Abstract
Background
Oridonin, an ingredient used in traditional Chinese medicine, has been demonstrated to play an important role in antitumour effects, but the mechanism underlying its antitumour properties is still not clear.
Methods
To verify the anti-cancer effects of oridonin via a miRNA-dependent mechanism, comprehensive miRNA expression profiling of oridonin-treated BxPC-3 human pancreatic cancer cells was performed using a miRNA microarray assay based on Sanger miR-Base Release 20, followed by a validation using real-time PCR. MicroRNA target prediction and Gene Ontology and KEGG pathway analysis were performed to investigate possible pathways involved.
Results
The results showed that 105 miRNAs were significantly differentially expressed (signal reading >500, p ≤ 0.01, |Log2-value| ≥1) in oridonin-treated BxPC-3 human pancreatic cancer cells.
Conclusions
Our data indicates that oridonin inhibits BxPC-3 cells probably through regulating the expression of miRNAs. Interruption of miRNA profiling may provide new therapeutic methods for the clinical treatment of pancreatic cancer.
Keywords: Oridonin, miRNA, microarray, BxPC-3 pancreatic cancer cell
Background
Oridonin, a natural ent-kaurane diterpenoid compound, is isolated from the Chinese medicinal herb Rabdosia rubescens as well as other plants, such as Isodon trichocarpus and Isodon shikokianus. Oridonin has many physiological and pharmacological effects, including anti-inflammation, anti-bacterial and anti-tumour effects, and shows no obvious side effects when used for the treatment of various human diseases. Concerning anti-tumour effects, previous studies have reported that oridonin can induce cell growth inhibition, promote apoptosis and inhibit migration and invasion in many cancers [1-3]. Nevertheless, the mechanisms underlying the antitumour activity of oridonin have not been completely delineated.
MicroRNAs (miRNAs) are a novel class of non-coding RNAs with lengths of 17–25 nucleotides (nt) that can regulate gene expression in eukaryotic organisms by pairing with target mRNAs to repress translation or cause degradation of multiple target mRNAs [4]. Recent studies have shown that miRNAs play crucial roles in many biological processes, such as development, cell growth, differentiation, apodosis and even tumouriogenesis [5,6]. Furthermore, miRNAs can function both as tumour suppressors and oncogenes and might be a potential therapeutic target in cancer. Recent publications have shown that correcting abnormal miRNAs in tumours can inhibit the function of the target mRNA in vivo in a mouse model [7,8].
Traditional Chinese medicines have become a popular topic in relation to their potential anti-tumour properties. However, there are no available reports on oridonin which inhibits pancreatic cancer via miRNA regulation. In this study, we establish a sensitive microarray chip for miRNA expression profiling in BxPC-3 pancreatic cancer cells treated with oridonin to verify our hypothesis that oridonin alters the miRNA expression profile in pancreatic cancer, and we show that miRNAs have potential applications in the future clinical treatment of tumours.
Methods
Cell culture
The BxPC-3 human pancreatic cancer cell line was provided by the Institute of Biochemistry and Cell Biology, Shanghai Institute of Biological Sciences, Chinese Academy of Sciences (ATCC® CRL1687™). The cells were cultured in RPMI 1640 (GIBCO, NY, United States) culture medium containing 10% foetal bovine serum (FBS, Gibco), 300 mg/L glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin in an incubator with 5% CO2 at 37°C. Cells in logarithmic growth phase were seeded in 60 mm dishes at a density of 4 × 104 cell/cm2 and incubated overnight. One group of these cells was subsequently treated with 87.8 μM oridonin (Gracia Chemical Technology Company, LTD, 98% purity, HPLC) dissolved in DMSO (final DMSO concentration in growth media is 0.1%), and another was used as a blank control group cultured in medium containing 0.1% DMSO for 24 hours. At least 3 independent experiments were performed.
RNA isolation and miRNA microarray
After 24 hours of treatment, total RNA (containing small RNAs) was extracted using the TRIzol LS reagent (Invitrogen Life Technologies) following the manufacturer’s protocol. The microarray assay (μParafloTM MicroRNA Microarray Assay) was performed by a service provider (LC Sciences), including quality control, labelling, chip hybridisation, signal amplification image acquisition and microarray data analysis. Hybridisation was performed overnight on a μParaflo microfluidic chip using a micro-circulation pump (Atactic Technologies) [9]. On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target microRNA (from miRBase, http://www.mirbase.org/) or other RNA (control or customer defined sequences) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes were made by in situ synthesis using PGR (photogenerated reagent) chemistry. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. Hybridization used 100 L 6xSSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25% formamide at 34°C. After RNA hybridization, tag-conjugating Cy3 dye were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Device) and digitized using Array-Pro image analysis software (Media Cybernetics). The data were analysed by first subtracting the background and then normalising the signals using a LOWESS filter (locally weighted regression). Then, the ratio of detected signals showing a log2 fold change [log2 (oridonin/control)] was used to define differentially expressed miRNAs, and Student’s t-test was employed to calculate P values.
MiRNA target prediction and Gene Ontology and KEGG pathway analysis
The prediction of miRNA targets was performed using the online software TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/cgi-bin/new_PicTar_vertebrate.cgi) and miRanda (http://www.microrna.org/microrna/home.do). The intersection of the results from these three types of software was taken as the final target genes of significantly differentially expressed miRNAs. Then, the target genes were analysed in terms of the annotation of their Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using Fisher’s exact test.
Reverse-transcription and Quantitative Real-time PCR
To validate the microarray data, total RNA from the same preparation used for microarray analysis was reverse-transcribed to cDNA in a Mycycler™ Thermal Cycler (Bio-Rad, USA), and quantitative real-time polymerase chain reaction (qPCR) was performed in a Real-Time PCR Detector (Bio-Rad, USA) using the PrimeScript™ miRNA qPCR Starter Kit Ver.2.0 (TaKaRa, Dalian, China), following the manufacturer’s protocol. Each reaction was performed in a final volume of 25 μl containing 1 μl cDNA, 0.4 μM of each primer and 1× SYBR Premix Ex TaqII. The amplification program was as follows: denaturation at 95°C for 10 sec, followed by 40 cycles of denaturation at 95°C for 5 sec and extension at 60°C for 20 sec, in which fluorescence was obtained. For quantification, RNU6B was used as the internal control, and expression levels of each mature miRNA were normalised using the 2-△△CT method [10]. All assays were performed in triplicate.
Statistical analysis
A log2 fold change [log2 (oridonin/control)] was used to define differentially expressed miRNAs, and Student’s t-test was employed to calculate P values. The target genes were analysed in terms of the annotation of their Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using Fisher’s exact test. Results of realtime RT-PCR experiments are expressed as means ± standard deviation (SD). Statistical comparisons were performed with the SPSS 17.0 software (Univariate Analysis of Variance) and statistical significance was considered for P values lower than 0.05.
Results
MiRNA expression was altered in BxPC-3 cells treated with oridonin
To study the responses of miRNAs to oridonin, microarray analysis of miRNA expression in BxPC-3 cells treated with oridonin was compared with the expression of miRNAs in DMSO treated cells. Only miRNAs showing significant expression among the oridonin treatments and their controls are reported (Table 1). As shown in Table 1, 105 reporters presented a strong response (signal reading >500, p ≤ 0.01, |Log2-valueb| ≥1) and significant regulation. Among these 105 miRNAs, 49 miRNAs were significantly down-regulated, whereas 56 were significantly up-regulated by oridonin. Among them, there are many new miRNAs whose function has been scarcely described in the literature.
Table 1.
miRNA regulation of oridonin in BxPC-3 cells
| Reporter name a | Log2-value b | p-Value | Reporter name a | Log2-value b | p-Value b |
|---|---|---|---|---|---|
| hsa-miR-513a-5p | 15.70 | 2.09E-66 | hsa-miR-27b-5p | −16.93 | 0.00E + 00 |
| hsa-miR-3661 | 12.73 | 0.00E + 00 | hsa-miR-205-3p | −16.63 | 0.00E + 00 |
| hsa-miR-4470 | 12.24 | 0.00E + 00 | hsa-miR-4262 | −16.47 | 0.00E + 00 |
| hsa-miR-409-3p | 10.68 | 0.00E + 00 | hsa-miR-499a-5p | −16.36 | 0.00E + 00 |
| hsa-miR-3197 | 10.32 | 0.00E + 00 | hsa-miR-3934-3p | −16.32 | 0.00E + 00 |
| hsa-miR-5096 | 10.29 | 0.00E + 00 | hsa-miR-193b-3p | −7.45 | 0.00E + 00 |
| hsa-miR-4267 | 10.13 | 0.00E + 00 | hsa-miR-421 | −6.93 | 0.00E + 00 |
| hsa-miR-466 | 8.90 | 0.00E + 00 | hsa-miR-10b-3p | −5.74 | 0.00E + 00 |
| hsa-miR-615-5p | 8.80 | 0.00E + 00 | hsa-miR-7641 | −5.53 | 0.00E + 00 |
| hsa-miR-7108-5p | 8.13 | 0.00E + 00 | hsa-miR-425-5p | −4.77 | 0.00E + 00 |
| hsa-miR-6791-5p | 7.92 | 0.00E + 00 | hsa-miR-125b-5p | −4.38 | 0.00E + 00 |
| hsa-miR-1246 | 6.86 | 0.00E + 00 | hsa-miR-200b-3p | −3.98 | 0.00E + 00 |
| hsa-miR-6807-5p | 6.57 | 1.38E-43 | hsa-miR-3960 | −3.95 | 0.00E + 00 |
| hsa-let-7f-5p | 6.41 | 0.00E + 00 | hsa-miR-132-3p | −3.55 | 0.00E + 00 |
| hsa-miR-1307-3p | 5.81 | 0.00E + 00 | hsa-miR-361-5p | −3.34 | 0.00E + 00 |
| hsa-miR-4514 | 5.36 | 1.44E-59 | hsa-miR-3178 | −3.31 | 0.00E + 00 |
| hsa-miR-4472 | 5.22 | 0.00E + 00 | hsa-miR-454-3p | −3.13 | 0.00E + 00 |
| hsa-miR-6126 | 5.04 | 0.00E + 00 | hsa-miR-320b | −2.71 | 0.00E + 00 |
| hsa-miR-6073 | 4.60 | 1.24E-43 | hsa-miR-455-3p | −2.62 | 0.00E + 00 |
| hsa-miR-4301 | 4.26 | 0.00E + 00 | hsa-miR-320e | −2.56 | 0.00E + 00 |
| hsa-miR-4484 | 3.79 | 3.65E-71 | hsa-miR-185-5p | −2.47 | 0.00E + 00 |
| hsa-miR-30c-1-3p | 3.65 | 0.00E + 00 | hsa-miR-320c | −2.38 | 0.00E + 00 |
| hsa-miR-4447 | 3.59 | 2.47E-42 | hsa-miR-4521 | −2.13 | 0.00E + 00 |
| hsa-miR-7977 | 3.56 | 0.00E + 00 | hsa-miR-320a | −2.10 | 0.00E + 00 |
| hsa-miR-5787 | 3.47 | 2.93E-51 | hsa-miR-193a-3p | −2.09 | 0.00E + 00 |
| hsa-miR-7150 | 3.44 | 2.24E-47 | hsa-miR-320d | −2.04 | 0.00E + 00 |
| hsa-miR-4516 | 3.16 | 0.00E + 00 | hsa-miR-92b-3p | −1.84 | 0.00E + 00 |
| hsa-miR-1273 g-3p | 3.06 | 0.00E + 00 | hsa-let-7i-5p | −1.81 | 0.00E + 00 |
| hsa-miR-6090 | 3.00 | 0.00E + 00 | hsa-miR-183-5p | −1.79 | 0.00E + 00 |
| hsa-miR-494-3p | 2.98 | 1.06E-72 | hsa-miR-365a-3p | −1.59 | 0.00E + 00 |
| hsa-miR-6786-5p | 2.87 | 1.26E-23 | hsa-miR-186-5p | −1.54 | 0.00E + 00 |
| hsa-miR-6727-5p | 2.81 | 0.00E + 00 | hsa-miR-125a-5p | −1.54 | 0.00E + 00 |
| hsa-miR-98-5p | 2.65 | 0.00E + 00 | hsa-miR-151a-3p | −1.43 | 0.00E + 00 |
| hsa-miR-668-3p | 2.64 | 6.84E-65 | hsa-miR-224-5p | −1.42 | 0.00E + 00 |
| hsa-miR-1275 | 2.59 | 1.69E-50 | hsa-miR-107 | −1.40 | 0.00E + 00 |
| hsa-miR-6125 | 2.53 | 1.03E-49 | hsa-miR-93-5p | −1.34 | 0.00E + 00 |
| hsa-miR-6087 | 2.50 | 0.00E + 00 | hsa-miR-3609 | −1.31 | 0.00E + 00 |
| hsa-miR-4505 | 2.31 | 1.03E-10 | sa-miR-103a-3p | −1.28 | 0.00E + 00 |
| ha-smiR-7110-5p | 2.26 | 3.03E-12 | hsa-miR-4286 | −1.22 | 0.00E + 00 |
| hsa-miR-1260a | 2.14 | 1.31E-09 | hsa-miR-3607-5p | −1.20 | 0.00E + 00 |
| hsa-miR-6803-5p | 2.13 | 1.94E-10 | hsa-miR-92a-3p | −1.19 | 0.00E + 00 |
| hsa-miR-29c-3p | 2.08 | 2.01E-13 | hsa-miR-429 | −1.17 | 0.00E + 00 |
| hsa-miR-4466 | 2.04 | 3.57E-06 | hsa-miR-20a-5p | −1.15 | 0.00E + 00 |
| hsa-miR-1973 | 1.93 | 7.49E-09 | hsa-miR-424-5p | −1.13 | 0.00E + 00 |
| hsa-miR-4739 | 1.86 | 1.47E-04 | hsa-miR-17-5p | −1.09 | 0.00E + 00 |
| hsa-miR-3196 | 1.85 | 1.80E-15 | hsa-miR-203a | −1.05 | 0.00E + 00 |
| hsa-miR-4497 | 1.76 | 1.16E-13 | hsa-miR-574-3p | −1.04 | 0.00E + 00 |
| hsa-miR-378 g | 1.76 | 2.93E-03 | hsa-miR-378c | −1.03 | 0.00E + 00 |
| hsa-miR-4459 | 1.73 | 5.05E-03 | hsa-miR-423-5p | −1.01 | 0.00E + 00 |
| hsa-miR-3665 | 1.70 | 2.46E-11 | |||
| hsa-miR-638 | 1.64 | 2.41E-04 | |||
| hsa-miR-7704 | 1.63 | 4.06E-08 | |||
| hsa-let-7 g-5p | 1.54 | 1.18E-05 | |||
| hsa-let-7e-5p | 1.52 | 3.00E-07 | |||
| hsa-miR-4508 | 1.04 | 2.30E-08 | |||
| hsa-miR-6089 | 1.03 | 2.17E-19 | |||
| Reporter name c | Log2-value b | p-Value | Reporter name c | Log2-value b | p-Value |
| hsa-miR-328-5p | 2.96 | 3.36E-18 | hsa-miR-3943 | −16.15 | 0.00E + 00 |
| hsa-miR-5194 | 6.18 | 8.69E-39 | hsa-miR-4536-3p | −16.08 | 0.00E + 00 |
| hsa-miR-6085 | 2.55 | 2.40E-12 | hsa-miR-1180-3p | −15.86 | 0.00E + 00 |
| hsa-miR-6880-5p | 2.69 | 1.08E-13 | hsa-miR-3188 | −15.82 | 0.00E + 00 |
| hsa-miR-4791 | 14.87 | 2.29E-39 | hsa-miR-1179 | −15.81 | 0.00E + 00 |
| hsa-miR-6124 | 3.00 | 1.28E-16 | hsa-miR-3169 | −9.50 | 0.00E + 00 |
| hsa-miR-1233-5p | 3.54 | 1.32E-20 | hsa-miR-15b-3p | −7.42 | 0.00E + 00 |
| hsa-miR-765 | 2.56 | 9.53E-10 | hsa-miR-301a-3p | −6.71 | 0.00E + 00 |
| hsa-miR-4463 | 2.34 | 1.77E-07 | hsa-miR-101-3p | −6.63 | 0.00E + 00 |
| hsa-miR-3065-5p | −6.07 | 0.00E + 00 | |||
| hsa-miR-625-5p | −3.97 | 0.00E + 00 | |||
| hsa-miR-24-2-5p | −3.06 | 0.00E + 00 | |||
| hsa-miR-128-3p | −2.86 | 0.00E + 00 | |||
| hsa-miR-4289 | −2.65 | 0.00E + 00 | |||
| hsa-miR-155-5p | −2.35 | 0.00E + 00 | |||
| hsa-miR-197-3p | −2.21 | 0.00E + 00 | |||
| hsa-miR-10a-5p | −2.08 | 0.00E + 00 |
aTranscripts showing strong signals (signal ≥ 500; |Log2-value| ≥ 1).
bOridonin/control.
cTranscripts showing weak signals (350 < signal < 500; |Log2-value| ≥ 2).
Previous studies related to miRNA expression in human pancreatic cancer are collected and summarised in Table 2 for comparison and discussion. Results showed that the expression of some miRNAs was changed dramatically after treatment with oridonin, as shown in Table 3 (20 miRNAs, including miR-205, miR-10b, miR-125b, miR-200b, miR-132, miR-320, miR-185, miR-424-5p, and miR-17-5p), which indicated that oridonin may influence BxPC-3 pancreatic cancer cells through regulating miRNAs, though verifying this hypothesis will require further investigation.
Table 2.
miRNA expression in pancreatic cancer
| miRNA | Regulation | Source | Reference |
|---|---|---|---|
| miR-17-5p | up | pancreatic cancer cell lines ( AsPC-1, KP-1 N, KP-3 and PANC-1 et al.) | [11,12] |
| miR-10a | up | 15 pancreatic cancer cell lines | [12] |
| miR-210 | up | pancreatic cancer patients | [13] |
| miR-214 | up | pancreatic cancer tissues | [14] |
| miR-15a | down | ||
| miR-107 | up | MiaPACA-2 and PANC-1 cells | [15,16] |
| miR-103 | up | ||
| miR-29a | up | ||
| miR-320 | up | ||
| miR-375 | down | Panc-1, SW1990, BxpC3 and Patu8988 | [17] |
| miR-483-3p | up | pancreatic cancer tissues | [18] |
| miR-21 | up | pancreatic cancer pecimens and 14 pancreatic cancer cell lines | [19,20] |
| miR-146a | down | Colo357 and Panc-1 | [21] |
| miR-424-5p | up | Human PDAC Tissues and PDAC Cell Lines | [22] |
| miR-155 | up | ||
| miR-221 | up | ||
| Let-7 | down | Pancreatic ductal adenocarcinoma samples | [23] |
| miR-126 | down | pancreatic tissue samples and cell lines | [24] |
| miR-132 | up | Pancreatic adenocarcinoma (PDAC) tissues | [25] |
| miR-212 | up | ||
| miR-96 | down | pancreatic cancer tissues and cell lines | [26] |
| miR-217 | down | PDAC tissues and cell lines | [27] |
| miR-494* | up | BxPC-3 cell | [28] |
| miR-140 | up | ||
| miR-148a* | up | ||
| miR-200b* | up | ||
| miR-564* | up | ||
| miR-195* | up | ||
| miR-637* | up | ||
| miR-34a | down | MIA PaCa-2 and AsPC-1 cells | [29] |
| miR-29c | down | normal pancreas and PDAC tissue | [30] |
| miR-494 | down | ||
| miR-615-5p | down | BxPC-3, CFPAC-1, SW1990, PANC-1 | [31] |
| 95 miRNA (let-7-family, miR-7, miR-92 and miR-93 et al.) | up | BxPC-3 cell | [16] |
*Passenger strand.
Table 3.
Differential expression of miRNAs in pancreatic cancer and pancreatic cancer induced by oridonin
| miRNA | Regulation | a Regulation reported in literature | Source in literature | Reference |
|---|---|---|---|---|
| miR-205 | down | up | BxPC-3 cell | [16] |
| miR-10b | down | up | BxPC-3 cell | [16] |
| miR-125b | down | up | BxPC-3 cell | [16] |
| miR-200b | down | up | BxPC-3 cell | [16] |
| miR-132 | down | up | BxPC-3 cell | [16] |
| miR-320 | down | up | MiaPACA-2 and PANC-1 cells | [15] |
| miR-185 | down | up | BxPC-3 cell | [16] |
| miR-92 | down | up | BxPC-3 cell | [16] |
| miR-183 | down | up | BxPC-3 cell | [16] |
| miR-186 | down | up | BxPC-3 cell | [16] |
| miR-125a | down | up | BxPC-3 cell | [16] |
| miR-151 | down | up | BxPC-3 cell | [16] |
| miR-224 | down | up | BxPC-3 cell | [16] |
| miR-107 | down | up | MiaPACA-2, PANC-1 and BxPC-3 cells | [15,16] |
| miR-93 | down | up | BxPC-3 cell | [16] |
| miR-103 | down | up | MiaPACA-2, PANC-1 and BxPC-3 cells | [15,16] |
| miR-20a | down | up | BxPC-3 cell | [16] |
| miR-424-5p | down | up | Human PDAC Tissues and PDAC Cell Lines | [22] |
| miR-17-5p | down | up | 14 pancreatic cancer cell lines (AsPC-1, KP-1 N, KP-3 and PANC-1 et al.) | [11] |
| miR-203 | down | up | BxPC-3 cell | [16] |
| miR-29c-3p | up | down | normal pancreas and PDAC tissue | [30] |
| miR-494 | up | down | ||
| miR-615-5p | up | down | BxPC-3, CFPAC-1, SW1990 and PANC-1 | [31] |
aRegulation reported in pancreatic cancer tissues/cells compared with normal pancreatic tissues/cells from the literature.
Target prediction and GO and KEGG pathway analyses
It has been demonstrated that one miRNA could target more than one gene, whereas some genes were targets of more than one miRNA. To predict the target mRNAs of the differentially expressed miRNAs, we performed target prediction for the differentially expressed miRNAs identified in the BxPC-3 cells using three different types of online software: TargetScan, PicTar and miRanda. The intersection of three software’s predictions was taken as the finally potential target genes.
GO and KEGG pathway analyses were performed on the target genes of the significantly differentially expressed miRNAs.
The enriched GO annotations are shown in Figure 1. The results revealed that the significantly enriched predicted target genes were involved mainly in the following categories: biological processes (e.g., signal transduction, regulation of transcription, DNA-dependent and multicellular organismal development), cellular components (e.g., cytoplasm, nucleus, membrane, integral to membrane and plasma membrane) and molecular functions (protein binding, metal ion binding and zinc ion binding).
Figure 1.
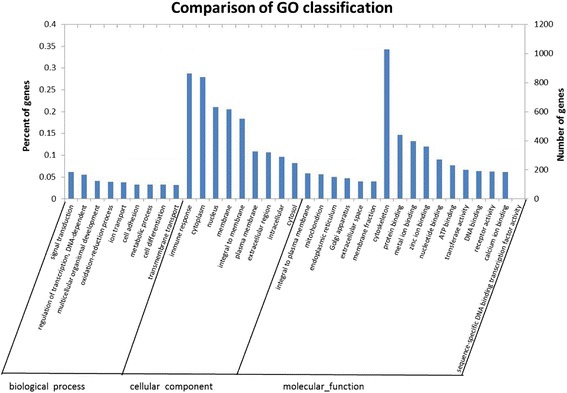
Distribution of GO categories for the predicted target genes of differentially expressed miRNAs identified in BxPC-3 cells treated with oridonin. The left vertical axis represents the percent of genes, the right vertical axis represents the number of genes and the horizontal axis represents the GO category, including biological processes, cellular components and molecular functions.
The KEGG pathway annotations of all of the target genes of the significantly differentially expressed miRNAs are shown in Table 4 (P ≥ 0.05). KEGG is a major public database of biological pathways, and significant enrichment in KEGG categories can identify differentially expressed genes involved in the main biochemical metabolic pathways and signal transduction pathways. The results presented in Table 4 revealed that the influence of BxPC-3 pancreatic cancer cells by oridonin may be related to neuroactive ligand-receptor interactions, pathways involved in cancer, MAPK signalling pathways, focal adhesion, calcium signalling pathways and other factors, prompting further study on the mechanism of pancreatic cancer inhibition by oridonin.
Table 4.
KEGG pathway annotation of the targets of differentially expressed miRNAs identified in BxPC-3 cells treated with oridonin
| Pathway Id | Pathway description | a S gene number | b TS gene number | c B gene number | d TB gene number | P value |
|---|---|---|---|---|---|---|
| 4080 | Neuroactive ligand-receptor interaction | 176 | 2158 | 206 | 2734 | 0.00899472 |
| 5200 | Pathways in cancer | 174 | 2158 | 204 | 2734 | 0.010837484 |
| 4010 | MAPK signaling pathway | 138 | 2158 | 155 | 2734 | 0.000542141 |
| 4510 | Focal adhesion | 123 | 2158 | 141 | 2734 | 0.00651097 |
| 4020 | Calcium signaling pathway | 118 | 2158 | 132 | 2734 | 0.000957212 |
| 4144 | Endocytosis | 92 | 2158 | 104 | 2734 | 0.00754512 |
| 4514 | Cell adhesion molecules (CAMs) | 79 | 2158 | 91 | 2734 | 0.03558939 |
| 4142 | Lysosome | 73 | 2158 | 79 | 2734 | 0.000942336 |
| 4640 | Hematopoietic cell lineage | 70 | 2158 | 76 | 2734 | 0.001553248 |
| 4670 | Leukocyte transendothelial migration | 69 | 2158 | 78 | 2734 | 0.0203194 |
| 4722 | Neurotrophin signaling pathway | 67 | 2158 | 76 | 2734 | 0.026182028 |
| 4660 | T cell receptor signaling pathway | 65 | 2158 | 74 | 2734 | 0.033536383 |
| 5414 | Dilated cardiomyopathy | 63 | 2158 | 68 | 2734 | 0.001810577 |
| 5410 | Hypertrophic cardiomyopathy (HCM) | 62 | 2158 | 66 | 2734 | 0.000696228 |
| 4350 | TGF-beta signaling pathway | 56 | 2158 | 62 | 2734 | 0.013956825 |
| 4512 | ECM-receptor interaction | 56 | 2158 | 63 | 2734 | 0.029353732 |
| 4912 | GnRH signaling pathway | 55 | 2158 | 62 | 2734 | 0.033519609 |
| 5215 | Prostate cancer | 52 | 2158 | 56 | 2734 | 0.004141376 |
| 5412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 52 | 2158 | 56 | 2734 | 0.004141376 |
| 4730 | Long-term depression | 47 | 2158 | 51 | 2734 | 0.009668211 |
| 5212 | Pancreatic cancer | 45 | 2158 | 50 | 2734 | 0.031740035 |
| 4720 | Long-term potentiation | 44 | 2158 | 48 | 2734 | 0.015812816 |
| 4662 | B cell receptor signaling pathway | 43 | 2158 | 48 | 2734 | 0.042316964 |
| 5211 | Renal cell carcinoma | 41 | 2158 | 45 | 2734 | 0.025499631 |
| 5220 | Chronic myeloid leukemia | 41 | 2158 | 45 | 2734 | 0.025499631 |
| 4115 | p53 signaling pathway | 39 | 2158 | 43 | 2734 | 0.034764924 |
| 5014 | Amyotrophic lateral sclerosis (ALS) | 37 | 2158 | 38 | 2734 | 0.001316211 |
| 5213 | Endometrial cancer | 32 | 2158 | 34 | 2734 | 0.015598239 |
| 520 | Amino sugar and nucleotide sugar metabolism | 26 | 2158 | 28 | 2734 | 0.046209141 |
| 51 | Fructose and mannose metabolism | 22 | 2158 | 23 | 2734 | 0.030443267 |
| 4330 | Notch signaling pathway | 20 | 2158 | 20 | 2734 | 0.008648864 |
aThe number of significantly differentially expressed genes matching KEGG pathways.
bThe total number of significantly differentially expressed genes.
cThe number of genes matching KEGG pathways.
dThe total number of genes.
Validation of miRNA microarray data via Quantitative RT-PCR
Among the significantly regulated miRNAs identified in the microarray assay, 4 miRNAs were selected for further validation via quantitative real-time PCR. The quantitative RT-PCR results showed that miR-409-3p was upregulated 2.04 times, miR-103a-3p was downregulated 1.85 times, miR-200b-3p was downregulated 2.22 times and miR-107 was downregulated 2.13 times in the oridonin treatment group compared with the control (Figure 2), which correlated well with the microarray results in Table 1.
Figure 2.
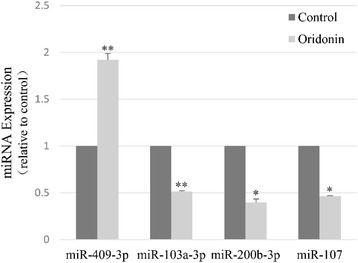
qPCR validation of a subset of miRNA microarray data. The horizontal axis represents the miRNAs, and the vertical axis represents the expression of miRNAs. The black bar represents the control group, and the grey bar represents the oridonin group. The data are expressed as the mean ± standard deviation (SD). **Significantly different from the control (p < 0.01); *different from the control (p < 0.05).
Discussion
With the discovery of miRNAs, it has been shown that miRNAs can function as endogenous posttranscriptional gene regulators through binding to the 3′ untranslated region of target mRNAs, and emerging evidence suggests that miRNAs play an important role in regulating diverse biological processes. Abnormal expression of miRNAs is associated with many diseases, such as nervous system diseases, cardiovascular disease and cancer. Several studies have demonstrated that aberrant miRNA expression is involved in pancreatic cancer (Table 2).
Pancreatic cancer is one of the most lethal malignancies, characterised by its highly metastatic potential, worst prognosis and strong resistance to chemotherapy and radiation therapy. The overall 5-year survival rate of pancreatic cancer is less than 5%. Chemotherapy and radiation therapy are the main therapeutic methods used to treat such cancers, however, these treatments produce deleterious side effects. Therefore, there is an urgent need to find safer treatments. Recently, traditional Chinese medicines have become a “hot spot” in relation to their potential anti-tumour properties, although the mechanisms of such anti-tumour effects are not clear. Some studies showed that the anti-cancer mechanisms of the active ingredients of traditional Chinese medicines may be associated with miRNAs, which can be treated as targets for cancer therapies [32-34]. Previous studies revealed that oridonin can cause cell cycle arrest, induce apoptosis and enhance the antitumour activity of gemcitabine in pancreatic cancer [35-37]. In this study, the miRNA expression was profiled in BxPC-3 human pancreatic cancer cells treated with oridonin. MicroRNA results showed that 105 miRNAs were significantly altered by oridonin treatment (Table 1). Among them, many have been reported to be associated with tumorigenesis or cancer progression. For instance, miR-424-5p (Table 3) is overexpressed in human pancreatic cancer. Down-regulation of miR-424-5p inhibits cell proliferation, migration and invasion and increases cell apoptosis in PANC-1 cells [22]. In addition, miR-17-5p, which is related to a poor prognosis, is overexpressed in pancreatic cancer [11]. Both miR-424-5p and miR-17-5p were found to be down-regulated by oridonin in our microarray data, implying that oridonin may inhibit pancreatic cancer cell proliferation, migration, invasion, and induce apoptosis by down-regulating miR-424-5p and miR-17-5p.
Four miRNAs (miR-409-3p, miR-103a-3p, miR-200b-3p and miR-107) were chosen to validate the microarray assay via quantitative real-time PCR. PCR results showed a well correlation with the microarray results, confirming the significant difference between oridonin treated and untreated cells. It has been reported that epigenetic silencing of miR-107 can regulate the expression of cyclin-dependent kinase 6 in pancreatic cancer [15]; while interfering miR-409-3p promotes tumour growth, the epithelial-to-mesenchymal transition (EMT) and bone metastasis [38]. miR-409-3p also suppresses the migration and invasion of bladder cancer T24 and 5,637 cells via targeting c-Met [39] and regulates cell proliferation and apoptosis by targeting PHF10 in SGC-7901 gastric cancer cells [40]. However, the effect of this miRNA on pancreatic cancer has rarely been described, similar to the situation for miR-103a-3p and miR-200b-3p. Based on the literature and our analysis on miRNA expression in cancer cells, we presume that these miRNAs likely play similar roles in pancreatic cancer, such as inhibiting cell proliferation, migration, invasion and inducing apoptosis. Thus, interruption of miRNA expression may be potential therapeutic targets for pancreatic cancer, although further studies are required to explore this possibility.
For further investigation, Gene Ontology analysis and KEGG pathway annotation were applied. GO enrichment analysis showed that the mRNA clusters were significantly enriched for the categories that are essential for cell survival. A total of 31 enrichment pathways for predicted target genes were listed in Table 4. Among them, the top 5 signaling pathways were neuroactive ligand-receptor interactions, Pathways in cancer, MAPK, focal adhesion and calcium signalling pathways. The results showed that 176 predicted target genes are associated with neuroactive ligand-receptor interactions, 138 genes are associated with MAPK signaling pathways, while 118 genes are associated with calcium signalling pathways. Data from previous research suggest that oridonin can enhance the antitumour activity of gemcitabine in pancreatic cancer through the MAPK-p38 signalling pathway [36] and inhibit BxPC-3 cell growth through caspase signaling pathways [41], which verified the results of KEGG pathway annotation. In conclusion, The KEGG pathway annotation revealed that BxPC-3 pancreatic cancer cells may be influenced by oridonin through these pathways and provided new research directions.
Conclusion
In conclusion, the results of the present study provide new insights into the general mechanisms underlying the suppression of BxPC-3 cells by oridonin treatment and may provide new therapeutic methods for pancreatic cancer.
Acknowledgements
This research was supported by the Zhejiang Province Project of Science Technology Department (NO.2014C33265 ), the Natural Science Foundation of Zhejiang Province (NO.LY14H160037 ) and the Natural Science Foundation of Jiangxi Province (NO. 20122BAB215042).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZG and JX Conceived and designed the experiments and drafted the manuscript. ZG and SL performed the experiments. ZG, XL and BX analysed the data. All authors read and approved the final manuscript.
Contributor Information
Zhifang Gui, Email: 934309858@qq.com.
Shuquan Li, Email: 1522716800@qq.com.
Xing Liu, Email: bjxj2005@163.com.
Bin Xu, Email: xubinmd@163.com.
Jian Xu, Email: xujian832002@163.com.
References
- 1.Ikezoe T, Chen SS, Tong X-J, Heber D, Taguchi H, Koeffler HP. Oridonin induces growth inhibition and apoptosis of a variety of human cancer cells. Int J Oncol. 2003;23(4):1187–93. [PubMed] [Google Scholar]
- 2.Wang S, Zhong Z, Wan J, Tan W, Wu G, Chen M, et al. Oridonin induces apoptosis, inhibits migration and invasion on highly-metastatic human breast cancer cells. Am J Chin Med. 2013;41(01):177–96. doi: 10.1142/S0192415X13500134. [DOI] [PubMed] [Google Scholar]
- 3.Chen RY, Xu B, Chen SF, Chen SS, Zhang T, Ren J, Xu J. Effect of oridonin-mediated hallmark changes on inflammatory pathways in human pancreatic cancer (BxPC-3) cells. World J Gastroenterol. 2014;20(40):14895–14903. doi: 10.3748/wjg.v20.i40.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharmacol Sci. 2006;101(4):267–70. doi: 10.1254/jphs.CPJ06013X. [DOI] [PubMed] [Google Scholar]
- 6.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6(1):60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 8.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102(50):18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Q, Hong A, Sheng N, Zhang X, Matejko A, Jun KY, et al. microParaflo biochip for nucleic acid and protein analysis. Methods Mol Biol. 2007;382:287–312. doi: 10.1007/978-1-59745-304-2_19. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–57. doi: 10.4161/cbt.10.8.13083. [DOI] [PubMed] [Google Scholar]
- 12.Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. 2012;19(7):2394–402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- 13.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, et al. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010;3(2):109–13. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9(3):293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Li M, Wang H, Fisher WE, Lin PH, Yao Q, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33(4):698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song S, Zhou J, He S, Zhu D, Zhang Z, Zhao H, et al. Expression levels of microRNA-375 in pancreatic cancer. Biomedical Rep. 2013;1(3):393–8. doi: 10.3892/br.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao J, Zhang S, Zhou Y, Hu X, Shao C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585(1):207–13. doi: 10.1016/j.febslet.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 19.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12(12):2171–6. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8(5):1067–74. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Vandenboom TG, 2nd, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70(4):1486–95. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu K, Hu G, He X, Zhou P, Li J, He B, et al. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19(4):739–48. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 23.Torrisani J, Bournet B, du Rieu MC, Bouisson M, Souque A, Escourrou J, et al. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20(8):831–44. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 24.Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, et al. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10(1):3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 25.Park JK, Henry JC, Jiang J, Esau C, Gusev Y, Lerner MR, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406(4):518–23. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70(14):6015–25. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 27.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31(10):1726–33. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 28.Hanoun N, Delpu Y, Suriawinata AA, Bournet B, Bureau C, Selves J, et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56(7):1107–18. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 29.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6(8):e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–52. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 31.Gao W, Gu Y, Li Z, Cai H, Peng Q, Tu M, et al. miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene. 2015;34(13):1629–1640. doi: 10.1038/onc.2014.101. [DOI] [PubMed] [Google Scholar]
- 32.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7(3):464–73. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 33.Bae S, Lee EM, Cha HJ, Kim K, Yoon Y, Lee H, et al. Resveratrol alters microRNA expression profiles in A549 human non-small cell lung cancer cells. Mol Cells. 2011;32(3):243–9. doi: 10.1007/s10059-011-1037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Hui L, Xu W, Shen H, Chen Q, Long L, et al. Triptolide modulates the sensitivity of K562/A02 cells to adriamycin by regulating miR-21 expression. Pharm Biol. 2012;50(10):1233–40. doi: 10.3109/13880209.2012.665931. [DOI] [PubMed] [Google Scholar]
- 35.Bu HQ, Liu DL, Wei WT, Chen L, Huang H, Li Y, et al. Oridonin induces apoptosis in SW1990 pancreatic cancer cells via p53- and caspase-dependent induction of p38 MAPK. Oncol Rep. 2014;31(2):975–82. doi: 10.3892/or.2013.2888. [DOI] [PubMed] [Google Scholar]
- 36.Bu HQ, Luo J, Chen H, Zhang JH, Li HH, Guo HC, et al. Oridonin enhances antitumor activity of gemcitabine in pancreatic cancer through MAPK-p38 signaling pathway. Int J Oncol. 2012;41(3):949–58. doi: 10.3892/ijo.2012.1519. [DOI] [PubMed] [Google Scholar]
- 37.Qi X, Zhang D, Xu X, Feng F, Ren G, Chu Q, et al. Oridonin nanosuspension was more effective than free oridonin on G2/M cell cycle arrest and apoptosis in the human pancreatic cancer PANC-1 cell line. Int J Nanomedicine. 2012;7:1793–804. doi: 10.2147/IJN.S29483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Josson S, Gururajan M, Hu P, Shao C, Chu GC, Zhau HE, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20(17):4636–46. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36(1):62–8. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Nie H, Wang M, Su L, Li J, Yu B, et al. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett. 2012;320(2):189–97. doi: 10.1016/j.canlet.2012.02.030. [DOI] [PubMed] [Google Scholar]
- 41.Xu B, Shen W, Liu X, Zhang T, Ren J, Fan Y, et al. Oridonin inhibits BxPC-3 cell growth through cell apoptosis. Acta Biochim Biophys Sin. 2015;47(3):164–73. doi: 10.1093/abbs/gmu134. [DOI] [PubMed] [Google Scholar]


