Abstract
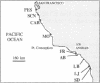
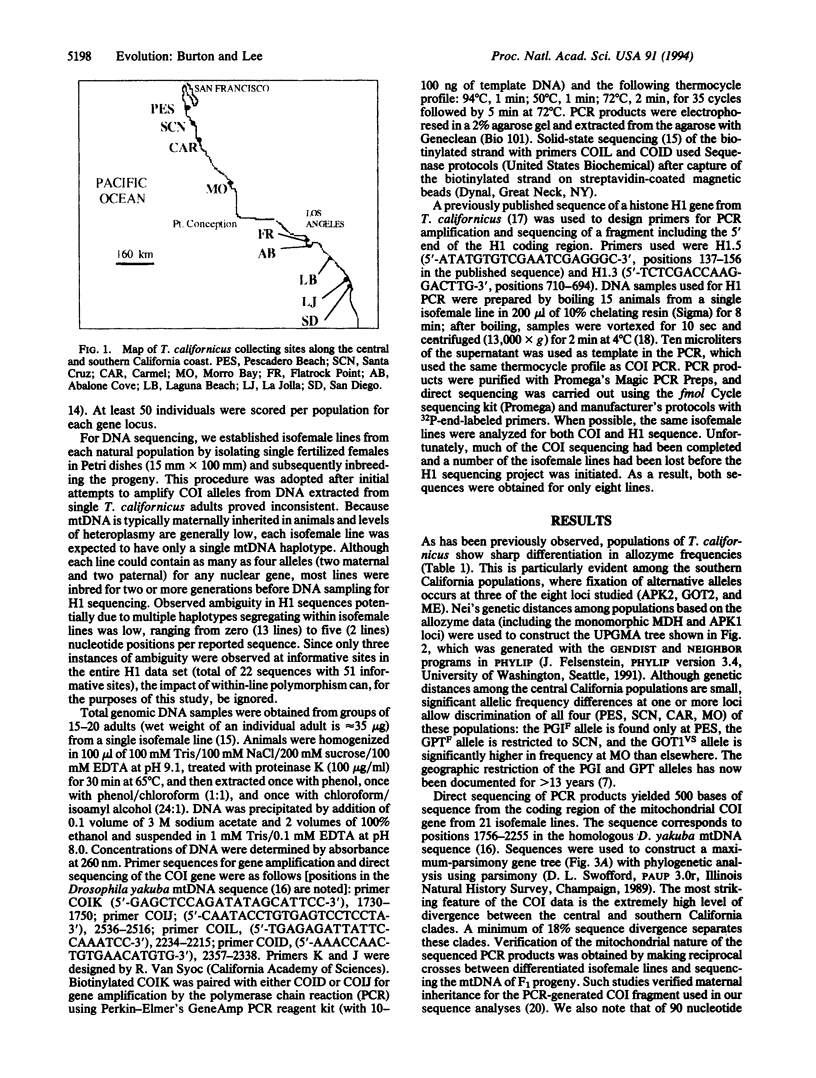
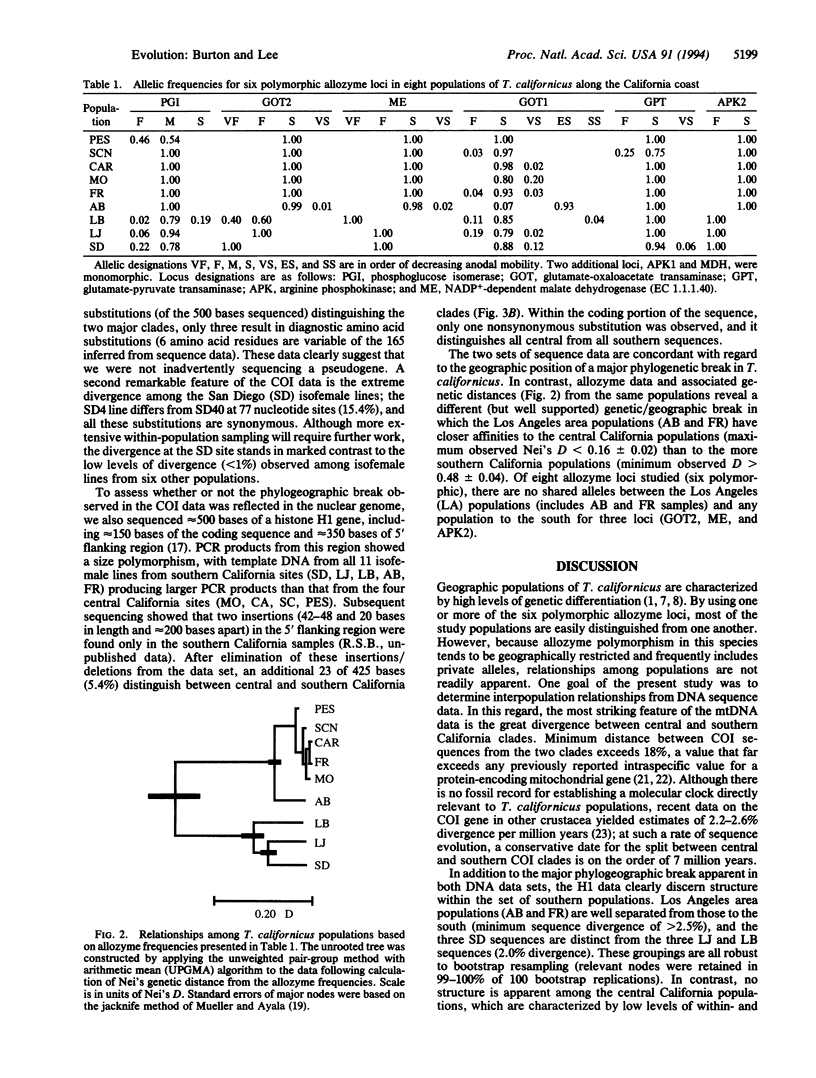
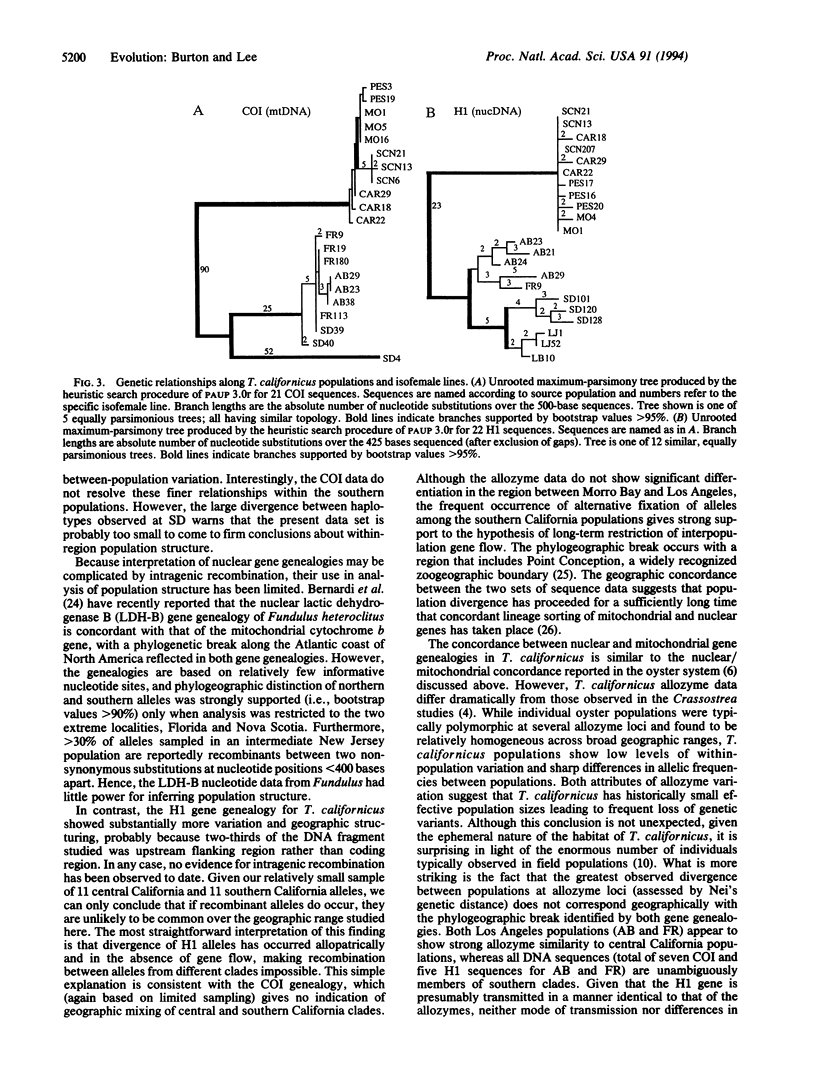
The genetic structure of natural populations is frequently inferred from geographic distributions of alleles at multiple gene loci. Surveys of allozyme polymorphisms in the tidepool copepod Tigriopus californicus have revealed sharp genetic differentiation of populations, indicating that gene flow among populations is highly restricted. Analysis of population structure in this species has now been extended to include nuclear and mitochondrial gene genealogies. DNA sequences of the mtDNA-encoded cytochrome-c oxidase subunit I gene from 21 isofemale lines derived from seven populations reveal a phylogeographic break between populations north and south of Point Conception, California, with sequence divergence across the break exceeding 18%, the highest level of mtDNA divergence yet reported among conspecific populations. Divergence between populations based on 22 sequences of the nuclear histone H1 gene is geographically concordant with the mitochondrial sequences. In contrast with previously studied nuclear genes in other sexually reproducing metazoans, the H1 gene genealogy from T. californicus shows no evidence of recombination. The apparent absence of intragenic recombinants probably results from the persistent lack of gene flow among geographically separated populations, a conclusion strongly supported by allozyme data and the mitochondrial gene genealogy. Despite strong population differentiation at allozyme loci, the phylogeographic break identified by the DNA sequences was not evident in the allozyme data.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi G., Sordino P., Powers D. A. Concordant mitochondrial and nuclear DNA phylogenies for populations of the teleost fish Fundulus heteroclitus. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9271–9274. doi: 10.1073/pnas.90.20.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D., Cook A., Wagner M., Wells D. Closely linked H2B genes in the marine copepod, Tigriopus californicus indicate a recent gene duplication or gene conversion event. DNA Seq. 1992;2(6):387–396. doi: 10.3109/10425179209020818. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Dill M. M., Burton R. S. Genetics of mitochondrial glutamate-oxaloacetate transaminase (GOT-2) in Tigriopus californicus. Biochem Genet. 1984 Apr;22(3-4):339–347. doi: 10.1007/BF00484232. [DOI] [PubMed] [Google Scholar]
- Karl S. A., Avise J. C. Balancing selection at allozyme loci in oysters: implications from nuclear RFLPs. Science. 1992 Apr 3;256(5053):100–102. doi: 10.1126/science.1348870. [DOI] [PubMed] [Google Scholar]
- Knowlton N., Weigt L. A., Solórzano L. A., Mills D. K., Bermingham E. Divergence in proteins, mitochondrial DNA, and reproductive compatibility across the isthmus of Panama. Science. 1993 Jun 11;260(5114):1629–1632. doi: 10.1126/science.8503007. [DOI] [PubMed] [Google Scholar]
- Mueller L. D., Ayala F. J. Estimation and interpretation of genetic distance in empirical studies. Genet Res. 1982 Oct;40(2):127–137. doi: 10.1017/s0016672300019005. [DOI] [PubMed] [Google Scholar]
- Reeb C. A., Avise J. C. A genetic discontinuity in a continuously distributed species: mitochondrial DNA in the American oyster, Crassostrea virginica. Genetics. 1990 Feb;124(2):397–406. doi: 10.1093/genetics/124.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P. S., Metzger D. A., Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991 Apr;10(4):506–513. [PubMed] [Google Scholar]