Abstract
Lipoic acid, an essential enzyme cofactor, is required in three domains of life. In the past 60 years since its discovery, most of the pathway for lipoic acid synthesis and metabolism has been elucidated. However, genetic control of lipoic acid synthesis remains unclear. Here, we report integrative evidence that bacterial cAMP-dependent signaling is linked to lipoic acid synthesis in Shewanella species, the certain of unique marine-borne bacteria with special ability of metal reduction. Physiological requirement of protein lipoylation in γ-proteobacteria including Shewanella oneidensis was detected using Western blotting with rabbit anti-lipoyl protein primary antibody. The two genes (lipB and lipA) encoding lipoic acid synthesis pathway were proved to be organized into an operon lipBA in Shewanella, and the promoter was mapped. Electrophoretic mobility shift assays confirmed that the putative CRP-recognizable site (AAGTGTGATCTATCTTACATTT) binds to cAMP-CRP protein with origins of both Escherichia coli and Shewanella. The native lipBA promoter of Shewanella was fused to a LacZ reporter gene to create a chromosome lipBA-lacZ transcriptional fusion in E. coli and S. oneidensis, allowing us to directly assay its expression level by β-galactosidase activity. As anticipated, the removal of E. coli crp gene gave above fourfold increment of lipBA promoter-driven β-gal expression. The similar scenario was confirmed by both the real-time quantitative PCR and the LacZ transcriptional fusion in the crp mutant of Shewanella. Furthermore, the glucose effect on the lipBA expression of Shewanella was evaluated in the alternative microorganism E. coli. As anticipated, an addition of glucose into media effectively induces the transcriptional level of Shewanella lipBA in that the lowered cAMP level relieves the repression of lipBA by cAMP-CRP complex. Therefore, our finding might represent a first paradigm mechanism for genetic control of bacterial lipoic acid synthesis.
Keywords: cAMP-receptor protein (CRP), LipA, LipB, Lipoic acid, lipoic acid synthesis, Shewanella
Introduction
Lipoic acid (6,8-dithiooctanoic acid, thioctic acid, or R-5-(1,2-dithiolan-3-yl) pentanoic acid), is a type of two-sulfur inserted eight-carbon fatty acid derivative and acts as a coenzyme widespread in three domains of life (Perham 2000; Cronan et al. 2005). This covalently bound cofactor is required for aerobic metabolism of 2-oxoacids in Escherichia coli and C1 metabolism in plants like Arabidopsis (Perham 2000; Cronan et al. 2005; Engel et al. 2007). In E. coli, the three well-known enzymes whose activities require lipoylation, the post-translational modification, include PDH (pyruvate dehydrogenase), OGDH (2-oxoglutarate dehydrogenase), and GCV (glycine cleavage system) system (Cronan et al. 2005; Hermes and Cronan 2009). All the three enzyme systems possess such subunits (the E2 subunits of both PDH and OGDH, and the H protein of GCV system) that contain no less one lipoyl domains (LD) featuring with a conserved structure of around 80 residues long (Reche 2000; Cronan et al. 2005). Generally, a specific/conserved lysine residue on these LDs is attached by lipoic acid via an amide bond (Perham 2000). Therefore, it seems likely that lipoic acid facilitates shuttle of the activated reaction intermediates amongst the active sites of the lipoate-dependent multi-enzyme systems (Perham 2000; Cronan et al. 2005).
Most of current knowledge of lipoic acid metabolisms comes from studies with E. coli (Zhao et al. 2003; Cronan et al. 2005). Two alternative strategies have been developed in E. coli to satisfy the trace physiological demand for lipoic acids. It includes de novo biosynthesis pathway and the scavenging route (Cronan et al. 2005; Hermes and Cronan 2009; Rock 2009; Christensen and Cronan 2010). The former pathway is constituted of two consecutive steps: the LipB (octanoyl-ACP: protein N-octanoyl-transferase) transfers the endogenously produced octanoyl moieties from octanoyl-ACP (an intermediate of the fatty acid biosynthesis) to lipoyl domains (Fig.1A) (Jordan and Cronan 2003; Zhao et al. 2003, 2005); in the second step the LipA (lipoyl synthase) uses S-adenosyl-l-methionine (SAM)-dependent radical chemistry to insert two sulfur atoms at carbons 6 and (Fig.1A) (Zhao et al. 2003; Cronan et al. 2005; Douglas et al. 2006; Christensen and Cronan 2010). The lipoyl protein ligase (LplA) plays a critical role in utilization of exogenous lipoic acids from environments in which the lipoyl-adenylate intermediate is required (Fig.1A) (Morris et al. 1994, 1995; Reed et al. 1994). Although the metabolic mechanism of lipoic aids that was initially discovered in the early of 1940s (Reed 2001) was extensively investigated (Reed 2001; Cronan et al. 2005; Hermes and Cronan 2009; Rock 2009; Christensen and Cronan 2010), its genetic regulation/control is poorly understood (Kaleta et al. 2010; Feng and Cronan 2014).
Figure 1.
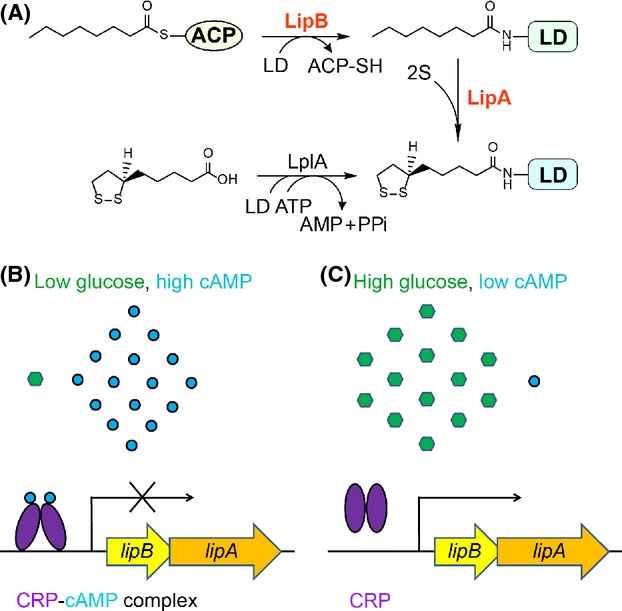
Working model for pathway for bacterial lipoic acid metabolism and its possible regulation. (A) A pathway proposed for lipoic acid synthesis and its scavenging in Shewanella. LipA, lipoic acid synthase; LipB, Octanoyl-ACP: protein ligase (N-octanoyltransferase); PdhR, pyruvate dehydrogenase operon repressor; LD, lipoyl domain (in light blue); ACP, acyl carrier protein (in white oval). Shewanella lipB/A expression is shut off by the cAMP-CRP complex on the condition of low glucose level (or high cAMP concentration) (B), whereas is induced upon high concentration of glucose is available (or cytosolic cAMP is limited) (C). Blue dots denote cAMP molecules, green regular hexagon represents glucose, and purple ovals indicate CRP protein. CRP, cAMP-receptor protein.
cAMP receptor protein (CRP, also called catabolic activator protein, CAP) is a type of global regulator, representing a classical model for bacterial gene regulation systems(Green et al. 2014). The paradigm version of CRP is E. coli crp protein product that modulates expression of hundreds of genes involved in a variety of bacterial physiological aspects such as energy metabolism (e.g., galactose catabolism) (Zheng et al. 2004; Green et al. 2014). Indeed, the activity of CRP requires the presence of its physiological ligand/effector, cyclic AMP (cAMP) (Zheng et al. 2004; Green et al. 2014). Upon the CRP protein is occupied by the cAMP small molecule, it proceeds to an allosteric alteration/structural rearrangement, allowing its acquisition of an ability to specifically bind a collection of specific target DNA sequences(Schultz et al. 1991; Green et al. 2014). As we know, the typical CRP box (cAMP-CRP binding site) is referred to the imperfect palindromic consensus sequence “N3TGTGAN6TCACAN3” (Zheng et al. 2004). In the similarity to the well-studied FadR regulator that has dual functions in fatty acid metabolism (Feng and Cronan 2009a,b, 2010, 2012), it appears that the dimeric CRP protein-mediated regulation also can exert two opposite roles, i.e., either activation (Hanamura and Aiba 1992; Ishizuka et al. 1994; Zheng et al. 2004) or repression (Aiba 1983; Hanamura and Aiba 1991; Ishizuka et al. 1994) in response to distinct external and/or internal stimuli/inputs (Green et al. 2014). Recently, comparative genomics-based reconstruction of bacterial regulatory networks RegPrecise (http://regprecise.lbl.gov/RegPrecise) by Rodionov's research group (Rodionov et al. 2011; Novichkov et al. 2013) predicted that a possible CRP box (AAGTGTGATCTATCTTACATTT) is present in front of lipBA operon (SO1162-S01161) of Shewanella oneidensis MR-1(a marine-borne species of γ-proteobacteria family) with considerable potential for the remediation of contaminated environments and application in microbial fuel cells (Fredrickson and Romine 2005; Fredrickson et al. 2008).
More importantly, it seemed likely that the predicted site reflects an evolutionally conserved regulatory mechanism in that it is found in nearly all the Shewanella species with known genome sequences and similar scenario were seen even with the two human pathogens Salmonella typhimurium and Klebsiella pneumonia. This might raise a possibility that cAMP signaling is linked to bacterial lipoic acid synthesis in certain species of γ-proteobacteria. However, this hypothesis requires further in vitro and in vivo experimental verification.
In this paper, we aimed to resolve this unanswered question. As anticipated, electrophoresis mobility shift assays (EMSA), we conducted and confirmed that the two CRP proteins of E. coli and Shewanella are functionally exchangeable and the predicted CRP sites of Shewanella are functional. Using the chromosome lipBA-lacZ transcriptional fusion in E. coli, we visualize that the removal of E. coli crp gene gave above fourfold increment of lipBA promoter-driven β-gal expression, which is almost identical to the scenario seen with Shewanella. Somewhat it is unexpected, but not without precedent that an addition of glucose into media effectively induces lipBA expression in Shewanella, in that the lowered cAMP level relieves the repression of lipBA by cAMP-CRP complex (Fig.1B and C). Therefore, our finding answered the long-term unresolved question in the field of lipoic acid metabolism and might represent a first paradigm illustrating the genetic control of bacterial lipoic acid synthesis by cAMP-dependent CRP signaling in certain species of γ-proteobacteria.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains used here were derivatives of both E. coli K-12 and S. oneidensis MR-1 (Table1) and cultivated aerobically at 37°C and 30°C, respectively. For the growth of E. coli, the following three media were utilized, including Luria-Bertani (LB) medium (10 g of tryptone, 5 g of yeast extract and 10 g of NaCl per liter; pH 7.5), rich broth (RB) medium (10 g of tryptone, 1 g of yeast extract, and 5 g of NaCl per liter), and M9 minimal medium with either 5 mmol/L sodium acetate or 0.4% glucose as the sole carbon source (Feng and Cronan 2009b, 2010). M1-defined medium containing 0.02% (w/v) of vitamin-free casamino acids and 15 mmol/L lactate as electron donor was used to cultivate S. oneidensis (Gao et al. 2008). If required, chemicals or antibiotics were added as follows: 2,6-diaminopimelic acid (DAP), 0.3 mmol/L; sodium ampicillin, 100 μg/mL; kanamycin sulfate, 25 μg/mL; and tetracycline, 15 μg/mL; gentamycin, 15 μg/mL.
Table 1.
Bacterial strains and plasmids in this study
| Bacteria or plasmids | Relevant characteristics | Refs or origins |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| BL21(DE3) | Engineered E. coli strain as an expression host for recombinant plasmids | Lab stock |
| MG1655 | Wild type of E. coli K-12 (F-, λ-, rph-1) | CGSC1, Lab stock |
| WM3064 | Donor strain for conjugation; ΔdapA | W. Metcalf, UIUC |
| BW25113 | A Δlac strain of E. coli K-12 (F-, λ-, rph-1, Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) Δ(rhaD-rhaB)568 hsdR514) | CGSC1, Baba et al. (2006) |
| JW5702-4 | (BW25113, Δcrp-765::kan) | CGSC1, Baba et al. (2006), Feng and Cronan (2010) |
| MC1061 | F-, λ-, Δ(araA-leu)7697, [araD139]B/r, Δ(codB-lacI)3, galK16, galE15(GalS), e14-, mcrA0, relA1, rpsL150(strR), spoT1, mcrB1, hsdR2 | Lab stock, Casadaban and Cohen (1980), Feng and Cronan (2009b) |
| RH77 | MC4100, ΔcyaA, Δcrp::Tn10 | Lab stock Feng and Cronan (2010, 2012) |
| DH5α (λ-pir) | An E. coli Δlac host for pAH125 and its derivatives | Feng and Cronan (2009a, 2012), Haldimann and Wanner (2001) |
| FYJ208 | Vibrio cholerae O395 | Jame Jun Zhu's lab |
| FYJ239 | BL21(DE3) carrying pET28-crp_ec | Feng and Cronan (2012), Goble et al. (2013) |
| FYJ426 | Salmonella enterica serovar Typhimurium 14028s | Slauch's lab |
| FYJ452 | DH5a(λ-pir) carrying pAH-PlipBA_she | This work |
| FYJ453 | MC4100 whose chromosome was integrated with the lipBA_she-lacZ transcriptional fusion at the λ phage site | This work |
| FYJ457 | MC1061, lipBA_she-lacZ transcriptional fusion | P1vir(FYJ453) × MC10612, This work |
| FYJ458 | MC4100, ΔcyaA, Δcrp::Tn10, lipBA_she-lacZ transcriptional fusion | P1vir(FYJ453) × RH772, This work |
| FYJ462 | Topo carrying pET28-crp_she | This work |
| FYJ463 | BL21(tuner) carrying pET28-crp_she | This work |
| S. oneidensis | ||
| MR-1 | Wild-type | Gao's lab |
| HG0624 | Δcrp derived from MR-1 | Gao et al. (2010) |
| HG1162-1 | ΔlipBA derived from MR-1 | This work |
| HG0424 | ΔaceE derived from MR-1 | This work |
| HG1329 | ΔcyaC derived from MR-1 | This work |
| Plasmids | ||
| pET28(a) | Commercial T7-driven expression vector, KmR | Novagen |
| pET28-crp_ec | pET28(a) carrying E. coli crp gene, KmR | Feng and Cronan (2012), Goble et al. (2013) |
| pAH125 | A promoter-less lacZ reporter plasmid in E. coli, KmR | Haldimann and Wanner (2001) |
| pHG101 | A promoter-less broad-host KmR vector | Wu et al. (2011) |
| pHGEI01 | An integrative lacZ reporter vector | Fu et al. (2014) |
| pAH-PlipBA_she | A pAH125 derivative encoding Shewanella oneidensis lipBA promoter region (∽350 bp) | This work |
| pET28-crp_she | pET28(a) encoding S. oneidensis crp gene, KmR | This work |
CGSC denotes Coli Genetic Stock Center, Yale University.
Selection for kanamycin resistance.
Plasmids and DNA manipulations
Using polymersase chain reaction (PCR) with primers crp_she-F plus crp_she-R (Table2), the S. oneidensis crp gene was amplified and inserted into the BamHI and XhoI sites of pET28a(+) expression vector, giving the recombinant plasmid pET28-crp_she (Table1). The promoter region of S. oneidensis lipBA (referred to PlipBA_she) covering the predicted CRP site (Table3) was amplified with a set of specific primers PlipBA-F plus PlipBA-R and cloned into the two cuts SalI and EcoRI of promoter-less plasmid pAH125 to give the recombinant plasmid pAH-PlipBA_she. Consequently, the pAH-PlipBA_she plasmid was transformed into MC4100 (Δlac), resulting in the LacZ reporter strain FYJ453 with PlipBA_she-lacZ transcriptional fusion on chromosome (Table1). The inserts introduced in the recombinant plasmids we generated were validated by both PCR assays and direct DNA sequencing (Feng and Cronan 2011a,b).
Table 2.
DNA oligonucleotide sequences used in this work
| Primers | Primer sequences | Purposes |
|---|---|---|
| crp_she-F (BamHI) | 5′-CG GGATCC ATG GCT CTG ATT GGT AAG CC-3′ | Gene cloning |
| crp_she-R (XhoI) | 5′-CCG CTCGAG TTA ACG GGT ACC ATA TAC CAC-3′ | |
| crp_she-ck1 | 5′-GTG AAT CCA GTG AGT TTG ACA-3′ | PCR detection for the crp mutant of Shewanella |
| crp_she-ck2 | 5′-CAG AGT TGA CTA ACG CCT TG-3′ | |
| PlipBA-F (SalI) | 5′-CCG GTCGAC GAT GAA CTG ATG GAG TTC CCC-3′ | PCR amplification and cloning of the lipBA promoter |
| PlipBA-R (EcoRI) | 5′-AACC GAATTC CAA GGG CAA CCT CTC CCC TA-3′ | |
| lipBA_she CRP site-F (43 bp) | 5′-CAA GGT CAT AAA GTG TGA TCT ATC TTACATTTA TGG CCA AGA G-3′ | Synthesis of the predicted CRP site of Shewanella lipBA |
| lipBA_she CRP site-R (43 bp) | 5′-CTC TTG GCC ATA AATGTA AGA TAG ATC ACACTT TAT GAC CTT G-3′ | |
| lipA_ec CRP site-F (42 bp) | 5′-ACG GAG TAA TAGATG TTA TCC GTA ATG CATTTT GAA AAA GTA-3′ | Synthesis of the suspected CRP site of E. coli lipA |
| lipA_ec CRP site-R (42 bp) | 5′-TAC TTT TTC AAA ATG CAT TAC GGA TAA CAT CTA TTA CTC CGT-3′ | |
| fadD_ec CRP site-F (48 bp) | 5′-GTA AAG ATA AAA ATA AAT AGT GAC GCG CTTCGC AACCTT TTC GTT GGG-3′ | Synthesis of the known CRP site of E. coli fadD |
| fadD_ec CRP site-R (48 bp) | 5′-CCC AAC GAA AAG GTT GCG AAG CGC GTC ACTATT TAT TTT TAT CTT TAC-3′ | |
| ybeD_ec CRP site-F(42 bp) | 5′-AAA CAC TTG AAAGTG TAA TTT CCG TCC CCATAT ACT AAG CAT-3′ | Synthesis of the anticipated CRP site of E. coli ybeD |
| ybeD_ec CRP site-R(42 bp) | 5′-ATG CTT AGT ATATGG GGA CGG AAA TTA CACTTT CAA GTG TTT-3′ | |
| ybeD_es CRP site-F(42 bp) | 5′-GAA CAC TTG AAA GTG TGA TTT CCA TCC CCA TAT ACT AGG TAT-3′ | Synthesis of the CRP site of ybeD gene from Enterobacter sp. 638 |
| ybeD_es CRP site-R(42 bp) | 5′-ATA CCT AGT ATA TGG GGA TGG AAA TCA CAC TTT CAA GTG TTC-3′ | |
| ybeD_kp CRP site-F(42 bp) | 5′-GAACACTTGAAA GTG TGA TTT CCA TCC CCA TATACTATTCAT-3′ | Synthesis of the CRP site of ybeD gene from Klebsiella pneumonia |
| ybeD_kp CRP site-R(42 bp) | 5′-ATG AAT AGT ATATGG GGA TGG AAA TCA CACTTT CAA GTG TTC-3′ | |
| ybeD_st CRP site1-F(42 bp) | 5′-GAA CGCTTGAAA GTG TGA TTT TCG TCC CCA TAT ACTATGCAT-3′ | Synthesis of the CRP site 1 of ybeD gene from Salmonella typhimurium LT2 |
| ybeD_st CRP site1-R(42 bp) | 5′-ATG CAT AGT ATA TGG GGA CGA AAA TCA CACTTT CAA GCG TTC-3′ | |
| ybeD_st CRP site2-F(42 bp) | 5′-CTG TGG CGG GAG TTG TTA TTT TTT TTA CGT AAT GCC GGA GCT-3′ | Synthesis of the CRP site 2 of the ybeD gene from Salmonella typhimurium LT2 |
| ybeD_st CRP site2-R(42 bp) | 5′-AGC TCC GGC ATTACG TAA AAA AAA TAACAACTC CCG CCA CAG-3′ | |
| ybeD_yp CRP site-F (42 bp) | 5′-ATT GGC CCC ATATTG TGA TTA ATC TTA TAT TGC AAA TAA GCT-3′ | Synthesis of the CRP site of the ybeD gene from Yersinia pestis |
| ybeD_yp CRP site-R (42 bp) | 5′-AGC TTA TTT GCA ATA TAA GAT TAA TCA CAA TAT GGG GCC AAT-3′ | |
| LacZ-R | 5′-CAG TGA ATC CGT AAT CAT GGT C-3′ | PCR assay for the lipBA-lacZ junction |
| crp_ec-F | 5′-CAG GTA GCG GGA AGC ATA TTT C-3′ | PCR assay for the E. coli crp |
| crp_ec-R | 5′-CAG CGT TTG TCG AAG TGC ATA G-3′ | |
| ybeD-F (19-39) | 5′-GAT GAA CTG ATG GAG TTC CCC-3′ | PCR (RT-PCR) assay for the S. oneidensis ybeD |
| ybeD-R (223-243) | 5′-GAT GTT GGC GAG CTC TGT GTA-3′ | |
| lipB-F (471-491) | 5′-CTG TGG ATC GTT GAA CAT CCA-3′ | PCR (RT-PCR) assay for the S. oneidensis lipB |
| lipB-R (760-780) | 5′-GAC CTA AGG AAG CCA CTT TGC-3′ | |
| lipA-F (1020-1040) | 5′-CTG AAC GTT TAC AAC CCG GAG-3′ | PCR (RT-PCR) assay for the S. oneidensis lipA |
| lipA-R (1253-1273) | 5′-CAT AAA GGT TGC TGT GCC GTG-3′ | |
| ybeD-lipB-F (208-229) | 5′-CAT ATC GAA ACC CTG TAC ACA G-3′ | PCR (RT-PCR) assay for the ybeD-lipB junction of S. oneidensis |
| ybeD-lipB-R (471-492) | 5′-GTG GAT GTT CAA CGA TCC ACA G-3′ | |
| lipB-lipA-F (910-930) | 5′-GCC CAC AAA CTG TGA TAG AAG-3′ | PCR (RT-PCR) assay for the lipA-lipB junction of S. oneidensis |
| lipB-lipA-R (1160-1181) | 5′-CTT GCT TAA TGT CGA GAA TGC G-3′ | |
| lipBA-Nest (769-789) | 5′-GGA TCC TAA GAC CTA AGG AAG-3′ | 5′-RACE of S. neidensis lipBA |
| lipBA-GSP (868-898) | 5′-CTG CAT TGC ACC ATT TCA AGG-3′ | |
| 16S_she-F | 5′-GAT AAC AGT TGG AAA CGA CTG-3′ | PCR (RT-PCR) assay |
| 16S_she-R | 5′-CTT TCC TCC CTA CTG AAA GTG-3′ |
The underlined italic letters represent restriction sites, and the bold letters denote the known (and/or predicted) CRP-binding sites. RT-PCR, reverse transcription-polymersase chain reaction; CRP, cAMP-receptor protein.
Table 3.
CRP binding sites in front of potential lipB/A operons from a variety of species amongst γ-proteobacteria
| Organisms | Gene | Loci | CRP site | Position1 | Score |
|---|---|---|---|---|---|
| Enterobacter sp. 638 | ybeD | Ent638_1166 | AAGTGTGATTTCCATCCCCATA | −90 | 4.4 |
| Escherichia coli MG1655 | ybeD | b0631 | AAGTGTAATTTCCGTCCCCATA | −94 | 3.6 |
| Cistrobacter koseri | ybeD | CKO_02527 | AAGTGTGATTTCCATCCCCATA | −91 | 4.4 |
| Klebsiella pneumonia | ybeD | KPN_00663 | AAGTGTGATTTCCATCCCCATA | −97 | 4.4 |
| Salmonella typhimurium LT2 | ybeD | STM0636 | AGTTGTTATTTTTTTTACGTAA AAGTGTGATTTTCGTCCCCATA |
−35 −93 |
3.9 4.2 |
| Yersinia pestis | ybeD | y1174 | TATTGTGATTAATCTTATATTG | −146 | 4.2 |
| Shewanella baltica | lipB | Sba_3281 | AAATGTGATCTGTCTTACATTT | −74 | 5.2 |
| S. halifaxensis | lipB | ShaI_3240 | AAATGTGATCCGTATTACATTT | −76 | 5.2 |
| S. loihica | lipB | Shew_2941 | AAATGTGATCTACCTTACATTT | −70 | 5.3 |
| S. oneidensis | lipB | SO1162 | AAGTGTGATCTATCTTACATTT | −68 | 5.1 |
| S. pealeana | lipB | Spea_3155 | AAATGTGATCCGTATTACATTT | −76 | 5.2 |
| S. piezotolerans | lipB | swp_3928 | AAATGTGATCTGTCTTACATTT | −69 | 5.2 |
| S. putrefaciens | lipB | Sputcn32_2875 | AAATGTGATCTATCTTACATTT | −69 | 5.3 |
| S. sediminis | lipB | Ssed_3491 | AAATGTGATCTAGCTTACATTT | −75 | 5.3 |
| S. woodyi | lipB | swoo_3714 | AAGTGTGATCTAGCTTACAATT | −74 | 5.1 |
| S. sp ANA-3 | lipB | Shewanan3_0989 | AAATGTGATCTGTCTTACATTT | −74 | 5.2 |
| S. sp MR-4 | lipB | Shewmr4_0985 | AAATGTGATCTGTCTTACATTT | −74 | 5.2 |
| S. sp MR-7 | lipB | Shewmr7_1050 | AAATGTGATCTGTCTTACATTT | −74 | 5.2 |
| S. sp W3-18-1 | lipB | Sputw3181_1028 | AAATGTGATCTATCTTACATTT | −75 | 5.3 |
CRP, cAMP-receptor protein.
The position is relative to the translation initiation site. All the information is sampled from the RegPrecise database (http://regprecise.lbl.gov/RegPrecise/search.jsp).
The lipBA promoter activity was assessed using an integrative lacZ reporter system as described recently (Fu et al. 2014). A fragment covering the sequence upstream of the lipB gene from −300 to +1 was amplified and cloned into the reporter vector pHGEI01, verified by sequencing, and the correct plasmid was then transferred into S. oneidensis strains by conjugation. Proper integration of the promoter fusion constructs was confirmed by PCR. To eliminate the antibiotic marker, the helper plasmid pBBR-Cre was transferred into the strains carrying the correct integrated construct. Colonies without the integrated antibiotic marker were screened and verified by PCR, and followed by the loss of pBBR-Cre as described previously (Fu et al. 2013).
In-frame mutant construction and complementation
In-frame deletion strains for S. oneidensis were constructed using the att-based Fusion PCR method as described previously (Jin et al. 2013). In brief, two fragments flanking gene of interest were amplified by PCR, which were linked together by a second round of PCR. The fusion fragments were introduced into plasmid pHGM1.0 by using Gateway BP clonase II enzyme mix (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instruction, resulting in mutagenesis vectors in E. coli WM3064, which were subsequently transferred into S. oneidensis MR-1 via conjugation. Integration of the mutational constructs into the chromosome was selected by resistance to gentamycin and confirmed by PCR. The verified transconjugants were grown in LB broth in the absence of NaCl and plated on LB supplemented with 10% sucrose. Gentamycin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the target gene. Mutants were verified by direct sequencing of the mutated regions.
Plasmids pHG101 and pHG102 were used in genetic complementation of mutants (Wu et al. 2011). For complementation of genes next to their promoter, a fragment containing the gene of interest and its native promoter was generated by PCR and cloned into pHG101. For the remaining genes, the gene of interest was amplified and inserted into MCS of pHG102 under the control of the arcA promoter, which is constitutively active (Gao et al. 2010). The resulting vectors were transferred into its corresponding mutant strain via conjugation and its presence was confirmed by plasmid purification and restriction enzyme digestion.
P1vir phage transductions
Following the protocol described by Miller (1992), we conducted the experiment of P1vir transduction. Transduction of strain MC1061 with a lysate grown on FYJ453 (PlipBA_she-lacZ) with selection for kanamycin resistance gave strain FYJ457 (MC1061, PlipBA_she-lacZ). Strain FYJ458 was constructed by transduction of strain RH77 (MC4100, Δcrp::Tn10) with a P1vir lysate grown on FYJ457 (MC1061, PlipBA_she-lacZ) with selection for kanamycin resistance (Table1). All the relevant genotypes were determined using PCR with a primer set (e.g., PlipBA-F plus LacZ-R, Table2), and the PCR products were confirmed by direct DNA sequencing (Feng and Cronan 2012).
RNA isolation and RT-PCR
Mid-log phase cultures of S. oneidensis MR-1 grown in RB media were collected for total bacterial RNA preparations. As we did before, the RNeasy bacterial RNA isolation kit (Qiagen, Hilden, Germany) was adopted (Feng and Cronan 2009b; Feng et al. 2013b). The quality of the acquired RNA samples was visualized using agarose gel electrophoresis. Using the general PCR assay in which the total RNA samples function as templates with primers 16S_she-F plus 16S_she-R (Table2), the possible contamination of trace genomic DNA in the RNA samples was routinely figured out as we described earlier (Feng and Cronan 2009b, 2010).
On the basis of above qualified RNA samples, we performed the reverse transcription (RT)-PCR experiments (Feng and Cronan 2009b, 2010). Briefly, 1 μg of total RNA was mixed with 0.5 μg of random primers (11 μL in total), denatured (70°C for 5 min), and then chilled on ice (5 min). The RT reaction mixture (20 μL total volume) comprised 10 μL of denatured RNA template, 1 μL of random primers, 4 μL of ImProm-II 5× reaction buffer, 2.5 μL of 1 mol/L MgCl2, 1 μL of deoxynucleoside triphosphate mix, 0.5 μL of the recombinant RNasin RNase inhibitor, and1 μL of ImProm-II reverse transcriptase (Feng and Cronan 2009b, 2011a). The program for RT reaction included the equilibration at 25°C for 5 min, an extension at 42°C for 60 min, and the inactivation of enzyme at 70°C for 15 min. As a result, the cDNA pool (1 μL) was used as the template to PCR-amplify the lipBA operon-related genes/DNA fragments.
Real-time quantitative RT-PCR
On the basis of SYBR Green dye method as we previously mentioned (Feng and Cronan 2009b, 2010), real-time quantitative RT-PCR (qRT-PCR) experiments were employed to evaluate the altered expression profile of S. oneidensis lipBA operon in the Δcrp mutant. qPCR reaction system (20 μL) contained 12.5 μL of iQ™ SYBR Green Supermix, 1 μL of each primer, 1 μL of the diluted cDNA sample, and 4.5 μL of sterile water. All the data were collected in triplicate on a Mastercycler® eprealplex (Eppendorf, Hauppauge, NY, USA), using the program of a denaturing cycle at 95°C for 15 min, 45 cycles comprising 94°C for 20 sec, 60°C for 20 sec, and 72°C for 20 sec, and a final step featuring with gradient temperature from 60°C to 90°C for dissociating double stranded DNA products. The reference gene was the16S_she rRNA-encoding gene (Table2) and water acted as blank control to monitor cross-contamination of various cDNA samples. The relative expression levels were calculated with the 2−ΔΔCT method developed by Livak and Schmittgen (2001).
5′-RACE
RLM-RACE (Ambicon, Grand Island, NY, USA), an improved version of 5′-RACE kit, was applied in mapping the transcription start site of S. oneidensis lipBA operon (Feng and Cronan 2011a,b). The nested PCR reactions were established using two sets of combined primers (Outer Primer plus lipBA-GSP and Inner Primer plus lipBA-Nest primer) (Table2). The PCR program was described with a denaturing cycle at 95°C for 5 min followed by 35 cycles comprising 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. The purified PCR products were sent for direct DNA sequencing. The transcriptional start site was assigned to first nucleotide adjacent to the RLM-RACE adaptor (Feng and Cronan 2009a,b; Feng et al. 2013b).
Enzymatic assays
For PDH assay, cells were grown at 30°C in 25 mL of LB containing the appropriate antibiotics until the beginning of the stationary phase, harvested, and washed twice with a 0.04 mol/L potassium phosphate buffer (pH 7.5). The resulting pellets were frozen rapidly and stored at −80°C. Cell extracts were prepared by resuspending the thawed pellets in 2 mL of the same buffer prior to sonication with a microtip in a Branson model 200 Sonifier (2 min total, with 40-sec pulses at 20-sec intervals). Cell debris was removed by centrifugation (10 min at 12,000g and 4°C), and the supernatants were used for assays at 25°C as described previously (Reed and Cronan 1993). Protein concentrations were determined by the bicinchoninic acid protein assay reagent (Pierce Chemical Co., Rockford, IL, USA).
To measure the β-galactosidase activities in E. coli, bacterial lysates from mid-log phase cultures grown in LB (or M9) media were prepared by treatment with sodium dodecyl sulfate-chloroform (Miller 1972; Feng and Cronan 2009b). Similarly, cells of S. oneidensis (mid-log phase under experimental settings) were pelleted for assaying its β-galactosidase activity with an assay kit as described previously (Wu et al. 2011).
Measurement of intracellular cAMP levels
Cells in mid-log phase cultures (∽0.3 of OD600) were collected by centrifugation and washed twice with charcoal-treated phosphate-buffered saline (PBS; pH 7.0). Both supernant and pellet fractions were applied to the cAMP assay using Cyclic AMP EIA kit (Cayman Chemical Co., Ann Arbor, Michigan, USA) according to the manufacturer's instruction.
Expression, purification and identification of two CRP proteins
To prepare the recombinant CRP protein in two versions (CRP_ec and CRP_she), the engineered E. coli strains carrying either pET28-crp_ec or pET28-crp_she (Table1) were induced with 0.3 mmol/L isopropyl β-d-1-thiogalactopyranoside (IPTG) at 30°C for 5 h(Feng and Cronan 2012). Following bacterial lysis by a French pressure cell, the clarified supernatants by centrifugation (30,966g, 30 min) were loaded onto a nickel chelate column (Qiagen). After removal of the contaminated protein by washing with 1×phosphate buffered saline (PBS) with 50 mmol/L imidazole, the interested CRP proteins (CRP_ec or CRP_she) were eluted using elution buffer containing 150 mmol/L imidazole. Finally, the protein was concentrated by ultrafiltration (30 kDa cut-off) and exchanged into 1× PBS (pH 7.4) containing 10% glycerol. The purity of the recombinant CRP proteins was judged by 12% sodiumdodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (Feng and Cronan 2009b, 2011b). To verify the identity of the acquired proteins, the de-stained (SDS-PAGE) gel slices were subjected to liquid chromatography quadrupole time-of-flight mass spectrometry using a Waters Q-Tof API-US Quad-ToF mass spectrometer linked to a Waters nanoAcquity UPLC(Feng and Cronan 2011a; Feng et al. 2013a,b).
Electrophoretic mobility shift assays
The function of the predicted CRP-binding site of Shewanella lipBA operon was assessed in vitro using electrophoretic mobility shift assays (EMSA) with little improvements (Feng and Cronan 2011a; Goble et al. 2013; Feng et al. 2014). In the EMSA tests, nine pieces of DNA probes were composed of seven suspected probes (lipBA_she, ybeD_ec, ybeD_es, ybeD_kp, ybeD_st1, ybeD_st2, and ybeD_yp) and the two control probes, the fadD_ec site with known function (the positive control) and the lipA_ec without any function (the negative control) (Table3). The digoxigenin (DIG)-labeled DNA probes were prepared in vitro through annealing two complementary oligonucleotides in TEN buffer (10 mmol/L Tris-HCl, 1 mmol/L EDTA, 100 mmol/L NaCl; pH 8.0) and then labeled by the terminal transferase with DIG-ddUTP (Roche, Indianapolis, IN, USA) (Feng et al. 2014).
In the presence/absence of cAMP (20 pmol), the various DIG-labeled DNA probes (0.2 pmol) were incubated with or without CRP protein in the binding buffer (Roche) at room temperature for around 20 min. Following the separation of the DNA-protein complexes with a native 7% PAGE gel, the chemiluminescent signals were further captured by the exposure to ECL film (GE Healthcare, Piscataway, NJ, USA) (Feng and Cronan 2011b, 2012).
Bioinformatic analyses
The alignments of DNA (and/or protein) sequences were conducted using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and final output was processed by the ESPript 2.2 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The lipBA regulons and the possible CRP-recognizable sites of γ-proteobacteria were collected from the RegPrecise database (Novichkov et al. 2010a) and were analyzed (Feng et al. 2013a) using RegPredict software (Novichkov et al. 2010b). The sequence logo for the CRP consensus palindrome was generated by WebLogo (http://weblogo.berkeley.edu/logo.cgi). The software of SPDBV_4.01 (http://spdbv.vital-it.ch/) was used for structure modeling.
Results
Shewanella lipBA is an operon
The paradigm pathway of lipoic acid synthesis is encoded by two genes lipB and lipA of E. coli. The two sequential steps of this pathway included LipB-catalyzed transfer of octanoyl moiety from octanoyl-ACP to lipoyl domains of the cognate enzymes and LipA-mediated insertion of sulfur atoms at C6 and C8 of LD-bound octanoyl moiety to give lipoate (Fig.1). Therefore, we are interested in examining the genetic context of the lipB and/or lipA in γ-proteobacteria using RegPredict software (Novichkov et al. 2010b) (Fig.2). In addition to the reference strains (e.g., E. coli, Salmonella enterica, Yersinia pestitis, etc.), all the other samples are focused on Shewanella species from the RegPrecise database (Novichkov et al. 2010a). We noted that the ybeD (SO1163) gene is constantly present upstream of the lipB gene (Fig.2), and YbeD protein of E. coli origin exhibits a striking structural homology to the allosteric regulatory domain of d-3-phosphoglycerate dehydrogenase (Kozlov et al. 2004).
Figure 2.
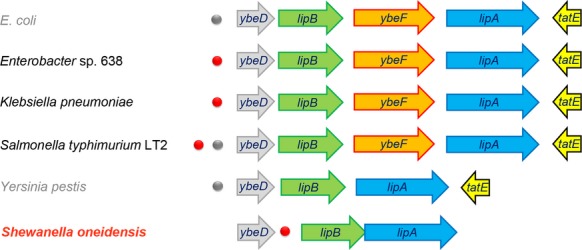
Gemomic context of the lipB/A operon/genes in the selected γ-proteobacteria. Blue arrows represent the lipA genes that encode the lipoic acid synthase catalyzing the last committed reaction of lipoic acid biosynthesis pathway, whereas green arrows indicate the octanoyl-protein ligase-encoding genes (lipB).The gray arrow upstream of the lipB gene refers to the ybeD gene of unknown function. In some cases, the tatE gene (Sec-independent protein translocase) downstream of lipA is shown with yellow arrow. In the four species (Escherichia coli, Enterobacter sp. 638, Klebsiella pneumoniae, and Salmonella typhimurium LT2), the ybeF gene encoding an LysR-type transcription factor (in orange) is located between lipA and lipB. The predicted CRP-binding palindromes are highlighted with dots (red dots represent the experimentally verified sites, whereas the gray ones are not experimentally validated). CRP, cAMP-receptor protein.
Unlike the scenario seen with E. coli that lipB and lipA are separated by a gene (ybeF) encoding a LysR-family transcription factor of unknown function (Feng and Cronan 2014) (Fig.2), it seemed likely that lipB and lipA constitutes an operon in most of species of Shewanella (Fig.1). Although physiological advantages for the co-transcription of these two genes are expected, experimental evidence is lacking. To address this hypothesis, the strain of S. oneidensis MR-1 was selected for our experiments. We established the combined PCR and RT-PCR assays using five pairs of specific primer pairs (Table2 and Fig.3A). The positive amplifications (1, 3 and 5) were obtained by both PCR and RT-PCR showed that all three genes (ybeD, lipB and lipA) are transcribed (Fig.3A). The fact that the primed amplicon (designated to 2) was observed only by PCR, but not by RT-PCR suggested that ybeD is not co-transcribed together with lipB (Fig.3A). As anticipated, the designed amplicon covering both lipB and lipA was positive in both PCR and RT-PCR assays, validating that lipB and lipA act as an operon (transcriptional unit) (Fig.3A).
Figure 3.
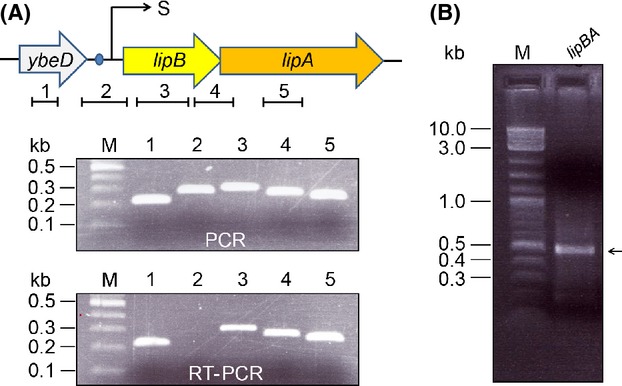
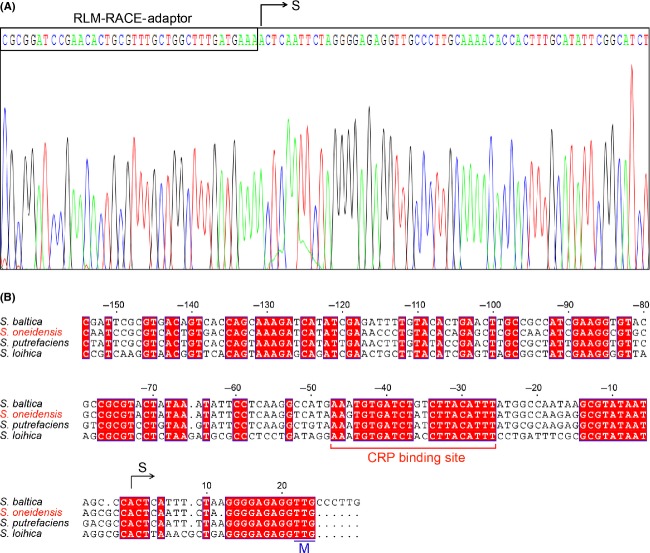
Determination of the Shewanella lipBA as an operon. (A) Genetic organization and transcriptional analyses of Shewanella lipBA operon. The three arrows represent ybeD (in gray), lipB (in yellow) and lipA (in orange), respectively. The numbered short lines (1, 2, 3, 4, and 5) indicate the specific PCR amplicons. The transcription start sites (S) is indicated with an arrow. The PCR and RT-PCR products were separated by the electrophoresis of 1.5% agarose gel. (B) Electrophoretic analyses for the 5′-RACE product of Shewanella lipBA operon 5′-RACE product were separated with 2.0% agarose gel and the expected size was highlighted with an arrow. kb, kilo-base pair; M, 100 bp DNA ladder (New England Bio-labs, Ipswich, MA, USA). RT-PCR, reverse transcription-polymersase chain reaction.
S. oneidensis lipBA promoter
DNA sequences recognized by CRP proteins of E. coli and S. oneidensis are predicted to be similar and the interaction depends on cAMP (Gao et al. 2010; Fu et al. 2013; Zhou et al. 2013). Given the fact that a predicted CRP-binding site (AAGTGTGATCTATCTTACATTT) is located in the intergenic region between the ybeD gene and the lipBA operon of S. oneidensis (Fig.2), we thus mapped the promoter by employing an improved method of 5′-RACE (RLM-RACE). As a result, we acquired the 5′-RACE products of approximately 450 bp in length (Fig.3B). The result of the direct DNA sequencing showed the 5′-end of the S. oneidensis lipBA transcript (i.e., transcription start site, A) is located 20 nucleotides upstream its translation initiation codon TTG (Fig.4C and D). Apparently, the assumed CRP-recognizable site appears to be 25 bp upstream of the transcription start site (Fig.4D). Furthermore, the multiple sequence alignment clearly indicated that the CRP binding sites of different origins are extremely conserved, in that 16 of 22 nucleotides are identical at least (if not all) in the examined Shewanella species (Fig. 6A and B). However, the function of these putative sites needs further experimental validation.
Figure 4.
Use of 5′-RACE analyses to map the Shewanella lipBA promoter. (A) Direct DNA sequencing of the RLM-RACE product of the Shewanella lipBA operon. (B) Sequence comparison of the promoter regions of the Shewanella lipBA operon. The multiple alignments were conducted using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the resultant output was processed by program ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) (Feng and Cronan 2009b, 2010; Feng et al. 2013a). Identical residues are indicated with white letters on a red background, similar residues are red letters on yellow, varied residues are in black letters, and dots represent missing residues. S, transcription start site; M, translational initiation site. The predicted CRP-recognizable palindrome is underlined. CRP, cAMP-receptor protein.
Figure 6.
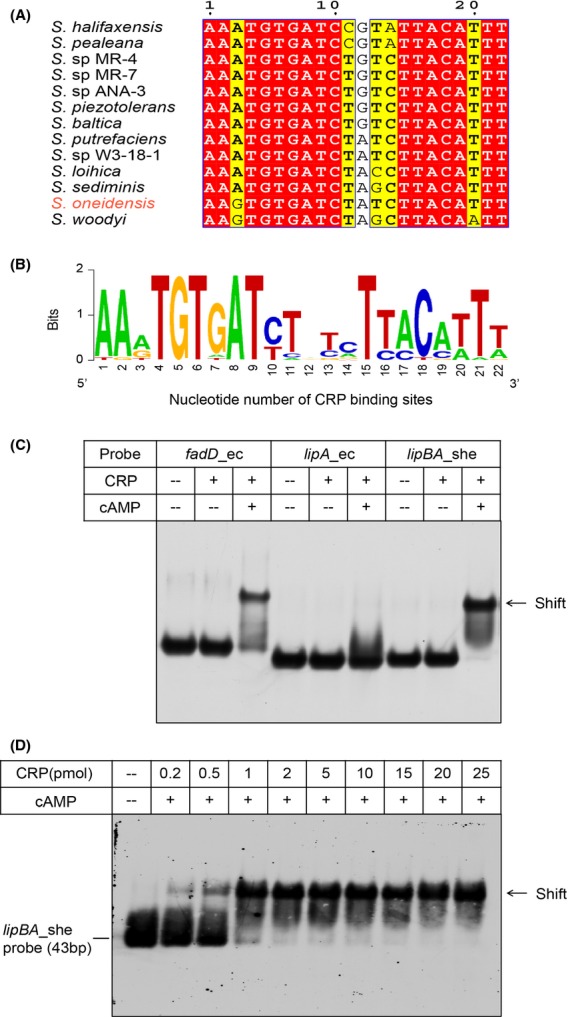
Binding of Shewanella lipBA to the cAMP-CRP functional complex. (A) Multiple sequence alignment of CRP-recognizable sites from Shewanella lipBA operon. Multiple sequence alignment was performed as described in Figure2. Identical residues are indicated with white letters on a red background, similar residues are black letters on yellow, and varied residues are in black letters. Totally, the CRP-binding sites are sampled from 13 different species of Shewanella. (B) Sequence logo for the CRP palindromic consensus sequences. The palindromic sequences used here are identical to those listed in (A), and the sequence logo was generated using WebLogo (http://weblogo.berkeley.edu/logo.cgi). (C) Escherichia coli CRP binds to Shewanella lipBA promoter, but not E. coli lipA promoter. The CRP site of E. coli fadD (fadD_ec) is used as positive control, while the possible CRP site of E. coli lipA (lipA_ec) is referred to negative control (Table2). The plus sign represents addition of the CRP protein and/or cAMP, whereas the minus sign denotes no addition of the CRP protein and/or cAMP. Designations: ec, E. coli; she, Shewanella. (D) Dose-dependent binding of E. coli CRP binds to Shewanella lipBA promoter. The level of CRP protein in (A) is 2 pmol, and the amount of cAMP is 20 pmol. The protein samples were incubated with 0.2 pmol of DIG-labeled lipBA_she probe (43 bp) in a total volume of 20 μL. A representative result from three independent gel shift assays (7% native PAGE) is given. CRP, cAMP-receptor protein.
Physiological requirement of protein lipoylation
It is reasonable that co-expression of LipB octanoyltransferase and LipA lipoate synthase assures the economical production of lipoic acid (an energy-expansive molecule) to effectively satisfy the metabolic/physiological requirement of protein lipoylation in organisms. Given the fact that both PDH and OGDH are proceeded such kind of post-translational modification, we thereby developed the anti-LA Western blot to detect this metabolic requirement in four γ-protebacteria species (E. coli, S. enterica, V. cholerae, and S. oneidensis). As expected, we did observe that lipoylation occurs in PDH and OGDH of Shewanella, which is in much similarity to the scenario seen with E. coli (Fig.5A). Because lipoylation is essential for the function of all characterized PDH and OGDH proteins, the result points out the metabolic significance of this common enzyme cofactor in S. oneidensis.
Figure 5.
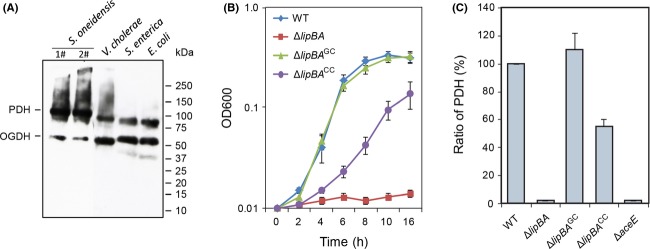
Physiological requirement of protein lipoylation in γ-proteobacteria. (A) Use of anti-LA Western blot to detect the requirement of protein lipoylation for γ-proteobacteria. Four species of γ-proteobacteria tested here include Escherichia coli, Salmonella enterica (S. enterica), Vibrio cholerae (V. cholerae) and Shewanella oneidensis (S. oneidensis). (B) Growth of the S. oneidensis lipBA mutant on lactate Complementation was carried out by either genetically (ΔlipBAGC, expressing a copy of the lipBA genes in trans) or chemically (ΔlipBACC, with the addition of lipoic acid of 3 pmol/mL). (C) Analyses for PDH activity The PDH dehydrogenase activities are given as micromoles of 3-acetylpyridine adenine dinucleotide reduced per milligram of protein per hour for extracts of the same number of cells estimated by OD600 readings. The relative activities (RA) were obtained by normalizing the values of other strains to the mean of wild-type values. In both (B) and (C), error bars represent standard deviations from at least three independent experiments. PDH, pyruvate dehydrogenase; OGDH, 2-oxoglutarate dehydrogenase; LA, Lipoic acid; kDa, kilo-dalton.
Subsequently, we constructed a lipBA null mutant from the S. oneidensis wild-type strain. The mutant was unable to grow on minimal medium unless lipoic acid was supplemented (Fig.5B), a phenotype observed from E. coli lip mutants (Reed and Cronan 1993). Additionally, the PDH assay revealed that this ΔlipBA strain contained no detectable dehydrogenase activities (Fig.5C). Importantly, the phenotypes resulting from the lipBA deletion were restored by their expression in trans, indicating that they are due to the intended mutation. These data, collectively, conclude that the lipBA genes are the only enzyme accountable for protein lipoylation in S. oneidensis.
Characterization of S. oneidensis CRP protein
S. oneidensis CRP and its counterpart of E. coli are highly homologous (Fig. S1A), and have been shown to be functionally equivalent/exchangeable in vivo (Saffarini et al. 2003). However, whether this is the case in vitro remains undefined. In addition to the E. coli CRP protein, an N-terminal hexahistidine fused S. oneidensis CRP protein was over-expressed, purified to homogeneity and gave a single protein band with an estimated molecular mass (∽24 kDa) (Fig. S2B). The tertiary structure of S. oneidensis CRP protein was modeled using SPDBV_4.01 software, which is highly similar to that of E. coli (Fig. S3C). Liquid chromatography mass spectrometry analyses of tryptic peptides of the recombinant CRP protein band excised from an SDS-PAGE gel validated its identity in that the peptides matched S. oneidensis CRP with 70% coverage of the expected peptides (Fig. S4D). The two versions of CRP proteins we prepared were subsequently used for functional analyses of the above predicted CRP-specific palindromic sites.
Shewanella lipBA binds the cAMP-CRP complex
To test the activity of the DNA probe derived from the S. oneidensis lipBA promoter (Fig.6A and B), we conducted EMSA assays. First, the positive control fadD_ec probe with a known function (Feng and Cronan 2012) binds well to E. coli CRP protein in the presence of cAMP effector molecule, whereas the negative control lipA_ec probe with a nonfunctional CRP site did not (Fig.6C). As expected, the lipBA_she probe exhibited the appreciably comparable activity of binding cAMP-CRP complex relative to the positive control. Apparently, our result is much consistent with previous observations with the CRP regulatory protein in the context of other metabolisms (Gao et al. 2010; Fu et al. 2013; Zhou et al. 2013), proving the prediction of Novichkov et al. (2013) is correct. Additionally, the specific binding of lipBA_she to cAMP-CRP complex seemed to be in a protein dose-dependent manner (Fig.6D).Not only does the CRP protein of E. coli origin interacts with E. coli fadD probe (Figs.6C and 7A) and Shewanella lipBA probe (Figs.6C and 7C), but also the CRP protein encoded by Shewanella binds to E. coli fadD probe (Fig.7B) and Shewanella lipBA probe (Fig.7D). It thus fully demonstrated that the two versions of CRP protein are functionally exchangeable in vitro.
Figure 7.
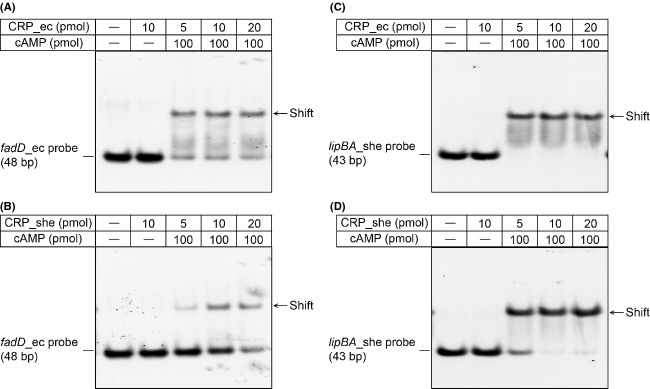
The two CRP proteins of Escherichia coli and Shewanella are functionally equivalent. (A) Binding of E. coli CRP to E. coli fadD probe. (B) E. coli CRP interacts with Shewanella lipBA promoter. (C) Interplay between Shewanella CRP and E. coli fadD probe. (D) Shewanella CRP binds Shewanella lipBA promoter. The CRP sites of E. coli fadD (fadD_ec) and Shewanella lipBA (lipBA_she) are listed in Table2. The plus sign denotes the addition of the CRP protein and/or cAMP, whereas the minus sign suggests no addition of the CRP protein and/or cAMP. Designations: ec, E. coli; she, Shewanella. When necessary in the EMSA tests, the level of cAMP is 20 pmol. The CRP protein samples were incubated with 0.2 pmol of DIG-labeled probe in a total volume of 20 μL. A representative result is shown from three independent gel shift assays (7% native PAGE). CRP, cAMP-receptor protein.
Similarly, we also tested a series of predicted CRP-binding sites located upstream of ybeD-lipB loci (Fig.2 and Table3) using EMSA tests with E. coli CRP protein. Unlike the lipBA_she probe (Fig. S2A), neither the E. coli ybeD probe (ybeD_ec, Fig. S2B) nor the Y. pestis ybeD probe (ybeD_yp, Fig. S2G) are functional for the cAMP-CRP complex. By contrast, the prediction in CRP-recognizable sites (ybeD_es and ybeD_kp) in front of the ybeD gene of both Enterobacter sp. 638 and Klebsiella pneumonia are correct in that both bind to the CRP protein (Fig. S2C and D). Of particular note, among the two CRP sites (ybeD_st1 and ybeD_st2) proposed for S. enteric ybeD gene, only the ybeD_st1 site is functional (Fig. S2E), while the other one was not (Fig. S2F).
A regulatory role for CRP in lipBA expression of S. oneidensis
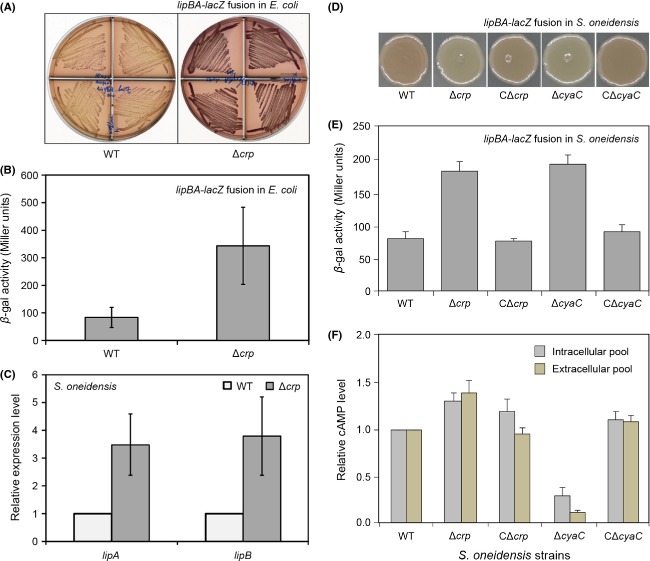
Two approaches (lipBA_she-lacZ transcriptional fusion and the real-time qRT-PCR) were used to examine the in vivo regulatory role of cAMP-CRP complex in expression of S. oneidensis lipBA operon encoding lipoic acid synthesis machinery. First, S. oneidensis lipBA promoter was fused to a LacZ reporter gene to allow direct assaying β-gal activity of lipBA_she-lacZ transcriptional fusion integrated into E. coli chromosome (Table1). In light of the functional equivalence of Shewanella CRP to E. coli CRP, we firstly compared the alteration of β-gal activity in the model organism E. coli (Δcrp mutant and its parental strain of E. coli). As anticipated, MacConkey plates-based experiments visualized that the lipBA_she promoter-driven β-gal activity is appreciably stronger (illustrated with purple) in the Δcrp mutant than that of the wild type E. coli (low activity denoted by yellow) (Fig.8A). Direct measurement of LacZ activity revealed that deletion of crp gene gave three- to fourfold increment of lipBA_she transcription level (Fig.8B). A similar lacZ reporter construct was also integrated into the chromosome of S. oneidensis wild-type and its Δcrp mutant strains (Fig.8D and E) (Fu et al. 2014). Consequently, the significant alteration/improvement of lipBA_she-lacZ expression level was detected upon the removal of the crp gene from S. oneidensis (Fig.8D and E). Second, the real-time qPCR-based analyses of transcriptional profile showed that no less threefold increment of lipA and/or lipB expression was observed in the Δcrp mutant of S. oneidensis in relative to the wild type strain (Fig.8C). Of particular note, repression of S. oneidensis lipBA expression by CRP depends on production of cyclic AMP (Fig.8E and F). Given the above combined in vitro and in vivo data, we concluded that the cAMP-CRP complex is a repressor for lipBA expression in S. oneidensis.
Figure 8.
In vivo effect of CRP-cAMP complex on lipBA expression of S. oneidensis. (A) MacConkey agar plate-based visualization for effect of Escherichia coli CRP on Shewanella lipBA promoter-driven lacZ transcription. The two E. coli strains with the lipBA-lacZ transcriptional fusion include FYJ457 (WT) and FYJ458 (Δcrp). To assay lipBA-lacZ expression, we used MacConkey agar plate with 0.4% lactose as a sole carbon source. The bacteria were maintained at 37°C for around 36 h. Purple denotes high level of β-gal activity, whereas yellow indicates low level of β-gal activity. (B) β-gal analyses for CRP-mediated regulation of lipBA_she transcription in model organism E. coli. Mid-log phase cultures in RB media were collected to test β-gal activity. The data are expressed in average ± standard deviation (SD), and error bars indicate SD. No less than three independent experiments were performed. The two E. coli strains are FYJ457 (WT) and FYJ458 (Δcrp), respectively. (C) Real-time quantitative PCR (qPCR) assays for altered expression profile of lipA and lipB upon the removal of crp gene from Shewanella. The two strains of Shewanella grown in RB media are MR-1 (S. oneidensis MR-1, WT) and HG0624 (S. oneidensis MR-1, Δcrp). Mig-log phase bacteria were collected for isolation of total RNA. The data are expressed as averages ± standard deviations (SD), and error bars mean SD. Three independent experiments were performed here. Colony comparison (D) and β-gal activity (E) of the S. oneidensis reporter strains carrying the chromosomal lipBA-lacZ fusions grown on minimal medium plates. (F) Direct measurement of bacterial cAMP level. The intracellular (pelleted cells) and extracellular (supernatant) level of bacterial cAMP pools were assayed after centrifugation. A standard curve with cAMP by values of OD450 was generated for each patch of samples. Relative levels were calculated by normalizing to the values of the wild-type, which was set to 1. Both CΔcrp and CΔcyaC strains were designed to express a single copy of the corresponding genes in trans. Error bars represent standard deviations from at least three independent experiments. CRP, cAMP-receptor protein.
Glucose improves the expression of S. oneidensis lipBA in the alternative model microorganism E. coli
It is well known that an addition of glucose into media can lower the level of cytosolic cAMP in E. coli, which might in turn impair at least partially CRP-mediated regulation. Somewhat it is unusual that not all the species of Shewanella genus can utilize/metabolize glucose in that the S. oneidensis glucose transporter-encoding gene glcP is a pseudo-gene with a frame-shift (Romine et al. 2008; Rodionov et al. 2010). Given the above technical problem, we therefore attempted to examine the so-called “glucose effect” with the engineered E. coli strain FYJ457 carrying the lipBA_she-lacZ transcriptional fusion (Fig. S3). As expected, we observed that the level of lipBA expression was induced by the addition of glucose (5 mmol/L) to about threefold higher than that grown in the M9 minimal media with acetate (5 mmol/L) as the sole carbon source (Fig. S3).
Together, we proposed for the first time that the global regulator, the cAMP-CRP complex represses bacterial lipoic acid synthesis in Shewanella, posing the relevance of the cAMP signaling to the production of the sulfur-containing C8 enzyme cofactor, lipoic acid (Fig.1A). This regulatory network can respond to the statue of glucose/cAMP level, i.e., the low glucose/high cAMP level shuts down lipBA expression (Fig.1B), whereas the high glucose/low cAMP level de-represses lipBA transcription (Fig.1C).
Discussion
Biotin and lipoic acid both are sulfur-containing fatty acid derivatives and act as enzyme cofactors required for central metabolism in the three domains of life. Unlike the fact that the regulation of bacterial biotin metabolism has been extensively investigated, the knowledge about genetic control of lipoic acid synthesis remains missing or lagged. Although a recent bioinformatics-based proposal was raised, that is, the PDH repressor involved in E. coli lipoic acid synthesis (Kaleta et al. 2010; Gohler et al. 2011), this prediction was not validated by the physiological evidence in vivo (Feng and Cronan 2014). Through revisiting this long-term unanswered issue, the data shown here might establish for the first time the link of cAMP signaling to bacterial lipoic acid metabolism (Fig.1). Somewhat this finding can resolve the puzzle/discrepancy. As we knew that cAMP-dependent CRP regulatory system is a common global regulator involved in a variety of physiological processes (such as sugar metabolism) in most of the bacterial species, our finding extended this regulatory network into the field of vitamin synthesis.
As an important second messenger, the pool of cAMP molecule is at least determined by the following three factors: First, The activity of cyclic adenylate cyclase (cyaA) is responsible for the formation of cAMP molecules (Of note, 90% of cAMP that is made by intracellular adenylyl cyclases) (Pastan and Perlman 1970; Hantke et al. 2011); Second, the cAMP phosphodiesterase (CpdA) has the opposite enzymatic activity to break a phosphodiester bond of cAMP (Imamura et al. 1996). Not only is the production of CyaA regulated by the CRP regulator at the transcriptional level (Aiba 1985; Qu et al. 2013; You et al. 2013), but was also controlled at the metabolic level by the phosphorylation of EIIAGlc, a core component of PTS system (Crasnier-Mednansky 2008; Deutscher 2008; Gorke and Stulke 2008; Narang 2009). Third, the TolC pump is found to export cAMP outside of E. coli cells in maintaining its sensitivity in the changing metabolic environment (Hantke et al. 2011). Given the fact that all the three homologs of cyaA (SO_4312), cpdA (SO_3901), and tolC (SO_3904) are encoded in Shewanella genomes, it might raise the possibility of unexpected complexity in the linking of the second messenger cAMP signaling to lipoic acid synthesis in the context of Shewanella physiology. More interestingly, we recently discovered a novel enzyme, cAMP deaminase (referred to CadD) from the human pathogen Leptospira interrogans (Goble et al. 2013), and established a new mechanism for quenching the cAMP-dependent signaling. In light that we failed to search a CadD-like homolog, thereby we are not quite sure whether it might be implicated into bacterial lipoic acid synthesis or not yet.
The fact that two critical genes of lipoic acid synthesis lipB and lipA are organized into an operon (Figs.2 and 3), is somehow what we expected, in that it acts as a physiological advantage for economic and effective production of this enzyme cofactor. It is well-known that the genus Shewanella (belonging to the γ-proteobacteria) inhabited in energy-rich, redox-fluctuating environments and in turn evolved to possess diverse metabolic capabilities, e.g., coupling the turnover of organic matter with anaerobic respiration of different electron acceptors (Fredrickson et al. 2008). To our surprise, an appreciably conserved CRP-binding site is constantly present in the promoter regions of lipBA operon from nearly all the Shewanella species with sequenced genomes (Fig.2). It is of no doubt that a common and novel regulatory mechanism for lipoic acid biosynthesis is present in the genus Shewanella.
What kind of selective pressure or evolution consequence does it reflect in adaption to its unique environmental niche? In the paradigm microorganism E. coli, CRP is a global regulator for catabolite repression (Aiba 1983; Schultz et al. 1991; Chandler 1992). However, the protein in S. oneidensis was initially characterized as a principal regulator controlling anaerobic respiration of many electron acceptors (Saffarini et al. 2003). In recent years, it has been repeatedly shown that the regulator in fact plays a more comprehensive role in the physiology, covering both aerobic and anaerobic respiration (Gao et al. 2010; Dong et al. 2012; Fu et al. 2013, 2014; Zhou et al. 2013). In contrast to the pathway-specific regulators BirA (Beckett 2007) and BioR (Feng et al. 2013a,b), both of which negotiate production of the other enzyme cofactor biotin, we believed that Shewanella genus have evolved an unknown strategy to share the cAMP-dependent CRP regulatory architecture with other biological processes to efficiently control lipoic acid synthesis. Given the fact that glucose can induce lipBA expressions (Fig.1B), together with the above information, we concluded that the logic for this kind of regulation does make sense. The reasons are described as follows: (1) the anaerobic growth environment preferred by Shewanella determines an entry of glucose into the glycolytic pathway, giving two pyruvate molecules each glucose; (2) in the Krebs cycle, the resulting pyruvate is catalyzed by PDH to give acetyl-CoA; (3) the full activity of PDH requires the lipoylation, a post-translational modification of protein (which is validated by the scenario seen in the Anti-LA Western blot, i.e., PDH is the prevalent protein form relative to OGDH, Figure5); (4) the protein lipoylation depends on the availability of lipoic acids; (5) de novo LipB-LipA synthesis pathway is necessary to be turned on in addition to the LplA-mediated scavenging route of lipoic acid; (6) de-repression of lipBA expression might facilitate meeting the physiological requirement for lipoic acid production in such situation (vice versa, Fig.1C).
Of particular note, we also detected functional CRP-binding sites ahead of ybeD with unkown function in limited species such as human pathogen S. enterica (Figs.2 and S2). Although that lipB gene is adjacent to ybeD (of note, we lacked evidence proving if they are co-transcribed or not), it required further experimental evidence for CRP regulate lipB or not in this case. It is of interest to test this hypothesis. In fact, it has already been being our research direction in aiming to answer/pursue this question. To the best of our knowledge, our findings reveal, for the first time, a new molecular mechanism for genetic control of bacterial lipoic acid synthesis.
Acknowledgments
This work was supported by a start-up package from Zhejiang University (Y. F.) and National Natural Science Foundation of China (31270097) (H. G). Y. F. is a recipient of the “Young 1000 Talents” Award.
Conflict of Interest
None declared.
Supporting Information
Characterization of Shewanella CRP protein. A. Sequence comparison of CRP proteins from three different organisms. As we described in Figures 2 and 4, the multiple alignments of CRP proteins were carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Identical residues are in white letters with red background, similar residues are in black letters with yellow background, varied residues arein black letters, and dots represent gaps. The predicted secondary structure was shown in top. α: α-helix; β: β-sheet; T: β-turns/coils. The three organisms used here are E. coli, V. cholerae, and S. oneidensis, respectively. (B) SDS-PAGE profile of the purified Shewanella CRP protein. The protein sample was separated with 4–20% gradient Mini-PROTEAN@ TGXTM Gel (Bio-Rad).The monomeric CRP protein with the estimated molecular weight of ∽24 kDa is indicated with an arrow. (C) Modeled structure of Shewanella CRP protein. Structure modeling was proceeded by the software of SPDBV_4.01 using E. coli CRP regulator with known structure (PDB: 2WC2) as structural template. N: N-terminus, C: C-terminus. (D) MS identification the recombinant Shewanella CRP protein. The peptide fragments that match Shewanella CRP protein are highlighted in bold and underlined type (70% coverage in total).
Figure S2. Diversity in binding of bacterial ybeD probes to CRP protein. (A) The CRP site of Shewanella lipBA gene (referred to lipBA_she) can interact with E. coli CRP protein. (B) The predicted CRP site in front of E. coli ybeD-lipB-ybeF-lipA operon (ybeD_ec) has no ability to bind to the CRP protein. The putative CRP sites of the ybeD-lipB-ybeF-lipA operon from Enterobacter sp. 638 (ybeD_es, C) and Klebsiella pneumonia (ybeD_kp, D) are functional. The predicted CRP site 1 of Salmonella enteric ybeD-lipB-ybeF-lipA operon is functional (E), whereas the site 2 is inactive (F). (G) No binding of the cAMP-CRP complex to the suspected CRP site in front of the ybeD-lipB-lipA operon of Yersinia pestis. All the EMSA experiments (7% native PAGE) were conducted as we described (Feng and Cronan 2012; Feng et al. 2013a) with a minor change. The level of cAMP added is 20 pmol. The E. coli CRP protein samples in various concentrations were incubated with 0.2 pmol of DIG-labeled probe in a total volume of 15 µL. A representative result is given. The sequences of all the DNA probes used here are listed in Tables 2 and 3. The minus sign denotes no addition of the CRP protein and/or cAMP molecule. Designations: she, Shewanella; ec, E. coli; es, Enterobacter sp. 638; kp, Klebsiella pneumonia; st, Salmonella typhimurium LT2, and yp,Yersinia pestis.
Figure S3. Induction of Shewanella lipBA expression by glucose in the alternative model E. coli. To test the effect of glucose on Shewanella lipBA expression, the E. coli strain carrying the lipBA_she-lacZ transcriptional fusion (FYJ457) was used here. Mid-log phase cultures in M9 media with acetate and/or glucose (5 mmol/L) as sole carbon source were sampled for assaying β-gal activity. The data from more than three independent experiments is expressed in average ± standard deviation (SD), and error bars indicate SD.
References
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983;32:141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Aiba H. Transcription of the Escherichia coli adenylate cyclase gene is negatively regulated by cAMP-cAMP receptor protein. J. Biol. Chem. 1985;260:3063–3070. [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D. Biotin sensing: universal influence of biotin status on transcription. Annu. Rev. Genet. 2007;41:443–464. doi: 10.1146/annurev.genet.41.042007.170450. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chandler MS. The gene encoding cAMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc. Natl. Acad. Sci. USA. 1992;89:1626–1630. doi: 10.1073/pnas.89.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen QH. Cronan JE. Lipoic acid synthesis: a new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry. 2010;49:10024–10036. doi: 10.1021/bi101215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasnier-Mednansky M. Is there any role for cAMP-CRP in carbon catabolite repression of the Escherichia coli lac operon? Nat. Rev. Microbiol. 2008;6:954. doi: 10.1038/nrmicro1932-c1. ; author reply 954. [DOI] [PubMed] [Google Scholar]
- Cronan JE, Zhao X. Jiang Y. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv. Microb. Physiol. 2005;50:103–146. doi: 10.1016/S0065-2911(05)50003-1. [DOI] [PubMed] [Google Scholar]
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Dong Y, Wang J, Fu H, Zhou G, Shi M. Gao H. A Crp-dependent two-component system regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS One. 2012;7:e51643. doi: 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Kriek M, Bryant P. Roach PL. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew. Chem. 2006;45:5197–5199. doi: 10.1002/anie.200601910. [DOI] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu U, Morgenthal K, Weckwerth W, Parnik T, et al. Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol. 2007;144:1328–1335. doi: 10.1104/pp.107.099317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 2009a;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. A new member of the Escherichia coli fad regulon: transcriptional regulation of fadMybaW. J. Bacteriol. 2009b;191:6320–6328. doi: 10.1128/JB.00835-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Overlapping repressor binding sites result in additive regulation of Escherichia coli FadH by FadR and ArcA. J. Bacteriol. 2010;192:4289–4299. doi: 10.1128/JB.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol. Microbiol. 2011a;80:195–218. doi: 10.1111/j.1365-2958.2011.07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. The Vibrio cholerae fatty acid regulatory protein, FadR, represses transcription of plsB, the gene encoding the first enzyme of membrane phospholipid biosynthesis. Mol. Microbiol. 2011b;81:1020–1033. doi: 10.1111/j.1365-2958.2011.07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Crosstalk of Escherichia coli FadR with global regulators in expression of fatty acid transport genes. PLoS One. 2012;7:e46275. doi: 10.1371/journal.pone.0046275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. PdhR, the pyruvate dehydrogenase repressor, does not regulate lipoic acid synthesis. Res. Microbiol. 2014;165:429–438. doi: 10.1016/j.resmic.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Xu J, Zhang H, Chen Z. Srinivas S. Brucella BioR regulator defines a complex regulatory mechanism for bacterial biotin metabolism. J. Bacteriol. 2013a;195:3451–3467. doi: 10.1128/JB.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang H. Cronan JE. Profligate biotin synthesis in α-proteobacteria - a developing or degenerating regulatory system? Mol. Microbiol. 2013b;88:77–92. doi: 10.1111/mmi.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Napier BA, Manandhar M, Henke SK, Weiss DS. Cronan JE. A Francisella virulence factor catalyses an essential reaction of biotin synthesis. Mol. Microbiol. 2014;91:300–314. doi: 10.1111/mmi.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson JK. Romine MF. Genome-assisted analysis of dissimilatory metal-reducing bacteria. Curr. Opin. Biotechnol. 2005;16:269–274. doi: 10.1016/j.copbio.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- Fu H, Chen H, Wang J, Zhou G, Zhang H, Zhang L, et al. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ. Microbiol. 2013;15:2198–2212. doi: 10.1111/1462-2920.12091. [DOI] [PubMed] [Google Scholar]
- Fu H, Jin M, Ju L, Mao Y. Gao H. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ. Microbiol. 2014;16:3181–3195. doi: 10.1111/1462-2920.12457. [DOI] [PubMed] [Google Scholar]
- Gao H, Wang X, Yang Z, Palzkill T. Zhou J. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genom. 2008;9:42. doi: 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang X, Yang ZK, Chen J, Liang Y, Chen H, et al. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS One. 2010;5:e15295. doi: 10.1371/journal.pone.0015295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goble AM, Feng Y, Raushel FM. Cronan JE. Discovery of a cAMP deaminase that quenches cyclic AMP-dependent regulation. ACS Chem. Biol. 2013;8:2622–2629. doi: 10.1021/cb4004628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohler AK, Kokpinar O, Schmidt-Heck W, Geffers R, Guthke R, Rinas U, et al. More than just a metabolic regulator–elucidation and validation of new targets of PdhR in Escherichia coli. BMC Syst. Biol. 2011;5:197. doi: 10.1186/1752-0509-5-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B. Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Green J, Stapleton MR, Smith LJ, Artymiuk PJ, Kahramanoglou C, Hunt DM, et al. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr. Opin. Microbiol. 2014;18:1–7. doi: 10.1016/j.mib.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A. Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A. Aiba H. Molecular mechanism of negative autoregulation of Escherichia coli crp gene. Nucleic Acids Res. 1991;19:4413–4419. doi: 10.1093/nar/19.16.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A. Aiba H. A new aspect of transcriptional control of the Escherichia coli crp gene: positive autoregulation. Mol. Microbiol. 1992;6:2489–2497. doi: 10.1111/j.1365-2958.1992.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Hantke K, Winkler K. Schultz JE. Escherichia coli exports cyclic AMP via TolC. J. Bacteriol. 2011;193:1086–1089. doi: 10.1128/JB.01399-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hermes FA. Cronan JE. Scavenging of cytosolic octanoic acid by mutant LplA lipoate ligases allows growth of Escherichia coli strains lacking the LipB octanoyltransferase of lipoic acid synthesis. J. Bacteriol. 2009;191:6796–6803. doi: 10.1128/JB.00798-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura R, Yamanaka K, Ogura T, Hiraga S, Fujita N, Ishihama A, et al. Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 1996;271:25423–25429. doi: 10.1074/jbc.271.41.25423. [DOI] [PubMed] [Google Scholar]
- Ishizuka H, Hanamura A, Inada T. Aiba H. Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 1994;13:3077–3082. doi: 10.1002/j.1460-2075.1994.tb06606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Jiang Y, Sun L, Yin J, Fu H, Wu G, et al. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One. 2013;8:e75610. doi: 10.1371/journal.pone.0075610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SW. Cronan JE., Jr The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J. Bacteriol. 2003;185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleta C, Gohler A, Schuster S, Jahreis K, Guthke R. Nikolajewa S. Integrative inference of gene-regulatory networks in Escherichia coli using information theoretic concepts and sequence analysis. BMC Syst. Biol. 2010;4:116. doi: 10.1186/1752-0509-4-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Elias D, Semesi A, Yee A, Cygler M. Gehring K. Structural similarity of YbeD protein from Escherichia coli to allosteric regulatory domains. J. Bacteriol. 2004;186:8083–8088. doi: 10.1128/JB.186.23.8083-8088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- Morris TW, Reed KE. Cronan JE., Jr Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- Morris TW, Reed KE. Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J. Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang A. cAMP does not have an important role in carbon catabolite repression of the Escherichia coli lac operon. Nat. Rev. Microbiol. 2009;7:250. doi: 10.1038/nrmicro1932-c3. [DOI] [PubMed] [Google Scholar]
- Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, et al. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010a;38:D111–D118. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, et al. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 2010b;38:W299–W307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. RegPrecise 3.0 – a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I. Perlman R. Cyclic adenosine monophosphate in bacteria. Science. 1970;169:339–344. doi: 10.1126/science.169.3943.339. [DOI] [PubMed] [Google Scholar]
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- Qu S, Zhang Y, Liu L, Wang L, Han Y, Yang R, et al. Cyclic AMP receptor protein is a repressor of adenylyl cyclase gene cyaA in Yersinia pestis. Can. J. Microbiol. 2013;59:304–310. doi: 10.1139/cjm-2012-0705. [DOI] [PubMed] [Google Scholar]
- Reche PA. Lipoylating and biotinylating enzymes contain a homologous catalytic module. Protein Sci. 2000;9:1922–1929. doi: 10.1110/ps.9.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ. A trail of research from lipoic acid to alpha-keto acid dehydrogenase complexes. J. Biol. Chem. 2001;276:38329–38336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- Reed KE. Cronan JE., Jr Lipoic acid metabolism in Escherichia coli: sequencing and functional characterization of the lipA and lipB genes. J. Bacteriol. 1993;175:1325–1336. doi: 10.1128/jb.175.5.1325-1336.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KE, Morris TW. Cronan JE., Jr Mutants of Escherichia coli K-12 that are resistant to a selenium analog of lipoic acid identify unknown genes in lipoate metabolism. Proc. Natl. Acad. Sci. USA. 1994;91:3720–3724. doi: 10.1073/pnas.91.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CO. Opening a new path to lipoic acid. J. Bacteriol. 2009;191:6782–6784. doi: 10.1128/JB.01151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Yang C, Li X, Rodionova IA, Wang Y, Obraztsova AY, et al. Genomic encyclopedia of sugar utilization pathways in the Shewanella genus. BMC Genom. 2010;11:494. doi: 10.1186/1471-2164-11-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Novichkov PS, Stavrovskaya ED, Rodionova IA, Li X, Kazanov MD, et al. Comparative genomic reconstruction of transcriptional networks controlling central metabolism in the Shewanella genus. BMC Genom. 2011;12(Suppl. 1):S3. doi: 10.1186/1471-2164-12-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romine MF, Carlson TS, Norbeck AD, McCue LA. Lipton MS. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl. Environ. Microbiol. 2008;74:3257–3265. doi: 10.1128/AEM.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarini DA, Schultz R. Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 2003;185:3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz SC, Shields GC. Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Wu L, Wang J, Tang P, Chen H. Gao H. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS One. 2011;6:e21479. doi: 10.1371/journal.pone.0021479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Miller JR, Jiang Y, Marletta MA. Cronan JE. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem. Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Zhao X, Miller JR. Cronan JE. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry. 2005;44:16737–16746. doi: 10.1021/bi051865y. [DOI] [PubMed] [Google Scholar]
- Zheng D, Constantinidou C, Hobman JL. Minchin SD. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Yin J, Chen H, Hua Y, Sun L. Gao H. Combined effect of loss of the caa 3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J. 2013;7:1752–1763. doi: 10.1038/ismej.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of Shewanella CRP protein. A. Sequence comparison of CRP proteins from three different organisms. As we described in Figures 2 and 4, the multiple alignments of CRP proteins were carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Identical residues are in white letters with red background, similar residues are in black letters with yellow background, varied residues arein black letters, and dots represent gaps. The predicted secondary structure was shown in top. α: α-helix; β: β-sheet; T: β-turns/coils. The three organisms used here are E. coli, V. cholerae, and S. oneidensis, respectively. (B) SDS-PAGE profile of the purified Shewanella CRP protein. The protein sample was separated with 4–20% gradient Mini-PROTEAN@ TGXTM Gel (Bio-Rad).The monomeric CRP protein with the estimated molecular weight of ∽24 kDa is indicated with an arrow. (C) Modeled structure of Shewanella CRP protein. Structure modeling was proceeded by the software of SPDBV_4.01 using E. coli CRP regulator with known structure (PDB: 2WC2) as structural template. N: N-terminus, C: C-terminus. (D) MS identification the recombinant Shewanella CRP protein. The peptide fragments that match Shewanella CRP protein are highlighted in bold and underlined type (70% coverage in total).
Figure S2. Diversity in binding of bacterial ybeD probes to CRP protein. (A) The CRP site of Shewanella lipBA gene (referred to lipBA_she) can interact with E. coli CRP protein. (B) The predicted CRP site in front of E. coli ybeD-lipB-ybeF-lipA operon (ybeD_ec) has no ability to bind to the CRP protein. The putative CRP sites of the ybeD-lipB-ybeF-lipA operon from Enterobacter sp. 638 (ybeD_es, C) and Klebsiella pneumonia (ybeD_kp, D) are functional. The predicted CRP site 1 of Salmonella enteric ybeD-lipB-ybeF-lipA operon is functional (E), whereas the site 2 is inactive (F). (G) No binding of the cAMP-CRP complex to the suspected CRP site in front of the ybeD-lipB-lipA operon of Yersinia pestis. All the EMSA experiments (7% native PAGE) were conducted as we described (Feng and Cronan 2012; Feng et al. 2013a) with a minor change. The level of cAMP added is 20 pmol. The E. coli CRP protein samples in various concentrations were incubated with 0.2 pmol of DIG-labeled probe in a total volume of 15 µL. A representative result is given. The sequences of all the DNA probes used here are listed in Tables 2 and 3. The minus sign denotes no addition of the CRP protein and/or cAMP molecule. Designations: she, Shewanella; ec, E. coli; es, Enterobacter sp. 638; kp, Klebsiella pneumonia; st, Salmonella typhimurium LT2, and yp,Yersinia pestis.
Figure S3. Induction of Shewanella lipBA expression by glucose in the alternative model E. coli. To test the effect of glucose on Shewanella lipBA expression, the E. coli strain carrying the lipBA_she-lacZ transcriptional fusion (FYJ457) was used here. Mid-log phase cultures in M9 media with acetate and/or glucose (5 mmol/L) as sole carbon source were sampled for assaying β-gal activity. The data from more than three independent experiments is expressed in average ± standard deviation (SD), and error bars indicate SD.