Abstract
It has become a current social trend for women to delay childbearing. However, the quality of oocytes from older females is compromised and the pregnancy rate of older women is lower. With the increased rate of delayed childbearing, it is becoming more and more crucial to understand the mechanisms underlying the compromised quality of oocytes from older women, including mitochondrial dysfunctions, aneuploidy and epigenetic changes. Establishing proper epigenetic modifications during oogenesis and early embryo development is an important aspect in reproduction. The reprogramming process may be influenced by external and internal factors that result in improper epigenetic changes in germ cells. Furthermore, germ cell epigenetic changes might be inherited by the next generations. In this review, we briefly summarise the effects of ageing on oocyte quality. We focus on discussing the relationship between ageing and epigenetic modifications, highlighting the epigenetic changes in oocytes from advanced-age females and in post-ovulatory aged oocytes as well as the possible underlying mechanisms.
Introduction
The average age of women bearing children has increased by ∼5 years in the past several decades (te Velde & Pearson 2002). However, advanced maternal age has deleterious effects on oocyte maturation and embryonic development, for which the decreasing oocyte quality with ageing may play a key role (Henderson & Edwards 1968, Tarin et al. 1998a , Sher et al. 2007, Simsek-Duran et al. 2013, Di Emidio et al. 2014, Rambags et al. 2014). These aspects have been well reviewed in our previous publications (Miao et al. 2009, Qiao et al. 2014); therefore, we summarise what is known on epigenetic alterations and relate changes in the epigenome with alterations in gene expression, mitochondrial dysfunction, nutrition, and hormonal homeostasis (Fig. 1).
Figure 1.
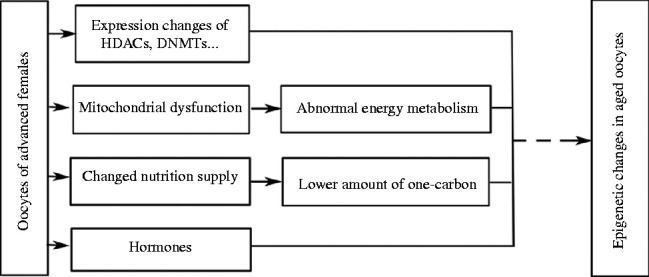
Schematics about relationship between epigenetic changes in oocytes and advanced maternal age. Advanced maternal age causes decrease in oocyte quality, including expression of HDACs and DNMTs, mitochondrial dysfunction, abnormal nutrition supply, changed levels of hormones and others. Thus, many pathways in oocytes may be disrupted, which may be involved in the process of establishing proper epigenetic modification in oocytes.
The risk of childlessness, stillbirth and multiple births for daughters born by aged mothers (≥40 years) is increased when compared with daughters born to young mothers (24–30 years of age) (Smits et al. 2002, Sekhon et al. 2014, Selemani et al. 2014). Several studies have indicated that maternal age is negatively correlated with the health of the offspring (Rocca et al. 1991, Kemkes-Grottenthaler 2004, Brion et al. 2008, Gale 2010). The Barker hypothesis suggests that the foetal development process is the origin of diseases in offspring (Barker 1995), and compromised pre-ovulation oocyte quality, especially related with epigenetic changes, may be crucial as well (Ge et al. 2014a ). Epigenetics, including DNA methylation, histone modification and non-coding RNA, regulate gene expression by changing the conformation of chromosomes other than changing DNA sequences. However, epigenetic modifications may be affected by external and internal factors and the epigenetic changes may be inherited by daughter cells or the offspring (Flanagan et al. 2006, Bird 2007, Goldberg et al. 2007). Therefore, establishing proper genomic imprinting is a crucial event during oocyte maturation and early embryonic development in humans and other mammalian species. In the mouse female germline, DNA methylation is re-methylated during oocyte maturation after birth and it is completely established at the germinal vesicle (GV) stage. This process is mainly catalysed by DNA methyltransferase 3s (DNMT3s, reviewed by Tomizawa et al. (2012)). In mammalian oocytes, the reprogramming process takes place during oogenesis for histone modification and non-coding RNAs, too which is well discussed in previous reviews (Gu et al. 2010, Hales et al. 2011). Thus, if the internal and/or external factors are changed during oogenesis, the epigenetic reprogramming process may be disturbed. For instance, in female mice DNA methylation of imprinted genes in oocytes is altered by maternal diabetes and obesity (Ge et al. 2013a , b , 2014b ). Furthermore, if one-carbon metabolism is abnormal, DNA methylation is changed in many tissues and it is correlated with diseases (Steegers-Theunissen et al. 2013).
Altered epigenetic modification may be an important factor for the complications seen in children of older mothers (Ge et al. 2014a , b ). In this paper, we review the evidence for epigenetic alterations that have been detected in oocytes and that could be responsible for the age-related effects and the potential mechanisms.
Ageing and epigenetic modifications in tissues
Damage to the reproductive and other systems increases with ageing in humans and in many other complex organisms, and there is a decrease in the adaptability and energy utilisation of the system. The accumulation of damage is caused by external and internal factors. Genetic factors only explain 20–30% of the variation in the human lifespan for twins and long-lived families, although these factors are crucial for survival to very old ages (Herskind et al. 1996, Mitchell et al. 2001, Poulsen et al. 2007). The other 70–80% of the variation may be caused by stochastic events, the environment and other non-genetic factors. Therefore, epigenetics, which is the link between the environment and genes and which regulates gene expression by mechanisms other than changes in the underlying DNA sequence (Goldberg et al. 2007), has been recognised as a possible contribution to the ageing phenotype (Wilson & Jones 1983, Fraga et al. 2005, McCauley & Dang 2015).
In mammals, epigenetic modifications are established during foetal development and most of them will be maintained throughout life by DNMT1. But the established epigenetic modifications in organs may be changed after birth if the external and/or internal environment is altered. The genome-wide result shows that global hypomethylation is associated with ageing (Liu et al. 2003, 2011). Genome-wide analysis shows that ageing is associated with a decrease in global genomic methylation, including CpG-poor promoters and tissue-specific genes (Heyn et al. 2012). Similar results have been obtained by other studies in blood and different tissues (Bjornsson et al. 2008, Moore et al. 2008, Bollati et al. 2009, Liu et al. 2009a , Kim et al. 2010, Gentilini et al. 2013, Wnuk et al. 2014). During the lifetime of monozygotic twins, the older monozygous twins exhibit remarkable differences not only in genomic distribution of 5-methylcytosine DNA but also in histone acetylation compared with the early years of life (Fraga et al. 2005). In the process of postnatal development and ageing of rhesus, dimethylation of histone 3 lysine 4 (H3K4me2) globally increases at promoters and enhancers (Han et al. 2012). Small non-coding RNA known as microRNA (miRNA) negatively modulates gene expression through binding to target mRNAs. The expression of miRNA is regulated by DNA methylation (Anwar & Lehmann 2014). Recently, the expression of miRNAs in ageing organs of mice and humans has also been observed (Pincus et al. 2011, Inukai et al. 2012, Ye et al. 2014). In older (>38 years) women, the miRNA profiling in the follicular fluid is clearly different compared with younger ones (<31 years) (Diez-Fraile et al. 2014). Although Anwar & Lehmann (2014) discussed that DNA methylation regulates the expression of miRNAs, the detailed mechanisms remained obscure. Besides DNA methylation, histone modifications and miRNAs, the chromatin structure and transposable elements in organisms are also altered with ageing in Caenorhabditis elegans, Drosophila and mouse model systems and the tissue culture-based replicative senescence model of cell ageing (Wood & Helfand 2013). Although the mechanism causing epigenetic changes in tissues after birth may be different from that in oocytes, the alterations in epigenetic modifications in different organs may indicate that epigenetic modifications in oocytes may be affected by ageing.
Epigenetic changes in ageing oocytes
Herein, ‘ageing oocyte’ is defined as oocyte ageing that occurs in the ovaries of females who show a progressive decline in oocyte number and poor oocyte quality in ovaries during reproductive ageing (Tatone et al. 2008). The pregnancy rate of older women is lower compared with that of younger women, and the lower pregnancy rate (Menken et al. 1986) may mainly be caused by compromised oocyte quality (Wang et al. 2012). The child born to an older mother tends to exhibit the onset of some diseases in adulthood (Rocca et al. 1991, Kemkes-Grottenthaler 2004). These observations prompted us to ponder the mechanisms underlying this phenomenon. External and internal environmental alterations may cause changes in epigenetic modifications in oocytes. In animal models, the establishment of epigenetic modifications in oocytes is affected by maternal diets (Steegers-Theunissen et al. 2013), non-communicable diseases (Ge et al. 2013b ) and other factors (Li et al. 2014). If the changes in epigenetic modifications occur in the germline, it would affect embryonic development and health of the offspring, and this effect may even extend to further generations (Ge et al. 2014b ). Therefore, we discuss the association of oocyte quality, highlighting the epigenetic modifications, with maternal ageing.
Ageing and oocyte quality
Many studies in the IVF–ET setting have demonstrated that ageing does not appear to affect the ability of oocytes to become fertilised, but compared with younger women, the implantation rates are lower and the spontaneous abortion rates are higher for older women (Romeu et al. 1987, Warburton et al. 1987, Lim & Tsakok 1997). Navot et al. (1991) reported that the age-related decline in female fertility is attributed to oocyte quality. For young mothers, about 20% of oocytes are aneuploid (Hassold et al. 2007), but the percentage increases to 50% or more in the oocytes of older mothers (Fragouli et al. 2011, Handyside et al. 2012). One reason may be that the level of cohesin, which is the key protein that regulates chromosome separation, falls below the level required to stabilise chiasmata and to hold sister centromeres tightly together in pre-ovulatory ageing oocytes (Lister et al. 2010, Jessberger 2012). Cohesion loss may be responsible for age-related meiotic segregation errors in mammalian oocytes (Petronczki et al. 2003, Revenkova et al. 2004, Tsutsumi et al. 2014). In humans, the expression of REC8 and SMC1B, the subunit of cohesin, in oocytes is decreased in 40-year-old women compared with 20-year-old women (Tsutsumi et al. 2014). There are other factors that may induce oocyte aneuploidy, which have been well reviewed recently (Jones & Lane 2013). Mitochondrial dysfunctions are observed in pre-ovulatory ageing oocytes (Dorland et al. 1998, Bentov et al. 2011, Eichenlaub-Ritter et al. 2011). As the amount of mitochondria decreases, the mutation of mitochondrial DNA may become increased and mitochondrial function is also affected in the oocytes of advanced-age female bovine, hamsters and mice (Eichenlaub-Ritter et al. 2011, Iwata et al. 2011, Simsek-Duran et al. 2013). Gene expression and chromatin structure (Wood & Helfand 2013) are also found to be affected by ageing. Hamatani et al. (2004) compared the difference in the expression profile at the transcript level of metaphase II (MII) oocytes of 5- to 6-week-old mice with that of 42- to 45-week-old mice. Among ∼11 000 genes whose transcripts are detected in oocytes, 5% showed obvious expression changes (Hamatani et al. 2004). Similar expression profiles in aged mouse oocytes have been reported in another study (Pan et al. 2008). An alteration in the expression profile of human MII oocytes is also associated with female ageing (Grondahl et al. 2010, Santonocito et al. 2013). In mouse oocytes, the mRNA and protein expression levels are altered with ageing, and dysfunctions of the ageing ovary may be a reason for the altered expression of mRNAs and proteins in pre-ovulatory ageing oocytes (Schwarzer et al. 2014, Tatone et al. 2014).
Ageing oocytes and epigenetics
Previous discussions indicate that the epigenetic modifications of oocytes may be affected by advanced maternal age because the expression of DNMTs and histone acetyltransferases (Myst1 (Kat8) and Mrgx (Morf412); Hamatani et al. 2004) is altered with ageing. Therefore, the changes in epigenetic modifications may partly explain why the child of an older woman is predisposed to the onset of hypertension, obesity and other diseases in adulthood (Aagesen et al. 1984, Malini & Ramachandra 2006).
Ageing oocytes and DNA methylation
If DNMT1, which maintains the DNA methylation patterns in oocytes and embryos (Mertineit et al. 1998), is deleted in mouse oocytes, the embryos show a loss of allele-specific expression and methylation at certain imprinted loci, and the foetuses of homozygous females die during the last third of gestation (Howell et al. 2001). In 5- to 6-week-old mouse oocytes, the gene expression profile is different from that in 42- to 45-week-old mouse oocytes. DNMT1, 3b and 3l are involved in the differential gene expression (Hamatani et al. 2004). Yue et al. (2012) found that the changes in genome-wide DNA methylation in oocytes and preimplantation embryos of 35- to 40-week-old mice were associated with decreased expression of DNMTs. The pregnancy rate of older Kunming mice (35–40 weeks old) is lower than that of younger mice, and the stillbirth and foetal malformation rate are higher in the older group compared with the younger group, which may be associated with abnormal DNA methylation in oocytes (Yue et al. 2012). In humans, TAP73 expression which is regulated by DNA methylation patterns is lower in the oocytes of women older than 38 years of age compared with the oocytes of women younger than 36 years of age (Guglielmino et al. 2011). There are still no direct proofs that the DNA methylation status in human oocytes is affected by ageing.
However, one study (Lopes et al. 2009) reports that the increase in resorption sites, morphological abnormalities and delayed development are related with the age of C57BL/6 mice (43–47 weeks old), but the monoallelic expression of the imprinted genes H19 and Snrpn is normal in the blastocysts of aged female mice and the DNA methylation patterns of the differentially methylated regions (DMRs) of Snrpn, Kcnq1ot1, U2af1-rs1 (Zrsr1), Peg1, Igf2r and H19 are not altered. By Restriction Landmark Genome Scanning, the investigators also did not find significant differences in genome-wide DNA methylation in embryos and placentas from aged female mice (Lopes et al. 2009). This is contradictory to previous reports (Hamatani et al. 2004). The authors propose that the contradiction may be related to the materials selected and the limitation of the technique utilised in their study (Lopes et al. 2009). During zebrafish ageing, two CpG island shores are hypomethylated in oocytes, but they are de novo methylated in fertilised eggs (Shimoda et al. 2014). This suggests that the loss of methylation might be reset after fertilisation because there is a de-methylation and re-methylation process during early embryonic development.
Although reports about DNA methylation changes in oocytes from advanced-age females are contradictory, the popular viewpoint proposes that the DNA methylation in oocytes may be changed by pre-ovulatory ageing.
Ageing oocytes and histone modifications
Histone modifications, including methylation, acetylation, ubiquitination and other modifications, represent another crucial and well-investigated epigenetic modification. During meiosis, histone is deacetylated globally at the MI and MII stages by histone deacetylase (HDAC) activity in mammalian oocytes as revealed by immunostaining (Kim et al. 2003, Akiyama et al. 2004, Reddy & Villeneuve 2004). Akiyama et al. (2006) reported that if meiotic histone deacetylation was inhibited, aneuploidy occurred in fertilised mouse oocytes and this resulted in embryonic death in the uterus at an early stage of development. HDAC is downregulated at transcript level in ageing mouse (42- to 45-week-old) oocytes (Hamatani et al. 2004) although histone still remains acetylated in the oocytes of 10-month-old female mice (Akiyama et al. 2006). This suggests that histone modification in pre-ovulatory ageing oocytes may be affected (Table 1) and during development it may result in embryonic death. Similar results were obtained by Manosalva & Gonzalez (2009) and Suo et al. (2010). The expression of Sirt2 which is related with the acetylation of histone H4K16 in the oocytes of aged mice is lower compared with that in younger mouse oocytes (Zhang et al. 2014a ).
Table 1.
Effects of advanced maternal age on epigenetics in oocytes.
| Species | Epigenetic changes | References |
|---|---|---|
| Mouse | Genome-wide DNA methylation is lower in 35- to 40-week-old mouse oocytes | Yue et al. (2012) |
| Mouse | DNMTs (DNMT1, 3a, 3b and 3l) expression is decreased in aged mouse oocytes | Hamatani et al. (2004) and Pan et al. (2008) |
| Mouse | Histone deacetylase is downregulated and histone remains acetylated in older mouse oocytes. Histone acetylation of H4K12 is affected in aged GV and MII oocytes | Hamatani et al. (2004), Akiyama et al. (2006), Manosalva & Gonzalez (2009) and Suo et al. (2010) |
| Mouse | Sirt2 expression is lower in the oocytes of old mice compared with young mice | Zhang et al. (2014a , b ) |
| Mouse | H3K9me3, H3K36me2, H3K79me2 and H4K20me2 are altered in aged oocytes | Manosalva & Gonzalez (2010) |
| Mouse | The expression of Cbx1 and Sirt1 is changed in the oocytes of older mice | |
| Mouse | Histone 3 lysine 4 methylation is changed in aged GV oocytes | Shao et al. (2014) |
| Bovine | Non-imprinted genes (SLC2A1, PRDX1, ZAR1 and BTS) are hypomethylated in the oocytes of adult cows compared with prepubertal calves | Diederich et al. (2012) |
| Zebrafish | Two CpG island shores hypomethylated in oocytes with ageing | Shimoda et al. (2014) |
| Human | The deacetylation of H4K12 in human MII oocytes is affected at an age-dependent manner | van den Berg et al. (2011) |
| Human | The expression of ubiquilin I, USP2, USP34 and USP42 is affected in the oocytes of women aged 37–39 years | Grondahl et al. (2010) |
| Human | MicroRNAs expression profiling of the follicular fluid of younger females is different from that of the follicular fluid of older females | Diez-Fraile et al. (2014) |
| Equine | Three miRNAs are expressed in significantly higher amounts in exosomes isolated from follicular fluid of old compared to young mares | da Silveira et al. (2012) |
Another study found that the histone methylation in mouse GV oocytes was affected by advanced maternal age (Manosalva & Gonzalez 2010) (Table 1). Concomitantly, the GV and MII oocytes of older females lack H3K9me3, H3K36me2, H3K79me2 and H4K20me2 compared with the GV and MII oocytes of younger females. Meanwhile, the expression of the histone methylation-related factors (Cbx1 and Sirt1) is changed in ageing GV oocytes. Histone 3 lysine 4 methylation in mouse GV oocytes is also changed by ageing (Shao et al. 2015).
In humans, the mRNA expression profile of MII oocytes is related with ageing. The differently expressed genes are involved in many biological processes, such as cell cycle, metabolism, apoptosis, protein modification and others (Grondahl et al. 2010). Recently, van den Berg et al. have shown that the histone acetylation staining of H4K5, H4K8, H4K12 and H4K16 was intensive in GV oocytes; however in MI and MII oocytes, chromatin was deacetylated in variable proportions. They also investigated the relationship between histone acetylation and maternal age. The results indicate that advanced maternal age negatively influences the deacetylation of H4K12 in human MII oocytes (van den Berg et al. 2011).
For women at aged 37–39 years, the gene for ubiquilin 1 (a ubiquitin-like protein) is downregulated in the oocytes, but three genes for the ubiquitin-specific peptidases USP2, USP34 and USP42 are upregulated (Grondahl et al. 2010). This suggests that the ubiquitination may be affected by age in human oocytes (Steuerwald et al. 2007). This indicates that histone ubiquitination might also be affected in oocytes by ageing, but there is still no solid evidence to confirm it.
Ageing oocytes and miRNA
miRNA is a kind of small non-coding RNA which functions in post-transcriptional regulation of gene expression upon recruitment into effector complexes (miRNA protein complexes or microRNPs; Truesdell et al. 2012). The post-transcriptional regulation may be particularly crucial for early mammalian development, from maturation of the germ line to initiation of gastrulation, because the genome is transcriptionally silent from the fully grown oocyte stage until zygotic genome activation (Abe et al. 2010). Small RNA is present in mouse oocytes, including miRNA (Tam et al. 2008), but Suh et al. (2010) suggest that miRNA function is globally suppressed during oocyte maturation and preimplantation development. However, loss of Dicer, which is crucial for the generation of endo-siRNA and miRNA, in mouse oocytes results in severe spindle and chromosomal segregation defects (Murchison et al. 2007), while loss of Dgcr8, which is essential only for miRNA processing, in the mouse has no effects on mRNA expression (Suh et al. 2010). Once Ago2 is knocked out in mouse oocytes, the phenotype is similar to that observed in Dicer-knockout mouse oocytes (Kaneda et al. 2009). These results suggest that miRNAs function may be suppressed in mouse oocytes. However, miRNA mediates mRNA translation activation by FXR1 in Xenopus laevis oocytes (Truesdell et al. 2012). In mouse oocytes, miRNA-335-5p could affect oocyte maturation by regulating cytoskeleton dynamics (Cui et al. 2013). A similar result that miRNA-27a activation is not suppressed is observed in porcine oocytes (Chen et al. 2012). Many studies also demonstrated that miRNAs were essential for follicle development in different species (Abramov et al. 2013, Sohel et al. 2013, Yang et al. 2013, Zhang et al. 2014b ). miRNAs expressed in oocytes could regulate bovine early embryogenesis (Tripurani et al. 2013). This contradiction has been well reviewed in previously published papers (Svoboda & Flemr 2010, Suh & Blelloch 2011).
It was shown that 79 miRNAs and 41 miRNAs existed in the microvesicles and exosomes isolated from equine follicular fluid respectively (Table 1), and three miRNAs are expressed significantly higher in exosomes isolated from follicular fluid of old mares compared with young ones (da Silveira et al. 2012). In humans, miRNAs are abundant in MII oocytes and cumulus cells and they may be essential for follicle development (Assou et al. 2013). Moreover, the miRNA expression profile in follicular fluid of women with polycystic ovary syndrome or premature ovarian failure is different from that in follicular fluid of unaffected women (Roth et al. 2014). In addition, miRNAs expression profiling of the follicular fluid of younger (<31 years) and older (>38 years) individuals was also investigated and the result showed that the expression of four miRNAs is different. These miRNAs are involved in carbohydrate digestion and absorption, p53 signalling and other biological processes that may be related with fertility. Therefore, this set of miRNAs and their respective targets should be evaluated in relationship with reproductive ageing (Diez-Fraile et al. 2014). However, the correlation between miRNA expression and oocyte quality during maternal ageing is still unknown.
Post-ovulatory ageing of oocytes and epigenetic modifications
After ovulation, when the arrested MII oocytes are not fertilised during the window of the optimal fertilisation time in vivo or in vitro, the unfertilised oocytes undergo a time-dependent decline concerning quality and this is called ‘post-ovulatory ageing of oocytes’ (Liang et al. 2012). Studies indicate that post-ovulatory ageing of mouse oocytes decreases the pregnancy rate, litter size and increases the percentage of male offspring compared with control females (Tarin et al. 1999, 2002, Kosubek et al. 2010, Liang et al. 2011). F1 offspring derived from post-ovulatory ageing oocytes are prone to the onset of growth retardation, delayed development of the righting reflex and emotionality (Tarin et al. 1999). Otherwise, post-ovulatory ageing of mouse oocytes decreases reproductive fitness and longevity of offspring (Tarin et al. 2002). This suggests that the epigenetic modification in post-ovulatory ageing of oocytes may be altered (Table 2). In our laboratory, we have analysed methylation patterns of imprinted genes in mouse oocytes during the post-ovulatory process. We examined methylation patterns of Snrpn and Peg1 in in vivo and in vitro oocytes at 13, 21 and 29 h of human chorionic gonadotrophin (hCG) injection, respectively, and loss of methylation was observed at 29 h of hCG injection (Liang et al. 2008). Imamura et al. (2005) also reported Peg1 lost methylation in oocytes during post-ovulatory ageing. However, only a small number of oocytes showed aberrant methylation in the DMR of Peg3 in offspring derived from post-ovulatory ageing mouse oocytes (Liang et al. 2011). Although the DNA methylation of some imprinted genes is influenced by oocyte ageing, whether the whole-genome methylation patterns and histone modification are affected by oocyte post-ovulatory ageing is unclear. The histone modifications in post-ovulatory ageing of mouse oocytes are changed at 19 h of hCG injection compared with that at 14 h of hCG injection. When extending the time to 24 h of hCG injection, the fluorescence signals of acetylation of H3K14 also increased in oocytes (Huang et al. 2007). Another study reported that histone acetylation of H3K14 and H4K12 increased in mouse oocytes during post-ovulatory ageing (Liu et al. 2009b ). This phenomenon is also observed in porcine oocytes during post-ovulatory ageing (Cui et al. 2011), although, the detailed underlying mechanism is still unknown.
Table 2.
Effects of post-ovulatory ageing of oocytes on epigenetic modifications.
| Species | Epigenetic changes | References |
|---|---|---|
| Mouse | DNA methylation patterns of Snrpn and Peg1 in oocytes is altered at 29 h after hCG injection | Liang et al. (2008) |
| Mouse | Peg1 loss of methylation in oocytes during post-ovulatory ageing | Imamura et al. (2005) |
| Mouse | Acetylation of H4K8, H4K12 and H3K14 is altered in ageing oocytes | Huang et al. (2007) and Liu et al. (2009b) |
| Porcine | Histone of H4K12 is changed in post-ovulatory ageing oocytes | Cui et al. (2011) |
Oocyte ageing and epigenetics: underlying mechanism(s)
Changes in enzymes related with epigenetic modifications
Epigenetic modifications are catalysed by numerous proteins, including DNMTs, ten-eleven-translocations (TETs), HDACs, ZFP57, Dicer, and KAP1/TRIM28. If their expression is affected in oocytes, the epigenetic modifications may be altered. DNMT1 and DNMT1o are crucial for maintaining proper methylation and DNMT3a, b and l are key de novo methylation enzymes. When the expression of DNMT1, which is necessary for maintaining DNA methylation, is disrupted in mouse oocytes, DNA methylation of imprinted genes is not maintained properly during early embryonic development (Hirasawa et al. 2008, Kurihara et al. 2008). The changed expression of DNMTs in oocytes from individuals of advanced maternal age (Hamatani et al. 2004, Grondahl et al. 2010) might be the direct reason for causing the DNA methylation alterations (Fig. 1). In MII oocytes of 35- to 40-week-old mice, the protein expression of DNMT1, DNMT3a, DNMT3b and DNMT3l is obviously lower than those in MII oocytes of 6- to 8-week-old mice (Yue et al. 2012). Anckaert et al. (2013) used 14-day in vitro follicle culture as a model to investigate pre-ovulatory intrafollicular oocyte ‘ageing’ and found that the mRNA levels of Dnmt3a, Dnmt3l and Zfp57 were altered compared with 12-day in vitro follicle culture in the mouse.
The expression of enzymes related with histone modification is also affected by advanced maternal ageing in oocytes. For example, Hdac2 is downregulated and the expression of histone acetyltransferases (Myst1 and Mrgx) decreases in aged mouse oocytes (Hamatani et al. 2004). In old mice, the expression of the histone methylation-related factors Cbx1 and Sirt2 was changed in GV oocyte (Manosalva & Gonzalez 2010). The protein and kinase activities of CDC2A decreased in the GV and MII oocytes of old mice (Manosalva & Gonzalez 2009). Although specific inhibitors of HDACs could delay post-ovulatory oocyte ageing in mice and pigs (Huang et al. 2007, Jeseta et al. 2008, Lee et al. 2013), it is still unclear whether the change in histone acetylation is caused by the altered level or activities of HDACs for these post-ovulatory ageing oocytes from young mice.
Other enzymes related with epigenetic modifications, for example TETs (Yamaguchi et al. 2012), Trim28 (Messerschmidt et al. 2012) and Dicer (Murchison et al. 2007), may also be critical for oocyte developmental potential. However, there are few studies to investigate whether their expression in oocytes is affected by ageing or not. The proteins of the TET family are not only involved in DNA demethylation during early embryo development (Tahiliani et al. 2009, Gu et al. 2011, Ma et al. 2012), they are also crucial for female germ cells to complete meiosis (Yamaguchi et al. 2012). If the expression of Tet3 is suppressed in oocytes, the paternal global demethylation process at the zygote stage is impaired (Gu et al. 2011). Although TETs are associated with ageing and diseases (van den Hove et al. 2012), a role for them in pre-ovulatory ageing oocyte and post-ovulatory ageing oocyte is still undetermined.
Changes in mitochondrial activity
Mitochondrial dysfunction may be another crucial factor inducing epigenetic changes in the oocytes of advanced females (Fig. 1). Oocyte maturation includes nuclear maturation and cytoplasmic maturation, and there are many biological events involved in these two processes, such as gene expression and histone and chromatin modifications (Eppig 1996). During oocyte maturation, energy (ATP) required is supplied by mitochondria (Torner et al. 2004). Histone and DNMTs use S-adenosyl-l-methionine (SAM) as a donor of methyl groups. SAM is biosynthesised using methionine and ATP (Igarashi & Katoh 2013). Wellen et al. (2009) demonstrated that ATP-citrate lyase was necessary for histone modification. Therefore, we conclude that if mitochondrial function is compromised by maternal age, the epigenetic modification may also be affected. In aged mouse oocytes, ATP and mitochondrial genomes are reduced to 38.4 and 44% respectively (Simsek-Duran et al. 2013). In bovine oocytes, the number of mitochondria and content of ATP are also affected by maternal age (Iwata et al. 2011). In post-ovulatory aged oocytes, reactive oxygen species (ROS) may be another factor inducing epigenetic changes. During in vitro ageing in porcine oocyte, H4K12 acetylation levels are related with ooplasmic ROS content (Cui et al. 2011). Glucose level is related with de novo purine and cAMP synthesis, which is associated with nuclear maturation of oocytes (Colton et al. 2003). The cumulus cells supply nutrition to oocytes in the final phase of oocyte maturation (Gilchrist et al. 2008). The function of cumulus cells is compromised by maternal age (Tatone & Amicarelli 2013). Thus, the energy transmitted from cumulus cells decreases, which may affect the establishment of epigenetic modifications in aged oocytes.
Nutrition effect
The changed nutrition supplies, especial one-carbon, may contribute to epigenetic changes in aged oocytes (Anckaert et al. 2010, Steegers-Theunissen et al. 2013; Fig. 1). For instance, the folate status declines with ageing, including decreased folate intake and altered folate availability (Jacob et al. 1998, Rampersaud et al. 2000). Therefore, the disruption of folate-mediated one-carbon metabolism by ageing may be another reason causing the abnormal DNA methylation in aged oocytes (Jacques et al. 2001).
Possible hormonal effects
A study demonstrated that oestrogen replacement therapy in menopause women reduced the total plasma homocystine concentration and increased genomic DNA methylation of mononuclear cells (Friso et al. 2007). In rodent brain, the methylation percent on the promoter of oestrogen receptor alpha is modulated by the expression of oestrogen (Schwarz et al. 2010). The DNA methylation is not only affected by hormones present in tissues, may be also in oocytes. In humans and mice, the methylation patterns of Peg1 and H19 are changed by hormones used for superovulation of growing oocytes (Sato et al. 2007). With ageing, the androgen level, which can be converted to oestrogen, is reduced for females (Blevins et al. 2013). The DNMT proteins and transcripts in the livers of 3-, 12-, and 24-month-old Ames dwarf mice are drastically reduced compared with WT siblings, and growth hormone appears to modulate the expression of DNMT1 and 3a (Armstrong et al. 2014). The amount of oestrogen which is important for follicular development decreases with ageing (Olsen & Kovacs 1996). Although there is no direct evidence showing that the epigenetic modifications are affected by hormones in pre- and post-ovulatory aged oocytes, it might contribute to the changes in epigenetic modifications in aged oocytes, which needs experimental validation (Fig. 1).
Conclusion(s) and perspectives
The above discussions suggest that advanced maternal age and post-ovulatory oocyte ageing are deleterious to oocyte quality, including oocyte maturation, chromosome segregation, epigenetic modifications and mitochondrial function, and the health of the offspring from advanced-age mothers may also be affected by compromised oocyte quality (Takeo et al. 2013a ). In this review, we mainly discussed the possible relationship between advanced maternal age and epigenetic modifications in oocytes and the potential underlying mechanisms. Two major problems are still unresolved: i) the detailed mechanisms underlying compromised oocyte quality including epigenetic changes caused by advanced maternal age and ii) the prevention of the adverse effects of oocyte ageing on epigenetic changes. As discussed previously, changes in enzymes including methyltransferases (DNMTs) and demethylases (TETs) may be the direct reasons for epigenetic alterations in aged oocytes, but whether/how ageing induces the changes in their expression needs further clarification. With the application of new technologies, it has been possible to test the transcriptome, global DNA methylation, histone modifications and proteome in limited number of cells or even in a single cell (Guo et al. 2014, Lovatt et al. 2014, Smallwood et al. 2014), which may help to address this issue. Another important study still to pursue is how to prevent the age-related deleterious effects on oocytes. If the disulphide-reducing agent dithiothreitol (DTT), an antioxidant, is supplemented to the culture medium, the negative effects of post-ovulatory ageing of mouse oocytes in vitro on fertilisation, cellular fragmentation at 24 h post-insemination and the potential of embryos for development until the blastocyst stage are prevented, at least in part (Tarin et al. 1998b ). N-acetyl-cysteine (NAC) supplemented to medium can also decrease ROS levels in post-ovulatory aged oocytes, but only resveratrol increased the fertilisation rate (Takeo et al. 2013b ). Whether the epigenetic changes in these oocytes are prevented is unknown. But another study demonstrated that if adult female mice were subjected to caloric restriction, they did not exhibit age-related increases in oocyte aneuploidy, meiotic spindle abnormalities or mitochondrial dysfunction, all of which occurred in the oocytes of age-matched controls (Selesniemi et al. 2011). This indicates that the epigenetic modification in aged oocytes may also be affected by calorie restricted diet because energy is crucial for epigenetic modifications. In medium supplemented with pyruvate, post-ovulatory oocyte ageing is prevented and changes in histone acetylation are corrected in the mouse oocyte (Liu et al. 2009b ). Some studies suggest that both DNA methylation and histone modification are associated with caloric restriction (Li et al. 2011, Chouliaras et al. 2012, Chen et al. 2013). One-carbon supplies methyl for methylation, so changing nutrients in diet might prevent abnormal methylation in oocytes. If the levels of methyl donor are lower in the medium during mouse follicle culture, the establishment of oocyte imprinting is affected (Anckaert et al. 2010). Steegers-Theunissen et al. (2013) reviewed the effects of one-carbon on reproduction and long-term health of offspring. These studies suggest that age-related effects, including epigenetic changes, on oocytes might be prevented by diets, medicine or other methods. However, until now we still cannot effectively prevent the age-related deleterious effects on oocytes.
Acknowledgements
The authors thank all the members of the Reproductive Medicine Center, Henan Provincial People's Hospital for their help with this work.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
Funding
This work is supported by the National Natural Science Foundation of China (3147055) and the National Basic Research Program of China (2012CB944404, 2011CB944501).
References
- Aagesen L, Grinsted J, Mikkelsen M. Advanced grandmaternal age on the mother's side – a risk of giving rise to trisomy 21. Annals of Human Genetics. 1984;48:297–301. doi: 10.1111/j.1469-1809.1984.tb00843.x. [DOI] [PubMed] [Google Scholar]
- Abe K, Inoue A, Suzuki MG, Aoki F. Global gene silencing is caused by the dissociation of RNA polymerase II from DNA in mouse oocytes. Journal of Reproduction and Development. 2010;56:502–507. doi: 10.1262/jrd.10-068A. [DOI] [PubMed] [Google Scholar]
- Abramov R, Fu G, Zhang Y, Peng C. Expression and regulation of miR-17a and miR-430b in zebrafish ovarian follicles. General and Comparative Endocrinology. 2013;188:309–315. doi: 10.1016/j.ygcen.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kim JM, Nagata M, Aoki F. Regulation of histone acetylation during meiotic maturation in mouse oocytes. Molecular Reproduction and Development. 2004;69:222–227. doi: 10.1002/mrd.20121. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. PNAS. 2006;103:7339–7344. doi: 10.1073/pnas.0510946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckaert E, Romero S, Adriaenssens T, Smitz J. Effects of low methyl donor levels in culture medium during mouse follicle culture on oocyte imprinting establishment. Biology of Reproduction. 2010;83:377–386. doi: 10.1095/biolreprod.109.082164. [DOI] [PubMed] [Google Scholar]
- Anckaert E, Sanchez F, Billooye K, Smitz J. Dynamics of imprinted DNA methylation and gene transcription for imprinting establishment in mouse oocytes in relation to culture duration variability. Biology of Reproduction. 2013;89:130. doi: 10.1095/biolreprod.113.111641. [DOI] [PubMed] [Google Scholar]
- Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World Journal of Gastroenterology. 2014;20:7894–7913. doi: 10.3748/wjg.v20.i24.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong VL, Rakoczy S, Rojanathammanee L, Brown-Borg HM. Expression of DNA methyltransferases is influenced by growth hormone in the long-living ames dwarf mouse in vivo and in vitro . Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 2014;69:923–933. doi: 10.1093/gerona/glt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Al-edani T, Haouzi D, Philippe N, Lecellier CH, Piquemal D, Commes T, Ait-Ahmed O, Dechaud H, Hamamah S. MicroRNAs: new candidates for the regulation of the human cumulus–oocyte complex. Human Reproduction. 2013;28:3038–3049. doi: 10.1093/humrep/det321. [DOI] [PubMed] [Google Scholar]
- Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995;311:171–174. doi: 10.1136/bmj.311.6998.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentov Y, Yavorska T, Esfandiari N, Jurisicova A, Casper RF. The contribution of mitochondrial function to reproductive aging. Journal of Assisted Reproduction and Genetics. 2011;28:773–783. doi: 10.1007/s10815-011-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg IM, Eleveld C, van der Hoeven M, Birnie E, Steegers EA, Galjaard RJ, Laven JS, van Doorninck JH. Defective deacetylation of histone 4 K12 in human oocytes is associated with advanced maternal age and chromosome misalignment. Human Reproduction. 2011;26:1181–1190. doi: 10.1093/humrep/der030. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, et al. Intra-individual change over time in DNA methylation with familial clustering. Journal of the American Medical Association. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins JK, Coxworth JE, Herndon JG, Hawkes K. Brief communication: Adrenal androgens and aging: female chimpanzees (Pan troglodytes) compared with women. American Journal of Physical Anthropology. 2013;151:643–648. doi: 10.1002/ajpa.22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mechanisms of Ageing and Development. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion MJ, Leary SD, Lawlor DA, Smith GD, Ness AR. Modifiable maternal exposures and offspring blood pressure: a review of epidemiological studies of maternal age, diet, and smoking. Pediatric Research. 2008;63:593–598. doi: 10.1203/PDR.0b013e31816fdbd3. [DOI] [PubMed] [Google Scholar]
- Chen L, Hu X, Dai Y, Li Q, Wang X, Xue K, Li Y, Liang J, Wang Y, Liu X, et al. MicroRNA-27a activity is not suppressed in porcine oocytes. Frontiers in Bioscience. 2012;4:2679–2685. doi: 10.2741/E574. [DOI] [PubMed] [Google Scholar]
- Chen PY, Ganguly A, Rubbi L, Orozco LD, Morselli M, Ashraf D, Jaroszewicz A, Feng S, Jacobsen SE, Nakano A, et al. Intrauterine calorie restriction affects placental DNA methylation and gene expression. Physiological Genomics. 2013;45:565–576. doi: 10.1152/physiolgenomics.00034.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DL, Kenis G, Keitel S, Hof PR, van Os J, Steinbusch HW, Schmitz C, Rutten BP. Prevention of age-related changes in hippocampal levels of 5-methylcytidine by caloric restriction. Neurobiology of Aging. 2012;33:1672–1681. doi: 10.1016/j.neurobiolaging.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton SA, Humpherson PG, Leese HJ, Downs SM. Physiological changes in oocyte–cumulus cell complexes from diabetic mice that potentially influence meiotic regulation. Biology of Reproduction. 2003;69:761–770. doi: 10.1095/biolreprod.102.013649. [DOI] [PubMed] [Google Scholar]
- Cui MS, Wang XL, Tang DW, Zhang J, Liu Y, Zeng SM. Acetylation of H4K12 in porcine oocytes during in vitro aging: potential role of ooplasmic reactive oxygen species. Theriogenology. 2011;75:638–646. doi: 10.1016/j.theriogenology.2010.09.031. [DOI] [PubMed] [Google Scholar]
- Cui XS, Sun SC, Kang YK, Kim NH. Involvement of microRNA-335-5p in cytoskeleton dynamics in mouse oocytes. Reproduction, Fertility, and Development. 2013;25:691–699. doi: 10.1071/RD12138. [DOI] [PubMed] [Google Scholar]
- Di Emidio G, Falone S, Vitti M, D'Alessandro AM, Vento M, Di Pietro C, Amicarelli F, Tatone C. SIRT1 signalling protects mouse oocytes against oxidative stress and is deregulated during aging. Human Reproduction. 2014;29:2006–2017. doi: 10.1093/humrep/deu160. [DOI] [PubMed] [Google Scholar]
- Diederich MI, Hansmann T, Heinzmann J, Barg-Kues B, Herrmann D, Aldag P, Baulain U, Reinhard R, Kues W, Weissberger C, Haaf T, Niemann H. DNA methylation and mRNA expression profiles in bovine oocytes derived from prepubertal and adult donors. Reproduction. 2012;144:319–330. doi: 10.1530/REP-12-0134. [DOI] [PubMed] [Google Scholar]
- Diez-Fraile A, Lammens T, Tilleman K, Witkowski W, Verhasselt B, De Sutter P, Benoit Y, Espeel M, D'Herde K. Age-associated differential microRNA levels in human follicular fluid reveal pathways potentially determining fertility and success of in vitro fertilization. Human Fertility. 2014;17:90–98. doi: 10.3109/14647273.2014.897006. [DOI] [PubMed] [Google Scholar]
- Dorland M, van Kooij RJ, te Velde ER. General ageing and ovarian ageing. Maturitas. 1998;30:113–118. doi: 10.1016/S0378-5122(98)00066-8. [DOI] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Wieczorek M, Luke S, Seidel T. Age related changes in mitochondrial function and new approaches to study redox regulation in mammalian oocytes in response to age or maturation conditions. Mitochondrion. 2011;11:783–796. doi: 10.1016/j.mito.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reproduction, Fertility, and Development. 1996;8:485–489. doi: 10.1071/RD9960485. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, et al. Intra- and interindividual epigenetic variation in human germ cells. American Journal of Human Genetics. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. PNAS. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Goodall NN, Sanchez-Garcia JF, Colls P, Wells D. The cytogenetics of polar bodies: insights into female meiosis and the diagnosis of aneuploidy. Molecular Human Reproduction. 2011;17:286–295. doi: 10.1093/molehr/gar024. [DOI] [PubMed] [Google Scholar]
- Friso S, Lamon-Fava S, Jang H, Schaefer EJ, Corrocher R, Choi SW. Oestrogen replacement therapy reduces total plasma homocysteine and enhances genomic DNA methylation in postmenopausal women. British Journal of Nutrition. 2007;97:617–621. doi: 10.1017/S0007114507433013. [DOI] [PubMed] [Google Scholar]
- Gale EA. Maternal age and diabetes in childhood. BMJ. 2010;340:c623. doi: 10.1136/bmj.c623. [DOI] [PubMed] [Google Scholar]
- Ge ZJ, Liang QX, Luo SM, Wei YC, Han ZM, Schatten H, Sun QY, Zhang CL. Diabetic uterus environment may play a key role in alterations of DNA methylation of several imprinted genes at mid-gestation in mice. Reproductive Biology and Endocrinology. 2013a;11:119. doi: 10.1186/1477-7827-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge ZJ, Liang XW, Guo L, Liang QX, Luo SM, Wang YP, Wei YC, Han ZM, Schatten H, Sun QY. Maternal diabetes causes alterations of DNA methylation statuses of some imprinted genes in murine oocytes. Biology of Reproduction. 2013b;88:117. doi: 10.1095/biolreprod.112.105981. [DOI] [PubMed] [Google Scholar]
- Ge ZJ, Zhang CL, Schatten H, Sun QY. Maternal diabetes mellitus and the origin of non-communicable diseases in offspring: the role of epigenetics. Biology of Reproduction. 2014a;90:139. doi: 10.1095/biolreprod.114.118141. [DOI] [PubMed] [Google Scholar]
- Ge ZJ, Luo SM, Lin F, Liang QX, Huang L, Wei YC, Hou Y, Han ZM, Schatten H, Sun QY. DNA methylation in oocytes and liver of female mice and their offspring: effects of high-fat-diet-induced obesity. Environmental Health Perspectives. 2014b;122:159–164. doi: 10.1289/ehp.1307047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentilini D, Mari D, Castaldi D, Remondini D, Ogliari G, Ostan R, Bucci L, Sirchia SM, Tabano S, Cavagnini F, et al. Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians’ offspring. Age. 2013;35:1961–1973. doi: 10.1007/s11357-012-9463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Human Reproduction Update. 2008;14:159–177. doi: 10.1093/humupd/dmm040. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Grondahl ML, Yding Andersen C, Bogstad J, Nielsen FC, Meinertz H, Borup R. Gene expression profiles of single human mature oocytes in relation to age. Human Reproduction. 2010;25:957–968. doi: 10.1093/humrep/deq014. [DOI] [PubMed] [Google Scholar]
- Gu L, Wang Q, Sun QY. Histone modifications during mammalian oocyte maturation: dynamics, regulation and functions. Cell Cycle. 2010;9:1942–1950. doi: 10.4161/cc.9.10.11599. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guglielmino MR, Santonocito M, Vento M, Ragusa M, Barbagallo D, Borzi P, Casciano I, Banelli B, Barbieri O, Astigiano S, et al. TAp73 is downregulated in oocytes from women of advanced reproductive age. Cell Cycle. 2011;10:3253–3256. doi: 10.4161/cc.10.19.17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Hales BF, Grenier L, Lalancette C, Robaire B. Epigenetic programming: from gametes to blastocyst. Birth Defects Research. Part A, Clinical and Molecular Teratology. 2011;91:652–665. doi: 10.1002/bdra.20781. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS. Age-associated alteration of gene expression patterns in mouse oocytes. Human Molecular Genetics. 2004;13:2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- Han Y, Han D, Yan Z, Boyd-Kirkup JD, Green CD, Khaitovich P, Han JD. Stress-associated H3K4 methylation accumulates during postnatal development and aging of rhesus macaque brain. Aging Cell. 2012;11:1055–1064. doi: 10.1111/acel.12007. [DOI] [PubMed] [Google Scholar]
- Handyside AH, Montag M, Magli MC, Repping S, Harper J, Schmutzler A, Vesela K, Gianaroli L, Geraedts J. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. European Journal of Human Genetics. 2012;20:742–747. doi: 10.1038/ejhg.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Human Molecular Genetics. 2007;16:R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- Henderson SA, Edwards RG. Chiasma frequency and maternal age in mammals. Nature. 1968;218:22–28. doi: 10.1038/218022a0. [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Human Genetics. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, et al. Distinct DNA methylomes of newborns and centenarians. PNAS. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes and Development. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hove DL, Chouliaras L, Rutten BP. The role of 5-hydroxymethylcytosine in aging and Alzheimer's disease: current status and prospects for future studies. Current Alzheimer Research. 2012;9:545–549. doi: 10.2174/156720512800618008. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/S0092-8674(01)00280-X. [DOI] [PubMed] [Google Scholar]
- Huang JC, Yan LY, Lei ZL, Miao YL, Shi LH, Yang JW, Wang Q, Ouyang YC, Sun QY, Chen DY. Changes in histone acetylation during postovulatory aging of mouse oocyte. Biology of Reproduction. 2007;77:666–670. doi: 10.1095/biolreprod.107.062703. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Katoh Y. Metabolic aspects of epigenome: coupling of S-adenosylmethionine synthesis and gene regulation on chromatin by SAMIT module. Sub-Cellular Biochemistry. 2013;61:105–118. doi: 10.1007/978-94-007-4525-4_5. [DOI] [PubMed] [Google Scholar]
- Imamura T, Kerjean A, Heams T, Kupiec JJ, Thenevin C, Paldi A. Dynamic CpG and non-CpG methylation of the Peg1/Mest gene in the mouse oocyte and preimplantation embryo. Journal of Biological Chemistry. 2005;280:20171–20175. doi: 10.1074/jbc.M501749200. [DOI] [PubMed] [Google Scholar]
- Inukai S, de Lencastre A, Turner M, Slack F. Novel microRNAs differentially expressed during aging in the mouse brain. PLoS ONE. 2012;7:e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Goto H, Tanaka H, Sakaguchi Y, Kimura K, Kuwayama T, Monji Y. Effect of maternal age on mitochondrial DNA copy number, ATP content and IVF outcome of bovine oocytes. Reproduction, Fertility, and Development. 2011;23:424–432. doi: 10.1071/RD10133. [DOI] [PubMed] [Google Scholar]
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, Henning SM, Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. Journal of Nutrition. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- Jacques PF, Bostom AG, Wilson PW, Rich S, Rosenberg IH, Selhub J. Determinants of plasma total homocysteine concentration in the Framingham Offspring cohort. American Journal of Clinical Nutrition. 2001;73:613–621. doi: 10.1093/ajcn/73.3.613. [DOI] [PubMed] [Google Scholar]
- Jeseta M, Petr J, Krejcova T, Chmelikova E, Jilek F. In vitro ageing of pig oocytes: effects of the histone deacetylase inhibitor trichostatin A. Zygote. 2008;16:145–152. doi: 10.1017/S0967199408004668. [DOI] [PubMed] [Google Scholar]
- Jessberger R. Age-related aneuploidy through cohesion exhaustion. EMBO Reports. 2012;13:539–546. doi: 10.1038/embor.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT, Lane SI. Molecular causes of aneuploidy in mammalian eggs. Development. 2013;140:3719–3730. doi: 10.1242/dev.090589. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Tang F, O'Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics & Chromatin. 2009;2:9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemkes-Grottenthaler A. Parental effects on offspring longevity – evidence from 17th to 19th century reproductive histories. Annals of Human Biology. 2004;31:139–158. doi: 10.1080/03014460410001663407. [DOI] [PubMed] [Google Scholar]
- Kim JM, Liu H, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. Journal of Cell Biology. 2003;162:37–46. doi: 10.1083/jcb.200303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS ONE. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosubek A, Klein-Hitpass L, Rademacher K, Horsthemke B, Ryffel GU. Aging of Xenopus tropicalis eggs leads to deadenylation of a specific set of maternal mRNAs and loss of developmental potential. PLoS ONE. 2010;5:e13532. doi: 10.1371/journal.pone.0013532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kawamura Y, Uchijima Y, Amamo T, Kobayashi H, Asano T, Kurihara H. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Developmental Biology. 2008;313:335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Lee AR, Kishigami S, Amano T, Matsumoto K, Wakayama T, Hosoi Y. Nicotinamide: a class III HDACi delays in vitro aging of mouse oocytes. Journal of Reproduction and Development. 2013;59:238–244. doi: 10.1262/jrd.2012-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Daniel M, Tollefsbol TO. Epigenetic regulation of caloric restriction in aging. BMC Medicine. 2011;9:98. doi: 10.1186/1741-7015-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang T, Qin XS, Ge W, Ma HG, Sun LL, Hou ZM, Chen H, Chen P, Qin GQ, et al. Exposure to diethylhexyl phthalate (DEHP) results in a heritable modification of imprint genes DNA methylation in mouse oocytes. Molecular Biology Reports. 2014;41:1227–1235. doi: 10.1007/s11033-013-2967-7. [DOI] [PubMed] [Google Scholar]
- Liang XW, Zhu JQ, Miao YL, Liu JH, Wei L, Lu SS, Hou Y, Schatten H, Lu KH, Sun QY. Loss of methylation imprint of Snrpn in postovulatory aging mouse oocyte. Biochemical and Biophysical Research Communications. 2008;371:16–21. doi: 10.1016/j.bbrc.2008.03.105. [DOI] [PubMed] [Google Scholar]
- Liang XW, Ge ZJ, Guo L, Luo SM, Han ZM, Schatten H, Sun QY. Effect of postovulatory oocyte aging on DNA methylation imprinting acquisition in offspring oocytes. Fertility and Sterility. 2011;96:1479–1484. doi: 10.1016/j.fertnstert.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Liang X, Ma J, Schatten H, Sun Q. Epigenetic changes associated with oocyte aging. Science China. Life Sciences. 2012;55:670–676. doi: 10.1007/s11427-012-4354-3. [DOI] [PubMed] [Google Scholar]
- Lim AS, Tsakok MF. Age-related decline in fertility: a link to degenerative oocytes? Fertility and Sterility. 1997;68:265–271. doi: 10.1016/S0015-0282(97)81513-0. [DOI] [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, et al. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Current Biology. 2010;20:1511–1521. doi: 10.1016/j.cub.2010.08.023. [DOI] [PubMed] [Google Scholar]
- Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mechanisms of Ageing and Development. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Tollefsbol TO. DNA methylation impacts on learning and memory in aging. Neurobiology of Aging. 2009a;30:549–560. doi: 10.1016/j.neurobiolaging.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Wu YG, Lan GC, Sui HS, Ge L, Wang JZ, Liu Y, Qiao TW, Tan JH. Pyruvate prevents aging of mouse oocytes. Reproduction. 2009b;138:223–234. doi: 10.1530/REP-09-0122. [DOI] [PubMed] [Google Scholar]
- Liu L, van Groen T, Kadish I, Li Y, Wang D, James SR, Karpf AR, Tollefsbol TO. Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clinical Epigenetics. 2011;2:349–360. doi: 10.1007/s13148-011-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes FL, Fortier AL, Darricarrere N, Chan D, Arnold DR, Trasler JM. Reproductive and epigenetic outcomes associated with aging mouse oocytes. Human Molecular Genetics. 2009;18:2032–2044. doi: 10.1093/hmg/ddp127. [DOI] [PubMed] [Google Scholar]
- Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S, Francis C, Spaethling JM, Wolf JA, Grady MS, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nature Methods. 2014;11:190–196. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JY, Liang XW, Schatten H, Sun QY. Active DNA demethylation in mammalian preimplantation embryos: new insights and new perspectives. Molecular Human Reproduction. 2012;18:333–340. doi: 10.1093/molehr/gas014. [DOI] [PubMed] [Google Scholar]
- Malini SS, Ramachandra NB. Influence of advanced age of maternal grandmothers on Down syndrome. BMC Medical Genetics. 2006;7:4. doi: 10.1186/1471-2350-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva I, Gonzalez A. Aging alters histone H4 acetylation and CDC2A in mouse germinal vesicle stage oocytes. Biology of Reproduction. 2009;81:1164–1171. doi: 10.1095/biolreprod.109.078386. [DOI] [PubMed] [Google Scholar]
- Manosalva I, Gonzalez A. Aging changes the chromatin configuration and histone methylation of mouse oocytes at germinal vesicle stage. Theriogenology. 2010;74:1539–1547. doi: 10.1016/j.theriogenology.2010.06.024. [DOI] [PubMed] [Google Scholar]
- McCauley BS, Dang W. Histone methylation and aging: lessons learned from model systems. Biochimica et Biophysica Acta. 2015 doi: 10.1016/j.bbagrm.2014.05.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menken J, Trussell J, Larsen U. Age and infertility. Science. 1986;233:1389–1394. doi: 10.1126/science.3755843. [DOI] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Miao YL, Kikuchi K, Sun QY, Schatten H. Oocyte aging: cellular and molecular changes, developmental potential and reversal possibility. Human Reproduction Update. 2009;15:573–585. doi: 10.1093/humupd/dmp014. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Hsueh WC, King TM, Pollin TI, Sorkin J, Agarwala R, Schaffer AA, Shuldiner AR. Heritability of life span in the Old Order Amish. American Journal of Medical Genetics. 2001;102:346–352. doi: 10.1002/ajmg.1483. [DOI] [PubMed] [Google Scholar]
- Moore LE, Pfeiffer RM, Poscablo C, Real FX, Kogevinas M, Silverman D, Garcia-Closas R, Chanock S, Tardon A, Serra C, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case–control study. Lancet Oncology. 2008;9:359–366. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes and Development. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navot D, Bergh PA, Williams MA, Garrisi GJ, Guzman I, Sandler B, Grunfeld L. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet. 1991;337:1375–1377. doi: 10.1016/0140-6736(91)93060-M. [DOI] [PubMed] [Google Scholar]
- Olsen NJ, Kovacs WJ. Gonadal steroids and immunity. Endocrine Reviews. 1996;17:369–384. doi: 10.1210/edrv-17-4-369. [DOI] [PubMed] [Google Scholar]
- Pan H, Ma P, Zhu W, Schultz RM. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Developmental Biology. 2008;316:397–407. doi: 10.1016/j.ydbio.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/S0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Pincus Z, Smith-Vikos T, Slack FJ. MicroRNA predictors of longevity in Caenorhabditis elegans . PLoS Genetics. 2011;7:e1002306. doi: 10.1371/journal.pgen.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatric Research. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- Qiao J, Wang ZB, Feng HL, Miao YL, Wang Q, Yu Y, Wei YC, Yan J, Wang WH, Shen W, et al. The root of reduced fertility in aged women and possible therapentic options: current status and future perspects. Molecular Aspects of Medicine. 2014;38:54–85. doi: 10.1016/j.mam.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Rambags BP, van Boxtel DC, Tharasanit T, Lenstra JA, Colenbrander B, Stout TA. Advancing maternal age predisposes to mitochondrial damage and loss during maturation of equine oocytes in vitro . Theriogenology. 2014;81:959–965. doi: 10.1016/j.theriogenology.2014.01.020. [DOI] [PubMed] [Google Scholar]
- Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. American Journal of Clinical Nutrition. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nature Cell Biology. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- Rocca WA, van Duijn CM, Clayton D, Chandra V, Fratiglioni L, Graves AB, Heyman A, Jorm AF, Kokmen E, Kondo K, et al. Maternal age and Alzheimer's disease: a collaborative re-analysis of case-control studies. EURODEM Risk Factors Research Group. International Journal of Epidemiology. 1991;20(Suppl 2):S21–S27. doi: 10.1093/ije/20.Supplement_2.S21. [DOI] [PubMed] [Google Scholar]
- Romeu A, Muasher SJ, Acosta AA, Veeck LL, Diaz J, Jones GS, Jones HW, Jr, Rosenwaks Z. Results of in vitro fertilization attempts in women 40 years of age and older: the Norfolk experience. Fertility and Sterility. 1987;47:130–136. doi: 10.1016/s0015-0282(16)49948-6. [DOI] [PubMed] [Google Scholar]
- Roth LW, McCallie B, Alvero R, Schoolcraft WB, Minjarez D, Katz-Jaffe MG. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. Journal of Assisted Reproduction and Genetics. 2014;31:355–362. doi: 10.1007/s10815-013-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santonocito M, Guglielmino MR, Vento M, Ragusa M, Barbagallo D, Borzi P, Casciano I, Scollo P, Romani M, Tatone C, et al. The apoptotic transcriptome of the human MII oocyte: characterization and age-related changes. Apoptosis. 2013;18:201–211. doi: 10.1007/s10495-012-0783-5. [DOI] [PubMed] [Google Scholar]
- Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Human Reproduction. 2007;22:26–35. doi: 10.1093/humrep/del316. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology. 2010;151:4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer C, Siatkowski M, Pfeiffer MJ, Baeumer N, Drexler HC, Wang B, Fuellen G, Boiani M. Maternal age effect on mouse oocytes: new biological insight from proteomic analysis. Reproduction. 2014;148:55–72. doi: 10.1530/REP-14-0126. [DOI] [PubMed] [Google Scholar]
- Sekhon LH, Gerber RS, Rebarber A, Saltzman DH, Klauser CK, Gupta S, Fox NS. Effect of oocyte donation on pregnancy outcomes in in vitro fertilization twin gestations. Fertility and Sterility. 2014;101:1326–1330. doi: 10.1016/j.fertnstert.2014.01.055. [DOI] [PubMed] [Google Scholar]
- Selemani M, Mwanyangala MA, Mrema S, Shamte A, Kajungu D, Mkopi A, Mahande MJ, Nathan R. The effect of mother's age and other related factors on neonatal survival associated with first and second birth in rural, Tanzania: evidence from Ifakara health and demographic surveillance system in rural Tanzania. BMC Pregnancy and Childbirth. 2014;14:240. doi: 10.1186/1471-2393-14-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selesniemi K, Lee HJ, Muhlhauser A, Tilly JL. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. PNAS. 2011;108:12319–12324. doi: 10.1073/pnas.1018793108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao GB, Wang J, Zhang LP, Wu CY, Jin J, Sang JR, Lu HY, Gong AH, Du FY, Peng WX. Aging alters histone H3 lysine 4 methylation in mouse germinal vesicle stage oocytes. Reproduction, Fertility, and Development. 2015 doi: 10.1071/RD13293. In press. [DOI] [PubMed] [Google Scholar]
- Sher G, Keskintepe L, Keskintepe M, Ginsburg M, Maassarani G, Yakut T, Baltaci V, Kotze D, Unsal E. Oocyte karyotyping by comparative genomic hybridization [correction of hybrydization] provides a highly reliable method for selecting “competent” embryos, markedly improving in vitro fertilization outcome: a multiphase study. Fertility and Sterility. 2007;87:1033–1040. doi: 10.1016/j.fertnstert.2006.08.108. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Izawa T, Yoshizawa A, Yokoi H, Kikuchi Y, Hashimoto N. Decrease in cytosine methylation at CpG island shores and increase in DNA fragmentation during zebrafish aging. Age. 2014;36:103–115. doi: 10.1007/s11357-013-9548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biology of Reproduction. 2012;86:71. doi: 10.1095/biolreprod.111.093252. [DOI] [PubMed] [Google Scholar]
- Simsek-Duran F, Li F, Ford W, Swanson RJ, Jones HW, Jr, Castora FJ. Age-associated metabolic and morphologic changes in mitochondria of individual mouse and hamster oocytes. PLoS ONE. 2013;8:e64955. doi: 10.1371/journal.pone.0064955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nature Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits LJ, Zielhuis GA, Jongbloet PH, Van Poppel FW. Mother's age and daughter's fecundity. An epidemiological analysis of late 19th to early 20th century family reconstitutions. International Journal of Epidemiology. 2002;31:349–358. doi: 10.1093/ije/31.2.349. [DOI] [PubMed] [Google Scholar]
- Sohel MM, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, Looft C, Rings F, Uddin MJ, Spencer TE, Schellander K, et al. Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: implications for bovine oocyte developmental competence. PLoS ONE. 2013;8:e78505. doi: 10.1371/journal.pone.0078505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegers-Theunissen RP, Twigt J, Pestinger V, Sinclair KD. The periconceptional period, reproduction and long-term health of offspring: the importance of one-carbon metabolism. Human Reproduction Update. 2013;19:640–655. doi: 10.1093/humupd/dmt041. [DOI] [PubMed] [Google Scholar]
- Steuerwald NM, Bermudez MG, Wells D, Munne S, Cohen J. Maternal age-related differential global expression profiles observed in human oocytes. Reproductive Biomedicine Online. 2007;14:700–708. doi: 10.1016/S1472-6483(10)60671-2. [DOI] [PubMed] [Google Scholar]
- Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current Biology. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo L, Meng QG, Pei Y, Yan CL, Fu XW, Bunch TD, Zhu SE. Changes in acetylation on lysine 12 of histone H4 (acH4K12) of murine oocytes during maternal aging may affect fertilization and subsequent embryo development. Fertility and Sterility. 2010;93:945–951. doi: 10.1016/j.fertnstert.2008.12.128. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Reports. 2010;11:590–597. doi: 10.1038/embor.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S, Goto H, Kuwayama T, Monji Y, Iwata H. Effect of maternal age on the ratio of cleavage and mitochondrial DNA copy number in early developmental stage bovine embryos. Journal of Reproduction and Development. 2013a;59:174–179. doi: 10.1262/jrd.2012-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S, Kawahara-Miki R, Goto H, Cao F, Kimura K, Monji Y, Kuwayama T, Iwata H. Age-associated changes in gene expression and developmental competence of bovine oocytes, and a possible countermeasure against age-associated events. Molecular Reproduction and Development. 2013b;80:508–521. doi: 10.1002/mrd.22187. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin JJ, Brines J, Cano A. Long-term effects of delayed parenthood. Human Reproduction. 1998a;13:2371–2376. doi: 10.1093/humrep/13.9.2371. [DOI] [PubMed] [Google Scholar]
- Tarin JJ, Ten J, Vendrell FJ, Cano A. Dithiothreitol prevents age-associated decrease in oocyte/conceptus viability in vitro . Human Reproduction. 1998b;13:381–386. doi: 10.1093/humrep/13.2.381. [DOI] [PubMed] [Google Scholar]
- Tarin JJ, Perez-Albala S, Aguilar A, Minarro J, Hermenegildo C, Cano A. Long-term effects of postovulatory aging of mouse oocytes on offspring: a two-generational study. Biology of Reproduction. 1999;61:1347–1355. doi: 10.1095/biolreprod61.5.1347. [DOI] [PubMed] [Google Scholar]
- Tarin JJ, Perez-Albala S, Perez-Hoyos S, Cano A. Postovulatory aging of oocytes decreases reproductive fitness and longevity of offspring. Biology of Reproduction. 2002;66:495–499. doi: 10.1095/biolreprod66.2.495. [DOI] [PubMed] [Google Scholar]
- Tatone C, Amicarelli F. The aging ovary – the poor granulosa cells. Fertility and Sterility. 2013;99:12–17. doi: 10.1016/j.fertnstert.2012.11.029. [DOI] [PubMed] [Google Scholar]
- Tatone C, Amicarelli F, Carbone MC, Monteleone P, Caserta D, Marci R, Artini PG, Piomboni P, Focarelli R. Cellular and molecular aspects of ovarian follicle ageing. Human Reproduction Update. 2008;14:131–142. doi: 10.1093/humupd/dmm048. [DOI] [PubMed] [Google Scholar]
- Tatone C, Eichenlaub-Ritter U, Amicarelli F. Dicarbonyl stress and glyoxalases in ovarian function. Biochemical Society Transactions. 2014;42:433–438. doi: 10.1042/BST20140023. [DOI] [PubMed] [Google Scholar]
- Tomizawa S, Nowacka-Woszuk J, Kelsey G. DNA methylation establishment during oocyte growth: mechanisms and significance. International Journal of Developmental Biology. 2012;56:867–875. doi: 10.1387/ijdb.120152gk. [DOI] [PubMed] [Google Scholar]
- Torner H, Brussow KP, Alm H, Ratky J, Pohland R, Tuchscherer A, Kanitz W. Mitochondrial aggregation patterns and activity in porcine oocytes and apoptosis in surrounding cumulus cells depends on the stage of pre-ovulatory maturation. Theriogenology. 2004;61:1675–1689. doi: 10.1016/j.theriogenology.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Tripurani SK, Wee G, Lee KB, Smith GW, Wang L, Jianboyao Y. MicroRNA-212 post-transcriptionally regulates oocyte-specific basic-helix–loop–helix transcription factor, factor in the germline alpha (FIGLA), during bovine early embryogenesis. PLoS ONE. 2013;8:e76114. doi: 10.1371/journal.pone.0076114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Scientific Reports. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi M, Fujiwara R, Nishizawa H, Ito M, Kogo H, Inagaki H, Ohye T, Kato T, Fujii T, Kurahashi H. Age-related decrease of meiotic cohesins in human oocytes. PLoS ONE. 2014;9:e96710. doi: 10.1371/journal.pone.0096710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velde ER, Pearson PL. The variability of female reproductive ageing. Human Reproduction Update. 2002;8:141–154. doi: 10.1093/humupd/8.2.141. [DOI] [PubMed] [Google Scholar]
- Wang YA, Farquhar C, Sullivan EA. Donor age is a major determinant of success of oocyte donation/recipient programme. Human Reproduction. 2012;27:118–125. doi: 10.1093/humrep/der359. [DOI] [PubMed] [Google Scholar]
- Warburton D, Kline J, Stein Z, Hutzler M, Chin A, Hassold T. Does the karyotype of a spontaneous abortion predict the karyotype of a subsequent abortion? Evidence from 273 women with two karyotyped spontaneous abortions. American Journal of Human Genetics. 1987;41:465–483. [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wnuk M, Lewinska A, Gurgul A, Zabek T, Potocki L, Oklejewicz B, Bugno-Poniewierska M, Wegrzyn M, Slota E. Changes in DNA methylation patterns and repetitive sequences in blood lymphocytes of aged horses. Age. 2014;36:31–48. doi: 10.1007/s11357-013-9541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Helfand SL. Chromatin structure and transposable elements in organismal aging. Frontiers in Genetics. 2013;4:274. doi: 10.3389/fgene.2013.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang S, Luo A, Ding T, Lai Z, Shen W, Ma X, Cao C, Shi L, Jiang J, et al. Expression patterns and regulatory functions of microRNAs during the initiation of primordial follicle development in the neonatal mouse ovary. Biology of Reproduction. 2013;89:126. doi: 10.1095/biolreprod.113.107730. [DOI] [PubMed] [Google Scholar]
- Ye Y, Li D, Ouyang D, Deng L, Zhang Y, Ma Y, Li Y. MicroRNA expression in the aging mouse thymus. Gene. 2014;547:218–225. doi: 10.1016/j.gene.2014.06.039. [DOI] [PubMed] [Google Scholar]
- Yue MX, Fu XW, Zhou GB, Hou YP, Du M, Wang L, Zhu SE. Abnormal DNA methylation in oocytes could be associated with a decrease in reproductive potential in old mice. Journal of Assisted Reproduction and Genetics. 2012;29:643–650. doi: 10.1007/s10815-012-9780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hou X, Ma R, Moley K, Schedl T, Wang Q. Sirt2 functions in spindle organization and chromosome alignment in mouse oocyte meiosis. FASEB Journal. 2014a;28:1435–1445. doi: 10.1096/fj.13-244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jiang X, Zhang Y, Xu B, Hua J, Ma T, Zheng W, Sun R, Shen W, Cooke HJ, et al. microRNA 376a regulates follicle assembly by targeting Pcna in fetal and neonatal mouse ovaries. Reproduction. 2014b;148:43–54. doi: 10.1530/REP-13-0508. [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a