Abstract
Assimilation and integration of “omics” technologies, including genomics, epigenomics, proteomics, and metabolomics has readily altered the landscape of medical research in the last decade. The vast and complex nature of omics data can only be interpreted by linking molecular information at the organismic level, forming the foundation of systems biology. Research in pulmonary biology/medicine has necessitated integration of omics, network, systems and computational biology data to differentially diagnose, interpret, and prognosticate pulmonary diseases, facilitating improvement in therapy and treatment modalities. This review describes how to leverage this emerging technology in understanding pulmonary diseases at the systems level –called a “systomic” approach. Considering the operational wholeness of cellular and organ systems, diseased genome, proteome, and the metabolome needs to be conceptualized at the systems level to understand disease pathogenesis and progression. Currently available omics technology and resources require a certain degree of training and proficiency in addition to dedicated hardware and applications, making them relatively less user friendly for the pulmonary biologist and clinicians. Herein, we discuss the various strategies, computational tools and approaches required to study pulmonary diseases at the systems level for biomedical scientists and clinical researchers.
Keywords: Genomics, Proteomics, RNA-Seq, ChIP-Seq, Epigenomics, Systems Biology, Network Biology, Interactomes, Transcriptomes, Diseasosomes
Introduction
Biological systems function as a network of interconnected and mutually dependent components, constituting the unified whole. Completion of the Human Genome Project reinforced this idea and signaled a paradigm shift in analyzing biological phenomena, introducing a holistic approach of systems biology a decade ago [1–6]. Increasingly, technology provides omic scale data, while systems-level models and measurements predict and validate biological phenomena [3, 7]. Thus, the pervasive nature of systems biology in molecular diagnostics, genetic and proteomic mapping of diseases is well recognized [7].
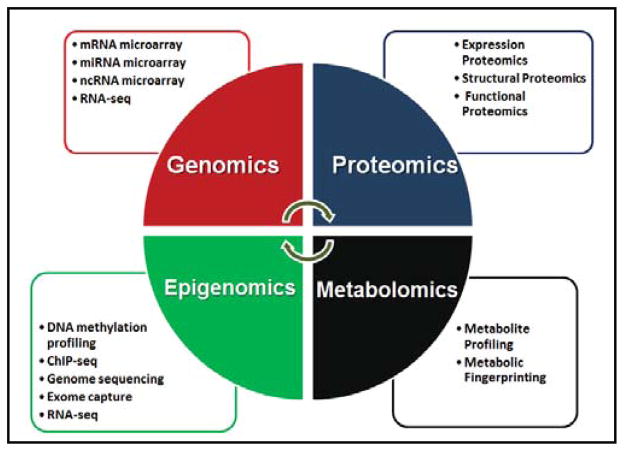
Systems biology integrates data from multiple sources into predictive molecular models analogous to the study of organ pathophysiology, including the lung [8]. Lung cancer, chronic obstructive pulmonary disease (COPD), asthma and idiopathic pulmonary fibrosis (IPF) are highly complex diseases with overlapping pathologies associated with lung remodeling and altered molecular and physiological functions. Genomics, epigenomics, proteomics and metabolomics are thus used to ascertain the molecular mechanisms underlying these functional alterations (Fig. 1). Attempts have been made to integrate data from these technologies using a systems biology approach in pulmonary medicine [8–10]. In the present review, we summarize and provide guidelines to integrate the different omics based technologies currently applied to pulmonary diseases. Detailed work flows and resources for analyzing data obtained from omics approaches provide an easy access and understanding for biologists and clinicians alike who are just entering in the field of systems biology.
Fig. 1.
Global screening strategies and techniques driving “omics” era. Genomics applications analyze mRNA, miRNA and other small RNA moieties, including lncRNA by microarray and RNA-seq technologies. These allow genome-wide interrogation of biological phenomena, as well as detect expression changes in the transcriptome. Proteomics approaches deal exclusively with changes at the proteome level. Existing proteomic techniques study structural and functional characteristics of proteins in addition to detecting changes in the global protein expression levels of an organism. In combination with genomics, proteomic analysis can offer detailed insights into the transcriptional and translational events occurring in a biological system. Detecting, identifying and quantifying small molecule metabolites that are central to living systems using metabolomic approaches is vital for a systems level understanding and can be integrated with other work flows. In order to truly understand gene regulation, analysis of epigenetic events that at the chromosomal level is indispensable. The integration of protocols epigenetic like DNA methylation and ChIP with genomic approaches like microarray and sequencing allow rapid and global analysis of the epigenome. Although, “omic” technologies are specific in terms of the processes they analyze, integration of one or more of these technologies is the cornerstone of systems approach to biological systems.
Genomics & Epigenomics in Pulmonary Biology
Genomics
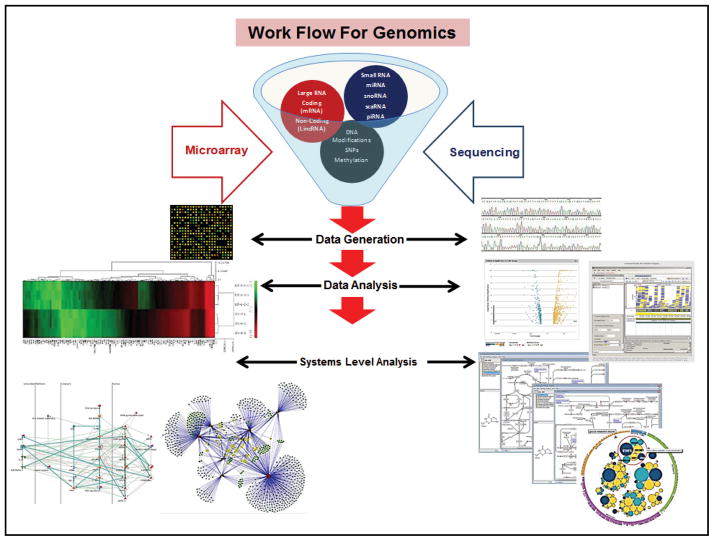
Large-scale genome sequencing and microarray projects have resulted in paradigm shifting approaches to understanding human diseases, including pulmonary disorders, as evidenced by a steady increase in publications (Fig. 2). Transcriptional profiles of disease phenotypes have contributed towards a better understanding of the molecular mechanisms driving pulmonary pathologies such as idiopathic pulmonary fibrosis (IPF), asthma, COPD, and pulmonary hypertension [11–18]. Currently established work flows for genomics experiments involve data generation, analysis and more recently systems level analysis (Fig. 3). Herein, we provide details of current and future technologies used in the area of genomics.
Fig. 2.
Emergence of Genomics in pulmonary biology. PubMed search for the co-occurrence of the terms “Genomics and pulmonary biology” identified 800 publications featuring these key words in the last decade. Interestingly, this number reflects a 20-fold increase in the number of publications with similar key words from the previous decade. The dramatic increase can be attributed to the availability of cheaper technologies, highly developed databases and analysis platforms with user friendly interface.
Fig. 3.
Workflow for genomics data obtained from microarray and RNA-seq. RNA or DNA extracted from different sources (cells/tissues) is the starting point for conventional genomics work flow. RNA/DNA is used for microarray or RNA-seq analysis as per recommended protocols. These methods allow users to study large and small RNA populations as well as DNA modifications. The second step of genomic workflow comprises of retrieving data using different sample processing software that are unique to a given application. Standard outputs from the software comprise of signal intensity values (microarray), sequence reads (RNA-seq), annotations and accession identifiers for the different genes that are being studied. This information serves as the input for the third step of the workflow, which is data analysis. The raw data obtained has to converted into biologically relevant information like differential gene expression data (fold change values), sequence alignment files (RNA-seq) and clusters of similarly affected targets (hierarchical clustering and volcano plots). The last and most important step of genomic work flows is systems level analysis. Using the information generated from the data analysis step, users can derive additional details about the alterations in biological pathways, networks and diseases that can be correlated to the biological phenomena being studied.
Microarray technology in pulmonary biology
DNA microarrays have defined genome-wide signatures of smoking and smoking cessation on bronchial and nasal epithelium [19–21], identifying novel molecules that serve as diagnostic markers in smokers with clinical suspicion of lung cancer [22, 23]. Genes regulating collagen synthesis/deposition were similarly defined for early detection of fibrotic changes in the lung [24, 25]. Likewise, prognostic gene signature during progression and resolution of acute lung injury (ALI) were identified from the microarray profiles of patient lungs [26, 27]. Metastasis signatures are similarly obtained from transcriptomic data on critical lung cancer genes [28–31]. A detailed account of genomics driven understanding of lung disorders has been published [32]. Genome wide association studies (GWAS) based on data from DNA microarray analysis have aided search for single-nucleotide polymorphisms (SNPs) within a population, which has successfully predicted pharmacogenetic factors associated with diseases like asthma and response to bronchodilators [33–37].
Microarray data analysis can be performed using a number of free and commercial tools (Table 1). Tools like Database for Annotation, Visualization and Integrated Discovery (DAVID) v.6.7 [38, 39], GenePattern 2.0 [40], Galaxy [41], Cytoscape [42], GenomeSpace (http://www.genomespace.org) and Bioconductor (http://www.bioconductor.org/) are free to use and have been routinely cited in published literature. Most cited commercial software tools include Ingenuity Pathway Analysis (IPA) (www.Ingenuity.com), GeneGO, MetaCore (http://portal.genego.com/) Partek 6.3 (www.partek.com). These enables biologists to discover visualize and explore therapeutically relevant transcriptional networks. Researchers must download, analyze and compare existing microarray datasets from public repositories like Gene Expression Omnibus [43, 44], ArrayExpress [45], and Stanford Microarray Database [46] to strengthen hypothesis and avoid redundant experimentation as shown by Sirota et al. [47], who created a set of 100 disease signatures from the Gene Expression Omnibus and identified many novel drug–disease gene pairs.
Table 1.
Resources for processing and analyzing genomics data
| Software tools | Microarray Analysis | |
|---|---|---|
| 1. | (DAVID) v.6.7 | http://david.abcc.ncifcrf.gov/ |
| 2. | GenePattern 2.0 | http://www.broadinstitute.org/cancer/software/genepattern/ |
| 3. | Galaxy Project | http://galaxyproject.org/ |
| 4. | GenomeSpace | http://www.genomespace.org |
| 5. | Bioconductor | http://www.bioconductor.org/ |
| 6. | Cytoscape | http://www.cytoscape.org/ |
| 7. | Ingenuity Pathways Analysis | www.Ingenuity.com |
| 8. | GeneGO, MetaCore | http://portal.genego.com/ |
| 9. | Partek 6.3 | www.partek.com |
|
| ||
| Software tools | RNA-seq Analysis | |
|
| ||
| 1. | Galaxy Project | http://galaxyproject.org/ |
| 2. | GenePattern 2.0 | http://www.broadinstitute.org/cancer/software/genepattern/ |
| 3. | R-workbench | http://www.ebi.ac.uk/Tools/rcloud/ |
| 4. | Palmapper | http://raetschlab.org/suppl/palmapper |
| 5. | mTim | http://raetschlab.org/suppl/mtim |
| 6. | rQuant | http://raetschlab.org/suppl/rquant |
| 7. | rDiff | http://cbio.mskcc.org/public/raetschlab/user/drewe/rdiff/ |
miRNA signature profiling in pulmonary diseases
A host of miRNAs have been identified as critical players in the development, progression and status of multiple lung pathologies [48–55] including in IPF [56, 57] and smokers lung [58]. These profiles are distinct from the miRNA profiles in chronic obstructive pulmonary disease (COPD), cancer, asthma and cardiovascular disease [59–65]. For example, targeting an miRNA let-7c has been proposed as it is a pathogenic link between COPD and lung cancer [66]. About 200 miRNAs are reported to be altered in steroid naïve asthmatic subjects, thereby establishing a potential link between inflammation and aberrant miRNA expression in patients with asthma [67–69]. Altered miRNAs in IPF have been extensively studied in pathogenesis of IPF and pulmonary hypertension [70–72]. While such molecular information is routinely generated, the precise mechanisms and roles of these miRNAs can only be ascertained by a combination of systems biology driving traditional experimental approaches [73].
Indeed, systems level analyses using miRNA expression profiles of lung cancer and interstitial lung diseases (ILDs) are being used to understand pathogenesis and progression of pulmonary disorders [9, 10, 74–79]. A number of computationally and experimentally derived databases of miRNA have been published [80–92] and are listed in Table 2. Approaches combining high-throughput experimental data together with sequence-based putative miRNA predictions have also been successfully applied [92–96]. Users can retrieve miRNA and mRNA gene expression profiles from NCI-60 (a set of 60 human cancer cell lines derived from diverse tissues) and use a tool, called CellMiner [97], to query these microarray datasets. Recently, transcriptome scale study on miRNA-lncRNA interactions have also added a new dimension to genomics analysis [98]. Such an approach has not been applied to study pulmonary disorders and will definitely contribute to a better understanding of disease regulation and progression. LncRNAs (long non coding RNAs) are a class of nonprotein coding RNAs that interact with other classes of RNAs including mRNAs and miRNAs and modulate their regulatory role via physical interactions [99]. Some of the more recent findings on the role of LncRNA in lung cancer have been reviewed recently [100]. The availability of high density LncRNA microarrays and network driven systems approach [101, 102] for studying the role of LncRNAs in human diseases makes it a promising approach for pulmonary biologists.
Table 2.
List of databases and tools for analyzing miRNA
| Databases | Web Links |
|---|---|
| 1. miRBase | http://www.mirbase.org/ |
| 2. miRNAmap | http://mirnamap.mbc.nctu.edu.tw/ |
| 3. miRWalk | http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html |
| 4. miRDB | http://mirdb.org/miRDB/ |
| 5. TarBase V.5c | http://diana.cslab.ece.ntua.gr/tarbase/ |
| 6. miRGen | http://www.diana.pcbi.upenn.edu/miRGen.html |
| 7. miROrtho | http://cegg.unige.ch/mirortho |
| 8. mir2disease | http://www.mir2disease.org/ |
| 9. miRanda | http://www.microrna.org/microrna/home.do |
| 10. TargetScan 6.2 | http://www.targetscan.org/ |
| 11. miRanda | http://www.ebi.ac.uk/enrightsrv/microcosm/htdocs/targets/v5/ |
| 12. DIANAmicroT | www.microrna.gr/microT |
| 13. PicTar | http://pictar.mdc-berlin.de/ |
| 14. miRecords | http://mirecords.umn.edu/miRecords/ |
| 15. mirTarBase | http://mirtarbase.mbc.nctu.edu.tw/ |
| 16. miRWalk | http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/ |
| 17. miRNAMap | http://mirnamap.mbc.nctu.edu.tw/ |
RNA-seq: Next generation technology for studying pulmonary disorders
RNA-seq is superior to previously developed transcriptomic methods. Unlike microarrays, RNA-seq detects known and unknown transcripts with low signal/noise ratio and high degree of sensitivity [103]. Recently, RNA-seq identified novel transcripts associated with smoking and lung cancer in small airway epithelium, and changes in RNA in peripheral lung in emphysema and IPF [104–107]. RNA-seq also identified novel and confirmative genes linked to asthma, indicating differences in biological processes in the airways of asthma patients [108]. In RNA-seq, differential gene expression data sets are analyzed by Software suites like GenePattern 2.0 [40], Galaxy [41] and Bioconductor [109]. The Bioconductor package ArrayExpressHTS [110] and R-workbench generates binary alignment (BAM) format [111], allowing users to process and analyze raw RNA-seq data. The Ensembl genome browser [112] can be used to display the BAM files containing the sequence alignment data. Currently ArrayExpress provides approximately 1200 BAM files for 125 RNA-seq experiments, from 14 different species [45]. Commercial software suites like NGS (Next Generation Sequencing; Genomatix) also provide a comprehensive promoter and transcript annotation database along with high end GUI (Graphic User Interface] for visualization and analysis of RNA-seq data (Table 1].
Epigenomics
Epigenetic alterations in idiopathic pulmonary fibrosis (IPF) and COPD have recently received considerable attention [113–120]. DNA-methylation profiles identified 870 genes and 625 CpG islands differentially methylated in IPF tissues [121, 122]. Surprisingly, a significant overlap was found between lung cancer and IPF, providing valuable insight into common pathways driving these two distinct diseases. Epigenetics approach also indentified changes in miRNAs and fibroblast signature for genes regulating extracellular matrix in IPF and lung cancer [123–125]. An epigenomic approach focusing on DNAmethylation to identify potential biomarkers for lung cancer was recently published [125]. While methylation of DNA is associated with decreased gene expression, acetylation of histones results in relaxation of chromatin facilitating gene transcription. Therefore, studying histone modifications becomes paramount to understand regulation of pulmonary diseases, as elegantly demonstrated in a recent finding that implicates histone modifications in decreased Fas expression and apoptosis resistance in fibrotic lung fibroblasts [126]. Further, the identification of distinct posttranslational histone modification patterns in histone H3 and histone H4 in lung cells [127], which may be considered as usable biomarkers for Cigarette Smoke-induced chronic lung diseases, supports the importance of epigenomics in pulmonary disorders. A more comprehensive summary of the latest experimental and translational epigenetic studies in pulmonary disorders can be found elsewhere [128].
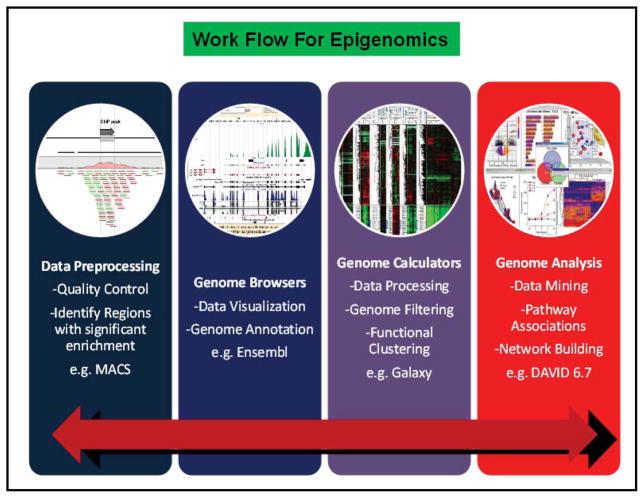
Epigenomic work flows for data generation and analysis are similar to genomic studies (Fig. 4]. Repitools, a part of Bio-conductor ver. 2.12, is used for enrichment-based epigenomic data analyses. Standard features include summarization and visualization of epigenomic data across promoters in relation to gene expression context, detection of regions of differential methylation/binding and, options for quantifying methylation. Epigenomix, a tool within Bio-conductor package performs integrative analysis for gene expression and histone modification data sets obtained by chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq). Methylated DNA immunoprecipitation (MeDIP) is a global purification technique used to enrich methylated DNA sequences [129]. MEDIPS is a Bioconductor package that is developed for analyzing data derived from methylated DNA immunoprecipitation (MeDIP) experiments followed by sequencing (MeDIP-seq). MEDIPS also provides several functionalities for the analysis including calculation of differential coverage between groups of samples as well as saturation and correlation analyses.
Fig. 4.
Work flow for processing and analyzing epigenomics data. Epigenomics shares a number of technologies and applications with genomics and proteomics, which allows users to overlap work flows for data generation and analysis. While initial steps of data retrieval and processing are highly similar to microarrays, there are additional steps like using genome browsers for alignment and annotations (similar to RNA-seq). Downstream analysis after data processing comprise of data clustering, pathway mining and network analysis to predict the most significantly altered candidates. For certain applications, software tools available on the Bioconductor package can integrate conventional genomic and proteomic workflow with output obtained from quantitative sequencing data (e.g. ChIP-seq, MBD-seq and CMS-seq). Given that the technology is still in early stages of development, users need to select different tools from different sources to put together user-defined work flows.
To predict absolute and relative methylation levels in MeDIP-microarray experiments a software package called MEDME is used. Bisulfite sequencing is another widely used technique that determines pattern of methylation following bisulphite treatment of DNA [130]. Data analysis for this follows a two-step analysis process, methylation calling and visualization. Bismark [131] is a program that can map bisulfite treated sequencing reads to a genome of interest and perform methylation calls in a single step, with output that can be viewed on genome viewer. SeqMonk, IGV and TABLET [132] are tools for visualization and analysis of high throughput mapped sequence data. For downstream analysis of epigenomics data, software like Galaxy, Database for Annotation, Visualization and Integrated Discovery (DAVID) v.6.7 [38, 39], Ingenuity Pathways Analysis (IPA) (www.Ingenuity.com), GeneGO, MetaCore (http://portal.genego.com/), Partek 6.3 (www.partek.com) allow users to correlate epigenomics data with pathway and biological networks. NCBI epigenomics is a curated resource for whole-genome epigenetic data sets that processes raw data maps to genomic coordinates that can be visualized using genome browsers or downloaded for local analysis [133, 134].
Proteomics & Metabolomics in Pulmonary Biology
Proteomics
Since most biological activities are driven by alterations in protein function proteomic approaches are increasingly used to understand pulmonary disorders [135–139]. A recent study using 2-dimensional difference in-gel electrophoresis (2D-DIGE) coupled with MALDI-ToF/ToF MS identified hyperoxia induced temporal alterations proteome, highlighting the role of proteins in translational regulation [140]. The human Bronchoalveolar Lavage Fluid (BALF) phosphoproteome recently indicated a diverse group of proteins, hitherto unknown for their biological functions [141]. Protein profiles specific to Ventilator-associated pneumonia (VAP) were readily identified in BALF, offering promising candidates for understanding the mechanism driving VAP [142]. Likewise, protein signatures of sarcoidosis, IPF, pulmonary langerhans cell histiocytosis, PF due to systemic sclerosis were highlighted, and using systems biology platforms, common and distinct pathways were identified [143]. Intriguingly, phosphoproteomics on lung tissues of lung cancer and COPD patients revealed that activation of the NF-κB pathway was the most relevant signaling link between the two diseases, providing clues to developing therapies [144–148].
Complex proteomic methodologies generate immense data, which requires multiple downstream analysis workflows (Fig. 5). Unlike genomics, the under-developed tools for proteomic analysis have many pitfalls [149–151]. However, use of databases such as NCBI, UniProtKB, Swiss-Prot, Protein Information resource (PIR) and OWL, has streamlined protein identification pipeline (Table 4). PROSITE [152] and SWISS-MODEL Repository [153] are used to define domains, families, and structures. Proteins are identified by matching empirically acquired information against a protein database using tools like Compute pI/Mw, ProtParam, PeptideCutter and ProtScale, which includes PTMs in mass calculations [154]. TagIdent [154–156], AACompIdent [154, 157, 158] and AACompSim [154, 157] are online tools that accept multiple data types for protein identification, calculating protein isoelectric point (pI), molecular weight and isoforms of interest, AA composition and sequence tag. Tools such as MOWSE can query user provided peptide or sequence data against the protein databases and rank the results according to a scoring algorithm [159].
Fig. 5.
Proteomic approaches and data analysis work flows. Proteomic analysis can be broadly divided into gel-based and shotgun-based approaches. The data obtained from either of these two methods can be analyzed in different ways depending on the biological properties being probed. Most proteomic analysis is highly sequential in nature and work flows have to be integrated by the user to customize data processing and analysis. Identification of proteins is the preliminary step for analyzing proteomic data. This can be done by querying known databases, analyzing and calculating the masses of peptides and analyzing known posttranslational modifications. Data obtained from high end technologies like mass spectrometry require synergistic tools which can generate databases, perform spectral matches, calculate false discovery rates (FDR) and quantify proteins.
Table 4.
Resources for protein sequence, structure and function
| Databases | Web Links |
|---|---|
| 1. NCBI | http://www.ncbi.nlm.nih.gov/protein/ |
| 2. Protein Information Resource (PIR) | http://pir.georgetown.edu/ |
3. UniProtKB
|
http://www.uniprot.org/ |
| 4. HPRD | http://www.hprd.org/query |
| 5. Human ProteinPedia | http://www.humanproteinpedia.org/index_html |
| 6. nExtProt 3.0 | http://www.nextprot.org/db |
| 7. PRIDE | http://www.ebi.ac.uk/pride/ |
While, Mascot, ProFound and SEQUEST are commercial suites [160–162], open source tools like Protein Prospector, X!Tandem 4 and the Open Mass Spectrometry Search Algorithm (OMSSA) [163, 164] interpret MS-MS, and ICAT data (http://prospector.ucsfedu/prospector/mshome.htm) in equally robust and reliable manner. For versatile tasks, including protein database generation, spectral reduction, peptide false discovery rate (FDR) analysis, peptide quantitation, protein parsimony, protein FDR analysis and protein quantitation proteomic work flows are not well developed. For instance, FDR analysis is critical for maximizing sensitivity while simultaneously controlling specificity. We suggest use of Coon OMSSA Proteomic Analysis Software Suite (COMPASS): a free and open-source software pipeline for high-throughput analysis of proteomics data, designed around the Open Mass Spectrometry Search Algorithm, which provide a viable alternative [165].
Researchers can also access data from exiting proteomics experiments through a number of online resources. Databases like PeptideAtlas [166], Global Proteome Machine Database (GPMDB) [167], MaxQB [168], provide a repository of proteomics data with varying options [169]. Most databases preprocess the data; therefore, to obtain raw data, the PRoteomics IDEntifications (PRIDE) database [170–173] is widely used. PRIDE stores protein and peptide identifications (IDs) and quantitative values (including PTMs), analyzed as mass spectra and the related technical and biological metadata. PRIDE also features several popular tools and allows easy integration of user data into different workflows. Repositories like Model Organism Protein Expression Database (MOPED), PaxDB [174, 175], is focused on protein expression information while MassBank [176] and neXtProt [177, 178] represent an extra layers of information on top of the experimental data from MS.
The rapid emergence of next generation technologies including new MS methods detect proteins with high sensitivity [179]. With triple quadruple approach, researchers can process samples by selected reaction monitoring (SRM), also known as multiple reaction monitoring (MRM) for targeted proteomic analysis [180, 181]. A major resource is SRMAtlas, which is a compendium of targeted proteomics assays run by the Institute for Systems Biology in Seattle, WA. A superior technique to SRM is called SWATH, which allows users to analyze more proteins and peptides as compared to the SRM approach and employs complex mass spectra generated by data-independent acquisition to query for the presence of specific peptides [182]. Recent developments and clinical application of proteomics can be found elsewhere [183–185]. In summation, the emergence of these high end proteomic platforms and development of sophisticated data analysis tools will greatly accelerate research in pulmonary biology.
Metabolomics
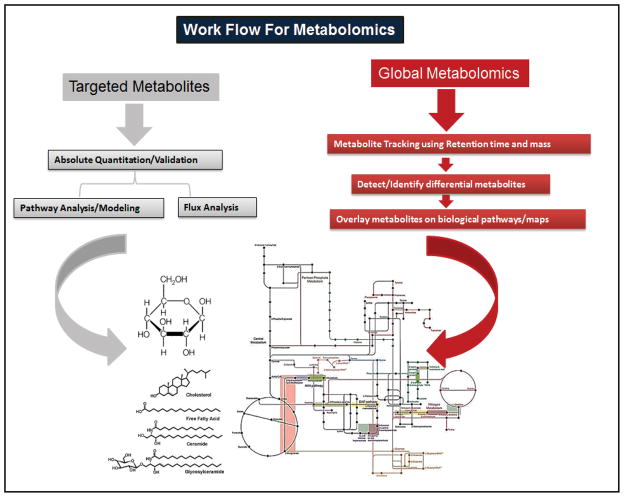
Metabolites from tissues and bio-fluids, includingBALF are identified and quantified using nuclear magnetic resonance (NMR) spectroscopy, Fourier transformed infrared spectroscopy (FT-IR], Fourier transformed ion cyclotron resonance mass spectroscopy (FTICR-MS) and mass spectrometry (MS) [186–191], gas chromatography mass spectrometry (GC–MS) and liquid chromatography based mass spectrometry (LC–MS) [192]. The information derived from these consists of metabolic intermediates, secondary metabolites, hormones, and other signaling molecules, forming the “metabolome” [193]. Analysis of metabolic responses to pharmacological interventions to diseases is defined as “metabonomics” [194], which aims to refine therapy and diagnosis as shown in lung cancer diagnosis [195]. However, these analysis are not trivial and necessitates the use of multiple techniques [196]. Metabolome is thus analyzed by targeted and global analysis (metabolite profiling and fingerprinting) (Fig. 6). The targeted approach being quantitative with a priori knowledge, while global approach generates qualitative profiles of all metabolites, identifying a unique pattern characterizing a snapshot or a fingerprint of the metabolism [190].
Fig. 6.
Targeted and global metabolomics approach for metabolite identification and quantification. Data analysis workflows for global metabolomics are comparable to genomics and proteomics since many of the techniques generate raw data that are of similar format (e.g. KEGG for pathway analysis). Software tools like MetaboAnalyst 2.0 can be used to integrate different “omics data” with metabolomics outputs for a systems level analysis.
Pattern recognition tools and discriminate analysis techniques together classify these fingerprints and associate it with a biological response [197]. This approaches have indeed characterized biochemical fingerprints of airway lining in childhood asthma patients and provided fingerprints of exhaled breath condensate in different clinical settings [198, 199]. Capillary Electrophoresis coupled to Time-of-Flight Mass Spectrometry (CE–TOF-MS) identified and validated metabolic biomarkers in ventilator-induced lung injury (VILI) [200]. Despite early success, sample collection, preparation, and analysis of NMR data remain challenging [201]. Metabolite profiling, an extension of functional genomics, assesses metabolic state of cells and aid in identifying disease genes where no apparent phenotype exists. For example, fingerprint of exhaled breath condensate (EBC) to discern severe asthma phenotype [193, 197, 202, 203].
Integration of metabolome to global omics identified novel dysregulated phospholipids as biomarkers that associate with asthma risk alleles [204]. Potential cancer drug targets have also been identified by integrating cancer gene co-expression network and metabolic networks [205]. Thus, users can integrate “omic” profiles to better understand pulmonary disorders by combining genomic, proteomic and metabolic signatures.
The sheer volume of data and complexity of metabolome requires a combination of mathematical, statistical and bioinformatical tools for analysis (Table 5) as reviewed previously [206, 207]. The Automated Mass Spectral Deconvolution, Identification System 2.63 (AMDIS) [208] and Mzmine [209] are the tools used to process raw data obtained by GC-MS and LC-MS. Recent additions include, MeltDB, a web-based software platform for the analysis and annotation of datasets from metabolomics experiments [210], while software suites like MetaboAnalyst 2.0 offer easy to use and visualize interface for researchers. MetaboAnalyst accepts a variety of metabolomics data types and consists of different functional modules - data processing, functional enrichment, statistical and metabolic pathway analysis [211]. XCMS software tool [212] is a unique tools that integrates peak detection, statistical analysis, and subsequent visualization of raw data for verification purposes and can be used for analyzing metabolite data from LC/MS, peptide digests, and GC/MS. Pathway analysis and data visualization tools form critical components of metabolomic data analysis. Prominent options include KEGG (http://www.genome.ad.jp/kegg/) and MetaCyc (http://metacyc.org/].
Table 5.
Software tools for epigenomics data processing and analyses
Databases that catalogue information about the human metabolome are vital in deducing biological interpretations from a metabolomics experiment. The human metabolome database (HMDB) [213–215] version 3.5 (http://www.hmdb.ca) contains) contains 41,469 metabolite entries including both water-soluble and lipid soluble metabolites as well as metabolites that would be regarded as either abundant (> 1 uM) or relatively rare (< 1 nM). Additionally, 5,688 protein (and DNA) sequences are linked to these metabolite entries. The database supports extensive text, sequence, chemical structure and relational query searches all displayed in interactive easy to comprehend viewing applets.
Conclusion
Different omics approaches attempt to unravel the high degree of complexity in biological systems and explain the onset and progression of diseases, revealing mechanisms that can be used for targeted interventional therapies. The sheer breadth of information generated by these technologies from different vantage points (RNA, DNA, proteins, chromatin, and metabolites) necessitates integration to arrive at a holistic picture of a given disease. Thus, the use of systems biology approach that integrates results obtained from high throughput technologies is fast becoming a central theme in medical research, especially in context of personalized medicine [216]. A “systomics” approach would combine data obtained from genomic, proteomic, epigenomic and metabolomic data to study and treat human diseases. For instance, systomic medicine can allow researchers and clinicians to study closely related pulmonary remodeling disorders like COPD, asthma, lung cancer and emphysema that have several overlapping phenotypes and symptoms (Fig. 7). Integration of data from the different omics platforms would allow the users to better understand the cause and effect relationships between molecular disruptions (e.g. gene mutations, alterations in protein expression and structure and fluctuating levels of metabolites) and help understand progression of irreversible lung remodeling diseases at the molecular level at a holistic level. This will allow us to identify common causative molecular events in these lung diseases, identifying novel drug targets and biomarkers that could pave the way for personalized medical treatment of pulmonary disorders based on individual genetics, age, environment and physiology.
Fig. 7.
Systomics: Integrative approach for pulmonary medicine. The confluence of genomics, proteomics, metabolomics and epigenomics drives systems driven approach for studying pulmonary disorders. Integration of work flows and data obtained from these will contribute to a more holistic understanding of the onset and progression of pulmonary diseases. This in turn will allow identification of novel disease phenotypes and assist drug designing strategies based on unique biomarkers. In summation, the integration of omics approaches will pave the way for personalized medicine in pulmonary medicine.
Table 3.
Protein-Protein Interactions Databases
| Databases | Web Links |
|---|---|
| l. STRING 9.05 | http://string-db.org |
| 2. Molecular Interaction Database (MINT) | http://mint.bio.uniroma2.it/mint/Welcome.do |
| 3. Database of Interacting Proteins (DIP) | http://dip.doe-mbi.ucla.edu/dip/Main.cgi |
| 4. IntAct | http://www.ebi.ac.uk/intact/ |
| 5. HPRD | http://www.hprd.org/ |
| 6. BioGRID 3.2 | http://thebiogrid.org/ |
| 7. PrePPT | http://bhapp.c2b2.columbia.edu/PrePPI/ |
Table 6.
Software tools for analyzing and interpreting metabolomics data
Acknowledgments
This work was supported by American Heart Association Grant. SDG-155-N (V.D.) and Moffitt Cancer Center Lung SPORE Career Development Grant (V.D.)
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Ideker T, Galitski T, Hood L. A new approach to decoding life: Systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 4.Kitano H. Systems biology: A brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 5.Aderem A. Systems biology: Its practice and challenges. Cell. 2005;121:511–513. doi: 10.1016/j.cell.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Kirschner MW. The meaning of systems biology. Cell. 2005;121:503–504. doi: 10.1016/j.cell.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheppard D. Aspen lung conference 2010: Systems biology of lung diseases--progress in the omics era. Proc Am Thorac Soc. 2011;8:199–202. doi: 10.1513/pats.201010-065MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auffray C, Adcock IM, Chung KF, Djukanovic R, Pison C, Sterk PJ. An integrative systems biology approach to understanding pulmonary diseases. Chest. 2010;137:1410–1416. doi: 10.1378/chest.09-1850. [DOI] [PubMed] [Google Scholar]
- 10.Kaminsky DA, Irvin CG, Sterk PJ. Complex systems in pulmonary medicine: A systems biology approach to lung disease. J Appl Physiol. 2011;110:1716–1722. doi: 10.1152/japplphysiol.01310.2010. [DOI] [PubMed] [Google Scholar]
- 11.Wu W, Kaminski N. Chronic lung diseases. Wiley Interdiscip Rev Syst Biol Med. 2009;1:298–308. doi: 10.1002/wsbm.23. [DOI] [PubMed] [Google Scholar]
- 12.Gosselink JV, Hayashi S, Elliott WM, Xing L, Chan B, Yang L, Wright C, Sin D, Pare PD, Pierce JA, Pierce RA, Patterson A, Cooper J, Hogg JC. Differential expression of tissue repair genes in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;181:1329–1335. doi: 10.1164/rccm.200812-1902OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel NM, Kawut SM, Jelic S, Arcasoy SM, Lederer DJ, Borczuk AC. Pulmonary arteriole gene expression signature in idiopathic pulmonary fibrosis. Eur Respir J. 2013;41:1324–1330. doi: 10.1183/09031936.00084112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mura M, Anraku M, Yun Z, McRae K, Liu M, Waddell TK, Singer LG, Granton JT, Keshavjee S, de Perrot M. Gene expression profiling in the lungs of patients with pulmonary hypertension associated with pulmonary fibrosis. Chest. 2012;141:661–673. doi: 10.1378/chest.11-0449. [DOI] [PubMed] [Google Scholar]
- 15.Drake JI, Gomez-Arroyo J, Dumur CI, Kraskauskas D, Natarajan R, Bogaard HJ, Fawcett P, Voelkel NF. Chronic carvedilol treatment partially reverses the right ventricular failure transcriptional profile in experimental pulmonary hypertension. Physiol Genomics. 2013;45:449–461. doi: 10.1152/physiolgenomics.00166.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XQ, Wang XM, Zhou TF, Dong LQ. Screening of differentially expressed genes and small molecule drugs of pediatric allergic asthma with DNA microarray. Eur Rev Med Pharmacol Sci. 2012;16:1961–1966. [PubMed] [Google Scholar]
- 17.Mariani TJ, Martinez F. Chronic obstructive pulmonary disease genomics: Yesterday, discovering population biomarkers; tomorrow, defining disease clusters. Am J Respir Crit Care Med. 2013;187:900–902. doi: 10.1164/rccm.201302-0340ED. [DOI] [PubMed] [Google Scholar]
- 18.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Rna-guided human genome engineering via cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biology. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Sebastiani P, Liu G, Schembri F, Dumas YM, Langer EM, Alekseyev Y, O’Connor GT, Brooks DR, Lenburg ME, Spira A. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 2010;41:1–8. doi: 10.1152/physiolgenomics.00167.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, Sridhar S, Beamis J, Lamb C, Anderson T, Gerry N, Keane J, Lenburg ME, Brody JS. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 23.Beane J, Sebastiani P, Whitfield TH, Steiling K, Dumas YM, Lenburg ME, Spira A. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res (Phila) 2008;1:56–64. doi: 10.1158/1940-6207.CAPR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaauboer ME, Emson CL, Verschuren L, van Erk M, Turner SM, Everts V, Hanemaaijer R, Stoop R. Novel combination of collagen dynamics analysis and transcriptional profiling reveals fibrosis-relevant genes and pathways. Matrix Biology. 2013;32:424–431. doi: 10.1016/j.matbio.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Lee TH, McKleroy W, Khalifeh-Soltani A, Sakuma S, Lazarev S, Riento K, Nishimura SL, Nichols BJ, Atabai K. Functional genomic screen identifies novel mediators of collagen uptake. Molecular biology of the cell. 2014 doi: 10.1091/mbc.E13-07-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank JA, Erle DJ. Progress toward a systems biology approach to acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2007;293:L290–291. doi: 10.1152/ajplung.00220.2007. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Shi JX, Pan XF, Feng J, Zhao H. DNA microarray-based screening of differentially expressed genes related to acute lung injury and functional analysis. Eur Rev Med Pharmacol Sci. 2013;17:1044–1050. [PubMed] [Google Scholar]
- 28.Singhal S, Miller D, Ramalingam S, Sun SY. Gene expression profiling of non-small cell lung cancer. Lung Cancer. 2008;60:313–324. doi: 10.1016/j.lungcan.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Ponten F, Konig A, Fernandes O, Karlsson M, Helenius G, Karlsson C, Rahnenfuhrer J, Hengstler JG, Micke P. Biomarker discovery in non-small cell lung cancer: Integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 30.Bourboulia D, Han H, Jensen-Taubman S, Gavil N, Isaac B, Wei B, Neckers L, Stetler-Stevenson WG. Timp-2 modulates cancer cell transcriptional profile and enhances e-cadherin/beta-catenin complex expression in a549 lung cancer cells. Oncotarget. 2013;4:166–176. doi: 10.18632/oncotarget.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C, Xiong M, Luo Y, Li J, Zhang Y, Dong Y, Zhu Y, Niu T, Wang Z, Duan L. Genome-wide transcriptional analysis of apoptosis-related genes and pathways regulated by h2ax in lung cancer a549 cells. Apoptosis. 2013;18:1039–1047. doi: 10.1007/s10495-013-0875-x. [DOI] [PubMed] [Google Scholar]
- 32.Herazo-Maya JD, Kaminski N. Evolving genomics of pulmonary fibrosis. Idiopathic Pulmonary Fibrosis. 2014:379–402. [Google Scholar]
- 33.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner AM. Fifty years on: Gwas confirms the role of a rare variant in lung disease. PLoS Genetics. 2013;9:e1003768. doi: 10.1371/journal.pgen.1003768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan QL, Gaume BR, Hawkins GA, Himes BE, Bleecker ER, Klanderman B, Irvin CG, Peters SP, Meyers DA, Hanrahan JP, Lima JJ, Litonjua AA, Tantisira KG, Liggett SB. Regulatory haplotypes in arg1 are associated with altered bronchodilator response. Am J Respir Crit Care Med. 2011;183:449–454. doi: 10.1164/rccm.201005-0758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu AC, Himes BE, Lasky-Su J, Litonjua A, Li L, Lange C, Lima J, Irvin CG, Weiss ST. Development of a pharmacogenetic predictive test in asthma: Proof of concept. Pharmacogenet Genomics. 2010;20:86–93. doi: 10.1097/FPC.0b013e32833428d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. Genepattern 2. 0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 41.Goecks J, Nekrutenko A, Taylor J. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: Ncbi gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. Ncbi geo: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, Kurbatova N, Malone J, Mani R, Mupo A, Pedro Pereira R, Pilicheva E, Rung J, Sharma A, Tang YA, Ternent T, Tikhonov A, Welter D, Williams E, Brazma A, Parkinson H, Sarkans U. Arrayexpress update--trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, Eisen MB, Spellman PT, Brown PO, Botstein D, Cherry JM. The Stanford microarray database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Science Trans Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB. Integrating the micrornome into the study of lung disease. Am J Resp Crit Care Med. 2009;179:4–10. doi: 10.1164/rccm.200807-1042PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomankova T, Petrek M, Kriegova E. Involvement of micrornas in physiological and pathological processes in the lung. Respir Res. 2010;11:159. doi: 10.1186/1465-9921-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou T, Garcia JGN, Zhang W. Integrating micrornas into a system biology approach to acute lung injury. Translat Res. 2011;157:180–190. doi: 10.1016/j.trsl.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oglesby IK, McElvaney NG, Greene CM. Micrornas in inflammatory lung disease--master regulators or target practice? Respir Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayed D, Abdellatif M. Micrornas in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 53.Pagdin T, Lavender P. Micrornas in lung diseases. Thorax. 2012;67:183–184. doi: 10.1136/thoraxjnl-2011-200532. [DOI] [PubMed] [Google Scholar]
- 54.Sittka A, Schmeck B. Micrornas in the lung. Adv Exp Med Biol. 2013;774:121–134. doi: 10.1007/978-94-007-5590-1_7. [DOI] [PubMed] [Google Scholar]
- 55.Sessa R, Hata A. Role of micrornas in lung development and pulmonary diseases. Pulmonary Circ. 2013;3:315–328. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandit KV, Milosevic J, Kaminski N. Micrornas in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 57.Kass DJ, Kaminski N. Evolving genomic approaches to idiopathic pulmonary fibrosis: Moving beyond genes. Clin Transl Sci. 2011;4:372–379. doi: 10.1111/j.1752-8062.2011.00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi K, Yokota SI, Tatsumi N, Fukami T, Yokoi T, Nakajima M. Cigarette smoking substantially alters plasma microrna profiles in healthy subjects. Toxicol Appl Pharmacol. 2013;272:154–160. doi: 10.1016/j.taap.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 59.Banerjee A, Luettich K. Micrornas as potential biomarkers of smoking-related diseases. Biomark Med. 2012;6:671–684. doi: 10.2217/bmm.12.50. [DOI] [PubMed] [Google Scholar]
- 60.Graff JW, Powers LS, Dickson AM, Kim J, Reisetter AC, Hassan IH, Kremens K, Gross TJ, Wilson ME, Monick MM. Cigarette smoking decreases global microrna expression in human alveolar macrophages. PLoS One. 2012;7:e44066. doi: 10.1371/journal.pone.0044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating micrornas as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138–1147. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]
- 62.Donaldson A, Natanek SA, Lewis A, Man WD, Hopkinson NS, Polkey MI, Kemp PR. Increased skeletal muscle-specific microrna in the blood of patients with copd. Thorax. 2013;68:1140–1149. doi: 10.1136/thoraxjnl-2012-203129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Madhavan D, Cuk K, Burwinkel B, Yang R. Cancer diagnosis and prognosis decoded by blood-based circulating microrna signatures. Front Genet. 2013;4:116. doi: 10.3389/fgene.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothschild SI. Epigenetic therapy in lung cancer - role of micrornas. Front Oncol. 2013;3:158. doi: 10.3389/fonc.2013.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of micrornas to hla-g and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin JI, Brusselle GG. Mechanistic links between copd and lung cancer: A role of microrna let-7? Nat Rev Cancer. 2014;14:70–70. doi: 10.1038/nrc3477-c1. [DOI] [PubMed] [Google Scholar]
- 67.Jardim MJ, Dailey L, Silbajoris R, Diaz-Sanchez D. Distinct microrna expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am J Respir Cell Mol Biol. 2012;47:536–542. doi: 10.1165/rcmb.2011-0160OC. [DOI] [PubMed] [Google Scholar]
- 68.Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, Nguyen C, Solon M, Barczak AJ, Zlock LT, Blagev DP, Finkbeiner WE, Ansel KM, Arron JR, Erle DJ, Woodruff PG. Airway epithelial mirna expression is altered in asthma. Am J Respir Crit Care Med. 2012;186:965–974. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plank M, Maltby S, Mattes J, Foster PS. Targeting translational control as a novel way to treat inflammatory disease: The emerging role of micrornas. Clin Exp Allergy. 2013;43:981–999. doi: 10.1111/cea.12170. [DOI] [PubMed] [Google Scholar]
- 70.Lino Cardenas CL, Kaminski N, Kass DJ. Micromanaging micrornas: Using murine models to study micrornas in lung fibrosis. Drug Discovery Today: Disease Models. doi: 10.1016/j.ddmod.2012.11.003. doi.org/10.1016/j.ddmod.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R, Denby L, Dempsie Y, Long L, Morrell NW, Baker AH. Dynamic changes in lung microrna profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscl Thrombo Vase Biol. 2010;30:716–723. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 72.Courboulin A, Paulin R, Giguere NJ, Saksouk N, Perreault T, Meloche J, Paquet ER, Biardel S, Provencher S, Cote J, Simard MJ, Bonnet S. Role for mir-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–548. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.White K, Loscalzo J, Chan SY. Holding our breath: The emerging and anticipated roles of microrna in pulmonary hypertension. Pulmon Circ. 2012;2:278–290. doi: 10.4103/2045-8932.101395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho JH, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, Dakhallah D, Price J, Bornman D, Zhang S, Marsh C, Galas D. Systems biology of interstitial lung diseases: Integration of mrna and microrna expression changes. BMC Med Genomics. 2011;4:8. doi: 10.1186/1755-8794-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 75.Weng C-W, Lee S-C, Lee Y-L, Ng K-L. Analysis of the nci-60 dataset for cancer-related microrna and mrna using expression profiles. Comput Biol Chem. 2013;44:15–21. doi: 10.1016/j.compbiolchem.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 76.Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, Shapiro CL, Huebner K. Integrated microrna and mrna signatures associated with survival in triple negative breast cancer. PLoS One. 2013;8:e55910. doi: 10.1371/journal.pone.0055910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhattacharya S, Mariani TJ. Systems biology approaches to identify developmental bases for lung diseases. Pediatr Res. 2013;73:514–522. doi: 10.1038/pr.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agusti A, Sobradillo P, Celli B. Addressing the complexity of chronic obstructive pulmonary disease: From phenotypes and biomarkers to scale-free networks, systems biology, and p4 medicine. Am J Respir Crit Care Med. 2011;183:1129–1137. doi: 10.1164/rccm.201009-1414PP. [DOI] [PubMed] [Google Scholar]
- 79.Silverman EK, Loscalzo J. Network medicine approaches to the genetics of complex diseases. Discov Med. 2012;14:143–152. [PMC free article] [PubMed] [Google Scholar]
- 80.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microrna. Org resource: Targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. Mirbase: Tools for microrna genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 83.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Microrna targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG. Diana-microt web server: Elucidating microrna functions through target prediction. Nucleic Acids Res. 2009;37:W273–276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microrna target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 87.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: A functional update of tarbase. Nucleic Acids Res. 2009;37:D155–158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. Mirecords: An integrated resource for microrna-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, Chien CH, Wu MC, Huang CY, Tsou AP, Huang HD. Mirtarbase: A database curates experimentally validated microrna-target interactions. Nucleic Acids Res. 2011;39:D163–169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dweep H, Sticht C, Pandey P, Gretz N. Mirwalk-database: Prediction of possible mirna binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. Mirnamap: Genomic maps of microrna genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microrna targets. Nat Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 93.Sales G, Coppe A, Bisognin A, Biasiolo M, Bortoluzzi S, Romualdi C. Magia, a web-based tool for mirna and genes integrated analysis. Nucleic Acids Res. 2010;38:W352–359. doi: 10.1093/nar/gkq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elkan-Miller T, Ulitsky I, Hertzano R, Rudnicki A, Dror AA, Lenz DR, Elkon R, Irmler M, Beckers J, Shamir R, Avraham KB. Integration of transcriptomics, proteomics, and microrna analyses reveals novel microrna regulation of targets in the mammalian inner ear. PLoS One. 2011;6:e18195. doi: 10.1371/journal.pone.0018195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beck D, Ayers S, Wen J, Brandl MB, Pham TD, Webb P, Chang CC, Zhou X. Integrative analysis of next generation sequencing for small non-coding rnas and transcriptional regulation in myelodysplastic syndromes. BMC Med Genomics. 2011;4:19. doi: 10.1186/1755-8794-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muniategui A, Pey J, Planes FJ, Rubio A. Joint analysis of mirna and mrna expression data. Brief Bioinform. 2013;14:263–278. doi: 10.1093/bib/bbs028. [DOI] [PubMed] [Google Scholar]
- 97.Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, Pommier Y, Weinstein JN. Cellminer: A relational database and query tool for the nci-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncrnamirna interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flintoft L. Non-coding rna: Structure and function for lncrnas. Nature reviews Genetics. 2013;14:598. doi: 10.1038/nrg3561. [DOI] [PubMed] [Google Scholar]
- 100.Enfield KSS, Pikor LA, Martinez VD, Lam WL. Mechanistic roles of noncoding rnas in lung cancer biology and their clinical implications. Genet Res Int. 2012;2012:16. doi: 10.1155/2012/737416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang X, Gao L, Guo X, Shi X, Wu H, Song F, Wang B. A network based method for analysis of lncrna-disease associations and prediction of lncrnas implicated in diseases. PLoS One. 2014;9:e87797. doi: 10.1371/journal.pone.0087797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu M-X, Chen X, Chen G, Cui Q-H, Yan G-Y. A computational framework to infer human disease-associated long noncoding rnas. PLoS One. 2014;9:e84408. doi: 10.1371/journal.pone.0084408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Z, Gerstein M, Snyder M. Rna-seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beyond the genome 2012 boston, ma USA 27–29 september 2012. Abstracts BMC Proc. 2012;6:O1–P54. [PubMed] [Google Scholar]
- 105.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO, Wang S, Levy S, Massion PP, Lenburg M, Spira A. Characterizing the impact of smoking and lung cancer on the airway transcriptome using rna-seq. Cancer Prev Res (Phila) 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hackett NR, Butler MW, Shaykhiev R, Salit J, Omberg L, Rodriguez-Flores JL, Mezey JG, Strulovici-Barel Y, Wang G, Didon L, Crystal RG. Rna-seq quantification of the human small airway epithelium transcriptome. BMC Genomics. 2012:13. doi: 10.1186/1471-2164-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou C, Chen H, Han L, Xue F, Wang A, Liang Y-j. Screening of genes related to lung cancer caused by smoking with rna-seq. Eur Rev Med Pharmacol Sci. 2014;18:117–125. [PubMed] [Google Scholar]
- 108.Yick CY, Zwinderman AH, Kunst PW, Grunberg K, Mauad T, Dijkhuis A, Bel EH, Baas F, Lutter R, Sterk PJ. Transcriptome sequencing (rna-seq) of human endobronchial biopsies: Asthma versus controls. The European respiratory journal. 2013;42:662–670. doi: 10.1183/09031936.00115412. [DOI] [PubMed] [Google Scholar]
- 109.Robinson MD, McCarthy DJ, Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goncalves A, Tikhonov A, Brazma A, Kapushesky M. A pipeline for rna-seq data processing and quality assessment. Bioinformatics. 2011;27:867–869. doi: 10.1093/bioinformatics/btr012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yang IV. Epigenomics of idiopathic pulmonary fibrosis. Epigenomics. 2012;4:195–203. doi: 10.2217/epi.12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Adcock IM, Ford P, Ito K, Barnes PJ. Epigenetics and airways disease. Respiratory Res. 2006;7:21. doi: 10.1186/1465-9921-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: Molecular mechanisms and implications for disease. Trends Mol Med. 2010;16:7–16. doi: 10.1016/j.molmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 116.Martino D, Prescott S. Epigenetics and prenatal influences on asthma and allergic airways disease. Chest. 2011;139:640–647. doi: 10.1378/chest.10-1800. [DOI] [PubMed] [Google Scholar]
- 117.Schwartz DA. Epigenetics and environmental lung disease. Proc Am Thorac Soc. 2010;7:123–125. doi: 10.1513/pats.200908-084RM. [DOI] [PubMed] [Google Scholar]
- 118.Sundar IK, Yao H, Rahman I. Oxidative stress and chromatin remodeling in chronic obstructive pulmonary disease and smoking-related diseases. Antioxid Redox Signal. 2013;18:1956–1971. doi: 10.1089/ars.2012.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rosas IO, Yang IV. The promise of epigenetic therapies in treatment of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:336–338. doi: 10.1164/rccm.201212-2272ED. [DOI] [PubMed] [Google Scholar]
- 120.Vucic EA, Chari R, Thu KL, Wilson IM, Cotton AM, Kennett JY, Zhang M, Lonergan KM, Steiling K, Brown CJ, McWilliams A, Ohtani K, Lenburg ME, Sin DD, Spira A, MacAulay CE, Lam S, Lam WL. DNA methylation is globally disrupted and associated with expression changes in copd small airways. Am J Respir Cell Mol Biol. 2013 doi: 10.1165/rcmb.2013-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, Bray M, Zhang K, Thannickal VJ, Hagood JS. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rabinovich EI, Kapetanaki MG, Steinfeld I, Gibson KF, Pandit KV, Yu G, Yakhini Z, Kaminski N. Global methylation patterns in idiopathic pulmonary fibrosis. PLoS One. 2012;7:e33770. doi: 10.1371/journal.pone.0033770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pandit KV, Milosevic J, Kaminski N. Micrornas in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 124.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Epigenetic regulation of mir-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Balgkouranidou I, Liloglou T, Lianidou ES. Lung cancer epigenetics: Emerging biomarkers. Biomark Med. 2013;7:49–58. doi: 10.2217/bmm.12.111. [DOI] [PubMed] [Google Scholar]
- 126.Huang SK, Scruggs AM, Donaghy J, Horowitz JC, Zaslona Z, Przybranowski S, White ES, Peters-Golden M. Histone modifications are responsible for decreased fas expression and apoptosis resistance in fibrotic lung fibroblasts. Cell Death Dis. 2013;4:e621. doi: 10.1038/cddis.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sundar IK, Nevid MZ, Friedman AE, Rahman I. Cigarette smoke induces distinct histone modifications in lung cells: Implications for the pathogenesis of copd and lung cancer. J Proteome Res. 2013;13:982–996. doi: 10.1021/pr400998n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu Y, Li H, Xiao T, Lu Q. Epigenetics in immune-mediated pulmonary diseases. Clinic Rev Allerg Immunol. 2013;45:314–330. doi: 10.1007/s12016-013-8398-3. [DOI] [PubMed] [Google Scholar]
- 129.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 130.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet--next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fingerman IM, Zhang X, Ratzat W, Husain N, Cohen RF, Schuler GD. Ncbi epigenomics: What’s new for 2013. Nucl Acids Res. 2013;41:D221–225. doi: 10.1093/nar/gks1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fingerman IM, McDaniel L, Zhang X, Ratzat W, Hassan T, Jiang Z, Cohen RF, Schuler GD. Ncbi epigenomics: A new public resource for exploring epigenomic data sets. Nucl Acids Res. 2011;39:D908–912. doi: 10.1093/nar/gkq1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hirsch J, Hansen KC, Burlingame AL, Matthay MA. Proteomics: Current techniques and potential applications to lung disease. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1–23. doi: 10.1152/ajplung.00301.2003. [DOI] [PubMed] [Google Scholar]
- 136.Wattiez R, Falmagne P. Proteomics of bronchoalveolar lavage fluid. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:169–178. doi: 10.1016/j.jchromb.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 137.Bowler RP, Ellison MC, Reisdorph N. Proteomics in pulmonary medicine. Chest. 2006;130:567–574. doi: 10.1378/chest.130.2.567. [DOI] [PubMed] [Google Scholar]
- 138.Wiktorowicz J, Jamaluddin M. Proteomic analysis of the asthmatic airway. In: Brasier AR, editor. Heterogeneity in asthma. Vol. 795. Springer; USA: 2014. pp. 221–232. [DOI] [PubMed] [Google Scholar]
- 139.Xu D-D, Deng D-F, Li X, Wei L-L, Li Y-Y, Yang X-Y, Yu W, Wang C, Jiang T-T, Li Z-J, Chen Z-L, Zhang X, Liu J-Y, Ping Z-P, Qiu Y-Q, Li J-C. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using itraq-coupled two-dimensional lc-ms/ms. Proteomics. 2014;14:322–331. doi: 10.1002/pmic.201300383. [DOI] [PubMed] [Google Scholar]
- 140.Konsavage WM, Umstead TM, Wu Y, Phelps DS, Shenberger JS. Hyperoxia-induced alterations in the pulmonary proteome of juvenile rats. Exp Lung Res. 2013;39:107–117. doi: 10.3109/01902148.2013.763871. [DOI] [PubMed] [Google Scholar]
- 141.Giorgianni F, Mileo V, Desiderio DM, Catinella S, Beranova-Giorgianni S. Characterization of the phosphoproteome in human bronchoalveolar lavage fluid. Int J Proteomics. 2012;2012:460261. doi: 10.1155/2012/460261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nguyen EV, Gharib SA, Palazzo SJ, Chow YH, Goodlett DR, Schnapp LM. Proteomic profiling of bronchoalveolar lavage fluid in critically ill patients with ventilator-associated pneumonia. PLoS One. 2013;8:e58782. doi: 10.1371/journal.pone.0058782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Landi C, Bargagli E, Bianchi L, Gagliardi A, Carleo A, Bennett D, Perari MG, Armini A, Prasse A, Rottoli P, Bini L. Towards a functional proteomics approach to the comprehension of idiopathic pulmonary fibrosis, sarcoidosis, systemic sclerosis and pulmonary langerhans cell histiocytosis. J Proteomics. 2013;83:60–75. doi: 10.1016/j.jprot.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 144.Wang YT, Tsai CF, Hong TC, Tsou CC, Lin PY, Pan SH, Hong TM, Yang PC, Sung TY, Hsu WL, Chen YJ. An informatics-assisted label-free quantitation strategy that depicts phosphoproteomic profiles in lung cancer cell invasion. J Proteome Res. 2010;9:5582–5597. doi: 10.1021/pr100394u. [DOI] [PubMed] [Google Scholar]
- 145.Lopez E, Cho WC. Phosphoproteomics and lung cancer research. Int J Mol Sci. 2012;13:12287–12314. doi: 10.3390/ijms131012287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sudhir PR, Hsu CL, Wang MJ, Wang YT, Chen YJ, Sung TY, Hsu WL, Yang UC, Chen JY. Phosphoproteomics identifies oncogenic ras signaling targets and their involvement in lung adenocarcinomas. PLoS One. 2011;6:e20199. doi: 10.1371/journal.pone.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Park JO, Choi DY, Choi DS, Kim HJ, Kang JW, Jung JH, Lee JH, Kim J, Freeman MR, Lee KY, Gho YS, Kim KP. Identification and characterization of proteins isolated from microvesicles derived from human lung cancer pleural effusions. Proteomics. 2013;13:2125–2134. doi: 10.1002/pmic.201200323. [DOI] [PubMed] [Google Scholar]
- 148.Pastor MD, Nogal A, Molina-Pinelo S, Carnero A, Paz-Ares L. Proteomic biomarkers in lung cancer. Clin Transl Oncol. 2013;15:671–682. doi: 10.1007/s12094-013-1034-0. [DOI] [PubMed] [Google Scholar]
- 149.Krueger KE, Srivastava S. Posttranslational protein modifications: Current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteom. 2006;5:1799–1810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 150.Vaudel M, Sickmann A, Martens L. Introduction to opportunities and pitfalls in functional mass spectrometry based proteomics. Biochim Biophys Acta. 2014;1844:12–20. doi: 10.1016/j.bbapap.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 151.Perez-Riverol Y, Hermjakob H, Kohlbacher O, Martens L, Creasy D, Cox J, Leprevost F, Shan BP, Perez-Nueno VI, Blazejczyk M, Punta M, Vierlinger K, Valiente PA, Leon K, Chinea G, Guirola O, Bringas R, Cabrera G, Guillen G, Padron G, Gonzalez LJ, Besada V. Computational proteomics pitfalls and challenges: Havanabioinfo 2012 workshop report. J Proteomics. 2013;87:134–138. doi: 10.1016/j.jprot.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 152.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at prosite. Nucl Acid Res. 2013;41:D344–347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The swiss-model repository and associated resources. Nucl Acid Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the expasy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 155.Wilkins MR, Gasteiger E, Sanchez JC, Appel RD, Hochstrasser DF. Protein identification with sequence tags. Curr Biol. 1996;6:1543–1544. doi: 10.1016/s0960-9822(02)70764-1. [DOI] [PubMed] [Google Scholar]
- 156.Wilkins MR, Gasteiger E, Tonella L, Ou K, Tyler M, Sanchez JC, Gooley AA, Walsh BJ, Bairoch A, Appel RD, Williams KL, Hochstrasser DF. Protein identification with n and c-terminal sequence tags in proteome projects. J Mol Biol. 1998;278:599–608. doi: 10.1006/jmbi.1998.1726. [DOI] [PubMed] [Google Scholar]
- 157.Wilkins MR, Pasquali C, Appel RD, Ou K, Golaz O, Sanchez JC, Yan JX, Gooley AA, Hughes G, Humphery-Smith I, Williams KL, Hochstrasser DF. From proteins to proteomes: Large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology (NY) 1996;14:61–65. doi: 10.1038/nbt0196-61. [DOI] [PubMed] [Google Scholar]
- 158.Wilkins MR, Ou K, Appel RD, Sanchez J-C, Yan JX, Golaz O, Farnsworth V, Cartier P, Hochstrasser DF, Williams KL, Gooley AA. Rapid protein identification using n-terminal “sequence tag” and amino acid analysis. Biochem Biophys Res Com. 1996;221:609–613. doi: 10.1006/bbrc.1996.0643. [DOI] [PubMed] [Google Scholar]
- 159.Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 160.Zhang W, Chait BT. Profound: An expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 161.Eng J, McCormack A, Yates J. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 162.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 163.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 164.Balgley BM, Laudeman T, Yang L, Song T, Lee CS. Comparative evaluation of tandem ms search algorithms using a target-decoy search strategy. Mol Cell Proteomics. 2007;6:1599–1608. doi: 10.1074/mcp.M600469-MCP200. [DOI] [PubMed] [Google Scholar]
- 165.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. Compass: A suite of pre- and post-search proteomics software tools for omssa. Proteomics. 2011;11:1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deutsch EW, Lam H, Aebersold R. Peptideatlas: A resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 168.Schaab C, Geiger T, Stoehr G, Cox J, Mann M. Analysis of high accuracy, quantitative proteomics data in the maxqb database. Mol Cell Proteomics. 2012;11:M111 014068. doi: 10.1074/mcp.M111.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mead JA, Bianco L, Bessant C. Recent developments in public proteomic ms repositories and pipelines. Proteomics. 2009;9:861–881. doi: 10.1002/pmic.200800553. [DOI] [PubMed] [Google Scholar]
- 170.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Perez-Riverol Y, Reisinger F, Rios D, Wang R, Hermjakob H. The proteomics identifications (pride) database and associated tools: Status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jones P, Martens L. Using the pride proteomics identifications database for knowledge discovery and data analysis. Methods Mol Biol. 2010;604:297–307. doi: 10.1007/978-1-60761-444-9_20. [DOI] [PubMed] [Google Scholar]
- 172.Jones P, Cote R. The pride proteomics identifications database: Data submission, query, and dataset comparison. Methods Mol Biol. 2008;484:287–303. doi: 10.1007/978-1-59745-398-1_19. [DOI] [PubMed] [Google Scholar]
- 173.Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. Pride: The proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 174.Kolker E, Higdon R, Haynes W, Welch D, Broomall W, Lancet D, Stanberry L, Kolker N. Moped: Model organism protein expression database. Nucleic Acids Res. 2012;40:D1093–1099. doi: 10.1093/nar/gkr1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, von Mering C. Paxdb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11:492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. Massbank: A public repository for sharing mass spectral data for life sciences. J Mass Spectromet. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 177.Gaudet P, Argoud-Puy G, Cusin I, Duek P, Evalet O, Gateau A, Gleizes A, Pereira M, Zahn-Zabal M, Zwahlen C, Bairoch A, Lane L. Nextprot: Organizing protein knowledge in the context of human proteome projects. J Proteome Res. 2013;12:293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- 178.Lane L, Argoud-Puy G, Britan A, Cusin I, Duek PD, Evalet O, Gateau A, Gaudet P, Gleizes A, Masselot A, Zwahlen C, Bairoch A. Nextprot: A knowledge platform for human proteins. Nucleic Acids Res. 2012;40:D76–83. doi: 10.1093/nar/gkr1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Method of the year 2012. Nat Meth. 2013;10:1–1. doi: 10.1038/nmeth.2329. [DOI] [PubMed] [Google Scholar]
- 180.Doerr A. Targeted proteomics. Nat Meth. 2011;8:43–43. [Google Scholar]
- 181.Doerr A. Mass spectrometry-based targeted proteomics. Nat Meth. 2013;10:23–23. doi: 10.1038/nmeth.2286. [DOI] [PubMed] [Google Scholar]
- 182.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the ms/ms spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol Cell Proteom. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Savaryn JP, Catherman AD, Thomas PM, Abecassis MM, Kelleher NL. The emergence of top-down proteomics in clinical research. Genome Med. 2013;5:1–8. doi: 10.1186/gm457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yassine H, Borges CR, Schaab MR, Billheimer D, Stump C, Reaven P, Lau SS, Nelson R. Mass spectrometric immunoassay and multiple reaction monitoring as targeted ms-based quantitative approaches in biomarker development: Potential applications to cardiovascular disease and diabetes. Proteomics Clin Appl. 2013;7:528–540. doi: 10.1002/prca.201200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Krastins B, Prakash A, Sarracino DA, Nedelkov D, Niederkofler EE, Kiernan UA, Nelson R, Vogelsang MS, Vadali G, Garces A. Rapid development of sensitive, high-throughput, quantitative and highly selective mass spectrometric targeted immunoassays for clinically important proteins in human plasma and serum. Clin Biochem. 2013;46:399–410. doi: 10.1016/j.clinbiochem.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kim HK, Choi YH, Erkelens C, Lefeber AW, Verpoorte R. Metabolic fingerprinting of ephedra species using 1h-nmr spectroscopy and principal component analysis. Chem Pharm Bull (Tokyo) 2005;53:105–109. doi: 10.1248/cpb.53.105. [DOI] [PubMed] [Google Scholar]
- 187.Choi HK, Choi YH, Verberne M, Lefeber AW, Erkelens C, Verpoorte R. Metabolic fingerprinting of wild type and transgenic tobacco plants by 1h nmr and multivariate analysis technique. Phytochemistry. 2004;65:857–864. doi: 10.1016/j.phytochem.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 188.Johnson HE, Broadhurst D, Kell DB, Theodorou MK, Merry RJ, Griffith GW. High-throughput metabolic fingerprinting of legume silage fermentations via fourier transform infrared spectroscopy and chemometrics. Appl Environ Microbiol. 2004;70:1583–1592. doi: 10.1128/AEM.70.3.1583-1592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gidman E, Goodacre R, Emmett B, Smith AR, Gwynn-Jones D. Investigating plant-plant interference by metabolic fingerprinting. Phytochemistry. 2003;63:705–710. doi: 10.1016/s0031-9422(03)00288-7. [DOI] [PubMed] [Google Scholar]
- 190.Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 191.Serkova NJ, Van Rheen Z, Tobias M, Pitzer JE, Wilkinson JE, Stringer KA. Utility of magnetic resonance imaging and nuclear magnetic resonance-based metabolomics for quantification of inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295:L152–161. doi: 10.1152/ajplung.00515.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wishart DS. Quantitative metabolomics using nmr. TrAC Trends in Analytical Chemistry. 2008;27:228–237. [Google Scholar]
- 193.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 194.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological nmr spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 195.Wen T, Gao L, Wen Z, Wu C, Tan CS, Toh WZ, Ong CN. Exploratory investigation of plasma metabolomics in human lung adenocarcinoma. Mol Bio Syst. 2013;9:2370–2378. doi: 10.1039/c3mb70138g. [DOI] [PubMed] [Google Scholar]
- 196.Weckwerth W. Metabolomics in systems biology. Annu Rev Plant Biol. 2003;54:669–689. doi: 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- 197.Raamsdonk LM, Teusink B, Broadhurst D, Zhang NS, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, Oliver SG. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 198.Carraro S, Rezzi S, Reniero F, Heberger K, Giordano G, Zanconato S, Guillou C, Baraldi E. Metabolomics applied to exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med. 2007;175:986–990. doi: 10.1164/rccm.200606-769OC. [DOI] [PubMed] [Google Scholar]
- 199.de Laurentiis G, Paris D, Melck D, Maniscalco M, Marsico S, Corso G, Motta A, Sofia M. Metabonomic analysis of exhaled breath condensate in adults by nuclear magnetic resonance spectroscopy. Eur Respir J. 2008;32:1175–1183. doi: 10.1183/09031936.00072408. [DOI] [PubMed] [Google Scholar]
- 200.Naz S, Garcia A, Rusak M, Barbas C. Method development and validation for rat serum fingerprinting with ce-ms: Application to ventilator-induced-lung-injury study. Anal Bioanal Chem. 2013;405:4849–4858. doi: 10.1007/s00216-013-6882-5. [DOI] [PubMed] [Google Scholar]
- 201.Emwas A-H, Salek R, Griffin J, Merzaban J. Nmr-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics. 2013;9:1048–1072. [Google Scholar]
- 202.Carraro S, Giordano G, Reniero F, Carpi D, Stocchero M, Sterk PJ, Baraldi E. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy. 2013;68:110–117. doi: 10.1111/all.12063. [DOI] [PubMed] [Google Scholar]
- 203.Jung J, Kim SH, Lee HS, Choi GS, Jung YS, Ryu DH, Park HS, Hwang GS. Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clin Exp Allergy. 2013;43:425–433. doi: 10.1111/cea.12089. [DOI] [PubMed] [Google Scholar]
- 204.Ried JS, Baurecht H, Stuckler F, Krumsiek J, Gieger C, Heinrich J, Kabesch M, Prehn C, Peters A, Rodriguez E, Schulz H, Strauch K, Suhre K, Wang-Sattler R, Wichmann HE, Theis FJ, Illig T, Adamski J, Weidinger S. Integrative genetic and metabolite profiling analysis suggests altered phosphatidylcholine metabolism in asthma. Allergy. 2013;68:629–636. doi: 10.1111/all.12110. [DOI] [PubMed] [Google Scholar]
- 205.Chen J, Ma M, Shen N, Xi JJ, Tian W. Integration of cancer gene co-expression network and metabolic network to uncover potential cancer drug targets. J Proteome Res. 2013;12:2354–2364. doi: 10.1021/pr400162t. [DOI] [PubMed] [Google Scholar]
- 206.Mehrotra B, Mendes P. Bioinformatics approaches to integrate metabolomics and other systems biology data. In: Saito K, Dixon R, Willmitzer L, editors. Plant metabolomics. Vol. 57. Springer; Berlin Heidelberg: 2006. pp. 105–115. [Google Scholar]
- 207.Shulaev V. Metabolomics technology and bioinformatics. Brief Bioinformat. 2006;7:128–139. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- 208.Stein SE. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J Am Soc Mass Spectrom. 1999;10:770–781. [Google Scholar]
- 209.Pluskal T, Castillo S, Villar-Briones A, Oresic M. Mzmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Neuweger H, Albaum SP, Dondrup M, Persicke M, Watt T, Niehaus K, Stoye J, Goesmann A. Meltdb: A software platform for the analysis and integration of metabolomics experiment data. Bioinformatics. 2008;24:2726–2732. doi: 10.1093/bioinformatics/btn452. [DOI] [PubMed] [Google Scholar]
- 211.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. Metaboanalyst 2. 0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40:W127–133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. Xcms: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 213.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. Hmdb 3. 0--the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. Hmdb: The human metabolome database. Nucleic Acids Res. 2007;35:D521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Hmdb: A knowledge base for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hood L. Systems biology and p4 medicine: Past, present, and future. Rambam Maimonides Med J. 2013;4:e0012. doi: 10.5041/RMMJ.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]