (Fig. 6, B and C; and Fig. S5 A). Expression of ciliopathy-associated Tmem231 variants in Tmem231−/− MEFs failed to restore ciliary Arl13b localization to the same extent as wild-type Tmem231 (Fig. 6, B and C; and Fig. S5 A). Each of these mutant forms of Tmem231 (p.Leu81Phe, p.Pro125Ala, p.Asn90Ile, and p.Ala216Pro) rescue ciliary Arl13b to a modest extent, suggesting that the OFD3- and MKS-associated mutations are hypomorphic and partially compromise TMEM231 function.
To test how the disease-associated mutations affect Tmem231 function, we tested whether mutant forms of Flag-tagged Tmem231 could associate with V5-tagged B9d1 by coimmunoprecipitation (Fig. 6 D). The four disease-associated Tmem231 mutant proteins retain their ability to interact with B9d1, but two mutations, p.Asn90Ile and p.Pro125Ala, compromised Tmem231 protein levels (Fig. 6 D).
To explore further how the disease-associated mutations in Tmem231 affect its function, we tested if they were able to support MKS complex organization at the TZ of Tmem231−/− MEFs. In the absence of Tmem231, B9d1 fails to localize to the TZ (Fig. 4 A; Fig. 6, E and F; and Fig. S5 F). Expression of wild-type Flag-Tmem231 in Tmem231−/− MEFs restored B9d1 localization to the TZ (Fig. 6, E and F; and Fig. S5 F). In contrast, all four disease-associated mutant forms of Tmem231 failed to restore B9d1 localization to the TZ to the same extent as wild-type Tmem231 (Fig. 6, E and F). Interestingly, p.Asn90Ile expression in wild-type MEFs mislocalized B9d1 away from the TZ to the centrosome, suggesting this mutant may interfere with the function of wild-type Tmem231 (Fig. 6, E and F; and Fig. S5 F). Thus, disease-associated mutations in Tmem231 disrupt formation of the MKS complex at the TZ, leading to defective ciliary membrane composition and ciliary signaling.
Discussion
We found that B9d1 and Tmem231 are evolutionarily conserved components of the TZ MKS complex required to control the composition of the ciliary membrane. In mice, B9d1 and Tmem231 are required for the TZ localization of each other and their interacting proteins, Tmem67 and Mks1. Similarly, in C. elegans, TMEM-231 localizes to the TZ and participates with other MKS complex proteins to build the TZ and prevent a nonciliary protein, TRAM-1a, from entering the cilium. Like B9d1, Tmem231 is required for mammalian development: Tmem231 mutant mice have developmental defects reminiscent of MKS patients, including corticomedullary kidney cysts, polydactyly, and hepatic ductal plate malformation. Consistent with a role in human disease, we identified hypomorphic and truncating TMEM231 mutations that segregate with OFD3 and MKS.
B9d1 and Tmem231 are essential for the organization of the MKS complex at the TZ. Each is required for the other, and Mks1 and Tmem67, to localize to the TZ. Additionally, an extracytosolic protein, Tctn1, is required for the TZ localization of B9d1 and Tmem231. Thus, the MKS complex is comprised of functionally integrated components that span the membrane and depend on extracytosolic (e.g., Tctn1), transmembrane (e.g., Tmem231), and cytoplasmic (e.g., B9d1) constituents for its organization. However, none of these components are necessary for formation of the entire TZ. For example, Tmem231, B9d1, and Tctn1 are required for Mks1 and Tmem67 TZ localization, but not Rpgrip1l or Nphp1 localization (this study; Garcia-Gonzalo et al., 2011). Tctn1 is required for Mks1, Tmem67, Tmem231, and B9d1 to localize to the TZ, but not for Cc2d2a, Rpgrip1l, or Tmem216 (this study; Garcia-Gonzalo et al., 2011). Tmem231 and B9d1 are needed for TZ localization of each other, Mks1, and Tmem67, and also Cc2d2a and Tmem17 (this study; Chih et al., 2012). Reciprocally, Cc2d2a and Tmem17 are necessary for Tmem231 and B9d1 to localize to the TZ (Chih et al., 2012). These data are consistent with at least two possible models of MKS complex assembly.
In hierarchical assemblies, a core component recruits a set of interactors that, in turn, recruit the peripheral proteins of the complex. A candidate for the initiating TZ component in C. elegans is MKS-5, the orthologue of mammalian Rpgrip1l and Rpgrip1 (Williams et al., 2011). In the absence of MKS-5, all other MKS complex components tested fail to localize to the TZ (Huang et al., 2011; Williams et al., 2011). Moreover, MKS-5 localization to the TZ is not dependent on the MKS complex components MKS-6 (orthologue of Ccd2a) or MKSR-2 (B9d2), raising the possibility that MKS-5 forms a core scaffold for TZ assembly in nematodes (Williams et al., 2011). Consistent with a central role for MKS-5 in TZ assembly, we found that it is critical for TMEM-231 localization to the TZ. One of the mammalian homologues of MKS-5, Rpgrip1l, behaves similarly in that Rpgrip1l does not require B9d1, Tmem231, or Tctn1 to localize to the TZ. However, whether Rpgrip1l is required for Tmem231 or B9d1 to localize to the TZ remains unclear. Furthermore, whether the second mammalian MKS-5 homologue, Rpgrip1, plays a semi-redundant role with Rpgrip1l in MKS complex formation remains unclear.
Alternatively, the mammalian TZ may not be assembled on any one protein, but might require multiple reciprocal interactions, as would occur if the complex were destabilized by the absence of any of several core constituents. Our finding that B9d1 and Tmem231 are reciprocally required for each other’s localization and mutually essential for TZ function suggests that this model may better describe mammalian TZ assembly. However, blended models involving both hierarchical and reciprocal dependencies can also account for the available data.
Components of the MKS complex are required to control the protein composition of the ciliary membrane (Garcia-Gonzalo et al., 2011; Dowdle et al., 2011; Chih et al., 2012). Consistent with their inclusion in the MKS complex, Tmem231 and B9d1 are necessary to localize certain membrane proteins to cilia, both in MEFs and in diverse mammalian tissues. For example, Arl13b, Adcy3, Pkd2, and Inpp5e fail to localize to cilia in Tmem231−/− MEFs, and Tmem231 and B9d1 are required for Arl13b ciliary localization in all mouse tissues examined. Interestingly, despite the varied functions of cilia in different tissues, we found that the primary cilia of varied cell types possess TZs of similar composition and Tmem231 is required to control ciliary composition in all of the diverse tissues examined.
Similar to mammalian Tmem231, C. elegans TMEM-231 controls protein entry into cilia, as TRAM-1a, a protein normally excluded from cilia, exhibits ciliary localization in tmem-231 mutant nematodes. This defect is comparable to that observed upon disruption of other TZ proteins in C. elegans, including the MKS module components MKSR-1 (B9d1), MKSR-2 (B9d2), and MKS-6 (Cc2d2a; Williams et al., 2011), and suggests that the TZ functions to promote the localization of certain membrane-associated proteins to the cilium and to prevent the ciliary entry of others.
Because of its roles in controlling ciliary composition, the TZ is essential for ciliary signaling and mammalian development (Reiter and Skarnes, 2006; Weatherbee et al., 2009; Dowdle et al., 2011; Garcia-Gonzalo et al., 2011; Sang et al., 2011; Chih et al., 2012). Similarly, Tmem231 is crucial for mouse development: Tmem231 mutant mice develop phenotypes that are hallmarks of MKS, including cystic kidneys, ductal plate malformations, and hindlimb polydactyly. The Tmem231 mutant phenocopies that of B9d1 mutants on a mixed genetic background, emphasizing their shared functions (Dowdle et al., 2011).
The two siblings that originally defined OFD3 are compound heterozygotes for missense mutations in TMEM231 predicted to produce p.Leu81Phe and p.Pro125Ala. The latter allele, encoding p.Pro125Ala, is homozygous in a MKS fetus and compound heterozygous with a separate heterozygous mutation (p.Asn90Ile, p.Gln182X, and c.664+4A>G) in three additional MKS families. Present in five different ciliopathy pedigrees (both OFD3 and MKS), it is unclear whether the hypomorphic p.Pro125Ala allele is a founder mutation that occurred in northern European populations or whether it is a mutational hotspot. Similarly, we identified a recurrent intronic c.664+4A>G mutation that disrupts the exon 5 splice donor site, resulting in aberrant splicing and premature termination. This change is present in four MKS pedigrees of varying ethnicity: two in homozygosity and two families with heterozygous c.664+4A>G changes with a second pathogenic heterozygous TMEM231 variant in trans. Together, our mutational data indicate that TMEM231 contributes to >5% of MKS cases, and the identification of two recurrent mutations will aid in the development of diagnostic tests for suspected ciliopathies.
To understand how nonsynonymous ciliopathy-associated TMEM231 mutations cause disease, we assessed if they disrupt the ability of Tmem231 to function with the MKS complex. The disease-associated mutant forms of Tmem231 are compromised in their ability to restore Arl13b to Tmem231−/− cilia. This phenotype is consistent with the failure of the disease-associated forms of Tmem231 to support formation of the MKS complex to the TZ: the mutant proteins fail to rescue B9d1 localization to the TZ in Tmem231−/− cilia. Interestingly, although two mutations destabilize Tmem231, all four mutant proteins interact with B9d1 similar to wild-type Tmem231, suggesting that the hypomorphic nature of these point mutants may be separate from their capacity to interact with other complex components.
We hypothesized that the OFD3-associated mutant proteins would be more functional than MKS-associated mutant proteins, given the greater severity of clinical ailments in MKS. However, one of the MKS-associated mutant proteins, p.Asn90Ile, displays more activity than the OFD3-associated mutant protein, p.Leu81Phe, in our cell-based assay. One possible explanation is that the two mutant TMEM231 alleles associated with OFD3 may partially complement each other. Partial intragenic complementation would indicate that Leu81 and Pro125 participate in separate aspects of TMEM231 function. Whereas p.Asn90Ile retains partial function in localizing Arl13b to cilia, this mutation interferes with other critical aspects of TMEM231 function such as B9d1 localization to the TZ. Given our inability to stratify TMEM231 allele pathogenicity by phenotype, we surmise that the MKS samples have an increased mutational burden in other ciliary or TZ genes in comparison to the OFD3-affected individuals, accounting, at least in part, for the different phenotypes of the two syndromes.
How might TMEM231 and the TZ control the composition of the ciliary membrane? The loss of ciliary proteins from Tmem231 mutant cells and the entrance of TRAM-1a into the cilia of C. elegans tmem-231 mutant animals suggest that the TZ acts as a membrane diffusion barrier, allowing ciliary proteins to enter and preventing entry of nonciliary proteins. Knockdown of B9d1 or Tmem231 can increase the entry rate of ciliary proteins into the cilium, suggesting that the MKS complex serves as an entry barrier (Chih et al., 2012).
Conversely, the MKS complex could act as an exit barrier, retaining certain membrane-associated proteins within cilia, while allowing exit of nonciliary membrane proteins. In this model, disruption of MKS complex function would not alter entry of ciliary membrane proteins, but would prevent their accumulation, resulting in decreased ciliary localization at steady state. We anticipate that assessing the role of the MKS complex in entry and exit of membrane proteins at the cilium will test these models.
Materials and methods
Plasmids
Mouse Tmem231 was cloned into the pGLAP5 destination vector (plasmid 19706, backbone: pEF5/FRT-V5, promotor: EF1a; Addgene) using Clonase II (Invitrogen) to produce pGLAP-Tmem231, as previously described (Dowdle et al., 2011). Similarly, mouse B9d1 and Mks1 were cloned in the pEF/V5/Frt (Invitrogen) destination vector using Clonase II to produce pEF/V5/Frt-B9d1 and pEF/V5/Frt-Mks1, as described previously (Dowdle et al., 2011). Mouse Tmem231 was cloned into a pCMV-Flag vector, such that the resulting construct expressed N-terminally flag-tagged mouse Tmem231 (pCMV-Flag-Tmem231). Mutant alleles were constructed from pCMV-Flag-Tmem231 using QuikChange site-directed mutagenesis with Herculase II DNA polymerase (Agilent Technologies). The four mutations are as follows: Tmem231p.L81F (c.241C>T), Tmem231p.N90I (c.269A>T), Tmem231p.P125A (c.373C>G), and Tmem231p.A216P (c.646G>C).
Immunoprecipitation and immunoblot
COS1 or 293T cells, grown on 15-cm-diameter plates, were transfected with pCMV-Flag-Tmem231 and pEF/V5/Frt-Mks1 or B9d1 using Lipofectamine 2000 (Life technologies) and lysed after 48 h in a buffer consisting of 50 mM Hepes, pH 7.4, 300 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10% glycerol, 0.3% NP-40, 0.5 mM DTT, and protease and phosphatase inhibitors. Lysates were cleared by centrifugation at 3,500 g for 20 min and incubated with FLAG-M2 beads (Sigma-Aldrich) for 4–6 h at 4°C. After 3× wash in lysis buffer, beads were resuspended in 6× SDS-PAGE loading buffer and denatured at 100°C for 5 min. Immunoprecipitations were resolved on 4–15% TGX gradient gels (Bio-Rad Laboratories) and transferred to PVDF membrane (EMD Millipore). Membranes were blocked with 5% nonfat dried milk in TBS with 0.1% Tween, and then incubated with primary antibodies followed by secondary antibodies in 5% nonfat dried milk in TBS with 0.1% Tween. Blots were developed with Western lightning enhanced chemiluminescence (PerkinElmer). Apparent molecular weights were determined using full-range rainbow prestained protein standards (GE Healthcare).
Mass spectrometry
IMCD3 cell lines harboring stable, single integrations of pGLAP5 vector or pGLAP5-Tmem231 were created using IMCD3 Flp-In cells (gift of M. Nachury, Stanford University, Palo Alto, CA). The IMCD3 Flp-In cells were created based on the Invitrogen Flp-In system. In brief, transfection of FRT site–bearing pGLAP5-based vectors and a Flp recombinase expression plasmid recombines the pGLAP-based vector at a genomic FRT site within the IMCD3 Flp-In genome. 50 15-cm-diameter plates each of pGLAP5 (control) and pGLAP5-Tmem231 stable cells were used for LAP purification, as described previously (Cheeseman and Desai, 2005). Cell lysates were cleared via centrifugation, GFP trap beads (ChromoTek) were used in the first capture, and after TEV cleavage, samples were further purified with S protein agarose. Purified LAP complexes were denatured in 3× SDS-PAGE sample buffer at 100°C for 10 min and resolved on a 4–15% Tris-glycine gradient gel (Bio-Rad Laboratories). After SYPRO Ruby (Invitrogen) staining, gel bands were excised and digested with trypsin and Protease Max (Promega). Acrylamide was used as the alkylation agent to produce a propanamide modification.
Samples were processed and interacting proteins were identified as previously described (Dowdle et al., 2011). In brief, nano-reverse phase HPLC using a 2D nanoLC (Eksigent) was performed with buffer A (0.1% formic acid in water), buffer B (0.1% formic acid in acetonitrile), and a self-packed fused silica column using duragel C18 matrix (Peeke). A linear gradient from 5 to 40% B over 60 min with a flow rate of 600 nl/min was used. A Michrom Advance source was interfaced with the nano-HPLC. An LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) in data-dependent acquisition mode was used to perform MS/MS in the ion trap of the top eight most intense ions. A full scan in the Orbitrap of the precursor ion was done with a resolution of 60,000 at 400 m/z. The .RAW files were then converted to mzXML format with msconvert script and searched with a Sorcerer (SageN) processor with Sequest. A precursor mass tolerance of 50 ppm was used to search the database. Positive interactions were defined as those with three or more identified peptides in the Tmem231-LAP purification and zero or one peptide identified in the control purification.
Mouse strains
The Institutional Animal Care and Use Committee at the University of California, San Francisco, approved all mouse protocols. B9d1+/− (B9d1tm1a(EUCOMM)Wtsi) mice have been described previously (Dowdle et al., 2011). In brief, the B9d1tm1a(EUCOMM)Wtsi gene trap was targeted to exon 3 of B9d1. These mice were outcrossed to CD1 mice (Charles River) and made conditionally ready by crossing to a mouse expressing to a ubiquitous FLPe recombinase-expressing mouse. Subsequently, the B9d1flox/flox mice were crossed to a ubiquitous Cre recombinase-expressing mouse to delete exon 3. The Tmem231+/− (B6N.129S5-Tmem231Gt(OST335874)Lex/Mmucd) strain was obtained from the Mutant Mouse Regional Resource Center at the University of California, Davis (Tang et al., 2010). The Tmem231 gene trap mutation was made in 129/SvEvBrd-derived embryonic stem cells. Chimeric animals were crossed to C57BL/6J albino mice (The Jackson Laboratory) to generate F1 heterozygous animals. F1 mice were intercrossed to produce F2 heterozygotes, which were backcrossed to C57BL/6NCrl (Charles River) to N11 before cryopreservation of spermatozoa. The Tmem231+/− strain was recovered at University of California, San Francisco, in a C56BL/6NCrl donor, and the mice were maintained by crossing to C56BL/6NCrl. The mixed background strain was created by outcrossing Tmem231+/− mice with CD1 mice (Charles River) for two generations.
Immunohistochemistry and histology
For tissue analysis, organs were fixed in 10% formalin overnight at 4°C, sectioned, and stained with hematoxylin and eosin according to standard protocols. Immunofluorescent staining of paraffin sections was performed as described previously (van Eyll et al., 2004). In brief, paraffin sections were rehydrated, permeabilized for 15 min with 0.3% Triton X-100 in PBS, blocked for 45 min with 3% milk/10% BSA/0.3% Triton X-100 in PBS at room temperature, and stained with primary and secondary antibodies. For B9d1 staining, an additional antigen retrieval was performed by incubating sections with 1% SDS dissolved in PBS for 5 min at room temperature followed by extensive rinses in PBS.
Antibodies
Primary antibodies used were mouse α-acetylated tubulin 6-11B-1 (TubAc; Sigma-Aldrich), rabbit α-Tmem231 (Sigma-Aldrich), rabbit α-Mks1 (Proteintech), rabbit α-Tmem67/Mks3 (Proteintech), rabbit α-Rpgrip1l (Proteintech), rabbit α-Nphp1 (gift of G. Pazour, University of Massachusetts Medical School, Worcester, MA; amino acids 1–209 [Benzing et al., 2001; Fliegauf et al., 2006]), rabbit α-Arl13b (Proteintech), mouse α-Arl13b (Proteintech), rabbit α-INPP5e (gift of S. Schurmans, Université de Liège, Liège, Belgium; antigen: N-terminal MLQGQPPNTEKKLIP peptide [Jacoby et al., 2009]), rabbit α-Adcy3 (Proteintech), rabbit α-IFT88 (Proteintech), rabbit α-Pkd2 (YCC; gift of S. Somlo, Yale School of Medicine, New Haven, CT; antigen: amino acids 687–962 [Wu et al., 1998]), rabbit goat α–γ-tubulin (Santa Cruz Biotechnology, Inc.), goat α-V5 (Abcam), and mouse α-FLAG-M2 (Sigma-Aldrich). Rabbit α-B9d1 antibody was generated by injecting a full-length mouse B9d1 protein fused to a His tag into rabbits (Pacific Immunology) and affinity purifying the antibody with recombinant B9d1-6×His. Secondary antibodies are as follows: donkey α-mouse 647, donkey α-mouse 488, donkey α-rabbit 488, donkey α-rabbit 647, and donkey α-goat 568 (Alexa Fluor).
Cell lines
MEFs were derived from E13.5 littermate-matched wild-type and B9d1 mutant or Tmem231 mutant embryos and grown in Advanced DMEM supplemented with 10% FBS, 1% GlutaMax, and 1% antibiotic-antimycotic (Invitrogen). COS-1 and 293T cells were grown in DMEM supplemented with 10% FBS and 1% GlutaMax.
C. elegans strains used and localization studies
All strains (Table S2) were maintained and cultured on NGM plates at 20°C. The tmem-231(tm5963), mks-5(tm3100), and nphp-4(tm925) mutations were obtained from the National Bioresource Project (Japan) and we previously generated the mks-2(nx111) mutant (Huang et al., 2011). All mutants were outcrossed to wild type (N2) at least five times.
The construct harboring GFP-tagged C. elegans TMEM-231 was generated by creating a translational fusion construct with its native promoter (1,620-bp upstream of the start codon) and all exons and introns fused in-frame to EGFP at the C-terminal end of the coding region together with a 3′ UTR from unc-54. Multiple transgenic lines for this construct were generated by coinjection of the rol-6(su1006) and screening for roller worms as described previously (Williams et al., 2011). Standard mating procedures were used to introduce the GFP-tagged TMEM-231 protein construct into mks-2, mks-5, and nphp-4 mutant backgrounds and fluorescently labeled protein constructs for TMEM-17, MKS-5, NPHP-1, and TRAM-1 into the tmem-231 mutant background. Single-worm PCR amplifications were used to genotype the various mutants. Information for all mutant strains used, namely mks-2(nx111) (3,573-bp deletion), mks-5(tm3100) (540-bp deletion), tmem-231(tm5963) (297-bp deletion), and nphp-4(tm925) (1,109-bp deletion), is available from WormBase.
For microscopy analyses, live worms were anaesthetized using 10 mM levamisole, mounted on 5% agarose pads, and visualized at 20°C by spinning-disc confocal microscopy performed on an inverted microscope (Zxio Observer; Carl Zeiss) equipped with a WaveFX spinning disc confocal system (Quorum Technologies) outfitted with a 9100 EMCCD camera (Hamamatsu Photonics) and 100× oil objective, 1.40 NA. Volocity (PerkinElmer) was used for image acquisition and subsequent visualization. The subcellular localization of the fluorescent proteins was assessed in both the wild-type animals and the indicated mutant backgrounds. Mislocalization phenotypes were confirmed by analyzing at least 30 animals for each strain.
Mutation analysis of ciliopathy patients
Full ethics approval was obtained by the Northern and Yorkshire Research Ethics Committee (UK), the University of Michigan, Necker Hospital (France), and Duke University Medical Center, and informed written consent was obtained from patients and their relatives. DNA was extracted from whole blood or fetal tissue using standard techniques. All coding exons and exon–intron boundaries of TMEM231 were amplified in 1,056 patients with nephronophthisis-related ciliopathies in a large-scale microfluidic multiplex PCR by using a Fluidigm Access Array system (48.48). By primer pooling and microfluidics, this approach allows for simultaneous generation of up to 600 amplicons for each of 48 patient samples at a time (Halbritter et al., 2012, 2013). In a second PCR reaction, amplicons were barcoded by adding a patient-specific 10-bp nucleotide sequence. Consecutively, generated amplicons from 1,056 patients were pooled and submitted to next generation sequencing, performed on a Genome Analyzer II platform (Illumina). Whole exome sequencing and variant filtering was conducted as described previously (Gordon et al., 2013). In brief, SureSelect libraries (Agilent Technologies) were prepared from 3 µg of genomic DNA sheared with an S2 Ultrasonicator (Covaris) according to the manufacturers’ instructions. Exome capture was performed with the 51-Mb SureSelect Human All Exon kit V5 (Agilent Technologies) via a multiplex approach with molecular barcodes for traceable identity of samples. Sequencing was performed on a pool of barcoded exome libraries with a HiSeq2500 (Illumina) machine. 76-bp paired-end reads were generated with the HiSeq2500. After demultiplexing, paired-end sequences were aligned to the reference human genome (hg19, UCSC Genome Browser) with the Burrows-Wheeler Aligner (Illumina). The mean depth of coverage obtained for each sample was >80×, and >90% of the exome was covered at least 15×.
Ciliary exome–targeted sequencing and bioinformatics filtering was conducted using custom SureSelect capture kits (Agilent Technologies) targeting 4.5 Mb of 20,168 exons (1,221 ciliary candidate genes), including TMEM231 (Thomas et al., 2012; Failler et al., 2014). In brief, SureSelect libraries were prepared from 3 µg of 300 genomic DNA samples sheared with an S2 Ultrasonicator, according to the manufacturers’ instructions. Precapture SOLiD libraries were prepared without any barcode. The SOLiD molecular barcodes for traceable ID of samples were added at the end of the capture step. The Ovation Ultralow System (NuGEN Technologies) was used to prepare HiSeq2500 precapture barcoded libraries. The ciliome capture by hybridization was performed on a pool of 10–16 barcoded precapture libraries. Sequencing was performed on pools of barcoded ciliome libraries (64 barcoded ciliome libraries per SOLiD FlowChip,\ and 16 ciliome libraries per HiSeq FlowCell lane) using SOLiD5500XL (Life Technologies) and HiSeq2500. Paired-end reads were generated (75 + 35 for SOLiD and 100 + 100 for HiSeq) and mapped on the human genome reference (NCBI build37/hg19 version) using Burrows-Wheeler Aligner or mapread (SOLiD). Downstream processing was performed with the Genome Analysis Toolkit, SAMtools, and Picard Tools, following documented best practices (Van der Auwera et al., 2013). All variants were annotated using a software system developed by the Paris Descartes University Bioinformatics platform. The mean depth of coverage obtained was >90× and >89% of the exome was covered at least 15×. Different filters were applied to exclude all variants located in nonexonic regions, pseudogenes, UTRs, or known polymorphic variants with a frequency ∼1% (i.e., present in databases such as dbSNP and 1000 Genome Projects, and all variants identified by in-house exome sequencing of 5,150 exomes and 1,020 ciliomes). The functional consequence of missense variants was predicted using the SIFT (http://sift.jcvi.org/www/SIFT_enst_submit.html) and PolyPhen2 programs (http://genetics.bwh.harvard.edu/pph2/).
For candidate gene analysis of 95 MKS samples, we conducted bidirectional Sanger sequencing of exons and splice junctions according to standard methodology. Mutations identified in all studies were confirmed using Sanger sequencing in patients and their parents to confirm segregation.
RT-PCR to evaluate splice-site mutations
We cultured primary skin fibroblasts from MKS fetus MKS-374 according to standard procedures and harvested total RNA in Trizol according to the manufacturer’s instructions. We generated oligodT-primed cDNA using Superscript III reverse transcription (Life Technologies), and conducted PCR using primers placed in exons 4 and 6 (TMEM231-4F-RT: 5′-ACCCTCGTGATGCAGAGC-3′; TMEM231-6R-RT: 5′-TTGGGATCATTCAGGACGGT-3′; TMEM231-6R2-RT: 5′-GAAATGACTTCCACAGGGTAT-3′). Resulting PCR products were sequenced directly using bidirectional Sanger sequencing.
Immunofluorescence
For cilia and TZ staining, MEFs were plated on glass coverslips, grown to confluence, and starved for 24–72 h in OptiMEM. For Mks1, Tmem67, and Inpp5e staining, cells were fixed in methanol for 3 min at −20°C. For B9d1, Tmem231, and Rpgrip1l, cells were fixed for 10 min at RT (20–25°C) in 4% methanol-free formaldehyde (FA) in PBS, followed by antigen retrieval for 5 min at RT in 1% SDS dissolved in PBS. After SDS antigen retrieval, cells were washed 3 × 10 min with PBS. Arl13b, Pkd2, Ift88, Nphp1, and Adcy3 cells were fixed for 10 min at RT with 5% FA in PBS, followed by methanol for 3 min at −20°C. After fixation, cells were blocked for 60 min at RT or overnight at 4°C in dPBS + 0.1% Triton X-100 + 2.5% BSA (IF block). Cells were incubated with primary antibodies diluted in IF block overnight at 4°C. Coverslips were washed with dPBS, and then incubated with Alexa-conjugated secondary antibodies raised in goat or donkey (Invitrogen) at RT. After three dPBS washes, coverslips were mounted using ProLong Gold antifade (Invitrogen) and sealed with nail polish. Cells were imaged with a TCS SP5 microscope (Leica). Images were processed using FIJI (ImageJ).
Rescue assays were modified and performed similar to previously described experiments (Garcia-Gonzalo et al., 2011). In brief, 2 × 105 MEFs were plated per coverslip in a 24-well plate in Advanced DMEM supplemented with 10% FBS, 1% GlutaMax, and 1% antibiotic-antimycotic, and transfected with 0.4 µg of empty FLAG, wild-type FLAG-Tmem231, or mutant FLAG-Tmem231 constructs using JetPrime (Polyplus Transfection). The transfection medium was replaced with full media at 24 h after transfection. At 48 h after transfection, cells were starved in OptiMEM for 48 h. Cells were fixed for 10 min at RT with 5% FA in PBS, followed by methanol for 3 min at −20°C and stained with one of the following combinations: mouse α-TubAc, rabbit α-Arl13b, and goat α-γ-tubulin; or mouse α-Arl13b, rabbit α-B9d1, and goat α-γ-tubulin. Samples were imaged on a TCS SPE DM5500Q confocal microscope (Leica) at room temperature, using an ACS Apochromat 63× objective (1.30 NA, oil) with the LAS AF acquisition software and Type F Immersion Liquid (Leica), taking care to use the same settings for each coverslip.
Quantitation
FIJI was used for image quantitation. To determine percent ciliogenesis, total cilia were counted manually and total nuclei were determined using the Watershed function. To quantify ciliary proteins that localize along the ciliary length (Arl13b, Pkd2, Adcy3, Inpp5e, and TubAc), a segmented line (width = 4 px) was drawn from the base to the tip of the cilium according to the TubAc channel, and area, length, and integrated density were measured. To quantify TZ proteins, a circle (1.02 × 1.02 µm) was used to measure the integrated density between the γ-tubulin and TubAc staining. Finally, a freehand selection around the γ-tubulin staining was used to measure the area and integrated density of the centrosome at the base of cilia. For each of these measurements, two background measurements were taken immediately adjacent the point of interest. The raw integrated density of the background measurements were divided by the area, and these were averaged for the corrected background (A.U./µm2). This was subtracted from the raw integrated density per micrometer squared of each experimental measurement to provide the corrected experimental arbitrary unit per micrometer squared. These ciliary intensity values were thresholded and plotted in GraphPad Prism with error bars representing the 95% confidence interval of the dataset. An unpaired Student’s t test with Welch’s correction was used to determine the statistical significance of the dataset. Finally, the data were normalized to the mean of the wild-type dataset. The thresholded dataset was used to determine the percentage of positive/negative cilia for each marker.
Online supplemental material
Fig. S1 illustrates the requirements of Tmem231 and B9d1 for ciliary membrane–associated protein localization and ciliary length control. Fig. S2 depicts the requirement of Tmem231 and B9d1 for the localization of other MKS complex components to the TZ. Fig. S3 is an alignment of the sequences of Tmem231 orthologues from diverse ciliated species. Fig. S4 shows sequences of the TMEM231 mutations identified in the OFD3- and MKS-affected individuals described in this study. Fig. S5 displays quantification of the rescue assays (related to Fig. 6). Table S1 shows mutations of TMEM231 associated with OFD3 and MKS. Table S2 shows the C. elegans strains used. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201411087/DC1.
Supplementary Material
Acknowledgments
We thank the patients and relatives for contributing to the study. Additionally, we thank Karen Fontaine Marchand, Muriel Poilpré, Anne-Lise Delezoide, Elisabeth Martin de Lassalle, Odile Boute, Martine Bucourt, Christine Muti, Tahar Yaacoubi, Soumaya Mougou-Zerelli, Martine Le Merrer, Frédérique Le Breton, Carine Abel, Stéphane Triau, and Agnès Guichet for referral of the MKS families for research studies. We also thank Christine Bole-Feysot (Genomics Platform) and Patrick Nitschke (Bioinformatics platform) at the Imagine Institute, Paris, for assistance with next-generation sequencing and analysis. Members of the Reiter laboratory provided valuable discussion. We thank the Cardiovascular Research Institute Imaging Core at the University of California, San Francisco, and Chris Adams at the Standford BioX mass spectrometry core. We thank Maxence Nachury, Stéphane Schurmans, Stefan Somlo, and Gregory Pazour for providing cell lines, plasmids, or antibodies.
This work was supported by grants from the Agence Nationale de la Recherche (ANR; 20100BLAN112202 to T. Attié-Bitach and S. Saunier) and investments for the future program (ANR-10-IAHY-01 to T. Attié-Bitach and S. Saunier), and the Fondation pour la Recherche Médicale (DEQ20130326532 to S. Saunier). M.R. Leroux acknowledges funding from the March of Dimes and Canadian Institutes of Health Research, and a senior scholar award from the Michael Smith Foundation for Health Research. E.C. Roberson acknowledges support from the Achievement Rewards for College Scientists Foundation. This work was also funded by US National Institutes of Health (NIH) grants DK072301 to N. Katsanis and E.E. Davis, HD042601 to N. Katsanis, NS039818 to A. Ashley-Koch, and DK-068306 to F. Hildebrandt; grant ANR 2010 FOETOCILPATH N° 1122 01 to T. Attié-Bitach; funding from EU seventh FP under GA nr. 241955, project SYSCILIA, to N. Katsanis and E.E. Davis; and NIH (AR054396 and GM095941), the Burroughs Wellcome Fund, the Packard Foundation, and the Sandler Family Supporting Foundation to J.F. Reiter. N. Katsanis is a distinguished Jean and George Brumley Professor. F. Hildebrandt is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and a Warren E. Grupe Professor.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Adcy3
- Adenylyl cyclase 3
- E
- embryonic day
- LAP
- localization and affinity purification
- MEF
- mouse embryonic fibroblast
- MKS
- Meckel syndrome
- NPHP
- Nephronophthisis
- OFD3
- orofaciodigital syndrome 3
- Pkd2
- Polycystic kidney disease 2
- Smo
- Smoothened
- TZ
- transition zone
References
- Arts H.H., Doherty D., van Beersum S.E.C., Parisi M.A., Letteboer S.J.F., Gorden N.T., Peters T.A., Märker T., Voesenek K., Kartono A., et al. 2007. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 39:882–888 10.1038/ng2069 [DOI] [PubMed] [Google Scholar]
- Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B., et al. 2007. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 80:186–194 10.1086/510499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraitser M.1986. The orofaciodigital (OFD) syndromes. J. Med. Genet. 23:116–119 10.1136/jmg.23.2.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzing T., Gerke P., Höpker K., Hildebrandt F., Kim E., and Walz G.. 2001. Nephrocystin interacts with Pyk2, p130Cas, and tensin and triggers phosphorylation of Pyk2. Proc. Natl. Acad. Sci. USA. 98:9784–9789 10.1073/pnas.171269898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas N.J., Inglis P.N., Li C., Robinson J.F., Parker J.D.K., Healey M.P., Davis E.E., Inglis C.D., Toivonen T., Cottell D.C., et al. 2009. Functional interactions between the ciliopathy-associated Meckel syndrome 1 (MKS1) protein and two novel MKS1-related (MKSR) proteins. J. Cell Sci. 122:611–624 10.1242/jcs.028621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S., Hori Y., Kaplan O.I., Kida K., Toivenon T., Foley-Fisher C., Cottell D., Katada T., Kontani K., and Blacque O.E.. 2010. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188:953–969 10.1083/jcb.200908133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1. [DOI] [PubMed] [Google Scholar]
- Chih B., Liu P., Chinn Y., Chalouni C., Komuves L.G., Hass P.E., Sandoval W., and Peterson A.S.. 2012. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14:61–72 10.1038/ncb2410 [DOI] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y.R., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., and Witman G.B.. 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190:927–940 10.1083/jcb.201006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.E., and Katsanis N.. 2012. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr. Opin. Genet. Dev. 22:290–303 10.1016/j.gde.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle W.E., Robinson J.F., Kneist A., Sirerol-Piquer M.S., Frints S.G.M., Corbit K.C., Zaghloul N.A., van Lijnschoten G., Mulders L., Verver D.E., et al. 2011. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am. J. Hum. Genet. 89:94–110 10.1016/j.ajhg.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failler M., Gee H.Y., Krug P., Joo K., Halbritter J., Belkacem L., Filhol E., Porath J.D., Braun D.A., Schueler M., et al. 2014. Mutations of CEP83 cause infantile nephronophthisis and intellectual disability. Am. J. Hum. Genet. 94:905–914 10.1016/j.ajhg.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Horvath J., von Schnakenburg C., Olbrich H., Müller D., Thumfart J., Schermer B., Pazour G.J., Neumann H.P.H., Zentgraf H., et al. 2006. Nephrocystin specifically localizes to the transition zone of renal and respiratory cilia and photoreceptor connecting cilia. J. Am. Soc. Nephrol. 17:2424–2433 10.1681/ASN.2005121351 [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., and Reiter J.F.. 2012. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J. Cell Biol. 197:697–709 10.1083/jcb.201111146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Corbit K.C., Sirerol-Piquer M.S., Ramaswami G., Otto E.A., Noriega T.R., Seol A.D., Robinson J.F., Bennett C.L., Josifova D.J., et al. 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43:776–784 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., Ocbina P.J.R., and Anderson K.V.. 2009. The primary cilium as a Hedgehog signal transduction machine. Methods Cell Biol. 94:199–222 10.1016/S0091-679X(08)94010-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorden N.T., Arts H.H., Parisi M.A., Coene K.L.M., Letteboer S.J.F., van Beersum S.E.C., Mans D.A., Hikida A., Eckert M., Knutzen D., et al. 2008. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet. 83:559–571 10.1016/j.ajhg.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.T., Petit F., Kroisel P.M., Jakobsen L., Zechi-Ceide R.M., Oufadem M., Bole-Feysot C., Pruvost S., Masson C., Tores F., et al. 2013. Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. Am. J. Hum. Genet. 93:1118–1125 10.1016/j.ajhg.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbritter J., Diaz K., Chaki M., Porath J.D., Tarrier B., Fu C., Innis J.L., Allen S.J., Lyons R.H., Stefanidis C.J., et al. 2012. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J. Med. Genet. 49:756–767 10.1136/jmedgenet-2012-100973 [DOI] [PubMed] [Google Scholar]
- Halbritter J., Bizet A.A., Schmidts M., Porath J.D., Braun D.A., Gee H.Y., McInerney-Leo A.M., Krug P., Filhol E., Davis E.E., et al. 2013. Defects in the IFT-B component IFT172 cause Jeune and Mainzer-Saldino syndromes in humans. Am. J. Hum. Genet. 93:915–925 10.1016/j.ajhg.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F., Benzing T., and Katsanis N.. 2011. Ciliopathies. N. Engl. J. Med. 364:1533–1543 10.1056/NEJMra1010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J.E., and Scott M.P.. 2005. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6:306–317 10.1038/nrm1622 [DOI] [PubMed] [Google Scholar]
- Hopp K., Heyer C.M., Hommerding C.J., Henke S.A., Sundsbak J.L., Patel S., Patel P., Consugar M.B., Czarnecki P.G., Gliem T.J., et al. 2011. B9D1 is revealed as a novel Meckel syndrome (MKS) gene by targeted exon-enriched next-generation sequencing and deletion analysis. Hum. Mol. Genet. 20:2524–2534 10.1093/hmg/ddr151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Milenkovic L., Jin H., Scott M.P., Nachury M.V., Spiliotis E.T., and Nelson W.J.. 2010. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 329:436–439 10.1126/science.1191054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Szymanska K., Jensen V.L., Janecke A.R., Innes A.M., Davis E.E., Frosk P., Li C., Willer J.R., Chodirker B.N., et al. 2011. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 89:713–730 10.1016/j.ajhg.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., and Anderson K.V.. 2005. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA. 102:11325–11330 10.1073/pnas.0505328102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert M.C., Weihbrecht K., Searby C.C., Li Y., Pope R.M., Sheffield V.C., and Seo S.. 2012. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA. 109:19691–19696 10.1073/pnas.1210916109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby M., Cox J.J., Gayral S., Hampshire D.J., Ayub M., Blockmans M., Pernot E., Kisseleva M.V., Compère P., Schiffmann S.N., et al. 2009. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet. 41:1027–1031 10.1038/ng.427 [DOI] [PubMed] [Google Scholar]
- Mougou-Zerelli S., Thomas S., Szenker E., Audollent S., Elkhartoufi N., Babarit C., Romano S., Salomon R., Amiel J., Esculpavit C., et al. 2009. CC2D2A mutations in Meckel and Joubert syndromes indicate a genotype–phenotype correlation. Hum. Mutat. 30:1574–1582 10.1002/humu.21116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E.A., Tory K., Attanasio M., Zhou W., Chaki M., Paruchuri Y., Wise E.L., Wolf M.T.F., Utsch B., Becker C., et al. 2009. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11). J. Med. Genet. 46:663–670 10.1136/jmg.2009.066613 [DOI] [PubMed] [Google Scholar]
- Reiter J.F., and Skarnes W.C.. 2006. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 20:22–27 10.1101/gad.1363606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani M., Micalizzi A., Kraoua I., Dotti M.T., Cavallin M., Sztriha L., Ruta R., Mancini F., Mazza T., Castellana S., et al. 2014. Mutations in B9D1 and MKS1 cause mild Joubert syndrome: expanding the genetic overlap with the lethal ciliopathy Meckel syndrome. Orphanet J. Rare Dis. 9:72 10.1186/1750-1172-9-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen R.1984. The Meckel syndrome: clinicopathological findings in 67 patients. Am. J. Med. Genet. 18:671–689 10.1002/ajmg.1320180414 [DOI] [PubMed] [Google Scholar]
- Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M., et al. 2011. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 145:513–528 10.1016/j.cell.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R., Faqeih E., Seidahmed M.Z., Sunker A., Alali F.E., AlQahtani K., and Alkuraya F.S.. 2011. A TCTN2 mutation defines a novel Meckel Gruber syndrome locus. Hum. Mutat. 32:573–578 10.1002/humu.21507 [DOI] [PubMed] [Google Scholar]
- Shaheen R., Ansari S., Mardawi E.A., Alshammari M.J., and Alkuraya F.S.. 2013. Mutations in TMEM231 cause Meckel-Gruber syndrome. J. Med. Genet. 50:160–162 10.1136/jmedgenet-2012-101431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.A., and Gardner-Medwin D.. 1993. Orofaciodigital syndrome type III in two sibs. J. Med. Genet. 30:870–872 10.1136/jmg.30.10.870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith U.M., Consugar M., Tee L.J., McKee B.M., Maina E.N., Whelan S., Morgan N.V., Goranson E., Gissen P., Lilliquist S., et al. 2006. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat. Genet. 38:191–196. [DOI] [PubMed] [Google Scholar]
- Srour M., Hamdan F.F., Schwartzentruber J.A., Patry L., Ospina L.H., Shevell M.I., Désilets V., Dobrzeniecka S., Mathonnet G., Lemyre E., et al. 2012. Mutations in TMEM231 cause Joubert syndrome in French Canadians. J. Med. Genet. 49:636–641 10.1136/jmedgenet-2012-101132 [DOI] [PubMed] [Google Scholar]
- Sugarman G.I., Katakia M., and Menkes J.. 1971. See-saw winking in a familial oral-facial-digital syndrome. Clin. Genet. 2:248–254 10.1111/j.1399-0004.1971.tb00285.x [DOI] [PubMed] [Google Scholar]
- Tang T., Li L., Tang J., Li Y., Lin W.Y., Martin F., Grant D., Solloway M., Parker L., Ye W., et al. 2010. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28:749–755 10.1038/nbt.1644 [DOI] [PubMed] [Google Scholar]
- Thomas S., Legendre M., Saunier S., Bessières B., Alby C., Bonnière M., Toutain A., Loeuillet L., Szymanska K., Jossic F., et al. 2012. TCTN3 mutations cause Mohr-Majewski syndrome. Am. J. Hum. Genet. 91:372–378 10.1016/j.ajhg.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuz K., Hsiao Y.C., Juárez O., Shi B., Harmon E.Y., Phelps I.G., Lennartz M.R., Glass I.A., Doherty D., and Ferland R.J.. 2013. The Joubert syndrome-associated missense mutation (V443D) in the Abelson-helper integration site 1 (AHI1) protein alters its localization and protein-protein interactions. J. Biol. Chem. 288:13676–13694 10.1074/jbc.M112.420786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 11:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eyll J.M., Pierreux C.E., Lemaigre F.P., and Rousseau G.G.. 2004. Shh-dependent differentiation of intestinal tissue from embryonic pancreas by activin A. J. Cell Sci. 117:2077–2086 10.1242/jcs.01067 [DOI] [PubMed] [Google Scholar]
- Weatherbee S.D., Niswander L.A., and Anderson K.V.. 2009. A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Hum. Mol. Genet. 18:4565–4575 10.1093/hmg/ddp422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.L., Li C., Kida K., Inglis P.N., Mohan S., Semenec L., Bialas N.J., Stupay R.M., Chen N., Blacque O.E., et al. 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192:1023–1041 10.1083/jcb.201012116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., D’Agati V., Cai Y., Markowitz G., Park J.H., Reynolds D.M., Maeda Y., Le T.C., Hou H. Jr, Kucherlapati R., et al. 1998. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell. 93:177–188 10.1016/S0092-8674(00)81570-6 [DOI] [PubMed] [Google Scholar]
- Yoder B.K., Hou X., and Guay-Woodford L.M.. 2002. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 13:2508–2516 10.1097/01.ASN.0000029587.47950.25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.