Summary
Breakpoint junctions of the chromosomal translocations that occur in human cancers display hallmarks of nonhomologous end-joining (NHEJ). In mouse cells, translocations are suppressed by canonical NHEJ (c-NHEJ) components, which include DNA ligase IV (LIG4), and instead arise from alternative NHEJ (alt-NHEJ). Here we used designer nucleases (ZFNs, TALENs, and CRISPR/Cas9) to introduce DSBs on two chromosomes to study translocation joining mechanisms in human cells. Remarkably, translocations were altered in cells deficient for LIG4 or its interacting protein XRCC4. Translocation junctions had significantly longer deletions and more microhomology, indicative of alt-NHEJ. Thus, unlike mouse cells, translocations in human cells are generated by c-NHEJ. Human cancer translocations induced by paired Cas9 nicks also showed a dependence on c-NHEJ, despite having distinct joining characteristics. These results demonstrate an unexpected and striking species-specific difference for common genomic rearrangements associated with tumorigenesis.
Introduction
Recurrent reciprocal chromosomal translocations are associated with oncogenesis (Mani and Chinnaiyan, 2010; Mitelman et al., 2007). Translocations frequently generate fusion genes with novel properties that drive oncogenesis; to date more than 300 genes have been associated with oncogenic translocations in both haematological malignancies and solid tumors. In addition to generating fusion genes, translocations can also enhance the expression of proto-oncogenes, the classic example of which results in c-Myc overexpression. Breakpoint junction analysis has demonstrated that oncogenic translocations typically arise by some form of non-homologous end-joining (NHEJ).
The canonical pathway of NHEJ (c-NHEJ) is required for cellular resistance to ionizing radiation as well as for immune system rearrangements and is active throughout the cell cycle (Deriano and Roth, 2013; Goodarzi and Jeggo, 2013; Pannunzio et al., 2014). Critical components of c-NHEJ include the Ku70/80 heterodimer, DNA-PKcs, DNA ligase IV (LIG4), and XRCC4. Loss of c-NHEJ components does not, however, completely abrogate NHEJ (Delacote et al., 2002; Kabotyanski et al., 1998; Liang and Jasin, 1996), suggesting that there are alternative ways to join ends, referred to as alt-NHEJ. Whether alt-NHEJ is a distinct, regulated pathway(s) or involves the co-opting of non-c-NHEJ proteins with some c-NHEJ components is a subject of debate (Deriano and Roth, 2013; Goodarzi and Jeggo, 2013; Pannunzio et al., 2014). Junctions that form by alt-NHEJ have more microhomology and longer deletions than junctions formed by c-NHEJ (Fattah et al., 2010; Guirouilh-Barbat et al., 2007; Kabotyanski et al., 1998; Oh et al., 2013; Simsek and Jasin, 2010; Smith et al., 2003). Proteins that promote alt-NHEJ include the end resection factor CtIP (Bennardo et al., 2008) and LIG3α (Wang et al., 2005).
Most studies analyzing translocation formation have been performed in mouse cells, in particular in lymphoid cells involving programmed DSBs and embryonic stem cells using I-SceI or zinc finger nuclease (ZFN)-generated breaks (Boboila et al., 2012a; Nussenzweig and Nussenzweig, 2010; Weinstock et al., 2007; Simsek et al., 2011a). These studies uniformly demonstrated that c-NHEJ suppresses translocation formation at nonhomologous sequences. Thus, in the absence of Ku, LIG4, or XRCC4, translocations are increased in frequency. Since alt-NHEJ proteins CtIP and LIG3 promote translocation formation (Zhang and Jasin, 2011; Simsek et al., 2011a) and translocation junction sequences in wild-type and c-NHEJ mutants have similar characteristics, it appears that translocations in mouse cells typically arise by alt-NHEJ.
In contrast to mouse cells, translocation junctions in human tumors do not always show significant lengths of microhomology (Gillert et al., 1999; Langer et al., 2003; Zucman-Rossi et al., 1998; Mattarucchi et al., 2008). Similarly, cancer and model translocations induced by nucleases in several human cell lines also show little or no microhomology at translocation junctions (Brunet et al., 2009; Piganeau et al., 2013). Studies in human cells deficient in c-NHEJ are limited. Ionizing radiation, a potent inducer of translocations in rodent cells, does not significantly induce translocations in a LIG4 mutant human cell line (Soni et al., 2014). In contrast, knockdown of c-NHEJ components did decrease androgen-induced translocations, although junction analysis was not reported (Lin et al., 2009).
To address the role of NHEJ pathways in the joining phase of chromosomal translocation formation, we took advantage of nucleases designed to introduce site-specific DSBs at endogenous loci in human cells (Gaj et al., 2013; Urnov et al., 2010) to induce translocations (Brunet et al., 2009; Piganeau et al., 2013). Using multiple cell lines and different nucleases to provoke DSBs, we found that the translocation frequency was often reduced in human cells in the absence of LIG4 or XRCC4, in stark contrast to results from c-NHEJ-deficient mouse cells. The translocations that were formed in human c-NHEJ mutants had frequent microhomologies and long deletions. Consistent with a requirement for c-NHEJ, loss of alt-NHEJ components did not affect translocation formation, unless c-NHEJ was also impaired. We also found that different types of end structures gave rise to different joining characteristics in wild-type cells. Translocations induced by wild-type Cas9 frequently had precisely joined ends, indicating that c-NHEJ can be highly accurate, whereas those induced by Cas9 nickase (nCas9) had more varied junctions; in either case, the absence of LIG4 led to greater inaccuracy in joining. Thus, our studies reveal an unexpected and striking species-specific difference in the generation of these oncogenic rearrangements.
Results
Intrachromosomal DSB repair is altered in c-NHEJ-deficient human cells
To analyze the repair of chromosomal DSBs in c-NHEJ-deficient human cells, we used LIG4 and XRCC4 mutant HCT116 cell lines (Oh et al., 2013; B.R. and E.A.H., in preparation). LIG4 null (L4−/−) cells are deficient in LIG4 but maintain XRCC4 expression; XRCC4 mutant (X4−/−) cells are deficient in both XRCC4 and LIG4, because LIG4 is unstable in the absence of XRCC4 (Bryans et al., 1999) (Figure 1A). We expressed ZFNp84, which cleaves the p84/AAVS1 locus (Brunet et al., 2009) and estimated the insertion/deletion (indel) frequency resulting from NHEJ at this site. In both X4−/− and L4−/− cells, the indel frequency was approximately half that of control cells (Figure S1A), suggesting reduced NHEJ. A similar reduction in indel frequency was obtained at a second locus with other nucleases (TALENs, Cas9, nCas9; see below, Figure 4C).
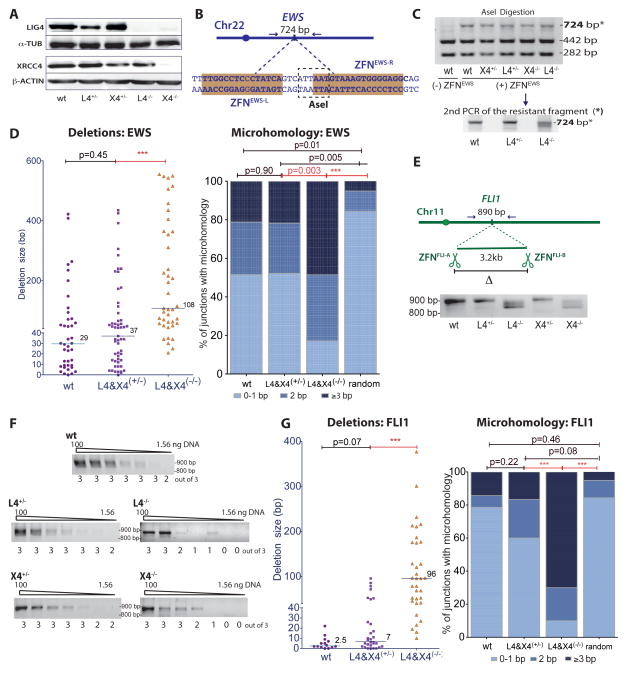
Figure 1. Intrachromosomal DSB repair in c-NHEJ deficient human cells is inefficient and shows a shift towards longer deletions and microhomology.
A. Absence of LIG4 in both L4−/− and X4−/− HCT116 cells.
B. The ZFNEWS cleavage site overlaps an AseI restriction site.
C. After ZFNEWS expression, AseI-resistant PCR products (*: 724bp) were purified and re-amplified (examples shown for wt, L4+/−, and L4−/− cells) for sequencing.
D. AseI-resistant NHEJ junctions at the ZFNEWS site from L4−/− and X4−/− cells demonstrate a shift towards longer deletions and increased microhomology (wt, n = 4; L4, n = 2; X4, n = 3). Junctions from L4 and X4 heterozygous or deficient cell lines are combined here and below, although results from individual cell lines were similar. The median deletion length is indicated, and each value represents the combined deletion from both ends of an individual junction. In the microhomology analysis, junctions that contain insertions were not included. Deletion and microhomology distributions here and below were compared by Mann-Whitney analysis. For random microhomology distribution, the probability that a junction will have microhomology by chance assumes an unbiased base composition (Roth et al. 1985). ***, p < 0.0001.
E. Two DSBs 3.2 kb apart were introduced by ZFNFLI-A and ZFNFLI-B in a FLI1 gene intron. To detect the 3.2-kb deletion, an ~890 bp fragment was PCR amplified using primers flanking the two DSBs.
F. Limiting dilution of genomic DNA to estimate the frequency of deletions after expression of ZFNFLI-A and ZFNFLI-B. PCR amplification of the 3.2-kb deletion product was performed with serial dilutions of genomic DNA (100, 50, 25, 12.5, 6.25, 3.125, and 1.56 ng). The number of times a PCR fragment was detected from 3 independent amplifications is indicated below the gel.
G. Junctions from joining two DSBs 3.2 kb apart at the FLI1 locus from L4−/− and X4−/− cells demonstrate a shift towards longer deletions and increased microhomology.
See also Figure S1.
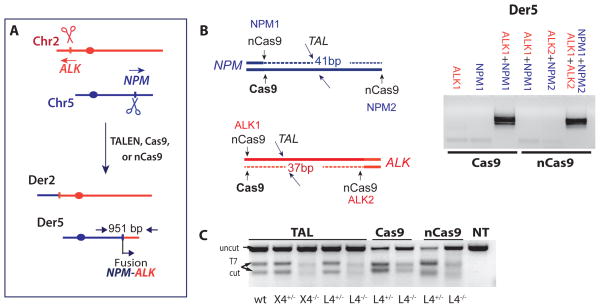
Figure 4. Cancer translocation induced by paired nicks and DSBs.
A. Induction of NPM-ALK cancer translocations with sequence specific nucleases. Der5 encodes the NPM-ALK fusion.
B. Der5 is detected only when DSBs or paired nicks are induced on both chromosomes. Wild-type Cas9 with gRNAs for NPM1 and ALK1 gives rise to translocations as does nCas9 with gRNAs NPM1+NPM2 and ALK1+ALK2. Relative positions of cleavage sites are indicated.
C. Indel formation at the ALK locus, as monitored by the T7-endonuclease assay.
See also Figure S4.
Junction characteristics were examined in the c-NHEJ mutants using ZFNEWS, which cleaves the EWS locus at an AseI restriction site (Piganeau et al., 2013) (Figure 1B, C). AseI-resistant junctions from X4−/− and L4−/− cells had longer deletions compared to control cells (p < 0.0001, Mann-Whitney test; Figures 1D, S1B). Microhomology at the junctions was also altered in the X4−/− and L4−/− cells, such that 49% of junctions had ≥ 3 bp microhomology in contrast to only ~20% of junctions in control cells (p = 0.003; Figure 1D). For both mutant and control cells, the microhomology distribution was different from that expected by the random occurrence of microhomology; however, the deviation from random was especially apparent for the mutant cells (p < 0.0001), providing strong evidence that microhomology is critical in the joining process in the absence of LIG4.
To determine if repair of two DSBs by deletion formation has the same dependency on c-NHEJ as the repair of a single DSB, we induced DSBs 3.2 kb apart within the FLI1 gene with ZFNs (Figure 1E). A fragment corresponding to the 3.2 kb deletion product was PCR amplified using primers that flanked the two DSBs. By limiting dilution, the deletion product was estimated to be ~4 to 8-fold less abundant in the mutant cells (Figure 1F). The PCR product showed minimal deletion from the two DNA ends in control cells, whereas it was frequently shorter in X4−/− and L4−/− cells (Figure 1E). Sequencing confirmed that many of the junctions from control cells occurred with minimal processing of the DNA ends, whereas long deletions were common in the mutant cells (median = 96 bp; Figures 1G, S1C). Further, ≥ 3 bp microhomology was infrequent at junctions from control cells but common at junctions from X4−/− and L4−/− cells (≤ 20% and ~70% of junctions, respectively). Thus, as with repair of a single DSB, intrachromosomal repair of two DSBs in human cells requires the LIG4:XRCC4 complex, similar to rodent cells (Guirouilh-Barbat et al., 2007; Simsek and Jasin, 2010).
Chromosomal translocations rely on c-NHEJ in human cells
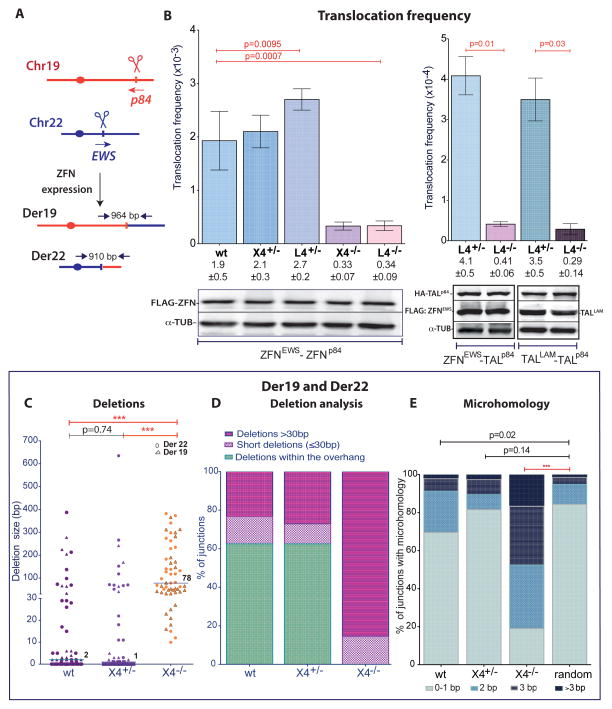
To determine the mechanism by which DSBs give rise to chromosomal translocations in human cells, we coexpressed ZFNp84 and ZFNEWS to induce concomitant DSBs on Chr19 and Chr22, respectively (Figure 2A). The translocation frequency was determined by PCR screening of small pools of cells (Brunet et al., 2009; Piganeau et al., 2013), amplifying a > 900 bp fragment for each derivative chromosome. Translocation frequencies were 5- to 6-fold lower for L4−/− and X4−/− cells compared to control cells (Figure 2B), indicating a reliance on c-NHEJ for efficient joining. Since ZFN protein levels were similar in all cell lines (Figure 2B), the lower translocation frequency was not due to reduced ZFN expression. To confirm that the reduced frequency was not specific to a particular ZFN pair, ZFNEWS was paired with a TALEN, TALp84, which cleaves close to the ZFNp84 site; TALp84 was also paired with another TALEN, TALLAM, which cleaves a locus on Chr1. Like ZFNs, TALENs generate 5′-overhangs, although the overhangs are more variable (Piganeau et al., 2013). Importantly, L4−/− cells also showed lower translocation frequencies with these nuclease pairs (Figure 2B).
Figure 2. c-NHEJ generates chromosomal translocations in HCT116 human cells.
A. Induction of chromosomal translocations with sequence-specific nucleases. Derivative chromosomes. Der19 and Der22 were detected by PCR using primers that flank the cleavage sites after ZFNp84 and ZFNEWS expression.
B. Translocation frequency is reduced in L4−/− and X4−/− HCT116 cells. Translocations were quantified as follows: ZFNEWS and ZFNp84, Der19 and Der22 (wt, n = 7; L4, n = 3; X4, n = 4); ZFNEWS and TALp84, Der22 (n = 4); TALLAM and TALp84, Der1 (n = 4). Der19 and Der22 were assessed in the same experiment and the frequencies averaged. Error bars, +/− SEM. Nuclease expression was detected 48 h after transfection.
C. – E. Translocation junction analysis from X4−/− cells demonstrates a shift towards longer deletions and an increased presence of microhomology. Der19 and Der22 junctions derived from ZFN expression were pooled. C, Deletion lengths from individual Der19 and Der22 junctions are indicated by the triangle and circle, respectively. D, Junctions are grouped according to whether the deletions were restricted to the overhang or were short (≤ 30 bp) or long (> 30 bp) deletions extending outside of the overhang. E, Microhomology distribution.
See also Figure S2.
We analyzed ~50 Der19 and Der22 junctions (Figure S2). In control cells, most deletions were short (≤ 2 bp median), such that more than half of junctions maintained all or part of the ZFN 5′-overhang (Figure 2C, D), presumably by fill-in synthesis. The median deletion length in X4−/− cells was much longer (78 bp, p < 0.001), and none of the deletions were restricted to the ZFN overhang. Instead, most junctions had deletions > 30 bp (85%), with many > 200 bp. Microhomology was also different in the c-NHEJ mutants. Breakpoint junctions recovered from control cells had microhomology distributions only marginally different from that expected from the chance joining of two random sequences (Figure 2E). In contrast, the microhomology distribution was significantly different from random joining in X4−/− cells (p < 0.0001), with ≥ 3 bp microhomology observed in 48% of junctions. Insertions were observed in a fraction of junctions from both control and mutant cells (Figure S2). Many insertions were a few bp, but some were longer and were derived from other chromosomes or from Chr19 or Chr22 sequences close to the breakpoints (see also, Brunet et al., 2009; Piganeau et al., 2013).
Chromosomal translocations in a human pre-B cell line depend on c-NHEJ
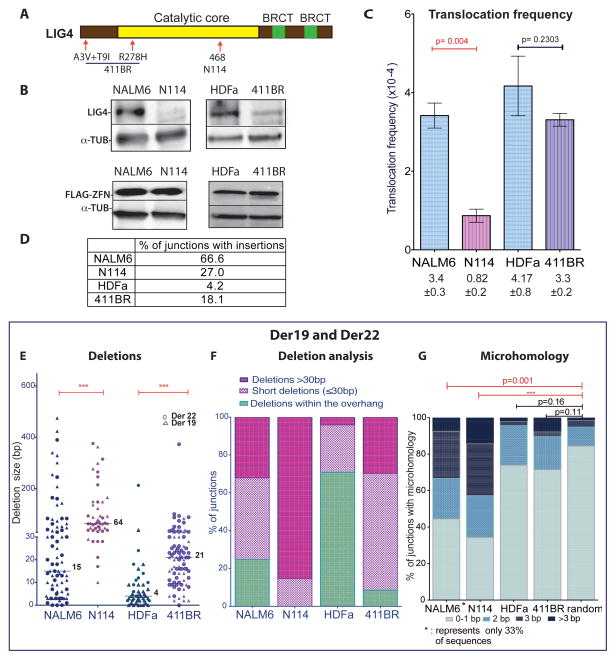
The dependence on c-NHEJ for translocation formation in human cells was quite surprising, given that c-NHEJ suppresses translocations in mouse cells. For comparison, we tested a second human LIG4 knockout cell line, N114, a derivative of the pre-B cell line, NALM6 (Grawunder et al., 1998) (Figure 3A, B). As with HCT116 L4−/− cells, indels were reduced at the ZFNp84 site in N114 cells compared to NALM6 cells (Figure S3A), consistent with a c-NHEJ defect in these cells. Notably, a substantial decrease in the ZFN-induced translocations was observed in N114 cells compared to parental cells (Figure 3C).
Figure 3. Chromosomal translocation formation is impaired in human LIG4 mutant cells.
A. Locations of LIG4 mutations in N114 pre-B cells and 411BR fibroblasts.
B. Western blotting for LIG4 and ZFNs.
C. LIG4-null N114 cells have a significantly reduced ZFN-induced translocation frequency (n = 3), while hypomorphic 411BR cells have only a mild reduction (n = 4). Der19 and Der22 formation is pooled. Error bars, +/− SEM.
D. High insertion frequency in Der19 and Der22 junctions from NALM6.
E. – G. Translocation junction analysis from LIG4 mutant cells. E, F, Junctions show longer deletions which always (N114) or mostly (411BR) extend beyond the ZFN overhang. G, Microhomology distribution at translocation junctions. Microhomology can only be determined at junctions without insertions (only 33% of junctions from wild-type NALM6 pre-B cells). Microhomology distributions from both pre-B cell lines was different from that expected by chance. By contrast, neither wild-type HDFa nor hypomorphic 411BR fibroblasts had microhomology distributions that differed from chance.
See also Figure S3.
In striking contrast to the HCT116 cells, most translocation breakpoint junctions from the parental pre-B cell line had short insertions (67% of junctions; Figures 3D, S3B), which likely arose from terminal deoxynucleotidyl transferase (TdT) activity in the pre-B cells, whereas only 27% of junctions from the mutant cells had insertions (p < 0.0001, Fisher’s exact test). The median deletion length in the translocation junctions from NALM6 cells was 15 bp (Figure 3E), and a substantial fraction were deleted only within the ZFN overhang (25%; Figure 3F). By contrast, in N114 cells deletions were significantly longer (median 64 bp; p < 0.0001) such that all extended beyond the ZFN overhang. Thus, longer deletions appear to be characteristic of translocation junctions in c-NHEJ deficient human cells.
Microhomology comparisons were more limited, given the high frequency of insertions in the junctions from the parental cells. In the 33% of junctions from NALM6 cells without insertions, ≥ 3 bp microhomology was over-represented compared to the chance joining of two random sequences (35%, p = 0.001; Figure 3G). In the residual translocation junctions from N114 cells, ≥ 3 bp microhomology was even more frequently observed (48% of junctions), such that they showed a strongly biased distribution compared to random (p < 0.0001; Figure 3G).
Patient-derived LIG4 mutant cells have an intermediate translocation phenotype
Hypomorphic LIG4 mutations have been reported in patients with a radiosensitive syndrome (O’Driscoll et al., 2001). The primary skin fibroblast cell line 411BR was derived from a patient containing homozygous LIG4 mutations in the catalytic domain and N terminus (O’Driscoll et al., 2001) (Figure 3A, B). The mutant LIG4 protein has substantially reduced catalytic activity but still interacts with XRCC4, and supports nearly normal levels of V(D)J recombination albeit with altered joining characteristics. Consistent with the V(D)J recombination assays, indels were not reduced at the ZFNp84 site in 411BR cells compared to a non-isogenic control skin fibroblast cell line, HDFa (Figure S3A). In addition, the translocation frequency was only mildly reduced and this reduction was not statistically significant (Figure 3C). As with HCT116 cells, only a fraction of translocation junctions recovered from HDFa and 411BR cells contained insertions (Figures 3D, S3C), reinforcing a role for TdT in generating the frequent insertions observed in pre-B cells. The median deletion length for 411BR cells was 21 bp, an increase relative to HDFa cells (4 bp; p < 0.0001), but not as much as for LIG4-null N114 cells (64 bp; p < 0.0001; Figure 3E, F). Microhomology distribution in junctions from 411BR cells did not differ from either control cells or a random distribution (Figure 3G). Thus, the hypomorphic 411BR cells presented an intermediate phenotype, suggesting that c-NHEJ is functional for translocation formation but altered such that longer deletions result.
NPM-ALK translocations induced by Cas9 DSBs and nCas9 paired nicks involve c-NHEJ
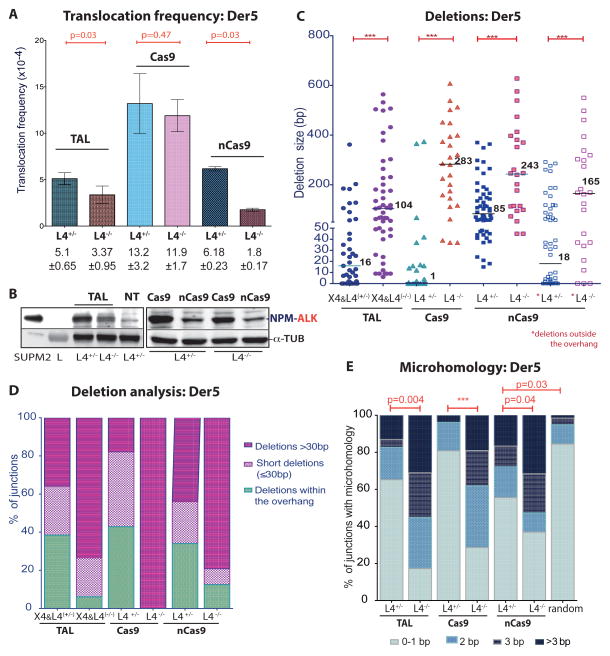
We previously induced the NPM-ALK translocation found in anaplastic large cell lymphoma using TALENs which target the NPM and ALK loci on Chr5 and Chr2, respectively (Piganeau et al., 2013) (Figures 4A, B, S4A). To examine the effect of c-NHEJ components on the formation of an oncogenic translocation, TALNPM and TALALK were expressed in the HCT116 mutants. As with ZFNp84, indel formation was reduced at the TALALK site in the absence of LIG4 or XRCC4 (Figures 4C, S4B). NPM-ALK translocations were also reduced in the L4−/− cells compared to L4+/− cells (1.5 fold, p = 0.03, Figure 5, with similar TALEN expression levels, Figure S5A), although not as dramatically as translocations induced at the EWS and p84 loci (Figure 2B). Correlating with the mild reduction in translocation frequency, the level of the NPM-ALK fusion protein was also somewhat reduced in the mutant cells (Figure 5B). As observed with ZFNs, TALEN-induced translocation junctions were dramatically altered by LIG4 or XRCC4 loss, such that deletions and microhomologies were significantly longer (Figures 5C–E, S5B).
Figure 5. Cancer translocations induced by paired nicks and DSBs.
A. – E. Der5 translocations induced by expression of the indicated nucleases in wild-type and mutant HCT116 cells. Nucleases: TAL, TALALK+TALNPM (n = 4); Cas9, Cas9+gRNAs (ALK1+NPM1) (n = 4); nCas9, nCas9+gRNAs (ALK1+ALK2 and NPM1+NPM2) (n = 3). A, Translocation frequency. Error bars, +/− SEM. B. The NPM-ALK fusion protein was detected 48 h after expression of the indicated nucleases. C-E, Translocation junction analysis. Junctions from L4−/− cells exhibited longer deletions and more microhomology. C, Deletion lengths from individual Der5 junctions. For wild-type Cas9, a large fraction of junctions from control L4+/− cells were blunt end ligations of the two ends but these were absent from L4−/− cells. For nCas9, the deletions are quite long, reflecting the long overhangs; however, in L4−/− cells, unlike L4+/− cells, most of the deletions extended well past the overhangs (see last two columns with asterisks for deletion lengths beyond the overhang). D, Junctions are grouped according to whether the deletions extend beyond the overhang (≤ 30 bp or > 30 bp) or are restricted to the overhang (or, in the case of wild-type Cas9, are 0 bp). E, Microhomology distribution. Microhomology in junctions from control L4+/− cells is distributed almost identically to that expected by chance with wild-type Cas9 but is greater with nCas9 which leaves long overhangs.
See also Figure S5.
We also tested Cas9-dependent induction of DSBs (Cong et al., 2013; Mali et al., 2013b) for translocation formation. Guide RNAs (gRNAs) were designed to result in DSBs near the TALEN cleavage sites upon Cas9 expression (NPM1 and ALK1, Figure 4B). Indel formation was increased at the CasALK1 site relative to the TALALK site, but importantly it was also reduced in L4−/− cells (Figures 4C, S4B).
Cas9-mediated translocation formation, which required both CasNPM1 and CasALK1 (Figure 4B), was more efficient than with the cognate TALEN pair (~3-fold; Figure 5A), consistent with the increased indel formation (Figure 4C). The presence of the NPM-ALK fusion protein confirmed the translocations (Figure 5B). Interestingly, only a slight reduction in translocation frequency was observed in the L4−/− cells (Figure 5A). Unlike ZFNs and TALENs, wild-type Cas9 generates blunt DNA ends or a short overhang in vitro (Jinek et al., 2012). Minimal processing of these DNA ends was observed in control cells, such that many junctions apparently arose by direct end ligation (Figures 5C–E, S5C). Overall, the median deletion length was 1 bp and the microhomology distribution was almost identical to that expected by chance (p = 0.81).
Translocation junctions were substantially altered in the L4−/− cells. None of the junctions arose by direct joining and the median deletion length was quite long (283 bp; Figures 5C–E, S5C). The microhomology distribution was significantly different from random (p < 0.0001), with 38% of the junctions having microhomology ≥ 3bp. Thus, NPM-ALK translocations induced by Cas9-generated DSBs arise by c-NHEJ in control cells with little processing of the DNA ends; in the absence of LIG4, alt-NHEJ gives rise to translocations with a remarkably different junction spectrum.
Patient-derived translocations often exhibit deletions and duplications at the breakpoint junctions (e.g., Zucman-Rossi et al., 1998), suggesting that DSBs with blunt DNA ends may not give rise to such translocations. nCas9, a Cas9 nickase due to a D10A mutation (Jinek et al., 2012), can generate DSBs with overhangs if two gRNAs are used that cleave opposite strands, i.e., paired nicks (Mali et al., 2013a; Ran et al., 2013). We tested gRNAs that would give rise to offset nicks at the NPM and ALK loci, leading to DSBs with 5′ overhangs of 41 and 37 bp, respectively (NPM1+NPM2 and ALK1+ALK2, Figure 4B). Indel formation from paired nicks directed to the ALK locus was efficient in control cells, consistent with DSB formation, and was reduced in L4−/− cells (Figures 4C, S4B).
Translocations were not recovered when a single nick was introduced on each chromosome but were recovered when paired nicks were introduced (Figure 4B), indicating that DSBs are required to drive translocation formation. Again, the NPM-ALK fusion protein was detected (Figure 5B). The translocation frequency was not as high as with Cas9-induced DSBs, possibly due to the requirement for four cleavage events by nCas9, rather than two by wild-type Cas9 (Figure 5A). In contrast to wild-type Cas9, a clear decrease in nCas9-induced translocations was observed in the L4−/− cells (Figure 5A).
Translocation junctions from paired nicks differed substantially from those from wild-type Cas9-induced DSBs. The median deletion length was 85 bp with the paired nicks in the control cells compared with 1 bp with wild-type Cas9 (Figures 5C, D, S5D). Many deletions involved only the ~40 base overhangs at each end (34%). Nevertheless, the median deletion length with the paired nicks was substantially longer in the L4−/− cells (243 bp, p = 0.0004), such that the majority of deletions extended beyond the overhangs into the double-stranded region (88%). Microhomology distribution for L4+/− cells was different from that expected by chance (Figure 5E), suggesting that the presence of long overhangs may promote more microhomology-mediated events even in the presence of LIG4. However, microhomology was much longer in the L4−/− cells (p < 0.04).
Alt-NHEJ does not affect translocations in human cells unless c-NHEJ is also deficient
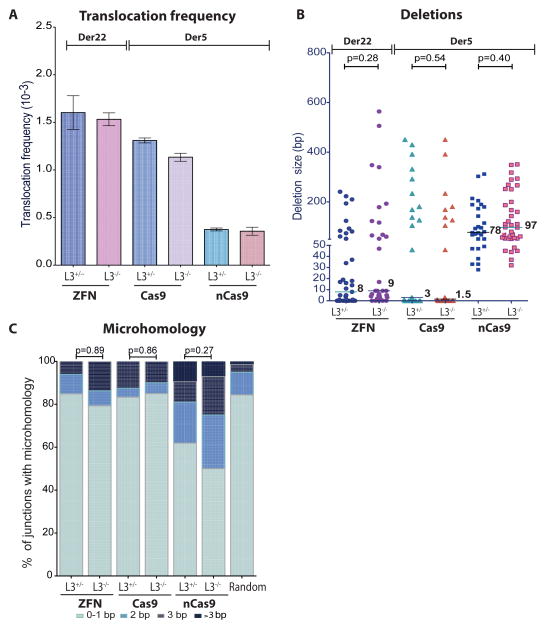
LIG3 is the major DNA ligase involved in translocation formation in mouse cells (Simsek et al., 2011a). To determine if LIG3 affects translocation formation in human cells, we tested LIG3-null HCT116 cells (Oh et al., 2014) (Figure S6A). LIG3 deficiency did not affect indel efficiency at ZFN, Cas9, or nCas9-induced DSBs (Figure S6A). Similarly, LIG3 deficiency did not significantly affect the recovery of translocations (Figure 6A). Translocation junctions were also not significantly altered for either deletions or microhomology (Figures 6B, C, S6B-D). Thus, LIG3 has either no role or a minor role in translocation formation in human cells.
Figure 6. LIG3 deficiency does not affect translocations in human cells.
A. – C. Neither translocation frequency nor junction characteristics are altered by LIG3 loss. Translocations are induced by expression of the indicated nucleases: ZFN, ZFNp84+ZFNEWS, generating Der22 (n = 3); Cas9, Cas9+gRNAs (ALK1+NPM1) or nCas9, nCas9+gRNAs (ALK1+ALK2 and NPM1+NPM2), generating Der5 (n = 2). B, Translocation frequency. Error bars, +/− SEM. C, Deletion lengths from individual Der22 (ZFNs) and Der5 (Cas9, nCas9) junctions. D, Microhomology distribution. Microhomology in junctions from control and LIG3-deficient cells was similar.
See also Figure S6.
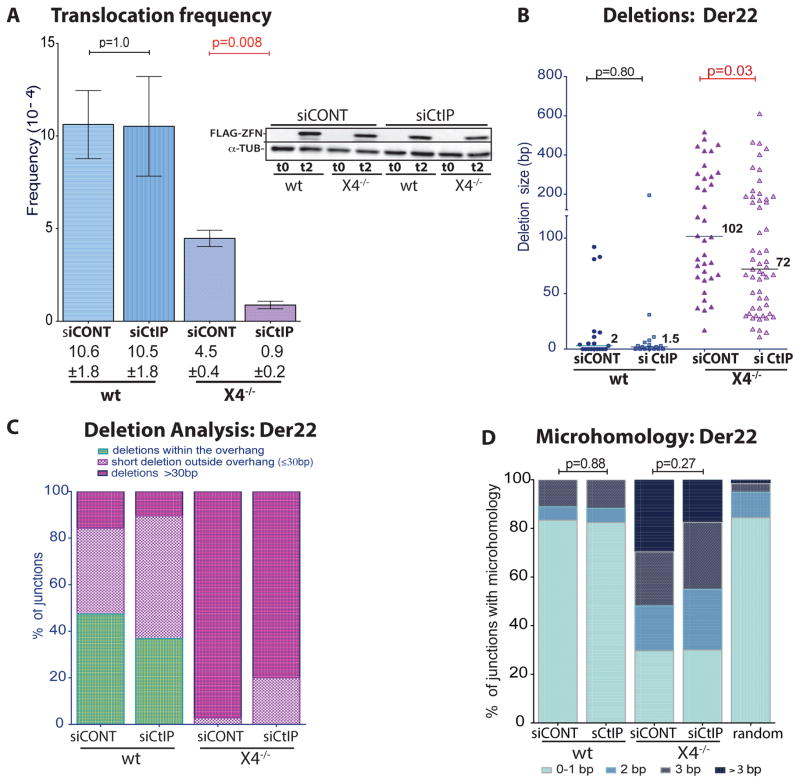
In mouse cells, translocations are thought to arise by CtIP-mediated resection of DNA ends, followed by annealing at microhomologies present in the resected DNA, since CtIP knockdown leads to smaller deletions and microhomologies at junctions (Zhang and Jasin, 2011). However, CtIP knockdowns had minimal effect on indel efficiency in wild-type and X4−/− HCT116 cells (Figure S7A). CtIP depletion also had no discernible effect in wild-type cells on ZFNp84-ZFNEWS-induced translocation frequency or junction characteristics (Figures 7, S7B). However, in X4−/− cells translocations were significantly reduced (Figure 7A, p = 0.008). Residual translocations showed smaller deletions (Figure 7B, C, S7C; p = 0.03), indicating that CtIP plays a role in resecting DNA ends in the absence of c-NHEJ components. Interestingly, microhomology was not significantly altered (Figure 7D). Thus, in human cells, CtIP appears to participate in translocations only in the absence of c-NHEJ.
Figure 7. CtIP affects translocations in XRCC4-deficient but not wild-type cells.
A. Translocation frequency is reduced with CtIP knockdown in X4−/− but not wild-type human cells (n = 3). Error bars, +/− SEM. FLAG-ZFN levels at the time of transfection (t0) and 48 h later (t2) is shown in the inset.
B. Deletion lengths from individual Der22 junctions are reduced in X4−/− but not wild-type cells with CtIP knockdown.
C. Fewer deletions extend ≤ 30 bp beyond the overhang in X4−/− cells with CtIP knockdown.
D. Microhomology in junctions from control and X4−/− cells is not affected by CtIP.
See also Figure S7.
Discussion
We demonstrate here that c-NHEJ is the predominant mechanism for joining DNA ends during translocation formation in human cells. In cells lacking the LIG4/XRCC4 complex, translocation junctions were altered with longer deletions and microhomologies, signatures of alt-NHEJ. Our results suggest that patient-derived translocations also occur primarily through c-NHEJ. Translocation frequency was often reduced in c-NHEJ mutants, although the efficiency with which joining was redirected to alt-NHEJ varied. In contrast, loss of alt-NHEJ components did not alter either translocation frequency or outcome unless c-NHEJ was also disrupted.
Different LIG4 mutants show subtly different joining characteristics
In each c-NHEJ mutant cell line tested, translocation junction characteristics differed from wild-type cells but with subtle differences. HCT116 c-NHEJ mutants had junctions with longer deletions and more microhomology regardless of the nuclease employed (ZFNs, TAL, Cas9, nCas9). Cas9-generated translocations from LIG4-null cells had the longest median deletion length of any of the translocations analyzed, implying that c-NHEJ is particularly critical for maintaining sequence information at these ends. Translocation frequency was lower in the HCT116 c-NHEJ mutants, but the reduction was greater at the EWS and p84 loci than at NPM and ALK, even when comparing the same type of nuclease (TALENs). These results suggest locus effects in the ability to switch from c-NHEJ to alt-NHEJ; alternatively, the oncogenic NPM-ALK fusion protein formed by the translocation could provide a slight growth advantage (Zhang et al., 2013). Interestingly, Cas9-induced NPM-ALK translocations were not significantly reduced in the absence of LIG4, suggesting that alt-NHEJ can efficiently act on these ends to generate translocations.
In patient 411BR cells, the translocation frequency was not affected, consistent with a report that low levels of LIG4 are sufficient for DSB repair in vitro (Windhofer et al., 2007), but deletion lengths were intermediate between control cells and other LIG4 mutants. The remaining LIG4 in these cells has residual catalytic activity (O’Driscoll et al., 2001), and it may also promote DNA-PKcs bridging activity (Cottarel et al., 2013) to protect ends from extensive degradation.
The pre-B cell line NALM6 showed a large number of junctions with small insertions, likely due to TdT activity (Smith et al., 2003). TdT is required for diversification of antigen receptors, and TdT-generated insertions at these and other DSBs are dependent on c-NHEJ components (Boubakour-Azzouz et al., 2012; Smith et al., 2003). Consistent with this dependence, N114 cells had fewer insertions. NALM6 junctions without insertions showed longer microhomology than is typical of wild-type cells, raising the possibility that some of the junctions that did not engage TdT did not join by c-NHEJ; alternatively, TdT may have created microhomology in some of these junctions (Lieber, 2010).
Translocations in human and mouse cells form by different mechanisms
The requirement for c-NHEJ components in chromosomal translocation formation in human cells is in direct opposition to findings from mouse studies. In mice and mouse cell lines, c-NHEJ suppresses translocations while alt-NHEJ promotes their formation. Mice with mutations in c-NHEJ components develop RAG recombinase-mediated pro-B lymphomas on a p53-deficient background with complex Myc translocations involving microhomology (Zhu et al., 2002). Similarly, c-NHEJ mutant B cells undergoing class switching have increased Myc translocations, implying that alt-NHEJ is a robust mechanism for generating translocations (Boboila et al., 2010). In addition to immune system-generated DSBs, nuclease-induced translocations in mouse embryonic stem cells form by alt-NHEJ, such that the translocation junctions show reduced microhomology and deletions in the absence of LIG3 or CtIP (Simsek et al., 2011a; Zhang and Jasin, 2011). Consistent with a c-NHEJ independent mechanism, junctions from wild-type, LIG4/XRCC4, and Ku70 mutant murine cells show similar characteristics (Simsek et al., 2011a; Simsek and Jasin, 2010; Weinstock et al., 2007). Thus, in mouse cells DSB repair leading to translocations differs from intrachromosomal repair of a single DSB (Simsek and Jasin, 2010; Weinstock et al., 2006), whereas in human cells both types of repair are similar and mediated by c-NHEJ.
Although NHEJ components are conserved, human cells have substantially higher DNA-PK activity than rodent cells (Finnie et al., 1995; Beamish et al., 2000; Meek et al., 2001). Another striking difference is that loss of the Ku protein is incompatible with human cell survival (Li et al., 2002) due to an essential role in telomere maintenance (Wang et al., 2009), whereas rodent cells and mice are viable in the absence of Ku (Nussenzweig et al., 1996; Taccioli et al., 1994; Zhu et al., 1996). The enhanced DNA-PK activity and differential Ku-requirements imply that basic DSB metabolism may be different between humans and rodents, a hypothesis supported by our studies.
Nuclease induction of oncogenic translocations: DSBs and paired nicks
Developing methodologies to induce oncogenic translocations has ramifications for understanding the role of translocations in tumor formation and their etiology. Here, we extended our previous studies in which cancer-relevant translocations were induced by ZFNs and TALENs (Piganeau et al., 2013) to Cas9 and nCas9 to generate the NPM:ALK translocation associated with lymphoma. Cas9 has also been used in HEK293 cells to induce a translocation found in lung tumors (Choi and Meyerson, 2014).
Cas9 has several advantages as a nuclease, in particular that it is easy to engineer by changing the gRNA sequence. Because mismatches to the gRNA can be tolerated, however, wild-type Cas9 has been associated with off-target mutagenesis (Fu et al., 2013). Paired nicks generated by nCas9 have the potential to substantially increase cleavage specificity, given that two gRNAs are required per DSB (Mali et al., 2013a; Ran et al., 2013), and we have determined that nCas9-generated paired nicks efficiently induce translocations.
Somewhat unexpectedly, we found that the DNA end structure markedly affected translocation junction characteristics. Cas9-induced DSBs are blunt or have a small overhang (Jinek et al., 2012), and many of the resultant translocation junctions from control cells appear to be simple c-NHEJ-dependent joining of the DNA ends. As translocation formation destroys the gRNA site, these events must arise from a single joining event, rather than iterative cycles of breakage and rejoining. The precise conservation of DNA end sequences in many translocations seems surprising, given that translocation formation results in a chromosome aberration. However, it is consistent with c-NHEJ protection of DNA ends and suggests that c-NHEJ is not an inherently error-prone process (Betermier et al., 2014).
The precise joining of Cas9-induced DSBs means that the translocation junctions often do not exhibit the small deletions typical of patient-derived translocations (Gillert et al., 1999; Langer et al., 2003; Reichel et al., 1998; Weinstock et al., 2006; Zhang et al., 2002; Zucman-Rossi et al., 1998), suggesting that cancer translocations may also arise from some other type of DNA end structure. nCas9-induced nicks provide flexibility in DNA end structures. The paired nicks we used to induce translocations were ~40 bp apart and thus led to relatively long 5′ overhangs. Overhang sequences are retained in some junctions, suggestive of fill-in synthesis, as observed at ZFN and TAL overhangs (this report; Brunet et al., 2009; Piganeau et al., 2013). But deletions are significantly longer than those generated from wild-type Cas9, often encompassing one or both overhangs. Offset nicks with large overhangs that are filled in, however, such as those generated here by nicking, have been proposed to lead to duplications of sequences found in some leukemic translocation junctions (Gillert et al., 1999; Reichel et al., 1998), indicating that the paired nicks may provide a good model to interrogate the factors involved in these translocations.
Insertions ranging from ~20 bp to several hundred bp were found in ~6% of translocation junctions (Figures S2, S5). These insertions were frequently derived from sequences close to one of the four DNA ends, as reported for mouse cells (Simsek and Jasin, 2010). However, unlike mouse cells, a significant fraction were also derived from other chromosomes, as reported for other human translocations (Piganeau et al., 2013; Sobreira et al., 2011; Zucman-Rossi et al., 1998). A plausible mechanism for their derivation is replication primed by one of the DNA ends using microhomology (Simsek and Jasin, 2010; Weinstock et al., 2006). Microhomology-primed duplication of sequences has similarities to what has been termed microhomology-mediated break induced replication, which has been postulated as a mechanism to give rise to copy number variants in the germline (Carvalho et al., 2013; Hastings et al., 2009).
Conclusions
Understanding mechanisms of genome rearrangements that contribute to the accumulation of cancer-driving mutations and secondary mutations resulting from cancer therapies is a critical area of research. The results presented here provide strong evidence that c-NHEJ is the primary pathway for chromosomal translocation formation in human cells. It is noteworthy in this regard that patients with LIG4 mutations (LIG4 syndrome) are not extremely cancer prone, unlike patients with mutations in genes in many other DNA repair pathways (Woodbine et al., 2014). While loss of c-NHEJ components may result in decreased DSB repair with a concomitant increase in apoptosis, our results here suggest that chromosomal translocations that would promote oncogenesis are also less likely to form, providing an alternative (or additional) explanation for the lack of a strong tumor predisposition. In conclusion, while the high level of c-NHEJ activity in human cells compared to rodents may provide a risk for oncogenic translocations, this is offset by the more accurate and efficient DSB repair from c-NHEJ that may restrain cancer incidence while permitting a longer lifespan.
Experimental Procedures
Additional experimental procedures are available online in the Supplemental Information. ZFNEWS, ZFNFLI1, ZFNP84, TALALK and TALNPM have been described (Brunet et al., 2009; Piganeau et al., 2013). (Note: p84 is encoded by the AAVS1 locus.) Cas9/gRNA target sequences are underlined with PAM sequences in bold:
NPM: CCTCGAACTGCTACTGGGTTCACCTCAGCCTCTGGAATAGCTAGAACTACAGG
ALK: CCTCAGGTAACCCTAATCTGATCACGGTCGGTCCATTGCATAGAGGAGG
Cells were nucleofected with nuclease expression vectors; genomic DNA was isolated 48 h later for all repair assays. Translocation frequency was calculated using nested PCR on small pools of cells to amplify translocation junctions (Brunet et al., 2009; Piganeau et al., 2013).
Supplementary Material
Acknowledgments
We thank Michael Lieber (USC) and Penny Jeggo (Sussex) for cell lines, Jean-Paul Concordet, Marine Charpentier, and Anne de Cian (MNHN) for discussions and technical assistance, Sangamo BioSciences, especially Fyodor Urnov and Lei Zhang, for providing ZFNs, and members of the Jasin laboratory, especially Francesca Cavallo and Gemma Regan-Mochrie. This work was supported by La Ligue Nationale contre le Cancer (H.G), Le Canceropole IDF (M.P.), an ANR grant ANR-12-JSV6-0005 (B.R.), and NIH grants to A.T. (GM047251, ES012512, NCI P30CA118100), E.A.H. (GM088351, CA15446), and M.J. (GM054668).
Footnotes
Author contributions
MJ, EB conceived the study; AT, EAH, CG, MJ, EB designed experiments; HG, MP, BR, JBR performed experiments; AS, BR, SO developed key reagents; MJ, EB wrote the paper with AT, EAH, CG.
References
- Beamish HJ, Jessberger R, Riballo E, Priestley A, Blunt T, Kysela B, Jeggo PA. The C-terminal conserved domain of DNA-PKcs, missing in the SCID mouse, is required for kinase activity. Nucleic Acids Res. 2000;28:1506–1513. doi: 10.1093/nar/28.7.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 2014;10:e1004086. doi: 10.1371/journal.pgen.1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012a;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Oksenych V, Gostissa M, Wang JH, Zha S, Zhang Y, Chai H, Lee CS, Jankovic M, Saez LM, et al. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1) Proc Natl Acad Sci USA. 2012b;109:2473–2478. doi: 10.1073/pnas.1121470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakour-Azzouz I, Bertrand P, Claes A, Lopez BS, Rougeon F. Terminal deoxynucleotidyl transferase requires KU80 and XRCC4 to promote N-addition at non-V(D)J chromosomal breaks in non-lymphoid cells. Nucleic Acids Res. 2012;40:8381–8391. doi: 10.1093/nar/gks585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci U S A. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, Belmont JW, Hastings PJ, Lupski JR. Replicative mechanisms for CNV formation are error prone. Nat Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel J, Frit P, Bombarde O, Salles B, Negrel A, Bernard S, Jeggo PA, Lieber MR, Modesti M, Calsou P. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol. 2013;200:173–186. doi: 10.1083/jcb.201203128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An Xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA, Jackson SP. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci U S A. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, Angermuller S, Borkhardt A, Harbott J, Lampert F, et al. A DNA damage repair mechanism is involved in the origin of chromosomal translocations t(4;11) in primary leukemic cells. Oncogene. 1999;18:4663–4671. doi: 10.1038/sj.onc.1202842. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo PA. The repair and signaling responses to DNA double-strand breaks. Adv Genet. 2013;82:1–45. doi: 10.1016/B978-0-12-407676-1.00001-9. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T, Metzler M, Reinhardt D, Viehmann S, Borkhardt A, Reichel M, Stanulla M, Schrappe M, Creutzig U, Ritter J, et al. Analysis of t(9;11) chromosomal breakpoint sequences in childhood acute leukemia: almost identical MLL breakpoints in therapy-related AML after treatment without etoposides. Genes Chrom Cancer. 2003;36:393–401. doi: 10.1002/gcc.10167. [DOI] [PubMed] [Google Scholar]
- Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci U S A. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet. 2010;11:819–829. doi: 10.1038/nrg2883. [DOI] [PubMed] [Google Scholar]
- Mattarucchi E, Guerini V, Rambaldi A, Campiotti L, Venco A, Pasquali F, Lo Curto F, Porta G. Microhomologies and interspersed repeat elements at genomic breakpoints in chronic myeloid leukemia. Genes Chrom Cancer. 2008;47:625–632. doi: 10.1002/gcc.20568. [DOI] [PubMed] [Google Scholar]
- Meek K, Kienker L, Dallas C, Wang W, Dark MJ, Venta PJ, Huie ML, Hirschhorn R, Bell T. SCID in Jack Russell terriers: a new animal model of DNA-PKcs deficiency. J Immunol. 2001;167:2142–2150. doi: 10.4049/jimmunol.167.4.2142. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- Oh S, Harvey A, Zimbric J, Wang Y, Nguyen T, Jackson PJ, Hendrickson EA. DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.04.015. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Wang Y, Zimbric J, Hendrickson EA. Human LIGIV is synthetically lethal with the loss of Rad54B-dependent recombination and is required for certain chromosome fusion events induced by telomere dysfunction. Nucleic Acids Res. 2013;41:1734–1749. doi: 10.1093/nar/gks1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio NR, Li S, Watanabe G, Lieber MR. Non-homologous end joining often uses microhomology: Implications for alternative end joining. DNA Repair (Amst) 2014;17:74–80. doi: 10.1016/j.dnarep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah G, Zhang L, Holmes M, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel M, Gillert E, Nilson I, Siegler G, Greil J, Fey GH, Marschalek R. Fine structure of translocation breakpoints in leukemic blasts with chromosomal translocation t(4;11): the DNA damage-repair model of translocation. Oncogene. 1998;17:3035–3044. doi: 10.1038/sj.onc.1202229. [DOI] [PubMed] [Google Scholar]
- Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. DNA Ligase III Promotes alternative nonhomologous end-Joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Riballo E, Kysela B, Baldeyron C, Manolis K, Masson C, Lieber MR, Papadopoulo D, Jeggo P. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 2003;31:2157–2167. doi: 10.1093/nar/gkg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira NL, Gnanakkan V, Walsh M, Marosy B, Wohler E, Thomas G, Hoover-Fong JE, Hamosh A, Wheelan SJ, Valle D. Characterization of complex chromosomal rearrangements by targeted capture and next-generation sequencing. Genome Res. 2011;21:1720–1727. doi: 10.1101/gr.122986.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni A, Siemann M, Grabos M, Murmann T, Pantelias GE, Iliakis G. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku298. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhofer F, Wu W, Iliakis G. Low levels of DNA ligases III and IV sufficient for effective NHEJ. J Cell Physiol. 2007;213:475–483. doi: 10.1002/jcp.21120. [DOI] [PubMed] [Google Scholar]
- Woodbine L, Gennery AR, Jeggo PA. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair. 2014;16:84–96. doi: 10.1016/j.dnarep.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wei F, Wang HY, Liu X, Roy D, Xiong QB, Jiang S, Medvec A, Danet-Desnoyers G, Watt C, et al. The potent oncogene NPM-ALK mediates malignant transformation of normal human CD4(+) T lymphocytes. Am J Pathol. 2013;183:1971–1980. doi: 10.1016/j.ajpath.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Strissel P, Strick R, Chen J, Nucifora G, Le Beau MM, Larson RA, Rowley JD. Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia. Proc Natl Acad Sci U S A. 2002;99:3070–3075. doi: 10.1073/pnas.042702899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J, Legoix P, Victor JM, Lopez B, Thomas G. Chromosome translocation based on illegitimate recombination in human tumors. Proc Natl Acad Sci USA. 1998;95:11786–11791. doi: 10.1073/pnas.95.20.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.