Abstract
Recent studies have suggested an association between alcoholism and DNA methylation, a mechanism that can mediate long-lasting changes in gene transcription. Here, we examined the contribution of DNA methylation to the long-term behavioral and molecular changes induced by a history of alcohol dependence. In search of mechanisms underlying persistent rather than acute dependence-induced neuroadaptations, we studied the role of DNA methylation regulating medial prefrontal cortex (mPFC) gene expression and alcohol-related behaviors in rats 3 weeks into abstinence following alcohol dependence. Postdependent rats showed escalated alcohol intake, which was associated with increased DNA methylation as well as decreased expression of genes encoding synaptic proteins involved in neurotransmitter release in the mPFC. Infusion of the DNA methyltransferase inhibitor RG108 prevented both escalation of alcohol consumption and dependence-induced downregulation of 4 of the 7 transcripts modified in postdependent rats. Specifically, RG108 treatment directly reversed both downregulation of synaptotagmin 2 (Syt2) gene expression and hypermethylation on CpG#5 of its first exon. Lentiviral inhibition of Syt2 expression in the mPFC increased aversion-resistant alcohol drinking, supporting a mechanistic role of Syt2 in compulsive-like behavior. Our findings identified a functional role of DNA methylation in alcohol dependence-like behavioral phenotypes and a candidate gene network that may mediate its effects. Together, these data provide novel evidence for DNA methyltransferases as potential therapeutic targets in alcoholism.
Keywords: alcoholism, DNA methylation, DNMT, epigenetics, neurotransmitter release, plasticity
Introduction
Alcoholism is a complex disorder that results from the interplay between genetic and environmental factors. Although it has been demonstrated that alcoholism is associated with long-lasting drug-induced changes in gene expression and neuronal plasticity (Tapocik et al., 2012), the molecular mechanisms modulating these changes and their maintenance are still unclear.
Substantial evidence suggests that epigenetic mechanisms are involved in the regulation of alcohol-related behaviors. For instance, the histone deacethylase (HDAC) inhibitor trichostatin A (TSA) decreases alcohol intake in the alcohol preferring P rats and prevents the development of alcohol withdrawal-related anxiety (Pandey et al., 2008; Sakharkar et al., 2012, 2014). Although several studies support a role of histone acetylation in regulation of alcohol-related behaviors (Tabakoff et al., 1986; Pandey et al., 2008; Agudelo et al., 2011; Sakharkar et al., 2012), much less is known about the possible role of DNA methylation. DNA methylation can lead to long-lasting and stable changes in gene expression, has the ability to change dynamically in response to external factors, and provides a mechanism through which the environment can influence gene expression and hence behavioral phenotypes of importance for addiction. Accordingly, environmental factors, including stress (Weaver et al., 2004; Murgatroyd et al., 2009) and exposure to drugs of abuse, regulate methylation patterns in the brain (Wong et al., 2011; Tian et al., 2012). Several studies also indicate that DNA methylation may influence reinforcing properties of drugs of abuse (Wong et al., 2011). For example, inhibition of DNA methyltransferase (DNMT) activity in the nucleus accumbens (NAc) increased, whereas hypermethylation induced by NAc-specific Dnmt3a overexpression attenuated, cocaine reward (LaPlant et al., 2010). Moreover, systemic inhibition of DNMT activity decreases excessive alcohol drinking and seeking behaviors in rodents (Warnault et al., 2013).
Although previous studies suggest a role of DNA methylation in alcohol related-behaviors, mechanisms through which DNA methylation contributes to long-term neuroadaptations in alcohol dependence are presently unknown. Here, we examined the possible contribution of DNA methylation to the long-term behavioral and molecular changes induced by a history of alcohol dependence. We focused on the mPFC because of its prominent role in drug-induced neuroadaptations associated with drug seeking and alcohol dependence (Tzschentke, 2000; Kalivas, 2008; Tapocik et al., 2012, 2014). Using our model of postdependent (PD) rats (Rimondini et al., 2002; Tapocik et al., 2012), we first measured DNA methylation levels in the mPFC 3 weeks into protracted abstinence from alcohol vapor. Next, we functionally assessed the role of DNA hypermethylation in alcohol-related behaviors. Furthermore, we performed whole transcriptome sequencing (WT seq) and pyrosequencing analysis to identify persistent alcohol-induced changes in gene expression that are driven by DNA methylation changes. Finally, using a lentiviral approach, we confirmed the role of Syt2, a gene identified in the WT seq and regulated by DNA methylation, in alcohol self-administration and aversion-resistant alcohol seeking, traits thought to be hallmarks of alcohol dependence.
Materials and Methods
Animals.
Male Wistar rats (200–225 g, Charles River) were housed under a reverse light cycle with food and water ad libitum and were habituated to the facility and handled before experiments. Testing took place during the dark phase. Procedures were approved by the National Institute on Alcohol Abuse and Alcoholism Animal Care and Use Committee.
Dependence induction.
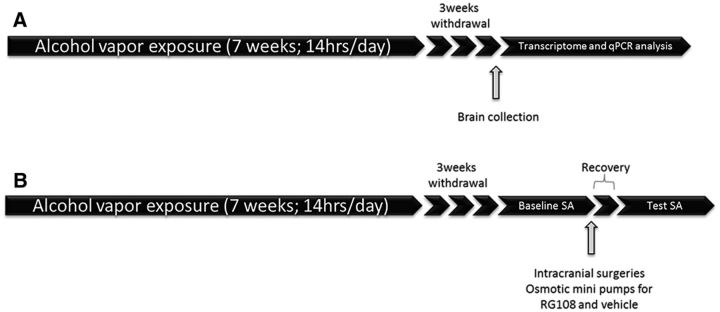
Dependence was induced using chronic intermittent alcohol vapor exposure as described previously (Rimondini et al., 2002). Briefly, rats were exposed to alcohol vapor for 14 h each day (on at 7:30 P.M., off at 9:30 A.M.) for 7 weeks, resulting in blood alcohol concentrations (BACs) between 150 and 300 mg/dl. Controls were kept in identical chambers with normal air flow. Once weekly, blood was collected from the lateral tail vein. BACs were assessed using quantitative gas chromatography (Tapocik et al., 2012). Molecular and behavioral tests were performed 3 weeks after the end of the exposure to assess persistent effects of alcohol exposure (Fig. 1).
Figure 1.
Experimental timeline: Rats are exposed to alcohol vapor for 7 weeks (14 h per day). A, Amg, Hipp, NAc, and mPFC were collected 3 weeks after the end of alcohol exposure. B, Alcohol self-administration (SA) was measured 3 weeks after alcohol exposure. Once SA was stable (baseline), cannulae connected to an osmotic mini-pump containing either RG108 or vehicle were implanted into mPFC of PD and control rats. The rats were tested for SA after 1 week recovery.
Surgery.
For the RG108 experiments, rats received continuous infusion of RG108 (100 μm dissolved in 5% 2-hydroxypropyl β-cyclodextrin (w/v)) into the lateral ventricle (coordinates relative to bregma: anteroposterior, −0.8 mm; mediolateral, ±1.5 mm; dorsoventral, −5.0 mm) or the mPFC (coordinates relative to bregma: anteroposterior, 2.5 mm; mediolateral, ±1.5 mm; dorsoventral, −3.5 mm; 10° angle). The rats underwent surgery 3 weeks after the end of alcohol exposure using the osmotic mini pumps 2002 (0.5 μl h−1; Alzet) and 2004 (0.25 μl h−1; Alzet) for lateral ventricle and mPFC infusion, respectively.
For the lentiviral microinjection, rats received 2 injections bilaterally (1 μl per injection; rate: 0.25 μl/min) directly into the mPFC (coordinates relative to bregma: anteroposterior, 2.5 and 3 mm; mediolateral, ±0.7 mm; dorsoventral, −3.5 mm) of a lentiviral vector containing a shRNA to Syt2 (CTTCTCTAAGCATGACATCAT; titer: 9.7 × 109 TU/ml; Sigma) and a scrambled control (titer: 2.9 × 109 TU/ml). Rats were subjected to behavioral studies after a 1 week recovery period.
Two-bottle free choice.
Rats had access to 2 bottles in their home cage. One bottle contained saccharin 0.2% and the second bottle contained saccharin 0.2% with increasing concentrations of alcohol (3%, 6%, and 8%). After 10 d of stable alcohol consumption at 8%, rats were separated into 2 groups (control and PD rats). PD rats were then exposed to alcohol vapor for 7 weeks. Beginning 3 weeks after rats were removed from alcohol vapor, cannulae connected to an osmotic mini pumps containing either RG108 or vehicle were implanted into lateral ventricle of PD and control rats. Rats were then tested for alcohol intake after 1 week recovery.
Alcohol self-administration.
Training and testing for operant self-administration of 10% alcohol in water were as described previously (Cippitelli et al., 2010). Once self-administration was stable at a fixed ratio 1 (FR1) (baseline), cannulae connected to osmotic mini pumps containing either RG108 or vehicle were implanted into mPFC of PD and control rats. LaPlant et al. (2010) have previously demonstrated that 100 μm of RG108 infused at a rate of 0.25 μl/min significantly decreased DNA methylation. The rats were tested for self-administration after 1 week recovery (Fig. 1).
Behavioral test after Syt2 inhibition.
Rats were trained to self-administer alcohol as described above. Once self-administration was stable, rats received a microinjection of Syt2 lentiviral vector or scrambled lentiviral vector directly into the mPFC and were allowed to recover for 1 week. Rats were then tested for alcohol self-administration after 1 week recovery. Following this test, rats were assessed for compulsivity-like behavior when alcohol was mixed with increasing concentration of quinine (0.005, 0.01, 0.025, 0.05, and 0.075 g/L) during alcohol-self administration.
DNA isolation and global DNA methylation analysis.
Cannulae connected to osmotic mini-pumps containing either RG108 or vehicle were implanted in lateral ventricles. Animals were decapitated 2 weeks after surgery (Fig. 1). Bilateral samples from the mPFC were dissected out as described previously (Björk et al., 2006) and stored at −80°C. DNA was extracted from mPFC using QIAamp DNA Mini Kit (QIAGEN) following the manufacturer's instructions.
Global DNA methylation was measured using Methylamp Global DNA Methylation Quantification Ultra Kit (Epigentek) following the manufacturer's instructions. Raw values were colorimetrically quantified, and total methylation level was estimated by generating a standard curve from Epigentek's methylated DNA standard. Values are represented as methylation percentage relative to vehicle control. For Tables 1 and 2, samples were dissected from rats that received vehicle microinfusion.
Table 1.
Global DNA methylation levelsa
| Brain region | Control | PD |
|---|---|---|
| mPFC | 100 ± 17.65 | 200.34 ± 38.45* |
| Amg | 100 ± 28.7 | 105.79 ± 29.2 |
| Hipp | 100 ± 13.08 | 92.51 ± 15.13 |
| NAc | 100 ± 17.0 | 144 ± 6.8* |
aMeasure of global DNA methylation relative to control (%) in mPFC, NAc, Amg, and Hipp of control and PD rats.
*p < 0.05, Control vs. PD.
Table 2.
Effect of RG108 on global DNA methylation levels in the mPFCa
| Drug treatment | Control | PD |
|---|---|---|
| Vehicle | 100 ± 17.65442 | 200.34 ± 38.45* |
| RG108 | 109.35 ± 11.36 | 119.62 ± 19.11** |
aMeasure of DNA methylation levels in the mPFC after RG108 treatment in control and PD rats. Data show % methylation relative to control/vehicle group.
*p < 0.05, Control vs. PD;
**p < 0.05, Vehicle vs. RG108.
For pyrosequencing analysis, DNA was sent to EpigenDx, a DNA methylation and pyrosequencing laboratory service, which designed and analyzed our pyrosequencing assays.
Western blot.
Tissue was homogenate in lysis buffer (RIPA buffer, DTT, Cell Signaling Technology). Supernatant containing proteins was collected after 10 min centrifugation (10,000 × g at 4°C). Protein concentration was assessed using the Pierce BCA protein assay kit (Thermo Scientific). Protein samples were denaturated at 70°C for 10 min and run on a 4%–12% Bis-Tris gel (NuPAGE Novex, Invitrogen) and then transferred to a PVDF membrane (Millipore). The membrane was blocked with 5% nonfat dry milk and incubated overnight with the primary antibody (rabbit anti-DNMT1, 1/1500; Epigentek), rabbit anti-DNMT3a (1/100; Santa Cruz Biotechnology), rabbit anti-DNMT3B (1/200; Santa Cruz Biotechnology), or rabbit anti-βtubulin (1/10,000; Abcam). The membrane was washed with TBST and then incubated with secondary antibody anti-rabbit HRP (1:10,000; Cell Signaling Technology) for 1 h. Detection and densitometric evaluations were performed using the ECL Western blotting detection reagent (GE Healthcare) and ImageJ software.
Immunohistochemistry.
Three weeks after completion of alcohol exposure, animals were intracardially perfused with 4% PFA-1× PBS. Brains were harvested, postfixed for 2 h, dehydrated in 30% sucrose solution, snap frozen in isopentane, and stored at −80°C. Sections were incubated in rabbit anti-DNMT1 (1 μg/ml; Abcam) or mouse anti-5Mec (1:300, Acris) and chicken anti-NeuN (1:500; EMD Millipore) for 48 h at 4°C and with the secondary antibody for 2 h at room temperature (donkey anti-rabbit 488 for DNMT1, donkey anti-mouse 555 for 5MeC, and donkey anti-chicken 633 for NeuN, 1:1000; Invitrogen). Cells stained positive for NeuN and DNMT1 or 5MeC were quantified using BioQuant Image Analyzer. DNMT1 and 5MeC expression was quantified on Bioquant by measuring the average density of expression per neuron. Average density represents DNMT1 and 5MeC total density divided by total number of neurons.
RNA isolation and whole transcriptome library preparation.
Total RNA was isolated (as described above) and then quantified on a Bioanalyzer (Agilent Technologies). Whole transcriptome sequencing libraries (4 samples/condition, 8 libraries in total) were prepared following the manufacturer's instructions for the Whole Transcriptome Sample Prep Kit (Illumina). Total RNA was ribosomal depleted, fragmented, hybridized, and ligated to 3′ and 5′ adaptor primers, followed by reverse transcription. cDNA was size selected and amplified. Each library template was hybridized to one lane in the flow cell and amplified to form clusters, followed by sequencing.
Whole transcriptome sequencing.
All FASTQ files were uploaded and stored on Simbiot (Umylny, 2012), and all processing was performed using Simbiot system (Genewiz). Quality check was performed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and all sequencing reads were mapped using tophat (Langmead et al., 2009; Trapnell et al., 2009) to Ensembl (Flicek et al., 2012) version 63 of the Rat genome. The mapped results were processed using cufflinks (Trapnell et al., 2012), producing Fragments per Kilobase of transcript per Million mapped reads data matrix, and with htseq-count (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html), producing the raw hit counts data matrix. The raw hit counts gene matrixes was transformed using DESeq (Anders and Huber, 2010) variance stabilization algorithm and normalized using quantile normalization function built into the Bioconductor (Gentleman et al., 2004) Limma (Smyth, 2005) package. The normalized data matrix was then analyzed using Limma and SAMR (Tusher et al., 2001) algorithms.
Pathway analyses of whole transcriptome sequencing data.
Normalized values for each gene were bioinformatically analyzed using Ingenuity Pathways software (IPA, QIAGEN). Methodological details on IPA can be found at http://www.ingenuity.com/science/knowledge_base.html. For IPA analysis, we selected genes from RNA-seq dataset with a 1.2-fold change cutoff and a p value <0.05.
Reverse transcription and qPCR.
cDNA was reverse transcribed from total RNA, and qPCRs were performed using standard TaqMan chemistry and a laser-equipped thermal cycler to detect changes in fluorescence in real time (Applied Biosystems). cDNA concentrations were calculated according to the ΔΔCt method, corrected for differences in PCR efficiency, and normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh).
Statistical analysis.
All data results were analyzed using ANOVA, with factors for the respective analysis indicated in conjunction with its results. When appropriate, post hoc comparisons were performed using the Newman–Keuls test. The accepted level of significance for all tests was p ≤ 0.05.
Results
A history of alcohol dependence leads to DNA hypermethylation in the mPFC
To determine whether history of alcohol dependence was associated with changes in DNA methylation, we first measured global DNA methylation in brain regions associated with addiction. We found that alcohol exposure increased DNA methylation in the mPFC and the nucleus accumbens (NAc) but not the amygdala (Amg) or the hippocampus (Hipp) (one-way ANOVA: F(1–9) = 5.32; p = 0.04; n = 6–8/group; Table 1). We chose to focus our study on the mPFC, which showed the more pronounced change in DNA methylation.
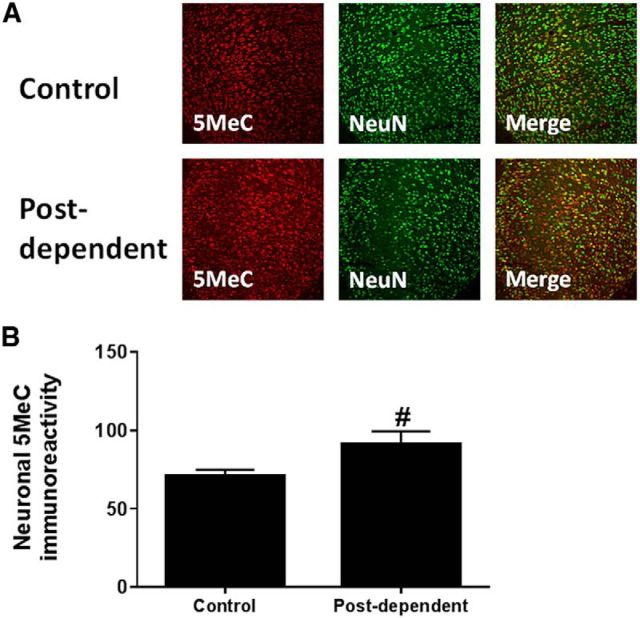
We measured protein expression levels of DNA methyltransferases DNMT1, DNMT3a, and DNMT3b in the mPFC. Western blot analysis showed no changes in protein expression after history of alcohol dependence (n = 8/group; Fig. 2A). However, using an immunohistochemistry approach that allows us to look at specific cell types, we found that DNMT1 was increased specifically in neurons (n = 6 or 7/group; p = 0.03; Fig. 2B,C). These results are consistent with previous studies suggesting that neurons are epigenetically more sensitive to environmental conditions than non-neuronal cells in the CNS (Feng et al., 2010; Iwamoto et al., 2011). Furthermore, we found that increased DNMT1 expression in neuronal cells of mPFC was associated with increased neuronal DNA methylation (n = 12 or 13/group, p < 0.05; Fig. 3).
Figure 2.
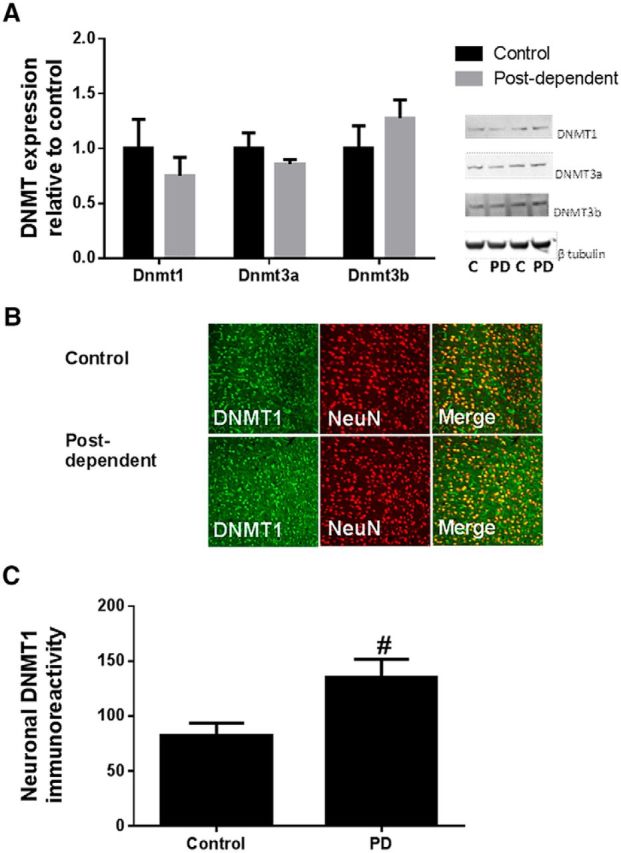
History of alcohol dependence increases DNMT1 expression in neuronal cells in mPFC. A, Western blot analysis shows no difference in DNMT1, DNMT3a, and DNMT3b protein expression. Black bars indicate mean values (± SEM) of control rats. Gray bars indicate mean values (± SEM) of PD rats. Protein expression is represented as DNMT/β-tubulin relative to control. B, C, Immunohistochemical detection of DNMT1 (green) and neuronal cells (red) in the mPFC of control (top) and PD rats (bottom). Right top and bottom, Merge DNMT1/NeuN. C, Average density of DNMT1 in neuronal cells. #p < 0.05, control versus PD rats.
Figure 3.
A, Immunohistochemical detection of 5MeC (red) and neuronal cells (green) in the mPFC of control (top) and PD (bottom). Right top and bottom, Merge 5MeC/NeuN. B, Average density of 5MeC in neuronal cells. #p < 0.05, control versus PD rats.
DNA methylation modulates alcohol intake
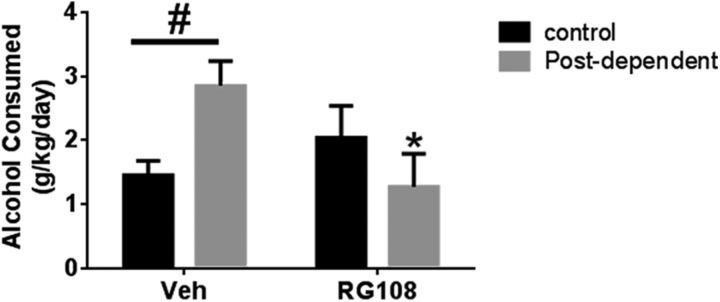
To understand the role of DNA methylation in alcohol-related behaviors, we pharmacologically decreased DNA methylation using the DNMT inhibitor RG108. Similar to what was found by Warnault et al. (2013), we showed that intracerebroventricular infusion of RG108 prevented escalated alcohol intake, which suggests a role of DNA methylation in alcohol consumption. Two-way ANOVA indicated a significant interaction between dependence history (control vs PD) and drug treatment (vehicle vs RG108) (F(1,21) = 8.28; p < 0.01). Post hoc tests showed a significant increase in alcohol consumption in PD compared with controls rats (p < 0.05) and a significant decrease in alcohol consumption in PD-RG108 compared with PD-vehicle rats (p < 0.05; n = 5–7/group; Fig. 4). In addition, RG108 treatment lowered the increased DNA methylation in the mPFC of PD rats but did not influence it in control rats. Two-way ANOVA showed a significant interaction between group and drug treatment (F(1–23) = 4.2; p ≤ 0.05). Post hoc tests showed a significant increase in DNA methylation in the mPFC of PD compared with control rats and a significant decrease in DNA methylation in the mPFC of PD-RG108 compared with PD-vehicle rats (p = 0.01; n = 6–8/group; Table 2).
Figure 4.
Intracerebroventricular infusion of RG108 prevents escalation in alcohol intake in PD rats. Black bars indicate mean values (± SEM) of control rats. Gray bars indicate mean values (± SEM) of PD rats. PD rats show escalated alcohol consumption that is prevented by intracerebroventricular infusion of RG108. #p < 0.05, control versus PD rats. *p < 0.05, vehicle versus RG108.
Escalation of alcohol self-administration following a history of alcohol dependence is in part mediated by DNA hypermethylation in the mPFC
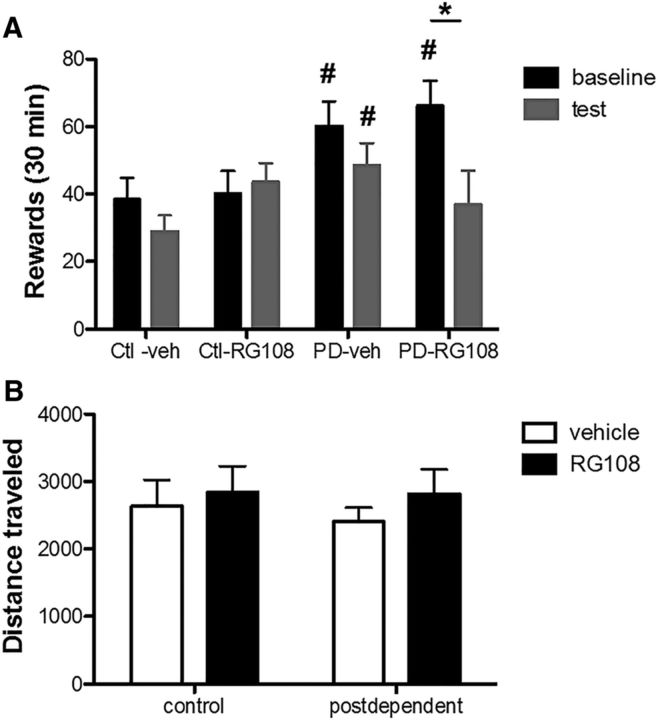
Because DNA methylation was found to be specifically increased in the mPFC and the NAc, we next wanted to determine whether hypermethylation in the mPFC alone is causally related to escalation of alcohol consumption. To address this question, rats received infusion of RG108 directly into the mPFC 3 weeks into protracted abstinence. The dose used for this experiment has previously been shown to significantly decrease DNA methylation (LaPlant et al., 2010). For this study, we assessed alcohol intake using operant self-administration because this methodology is thought to more directly gauge reinforcing properties of alcohol and allows examination of alcohol-taking as well as alcohol-seeking behavior that is in part driven by the mPFC (Koros et al., 1999; Dayas et al., 2007). A history of alcohol dependence resulted in a significant escalation of operant alcohol self-administration (F(1–17) = 0.003; n = 10/group; Fig. 5A) as determined by rewards received during a 30 min session. Baseline was calculated as the last 3 d of self-administration before surgery. Infusion of RG108 directly into mPFC prevented the escalation of alcohol self-administration observed in PD rats but did not influence self-administration rates in control rats. ANOVA indicated a main effect of group (control vs PD: F(1,15) = 5.9; p = 0.02); and an interaction between time (baseline vs test) × group × drug (RG108 vs vehicle: F(1,15) = 5.9; p = 0.03; n = 5/group). Post hoc tests showed a significant decrease in alcohol self-administration in PD-RG108 treated rats compared with PD-vehicle rats but no effect of RG108 on control rats (p < 0.001; Fig. 5A). RG108 treatment did not modify locomotor activity, making it unlikely that nonspecific motor impairment or sedation would account for the findings, and supporting a specific role of RG108 in alcohol consumption (Fig. 5B).
Figure 5.
Infusion of RG108 directly into mPFC prevents escalation in alcohol intake in PD rats. A, Black bars indicate mean values (± SEM) of rats before drug injection. Gray bars indicate mean values (± SEM) of rats after drug injection. Ctl refers to Control rats. Infusion of RG108 into mPFC prevents the increased alcohol consumption observed in the PD compared with control rats. B, RG108 does not modify locomotor activity. White bars indicate mean values (± SEM) of rats treated with vehicle. Black bars indicate mean values (± SEM) of rats treated with RG108. #p < 0.05, control versus PD rats. *p < 0.05, vehicle versus RG108.
Chronic intermittent alcohol exposure regulates expression of genes involved in neurotransmitter release
Whole transcriptome sequencing analysis identified 784 genes with nominally significant changes in expression within the mPFC after a history of alcohol dependence (p < 0.05, no false discovery rate correction; n = 4/group). Full list of gene expression changes between control and PD rats can be found at: http://www.hu.liu.se/ike/forskning/cellbiologi/thorsell-annika?l=sv (click on raw sequencing data). Within this list, bioinformatics analysis identified two overrepresented categories of genes, related to gene expression and neurotransmission (Table 3). Based on a bioinformatics network analysis and the crucial role of neurotransmission for addiction, we selected a subset of 7 genes coding for synaptic proteins for confirmation by qPCR (n = 8/group; Fig. 6A,B). qPCR confirmed that these genes were significantly downregulated in the mPFC of PD rats compared with control rats (t test; p < 0.05). Decreased expression of these genes was specific to the mPFC, as their expression did not change or were increased in the Amg, NAc, or Hipp 3 weeks after chronic alcohol exposure (n = 10/group; Table 4).
Table 3.
Top 6 GO functional enrichment analysis for the list of 784 significantly altered mRNAsa
| Category | Term | Function | Term description | Genes | Count | List total | Pop hits | Pop total | Fold enrichment | p value | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | GO:0006836 | Neurotransmission* | Neurotransmitter transport | SYT1, RAB3C, NOS1AP, SLC6A1, SLC6A11, SYT2, STX1B, RIMS1, PARK7, LIN7A, SLC1A3, SYN1, TRIM9, CACNA1A, NSF, LPHN1 | 16 | 395 | 100 | 12,092 | 4.898025316 | 7.68E-07 | 0.002 | 0.002 | 0.001* |
| GOTERM_BP_FAT | GO:0007269 | Neurotransmitter secretion | SYT1, RAB3C, NOS1AP, SYN1, TRIM9, SYT2, NSF, CACNA1A, LPHN1, LIN7A | 10 | 395 | 52 | 12,092 | 5.887049659 | 4.01E-05 | 0.097 | 0.050 | 0.071 | |
| GOTERM_CC_FAT | GO:0044451 | Regulation of gene expression* | Nucleoplasm part | NR6A1, HR, WBP11, TBP, DMAP1, MEIS1, POLR2B, PNN, ARNT, ERCC5, BRPF1, GTF3C6, GATAD2A, EWSR1, CHD4, MYST3, ATPAF2, RARG, SMAD9, SMAD7, YWHAB, ECSIT, GPS2, MED4, EP300, SMARCE1, RBPSUH, PIAS3, HIPK2, PAF1, HDAC7, NCOR2, MED1 | 33 | 354 | 430 | 10,776 | 2.336145053 | 1.20E-05 | 0.005 | 0.005 | 0.017* |
| GOTERM_CC_FAT | GO:0070013 | Intracellular organelle lumen | NR6A1, HR, TBP, DMAP1, PNN, RBM4B, FAR2, BRPF1, IDH3G, P4HA3, MRPL36, MYST3, ATPAF2, RARG, POLR1E, ERP29, CDK9, FGF22, ECSIT, MED4, EP300, SMARCE1, PIAS3, HIPK2, ATP5C1, RANGRF, PAF1, TGFB1I1, MED1, TXN2, NFKBIE, GLUD1, WBP11, MEIS1, POLR2B, ARNT, LOC684557, ERCC5, TAP1, GTF3C6, GATAD2A, ETV6, NSUN2, CHD4, DNAJA3, EWSR1, LYZ2, SHMT2, SMAD9, SMAD7, PNO1, FDXR, YWHAB, IDH3B, ILF3, GPS2, PEO1, MRPL23, DDX56, PHF2, RBPSUH, PES1, HDAC7, NCOR2, DAP3 | 65 | 354 | 1178 | 10,776 | 1.679663895 | 2.79E-05 | 0.011 | 0.006 | 0.039* | |
| GOTERM_MF_FAT | GO:0003712 | Transcription cofactor activity | DCC, HR, YWHAB, DMAP1, PPARGC1A, ARNT, GPS2, MED4, NCOA2, EP300, ATN1, ZMIZ2, WDR77, BCL11A, HIPK2, TGFB1I1, MKL1, HDAC7, NCOR2, MED1 | 20 | 387 | 164 | 11,963 | 3.769773744 | 1.38E-06 | 0.001 | 0.001 | 0.002* | |
| GOTERM_MF_FAT | GO:0008134 | Transcription factor binding | DCC, HR, TBP, HSPA1A, DMAP1, ARNT, ATN1, WDR77, BCL11A, MKL1, CHD4, MYST3, RARG, YWHAB, PPARGC1A, GPS2, MED4, NCOA2, EP300, RBPSUH, ZMIZ2, HIPK2, TGFB1I1, NCOR2, HDAC7, MED1 | 26 | 387 | 309 | 11,963 | 2.601021884 | 2.28E-05 | 0.016 | 0.008 | 0.034* |
ap < 0.05 (B-H, FDR < 0.2). B-H, Benjamini-Hochberg method; FDR, false discovery rate; GO, gene ontology.
*GO category that reached significance with p < 0.05.
Figure 6.
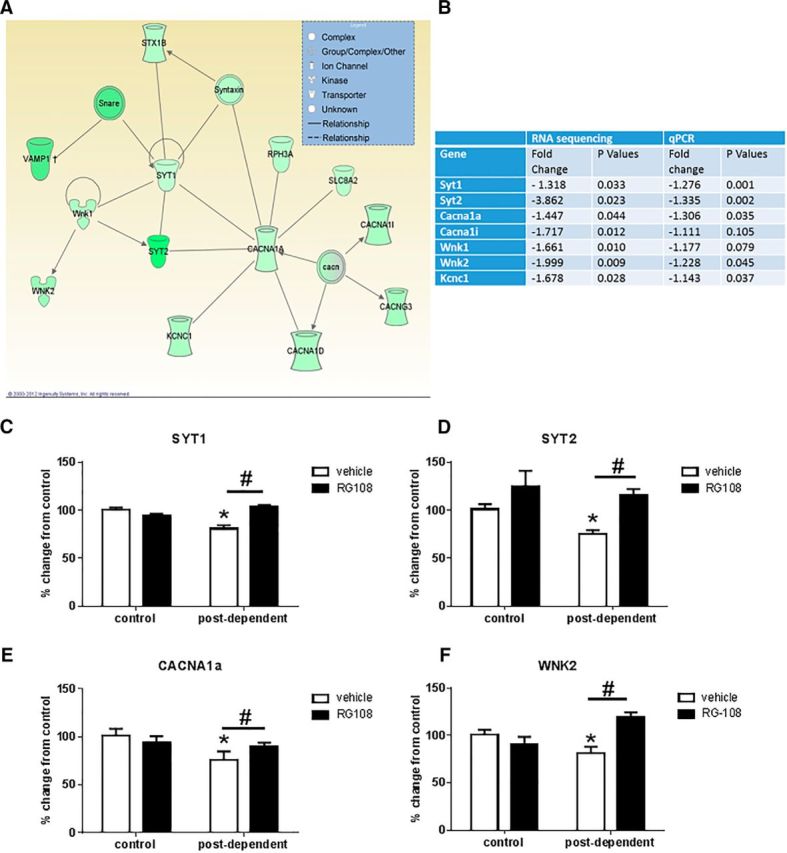
RG108 prevents gene expression changes induced by a history of alcohol dependence. A, Figure from Ingenuity Pathway Analysis showing one of the top gene networks that is downregulated by chronic alcohol exposure. This network includes genes involved in calcium release and exocytosis, two processes critical for synaptic transmission. B, Table shows the fold change in gene expression that is obtained by RNA sequencing and qPCR validation. RG108 prevents alcohol-induced decrease expression of syt1 (C), syt2 (D), cacna1a (E), and wnk2 (F). White bars indicate mean values (± SEM) of rats treated with vehicle. Black bars indicate mean values (± SEM) of rats treated with RG108. #p < 0.05, control versus PD rats. *p < 0.05, vehicle versus RG108.
Table 4.
RG108 treatment does not regulate the expression cacna1a, syt1, syt2, cacna1l, Slc8a2, and wnk2 in the Amg, NAc, and Hippa
| CTL/vehicle |
CTL/RG108 |
PD/vehicle |
PD/RG108 |
|||||
|---|---|---|---|---|---|---|---|---|
| Fold change | SEM | Fold change | SEM | Fold change | SEM | Fold change | SEM | |
| Amg | ||||||||
| Cacna1a | 1.01 | 0.05 | −1.02 | 0.07 | −1.13 | 0.04 | −1.12 | 0.05 |
| Syt1 | 1.01 | 0.04 | 1.01 | 0.04 | −1.05 | 0.05 | −1.05 | 0.05 |
| Syt2 | 1.02 | 0.07 | −1.09 | 0.09 | −1.24 | 0.11 | −1.12 | 0.05 |
| Cacna1l | 1.01 | 0.06 | 1.10 | 0.06 | 1.10 | 0.12 | 1.05 | 0.06 |
| Slc8a2 | 1.02 | 0.08 | 1.19 | 0.10 | 1.56 | 0.23 | 1.26 | 0.11 |
| wnk2 | 1.01 | 0.05 | 1.08 | 0.06 | 1.19* | 0.05 | 1.32* | 0.14 |
| Hipp | ||||||||
| Cacna1a | 1.01 | 0.06 | 1.03 | 0.05 | 1.07 | 0.05 | 1.24 | 0.11 |
| Syt1 | 1.01 | 0.04 | 1.15 | 0.03 | 1.25 | 0.05 | 1.09 | 0.08 |
| Syt2 | 1.02 | 0.08 | 1.32 | 0.15 | 1.52* | 0.13 | 1.50* | 0.14 |
| Cacna1l | 1.01 | 0.07 | 1.00 | 0.06 | 1.11 | 0.11 | 1.18 | 0.11 |
| Slc8a2 | 1.02 | 0.08 | 1.05 | 0.06 | 1.20* | 0.06 | 1.17* | 0.05 |
| Wnk2 | 1.01 | 0.06 | 0.81 | 0.04 | 1.29* | 0.16 | 1.01 | 0.05 |
| NAc | ||||||||
| Cacna1a | 1.12 | 0.19 | 1.29 | 0.12 | 1.30 | 0.15 | 1.19 | 0.13 |
| Syt1 | 1.09 | 0.17 | 1.19 | 0.09 | 1.12 | 0.09 | 1.02 | 0.07 |
| Syt2 | 1.24 | 0.38 | 1.53 | 0.23 | 1.85 | 0.49 | 1.42 | 0.26 |
| Cacna1l | 1.02 | 0.08 | −1.06 | 0.08 | 1.00 | 0.10 | 0.88 | 0.09 |
| Slc8a2 | 1.02 | 0.07 | −1.07 | 0.07 | −1.03 | 0.08 | −1.22 | 0.07 |
| Wnk2 | 1.01 | 0.06 | −1.02 | 0.06 | 1.02 | 0.06 | −1.19 | 0.05 |
amRNA levels (fold change) of cacna1a, syt1, syt2, cacna1l, Slc8a2, and wnk2 in the Amg, NAc, and Hipp of control and PD rats treated with vehicle or RG108.
*p < 0.05, control versus PD rats.
Synaptic transmission genes repressed by chronic intermittent alcohol exposure are rescued by DNMT inhibition using RG108
Next, to assess whether DNA hypermethylation accounts for the expression changes of the qPCR validated differentially expressed genes, we investigated whether RG108 treatment into mPFC reversed the persistent gene expression changes (Fig. 6C–F). RG108 treatment prevented the downregulation of 4 of the 7 genes (Syt1, Syt2, Wnk2, and Cacna1a; n = 7 or 8/group) that were confirmed by qPCR. Two-way ANOVA showed a main effect of drug (vehicle vs RG108) on Syt2 (F(1–25) = 14.16; p < 0.001), Syt1 (F(1–28) = 9.72; p < 0.005), and Wnk2 (F(1–28) = 4.01; p = 0.05) transcripts. A significant interaction (treatment × drug) was found for Cacna1a (F(1–28) = 3.988; p = 0.05), Wnk2 (F(1–28) = 10.30; p = 0.003), and Syt1 (F(1–28) = 25.65; p < 0.0001). Post hoc analysis showed a significant difference in Wnk2, Syt1, and Cacna1 mRNA levels between vehicle- and RG108-treated PD rats. In contrast, RG108 had no effect in controls.
Intracerebroventricular infusion of RG108 did not influence the expression of these transcripts in the Amg, NAc, and or Hipp (Table 4), suggesting a specific effect of RG108 in the mPFC. Together, these findings suggest that chronic intermittent exposure to alcohol persistently decreases the expression of these synaptic genes through increased DNA methylation.
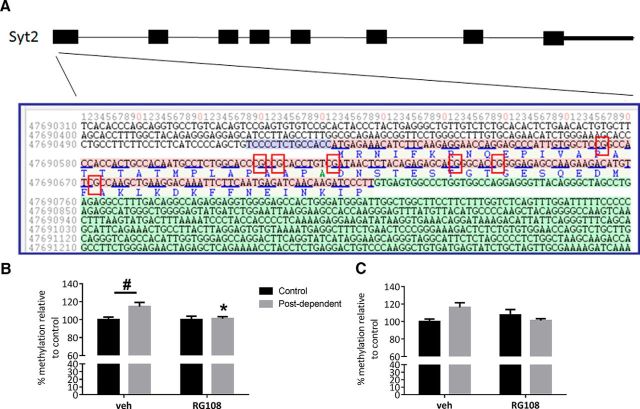
RG108 treatment prevented hypermethylation on exon 1 of Syt2 induced by chronic intermittent alcohol exposure
To determine whether alcohol exposure directly decreases the expression of these synaptic transmission genes through increased DNA methylation, we measured DNA methylation levels on the promoter region and exon 1 of Syt2 and on the promoter region of Cacna1a in the mPFC by pyrosequencing (n = 6–8). We found that CpG#5 on exon 1 of Syt2 was significantly hypermethylated in PD rats compared with control rats and that this hypermethylation was prevented by RG108 treatment (Fig. 7A,B). Two-way ANOVA showed a significant interaction of group × drug (F(1–23) = 4.31; p < 0.05). Post hoc tests showed a significant increase in DNA methylation in the mPFC of PD rats compared with control rats (p = 0.02) and a significant decrease of DNA methylation in PD-RG108 rats compared with PD-vehicle rats (p = 0.007). Interestingly, we found similar results for CpG site 6, which is next to CpG#5 (Fig. 7C). Two-way ANOVA showed a significant interaction group × drug (F(1–23) = 4.31; p = 0.03). Post hoc tests showed an increase in DNA methylation in mPFC of PD compared with control rats that is close to significance (p = 0.06). Like promoter-silencing hypermethylation, increased DNA methylation on exon 1 is associated with gene silencing (Brenet et al., 2011). Therefore, these results suggest that alcohol consumption may decrease the expression of Syt2 through increased DNA methylation on its exon 1. In contrast, DNA methylation levels on the promoter region of Cacna1a were similar between control and PD rats, suggesting that RG108 regulates Cacna1a expression through indirect mechanisms (data not shown).
Figure 7.
History of alcohol dependence increases DNA methylation on exon 1 of syt2. A, Figure shows sequence of exon 1 of syt2 and the location of the 7 CpG sites. Bar graph represents DNA methylation level (%) at CpG#5 (B) and at CpG#6 (C) on exon 1. Black bars indicate mean values (± SEM) of control rats. Gray bars indicate mean values (± SEM) of PD rats. #p < 0.05, control versus PD rats. *p < 0.05 vehicle versus RG108.
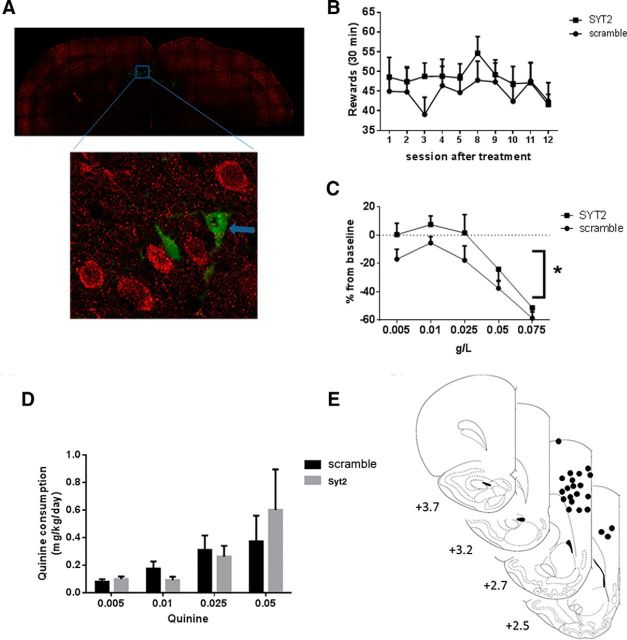
SYT2 knockdown contributes to compulsive-like drinking
To determine whether decreased expression of Syt2 plays a functional role in alcohol-related behaviors induced by a history of alcohol dependence, we inhibited Syt2 expression specifically in the mPFC of naive rats using a lentiviral vector. Cells infected by our “shRNA Syt2 lentiviral vector” show no expression of SYT2, confirming that it inhibited its expression (Fig. 8A). Syt2 inhibition did not modify alcohol self-administration rates (n = 7–9; Fig. 8B). However, cortical Syt2 inhibition resulted in tolerance to quinine adulteration, suggesting a compulsive-like behavior similar to that observed in alcohol dependence (Vendruscolo et al., 2012). Repeated-measures ANOVA showed a main effect of treatment (F(1,56) = 4.57; p ≤ 0.05; Fig. 8C). Cortical Syt2 inhibition did not affect taste perception as rats with Syt2 inhibition drank similar amounts of quinine solution as rats injected with the scrambled virus (Fig. 8D,E).
Figure 8.
Syt2 inhibition increases tolerance to quinine adulteration. A, Immunohistochemical detection of syt2 (red) and cell infected by shRNA lentiviral vector specific to syt2. The figure shows that cells infected by lentivirus do not express SYT2 (green). B, Number of reward for alcohol in rats that received injection of shRNA lentiviral vector specific to Syt2 (square) and rats that received the scrambled lentiviral vector. C, Compulsive-like drinking (i.e., persistent alcohol drinking despite the aversive bitter taste of quinine added to the alcohol solution). The data represent the percentage change from baseline (i.e., lever presses for alcohol alone before adulteration with quinine). *p < 0.05; scramble vs. Syt2. D, Syt2 inhibition does not modify quinine consumption. Bar graph represents quinine consumption as measured by two-bottle free choice. E, Graph shows mapping of viral injection sites within the mPFC (•).
Discussion
In this study we sought to determine the epigenetic events that occur during protracted abstinence from alcohol dependence a stage that in human alcoholics is associated with the emergence of alcohol craving and high relapse risk (Heilig et al., 2010). We showed that a history of alcohol dependence is associated with increased DNA methylation specifically in the mPFC. Importantly, we demonstrated that DNA methylation is causally related to alcohol intake and seeking behaviors. Furthermore, our gene expression analysis suggests that DNA methylation regulates alcohol-induced neurotransmission-related gene expression changes. For instance, Syt2, one of the neurotransmission-related genes found to be altered, had increased DNA methylation at 2 CpG sites in exon 1. RG108 treatment was able to restore both Syt2 expression and DNA methylation levels, suggesting a direct role of DNA methylation for Syt2 silencing. Finally, we identified a causal role of Syt2 in compulsivity-like behavior, a hallmark of alcohol dependence.
Convergent evidence suggests that upregulated DNMT1 contributes to the maintenance of alcohol-induced DNA hypermethylation. First, RG108 has been designed to selectively target the catalytic domain of DNMT1 (Brueckner et al., 2005). Second, DNMT1 was upregulated in the mPFC after repeated cycles of alcohol intoxication. Third, increased DNMT1 expression was associated with increased DNA methylation in neuronal cell. Finally, DNMT1 is the main enzyme that maintains DNA methylation, the mechanisms that we inhibited with RG108. Rather than inducing de novo methylation, the main role of DNMT1 is to maintain DNA methylation already in place. We therefore do not believe that elevated activity of DNMT1 caused DNA hypermethylation as such in our study. Instead, it is more likely that it contributed to maintaining alcohol-induced DNA hypermethylation, which was already in place 3 weeks after alcohol exposure, before RG108 was administered. Our hypothesis is that repeated cycles of alcohol intoxication followed by protracted abstinence induced DNA hypermethylation, which was then maintained by DNMTs and specifically DNMT1. However, we cannot rule out a possible interaction between DNMT1 and the other DNMT enzymes, specifically DNMT3a, which can interact with DNMT1 to maintain DNA methylation (Feng et al., 2010; Iwamoto et al., 2011). Furthermore, although RG108 was designed to inhibit DNMT, it is unclear whether it may also affect DNMT3a and DNMT3b activity.
To identify the molecular mechanisms through which DNA methylation might affect alcohol consumption, we investigated transcriptome changes induced by a history of alcohol dependence and assessed whether some of these changes may be driven by DNA hypermethylation. Because the magnitude of long-term neuronal gene expression changes is frequently small and easily lost in attempts to apply transcriptome-wide false discovery rate corrections, we relied on nominal expression values and bioinformatics pathway analysis for our discovery effort and validated the functionally relevant candidate hits by qPCR.
Using this strategy, we found that a history of alcohol dependence had a global effect in the mPFC on a gene network that includes proteins regulating synaptic vesicle formation and function, such as Syt1, Syt2, Wnk2, and Cacna1a. These data suggest a persistent dysregulation of synaptic transmission in PD mPFC neurons that may be responsible for the escalated alcohol consumption we observed following a history of alcohol dependence. These genes regulate Ca2+-evoked neurotransmitter release (Chapman, 2008) and may therefore be involved in changes in neurotransmitter release observed after alcohol exposure. Consistent with our data, Worst et al. (2005) observed a downregulation of Syt1 expression in the prefrontal cortex of Alko alcohol accepting rats compared with their alcohol nonaccepting counterparts. Interestingly, a recent study from Varodayan et al. (2011) showed that the expression of Syt1 is upregulated in mouse cortical neurons following a single acute alcohol exposure. If these tissue culture findings reflect processes that occur in vivo, these observations collectively suggest the possibility of an allostatic process, in which acute upregulation of Syt1 expression and function by alcohol are followed by a persistent downregulation to a new set point.
Importantly, we demonstrated that RG108 can reverse >50% of the persistent gene expression changes, supporting that they are mediated by DNA hypermethylation. The observation that RG108 did not uniformly rescue all gene expression changes is unsurprising and suggests that DNA methylation does not account for all alcohol-induced neuroadaptations. However, DNA methylation can have a significant impact on genes involved in important neuronal functions, such as neurotransmitter release. Because of the complexity of how DNA methylation is regulated, we chose to first focus on the role of DNA methylation on select genes to begin elucidating the possible functional contribution of at least some of them.
We show two observations likely pointing to two different mechanisms through which DNA methylation can regulate gene expression in the PD state. For instance, we found that repression of cacna1a is likely to be indirectly mediated by increased DNA methylation. Indeed, although RG108 treatment restored cacna1a expression levels, DNA methylation profile of cacna1a was not altered by alcohol exposure. In contrast, Syt2 seems to be directly regulated by DNA methylation. Our pyrosequencing data showed that repeated alcohol intoxication induced hypermethylation on exon 1 of Syt2. Similar to promoter hypermethylation, DNA methylation on exon 1 is associated with gene silencing (Brenet et al., 2011). Furthermore, RG108 infusions restored both Syt2 and DNA methylation levels, suggesting that repeated cycles of alcohol intoxication followed by protracted abstinence downregulated Syt2 through increased DNA methylation.
Recent publications suggest that DNA can be actively demethylated (Wu and Zhang, 2014). It is possible that, when DNMT enzymes are inhibited, active demethylation reverses alcohol-induced hypermethylation and therefore restores gene expression levels. Additional research will be required to examine this hypothesis.
Furthermore, we found that Syt2 has a mechanistic role in alcohol dependence-induced behaviors. Inhibition of Syt2 directly in the mPFC induced compulsivity-like behavior, thought to be one of the hallmarks of alcohol dependence. In contrast, Syt2 inhibition did not modify alcohol self-administration, suggesting that Syt2 alone may not be sufficient to cause an escalation in alcohol consumption after alcohol dependence. Principal component analysis of behaviors assessed PD animals from a prior study (Vendruscolo et al., 2012) indicates that compulsivity-like, quinine-resistant alcohol consumption and escalation of self-administration rates load on separate factors (data not shown). Overall, interaction between Syt2 and other synaptic transmission genes may be necessary to alter alcohol consumption in PD rats. This points to the possibility that the complex behavioral phenotype induced by a history of alcohol dependence is driven by multiple factors and that therapeutics may have better prospect of being effective if they target pathways rather than individual molecular targets. In that context, DNMT inhibition may offer an attractive therapeutic mechanism as it can simultaneously control the expression of multiple genes. Gene expression regulation by DNA methylation is very complex, and future experiments investigating genome-wide changes in alcohol-induced DNA methylation will be needed to better understand the mechanisms through which DNA methylation regulates alcohol-induced neuroadaptations.
In conclusion, we found that DNMT inhibition prevented both escalated alcohol intake and gene expression changes induced by a history of alcohol dependence. Thus, DNMT inhibitors may have a potential to be pharmacotherapies for alcohol dependence. DNMT inhibitors, such as 5-azacytidine and 5-aza-2′-deoxycytidine, are currently FDA approved for the treatment of myelodysplastic syndrome (Fandy, 2009). Our findings provide an initial rationale for exploring the potential of these or other DNMT inhibitors as a treatment for alcohol dependence.
Notes
Supplemental material for this article is available at http://www.hu.liu.se/ike/forskning/cellbiologi/thorsell-annika?l=sv. This material has not been peer reviewed.
Footnotes
We thank Karen Smith and Juan Rivas for assistance in preparing the manuscript; and Lauren Bell and Michelle Zook for help with the animal behavior study.
The authors declare that, except for income received from our primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service, and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
References
- Agudelo M, Gandhi N, Saiyed Z, Pichili V, Thangavel S, Khatavkar P, Yndart-Arias A, Nair M. Effects of alcohol on histone deacetylase 2 (HDAC2) and the neuroprotective role of trichostatin A (TSA) Alcohol Clin Exp Res. 2011;35:1550–1556. doi: 10.1111/j.1530-0277.2011.01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk K, Saarikoski ST, Arlinde C, Kovanen L, Osei-Hyiaman D, Ubaldi M, Reimers M, Hyytiä P, Heilig M, Sommer WH. Glutathione-S-transferase expression in the brain: possible role in ethanol preference and longevity. FASEB J. 2006;20:1826–1835. doi: 10.1096/fj.06-5896com. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- Cippitelli A, Karlsson C, Shaw JL, Thorsell A, Gehlert DR, Heilig M. Suppression of alcohol self-administration and reinstatement of alcohol seeking by melanin-concentrating hormone receptor 1 (MCH1-R) antagonism in Wistar rats. Psychopharmacology (Berl) 2010;211:367–375. doi: 10.1007/s00213-010-1891-y. [DOI] [PubMed] [Google Scholar]
- Dayas CV, Liu X, Simms JA, Weiss F. Distinct patterns of neural activation associated with ethanol seeking: effects of naltrexone. Biol Psychiatry. 2007;61:979–989. doi: 10.1016/j.biopsych.2006.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16:2075–2085. doi: 10.2174/092986709788612738. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri AK, Keefe D, Keenan S, Kinsella R, Komorowska M, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M, Crabbe JC, Becker HC. Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addict Biol. 2010;15:169–184. doi: 10.1111/j.1369-1600.2009.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, Saito T, Geschwind DH, Kato T. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW. Addiction as a pathology in prefrontal cortical regulation of corticostriatal habit circuitry. Neurotox Res. 2008;14:185–189. doi: 10.1007/BF03033809. [DOI] [PubMed] [Google Scholar]
- Koros E, Kostowski W, Bienkowski P. Operant responding for ethanol in rats with a long-term history of free-choice ethanol drinking. Alcohol Alcohol. 1999;34:685–689. doi: 10.1093/alcalc/34.5.685. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. FASEB J. 2002;16:27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36:61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Baxstrom K, Shi G, Moonat S, Pandey SC. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: a role in anxiety-like and alcohol-drinking behaviours. Int J Neuropsychopharmacol. 2014;17:1207–1220. doi: 10.1017/S1461145714000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Bioinformatics and computational biology solutions using R and bioconductor. Springer: New York; 2005. Limma: linear models for microarray data. [Google Scholar]
- Tabakoff B, Cornell N, Hoffman PL. Alcohol tolerance. Ann Emerg Med. 1986;15:1005–1012. doi: 10.1016/S0196-0644(86)80119-6. [DOI] [PubMed] [Google Scholar]
- Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR, Schwandt M, Sommer WH, Heilig M. Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J. 2012;13:286–296. doi: 10.1038/tpj.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A, Sun H, Schank JR, King C, Heilig M. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci. 2014;34:4581–4588. doi: 10.1523/JNEUROSCI.0445-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Zhao M, Li M, Song T, Zhang M, Quan L, Li S, Sun ZS. Reversal of cocaine-conditioned place preference through methyl supplementation in mice: altering global DNA methylation in the prefrontal cortex. PLoS One. 2012;7:e33435. doi: 10.1371/journal.pone.0033435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschentke TM. The medial prefrontal cortex as a part of the brain reward system. Amino Acids. 2000;19:211–219. doi: 10.1007/s007260070051. [DOI] [PubMed] [Google Scholar]
- Umylny BWSJ. Beyond the pipelines: Clous computing facilitates management, distribution, security and analysis of high speed sequencer data. In: Harbers M, Kahl G, editors. Tag-based next generation sequencing. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2012. [Google Scholar]
- Varodayan FP, Pignataro L, Harrison NL. Alcohol induces synaptotagmin 1 expression in neurons via activation of heat shock factor 1. Neuroscience. 2011;193:63–71. doi: 10.1016/j.neuroscience.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo LF, Barbier E, Schlosburg JE, Misra KK, Whitfield TW, Jr, Logrip ML, Rivier C, Repunte-Canonigo V, Zorrilla EP, Sanna PP, Heilig M, Koob GF. Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. J Neurosci. 2012;32:7563–7571. doi: 10.1523/JNEUROSCI.0069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling: a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wong CC, Mill J, Fernandes C. Drugs and addiction: an introduction to epigenetics. Addiction. 2011;106:480–489. doi: 10.1111/j.1360-0443.2010.03321.x. [DOI] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, Vrana KE. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80:529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]