Abstract
The gamma(1)34.5 gene of herpes simplex virus is dispensable in some cell lines (e.g., Vero). In others (e.g., human neuroblastoma cell line SK-N-SH), the gamma(1)34.5- deletion mutant triggers a premature total shutoff of all protein synthesis, thereby rendering the cell nonviable and reducing drastically viral yields. The inability to prevent the cellular stress response that causes the infected cell to die may be responsible for the inability of the deletion mutant to multiply and cause pathology in the central nervous system of mice. The gamma(1)34.5 gene consists of an amino-terminal domain, a variable linker sequence consisting of 3 amino acids repeated 5-10 times, and a carboxyl-terminal domain homologous to the corresponding domain of MyD116, a gene expressed in myeloid leukemia cells induced to differentiate by interleukin 6, and growth arrest and DNA damage gene 34 (GADD34), a gene induced by growth arrest and DNA damage. We have constructed several viral mutants from which various domains of the gamma(1)34.5 gene had been deleted or rendered mute by the insertion of a stop codon. Studies on those mutants show that the domain of the gamma(1)34.5 gene necessary to preclude the total shutoff of protein synthesis corresponds to the carboxyl-terminal domain of the gamma(1)34.5 gene homologous to the corresponding coding domain of the MyD116 and GADD34 genes.
Full text
PDF

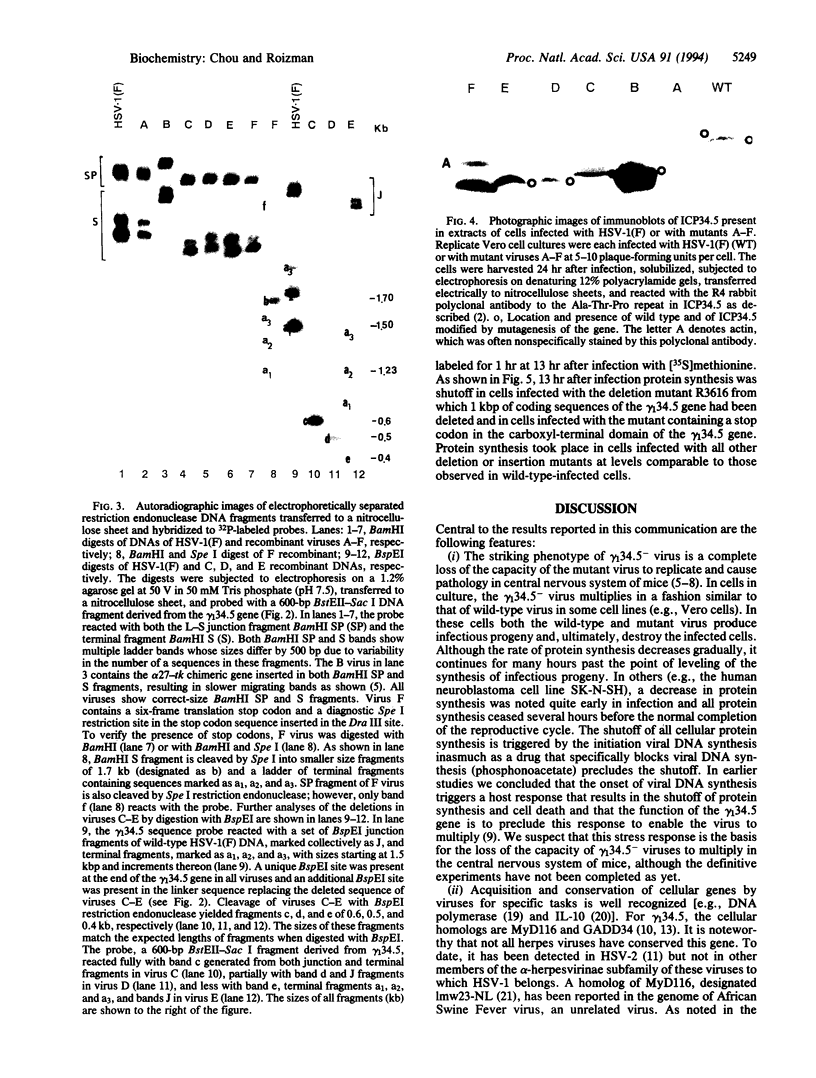
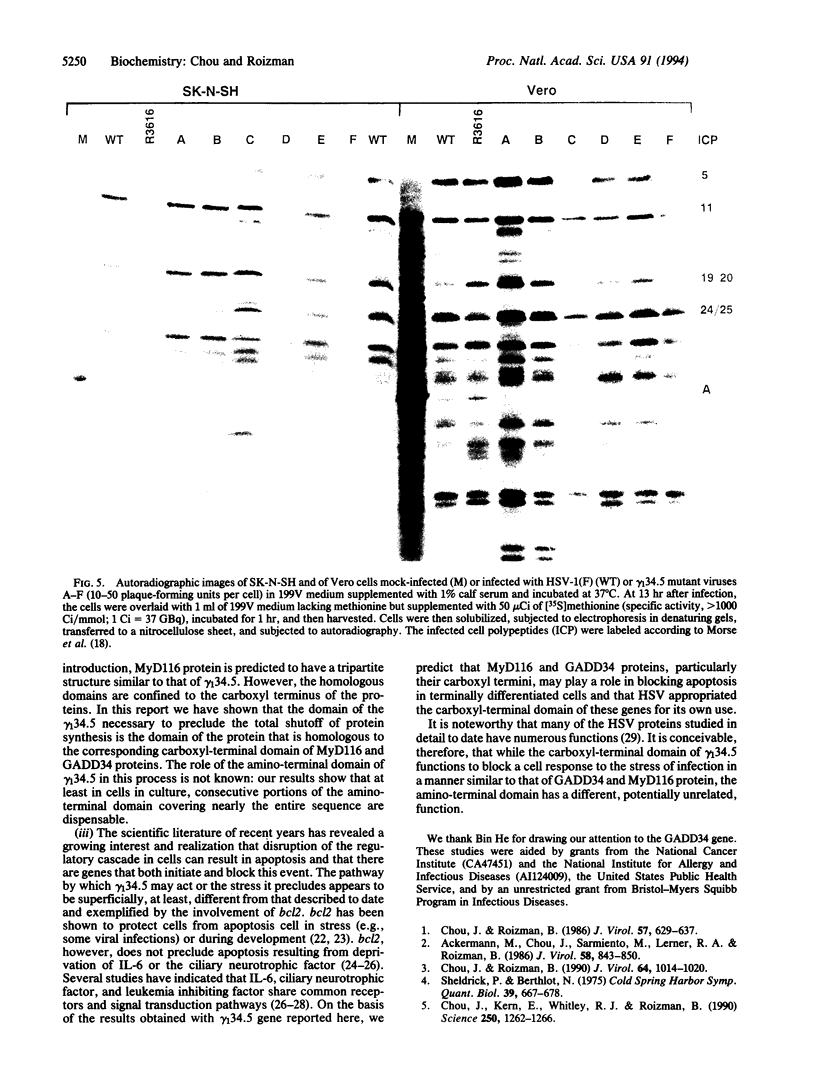

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann M., Chou J., Sarmiento M., Lerner R. A., Roizman B. Identification by antibody to a synthetic peptide of a protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J Virol. 1986 Jun;58(3):843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp T. E., Wyatt S., Paterson H. F., Davies A. M. The proto-oncogene bcl-2 can selectively rescue neurotrophic factor-dependent neurons from apoptosis. Cell. 1993 Apr 23;73(2):295–307. doi: 10.1016/0092-8674(93)90230-n. [DOI] [PubMed] [Google Scholar]
- Chou J., Kern E. R., Whitley R. J., Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990 Nov 30;250(4985):1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J Virol. 1990 Mar;64(3):1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol. 1986 Feb;57(2):629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Nebert D. W., Hollander M. C., Luethy J. D., Papathanasiou M., Fargnoli J., Holbrook N. J. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989 Oct;9(10):4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing D. P., Comeau M. R., Friend D. J., Gimpel S. D., Thut C. J., McGourty J., Brasher K. K., King J. A., Gillis S., Mosley B. The IL-6 signal transducer, gp130: an oncostatin M receptor and affinity converter for the LIF receptor. Science. 1992 Mar 13;255(5050):1434–1437. doi: 10.1126/science.1542794. [DOI] [PubMed] [Google Scholar]
- Ip N. Y., Nye S. H., Boulton T. G., Davis S., Taga T., Li Y., Birren S. J., Yasukawa K., Kishimoto T., Anderson D. J. CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell. 1992 Jun 26;69(7):1121–1132. doi: 10.1016/0092-8674(92)90634-o. [DOI] [PubMed] [Google Scholar]
- Javier R. T., Thompson R. L., Stevens J. G. Genetic and biological analyses of a herpes simplex virus intertypic recombinant reduced specifically for neurovirulence. J Virol. 1987 Jun;61(6):1978–1984. doi: 10.1128/jvi.61.6.1978-1984.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987 Jan;6(1):169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord K. A., Abdollahi A., Thomas S. M., DeMarco M., Brugge J. S., Hoffman-Liebermann B., Liebermann D. A. Leukemia inhibitory factor and interleukin-6 trigger the same immediate early response, including tyrosine phosphorylation, upon induction of myeloid leukemia differentiation. Mol Cell Biol. 1991 Sep;11(9):4371–4379. doi: 10.1128/mcb.11.9.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord K. A., Hoffman-Liebermann B., Liebermann D. A. Sequence of MyD116 cDNA: a novel myeloid differentiation primary response gene induced by IL6. Nucleic Acids Res. 1990 May 11;18(9):2823–2823. doi: 10.1093/nar/18.9.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Barnett B. C. Neurovirulence factor. Nature. 1991 Oct 17;353(6345):609–609. doi: 10.1038/353609b0. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Cunningham C., McIntyre G., Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J Gen Virol. 1991 Dec;72(Pt 12):3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- Moore K. W., Vieira P., Fiorentino D. F., Trounstine M. L., Khan T. A., Mosmann T. R. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. 1990 Jun 8;248(4960):1230–1234. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez G., London L., Hockenbery D., Alexander M., McKearn J. P., Korsmeyer S. J. Deregulated Bcl-2 gene expression selectively prolongs survival of growth factor-deprived hemopoietic cell lines. J Immunol. 1990 May 1;144(9):3602–3610. [PubMed] [Google Scholar]
- Post L. E., Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981 Jul;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Raff M. C., Barres B. A., Burne J. F., Coles H. S., Ishizaki Y., Jacobson M. D. Programmed cell death and the control of cell survival: lessons from the nervous system. Science. 1993 Oct 29;262(5134):695–700. doi: 10.1126/science.8235590. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sussman M. D., Lu Z., Kutish G., Afonso C. L., Roberts P., Rock D. L. Identification of an African swine fever virus gene with similarity to a myeloid differentiation primary response gene and a neurovirulence-associated gene of herpes simplex virus. J Virol. 1992 Sep;66(9):5586–5589. doi: 10.1128/jvi.66.9.5586-5589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha M. Y., Clements G. B., Brown S. M. The herpes simplex virus type 2 (HG52) variant JH2604 has a 1488 bp deletion which eliminates neurovirulence in mice. J Gen Virol. 1989 Nov;70(Pt 11):3073–3078. doi: 10.1099/0022-1317-70-11-3073. [DOI] [PubMed] [Google Scholar]
- Thompson R. L., Rogers S. K., Zerhusen M. A. Herpes simplex virus neurovirulence and productive infection of neural cells is associated with a function which maps between 0.82 and 0.832 map units on the HSV genome. Virology. 1989 Oct;172(2):435–450. doi: 10.1016/0042-6822(89)90186-4. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. T. Programmed cell death: apoptosis and oncogenesis. Cell. 1991 Jun 28;65(7):1097–1098. doi: 10.1016/0092-8674(91)90002-g. [DOI] [PubMed] [Google Scholar]