SUMMARY
In addition to the ever-present concern of medical professionals about epidemics of infectious diseases, the relative ease of access and low cost of obtaining, producing, and disseminating pathogenic organisms or biological toxins mean that bioterrorism activity should also be considered when facing a disease outbreak. Utilization of whole-genome sequencing (WGS) in outbreak analysis facilitates the rapid and accurate identification of virulence factors of the pathogen and can be used to identify the path of disease transmission within a population and provide information on the probable source. Molecular tools such as WGS are being refined and advanced at a rapid pace to provide robust and higher-resolution methods for identifying, comparing, and classifying pathogenic organisms. If these methods of pathogen characterization are properly applied, they will enable an improved public health response whether a disease outbreak was initiated by natural events or by accidental or deliberate human activity. The current application of next-generation sequencing (NGS) technology to microbial WGS and microbial forensics is reviewed.
INTRODUCTION
The term next-generation sequencing (NGS) as used here includes not only the physical process for the acquisition of sequence data but also novel methods for sample preparation and powerful new informatic approaches for handling and analyzing the resultant data (Fig. 1). The advent of inexpensive, ultra-high-throughput DNA sequencing instrumentation has transformed microbial whole-genome sequencing (WGS) from a million-dollar, team science enterprise into a routine exercise in molecular biology. Benchtop sequencers are now capable of producing up to 35 Mb (454 GS Junior), 2 Gb (Ion Torrent), or 15 Gb (Illumina MiSeq) of sequence per run and have a more-than-sufficient capacity for single-organism sequencing. While the de novo assembly of genomes remains challenging, the volume of sequence data produced by NGS technology is in fact usually high enough that several relatively small bacterial genomes can be sequenced in one run without sacrificing coverage depth. In addition to developments in the technology for acquiring the actual DNA sequence, there are also new methods that greatly simplify the preparation of NGS libraries (1–3). Enzyme-based technologies such as Nextera-Illumina transposition, which combines enzyme-based fragmentation with the addition of Illumina adaptors, can efficiently produce a sequence-ready library from <50 ng of DNA (4). Alternative enzyme-based fragmentation protocols have also been developed to prepare libraries for Ion Torrent sequencing (5).
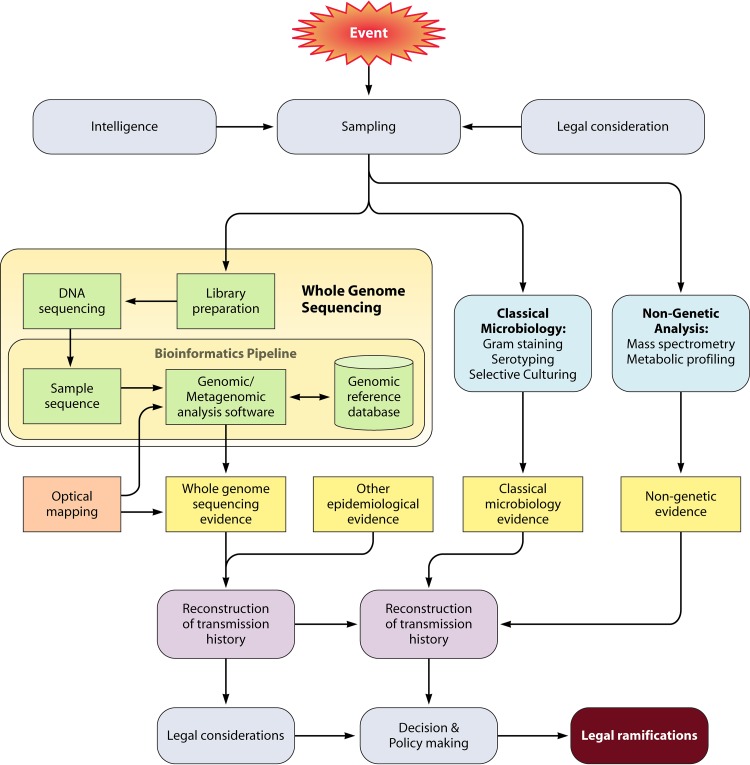
FIG 1.
Microbial forensics analysis diagram illustrating where NGS can fit within microbial forensic analysis. Legal considerations such as patient privacy and/or the rights of defendants/perpetrators are important to consider at the time of sample collection. WGS analysis requires both NGS and bioinformatic capacity. Legal requirements are also a factor in the possible uses of WGS information.
These remarkable advancements in methods and instrumentation have turned NGS into a resource that can be accessed and utilized relatively rapidly, cheaply, and easily, nearly anywhere in the world (6, 7). The process of generating data by WGS is therefore no longer a time-limiting step, and remaining bottlenecks lie in three main areas.
The first bottleneck is the absence of standard operational protocols for the acquisition of samples in conjunction with their contextual epidemiological information. These protocols should be formulated not only with an eye to the best scientific standards but also for the application of the appropriate legal safeguards to both respect individual privacy and permit future independent verification of the WGS results. Given that international travel increases the potential spread of infectious diseases, these protocols should be globally relevant and applicable. A promising approach to dealing with these problems is the Global Biosurveillance Technology Initiative (GBTI), which aims to implement the same standard NGS protocols within the Global Emerging Infections Surveillance and Response System Operations (GEIS) Partner Laboratory Network, which in turn is part of the U.S. National Strategy for Biosurveillance and the Global Health Security Agenda. Due to the limitations of space in this review, we refer readers to the other work for more in-depth information on the approach being implemented by these workgroups (8, 9).
The second bottleneck is the critical evaluation of raw sequence quality and performance of the bioinformatic methods used to map and identify sequence variants. Currently, quality indicators are often based on achieving the best that can be managed by using a particular sequencing technology. This is a rapidly changing field, and there is a need for common metrics to be developed that can be applied across different sequencing platforms and that encapsulate the quality of the sequence information used in the analysis. Harmonization of study results can be achieved by using different study centers and technologies to sequence a DNA sample; some samples, however, may be available in very limited quantities (10), and there are also barriers to the shipment of material that may contain infectious agents (11). This potential limitation has been addressed through the development by the Human Microbiome Project of an artificial DNA sample prepared from a “mock” microbial community to calibrate the results obtained from different sequencing centers and machines.
The third bottleneck involves the improvement in data analysis procedures to convert floods of data into actionable results. Accurate base calling is essential for mutation detection in WGS and most forensic applications. An in-depth discussion of the technological approaches and scientific details of NGS chemistry (specifically comparing the most popular variants Roche 454 GS Junior, Life Technologies Ion Torrent Personal Genome Machine [PGM], and Illumina MiSeq) is beyond the scope of this report and has been reviewed in depth elsewhere (12–14). In general, NGS has higher error rates than Sanger sequencing, but these higher rates are mitigated by repeated sequencing to establish a sequence consensus and data filtering (based on quality and error probability scores such as Q20 [quality score in which 1 error is estimated to occur in every 100 bases]); for NGS, there are techniques for the generation of FASTQ files that include quality assessment such as artificial Phred scores so that cross-platform results from different sequencing pipelines can be compared (15, 16). In tests, sequences generated by the Ion Torrent, MiSeq, and Pacific Biosciences technologies all displayed near-perfect coverage behavior on GC-rich, neutral, and moderately AT-rich genomes, but a profound bias was observed upon sequencing of the extremely AT-rich genome of Plasmodium falciparum on the Ion Torrent PGM platform, resulting in no useful data (12, 14).
Recently, single molecule, real-time (SMRT) sequencing, a fourth high-throughput sequencing technology, has begun to be used more frequently in WGS applications. While the Pacific Biosciences PacBio RS instrument is more error prone than other NGS platforms, it is the first of its kind to utilize SMRT sequencing, which produces very long read lengths (>20,000 bp, with an average length of ∼3,000 bp) that are 30 to 200 times longer than those produced by other NGS instruments and 4 times longer than those produced by the original instrument released in 2011 (17). The advantages of SMRT are that library preparation is minimal, and its long read lengths allow rapid assembly of the WGS (18). When SMRT sequencing was combined with circular library preparation, the consensus sequence error rate dropped to within the limits achieved by other NGS technologies (19). Recently, a device from Oxford Nanopore Technologies, while still under development and exhibiting sequencing bias, has also demonstrated the capacity to produce long sequence reads (10,000 bp) that facilitate de novo WGS assembly (20, 21).
There are two main approaches to genome assembly, reference guided and de novo, which have been discussed in depth elsewhere (22–24). The reference-guided method, which refers to alignment of sequencing reads to each other and to a selected reference genome, is helpful for rapidly assembling and annotating newly sequenced genomes. There is, however, a risk of not properly assembling genomic regions or annotating genes if the new sequences are too divergent from that of the chosen reference genome. Errors in genome assembly, especially in the context of microbial forensics dealing with novel strains or pathogens, can have serious consequences. If WGS data are misassembled, this can lead to the omission of critical elements of a genome, such as plasmids containing virulence factors, antibiotic resistance genes, or bioengineered DNA originating from different organisms. For WGS, a preferable option is the de novo approach, which allows the assembly of short reads into longer contigs and scaffolds without a reference. Many tools, including Velvet (25), SOAPdenovo (26), ABySS (27), and ALLPATHS-LG (28), are now available for de novo genome assembly (Fig. 2). However, during recent genome assembly “competitions,” such as GAGE-B (29) and the Assemblathons (24, 30), it has been noted that no single assembler outperforms others in all assembly metrics for all organisms. Specifically, both of the competitions mentioned above found that (i) on a single organism, different assemblers were ranked completely differently depending on which metric was being used to judge the quality of the assembly and (ii) for the same metric, assemblers were ranked differently for different organisms. A detailed review of genome assembly can be found elsewhere (31).
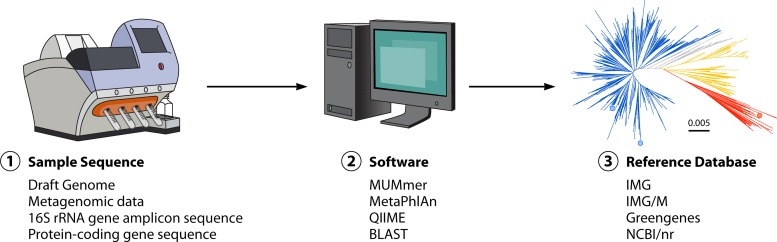
FIG 2.
Conceptual representation of a microbial forensics bioinformatic pipeline. There are three components necessary for microbial forensic analysis with NGS data. First, a DNA sequence of the sample is needed (1), which could be a completed draft genome assembly, shotgun metagenomic sequences, or amplicon sequences such as 16S rRNA or protein-encoding genes. Algorithm/software implementation (2) is the necessary link between the sample sequence data (1) and a comprehensive reference database (3) consisting of all available draft and complete genomes, metagenomes, 16S rRNA gene sequences, or other protein-encoding gene sequences.
Standard tools for whole-genome alignment, such as MUMmer (32), can be used to compare assembled genomes to databases of both finished and draft microbial genomes, which are available from sources such as the NCBI and the Integrated Microbial Genomes (IMG) database (33). To evaluate the evolution of strains during an outbreak, it is also useful to carry out phylogenetic analysis to compare the finished assembly and trace genetic changes, not only in reference to the current epidemic but also in the broader global context. For an in-depth discussion of the strengths and weaknesses of these programs, we direct readers to several excellent publications on this topic (34–37). In brief, the programs that are most helpful in correctly identifying the most probable taxonomic lineages and assisting in tracing the transmission path of an epidemic can incorporate relevant epidemiologic data (such as spatial overlaps) and the natural history of the organism (e.g., the average frequency with which mutations occur in a given species) during the building of phylogenetic trees (38–40).
Advances in NGS have resulted in a situation in which rapid WGS of the causative microbe during an outbreak is practical (41). As a result, first responders (such as medical, law, and fire service personnel who might be providing patient care in the field) should be aware of the power and the limitations of WGS so that they know to obtain appropriate samples and contextual information, which will complement WGS in building the models needed to predict the course, or define the origin, of an epidemic.
THE CASE FOR APPLYING WGS TO DETERMINE WHETHER AN OUTBREAK IS DUE TO DELIBERATE, ACCIDENTAL, OR NATURAL CAUSES
Genomic microbiology can give detailed information on biological samples, and the rapid advances in molecular technology allow WGS to be used as a routine investigatory tool. From a policy- and decision-making standpoint, it is critical to determine whether an outbreak is due to natural circumstances, an accidental release of a cultured or engineered organism, or a deliberate introduction of a known pathogenic organism. Under these circumstances, the ultimate goals are identification of the agent responsible for the outbreak and attribution of both its biological and geopolitical origins, with a high degree of certainty. The resultant information is central to making correct decisions that are required in response to the outbreak, such as whether a public health emergency should be declared, what scientific and legal mechanisms should be used to limit transmission and contain the outbreak, and even whether to trigger a response under the Biological and Toxin Weapons Convention (BTWC) (42).
Prior to the application of WGS to forensics and bioterrorism, identification of the “unique signature” of a deliberately engineered organism was not possible (43). During the Amerithrax investigation of the 2001 mailing of anthrax spores, for example, microbiological characterization of strains was by done by WGS (10, 44). Upon compilation of appropriate reference databases, the Bacillus anthracis organisms used in the attack were identified as the laboratory Ames strain, and the material was defined further as a B. anthracis Ames ancestor strain (45–47). At that time, however, WGS was slow and prohibitively expensive and required 5 μg of genomic DNA; as a result, some samples were never tested, and the use of multiple sequencing runs for validation of the single-nucleotide variants in the sequenced genomes was limited (10). On-site WGS was not possible, and the long and complex “chain of custody” resulted in challenges to the DNA evidence. In addition, sampling was carried out by individuals who were the holders of anthrax stocks and who turned out to be potential defendants/perpetrators. This situation may have significantly biased the sample collection, especially once it was known that genetic analysis was a key component of the investigation. Inconsistencies in sampling methods and the fact that samples were cultured prior to sequencing may also have obscured genetic diversity among the original samples. The collection of only two samples from each source limited their availability for subsequent validation using newer WGS technologies. Little information was available on the provenance of the anthrax samples and contextual data on the genetic interrelatedness, thus restricting the possibility for tracking their movement between laboratories. These limitations weakened the investigation as a whole, leading the National Research Council Committee, which reviewed the scientific evidence from the investigation, to conclude that despite considerable data indicating a specific perpetrator, “it was not possible to reach a definitive conclusion about the origins of the B. anthracis in the mailings” (48).
WGS now offers the potential for providing more accurate and rapid information, thus enhancing the likelihood of meeting criteria described in the International Health Regulations (IHRs) (2005), which state that parties are required to assess reports of urgent events within 48 h to determine whether an event is “notifiable” and to have the capacity for rapid determination of control measures required to prevent domestic and international spread. U.S. and the European Union laws and World Trade Organization (WTO) regulations require quick responses, with a scientific basis, to avoid needless disruption of trade and travel relating to a potential public health emergency of international concern (PHEIC).
Biosurveillance Information
Indicators of the release of an infectious agent include the occurrence of disease in humans as well as detection of pathogens through surveillance systems. There is a need to coordinate public health, law enforcement, and national security activities under a common umbrella and to employ the recent advances in WGS as applied to microbial forensics during this concerted action. It is essential that the details obtained during biosurveillance be integrated into the analysis of samples in order to obtain the maximum amount of useful information.
The current U.S. strategy for biosurveillance is an “all-hazards” approach so that potentially dangerous incidents are monitored through the same surveillance systems regardless of whether they are believed to result from a deliberate or accidental release or a natural occurrence (9). Since 2001, environmental and informational surveillance systems have been greatly expanded in type and geographic distribution such that outbreaks can be detected by using biological agent detectors. An example of these surveillance systems is the Bioagent Autonomous Networked Detector (BAND) project, which involves air sampling in at-risk urban areas at least once every 3 h (49). The BAND project focuses on pathogens such as bioterrorism category A agents, including anthrax, botulism, plague, smallpox, tularemia, or viral hemorrhagic fever (50). These surveillance systems involve several agencies that have cooperated to provide sample libraries for detecting potential threats and ruling out background material (49). They employ protocols consistent with microbial forensic applications, have sophisticated sampling algorithms, and involve multiple national security and law enforcement agencies. Programs to increase water sampling have been initiated but are technologically much more difficult (49). Despite increasing cooperation between state and federal agencies and among international agencies, the limits of physical surveillance methods make it likely that an “attack” will first be detected through epidemiological indicators rather than environmental sensors (51).
Electronic health records (EHRs) throughout the United States can be used in informational surveillance to identify epidemics in their early stages. This surveillance includes new reporting requirements and coordination both nationally, within state and federal frameworks, and internationally (52–54). While the Privacy Rule permits disclosure of private health information without authorization for public health surveillance, investigations, and interventions, the disclosure of this information has legal and logistical consequences, since criminal prosecution and national security interests are not always the same as those of public health (55). Most developed states and countries have privacy statutes that dictate how patient records may be used (56, 57). These laws, which differ considerably among states and countries, need to be taken into consideration prior to sample collection. Since 2001, a number of protocols have been developed/proposed to deal with these potential conflicts, but this has not been addressed in many states. Even in those states and countries that have dealt with these issues, it is important to recognize the impact of new technologies (55).
Issues for consideration in the use of U.S. patient records.
In the United States, HIPAA (the Health Insurance Portability and Accountability Act) allows free access to patient records held by “covered entities” (which include various health plans, health care clearinghouses, and health care providers that transmit health information electronically) for legitimate public health purposes but more limited access for judicial proceedings. For the latter, a court order or other legal order (e.g., a subpoena) is necessary for the release of records, and notice must be provided to the person whose records are sought (58, 222). If an environmental sample could include evidence to incriminate an individual whose health records are sought, additional state and federal criminal evidence safeguards may exist.
There is a potential loophole in that HIPAA does not block public health officials from sharing patient records with law enforcement officials (because public health entities are not “covered entities” under HIPAA), but there could be serious political and public trust consequences if there was a perception that public health officials were being used to “spy” on individuals for law enforcement. The full scope of that action will depend on the national security protocol that is invoked, and although in this setting, a covered entity may disclose patient records, it is not required to do so.
As authorized by the National Security Act (59) and the USA Patriot Act (60) or Executive Order (222), covered entities can release patient records to authorized federal officials for the conduct of lawful intelligence, counterintelligence, and other national security proceedings. A full debate on the limits of emergency powers and how they may affect patient privacy has not occurred, and there are significant gaps in our understanding of how patient records may be used in a public health emergency. The initial discussions on HIPAA exceptions have focused on the patient records of the potential perpetrator, and while discussions on the creation of large bioterrorism databases of patient records have begun, these debates have not yet resulted in any clear directives. Large-scale, deidentified “biobanks” of DNA isolated from residual clinical samples have been established at several institutions. There are concerns, however, about the risk to patients' genomic privacy posed by these collections, as exemplified by the recent reidentification of several participants in the 1000 Genomes project (61–64). HIPAA regulations permit disclosures to law enforcement agencies investigating potential acts of bioterrorism, but not disclosures of DNA sequence data [Code of Federal Regulations Title 45 §164.512(a)(2)(i) (58)]. Although this precluded DNA analysis, it is likely best interpreted as referring to human DNA; the DNA of microbes can also have identifying information associated with it. For example, such data may reveal details about an individual's movement and travel.
As mentioned above, state as well as federal laws may apply to patient record access. Health systems throughout the country have implemented new EHRs, but the level of detail and laboratory results included in the record vary from system to system. Many records would not include WGS data even if this level of testing were taking place. “Meaningful use” criteria dictated for EHRs by the Centers for Medicare and Medicaid Services do not address this use.
If a public health emergency has been declared, states that have passed legislation incorporating the Model State Emergency Health Powers Act (a majority) have sweeping powers to deal with such a situation. This would allow significant access to patient records by authorized personnel, but as states may have different limitations and definitions of a public health emergency, access would need to be dealt with on a case-by-case basis. The full extent of coordination among public health officials, law enforcement agencies, and national security officials will be context dependent and has not yet been fully resolved. For example, it may be too late to start gathering identifiable data after an emergency is declared, and routine collection of identifiable and linked data may be necessary. It is also not clear what might happen if a public health emergency spans several states with different emergency criteria and regulations. Nonetheless, it is unlikely that these problems will be addressed legislatively. States want to retain their powers in emergency situations, and even at the federal level, there has been unproductive interagency competition. Thus, protocols that model potential scenarios need to be developed so that gaps and overlaps in real-time legal decision-making can be exposed and addressed. In addition to the challenges of patient record access, their usefulness in routine surveillance will be affected by heterogeneous institutional facilities, resources, and reimbursement agreements as well as by differences in clinical decision-making, which may result in inconsistent sampling (65). Participants in a recent World Cohort Integration workshop listed training in biobank management as one of their top priorities (66).
The all-hazards approach may be an adaptable strategy to safeguard the public health of the nation. In the case of natural pandemics or the release of bioterror agents, the initial casualties may first be seen in hospitals or other health care settings. Information indicative of the early phase of an “outbreak” will be available initially to public health officials who are trained to gather epidemiological evidence in order to contain disease and treat affected individuals; however, they are often neither knowledgeable about nor focused on determining the source of the agent, identifying a possible perpetrator, or collecting and preserving evidence for legal proceedings. It is therefore essential that leaders in this area have the knowledge and experience to recognize the potential for an accidental or deliberate release and have the authority to initiate an alert and the associated response. Access to the current technologies used for microbial forensics, such as WGS, would be beneficial in an investigation of a routine outbreak as well as one that involves criminal activity and/or national security.
Current Epidemiological Approaches to the Detection of Deliberate or Accidental Outbreaks
A number of authors have described epidemiological indications that an outbreak may be caused by bioterrorism (67–71). Dembek et al. list 11 “clues” supporting the likelihood of an intentional attack: (i) a highly unusual event with large numbers of casualties, which occurs with no plausible natural explanation; (ii) a higher rate of morbidity or mortality than expected; (iii) an uncommon disease; (iv) a point source outbreak; (v) multiple epidemics; (vi) lower attack rates for individuals protected from biological incident or attack by vaccination, antibiotics, wearing personal protective equipment, or living in an air-water barrier isolation unit; (vii) dead animals; (viii) reverse spread (people to animals); (ix) an unusual disease manifestation; (x) a disease pattern consistent with the discharge of the pathogen from a single point followed by its subsequent distribution by air or water currents; and (xi) direct evidence (70). The appearance of any of these clues during an outbreak should prompt consideration that an accidental or deliberate release of a bioweapon may have occurred. Grunow and Finke as well as Radosavljevic and Belojevic have developed epidemiological approaches that yield a quantitative assessment of whether an outbreak is intentional, accidental, or natural (69, 71). Since these algorithms are based on incomplete evidence and qualitative data, they are necessarily imprecise. Neither of these approaches has been fully validated or applied in the course of an outbreak, but both approaches could provide guidance to public health officials in the early stages (or in a later retrospective assessment) of an event, in order to make decisions such as whether and when to include law enforcement in the investigation. Dembek et al. applied the test described by Grunow and Finke retrospectively to a number of case scenarios and found that it accurately identified the situations most likely to have resulted from bioterrorism but failed to identify deliberate attacks that involved common pathogens (70). Moreover, the difficulty with this tool is that considerable investigation may be required before a valid assessment can be made, and by then, crucial leads may be lost or new events may initiated.
In that the approach of Grunow and Finke is felt to be too slow for use in attacks that involve mass casualties, Radosavljevic and Belojevic created an alternative method that uses similar epidemiological clues but weighs factors differently, enabling, they believe, a more rapid assessment of the nature and etiology of an outbreak. Application of the test by Radosavljevic and Belojevic to the North American outbreak of swine flu in 2009, the Kosovo tularemia outbreak of 2000, and the Sverdlovsk anthrax release of 1978 yielded conclusions that were consistent with those obtained after final analyses of these events (71). Applying a tool retrospectively is different, however, than using it in real time. The difficulty with the tools of both Grunow and Finke and Radosavljevic and Belojevic is that it is difficult to anticipate the availability and accuracy of data.
In reality, public health officials often need to act long before all of the information can be assembled. Thus, although epidemiological tools and other indicators will continue to play a crucial role in the characterization of a suspicious outbreak, concurrent application of WGS and other methods of microbial forensics will generally yield more rapid and accurate results. Radosavljevic and Belojevic found that of all the variables that they examined, identification of the etiologic pathogen was the most important component of the scoring system and concluded that WGS should be applied as soon as any suspicious indicator is identified. Challenges that need to be addressed for WGS to be of use during an outbreak are the legal issues (discussed above) and the practicality of comprehensive sample collection and storage (discussed in depth in the context of the German Escherichia coli O104:H4 outbreak, below).
Advantages and Limitations of Approaches Other than WGS in the Characterization of Infectious Disease Outbreaks
Beginning with the pioneering work of Ferdinand Cohn (1828 to 1898), the growth characteristics of microorganisms have been used to identify and classify them. One of the key indicators of an engineered outbreak is the presence of a rare or out-of-context pathogen (i.e., anthrax spores in mail). “Classical” techniques are the current mainstay of microbial identification, but they usually require specialized skills that cannot be rapidly acquired during an emergency when the ability to process a large number of samples is paramount. Currently, automated identification systems (e.g., BD Phoenix 100, bioMérieux Vitek, and Biolog, to name a few) are available for high-throughput, phenotypic characterization of some prokaryotic microorganisms, based on growth under different culture conditions (72–75). In an outbreak situation, antibiograms (the ability of an isolate to proliferate, or not, in the presence of different drugs) have been used to both track and provide information on the drug susceptibility of target organisms. This information can be vital to control an outbreak and minimize the number of fatalities (76).
Careful examination and analysis of bacterial colony morphology are powerful tools in the hands of experienced microbiologists, as illustrated in the study of the anthrax formulation used in the Amerithrax attacks. Key morphological variants were identified in samples gathered from the incident premises (10). This knowledge, combined with WGS of the relevant isolates, provided a highly distinctive genetic “fingerprint” for investigators to trace the original source of the anthrax spores (10). Culture-based approaches share two major limitations: the organism must be both (i) “culturable” and (ii) able to be isolated from the initial biological mixture in which it is collected, i.e., in a monoculture. It is estimated, however, that >99% of bacteria cannot be cultured in the laboratory under any known conditions (77–79). As, historically, the inability to culture a biological organism is not a limitation to its use in warfare, in cases of suspected bioterrorism, techniques that are not limited to cultured organisms should be used (80, 81).
Nevertheless, when available, culture-based techniques are the mainstay of classical microbial identification and can provide the foundation needed for WGS and proteomic-based approaches. A variety of techniques, including elemental analysis and infrared spectroscopy of proteins, are being used to characterize biological samples in forensic investigations; mass spectrometry (MS) has become the most commonly used and one of the most powerful (82). This technology generates a proteomic fingerprint of biological material, using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS. Comparison of data from a bacterial culture to data in a reference database of MALDI-TOF MS spectra enables identification of the genus and species. It is likely that recent advances in this technology will allow not only the identification of the cultured strain but also the determination of whether it expresses specific effector molecules (toxins and drug efflux pumps, etc.) (83–86).
Amplification of “signature” DNA in samples by PCR can bypass the requirement for an organism to be isolated and grown in culture. Identification of repetitive-unit variable-number tandem repeats (MLVA [multiple-locus variable-number tandem-repeat analysis]) has been used to genotype bacterial pathogens (87–89). These DNA repeat regions are prone to strand slippage occurring during DNA replication and also to expansion and contraction in numbers during DNA recombination and are therefore frequently polymorphic (90, 91). The variation in the number of repetitive units is often easily detected following PCR amplification with genus- or species-specific primers, size fractionation in agarose gels, and subsequent DNA staining. The two major drawbacks of this genotyping method are that (i) the evolutionary relationship between strains cannot be inferred (due to the potential for both expansion and contraction in the number of repeats) and (ii) small changes that could differentiate closely related strains may not be detected.
Multilocus sequence typing (MLST) is a second PCR-based approach, but this approach targets DNA sequences under evolutionary pressure, to type microbial pathogens at the sequence level (92–95). The investigation of the source of outbreaks of nontyphoidal Salmonella in the Gambia provides an example of the application of MLST. Seven genes were sequenced in multiple isolates, in order to type Salmonella isolates causing infections in children and their companion animals. The salmonella strains from each host species were distinct, suggesting primarily human-to-human transmission of Salmonella in this population (96). Uneven evolutionary selection among the MLST targets and an accompanying discovery bias may give the misleading impression that a pathogen is genetically monomorphic, when this is not the case (97). Careful selection of target genes is essential so that MLST is able to discriminate among closely related strains. When this is not possible due to inadequate a priori data, in-depth sequencing of selected isolates can be used to identify appropriate MLST targets.
In this presentation, we evaluate the advantages and limitations of WGS in the investigation of infectious disease outbreaks and its ability to identify and characterize the outbreak source. To do this, we have analyzed several paradigmatic cases in which genomic microbiology has been used to identify the relevant technical, scientific, and legal challenges. We have summarized recent examples that made use of current WGS technologies with the goal of pointing out the most effective approaches in their deployment.
OUTBREAK OF ENTEROHEMORRHAGIC ESCHERICHIA COLI O104:H4 IN GERMANY (2011)
E. coli strains are ordinarily commensal, enteric organisms, but several, such as enteroaggregative E. coli (EAEC) strains and Shiga toxin-producing enterohemorrhagic E. coli (EHEC) strains, possess virulence factors that enable them to be pathogenic. Shiga toxin can both cause local damage in the gut (resulting in bloody diarrhea) and, after entry through the gut wall, damage the kidney, resulting in hemolytic-uremic syndrome (HUS) (98).
In May to June 2011, >4,000 cases of bloody diarrhea and 850 cases of HUS, including 50 deaths, were reported in Germany. These illnesses and fatalities were subsequently determined to be caused by a strain of Shiga toxin-producing EHEC serotype O104:H4 that also contained a plasmid characteristic of EAEC and a plasmid encoding an extended-spectrum beta-lactamase (99, 100). Historically, the O104:H4 strain has not been associated with a high rate of HUS (101, 102), but the proportion of infected patients who went on to develop HUS and other complications in this outbreak was unexpectedly high (103, 104). Isolates of E. coli O104:H4 from these cases, as well as some from France in June 2011, were indistinguishable from previous strains by the current conventional molecular epidemiological methods of characterization (serotyping, multilocus sequence typing, repetitive extragenic palindromic PCR [REP-PCR], pulsed-field gel electrophoresis, optical mapping, and antimicrobial susceptibility testing) (100, 105). Rohde et al. used a rapid benchtop sequencing technology, open-source data release, and crowd-sourced analyses to reveal in <1 week that the strain responsible for this outbreak belonged to an EAEC lineage and had acquired genes for Shiga toxin and antibiotic resistance (106).
Virulence Characteristics of the Causal Organism and Distinguishing between Accidental/Deliberate Release and Natural Transmission
Mellmann et al. refined the initial draft genome and characterized the Shiga toxin gene carried by the German strains as being most similar to those of EHEC lineages, although the heat-stable enterotoxin gene astA, in addition to TEM-1 and CTX-M-14 beta-lactamase antibiotic resistance genes, was EAEC-like. This work suggested an evolutionary model whereby the hybrid outbreak strain arose in an EAEC genetic background by the acquisition of Shiga toxin followed by several gene insertion events, which added both astA and beta-lactamase antibiotic resistance (40). The combination of the virulence traits from both EAEC and EHEC strains seemed to have produced a more virulent strain.
The genome sequences of the O104:H4 outbreak strain, seven other EAEC O104:H4 strains, and four reference strains were obtained as one component of the investigation to find the source of the outbreak (107). Rasko et al. found Shiga toxin-encoding prophage elements as well as structural variation around regions harboring virulence factor genes and pathogenicity islands (107). Phylogenetic trees, drawn on the basis of the whole-genome sequences obtained from the O104:H4 outbreak and related strains, were in agreement with the high likelihood that the Shiga toxin-producing outbreak strains evolved from an EAEC O104:H4 strain that recently acquired the toxin-encoding phage via natural lateral gene transfer. Therefore, the most plausible interpretation of the accumulated data regarding this episode is that the source was a Shiga toxin-producing O104:H4 strain, which resulted from a naturally occurring exchange of genetic material and not a deliberate or accidental release of an engineered bioweapon. A deliberate introduction of a Shiga toxin-encoding prophage would likely have shown other features, such as the presence of cloning vectors (containing known restriction sites), in the genomic analysis or topographical changes in the genome organization, which might be the hallmarks of scarless genetic engineering. In future infectious disease outbreaks, WGS will be able to supply this type of information in real time and hopefully will lead to a more rapid optimization of the mitigation strategies being employed.
Tracing the Origins of This Outbreak
Foodborne pathogens have been used in the past as bioweapons, and when their clinical presentation is similar to those of other diseases that might be expected in the target population, the outbreak may not receive the appropriate level of attention by public health authorities (67, 70, 108). Importantly, this outbreak of illness caused by O104:H4 met several of the criteria for a biological attack, as discussed by Grunow and Finke as well as Radosavljevic and Belojevic (i.e., presentation of greater virulence due to a new combination of antibiotic resistance and toxin genes) (69, 71). However, classical epidemiology linked the EHEC O104:H4 strain to a 15,000-kg shipment of fenugreek seed from Egypt that was sent to Germany in December 2009. Some of the seeds were also sent to France and caused a smaller outbreak there (100). These two episodes were ultimately linked by WGS, but due to difficulties in detecting and culturing this organism directly from the environment, neither samples nor WGS evidence was available to trace the E. coli O104:H4 isolate back to the seeds imported from the Egyptian supplier. It remains to be determined whether metagenomics (discussed below) will be useful in this role of tracing and characterizing virulent strains, sans culture, in such environmental samples (109). The EHEC O104:H4 outbreak demonstrates the vulnerability of our food chain and the potential health and economic consequences that can result from even limited product contamination. It also illustrates why unusual outbreaks involving endemic microbes must be taken as seriously as those involving potential agents of bioterrorism. Although there did not appear to be criminal intent involved then, the complicated economic and legal issues that came into play document the importance of having and using sophisticated microbial forensic techniques to aid in such an investigation and the subsequent legal proceedings.
Rasko et al. sequenced an additional outbreak strain and other historical EAEC O104:H4 strains to place the outbreak within a global context (107). In these comparisons, the core genome of the EHEC outbreak strains were closely grouped with EAEC O104:H4 strains. Only 238 sequence differences were observed between the outbreak strains C227-11 and TY2482 (107). As these NGS data were obtained by using a PacBio SMRT sequencer, which provides long sequence reads, facilitating de novo genome assembly at the cost of sequence accuracy, Grad et al. resequenced the second outbreak strain and three additional German outbreak strains using the more accurate Illumina instrument and by doing so illustrate the current strengths and limitations of each technology. Gire et al. also found during sequencing of Ebola virus (EBOV) during the 2014 outbreak that the accuracy of Illumina sequencing was advantageous (111). However, as discussed above, the PacBio technology offers high-speed sequencing and assembly at the cost of an increased incidence of in-sequence errors. Library sequencing using the Illumina machine produces a less error-prone sequence, but its shorter reads require greater effort during alignment to a reference genome or for assembly into a new whole-genome sequence de novo. Using the Illumina sequence output, Grad et al. were able to confirm the close relationships among the outbreak strains (which they found to differ, however, by only two single nucleotide polymorphisms [SNPs]) (39). The number of strains available for sequencing is critical for WGS to be a useful adjunct technology to the epidemiological study of transmission pathways. Sequencing of 11 EHEC O104:H4 isolates from the small number of French cases and identification of 19 divergent SNPs allowed a more nuanced perspective on this entire episode (39).
Although the outbreak was first recognized in relation to salad products eaten in northern Germany (and later in a separate but related outbreak in France), it quickly involved most of Europe. Within 2 weeks, 12 European nations had reported outbreaks that might constitute a “public health emergency of international concern” (PHEIC) pursuant to the requirements of WHO International Health Regulations (53, 112). An outbreak is considered a PHEIC if two of the following four factors are present: (i) serious public health impact, (ii) unusual or unexpected circumstances, (iii) risk of international spread, and (iv) significant risk of international or trade restrictions (53). The EHEC O104:H4 outbreak clearly met these standards, and while the WHO never formally declared a public health emergency, the outbreak produced significant concern and had serious international economic effects. None of the affected countries attempted to hide the outbreaks, and this certainly aided in the investigation and facilitated international cooperation. The government of Germany, the epicenter of the outbreak, made every effort to be transparent and provided considerable information to the public during the course of the investigation. It appears, however, that this open approach also had negative economic effects. When the outbreak was first reported, Spanish cucumbers were suggested to be a potential source. It was not until a week later, after a more sophisticated epidemiologic investigation was conducted, that sprout seeds imported from Egypt were implicated as the likely source (113). In the meantime, many countries acted proactively to remove cucumbers and tomatoes from stores and production, Russia issued a trade ban on Spanish cucumbers, and the Spanish cucumber and vegetable market suffered significant losses (estimated to be €200 million a week) (114). Some of these actions may have violated European law and/or constituted WTO violations (112). Spain sought compensation for the economic losses that were incurred, but legal mechanisms for obtaining such funds were unclear (112). Ultimately, the European Union proposed several measures to support the promotion of fresh fruit and vegetables following the E. coli crisis in member states, which included Spain (115).
The IHRs (2005) require parties “to avoid responses to disease events that lack a scientific and public health basis and that impose unnecessary restrictions on international trade and travel” (53). Similar obligations exist under European Union and WTO rules (112). The difficulty with this requirement is that there is no real standard for what constitutes such a basis, and it can be difficult to balance this requirement with the need to protect public health. What is clear is that the resulting response was neither rapid enough nor accurate enough to prevent social and economic consequences. Classical epidemiologic approaches are useful but may be slow and inaccurate. In the EHEC O104:H4 outbreak, DNA sequencing was constrained by legal and scientific limitations. Patient samples were not necessarily collected because German health insurance limits laboratory testing to the minimum that is needed for establishing a diagnosis (116). Even more importantly, although 10,000 samples were taken, EHEC O104:H4 contamination was never detected in any food sample, due at least in part to the fact that there was no standardized method for collection and handling.
The issue of test validation remains a major legal challenge (117). Most laboratories are unable to accumulate enough samples to validate an assay to detect a rare pathogen prior to an outbreak, and when an outbreak occurs, it is too late to focus on validation. Even when it is theoretically possible to carry out validation testing, there is a question of what level is necessary, depending on the context of the situation. A test may not meet criminal prosecution standards but could provide adequate evidence for a public health decision during a rapidly progressing outbreak. If, however, a test or technology has not been approved by the relevant government agencies, even if scientifically validated, it may not be used. An accelerated process is needed for the evaluation and approval of potentially useful tools in an outbreak. To accomplish this would require decisions to be reached about what minimum preliminary evidence is needed to deploy techniques that, while not fully validated, offer significant potential benefits in outbreak detection and control.
Retrospective Metagenomics Study
NGS methods have been used extensively for single-organism, whole-genome analysis, but only recently have methods become available for characterizing whole populations of microorganisms or for forensic use in decision-making. Dramatic improvements in NGS technology and methods for data handling and analysis over the last several years (118) have enabled metagenomics, the application of sequencing to study DNA collected directly from environmental samples (without purification and amplification such as that achieved by selective culture) such as soil (109), seawater (119, 120), hospitals (121), and human-associated habitats (122). Thus, a metagenome is the complete collection of genetic material recovered directly from an environmental sample, representative of the organisms present in that sample (bacterial, viral, human, animal, or otherwise). The National Research Council recently compared advancements in metagenomics to “a reinvention of the microscope in the expanse of research questions it opens to investigation” (123).
One of the important lessons to be learned from this outbreak is the necessity of archiving residual patient samples in specific biobanks. In order to accomplish this, however, it is necessary to have in place ahead of time adequate funding and standard protocols for both the collection of samples and the accurate recording and archiving of associated epidemiological data and to ensure patient privacy, as discussed above (62, 124). Currently, there are no such prospective protocols for the archiving of samples to be used in WGS or for the education of the public on the value of patient records and microbiological samples (isolates) collected during an epidemic for basic and clinical biomedical research (62, 125). In a retrospective study, samples collected during the 2011 Shiga-toxigenic E. coli (STEC) O104:H4 outbreak in Germany were sequenced in depth and analyzed by using a metagenomic approach. The resulting data highlight the limits of current metagenomic approaches and point out areas, such as sample collection and sequencing depth, that need to be further developed in order for this approach to play a key role in the genetic characterization of a pathogen during an outbreak (126). Loman et al. used total DNA isolated directly from clinical samples to prepare sequencing libraries, which were then run on both the Illumina MiSeq and multiplexed HiSeq instruments (126). Encouragingly, those researchers were able to assemble a full draft genome of the pathogen (STEC) out of this mass of data. Another arguable success is that those researchers were able to detect sequences from the pathogen in 67% of the samples. However, a sensitivity of 67% is low for a clinical diagnosis and may yet be too low for forensic attribution purposes. Notwithstanding these results, this study found many potential pathogens in any particular sample. To narrow down the list of potential causative agents, those researcher had to determine which species were present by using only microbial contigs that were present in at least 20 patients but not present in any healthy controls. While Loman et al. concluded from their studies that metagenomic data can be used to infer a causal link by detection of a pathogen, in fact, a single sample and the coincident occurrence of disease are not definitive evidence of causality (127). It is, as Loman et al. themselves note, essential that multiple “suspect” and “background” samples be acquired to determine if there is a statistically significant link (126).
OUTBREAKS OF METHICILLIN-RESISTANT STAPHYLOCOCCUS AUREUS IN THE UNITED KINGDOM (2009 AND 2011) AND GERMANY (2010)
The emergence of community-acquired and methicillin-resistant Staphylococcus aureus (MRSA) has posed important new public health risks. Until the 1990s, infections with MRSA were limited largely to health care settings, but in the late 1990s, community-associated strains were identified in children, sports teams, and men who have sex with men (128). These strains are particularly dangerous because they combine antibiotic resistance, exceptional virulence, and person-to-person transmissibility. MRSA infections, which are quickly approaching pandemic levels, are creating problems in the community and rapidly displacing hospital-acquired strains. It is therefore no longer relevant to distinguish between hospital- and community-acquired strains. The public health importance of the increasing prevalence of antibiotic-resistant, virulent MRSA may be second only to that of HIV/AIDS (128). The evolution of these strains appears to involve naturally occurring horizontal acquisition of elements conferring methicillin resistance and other changes that are not yet fully understood but which also affect virulence.
Pathogens such as MRSA, which are spread by person-to-person contact, are less contagious than those transmitted by aerosol. S. aureus, however, resists decontamination protocols (persisting for up to 7 months on dry surfaces), making it difficult to remove from inanimate objects. In addition, the ease by which the S. aureus genome can be manipulated, its capacity to grow in large quantities in culture, the high morbidity and mortality rates associated with this organism, and, finally, the absence of an effective vaccine collectively are reasons why this organism is more of a threat (129, 130).
Virulence Characteristics of the Causal Organism
The MRSA strain (designated sequence type 2371 [ST2371]), which was isolated in Cambridge, United Kingdom, was clearly derived from ST22 (the MRSA sequence type that is the cause of ∼80% of United Kingdom hospital-associated MRSA infections) but also had acquired the gene for the exotoxin Panton-Valentine leucocidin (PVL) (76, 131). The prophage carrying the genes encoding PVL had independently integrated into S. aureus genomes on several different occasions, and it is therefore probable that the acquisition of these toxin genes by the ST22 strain occurred by this mechanism (132). SNPs previously associated with resistance to ciprofloxacin and rifampin were also identified within the ST2371 sequence (133–135). Data from WGS have provided evidence for the potential acquisition of these traits by chance and suggested a plausible explanation for this particular outbreak.
The use of surveillance to detect and monitor outbreaks caused by exceptionally virulent MRSA strains (represented by a relatively small number of clones and asymptomatic carriage by a large proportion of the population) has become a central element in controlling this organism. In light of this situation, the identification of new, virulent strains can be quite challenging. Strategies to contain the spread of and eradicate novel MRSA strains require rapid identification of both symptomatic and asymptomatic carriers. One obstacle to accomplishing this goal is that MRSA isolates belonging to a single lineage may be impossible to distinguish by using clinically available typing techniques. In contrast, WGS provides the necessary resolution for identifying individual strains and thus defining transmission pathways. Recently, Köser et al., Harris et al., and Nübel et al. employed WGS to characterize strains of methicillin-resistant Staphylococcus aureus, which appear to have emerged from closely related strains undistinguishable by current genotyping assays (76, 131, 136).
Tracing the Origins of These Outbreaks
Gray et al. and Harris et al. performed WGS of S. aureus isolates and suggested that the rate of evolutionary change would provide a sufficient phylogenetic signal to trace the transmission pathway of the bacterium during an epidemiological study (137, 138). At the Cambridge University Hospitals, samples were collected from neonates at the time of admission and twice weekly thereafter, and S. aureus isolation was routinely carried out by culture on selective medium (76, 131, 136). An archive of local surveillance samples was therefore available for in-depth analysis, when an outbreak was suspected. Bacterial DNA extracted from archived and current samples was used to prepare bar-coded sequencing libraries (76, 131, 136). In the groundbreaking study performed by Köser et al., antibiograms were used to identify an unusual resistance profile and, in combination with WGS, enabled tracking of the course of transmission (76).
MRSA carried by asymptomatic individuals is a real challenge to the prevention of virulent strain reintroduction during outbreak control by classical epidemiology. By using WGS, Harris et al. documented 26 cases in which there was carriage of the MRSA outbreak strain. Those researchers also identified transmission among patients in the special baby care unit, their mothers, and members of the community. More significantly, they discovered that a staff member carrying MRSA had acted as a reservoir, thus causing the outbreak to persist even after deep cleaning of the special baby care unit (131). Building on this work, Nübel et al. performed a similar study at a tertiary care hospital in Berlin, Germany, and also identified transmission occurring via a staff member. However, a more frequent cause of new transmissions in their study was infected infants prior to their diagnosis, suggesting that increased surveillance was needed in this hospital to identify affected patients and institute contact isolation protocols (136).
These examples demonstrate the power of WGS technologies, especially when used in conjunction with standard methods, for identifying an outbreak. The establishment of a “causal” link required classic epidemiological surveys to identify possible asymptomatic carriers, thus reducing the number of S. aureus isolates for deep sequencing. The isolation of S. aureus DNA also involved routine, selective, automated culture of S. aureus from a large number of clinical specimens prior to the outbreak and the maintenance of an archive of S. aureus samples. These resources enabled retrospective WGS analysis to trace the transmission pathway in the outbreak. This study went beyond standard epidemiology and involved outreach to obtain additional samples from family doctors, other hospital departments, and screening of staff members. The multiplexing of WGS in these studies allowed large numbers of samples to be analyzed economically. This study also highlights the need for epidemiological information to be gathered to maximize the impact of WGS data.
Asymptomatic members of the community who carry the MRSA strain in question may increase sample noise, which can be reduced by the implementation of an investigatory “filter.” The risks and benefits of identification (by WGS and surveillance) and decolonization of outbreak strains from healthy individuals need to be considered. Moreover, the added resolution provided by WGS in this context raises additional ethical and legal considerations. These questions include “who should be tested” and “what controls should be implemented to prevent further transmission,” once an individual tests positive. For example, testing in the community would normally focus on only symptomatic individuals (139). The studies by Köser et al. and Harris et al. indicate, however, at least in the case of MRSA, that in settings where the disease is endemic, testing of asymptomatic individuals is warranted, and this may well be the case for several other diseases. In a recent Oxford University Hospital study of a Clostridium difficile outbreak, disease transmission from asymptomatic carriers was implicated as the source of infection in the majority of cases (140). Public health authorities will need to determine the extent of testing that is necessary or legally permissible, and this decision may also depend on the characteristics of the MRSA strain involved. A particularly virulent or resistant strain may represent a public health emergency (52) that would justify further intrusion and mandatory treatment. A less virulent strain may not justify such controls. Resolving this question would require evidence, which can be provided by WGS, that a full investigation of a pathogen needs to be conducted as quickly as possible before an outbreak is out of control.
Similarly, these studies raise questions of whether asymptomatic health care workers who carry the organism should be permitted to work with patients or even be in the hospital. Typically, health care workers who are infected with blood-borne pathogens, such as hepatitis (hepatitis B virus [HBV] or hepatitis C virus [HCV]) or HIV are permitted to continue patient care as long as they adopt additional, individual preventive strategies (e.g., double gloving) (141, 142). The skin-to-skin transmission of MRSA may pose an additional risk. Nonetheless, B. A. Diep, who commented on the study by Harris et al., cautioned against placing large numbers of asymptomatic carriers (who could potentially be discovered during surveillance using high-throughput sequencing) under contact precautions unless a clear benefit to such contact precautions can be identified (143). The perceived risk to job security, treatment delay, and expense of antibiotic purging of MRSA, combined with the risk of triggering the development of further resistance to the antibiotic used in healthy individuals, reduce the enthusiasm for prospective molecular epidemiological studies.
Protocols and policies to deal with all of these questions need to be circulated and discussed prior to an MRSA outbreak.
OUTBREAKS OF MYCOBACTERIUM TUBERCULOSIS IN GERMANY (1997 TO 1998), CANADA (2006 TO 2008), AND THE UNITED KINGDOM (1994 TO 2011)
Mycobacterium tuberculosis is the causative agent of human tuberculosis (TB). Currently, in low-incidence areas such as the United States, characterization of a new infection is done only in the setting of a cluster of new cases or for epidemiologic research into the factors involved in tuberculosis spread (144, 145). Since M. tuberculosis is slow growing, a recent transmission event is considered to be one that occurred within the last 2 years, but it may be detected only upon reactivation years after the original exposure. The main tool employed to contain tuberculosis outbreaks (>3 culture-confirmed tuberculosis cases) is an epidemiologic questionnaire that focuses on the characterization of the chain of transmission and subsequent chemotherapy for identified patients (146–148).
Virulence Characteristics of the Causal Organism and Distinguishing between Accidental/Deliberate Release and Natural Transmission
While M. tuberculosis is not typically regarded as an effective agent of bioterrorism (transmission is difficult, and it has a long latency period), it is nonetheless one of a number of global infectious agents (e.g., severe acute respiratory syndrome [SARS], SARS-like virus, and swine flu) that are transmitted by aerosol and have the potential to cause massive disruption to trade and travel (149–153). M. tuberculosis can thus be considered a potential instrument of bioterrorism, even though it may not have as rapid an impact as other bioweapons. The ability of M. tuberculosis to disrupt trade and air transit was highlighted in 2007 when Andrew “Drew” Speaker traveled extensively by commercial airlines while infected with multidrug-resistant TB (MDR-TB) (154, 155). The Georgia lawyer was diagnosed in the United States, but against the advice of health authorities, he subsequently flew to Europe for his wedding and honeymoon. Speaker disregarded requests to contact Italian health officials at his European destination and was served with an isolation order only upon his return to the United States. The recent emergence of totally drug-resistant tuberculosis in South Africa only exacerbates concern for those unknowingly exposed to this bacterium (156–158). This example illustrates the difficulty of categorizing any individual incident as accidental/deliberate exposure or a natural disaster (149–153, 159).
Over a 3-year period, a relatively large number of tuberculosis cases, which involved the same M. tuberculosis mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) genotype, occurred in a medium-sized community in Canada. Walker et al. had previously collected samples from tuberculosis patients and infected household contacts and analyzed them by WGS. From these data, those authors estimated that Mycobacterium accumulates 0.5 single nucleotide polymorphisms per genome per year (144). Mycobacterium appears to be evolutionarily stable, and Casali et al. found little evidence for the occurrence of any backward mutations, suggesting that high confidence could be placed on even a small number of SNPs (in contrast to RNA viruses, described below) (160). Gardy et al. used WGS to discern differences in the bacterium isolated during the Canadian outbreak, which were then combined with results from an epidemiologic investigation (145). Those researchers discovered that this cluster was due to two independent but coincident events which inflated the number of tuberculosis cases occurring in this outbreak. The upsurge in tuberculosis cases in this particular location was due to an increase in illicit drug use (occurring within the population) that resulted in an inadvertent increase in the number of hosts who were highly vulnerable to infection rather than the emergence of a hypervirulent M. tuberculosis strain (161).
Retrospective United Kingdom and German studies examined the utility of WGS for the identification of factors resulting in increased tuberculosis transmission (144, 162). The increased genetic resolution allowed the determination of the direction of transmission in outbreaks, reconstruction of the infection timeline, and identification of superspreaders (the treatment of whom would be essential to prevent future transmission) (144, 162).
Tracing the Origins of Tuberculosis Outbreaks
The emergence of drug resistance in M. tuberculosis and carriage by asymptomatic individuals have created a worldwide public health emergency. It will be necessary for public health officials to make decisions about whether to allow entry to people exposed to drug-resistant TB. Similarly, should an outbreak occur, public health officials will need to make decisions about possible vaccination programs and isolation of affected individuals. Infectious tuberculosis is already a quarantinable communicable disease, but as the Speaker case demonstrated in 2007, the Secretary of Health and Human Services cannot exercise legal authority without causing panic and disruption of trade and travel (163). It is essential that there be evidence-based tools for use by the highest levels of authority. Measures as extreme as the detention of affected individuals should be employed only when there is considerable certainty that the correct subjects have been identified. The use of WGS in this setting can provide critical information.
The determination of M. tuberculosis drug sensitivity can be extremely challenging, but WGS can also provide information on mutations known to be associated with drug resistance (160, 164). Both PCR/hybridization and multilocus sequencing schema have been used to identify mutations in genes known to be involved in first- and second-line tuberculosis drug resistance (165). However, as WGS becomes cheaper and faster and as the analysis pipeline becomes more turnkey, it could supplant these methods, as it can provide information not only on drug resistance but also on strain evolution and the chain of transmission during tuberculosis outbreaks. This in turn raises new questions of potential legal consequences when an individual does not comply with treatment requirements. Heretofore, causation in deliberate or negligent transmission has been difficult to prove, but the greater sensitivity of WGS may help overcome this problem.
Althomsons et al. were able to identify small tuberculosis clusters at high risk of metastasizing into larger outbreaks in low-incidence areas (166). In these cases, where increased contact surveillance is warranted but has limitations, WGS may provide useful assistance to epidemiologists to recognize and link cases in ambiguous outbreaks and to determine whether there are undiagnosed patients in need of treatment (144, 160, 164, 167, 168). In an era of increasing global travel, genome sequence information may also provide a valuable clue to identify the source of an outbreak of extremely drug-resistant TB (XDR-TB). MLST DNA sequencing projects suggest that the geographical variation in human M. tuberculosis is higher than was expected for this slow-growing and evolving bacterium (169).
OUTBREAKS OF FOOT-AND-MOUTH DISEASE VIRUS IN THE UNITED KINGDOM (2001 AND 2007) AND BULGARIA (2011)
WGS was used to analyze outbreaks of foot-and-mouth disease (caused by foot-and-mouth disease virus [FMDV]) in the United Kingdom (2001 and 2007) and later in Bulgaria (2011). This technology allows the examination of the transmission pathways that are key factors in the control of an easily transmitted RNA virus in the context of both an accidental release and its transmission from a population in which it is well established. Although eradicated in most western countries, FMDV is endemic to many developing nations.
FMDV, a small-genome RNA virus, is an excellent candidate for WGS analysis by conventional Sanger sequencing; although the genome is small, the high error rate inherent in its mode of replication ensures that sufficient mutations occur to differentiate viruses from closely related sources. This mutation rate has been attributed to the error-prone RNA-dependent RNA polymerase used during virus replication (170, 171). This high rate of change, however, results in a paraphyletic constellation of viruses in infected individuals similar to that occurring in human individuals infected with the RNA virus HIV, which also has a high error rate during virus replication (172). The recent outbreaks of infections with human RNA viruses (such as Ebola virus, SARS coronavirus [SARS-CoV], Middle East respiratory syndrome [MERS], and HIV/AIDS) document the importance of computer modeling for this type of WGS data in epidemics (111, 151, 173–175). The experience of using WGS for FMDV in the 2001, 2007, and 2011 outbreaks illustrates the advantage of gathering additional background information during sample collection. This point was especially important when dealing with rapidly mutating FMDV, as it was helpful to obtain optimum results when tracking and tracing the source and trajectory of FMDV epidemics to place the WGS data in context when using bioinformatic tools.
FMDV spreads quickly, and many farms contain tens of thousands (if not far more) animals; thus, information on the movement of most animals during the United Kingdom and Bulgarian epidemics was readily available (176). In epidemiological studies, phylogenetic analysis of WGS is the technique most frequently used to scrutinize these data (38–40). Due to the high mutation rate of FMDV, the identity between sequentially isolated viruses can be as low as 80%; some mutations result in a loss of infectivity, thereby causing a number of genomic “bottlenecks” at the time of transmission. As mentioned above, an extensive database of epidemiological information collected during the 2001 FMDV outbreak was available for computer modeling of RNA virus evolution. In cases where both the time of initial infection and the number of cases or infected premises are unknown, this could be used to decrease the number of possible scenarios used to identify the most probable sequence of events (177). The variation occurring within the host animal which results in tissue-specific and time-dependent differences in virus sampling could otherwise have greatly decreased the ability of WGS to be as informative in RNA virus outbreaks (177). In FMDV outbreaks, it is especially important that contextual sample information and spatial, temporal, and genetic data are considered simultaneously along with information on the latency of the disease in order to obtain the most accurate information to plot the transmission route of infection and identify the most effective intervention strategies (178).
Virulence Characteristics of the Causal Organism and Distinguishing between Accidental/Deliberate Release and Natural Transmission
During the 2001 outbreak in the United Kingdom, samples were collected from the majority of the 2,030 infected premises and stored at the Institute of National Health Pirbright Laboratory. Additional strains from enzootic areas of the globe have also been sequenced and are available for comparison (179–182). Cottam et al. analyzed the FMDV nucleotide sequences obtained from samples gathered at 21 infection sites over the 7 months of the 2001 United Kingdom outbreak (183). They determined the “consensus sequence” of the virus population occurring in both infected animals and premises and determined that ∼2 × 10−5 substitutions occurred at each site every day. This high mutation rate resulted in an average of 1.5 nucleotide changes occurring at each infected farm (183).
In the 2007 epidemic, again in the United Kingdom, WGS suggested that within 24 h of receipt of the first samples, this outbreak was due to an accidental release (184). The FMDV O1/BFS 1860/UK/67 strain was in culture at both the Institute of Animal Health Laboratories (sample IAH2) and Merial Animal Health Limited (sample MAH) at Pirbright. The isolates from the infected premises adjacent to Pirbright were 99.84% identical to the O1/BFS 1860/UK/67 reference strain, indicating (for an RNA virus) an extremely close evolutionary link. In addition, growth of FMDV in culture had left a unique molecular signature within the genome of the epidemic strain. When grown in vitro, the FMDV strain acquires positively charged amino acid residues on the virus coat to better attach to heparin sulfate receptors on the cell. These are, however, a maladaptation in vivo and result in faster clearing from circulation (185). The early outbreak isolates contained both culture-specific mutations viral protein 3 amino acid 56 (VP356; histidine to arginine) and VP360 (aspartic acid to glycine), which favor heparin sulfate receptor binding. This supported the premise that the 2007 United Kingdom epidemic was caused by an accidental release of a cultured virus from Merial Animal Health Limited or the Institute of Animal Health Laboratories (186).
Tracing the Origins of FMDV Outbreaks
In January 2011, an FMDV-infected wild boar was discovered near the border between Turkey and Bulgaria. Sequencing of the FMDV VP1 gene identified it as belonging to an ANT-10 lineage, which was discovered in Iran and found to be spreading through the Middle East. Subsequently, ∼2,230 animals were slaughtered to eradicate the disease from Bulgaria. Information on the locations and dates of sample collection was gathered and analyzed in conjunction with FMDV sequence data to trace the source and course of this epidemic (187).
During the 2007 outbreak, temporal information and the rapidity of the sequencing effort were used to identify the existence of a “missing link” in the infection chain and redirect efforts during the eradication program (184). As in the 2007 outbreak in the United Kingdom, WGS data were used to determine the number of changes between farms and provided the resolution necessary to identify greater-than-expected evolutionary “jumps” (184, 187). The Bulgarian epidemiologists were able to use the information discovered during the contemporaneous whole-genome analysis to identify infected premises that were previously unrecognized. Those authors, however, concluded that sequence data alone were not sufficient in their models to determine the infection origin, but when combined with spatiotemporal data, they were extremely useful in determining the dynamics of their epidemic (187).
Determining unambiguously the country from which an epidemic strain arose is challenging and, as mentioned above, requires an extensive WGS database. Comparison, for instance, of the 2001 FMDV isolates with global strains suggested that a strain then circulating in South Africa was the most probable source of the United Kingdom outbreak, based on the similarity of the consensus sequenced genomes (179). The 2011 Bulgaria outbreak viruses were all closely related to viruses collected in Turkey during 2010, but phylogenetic analysis based on the identified SNPs suggested that it was more distantly related to those in circulation in 2011. It is possible that the wild boar population in the transborder regions between these two countries could act as a FMDV reservoir; however, it is equally possible that the initial identification of FMDV in a wild animal represented an incorrect disposal of domestic meat during the Kurban Bayram Festival or even interactions of wild and domestic animals occurring close to the location where the first identified case was discovered. The authors of that study therefore conclude that the immediate source of the 2011 outbreak had not been sampled and could not be determined (187). The identity of the FMDV reservoir is therefore in doubt, and extensive sampling of cloven-hoofed animals at the border between Turkey and Bulgaria would have been needed to truly determine the source of the 2011 outbreak.
IMPLICATIONS
Based on the cases presented here, it is clear that in order to identify virulence factors within whole-genome sequences, an extensive knowledge of the genetic basis of virulence needs to be available. Information on the natural history of the disease and its latency, as well as the rate of mutation accumulation in the causal microbe, is essential to place data from WGS into context.
When using information from WGS to build models to predict the course of an epidemic, information such as the location and timing of sample collection is critical. Based on these models, breaks in containment can be identified, but obviously, this information is useful to first responders only if provided in real time (6). For WGS information to be useful in a fast-moving epidemic, both sequencing and analysis must be completed and made available in a timely manner.
In situations where an accidental or deliberate release of pathogens is suspected, sequence data need to be examined for genetic signatures left by “enabling” manufacturing processes (such as cell culture in the case of FMDV or heat attenuation in anthrax strains) (10, 185). To identify and prevent “natural” outbreaks, the evidence left by the mechanisms that led to the lateral transfer of virulence genes needs to be identified and compiled.
Limitations of WGS in Infectious Disease Outbreaks
Preanalytical limitation.
One of the limitations of the current WGS approaches is the need for simplification of biological samples by selective culture prior to sequencing. This limits the species that are able to be sequenced, and if the initial sample contains a mixture of genotypes, it will likely result in the sequencing of only the strains that grow best in culture (40, 76, 106, 107, 109, 122, 131, 144, 145, 178, 183, 184, 187). Recent advances in metagenomic sequencing have been made, as discussed above, but currently, these methods require a large investment of resources and are not without their own limitations (126). Currently, a major challenge to the use of metagenomic samples in pathogen identification and characterization is the fact that parasites are typically present at only low copy numbers in the clinical and environmental samples available for analysis. In epidemiologic studies, data from populations rather than from individuals are required. Sequencing at the depth required to obtain pathogen WGS from metagenomic samples is often difficult and incompatible with efficient multiplex sample sequencing.
New strategies for high-throughput enrichment to be used in combination with metagenomic methods could lead, even in large studies, to rapid culture-free NGS. Approaches that limit the amount of eukaryotic (host) CpG-methylated DNA combined with metagenomic sequencing enhance the detection of infecting bacteria (188–190). For example in epidemiological studies involving M. tuberculosis, in which obtaining material from slowly replicating organisms is a major limitation, enrichment of bacterial DNA in sputum (1 ml of a smear-positive sputum sample typically contains 104 CFU of M. tuberculosis) combined with sequencing may be an efficient approach to provide WGS data in a timely manner.
Alternative approaches might be useful for other more complex sample sources. Fluorescence in situ hybridization-flow cytometry-cell sorting is a technique with the potential to preferentially isolate any target bacterium from a soil sample for NGS (191). This approach could permit WGS of select species (such as MRSA) in environmental samples (192). Selective whole-genome amplification methods like those already practical for the small genomes of viruses such as FMDV are also undergoing rapid development for larger genomes and will soon also offer the option of culture-free sequencing of targeted genomes (193, 221). This is, however, a rapidly moving field, and while there is no guarantee that the current growth rate will continue indefinitely, the cost of sequencing has fallen precipitously in recent years (194). When newer sequencing technologies (discussed above) enter the commercial marketplace, they will likely facilitate sample sequencing at great depth (195). It is also feasible, therefore, that new developments in technology and bioinformatics will allow multiplex sample processing without enrichment, for the large numbers of samples needed for epidemiological studies (126).
Analytical limitations.
Another limitation of WGS for the identification and tracking of the source of an infectious disease outbreak is that of up-to-date, large-scale informatics databases. This problem is illustrated by the dearth of contemporary (circulating) Vibrio cholerae strains that have been or are available for WGS; such a collection would have provided a useful context for the strains sequenced during the investigation into the source of the 2010 Haitian cholera epidemic (196). Although facilitated by the availability of rapid and inexpensive WGS technology, analysis of this outbreak was constrained by the lack of a comprehensive database containing sequences of contemporarily circulating strains from different locations (197, 198). Nevertheless, even in this example, comparison of the sequences obtained from the Haitian epidemic with data on those circulating in Latin America, Africa, Bangladesh, and Nepal suggested an Asian origin (199–201). Similarly, during the Amerithrax investigation, a reference library of samples was gathered only after the fact and with the assistance of potential defendants. Furthermore, sampling was not uniform and was not carried out with consideration of repeated sequencing and verification of the initial results (9). These problems could have been avoided if a preexisting, verified reference database/library of WGS strains was already available.
Development of reference databases.
A plan for microbial forensics analysis generally includes the following elements: the acquisition of DNA sequence reads and the use of software to assign reads (or assemblies) to taxa in a reference database (Fig. 1). Any software used to assign a read or assembled contig to a known organism is ultimately dependent on the availability of a high-quality reference genome database. All cases are variations on a theme: a software algorithm is used to match the WGS of an isolate sequence of known origin in a comprehensive database. In the case of a single genome, amplified segments or whole assemblies are aligned against databases containing sequences of known origin in order to make a proper identification (32, 202). In environmental metagenomic studies, reads or contigs are searched against databases in order to classify reads taxonomically for both species identification and abundance estimation (when a sample has a plurality of genomes). Individual algorithms employ different techniques for reducing the computational burden of this process. A tool used by the Human Microbiome Consortium starts with the IMG database of microbial genomes (33) and creates a customized reduced-representation database containing only “perfect” marker genes that are both unique and universal within a clade (203). Other read-based tools employ different methods, such as manually curing lists of lineage-specific versions of near-universal genes (204) or constructing Burrows-Wheeler indices upon which to align sequences using fast, short-read alignment algorithms (205).
The greatest limitation of WGS analysis is often not the software or the sequencing technology but the completeness and correctness of the reference database being used. None of the methods discussed here will perform well when used to match a sequence read or assembly to a database that does not contain a labeled sequence corresponding to the source organism for the query sequence. From a forensic attribution perspective, the utility of interpretation software is dependent on a continuously updated database that not only would encompass pathogen genomes to detect the presence of a species (or lack thereof) but also would contain a catalog of point mutations or genes that account for drug resistance, virulence, or marks of genetic engineering (206). Furthermore, detection of genetic engineering is challenging not only due to the large number of cloning vectors that may closely resemble the sequence of the organism from which they are derived (e.g., E. coli) but also because of the availability of numerous scarless genetic engineering procedures (207–210). Each day, new organisms are being sequenced, new resistance mechanisms are being identified/recognized, and new association studies are finding genetic determinants that affect virulence. Maintaining constantly up-to-date databases comprising all this information is difficult but is an essential element in NGS.
It is important to recognize that the relevant reference population will not always be available in the standard databases (108). In general, algorithms that map sequence reads to reference databases do so probabilistically, accounting for both the degree of similarity and the size of the reference database. For example, the E value in a BLAST search describes the number of random hits one can “expect” by chance, when searching a database of a particular size. Thus, the probability of an apparent match depends greatly on the size of the reference database, which may include only a portion of the total genetic data available. The size of this portion varies globally, regionally, and locally. Evaluation of processes for the selection of an appropriate “background” or reference population is outside the scope of this review, but it is important that the chosen software method be limited to searching a reduced or customized database.
Rapid and Validated Discrimination of Accidental and Deliberate Infectious Disease Outbreaks by Using WGS
The development of a bioweapon does not require genetic engineering, as bacteria have already acquired an arsenal of virulence factors. Today, nearly all countries have the technological potential to produce large amounts of pathogenic microorganisms safely. Mass production of biological agents may, however, result in characteristic genetic changes, which can later be recognized within the sequenced genome. Common pathogens can be genetically manipulated to cause higher morbidity or mortality rates, survive longer in the environment, be delivered with greater ease, and be resistant to common antibiotics. Deliberate action or accidental release may not be initially considered or easily recognized in outbreaks caused by these organisms. WGS can be used to identify regions of topological discordance, hallmarks of either lateral transfer of genes or scarless genetic engineering. Identification of any virulence and antibiotic resistance genes that have potentially been transferred horizontally between prokaryotic species is of major importance in the characterization of an outbreak strain and identification of a new pathogen arising from a previously commensal species (211). WGS has been used very effectively to characterize the interspecies or interstrain exchange of virulence factors (e.g., acquisition of the cytolytic toxin Panton-Valentine leucocidin by S. aureus sequence type 22 and Stx by Escherichia coli serotype O104:H4, as discussed previously [40, 76, 138]). In these outbreaks, a key consideration was the speed with which WGS can be used to identify virulence factors and provide results. As discussed above, the identification of these events is greatly facilitated by de novo microbial genome assembly rather than the simpler reference-guided genome assembly. The recent advances in third-generation sequencing technology discussed above, however, will lead to future improvements in the rapid assembly of finished de novo microbial genome assemblies (18).
After sequence reads have been assembled, standard sequence similarity tools can be used to compare them to databases of both finished and draft microbial genomes (32), which are available from sources such as the NCBI and IMG (33). Comparison of an outbreak strain with its nearest evolutionary neighbor can identify where a breakdown in the synteny between the two strains occurs. Boundaries of the novel DNA can then be closely examined for evidence of unnatural genetic changes (suggesting that the bacterium has been genetically manipulated in order to create a bioweapon) or for evidence that a naturally occurring mobile and/or invasive DNA element had horizontally transferred virulence genes (131). In the examples described in this review (acquisition of the cytolytic toxin Panton-Valentine leucocidin by S. aureus sequence type 22 and Stx by Escherichia coli serotype O104:H4), the WGS evidence suggested that the toxins were naturally acquired by the bacteria rather than generated deliberately or even accidentally by human activity (40, 76, 138).
A deliberate or accidental release of an organism may also be suspected if the genetic signature left by mass production, such as in vitro growth in culture, can be detected within the whole genome sequence. Recognition of this phenomenon requires that analysts understand the potential consequences of production technology on the pathogen and can recognize the resulting molecular signature within WGS data. As discussed in our case studies, such information was available during the WGS analysis of the outbreak FMDV isolate from the United Kingdom in 2007 (212). It allowed the identification of the mutations that are selected only during the in vitro culture of the virus in the outbreak virions and provided supporting evidence that this outbreak was due to an accidental or deliberate release (184, 212, 213).
If WGS reveals that genetically identical or highly related material is the source of multiple sequential or coincident cases/epidemics, the conclusion is that there is a reservoir and that deliberate or accidental reintroduction of the pathogen is occurring. Tracing the causal organism along the epidemic pathway not only allows characterization of the genetic signature of the source material but also indicates whether the epidemic was due to the synchronous introduction of the pathogen at multiple locations, which may be an indication of deliberate activity. Rapid sequencing of 99 Ebola virus (EBOV) isolates involved in the 2014 outbreak showed that they arose from a single-crossover event from the natural host, which then spread through the human population (111). This then permitted the focus to remain on containing human-to-human EBOV transmission in the current outbreak rather than on the identification of possible zoonotic sources of new transmissions (214). During the Canadian epidemiologic investigation of a Mycobacterium tuberculosis outbreak, WGS was used to both identify the index cases and determine that this epidemic was due to two coincident outbreaks of genetically dissimilar strains (161). WGS has also been successfully used in evaluations of nosocomial cases of MRSA to identify and eliminate the accidental source of repeated MRSA outbreaks (76, 131, 136).
During the United Kingdom outbreaks in 2001 and 2007 and the Bulgarian outbreak in 2011, the library of FMDV sequences allowed WGS to be used to identify the probable country of origin of the infection (179–183). However, while WGS was able to link the French and German 2010 EHEC O104:H4 outbreaks and to place this outbreak within its historical context (100, 107), an inadequate number of isolates was sequenced to test the epidemiological hypothesis that implicated fenugreek seed from Egypt as the outbreak source (114).
In conclusion, WGS can be helpful in identifying a bioterrorism event if there is evidence of genetic signatures left by an enabling manufacturing process or if the hallmarks of genetic manipulation are left within the genomic sequence. It is also suspicious if multiple epidemics seem to arise from genetically identical material or if no natural explanation for the acquisition of virulence factors can be inferred from the WGS data.
Legal Framework/Validation
New technologies require new legal responses. Although the improved resolution offered by WGS and related technologies in microbial forensics may provide greater clarity in many contexts, this “improvement” may come into conflict with existing legal rules and protocols and create new gaps. Assuming that reference databases are developed, ongoing sampling, both patient and environmental, needs to be done in a consistent fashion to ensure the continued utility of these reference databases. Since it is impossible to predict when clinical samples might be needed, the overall best practices for WGS use may be that samples should be archived in a manner compatible with future NGS (215–217). This may require the coordination of clinical diagnostics and compatible systems across states. There are no laws or regulations, however, that could require such action. New or updated protocols would be useful to allow the coordination of law enforcement, public health, and national security officials; however, at a minimum, attempts should be made to establish voluntary protocols at reference laboratories, such as the Global Microbial Identifier (GMI) Initiative (218). However, there are questions regarding the source of funding for large databases of pathogen DNA samples and whether these samples might in themselves constitute a risk to patient privacy, which at a minimum would require both oversight and an adjustment of state and federal privacy laws. To be most effective, these samples should consist of not only those collected at tertiary reference laboratories during outbreaks but also those collected during routine surveillance before an outbreak occurs (117). The Human Microbiome Project, however, has just begun the formidable task of identifying the microbiota of humans; continual sampling of the microorganisms in circulation and archiving these samples suitably for future WGS studies would be a massive undertaking (219).
Ironically, WGS may cause additional forensic headaches because it lends additional clarity (172, 220). When such clarity was not possible, the legal system tolerated decisions made on conjectures and gaps in evidence. It is likely that this will no longer be the case. On a related note, many laboratories are currently using laboratory assays that were developed for research and have never been validated for forensic applications (117). These tests may provide answers but not ones that would be admissible in a court of law. As a result, it is important that protocols be expanded or new ones be developed to facilitate cooperative efforts between public health and law enforcement agencies for the purpose of addressing these unprecedented situations.
There will also be new privacy concerns and questions about individual liberties. For example, during an outbreak, it is now possible to identify people who are the unintentional source or propagators of the outbreak. What should the legal response be to inadvertent endangerment by bodily substance in this context? Can authorities legally detain an asymptomatic index patient in an outbreak and/or limit their activities if they are still infectious? Can the criminal justice system use evidence based on microbial forensic WGS technologies to elucidate an individual's incriminating travel and movements? What are the limits to patient expectations of privacy and confidentially in the context of microbial WGS?
CONCLUSION
Guidelines and standard operating procedures are needed in order to gather and integrate WGS data with other forms of evidence (epidemiological, nongenetic, and classical forensics) and to integrate a combined and properly weighted WGS analysis with other information in order to ensure the most appropriate public health, legal, or policy decision. In principle, a microorganism's genome sequence contains all this information, and rapid, inexpensive NGS has the potential to replace many of the above-mentioned labor-intensive procedures currently used in microbial forensics attributions.
ACKNOWLEDGMENTS
We thank E. Houpt and A. Mathers for expert advice and informative discussions on Mycobacterium tuberculosis and methicillin-resistant Staphylococcus aureus and C. Shaw for expert advice and informative discussions in regard to the United Kingdom outbreaks of foot-and-mouth disease virus. We also acknowledge the thoughtful suggestions on this report made by A. Mackey and the careful reading of the manuscript by G. Ramakrishnan. Finally, we acknowledge the administrative and organizational support provided by Pam Schaefer in the Division of Infectious Diseases and International Health, Department of Medicine.
This work was supported by a grant from the Department of Defense to E.L.H. and by grant AI103536-01 to C.A.G.
Biographies

Carol A. Gilchrist, Ph.D., received her degree at the University of Western Ontario and is currently an Assistant Professor of Research in the Division of Infectious Diseases, University of Virginia. Dr. Gilchrist's research has focused on using molecular methods to understand the biology and pathogenic phenotype of enteric parasites. She is coauthor of the paper describing the sequenced Entamoeba histolytica genome in 2005 and devised the current E. histolytica multilocus sequence typing system. She is part of a collaboration currently focused on obtaining additional WGS of E. histolytica and Cryptosporidium hominis protozoan pathogens to understand the genetic variability inherent to these human parasites.

Stephen D. Turner received his B.S. in biology at James Madison University, M.S. in applied statistics at Vanderbilt University, and Ph.D. in human genetics at Vanderbilt University, where he developed and applied new methods for detecting gene-gene interactions in large-scale human genetics studies. He did a postdoctoral fellowship in genetic epidemiology at the University of Hawaii Cancer Center before joining the University of Virginia faculty in the Department of Public Health Sciences. Dr. Turner established UVA's bioinformatics core facility in 2011 to provide a centralized resource for expert bioinformatics consulting and data analysis as a service.

Margaret F. Riley is Professor of Law at the University of Virginia School of Law, where she teaches in the areas of Bioethics, Food and Drug Law, Health Law, Animal Law, and Public Health Law. She also has secondary appointments in the Department of Public Health Sciences at the University of Virginia School of Medicine and in the Batten School of Leadership and Public Policy. She is a graduate of Duke University and Columbia University Law School and was a litigation associate at Rogers & Wells in New York, NY, and Pepper Hamilton & Scheetz in Philadelphia, PA, prior to joining the faculty at the University of Virginia. Her areas of interest include health institutions and reform, biomedical ethics and research, food and drug law, genomics, reproductive technologies, stem cell research, biotechnology, health disparities, and chronic disease.

William A. Petri, Jr., M.D., Ph.D., is Chief of the Division of Infectious Diseases and International Health at the University of Virginia. He has an active clinical practice of internal medicine and infectious diseases and conducts research in microbial pathogenesis and immunity at the University and in Bangladesh.

Erik L. Hewlett received his M.D. from the Johns Hopkins School of Medicine and did his internal medicine training at Cornell-NY Hospital. He then completed postdoctoral training at the NIH in the National Institute of Arthritis, Metabolism, and Digestive Diseases, during which time he began his current research on Bordetella pertussis and adenylate cyclase toxin. He joined the faculty of the University of Virginia School of Medicine in Geographic Medicine and Clinical Pharmacology in 1980 and is now Professor of Medicine in the Division of Infectious Diseases and International Health and Professor of Microbiology, Immunology, and Cancer Biology. The focus of his research program is the molecular pathogenesis of infectious diseases, including pertussis, anthrax, and C. difficile-associated diarrhea. He is director of the UVA Traveler's Clinic, teaches medical and graduate students, and is chair of the Board of Directors of the UVA Licensing and Ventures Group, which handles UVA intellectual property.
REFERENCES
- 1.Oyola SO, Otto TD, Gu Y, Maslen G, Manske M, Campino S, Turner DJ, Macinnis B, Kwiatkowski DP, Swerdlow HP, Quail MA. 2012. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics 13:1. doi: 10.1186/1471-2164-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. 2009. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods 6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knierim E, Lucke B, Schwarz JM, Schuelke M, Seelow D. 2011. Systematic comparison of three methods for fragmentation of long-range PCR products for next generation sequencing. PLoS One 6:e28240. doi: 10.1371/journal.pone.0028240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lienau EK, Strain E, Wang C, Zheng J, Ottesen AR, Keys CE, Hammack TS, Musser SM, Brown EW, Allard MW, Cao G, Meng J, Stones R. 2011. Identification of a salmonellosis outbreak by means of molecular sequencing. N Engl J Med 364:981–982. doi: 10.1056/NEJMc1100443. [DOI] [PubMed] [Google Scholar]
- 7.Long SW, Williams D, Valson C, Cantu CC, Cernoch P, Musser JM, Olsen RJ. 2013. A genomic day in the life of a clinical microbiology laboratory. J Clin Microbiol 51:1272–1277. doi: 10.1128/JCM.03237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellerin C. 5 June 2013. Global force's needs shape DOD biosurveillance. U.S. Department of Defense, Washington, DC: http://www.defense.gov/news/newsarticle.aspx?id=120224. [Google Scholar]
- 9.Obama B. 31 July 2012. National strategy for biosurveillance. https://www.whitehouse.gov/sites/default/files/National_Strategy_for_Biosurveillance_July_2012.pdf.
- 10.Rasko DA, Worsham PL, Abshire TG, Stanley ST, Bannan JD, Wilson MR, Langham RJ, Decker RS, Jiang L, Read TD, Phillippy AM, Salzberg SL, Pop M, Van Ert MN, Kenefic LJ, Keim PS, Fraser-Liggett CM, Ravel J. 2011. Bacillus anthracis comparative genome analysis in support of the Amerithrax investigation. Proc Natl Acad Sci U S A 108:5027–5032. doi: 10.1073/pnas.1016657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Code of Federal Regulations. 2013. Title 42. Public health. Chapter I. Public Health Service, Department of Health and Human Services. Subchapter F. Quarantine, inspection, licensing. Part 71. Foreign quarantine. Subpart F. Importations. §71.54. Import regulations for infectious biological agents, infectious substances, and vectors. [Google Scholar]
- 12.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 13.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 14.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 15.Frampton M, Houlston R. 2012. Generation of artificial FASTQ files to evaluate the performance of next-generation sequencing pipelines. PLoS One 7:e49110. doi: 10.1371/journal.pone.0049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Su X, Wang A, Xu J, Ning K. 2013. QC-Chain: fast and holistic quality control method for next-generation sequencing data. PLoS One 8:e60234. doi: 10.1371/journal.pone.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts RJ, Carneiro MO, Schatz MC. 2013. The advantages of SMRT sequencing. Genome Biol 14:405. doi: 10.1186/gb-2013-14-6-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 19.Lou DI, Hussmann JA, McBee RM, Acevedo A, Andino R, Press WH, Sawyer SL. 2013. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A 110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennisi E. 2014. Genomics DNA sequencers still waiting for the nanopore revolution. Science 343:829–830. doi: 10.1126/science.343.6173.829. [DOI] [PubMed] [Google Scholar]
- 21.Koren S, Phillippy AM. 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23C:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Narzisi G, Mishra B. 2011. Comparing de novo genome assembly: the long and short of it. PLoS One 6:e19175. doi: 10.1371/journal.pone.0019175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Chen J, Yang Y, Tang Y, Shang J, Shen B. 2011. A practical comparison of de novo genome assembly software tools for next-generation sequencing technologies. PLoS One 6:e17915. doi: 10.1371/journal.pone.0017915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Earl D, Bradnam K, St John J, Darling A, Lin D, Fass J, Yu HOK, Buffalo V, Zerbino DR, Diekhans M, Nguyen N, Ariyaratne PN, Sung W-K, Ning Z, Haimel M, Simpson JT, Fonseca NA, Birol İ, Docking TR, Ho IY, Rokhsar DS, Chikhi R, Lavenier D, Chapuis G, Naquin D, Maillet N, Schatz MC, Kelley DR, Phillippy AM, Koren S, Yang S-P, Wu W, Chou W-C, Srivastava A, Shaw TI, Ruby JG, Skewes-Cox P, Betegon M, Dimon MT, Solovyev V, Seledtsov I, Kosarev P, Vorobyev D, Ramirez-Gonzalez R, Leggett R, MacLean D, Xia F, Luo R, Li Z, Xie Y, et al. . 2011. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res 21:2224–2241. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A 108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magoc T, Pabinger S, Canzar S, Liu X, Su Q, Puiu D, Tallon LJ, Salzberg SL. 2013. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics 29:1718–1725. doi: 10.1093/bioinformatics/btt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, Chitsaz H, Chou W-C, Corbeil J, Del Fabbro C, Docking TR, Durbin R, Earl D, Emrich S, Fedotov P, Fonseca NA, Ganapathy G, Gibbs RA, Gnerre S, Godzaridis E, Goldstein S, Haimel M, Hall G, Haussler D, Hiatt JB, Ho IY, Howard J, Hunt M, Jackman SD, Jaffe DB, Jarvis ED, Jiang H, Kazakov S, Kersey PJ, Kitzman JO, Knight JR, Koren S, Lam T-W, Lavenier D, Laviolette F, Li Y, Li Z, Liu B, Liu Y, Luo R, Maccallum I, et al. . 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker S, Joecker A, Church G, Snyder M, West J, Salzberg S, Worthey E, Smith T, Wang J, Reid JG. 2012. Genome interpretation and assembly—recent progress and next steps. Nat Biotechnol 30:1081–1083. doi: 10.1038/nbt.2425. [DOI] [PubMed] [Google Scholar]
- 32.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter 10:Unit 10.3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 33.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- 35.Chan CX, Ragan MA. 2013. Next-generation phylogenomics. Biol Direct 8:3. doi: 10.1186/1745-6150-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin AP. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol 68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei M, Sudhir K. 2000. Molecular evolution and phylogenetics, 1st ed Oxford University Press, New York, NY. [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, Godfrey P, Haas BJ, Murphy CI, Russ C, Sykes S, Walker BJ, Wortman JR, Young S, Zeng Q, Abouelleil A, Bochicchio J, Chauvin S, Desmet T, Gujja S, McCowan C, Montmayeur A, Steelman S, Frimodt-Møller J, Petersen AM, Struve C, Krogfelt KA, Bingen E, Weill F-X, Lander ES, Nusbaum C, Birren BW, Hung DT, Hanage WP. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci U S A 109:3065–3070. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croucher NJ, Didelot X. 2015. The application of genomics to tracing bacterial pathogen transmission. Curr Opin Microbiol 23C:62–67. doi: 10.1016/j.mib.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 42.United Nations. 1972. Biological and Toxin Weapons Convention. United Nations, Washington, DC. [Google Scholar]
- 43.Dando M, Pearson G, Kriz B (ed). 2001. Scientific and technical means of distinguishing between natural and other outbreaks of disease. Proceeding of the NATO Advanced Research Workshop, vol 35 Springer, New York, NY. [Google Scholar]
- 44.CDC. 2001. Update: investigation of anthrax associated with intentional exposure and interim public health guidelines, October 2001. MMWR Morb Mortal Wkly Rep 50:889–893. [PubMed] [Google Scholar]
- 45.Read TD, Salzberg SL, Pop M, Shumway M, Umayam L, Jiang L, Holtzapple E, Busch JD, Smith KL, Schupp JM, Solomon D, Keim P, Fraser CM. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028–2033. doi: 10.1126/science.1071837. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmaster AR, Fitzgerald CC, Ribot E, Mayer LW, Popovic T. 2002. Molecular subtyping of Bacillus anthracis and the 2001 bioterrorism-associated anthrax outbreak, United States. Emerg Infect Dis 8:1111–1116. doi: 10.3201/eid0810.020394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolonay JF, Beanan MJ, Dodson RJ, Brinkac LM, Gwinn M, DeBoy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pop M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang L, Hance IR, Weidman JF, Berry KJ, Plaut RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Koehler TM, Hanna PC, Kolstø A-B, Fraser CM. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 48.Committee on Review of the Scientific Approaches Used During the FBI's Investigation of the 2001 Bacillus Anthracis Mailings, National Research Council. 2011. Review of the scientific approaches used during the FBI's investigation of the 2001 anthrax letters. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 49.Ervin A, Hultgren A, Rhyne E, Ward K. 2010. Sensing dispersal of chemical and biological agents in urban environments, p 423–434. In Voeller JG. (ed), Wiley handbook of science and technology for homeland security. Wiley, Hoboken, NJ. [Google Scholar]
- 50.CDC. 2015. CDC bioterrorism agents/diseases by category. CDC, Atlanta, GA. [Google Scholar]
- 51.Lipsitch M, Finelli L, Heffernan RT, Leung GM, Redd SC. 2011. Improving the evidence base for decision making during a pandemic: the example of 2009 influenza A/H1N1. Biosecur Bioterror 9:89–115. doi: 10.1089/bsp.2011.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Public Health Law Program, Centers for Disease Control and Prevention. 2009. Selected federal legal authorities pertinent to public health emergencies. CDC, Atlanta, GA. [Google Scholar]
- 53.World Health Organization. 2008. International health regulations (2005), 2nd ed World Health Organization, Geneva, Switzerland. [Google Scholar]
- 54.OIE. 2009. International trade: rights and obligations of OIE members. OIE, Paris, France. [Google Scholar]
- 55.Sutton V. 2005. Dual purpose bioterrorism investigations in law enforcement and public health protection: how to make them work consistent with the rule of law. Houst J Health Law Policy 6:151–170. [Google Scholar]
- 56.European Parliament, Council of the European Union. 1995. Directive 95/46/EC of the European Parliament and of the Council of 24 October 1995 on the protection of individuals with regard to the processing of personal data and on the free movement of such data, articles 3 and 13. Official Journal L 281. [Google Scholar]
- 57.US Congress. 1996. Health Insurance Portability and Accountability Act. 42 US Code 1320. [Google Scholar]
- 58.Code of Federal Regulations. Title 45. Public welfare. Subtitle A. Department of Health and Human Services. Subchapter C. Administrative data standards and related requirements. Part 164. Security and privacy. Subpart E. Privacy of individually identifiable health information. §164.512. Uses and disclosures for which an authorization or opportunity to agree or object is not required. [Google Scholar]
- 59.US Congress. 1947. National Security Act of 1947, 50 U.S.C.A. §§ 3001-3234. Accessible from West, Westlaw through Public Law (P.L.) 113-296 (excluding P.L. 113-235, 113-287, and 113-291).
- 60.US Congress. 2001. Uniting and Strengthening America by Providing Appropriate Tools Required (USA Patriot Act) Act of 2001. Public Law 107-56. [Google Scholar]
- 61.Iverson E, Celious A, Shehane E, Oerke M, Warren V, Eastman A, Kennedy CR, Freeman BD. 2013. Critical illness research involving collection of genomic data: the conundrum posed by low levels of genomic literacy among surrogate decision makers for critically ill patients. J Empir Res Hum Res Ethics 8:53–57. doi: 10.1525/jer.2013.8.3.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. 2008. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korf BR. 2013. Genomic privacy in the information age. Clin Chem 59:1148–1150. doi: 10.1373/clinchem.2013.205260. [DOI] [PubMed] [Google Scholar]
- 64.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. 2013. Identifying personal genomes by surname inference. Science 339:321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 65.Moulson G, Cheng M. 2011. German shortcomings in focus as outbreak wanes. NBCNEWS.com. http://www.nbcnews.com/id/43384471/ns/health-food_safety/t/german-shortcomings-focus-outbreak-wanes/#.VQr_j7l0w5s.
- 66.Paulus JK, Santoyo-Vistrain R, Havelick D, Cohen A, Kalyesubula R, Ajayi IO, Mattsson JG, Adami H-O, Dalal S. 2012. Global teaching and training initiatives for emerging cohort studies. J Epidemiol Glob Health 2:125–133. doi: 10.1016/j.jegh.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treadwell TA, Koo D, Kuker K, Khan AS. 2003. Epidemiologic clues to bioterrorism. Public Health Rep 118:92–98. doi: 10.1016/S0033-3549(04)50224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rotz LD, Hughes JM. 2004. Advances in detecting and responding to threats from bioterrorism and emerging infectious disease. Nat Med 10:S130–S136. doi: 10.1038/nm1152. [DOI] [PubMed] [Google Scholar]
- 69.Grunow R, Finke E-J. 2002. A procedure for differentiating between the intentional release of biological warfare agents and natural outbreaks of disease: its use in analyzing the tularemia outbreak in Kosovo in 1999 and 2000. Clin Microbiol Infect 8:510–521. doi: 10.1046/j.1469-0691.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- 70.Dembek ZF, Kortepeter MG, Pavlin JA. 2007. Discernment between deliberate and natural infectious disease outbreaks. Epidemiol Infect 135:353–371. doi: 10.1017/S0950268806007011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Radosavljevic V, Belojevic G. 2012. Unusual epidemic events: a new method of early orientation and differentiation between natural and deliberate epidemics. Public Health 126:77–81. doi: 10.1016/j.puhe.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 72.Bailey AM, Paulsen IT, Piddock LJV. 2008. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob Agents Chemother 52:3604–3611. doi: 10.1128/AAC.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberhardt MA, Palsson BØ, Papin JA. 2009. Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oberhardt MA, Puchałka J, Martins dos Santos VAP, Papin JA. 2011. Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis. PLoS Comput Biol. 7:e1001116. doi: 10.1371/journal.pcbi.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lagier J-C, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape J-F, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 76.Köser CU, Holden MTG, Ellington MJ, Cartwright EJP, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rappé MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu Rev Microbiol 57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 78.Tringe SG, Rubin EM. 2005. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet 6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 79.Wooley JC, Godzik A, Friedberg I. 2010. A primer on metagenomics. PLoS Comput Biol 6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wheelis M. 2002. Biological warfare at the 1346 siege of Caffa. Emerg Infect Dis 8:971–975. doi: 10.3201/eid0809.010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keremidis H, Appel B, Menrath A, Tomuzia K, Normark M, Roffey R, Knutsson R. 2013. Historical perspective on agroterrorism: lessons learned from 1945 to 2012. Biosecur Bioterror 11(Suppl 1):S17–S24. doi: 10.1089/bsp.2012.0080. [DOI] [PubMed] [Google Scholar]
- 82.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier P-E, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 83.Gagnaire J, Dauwalder O, Boisset S, Khau D, Freydière A-M, Ader F, Bes M, Lina G, Tristan A, Reverdy M-E, Marchand A, Geissmann T, Benito Y, Durand G, Charrier J-P, Etienne J, Welker M, Van Belkum A, Vandenesch F. 2012. Detection of Staphylococcus aureus delta-toxin production by whole-cell MALDI-TOF mass spectrometry. PLoS One 7:e40660. doi: 10.1371/journal.pone.0040660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adrait A, Lebert D, Trauchessec M, Dupuis A, Louwagie M, Masselon C, Jaquinod M, Chevalier B, Vandenesch F, Garin J, Bruley C, Brun V. 2012. Development of a protein standard absolute quantification (PSAQ) assay for the quantification of Staphylococcus aureus enterotoxin A in serum. J Proteomics 75:3041–3049. doi: 10.1016/j.jprot.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 85.Jungo F, Bougueleret L, Xenarios I, Poux S. 2012. The UniProtKB/Swiss-Prot Tox-Prot program: a central hub of integrated venom protein data. Toxicon 60:551–557. doi: 10.1016/j.toxicon.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Picotti P, Bodenmiller B, Aebersold R. 2013. Proteomics meets the scientific method. Nat Methods 10:24–27. doi: 10.1038/nmeth.2291. [DOI] [PubMed] [Google Scholar]
- 87.Ali IKM, Zaki M, Clark CG. 2005. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol 43:5842–5847. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, Okinaka R, Jackson PJ, Hugh-Jones ME. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol 182:2928–2936. doi: 10.1128/JB.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scott AN, Menzies D, Tannenbaum T-N, Thibert L, Kozak R, Joseph L, Schwartzman K, Behr MA. 2005. Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis. J Clin Microbiol 43:89–94. doi: 10.1128/JCM.43.1.89-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. 2005. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res 33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chi LM, Lam SL. 2005. Structural roles of CTG repeats in slippage expansion during DNA replication. Nucleic Acids Res 33:1604–1617. doi: 10.1093/nar/gki307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Pontén TS, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS, Acosta N, Bhattacharyya T, Diosque P, Carrasco HJ, Miles MA. 2011. Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis 5:e1049. doi: 10.1371/journal.pntd.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aanensen DM, Spratt BG. 2005. The Multilocus Sequence Typing Network: mlst.net.Nucleic Acids Res 33:W728–W733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baker S, Hanage WP, Holt KE. 2010. Navigating the future of bacterial molecular epidemiology. Curr Opin Microbiol 13:640–645. doi: 10.1016/j.mib.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dione MM, Ikumapayi UN, Saha D, Mohammed NI, Geerts S, Ieven M, Adegbola RA, Antonio M. 2011. Clonal differences between non-typhoidal Salmonella (NTS) recovered from children and animals living in close contact in the Gambia. PLoS Negl Trop Dis 5:e1148. doi: 10.1371/journal.pntd.0001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gibbons HS, Krepps MD, Ouellette G, Karavis M, Onischuk L, Leonard P, Broomall S, Sickler T, Betters JL, McGregor P, Donarum G, Liem A, Fochler E, McNew L, Rosenzweig CN, Skowronski E. 2012. Comparative genomics of 2009 seasonal plague (Yersinia pestis) in New Mexico. PLoS One 7:e31604. doi: 10.1371/journal.pone.0031604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 99.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 100.Gault G, Weill FX, Mariani-Kurkdjian P, Jourdan-da Silva N, King L, Aldabe B, Charron M, Ong N, Castor C, Mace M, Bingen E, Noel H, Vaillant V, Bone A, Vendrely B, Delmas Y, Combe C, Bercion R, d'Andigne E, Desjardin M, de Valk H, Rolland P. 2011. Outbreak of haemolytic uraemic syndrome and bloody diarrhoea due to Escherichia coli O104:H4, south-west France, June 2011. Euro Surveill 16(26):pii=19905 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19905. [DOI] [PubMed] [Google Scholar]
- 101.Bae WK, Lee YK, Cho MS, Ma SK, Kim SW, Kim NH, Choi KC. 2006. A case of hemolytic uremic syndrome caused by Escherichia coli O104:H4. Yonsei Med J 47:437–439. doi: 10.3349/ymj.2006.47.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mellmann A, Bielaszewska M, Köck R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg Infect Dis 14:1287–1290. doi: 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Müller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 104.Jansen A, Kielstein JT. 2011. The new face of enterohaemorrhagic Escherichia coli infections. Euro Surveill 16(25):pii=19898 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19898. [PubMed] [Google Scholar]
- 105.Mariani-Kurkdjian P, Bingen E, Gault G, Jourdan-Da Silva N, Weill F-X. 2011. Escherichia coli O104:H4 south-west France, June 2011. Lancet Infect Dis 11:732–733. doi: 10.1016/S1473-3099(11)70266-3. [DOI] [PubMed] [Google Scholar]
- 106.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 107.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin C-S, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Møller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Budowle B, Schutzer SE, Breeze RG, Keim PS, Morse SA (ed). 2011. Microbial forensics, 2nd ed Elsevier, San Diego, CA. [Google Scholar]
- 109.Daniel R. 2005. The metagenomics of soil. Nat Rev Microbiol 3:470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- 110.Reference deleted.
- 111.Gire SK, Goba A, Andersen KG, Sealfon RSG, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang P-P, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JLB, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, et al. . 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fidler D. 2011. International law and the E. coli outbreaks in Europe. Am Soc Int Law Insights 15:1–6. http://www.asil.org/insights/volume/15/issue/14/international-law-and-e-coli-outbreaks-europe. [Google Scholar]
- 113.Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, Wilking H, Deleré Y, an der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Höhle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kühne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365:1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 114.Tremlett G, Pidd H. 31 May 2011. Germany admits Spanish cucumbers are not to blame for E coli outbreak. Guardian. http://www.theguardian.com/uk/2011/may/31/e-coli-deaths-16-germany-sweden. [Google Scholar]
- 115.European Commission. 18 November 2011. €17 million additional EU support for the promotion of fresh fruit and vegetables following the E-coli crisis. Press release European Commission, Brussels, Belgium: http://europa.eu/rapid/press-release_IP-11-1373_en.htm?locale=en. [Google Scholar]
- 116.Turner M. 2011. Microbe outbreak panics Europe. Nature 474:137. doi: 10.1038/474137a. [DOI] [PubMed] [Google Scholar]
- 117.Budowle B, Connell ND, Bielecka-Oder A, Colwell RR, Corbett CR, Fletcher J, Forsman M, Kadavy DR, Markotic A, Morse SA, Murch RS, Sajantila A, Schmedes SE, Ternus KL, Turner SD, Minot S. 2014. Validation of high throughput sequencing and microbial forensics applications. Investig Genet 5:9. doi: 10.1186/2041-2223-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wetterstrand KS. 2013. DNA sequencing costs: data from the NHGRI large-scale genome sequencing program. NIH, Bethesda, MD. [Google Scholar]
- 119.DeLong EF. 2005. Microbial community genomics in the ocean. Nat Rev Microbiol 3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- 120.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y-H, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJ, Brown GZ, Green JL. 2012. Architectural design influences the diversity and structure of the built environment microbiome. ISME J 6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.National Research Council Committee on Metagenomics: Challenges and Functional Applications. 2007. The new science of metagenomics: revealing the secrets of our microbial planet. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 124.Giesbertz NAA, Bredenoord AL, van Delden JJM. 2012. Inclusion of residual tissue in biobanks: opt-in or opt-out? PLoS Biol 10:e1001373. doi: 10.1371/journal.pbio.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.WHO. 2000. Guidelines for the collection of clinical specimens during field investigation of outbreaks. WHO/CDS/CSR/EDC/2000.4 World Health Organization, Geneva, Switzerland. [Google Scholar]
- 126.Loman NJ, Constantinidou C, Christner M, Rohde H, Chan J, Quick J, Weir J, Quince C, Smith G, Betley J, Aepfelbacher M, Pallen M. 2013. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA 309:1502–1510. doi: 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- 127.Taniuchi M, Sobuz SU, Begum S, Platts-Mills JA, Liu J, Yang Z, Wang X-Q, Petri WA Jr, Haque R, Houpt ER. 2013. Etiology of diarrhea in Bangladeshi infants in the first year of life using molecular methods. J Infect Dis 208:1794–1802. doi: 10.1093/infdis/jit507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Otto M. 2012. MRSA infection is associated with increased mortality. Future Microbiol 7:189–191. doi: 10.2217/fmb.11.156. [DOI] [PubMed] [Google Scholar]
- 129.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.CDC. 2007. Active Bacterial Core Surveillance (ABCs) report, Emerging Infections Program Network, methicillin-resistant Staphylococcus aureus, 2007. CDC, Atlanta, GA. [Google Scholar]
- 131.Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Boakes E, Kearns AM, Ganner M, Perry C, Warner M, Hill RL, Ellington MJ. 2011. Molecular diversity within clonal complex 22 methicillin-resistant Staphylococcus aureus encoding Panton-Valentine leukocidin in England and Wales. Clin Microbiol Infect 17:140–145. doi: 10.1111/j.1469-0691.2010.03199.x. [DOI] [PubMed] [Google Scholar]
- 133.Dinges MM, Orwin PM, Schlievert PM. 2000. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 13:16–34. doi: 10.1128/CMR.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tanaka M, Wang T, Onodera Y, Uchida Y, Sato K. 2000. Mechanism of quinolone resistance in Staphylococcus aureus. J Infect Chemother 6:131–139. doi: 10.1007/s101560070010. [DOI] [PubMed] [Google Scholar]
- 135.Wichelhaus TA, Schäfer V, Brade V, Böddinghaus B. 2001. Differential effect of rpoB mutations on antibacterial activities of rifampicin and KRM-1648 against Staphylococcus aureus. J Antimicrob Chemother 47:153–156. doi: 10.1093/jac/47.2.153. [DOI] [PubMed] [Google Scholar]
- 136.Nübel U, Nachtnebel M, Falkenhorst G, Benzler J, Hecht J, Kube M, Bröcker F, Moelling K, Bührer C, Gastmeier P, Piening B, Behnke M, Dehnert M, Layer F, Witte W, Eckmanns T. 2013. MRSA transmission on a neonatal intensive care unit: epidemiological and genome-based phylogenetic analyses. PLoS One 8:e54898. doi: 10.1371/journal.pone.0054898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gray RR, Tatem AJ, Johnson JA, Alekseyenko AV, Pybus OG, Suchard MA, Salemi M. 2011. Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a Bayesian framework. Mol Biol Evol 28:1593–1603. doi: 10.1093/molbev/msq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barton M, Hawkes M, Moore D, Conly J, Nicolle L, Allen U, Boyd N, Embree J, Van Horne L, Le Saux N, Richardson S, Moore A, Tran D, Waters V, Vearncombe M, Katz K, Weese JS, Embil J, Ofner-Agostini M, Ford-Jones EL. 2006. Guidelines for the prevention and management of community-associated methicillin-resistant Staphylococcus aureus: a perspective for Canadian health care practitioners. Can J Infect Dis Med Microbiol 17:4C–24C. [PMC free article] [PubMed] [Google Scholar]
- 140.Eyre DW, Griffiths D, Vaughan A, Golubchik T, Acharya M, O'Connor L, Crook DW, Walker AS, Peto TEA. 2013. Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS One 8:e78445. doi: 10.1371/journal.pone.0078445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Henderson DK, Dembry L, Fishman NO, Grady C, Lundstrom T, Palmore TN, Sepkowitz KA, Weber DJ. 2010. SHEA guideline for management of healthcare workers who are infected with hepatitis B virus, hepatitis C virus, and/or human immunodeficiency virus. Infect Control Hosp Epidemiol 31:203–232. doi: 10.1086/650298. [DOI] [PubMed] [Google Scholar]
- 142.Centers for Disease Control and Prevention. 2012. Updated CDC recommendations for the management of hepatitis B virus-infected health-care providers and students. MMWR Recommend Rep 61(RR3):1–12. [PubMed] [Google Scholar]
- 143.Diep BA. 2013. Use of whole-genome sequencing for outbreak investigations. Lancet Infect Dis 13:99–101. doi: 10.1016/S1473-3099(12)70276-1. [DOI] [PubMed] [Google Scholar]
- 144.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 146.National TB Controllers Association on Genotyping Tuberculosis, CDC Advisory Group. 2004. Guide to the application of genotyping to tuberculosis prevention and control. CDC, Atlanta, GA. [Google Scholar]
- 147.Mitruka K, Oeltmann JE, Ijaz K, Haddad MB. 2011. Tuberculosis outbreak investigations in the United States, 2002-2008. Emerg Infect Dis 17:425–431. doi: 10.3201/eid1703.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McElroy PD, Rothenberg RB, Varghese R, Woodruff R, Minns GO, Muth SQ, Lambert LA, Ridzon R. 2003. A network-informed approach to investigating a tuberculosis outbreak: implications for enhancing contact investigations. Int J Tuberc Lung Dis 7:S486–S493. [PubMed] [Google Scholar]
- 149.Girard MP, Tam JS, Assossou OM, Kieny MP. 2010. The 2009 A (H1N1) influenza virus pandemic: a review. Vaccine 28:4895–4902. doi: 10.1016/j.vaccine.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 150.Morse SS, Mazet JAK, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Institute of Medicine Forum on Microbial Threat. 2004. Learning from SARS: preparing for the next disease outbreak. Workshop summary. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 152.Van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus ADME, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RAM. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chan JFW, Li KSM, To KKW, Cheng VCC, Chen H, Yuen K-Y. 2012. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect 65:477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.US Department of Health and Human Services. 2007. Update on CDC investigation into people potentially exposed to patient with extensively drug-resistant TB. CDC Newsroom press release. CDC, Atlanta, GA. [Google Scholar]
- 155.CNN. com. 2007. Doctors: TB traveler's diagnosis more treatable than thought. CNN.com. http://edition.cnn.com/2007/HEALTH/07/03/tb.speaker/index.html.
- 156.Klopper M, Warren RM, Hayes C, Gey van Pittius NC, Streicher EM, Müller B, Sirgel FA, Chabula-Nxiweni M, Hoosain E, Coetzee G, van Helden PD, Victor TC, Trollip AP. 2013. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg Infect Dis 19:449–455. doi: 10.3201/eid1903.120246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gupta JK, Lin C-H, Chen Q. 2012. Risk assessment of airborne infectious diseases in aircraft cabins. Indoor Air 22:388–395. doi: 10.1111/j.1600-0668.2012.00773.x. [DOI] [PubMed] [Google Scholar]
- 158.Martinez L, Blanc L, Nunn P, Raviglione M. 2008. Tuberculosis and air travel: WHO guidance in the era of drug-resistant TB. Travel Med Infect Dis 6:177–181. doi: 10.1016/j.tmaid.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 159.Ruger JP. 2010. Control of extensively drug-resistant tuberculosis (XDR-TB): a root cause analysis. Glob Heal Gov 3:1–20. [PMC free article] [PubMed] [Google Scholar]
- 160.Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, Bentley SD, Parkhill J, Nejentsev S, Hoffner SE, Horstmann RD, Brown T, Drobniewski F. 2012. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res 22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Deiss RG, Rodwell TC, Garfein RS. 2009. Tuberculosis and illicit drug use: review and update. Clin Infect Dis 48:72–82. doi: 10.1086/594126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bush GW. 2003. Executive Order 13295: revised list of quarantinable communicable diseases. [Google Scholar]
- 164.Nachega JB, Chaisson RE. 2003. Tuberculosis drug resistance: a global threat. Clin Infect Dis 36:S24–S30. doi: 10.1086/344657. [DOI] [PubMed] [Google Scholar]
- 165.Daum LT, Rodriguez JD, Worthy SA, Ismail NA, Omar SV, Dreyer AW, Fourie PB, Hoosen AA, Chambers JP, Fischer GW. 2012. Next-generation Ion Torrent sequencing of drug resistance mutations in Mycobacterium tuberculosis strains. J Clin Microbiol 50:3831–3837. doi: 10.1128/JCM.01893-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Althomsons SP, Kammerer JS, Shang N, Navin TR. 2012. Using routinely reported tuberculosis genotyping and surveillance data to predict tuberculosis outbreaks. PLoS One 7:e48754. doi: 10.1371/journal.pone.0048754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li J, Driver CR, Munsiff SS, Fujiwara PI. 2003. Finding contacts of homeless tuberculosis patients in New York City. Int J Tuberc Lung Dis 7:S397–S404. [PubMed] [Google Scholar]
- 168.Lowther SA, Miramontes R, Navara B, Sabuwala N, Brueshaber M, Solarz S, Haddad MB, Sodt D, Lynfield R. 2008. Outbreak of tuberculosis among Guatemalan immigrants in rural Minnesota. Public Health Rep 126:726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S. 2008. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A 96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Domingo E. 2000. Viruses at the edge of adaptation. Virology 270:251–253. doi: 10.1006/viro.2000.0320. [DOI] [PubMed] [Google Scholar]
- 172.Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML. 2010. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci U S A 107:21242–21247. doi: 10.1073/pnas.1015673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Förster-Waldl E, Sadeghi K, Tamandl D, Gerhold B, Hallwirth U, Rohrmeister K, Hayde M, Prusa AR, Herkner K, Boltz-Nitulescu G, Pollak A, Spittler A. 2005. Monocyte Toll-like receptor 4 expression and LPS-induced cytokine production increase during gestational aging. Pediatr Res 58:121–124. doi: 10.1203/01.PDR.0000163397.53466.0F. [DOI] [PubMed] [Google Scholar]
- 174.Memish ZA, Cotten M, Watson SJ, Kellam P, Zumla A, Alhakeem RF, Assiri A, Rabeeah AA, Al-Tawfiq JA. 2014. Community case clusters of Middle East respiratory syndrome coronavirus in Hafr Al-Batin, Kingdom of Saudi Arabia: a descriptive genomic study. Int J Infect Dis 23:63–68. doi: 10.1016/j.ijid.2014.03.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. The early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Monke J. 2007. CRS report for congress. Agroterrorism: threats and preparedness. Congressional Research Service, Washington, DC. [Google Scholar]
- 177.Haydon DT, Samuel AR, Knowles NJ. 2001. The generation and persistence of genetic variation in foot-and-mouth disease virus. Prev Vet Med 51:111–124. doi: 10.1016/S0167-5877(01)00210-0. [DOI] [PubMed] [Google Scholar]
- 178.Morelli MJ, Thébaud G, Chadœuf J, King DP, Haydon DT, Soubeyrand S. 2012. A Bayesian inference framework to reconstruct transmission trees using epidemiological and genetic data. PLoS Comput Biol 8:e1002768. doi: 10.1371/journal.pcbi.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mason PW, Pacheco JM, Zhao Q-Z, Knowles NJ. 2003. Comparisons of the complete genomes of Asian, African and European isolates of a recent foot-and-mouth disease virus type O pandemic strain (PanAsia). J Gen Virol 84:1583–1593. doi: 10.1099/vir.0.18669-0. [DOI] [PubMed] [Google Scholar]
- 180.Mohapatra JK, Sanyal A, Hemadri D, Tosh C, Biswas S, Knowles NJ, Rasool TJ, Bandyopadhyay SK, Pattnaik B. 2008. Comparative genomics of serotype Asia 1 foot-and-mouth disease virus isolates from India sampled over the last two decades. Virus Res 136:16–29. doi: 10.1016/j.virusres.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 181.Carrillo C, Tulman ER, Delhon G, Lu Z, Carreno A, Vagnozzi A, Kutish GF, Rock DL. 2005. Comparative genomics of foot-and-mouth disease virus. J Virol 79:6487–6504. doi: 10.1128/JVI.79.10.6487-6504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Belsham GJ, Jamal SM, Tjørnehøj K, Bøtner A. 2011. Rescue of foot-and-mouth disease viruses that are pathogenic for cattle from preserved viral RNA samples. PLoS One 6:e14621. doi: 10.1371/journal.pone.0014621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cottam EM, Haydon DT, Paton DJ, Gloster J, Wilesmith JW, Ferris NP, Hutchings GH, King DP. 2006. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J Virol 80:11274–11282. doi: 10.1128/JVI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cottam EM, Wadsworth J, Shaw AE, Rowlands RJ, Goatley L, Maan S, Maan NS, Mertens PPC, Ebert K, Li Y, Ryan ED, Juleff N, Ferris NP, Wilesmith JW, Haydon DT, King DP, Paton DJ, Knowles NJ. 2008. Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathog 4:e1000050. doi: 10.1371/journal.ppat.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhu W, Li J, Liang G. 2011. How does cellular heparan sulfate function in viral pathogenicity? Biomed Environ Sci 24:81–87. doi: 10.3967/0895-3988.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 186.Spratt BG. 2007. Independent review of the safety of UK facilities handling foot-and-mouth disease virus. DEFRA, London, United Kingdom. [Google Scholar]
- 187.Valdazo-González B, Polihronova L, Alexandrov T, Normann P, Knowles NJ, Hammond JM, Georgiev GK, Ozyörük F, Sumption KJ, Belsham GJ, King DP. 2012. Reconstruction of the transmission history of RNA virus outbreaks using full genome sequences: foot-and-mouth disease virus in Bulgaria in 2011. PLoS One 7:e49650. doi: 10.1371/journal.pone.0049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Oyola SO, Gu Y, Manske M, Otto TD, O'Brien J, Alcock D, Macinnis B, Berriman M, Newbold CI, Kwiatkowski DP, Swerdlow HP, Quail MA. 2013. Efficient depletion of host DNA contamination in malaria clinical sequencing. J Clin Microbiol 51:745–751. doi: 10.1128/JCM.02507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lister R, Ecker JR. 2009. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res 19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ballestar E, Wolffe AP. 2001. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem 268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 191.Kalyuzhnaya MG, Zabinsky R, Bowerman S, Baker DR, Lidstrom ME, Chistoserdova L. 2006. Fluorescence in situ hybridization-flow cytometry-cell sorting-based method for separation and enrichment of type I and type II methanotroph populations. Appl Environ Microbiol 72:4293–4301. doi: 10.1128/AEM.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Van Rijen MML, Kluytmans-van den Bergh MFQ, Verkade EJM, Ten Ham PBG, Feingold BJ, Kluytmans JAJW. 2013. Lifestyle-associated risk factors for community-acquired methicillin-resistant Staphylococcus aureus carriage in the Netherlands: an exploratory hospital-based case-control study. PLoS One 8:e65594. doi: 10.1371/journal.pone.0065594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hupalo DN, Bradic M, Carlton JM. 2015. The impact of genomics on population genetics of parasitic diseases. Curr Opin Microbiol 23C:49–54. doi: 10.1016/j.mib.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hall N. 2013. After the gold rush. Genome Biol 14:115. doi: 10.1186/gb-2013-14-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Eisenstein M. 2012. Oxford Nanopore announcement sets sequencing sector abuzz. Nat Biotechnol 30:295–296. doi: 10.1038/nbt0412-295. [DOI] [PubMed] [Google Scholar]
- 196.Pan American Health Organization. 2011. Cholera and post-earthquake response in Haiti. Pan American Health Organization, Washington, DC. [Google Scholar]
- 197.Keim PS, Aarestrup FM, Shakya G, Price LB, Hendriksen RS, Engelthaler DM, Pearson T. 2011. Reply to “South Asia instead of Nepal may be the origin of the Haitian cholera outbreak strain.” mBio 2(6):e00245-11 (Reply.) doi: 10.1128/mBio.00245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pun SB. 2011. South Asia instead of Nepal may be the origin of the Haitian cholera outbreak strains. mBio 2(5):e00219-11. doi: 10.1128/mBio.00219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P. 2011. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis 17:2113–2121. doi: 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Bortolaia V, Pearson T, Waters AE, Upadhyay BP, Shrestha SD, Adhikari S, Shakya G, Keim PS, Aarestrup FM. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2(4):e00157-11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 203.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu B, Gibbons T, Ghodsi M, Treangen T, Pop M. 2011. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics 12(Suppl 2):S4. doi: 10.1186/1471-2164-12-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Davenport CF, Neugebauer J, Beckmann N, Friedrich B, Kameri B, Kokott S, Paetow M, Siekmann B, Wieding-Drewes M, Wienhöfer M, Wolf S, Tümmler B, Ahlers V, Sprengel F. 2012. Genometa—a fast and accurate classifier for short metagenomic shotgun reads. PLoS One 7:e41224. doi: 10.1371/journal.pone.0041224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Köser C, Ellington M, Cartwright E. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 8:e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Blank K, Hensel M, Gerlach RG. 2011. Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One 6:e15763. doi: 10.1371/journal.pone.0015763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fehér T, Karcagi I, Gyorfy Z, Umenhoffer K, Csörgo B, Pósfai G. 2008. Scarless engineering of the Escherichia coli genome. Methods Mol Biol 416:251–259. doi: 10.1007/978-1-59745-321-9_16. [DOI] [PubMed] [Google Scholar]
- 209.Liang R, Liu J. 2010. Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiol 10:209. doi: 10.1186/1471-2180-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sun W, Wang S, Curtiss R. 2008. Highly efficient method for introducing successive multiple scarless gene deletions and markerless gene insertions into the Yersinia pestis chromosome. Appl Environ Microbiol 74:4241–4245. doi: 10.1128/AEM.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AKC, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2(6):e00204-11. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sa-Carvalho D, Rieder E, Baxt B, Rodarte R, Tanuri A, Mason PW. 1997. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J Virol 71:5115–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Juleff N. 2013. Accumulation of nucleotide substitutions occurring during experimental transmission of foot-and-mouth disease virus. J Gen Virol 94:108–119. doi: 10.1099/vir.0.046029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Associated Press. 2014. Ebola scares West Africans away from bush meat. CBSNEWS.com. http://www.cbsnews.com/news/ebola-virus-scares-west-africans-away-from-bush-meat/.
- 215.Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. 2013. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J Clin Microbiol 51:1396–1401. doi: 10.1128/JCM.03332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Logan G, Freimanis GL, King DJ, Valdazo-González B, Bachanek-Bankowska K, Sanderson ND, Knowles NJ, King DP, Cottam EM. 2014. A universal protocol to generate consensus level genome sequences for foot-and-mouth disease virus and other positive-sense polyadenylated RNA viruses using the Illumina MiSeq. BMC Genomics 15:828. doi: 10.1186/1471-2164-15-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bertelli C, Greub G. 2013. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin Microbiol Infect 19:803–813. doi: 10.1111/1469-0691.12217. [DOI] [PubMed] [Google Scholar]
- 218.Work Group 4. 2013. News from Work Group 4—launch of pilot proficiency test. Global Microbial Identifier Initiative, Lyngby, Denmark. [Google Scholar]
- 219.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Metzker ML, Mindell DP, Liu X-M, Ptak RG, Gibbs RA, Hillis DM. 2002. Molecular evidence of HIV-1 transmission in a criminal case. Proc Natl Acad Sci U S A 99:14292–14297. doi: 10.1073/pnas.222522599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Leichty AR, Brisson D. 2014. Selective whole genome amplification for resequencing target microbial species from complex natural samples. Genetics 198:473–481. doi: 10.1534/genetics.114.165498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Reagan R. 1981. Executive Order 12333: United States intelligence activities. [Google Scholar]