Abstract
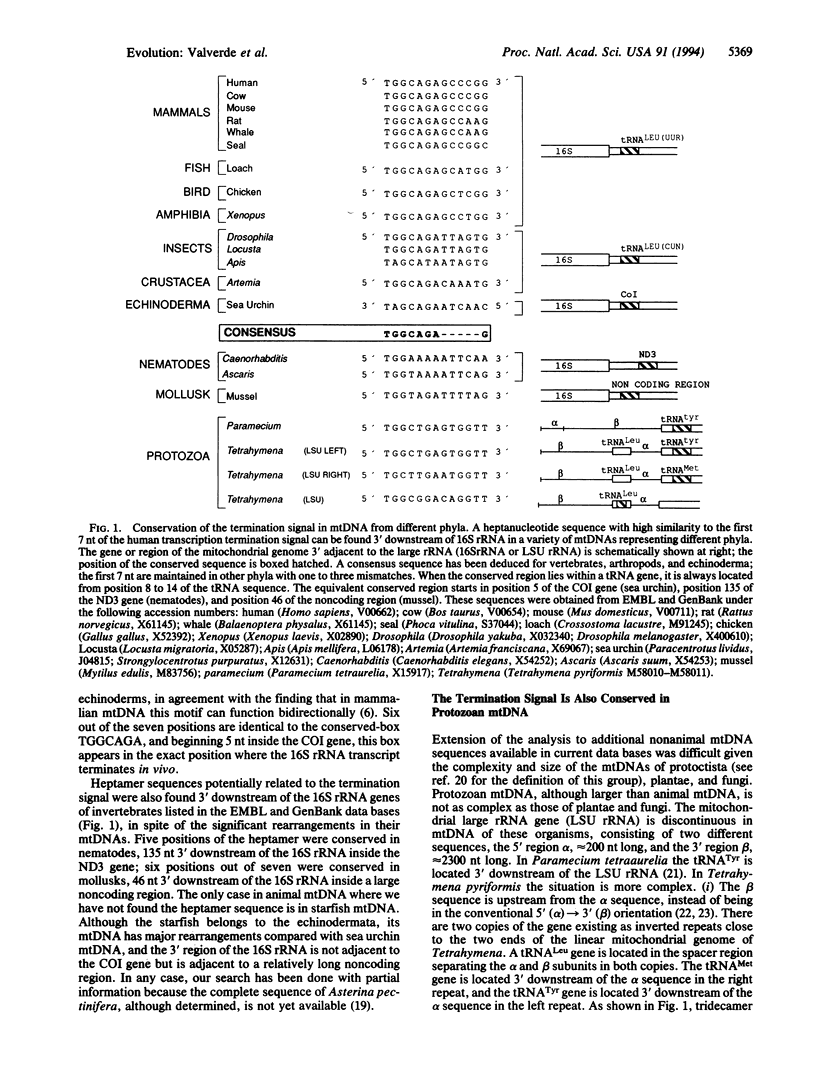
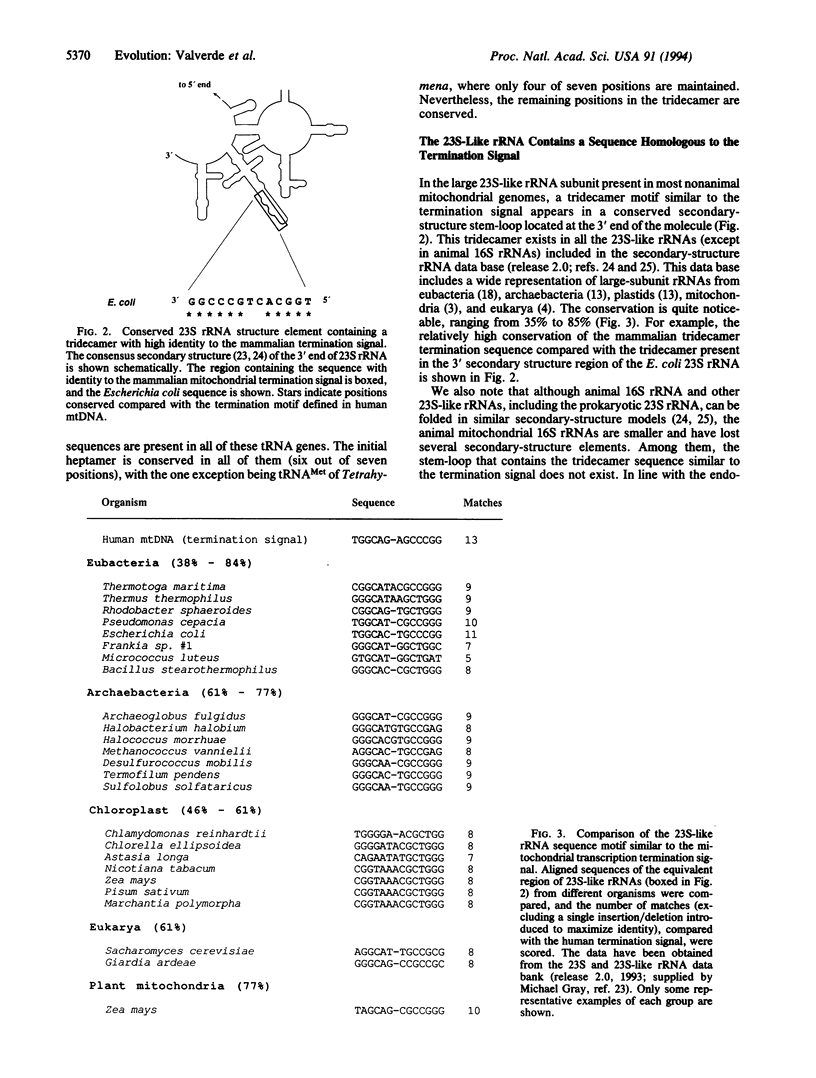
A search of sequence data bases for a tridecamer transcription termination signal, previously described in human mtDNA as being responsible for the accumulation of mitochondrial ribosomal RNAs (rRNAs) in excess over the rest of mitochondrial genes, has revealed that this termination signal occurs in equivalent positions in a wide variety of organisms from protozoa to mammals. Due to the compact organization of the mtDNA, the tridecamer motif usually appears as part of the 3' adjacent gene sequence. Because in phylogenetically widely separated organisms the mitochondrial genome has experienced many rearrangements, it is interesting that its occurrence near the 3' end of the large rRNA is independent of the adjacent gene. The tridecamer sequence has diverged in phylogenetically widely separated organisms. Nevertheless, a well-conserved heptamer--TGGCAGA, the mitochondrial rRNA termination box--can be defined. Although extending the experimental evidence of its role as a transcription termination signal in humans will be of great interest, its evolutionary conservation strongly suggests that mitochondrial rRNA transcription termination could be a widely conserved mechanism in animals. Furthermore, the conservation of a homologous tridecamer motif in one of the last 3' secondary loops of nonmitochondrial 23S-like rRNAs suggests that the role of the sequence has changed during mitochondrial evolution.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asakawa S., Kumazawa Y., Araki T., Himeno H., Miura K., Watanabe K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol. 1991 Jun;32(6):511–520. doi: 10.1007/BF02102653. [DOI] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Cantatore P., Roberti M., Rainaldi G., Gadaleta M. N., Saccone C. The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial genome of Paracentrotus lividus. J Biol Chem. 1989 Jul 5;264(19):10965–10975. [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Clayton D. A. A tridecamer DNA sequence supports human mitochondrial RNA 3'-end formation in vitro. Mol Cell Biol. 1988 Oct;8(10):4502–4509. doi: 10.1128/mcb.8.10.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Clayton D. A. In vitro transcription of human mitochondrial DNA: accurate termination requires a region of DNA sequence that can function bidirectionally. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6277–6281. doi: 10.1073/pnas.83.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Cummings D. J. Mitochondrial genomes of the ciliates. Int Rev Cytol. 1992;141:1–64. doi: 10.1016/s0074-7696(08)62062-8. [DOI] [PubMed] [Google Scholar]
- Daga A., Micol V., Hess D., Aebersold R., Attardi G. Molecular characterization of the transcription termination factor from human mitochondria. J Biol Chem. 1993 Apr 15;268(11):8123–8130. [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Elliott D. J., Jacobs H. T. Mutually exclusive synthetic pathways for sea urchin mitochondrial rRNA and mRNA. Mol Cell Biol. 1989 Mar;9(3):1069–1082. doi: 10.1128/mcb.9.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta G., Pepe G., De Candia G., Quagliariello C., Sbisà E., Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989 Jun;28(6):497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- Garesse R. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics. 1988 Apr;118(4):649–663. doi: 10.1093/genetics/118.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. The evolutionary origins of organelles. Trends Genet. 1989 Sep;5(9):294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Gray M. W., Schnare M. N. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993 Jul 1;21(13):3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Schnare M. N., Gray M. W. A compilation of large subunit (23S- and 23S-like) ribosomal RNA structures. Nucleic Acids Res. 1992 May 11;20 (Suppl):2095–2109. doi: 10.1093/nar/20.suppl.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen T. Y., Schnare M. N., Gray M. W. Sequence heterogeneity in the duplicate large subunit ribosomal RNA genes of Tetrahymena pyriformis mitochondrial DNA. J Biol Chem. 1990 Dec 25;265(36):22336–22341. [PubMed] [Google Scholar]
- Heinonen T. Y., Schnare M. N., Young P. G., Gray M. W. Rearranged coding segments, separated by a transfer RNA gene, specify the two parts of a discontinuous large subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J Biol Chem. 1987 Feb 25;262(6):2879–2887. [PubMed] [Google Scholar]
- Hess J. F., Parisi M. A., Bennett J. L., Clayton D. A. Impairment of mitochondrial transcription termination by a point mutation associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1991 May 16;351(6323):236–239. doi: 10.1038/351236a0. [DOI] [PubMed] [Google Scholar]
- Kruse B., Narasimhan N., Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989 Jul 28;58(2):391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- Montoya J., Gaines G. L., Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983 Aug;34(1):151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Bird J. W., Clayton D. A. Identification of the 3'-ends of the two mouse mitochondrial ribosomal RNAs. The 3'-end of 16 S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J Biol Chem. 1983 Aug 25;258(16):10104–10110. [PubMed] [Google Scholar]
- Yoza B. K., Bogenhagen D. F. Identification and in vitro capping of a primary transcript of human mitochondrial DNA. J Biol Chem. 1984 Mar 25;259(6):3909–3915. [PubMed] [Google Scholar]
- de Bruijn M. H. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature. 1983 Jul 21;304(5923):234–241. doi: 10.1038/304234a0. [DOI] [PubMed] [Google Scholar]


