Abstract
Polyploidy or whole-genome duplication occurs in some animals and many flowering plants, including many important crops such as wheat, cotton and oilseed rape. The prevalence of polyploidy in the plant kingdom suggests it as an important evolutionary feature for plant speciation and crop domestication. Studies of natural and synthetic polyploids have revealed rapid and dynamic changes in genomic structure and gene expression after polyploid formation. Growing evidence suggests that epigenetic modifications can alter homoeologous gene expression and reprogram gene expression networks, which allows polyploids to establish new cytotypes, grow vigorously and adapt in local environments. Sequence and gene expression changes in polyploids have been well documented and reviewed elsewhere. This review is focused on developmental regulation and epigenetic changes including DNA methylation and histone modifications in polyploids.
Keywords: DNA methylation, polyploidy, gene expression, small RNAs, epigenetics, transposons, development
Introduction
Polyploidy or whole-genome duplication (WGD) describes an organism or cell that contains two or more sets of chromosomes. Since the report of polyploid evening primrose (Oenothera lamarkiana) in 1907 [1], polyploidy has been extensively studied from genetics and evolution [2] to genomics and function [3,4]. Polyploidy is widespread and is particularly common in angiosperms [5-11] and is a key driver of macro-evolutionary success [12]. Estimates indicate two rounds of ancestral WGDs occurred before the divergence of extant seed plants and angiosperms, giving rise to the diversification of genes and pathways important to seed and flower development and eventually the dominance of angiosperms on the planet [13,14]. To date, more than 70% of flowering plants are considered to be polyploids [15]. In addition, many plant species, such as soybean and maize, are paleopolyploids (ancient polyploids) [16,17]. However, polyploidy was traditionally considered an evolutionary dead end [11,12,18]. Polyploidy and aneuploidy often lead to carcinogenesis or birth defects in humans [19], and aneuploidy impairs proliferation and alters metabolic properties in mouse cell lines [20] and induces proteomic changes and phenotypic variation in yeast [21]. Aneuploids generally have larger changes than polyploids because dosage imbalance affects the stability and interaction of a protein in a regulatory complex [22].
Polyploids can be divided to autopolyploids and allopolyploids, according to their origin and composition of chromosomes [8,18]. The former are formed via duplication of a single genome, whereas the latter result from merging two or more divergent genomes (Figure 1). The distinction between them is unclear in some polyploids such as sugarcane [23]. As the frequency of intraspecific mating is much higher than interspecific crossing, autopolyploidy is predicted to be more common than allopolyploidy [24]. However, allopolyploids are more common than autopolyploids in nature [9,25]. This is probably because both heterozygosity and genetic redundancy are fixed in allopolyploids, leading to novel traits and improved adaptation to compete with progenitors and occupy new habitats [26,27]. However, relative contributions of ploidy and hybridity to physiology and phenotypes have not been tested until recently in maize hybrids [28] and Arabidopsis polyploids [29] at various ploidy levels. While ploidy and hybridization affect cell size and biomass, respectively, both ploidy and hybridization change seed size and weight [29], suggesting distinctive roles of ploidy and hybridization in promoting vegetative and reproductive traits.
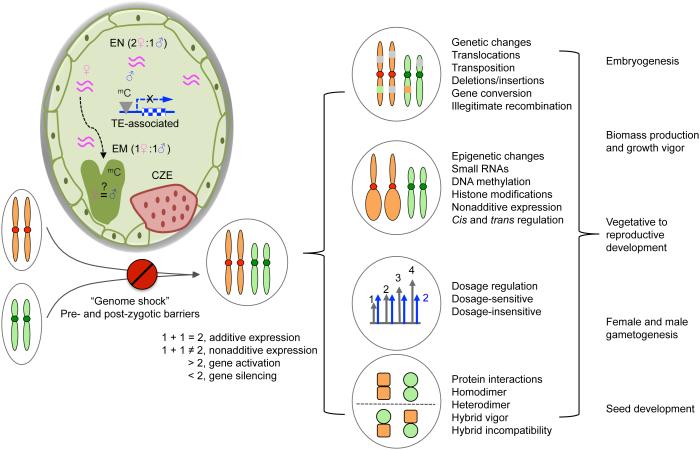
Figure 1.
Genetic and epigenetic mechanisms for gene expression and developmental changes in polyploids and hybrids. A bottleneck for allopolyploid formation is prezygotic and postzygotic barriers, the latter of which is related to hybrid incompatibility and ‘genome shock’ in the endosperm (EN) and embryo (EM). The hybrid incompatibility is partly mediated by small RNAs that can sense the dosage and divergence between maternal and paternal genomes in endosperm that contain 2:1 (maternal:paternal) genome ratio. These siRNAs (short-waved lines) can induce methylation (mC) of TE-associated genes and alter their expression in the endosperm including chalazal endosperm (CZE) that controls the endosperm cellularization process. Whether or not siRNAs in the endosperm are transferred into the embryo remains to be tested. After overcoming the hybrid incompatibility, the newly formed allopolyploids may undergo (1) genetic changes including various genomic rearrangements, (2) epigenetic changes that affect gene expression through histone modifications, DNA methylation and small RNAs, (3) dosage regulation and (4) orthologous/homoeologous protein interactions. Some of these changes are not mutually exclusive. A consequence is additive and nonadditive (deviated from the mid-parent value) gene expression, including gene activation and silencing. These induced genetic and epigenetic changes in homoeologous genes can affect embryogenesis, growth vigor, transition from vegetative to reproductive development, gametogenesis and seed development in response to developmental and environmental cues, some of which can be selected and maintained in the new allopolyploid species.
Polyploidization leads to instantaneous WGD, which is often followed by a diploidization process characterized by rapid genomic reorganization and massive gene loss [5,9-11] (Figure 1). This phenomenon has been observed in newly synthesized allopolyploid wheat [30], Brassica [31] and Tragopogon [32]. Some other allopolyploids, such as cotton [33] and Arabidopsis [34], do not show many changes in their genomic sequences. Recent sequencing results revealed relatively stable genome organization in domesticated allohexaploid wheat [35] and allotetraploid Brassica napus [36]. This discrepancy of genome stability between resynthesized and domesticated allopolyploids could be explained by the fact that the exact progenitors of allopolyploid crops no longer exist. Alternatively, natural selection has eliminated those early allopolyploids associated with many genomic rearrangements and epigenetic changes.
Polyploidy causes nuclear enlargement, chromosomal rearrangement and epigenetic changes, leading to reprogramming of transcriptome, proteome and metabolome networks [5]. Although these changes offer evolutionary flexibility and phenotypic diversity for new polyploids, they also invoke disadvantages, including increasing errors of chromosome pairing, reducing mating opportunity, and altering optimized patterns of gene expression and epigenetic modifications that are inherited from the parents [6,9]. Few mammals are polyploids, probably due to these disadvantages including disruption of imprinting gene expression and sex chromosome balance [8,37,38]. However, ploidy variation is often tolerated in plants, likely because their plastic genomes have evolved some mechanisms to mediate these deleterious changes [5]. The plasticity in plants is not only reflected in the genome structure and function but also in developmental regulation (Figure 1). Every plant cell is totipotent (equivalent to stem cells in animals) and has the potential to develop a new plant [39]. This genomic and developmental plasticity is predicted to associate with epigenetic changes in polyploid plants, which could promote growth, development and adaptation in response to myriad environmental cues or stress. In this review, we update views and perspectives of epigenetic (DNA methylation and histone modification) and developmental regulation in polyploidy (Figure 1).
DNA methylation changes in polyploids
DNA methylation affects molecular processes of plant and animal development, including transposon silencing, virus defense and gene imprinting [40-42]. In plants, DNA methylation occurs in CG, CHG and CHH (H = A, T or C) contexts through distinct pathways. In Arabidopsis, METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) are responsible for maintenance of CG and CHG methylation, respectively [43-45]. CHH methylation is established de novo by DOMAINS REARRANGED METHYLTRANSFERASE 1 and 2 (DRM1 and DRM2) guided by 24-nt small interfering RNAs (siRNAs) and CHROMOMETHYLASE 2 (CMT2) through interacting with DECREASE IN DNA METHYLATION1 (DDM1) [46-48]. In addition to the establishment and maintenance, DNA methylation can be removed by DNA demethylases DEMETER (DME) [49], REPRESSOR OF SILENCING 1 (ROS1) [50] and DEMETER-LIKE 2 and 3 (DML2 and DML3) [49,51]. DNA methylation may change, inducing activation or repression of transposons and genes, in response to developmental and environmental cues.
Intergenomic interactions between progenitor genomes in allopolyploids are predicted to induce epigenetic changes including DNA methylation and histone modifications [8]. DNA methylation variation between allopolyploids and their progenitors has been documented in many plants [52-57]. Methylation-sensitive amplification polymorphism analysis in Arabidopsis allotetraploids shows 8.3% of fragments examined are differentially methylated between F3 synthetic allotetraploids and their parents, A. thaliana and A. arenosa [52]. DNA methylation change (1.4–7%) is also observed in synthetic allopolyploid Brassica napus [53,58]. Gene silencing is stochastic during selfing generations in resynthesized Arabidopsis allotetraploids, and at least several genes are reactivated in both ddm1- and met1-RNAi lines [57]. In addition, demethylation of centromeric repeats and gene-specific regions are associated with transposable element (TE) activation in met1-RNAi lines of Arabidopsis allopolyploids [59]. In Arabidopsis allotetraploids, A. thaliana homoeologs of teosinte-branched1 and cycloidea3 (TCP3) and its neighboring genes are hypermethylated and silenced, which can be reactivated by blocking DNA methylation, suggesting a causal role for DNA methylation in silencing homoeologous genes in allopolyploids [55]. However, in a natural allotetraploid A. suecica, the A. arenosa MIR172b locus but not that of A. thaliana is hypermethylated and associated with low expression levels of the A. arenosa allele. These data indicate genome-specific changes of DNA methylation between homoeologous sequences in allopolyploids [54] (Figure 2). These homoeologous sequences coincide with genomic rearrangements between A. thaliana and A. arenosa, suggesting a correlation between genetic and epigenetic variation.
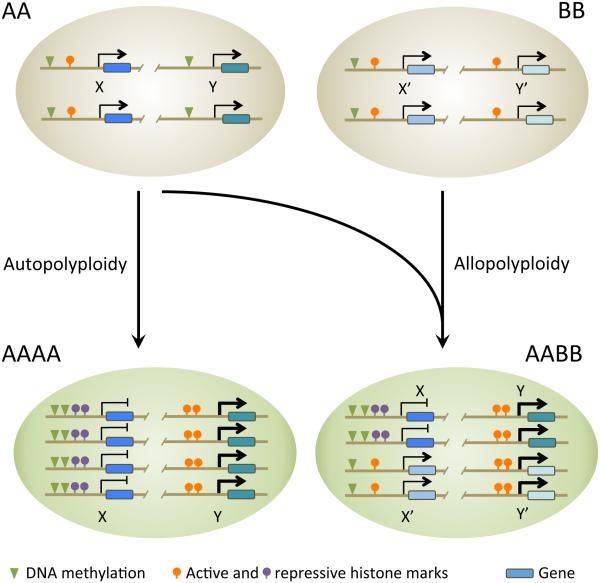
Figure 2.
A model for epigenetic regulation of gene expression in polyploidy. When a diploid (AA) becomes an autotetraploid (AAAA) through genome doubling, the expression of homologous genes can be affected. For example, gene X is silenced due to DNA hypermethylation and repressive histone markers in promoters, whereas gene Y is upregulated by loss of DNA methylation in the promoter. In allotetraploid (AABB), derived from hybridization between two diploids (AA and BB), epigenetic modifications contribute to nonadditive expression of homoeologous genes. Gene X of AA parent is repressed by DNA hypermethylation and histone modifications in the promoter, but the expression of homoeologous gene X’ of BB parent is not affected in the allotetraploid. Other homoeologous genes (Y and Y’) are both expressed because of DNA hypomethylation and active histone marks. DNA methylation can be induced via small RNA-dependent and independent pathways. The nonadditive expression of homoeologous genes (X and X’) and activation of both homoeologous genes (Y and Y’) could help newly formed allotetraploids establish new cytotypes, develop more vigorously and adapt in local environments. Epigenetic modifications of homoeologous loci could be regulated by environmental stress and developmental signals (see Figure 1). Arrow thickness corresponds to gene expression level. AA and BB indicate parental genomes (species).
Much of this genetic variation is partly related to TEs that have diverged between species [60-62]. In allopolyploid wheat, the reduced number of siRNAs corresponding to TEs correlates with decreased CG methylation in a retrotransposon family named Veju [56]. In another study, CCGG sites surrounding Stowaway-like Miniature Inverted–Repeat Transposable Elements are massively hypermethylated in allohexaploid wheat [63]. These results suggest that hypermethylation and hypomethylation of transposons can spontaneously occur in different genomic regions in polyploids. In addition, recent allopolyploid Spartina anglica undergoes little genetic variation, but 30% of the parental methylation patterns are altered in the allopolyploids, suggesting that DNA methylation change but not genetic variation contributes to phenotypic diversity in Spartina allopolyploids [64,65].
Although DNA methylation changes are primarily observed in allopolyploids [8,34], studies from autopolyploid Arabidopsis and Cymbopogon also reveal DNA methylation variation between autopolyploids and their isogenic diploid parents [66,67]. In situ immunodetection of 5-methylcytosine reveals enhanced genome-wide DNA methylation in autotetraploids compared with diploid Cymbopogon, which is thought to regulate native secondary metabolites and body size [66]. Gene expression changes in Arabidopsis autopolyploids are very limited in some ecotypes such as Ler-0 [34] but more common in other ecotypes [67], suggesting genotype-dependent regulation [67]. In the autotetraploid Col-0, MRD1 [67] is expressed more abundantly, which correlates with hypomethylation in 3'-flanking sequences of MRD1 (Figure 2). An interesting phenomenon of epigenetic regulation in autopolyploids is associated with an unusual epiallele of a transgene locus after tetraploidization [68,69]. The diploid A. thaliana, transformed with the hygromycin phosphotransferase (HPT) gene, shows stable hygromycin resistance over several generations of self-pollination or after outcrossing with diploid wildtype plants [69]. However, outcrossing to tetraploid plants leads to reduced HPT activity, and autotetraploid derivatives generate progeny with silenced HPT. This phenomenon is termed polyploidy-associated transcriptional gene silencing (paTGS). Further study indicates that paTGS is regulated by both DNA methylation and histone methylation [68].
Interploidy crosses within species often result in nonviable progeny [25,70]. This reproductive barrier limits gene flow between newly formed polyploids and parents, and also decreases a survival probability of polyploids because mating opportunities with other polyploids are rare [9]. However, this reproductive barrier promotes polyploid speciation and is established in endosperm and is also known as triploid block [71]. Interploidy crosses affect the ratio of maternal to paternal genome in the endosperm and changes timing of endosperm cellularization, eventually causing seed abortion [25]. Studies show that target genes of endosperm-specific AGAMOUS-LIKE (AGL) and Polycomb Repressive Complex 2 (PRC2) are deregulated by interploidy hybridization [72-74]. PRC2 comprises histone methyltransferase and regulates gene expression by increasing H3K27 methylation. Inactivation of MEDEA (MEA) or FERTILIZATION INDEPENDENT SEED 2 (FIS2), two subunits of PRC2, leads to endosperm over-proliferation and seed abortion [75,76]. A recent study in A. thaliana found that hypomethylated pollen derived from the met1 mutant could bypass the interploidy hybridization barrier [77]. A hypomethylated pollen genome induces de novo CHG methylation in FIS2-PRC2 target genes to suppress their expression. The gain of CHG methylation in these genes compensates the repression function of FIS2-PRC2, whose expression is altered by interploidy hybridization. These findings provide new insights into overcoming interploidy hybridization barriers in plant breeding (Figure 1).
Histone modifications in polyploids
In eukaryotic cell nuclei, DNA is packaged into a highly compacted structure known as chromatin through interacting with histones [78]. In response to developmental and environmental changes, the accessibility of chromatin is dynamically regulated by a suite of histone modifications, dubbed ‘histone code’ [79]. Core histones (H2A, H2B, H3 and H4) can be covalently modified at different positions of amino-terminal tails by different modifications, including acetylation, methylation, ubiquitination, phosphorylation, glycosylation, carbonylation, ADP ribosylation, sumoylation and biotinylation [79-81]. These modifications alone or in combination can change the accessibility of chromatin structures in the vicinity of genes to transcription machinery, leading to transcriptional activities. Among these modifications, histone methylation and acetylation have been most extensively studied and mainly occur in lysine and arginine residues on histone tails. Histone methylation and acetylation can be established and removed by corresponding enzymes, providing dynamic mechanisms for transcriptional regulation. Different histone modifications have different effects on gene expression. Histone modifications such as histone H3K9ac and H3K4me3 are usually found at euchromatin and are associated with active transcription, whereas marks such as H3K9me2 and H3K27me3 are generally related to gene repression and are found at heterochromatin [79,81,82].
Synthetic Arabidopsis allopolyploids flower later than the progenitors autotetraploid A. thaliana (Ler) and A. arenosa, and the natural allotetraploid A. suecica flowers even later than synthetic lines [34]. The flowering-time variation in these lines is correlated with different expression levels of FLC, which inhibits early flowering. The upregulation of FLC in Arabidopsis allopolyploids relative to the progenitors is mediated by increased levels of H3K9 acetylation and H3K4 dimethylation in the promoters [34] (Figure 2). In addition, compared with the parents A. thaliana and A. arenosa, Arabidopsis allotetraploids show morphological vigor, including a larger rosette and increased biomass. Genome-wide and biochemical analyses reveal that central circadian clock genes (CCA1, LHY, TOC1 and GI) mediate expression changes in downstream genes to produce more chlorophyll and starch in allotetraploids [83]. Repression of CCA1 and LHY in the allotetraploids correlates with lower levels of H3K9 acetylation and H3K4 methylation in promoters; and upregulation of TOC1 and GI is associated with increased levels of H3K9 acetylation and H3K4 methylation. These results show histone modifications play roles in diurnal regulation of circadian rhythms, which in turn increase expression levels of the genes related to starch metabolism and photosynthesis, leading to hybrid vigor in allopolyploids and hybrids (Figure 1).
Nucleolar dominance is an epigenetic phenomenon that describes uniparental ribosomal RNA (rRNA) gene silencing and is observed in interspecific hybrids and allopolyploids of many plants and animals [84-86]. Tandem rDNA repeats are recognized as nucleolus organizing regions (NORs). In allotetraploid A. suecica, endogenous A. thaliana rRNA genes are silenced but both A. thaliana and A. arensoa rRNA genes transfected in A. suecica protoplasts are equally expressed [85]. Moreover, the silenced A. thaliana rRNA genes can be reactivated by backcrossing synthetic A. suecica to tetraploid A. thaliana. These data argue against the hypothesis that dominant rRNA genes have a superior binding affinity for a limiting transcription factor [85]. Another study from allopolyploid B. napus shows inhibiting histone deacetylation or cytosine methylation derepresses silenced B. oleracea rRNA genes [84]. Further analysis showed that concerted changes in DNA methylation density and specific histone modifications in the promoters mediate the expression of rRNA genes, which involves a plant-specific histone deacetylase (HDT1) that is required for H3K9 deacetylation and subsequent H3K9 di-methylation (H3K9me2) [86]. In allohexaploid wheat (Triticum aestivum, AABBDD), rDNA loci from the A and D genomes are largely lost during allohexaploid evolution. A recent study shows that NORs from the A and D genomes start to disappear in the fourth generation and are completely eliminated by the seven generation in synthetic tetraploids [87]. The elimination of NORs is associated with increased DNA methylation (CHG and CHH methylation) and histone methylation (H3K27me3 and H3K9me2). These results suggest histone modification and DNA methylation are responsible for uniparental rRNA gene silencing or elimination in allopolyploids and interspecific hybrids.
Developmental regulation in polyploids
In response to genome shock in interspecific hybrids or after polyploid formation, newly formed polyploids must overcome genetic redundancy and reestablish new genomic composition and cytotype, accompanied with orchestration of novel and complex gene expression networks through genetic and epigenetic modifications [88]. Many duplicated genes may undergo progressive loss, pseudogenization (loss of function), subfunctionalization (partitioning of ancestral functions between duplicates) and neofunctionalization (evolving new function in one of the duplicated genes) [89].
Developmental regulation in allopolyploids is also observed in nucleolar dominance of B. napus, where B. oleracea rRNA genes are silenced and B. rapa rRNA genes are transcriptionally active [90]. However, the silenced rRNA genes are reactivated in reproductive tissues including flower buds, petals, sepals, anthers and siliques, suggesting a developmental role for reversal of gene silencing in interspecific hybrids and allopolyploids. Consistent with this finding, tissue-specific gene activation and silencing have also been described in many allopolyploids, including Arabidopsis [34], cotton [91,92] and Tragopogon [93]. Study of allotetraploid cotton using 63 gene pairs reveals that nine gene pairs show tissue-specific subfunctionalization, and 15 gene pairs exhibit tissue-specific neofunctionalization [92]. These data suggest that nonadditive expression of genes and reciprocal silencing of homoeologous alleles can occur in a tissue-specific manner and in resynthesized or natural allopolyploids, suggesting a general role of developmental regulation in gene expression during polyploidy formation and evolution.
Arabidopsis autotetraploids show similar phenotypes to their diploid progenitors, but the analysis of element components in leaves reveals higher potassium uptake in all Arabidopsis autotetraploids tested [94]. The Arabidopsis autotetraploids also have enhanced salinity tolerance, which is associated with elevated potassium and reduced sodium accumulation in leaves. Gene expression changes in roots but not in leaves of autopolyploids may be responsible for the enhanced potassium uptake. In another study, expression of circadian clock regulators in leaves of Arabidopsis allotetraploids was altered to promote expression of downstream genes in photosynthesis and carbohydrate metabolism, which consequently produced more chlorophyll and starch in the allotetraploids than the parents [83]. Compared to A. arenosa homoeologs, circadian genes of A. thaliana origin are more sensitive to expression changes in the allotetraploids. When rootstocks of diploid and autotetraploid Citrus limonia are grafted with diploid C. sinensis scions [95], the autotetraploid plants are much more tolerant to water deficit than diploid plants, which is mediated by upregulation of abscisic acid signaling genes in roots. These data suggest that homoeologous genes could be differentially regulated in different tissues or developmental stages in response to developmental and environmental signals.
The current view is that at the genome-wide level there is no obvious bias of homoeolog expression. However, in any given tissue or developmental stage, there can be expression bias towards homoeologous loci. This notion is supported by recent genome sequencing analyses in B. napus [36] and hexaploid wheat [35]. In B. napus, overall expression levels of homoeologous genes are relatively equal, but ~40% of homoeologs are expressed differently in roots and leaves [36]. Artificial selection of B. napus promotes optimization of seed oil biosynthesis, decrease of undesirable glucosinolates and enhancement of pathogen resistance through gene loss and subfunctionalization. In wheat, RNA sequencing using different cell types of endosperm at three different developmental stages reflects cell type- and stage-dependent genome dominance and asymmetric expression of homoeologs [96]. Although three wheat homoeologous genomes contribute relatively equally to the number of expressed genes in the endosperm, only 28% of the homoeologous triplet loci have the same expression patterns in different cell types or developmental stages. When homoeologs are clustered into 25 coexpression network modules in the endosperm, 23 (92%) modules show subgenomic dominance, but no single homoeologous genome is overly dominant. Understanding how homoeologs are differentially expressed during growth and development and how artificial selection affects epigenetic changes in polyploid crops will facilitate plant breeding and crop improvement.
Conclusions and prospects
A general feature of polyploidy is genetic redundancy and intergenomic interactions, which induce genetic and epigenetic changes, leading to reorganization of genome and regulatory networks that allow polyploids to establish new species and adapt in ecological niches. Many conclusions about epigenetic and developmental changes in polyploids have been obtained from results using a few genes and limited methodologies and techniques. Understanding the complexity of polyploid genomes and epigenomes requires the sequencing of many polyploid genomes. To date, few polyploid genomes have been sequenced, and using next-generation sequencing platforms alone often cannot effectively discriminate homoeologous genes and genomes in allopolyploids. The sequences of extant ‘progenitor’ genomes cannot resolve de novo changes after polyploid formation and within polyploid species. As more polyploid genomes are sequenced, more detailed mechanisms and functions for gene expression and morphological changes will be revealed. Furthermore, each polyploid cell contains more DNA molecules and likely transcribes more RNA molecules than a diploid cell. Conventional transcriptome analysis can only estimate relative transcript abundance for each gene in a given tissue or cell type, which underestimates the differences of total mRNA amount on the per-cell basis. This could under- or over-estimate upregulated or downregulated genes in polyploid cells. Developing high-throughput sequencing in single cells will provide new insights into genetic and epigenetic changes and developmental regulation of polyploidy, which could help develop tools to effectively improve production of many polyploid plants and crops to meet the growing demand for food, feed, biofuel and a better environment.
Highlights.
Polyploidy is a common evolutionary feature in all eukaryotes especially in plants
Ploidy and intergenomic hybridization induce genetic and epigenetic changes
DNA methylation and histone modifications affect homoeologous gene expression
Gene expression networks are reprogrammed by environmental and developmental cues
Studies of polyploidy at sequence and single-cell levels will reveal mechanistic insights
Acknowledgements
The work cited from the Chen laboratory was supported in part by grants from the U.S. National Institutes of Health, National Science Foundation (GM067015, ISO0733857, ISO1025947, ISO1238048 and MCB1110957), and the Cotton Incorporated (07-161). We apologize for omitting some references owing to the space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lutz AM. A preliminary note on the chromosomes of Oenothera Lamarckiana and one of its mutants, O. gigas. Science. 1907;26:151–152. doi: 10.1126/science.26.657.151. [DOI] [PubMed] [Google Scholar]
- 2.Lewis WH. Polyploidy: Biological Relevance. Plenum; New York: 1980. [Google Scholar]
- 3.Chen ZJ, Birchler JA. In: Polyploid and Hybrid Genomics. Chen ZJ, Birchler JA, editors. Wiley-Blackwell; New York: 2013. [Google Scholar]
- 4.Soltis P, Soltis DE. Polyploidy and Genome Evolution. Springer; Berlin Heidelberg: 2012. [Google Scholar]
- 5.Leitch AR, Leitch IJ. Genomic plasticity and the diversity of polyploid plants. Science. 2008;320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- • 6.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. This article reviews possible advantages and disadvantages of polyploid formation, including gene redundancy and asexual reproduction. It also shows constraints of being polyploid, including disrupting cellular architecture, reorganization of gene expression, unstable mitosis and meiosis. [DOI] [PubMed] [Google Scholar]
- 7.Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- • 8.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. This review summarizes the mechanisms of gene expression changes in polyploids, with a focus on effects of genomic change, transcriptional and post-transcriptional regulation on phenotypic variation in polyploids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hegarty MJ, Hiscock SJ. Genomic clues to the evolutionary success of polyploid plants. Curr Biol. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. [DOI] [PubMed] [Google Scholar]
- 10.Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 11.Soltis DE, Visger CJ, Soltis PS. The polyploidy revolution then…and now: Stebbins revisited. Am J Bot. 2014;101:1057–1078. doi: 10.3732/ajb.1400178. [DOI] [PubMed] [Google Scholar]
- 12.Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP. Recently formed polyploid plants diversify at lower rates. Science. 2011;333:1257. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- 13.Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- • 14.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nature Reviews Genetics. 2009;10:725–732. doi: 10.1038/nrg2600. The authors argue that ancient genome doubling could probably survive only under special conditions and then contribute to species diversification. [DOI] [PubMed] [Google Scholar]
- 15.Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biol. 2010;8:e1000409. doi: 10.1371/journal.pbio.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoemaker RC, Schlueter J, Doyle JJ. Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol. 2006;9:104–109. doi: 10.1016/j.pbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Stebbins GL. Variation and Evolution in Plants. Columbia University Press; New York: 1950. [Google Scholar]
- 19.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 20.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: biological implications. Trends in Genetics. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Premachandran MN, Prathima PT, Lekshmi M. Sugarcane and polyploidy: a review. J. Sugarcane Research. 2011;1:1–15. [Google Scholar]
- 24.Stebbins GL. Chromosomal Evolution in Higher Plants. Edward Arnold; London: 1971. [Google Scholar]
- 25.Schatlowski N, Kohler C. Tearing down barriers: understanding the molecular mechanisms of interploidy hybridizations. J Exp Bot. 2012;63:6059–6067. doi: 10.1093/jxb/ers288. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Ann Rev Ecol Syst. 1998;29:467–501. [Google Scholar]
- 27.Levin DA. Polyploidy and novelity in flowering plants. Amer. Nat. 1983;122:1–25. [Google Scholar]
- 28.Riddle NC, Jiang H, An L, Doerge RW, Birchler JA. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theor Appl Genet. 2010;120:341–353. doi: 10.1007/s00122-009-1113-3. [DOI] [PubMed] [Google Scholar]
- •• 29.Miller M, Zhang C, Chen ZJ. Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 (Bethesda) 2012;2:505–513. doi: 10.1534/g3.112.002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman M, Liu B, Segal G, Abbo S, Levy AA, Vega JM. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. This paper analyzed the effects of ploidy and hybridity on seed size, biomass and gene expression change using diploid, triploid and tetraploid hybrids and their respective parents in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell. 2007;19:3403–3417. doi: 10.1105/tpc.107.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buggs RJ, Chamala S, Wu W, Tate JA, Schnable PS, Soltis DE, Soltis PS, Barbazuk WB. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr Biol. 2012;22:248–252. doi: 10.1016/j.cub.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Liu B, Brubaker G, Cronn RC, Wendel JF. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome. 2001;44:321–330. [PubMed] [Google Scholar]
- •• 34.Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, Madlung A, Osborn TC, Doerge RW, Comai L, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. This paper provided a comprehensive view of genome-wide nonadditive gene expression in synthetic Arabidopsis allotetraploids, providing a molecular basis for de novo variation in resynthesized allopolyploids. The data lead to several new research directions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Wheat Genome Sequencing C: A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- • 36.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. This paper described the genome sequence of Brassica napus and studied consequences of genome duplication and hybridization. They found abundant homoeologous exchanges and subtle structural, functional, and epigenetic cross-talk in B. napus. [DOI] [PubMed] [Google Scholar]
- 37.Muller HJ. Why polyploidy is rarer in animals than in plants. Amer Nat. 1925;59:346–353. [Google Scholar]
- 38.Mable BK. 'Why polyploidy is rarer in animals than in plants': myths and mechanisms. Biol J Linn Soc Lond. 2004;82:453–466. [Google Scholar]
- 39.Steward FC, Mapes MO, Mears K. Growth and organized development of cultured cells. II. Organization in cultures grown from freely suspended cells. Am J Bot. 1958;45:705–708. [Google Scholar]
- • 40.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. This paper reviews DNA methylation pathways and their inheritance in plants and animals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 42.Richards EJ. DNA methylation and plant development. Trends Genet. 1997;13:319–323. doi: 10.1016/s0168-9525(97)01199-2. [DOI] [PubMed] [Google Scholar]
- 43.Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 45.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- • 48.Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. This article showed that in addition to the RdDM pathway, CHH methylation could also be catalyzed by CMT2 through genetic interaction with DDM1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 50.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 51.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci U S A. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 2002;129:733–746. doi: 10.1104/pp.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lukens LN, Pires JC, Leon E, Vogelzang R, Oslach L, Osborn T. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 2006;140:336–348. doi: 10.1104/pp.105.066308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian L, Li X, Ha M, Zhang C, Chen ZJ. Genetic and epigenetic changes in a genomic region containing MIR172 in Arabidopsis allopolyploids and their progenitors. Heredity. 2014;112:207–214. doi: 10.1038/hdy.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee HS, Chen ZJ. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc Natl Acad Sci USA. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, Levy AA. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics. 2011;188:263–272. doi: 10.1534/genetics.111.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics. 2004;167:1961–1973. doi: 10.1534/genetics.104.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y, Zhong L, Wu X, Fang X, Wang J. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta. 2009;229:471–483. doi: 10.1007/s00425-008-0844-8. [DOI] [PubMed] [Google Scholar]
- •• 59.Chen M, Ha M, Lackey E, Wang J, Chen ZJ. RNAi of met1 reduces DNA methylation and induces genome-specific changes in gene expression and centromeric small RNA accumulation in Arabidopsis allopolyploids. Genetics. 2008;178:1845–1858. doi: 10.1534/genetics.107.086272. This paper studied the function of DNA methylation in Arabidopsis allopolyploids using RNAi-mediated downregulation of MET1, which could induce genome-specific changes in gene expression in allotetraploids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB, Chacko B, Chen NW, Couloux A, Dalwani A, Denny R, et al. Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 2008;148:1740–1759. doi: 10.1104/pp.108.127902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yaakov B, Kashkush K. Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol Biol. 2012;80:419–427. doi: 10.1007/s11103-012-9957-3. [DOI] [PubMed] [Google Scholar]
- 64.Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien MA, Ainouche M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 2009;184:1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x. [DOI] [PubMed] [Google Scholar]
- 65.Salmon A, Ainouche ML, Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Mol Ecol. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- 66.Lavania UC, Srivastava S, Lavania S, Basu S, Misra NK, Mukai Y. Autopolyploidy differentially influences body size in plants, but facilitates enhanced accumulation of secondary metabolites, causing increased cytosine methylation. Plant J. 2012;71:539–549. doi: 10.1111/j.1365-313X.2012.05006.x. [DOI] [PubMed] [Google Scholar]
- 67.Yu Z, Haberer G, Matthes M, Rattei T, Mayer KF, Gierl A, Torres-Ruiz RA. Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:17809–17814. doi: 10.1073/pnas.1000852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baubec T, Dinh HQ, Pecinka A, Rakic B, Rozhon W, Wohlrab B, von Haeseler A, Mittelsten Scheid O. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic States in Arabidopsis. Plant Cell. 2010;22:34–47. doi: 10.1105/tpc.109.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •• 69.Mittelsten Scheid O, Afsar K, Paszkowski J. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat Genet. 2003;34:450–454. doi: 10.1038/ng1210. This paper showed a new phenomenon which is termed as polyploid-associated transcriptional gene silencing (paTGS), similar to paramutation-like effects. [DOI] [PubMed] [Google Scholar]
- • 70.Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. This article studied the effects of interploidy crosses on seed development. The maternal and paternal excess crosses led to different effects on endosperm development. [DOI] [PubMed] [Google Scholar]
- 71.Kohler C, Mittelsten Scheid O, Erilova A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010;26:142–148. doi: 10.1016/j.tig.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 72.Erilova A, Brownfield L, Exner V, Rosa M, Twell D, Mittelsten Scheid O, Hennig L, Kohler C. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 2009;5:e1000663. doi: 10.1371/journal.pgen.1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jullien PE, Berger F. Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet. 2010;6:e1000885. doi: 10.1371/journal.pgen.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu J, Zhang C, Baulcombe DC, Chen ZJ. Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci USA. 2012;109:5529–5534. doi: 10.1073/pnas.1203094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB, et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- •• 77.Schatlowski N, Wolff P, Santos-Gonzalez J, Schoft V, Siretskiy A, Scott R, Tamaru H, Kohler C. Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell. 2014;26:3556–3568. doi: 10.1105/tpc.114.130120. This paper demonstrated that in A. thaliana, hypomethylated pollen derived from met1 mutant can bypass the interploidy hybridization through inducing de novo CHG methylation in FIS2-PRC2 target genes to suppress their expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 79.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 80.Chen ZJ, Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim Biophys Acta. 2007;1769:295–307. doi: 10.1016/j.bbaexp.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 82.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- •• 83.Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. This work discovered altered expression of circadian clock genes by histone modifications that in turn mediated expression changes in the downstream genes to promote photosynthesis and starch metabolism in allotetraploids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen ZJ, Comai L, Pikaard CS. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci U S A. 1998;95:14891–14896. doi: 10.1073/pnas.95.25.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- • 87.Guo X, Han F. Asymmetric Epigenetic Modification and Elimination of rDNA Sequences by Polyploidization in Wheat. Plant Cell. 2014 doi: 10.1105/tpc.114.129841. This paper demonstrated a role for DNA methylation and histone modification in the elimination of rDNA sequences in polylploidization in wheat. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15:57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen ZJ, Pikaard CS. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA. 1997;94:3442–3447. doi: 10.1073/pnas.94.7.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chaudhary B, Flagel L, Stupar RM, Udall JA, Verma N, Springer NM, Wendel JF. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (gossypium) Genetics. 2009;182:503–517. doi: 10.1534/genetics.109.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buggs RJ, Zhang L, Miles N, Tate JA, Gao L, Wei W, Schnable PS, Barbazuk WB, Soltis PS, Soltis DE. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr Biol. 2011;21:551–556. doi: 10.1016/j.cub.2011.02.016. [DOI] [PubMed] [Google Scholar]
- •• 94.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. This paper showed that Arabidopsis autotetraploids exhibited higher potassium uptake due to gene expression changes in roots, although Arabidopsis autotetraploids showed similar phenotype to their diploid progenitors. [DOI] [PubMed] [Google Scholar]
- 95.Allario T, Brumos J, Colmenero-Flores JM, Iglesias DJ, Pina JA, Navarro L, Talon M, Ollitrault P, Morillon R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 2013;36:856–868. doi: 10.1111/pce.12021. [DOI] [PubMed] [Google Scholar]
- 96.Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, International Wheat Genome Sequencing C. Mayer KF, Olsen OA. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345:1250091. doi: 10.1126/science.1250091. [DOI] [PubMed] [Google Scholar]