Abstract
EZH2 is the catalytic subunit of Polycomb Repressor Complex 2 (PRC2) which catalyzes methylation of histone H3 at lysine 27 (H3K27me) and mediates gene silencing of target genes via local chromatin reorganization. Numerous evidences show that EZH2 plays a critical role in cancer initiation, progression and metastasis, as well as in cancer stem cell biology. Indeed, EZH2 dysregulation alters gene expression programs in various cancer types. The molecular mechanisms responsible for EZH2 alteration appear to be diverse and depending on the type of cancer. Furthermore, accumulating evidences indicate that EZH2 could also act as a PRC2-independent transcriptional activator in cancer. In this review, we address the current understanding of the oncogenic role of EZH2, including the mechanisms of EZH2 dysregulation in cancer and progresses in therapeutic approaches targeting EZH2.
Keywords: EZH2, polycomb repression, chromatin modification, histone lysine methylation, cancer, cancer stem cells
Introduction
During embryogenesis, the fertilized egg develops into a complex organism composed of many differentiated cell types. The maintenance of the differentiation status of these cell types requires a cellular memory system responsible for the stable inheritance of gene expression programs. The Polycomb group (PcG) and trithorax group (trxG) genes were discovered in Drosophila melanogaster as part of such a memory system [1,2]. They have been identified as repressors (PcG) and activators (trxG) of genetic programs, respectively. Mutations in the PcG and trxG genes result in pleiotropic defects, of which homeotic transformations are the most apparent [3]. In vertebrates, the PcG and trxG proteins have similar roles in the maintenance of homeotic gene expression patterns. Indeed, changes in the body plan have been observed in PcG and trxG gene homolog mouse mutants [4-7]. Although primarily known for their involvement in the control of homeotic genes during the establishment of the body plan, PcG and trxG members have also been shown to be implicated in the control of various cellular processes, such as stem cell renewal and differentiation, cell fate decisions, senescence, chromosome X-inactivation in mammals, or tumorigenesis and neoplastic development [8-10].
Perturbations in local chromatin structure cause inappropriate gene expression and genomic instability, resulting in cellular transformation and malignant outgrowth. Therefore, proteins involved in chromatin organization, including Polycomb group (PcG) proteins constitute fundamental players in cancer pathogenesis [11-13].
Polycomb repression and EZH2 activity
PcG proteins have been found to interact with each other to form multimeric, chromatin-associated protein complexes of two general types: the Polycomb Repressive Complex 1 (PRC1) and PRC2 [14,15]. These complexes post-translationally modify histone tails and are believed to cooperate in transcriptional repression of target genes by altering local, higher order chromatin structure. The PRC2 protein complex contains EZH2, a histone methyltransferase that catalyzes trimethylation of histone H3 lysine 27 (H3K27me3) [16,17]. In addition to EZH2, core PRC2 is composed of EED, SUZ12 and RBBP4/RbAp48/NURF55 which are required for full EZH2 histone methyltransferase activity [18,19]. The PRC1 protein complex binds to PRC2-modified residues (H3K27me3) and monoubiquitinylates histone H2A at lysine 119 (H2AK119ub1) [20,21]. These modifications (H3K27me3 and H2AK119ub1) cause in turn local chromatin compaction and transcriptional silencing.
EZH2 is the catalytic subunit of the PRC2 protein complex, and its C-terminal SET domain exhibits the H3K27 methyltransferase function. However, EZH2 by itself lacks enzymatic activity. Two other PRC2 components, the zinc-finger-containing protein SUZ12 and the WD40-repeat protein EED are required to maintain the integrity of the PRC2 complex and for EZH2 robust methyltransferase activity [22-24]. The fourth PRC2 core subunit, RBBP4/RbAp48 also contributes to PRC2 function but subcomplexes lacking this component retain substantial enzyme activity [18,19]. Additional PRC2 components, such as AEBP2, PCL (PHF) and JARID2 may function as accessory factors regulating the activity of the PRC2 protein complex. However, their role appears to be modulatory rather than essential [17,25-28].
In the context of the PRC2 protein assembly, EZH2 SET domain performs three successive methyl transfer reactions, producing ultimately H3K27me3. This contrasts with other SET-domain protein methyltransferases whose capacity for methyl transfer appears more limited. For example, SET7/9 produces only monomethylated products (H3K4me1), whereas G9a/EHMT2 catalyzes mono- and dimethylation (H3K9me1 and H3K9me2) and SUV39H2 can di- and trimethylate monomethylated substrates (H3K9me1 methylation into H3K9me2 and H3K9me3) [29,30].
EZH1 is a paralog of EZH2; PRC2-EZH2 and PRC2-EZH1-containing complexes control overlapping sets of target genes but act differently to maintain the repressed chromatin state [31]. Furthermore, EZH2 is mainly expressed in proliferating tissues, whereas EZH1 expression is found in dividing and differentiated cells [31,32]. This observation suggests that EZH2-containing PRC2 complexes might establish the H3K27me3 repressive marks, whereas EZH1-containing PRC2 complexes might contribute to the restoration of the H3K27me3 methylation profile after histone demethylation or histone exchanges.
Involvement of EZH2 in cancer
The genome-wide mapping of PcG target genes revealed more than 2,000 sites in the mouse embryonic stem cell genome [8,33,34]. Interaction of PcG proteins with chromatin at these loci is associated with increased levels of H3K27me3 repressive marks and Polycomb repression affects numerous genes encoding key developmental regulators and signaling proteins. These genomic studies point on the widespread roles of PRC2 and H3K27 methylation in developmental and differentiation processes of multicellular organisms, and on their implication in fundamental chromatin mechanisms that underlie stem cell regulatory circuits and cancer progression. Thus, it is not surprising that increasing evidences indicate that EZH2 deregulation is frequently observed in a variety of cancers (Table 1). Interestingly, different ways by which the function of EZH2 may be impaired in tumors have been described (Figure 1). It also appears that the type of EZH2 dysregulation often correlates with the malignancy. EZH2 overexpression is mainly found in solid tumors, whereas activating or inactivating mutations are identified in hematologic malignancies. A missense mutation Lys27Met (K27M) in the gene encoding histone H3.3 (H3F3A) is present at high frequencies in pediatric gliomas. This mutation behaves as a potent inhibitor of EZH2 activity. Thus, although mechanisms could be different, misregulation of H3K27 methylation is common in tumorigenesis and EZH2 appears to have both oncogenic and tumor suppressive functions.
Table 1.
EZH2 alterations and cancers
| Alteration | Cancer type | References |
|---|---|---|
| Overexpression | Prostate, breast, bladder, ovarian, renal carcinoma, lung, liver, brain, gastric, esophageal, pancreatic, melanoma | [35,38,39-56,166] |
| Activating mutations | Large follicular and B-cell lymphomas | [99,100,102,103] |
| Inactivating/hypomorphic mutations | Myeloproliferative neoplasms Pediatric cancers | [115,116,118-120] |
| H3.3K27M-mediated EZH2 inhibition | Pediatric gliomas | [121-124] |
Figure 1.
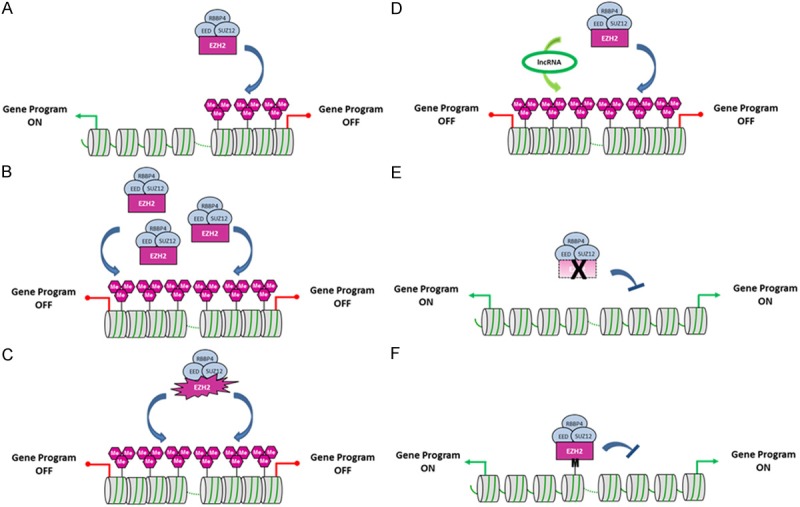
Schematic representation of the diverse EZH2 dysregulations found in cancer. A. The histone lysine methyltransferase EZH2 catalyzes H3K27 methylation at defined target genes and silences their expression. B. Overabundance of EZH2 is responsible for an increase in H3K27me3 repressive mark levels leading to the silencing of tumor suppressor genes in cancer cells. C. EZH2 bearing activating mutations at residues Y641, A677 or A687 possesses an enhanced activity leading to an increase in H3K27me3 levels. D. Overexpression of EZH2-interacting partners such as specific lncRNAs enhances recruitment of EZH2 to targets and increases H3K27me3 levels. E. EZH2 harboring an inactivating mutation or EZH2 gene deletion leads to a decrease in H3K27me3 levels and activation of EZH2 target gene programs in cancer. F. A lysine to methionine substitution at position 27 (K27M) in the gene encoding histone H3.3 (H3F3A) inhibits EZH2 activity and leads to nearly undetectable H3K27me3 repressive mark levels in pediatric gliomas. Purple hexagons represent H3K27me epigenetic marks, and M illustrates H3.3K27M mutant histones.
EZH2 overabundance in solid cancer
Overexpression of EZH2 was first found in prostate and breast cancer in microarray studies [35,36]. Furthermore, EZH2 overexpression is linked to aggressive and advanced metastatic stages of the disease and is strongly associated with poor clinical outcome and prognosis [36-38]. Overexpression of EZH2 has also been reported in a large number of other solid tumors such as bladder cancers [39-42], ovarian cancers [43,44], renal carcinomas [45], small cell and non-small cell lung cancers [46-49], hepatocellular carcinomas [50], brain tumors [51], kidney cancers [52], gastric tumors [53], esophageal cancers [54], pancreatic cancers [55] or melanomas [38,56]. EZH2 has been shown to promote cell proliferation, migration, and invasion in different in vitro cancer cell models [32,36,43,57-59]. EZH2 overexpressing cells are also tumorigenic when injected into the mammary fat pads of nude mice [60], while overexpression of wild-type EZH2 in mammary epithelial cells in vivo results in epithelial hyperplasia and promotes mammary tumor initiation [61,62]. Furthermore, the oncogenic property of EZH2 in mice correlates with its H3K27 methyltransferase activity [63].
Overabundance of EZH2 in tumor cells may result from different mechanisms. In some cases, EZH2 up-regulation is associated to gene amplification [35,64]. Using FISH techniques, Sramaki and colleagues studied the copy number of the EZH2 gene in several prostate cancer cell lines such as LNCaP, DU145, PC-3, 22Rv1, in xenografts and in clinical tumors [64]. In contrast to early prostate cancer, in late stage tumor samples, EZH2 gene amplification was shown to correlate with its overexpression.
Increased EZH2 levels in cancer may also be caused by a variety of transcriptional signals and pathways, some of them common for different cancer types while others may be more specific or limited to specific malignancies (Figure 2, Table 2). The MEK-ERK-ELK1 pathway, which is often activated in cancer, has been demonstrated to be responsible for EZH2 overexpression in triple-negative and ERBB2-overexpressing subtypes of breast cancer [65]. Upon phosphorylation ELK1 binds to three ELK1-binding motifs located within the EZH2 gene promoter and activates EZH2 transcription. The pRb-E2F signaling is another pathway involved in numerous tumors. Upon pRb/RB1 phosphorylation, E2F dissociates from the pRb-E2F complex and the activated E2F transcription factor binds to E2F-binding sites located in the EZH2 promoter to activate its transcription [32]. Overexpression of E2F or deregulation of the pRb-E2F pathway correlate with activated EZH2 expression in breast, bladder and small-cell lung cancer [32,66-68]. A number of transcription factors involved in tumorigenesis directly bind to the EZH2 promoter and activate its mRNA expression in different cancer models. In particular, MYC and ETS transcription factors directly regulate EZH2 transcription in prostate cancer [69,70], whereas NF-YA, STAT3 and the co-activator of the androgen receptor ATAD2/ANCCA regulate EZH2 expression in epithelial ovarian, colorectal, and breast cancer cells, respectively [71-73]. In Ewing’s sarcoma, the fusion oncoprotein EWS-FLI1 induces EZH2 expression which has a key role in endothelial/neuroectodermal differentiation and tumor growth [74]. Also, expression of EZH2 was found profoundly affected by the BRAF (V600E) mutation in melanoma although the precise mechanism of regulation is not yet fully understood [75].
Figure 2.
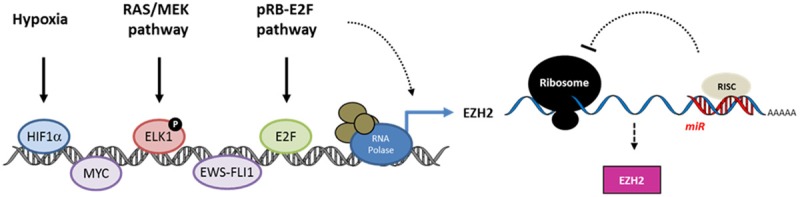
Schematic representation of EZH2 overexpression controlling levels. EZH2 overexpression in cancer cells is achieved at the transcriptional level through the binding of transcription factors to its promoter or at the post-transciptional level via the alteration of the micro-RNA regulation.
Table 2.
Regulators of EZH2 transcription
| Factor/Pathway | Cancer type | References |
|---|---|---|
| MYC | Prostate | [69] |
| ETS/ERG | Prostate | [70] |
| E2F (pRB-E2F pathway) | Breast | [32] |
| Lung | [68] | |
| Bladder | [66,67] | |
| ELK1 (MEK-ERK pathway) | Breast | [65,76] |
| ATAD2/ANCCA | Breast | [73] |
| NF-YA | Ovarian | [71] |
| STAT3 | Colorectal | [72] |
| BRAF (V600E) | Melanoma | [75] |
| EWS-FLI1 | Ewing’s sarcoma | [74] |
| HIF1α (Hypoxia) | Breast | [76] |
Hypoxia in solid tumors represents another pathway directly regulating EZH2 transcription. Chang and colleagues identified a consensus sequence for hypoxia-inducible factor-1α (HRE) within the EZH2 promoter [76]. Hypoxic microenvironment in tumors induces HIF1α binding to the HRE and transactivates EZH2 to promote breast tumor expansion (Table 2).
In addition, EZH2 abundance is controlled at the post-translational level by multiple micro-RNAs (miRs). The miR-25, -26a, -30d, -98, -101, -124, -137, -138, -144, -214 and let-7 interact with defined sequences within the EZH2 3’UTR and directly downregulate EZH2 protein abundance. The loss of control by these miR results in the up-regulation of EZH2 and appears to be involved in the aggressiveness of various cancers [57,77-98] (Figure 2, Table 3).
Table 3.
EZH2 post-transcriptional regulation by miR
| MiR | Cancer Type | References |
|---|---|---|
| miR-25, miR-30d | Thyroid cancer | [77] |
| miR-26a | Lymphoma, nasopharyngeal carcinoma, breast cancer, prostate cancer | [78-80] |
| miR-98 | Nasopharyngeal carcinoma, gastric cancer | [81,82] |
| miR-101 | Nasopharyngeal carcinoma, glioblastoma multiform, prostate cancer, bladder cancer, head and neck cancer, non-small cell lung cancer, melanoma | [57,83-88] |
| miR-124 | Hepatocellular carcinoma, gastric cancer | [89,90] |
| miR-137 | Melanoma | [91] |
| miR-138 | Head and neck cancer, glioblastoma multiform, non-small cell lung cancer | [92-94] |
| miR-144 | Bladder cancer | [98] |
| miR-214 | Gastric cancer, hepatocellular carcinoma | [81,95] |
| Let-7 | Prostate cancer, nasopharyngeal carcinoma | [96,97] |
Altogether, different studies suggest that the diverse mechanisms involved in EZH2 overabundance depend on the cell context. However, EZH2 functions as an oncogenic factor in the majority of solid tumors and regardless of the molecular mechanisms involved, EZH2 overabundance leads to higher levels of H3K27me3 repressive epigenetic marks that would be responsible in turn, for the silencing of tumor suppressor genes in cancer cells (Figure 1B).
EZH2 activating mutations in lymphoma
In about 7% of large follicular lymphomas and 22% of diffuse B-cell lymphomas, recurrent somatic mutations were identified at tyrosine 641 (Y641) within the catalytic SET domain of EZH2 [99]. Initially reported to be a loss-of-function mutation [99], it has been demonstrated that these mutations shift the methylation capacity of EZH2 [100]. Indeed, EZH2-Y641 mutants have reduced H3K27 mono- and dimethylation activities, but aberrantly elevated trimethyltransferase activity. Because of their reduced H3K27me1/2 activities, EZH2-Y641 mutant alleles are invariably found at heterozygous forms together with the wild-type EZH2 allele in lymphomas. In agreement with these observations, transgenic mice expressing the EZH2-Y641F mutant in lymphocytes displayed a global increase in trimethylated H3K27 in spleen cells and developed lymphomas when combined with Eµ-Myc expression [101]. Similar to the Y641 mutation, mutations within the EZH2 SET domain at residues Alanine 677 (A677G) and Alanine 687 (A687V) are drivers of H3K27 hypertrimethylation [102-104]. Thus, EZH2-Y641, EZH2-A677 and EZH2-A687 are oncogenic mutants responsible for an excess of H3K27me3 repressive marks impairing gene expression programs in lymphomas (Figure 1C).
Increased recruitment of EZH2 at chromatin in cancer
An excess of H3K27me3 repressive marks in cancer can also result from an increase of PRC2 recruitment at chromatin (Figure 1D). In particular, long non-coding RNAs (lncRNAs) have emerged as potential factors involved in PRC2 recruitment. HOTAIR is one of these lncRNAs interacting with EZH2 and playing an oncogenic role in cancer [105-109]. Overexpression of HOTAIR increases invasiveness and metastatic potential of epithelial cancer cells and induces relocalization of the PRC2 complex which binds to target genes in a pattern similar to that observed in embryonic fibroblasts, whereas HOTAIR knockdown decreases cancer invasiveness, especially in cells expressing high levels of PRC2 proteins [106]. Other lncRNAs, such as HEIH, PCAT-1, H19 or linc-UBC1 have been shown to interact with EZH2 and to be involved in cancer [110-113].
EZH2 null-mutations in malignant myeloid disorders
Mutations in EZH2 have been identified in about 10%-23% of various subtypes of myelodysplastic syndromes and myeloproliferative neoplasms, as well as in 13% of myelofibrosis [114-118]. Both monoallelic and biallelic mutations were identified, and almost all are predicted to inactivate the methyltransferase activity of EZH2. In particular mutations found in myeloid disorders include missense mutations, frameshift mutations, premature stop codons, and spliceosomal mutations. In addition to EZH2 mutation, decreased EZH2 expression was found to be associated with hemizygous deletion (7/del7q) involving the EZH2 gene. EZH2 loss-of-function mutations are associated with reduced H3K27me3 methylation and derepression of EZH2 target genes, which may contribute to leukemogenesis (Figure 1E). Consistent with these findings, deletion of Ezh2 alone is sufficient to induce myelodysplastic syndrome/myeloproliferative neoplasm-like diseases in mice [119].
Furthermore, EZH2-inactivating mutations are also primarily detected in pediatric cancers including core binding factor acute myeloid leukemia and T-lineage acute lymphoblastic leukemia [120].
Altogether, these studies outline the tumor suppressor function of EZH2 in myeloid malignancies, in addition to its role as an oncogene in other cancer types.
H3.3K27M mutation-dependent inhibition of EZH2 in high-grade pediatric gliomas
Sequencing studies of high-grade pediatric gliomas, including glioblastoma multiforms (GBM) and diffuse intrinsic pontine gliomas (DIPG) identify recurrent heterozygous Lysine to Methionine substitutions at position 27 (K27M) in the gene encoding histone H3.3 (H3F3A) [121,122]. H3.3K27M mutation occurs in 70-80% of midline GBM and DIPG in young children and confers a dismal prognosis [122-124]. The mutation also leads to nearly undetectable H3K27me3 repressive mark levels in gliomas [125-128]. Furthermore, it has been shown that H3.3K27M peptides bind to EZH2 and interfere with its methyltransferase activity, potentially as methionine mimics the structure of monomethyl lysine [127] (Figure 1F). Since H3.3 is a variant of canonical histone H3 which contributes to a minority of total histone H3 in gliomas, H3.3K27M behaves like a powerful dominant negative inhibitor of EZH2 activity. Interestingly, EZH2 protein and remaining H3K27me3 repressive marks were also shown to be locally increased at hundreds of gene loci in glioma cells expressing H3.3K27M [128]. Therefore, H3.3K27M mutation alters gene expression programs and the global epigenetic landscape which may drive to tumorigenesis in gliomas.
PRC2-independent function of EZH2 in cancer
EZH2 is primarily known for its role in gene silencing through H3K27 trimethylation. However, several studies reveal that EZH2 may also function as a transcriptional activator in different cancer models [129-131] (Figure 3).
Figure 3.
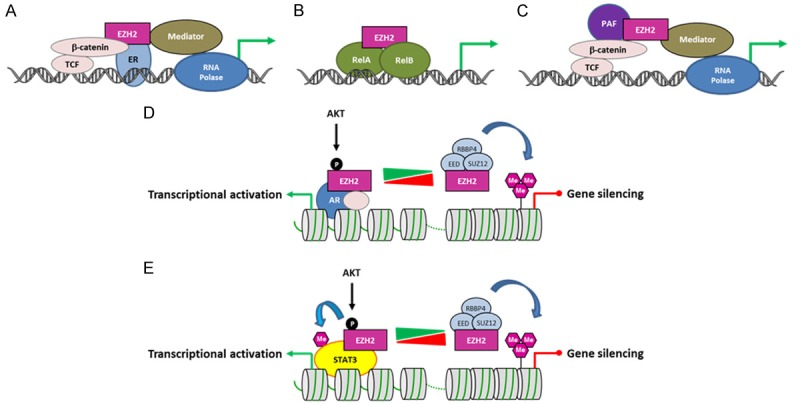
PRC2-independent transcriptional activation by EZH2 in cancer. A. In ER-positive breast cancer cells, EZH2 interacts with β-catenin and ER, and functionally enhances gene expression. B. In ER-negative breast cancer cells, EZH2 interacts with RELA/RELB to stimulate NF-κB target gene expression. C. In colorectal cancer cells, EZH2 forms a complex with β-catenin and PAF to promote transcription. D. In castration-resistant prostate cancer, AKT1-mediated phosphorylation of EZH2 at serine 21 allows EZH2 to interact with the AR at target genes to activate transcription. The AKT pathway then acts as a molecular switch changing EZH2 function from a chromatin silencer to a transcriptional co-activator of the AR. This transcriptional activation function is methyltransferase activity-dependent. E. AKT1-mediated phosphorylation of EZH2 at serine 21 also facilitates STAT3 methylation and activation in glioblastoma stem cells. ER: estrogen receptor; TCF: T-cell factor; AR: androgen receptor; PAF: PCNA-associated factor; RNA Polase: RNA polymerase II.
In breast cancer cells, EZH2 has been reported to act as a transcriptional activator but its mechanism of action depends on the cell type [129,130]. In estrogen receptor-positive, luminal-like MCF-7 breast cancer cells, EZH2 links physically the estrogen receptor α to the Wnt signaling components β-catenin and TCF at target gene promoters. By this way, EZH2 activates Cyclin D1 (CCND1) and MYC transcription, independently of its methyltransferase activity [129] (Figure 3A). By contrast, in estrogen receptor-negative, basal like MDA-MB-231 breast cancer cells, EZH2 interacts with the NF-κB components RELA and RELB to activate transcription of several NF-κB target genes, such as the TNF and IL6 genes [130] (Figure 3B). Thus, EZH2 could act as a transcriptional repressor via its H3K27 histone methyltransferase activity or, as a transcriptional activator through different molecular mechanisms, in promoting breast tumorigenesis.
The PCNA-associated factor PAF (KIAA0101) is overexpressed in colon cancers and is required for cancer cell proliferation via Wnt signaling activation. Jung and colleagues [131] identified a PAF-EZH2-β-catenin protein complex involved in Wnt target gene transactivation in colon cancer cells. Upon Wnt signaling activation, PAF dissociates from PCNA, binds to β-catenin and recruits EZH2 at Wnt target genes to induce their expression independently of EZH2’s enzymatic activity (Figure 3C).
EZH2 is oncogenic and functions as a transcriptional activator in castration-resistant prostate cancer (CRPC) [132]. However in contrast to what was described in breast cancer cells and in colon cancer cells, the transcriptional properties of EZH2 in CRPC rely on its methyltransferase activity, but do not require the other PRC2 components. It has been suggested that EZH2-mediated transcriptional activation may occur through the methylation of the androgen receptor or other associated proteins. Furthermore, Xu et al. [132] reported that AKT1-mediated phosphorylation of EZH2 at serine 21 (S21) allows the methyltransferase to interact with the androgen receptor at many target genes. Thus, the AKT pathway acts as a molecular switch changing EZH2 function from a chromatin silencer to a transcriptional co-activator of the androgen receptor (Figure 3D). A similar situation has been described in glioblastoma where AKT1-mediated phosphorylation of EZH2 at S21 enhances STAT3 activation via its EZH2-mediated trimethylation at lysine 180 [133] (Figure 3E).
The methyltransferase activity of EZH2 is required for EZH2-dependent gene activation in CRPC and in glioblastoma, indicating that EZH2 can methylate non-histone substrates. In this regard, it is worth noting that previous studies showed that EZH2 indeed methylates non-histone proteins such as the transcription factors GATA4 and RORα, although the role of these methylations in cancer remains to be explored [134,135].
Post-translational modifications of EZH2 in cancer
The discovery that AKT1-mediated EZH2 phosphorylation at S21 converts EZH2 silencing activity into an activating effector [60,132,133] underlines the important role cell signaling may have on EZH2 activity control. There is growing evidence showing that EZH2 activity and stability are tightly regulated by multiple post-translational modifications.
In addition to phosphorylation at S21, EZH2 could be phosphorylated at multiple threonine residues [136-139]. In particular, cyclin-dependent kinases (CDK1/2) phosphorylate EZH2 at T350 and T492. T350 phosphorylation promotes the interaction between EZH2 and lncRNAs such as HOTAIR [137]. Thus, CDK-mediated phosphorylation positively impacts EZH2 action by enhancing its recruitment at chromatin. In contrast, T492 phosphorylation reduces the methyltransferase activity of EZH2 by disrupting PRC2 assemblies [138]. However, phosphomimic change at the corresponding mouse Ezh2 residue (T487D) does not seem to have an effect on methyltransferase activity nor on PRC2 assembly [137]. In another study, Wu and Zhang [139] reported that CDK1-mediated phosphorylation at T350 and T492 promotes EZH2 ubiquitinylation and subsequent degradation of the protein by the proteasome. These discrepancies might reflect that distinct regulatory mechanisms control EZH2 activity in different cell types.
Finally, recent experiments suggest that glycosylation may also affect EZH2 stability and H3K27me3 levels [140]. EZH2 interacts with O-linked N-acetylglucosamine (GlcNAc) transferase (OGT) and the methyltransferase is O-GlcNAcylated at serine 75 (S75) in vivo. Furthermore, OGT knockdown specifically downregulates EZH2 protein stability and greatly reduces H3K27me3 levels. EZH2-S75A mutants also exhibit a reduction in stability. This OGT-EZH2 axis might then explain in part how the dysregulation of OGT is implicated in cancer.
Involvement of EZH2 in cancer stem cell biology
According to the cancer stem cell (CSC) hypothesis, the CSCs which represent a small fraction of the tumor population are the only tumor-initiating clones. CSCs have unlimited self-renewal capacities and the ability to differentiate in many cancer cell types. They are resistant to chemotherapy [141] and thought to be responsible for metastatic spreading [142]. Thus, deciphering the cellular regulations involved in CSC biology is central to the development of efficient anti-cancer opportunities.
EZH2 is a crucial factor playing a role in the maintenance of self-renewal of adult and em-bryonic stem cells [13,143,144], while EZH2 expression is reduced in differentiated cells. Since EZH2 overexpression is linked to cancer initiation and metastasis and considering that high-grade tumors are enriched with a high content of CSCs, it is proposed that EZH2 expression could favor the transition of dormant progenitors and/or differentiated cells into a more aggressive stem-cell like phenotype. Indeed, EZH2 expression has been shown to be involved in breast CSCs formation and expansion that promote cancer progression [76]. Similarly, EZH2 expression is crucial for glioblastoma CSC self-renewal and tumor-initiating capacity [145-147].
Anti-cancer strategies targeting EZH2
Numerous experimental evidences indicate that EZH2 plays a prominent role in carcinogenesis, from tumor initiation to metastasis, in various cancer types. EZH2 has thus emerged as a potential cancer therapeutic target. Indeed, knock-down of EZH2 inhibits cancer cell proliferation and decreases in vivo tumor growth in xenograft models [148,149].
The search for pharmacological inhibitors of EZH2 activity also yielded several promising molecules. Among them, 3-deazaneplanocin A (DNZep) has been shown to exhibit significant anti-tumor activity against different cancer models, such as breast, prostate, lung, brain, colorectal and liver cancer cells [150-152]. Indeed, DNZep treatment reduces EZH2 protein and H3K27me3 levels, reactivates PRC2 target genes and is responsible for apoptosis of cancer cells, but not of normal cells [150]. Interestingly, DNZep has also been shown to abrogate self-renewal and tumor-initiating capacities of glioblastoma cancer stem cells, ovarian cancer stem cell-like populations and prostate cancer stem cells [59,146,153,154]. However, DNZep is an S-adenosylhomocysteine (SAH) hydrolase inhibitor causing intracellular SAH levels to increase. Accumulation of SAH subsequently inhibits diverse methyltransferases including EZH2, by a feed-back loop mechanism (Figure 4). DNZep is therefore not selective and specific to EZH2 [155,156].
Figure 4.
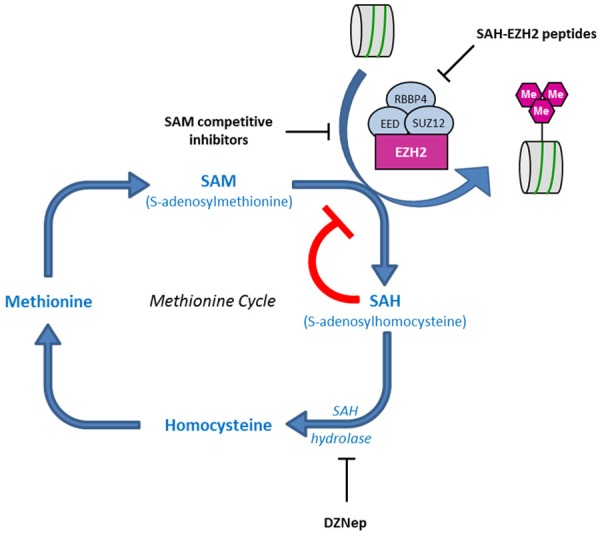
Methionine cycle and mode of action of several EZH2 inhibitors. DNZep is an inhibitor of SAH hydrolase leading to an accumulation of SAH which in turn inhibits EZH2 activity, as well as other cellular methltransferases. SAM competitors such as EPZ005687, EPZ-6438, EI1, UNC1999 and GSK126 bind to the SAH-binding pocket of EZH2 to prevent the recruitment of the SAM methyl donor. SAH-EZH2 peptides disrupt the PRC2 assembly required for full EZH2 activity.
More recently, high-throughput screens performed with the PRC2 complex led to the discovery of potent compounds that selectively inhibit EZH2 enzymatic activity [157-161]. These molecules including EPZ005687, EPZ-6438, EI1, UNC1999 and GSK126, all act as S-adenosylmethionine (SAM) competitive inhibitors (Figure 4).
The compound EPZ005687 has an equilibrium dissociation constant (Ki) value of 24 nM and has a 50-fold selectivity for EZH2 against the closely related EZH1 enzyme and more than 500-fold selectivity against 15 other protein methyltransferases [157]. Interestingly, EPZ005687 can also inhibit the H3K27 methylation activities of lymphoma cells harboring heterozygous EZH2-Y641 and EZH2-A677 mutations, killing these cells with minimal effects on the proliferation of wild-type cells [157]. EPZ-6438, with a Ki value of 2.5 nM, possesses a superior potency and drug-like properties, including good oral bioavailability in EZH2-mutant xenograft mice models [158]. The molecule is currently undergoing a phase I clinical trial in patients with advanced solid tumors or refractory B-cell lymphoma [158].
EI1 (Ki of about 13 nM), UNC1999 an orally bioavailable compound in mice, but non-selective for EZH1 and GSK126, the most potent EZH2 inhibitor (Ki of about 0.5-3 nM) [159-161], as well as EPZ005687 and EPZ-6438, all bind to the SAM pocket of the EZH2 catalytic SET domain and selectively inhibit its H3K27 methyltransferase activity. However, the inhibitors are only effective in killing cell lines harboring gain-of-function EZH2 mutations (EZH2-Y641, EZH2-A677), although all of these compounds induce a decrease in H3K27me3 levels in both EZH2-mutated and wild-type cancer cells. This indicates that the SAM competitive inhibitors may be more beneficial to patients with lymphoma rather than with other cancer types.
Since the enzymatic activity of EZH2 requires its association with other components of the PRC2 complex, such as EED and SUZ12, a strategy disrupting PRC2 assembly has been successfully developed to inhibit EZH2 methyltransferase activity [162]. A hydrocarbon-stapled peptide that mimics the α-helical EED-binding domain of EZH2 (SAH-EZH2 peptide) was shown to disrupt the interaction between EZH2 and EED (Figure 4). Indeed, the SAH-EZH2 peptide reduces the H3K27 methyltransferase activity of the PRC2 complex and leads to growth arrest and differentiation of MLL-AF9 leukemia cells which are dependent on PRC2 activity. The antiproliferative activity of SAH-EZH2 was also extended to EZH2-dependent B-cell lymphomas; the inhibitory peptide reduces H3K27 trimethylation, EZH2 protein levels, as well as cancer cell viability. Thus, disruption of the PRC2 complex may represent an alternative and complementary strategy for selectively arresting the proliferation of at least some EZH2-dependent cancers. It is also worth noting that the SAH-EZH2 peptide also dissociates EZH1-EED complexes, in addition to EZH2-EED assemblies [162].
In addition, some natural chemopreventive agents have been shown to effectively inhibit EZH2 and reactivate PRC2-silenced target genes in various cancer models. These compounds include epigallocatechin-3-gallate (EGCG), a major component of green tea and dietary omega-3 polyunsaturated fatty acids (omega-3 PUFAs) which both reduce EZH2 levels by increasing proteasomal degradation [163,164]. Curcumin, a natural ingredient of turmeric also decreases cancer proliferation by modulating EZH2 levels, but in that case downregulation of EZH2 expression is mediated by the MAPK pathway rather than by the increase of protein degradation [165].
Conclusion
A large set of experimental data have established that EZH2 acts as a key player in tumorigenesis. However, the molecular mechanisms involved in EZH2 dysregulation in cancer appear to be diverse. EZH2 overexpression is mainly found in solid tumors and activating mutations are found in B-cell lymphomas while inactivating mutations are often identified in myelodysplastic syndromes and myeloproliferative neoplasms. Finally a missense mutation in the gene encoding histone H3.3 (H3F3A) inhibiting EZH2 activity is present at high frequencies in pediatric gliomas. Thus, EZH2 functions as an oncogene in solid tumors and lymphomas whereas it behaves like a tumor suppressor gene in myeloid disorders and in pediatric glioblastomas. The oncogenic role of EZH2 mainly depends on its ability to repress gene expression programs via H3K27 methylation and chromatin compaction. However, studies in certain cancers revealed that the oncogenic function of EZH2 could also result of its action as a PRC2-independent transcriptional activator. Then, EZH2 could be involved in cancer through multiple mechanisms and could be regulated by different pathways depending on cellular context and cancer type. A better understanding of the regulatory network involving EZH2 is consequently required for the development of novel anti-cancer therapeutic strategies.
Acknowledgements
This work was supported by Inserm, the University of Lille, the CNRS and grants from the Ligue Contre le Cancer and the Cancéropole Nord-Ouest. BD is supported by a fellowship from the Région Nord-Pas de Calais and the University of Lille 1.
Disclosure of conflict of interest
None to disclose.
References
- 1.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 2.Schuettenbruger B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by Polycomb and Trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Jürgens G. A group of genes controlling the special expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155. [Google Scholar]
- 4.Yu BD, Hess JL, Horning SE, Brown GAJ, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 5.Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 6.van der Lugt NM, Alkema M, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev. 1996;58:153–164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- 7.Coré N, Bel S, Gaunt SJ, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 8.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 9.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83:184–193. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 13.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 14.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–213. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 16.Völkel P, Angrand PO. The control of histone lysine methylation in epigenetic regulation. Biochimie. 2007;89:1–20. doi: 10.1016/j.biochi.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of the Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 18.Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Köcher T, Cohen A, Stunnenberg HG, Wilm M, Müller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Wang LJ, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–978. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 22.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 25.Montgomery ND, Yee D, Montgomery SA, Magnuson T. Molecular and functional mapping of EED motifs required for PRC2-dependent histone methylation. J Mol Biol. 2007;374:1145–1157. doi: 10.1016/j.jmb.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, Plotnikov AN, Schapira M. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 37.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collett K, Eide GE, Arnes J, Stefansson IM, Eide J, Braaten A, Aas T, Otte AP, Akslen LA. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. J. Clin. Oncol. 2006;24:268–273. doi: 10.1158/1078-0432.CCR-05-1533. [DOI] [PubMed] [Google Scholar]
- 39.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP, Akslen LA. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 40.Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- 41.Hinz S, Kempkensteffen C, Christoph F, Hoffmann M, Krause H, Schrader M, Schostak M, Miller K, Weikert S. Expression of the polycomb group protein EZH2 and its relation to outcome in patients with urothelial carcinoma of the bladder. J Cancer Res Clin Oncol. 2008;134:331–336. doi: 10.1007/s00432-007-0288-8. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Albadine R, Magheli A, Guzzo TJ, Ball MW, Hinz S, Schoenberg MP, Netto GJ, Gonzalgo ML. Increased EZH2 protein expression is associated with invasive urothelial carcinoma of the bladder. Urol Oncol. 2012;30:428–433. doi: 10.1016/j.urolonc.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Weikert S, Christoph F, Köllermann J, Müller M, Schrader M, Miller K, Krause H. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int J Mol Med. 2005;16:349–353. [PubMed] [Google Scholar]
- 44.Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, Zeng YX, Kung HF, Xie D. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–1583. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Cai Q, Godwin AK, Zhang R. Enhancer of zeste homolog 2 promotes the proliferation and invasion of epithelial ovarian cancer cells. Mol Cancer Res. 2010;8:1610–1618. doi: 10.1158/1541-7786.MCR-10-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HW, Choe M. Expression of EZH2 in renal cell carcinoma as a novel prognostic marker. Pathol Int. 2012;62:735–741. doi: 10.1111/pin.12001. [DOI] [PubMed] [Google Scholar]
- 47.Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, Riquelme E, Galindo H, Moran CA, Kalhor N, Swisher SG, Simon GR, Stewart DJ, Lee JJ, Wistuba II. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res. 2013;19:6556–6565. doi: 10.1158/1078-0432.CCR-12-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, Yamanaka R, Tanaka Y, Nukiwa T, Marquez VE, Ishikawa Y, Ichinose M, Aburatani H. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Konishi J, Oizumi S, Kaga K, Matsuno Y, Nishimura M, Dosaka-Akita H. Distinctive expression of the polycomb group proteins Bmi1 polycomb ring finger oncogene and enhancer of zeste homolog 2 in nonsmall cell lung cancers and their clinical and clinicopathologic significance. Cancer. 2010;116:3015–3024. doi: 10.1002/cncr.25128. [DOI] [PubMed] [Google Scholar]
- 50.Lv Y, Yuan C, Xiao X, Wang X, Ji X, Yu H, Wu Z, Zhang J. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncol Rep. 2012;28:147–154. doi: 10.3892/or.2012.1787. [DOI] [PubMed] [Google Scholar]
- 51.Kondo Y, Shen L, Suzuki S, Kurokawa T, Masuko K, Tanaka Y, Kato H, Mizuno Y, Yokoe M, Sugauchi F, Hirashima N, Orito E, Osada H, Ueda R, Guo Y, Chen X, Issa JP, Sekido Y. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol Res. 2007;37:974–983. doi: 10.1111/j.1872-034X.2007.00141.x. [DOI] [PubMed] [Google Scholar]
- 52.Crea F, Hurt EM, Farrar WL. Clinical significance of Polycomb gene expression in brain tumors. Mol Cancer. 2010;9:265. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu B, Abourbih S, Sircar K, Kassouf W, Mansure JJ, Aprikian A, Tanguay S, Brimo F. Enhancer of zeste homolog 2 expression is associated with metastasis and adverse clinical outcome in clear cell renal cell carcinoma: a comparative study and review of the literature. Arch Pathol Lab Med. 2013;137:1326–1336. doi: 10.5858/arpa.2012-0525-OA. [DOI] [PubMed] [Google Scholar]
- 54.Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada A, Fujii S, Daiko H, Nishimura M, Chiba T, Ochiai A. Aberrant expression of EZH2 is associated with a poor outcome and P53 alteration in squamous cell carcinoma of the esophagus. Int J Oncol. 2011;38:345–353. doi: 10.3892/ijo.2010.868. [DOI] [PubMed] [Google Scholar]
- 56.Ougolkov AV, Bilim VN, Billadeau DD. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin Cancer Res. 2008;14:6790–6796. doi: 10.1158/1078-0432.CCR-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Pathol. 2007;34:597–600. doi: 10.1111/j.1600-0560.2006.00678.x. [DOI] [PubMed] [Google Scholar]
- 58.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, Noske DP, Tannous BA, Würdinger T. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, Zhang HB, Liao YJ, Zheng F, Zhu W, Liu TH, Bian XW, Guan XY, Lin MC, Zeng MS, Zeng YX, Kung HF, Xie D. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 60.Crea F, Hurt EM, Mathews LA, Cabarcas SM, Sun L, Marquez VE, Danesi R, Farrar WL. Pharmacologic disruption of Polycomb Repressive Complex 2 inhibits tumorigenicity and tumor progression in prostate cancer. Mol Cancer. 2011;10:40. doi: 10.1186/1476-4598-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, Kidwell KM, Kleer CG. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Gonzalez ME, Toy K, Filzen T, Merajver SD, Kleer CG. Targeted overexpression of EZH2 in the mammary gland disrupts ductal morphogenesis and causes epithelial hyperplasia. Am J Pathol. 2009;175:1246–1254. doi: 10.2353/ajpath.2009.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croonquist PA, Van Ness B. The polycomb group protein enhancer of zeste homolog 2 (EZH 2) is an oncogene that influences myeloma cell growth and the mutant ras phenotype. Oncogene. 2005;24:6269–6280. doi: 10.1038/sj.onc.1208771. [DOI] [PubMed] [Google Scholar]
- 65.Saramaki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 66.Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, Ochiai A. MEK–ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene. 2011;30:4118–4128. doi: 10.1038/onc.2011.118. [DOI] [PubMed] [Google Scholar]
- 67.Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, Kovacs G, Dennis N, Fisher C, Wooster R, Huddart R, Foster CS, Cooper CS. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23:1627–1630. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 68.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–5623. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 69.Coe BP, Thu KL, Aviel-Ronen S, Vucic EA, Gazdar AF, Lam S, Tsao MS, Lam WL. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One. 2013;8:e71670. doi: 10.1371/journal.pone.0071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–683. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, Albertini V, Malek A, Chiorino G, Catapano CV, Carbone GM. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garipov A, Li H, Bitler BG, Thapa RJ, Balachandran S, Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Mol Cancer Res. 2013;11:360–369. doi: 10.1158/1541-7786.MCR-12-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin YW, Ren LL, Xiong H, Du W, Yu YN, Sun TT, Weng YR, Wang ZH, Wang JL, Wang YC, Cui Y, Sun DF, Han ZG, Shen N, Zou W, Xu J, Chen HY, Cao W, Hong J, Fang JY. Role of STAT3 and vitamin D receptor in EZH2-mediated invasion of human colorectal cancer. J Pathol. 2013;230:277–290. doi: 10.1002/path.4179. [DOI] [PubMed] [Google Scholar]
- 74.Kalashnikova EV, Revenko AS, Gemo AT, Andrews NP, Tepper CG, Zou JX, Cardiff RD, Borowsky AD, Chen HW. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010;70:9402–9412. doi: 10.1158/0008-5472.CAN-10-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, Hotfilder M, Lowel D, von Luettichau I, Mossbrugger I, Quintanilla-Martinez L, Kovar H, Staege MS, Muller-Tidow C, Burdach S. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hou P, Liu D, Dong J, Xing M. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle. 2012;11:286–295. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, Fusco A. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab. 2012;97:E710–718. doi: 10.1210/jc.2011-3068. [DOI] [PubMed] [Google Scholar]
- 79.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Möller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 80.Salvatori B, Iosue I, Djodji Damas N, Mangiavacchi A, Chiaretti S, Messina M, Padula F, Guarini A, Bozzoni I, Fazi F, Fatica A. Critical role of c-Myc in acute myeloid leukemia involving direct regulation of miR-26a and histone methyltransferase EZH2. Genes Cancer. 2011;2:585–592. doi: 10.1177/1947601911416357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, Li XP. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 82.Huang SD, Yuan Y, Zhuang CW, Li BL, Gong DJ, Wang SG, Zeng ZY, Cheng HZ. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer. 2012;11:51. doi: 10.1186/1476-4598-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alajez NM, Shi W, Hui AB, Bruce J, Lenarduzzi M, Ito E, Yue S, O'Sullivan B, Liu FF. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis. 2010;1:e85. doi: 10.1038/cddis.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 86.Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, Ye ZY, Tao HQ. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46:2295–2303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 87.Banerjee R, Mani RS, Russo N, Scanlon CS, Tsodikov A, Jing X, Cao Q, Palanisamy N, Metwally T, Inglehart RC, Tomlins S, Bradford C, Carey T, Wolf G, Kalyana-Sundaram S, Chinnaiyan AM, Varambally S, D’Silva NJ. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30:4339–4349. doi: 10.1038/onc.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–678. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 89.Luo C, Merz PR, Chen Y, Dickes E, Pscherer A, Schadendorf D, Eichmüller SB. MiR-101 inhibits melanoma cell invasion and proliferation by targeting MITF and EZH2. Cancer Lett. 2013;341:240–247. doi: 10.1016/j.canlet.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 90.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, Kung HF, Xie D. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 91.Xie L, Zhang Z, Tan Z, He R, Zeng X, Xie Y, Li S, Tang G, Tang H, He X. microRNA-124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol Cell Biochem. 2014;392:153–159. doi: 10.1007/s11010-014-2028-0. [DOI] [PubMed] [Google Scholar]
- 92.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, Diederichs S, Eichmüller SB. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133:768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 93.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, Dai Y, Zhou X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, Peng Y. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta. 2013;1832:1697–1707. doi: 10.1016/j.bbadis.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H, Zhang H, Zhao M, Lv Z, Zhang X, Qin X, Wang H, Wang S, Su J, Lv X, Liu H, Du W, Zhou W, Chen X, Fei K. MiR-138 inhibits tumor growth through repression of EZH2 in nonsmall cell lung cancer. Cell Physiol Biochem. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- 96.Xia H, Ooi LL, Hui KM. MiR-214 targets beta-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7:e44206. doi: 10.1371/journal.pone.0044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kong D, Heath E, Chen W, Cher ML, Powell I, Heilbrun L, Li Y, Ali S, Sethi S, Hassan O, Hwang C, Gupta N, Chitale D, Sakr WA, Menon M, Sarkar FH. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One. 2012;7:e33729. doi: 10.1371/journal.pone.0033729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cai K, Wan Y, Sun G, Shi L, Bao X, Wang Z. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncol Rep. 2012;28:2101–2106. doi: 10.3892/or.2012.2027. [DOI] [PubMed] [Google Scholar]
- 99.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280:4531–4538. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 100.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, Marquez VE, Marra MA, Gascoyne RD, Humphries RK, Arrowsmith CH, Morin GB, Aparicio SA. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Berg T, Thoene S, Yap D, Wee T, Schoeler N, Rosten P, Lim E, Bilenky M, Mungall AJ, Oellerich T, Lee S, Lai CK, Umlandt P, Salmi A, Chang H, Yue L, Lai D, Cheng SW, Morin RD, Hirst M, Serve H, Marra MA, Morin GB, Gascoyne RD, Aparicio SA, Humphries RK. A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis. Blood. 2014;123:3914–3924. doi: 10.1182/blood-2012-12-473439. [DOI] [PubMed] [Google Scholar]
- 103.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, Chen SB, Della Pietra A 3rd, Dul E, Hughes AM, Gilbert SA, Thrall SH, Tummino PJ, Kruger RG, Brandt M, Schwartz B, Creasy CL. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, Smith JJ, Moyer MP, Richon VM, Copeland RA, Wigle TJ. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012;586:3448–3451. doi: 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 105.Ott HM, Graves AP, Pappalardi MB, Huddleston M, Halsey WS, Hughes AM, Groy A, Dul E, Jiang Y, Bai Y, Annan R, Verma SK, Knight SD, Kruger RG, Dhanak D, Schwartz B, Tummino PJ, Creasy CL, McCabe MT. A687V EZH2 Is a Driver of Histone H3 Lysine 27 (H3K27) Hypertrimethylation. Mol Cancer Ther. 2014;13:3062–3073. doi: 10.1158/1535-7163.MCT-13-0876. [DOI] [PubMed] [Google Scholar]
- 106.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 109.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, Shang JL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 112.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 114.He W, Cai Q, Sun F, Zhong G, Wang P, Liu H, Luo J, Yu H, Huang J, Lin T. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta. 2013;1832:1528–1537. doi: 10.1016/j.bbadis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 115.Cardoso C, Mignon C, Hetet G, Grandchamps B, Fontes M, Colleaux L. The human EZH2 gene: genomic organisation and revised mapping in 7q35 within the critical region for malignant myeloid disorders. Eur J Hum Genet. 2000;8:174–180. doi: 10.1038/sj.ejhg.5200439. [DOI] [PubMed] [Google Scholar]
- 116.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 117.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tönnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 118.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, Visconte V, Sugimoto Y, Prince C, O’Keefe C, Hsi ED, List A, Sekeres MA, Rao A, McDevitt MA, Maciejewski JP. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Makishima H, Jankowska AM, Tiu RV, Szpurka H, Sugimoto Y, Hu Z, Saunthararajah Y, Guinta K, Keddache MA, Putnam P, Sekeres MA, Moliterno AR, List AF, McDevitt MA, Maciejewski JP. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24:1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 120.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, Nagae G, Nakaseko C, Yokote K, Shimoda K, Koseki H, Suzuki Y, Sugano S, Aburatani H, Ogawa S, Iwama A. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, Ma J, Edmonson MN, Hedlund EK, Rusch MC, Shurtleff SA, Mulder HL, Boggs K, Vadordaria B, Cheng J, Yergeau D, Song G, Becksfort J, Lemmon G, Weber C, Cai Z, Dang J, Walsh M, Gedman AL, Faber Z, Easton J, Gruber T, Kriwacki RW, Partridge JF, Ding L, Wilson RK, Mardis ER, Mullighan CG, Gilbertson RJ, Baker SJ, Zambetti G, Ellison DW, Zhang J, Downing JR. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tönjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Co- llins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 123.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome Project. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, Bourgey M, Bourque G, Montpetit A, Bourret G, Lepage P, Fleming A, Lichter P, Kool M, von Deimling A, Sturm D, Korshunov A, Faury D, Jones DT, Majewski J, Pfister SM, Jabado N, Hawkins C. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Frühwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Dürken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 126.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Højfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Seker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu CM, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho YJ, Pfister SM. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 127.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, Sun L, Zhang Y, Chen Y, Li R, Zhang Y, Hong M, Shang Y. Integration of estrogen and Wnt signaling circuits by the Polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27:5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, Liou YC, Yu Q. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 132.Jung HY, Jun S, Lee M, Kim HC, Wang X, Ji H, McCrea PD, Park JI. PAF and EZH2 induce Wnt/beta-catenin signaling hyperactivation. Mol Cell. 2013;52:193–205. doi: 10.1016/j.molcel.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP, Liu XS, Brown M. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam DH, Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, Chung CH, Seo SB, Yoon JB, Ko E, Noh DY, Kim KI, Kim KK, Baek SH. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 137.Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, Simon JA, Huang H. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286:28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, Kang ML, Wong CH, Juan LJ. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Crea F, Danesi R, Farrar WL. Cancer stem cell epigenetics and chemoresistance. Epigenomics. 2009;1:63–79. doi: 10.2217/epi.09.4. [DOI] [PubMed] [Google Scholar]
- 143.Hurt EM, Farrar WL. Cancer stem cells: the seeds of metastasis? Mol Interv. 2008;8:140–142. doi: 10.1124/mi.8.3.7. [DOI] [PubMed] [Google Scholar]
- 144.Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011;15:243–250. [PMC free article] [PubMed] [Google Scholar]
- 145.Chen YH, Hung MC, Li LY. EZH2: a pivotal regulator in controlling cell differentiation. Am J Transl Res. 2012;4:364–375. [PMC free article] [PubMed] [Google Scholar]
- 146.Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, Kim M, Totonchy M, Cusack T, Ene C, Ma H, Su Q, Zenklusen JC, Zhang W, Maric D, Fine HA. Epigenetic-mediated dysfunction of the bone morphogenetic protein pathway inhibits differentiation of glioblastoma-initiating cells. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suvà ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, Marquez VE, Clément V, Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 148.Natsume A, Ito M, Katsushima K, Ohka F, Hatanaka A, Shinjo K, Sato S, Takahashi S, Ishikawa Y, Takeuchi I, Shimogawa H, Uesugi M, Okano H, Kim SU, Wakabayashi T, Issa JP, Sekido Y, Kondo Y. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in Glioblastoma. Cancer Res. 2013;73:4559–4570. doi: 10.1158/0008-5472.CAN-13-0109. [DOI] [PubMed] [Google Scholar]
- 149.Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, Ljungman M, Merajver SD, Kleer CG. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–853. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fan T, Jiang S, Chung N, Alikhan A, Ni C, Lee CC, Hornyak TJ. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Mol Cancer Res. 2011;9:418–429. doi: 10.1158/1541-7786.MCR-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Puppe J, Drost R, Liu X, Joosse SA, Evers B, Cornelissen-Steijger P, Nederlof P, Yu Q, Jonkers J, van Lohuizen M, Pietersen AM. BRCA1-deficient mammary tumor cells are dependent on EZH2 expression and sensitive to Polycomb Repressive Complex 2-inhibitor 3-deazaneplanocin A. Breast Cancer Res. 2009;11:R63. doi: 10.1186/bcr2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rao M, Chinnasamy N, Hong JA, Zhang Y, Zhang M, Xi S, Liu F, Marquez VE, Morgan RA, Schrump DS. Inhibition of histone lysine methylation enhances cancer-testis antigen expression in lung cancer cells: implications for adoptive immunotherapy of cancer. Cancer Res. 2011;71:4192–4204. doi: 10.1158/0008-5472.CAN-10-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rizzo S, Hersey JM, Mellor P, Dai W, Santos-Silva A, Liber D, Luk L, Titley I, Carden CP. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol Cancer Ther. 2011;10:325–335. doi: 10.1158/1535-7163.MCT-10-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, Jerva LF, Scott MP, Copeland RA. Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011;78:199–210. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 157.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, Scott MP, Jin L, Smith JJ, Olhava EJ, Chesworth R, Moyer MP, Richon VM, Copeland RA, Keilhack H, Pollock RM, Kuntz KW. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 158.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Waters NJ, Smith JJ, Porter-Scott M, Chesworth R, Moyer MP, Copeland RA, Richon VM, Uenaka T, Pollock RM, Kuntz KW, Yokoi A, Keilhack H. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 159.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang JH, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 162.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Choudhury SR, Balasubramanian S, Chew YC, Han B, Marquez VE, Eckert RL. (-)-Epigallocatechin-3-gallate and DZNep reduce polycomb protein level via a proteasome-dependent mechanism in skin cancer cells. Carcinogenesis. 2011;32:1525–132. doi: 10.1093/carcin/bgr171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dimri M, Bommi PV, Sahasrabuddhe AA, Khandekar JD, Dimri GP. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis. 2010;31:489–495. doi: 10.1093/carcin/bgp305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hua WF, Fu YS, Liao YJ, Xia WJ, Chen YC, Zeng YX, Kung HF, Xie D. Curcumin induces down-regulation of EZH2 expression through the MAPK pathway in MDA-MB-435 human breast cancer cells. Eur J Pharmacol. 2010;637:16–21. doi: 10.1016/j.ejphar.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 166.Alford SH, Toy K, Merajver SD, Kleer CG. Increased metastasis in patients with familial early-stage breast cancer and high EZH2 expression. Breast Cancer Res Treat. 2012;132:429–437. doi: 10.1007/s10549-011-1591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


