Abstract
We identified single nucleotide polymorphism (SNP) markers in the laccase gene to establish a line-diagnostic system for shiitake mushrooms. A total of 89 fungal isolates representing four lines, including Korean registered, Korean wild type, Chinese, and Japanese lines, were analyzed. The results suggest that SNP markers in the laccase gene can be useful for line typing in shiitake mushrooms.
Keywords: Laccase gene, Lentinula edodes, Mushroom, Shiitake, SNP
Shiitake mushrooms (Lentinula edodes) are wood-decaying basidiomycetes that have been cultivated widely and are consumed as food in East Asia, including Korea, China, and Japan. They have become the most popular edible mushroom in these countries. In addition to its culinary significance, the Shitake has received further attention recently because of its potential for use in medicine and pharmacology [1, 2]. Despite the economic value of L. edodes, typing and identification of its various lines (including commercial lines) still remain ambiguous [3, 4]. Development of reliable line typing of the shiitake mushroom might be of great significance for its cultivar development [5].
Laccases (EC 1.10.3.2; benzenediol:oxygen oxidoreductases), found in all basidiomycetes, are enzymes with possible applications in the pulp and paper manufacturing industries because they play a role in the biodegradation of lignin [6]. Laccase (p-diphenol oxidase) is a blue oxidase containing several copper ions that oxidizes ortho- and para-diphenols and aromatic amines by removing an electron and a proton from each hydroxyl group to form a free radical [7]. However, the full extent of their functions remain unclear; to date the laccases have been implicated in different biological processes such as sporulation, pigment production, rhizomorph formation, and lignin degradation [8, 9, 10]. Previous studies of the laccase gene characterized and identified its molecular structure and function in some species of basidiomycetes [11, 12, 13, 14]. Nevertheless, the possibility of the laccase gene providing genetic markers for screening/typing of the shiitake lines has not yet been tested. In this study, we aim to (1) develop single nucleotide polymorphism (SNP) markers in the laccase gene to establish an effective system for the identification of different lines of shiitake and (2) investigate whether the observed SNP polymorphisms translate into changes in amino acid sequences, potentially affecting its phenotype.
In order to explore SNPs in the laccase gene, we used four different lines of shiitake, that is, Korean registered, Korean wild type, Japanese, and Chinese lines. Twenty shiitake lines registered in Korea (Korean registered lines) were used: 10 lines from Korea Forest Research Institute (KFRI), including KFRI No. 401 (Sanlim 1Ho) to KFRI No. 169 (Sanlim 10Ho), nine lines from Forest Mushroom Research Institute (FMRI, Korea), including KFRI No. 1 (Sanjo 101Ho) to KFRI No. 689 (Sanjo 109Ho), and one line from Rural Development Administration (RDA, Korea), including KFRI No. 192 (Nonggi 3Ho). Twenty-nine wild type lines grown in Korea, including KFRI No. 36 (Korean wild type lines), were also used. Furthermore, 20 Japanese lines (e.g., KFRI No. 751) and the same number of Chinese lines (e.g., KFRI 478) were examined. The name of each line used in this study was based on voucher numbers in the list of lines maintained in KFRI. Cultures of mycelia from the shiitake lines were maintained on potato dextrose agar slopes according to the Difco manual (Difco Laboratories, Detroit, MI, USA). Liquid cultures of the fungi using potato dextrose broth were incubated at 25℃ for about 30 days, after which the mycelia were harvested from the medium.
Genomic DNA was isolated from a small piece of shiitake mycelia using the phenol-chloroform method or a plant genomic DNA kit (Bionics, Seoul, Korea). Total DNA was quantified using a DNA spectrometer (Nano-Drop ND-1000; NanoDrop Technologies, Wilmington, DE, USA). The laccase gene was then amplified with our newly developed primers using PCR conditions as follows: 3min denaturation at 95℃, 30 cycles of 1min denaturation at 94℃, 1 min annealing at 52℃ (Ta), and 1 min extension at 72℃, and finally, 5 min extension at 72℃. We designed primers using PRIMER 3 ver. 0.4.0, based on the sequence data deposited in GenBank (accession No. FJ473386) (Table 1). PCR was performed in a total reaction volume of 20 µL containing 400 mM KCl, 100 mM Tris-HCl pH 8.3, 15 mM MgCl2, 10 mM DTT, 5 µg/mL acetylated bovine serum albumin, 2.5mM dNTP, 1.0 U FR-Taq polymerase (Bionics), 10~20 ng/µL template DNA, and 200 nM of each of the forward and reverse primers. Each PCR product was then electrophoresed in a 1% agarose gel stained with 2-µL ethidium bromide in 1× TAE buffer.
Table 1. PCR primer pairs and sequencing primer pairs for the Letinula edodes laccase gene.

aTa is annealing temperature.
bThe primer pairs designed to amplify the LELCC gene were based on its open reading frame.
Amplified PCR products were purified enzymatically with Exonuclease I (Fermentas, Burlington, ON, Canada) and Fast Shrimp Alkaline Phosphatase (SAP; Fermentas), following the manufacturer's instructions. The purified laccase gene fragments were subject to direct sequencing in the forward and reverse directions using our newly designed internal primers and the same forward and reverse primers as in the PCR, using a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The DNA sequences were edited using BIOEDIT ver. 7.0.5.3 [15], then aligned using CLUSTAL X ver. 1.83 [16].
Different alleles of the LELCC (Letinula edodes laccase) gene were assessed by their SNPs using a TaqMan probe assay (Applied Biosystems) (Table 2). Each 10 µL reaction volume contained 4 µL of diluted template DNA (2 ng/µL), 5 µL of 2× TaqMan Universal PCR Master Mix (Applied Biosystems), 0.25 µL of each 40× SNP genotyping assay mix, and 0.75 µL of sterile-filtered water. SNPs were genotyped in an ABI StepOnePlus Real-Time PCR System (Applied Biosystems) using the following conditions: an initial denaturation of 10 min at 95℃, and 40 cycles of 92℃ for 15 sec and 60℃ for 1 min. StepOnePlus software ver. 2.0 (Applied Biosystems) was used to detect the fluorescence generated from each sample and to perform an automatic or a manual determination of SNP type for each assay.
Table 2. TaqMan probes for SNP genotyping of the Letinula edodes laccase gene.

Each allele specific TaqMan probe is labeled with a reporter dye at the 5' end.
aVIC dye is linked to the 5' end of the allele 1 probe.
bFAM dye is linked to the 5' end of the allele 2 probe.
In order to quantify SNP variation within four shiitake lines, we calculated genetic parameters such as expected heterozygosity (HE), observed heterozygosity (HO), polymorphism information content (PIC) [17], and Hardy-Weinberg equilibrium (HWE) p-values using PowerMarker ver. 3.25 [18]. The p-values for HWE were adjusted using the sequential Bonferroni correction [19]. Allelic and genotypic frequencies were calculated for the samples analyzed. The genetic variability of the sample as a whole was estimated as described for each of the four lines.
To examine if the observed SNPs represent synonymous (silent) or non-synonymous (replacement) substitutions, the full-length sequence of the LELCC gene for each shiitake line was prepared in a single contig created according to the international unit base (IUB) code by multiple sequence alignment using Clustal W. We also translated the nucleotide sequences of the LELCC gene contigs into amino acid sequences in accordance with open reading frame identification.
We successfully recovered a total of 2,249 bp sequences of the partial LELCC gene from all 89 samples of the four shiitake lines (GenBank accession No. HQ662226~HQ662271). From our alignment of the sequences from all shiitake lines, we identified the nucleotide positions of SNPs (Supplementary Fig. 1). Although our analyses yielded a total of 48 SNPs, we restricted to only 10 SNP markers (SNP 1 to SNP 10) based on the accuracy, amount of within-line polymorphism (i.e., frequency), and designed TaqMan probes.
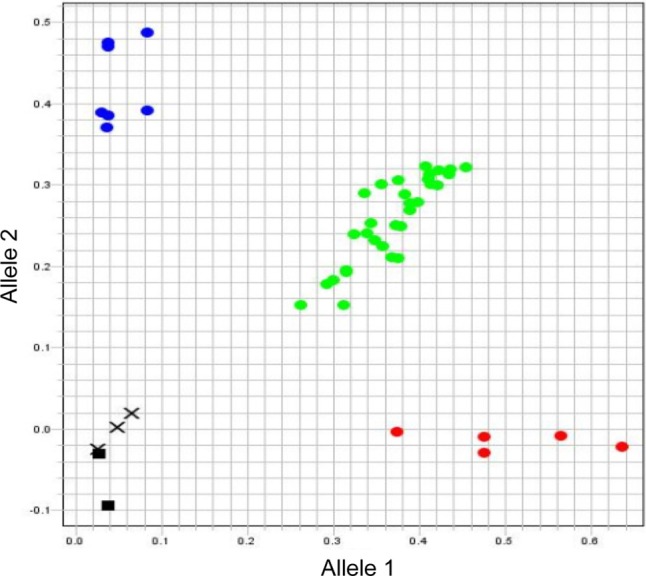
Allelic discrimination was accomplished using real-time PCR. The probes for SNP 8 and SNP 10 were excluded from this analysis owing to technical issues (e.g., low reproducibility of PCR). An example of SNP genotyping using the SNP 2 probe is shown in Fig. 1. The red color indicates the GG homozygote, which was labeled by fluorescent VIC, the blue color denotes the CC homozygote, labeled by FAM, and the green color indicates the GC heterozygote, fluorescently labeled by both VIC and FAM. All eighty-nine shiitake lines were genotyped with the eight SNP markers (Supplementary Table 1). The SNP markers we developed allowed the discrimination of 81 out of 89 lines, with only eight lines that could not be distinguished (KFRI 401 = FMRI137, China 482 = Japan 754, China 488 = China 494, China 490 = Japan 812) (see Supplementary Table 1).
Fig. 1. Allelic discrimination plot using allele specific probes for the Letinula edodes laccase gene. Red color indicates a GG homozygote that was labeled with fluorescent VIC, blue color indicates a CC homozygote that was labeled with fluorescent FAM, and green color indicates a GC heterozygote that was labeled with both fluorescent VIC and FAM. ▪, negative control; Χ, undetermined.
We performed statistical analyses on each of the four lines separately (Korean registered lines, Korean wild type lines, Japanese lines, and Chinese lines). The analysis classified genotypes of the LELCC gene by SNP markers and calculated the frequency of each genotype and the PIC values (Table 3). According to this analysis, the mean frequency of heterozygote individuals was 0.76 (Chinese) > 0.68 (Korean registered) > 0.55 (Japanese) > 0.29 (Korean wild type). However, the mean PIC values, which indicate the number of polymorphic loci/total number of loci analyzed, were similar between lines, ranging from 0.32 to 0.37.
Table 3. Genotype frequencies at eight SNP markers in four different lines of shiitake (Lentinula edodes).
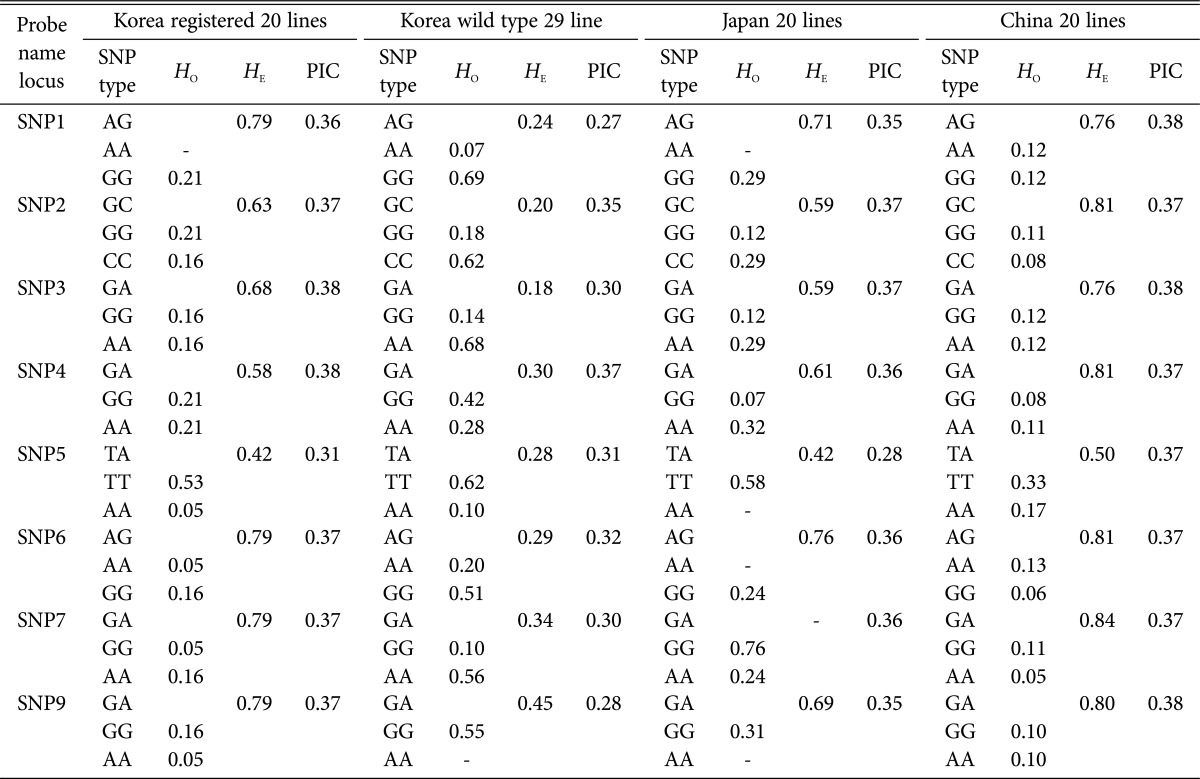
HO, homozygote frequency; HE, heterozygote frequency; PIC, polymorphism information content: number of polymorphic loci/total number of loci analyzed; SNP, single nucleotide polymorphism; -, value zero.
To assess genetic diversity across the shiitake lines, we estimated observed heterozygosity (HO), expected heterozygosity (HE), HWE p-values, and the amount of inbreeding-like effects within subpopulation (FIS), among subpopulations (FST), and within the entire population (FIT). The Ho and He of all samples pooled were 0.5906 and 0.4699, respectively. The genotypes at four out of eight SNP markers (SNPs 1, 6, 7, 9) were significantly different from Hardy-Weinberg expectations (Table 4), which suggests non-random mating at these markers.
Table 4. Summary statistic of SNP markers from pooled lines of shiitake.
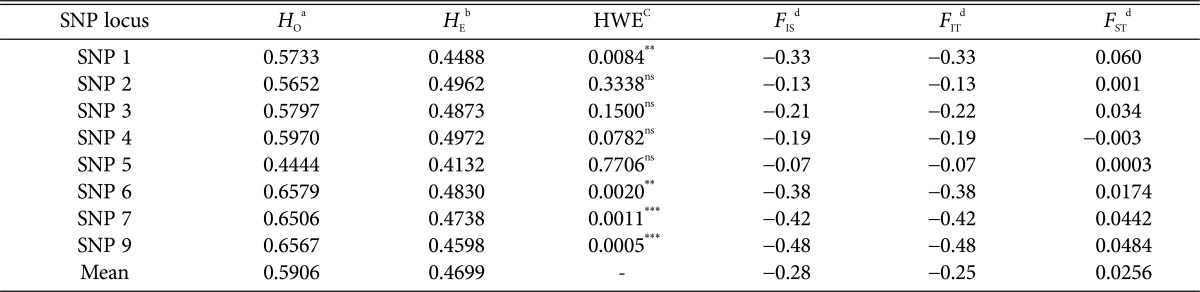
SNP, single nucleotide polymorphism.
aHO: observed heterozygosity is the proportion of heterozygous individuals in the population.
bHE: expected heterozygosity is defined as the probability that two randomly chosen alleles from the population are different.
cHWE: probability estimated from likelihood ratio (G2) tests for Hardy-Weinberg equilibrium at each locus. Significance levels at e = 0.05, 0.01, and 0.001 are indicated by ** and ***, respectively. ns, non-significance.
dF-statistics describe the amount of inbreeding-like effects within subpopulation (FIS), among subpopulations (FST), and within the entire population (FIT).
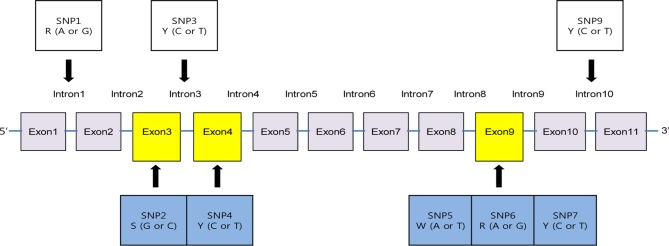
We determined the structure of the LELCC gene by identifying 11 exons and 10 introns, based on the splicing sites (AG/GT) in the sequences (Fig. 2). Among the observed eight SNP markers, SNPs 1, 3, and 9 were located within introns and the other five within exons (Fig. 2). We suggest that SNP 2 located in exon 3, SNP 4 in exon 4, and SNPs 5, 6, and 7 in exon 9 can be good probe candidates for the identification of specimens in the four shiitake lines. GGG (glycine: nonpolar) was converted to GCG (alanine: nonpolar) in SNP 2, and AAT (asparagine: neutral) was converted to TAT (tyrosine: aromatic) in SNP 5. In both cases a purine was substituted by a pyrimidine (G or A to C or T) and the corresponding amino acid was altered, i.e., replacement (nonsynonymous) polymorphisms. In contrast, the remaining polymorphisms were synonymous: GAC (aspartate: negative) converted to GAT (aspartate) in SNP 4, GTG (valine: nonpolar) converted to GTA (valine) in SNP 6, and TTG (leucine: nonpolar) converted to CTG (leucine) in SNP 7.
Fig. 2. A contig map of the Letinula edodes laccase gene showing the locations of eight single nucleotide polymorphisms (SNPs) that were identified in this study. Note that SNPs 2 and 5 are replacement polymorphisms, while SNPs 4, 6, and 7 are synonymous polymorphisms.
Until recently, line typing and breeding of shiitake (Lentinula edodes) cultivars have been depended on traditional DNA markers like restriction fragment length polymorphism [20], randomly amplified polymorphism DNA [21], amplified fragment length polymorphism (AFLP) [4], and microsatellites [22]. However, these markers have limitations in that they often show low levels of reproducibility and usually lack specific genetic information (e.g., AFLP; genetic loci showing DNA polymorphisms are usually unknown). Kim et al. [22] used co-dominant microsatellite DNA markers (GenBank accession No. DQ231475~DQ231479) to discriminate 89 lines of shiitake from East Asia, including Korea, China, and Japan, but they found that precise cultivar typing was difficult because of insufficient genetic information. The SNP markers that we developed here from the laccase gene, a gene known to be functionally important in basidiomycetes, can complement these limitations and allow for more reliable typing of shiitake mushrooms and possibly for cultivar development through marker association selection. It is worth noting that a combined analysis of SNP markers (that were developed for this study) and microsatellites (that were developed in our previous study [22]) allows all 89 shiitake lines to be distinguished.
Moreover, it will be interesting to examine the possible relationships between SNP markers and phenotypic traits, particularly shiitake pigmentation. We found two motifs using MotifFinder software (http://www.genome.jp/tools/motif/). First, the position of serpin was located between 120 bp and 130 bp at nucleotide sequences and was registered as PS00284 in PROSITE. Second, the position of multicopper oxidase 1 was between 137 bp and 157 bp and was registered as PS00079 in PROSITE. Serpins are a group of proteins with similar structures and were first identified as a set of protease inhibitors. The acronym "serpin" was coined because many serpins inhibit chymotrypsin-like serine proteases (serine protease inhibitors) [23]. A single fungal serpin has been characterized to date: celpin from Piromyces spp. line E2. Piromyces is an anaerobic fungus found in ruminant guts and is important for digesting plant material. Celpin is predicted to be an inhibitory molecule and contains two N-terminal dockerin domains in addition to the serpin domain. Dockerins are commonly found in proteins that localize to the fungal cellulosome, a large extracellular multiprotein complex that breaks down cellulose [23]. It is therefore suggested that celpin protects the cellulosome against plant proteases. Interestingly, certain bacterial serpins also localize to the cellulosome. Laccases are copper-containing oxidases that are found in many plants, fungi, and microorganisms. Our analysis of SNPs in shiitake laccases is expected to be useful in future genotyping or protein function studies.
ACKNOWLEDGEMENTS
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01118805)" Rural Development Administration, Republic of Korea, supported by grant (Code 306008-5) from Technology Development Program for Agriculture and Forestry, Ministry of Agriculture, Forestry and Fisheries, Republic of Korea, and supported by the Sangji University Research Fund, 2013 (Project No. 2013-0081) to Hyuk Je Lee.
ELECTRONIC SUPPLEMENTARY MATERIAL
Supplementary data including one table and one figure can be found with this article online at http://www.mycobiology.or.kr/src/sm/mb-43-75-s001.pdf.
Multiple sequence alignment of the LELCC (Letinula edodes laccase) gene sequences (2,249 bp) of 89 shiitake lines, illustrating locations of the single nucleotide polymorphism polymorphisms identified. R = A or G, Y = C or T, K = G or T, M= A or C, S = G or C, and W = A or T.
Genotyping at eight SNPs in the LELCC (Letinula edodes laccase) gene from each shiitake line
References
- 1.Oba K, Kobayashi M, Matsui T, Kodera Y, Sakamoto J. Individual patient based meta-analysis of lentinan for unresectable/recurrent gastric cancer. Anticancer Res. 2009;29:2739–2745. [PubMed] [Google Scholar]
- 2.Bisen PS, Baghel RK, Sanodiya BS, Thakur GS, Prasad GB. Lentinus edodes: a macrofungus with pharmacological activities. Curr Med Chem. 2010;17:2419–2430. doi: 10.2174/092986710791698495. [DOI] [PubMed] [Google Scholar]
- 3.Chiu SW, Ma AM, Lin FC, Moore D. Genetic homogeneity of cultivated strains of shiitake (Lentinula edodes) used in China as revealed by the polymerase chain reaction. Mycol Res. 1996;100:1393–1399. [Google Scholar]
- 4.Terashima K, Matsumoto T. Strain typing of shiitake (Lentinula edodes) cultivars by AFLP analysis, focusing on a heat-dried fruiting body. Mycoscience. 2004;45:79–82. [Google Scholar]
- 5.Hibbett DS, Donoghue MJ. Implications of phylogenetic studies for conservation of genetic diversity in Shiitake mushrooms. Conserv Biol. 1996;10:1321–1327. [Google Scholar]
- 6.Madhavi V, Lele SS. Laccase: properties and applications. BioResources. 2009;4:1694–1717. [Google Scholar]
- 7.Messerschmidt A, Huber R. The blue oxidase, ascorbate oxidase, laccase and ceruloplasmin. Eur J Biochem. 1990;187:341–352. doi: 10.1111/j.1432-1033.1990.tb15311.x. [DOI] [PubMed] [Google Scholar]
- 8.Clutterbuck AJ. The genetics of conidiophore pigmentation in Aspergillus nidulans. J Gen Microbiol. 1990;136:1731–1738. doi: 10.1099/00221287-136-9-1731. [DOI] [PubMed] [Google Scholar]
- 9.Leatham GF, Stahmann MA. Studies on the laccase of Lentinus edodes: specificity, locallization and association with the development of fruiting bodies. J Gen Microbiol. 1981;125:147–157. [Google Scholar]
- 10.Worrall JJ, Chet I, Hüttermann A. Association of rhizomorph formation with laccase activity in Armillaria spp. J Gen Microbiol. 1986;132:2527–2533. [Google Scholar]
- 11.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai S, Iimura Y, Takenouchi K, Katayama Y, Morohoshi N. Cloning and sequence analysis of laccase genes and construction of host vector system in Coriolus versicolor. J Cell Biochem Suppl. 1993;17:C192. [Google Scholar]
- 13.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the whiterot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 14.Yaver DS, Xu E, Golightly EJ, Brown KM, Brown SH, Rey MW, Schneider P, Halkier T, Mondorf K, Dalboge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1997;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 18.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 19.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni RK. DNA polymorphisms in Lentinula edodes, the Shiitake mushroom. Appl Environ Microbiol. 1991;57:1735–1739. doi: 10.1128/aem.57.6.1735-1739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Molina FI. Strain typing of Lentinula edodes by random amplified polymorphic DNA assay. FEMS Microbiol Lett. 1995;131:17–20. doi: 10.1111/j.1574-6968.1995.tb07747.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim KH, Kim YY, Ka KH, Lee HS, Bak WC, Jeong SJ, Seong JY, Suh DS. Microsatellite Markers for Population-Genetic Studies of Shiitake (Lentinula Edodes) Strains. Gene Genomics. 2009;31:403–411. [Google Scholar]
- 23.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of the LELCC (Letinula edodes laccase) gene sequences (2,249 bp) of 89 shiitake lines, illustrating locations of the single nucleotide polymorphism polymorphisms identified. R = A or G, Y = C or T, K = G or T, M= A or C, S = G or C, and W = A or T.
Genotyping at eight SNPs in the LELCC (Letinula edodes laccase) gene from each shiitake line


