Abstract
Leishmania donovani, a protozoan parasite, is the causative agent of visceral leishmaniasis. It lives and multiplies within the harsh environment of macrophages. In order to investigate how intracellular parasite manipulate the host cell environment, we undertook a quantitative proteomic study of human monocyte-derived macrophages (THP-1) following infection with L. donovani. We used the isobaric tags for relative and absolute quantification (iTRAQ) method and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to compare expression profiles of noninfected and L. donovani-infected THP-1 cells. We detected modifications of protein expression in key metabolic pathways, including glycolysis and fatty acid oxidation, suggesting a global reprogramming of cell metabolism by the parasite. An increased abundance of proteins involved in gene transcription, RNA splicing (heterogeneous nuclear ribonucleoproteins [hnRNPs]), histones, and DNA repair and replication was observed at 24 h postinfection. Proteins involved in cell survival and signal transduction were more abundant at 24 h postinfection. Several of the differentially expressed proteins had not been previously implicated in response to the parasite, while the others support the previously identified proteins. Selected proteomics results were validated by real-time PCR and immunoblot analyses. Similar changes were observed in L. donovani-infected human monocyte-derived primary macrophages. The effect of RNA interference (RNAi)-mediated gene knockdown of proteins validated the relevance of the host quantitative proteomic screen. Our findings indicate that the host cell proteome is modulated after L. donovani infection, provide evidence for global reprogramming of cell metabolism, and demonstrate the complex relations between the host and parasite at the molecular level.
INTRODUCTION
Visceral leishmaniasis (VL) is a vector-borne neglected tropical disease caused by an obligate intracellular protozoan parasite, Leishmania donovani (1). Infective metacyclic promastigotes infect cells of the monocyte/macrophage lineage, where they survive and multiply as intracellular amastigotes in the parasitophorous vacuole (PV), which acts as a safe haven (2).
Leishmania has developed an intricate relationship with its host, primarily cells of the monocyte/macrophage lineage, where it exploits and subverts the host immune system by either inducing immunosuppression or promoting proparasitic host factors to ensure its survival and growth in an otherwise harsh milieu (3). Hijacking of innate immune functions of macrophages by Leishmania appears to be a multifarious event, as macrophages have inherently evolved to defend the host against invading pathogens by a myriad of effectors rather than providing a favorable environment to the pathogen. The chief molecular mechanisms by which Leishmania is known to inhibit the activation of macrophages toward its own benefit include suppression of deadly antimicrobial free radicals such as nitric oxide (NO), faulty antigen presentation, selective induction and suppression of host cell apoptosis, inhibition of cytokine production and hence cytokine-inducible macrophage function, and activation of T cells (4–8). Leishmania has evolved sophisticated mechanisms to alter the physiological program and activation of adaptive immune responses of host cells by exploiting host cell signaling mechanisms such as the downregulation of Ca2+-dependent classical protein kinase C (PKC) activity and extracellular signal-regulated kinase (ERK) phosphorylation and activity (9, 10). Using mainly host tyrosine phosphatases, Leishmania is known to deactivate mitogen-activated protein kinases (MAPKs) in infected macrophages (5). Extensive manipulations of host cell effector (innate and adaptive) functions by pathogens must be reflected at the levels of transcripts as well as proteins. Enormous efforts made in the field of host gene expression profiling using different (murine and/or human) cell types and different species of Leishmania provide key insights into an extensive modulation of gene function and contribute to a better understanding of the dynamics of gene expression postinfection (11–14). This type of transcriptome-based analysis has major limitations, as it does not represent the true effectors of cellular functions—the proteins.
A recent study based on a comparative proteome analysis revealed differentially expressed proteins in CBA (inbred strain of mouse) macrophages infected with Leishmania amazonensis or L. major in an effort to identify key proteins likely to play a crucial role in determining the course of infection (15). In the present study, we have adopted a quantitative proteomics-based approach to gain insight into the reprogramming of the THP-1 cell line, an acute monocytic leukemia-derived human cell line, exposed to L. donovani for different time periods. Activated THP-1 cells have long been used as a versatile model system to study inflammatory responses, host cell apoptosis, and autophagy behavior in response to intracellular pathogens (16, 17). This model may not completely replicate the in vivo conditions after infection, but it is a well-established in vitro model system to study the host-Leishmania interface (18).
In the present study, quantitative changes in the dynamics of the host proteome status at intervals of 12, 24, and 48 h after infection with L. donovani were tracked by using isobaric tags for relative and absolute quantification (iTRAQ) followed by high-resolution mass spectroscopy (MS). Our data not only provide corroborating evidence for some previously identified specific proteins but also are indicative of a global reprogramming of host metabolic and regulatory events by L. donovani.
MATERIALS AND METHODS
Parasites.
L. donovani strain AG83 (MHOM/IN/1983/AG83), used in the present study, was routinely maintained in BALB/c mice by repeated passage to maintain its virulence. Amastigotes were routinely isolated from the spleen of infected BALB/c mice and were made to undergo transformation from amastigotes to promastigotes, prior to infection, as reported previously (19, 20). Promastigotes of L. donovani AG83 were routinely cultured in modified M199 medium (Sigma, St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco/BRL, Life Technologies, Scotland, United Kingdom) and 0.13 mg/ml penicillin-streptomycin at 26°C.
Cell culture and infection.
The THP-1 cell line, an acute monocytic leukemia-derived human cell line (ATCC TIB-202TM), was cultivated in suspension at a density of 105 to 106 cells/ml in RPMI 1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% heat-inactivated FBS and 1% streptomycin-penicillin at 37°C in 5% CO2. PMA (phorbol 12-myristate 13-acetate; Sigma, St. Louis, MO, USA)-treated and subsequently differentiated THP-1 cells act like macrophages and are widely used to study monocyte-derived macrophage (MDM) behavior. The differentiation of THP-1 cells into macrophages was induced by incubating cells for 48 h with 50 ng/ml PMA at 37°C in 5% CO2 in flat-bottom 6-well tissue culture plates (Greiner Bio-One, Germany).
Human peripheral blood mononuclear cells (PBMCs) were isolated by using a Ficoll Paque density gradient (Sigma, St. Louis, MO, USA) on whole blood collected from healthy donors. MDMs were isolated as described previously (21). Briefly, isolated monocytes were cultured overnight in 6-well plates at a density of 2 × 106 cells per well in the presence of macrophage colony-stimulating factor (M-CSF) (300 ng/ml) in RPMI medium with 10% fetal calf serum (FCS). Nonadherent cells were repeatedly washed, and cells were allowed to differentiate into MDMs in the presence of M-CSF and 10% FBS. Differentiated MDMs were harvested after 96 h.
For infection experiments, THP-1 cells as well as MDMs were harvested and distributed into six-well plates at 0.5 × 106 cells/well. Uninfected cells were used as controls. Stationary-phase promastigotes were added to confluent cells at an infection ratio of 20:1 to initiate infection. Excess noninfective promastigotes were removed by several washes with phosphate-buffered saline (PBS). An infection ratio of 20 promastigotes per macrophage was consistently found to be optimal, with an average infection ratio of almost 8 parasites per macrophage at 48 h postinfection. This infection ratio was hence selected for the following experiments. Infected cells were incubated for different time periods to allow the establishment of infection and the transformation and multiplication of the intracellular parasites.
Cell harvest and protein extraction.
Uninfected and infected THP-1 cells were harvested by the addition of 300 μl of 0.05% EDTA per well. Proteins were extracted from cells by frequent washing in ice-cold PBS followed by lysis in membrane lysis buffer (Invitrogen, Carlsbad, CA, USA). Briefly, Halt protease inhibitor cocktail (Thermo Fisher Scientific, USA), phenylmethylsulfonyl fluoride (Sigma, St. Louis, MO, USA), and the nuclease inhibitor benzonase (Sigma, St. Louis, MO, USA) were added to the lysis buffer. Several freeze-thaw cycles were performed in order to completely lyse the cells. The cell lysate was centrifuged at 13,000 × g for 20 min at 4°C, the supernatant was collected, and the protein concentration was determined by using a Bio-Rad protein assay kit (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Quantitative proteomics using iTRAQ labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS).
iTRAQ labeling was performed according to the manufacturer's protocol (AB Sciex, Foster City, CA). Briefly, equal amounts of protein (100 μg) from control cells (uninfected THP-1 cells) and infected THP-1 cells were precipitated with acetone. Proteins from each sample were dissolved in 20 μl of dissolution buffer (0.5 M trimethylammonium bicarbonate) and 1 μl of denaturant reagent (2% SDS) and reduced for 1 h at 60°C with 2 μl reducing agent. Cysteine blocking was performed by using 1 μl of cysteine blocking reagent for 10 min at room temperature. Tryptic digestion was performed by the addition of 10 μl of a trypsin solution (Sigma, USA) (1 μg/μl in double-distilled water), followed by incubation for 16 h at 37°C. The peptides were labeled by using appropriate iTRAQ labels (−114, −115, −116, and −117) (see Table S1 in the supplemental material) according to the iTRAQ reagent multiplex kit protocol. Differentially labeled samples were pooled and fractionated by using strong-cation-exchange (SCX) chromatography.
The labeled peptide mixture was separated by SCX chromatography using a high-performance liquid chromatography (HPLC) system (1200 series, quaternary pumps; Agilent) with a UV detector. The labeled samples were resuspended in SCX low-ionic-strength buffer (5 mM ammonium formate, 30% acetonitrile [ACN]) and loaded onto a PolyLC Polysulfoethyl A Zorbax-A SCX column (5 μm [2.1 mm by 150 mm]; Agilent). Peptide elution was performed by increasing the salt concentration (5 to 500 mM ammonium formate) and was detected at an absorbance of 214 nm. Initially, a total of 60 SCX fractions were collected, which in turn were pooled to obtain 40 fractions. The concatenation (pooling of equal-interval fractions) strategy was applied to reduce the peptide overlap among SCX fractions.
After SCX fractionation, the fractions were desalted by using desalting columns (ZipTip; Millipore) manually. The eluates were finally concentrated by using a speedVac concentrator (Thermo Scientific) prior to analysis by MS. Eluted fractions were further separated by reverse-phase chromatography using a Nano-LC column (Agilent Technologies, Germany). Mass spectrometric analysis of the column eluate was done by using a hybrid quadrupole LIT (linear ion trap) mass spectrometer (4000 Q Trap LC-MS/MS system; AB Sciex). For collision-induced dissociation (CID), the curtain gas was set at 15 lb, nitrogen was used as the collision gas, and the ionization tip voltage was 2,000 V. The information-dependent acquisition (IDA) mode of operation was used for the acquisition of survey scans from 400 to 1,600 atomic mass units (amu), followed by CID of the three most intense ions.
The peptide MS/MS spectra were extracted by using Analyst 1.4.2 software (AB Sciex). The MS/MS spectral data were analyzed for protein identification and quantification by using Protein Pilot v.3 software, revision 114732 (AB Sciex). The Paragon searching algorithm from Protein Pilot was set up to search iTRAQ 4-plex samples as variable modifications and with methyl methane thiosulfonate (MMTS) as a fixed modification. Detailed information about the Paragon algorithm was reported previously (22). The raw peptide identification results from the Paragon algorithm (AB Sciex) searches were further processed by the Pro Group algorithm (AB Sciex) within the Protein Pilot software before display. The human reference proteome (uniprot_sprot_20081216+Contams.fasta) and the Leishmania infantum database (release 4.0; http://tritrypdb.org/) were used for protein identification. The Pro Group algorithm (AB Sciex) within the Protein Pilot software was used to perform automatic bias correction to remove variations imparted due to unequal mixing of differentially labeled peptide samples (22). Search effort, through the Pro Group algorithm in the Protein Pilot software, automatically calculated the reporter peak area, effort factor (EF), and P value. The Unused-Prot score is Protein Pilot's measurement of protein identification confidence taking into account all peptide evidence for a protein, excluding any evidence that is better explained by a higher-ranking protein. The Proteomics System Performance Evaluation Pipeline incorporated directly into Protein Pilot was employed to conduct target/decoy searches to facilitate false-positive discovery rate (FDR) estimation. A FDR threshold of 5% was employed for identification and quantification.
The parameters that were used for identification and quantification of differentially abundant proteins included (i) proteins identified as having ≥95% confidence (Unused-Prot score, >1.3) and a minimum of two peptides, (ii) proteins with fold differences of ≥1.5 or ≤0.67, and (iii) proteins detected in a minimum of two technical replicates.
However, a large number of proteins that were significantly modulated in a minimum of 2 to 3 replicates but identified with a single peptide (having ≥95% confidence) were also included in the proteome screen. The correctness of identification was ascertained by using the Proteomics System Performance Evaluation Pipeline incorporated directly into Protein Pilot to conduct the false-positive discovery rate estimation for some of the proteins that were identified with a minimum of 1 representative peptide to authenticate the quantification. Although these proteins pass the statistical filter of Protein Pilot, their differential expression would be tentative.
Furthermore, at each time point, three biological replicates originating from three different cultures were used to perform quantitative proteomic experiments. Each biological replicate in turn had two different technical replicates represented by THP-1 cells infected with either clone 1 (C1) or clone 2 (C2) of the AG83 strain. The ratios of peak areas for each of the signature ions were obtained and bias corrected, according to the manufacturer's instructions, to account for isotopic overlap (22, 23). ProGROUP software (AB Sciex) was used to pool the data from all the experiments. A single protein identification was assigned to each protein, thus allowing comparison of replicate data sets.
Protein identification, functional classification, and determination of possible subcellular origins for each protein were performed by using Gene Ontology (GO) annotations (http://www.geneontology.org/). GO provides a set of hierarchical controlled vocabulary split into 3 categories, biological process, cellular component, and molecular function, that can be manually or electronically assigned to a UniProt KB entry.
RNA preparation and real-time PCR analysis.
Total RNA was isolated from THP-1 cells and human MDMs, infected or not infected with L. donovani, by using Tri reagent (Sigma, St. Louis, MO, USA) and subsequently treated with RNase-free DNase (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturers' instructions. The concentration and purity of RNA were determined by using a Nanodrop instrument (Thermo Fisher Scientific, USA). RNA (500 ng) was reverse transcribed by using a first-strand cDNA synthesis kit (Thermo Scientific, MA, USA). Controls having the same amount of RNA but lacking reverse transcriptase or the template were used to rule out the presence of DNA or any other contamination. Real-time PCR (RT-PCR) with gene-specific primers was performed on the resulting cDNA by using SYBR Fast green double-stranded DNA binding dye (Applied Biosystems, CA, USA) and the ABI Prism 700 sequence detection system instrument (Applied Biosystems, CA, USA). NCBI gene identifiers used in the study, primer sequences, and product sizes are given in the Table S2 in the supplemental material. The following program was used for PCR amplification: 50°C for 2 min, followed by 40 cycles at 95°C for 30 s, 62°C for 1 min, and 72°C for 20 s. The efficiency of primers was assessed by performing 10-fold dilution series experiments with cDNA to obtain the average slope of each primer pair (data not shown). The generation of specific PCR products was confirmed by melting curve analysis. All sample analysis was performed in triplicate. Amplification of U6 small nuclear RNA (RNU6A) was used as an internal control. To quantify gene expression, the comparative threshold cycle method for relative quantification (2−ΔΔCT = n-fold) was used.
RNA interference (RNAi) and intracellular amastigote screening.
Gene knockdown was performed to validate the proteins whose proteomics identification pattern suggested differential expression in uninfected and L. donovani-infected THP-1 cells.
Gene knockdown.
THP-1 cells were seeded into three 384-well plates at a confluence of 15,000 cells per well in 50 μl of RPMI supplemented with 10% FBS (Gibco, USA) and 50 ng/ml of PMA (Sigma, St. Louis, MO, USA) and incubated for 48 h at 37°C with 5% CO2. Once differentiated THP-1 cells attached to the bottom of the well, the cells were washed twice with PBS, followed by transfection with small interfering RNA (siRNA; Dharmacon, USA) (see Table S3 in the supplemental material). Briefly, transient transfection with siRNAs was carried out by using RNAi Max (Invitrogen). For each well, 3.9 μl of serum-free Opti-MEM (Invitrogen) and 0.1 μl of RNAi Max were preincubated for 5 min at room temperature. At the same time, 3.5 μl of serum-free Opti-MEM was mixed with 0.5 μl of each siRNA (5 μM) and also incubated for 5 min at room temperature. These two mixtures were combined and incubated for 20 min at room temperature for complex formation. After the addition of 42 μl of complete Dulbecco's modified Eagle's medium (DMEM) to the mixture, the entire solution was added to the cells in each well, resulting in a final concentration of 50 nM siRNAs. After transfection, cells were incubated for 48 h to allow gene silencing. The wells were aspirated, and 3 × 105 Leishmania parasites obtained from stationary-phase culture (5 days after seeding) were added to each well in 50 μl of RPMI supplemented with 10% FBS.
Intracellular amastigote screening.
The plates were fixed at 12, 24, and 48 h postinfection by the addition of 25 μl of a 2% formaldehyde solution. After incubation for 4 h, the wells were washed 5 times with PBS (using a BioTek 406 automated liquid handler), and finally, 10 μl of 5 μM Draq5 staining solution (catalog no. DR50200; Biostatus) was added. The plates were incubated with the staining solution for at least 12 h prior to reading in an Operetta automated fluorescence microscope (PerkinElmer) with a 635-nm filter at a ×20 lens magnification. Each well was imaged 5 times, and the images were analyzed by using LeishPlugin, designed by the IPK-Image Mining software suite. LeishPlugin was able to detect and quantify both host cells and intracellular parasites (24).
SDS-PAGE and Western blotting.
Cell lysis was carried out by using cell lysis buffer (0.125 mol/liter Tris, 4% SDS, 20% glycerol, and 10% 2-mercaptoethanol). A CB XTM protein assay kit (G Biosciences, USA) was used to determine the protein concentration. The cell lysate (30 to 60 μg) was electrophoresed in 12% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes at 4°C. Membrane blocking was performed in 5% fat-free milk in 0.05% PBS-Tween 20 (PBST) overnight at 4°C. The membranes were incubated with primary antibodies against hnRNP K (R332; Cell Signaling) (1:1,000), mitochondrial antiviral signaling protein (MAVS) (E-3; Santa Cruz Biotechnology) (1:1,000), protein tyrosine phosphatase nonreceptor type 6 (PTPN6)/Src homology region 2 domain-containing phosphatase 1 (SHP-1) (C-19; Santa Cruz Biotechnology) (1:1,000), the 78-kDa glucose-regulated protein (GRP78) (A-10; Santa Cruz Biotechnology) (1:1,000), high-mobility-group protein I (HMG-I) (EPR7839; Abcam) (1:10,000), histone H4 (L64C1; Cell Signaling) (1:1,000), and beta-tubulin (Thermo Scientific, USA) (1:5,000) prepared in PBST at room temperature for 1 h. The blots were subsequently incubated with the secondary antibody conjugated to horseradish peroxidase in PBST for 1 h at room temperature. Protein bands were observed on X-ray film by using an enhanced chemiluminescence kit (G Biosciences, USA) (25).
Ethical statement.
All experiments performed on human MDMs were approved by the Institutional Human Ethical Committee of the National Institute of Immunology under project no. IHEC#84/14.
Statistical analysis.
All proteomics results were analyzed and expressed as means ± standard errors of means (SEM). Proteins detected and quantified in a minimum of two replicates at each time point were considered for analysis. One-way analysis of variance (ANOVA) was applied to analyze the protein fold abundance at different time points. Student's t test was applied to analyze the real-time PCR data and the effect of knockdown on the infection ratio and intracellular amastigotes. Western blot results are representative of three independent experiments. P values of <0.05 were considered statistically significant. All statistical analyses were performed by using GraphPad Prism version 5.01.
Public availability and accessibility of proteomic data.
To make our findings publicly available and accessible to the research community, we have submitted our quantification data and the list of proteins and peptides identified to the Human Proteinpedia (HUPA) (http://www.humanproteinpedia.org/) (26, 27). The data (accession number HuPA_00807) can be freely downloaded from the following links: http://www.humanproteinpedia.org/data_display?exp_id=00807 for experiment details and http://www.humanproteinpedia.org/download_data?exp_id=00807 for download data.
RESULTS AND DISCUSSION
Identification and relative abundance of total proteins.
A quantitative proteomic approach using isobaric tagging and high-resolution mass spectrometry was used to compare differential host proteome abundances in response to L. donovani infection. A proteome comparison of infected and uninfected cells was made at 12, 24, and 48 h postinfection. The major limitation of performing differential host proteome modulation studies where the intracellular eukaryotic pathogen is still multiplying inside parasitophorous vacuoles (PVs) is contaminants originating from pathogenic peptides. To circumvent this issue, an equal amount of digested proteins isolated from Leishmania promastigotes that were used to infect macrophages was mixed with host proteins (uninfected control sample) just prior to peptide labeling using isobaric tags. This kind of approach has previously been used to normalize any signal related to the pathogen proteome (28). Moreover, parasite proteins represented a relatively small proportion of the total proteins, and any proteins that were of parasite origin were easily detected by MS identification, thereby ensuring that they were not erroneously included in the subsequent analysis.
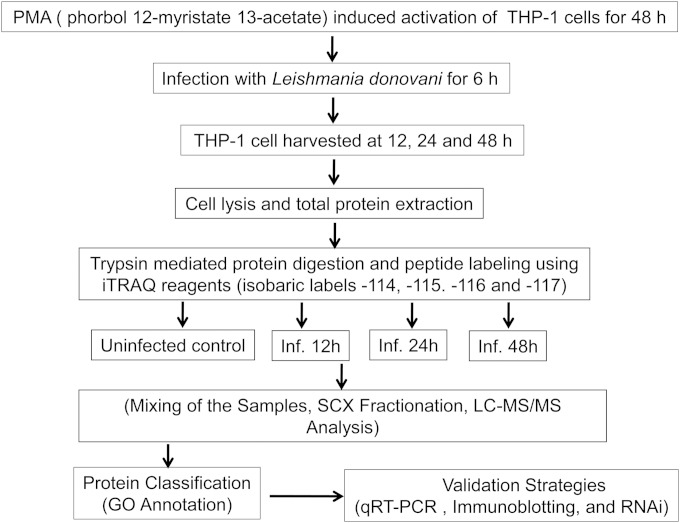
Figure 1 represents the workflow of the complete experimental strategy used in this study. Three independent biological replicate experiments were performed to increase the reproducibility and coverage of the total proteins identified. Sets A, B, and C represent three biological replicates at each time point (see Table S4 in the supplemental material). The technical replicates for each set are in turn represented as groups I and II (see Table S4 in the supplemental material). A total number of 1,624 proteins were identified at all three points.
FIG 1.
Workflow of the experimental strategy used in this study. Inf, infection.
Total numbers of nonredundant proteins identified at 12 h postinfection in sets A, B, and C were 192, 153, and 155, respectively (see Table S4A in the supplemental material). Furthermore, a total number of 329 nonredundant proteins (merged data from sets A, B, and C) were identified in THP-1 cells at 12 h postinfection (see Table S4B in the supplemental material). Total numbers of proteins identified in THP-1 cells at 24 h postinfection in set D, E, and F were 263, 197, and 221, respectively (see Table S4C in the supplemental material). A total number of 441 nonredundant proteins (merged data from sets D, E, and F) were identified, 32 of which were not quantified, while 12 proteins had unknown function (see Table S4D in the supplemental material). At 48 h postinfection, total numbers of proteins identified in host cells were 83, 167, and 193 in sets G, H, and I, respectively (see Table S4E in the supplemental material). A total number of 310 nonredundant proteins (merged data from sets G, H, and I) were identified, 29 of which were not quantified, while 4 proteins had unknown function (see Table S4F in the supplemental material). The total number of nonredundant proteins identified in this study was 637 (see Table S4G in the supplemental material). The low number of nonredundant proteins identified in the present proteome screen could be attributed to the AB Sciex 4000 Q Trap instrument, which has a relatively lower scan speed (4,000 amu/s) than those of new-generation high-end mass spectrometers.
Identification and functional annotation of differentially abundant protein signals.
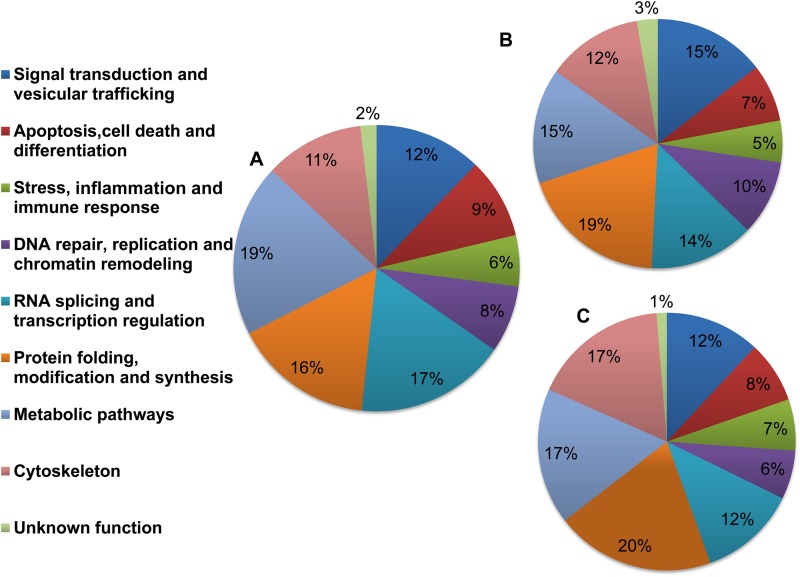
Using Gene Ontology (GO) annotations (http://www.geneontology.org/), 9 major functional classes were identified, namely, signal transduction and vesicular trafficking; apoptosis, cell death, and differentiation; stress, inflammation, and immune response; DNA repair, replication, and chromatin remodeling; RNA splicing and transcription regulation; protein folding, modification, and synthesis; metabolic pathways; cytoskeleton; and proteins with unknown function. The pie diagrams in Fig. 2A to C show the percent distributions of functionally annotated proteins at 12, 24, and 48 h post-Leishmania infection.
FIG 2.
Pie diagrams showing functional annotations and relative distributions of the total proteins detected in THP-1 cells infected with L. donovani. Nine major functional classes were identified based on their cellular function by using iTRAQ methodology as described in Materials and Methods. Percent distributions of the functionally annotated host proteins at 12 h (A), 24 h (B), and 48 h (C) after Leishmania infection are shown.
Pattern of differentially abundant proteins.
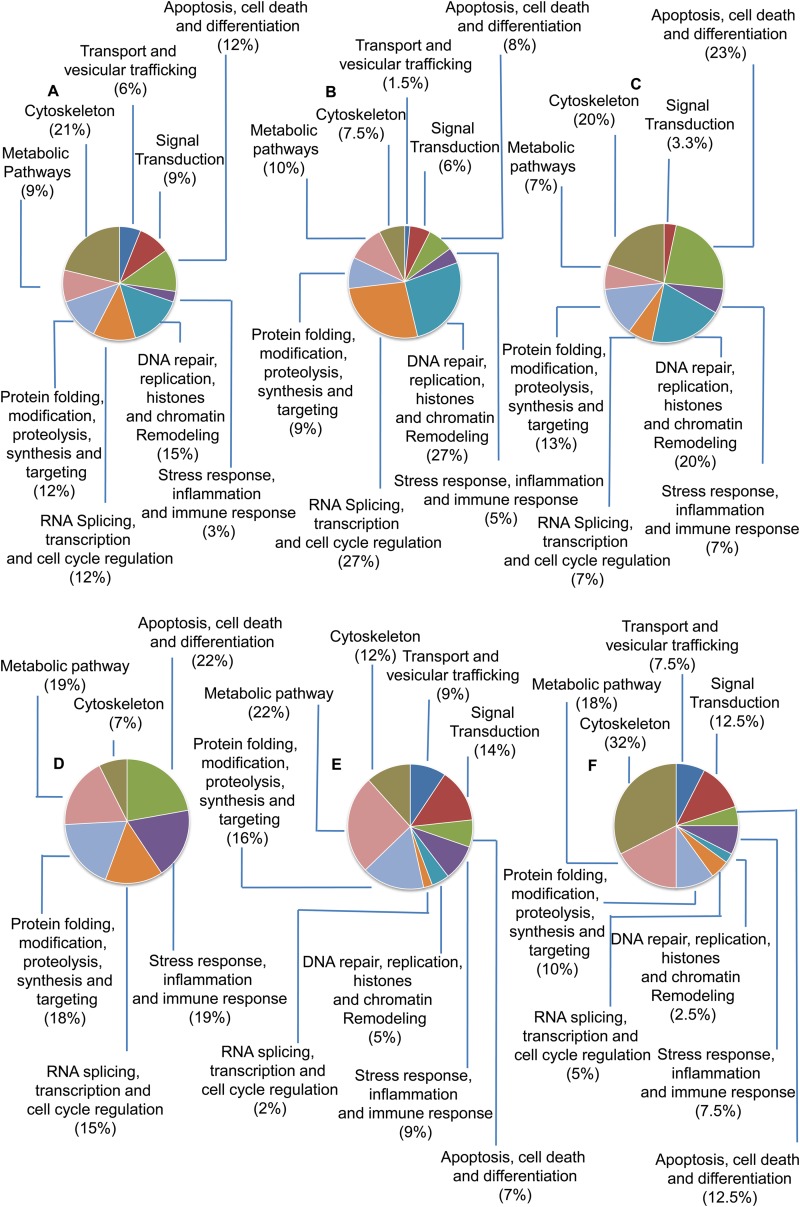
A list of differentially modulated proteins in THP-1 cells infected with L. donovani is shown in Table S5 in the supplemental material. A total of 199 host proteins were modulated at 12, 24, and 48 h post-Leishmania infection. Table S5 in the supplemental material represents some of the important proteins involved in signal transduction, cell survival responses, immune responses, chromatin remodeling, and RNA splicing that were critically modulated in macrophages after infection. The numbers of proteins that showed increased abundance at 12, 24, and 48 h in infected THP-1 cells compared to uninfected cells were 35, 69, and 30, respectively (see Table S5 in the supplemental material). The numbers of proteins that were found in low abundance were 27, 45, and 40 at 12, 24, and 48 h, respectively (see Table S5 in the supplemental material). The functional distribution of differentially modulated host proteins at 12, 24, and 48 h post-Leishmania infection is shown in Fig. 3. Nuclear proteins constituted >50% of the total proteins and were found in high abundance at 24 h post-Leishmania infection (Fig. 3B). Proteins involved in RNA splicing (hnRNPs) and transcription regulation were substantially more abundant at 24 h postinfection, and this probably represents the most intense phase of transcript-based information reprogramming in the host cells. Histones and proteins involved in chromatin remodeling were highly abundant at all three time points (Fig. 3A to C). This could possibly be an outcome of selective pressure of higher gene expression levels in infected cells, in order to cater to the need of increased metabolic events. Percentages of less abundant proteins at 12, 24, and 48 h are shown in Fig. 3D to F, respectively.
FIG 3.
Functional distribution of differentially modulated host proteins (more and less abundant proteins) at 12, 24, and 48 h after Leishmania infection. (A to C) Percentages of more abundant proteins at 12 h (A), 24 h (B), and 48 h (C) postinfection. (D to F) Percentages of less abundant proteins at 12 h (D), 24 h (E), and 48 h (F) postinfection.
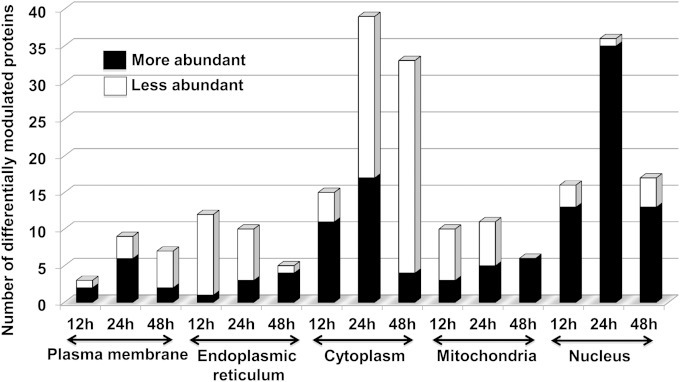
A further classification of differentially abundant proteins based on their origin of subcellular location, based chiefly on GO annotations (http://www.geneontology.org/), indicated a critical involvement of the endoplasmic reticulum (ER), mitochondria, and nucleus at different time points during the course of infection and parasite multiplication. Mitochondrial and ER-resident proteins were in abundance at 48 h postinfection (Fig. 4). The majority of nuclear proteins were abundant at all time points; however, the number was highest at 24 h after Leishmania infection (Fig. 4). In the present study, transcriptional events represent some of the most important cellular events deregulated at 24 h postinfection.
FIG 4.
Gene Ontology-based classification of differentially abundant proteins present in different intracellular compartments.
A summary list of differentially modulated proteins in THP-1 cells infected with L. donovani compared to uninfected cells at 12 h, 24 h, and 48 h is shown in Table 1. These proteins were further classified into the following categories: signal transduction, apoptosis, cell death and differentiation, immune and inflammatory responses, chromatin remodeling, and RNA splicing. We selected representative proteins from each group for further validation by quantitative real-time PCR and Western blotting.
TABLE 1.
List of selected differentially modulated proteins in THP-1 cells infected with Leishmania donovani compared to uninfected cells at 12 h, 24 h, and 48 ha
| Category and protein name | UniProt accession no. | Biological process(es) | 12 h |
24 h |
48 h |
|||
|---|---|---|---|---|---|---|---|---|
| Mean fold change in abundance (±SD) | No. of replicate expts | Mean fold change in abundance (±SD) | No. of replicate expts | Mean fold change in abundance (±SD) | No. of replicate expts | |||
| Signal transduction | ||||||||
| Adenylyl cyclase-associated protein | Q01518 | Activation of adenylate cyclase activity | 0.93 (±0.01) | 3 | 0.44 (±0.21) | 3 | 0.25 (±0.01) | 3 |
| cAMP-responsive element binding protein 3-like protein 4 | Q8TEY5 | Positive regulation of transcription from RNA polymerase II promoter | 15.3 (±1.98) | 3 | ||||
| Coronin 1C | Q9ULV4 | Actin cytoskeleton organization | 2.28 (±1.03) | 2 | 0.84 (±0.06) | 2 | ||
| Receptor-interacting serine/threonine protein kinase 1 | Q13546 | TRIF-dependent Toll-like receptor signaling pathway | 3.06 (±0.94) | 2 | ||||
| Protein tyrosine phosphatase nonreceptor type | P29350 | JAK-STAT cascade involved in growth hormone signaling pathway | 0.22 (±0.005) | 2 | ||||
| Apoptosis, cell death, and differentiation | ||||||||
| 78-kDa glucose-regulated protein | P11021 | ER overload response | 0.43 (±0.04) | 3 | 1.03 (±0.16) | 3 | 1.95 (±0.38) | 3 |
| 10-kDa heat shock protein, mitochondrial | P61604 | Activation of cysteine-type endopeptidase activity involved in the apoptotic process | 0.59 (±0.05) | 3 | 0.84 (±0.12) | 3 | 1.9 (±0.88) | 3 |
| Vimentin | P08670 | Cellular component disassembly involved in the execution phase of apoptosis | 0.58 (±0.16) | 3 | 1.74 (±0.55) | 3 | 2.19 (±0.10) | 3 |
| High-mobility-group protein HMG-I/HMG-Y | P17096 | DNA unwinding involved in replication | 1.56 (±0.07) | 3 | 3.54 (±0.1) | 3 | 5.74 (±0.34) | 3 |
| Annexin A5 | P08758 | Negative regulation of the apoptotic process | 0.34 (±0.13) | 3 | 0.35 (±0.12) | 2 | ||
| Immune and inflammatory responses | ||||||||
| Mitochondrial antiviral signaling protein | Q7Z434 | Activation of innate immune response | 7.86 (±3.55) | 2 | ||||
| Interleukin-1β | P01584 | Cytokine-mediated signaling pathway | 0.75 (±0.45) | 2 | 0.55 (±0.03) | 2 | ||
| CD166 antigen | Q13740 | Cell adhesion | 1.71 (±0.12) | 2 | ||||
| Chromatin remodeling and RNA splicing | ||||||||
| Histone H4 | P62805 | CENP-A containing nucleosome assembly at the centromere | 1.62 (±0.28) | 3 | 5.51 (±2.94) | 3 | 13.6 (±3.23) | 3 |
| Nucleolin | P19338 | Transcription regulation | 1.46 (±0.43)* | 3 | 3.23 (±2.3) | 3 | 1 (±0.02) | 3 |
| Heterogeneous nuclear ribonucleoprotein A2/B1 | P22626 | RNA transport and mRNA splicing via the spliceosome | 0.89 (±0.04) | 3 | 2.96 (±0.49) | 3 | 1.0 (±0.029) | 3 |
| Heterogeneous nuclear ribonucleoprotein H | P31943 | mRNA splicing via the spliceosome | 0.98 (±0.24) | 2 | 2 (±0.54) | 3 | 0.99 (±0.108) | 3 |
| Heterogeneous nuclear ribonucleoprotein D0 | Q14103 | mRNA splicing via the spliceosome | 0.71 (±0.08) | 3 | 3.07 (±1.64) | 3 | 1.08 (±0.09) | 3 |
| Heterogeneous nuclear ribonucleoprotein A1 | P09651 | RNA export from the nucleus and mRNA splicing via the spliceosome | 1.17 (±0.26) | 3 | 2.57 (±0.25) | 2 | ||
| Heterogeneous nuclear ribonucleoprotein K | P61978 | mRNA splicing via the spliceosome | 0.46 (±0.23) | 3 | 2.64 (±0.34) | 3 | 0.88 (±0.28) | 3 |
| Heterogeneous nuclear ribonucleoprotein C1/C2 | P07910 | mRNA splicing via the spliceosome | 1.13 (±0.10) | 3 | 2.28 (±1.04) | 3 | 1.37 (±0.24) | 3 |
| Heterogeneous nuclear ribonucleoprotein A3 | P51991 | mRNA splicing via the spliceosome | 0.69 (±0.13) | 2 | 2.79 (±1.16) | 3 | 1.16 (±0.15) | 3 |
The parameters used for identification and quantification of differentially modulated proteins included (i) proteins having ≥95% confidence (Unused-Prot score of >1.3); (ii) proteins with fold differences of ≥1.5 or ≤0.67, which were considered more abundant or less abundant, respectively; and (iii) proteins detected in a minimum of two technical replicate experiments. Fold changes represent the fold changes in abundance (higher/lower) in infected versus uninfected THP-1 cells. TRIF, TIR domain-containing adapter-inducing IFN-β; CENP-A, centromere protein A. *, nearest fold ratio for threshold cutoff is threshold cutoff ± 0.05.
Leishmania infection modulates hnRNPs in host cells.
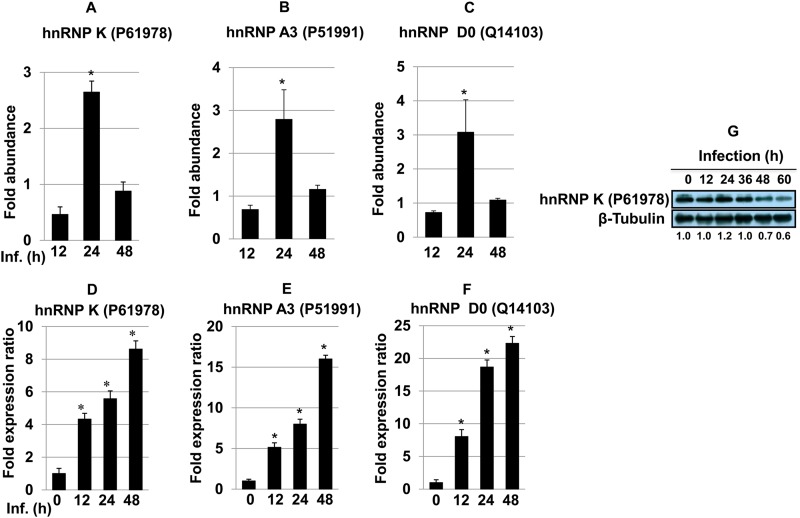
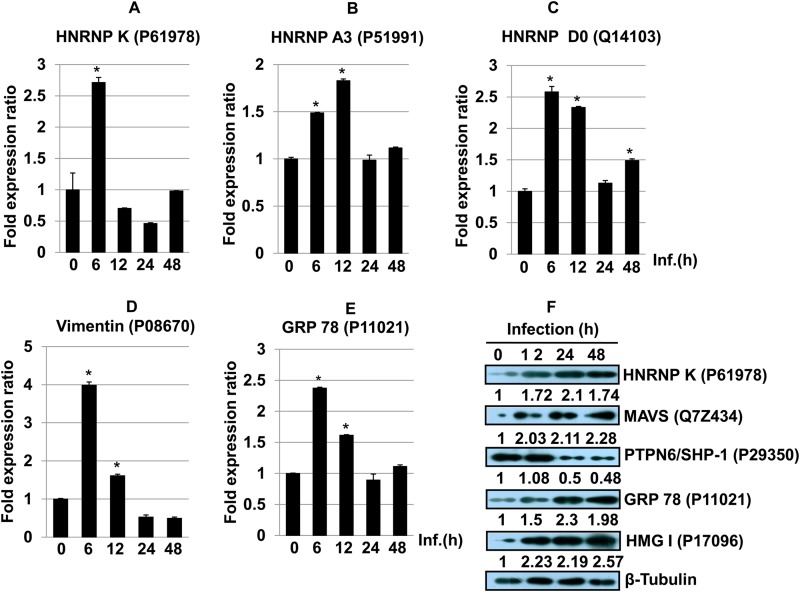
Regulatory factors involved in alternative splicing include trans-acting proteins (repressors and activators) and corresponding cis-regulatory sites (silencers and enhancers) on the pre-mRNA (29). hnRNPs are repressors of RNA splicing and act by blocking the access of the spliceosome to the polypyrimidine tract of RNA (30). Apart from their role in splicing, these proteins are also involved in mRNA transport, regulating the secondary structure of pre-mRNA molecules and hence determining their interaction with RNA binding proteins, ultimately generating RNA base information diversity (29). We observed increased abundances of several hnRNPs, such as hnRNP K (UniProt KB accession no. P61978) (∼2.6-fold change at 24 h) (Fig. 5A), hnRNP A3 (accession no. P51991) (∼2.8-fold) (Fig. 5B), and hnRNP D0 (accession no. Q14103) (∼3.0-fold) (Fig. 5C), at 24 h postinfection (Table 1). The relative gene expression patterns of hnRNP K (Fig. 5D), hnRNP A3 (Fig. 5E), and hnRNP D0 (Fig. 5F) in infected THP-1 cells were confirmed by using quantitative real-time PCR. Consistent and increased expression was observed at all time points, although it did not reflect the exact protein abundance pattern identified in the proteomics screen. This could be due to an increased stability of the corresponding transcripts. Immunoblot analysis was done to further validate the protein expression of hnRNP K in THP-1 cells after Leishmania infection (Fig. 5G). The maximum expression level of hnRNP K was observed at 24 h, followed a by time-dependent decrease in expression, thereby further validating the proteomics screen results.
FIG 5.
(A to C) Bar diagrams showing differential abundances of hnRNP K (A), hnRNP A3 (B), and hnRNP D0 (C) in host macrophages after Leishmania infection at different time points. One-way ANOVA was applied to proteomics data to analyze the fold abundances. (D to F) The expression of hnRNP K (D), hnRNP A3 (E), and hnRNP D0 (F) was assessed by qRT-PCR. Real-time PCR data represent means from three independent experiments. Student's t test was applied to analyze the data. U6 small RNA (RNU6A) was used as an internal control. Data analysis was performed by using the 2−ΔΔCT method. *, P < 0.05. Inf., infection. (G) Western blot showing time-dependent expression of hnRNP K in THP-1 cells infected with L. donovani. Beta-tubulin was used as a loading control. Densitometric analysis shows the fold change in expression in infected THP-1 cells with respect to the untreated control group. The data are representative of three independent experiments. UniProt accession numbers are shown in parentheses.
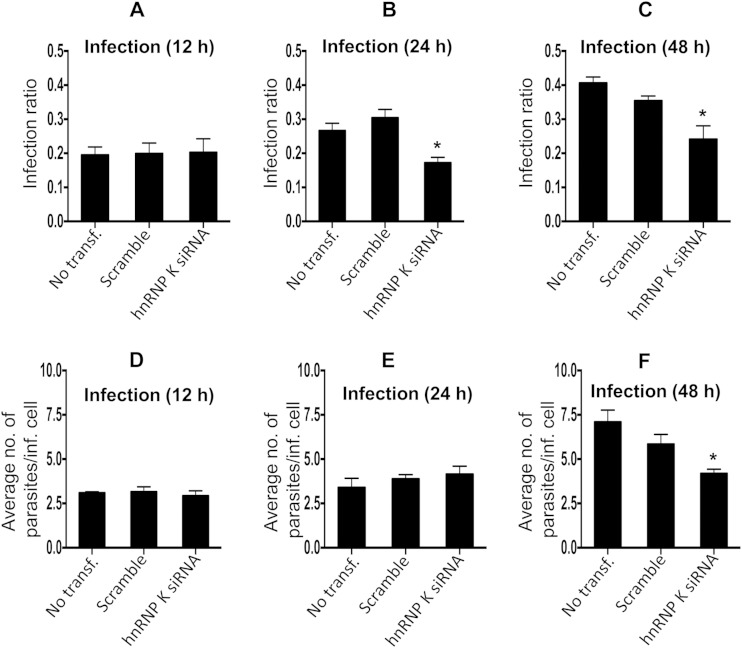
Heterogeneous nuclear ribonucleoprotein K, besides being a transcription factor, has diverse roles in pre-mRNA splicing, chromatin remodeling, transcription, and translation (31). hnRNP K is a substrate for granzyme A (GzmA) and caspases and is cleaved during cell death, and its inactivation is a common attribute during apoptosis (32). hnRNP K suppresses apoptosis by suppressing effector caspase-3 and -7 activities by activating caspase inhibitors (33). The role of hnRNP K in host-pathogen interactions is well documented for hepatitis B virus replication and hepatitis C virus pathogenesis (34, 35). We further validated the role of hnRNP K by RNAi and assessed the effect of knockdown on the parasite infection ratio and proliferation of amastigotes. We performed siRNA-mediated knockdown of the gene corresponding to hnRNP K in uninfected THP-1 cells, followed by challenge with Leishmania. Quantification of the number of intracellular amastigotes postinfection suggested reduced infection ratios (Fig. 6C) and parasite loads (Fig. 6F) at 48 h postinfection, further suggesting the supportive role of hnRNP K in Leishmania infection and proliferation. Microscopy-based images showing the effect of the knockdown of hnRNP K on the infection ratio and intracellular amastigotes are shown in Fig. S1 in the supplemental material. The increased abundance of this protein appears to be a proparasitic response; however, the exact molecular mechanism remains to be elucidated.
FIG 6.
(A to C) Bar diagrams showing the effect of gene knockdown of hnRNP K on the variation of the infection ratio at 12 h (A), 24 h (B), and 48 h (C). (D to F) Average number of parasites/infected cell at 12 h (D), 24 h (E), and 48 h (F). Results are representative of data from three separate experiments. Student's two-tailed t test was applied to analyze the data. *, P < 0.05. inf., infected; transf, transfection.
Proteins involved in immune responses.
Cells of the monocyte-macrophage lineage are the chief source of interleukin-1 (IL-1), the principal mediator of the host immune response. Leishmania averts the initiation of an effective immune response by suppressing the production of cytokines and chemokines involved in the proinflammatory response (IL-1 and tumor necrosis factor alpha [TNF-α]) or in T-cell activation (IL-12) (3, 36). Leishmania cell surface lipophosphoglycan (LPG) is known to suppress IL-1β transcription by acting through a promoter repression sequence (37). We found a lower abundance of IL-1β (UniProt KB accession no. P01584) (∼0.55-fold change at 24 h) in this screen, further validating the findings of previously reported in vitro experiments (37). Chitinase-3-like protein 1 (accession no. P36222) (∼0.5-fold at 24 h) (see Table S5 in the supplemental material) is secreted by various cell types, including macrophages (acts as a Th2-promoting cytokine), and is known to play an important role in the inflammatory response and activation of the AKT prosurvival signaling pathway (38, 39). CD166 antigen (accession no. Q13740), also known as activated leukocyte cell adhesion molecule, which is a transmembrane glycoprotein and is expressed in activated monocytes (40), showed higher levels (∼1.7-fold) (see Table S5 in the supplemental material) at 48 h postinfection, possibly promoting cell-cell adhesion and most likely contributing to information exchange in the form of signaling molecules (see Table S5 in the supplemental material).
Mitochondrial antiviral signaling protein (MAVS) is the first mitochondrial protein known to activate the nuclear factor kappa light chain enhancer of activated B cells (NF-κB) and interferon (IFN) regulatory factors (IRF3 and IRF7) responsible for the synthesis of type I interferons (IFN-α and IFN-β), an important component of antiviral signaling. Silencing of endogenous MAVS expression by RNAi prevents the activation of NF-κB, IRF3, and IRF7, thus blocking interferon production and promoting viral infection (41).
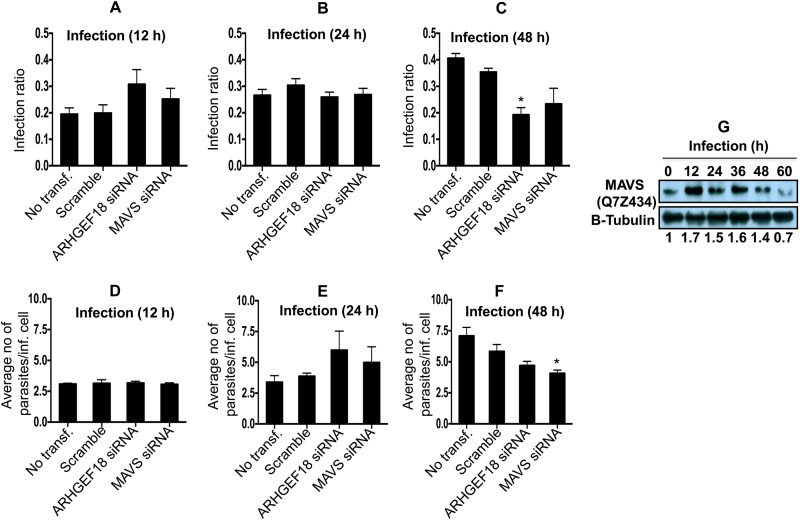
Our proteomic screen showed that MAVS (UniProt KB accession no. Q7Z434) (∼7.9-fold change at 12 h) was significantly abundant in host cells after Leishmania infection (Table 1). In order to ascertain the correctness of identification of a protein having a single peptide, we carried out validation by Western blotting and RNAi. Immunoblot analyses were done to further validate the protein expression of MAVS in THP-1 cells after Leishmania infection. An increase in the MAVS expression level was evidenced by Western blotting during progressive infection and proliferation of parasites (Fig. 7G). We found a gradual decline in the protein level of MAVS after 48 h of infection. The RNAi experiment for MAVS indicates a critical proparasitic role for this protein during Leishmania-macrophage interactions, as the knockdown of MAVS was adequate for the reduction of the average number of parasites per infected cell only at 48 h postinfection (Fig. 7F). Microscopy-based images showing the effect of the knockdown of MAVS on the infection ratio and intracellular amastigotes are shown in Fig. S2 in the supplemental material.
FIG 7.
(A to C) Bar diagrams showing the effect of ARHGEF18 (Rho guanine nucleotide exchange factor 18) and MAVS (mitochondrial antiviral signaling protein) gene knockdown on the variation of the infection ratio at 12 h (A), 24 h (B), and 48 h (C). (D to F) Average number of parasites/infected cell at 12 h (D), 24 h (E), and 48 h (F). Results are representative of data from three separate experiments. Student's two-tailed t test was applied to analyze the data. *, P < 0.05. inf., infected; transf, transfection. (G) Western blot showing time-dependent expression of MAVS in THP-1 cells infected with L. donovani. Densitometric analysis shows the fold change in expression in infected THP-1 cells with respect to the untreated control group. The data are representative of three independent experiments.
The possible cross talk of MAVS with the components of the NF-κB and IRF signaling pathways for proinflammatory cytokine and type I IFN production makes it an important regulator of immune responses for nonviral pathogens (42). Type I IFNs exhibit enhanced expression of class I major histocompatibility complex molecules, activation of natural killer cells, production of cytokines, and promotion of T cells toward a T helper 1 (Th1) phenotype (43). The role of type I IFN (IFN-α and IFN-β) in host macrophages infected with L. major has been well documented (44). IFN-α/β has been demonstrated to be responsible for the early induction of NOS2 (nitric oxide synthase 2) in L. major-infected mouse macrophages. L. major-infected dendritic cells (DCs) exhibit type I IFN (IFN-α and IFN-β)-associated upregulation of IRF2, IRF9, STAT1/2, and IFNAR (IFN-α/β receptor), leading to an enhanced production of IL-12. However, L. major, but not L. donovani, was found to induce the expression of IRF2, IRF7, and IFIT5 (IFN-induced protein with tetratricopeptide repeats 5), thereby ruling out the possible involvement of type I IFN (IFN-α and IFN-β) signaling pathways as factors mediating the production of IL-12 in L. donovani-infected human myeloid-derived DCs (45). Our findings further indicate that the proparasite effect of MAVS is probably independent of type I IFN (IFN-α and IFN-β) signaling pathways in the case of L. donovani-infected macrophages. MAVS has been shown to have a multitude of functions depending upon the cellular context. Alteration of the expression of MAVS may have a differential outcome depending upon its interaction with diverse mitochondrial and/or nonmitochondrial proteins, as MAVS functions are subject to fine-tuning by its interactors (46).
Host proteins are involved in signal transduction.
Leishmania has evolved sophisticated mechanisms by which it is able to sabotage host defense responses. We found that different signaling proteins have altered expression and possibly play a significant role in Leishmania infection and multiplication (see Table S5 in the supplemental material).
Members of the Rho family of GTPases (Cdc42, Rac1, and RhoA), a subfamily of the Ras superfamily, are known to organize the actin cytoskeleton and regulate the phagocytic oxidative burst in macrophages (47). Rac1 and RhoA are associated with phagosomes harboring Leishmania and act as an important component in cytoskeleton rearrangement during phagosome assembly (48, 49). Like all GTPases, Rho proteins act as binary switches, oscillating between inactive (GDP-bound) and active (GTP-bound) conformations. GEFs (guanine nucleotide exchange factors) and GAPs (GTPase-activating proteins) are involved in activating and inactivating this switch and thus are important regulators of cellular events during phagocytosis and pathogen establishment (48). SH3 domain binding protein 1 (SH3BP) (UniProt KB accession no. Q9Y3L3) demonstrates GAP activity that was more abundant in infected cells at 12 h (∼1.6-fold), thus repressing this switch to facilitate parasite survival. Interestingly, Rab GDP dissociation inhibitor beta (accession no. P50395) (∼0.6-fold at 24 h and ∼0.5-fold at 48 h) was found in low abundance in the proteomics screen. Rab GDP dissociation inhibitor beta regulates the GDP/GTP exchange reaction of most Rab proteins and activates the binary switch. These modulations of components of GTPase-mediated signaling point toward a downregulation of host defense responses against Leishmania infection and proliferation. GEFs, including Rho guanine nucleotide exchange factor 18 (ARHGEF18) (accession no. Q6ZSZ5), are also involved in actin cytoskeleton reorganization and act as a guanine nucleotide exchange factor for Rac1, inducing the production of reactive oxygen species (ROS) (49). This protein was highly abundant (∼3.6-fold) at 12 h postinfection (Table 1), further suggesting a role of Rho family GTPases in host-Leishmania interactions. The knockdown of the gene corresponding to ARHGEF18 suggests the essentiality of this gene during Leishmania infection, as the infection ratio as well as the total number of amastigotes were reduced at 48 h postinfection in ARHGEF18-depleted THP-1 cells (Fig. 7C).
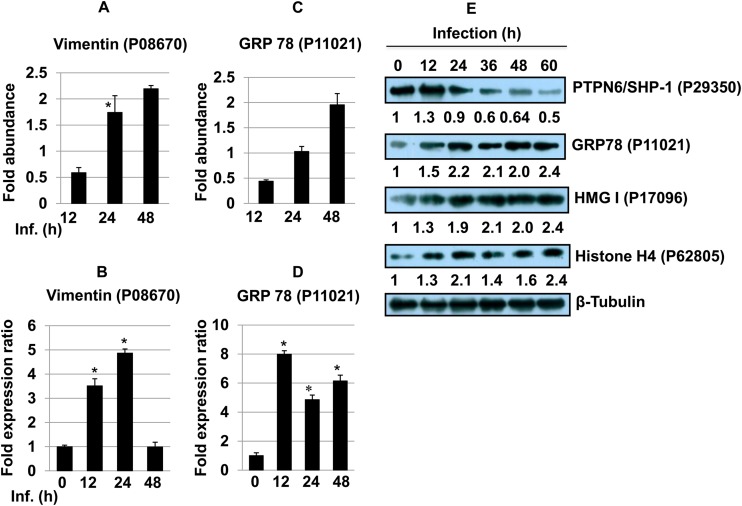
Protein tyrosine phosphatases (PTPs) dephosphorylate a wide variety of phosphoproteins involved in a myriad of signaling events involved in cell growth, differentiation, and cell death (50). Protein tyrosine phosphatase nonreceptor type 6 (PTPN6), also known as Src homology region 2 domain-containing phosphatase 1 (SHP-1) (UniProt KB accession no. P29350), showed a lower abundance at 48 h (∼0.22-fold) (Table 1) (51). SHP-1 reversibly associates with the IFN-α receptor complex upon IFN stimulation and selectively inhibits the JAK/STAT signaling pathway (3). SHP-1 is also known to have an inhibitory effect on JAK2, the Erk1/Erk2 MAPK, NF-κB, IRF-1, and AP-1, which in turn inhibit IFN-γ-inducible macrophage function (5, 52). The pattern of abundance of SHP-1 was observed in two of the replicate experiments and was identified based on a single peptide in the proteomic screen. The expression pattern of SHP-1 was further validated by immunoblot analyses (Fig. 8E). Western blot analysis showed increased expression of SHP-1 12 h after infection. However, a time-dependent decrease in SHP1 expression was observed from 24 to 60 h. It was interesting to find the downregulation of SHP-1 as a delayed response against Leishmania infection in host cells.
FIG 8.
(A and C) Bar diagrams showing differential abundances of vimentin (A) and the 78-kDa glucose-regulated protein (C) at different time points post-Leishmania infection. (B and D) The expression of vimentin (B) and 78-kDa glucose-regulated protein (D) was assessed by qRT-PCR. One-way ANOVA was applied to proteomics data to analyze the fold abundance. Real-time PCR data represent means from three independent experiments. Student's t test was applied to analyze the data. RNU6A was used as an internal control. Data analysis was performed by using the 2−ΔΔCT method. *, P < 0.05. Inf., infection. (E) Western blot analysis of PTPN6/SHP-1, GRP78, HMG-I, and histone H4 proteins across different time points in THP-1 cells infected with L. donovani. Beta-tubulin was used as a loading control. Densitometric analysis shows fold changes in expression in infected THP-1 cells with respect to the untreated control group. The data are representative of three independent experiments.
Necroptosis, an alternative form of programmed cell death, depends on two structurally related kinases, receptor-interacting serine/threonine kinase 1 (RIPK1) and RIPK3. The activation of RIPK1 begins by the oligomerization of Fas-associated protein with death domain (FADD) and TNF receptor-associated death domain (TRADD), which is triggered by TNF-α (53). The increased abundance of RIPK1 (UniProt KB accession no. Q13546) (∼3.0-fold at 24 h) suggests a possible involvement of this phenomenon in a subset of macrophages infected with Leishmania.
Leishmania promastigotes initially exploit complement or the mannose-fucose receptor to enter macrophages, a process assisted by Leishmania LPG (3). Newly formed phagosomes harboring Leishmania are partially covered by periphagosomal F-actin that prevents the interaction of early phagosomes with the late endocytic-lysosomal compartment (54). A similar mechanism has been reported for mycobacterial phagosomes, where coronin 1 (F-actin-interacting protein expressed exclusively in mammalian leukocytes) is involved in preventing the fusion of early phagosomes with late endosomes by F-actin recruitment (54, 55). Early phagosomes that retain coronin 1 are resistant to acidification and hence increase pathogen fitness (56). The increased abundance of coronin 1C (UniProt KB accession no. Q9ULV4) (∼2.3-fold at 12 h) during the initial phase of the host-Leishmania interaction is an indication of a similar mechanism possibly operating in early phagosomes retaining Leishmania.
The transcriptional activator cyclic AMP (cAMP)-responsive element binding protein 3-like protein 4 (CREB-4) (UniProt KB accession no. Q8TEY5), in combination with NF-κB, C/EBPδ, and NFAT, is involved in the transcriptional upregulation of genes to counter stress-induced damage (57). The CREB-4 level was 15.0-fold higher at an earlier time point (12 h) after infection, and this is a possible adaptation mechanism in Leishmania-infected macrophages against oxidative stress. No detectable levels of CREB-4 were observed at 24 and 48 h postinfection.
Proteins involved in cell survival.
While programmed cell death or apoptosis is an indispensable mechanism of host cells to counter intracellular pathogens, these pathogens, including Leishmania, are known to modulate and selectively suppress apoptotic machinery and successfully multiply inside the parasitophorous vacuole (58).
Perforin (UniProt KB accession no. P14222) induces cell death in association with granzymes (GzmA and GzmB) by forming a pore on the target cell. Subsequently, granzymes can trigger apoptosis by an indirect effector caspase activation mechanism by activating proapoptotic BH3-interacting domain death agonist (BID) (59). Perforin was more abundant (∼1.75-fold) at 24 h postinfection. Annexins can bind to certain membrane phospholipids in a Ca2+-dependent manner, and they are a link between Ca2+ signaling and membrane function. Annexin A5 (accession no. P08758) is a ligand highly specific for phosphatidylserine, which, once exposed to the outer surface of the plasma membrane, acts as an “eat me” signal for phagocytosis and necrotic cells (60). Regardless of the higher level of perforin at 24 h, a consistent low abundance of annexin A5 (∼0.34-fold at 24 h and ∼0.35-fold at 24 h) clearly indicates the containment of apoptosis in Leishmania-infected cells. Upregulation of proapoptotic perforin and downregulation of annexin A5 suggest antagonism between proapoptotic and antiapoptotic effectors in Leishmania-infected macrophages.
Vimentin (UniProt KB accession no. P08670) is an organizer of critical proteins involved in attachment, migration, cell signaling, inflammation, and apoptosis. Caspase-mediated cleavage of vimentin disrupts the association of intermediate filaments, which coincides transiently with nuclear fragmentation and facilitates apoptosis (61). Activated human macrophages are known to secrete vimentin into the extracellular space. Proinflammatory cytokines such as TNF-α can trigger its secretion, while IL-10 (Th2 cytokine) is known to block its secretion in a PKC-dependent manner (62). Vimentin is essential for the replication of foot-and-mouth disease virus, and it has been deregulated during Toxoplasma infection (28, 63). An early low abundance (∼0.58-fold at 12 h) followed by an augmentation of its abundance (∼1.74-fold at 24 h and ∼2.2-fold at 48 h) and gene expression (Fig. 8A and B) suggests its participation in the host response to Leishmania infection and multiplication.
We found increased abundances of several high-mobility-group (HMG) proteins (Table 1). HMG proteins act as “antirepressor” molecules that outcompete the binding of inhibitory proteins (of transcription) to scaffold attachment region (SAR) sequences (64). HMG-I/HMG-Y (UniProt KB accession no. P17096) showed an increased abundance at all time points (∼1.56-fold at 12 h, 3.54-fold at 24 h, and 5.74-fold at 48 h). These proteins are critical regulators of several genes involved in the immune response and signal transduction (most importantly CD44, NOS2, IL-2, TNFB, IFNB, COX2, and PKCG) and physically interact with a large number of transcription factors (namely, AP-1, IRF-1, NF-κB, C/EBPβ, and NF-AT) (64) that are known to play a significant role in host-Leishmania interactions.
Mammalian heat shock proteins (HSPs) and glucose-regulated proteins (GRPs) are molecular chaperones essentially involved in protein folding, stabilization, and translocation under normal and adverse conditions such as heat and oxidative stress, including infection. The level of the ER-resident chaperone 78-kDa glucose-regulated protein (GRP78) (UniProt KB accession no. P11021) is elevated during glucose starvation, Ca2+ depletion from the ER, accumulation of misfolded proteins, and reductive stress. The rapid elevation in the cytosolic level of Ca2+ as a result of the depletion of Ca2+ from the ER during Leishmania infection is a widely known phenomenon (3). The initially low abundance (∼0.4-fold at 12 h), possibly as a result of decreased production of proinflammatory cytokines, followed by the increased abundance (∼1.9-fold at 48 h) (Fig. 8C), which can be a direct consequence of increased stress conditions, including the unfolded-protein response (UPR) and further depletion of Ca2+ levels, further mirrors these events as reported above. Furthermore, validation of GRP78 gene expression was done by quantitative RT-PCR (qRT-PCR) of macrophages infected with Leishmania for 12, 24, and 48 h (Fig. 8D). Immunoblot analyses were done to validate the protein expression of HMG-I, vimentin, and GRP78 in THP-1 cells after Leishmania infection (Fig. 8E). Time-dependent increases in HMG-I and GRP78 protein expression levels were observed in Leishmania-infected THP-1 cells (Fig. 8E).
Increased abundance of proteins involved in translation.
Modulation of host protein synthesis is a widely known event in Toxoplasma-infected human fibroblast cells (28). Two important proteins, elongation factor 1 alpha 1 (UniProt KB accession no. P68104) (∼2.29-fold at 12 h and ∼1.55-fold at 24 h) and elongation factor 1 alpha 2 (accession no. Q05639) (∼2.27-fold at 12 h) were more abundant in infected macrophages. Notably, elongation factor 1α, in association with poly(ADP-ribose) polymerase 1 (PARP1) and protein tyrosine kinase (TXK), forms a complex that acts as a T helper 1 (Th1) cell-specific transcription factor to promote IFN-γ transcription (65). Several ribosomal proteins (both 60S and 40S ribosomal subunits) were found in high abundance across different time points after Leishmania infection. The relatively higher abundance of proteins involved in translation appears to be an adaptive mechanism to meet the increasing demand for metabolites and proteins during extended periods of parasite multiplication inside the host cell.
Histones are highly abundant in infected host cells.
We detected a significantly higher level of histone proteins, which correlates with an increased number of transcriptional events after Leishmania infection. Histone H2A is composed of several nonallelic variants whose composition varies during cell differentiation. The levels of several histone H2A and H2B proteins were significantly and consistently higher at different time points (Table 1). Histone H3.3 (UniProt KB accession no. P84243) (∼1.87-fold at 24 h) and histone H3.2 (accession no. Q71DI3) (∼3.45-fold at 24 h) were the only histone H3 variants that were more abundant in infected macrophages. Histone H1x (accession no. Q92522) was the only variant of linker histone H1 that was identified in this study (∼2.0-fold at 24 h). We also found a strong and consistent induction of histone H4 (accession no. P62805). Immunoblot analysis showed a time-dependent increase in the expression of histone H4 in infected THP-1 macrophages (Fig. 8E).
Altered expression patterns of identified proteins in L. donovani-infected human monocyte-derived macrophages.
Since our proteomic screen was done using infected THP-1 cells that were differentiated into macrophages by treatment with PMA, we decided to validate some of the proteins identified in our proteome screen using human monocyte-derived macrophages (MDMs). Real-time PCR analyses of hnRNP K (Fig. 9A), hnRNP A3 (Fig. 9B), hnRNP D0 (Fig. 9C), vimentin (Fig. 9D), and GRP78 (Fig. 9E) in human monocyte-derived macrophages infected with L. donovani across different time points was done. Significant increases in the expression levels of hnRNP K (Fig. 9A), hnRNP A3 (Fig. 9B), and hnRNP D0 (Fig. 9C) were observed at early time points (6 to 12 h) in L. donovani-infected human MDMs. Immunoblot analyses were done to further compare and validate the protein expressions of hnRNP K, MAVS, HMG-I, vimentin, SHP-1, and GRP78 in human monocyte-derived macrophages after Leishmania infection (Fig. 9). Time-dependent increases in the expression levels of hnRNP K, MAVS, HMG-I, and GRP78 were observed in human MDMs (Fig. 9F) after Leishmania infection, suggesting a possible involvement of these proteins during the course of infection and proliferation of parasites. The pattern of abundance of SHP-1 was also validated by immunoblot analyses (Fig. 9F). The level of SHP-1 showed an initial increase at 12 h and then decreased at later time points. Real-time PCR and Western analysis using MDMs clearly confirm the data obtained with infected THP-1 cells.
FIG 9.
(A to E) Real-time PCR analyses of hnRNP K (A), hnRNP A3 (B), hnRNP D0 (C), vimentin (D), and GRP78 (E) across different time points in human monocyte-derived macrophages infected with L. donovani. The data are representative of three different biological experimental replicates. Student's t test was applied to analyze the data. RNU6A was used as an internal control. Data analysis was performed by using the 2−ΔΔCT method. *, P < 0.05. (F) Western blot analysis of hnRNP K, MAVS, PTPN6/SHP-1, GRP78, and HMG-I. Beta-tubulin was used as a loading control. Densitometric analysis shows fold changes in expression in infected THP-1 cells with respect to the untreated control group. The data are representative of three independent experiments.
Altered expression patterns of metabolic enzymes and their functional significance.
In order to survive in host macrophages, intracellular parasites require highly adaptable physiological and metabolic systems. Previous reports showed a decrease in the glycolytic capacity of L. donovani during the course of differentiation (66). At the same time, beta-oxidation, amino acid catabolism, tricarboxylic acid (TCA) cycle, mitochondrial respiration chain, and oxidative phosphorylation capacities are all upregulated. These results indicate that the differentiating parasite shifts from glucose to fatty acids and amino acids as its main energy source (66). Leishmania promastigotes have also been reported to counter ROS and reactive nitrogen species (RNS) effects during the initial stage of infection, as observed by changes in the enzymatic machinery of pathways involved in maintaining redox homeostasis, trypanothione metabolism, oxidative phosphorylation, superoxide metabolism, the mitochondrial respiration process, and other essential metabolic pathways (67).
Metabolomics together with proteomics approaches could form one of the most important postgenomic analyses to reveal changes in metabolic fluxes and identify biomarkers during the course of pathogenic challenge inside macrophages. A recent study using 1H NMR (nuclear magnetic resonance)-based metabolic profiling of classically activated macrophages (caMϕs), alternatively activated macrophages (aaMϕs), and nonactivated macrophages (naMϕs), with or without infection with L. major revealed clear metabolic differences between aaMϕs, caMϕs, and naMϕs in the infected and uninfected states (68). The internal and external metabolome of L. major-infected macrophages showed elevated levels of acetate, alanine, pyruvate, succinate, and lactate, which appeared to be the main end products of intramacrophage parasites. The enhanced levels of creatine (aaMϕs, caMϕs, and naMϕs) and creatine phosphate (aaMϕs and naMϕs) in infected macrophages on the other hand reflected the higher energy demand of the macrophages in order to cope with infection, since creatine phosphate represents a major cellular short-term energy reserve for ATP generation.
A similarly enhanced metabolic status was reflected in the present proteomics screen, where a large number of enzymes involved in metabolic pathways were differentially modulated in macrophages in response to infection by L. donovani. The majority of differentially modulated enzymes were involved in glycolysis, the Krebs cycle, fatty acid oxidation, and electron transport, reflecting the altered metabolic needs of the infected macrophages (see Table S5 in the supplemental material).
Glycolysis is critically modulated inside Leishmania-infected macrophages.
Apart from acting as a safe haven for parasites, the PV provides essential nutrients, cations, and carbon sources for a successful infection. On the other hand, Leishmania is known to salvage low-molecular-weight nutrients (hexoses, amino acids, polyamines, purines, and vitamins) through its membrane transporters while multiplying inside PVs (69). Larger macromolecules such as proteins, carbohydrates, DNA, and RNA are taken up by parasites directly through endocytosis. Fructose-bisphosphate aldolase A (UniProt KB accession no. P04075) is known to bind and sequester Fe(II), a soluble form of iron available in the cellular pool (70), while Leishmania is an excellent iron miner, and the uptake of Fe(II) by the parasite plays a key role in amastigote differentiation in the PV (71). The lower abundance of this enzyme (∼0.3- and 0.5-fold at 24 h and 48 h, respectively) appears to be a host cell adaptation mechanism to deplete the intracellular pool of iron available to parasites to further resist their proliferation. Nevertheless, a lower level of fructose-bisphosphate aldolase A can promote the entry of the substrate of the enzyme into the pentose phosphate pathway, thus generating reducing equivalents (NADPH) and pentoses.
Pyruvate kinase M1/M2 (PKM1/M2), besides catalyzing the transfer of a phosphoryl group from phosphoenolpyruvate (PEP) to ADP and generating ATP, is also involved in caspase-independent cell death of tumor cells (72). Development of hypoxic conditions during infections and inflammatory events is a normal phenotype in cancer tumor and immune effector cells. Leishmania is known to activate the mammalian oxygen-sensing transcription factor hypoxia-inducible factor l (HIF-1) for its survival and multiplication inside macrophages (73). PKM2 is known to act as coactivator that stimulates HIF-1 transactivation of target genes involved in glucose uptake and cell survival (74). A sustained lower level of PKM1/M2 appears to be one of the antagonistic mechanisms against infection inside host cells. The lower abundance of another glycolytic enzyme, α-enolase (0.4- and 0.5-fold at 24 and 48 h, respectively), during the payoff phase of the reaction suggests the rerouting the glycolytic reaction intermediates in anabolic pathways to supplement the sustained need for metabolites inside persistently infected cells.
Higher abundances of other key glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase (UniProt KB accession no. P04406) (∼1.66-fold) and triose phosphate isomerase (accession no. P60174) (∼1.5-fold), were also observed.
Modulation of enzymes in the Krebs cycle.
The Krebs cycle provides precursors of amino acid biosynthesis as well as the reducing agent NADH used in various biochemical reactions. Mitochondrial fumarate hydratase (UniProt KB accession no. UP07954) was found in high abundance (∼1.8-fold at 24 h and 1.3-fold at 48 h). Malate dehydrogenase was initially found in lower abundance, but it was induced as infection progressed (∼0.5-fold at 12 h and ∼1.7-fold at 48 h) and host cells underwent a metabolic burden to synthesize a larger amount of the amino acid precursor needed to support parasite proliferation and auxotrophy for several metabolic needs. Another enzyme, the mitochondrial NAD-dependent malic enzyme (accession no. P23368) (∼1.9-fold at 24 h), acts on malate to form pyruvate, which is accompanied by the production of NADPH, an essential cofactor for fatty acid biosynthesis, along with acetyl coenzyme A (acetyl-CoA), through an auxiliary pathway. The increased abundances of these enzymes reflect the need to generate reaction intermediates and reducing power to fuel amino acid biosynthesis and other anabolic pathways.
Enzymes involved in β-oxidation of fatty acids are differentially upregulated.
Fatty acid oxidation inside mitochondria generates acetyl-CoA, which enters the TCA cycle, while NADH and reduced flavin adenine dinucleotide (FADH2) are used in the electron transport chain. Key enzymes of this pathway, such as acyl-CoA dehydrogenase family member 10 (UniProt KB accession no. Q6JQN1) (∼7.0-fold at 12 h), mitochondrial trifunctional enzyme subunit alpha (accession no. P40939) (∼1.56-fold at 24 h), and mitochondrial 2,4-dienoyl-CoA reductase (accession no. Q16698) (∼2.0-fold at 12 h) were found in higher abundance. This would provide an increased energy (ATP) supply and acetyl-CoA, which, in conjunction with oxaloacetate, can fuel the TCA cycle and complement the ever-increasing demand for reaction intermediates and reducing power (NADPH).
Enzymes involved in electron transport and mitochondrial respiration are upregulated.
The shift in the pattern of energy production was visible for electron transport, where mitochondrial ATP synthase subunit beta (UniProt KB accession no. P06576) (∼0.47-fold at 12 h and ∼3.1-fold at 48 h) and mitochondrial cytochrome b-c1 complex subunit 2 (accession no. P22695) (∼1.71-fold at 24 h) showed consistently higher abundances in infected host cells. This overabundance of some of the key enzymes in mitochondrial respiration indicates the active involvement of host mitochondrial machinery of oxidative phosphorylation in Leishmania infection.
In conclusion, our data provide the first comprehensive mechanistic insight into host proteome dynamics and the manipulation of gene regulatory events as one of the mechanisms by which Leishmania is known to command host defense responses for its own benefit. The host quantitative proteomics modulation screen in combination with the RNAi approach has the potential to identify critical host factors involved in Leishmania infection and multiplication. Future strategies will involve innovative genomics and proteomics approaches to identify potential targetable host factors against Leishmania using different host backgrounds. These studies can become the backbone for intuitive approaches for targeting host components, particularly signaling molecules, which may be important for parasite clearance from the infected host. Since leishmaniasis is a complex of diseases (VL is one among them) with a greater diversity of pathologies caused by different species throughout diverse geographical locations, using a single strain or species to identify host factors and altered mechanisms in a single cell type will not represent the true picture of host-Leishmania interaction. A larger number of studies involving actual patient samples and clinical isolates are needed to analyze the host, parasitoproteome, and metabolome in order to obtain the true picture of the host-parasite interface.
Supplementary Material
ACKNOWLEDGMENTS
Rentala Madhubala is a J. C. Bose National Fellow and is supported by a grant from the Department of Science and Technology, Government of India. Alok K. Singh is a Senior Research Fellow of the Council for Scientific and Industrial Research, Government of India.
We acknowledge the use of the Central Instrumentation Facility (CIF) at the School of Life Sciences, Jawaharlal Nehru University. We acknowledge the technical support provided for data submission to the Human Proteinpedia by Dipankar Malakar (AB Sciex, India), Annu Uppal (AB Sciex, India), T. S. Keshava Prasad (Institute of Bioinformatics, Bangalore, India), and Jayshree Advani (Institute of Bioinformatics, Bangalore, India).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02833-14.
REFERENCES
- 1.Herwaldt BL. 1999. Leishmaniasis. Lancet 354:1191–1199. [DOI] [PubMed] [Google Scholar]
- 2.Bogdan C, Röllinghoff M. 1999. How do protozoan parasites survive inside macrophages? Parasitol Today 15:22–28. doi: 10.1016/S0169-4758(98)01362-3. [DOI] [PubMed] [Google Scholar]
- 3.Olivier M, Gregory DJ, Forget G. 2005. Subversion mechanisms by which Leishmania parasites can escape the host immune response: a signaling point of view. Clin Microbiol Rev 18:293–305. doi: 10.1128/CMR.18.2.293-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proudfoot L, O'Donnell CA, Liew FY. 1995. Glycoinositolphospholipids of Leishmania major inhibit nitric oxide synthesis and reduce leishmanicidal activity in murine macrophages. Eur J Immunol 25:745–750. doi: 10.1002/eji.1830250318. [DOI] [PubMed] [Google Scholar]
- 5.Forget G, Siminovitch KA, Brochu S, Rivest S, Radzioch D, Olivier M. 2001. Role of host phosphotyrosine phosphatase SHP-1 in the development of murine leishmaniasis. Eur J Immunol 31:3185–3196. doi:. [DOI] [PubMed] [Google Scholar]
- 6.Moore KJ, Matlashewski G. 1994. Intracellular infection by Leishmania donovani inhibits macrophage apoptosis. J Immunol 152:2930–2937. [PubMed] [Google Scholar]
- 7.Carrera L, Gazzinelli RT, Badolato R, Hieny S, Muller W, Kuhn R, Sacks DL. 1996. Leishmania promastigotes selectively inhibit interleukin 12 induction in bone marrow-derived macrophages from susceptible and resistant mice. J Exp Med 183:515–526. doi: 10.1084/jem.183.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiner NE. 1987. Parasite accessory cell interactions in murine leishmaniasis. I. Evasion and stimulus-dependent suppression of the macrophage interleukin 1 response by Leishmania donovani. J Immunol 138:1919–1925. [PubMed] [Google Scholar]
- 9.Descoteaux A, Matlashewski G, Turco SJ. 1992. Inhibition of macrophage protein kinase C-mediated protein phosphorylation by Leishmania donovani lipophosphoglycan. J Immunol 149:3008–3015. [PubMed] [Google Scholar]
- 10.Martiny A, Meyer-Fernandes JR, de Souza W, Vannier-Santos MA. 1999. Altered tyrosine phosphorylation of ERK1 MAP kinase and other macrophage molecules caused by Leishmania amastigotes. Mol Biochem Parasitol 102:1–12. doi: 10.1016/S0166-6851(99)00067-5. [DOI] [PubMed] [Google Scholar]
- 11.Buates S, Matlashewski G. 2001. General suppression of macrophage gene expression during Leishmania donovani infection. J Immunol 166:3416–3422. doi: 10.4049/jimmunol.166.5.3416. [DOI] [PubMed] [Google Scholar]
- 12.Gregory DJ, Sladek R, Olivier M, Matlashewski G. 2008. Comparison of the effects of Leishmania major or Leishmania donovani infection on macrophage gene expression. Infect Immun 76:1186–1192. doi: 10.1128/IAI.01320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerfali FZ, Laouini D, Guizani-Tabbane L, Ottones F, Ben-Aissa K, Benkahla A, Manchon L, Piquemal D, Smandi S, Mghirbi O, Commes T, Marti J, Dellagi K. 2008. Simultaneous gene expression profiling in human macrophages infected with Leishmania major parasites using SAGE. BMC Genomics 9:238. doi: 10.1186/1471-2164-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramírez C, Díaz-Toro Y, Tellez J, Castilho TM, Rojas R, Ettinger NA, Tikhonova I, Alexander ND, Valderrama L, Hager J, Wilson ME, Lin A, Zhao H, Saravia NG, McMahon-Pratt D. 2012. Human macrophage response to L. (Viannia) panamensis: microarray evidence for an early inflammatory response. PLoS Negl Trop Dis 6:e1866. doi: 10.1371/journal.pntd.0001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menezes JPB, Almeida TF, Petersen ALOA, Guedes CES, Mota MSV, Lima JGB, Palma LC, Buck GA, Krieger MA, Probst CM, Veras PST. 2013. Proteomic analysis reveals differentially expressed proteins in macrophages infected with Leishmania amazonensis or Leishmania major. Microbes Infect 15:579–591. doi: 10.1016/j.micinf.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Rose SJ, Bermudez LE. 2014. Mycobacterium avium biofilm attenuates mononuclear phagocyte function by triggering hyperstimulation and apoptosis during early infection. Infect Immun 82:405–412. doi: 10.1128/IAI.00820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas D, Qureshi OS, Lee W-Y, Croudace JE, Mura M, Lammas DA. 2008. ATP-induced autophagy is associated with rapid killing of intracellular mycobacteria within human monocytes/macrophages. BMC Immunol 9:35. doi: 10.1186/1471-2172-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheekatla SS, Aggarwal A, Naik S. 2012. mTOR signaling pathway regulates the IL-12/IL-10 axis in Leishmania donovani infection. Med Microbiol Immunol 201:37–46. doi: 10.1007/s00430-011-0202-5. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee A, Padmanabhan PK, Singh S, Roy G, Girard I, Chatterjee M, Ouellette M, Madhubala R. 2007. Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J Antimicrob Chemother 59:204–211. doi: 10.1093/jac/dkl494. [DOI] [PubMed] [Google Scholar]
- 20.Sharma A, Madhubala R. 2009. Ubiquitin conjugation of open reading frame F DNA vaccine leads to enhanced cell-mediated immune response and induces protection against both antimony-susceptible and -resistant strains of Leishmania donovani. J Immunol 183:7719–7731. doi: 10.4049/jimmunol.0900132. [DOI] [PubMed] [Google Scholar]
- 21.Alanne-Kinnunen M, Lappalainen J, Öörni K, Kovanen PT. 2014. Activated human mast cells induce LOX-1-specific scavenger receptor expression in human monocyte-derived macrophages. PLoS One 9:e108352. doi: 10.1371/journal.pone.0108352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. 2008. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer's mice: understanding the interface between physiology and disease. PLoS One 3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siqueira-Neto JL, Moon S, Jang J, Yang G, Lee C, Moon HK, Chatelain E, Genovesio A, Cechetto J, Freitas-Junior LH. 2012. An image-based high-content screening assay for compounds targeting intracellular Leishmania donovani amastigotes in human macrophages. PLoS Negl Trop Dis 6:e1671. doi: 10.1371/journal.pntd.0001671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenedy MR, Vuppala SR, Siegel C, Kraiczy P, Akins DR. 2009. CspA-mediated binding of human factor H inhibits complement deposition and confers serum resistance in Borrelia burgdorferi. Infect Immun 77:2773–2782. doi: 10.1128/IAI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandasamy K, Keerthikumar S, Goel R, Mathivanan S, Patankar N, Shafreen B, Renuse S, Pawar H, Ramachandra YL, Acharya PK, Ranganathan P, Chaerkady R, Keshava Prasad TS, Pandey A. 2009. Human Proteinpedia: a unified discovery resource for proteomics research. Nucleic Acids Res 37:D773–D781. doi: 10.1093/nar/gkn701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad TSK, Kandasamy K, Pandey A. 2009. Human Protein Reference Database and Human Proteinpedia as discovery tools for systems biology. Methods Mol Biol 577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- 28.Nelson MM, Jones AR, Carmen JC, Sinai AP, Burchmore R, Wastling JM. 2008. Modulation of the host cell proteome by the intracellular apicomplexan parasite Toxoplasma gondii. Infect Immun 76:828–844. doi: 10.1128/IAI.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black DL. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 30.Tazi J, Bakkour N, Stamm S. 2009. Alternative splicing and disease. Biochim Biophys Acta 1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol 16:2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Domselaar R, Quadir R, van der Made AM, Broekhuizen R, Bovenschen N. 2012. All human granzymes target hnRNP K that is essential for tumor cell viability. J Biol Chem 287:22854–22864. doi: 10.1074/jbc.M112.365692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Z, Ko HL, Goh EH, Wang B, Ren EC. 2013. hnRNP K suppresses apoptosis independent of p53 status by maintaining high levels of endogenous caspase inhibitors. Carcinogenesis 34:1458–1467. doi: 10.1093/carcin/bgt085. [DOI] [PubMed] [Google Scholar]
- 34.Ng LFP, Chan M, Chan S-H, Cheng PC-P, Leung EH-C, Chen W-N, Ren E-C. 2005. Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS Med 2:e163. doi: 10.1371/journal.pmed.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan B, Lu K-Y, Reymond Sutandy FX, Chen Y-W, Konan K, Zhu H, Kao CC, Chen C-S. 2014. A human proteome microarray identifies that the heterogeneous nuclear ribonucleoprotein K (hnRNP K) recognizes the 5′ terminal sequence of the hepatitis C virus RNA. Mol Cell Proteomics 13:84–92. doi: 10.1074/mcp.M113.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiner SL, Zheng S, Wang ZE, Stowring L, Locksley RM. 1994. Leishmania promastigotes evade interleukin 12 (IL-12) induction by macrophages and stimulate a broad range of cytokines from CD4+ T cells during initiation of infection. J Exp Med 179:447–456. doi: 10.1084/jem.179.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatzigeorgiou DE, Geng J, Zhu B, Zhang Y, Liu K, Rom WN, Fenton MJ, Turco SJ, Ho JL. 1996. Lipophosphoglycan from Leishmania suppresses agonist-induced interleukin 1 beta gene expression in human monocytes via a unique promoter sequence. Proc Natl Acad Sci U S A 93:14708–14713. doi: 10.1073/pnas.93.25.14708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawada M, Hachiya Y, Arihiro A, Mizoguchi E. 2007. Role of mammalian chitinases in inflammatory conditions. Keio J Med 56:21–27. doi: 10.2302/kjm.56.21. [DOI] [PubMed] [Google Scholar]
- 39.Chen C-C, Llado V, Eurich K, Tran HT, Mizoguchi E. 2011. Carbohydrate-binding motif in chitinase 3-like 1 (CHI3L1/YKL-40) specifically activates Akt signaling pathway in colonic epithelial cells. Clin Immunol 140:268–275. doi: 10.1016/j.clim.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lunter PC, van Kilsdonk JWJ, van Beek H, Cornelissen IMHA, Bergers M, Willems PHGM, van Muijen GNP, Swart GWM. 2005. Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a novel actor in invasive growth, controls matrix metalloproteinase activity. Cancer Res 65:8801–8808. doi: 10.1158/0008-5472.CAN-05-0378. [DOI] [PubMed] [Google Scholar]
- 41.Seth RB, Sun L, Ea C-K, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Buss C, Opitz B, Hocke AC, Lippmann J, van Laak V, Hippenstiel S, Krüll M, Suttorp N, Eitel J. 2010. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-beta response and control of bacterial replication in human endothelial cells. J Immunol 184:3072–3078. doi: 10.4049/jimmunol.0902947. [DOI] [PubMed] [Google Scholar]
- 43.Belardelli F, Gresser I. 1996. The neglected role of type I interferon in the T-cell response: implications for its clinical use. Immunol Today 17:369–372. doi: 10.1016/0167-5699(96)10027-X. [DOI] [PubMed] [Google Scholar]
- 44.Mattner J, Schindler H, Diefenbach A, Röllinghoff M, Gresser I, Bogdan C. 2000. Regulation of type 2 nitric oxide synthase by type 1 interferons in macrophages infected with Leishmania major. Eur J Immunol 30:2257–2267. doi:. [DOI] [PubMed] [Google Scholar]
- 45.Favila MA, Geraci NS, Zeng E, Harker B, Condon D, Cotton RN, Jayakumar A, Tripathi V, McDowell MA. 2014. Human dendritic cells exhibit a pronounced type I IFN signature following Leishmania major infection that is required for IL-12 induction. J Immunol 192:5863–5872. doi: 10.4049/jimmunol.1203230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belgnaoui SM, Paz S, Hiscott J. 2011. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol 23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Etienne-Manneville S, Hall A. 2002. Rho GTPases in cell biology. Nature 420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 48.Morehead J, Coppens I, Andrews NW. 2002. Opsonization modulates Rac-1 activation during cell entry by Leishmania amazonensis. Infect Immun 70:4571–4580. doi: 10.1128/IAI.70.8.4571-4580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya T. 2003. G protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: regulation of cell shape and reactive oxygen species production. Circ Res 93:848–856. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 50.Zhang ZY. 2001. Protein tyrosine phosphatases: prospects for therapeutics. Curr Opin Chem Biol 5:416–423. doi: 10.1016/S1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- 51.Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L. 2000. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- 52.Contreras I, Gómez MA, Nguyen O, Shio MT, McMaster RW, Olivier M. 2010. Leishmania-induced inactivation of the macrophage transcription factor AP-1 is mediated by the parasite metalloprotease GP63. PLoS Pathog 6:e1001148. doi: 10.1371/journal.ppat.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shindo R, Kakehashi H, Okumura K, Kumagai Y, Nakano H. 2013. Critical contribution of oxidative stress to TNFα-induced necroptosis downstream of RIPK1 activation. Biochem Biophys Res Commun 436:212–216. doi: 10.1016/j.bbrc.2013.05.075. [DOI] [PubMed] [Google Scholar]
- 54.Lodge R, Descoteaux A. 2005. Modulation of phagolysosome biogenesis by the lipophosphoglycan of Leishmania. Clin Immunol 114:256–265. doi: 10.1016/j.clim.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 55.Pieters J. 2008. Coronin 1 in innate immunity. Subcell Biochem 48:116–123. doi: 10.1007/978-0-387-09595-0_11. [DOI] [PubMed] [Google Scholar]
- 56.Seto S, Tsujimura K, Koide Y. 2012. Coronin-1a inhibits autophagosome formation around Mycobacterium tuberculosis-containing phagosomes and assists mycobacterial survival in macrophages. Cell Microbiol 14:710–727. doi: 10.1111/j.1462-5822.2012.01754.x. [DOI] [PubMed] [Google Scholar]
- 57.Gerlo S, Kooijman R, Beck IM, Kolmus K, Spooren A, Haegeman G. 2011. Cyclic AMP: a selective modulator of NF-κB action. Cell Mol Life Sci 68:3823–3841. doi: 10.1007/s00018-011-0757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heussler VT, Küenzi P, Rottenberg S. 2001. Inhibition of apoptosis by intracellular protozoan parasites. Int J Parasitol 31:1166–1176. doi: 10.1016/S0020-7519(01)00271-5. [DOI] [PubMed] [Google Scholar]
- 59.Trapani JA, Smyth MJ. 2002. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol 2:735–747. doi: 10.1038/nri911. [DOI] [PubMed] [Google Scholar]
- 60.Gerke V, Creutz CE, Moss SE. 2005. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 61.Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. 2001. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ 8:443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- 62.Mor-Vaknin N, Punturieri A, Sitwala K, Markovitz DM. 2003. Vimentin is secreted by activated macrophages. Nat Cell Biol 5:59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- 63.Gladue DP, O'Donnell V, Baker-Branstetter R, Holinka LG, Pacheco JM, Fernández Sainz I, Lu Z, Ambroggio X, Rodriguez L, Borca MV. 2013. Foot-and-mouth disease virus modulates cellular vimentin for virus survival. J Virol 87:6794–6803. doi: 10.1128/JVI.00448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reeves R. 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277:63–81. doi: 10.1016/S0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 65.Maruyama T, Nara K, Yoshikawa H, Suzuki N. 2007. Txk, a member of the non-receptor tyrosine kinase of the Tec family, forms a complex with poly(ADP-ribose) polymerase 1 and elongation factor 1alpha and regulates interferon-gamma gene transcription in Th1 cells. Clin Exp Immunol 147:164–175. doi: 10.1111/j.1365-2249.2006.03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. 2008. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB J 22:590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- 67.Sardar AH, Kumar S, Kumar A, Purkait B, Das S, Sen A, Kumar M, Sinha KK, Singh D, Equbal A, Ali V, Das P. 2013. Proteome changes associated with Leishmania donovani promastigote adaptation to oxidative and nitrosative stresses. J Proteomics 81:185–199. doi: 10.1016/j.jprot.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 68.Lamour SD, Choi B-S, Keun HC, Müller I, Saric J. 2012. Metabolic characterization of Leishmania major infection in activated and nonactivated macrophages. J Proteome Res 11:4211–4222. doi: 10.1021/pr3003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McConville MJ, de Souza D, Saunders E, Likic VA, Naderer T. 2007. Living in a phagolysosome; metabolism of Leishmania amastigotes. Trends Parasitol 23:368–375. doi: 10.1016/j.pt.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 70.Spasojević I, Mojović M, Blagojević D, Spasić SD, Jones DR, Nikolić-Kokić A, Spasić MB. 2009. Relevance of the capacity of phosphorylated fructose to scavenge the hydroxyl radical. Carbohydr Res 344:80–84. doi: 10.1016/j.carres.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 71.Mittra B, Andrews NW. 2013. IRONy OF FATE: role of iron-mediated ROS in Leishmania differentiation. Trends Parasitol 29:489–496. doi: 10.1016/j.pt.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steták A, Veress R, Ovádi J, Csermely P, Kéri G, Ullrich A. 2007. Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res 67:1602–1608. doi: 10.1158/0008-5472.CAN-06-2870. [DOI] [PubMed] [Google Scholar]
- 73.Singh AK, Mukhopadhyay C, Biswas S, Singh VK, Mukhopadhyay CK. 2012. Intracellular pathogen Leishmania donovani activates hypoxia inducible factor-1 by dual mechanism for survival advantage within macrophage. PLoS One 7:e38489. doi: 10.1371/journal.pone.0038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. 2011. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.