Abstract
The bioengineering of a thermophilic enzyme starting from a mesophilic scaffold has proven to be a significant challenge as several stabilizing elements have been proposed to be the foundation of thermal stability including disulfide bridges, surface loop reduction, ionic pair networks, proline substitutions, and aromatic clusters. This study emphasizes the impact of increasing the rigidity of human carbonic anhydrase II (HCA II) via incorporation of proline residues at positions 170 and 234, which are located in surface loops that are able to accommodate restrictive main-chain conformations without rearrangement of the surrounding peptide backbone. Additionally, the effect of compactness of HCA II was examined by way of deletion of a surface loop (residues 230 through 240), which had been previously identified as a possible source of thermal stability for the hyperthermophilic CA isolated from the bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Differential scanning calorimetry analysis of these HCA II variants revealed that these structural modifications had a minimum effect on the thermal stability of the enzyme while kinetic studies showed unexpected effects on the catalytic efficiency and proton transfer rates. X-ray crystallographic analysis of these HCA II variants showed the electrostatic potential and configuration of the highly acidic loop (residues 230 and 240) plays an important role in its high catalytic activity. Based on these observations and the literature, a picture is emerging of the various components within the general structural architecture of HCA II that are key to stability. These elements could provide the blueprints for the rational thermal stability engineering of other enzymes.
Keywords: Carbonic anhydrase, protein thermal stability, differential scanning calorimetry, protein crystallography, bioengineering
Introduction
Human carbonic anhydrase II (HCA II) is zinc-containing metalloenzyme that catalyzes the reversible hydration of carbon dioxide (CO2) into bicarbonate (HCO3−) and a proton (H+) via a two-step ping-pong mechanism [1, 2]:
| (1) |
| (2) |
In the first step of the reaction, in the hydration direction, CO2 binds in a hydrophobic pocket at the base of the active site, with the carbon positioned for nucleophilic attack by a zinc-bound hydroxide (Zn-OH−) leading to the formation of HCO3−. The HCO3− is then displaced from the zinc and leaves the active site with water taking its place to form a zinc-bound water (Zn-H2O) (Eq. 1). The second step of the reaction is the transfer of a proton from Zn-H2O to the bulk solvent (B) to regenerate the Zn-OH− for the next catalytic cycle, this is achieved via a network of ordered waters in the active site (proton wire) and a proton shuttling residue His64, at the entrance of the active site cavity [3–6]. This proton transfer (PT) occurs at the rate of 106 s−1 and is the rate-limiting step of the overall maximum velocity of catalysis [6, 7].
The high catalytic activity, ease of expression, and purification of HCA II has generated interest in its use in carbon sequestration systems to reduce the amount of atmospheric CO2 emissions from the burning of fossil fuels [8, 9]. However, the denaturation of the enzyme at high temperatures and acidic pH is a resultant of the process, and therefore limits the usefulness of wild-type HCA II in these systems. As such, there is a wide interest in the bioengineering of a thermal stable HCA II variant that can withstand these harsh industrial settings [10, 11].
The rational design of thermal stable enzymes has proven to be a significant challenge in computational approaches because the proposed stabilizing elements such as core packing, surface electrostatics, and rigidity vary widely among different proteins [12–14]. Comparison of proteins isolated from thermophilic organisms to their mesophilic counterparts often reveals a sequence identity of 40 – 85% [15, 16], superimposable tertiary structure [17–23], and small differences (5 – 20 kcal mol−1) in the free energy of stabilization (ΔG) between the folded states [24]. In addition, stability studies of enzyme variants revealed that differences in ΔG as small as 3.0 – 6.5 kcal mol−1 can account for an increase in melting temperature (TM) by 12 °C [25, 26]. These small energy differences are encouraging for the design of thermal stable variants via site-directed mutagenesis, but caution must also be taken as unforeseen effects on the catalytic activity, folding, and solubility of the enzyme can occur [27, 28].
One current working hypothesis is that hyperthermophilic enzymes are more rigid than their mesophilic homologues and have a lower entropy of unfolding [27]. This is supported by numerous experimental data including frequency domain fluorometry and anisotropy decay [29], hydrogen-deuterium exchange [30–32], and tryptophan phosphorescence [33]. Proline, which has limited conformation geometry within the main-chain of a polypeptide chain and restricts the conformation of the preceding residues [34], has the lowest entropy of all the amino acids. There are a number of examples of engineered thermophilic enzymes that have utilized the concept of proline incorporation with increased stability [34–37]. Residue positions Lys170 and Glu234 in HCA II are excellent candidates for proline incorporation because they are located in β-turns, and therefore naturally adopt the restricted φ angle of proline. Thus, these sites contain the required dihedral angles seen in the i+1 position for Type I and II β-turns [38].
Additionally, loop truncation of several mesophilic proteins have been shown to increase the TM [39, 40]. HCA II has an extended loop region between residues 230–240 that is a good candidate for such a truncation. In the HCA II crystal structure this loop has a higher average Cα B-factor (thermal fluctuation) than the average for the enzyme and contains two solvent-exposed hydrophobic residues (Phe231 and Leu240) which have been previously shown to have detrimental effects on the thermal stability [11]. Chain annealing after deletion of the 230–240 loop would not be detrimental to the protein backbone, as the carbonyl of Leu229 would be sufficiently close to the amine of Met241 (2.8 Å) for ligation. Furthermore, the structural comparison of the hyperthermophilic CA isolated from the bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 (SspCA) with HCA II revealed this loop to be absent in SspCA [41].
This study describes the rigidification of HCA II via proline substitutions and surface loop deletion. These elements have only minor effects on the thermal stability, but do have a significant impact on the catalytic activity of the enzyme.
Results
Differential Scanning Calorimetry
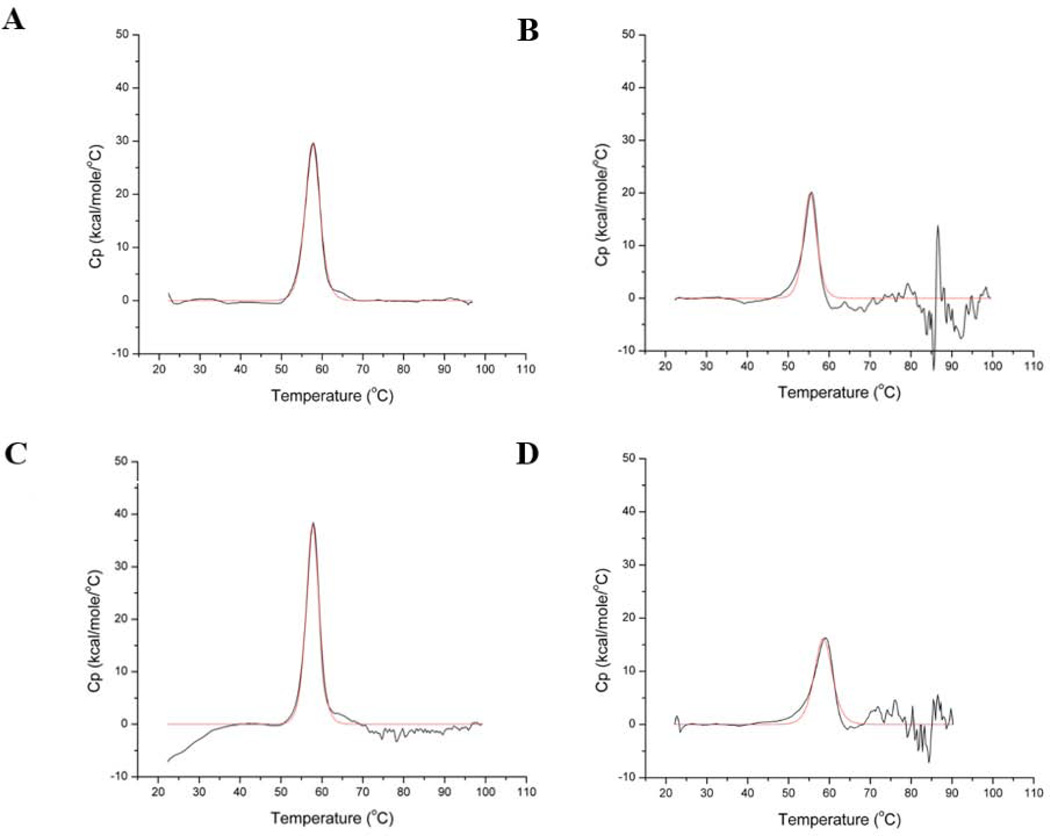
The thermograms for wild-type, K170P, E234P and Δ230–240 HCA II were obtained in triplicate at pH 7.8 before being averaged, buffer-subtracted and baseline corrected. The major transition peak for all variants displayed a single unfolding dynamic (Fig. 1). The transitions were calculated to be endothermic from Eq. 3 and were centered at the TM, with a maximum heat capacity (CP) occurring at the midpoint of the peak. A summary of the calculated TM values is given in Table 1. The calorimetric enthalpy at the transition midpoint (ΔH°M) was calculated via integration of the area under the unfolding peak and normalized to the protein molar concentrations. The identity of the van’t Hoff enthalpies (ΔHvH) to that of ΔH°M (~230 kcal mol−1) confirmed that the unfolding process is a reversible two-state transition for all variants.
Figure 1.
Thermograms of the unfolding transition for A) wild-type; B) K170P; C) E234P; and D) Δ230–240 HCA II at pH 7.8. The buffer subtracted, baseline-corrected data are shown in black with the fit to the data shown in red.
Table 1.
Thermal Stability and Catalytic Measurements
| Enzyme | TM (°C)a |
kcatex/KeffS (M−1µs−1)b |
kB (µs−1)c |
pKa ZnOH−c | pKa His64c |
|---|---|---|---|---|---|
| wild-type HCA II | 57.1 (± 0.1) | 120 (± 5) | 1.2 (±0.2) | 6.9 (± 0.1) | 7.2 (± 0.1) |
| K170P | 55.7 (± 0.1) | 63 (± 3) | 1.4 (±0.3) | 6.7 (± 0.1) | 6.4 (± 0.1) |
| E234P | 57.9 (± 0.1) | 56 (± 2) | 3.1 (± 0.6) | 7.2 (± 0.1) | 6.2 (± 0.1) |
| Δ230–240 | 58.9 (± 0.2) | 66 (± 3) | 1.0 (± 0.6) | 6.7 (± 0.2) | 5.8 (± 0.2) |
There were only modest measured increases in the TM for the E234P and Δ230–240 HCA II (TM ≈ 58 and 59 °C, respectively; Figs. 1C and D) compared to wild-type HCA II (TM ≈ 57 °C; Fig. 1A). Surprisingly, K170P HCA II, showed a slight lowering in stability with a TM ≈ 56 °C (Fig. 1B). The minor increase of E234P and decrease of K170P HCA II TMs do not follow the value reported with a proline substitution at residue L240P HCA II, which increased the thermal stability to 60 °C [42].
Catalytic Activity
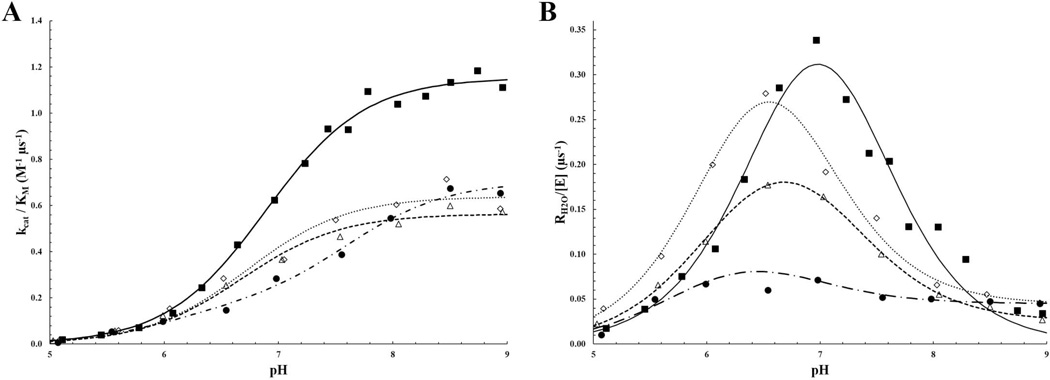
The pH profiles for the two rate constants, kcat/KM and RH2O, were determined by measuring the exchange of 18O-label between CO2 and water using mass spectrometry (Eqs. 6 and 7). The rates of catalyzed interconversion between CO2 and HCO3−, as measured by kcat/KM (Eq. 8) for K170P, E234P and Δ230–240 HCA II were all measured to be ~60 M−1 µs−1, an approximate 2-fold decrease compared to wild-type HCA II (Fig. 2A; Table 1). The measured rate constant for PT, kB, from the ZnH2O through the water network to His64 was determined by a fit from Fig. 2B curve to Eq. 9 and showed that K170P HCA II PT was comparable to that of wild-type HCA II (~1.4 µs−1), whereas E234P had a ~2.5 fold increase in kB (~3.1 µs−1) (Table 1). Interestingly, the pH profile for Δ230–240 displayed a broad curve that is indicative of a double-ionization fit that describes a kB rate that is similar to wild-type HCA II, but with larger uncertainty in the maximum rate (Table 1). All three variants revealed a pKa for the ZnOH− similar to wild-type HCA II (~7.0), but the pKa range for the proton shuttle residue, His64, is significantly different (5.8 – 6.4 for the variants compared to 7.2 for wild-type HCA II; Table 1).
Figure 2.
pH profiles for A) catalytic efficiency (kcat/KM) and B) proton transfer rate (RH2O/[E]) for the HCA II variants. Wild-type is shown as a closed square, K170P as an open diamond, E234 as an open triangle and Δ230–240 HCA II as a closed circle.
X-ray Crystallography
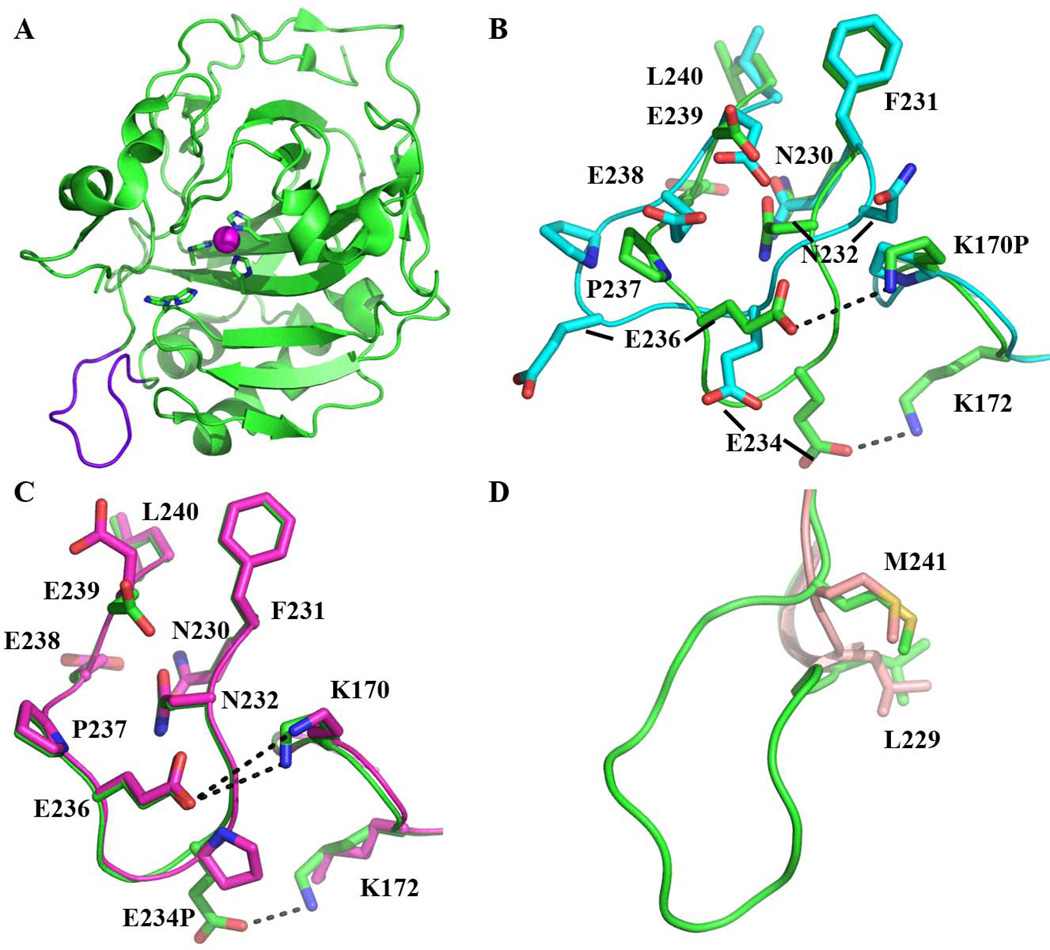
The K170P and Δ230–240 HCA II crystallized in conditions similar to that of the wild-type HCA II [43], but the crystal growth took longer (1 – 3 months) and formed in the orthorhombic P212121 space group, whereas wild-type HCA II crystals usually grow in one week and form in a monoclinic P21 space group. The K170P crystals grew using a protein concentration of 10 mg/mL (the same as wild-type), whereas Δ230–240 required a 5-fold higher concentration of 55 mg/mL. The change in space group of the K170P and Δ230–240 HCA II is most likely a direct consequence of a conformation change in the 230–240 loop in K170 HCA II and its deletion in the Δ230–240 HCA II, as in the wild-type HCA II monoclinic P21 structure this loop is involved in a crystallographic contact (Fig. 3A). The E234P variant crystallized in similar conditions, concentration, time, and space group as compared to wild-type HCA II, and also had no effect on the conformation of the 230–240 loop. The HCA II variants diffracted to a maximum resolutions of 1.35 Å for Δ230–240, 1.52 Å for E234P and 1.60 Å for K170P HCA II. A summary of the collected diffraction and final refinement statistics is shown in Table 2.
Figure 3.
Topology of the 230–240 loop in the HCA II variants. A) Cartoon view of wild-type HCA II (green) with the 230–240 loop highlighted in purple. The Zn2+ (magenta sphere) with the coordinating His residues and the proton shuttle residue, His64 shown in stick. B) Comparison of the 230–240 loop in the K170P (blue) to wild-type (green) HCA II. Side-chains are shown in stick and are as labeled. Residues G233 and G235 are not labeled for clarity. C) Comparison of the 230–240 loop in E234P (light purple) to wild-type (green) HCA II. Residues are as labeled with potential salt bridge interactions shown as a dashed line. D) Ligation between residues L229 and M241 in Δ230–240 (light pink) showing the deletion of the loop compared to wild-type HCA II (green) HCA II.
Table 2.
X-ray crystallographic data set and refinement statistics for K170P, E234P and Δ230–240 HCA II.
| K170P | E234P | Δ230–240 | |
|---|---|---|---|
| PDB Accession Number | 4QK1 | 4QK2 | 4QK3 |
| Wavelength (Å) | 1.5418 | 1.5418 | 0.9177 |
| Spacegroup | P212121 | P21 | P212121 |
| Unit cell parameters (Å; °) | a = 42.0, b = 69.0, c = 73.6; β = 90 | a = 42.2, b = 41.1, c = 71.9; β = 104.3 | a = 42.1, b = 69.0, c = 74.2; β = 90 |
| Resolution | 20.0 – 1.60 (1.66 – 1.60)* | 20.0 – 1.52 (1.57 – 1.52) | 20.0 – 1.35 (1.40 – 1.35) |
| Total number of measured reflections | 179331 | 259705 | 168087 |
| Total number of unique reflections | 27184 | 33910 | 44710 |
| Rsyma(%) | 10.4 (43.9) | 8.2 (24.2) | 12.0 (48.0) |
| I / σ I | 11.4 (3.2) | 25.8 (4.4) | 11.2 (2.0) |
| Completeness (%) | 93.9 (99.8) | 93.4 (89.1) | 91.9 (93.8) |
| Redundancy | 6.6 (6.8) | 7.7 (7.7) | 3.8 (3.3) |
| Rcryst (%) | 17.7 | 15.9 | 21.7 |
| Rfree (%) | 22.3 | 17.8 | 25.0 |
| Residue Number | 256 | 257 | 246 |
| Number of water molecules | 195 | 275 | 167 |
| r.m.s.d.: Bond lengths (Å); angles (°) | 0.011; 1.486 | 0.008; 1.215 | 0.010; 1.440 |
| Coordinate error (Å) | 0.3 | 0.2 | 0.2 |
| Ramachandran statistics (%): Most favored; allowed | 94.9; 5.1 | 96.9; 3.1 | 94.6; 5.4 |
| Average B-factors (Å2): All; main-; side-chain; solvent | 28.4; 25.4; 30.2; 32.9 | 17.0; 13.0; 17.4; 29.5 | 21.9; 17.6; 23.6; 34.5 |
Rsym is defined as ∑hkl ∑i |(Ii(hkl)−<Ii(hkl) >|/ ∑hkl∑iIi(hkl) × 100, where Ii(hkl)is the intensity of an individual reflection and <Ii(hkl) > is the average intensity for this reflection; the summation is over all intensities.
Rcryst = (∑|Fo − Fc|/∑ |Fo|) × 100
Rfree is calculated in the same way as Rcryst except it is for data omitted from refinement (5% of reflections for all data sets).
Values in parentheses are for the highest resolution shell.
The initial mFo – DFc omit maps revealed positive density (>4σ) around the K170 and E234 amino acid sites and after building as proline side chains, the refined 2mFo – DFc maps unambiguously confirmed the incorporation of a proline residues at positions 170 and 234, with a maximum electron density of 0.32 and 0.46 e/Å3, respectively. Likewise, the deletion of the surface loop between residues 230 and 240 was seen as negative density (>4σ) in the initial mFo – DFc omit map in the region between Leu229 and Met241, confirming the Δ230–240 HCA II variant. After the deletion of the loop in the model and subsequent rounds of refinements, excellent electron density in the final 2mFo – DFC map for the ligation of Leu229 and Met241 was observed with a maximum electron density of 0.55 e/Å3, comparable to that of the surrounding peptide backbone.
Superimposition of the wild-type HCA II (PDB: 3KS3; [43]) onto E234P HCA II (Fig. 3C) revealed an r.m.s.d. of main-chain of 0.2 Å, indicating no change in the main-chain conformation. However, K170P HCA II (Fig. 3B) exhibited an alternative 230–240 loop main-chain conformer with a mean r.m.s.d. of 2.2 Å. This conformational loop change is most likely a direct consequence of the K170P substitution, as the basic lysine residue is involved in electrostatic interactions with the 230–240 acidic loop in wild-type HCA II. This would also explain why the observed space group changes from monoclinic to orthorhombic. All three structures, however, displayed an overall topology similar to that of the wild-type HCA II with an r.m.s.d. of ~0.2, 0.3 and 0.7 Å for the E234P, Δ230–240 and K170P HCA II Cα atoms, respectively. The incorporation of a proline residue at position 234 increased the overall rigidity of the 230–240 loop by ~9% compared to the wild-type HCA II structure as measured by the relative B-factors and this correlates well to the enhanced thermal stability (Fig. 1; Table 1). For comparison, the relative B-factors of the Cα backbone in the 230–240 loop for the K170P HCA II variant increased by ~33% compared to wild-type HCA II which also correlates to its apparent increased flexibility and loss in thermal stability. The proline substitution at residue 170 did have a slight effect on the rigidity (~2% decrease in the relative Cα backbone B-factors compared to wild-type) in the region between residues 167 and 173.
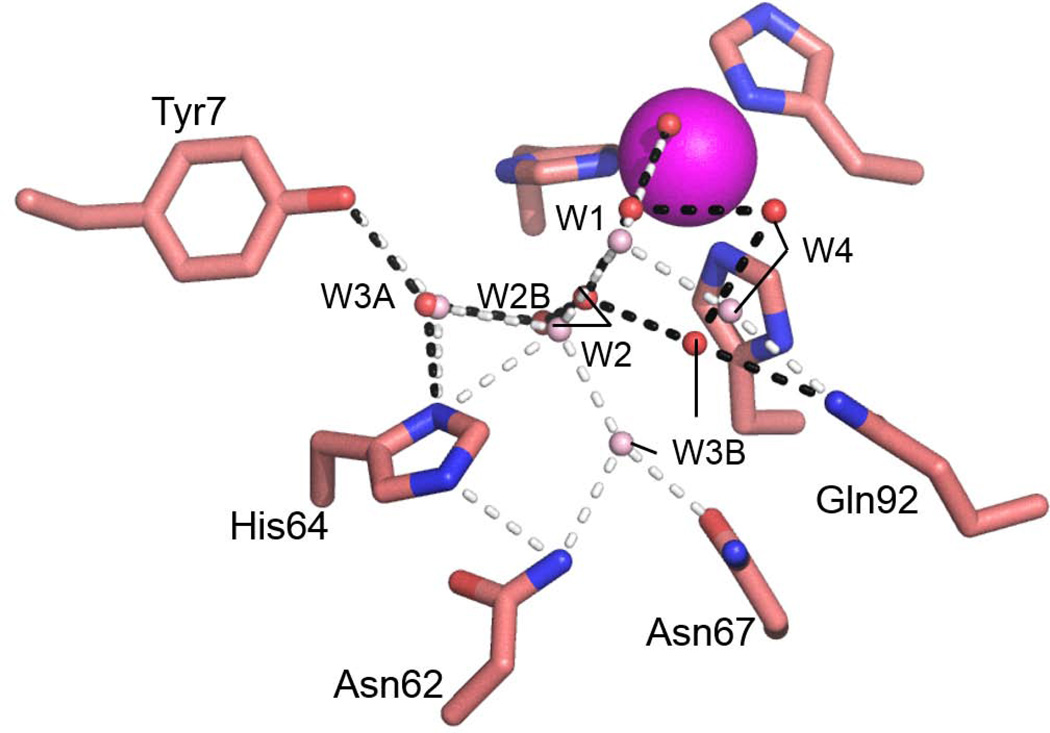
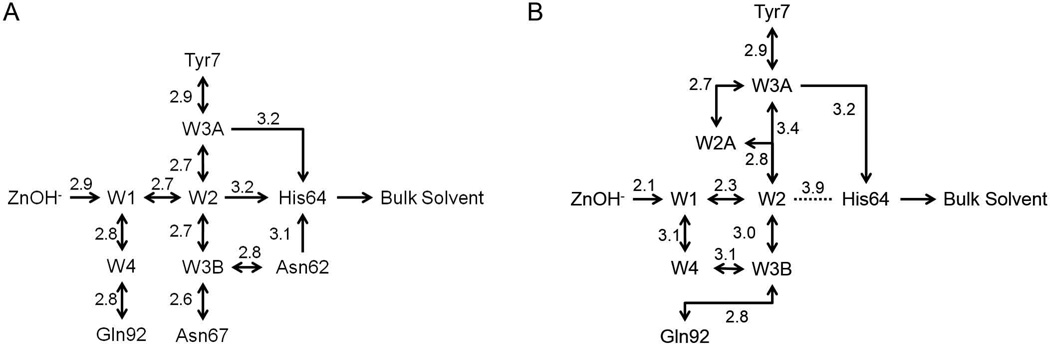
Comparison of the active site residues and water molecules involved in the PT network for K170P and E234P HCA II revealed very little deviation in their positioning relative to wild-type HCA II, and therefore provided no structural evidence for their altered catalytic efficiencies and PT rates. The Δ230–240 HCA II active site water molecules W1 and W2 were in approximate identical locations (0.2 Å) compared to wild-type (Fig. 4). There was a significant shift in the positioning of W3B (2.8 Å), which interacts with Asn62 and Asn67 in wild-type HCA II, to form interactions with Gln92 and W4 in the Δ230–240 HCA II active site. W4 was also repositioned by 2.2 Å resulting in loss of a hydrogen bond with Gln92 as compared to wild-type HCA II. Interestingly, the shifts seen for W1 and W2 in the Δ230–240 HCA II active site places W2 too far away from His64 for a potential hydrogen bond (3.9 Å; Fig. 5).
Figure 4.
Active site water network configuration in Δ230–240 (red spheres) overlaid with the wild-type (light pink spheres) HCA II. Potential hydrogen bond interactions for Δ230–240 are shown as black dashed lines whereas the white dashed lines represent the hydrogen bonds observed in wild-type HCA II Residues and water molecules are as labeled.
Figure 5.
Schematic of the proton transfer network for A) wild-type and B) Δ230–240 HCA II. Hydrogen bond interactions (not drawn to scale) are depicted as arrows with the distance between atoms label in Å.
Discussion
Previous studies have proposed that thermophilic enzymes owe their stability in part to the overall compactness and rigidity compared to their mesophilic homologues. For HCA II, this was shown to be true when a proline at position 240, L240P, was engineered into the enzyme and enhanced the thermal stability (ΔTM ≈ 3 °C) compared to wild-type HCA II [11, 42]. Additionally, the recently determined X-ray crystallographic structure of SspCA [41] implied that the absence of the 230–240 loop (HCA II numbering) may be linked to its extreme thermal stability (TM > 100 °C). The modest ~2 °C increase in melting temperature of Δ230–240 HCA II does support the hypothesis that thermophilic proteins display a more compact topology. However, this cannot account for the extreme thermal stability reported for SspCA (TM > 100 °C; [41]), but it may arise from other elements including the incorporation of ionic clusters on the surface and an extensive dimeric interface (HCA II being a monomer). Furthermore, the increase of the Δ230–240 HCA II stability is unremarkable when compared to previously reported HCA II variants which increased the TM by decreasing the surface hydrophobicity (ΔTM ≈ 7 °C) [11], and the incorporation of a conserved disulfide bridge (ΔTM ≈ 14 °C) [10].
The DSC studies of the E234P variant revealed only a modest 1 – 2 °C increase of TM compared to wild-type HCA II (Fig. 1; Table 1). The enhanced thermal stability is consistent with previous studies [11, 42]), but may imply that the greatest stabilizing effect may arise if the target residue is nonpolar. In contrast, the K170P variant has a TM compared to wild-type HCA II (Fig. 1; Table 1) despite a decreased thermal fluctuation in the 167–173 loop, as determined from B-factor analysis. The loss in thermal stability of K170P HCA II can be associated with the increase in flexibility of the 230–240 loop via loss of the tethering to the acidic loop by the basic residue (Fig. 3B). Site-directed mutagenesis of Lys172 may verify this hypothesis as it seems to provide potential electrostatic interactions with the 230–240 loop in addition to Lys170. Taken together, while it seems that the thermal stability of HCA II can be enhanced via rigidification and compression of the accessible surface area, there also seems to be some dependence on the surface polarity.
Measurement of the catalytic activity via 18O mass spectrometry of the HCA II variants revealed a two-fold decrease in kcat/KM compared to wild-type HCA II (~60 compared to 120 M−1 µs−1, respectively) with varying rates in PT (Fig. 2; Table 1). Additionally, the pKa of the active site was measured and shown to be altered in the variants as compared to wild-type HCA II, presumably as a direct effect of either loss in the 230–240 loop acidity in the E234P and Δ230–240 variants or reconfiguration of the loop around the peripheral of the active site with the K170P variant. These altered ionization constants for His64 could explain the increased PT rates for E234P HCA II, as a deprotonated His64 would be better suited to accept a proton from the water network. The lowered catalytic efficiency of the HCA II variants may be associated with these lowered pKa values for His64 by having an effect on the binding affinity and/or coordination of substrate molecules in the active site. While this effect is minimum in the positioning of the water molecules within the active site of the K170P and E234P variants, deletion of the acidic loop between residues 230 and 240 resulted in a distorted PT water network that included shifts in the positioning of W1, W2, W3B and W4, as well as the inclusion of an additional water molecule (W2A; Fig. 4). The W2A molecule shortens the hydrogen bond distance between W2 and W3A from 3.4 Å in wild-type HCA II (Fig. 5A) to ~2.8 Å in Δ230–240 HCA II (Fig. 5B). PT between W3A and His64 can then potentially occur at a distance (3.2 Å) similar to that observed in wild-type HCA II (Fig. 5). As seen in the RH2O pH profile (Fig. 2B), the rate of PT in the Δ230–240 HCA II variant has less of a dependence on the pH of the solvent and may reconfigure its PT water network in accordance with its environment. Further crystallographic studies at varying pH are needed to elucidate this unique water network.
Conclusions
The bioengineering of enzymes to meet specific criteria can be a valuable asset in a variety of industrial and medical systems. The ever increasing demand for a carbon sequestration agent to help combat global warming and the need for a reliable biomedical catalyst has fueled research into the rational design of a thermal stable variant of HCA II without detriment to high catalytic activity. Through superimposition of thermophilic proteins onto their mesophilic counterparts, several elements have been proposed to provide stability including a more rigid and compact surface, increased ionic networks on the enzyme surface and the inclusion of disulfide bridges and aromatic clusters within the enzyme core [12–14, 44, 45]. The extent to which these stabilizing elements contribute to the overall stability, however, fluctuates among different proteins. This study suggests that the stability of HCA II can be enhanced by rigidification of surface loops via incorporation of proline residues as well as increasing the surface compactness via loop deletion. The positioning of these alterations, however, may have unforeseen effects on the catalytic activity of HCA II as the acidic loop between residues 230 and 240 seems to modulate the pKa of the active site. Reduction of the electrostatic potential in this loop via the E234P substitution and reconfiguration of the loop through the K170P substitution resulted in decreased catalytic efficiency and altered PT rates whereas deletion of the 230–240 loop resulted in a complete reorganization of the active site PT water network. The successful biophysical analysis of HCA II variants containing the aforementioned thermal stabilizing elements could provide valuable insight into protein stability and promote significant developments in bioengineering computational techniques.
Materials & Methods
Protein Expression and Purification
HCA II cDNA containing the K170P and E234P mutations were prepared from an expression vector containing the enzyme coding region [46] via site-directed mutagenesis using the Stratagene QuikChange II kit and primers designed from Invitrogen. The plasmid cDNA expressing the loop deletion between residues 230 and 240 in HCA II (Δ230–240 HCA II) was created utilizing the PCR-mediated DNA deletion method [47]. In short, two primers (Primers A and B) were designed such that Primer A is the reverse complement of a sequence corresponding to 20 bases upstream of the sequence to be deleted followed by a reverse complimentary sequence 20 bases downstream of the deletion site. Primer B was designed in the same fashion but corresponding to the complementary strand; that is, the sequence of Primer B was identical to the plasmid primary lacking the sought deletion. The corresponding cDNA for each variant was transformed into E. coli XL1-Blue super-competent cells, which were then confirmed by DNA sequencing of the entire coding region.
The verified cDNA for each HCA II variant was transformed into E. coli BL21(DE3) cells in 1L of 2 × Luria broth containing ~0.1 mg/mL ampicillin and grown at 37 °C until a turbidity of ~0.6 measured at 600 nm was reached. Protein production was induced via the addition of ~0.1 mg/mL isopropyl β-D-1-thiogalactopyranoside (IPTG) and metal inclusion in the enzyme active site was insured via addition of ~1 mM zinc sulfate (final concentrations). The cells were incubated for an additional three hours and harvested by centrifugation for 12 minutes at 4000 rpm in a JA-10 rotor. The resulting pellet was suspended in 200 mM sodium sulfate, 100 mM Tris-HCl, pH 9.0 and lysed by addition of hen egg white lysozyme. Cellular debris was removed via addition of DNaseI followed by centrifugation for 75 mins at 15000 rpm in a JA-20 rotor. The resulting lysate was filtered using a 0.8 µm syringe filter (Amicon) and purified on an affinity column containing an agarose resin coupled with p-(aminomethyl)-benzene-sulfonamide, a tight-binding inhibitor of HCA II [48]. The bound HCA II variants were then eluted with 400 mM sodium azide, 100 mM Tris-HCl, pH 7.0 followed by concentration and buffer-exchanging in 50 mM Tris-HCl, pH 7.8 using Amicon Ultra centrifuge filters (10 kDa MWCO) at 8000 rpm in 12 min integrals using a JA-20 rotor.
Differential Scanning Calorimetry
The DSC experiments were performed to assess the thermal stability of the HCA II variants under near-physiological conditions using a VP-DSC calorimeter (Microcal, Inc., North Hampton, MA) with a cell volume of ~0.5 mL. The HCA II samples (6–10 µM) were extensively buffer-exchanged into 50 mM Tris-HCl, pH 7.8 and then degassed, while stirring, at 16 °C for 20 minutes prior to data collection. The DSC thermograms were collected in triplicate from 20 to 100 °C with a scan rate of 60 °C/h. The calorimetric enthalpies of unfolding were calculated by integrating the area under the peaks in the thermograms after adjusting the pre- and post-translation baselines. The thermograms were fit to a two-state reversible unfolding model to obtain van't Hoff enthalpies (ΔHvH) of unfolding.
The HCA II variant melting temperature (TM) values were obtained from the midpoints of the DSC curves, indicating a two-state transition. The difference in Gibbs free energy (ΔG°) at a given temperature, T, was calculated via [49]:
| (3) |
Where ΔH°M is the calorimetric enthalpy at TM and ΔCP is the observed change in heat capacity between the folded and unfolded states. The denaturation enthalpies (ΔH°) and entropies (ΔS°) were calculated at a given temperature using the Kirchoff's law equations [43]:
| (4) |
| (5) |
Thermograms for all samples were obtained in triplicate and then averaged to obtain the final profile, which was then used for reference subtraction and data analysis.
Kinetic Studies
The 18O exchange method is based on mass spectrometric measurements using a membrane inlet of the depletion 18O from CO2 [50]. The isotopic content of CO2 in solution is measured when it passes across a membrane and into an Extrel EXM-200 mass spectrometer. The measured variable is the atom fraction of 18O in CO2. The first step of catalysis has a probability of reversibly labeling the ZnOH− with 18O (Eq. 6). During the next step the 18OH− can be protonated and results in the release of H218O to the bulk solvent where it is essentially infinitely diluted by H216O (Eq. 7). In this process, His64 acts as a proton shuttle [5].
| (6) |
| (7) |
The 18O-exchange method obtains two different rates at chemical equilibrium [50]: R1, which is the rate of exchange of CO2 and HCO3− (Eq. 8); and RH2O, which is the rate of release of H218O from the enzyme (Eq. 9). In Eq. 8, kcatex is the rate constant for maximal conversion between substrate and product while KeffS is the effective binding constant of substrate ([S] is the concentration); [S] can be either CO2 or HCO3− depending on the direction of the reaction. The ratio expressed in Eq. 8 of kcatex/KeffS is in principle the same as kcat/KM obtained under steady state conditions.
| (8) |
In the second step of catalysis, the rate RH2O is the part of 18O exchange that is dependent on the rate of proton transfer from His64 to the labeled ZnOH− (i.e. in the dehydration direction) [5]. Eq. 9 shows the relationship between kB, the rate constant for proton transfer to ZnOH− and (Ka)donor and (Ka)ZnH2O that are the ionization constants for the proton donor and ZnH2O, respectively.
| (9) |
All enzyme kinetic measurements were done at 25 °C in the absence of buffer using a total substrate concentration (all species of CO2) of 25 mM. Kinetic constants and ionization constants shown in Eqs. 8 and 9 were determined through nonlinear least squares methods (Enzfitter, Biosoft).
Crystallization and Diffraction Collection
Crystal trays were prepared for the K170P and E234P HCA II variant with a 1:1 ratio of the reservoir solution and the protein sample (10 mg/mL) in 5 µL drops utilizing the hanging drop vapour diffusion method. The reservoir solutions contained 500 µL of 1.4 M sodium citrate, 50 mM Na2HPO4, pH 8.0 for the K170P HCA II variant and 1.4 M sodium citrate, 50 mM Tris-HCl, pH 7.8 for the E234P variant. Crystal formation for K170P HCA II appeared after 3 months, whereas crystals for E234P HCA II appeared within one week. Crystal trays were prepared for Δ230–240 HCA II loop deletion variant in 1:1 ratio of the reservoir solution containing 1.6 M sodium citrate, 100 mM Tris-HCl, pH 7.8 and protein sample at 50 mg/mL in 5 µL drops utilizing the hanging drop vapour diffusion method. Crystal formation occurred within one week.
Diffraction data for K170P and E234P HCA II were collected on an in-house Rigaku R-Axis IV++ image plate detector with a RU-H3R rotating Cu anode (Kα = 1.5418 Å) operating at 50 kV and 22 mA. The X-rays were focused using Osmic optics, followed by a helium-purged beam path. The crystal-to-detector distance was 80 mm for K170P and E234P HCA II. Each image was collected for five minutes with 1° oscillations. HKL2000 [51] was used to integrate, merge and scale the data sets. K170P HCA II crystallized in the orthorhombic space group P212121 whereas E234P HCA II crystallized in the monoclinic space group P21. The crystals diffracted to 1.60 and 1.52 Å resolution for K170P and E234P, respectively. The diffraction and refinement statistics for K170P and E234P HCA II are summarized in Table 2.
Diffraction data for Δ230–240 HCA II was collected on the F1 beamline at Cornell High Energy Synchrotron Source (CHESS F1; λ = 0.9177 Å) on an ADSC Q-270 detector using the microfocused beam. The crystal-to-detector distance was 150 mm. Each image was collected for 10 seconds with 0.5° oscillations. HKL2000 [51] was used to integrate, merge and scale the data sets. Δ230–240 HCA II crystallized in the orthorhombic space group P212121 and diffracted to a h 1.35 Å resolution. The diffraction and refinement statistics for Δ230–240 HCA II are summarized in Table 2.
Phase Determination and Structure Refinement
Initial phases for K170P and E234P HCA II were calculated using molecular replacement via Phaser [52] using the wild-type HCA II structure, with the residues at 170 and 234 replaced with alanine, respectively (PDB: 3KS3; [43]). 5% of the reflections were set for Rfree calculations. Initial phases for Δ230–240 HCA II variant were determined using wild-type HCA II. PHENIX.REFINE [53] was used in cycles of restrained refinement of the molecular model, alternating with manual building using COOT [54]. Initial mFo - DFc phase maps revealed positive density at position 170 and 234 sidechain deletions, while the Δ230–240 loop deletion variant revealed negative density for the deleted residues 229 and 241. Subsequent refinements showed excellent electron density in the 2mFo - DFc maps for the K170P and E234P HCA II (Fig. 1A and B) as well as for the newly formed peptide linkage in Δ230–240 HCA II. The final Rcryst and Rfree values are shown in Table 2. MOLPROBITY [55] was used to assess the quality of the final model. All structural figures were created in PyMOL [56].
B-factor Analysis
The thermal fluctuations of the loop regions encompassing residues 167–173 and 230–240 in all variants were compared via normalization to the corresponding residue with the highest Cα B-factor in each region. The B-factors of the remaining residues were converted to the percent of rigidification compared to the most flexible residue and the mean percentage was then calculated for each variant. These rigidification percentages of each variant was compared to HCA II in either the monoclinic spacegroup (PDB: 2ILI; [57]) or the orthorhombic spacegroup (PDB: 1UGG; [58]) for E234P and K170P HCA II, respectively, to minimize B-factors differences arising from resolution and spacegroup dissimilarities.
Acknowledgements
This research has partially been funded by the NIH (GM25154). The authors would like to thank the Center of Structural Biology for support of the X-ray facility at UF and the beamline scientists at the F1 station at CHESS for their technical support during data collection.
Abbreviations
- CO2
carbon dioxide
- Cp
specific heat
- DSC
differential scanning calorimetry
- H+
proton
- HCA II
human carbonic anhydrase II
- HCO3−
bicarbonate
- SspCA
Sulfurihydrogenibium yellowstonense YO3AOP1 carbonic anhydrase
- TM
melting temperature
- Zn-H2O
zinc-bound water
- Zn-OH−
zinc-bound hydroxide
- ΔG
Gibb’s free energy
Footnotes
Enzymes
Human carbonic anhydrase II (EC 4.2.1.1)
Database
Structural data are available in the PDB database under the accession numbers 4QK1, 4QK2 and 4QK3.
References
- 1.Aggarwal M, Boone CD, Kondeti B, McKenna R. Structural annotation of human carbonic anhydrases. J Enzyme Inhib Med Ch. 2013;28:267–277. doi: 10.3109/14756366.2012.737323. [DOI] [PubMed] [Google Scholar]
- 2.Boone CD, Pinard M, McKenna R, Silverman D. Catalytic Mechanism of α-Class Carbonic Anhydrases: CO2 Hydration and Proton Transfer. In. In: Frost SC, McKenna R, editors. Carbonic Anhydrase: Mechanism, Regulation, Links to Disease, and Industrial Applications. New York: Springer; 2014. pp. 31–52. [DOI] [PubMed] [Google Scholar]
- 3.Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase I. Stop-flow kinetic studies on the native human isoenzyme B and C. J Biol Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 4.Silverman DN, Lindskog S. The catalytic mechanism of carbonic anhydrase: implications of a rate-limiting protolysis of water. Acc Chem Res. 1988;21:30–36. [Google Scholar]
- 5.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28:7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 6.Steiner H, Jonsson BH, Lindskog S. Catalytic Mechanism of Carbonic-Anhydrase - Hydrogen-Isotope Effects on Kinetic-Parameters of Human C Isoenzyme. Eur J Biochem. 1975;59:253–259. doi: 10.1111/j.1432-1033.1975.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 7.Silverman DN, McKenna R. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc Chem Res. 2007;40:669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 8.Boone CD, Gill S, Habibzadegan A, McKenna R. Carbonic anhydrases and their industrial applications. Curr Topics Biochem Res. 2013;14:1–10. [Google Scholar]
- 9.Boone CD, Gill S, Habibzadegan A, McKenna R. Carbonic Anhydrase: An Efficient Enzyme with Possible Global Implications. Int J Chem Eng. 2013;2013:1–6. [Google Scholar]
- 10.Boone CD, Habibzadegan A, Tu C, Silverman DN, McKenna R. Structural and catalytic characterization of a thermally stable and acid-stable variant of human carbonic anhydrase II containing an engineered disulfide bond. Acta Crystallogr D. 2013;69:1414–1422. doi: 10.1107/S0907444913008743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher Z, Boone CD, Biswas SM, Venkatakrishnan B, Aggarwal M, Tu C, Agbandje-McKenna M, Silverman D, McKenna R. Kinetic and structural characterization of thermostabilized mutants of human carbonic anhydrase II. Protein Eng Des Sel. 2012;25:347–355. doi: 10.1093/protein/gzs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filikov AV, Hayes RJ, Luo P, Stark DM, Chan C, Kundu A, Dahiyat BI. Computational stabilization of human growth hormone. Protein Sci. 2002;11:1452–1461. doi: 10.1110/ps.3500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Permyakov SE, Makhatadze GI, Owenius R, Uversky VN, Brooks CL, Permyakov EA, Berliner LJ. How to improve nature: study of the electrostatic properties of the surface of alpha-lactalbumin. Protein Eng Des Sel. 2005;18:425–433. doi: 10.1093/protein/gzi051. [DOI] [PubMed] [Google Scholar]
- 14.Strickler SS, Gribenko AV, Keiffer TR, Tomlinson J, Reihle T, Loladze VV, Makhatadze GI. Protein stability and surface electrostatics: a charged relationship. Biochemistry. 2006;45:2761–2766. doi: 10.1021/bi0600143. [DOI] [PubMed] [Google Scholar]
- 15.Davies GJ, Gamblin SJ, Littlechild JA, Watson HC. The structure of a thermally stable 3-phosphoglycerate kinase and a comparison with its mesophilic equivalent. Proteins. 1993;15:283–289. doi: 10.1002/prot.340150306. [DOI] [PubMed] [Google Scholar]
- 16.Vieille C, Hess JM, Kelly RM, Zeikus JG. xylA cloning and sequencing and biochemical characterization of xylose isomerase from Thermotoga neapolitana. Appl Environ Microb. 1995;61:1867–1875. doi: 10.1128/aem.61.5.1867-1875.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auerbach G, Ostendorp R, Prade L, Korndörfer I, Dams T, Huber R, Jaenicke R. Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization. Structure. 1998;6:769–781. doi: 10.1016/s0969-2126(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 18.Chi YI, Martinez-Cruz LA, Jancarik J, Swanson RV, Robertson DE, Kim SH. Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability. FEBS Lett. 1999;445:375–383. doi: 10.1016/s0014-5793(99)00090-3. [DOI] [PubMed] [Google Scholar]
- 19.Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. P Natl Acad Sci USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isupov MN, Fleming TM, Dalby AR, Crowhurst GS, Bourne PC, Littlechild JA. Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus. J Mol Biol. 1999;291:651–660. doi: 10.1006/jmbi.1999.3003. [DOI] [PubMed] [Google Scholar]
- 21.Maes D, Zeelen JP, Thanki N, Beaucamp N, Alvarez M, Thi MH, Backmann J, Martial JA, Wyns L, Jaenicke R, Wierenga RK. The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparitive thermostability structural analysis of ten different TIM structures. Proteins. 1999;37:441–453. [PubMed] [Google Scholar]
- 22.Russell RJ, Ferguson JM, Hough DW, Danson MJ, Taylor GL. The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolutoin. Biochemistry. 1997;36:9983–9994. doi: 10.1021/bi9705321. [DOI] [PubMed] [Google Scholar]
- 23.Tahirov TH, Oki H, Tsukihara T, Ogasahara K, Yutani K, Ogata K, Izu Y, Tsunasawa S, Kato I. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J Mol Biol. 1998;284:101–124. doi: 10.1006/jmbi.1998.2146. [DOI] [PubMed] [Google Scholar]
- 24.Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974;86:665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- 25.Kuwajima K, Ikeguchi M, Sugawara M, Hiraoka Y, Sugai S. Kinetics of disulfide bond reduction in α-lactalbumin by dithiothreitol and molecular basis of superreactivity of the Cys6-Cys120 disulfide bond. Biochemistry. 1990;29:8240–8249. doi: 10.1021/bi00488a007. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 27.Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng Des Sel. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 29.Manco G, Giosuè E, D'Auria S, Herman P, Carrea G, Rossi M. Cloning, overexpression, and properties of a new thermophilic and thermostable esterase with sequence similarity to hormone-sensitive subfamily from the archaeon Archaeoglobus fulgidus. Arch Biochem Biophys. 2000;373:182–192. doi: 10.1006/abbi.1999.1497. [DOI] [PubMed] [Google Scholar]
- 30.Bonisch H, Backmann J, Kath T, Naumann D, Schafer G. Adenylate kinase from Sulfolobus acidocaldarius: expression in Escherichia coli and characterization by Fourier transform infrared spectroscopy. Arch Biochem Biophys. 1996;333:75–84. doi: 10.1006/abbi.1996.0366. [DOI] [PubMed] [Google Scholar]
- 31.Jaenicke R, Bohm G. The stability of proteins in extreme environments. Curr Opin Struc Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 32.Závodszky P, Kardos J, Svingor, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA. 1998;95:406–411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gershenson A, Schauerte JA, Giver L, Arnold FH. Tryptophan phophorescence study of enzyme flexibility and unfolding in laboratory-evolved thermostable esterases. Biochemistry. 2000;39:4658–4665. doi: 10.1021/bi992473s. [DOI] [PubMed] [Google Scholar]
- 34.Sriprapundh D, Vieille C, Zeikus JG. Molecular determinants of xylose isomerase thermal stability and activity: analysis of thermozymes by site-directed mutagenesis. Protein Eng. 2000;13:259–265. doi: 10.1093/protein/13.4.259. [DOI] [PubMed] [Google Scholar]
- 35.Nakai T, Okada K, Akutsu S, Miyahara I, Kawaguchi S, Kato R, Kuramitsu S, Hirotsu K. Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate. Biochemistry. 1999;38:2413–2424. doi: 10.1021/bi9819881. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Heatwole J, Soelaiman S, Shoham M. Crystal structure of a thermophilic alcohol dehydrogenase substrate complex suggests determinants of substrate specficity and thermostability. Proteins. 1999;37:619–627. doi: 10.1002/(sici)1097-0134(19991201)37:4<619::aid-prot12>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe K, Masuda T, Ohashi H, Mihara H, Suzuki Y. Multiple proline substitutions cumulatively thermostabilze Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur J Biochem. 1994;226:277–283. doi: 10.1111/j.1432-1033.1994.tb20051.x. [DOI] [PubMed] [Google Scholar]
- 38.Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- 39.Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol R. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villbrandt B, Sagner G, Schomburg D. Investigations on the thermostability and function of truncated Thermus aquaticus DNA polymerase fragments. Protein Eng. 1997;10:1281–1288. doi: 10.1093/protein/10.11.1281. [DOI] [PubMed] [Google Scholar]
- 41.Di Fiore A, Capasso C, De Luca V, Monti SM, Carginale V, Supuran CT, Scozzafava A, Pedone C, Rossi M, De Simone G. X-ray structure of the first `extremo-alpha-carbonic anhydrase', a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D. 2013;69:1150–1159. doi: 10.1107/S0907444913007208. [DOI] [PubMed] [Google Scholar]
- 42.Daigle R, Desrochers M. Carbonic Anhydrase Having Increased Stability Under High Temperature Conditions. 7521217. US: CO2 Solution, Inc; Patent, U. S. 2009
- 43.Avvaru BS, Kim CU, Sippel KH, Gruner SM, Agbandje-McKenna M, Silverman DN, McKenna R. A short, strong hydrogen bond in the active site of human carbonic anhydrase II. Biochemistry. 2010;49:249–251. doi: 10.1021/bi902007b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mårtensson L-G, Karlsson M, Carlsson U. Dramatic stabilization of the native state of human carbonic anhydrase II by an engineered disulfide bond. Biochemistry. 2002;41:15867–15875. doi: 10.1021/bi020433+. [DOI] [PubMed] [Google Scholar]
- 45.Boone CD, Gill S, Tu C, Silverman DN, McKenna R. Structural, catalytic and stabilizing consequences of aromatic cluster variants in human carbonic anhydrase II. Arch Biochem Biophys. 2013;539:31–37. doi: 10.1016/j.abb.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forsman C, Behravan G, Osterman A, Jonsson BH. Production of active human carbonic anhydrase II in E. coli. Acta Chem Scand B. 1988;42:314–318. doi: 10.3891/acta.chem.scand.42b-0314. [DOI] [PubMed] [Google Scholar]
- 47.Hansson MD, Rzeznicka K, Rosenback M, Hansson M, Sirijovski N. PCR-mediated deletion of plasmid DNA. Anal Biochem. 2008;375:373–375. doi: 10.1016/j.ab.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Khalifah RG, Strader DJ, Bryant SH, Gibson SM. Carbon-13 nuclear magnetic resonance probe of active-site ionizations in human carbonic anhydrase B. Biochemistry. 1977;16:2241–2247. doi: 10.1021/bi00629a031. [DOI] [PubMed] [Google Scholar]
- 49.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 50.Silverman DN. Carbonic anhydrase: oxygen-18 exchange catalyzed by an enzyme with rate-contributing proton-transfer steps. Methods Enzymol. 1982;87:732–752. doi: 10.1016/s0076-6879(82)87037-7. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 57.Fisher SZ, Maupin CM, Budayova-Spano M, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: insights into the proton transfer mechanism. Biochemistry. 2007;46:2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 58.Scolnick LR, Christianson DW. X-ray crystallographic studies of alanine-65 variants of carbonic anhydrase II reveal the structural basis of compromised proton transfer in catalysis. Biochemistry. 1996;35:16429–16434. doi: 10.1021/bi9617872. [DOI] [PubMed] [Google Scholar]