Abstract
Here we show that p38 mitogen activated protein kinase (p38 MAPK) phosphorylates spliced form of X-Box Binding Protein 1 (XBP1s) on Thr48 and Ser61 residues and greatly enhances nuclear migration of XBP1s. Mutation of Thr48 and Ser61 to alanine dramatically reduces nuclear translocation of XBP1s and activity. We also demonstrate that p38 MAPK activity is markedly reduced in the livers of obese mice and that activation of p38 MAPK by expression of constitutively active MAP Kinase Kinase 6 (MKK6Glu) greatly enhances nuclear translocation of XBP1s, reduces ER stress and establishes euglycemia in the severely obese and diabetic mice. Hence, our results define a crucial role for Thr48 and Ser61 phosphorylations of XBP1s in maintenance of glucose homeostasis in obesity and indicate that p38 MAPK activation in the livers of obese mice may provide a novel therapeutic approach for treatment of type 2 diabetes.
INTRODUCTION
The endoplasmic reticulum (ER) is responsible for the synthesis of secretory and membrane proteins, which acquire their lowest energy three-dimensional structures within this organelle. Perturbations of ER homeostasis can create a condition referred to as ER stress, and lead to activation of a group of complex signaling pathways, called the unfolded protein response (UPR)1–4. Two type-I transmembrane kinases (the PKR-like ER kinase - PERK and the inositol requiring enzyme 1 - IRE1), plus a specific type-II transmembrane protein (the activating transcription factor 6 - ATF6), have major roles in initiating UPR signaling1–4.
Of these three transmembrane proteins, IRE1 is most conserved evolutionarily. In addition to its kinase activity, IRE1 also has endoribonuclease activity5,6. X-Box Binding Protein 1 (XBP1) belongs to the CREB/ATF family of transcription factors and is a basic region–leucine zipper transcription factor7–10. IRE1 cleaves the full length XBP1 mRNA to initiate removal of a 26bp intron, converting the unspliced XBP1 protein (XBP1u) to the highly active spliced XBP1 (XBP1s), a transcription factor that functions as a master regulator of ER folding capacity1–4,11–14. Despite the critical importance of XBP1s in several different cellular processes, and despite its association with a number of diseases, little is known about how this transcription factor is regulated3,4,15–23.
We and others have previously shown that obesity leads to the development of ER stress and activates the UPR in the liver, adipose tissue and brain15–18,24,25, in turn contributing to the development of insulin resistance, type-2 diabetes and leptin resistance. We have also shown that nuclear translocation of XBP1s is severely reduced under conditions of obesity, due to loss of interactions between XBP1s and the p85 regulatory subunits26. These results indicate that obesity actually creates an XBP1-deficient condition in the liver. If XBP1s is reactivated in the liver by forced ectopic expression in severely obese and diabetic mice, blood glucose levels are reduced to euglycemia27.
p38 mitogen activated protein kinases (MAPKs) belong to a family of evolutionarily conserved serine/threonine MAPKs that link extracellular signals to the intracellular machinery that regulates a plethora of cellular processes28–31. Along with c-Jun amino terminal kinase (JNK), p38 MAPK is described as stress activated protein kinases (SAPKs). JNK and p38 MAPKs are both activated by environmental and genotoxic stress, and both have key roles in converting extracellular stimuli into a wide range of cellular responses28–31.
ER stress and UPR signaling also activate SAPKs, including JNK32 and p38 MAPK33; both signaling pathways are considered to be detrimental arms of UPR signaling, and both participate in the development of ER-stress-related pathologies34, especially under conditions when the cell can no longer cope with ER stress. The main aim of UPR signaling is to re-establish ER homeostasis. The ER is a dynamic organelle, with the capacity to respond rapidly to changes in the environment. For example UPR transcription factors such as ATF6 and XBP1 or the ATF4 are rapidly activated to re-establish equilibrium within the ER, under conditions that perturb the ER system. Of interest in the context of the current study, however, is that the time frame of activation of these “detrimental” arms is similar as for activation of the “protective” arms. Importantly, initiation of apoptosis occurs much later than this initial activation of UPR signaling parameters35. This temporal overlap between the activation of the SAPKs and that of the protective pathways, and occurrence of cellular pathology in a later time frame prompted us to investigate whether SAPK signaling also has positive effects on ER homeostasis, via regulation of UPR signaling elements.
RESULTS
SAPK signaling increases XBP1s activity
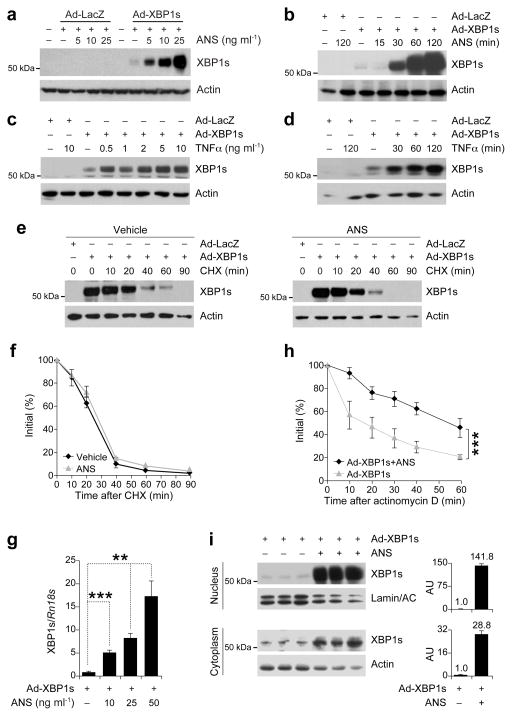
To investigate whether activation of JNK or p38 MAPK affects XBP1s activity, we expressed XBP1s in MEFs by infecting them with XBP1s-expressing adenovirus (Ad-XBP1s), or with LacZ-expressing adenovirus (Ad-LacZ; control). Subsequently, the cells were treated with increasing doses of anisomycin, an agent that activates JNK and p38 MAPK. Anisomycin treatment led to a robust increase in XBP1s protein levels (Fig. 1a), but only in cells infected with Ad-XBP1s. No upregulation of XBP1s proteins was noted in Ad-LacZ-infected control cells treated with up to 25 ng ml−1 anisomycin, showing that anisomycin by itself does not create ER stress nor does it induce XBP1 splicing (Fig. 1a). A time course experiment demonstrated that exposure of the XBP1s-expressing cells to anisomycin leads to upregulation of XBP1s protein levels within the first 30 min (Fig. 1b). To determine whether an alternative way of activating SAPKs would also increase XBP1s protein levels, we treated the XBP1s-expressing cells with increasing doses of TNFα: as with the anisomycin, TNFα stimulation markedly increased XBP1s protein levels (Fig. 1c); time course study revealed that TNFα also upregulates XBP1s levels within 30 min (Fig. 1d). And finally, undertaking the same experiment in Fao cells produced the same results (Supplementary Fig. 1a,b).
Figure 1.
SAPK signaling increases XBP1s mRNA stability and nuclear translocation. (a) XBP1s protein levels in the MEFs infected with adenoviruses expressing LacZ (Ad-LacZ) or XBP1s (Ad-XBP1s) and treated with anisomycin (ANS) at indicated concentrations for 2h. (b) XBP1s protein levels from MEFs infected with Ad-LacZ or Ad-XBP1s and treated with ANS (25 ng ml−1) for 15, 30, 60 and 120 min. (c–d) XBP1s immunoblotting in MEFs infected with Ad-LacZ or Ad-XBP1s and treated with TNFα (c) for 2 h at increasing concentrations or (d) at 10 ng ml−1 concentration for 30, 60 and 120 min. (e) XBP1s protein degradation rate in MEFs infected with Ad-LacZ or Ad-XBP1s, treated with vehicle or ANS (25 ng ml−1) for 2 h then subjected to cycloheximide (CHX) (10 μg ml−1) for indicated time points. XBP1s protein levels were determined by direct immunoblotting of the whole cell lysates. (f) XBP1s protein/actin ratio before and at indicated times after CHX treatment. (g) XBP1s mRNA levels after ANS stimulation. MEFs were infected with Ad-XBP1s and subsequently treated with ANS for 1 h at indicated concentrations. (h) XBP1s mRNA levels at indicated time points after addition of actinomycin D (10 μg ml−1) to the MEFs infected with Ad-XBP1s and stimulated with ANS (25 ng ml−1) for 1 h. (i) Cytoplasmic and nuclear XBP1s protein levels from MEFs infected with Ad-XBP1s and exposed to ANS (25 ng ml−1) for 2 h. Graph adjacent to each blot depicts the ratio of XBP1s in ANS- versus vehicle-treated cells. Each experiment was independently reproduced three times. Error bars are ±S.E.M. Significance was determined by two-way analysis of variance (ANOVA) with Bonferroni multiple-comparison analysis (Figures 1f and 1h) or Student’s t-test (Figure 1g) (**P<0.01, ***P<0.001).
We next investigated whether upregulation of XBP1s levels is due to an increase in the protein stability. Cells were infected with Ad-XBP1s, subsequently treated with vehicle or anisomycin, then further treated with cycloheximide to inhibit translation initiation; XBP1s levels were determined before and after addition of cycloheximide. To analyze the degradation rate of XBP1s in vehicle- or anisomycin-treated cells correctly, we specifically exposed blots from vehicle-treated cells longer and anisomycin-treated cells shorter to have same level of XBP1s signal at 0 time points of both groups. We observed no differences in the degradation rate of XBP1s protein in the vehicle- or anisomycin-treated cells (Fig. 1e,f). These results exclude the possibility that anisomycin increases XBP1s protein levels by increasing its stability.
However, we noted a dramatic increase in mRNA of XBP1s in the anisomycin-treated cells (Fig. 1g). To evaluate the possibility that anisomycin treatment increases the stability of XBP1s mRNA, we pretreated the XBP1s expressing cells with anisomycin and examined the degradation pattern of XBP1s mRNA after inhibition of transcription with actinomycin D. Anisomycin extended the half-life of the mRNA from around 15 min to 60 min (Fig. 1h). Furthermore, we also found that nuclear levels of XBP1s in anisomycin-treated cells were 141.8 times higher than in vehicle-treated cells, but cytoplasmic levels were only increased 28.8 times (Fig. 1i). Under normal conditions, the fold increase in XBP1s should be similar in the cytoplasmic and nuclear compartments. The five-fold discrepancy in these values prompted us to consider that perhaps the anisomycin increases the efficiency of nuclear translocation of XBP1s. When taken together, the results presented to this point suggest that SAPK signaling increases the mRNA stability of XBP1s and leads to its nuclear translocation with a higher efficiency.
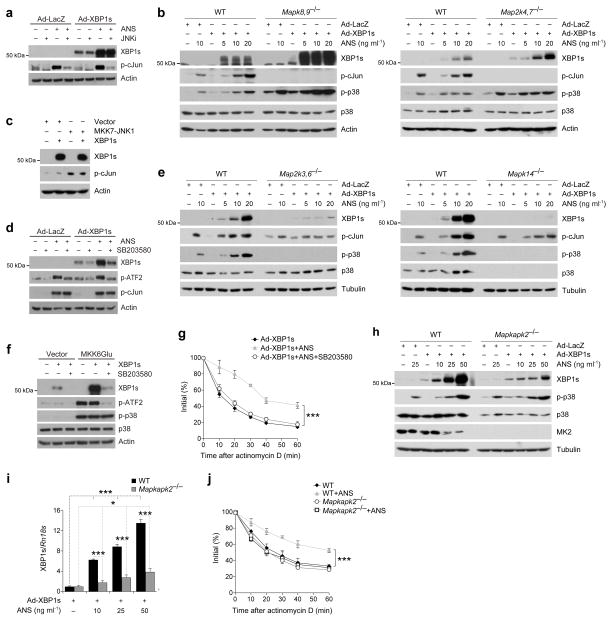
To unravel the molecular mechanisms underlying the above observations, we took several different approaches to explore whether activation of JNK, which is one of the most dominant signaling elements in SAPK signaling, affects XBP1s levels and nuclear translocation. First, Ad-LacZ- and Ad-XBP1s-infected cells were treated with a JNK inhibitor and then exposed to anisomycin. The JNK inhibitor was effective in completely abolishing anisomycin-induced JNK activation (Fig. 2a), but had no effect on anisomycin-mediated upregulation of XBP1s protein levels (Fig. 2a). As the second approach we used JNK1,2-deficient cells (Mapk8,9−/−) and showed that stimulation of Mapk8,9−/− cells with anisomycin also led to a robust increase in XBP1s protein (Fig. 2b, left), which was markedly higher than that in anisomycin-treated wild-type (WT) cells (Fig. 2b, left). While no JNK activity was detectable in the Mapk8,9−/− cells that were treated with anisomycin, we did note a marked increase in p38 MAPK phosphorylation (Fig. 2b, left). It is known that MAP kinase kinase (MKK) 4 and 7 are upstream kinases responsible for JNK activation36,37 and anisomycin or TNFα cannot activate JNK in MKK4 and 7 double knockout cells (Map2k4,7−/−)36,38. We thus assessed whether XBP1s can be regulated by anisomycin treatment in Map2k4,7−/− cells; these cells, along with their WT cells, were treated with anisomycin after XBP1s expression. We found that, as for the Mapk8,9−/− cells, anisomycin stimulation dramatically increased XBP1s levels in Map2k4,7−/− cells, to a level that was higher than that in WT cells (Fig. 2b, right). JNK activity was not detectable in the Map2k4,7−/− cells, whereas p38 MAPK activation was markedly increased (Fig. 2b, right). Furthermore, we repeated the experiments in Mapk8,9−/− and Map2k4,7−/− cells with TNFα: as with anisomycin, treatment of Mapk8,9−/− and Map2k4,7−/− cells with TNFα also led to a robust increase in XBP1s levels (Supplementary Fig. 2a,b). Finally, we utilized an MKK7-JNK1 fusion protein, previously shown to be a specific activator of JNK pathway39, and investigate whether activation of JNK alone can have effect on XBP1s. MKK7-JNK1 expression in the cells dramatically increased JNK activity (Fig. 2c). However, co-expression of MKK7-JNK1 with XBP1s did not increase protein levels of XBP1s (Fig. 2c), indicating that the process is independent of JNK activation.
Figure 2.
p38 MAPK increases mRNA stability of XBP1s through activation of MK2. (a) XBP1s, phospho-c-Jun (p-cJun) and actin levels from Ad-LacZ- or Ad-XBP1s-infected MEFs that were pre-treated with JNK inhibitor VIII (JNKi, 10 μM) for 30 min and subsequently subjected to ANS (25 ng ml−1) for 2 h. (b) Ad-LacZ- or Ad-XBP1s-infected (left) wild-type (WT) and JNK1,2-deficient (Mapk8,9−/−) or (right) WT and MKK4,7-deficient (Map2k4,7−/−) MEF cells were treated with ANS at indicated concentrations for 2 h. Whole cell lysates were immunoblotted with XBP1s, p-cJun, phospho-p38 MAPK (p-p38), p38 MAPK and actin antibodies. (c) HEK293 cells were transfected with XBP1s or co-transfected with XBP1s and MKK7-JNK1 plasmids (Empty vector was used for normalization of DNA transfection amount). XBP1s, p-cJun and actin levels 24 h after the transfections. (d) Ad-LacZ- or Ad-XBP1s-infected MEFs were pretreated with SB203580 (10 μM), a specific p38 MAPK inhibitor for 30 min and then subjected to ANS (25 ng ml−1) treatment for 2 h. XBP1s, phospho-ATF2 (p-ATF2), p-cJun and actin levels were determined in whole cell lysates. (e) Ad-LacZ- or Ad-XBP1s-infected (left) WT and MKK3,6-deficient (Map2k3,6−/−) or (right) WT and p38α-deficient (Mapk14−/−) cells were treated with ANS at indicated concentrations for 2 h. (f) HEK293 cells were transfected with XBP1s or MKK6Glu, or co-transfected with MKK6Glu and XBP1s together (Empty vector was used for normalization of DNA transfection amount). Transfected cells were either treated with SB203580 (10 μM) or vehicle for 24 h. (g) XBP1s mRNA levels from Ad-XBP1s-infected MEFs that are pretreated with SB203580 (10 μM) for 30 min, stimulated with ANS (25 ng ml−1) for additional 1 h and subjected to actinomycin D (10 μM) for indicated time points. (h) WT and MK2-deficient (Mapkapk2−/−) cells were infected with Ad-LacZ or Ad-XBP1s and subsequently treated with ANS for 2 h at indicated concentrations. (i) mRNA levels of XBP1s from WT and Mapkapk2−/− cells infected with Ad-XBP1s and treated with ANS at different concentrations for 1 h. (j) XBP1s mRNA levels from Ad-XBP1s-infected WT and Mapkapk2−/− MEFs stimulated either with vehicle or ANS (25 ng ml−1) for 1 h and then treated with actinomycin D (10 μg ml−1) for indicated time points. Each experiment was independently performed three times. Error bars are ±S.E.M. Significance was determined by two-way ANOVA (figure 2g), three-way ANOVA (figure 2j) with Bonferroni multiple-comparison analysis, or Student’s t-test (figure 2i) (*P<0.05, ***P<0.001).
p38 MAPK is another critical SAPK signaling element that is activated during ER stress conditions; also, in the absence of JNK activation, higher levels of p38 MAPK activation were noted (Fig. 2b and Supplementary Fig. 2a,b). We thus tested the possibility that p38 MAPK activation mediates the effects of anisomycin and TNFα on XBP1s. Ad-XBP1s-infected cells were treated with a specific p38 MAPK inhibitor (SB203580), then stimulated with anisomycin. The dramatic increase in the XBP1s levels was completely abolished by pretreatment of the cells with a p38 MAPK inhibitor (Fig. 2d). These findings suggest that anisomycin-induced upregulation of XBP1s levels is mediated via the p38 MAPK pathway.
MKK3 and 6 are required for p38 MAPK activation, and in fact, p38 MAPK cannot be activated in MKK3,6-deficient (Map2k3,6−/−) cells40. In light of this, we reasoned that anisomycin should not induce an increase in XBP1s levels in Map2k3,6−/− cells – and indeed, no increase in XBP1s levels was noted when Ad-XBP1s-infected Map2k3,6−/− cells were treated with anisomycin, relative to levels in anisomycin-stimulated WT cells (Fig. 2e, left): and, as previously reported40, p38 MAPK activation was dramatically reduced in Map2k3,6−/− cells (Fig. 2e, left). We also showed that Ad-XBP1s-infected p38α-deficient (Mapk14−/−) cells lack the ability to upregulate XBP1s levels in response to anisomycin stimulation (Fig. 2e, right).
A constitutively active form of MKK6 (MKK6Glu), one of the upstream kinases of p38 MAPK, was previously generated by mutation of Ser207 and Ser211 to glutamate. Expression of MKK6Glu specifically activates p38 MAPK41. We used MKK6Glu to assess whether activation of p38 MAPK alone is sufficient to increase XBP1s protein levels. Cells were first transfected with MKK6Glu and/or XBP1s at the presence or absence of p38 MAPK inhibitor. Expression of MKK6Glu markedly increased XBP1s protein levels and inhibition of p38 MAPK activity blocked the effect of MKK6Glu on XBP1s (Fig. 2f).
To investigate whether the increased stability of XBP1s mRNA was indeed mediated via the p38 MAPK pathway, we pretreated cells first with a p38 MAPK inhibitor, then treated them with anisomycin, and finally incubated with actinomycin D. Inhibition of p38 MAPK completely blocked the prolongation of the half-life of XBP1s mRNA induced by anisomycin stimulation (Fig. 2g).
p38 MAPK signaling has also been implicated in the regulation of stability of several different mRNAs42. One of the main molecules that links p38 MAPK activation to regulation of mRNA stability is MAPK-activated protein kinase 2 (MK2)42. MK2 was previously shown to regulate activation of several RNA-binding proteins that are involved in stabilization of mRNA. These include tristetraprolin (TTP), human antigen R (HuR) or ARE/poly(U)-binding degradation factor (AUF1). In addition p38 MAPK can also regulate RNA stability by directly phosphorylating KH-domain splicing regulatory protein (KSRP)42.
To investigate that the p38 MAPK-mediated increase in mRNA stability of XBP1s is due to MK2 activation, we stimulated Ad-XBP1s-infected WT and MK2-deficient (Mapkapk2−/−) cells with anisomycin: XBP1s protein levels were dramatically reduced in Mapkapk2−/− cells, indicating that increased mRNA stability is primarily mediated via MK2 activation (Fig. 2h). We next examined the mRNA levels of XBP1s in WT and Mapkapk2−/− cells that were infected with Ad-XBP1s and stimulated with anisomycin. We found that anisomycin increases the mRNA levels of XBP1s in a dose dependent manner in WT cells (Fig. 2i), but the increase is significantly lower (P<0.001) in the Mapkapk2−/− cells (Fig. 2i), supporting a dominant role for MK2 in p38 MAPK-mediated upregulation of XBP1s mRNA stability. Furthermore, an analysis of degradation rate of XBP1s mRNA in WT and Mapkapk2−/− cells that were either treated with vehicle or anisomycin documented that there were no differences in the degradation rate of XBP1s mRNA between vehicle-treated WT and Mapkapk2−/− and anisomycin-treated Mapkapk2−/− cells (Fig. 2j).
To investigate whether TTP might have a role in stabilizing XBP1s mRNA, Ad-XBP1s-infected TTP knockout (Zfp36−/−) cells were treated with anisomycin, then analyzed for mRNA degradation after adding actinomycin D. Results show that anisomycin is capable of increasing XBP1s mRNA stability even in Zfp36−/− cells (Supplementary Fig. 3a,b).
To investigate the role of AUF1 and KSRP in stabilizing XBP1s mRNA, we chose the most efficient shRNA for AUF1 and KSRP (Supplementary Fig. 3c,e) and showed that depletion of AUF1 or KSRP does not affect anisomycin-induced upregulation of XBP1s levels (Supplementary Fig. 3d,f). And finally, we sought to investigate the role of HuR in this process, but we could not detect expression of HuR in MEF cells (data not shown).
Despite these major efforts, we were unable to identify the RNA binding protein that is regulated by MK2 and is responsible for increasing the stability of XBP1s mRNA, leading us to conclude that another RNA binding protein, which is an (as yet) unknown target of MK2, should be responsible for stabilization of XBP1s mRNA and further investigation is needed for determination of this factor.
p38 MAPK phosphorylates XBP1s on Thr48 and Ser61
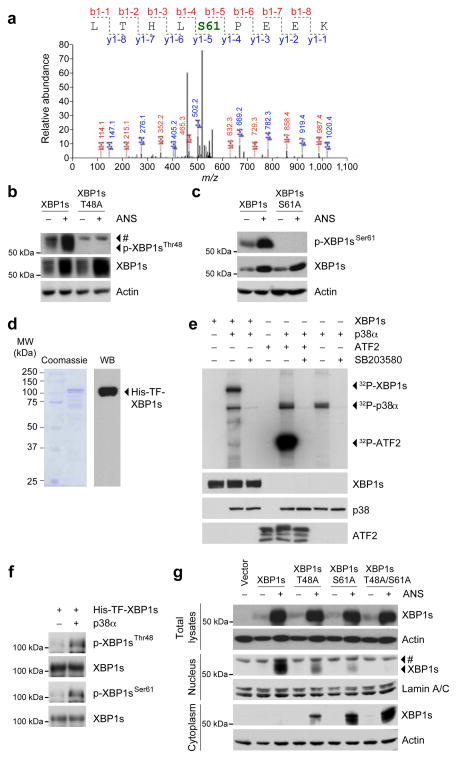
The dramatic increase in the efficiency of migration of XBP1s to the nucleus when p38 MAPK is activated was a surprising result (Fig. 1i), and prompted the hypothesis that p38 MAPK directly phosphorylates XBP1s and thus triggers this enhanced translocation. To test this hypothesis, we expressed a flag-tagged-XBP1s in cells and stimulated them with anisomycin. MS/MS analysis demonstrated that anisomycin stimulation increases phosphorylation of XBP1s at Ser61, which is a conserved phosphorylation site (LSPE) for p38 MAPK (Fig. 3a). One other phosphorylation site (Thr48) on XBP1s is also a conserved p38 MAPK phosphorylation site, although it was not detected with the MS/MS analysis. To convincingly demonstrate that Ser61 is indeed phosphorylated after anisomycin stimulation or via activation of p38 MAPK, and also to test whether Thr48 is also a phosphorylation site, we developed a phospho-specific antibody against phosphorylated XBP1s on Ser61 or on Thr48. We found that, in fact, stimulation of XBP1s-expressing cells with anisomycin greatly increased phosphorylation of the Thr48 residue (Fig. 3b). To unravel the role of this phosphorylation site we mutated the Thr48 residue to alanine (T48A), and showed that this completely eliminated the anisomycin-induced Thr48 phosphorylation of XBP1s, but did not affect the upregulation in XBP1s protein levels (Fig. 3b). Similar results were obtained from XBP1s Ser61 phosphorylation: anisomycin stimulation strongly induced phosphorylation of XBP1s at Ser61, but mutation of Ser61 did not alter the anisomycin-induced upregulation of XBP1s protein levels (Fig. 3c).
Figure 3.
p38 MAPK phosphorylates XBP1s at Thr48 and Ser61. (a) MS/MS analysis of XBP1s after 2-h anisomycin (25 ng ml−1) stimulation (see online methods for experimental details). (b–c) HEK293 cells were transfected with WT or mutant XBP1s; (b) XBP1s-T48A or (c) XBP1s-S61A. Subsequently, cells were stimulated with ANS (25 ng ml−1) for 2 h. p-XBP1sThr48, p-XBP1sSer61, XBP1s and actin levels were determined via direct immunoblotting. (d) Coomassie blue staining (left panel) and western blotting (right panel) for recombinant His-TF-XBP1s protein. (e) In vitro kinase assay of p38 MAPK for XBP1s and ATF2 at the presence or absence of SB203580 (10 μM). Total XBP1s, p38 and ATF2 protein levels were detected by western blotting, and phospho-XBP1s, phospho-p38 and phospho-ATF2 were detected by autoradiography. (f) p-XBP1sThr48, p-XBP1sSer61 and XBP1s protein levels on His-TF-XBP1s subjected to p38 MAPK in vitro kinase assay. (g) XBP1s protein levels in whole cell, cytoplasmic and nuclear extracts of HEK293 cells that were transfected with indicated XBP1s plasmids and further stimulated with vehicle or ANS (25 ng ml−1) for 2 h. Each experiment was independently reproduced three times. #: non-specific band.
To determine whether p38 MAPK can directly phosphorylate XBP1s, we produced a XBP1s fusion protein (Fig. 3d) and performed an in vitro kinase assay with an already activated recombinant p38 MAPK protein. Incubation of XBP1s with activated recombinant p38 MAPK markedly increased phosphorylation of XBP1s (Fig. 3e), however, addition of a p38 MAPK inhibitor to the kinase assay buffer completely reversed the effect of activated p38 MAPK on XBP1s. As a control, the p38 MAPK kinase assay was also done with ATF2 (Fig. 3e). In addition, an in vitro kinase assay was aimed at understanding whether Thr48 and Ser61 are indeed directly phosphorylated by p38 MAPK: direct immunoblotting with use of the phospho-specific antibodies for p-XBP1sThr48 and p-XBP1sSer61 demonstrated that p38 MAPK directly phosphorylates XBP1s on Thr48 and Ser61 residues in the in vitro kinase assay (Fig. 3f). To understand the role of these phosphorylations, and to exclude a possible overlapping function in regulation of XBP1s activity and nuclear translocation, we created mutant XBP1s at Thr48 and/or Ser61 residues by converting to alanine. We then investigated the contribution of each phosphorylation site on the nuclear translocation of XBP1s. WT, T48A, S61A and T48A/S61A mutant XBP1s were expressed in the cells, anisomycin was added, and total levels of cytoplasmic and nuclear XBP1s proteins were determined. Mutation of Thr48, of Ser61, or of both sites together, did not affect the anisomycin-stimulated upregulation of XBP1s protein levels (Fig. 3g). However, mutation of the Thr48 residue decreased XBP1s nuclear translocation by about 49%, mutation of the Ser61 residue by about 79%, while mutation of both sites dramatically decreased nuclear translocation of XBP1s (Fig. 3g). In the mean time, mutation of Thr48 and Ser61 led to accumulation of XBP1s in the cytoplasm indicating that the double mutant XBP1s, despite the dramatic increase in the protein levels can not migrate to the nucleus. These findings indicate that Thr48 and Ser61 phosphorylation is crucial for nuclear translocation of XBP1s (Fig. 3g).
p38 MAPK is crucial for XBP1s activity during refeeding
We have previously shown that XBP1s is generated in the liver when the animal is refed after a fasting period26. To create a metabolic overloading model in cells, following a 16-h starvation in medium without fetal bovine serum (FBS), cells were reincubated with medium containing 10% FBS. There was a marked increase in XBP1s splicing and nuclear translocation, as well as in activation of p38 MAPK after restimulation with 10% FBS (Supplementary Fig. 4a). However, inhibition of p38 MAPK activity by pretreatment of the starved cells with p38 MAPK inhibitor before addition of 10% FBS strongly reduced XBP1s nuclear translocation without altering XBP1s splicing, or without affecting the total amount of XBP1s that is produced in the cells (Supplementary Fig. 4a). We then re-did the same experiment, but in Mapk14−/− and Map2k3,6−/− cells: FBS-induced XBP1s nuclear translocation is dramatically reduced in Mapk14−/− and Map2k3,6−/− cells compared to WT cells, despite the fact that XBP1s splicing and protein levels in both cell lines were similar to that in their WT cells (Supplementary Fig. 4b,c).
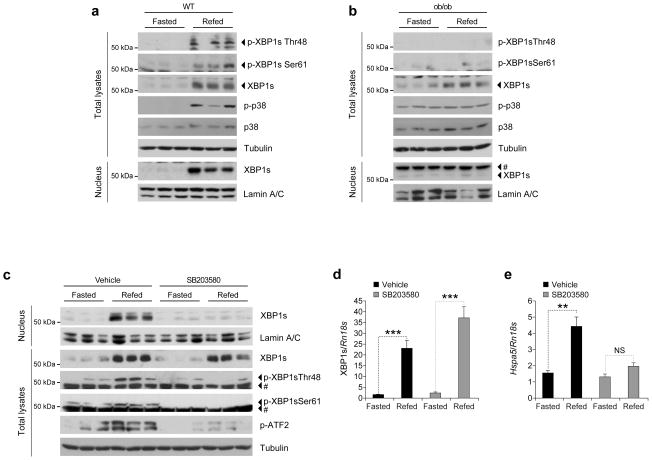
The next step was to determine whether refeeding after a fasting period induces p38 MAPK activation and XBP1s phosphorylation in the liver of lean WT mice. We fasted WT lean mice for 24 h and refed for 1 h: p38 MAPK phosphorylation was greatly increased in the liver following refeeding, and XBP1s total proteins were upregulated (Fig. 4a); also, phosphorylation of XBP1s at Thr48 and Ser61 was markedly increased after refeeding, indicating that XBP1s is phosphorylated at these sites during the refeeding process (Fig. 4a).
Figure 4.
Inhibition of p38 MAPK blocks XBP1s nuclear translocation. (a–b) (a) Eight-week-old WT lean male mice or (b) age-matched ob/ob males were fasted for 24 h and refed for 1 h. p-XBP1sThr48, p-XBP1sSer61, XBP1s, p-p38, p38 and tubulin levels in total liver extracts. Liver nuclear extracts were used to investigate XBP1s and lamin A/C protein levels. (c–e) Eight-week-old WT lean male mice were intraperitoneally injected with SB203580 (2 mg kg−1 per day) for 3 d. Subsequently, mice were fasted for 24 h and refed for 1 h. (c) Total lysates, nuclear and cytoplasmic protein fractions were prepared from the liver and indicated parameters were analyzed via western blotting. (d–e) (d) XBP1s and (e) Hspa5 mRNA levels at fasting and refed conditions in the livers of the mice either treated with vehicle or SB203580. Each experiment was independently reproduced three times. Error bars are ±S.E.M. Significance was determined by Student’s t-test (**P<0.01, ***P<0.001). NS: Non-significant.
We also wanted to know how p38 MAPK signaling is regulated in obese mice, since we have previously shown that XBP1s cannot migrate to the nucleus in the liver of obese mice during refeeding26. We found that refeeding of ob/ob mice after the fasting period does not increase p38 MAPK phosphorylation (Fig. 4b). XBP1s levels in total lysates are increased, but there is no detectable phosphorylations of Thr48 and Ser61 in XBP1s, and no detectable XBP1s protein in the nucleus (Fig. 4b).
To examine whether inhibition of p38 MAPK signaling alone blocks XBP1s nuclear translocation in the mouse, we treated 8-week-old WT lean male mice for 3 d either with vehicle or with the p38 MAPK inhibitor, after which the mice were starved for 24 h and refed for 1 h. XBP1s nuclear levels greatly increased after refeeding in vehicle-treated control group. However, XBP1s nuclear translocation in the liver of SB203580-treated mice after refeeding was dramatically reduced (Fig. 4c). Analysis of ATF2 phosphorylation revealed a marked reduction, indicating that SB203580 successfully blocked activation of p38 MAPK (Fig. 4c). XBP1s mRNA levels in the liver of refed mice that were either treated with vehicle or with the inhibitor were significantly increased in both groups (P<0.001) (Fig. 4d). Further analysis of mRNA levels of Hspa5 (encoding GRP78) revealed that Hspa5 expression was significantly increased in the vehicle-treated (P<0.01), but not in the inhibitor-treated group (Fig. 4e).
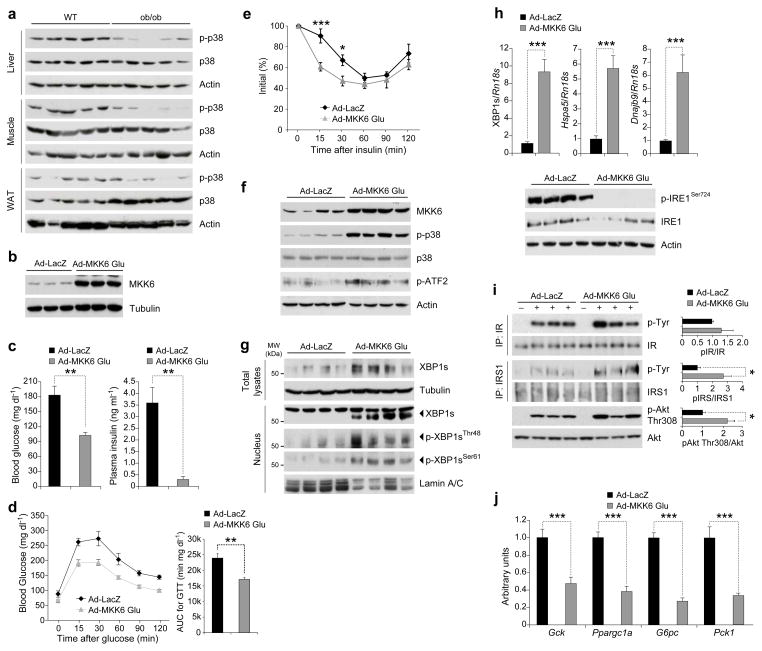
In addition to comparing fast and refeeding states, we also investigated the basal p38 MAPK activity in the liver, muscle and white adipose tissues of lean and obese mice. We found that p38 MAPK phosphorylation was markedly reduced in the liver, muscle and adipose tissues of ob/ob and high fat diet-fed obese mice when compared to their lean counterparts (Fig. 5a and Supplementary Fig. 5). Previous observations also indicated that p38 MAPK phosphorylation was reduced in the liver tumors of the mice that were obese43.
Figure 5.
Reactivation of p38 MAPK in the liver of ob/ob mice greatly enhances XBP1s nuclear translocation. (a) p-p38, p38 and actin levels in the liver, muscle and adipose tissues of eight-week-old WT and ob/ob mice. (b) MKK6 and tubulin protein levels in MEFs infected with Ad-LacZ or Ad-MKK6Glu. (c–j) Eight-week-old male ob/ob mice were infected with 8 x 106 plaque-forming units (PFU) g−1 of Ad-LacZ (n=5) or Ad-MKK6Glu (n=5) via tail vein injection. (c) (left) Six-hour fasted blood glucose (mg dl−1) levels on day 3 and (right) circulating insulin levels at six-hour fasted state on day 7 after virus injections. (d–e) (d) Glucose tolerance test (GTT) on day 5 and (e) insulin tolerance test (ITT) on day 3 after virus injections. (f–j) On post-injection day 7, mice were sacrificed after 6-h fasting and liver tissues were collected. (f) MKK6, p-p38, p38, p-ATF2 and actin levels, (g) total XBP1s and tubulin as well as nuclear XBP1s, p-XBP1sThr48, p-XBP1sSer61 and lamin A/C levels. (h) (top) mRNA levels of XBP1s, Hspa5 and Dnajb9, (bottom) p-IRE1Ser724, total IRE1 and actin protein levels. (i) IR and IRS1 tyrosine and Akt Thr308 phosphorylations together with their total protein levels after insulin infusion (0.75 IU kg−1) through the portal vein on post-injection day 7. Graphs next to the blots depict the ratio between phospho- and total-protein. (j) mRNA levels of Gck, G6pc, Pck1 and Ppargc1a in the livers. Three independent groups of mice (n=15 total in each group) were used in the experiments. Error bars are ±S.E.M. Significance was determined by two-way ANOVA with Bonferroni multiple-comparison analysis (figure 5e) or Student’s t-test (*P<0.05, **P<0.01, ***P<0.001). AUC: Area under a curve.
Activation of p38 MAPK in the liver regulates blood glucose
To determine whether reactivation of p38 MAPK in the liver of obese and diabetic mice would increase XBP1s phosphorylation, reduce ER stress, increase glucose tolerance and reduce blood glucose levels, we produced MKK6Glu-expressing adenovirus (Ad-MKK6Glu) (Fig. 5b). Next, eight-week-old ob/ob mice were injected through the tail vein with Ad-MKK6Glu or Ad-LacZ. Six-hour fasting blood glucose levels were significantly reduced (P<0.01) in the Ad-MKK6Glu-injected group compared with the Ad-LacZ-injected control mice (Fig. 5c, left). A significant reduction (P<0.01) was noted in the circulating insulin levels of the MKK6Glu overexpressing ob/ob mice, indicating that insulin sensitivity is increased (Fig. 5c, right). Moreover, as documented by glucose tolerance test (GTT), the glucose disposal rate in the Ad-MKK6Glu-injected group was markedly enhanced when compared with the Ad-LacZ-injected group (Fig. 5d). Performance of insulin tolerance test (ITT) also revealed increased insulin sensitivity in the Ad-MKK6Glu-injected group (Fig. 5e).
Furthermore, analysis of liver samples from Ad-LacZ- and Ad-MKK6Glu-injected ob/ob mice documented that p38 MAPK phosphorylation and downstream signaling were dramatically increased in the MKK6Glu expressing ob/ob livers (Fig. 5f). XBP1s total protein levels, nuclear protein levels, and Thr48 and Ser61 phosphorylations were markedly upregulated when MKK6Glu was expressed in the liver of ob/ob mice (Fig. 5g). mRNA levels of XBP1s, and expression of XBP1s-target genes, Hspa5 and Dnajb9 (ERdj4), were also significantly elevated (P<0.001), indicating increased XBP1s mRNA stability and activity (Fig. 5h, top). We next analyzed IRE1α phosphorylation levels as indicative of ER stress, and demonstrated that IRE1α phosphorylation was dramatically reduced in MKK6Glu-expressing ob/ob livers (Fig. 5h, bottom).
To investigate the insulin sensitivity, we infused insulin into the livers of the Ad-LacZ- and Ad-MKK6Glu-injected ob/ob mice and analyzed the activation of insulin receptor (IR), insulin receptor substrate 1 (IRS1) and Akt. No differences were seen between the two groups in terms of insulin-stimulated tyrosine phosphorylation of IR but there were significant increases in IRS1 tyrosine and Akt Thr308 phosphorylations (P<0.05) (Fig. 5i). Furthermore, expression levels of genes such as glucokinase (Gck), glucose-6-phosphatase (G6pc), phosphoenolpyruvate carboxykinase 1 (Pck1) and peroxisome proliferator-activated receptor-γ coactivator 1-α (Ppargc1a), which are involved in the glucose homeostasis, revealed a significant down regulation (P<0.001) when MKK6Glu was expressed in the livers of the ob/ob mice (Fig. 5j).
We also determined the extent of MKK6Glu expression in other tissues such as muscle and white adipose tissue (WAT) after tail vein injection of the adenoviruses. In addition, LacZ expression in the liver by tail vein injection of Ad-LacZ did not affect the blood glucose levels of ob/ob mice. Furthermore, tail vein injection of either Ad-LacZ or Ad-MKK6Glu did not alter the bodyweight and also did not lead to increase in liver function tests such as AST and ALT (Supplementary Fig. 6a–d)
To determine the role of XBP1s in MKK6Glu-mediated enhancement of glucose homeostasis we overexpressed MKK6Glu in the liver of 16-week high fat diet (HFD)-fed Xbp1flox/flox mice, while simultaneously depleting XBP1 with use of an adenovirus that expresses Cre recombinase (Ad-Cre). The first group of mice were injected with Ad-LacZ, second group with Ad-MKK6Glu plus Ad-LacZ and the third group with Ad-MKK6Glu and Ad-Cre. In parallel with the results obtained from the experiments on the ob/ob mice, injection of Ad-MKK6Glu into the Xbp1flox/flox mice led to a significant decrease in blood glucose levels (P<0.05) (Supplementary Fig. 7a). However, injection of Ad-Cre together with Ad-MKK6Glu completely eliminated the effect of MKK6Glu on blood glucose levels, indicating that XBP1s plays a major role in the improved glucose homeostasis mediated by MKK6Glu expression in the liver (Supplementary Fig. 7a). Next we performed a GTT; glucose disposal from circulation was markedly enhanced in the MKK6Glu-expressing Xbp1flox/flox mice, while depletion of XBP1 in the MKK6Glu-expressing mice reduced this effect (Supplementary Fig. 7b). We also analyzed p38 MAPK activation and MKK6 levels and documented that p38 MAPK was greatly activated in all the MKK6Glu expressing groups (Supplementary Fig. 7c). XBP1s nuclear translocation was markedly increased in the MKK6Glu expressing group, but completely eliminated in the MKK6Glu plus Cre recombinase-expressing group (Supplementary Fig. 7c). Furthermore, analysis of IRE1 phosphorylation revealed a significant down regulation (P<0.05) in the MKK6Glu expressing group, and this downregulation was abolished in the MKK6Glu plus Cre recombinase expressing group (Supplementary Fig. 7d). Next, we determined the expression of chaperones: MKK6Glu expression led to a significant upregulation in the expression of Hspa5 (P<0.001) and Dnajb9 (P<0.05) but this effect was diminished in the XBP1s-depleted group (Supplementary Fig. 7e). Finally, analysis of G6pc, Pck1 and Ppargc1a mRNA levels revealed that MKK6-mediated down regulation of these genes were lost in the XBP1s-depleted mice (Supplementary Fig. 7f).
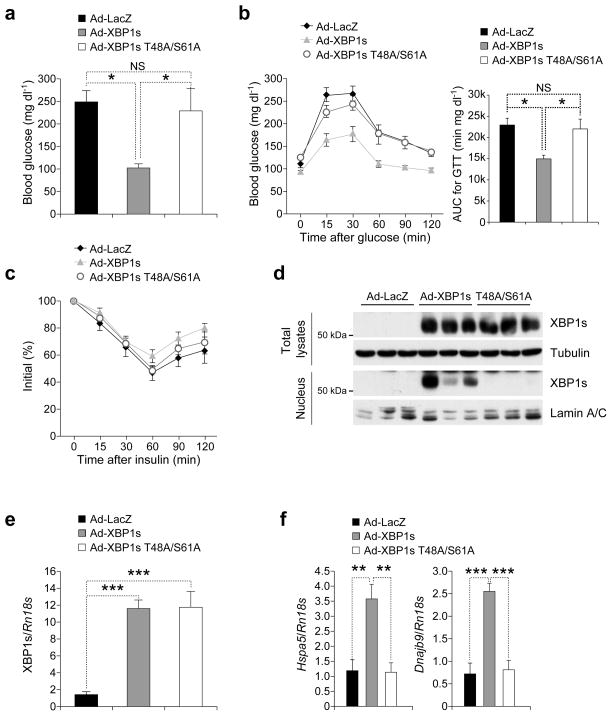
Based on the results that we have obtained to this point, the T48A/S61A double mutant XBP1s, when expressed in the liver of obese mice, should not translocate to the nucleus and should not have any effect on glucose homeostasis. To investigate whether this postulate is true, eight-week-old male ob/ob mice were injected either with Ad-LacZ or Ad-XBP1s or Ad-XBP1s T48A/S61A. Analysis of blood glucose levels showed that blood glucose of Ad-XBP1s-injected ob/ob mice were at euglycemia levels (Fig. 6a). However, expression of T48A/S61A mutant XBP1s had no effect on blood glucose levels (Fig. 6a). Performance of GTT revealed that mutation of Thr48 and Ser61 diminished the ability of XBP1s to enhance glucose tolerance (Fig. 6b). In parallel with our recent observations27 medium levels of XBP1s expression in the liver did not alter insulin tolerance (Fig. 6c).
Figure 6.
XBP1s-T48A/S61A cannot migrate to the nucleus in the liver and regulate glucose homeostasis. (a–f) Eight-week-old male ob/ob mice were injected with Ad-LacZ (n=5) or Ad-XBP1s (n=5) or Ad-XBP1s-T48A/S61A (n=5) (3.6 x 107 PFU g−1 for each) via the tail vein. (a–c) (a) Six-hour fasting blood glucose levels on day 3, (b) GTT on day 5 and (c) ITT on day 7 after adenovirus injections. (d) On post-injection day 9, mice were sacrificed after 6-h fasting. XBP1s protein levels were determined both in the whole liver lysates and in nuclear extracts. (e) XBP1s and (f) Hspa5 and Dnajb9 mRNA levels in the liver. Three independent groups of mice (n=15 total in each group) were used in the experiments. Error bars are ±S.E.M. Significance was determined by one-way ANOVA except figure 6c (two-way ANOVA) with Bonferroni multiple-comparison analysis (*P<0.05, **P<0.01, ***P<0.001).
Nuclear levels of XBP1s were markedly increased in the livers of Ad-XBP1s-injected group, but there were no detectable mutant XBP1s in the nuclear fractions of the livers of Ad-XBP1s T48A/S61A-injected group (Fig. 6d). Analysis of total protein and mRNA levels of XBP1s and XBP1s T48A/S61A demonstrated that both XBP1s were expressed at similar levels in the liver (Fig. 6d,e). Furthermore, upregulation of the chaperones, such as Hspa5 and Dnajb9, seen after XBP1s expression, were diminished in the livers of ob/ob mice that were expressing the mutant XBP1s (Fig. 6f). These results provide further evidence that similar to in vitro systems, phosphorylation of XBP1s on T48/S61 is imperative for its function in vivo.
DISCUSSION
In recent years we and others have shown that increased ER stress in obesity is a major link between obesity and type 2 diabetes15–18 and have identified XBP1 as a crucial factor for the maintenance of glucose homeostasis in obesity17,26. We have recently documented that a severe defect in the ability of XBP1s to migrate to the nucleus is a central pathology for the development of ER stress in obesity26 and forced ectopic expression of XBP1s in the liver of obese and diabetic mice greatly decreases ER stress, enhances glucose tolerance, and reduces the blood glucose levels to euglycemia27. Together, these findings indicate that approaches that will increase XBP1s activity could be an attractive therapeutic approach for the treatment of type 2 diabetes.
Our current work demonstrates that p38 MAPK, a well-known SAPK, is an important regulator of XBP1s nuclear translocation and activity. p38 MAPK increases the stability of XBP1s mRNA, and leads to marked upregulation in the protein levels. Furthermore, by phosphorylating XBP1s on Thr48 and Ser61, p38 MAPK greatly enhances the nuclear translocation of this transcription factor. Mutation of the two phosphorylation sites leads to a dramatic reduction in nuclear translocation of XBP1s, in vitro and in vivo. While it is remarkable that phosphorylation of two residues can create such a pronounced effect on the efficiency of XBP1s, the finding that preventing phosphorylation of XBP1s almost completely blocks its activity was quite unexpected. Considering this potent effect of Thr48 and Ser61 phosphorylation on XBP1s activity, and the role of p38 MAPK in this process, we propose that p38 MAPK is a crucial element of UPR signaling, and is required for maintenance of ER homeostasis.
Furthermore, we have also documented that deficiency of JNK in the cells leads to enhanced p38 MAPK activation. Considering the fact that p38 MAPK activation leads to a great enhancement in glucose homeostasis, it is also possible that improvement seen in metabolic homeostasis in JNK-deficient mice could also be contributed by increased p38 MAPK signaling and consequently upregulated ER folding capacity.
The only known physiological condition when XBP1s is activated in the liver is during the refeeding (metabolic load) of mice after a fasting period. Using this condition as an experimental model, we have demonstrated inhibition of p38 MAPK with the use of different approaches (pharmaceutical or genetic) blocks the nuclear translocation of XBP1s, without affecting its splicing or total protein levels. We further demonstrated that XBP1s is phosphorylated on Thr48 and Ser61 in the liver of lean mice during refeeding, and is then able to migrate to the nucleus. However, when obese mice are subjected to the same fasting and refeeding protocol, no such phosphorylation is evident, and no XBP1s is found in the nucleus. It is important to note that p38 MAPK activation markedly increases in the livers of lean mice after refeeding, but that increased in this phosphorylation is not observed in obese mice. Putting this information together with the data showing that inhibition of p38 MAPK in lean mice blocks XBP1s Thr48/Ser61 phosphorylation and nuclear translocation, we suggest that p38 MAPK activity is important for XBP1s activity and its nuclear translocation.
Indeed, increasing p38 MAPK activity by expressing MKK6Glu in the liver of obese and diabetic mice greatly increases XBP1s phosphorylation and nuclear translocation, and markedly enhances glucose tolerance. In other words, activation of p38 MAPK converts the hyperglycemic state of ob/ob mice to a euglycemic state. This is an unexpected observation because the general view in the obesity and diabetes field leans towards the idea that in general SAPK signaling is detrimental to metabolic homeostasis. The role of inflammation in development of insulin resistance and type 2 diabetes is currently an accepted hypothesis44–48. However, our results indicate that not all the inflammatory signaling pathways worsen glucose homeostasis in obesity. Despite the presence of an increased inflammatory state in obesity and increased activity of other inflammatory signaling cascades such as JNK pathway49, we find that p38 MAPK activity in the liver of obese mice is reduced, raising the possibility that p38 MAPK signaling pathway becomes resistant to cytokine signaling in obesity, since p38 MAPK pathway is normally activated with various inflammatory signals including TNFα. It is possible that this unbalanced activation of inflammatory signaling pathways or development of cytokine resistance could be playing an important role in development of various pathologies related to glucose homeostasis in obesity.
Results presented in this study provide indications that specific activators of p38 MAPK might help in re-establishment of ER homeostasis and treatment of type 2 diabetes in obesity. The importance of Thr48 and Ser61 phosphorylation suggests that inhibition of the phosphatase(s) that dephosphorylates XBP1s could be a novel approach for increasing XBP1s activity. Furthermore, the link that we have identified between p38 MAPK and XBP1s also has implications for diseases other than obesity and type 2 diabetes. For example, inhibition of XBP1 activity in cancer cells that exhibit high levels of protein synthesis could be a possible chemotherapeutic tool. Inhibition of XBP1 splicing by proteasome inhibitors has already been shown to be advantageous in multiple myeloma cells, since these cells have a high level of protein synthesis50. Our results suggest that inhibition of p38 MAPK activity (inhibitors for p38 MAPK already exist) could also be beneficial in the cancers that have high protein synthesis. Our findings also add another dimension to the use of p38 inhibitors: p38 MAPK inhibition is considered in the treatment of some inflammatory diseases such as Crohn’s disease, psoriasis, or asthma28. These therapeutic approaches should, however, be evaluated within the context of our results, and in light of the possibility that inhibition of XBP1s activity also decreases the ability of the cell to cope with the inflammatory conditions19,20.
ONLINE METHODS
Recombinant XBP1s
XBP1s CDS together with 6 × His-TF (Trigger Factor) tag at N-terminal was cloned into pGSC1 plasmid. The sequences of His-TF tag (6 × His + TF tag + 3C protease cleavage site) are as follow: MNHKVHHHHHHMQVSVETTQGLGRRVTITIAADSIETAVKSELVNVAKKVRIDG FRKGKVPMNIVAQRYGASVRQDVLGDLMSRNFIDAIIKEKINPAGAPTYVPGEY KLGEDFTYSVEFEVYPEVELQGLEAIEVEKPIVEVTDADVDGMLDTLRKQQATW KEKDGAVEAEDRVTIDFTGSVDGEEFEGGKASDFVLAMGQGRMIPGFEDGIKG HKAGEEFTIDVTFPEEYHAENLKGKAAKFAINLKKVEERELPELTAEFIKRFGVE DGSVEGLRAEVRKNMERELKSAIRNRVKSQAIEGLVKANDIDVPAALIDSEIDVL RRQAAQRFGGNEKQALELPRELFEEQAKRRVVVGLLLGEVIRTNELKADEERV KGLIEEMASAYEDPKEVIEFYSKNKELMDNMRNVALEEQAVEAVLAKAKVTEKE TTFNELMNQQASAGLEVLFQGP. For producing recombinant XBP1s protein, 5 ng pGSC1 plasmid containing XBP1s and His-TF tag was transformed into ArcticExpress TM (DE3) RP host stain E. coli (Agilent Technologies). The total protein was extracted and purified by Ni-affinity resin.
Mass Spectrometric Analysis by LC-MS/MS
MEFs were infected with flag-tagged XBP1s expressing adenovirus and treated with anisomycin (25 ng ml−1) for 2 h. Cells were washed with ice-cold PBS and lysed with RIPA buffer (50 mM Tris-HCl, pH 7.5; 2 mM EGTA; 0.3% CHAPS; 100 mM NaF; 10 mM Na4P2O7; 1 mM Na3VO4; 10 μg ml−1 Leupeptin; 10 μg ml−1 Aprotinin; 2 mM PMSF and 20 nM Okadaic acid). After overnight incubation with anti-flag antibody, Protein A sepharose beads were added for additional 2 h at 4°C. Immunoprecipitates were washed three times with RIPA buffer containing 150 mM NaCl and boiled for 5 min in 2× Laemmli buffer for elution of immunoprecipitated XBP1s. Samples were resolved in SDS-PAGE and stained with Coomassie blue (Bio-Rad). Protein from Coomassie-stained gel bands was digested with trypsin and the resulting peptide mixtures were subjected to microcapillary liquid chromatography tandem mass spectrometry (LC-MS/MS). MS/MS spectra were assigned by searching them with the XBP1s protein sequence using the SEQUEST algorithm.
In vitro Kinase Assay
5 μg of recombinant XBP1s was incubated with activated recombinant p38α MAPK (Cell Signaling) in kinase assay buffer (25 mM Tris-HCl, pH 7.5; 5 mM β-glycerophosphate; 2 mM dithiothreitol; 0.1 mM Na3VO4, 10 mM MgCl2) containing 5 μCi of [γ-32P]ATP (PerkinElmer) and 10 μM cold-ATP for 30 min at 30°C in the presence or absence of 10 μM SB203580 (Calbiochem). GST-ATF2 fusion protein (Cell Signaling) was used as a positive control. The reactions were terminated by addition of Laemmli buffer and samples were separated by SDS-PAGE and transferred to PVDF membranes. Total protein levels were detected by immunoblotting and incorporated 32P (phosphorylation) was visualized by autoradiography.
Analysis of in vivo Insulin Signaling
For in vivo insulin signaling analysis, mice were anaesthetized with xylazine/ketamine after 6 h of fasting. Insulin (0.75 IU kg−1) or saline was infused into the liver via the portal vein. Five minutes after infusion, livers were excised quickly and frozen in liquid nitrogen immediately and stored at −80°C until use.
Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT)
For GTT analysis, mice were intraperitoneally injected with D-glucose (0.5 g kg−1 body weight for ob/ob, 2 g kg−1 body weight for Xbp1flox/flox mouse) after an overnight fasting. Tail vein blood was collected at 0, 15, 30, 60, 90 and 120 min following glucose injection and blood glucose was measured with a glucose meter from Bayer. For ITT analysis, mice were fasted for 6 h (from 8 am to 2 pm) and intraperitoneally injected with recombinant human insulin (2 IU kg−1 for ob/ob mice) from Eli Lilly (Indianapolis, IN). Blood was taken from tail vein at 0, 15, 30, 60, 90 and 120 min after insulin injection and blood glucose was measured.
Statistical Analysis
Data are presented as means ± standard error of the mean (SEM). Statistical significance was calculated by Student’s t test or by multifactor analysis of variance (ANOVA), with factors of time, treatment, and in some cases, genotype. Non-significant interaction terms involving time were taken as an indication that treatment contrasts could be pooled across time. When ANOVA indicated a significant difference among the groups, we compared the groups using a stricter criterion for statistical significance according to the Bonferroni rule (corrected p value = pair-wise p value x number of comparisons). Significance was accepted at the level of P<0.05 (*), P<0.01 (**), or P<0.001 (***).
Supplementary Material
Acknowledgments
We thank to members of the Ozcan laboratory for their help during the execution of the experiments. We thank to L. Glimcher (Harvard School of Public Health) for providing us with the Xbp1flox/flox mouse strain. We thank to P. Blackshear (National Institute of Environment Health Sciences) for kindly providing us with Zfp36−/− cells and M. Gaestel (Hannover Medical School, Germany) for generously providing Mapkapk2−/− cells. We are grateful to H. Feldman (Harvard Medical School) for providing us help with the statistical analysis. This study was supported by the junior faculty start-up funds provided to U.O. by Children’s Hospital Boston, an RO1 grant (R01DK081009) and R56 grant (R56DK089111) provided to U.O. by US National Institutes of Health and the Timothy Murphy funds provided to Division of Endocrinology, Children’s Hospital Boston.
Footnotes
Additional methods. Detailed methodology is described in the Supplementary Methods.
Authors do not have any conflict of interest.
AUTHOR CONTRIBUTIONS
C.S. and J.L. came up with the hypothesis, designed and performed the experiments, analyzed the data and wrote the manuscript. Y.J., J.L., D.G., H.H., S.W.P. performed the experiments. R.J.D. provided reagents and advice through out the project. U.O. came up with the hypothesis, designed experiments, analyzed the data and wrote the manuscript.
References
- 1.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiological reviews. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 2.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 3.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 4.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annual review of cell and developmental biology. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 5.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes & development. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 10.Clauss IM, Chu M, Zhao JL, Glimcher LH. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic acids research. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular and cellular biology. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sriburi R, et al. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 14.Fagone P, et al. Phospholipid biosynthesis program underlying membrane expansion during B-lymphocyte differentiation. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M608175200. [DOI] [PubMed] [Google Scholar]
- 15.Ozcan U, et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Molecular cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozcan U, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science (New York NY. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science (New York, N Y. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 18.Ozcan L, et al. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell metabolism. 2009;9:35–51. doi: 10.1016/j.cmet.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nature immunology. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sado M, et al. Protective effect against Parkinson’s disease-related insults through the activation of XBP1. Brain research. 2009;1257:16–24. doi: 10.1016/j.brainres.2008.11.104. [DOI] [PubMed] [Google Scholar]
- 22.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koong AC, Chauhan V, Romero-Ramirez L. Targeting XBP-1 as a novel anti-cancer strategy. Cancer biology & therapy. 2006;5:756–759. doi: 10.4161/cbt.5.7.2973. [DOI] [PubMed] [Google Scholar]
- 24.Nakatani Y, et al. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. The Journal of biological chemistry. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 25.Ozawa K, et al. The endoplasmic reticulum chaperone improves insulin resistance in type 2 diabetes. Diabetes. 2005;54:657–663. doi: 10.2337/diabetes.54.3.657. [DOI] [PubMed] [Google Scholar]
- 26.Park SW, et al. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nature medicine. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, et al. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nature medicine. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulthard LR, White DE, Jones DL, McDermott MF, Burchill SA. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends in molecular medicine. 2009;15:369–379. doi: 10.1016/j.molmed.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annual review of cell and developmental biology. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 30.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 31.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annual review of immunology. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 32.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science (New York, NY. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 33.Matsuzawa A, Nishitoh H, Tobiume K, Takeda K, Ichijo H. Physiological roles of ASK1-mediated signal transduction in oxidative stress- and endoplasmic reticulum stress-induced apoptosis: advanced findings from ASK1 knockout mice. Antioxidants & redox signaling. 2002;4:415–425. doi: 10.1089/15230860260196218. [DOI] [PubMed] [Google Scholar]
- 34.Park SK, Sanders BG, Kline K. Tocotrienols induce apoptosis in breast cancer cell lines via an endoplasmic reticulum stress-dependent increase in extrinsic death receptor signaling. Breast cancer research and treatment. 2010;124:361–375. doi: 10.1007/s10549-010-0786-2. [DOI] [PubMed] [Google Scholar]
- 35.Lin JH, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science (New York, NY. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tournier C, et al. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes & development. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiological reviews. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 38.Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Molecular and cellular biology. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei K, et al. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Molecular and cellular biology. 2002;22:4929–4942. doi: 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brancho D, et al. Mechanism of p38 MAP kinase activation in vivo. Genes & development. 2003;17:1969–1978. doi: 10.1101/gad.1107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Molecular and cellular biology. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark A, Dean J, Tudor C, Saklatvala J. Post-transcriptional gene regulation by MAP kinases via AU-rich elements. Front Biosci. 2009;14:847–871. doi: 10.2741/3282. [DOI] [PubMed] [Google Scholar]
- 43.Park EJ, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annual review of physiology. 72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 45.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. The Journal of clinical investigation. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solinas G, Karin M. JNK1 and IKKbeta: molecular links between obesity and metabolic dysfunction. Faseb J. 2010;24:2596–2611. doi: 10.1096/fj.09-151340. [DOI] [PubMed] [Google Scholar]
- 47.Arkan MC, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nature medicine. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 48.Solinas G, et al. JNK1 in hematopoietically derived cells contributes to diet-induced inflammation and insulin resistance without affecting obesity. Cell metabolism. 2007;6:386–397. doi: 10.1016/j.cmet.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Hirosumi J, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 50.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.