Background: XMAP215 family members contain arrayed TOG domains that promote microtubule polymerization.
Results: TOG3 has a unique architecture that positions conserved residues in a TOG1- and TOG2-like tubulin-binding mode.
Conclusion: TOG1, TOG2, and TOG3 have similar architectures and predicted tubulin-binding modes that contrast with TOG4.
Significance: XMAP215 family members use a structurally diverse, polarized TOG domain array to promote microtubule polymerization.
Keywords: Cell Division, Crystal Structure, Microtubule, Microtubule-associated Protein, Structural Biology, Tubulin, Msps, TOG, XMAP215, Microtubule Dynamics
Abstract
Microtubule-associated proteins regulate microtubule (MT) dynamics spatially and temporally, which is essential for proper formation of the bipolar mitotic spindle. The XMAP215 family is comprised of conserved microtubule-associated proteins that use an array of tubulin-binding tumor overexpressed gene (TOG) domains, consisting of six (A–F) Huntingtin, elongation factor 3, protein phosphatase 2A, target of rapamycin (HEAT) repeats, to robustly increase MT plus-end polymerization rates. Recent work showed that TOG domains have differentially conserved architectures across the array, with implications for position-dependent TOG domain tubulin binding activities and function within the XMAP215 MT polymerization mechanism. Although TOG domains 1, 2, and 4 are well described, structural and mechanistic information characterizing TOG domains 3 and 5 is outstanding. Here, we present the structure and characterization of Drosophila melanogaster Mini spindles (Msps) TOG3. Msps TOG3 has two unique features as follows: the first is a C-terminal tail that stabilizes the ultimate four HEAT repeats (HRs), and the second is a unique architecture in HR B. Structural alignments of TOG3 with other TOG domain structures show that the architecture of TOG3 is most similar to TOG domains 1 and 2 and diverges from TOG4. Docking TOG3 onto recently solved Stu2 TOG1· and TOG2·tubulin complex structures suggests that TOG3 uses similarly conserved tubulin-binding intra-HEAT loop residues to engage α- and β-tubulin. This indicates that TOG3 has maintained a TOG1- and TOG2-like TOG-tubulin binding mode despite structural divergence. The similarity of TOG domains 1–3 and the divergence of TOG4 suggest that a TOG domain array with polarized structural diversity may play a key mechanistic role in XMAP215-dependent MT polymerization activity.
Introduction
Microtubules (MTs)2 are polar cytoskeletal filaments composed of αβ-tubulin that facilitate cargo trafficking, enable cell movement, establish cell polarity, and form the bipolar mitotic spindle. The ability of MTs to regulate multiple cellular processes relies on polymer dynamics conferred by GTP hydrolysis-dependent gain and loss of αβ-tubulin at MT ends (1–3). Although tubulin has an inherent ability to switch between phases of polymerization, depolymerization, and pause, cellular MT dynamics are regulated in space and time by a host of plus-end tracking proteins (+TIPs), including a family of MT polymerases defined by the Xenopus member, XMAP215 (4, 5). XMAP215 was originally identified in Xenopus laevis egg extracts and found to dramatically increase MT plus-end polymerization rates (6). XMAP215 orthologs have been discovered and characterized in species ranging from yeast to humans, with depletion of these proteins yielding decreased MT polymerization rates and aberrant spindle phenotypes (7–27).
The XMAP215 family has a conserved domain architecture; an N-terminal array of tumor overexpressed gene (TOG) domains bind and add tubulin heterodimers processively to MT plus-ends; and a C-terminal domain (CTD) that interacts with other microtubule-associated proteins to afford proper XMAP215 localization (28–34). Yeast family members contain two N-terminal TOG domains followed by a C-terminal coiled-coil homodimerization domain, which functionally yields a tetrameric TOG domain array (35, 36). This contrasts with higher eukaryotic XMAP215 family members, which have a pentameric TOG domain array and function as monomers (6, 29, 31, 37). Long flexible linkers connect TOG domains and promote MT lattice association in cell culture via stretches of basic residues (15, 29).
TOG domains form an oblong paddle-like structure consisting of six (A–F) HEAT repeats (HRs) (37–39). Intra-HEAT loops are positioned along one face of the TOG domain and form a 60-Å stretch that binds a single αβ-tubulin heterodimer (37, 40, 41). When conserved residues in the TOG domain's intra-HEAT loops are mutated, TOG-tubulin interactions are ablated, and XMAP215 MT polymerase activity is compromised (37–39). Mutating the tubulin binding activity of different TOG domains in the pentameric TOG domain array has differential effects on XMAP215 MT polymerization activity and spindle length. Mutations in the TOG1 or TOG2 intra-HEAT tubulin-binding loops compromise XMAP215-mediated MT polymerization activity more dramatically than homologous mutations in TOG3 or TOG4 (8, 16, 17, 20, 29, 42, 43). Mutating the tubulin-binding determinants in TOG5 does not appear to significantly affect the rate of MT polymerization (16, 29).
The ability of TOG domains to differentially promote MT polymerization rates may be partially explained by variation in TOG domain architectures. Structures to date of Saccharomyces cerevisiae TOG1 and TOG2, Drosophila melanogaster Mini spindles (Msps) TOG2 and TOG4, Caenorhabditis elegans ZYG-9 TOG3, and Homo sapiens ch-TOG TOG4 show that TOG domains have different architectural features that are dependent on the position of TOG domains within the array (16, 38–41). Specifically, TOG domains 1 and 2 are structurally similar to each other within and across species, whereas the structure of TOG4 is conserved across species but diverges from TOG1 and TOG2. The ZYG-9 TOG3 structure is unique among TOG domains solved to date, having an additional N-terminal HR positioned orthogonal to the other HRs in the TOG domain that interacts with HRs A–C (39). Studies hypothesize that ZYG-9 TOG3, which is the ultimate TOG domain in the ZYG-9 trimeric TOG domain array, is most similar to TOG5 from pentameric XMAP215 family members, but structural evidence for this observation is still at large (39, 44).
Two distinct questions arise from the determination that TOG domains have differential architectures and abilities to promote MT plus-end polymerization; the first concerns whether TOG domains in the pentameric array have a similar or distinct function, and the second concerns whether all the TOG domains bind free tubulin or a subset selectively binds MT lattice-incorporated tubulin. The latter suggests that XMAP215 TOG domain arrays are composed of position-specific TOG domain architectures that recognize different tubulin structural states, which may play a key mechanistic role in XMAP215-mediated MT polymerization. The divergent, yet positionally conserved architecture of TOG4, supports this hypothesis (16). Although structures of TOG domains 1, 2, and 4 are well described, structural information characterizing TOG3 and TOG5 from pentameric XMAP215 family members is lacking and is required to move toward a full understanding of the XMAP215-dependent MT polymerization mechanism.
Here, we present the x-ray crystal structure of D. melanogaster Msps TOG3, the first TOG3 structure reported from a pentameric XMAP215 family member. Msps TOG3 has unique structural elements that deviate from other TOG domain structures determined to date, including an extended HR B intra-HEAT loop and a stabilizing C-terminal tail that binds along HRs C–F. Although Msps TOG3 has unique structural features, modeling suggests that TOG3 has preserved the position of conserved tubulin-binding intra-HEAT loop residues in arrangements that mimic those observed in the Stu2 TOG1·αβ-tubulin and TOG2·αβ-tubulin complex structures. Our findings suggest that TOG3 binds αβ-tubulin in a mode similar to TOG1 and TOG2 but different from TOG4, highlighting the polarized nature of architecturally distinct TOG domains in the pentameric XMAP215 family TOG domain array, which is collectively used to promote the rate of MT polymerization.
EXPERIMENTAL PROCEDURES
Multiple Species Alignment
Proteins used to determine conservation included Msps (D. melanogaster), ch-TOG (Homo sapiens), CKAP5 (Bos taurus), CKAP5 (Gallus gallus), CKAP5 (Pan troglodytes), CKAP5 (Mus musculus), XMAP215 (X. laevis), MOR1 (Arabidopsis thaliana), TMBP200 (Nicotiana tabacum), CP224 (Dictyostelium discoideum), ZYG-9 (Caenorhabditis elegans), and Stu2 (S. cerevisiae). Clustal Omega was used to produce preliminary alignments that were then adjusted manually.
Construct Cloning and Expression
D. melanogaster Msps TOG3 (residues 582–825) was subcloned into pET28 (Novagen), engineering an N-terminal His6 tag. Msps TOG3 R816A was generated using a single round of QuikChange mutagenesis (Agilent Technologies, Cary, NC). pET28-TOG3 constructs were transformed into BL21 DE3 (pLysS) Escherichia coli for recombinant expression. Cultures (6 liters) were grown at 37 °C under kanamycin selection. Upon reaching an optical density of 0.6 (600 nm), protein expression was induced with 0.125 mm isopropyl 1-thio-β-d-galactopyranoside. The temperature was reduced to 18 °C, and cultures were grown for 20 h. Cells were harvested by centrifugation at 2,100 × g for 8 min. Pellets were resuspended in 200 ml of buffer A (25 mm Tris, pH 8.0, 300 mm NaCl, 10 mm imidazole, and 0.1% β-mercaptoethanol) and stored at −20 °C.
Selenomethionine (SeMet)-substituted Msps TOG3 expression and purification followed the above protocol with the following alterations: pET28-TOG3 was transformed into B834 DE3 auxotrophic E. coli and grown in 6 liters of minimal media (Molecular Dimensions) supplemented with l-SeMet (15 μg/ml). At an optical density (600 nm) of 0.4, protein expression was induced, and cells were grown at 18 °C for 12 h (45).
Protein Purification
Native, R816A, and SeMet-substituted Msps TOG3 were purified using affinity and ion exchange chromatography. Pellets were thawed, 1 mm phenylmethylsulfonyl fluoride was added, and cells were lysed by sonication at 4 °C. Lysate was cleared by centrifugation at 23,000 × g for 45 min at 4 °C. The supernatant was loaded onto a 10-ml Ni2+-nitrilotriacetic acid column (Qiagen) and subsequently washed with buffer A. A 250-ml linear gradient between buffer A and buffer B (25 mm Tris, pH 8.0, 300 mm NaCl, 300 mm imidazole, and 0.1% β-mercaptoethanol) was used to elute TOG3. Protein-containing fractions were pooled and exchanged into buffer A, and the His6 tag was cleaved by incubation with thrombin (Haematologic Technologies) for 16 h at 4 °C. The digested protein was filtered over a benzamidine-Sepharose (GE Healthcare) column to remove thrombin and successively exchanged into buffer Q (25 mm Tris, pH 9.1, and 0.1% β-mercaptoethanol). Protein was loaded onto a 10-ml Q-Sepharose fast flow (GE Healthcare) column and washed with buffer Q. Protein was eluted using a 250-ml linear gradient between buffer Q and buffer HQ (25 mm Tris, pH 9.1, 1 m NaCl, and 0.1% β-mercaptoethanol). Fractions deemed >98% pure by SDS-PAGE analysis were buffer-exchanged into storage buffer (10 mm Hepes, pH 7.0, 150 mm NaCl, and 0.1% β-mercaptoethanol), concentrated to 15 mg/ml, flash-frozen in liquid nitrogen, and stored at −80 °C.
Crystallization
Crystallization of native and SeMet-substituted Msps TOG3 followed the hanging drop diffusion method. After 1 day of equilibration at 20 °C, native Msps TOG3 rod-shaped crystals appeared in drops containing 2 μl of protein at 15 mg/ml and 2 μl of a 500-μl well solution (33% PEG 4000 and 27 mm potassium phosphate monobasic). SeMet-substituted TOG3 crystallization followed the native protein parameters, except rod-shaped crystals were grown using a well solution containing 38% PEG 4000, 175 mm ammonium acetate, and 25 mm sodium acetate trihydrate, pH 4.6. Native and SeMet-substituted TOG3 crystals were transferred into a cryoprotection solution containing the mother liquor supplemented with 35 and 30% glycerol, respectively, and flash-frozen in liquid nitrogen.
Data Collection, Structural Determination, and Refinement
Isomorphous native and SeMet-substituted single-wavelength anomalous diffraction (SAD) Msps TOG3 single crystal diffraction datasets were collected at the Advanced Photon Source SER-CAT 22-ID beamline at a wavelength of 0.97723 Å. The native TOG3 dataset was taken with 1° oscillations for a total of 180 frames. A SAD dataset was collected on a SeMet-substituted TOG3 crystal, collecting 0.5° oscillations across 125° plus inverse beam collected in 5° wedges. Data were indexed, integrated, and scaled using HKL2000 to resolutions of 2.3 and 2.6 Å for the native and SeMet datasets, respectively (46). Selenium sites were identified, phases were determined, and experimental electron density was calculated using PHENIX AutoSol. An initial model was built using PHENIX AutoBuild (47). Refinement cycles involved iterative manual building in Coot followed by refinement runs using PHENIX phenix.refine (47, 48). The SeMet TOG3 structure was built and refined against a maximum-likelihood Hendrickson-Lattman target function to 2.6 Å. The resulting structure was subsequently used as a molecular replacement search model to phase the native dataset using the AutoMR molecular replacement method in PHENIX. The native TOG3 structure was refined against a maximum likelihood target function using an Rfree based on 10% of the data randomly excluded from the refinement. Electrostatics were calculated using APBS, and pairwise alignments and r.m.s.d. values were calculated using the Dali pairwise alignment server (49, 50).
Circular Dichroism
Native and R816A mutant Msps TOG3 stocks were buffer-exchanged and diluted to 0.13 mg/ml in circular dichroism (CD) buffer (10 mm sodium phosphate, pH 7.5, and 50 mm sodium fluoride). CD spectra were collected using a Chirascan-plus CD spectrometer (Applied Photophysics, Leatherhead, UK) at room temperature using a 1-mm path length cuvette. Spectra were acquired from 260 to 185 nm with a step size of 0.5 nm. A baseline CD buffer spectrum was subtracted from each TOG3 construct spectrum. By monitoring CD at 208 and 222 nm while heating samples from 20 to 94 °C in a step size of 1 °C, the thermal melting curves of the native and R816A TOG3 constructs were obtained. Inflection points for melt curves were determined by taking the second derivative of each curve. All CD spectra were smoothed using the Chirascan-plus software.
RESULTS
TOG3 Is Conserved in XMAP215 Family Members with Pentameric TOG Arrays
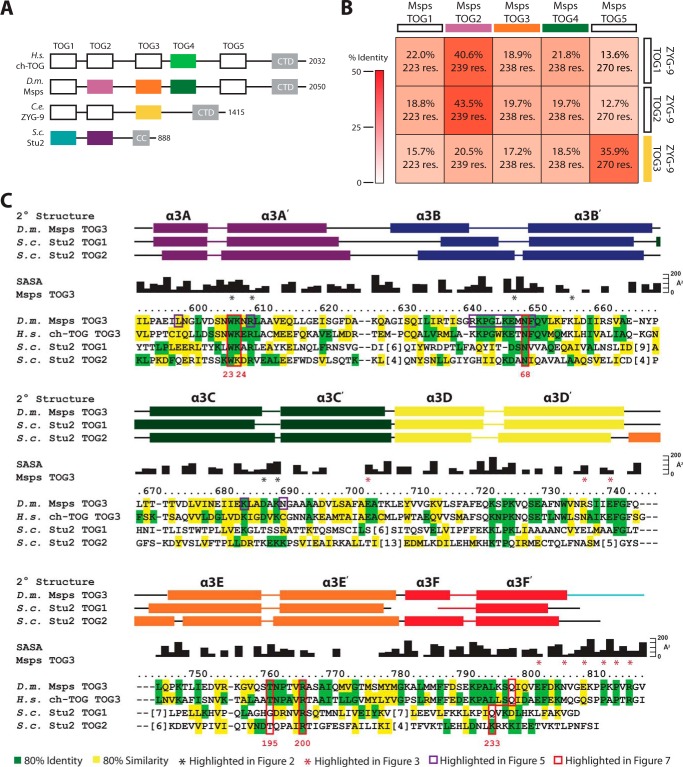
XMAP215 family members have TOG domain arrays of different lengths. Higher eukaryotic family members, including D. melanogaster Msps, contain pentameric TOG domain arrays, which contrasts with the dimeric and trimeric arrays of S. cerevisiae Stu2 and C. elegans ZYG-9, respectively (Fig. 1A). To date, TOG domain structures have been solved across multiple XMAP215 family members including Stu2 TOG1 and TOG2, ZYG-9 TOG3, Msps TOG2 and TOG4, and ch-TOG TOG4 (Fig. 1A, colored boxes) (16, 38–41). These structures demonstrate that XMAP215 TOG domain architectures are differential and position-dependent across the TOG domain array.
FIGURE 1.
TOG3 is conserved across pentameric XMAP215 family members. A, XMAP215 family domain architecture consists of arrayed TOG domains that interact with αβ-tubulin and a C-terminal domain (CTD) or coiled-coil (CC) that associate with other microtubule-associated proteins. XMAP215 family TOG domain structures solved to date are shown in color. B, sequence identity matrix comparing Msps and ZYG-9 TOG domains with the number of aligned residues indicated. C, sequence alignment of Msps TOG3, ch-TOG TOG3, Stu2 TOG1, and Stu2 TOG2. Conservation is mapped based on multiple species alignments within individual TOG domains with residues containing 80% identity and 80% similarity across species highlighted in green and yellow, respectively. Black residue numbers and solvent-accessible surface area (SASA) correspond to the Msps TOG3 structure. Red numbers below the alignment represent Stu2 TOG1 residues highlighted in Fig. 7. Secondary structure for Msps TOG3, Stu2 TOG1, and Stu2 TOG2 are presented above the alignment based on Protein Data Bank entries 4FFB, 4U3J, and 2QK1.
Although previously solved TOG domain structures helped elucidate the polarized nature of TOG domain arrays, the architecture and function of TOG domains 3 and 5 from pentameric family members are largely unknown. Thus, we set out to determine the structure of Msps TOG3. ZYG-9 TOG3 is the only TOG3 structure solved to date (39). To determine whether ZYG-9 TOG3 is a good model to predict the structure and function of Msps TOG3, we aligned and determined the pairwise sequence identities between TOG domains from Msps and ZYG-9. ZYG-9 TOG3 and Msps TOG3 showed only 17.2% sequence identity, suggesting that ZYG-9 TOG3 is not a good model for Msps TOG3. In contrast, ZYG-9 TOG3 is highly similar to Msps TOG5 with a sequence identity of 35.9% (Fig. 1B). The similarity between TOG5 domains from pentameric XMAP215 family members and ZYG-9 TOG3 has been noted previously, although no structural evidence to support the similarity has been presented (39, 44). The only pairwise identity that exceeds that between Msps TOG5 and ZYG-9 TOG3 is that between Msps TOG2 and ZYG-9 TOG1 (40.6% identity) and Msps TOG2 and ZYG-9 TOG2 (43.5% identity) (Fig. 1B). These analyses suggest ZYG-9 may have a TOG array architecture akin to a permuted Msps TOG2-TOG2-TOG5 array.
As Msps TOG3 and ZYG-9 TOG3 have comparatively low sequence identity (17.2%, Fig. 1B), we inquired whether TOG3 has a higher degree of conservation across XMAP215 family members with pentameric TOG arrays. Additionally, we asked how conserved regions in TOG3 compared with corresponding regions in other TOG domains in the array (i.e. TOG1 and TOG2). We created individual alignments for TOG1, TOG2, and TOG3 across 10 species ranging from S. cerevisiae and D. discoideum to H. sapiens, mapped residues that were 80% identical (green) or 80% similar (yellow) within a specific TOG domain, and then reduced this information into the alignment presented in Fig. 1C showing D. melanogaster, Msps TOG3, H. sapiens ch-TOG TOG3, S. cerevisiae, Stu2 TOG1, and S. cerevisiae Stu2 TOG2. TOG domains contain six HRs (A–F) with tubulin-binding residues located in the intra-HEAT loops that line one face of the domain (37–39). TOG3 is highly conserved across species with 35% of the residues in the domain having 80% cross-species identity. The highest areas of conservation map to the intra-HEAT loops. TOG3 intra-HEAT loops also display conservation with Stu2 TOG domains 1 and 2 (Fig. 1C), as we will describe in detail. These results indicate that TOG3 is highly conserved across pentameric XMAP215 family members and that the predicted TOG3 intra-HEAT tubulin-binding residues are conserved with the Stu2 TOG1 and TOG2 determinants known to engage αβ-tubulin.
Drosophila Msps TOG3 Is a Canonical TOG Domain with a Stabilizing C-terminal Extension
TOG domain structures of Stu2 TOG1 and TOG2, Msps TOG2 and TOG4, ZYG-9 TOG3, and ch-TOG TOG4 show that TOG domains have position-dependent architectures within the TOG domain array (16, 38–41). TOG domains 1 and 2 are similar within and across species, whereas the structure of TOG4 diverges yet is conserved across species. To further examine the structural differences within the pentameric TOG domain array, we screened TOG3 domains from pentameric XMAP215 family members and obtained crystals of Msps TOG3. Isomorphous native and SAD datasets were collected from single native and selenomethionine (SeMet)-substituted Msps TOG3 crystals, respectively. Diffraction data were indexed, integrated, and scaled to a resolution of 2.3 Å for the native dataset and 2.6 Å for the SeMet SAD dataset. The SeMet-substituted dataset was refined using experimental phase restraints against a maximum-likelihood Hendrickson-Lattman target to R and Rfree values of 0.18 and 0.23, respectively. The resulting SeMet TOG3 structure was used to perform molecular replacement on the native dataset. The resulting native model was refined to an R value of 0.18 and an Rfree value of 0.24 using a maximum likelihood refinement model. The Msps TOG3 structure has one molecule in the asymmetric unit that includes residues 592–818 and 115 water molecules. Residues 582–591 and 819–825 are omitted from the model due to lack of electron density, presumably because of high linker flexibility. Resulting crystallographic diffraction, phasing, and refinement statistics for the native and SeMet-substituted TOG3 structures are presented in Table 1.
TABLE 1.
Crystallographic data, phasing, and refinement statistics
Values in parentheses are for the highest resolution shells unless otherwise denoted.
| Crystal | Native | SeMet |
|---|---|---|
| PDB code | 4Y5J | |
| Data Collection | ||
| Beamline | APS 22-ID | APS 22-ID |
| Wavelength (nm) | 0.97723 | 0.97723 |
| Space group | P3212 | P3212 |
| Resolution (Å) | 50.0–2.30 (2.38–2.30) | 50.0–2.54 (2.63–2.54) |
| Unit cell dimensions: a, b, c (Å) | 49.8, 49.8, 205.0 | 49.8, 49.8, 205.4 |
| No. of reflections: measured/unique | 144,181 (10,580)/13,396 (1263) | 174,442 (12,493)/18,862 (1853) |
| Mean redundancy | 10.8 (8.9) | 9.2 (6.6) |
| Completeness (%) | 99.5 (95.0) | 99.7 (97.0) |
| I/σ | 33.4 (5.1) | 21.2 (3.6) |
| Rsym (%)a | 0.08 (0.37) | 0.10 (0.45) |
| Resolution at which anomalous completeness >85% for I/σ >5, >3, >2 (Å) | 3.4, 3.0, 2.9 | |
| Figure of meritb: all/centrics/acentrics | 0.44/0.12/0.49 | |
| Figure of meritb after density modification: all/centrics/acentrics | 0.75/0.65/0.76 | |
| Refinement | ||
| Resolution (Å) | 34.2–2.30 (2.39–2.30) | |
| No. of protein/solvent atoms | 1759/115 | |
| r.m.s.d. bond angles (°) | 0.008 | |
| r.m.s.d. bond lengths (Å) | 1.026 | |
| Ramachandran favored/allowed (%) | 98.2/1.8 | |
| Mean B (Å2) | 29.5 | |
| Phase error (°) | 24.3 | |
| Rc/Rfreed value (%) | 17.6 (20.3)/24.1 (28.7) | |
a Rsym = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h)] is the integrated intensity of the ith measurement, and 〈I(h)〉 is the mean of all measurements of Ih for the Miller indices h.
b Figure of merit is the weighted mean of the cosine of the deviation from αbest.
c R value = Σ(|Fobs| − k|Fcalc|)/Σ|Fobs|.
d Rfree was calculated using a 10% subset of the data that was removed randomly from the original data and excluded from refinement.
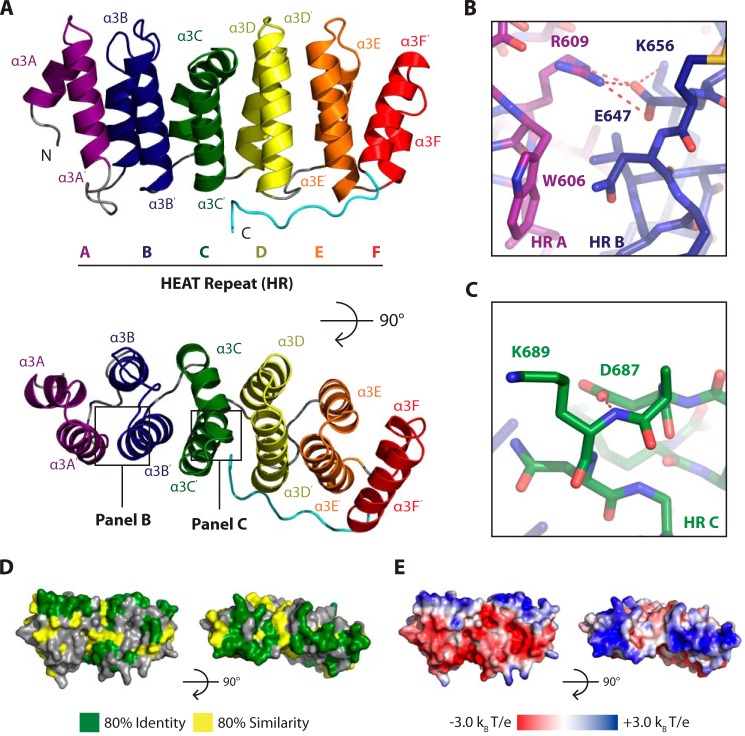
Msps TOG3, like other known TOG domain structures, is elongated and composed of six HRs that pack linearly against one another (Fig. 2A). Each HR consists of two antiparallel helices that are denoted by X and X′ (38). HRs are labeled A–F and can be grouped into two characteristic triads that have slight twists, HRs A–C and D–F (51, 52). Specifically, the HR A–C triad has a right-handed twist, whereas the HR D–F triad has a right-handed twist between HRs D and E and a left-handed twist between HRs E and F.
FIGURE 2.
Msps TOG3 is a canonical TOG domain with conserved solvent-exposed tubulin-binding residues. A, Msps TOG3 consists of six (A–F) HRs. B and C, TOG3 has solvent-exposed conserved tubulin-binding residues including a tryptophan (Trp-606) in HR A (B) and a lysine (Lys-689) in HR C (C). D, residue conservation, contoured as in Fig. 1C, mapped on the Msps TOG3 structure and oriented as in A. E, electrostatic potential mapped on Msps TOG3, oriented as in A and D.
Recent Stu2 TOG1· and TOG2·tubulin complex structures show that the TOG domain face formed by intra-HEAT loops contains conserved residues that are important for tubulin binding (40, 41). Specifically, the intra-HEAT loops in HRs A–D and in HRs E and F position residues to interact with β- and α-tubulin, respectively. Previous studies have shown that mutating residues on the intra-HEAT loop face abrogates the ability of TOG domains to interact with αβ-tubulin (37–39). Msps TOG3 has solvent-accessible residues, including a tryptophan (Trp-606) in intra-HEAT loop A and a lysine (Lys-689) in intra-HEAT loop C, that are positioned to interact with β-tubulin in a Stu2 TOG1- and TOG2-like binding mode (Fig. 2, B and C). We utilized the contours from Fig. 1C to map TOG3-specific conservation onto the Msps TOG3 structure and found that the highest area of similarity and identity are in the intra-HEAT loops, as expected by the placement of important tubulin-binding residues (Fig. 2D). This surface has a net positive charge, which complements the negative exterior surface charge of MTs (Fig. 2E) (49). Together, these data suggest that TOG3, like TOG1 and TOG2, can bind a sub-region of the αβ-tubulin heterodimer that upon polymerization is exposed on the MT exterior surface.
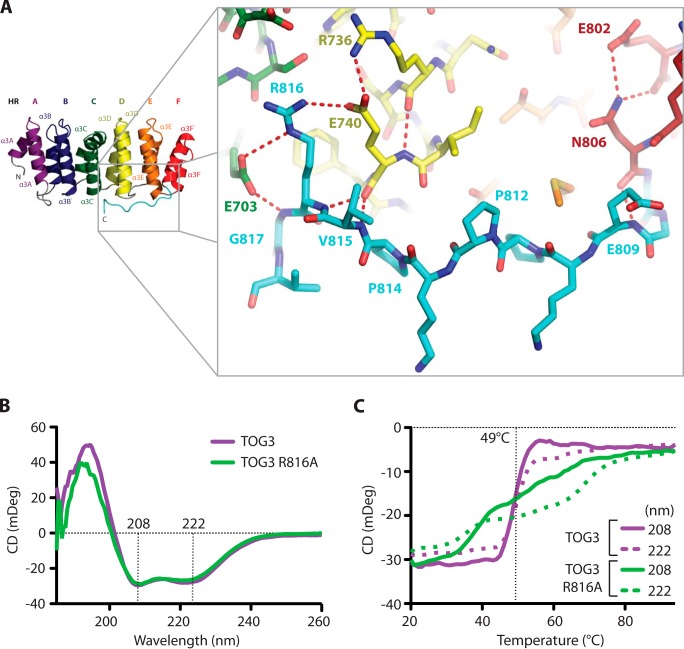
TOG3 has a unique, conserved C-terminal extension that binds along HRs C–F (see Figs. 1C and 2A). The C-terminal Msps TOG3 extension makes multiple conserved contacts with two main areas. The first area is with HR F, and the second area is with HRs C and D (see Figs. 1C, 2D, and 3A). In HR F, a hydrogen bond is formed between the Asn-806 backbone carbonyl and the backbone amide of Glu-809. The interaction between Asn-806 and Glu-809 is stabilized through buttressing interactions between the Asn-806 side chain and Glu-802 (Fig. 3A). The TOG3 C-terminal extension is further stabilized by a highly conserved arginine, Arg-816, which forms salt bridges with HR C Glu-703 and HR D Glu-740. The Glu-740 salt bridge is buttressed by a large hydrogen-bonding network involving HR D Arg-736, as well as the Val-815 backbone (Fig. 3A). The presence of multiple contacts between the C-terminal extension and the ultimate four HRs suggests that the C-terminal extension could promote TOG3 stability. We created two TOG3 constructs to test this hypothesis. The first removed the entire C-terminal extension and the second was an arginine to alanine mutation at position 816 (R816A). The construct lacking the C-terminal extension showed limited solubility and precipitated out of solution during purification, suggesting that the extension is necessary to maintain TOG3 domain stability. The R816A construct was stable enough to be purified, and we performed comparative circular dichroism (CD) studies on the wild type and R816A constructs to test the role of Arg-816 in promoting TOG3 domain stability. Both the wild type and R816A constructs produced CD spectra with minima at 208 and 222 nm at 20 °C, which is characteristic of α-helical proteins (Fig. 3B). We then performed thermal melts on each construct while monitoring CD at 208 and 222 nm to test domain stability. The CD signal of the native TOG3 construct began to change at 44 °C and showed a cooperative unfolding curve with an inflection point at 49 °C. This thermal unfolding profile contrasts with that of the R816A construct, whose CD signal began to change at 33 °C and continued to slowly change signal as the temperature was increased (Fig. 3C). Together, these results suggest that although the Arg-816 side chain is not necessary to preserve the overall helical domain architecture of TOG3, the interactions observed between the Arg-816 side chain with HRs C and D in our crystal structure are important to maintain domain stability, as shown through a noncooperative thermal melting curve.
FIGURE 3.
A conserved C-terminal extension stabilizes Msps TOG3. A, Msps TOG3 has a 12-residue C-terminal extension that makes multiple contacts with the ultimate four HRs. B, CD spectra of native (green) and point mutant R816A (purple) Msps TOG3 constructs indicate α-helical character at 20 °C, as indicated by signature minima at 208 and 222 nm. C, CD thermal melts of native and R816A TOG3 were taken in 1 °C steps from 20 to 94 °C. Native TOG3 has a cooperative unfolding profile with an inflection point at 49 °C. Msps TOG3 R816A has a noncooperative unfolding curve, which suggests that the Arg-816 side chain in the conserved C-terminal extension promotes domain stabilization though its interactions with HRs C and D.
Msps TOG3 Is Structurally Unique from ZYG-9 TOG3
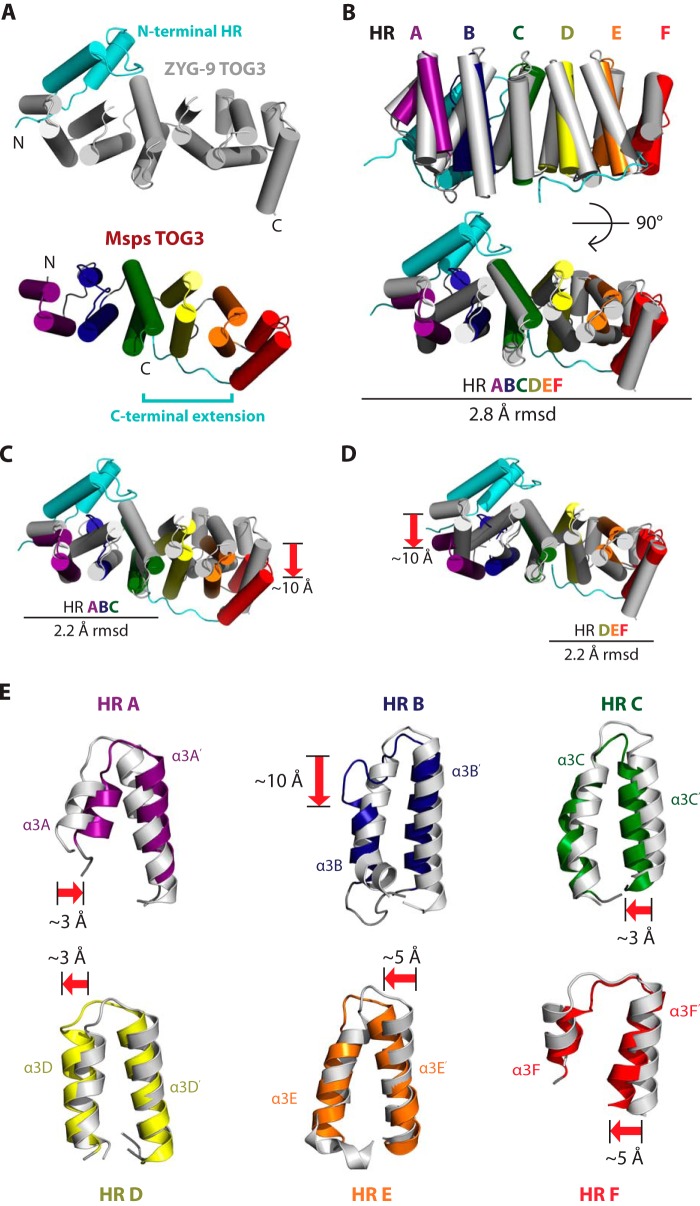
We next inquired how Msps TOG3 compares with other TOG structures solved to date. Of particular interest is how Msps TOG3 compares with the structure of C. elegans ZYG-9 TOG3 (39). In contrast to the pentameric TOG domain array present in higher eukaryotic XMAP215 family members, ZYG-9 contains a trimeric TOG domain array. ZYG-9 TOG3 is thought to be most similar to TOG5 domains from pentameric XMAP215 family members and has a unique extra N-terminal HR that is positioned orthogonal to HRs A–F (39, 44). We compared Msps TOG3 and ZYG-9 TOG3 to determine the degree of inter-species structural homology between these domains (Fig. 4A). Surprisingly, Msps TOG3 and ZYG-9 TOG3 align well over HRs A–F with a root mean square deviation (r.m.s.d.) of 2.8 Å across 207 Cα atoms (Fig. 4B). Structural alignment of respective HR triads HRs A–C and HRs D–F each produced r.m.s.d. values of 2.2 Å, indicating that neither HR triad is more structurally similar (Fig. 4, C and D).
FIGURE 4.
ZYG-9 TOG3 and Msps TOG3 have different domain architectures. A, ZYG-9 TOG3 and Msps TOG3 have unique features (colored in cyan), including an N-terminal HR (ZYG-9 TOG3) and a C-terminal extension (Msps TOG3). Based on sequence analysis, ZYG-9 TOG3 lacks determinants homologous to those found in the Msps TOG3 C-terminal extension and Msps TOG3 lacks determinants homologous to the ZYG-9 TOG3 extra N-terminal HR. B, Msps TOG3 and ZYG-9 TOG3 structures align well over HRs A–F with r.m.s.d. of 2.8 Å. C, Msps TOG3 and ZYG-9 TOG3 align over the HR A–C triad region with r.m.s.d. of 2.2 Å. The alignment yields an ∼10 Å relative displacement in HR F. D, Msps TOG3 and ZYG-9 TOG3 align equally well over the HR D–F triad (2.2 Å r.m.s.d.), suggesting that neither triad has a higher structural similarity. E, comparison of individual HRs from the Msps TOG3 and ZYG-9 TOG3 structural alignment presented in B. HRs A, C, and D show little structural deviation with shifts of ∼3 Å, while HRs E and F show slightly greater positional deviation with shifts of ∼5 Å. The greatest structural divergence occurs in HR B. Msps TOG3 HR B has a truncated α3B helix and has a longer serpentine intra-HEAT loop that bridges the ∼10 Å gap created by the comparatively truncated α3B helix.
Although Msps TOG3 and ZYG-9 TOG3 align well across HRs A–F, each domain has distinct features that distinguish them and suggest that ZYG-9 TOG3 is most similar to TOG5 from pentameric XMAP215 members. Specifically, 1) Msps TOG3 has a conserved C-terminal tail that is not present in the sequence flanking ZYG-9 TOG3 nor are the conserved Msps TOG3 determinants in HRs C–F that engage the C-terminal tail conserved in ZYG-9 TOG3. 2) The additional N-terminal HR in ZYG-9 TOG3 is not predicted in Msps TOG3. In contrast, the ZYG-9 N-terminal HR shows high similarity (61%) to a predicted HR at the N-terminal region flanking TOG5 domains from pentameric XMAP215 family members. Thus, we hypothesize that ZYG-9 TOG3 is more structurally similar to TOG5 than to TOG3. Additionally, ZYG-9 TOG3 lacks conserved structural determinants found in Msps TOG3 HR B, as discussed below.
Msps TOG3 Has a Truncated α3B Helix and an Extended Intra-HR B Loop
Based on the alignment of Msps TOG3 and ZYG-9 TOG3 across HRs A–F (see Fig. 4B), we next asked how each individual HR compared. Comparative examination of individual HRs shows that there is little structural deviation in HRs A, C, and D, with maximum positional shifts of ∼3 Å. Slightly larger deviations are present in HRs E and F, where structural elements are comparatively shifted ∼5 Å (Fig. 4E). The largest variation between ZYG-9 TOG3 and Msps TOG3 is in HR B. Msps TOG3 has a 5-residue C-terminal truncation of its α3B helix that is replaced by an extended intra-HR loop. The 5-residue difference between α3B helices contributes to an ∼10 Å shift in the C-terminal position of Msps TOG3 α3B (Fig. 4E). It is of interest to note that although there are significant architectural changes in Msps TOG3 HR B, tubulin-binding determinants in the intra-HEAT loop are positioned as observed in ZYG-9 TOG3.
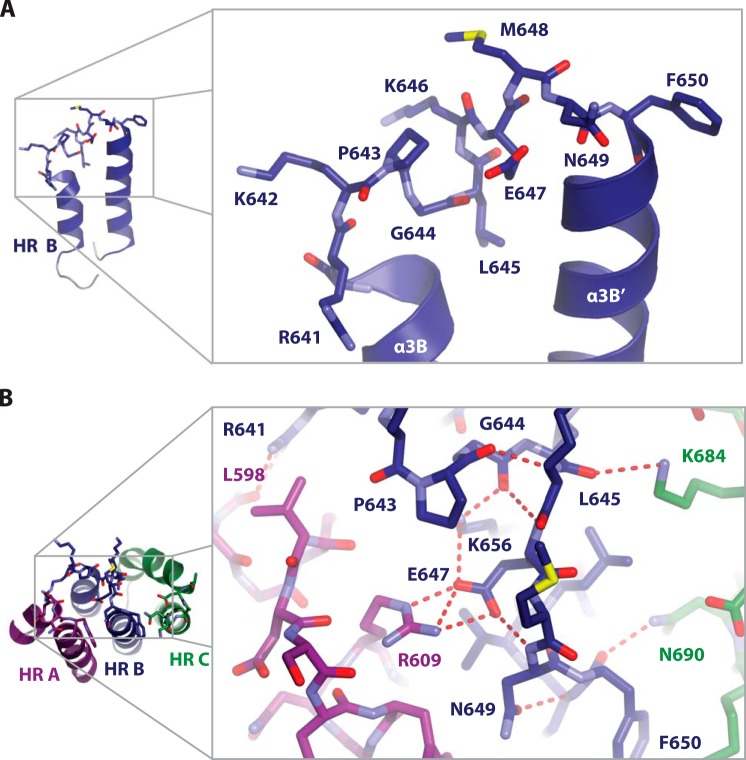
The extended Msps TOG3 HR B intra-HEAT loop consists of highly conserved residues Arg-641 to Phe-650 (Figs. 5A and see 1C). A salt bridge between HR A Arg-609 and Glu-647 supports the position of the HR B intra-HEAT loop (Fig. 5B). This salt bridge is conserved in all other TOG domain structures solved to date (16, 38–41, 51). In the context of the first HR triad, other unique intra- and inter-HR interactions support the position of HR B. There is an extensive intra-HR B hydrogen-bonding network that includes Pro-643, Gly-644, Glu-647, Asn-649, and Lys-656. Unique inter-HR contacts include a hydrogen bond between the HR B intra-HEAT loop Arg-641 guanidinium group and the HR A Leu-598 backbone carbonyl as well as interactions between HR C Lys-684 and Asn-690, and HR B intra-HEAT loop residues Leu-645 and Phe-650, respectively (Fig. 5B). These contacts appear to support the Msps TOG3 HR B intra-HEAT loop in an optimal conformation to interact with tubulin.
FIGURE 5.
TOG3 HR B has a unique serpentine intra-HR loop architecture that buttresses the position of tubulin-binding residues. A, The Msps TOG3 α3B helix is truncated relative to αB helices in other XMAP215 family TOG domains. A longer intra-HR loop (residues Arg-641 to Phe-650) is required to span the distance to α3B′ and optimally position HR B loop β-tubulin binding determinants. The TOG3-specific conserved determinants Lys-642, Pro-643, Gly-644, and a hydrophobic residue at position 645 (Leu-645) create a unique jog in the backbone that buttresses the HR B β-tubulin binding determinants. B, position of β-tubulin binding determinants in the HR B loop is maintained by multiple intra- and inter-HR interactions involving Leu-598, Arg-609, Arg-641, Pro-643, Gly-644, Leu-645, Glu-647, Asn-649, Phe-650, Lys-656, Lys-684, and Asn-690. The Arg-609 to Glu-647 salt bridge is conserved across TOG domain structures determined to date. Additional hydrogen-bonding networks, including an intra-HR B network and inter-HR interactions between HR B and HRs A and C, support the position of the HR B loop.
TOG3 Is Most Structurally Similar to Stu2 TOG1 and Has a Conserved TOG1- and TOG2-like Tubulin-binding Mode
Stu2 TOG1 and TOG2, ZYG-9 TOG3, Msps TOG2 and TOG4, and ch-TOG TOG4 constitute the XMAP215 family TOG domain structures solved and published to date (16, 38–41). These structures show that TOG1 and TOG2 have similar architectures but are divergent from TOG4. We performed pairwise alignments of Msps TOG3 over HRs A–F with each of the other TOG domain structures to determine their structural similarity (Fig. 6). Msps TOG3 is least similar to TOG4 structures with r.m.s.d. values of 4.6 and 4.8 Å for ch-TOG TOG4 and Msps TOG4, respectively (Fig. 6). Pairwise alignment of Msps TOG3 and Msps TOG4 across the HR triads A–C and D–F produce r.m.s.d. values of 2.3 and 2.9 Å, respectively, suggesting that the first triad is the most similar between these domains. When the HR A–C triads of TOG3 and TOG4 are aligned, there is an ∼18 Å shift in the relative positioning of HR F suggesting that this region of TOG4 may bind a distinct site on α-tubulin (16). The structural divergence of TOG4 contrasts with the lower r.m.s.d. values obtained when comparing Msps TOG3 to structures of Stu2 TOG1 (2.4 Å r.m.s.d.), Stu2 TOG2 (3.0 Å r.m.s.d.), and Msps TOG2 (3.2 Å r.m.s.d.) (Fig. 6). These data suggest that TOG domains 1–3 have similar architectures that differ significantly from TOG4 and that TOG3 may engage αβ-tubulin in a mode that parallels that of TOG1 and TOG2.
FIGURE 6.
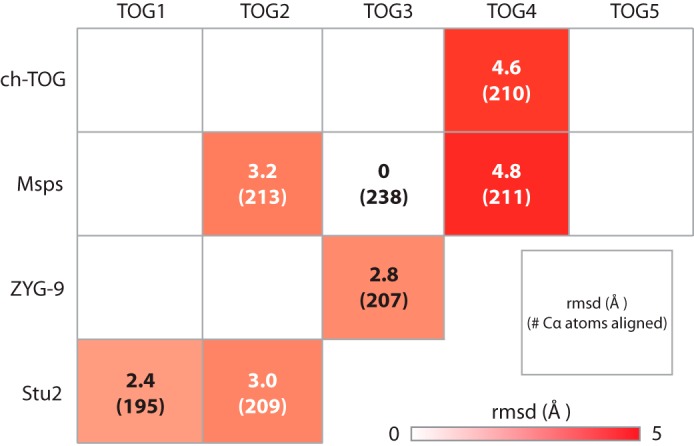
TOG3 is structurally similar to TOG1 and divergent from TOG4. Shown are pairwise structural comparisons of Msps TOG3 with XMAP215 family TOG structures determined to date that include: Stu2 TOG1 (4FFB) and TOG2 (2QK1); ZYG-9 TOG3 (2OF3); Msps TOG2 (2QK2) and TOG4 (4QMH); and ch-TOG TOG4 (4QMI). Msps TOG3 is most similar to Stu2 TOG1 with an r.m.s.d. value of 2.4 Å. Structural alignment with Stu2 TOG2, Msps TOG2, and ZYG-9 TOG3 yield an r.m.s.d. values ranging from 2.8 to 3.2 Å. The structure of TOG3 diverges from TOG4 structures with r.m.s.d. values of 4.6 and 4.8 Å for ch-TOG and Msps, respectively.
To assess how Msps TOG3 might interact with tubulin, we aligned TOG3 to Stu2 TOG1 from the Stu2 TOG1·αβ-tubulin complex structure and examined potential TOG3-tubulin interactions (Fig. 7, A–C). Stu2 TOG1 was recently solved in complex with curved αβ-tubulin, a conformation observed in nonpolymerized tubulin (40, 53–55). In the Stu2 TOG1·αβ-tubulin complex structure, HRs A–D contact β-tubulin, and HRs E and F contact α-tubulin (Fig. 7A) (40). When the TOG1 structure is modeled on the structure of straight αβ-tubulin, as found in polymerized MTs, a gap is formed between HRs E and F and α-tubulin (Fig. 7A, red arrow) (56, 57). These data suggest that TOG1 binds the natively curved conformation of the free αβ-tubulin heterodimer (40). Modeling Msps TOG3 onto the Stu2 TOG1·αβ-tubulin structure suggests that intra-HEAT loops in Msps TOG3 HRs A–D and HRs E and F are well positioned to contact β- and α-tubulin, respectively, through interactions homologous to those observed between Stu2 TOG1 and αβ-tubulin (Fig. 7, B and C). Similar results were obtained when TOG3 was docked on the structure of the Stu2 TOG2·αβ-tubulin complex (41). Interestingly, a gap is formed between TOG3 and tubulin when TOG3 is modeled on straight αβ-tubulin suggesting that TOG3, like TOG1 and TOG2, preferentially binds a curved tubulin conformation (Fig. 7B, red arrow).
FIGURE 7.
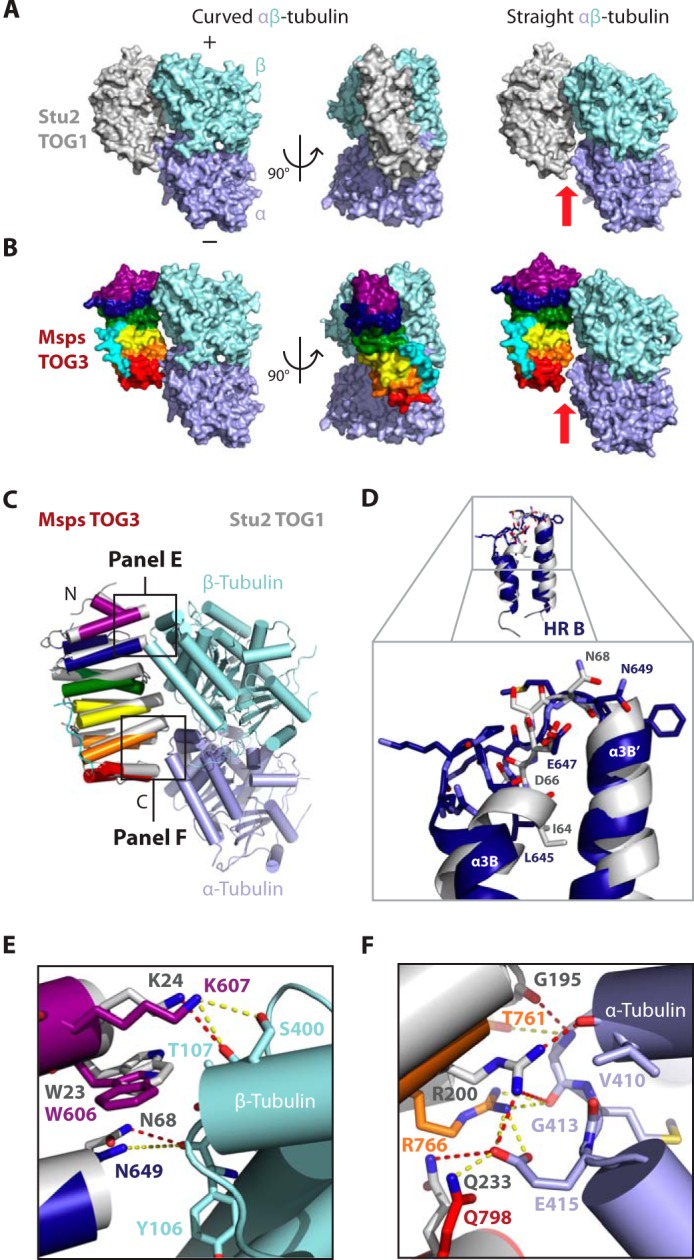
Despite structural divergence, Msps TOG3 has conserved the position of important tubulin-binding residues in a Stu2 TOG1- and TOG2-like tubulin-binding mode. A, Stu2 TOG1 bound to a curved (left, 4FFB) and straight (right, modeled) αβ-tubulin heterodimer. When modeled on straight tubulin (1JFF), a gap is formed between TOG1 HR F and α-tubulin (see red arrow). B, models of Msps TOG3 bound to curved and straight αβ-tubulin were generated by structurally aligning Msps TOG3 to the Stu2 TOG1 coordinate file used in A. TOG3 docks well onto curved αβ-tubulin. A gap is formed between TOG3 and α-tubulin when TOG3 is modeled on straight αβ-tubulin (see red arrow). C, cartoon model of Msps TOG3 (color) superimposed on the Stu2 TOG1 (gray) αβ-tubulin complex structure (4FFB). D–F, zoom views of the alignments shown in C. D, despite structural divergence, Msps TOG3 intra-HR loop B maintains the position of important, conserved tubulin-binding residues, including Asn-649, which is positioned similar to Stu2 TOG1 Asn-68. E and F, Msps TOG3 is predicted to interact with αβ-tubulin in a Stu2 TOG1 tubulin-binding mode. E, Msps TOG3 residues Trp-606, Lys-607, and Asn-649 are positioned similar to Stu2 TOG1 residues Trp-23, Lys-24, and Asn-68, respectively, which interact with β-tubulin residues Thr-107, Ser-400, and Tyr-106 (residue numbers are from S. cerevisiae β-tubulin; these residues are identical to Drosophila β-tubulin counterparts, with the exception of Ser-400 corresponding to Gly-400 in Drosophila). F, Msps TOG3 HRs E and F are predicted to make Stu2 TOG1-like contacts with α-tubulin, including a conserved network of salt bridges. Interactions observed in the Stu2 TOG1·αβ-tubulin complex are shown as red dashed lines, and potential homologous interactions between Msps TOG3 and tubulin are shown as yellow dashed lines.
Although our modeling indicates that Msps TOG3 may bind curved αβ-tubulin in a mode similar to Stu2 TOG1, we have shown that Msps TOG3 has a truncated α3B helix that diverges from other known TOG domain structures. We next examined how the different architecture of TOG3 HR B may affect the ability of TOG3 to bind αβ-tubulin. Interestingly, even though Msps TOG3 HR B has a unique architecture, HR B intra-HEAT loop residues are positioned nearly identical to the homologous Stu2 TOG1 tubulin-binding residues (Fig. 7, D–F). For example, the Msps Asn-649 and Stu2 Asn-68 side chains are equivalently positioned to interact with the β-tubulin Tyr-106 backbone carbonyl (Fig. 7, D and E). Thus, TOG3 has the potential to form similar interactions with β-tubulin using intra-HEAT loop B as Stu2 TOG1 does. In general, Msps TOG3 intra-HEAT loop tubulin-binding determinants are positioned similar to the conformation of Stu2 TOG1 residues that contact tubulin. The position of Msps TOG3 HR A residues Lys-607 and Trp-606 closely mimic those of Stu2 TOG1 HR A residues Lys-24 and Trp-23, which interact with β-tubulin Thr-107 and Ser-400 (Fig. 7E). Conserved tubulin-binding interactions continue into the second HR triad (Fig. 7F). Stu2 TOG1 HR E residue Arg-200 forms a conserved salt bridge with α-tubulin Glu-415. The homologous Msps TOG3 residue Arg-766 is similarly positioned to interact with α-tubulin Glu-415, as well. Additionally, the backbone carbonyl of Msps TOG3 HR E Thr-761 is arranged similar to Stu2 TOG1 Gly-195, which interacts with the backbone amide of α-tubulin Gly-413. Msps TOG3 Gln-798 and Stu2 TOG1 Gln-233, although located on different turns on the α3F′ helix, are both positioned to interact with α-tubulin Glu-415 (Fig. 7F and see 1C). Taken together, these findings suggest that although Msps TOG3 HRs have structurally diverged from HRs in XMAP215 family TOG1 and TOG2 structures, with an emphasis on HR B, TOG3 intra-HEAT loop tubulin-binding determinants have maintained a structural organization that predicts a tubulin-binding mode similar to how Stu2 TOG1 and TOG2 engage the curved αβ-tubulin heterodimer.
DISCUSSION
Implications for XMAP215 Arrangement at the MT Plus-end
Spatiotemporal regulation of MT polymerization by XMAP215 family members is essential for proper cellular function, with depletion of XMAP215 or its orthologs resulting in reduced MT polymerization rates and abnormal spindle phenotypes (7–11, 14, 17–27). Family members utilize characteristic domains to localize to MT plus-ends and promote MT polymerization. XMAP215 family members have a conserved C-terminal domain that binds SLAIN2 (H. sapiens)/Sentin (D. melanogaster), which confers EB1-dependent MT plus-end localization (33, 34). EB1 fenestrates the MT lattice, binding between four tubulin heterodimers where it recognizes tubulin in a GTP hydrolysis transition/post-hydrolysis state mimicked by the GTP analog GTPγS (58–60). Thus, EB1 binds just distal to the polymerizing MT GTP-cap and via SLAIN2/Sentin localizes the XMAP215 C terminus to this zone (33, 34, 61, 62). In contrast, the XMAP215 N-terminal TOG domain array binds and incorporates αβ-tubulin into the growing MT plus-end (28–31, 38, 40, 41). The differential positioning of the XMAP215 N- and C-terminal domains at the MT plus-end likely results in a dynamic/polarized arrangement on the MT and requires an extended XMAP215 configuration (31, 62, 63). How each TOG domain in the pentameric array is positioned along the growing MT plus-end remains to be determined. Studies to date suggest that TOG1 and TOG2 bind and tether tubulin heterodimers proximal to the MT plus-end to promote their incorporation into the lattice (40, 41). This raises the following question. What role do TOG domains 3–5 play in the polymerization mechanisms? Do they function like TOG1 and TOG2 or are they designed to bind the MT lattice in the space spanning the GTP cap and the EB1-binding zone?
A polarized arrangement of XMAP215 at the MT plus-end suggests that TOG domain structures along the array may be differentially honed to perform unique functions, either to bind and deliver free tubulin to the MT plus-end or bind and recognize different tubulin states along the polymerizing MT (different tubulin-nucleotide states and/or curved versus straight tubulin). We know from recent structures of individual Stu2 TOG domains in complex with tubulin that TOG1 and TOG2 bind a curved αβ-tubulin conformation predicted to be similar to the structural state of tubulin free in solution (40, 41). TOG4 reveals a conserved architecture distinct from structures of TOG1 and TOG2 that predict binding to a curved tubulin heterodimer, albeit in a mode that recognizes a unique site on α-tubulin (16). Additionally, TOG5 has the lowest degree of intra-TOG sequence similarity with other TOG domains in the array, suggesting that TOG5 may have unique structural determinants and tubulin-binding properties (16, 29, 39, 44). Although the structure of TOG5 is unknown, we predict that it is similar to the structure of ZYG-9 TOG3. ZYG-9 TOG3 is also predicted to engage tubulin in a curved conformation; however, ZYG-9 TOG3 has an additional N-terminal HR that is positioned orthogonal to the TOG domain's six core HRs, extends the TOG domain's lateral reach, and may facilitate binding across neighboring protofilaments during polymerization. In support of lateral tubulin binding, a linker-TOG5 construct has been shown to localize along the lengths of MTs in cell culture (15).
In this study, we asked whether a TOG3 domain from a pentameric XMAP215 family member has a unique architecture or whether TOG3 is structurally similar to either TOG1 and TOG2 or TOG4. Our analyses show that although TOG3 has unique structural features, including a C-terminal extension that stabilizes HRs C–F and a truncated α3B helix followed by an extended intra-HEAT loop B, overall the TOG3 structure closely resembles that of TOG1 and TOG2 and is divergent from TOG4. Despite significant differences in the architecture of TOG3 HR B, the TOG3 intra-HEAT loops that compose the domain's tubulin-binding surface are spatially positioned to engage tubulin, as observed in the Stu2 TOG1·tubulin and TOG2·tubulin complexes. This suggests that TOG3 has a tubulin-binding mode similar to TOG1 and TOG2 and uses its intra-HEAT loops to engage a curved conformation of the αβ-tubulin heterodimer.
Although we predict that TOG3 adopts a TOG1- and TOG2-like tubulin-binding mode, the question why TOG3 has divergent structural components (an elongated intra-HR B loop and an ordered C-terminal tail that binds alongside the domain) remains unknown. Three potential explanations for the conserved unique structural features of TOG3 are as follows. 1) These components may regulate domain flexibility, preventing or promoting TOG domain movement as tubulin transitions from a curved to a straight conformation during polymerization. 2) These structural components may allosterically affect tubulin binding. 3) TOG3's unique structural features may be used to engage other factors in trans or other Msps domains or linker regions in cis to control MT polymerization activity. Whether these unique determinants regulate domain flexibility, tubulin binding, or binding to other factors remains to be determined.
How does the structure of TOG domains across the pentameric array correlate with function? Studies show that Msps and XMAP215 TOG1–2 constructs can bind tubulin heterodimers over gel filtration, correlating with the ability of Stu2 TOG1 and TOG2 to individually bind tubulin heterodimers in a curved tubulin conformation (16, 29, 38, 40, 41). In contrast, an Msps TOG3-4 construct did not show robust tubulin binding over gel filtration, although it did increase MT polymerization activity, indicating that TOG3-4 forms transient interactions with tubulin heterodimers (16). Additionally, a full-length XMAP215 construct containing mutations that ablated the tubulin binding activity of TOG domains 1 and 2 retained some tubulin binding activity over gel filtration that was not evident when all five TOG domains were mutated (29). This suggests that all, or a subset of the ultimate three TOG domains, can bind free tubulin heterodimers. However, data also suggest that TOGs 3–5 can bind the straight conformation of tubulin found in the MT lattice. An Msps TOG3-4 construct when overexpressed in cell culture localized along the length of MTs, and this activity could be diminished when either the conserved TOG3 or TOG4 HR A loop tryptophan used to bind tubulin was mutated to glutamate (15, 16). Similarly, an XMAP215 TOG1-4 construct showed weak MT lattice binding in vitro that was not observed with an XMAP215 TOG1-2 construct (29). These published observations indicate that TOG domains have different tubulin-binding affinities and suggest that they may have differential capacities to recognize curved or straight tubulin. However, the XMAP215 family TOG domain structures determined to date (TOGs 1–4, as well as ZYG-9 TOG3, which we predict is homologous to the structure of TOG5) all have a tubulin-binding surface that would best complement the tubulin heterodimer's curved architecture. How then can TOGs 3–5 contribute to MT lattice binding, which indicates an ability to interact with straight tubulin? To accommodate both curved and straight tubulin binding, TOG domains 3–5 may remain structurally static and use only a portion of their tubulin-binding surface to interact with straight tubulin (as modeled in Fig. 7B, at right), or they may bend, undergoing a conformational change that facilitates binding to straight tubulin across the length of the TOG domain. Similarly, these TOG domains may bind free tubulin heterodimers through cooperative action with TOG domains 1 and 2 and dynamically change shape with tubulin as it transitions from its curved to straight conformation. Although data suggest that Stu2 TOG1 and TOG2 are not cooperative in their tubulin binding activity (41), cooperative tubulin binding activity between TOGs 1 and 2 with TOGs 3–5 has not been examined.
Organization Diversity of TOG Domains across the XMAP215 Family
The S. cerevisiae XMAP215 member Stu2 has only two TOG domains, but it drives MT polymerization as a homodimer (22, 24, 27, 35, 36, 40, 41). Thus, Stu2 functionally operates with four TOG domains (TOG1-TOG2)2. XMAP215 pentameric TOG array members are monomeric, and although TOG domains 1–3 bear structural similarity to Stu2 TOG domains 1 and 2, their spatial arrangement at the MT plus-end is likely divergent; a parallel 2 × 2 TOG arrangement in yeast that may function to enhance longitudinal and lateral tubulin interactions is likely to differ significantly from the potential linear 5 × 1 TOG domain arrangement of XMAP215 members with a pentameric TOG array. Additionally, the unique architecture of TOG4 and the sequence divergence of TOG5 predict novel functional attributes for these TOG domains that further distinguish the polymerization mechanism of pentameric TOG domain XMAP215 members from Stu2.
Distinct TOG Array Protein Families Differentially Regulate MT Dynamics
The use of architecturally distinct TOG domains in an array to differentially regulate MT dynamics is an emerging paradigm. CLASP is predicted to contain a trimeric TOG domain array that functions to promote MT pause (64–68). The structure of D. melanogaster CLASP (MAST) TOG1 reveals a XMAP215 TOG1-like architecture (69). In contrast, the structure of H. sapiens CLASP1 TOG2 is architecturally distinct from XMAP215 TOG domains 1–3 (51). When the first HR triad of Msps TOG3 and CLASP TOG2 was aligned, CLASP TOG2 HR F was displaced ∼11 Å relative to Msps TOG3 HR F due to a dramatic bend across the CLASP TOG2 tubulin-binding surface, which is predicted to underlie CLASP-dependent MT pause (51). The differential architectures of TOG domains between and within the XMAP215 and CLASP TOG arrays likely underlie each family's specific MT regulatory mechanism.
Characterizing how TOG domain arrays differentially regulate MT dynamics is dependent on understanding how individual TOG domains function collectively. In the case of the XMAP215 family members, mutating tubulin binding activity in any subset of TOG domains within the array affects tubulin binding and MT polymerization activity with concomitant effects on mitotic spindle size (8, 16, 17, 20, 29, 42, 43). The key to understanding the XMAP215 TOG array MT polymerization mechanism is determining how many tubulin heterodimers the array binds, whether the pentameric TOG array simply tethers tubulin heterodimers or actively potentiates longitudinal or lateral tubulin-tubulin interactions, and what nucleotide and/or conformational state of tubulin each TOG domain in the array preferentially binds.
The structure of Msps TOG3 furthers our understanding of the XMAP215 pentameric TOG array, highlighting that the first three TOG domains in the array have a common tubulin-binding surface and likely engage similar tubulin conformational states. Determining the structure of TOG5, the most sequence divergent XMAP215 family TOG domain, and understanding how all five TOG domains function collectively to drive MT polymerization are key next steps in understanding the XMAP215 MT polymerization mechanism.
Acknowledgment
We thank Ashutosh Tripathy for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants T32GM008570 (to the Program in Molecular and Cellular Biophysics, University of North Carolina at Chapel Hill) and Grant R01GM094415. This work was also supported by March of Dimes Grant FY14-247 (to K. C. S.).
The atomic coordinates and structure factors (code 4Y5J) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MT
- microtubule
- TOG
- tumor overexpressed gene
- HEAT
- Huntingtin, elongation factor 3, protein phosphatase 2A, target of rapamycin
- HR
- HEAT repeat(s)
- +TIPs
- plus-end tracking proteins
- XMAP215
- Xenopus microtubule-associated protein 215
- CTD
- C-terminal domain
- Msps
- Mini spindles
- SeMet
- selenomethionine
- SAD
- single-wavelength anomalous diffraction
- r.m.s.d.
- root mean square deviation
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Desai A., Mitchison T. J. (1997) Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 [DOI] [PubMed] [Google Scholar]
- 2. Gelfand V. I., Bershadsky A. D. (1991) Microtubule dynamics: mechanism, regulation, and function. Annu. Rev. Cell Biol. 7, 93–116 [DOI] [PubMed] [Google Scholar]
- 3. Wang H.-W., Nogales E. (2005) Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature 435, 911–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akhmanova A., Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 5. Galjart N. (2010) Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20, R528–R537 [DOI] [PubMed] [Google Scholar]
- 6. Gard D. L., Kirschner M. W. (1987) A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 105, 2203–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gard D. L., Becker B. E., Josh Romney S. (2004) MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 239, 179–272 [DOI] [PubMed] [Google Scholar]
- 8. Al-Bassam J., Kim H., Flor-Parra I., Lal N., Velji H., Chang F. (2012) Fission yeast Alp14 is a dose-dependent plus-end-tracking microtubule polymerase. Mol. Biol. Cell 23, 2878–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawamura E., Wasteneys G. O. (2008) MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J. Cell Sci. 121, 4114–4123 [DOI] [PubMed] [Google Scholar]
- 10. Matthews L. R., Carter P., Thierry-Mieg D., Kemphues K. (1998) ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cullen C. F., Deák P., Glover D. M., Ohkura H. (1999) mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol. 146, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charrasse S., Mazel M., Taviaux S., Berta P., Chow T., Larroque C. (1995) Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur. J. Biochem. 234, 406–413 [DOI] [PubMed] [Google Scholar]
- 13. Charrasse S., Schroeder M., Gauthier-Rouviere C., Ango F., Cassimeris L., Gard D. L., Larroque C. (1998) The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 111, 1371–1383 [DOI] [PubMed] [Google Scholar]
- 14. Goshima G., Wollman R., Stuurman N., Scholey J. M., Vale R. D. (2005) Length control of the metaphase spindle. Curr. Biol. 15, 1979–1988 [DOI] [PubMed] [Google Scholar]
- 15. Currie J. D., Stewman S., Schimizzi G., Slep K. C., Ma A., Rogers S. L. (2011) The microtubule lattice and plus-end association of Drosophila Mini spindles is spatially regulated to fine-tune microtubule dynamics. Mol. Biol. Cell 22, 4343–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fox J. C., Howard A. E., Currie J. D., Rogers S. L., Slep K. C. (2014) The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol. Biol. Cell 25, 2375–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brittle A. L., Ohkura H. (2005) Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 24, 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Popov A. V., Severin F., Karsenti E. (2002) XMAP215 Is required for the microtubule-nucleating activity of centrosomes. Curr. Biol. 12, 1326–1330 [DOI] [PubMed] [Google Scholar]
- 19. Cassimeris L., Becker B., Carney B. (2009) TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motil. Cytoskeleton 66, 535–545 [DOI] [PubMed] [Google Scholar]
- 20. Whittington A. T., Vugrek O., Wei K. J., Hasenbein N. G., Sugimoto K., Rashbrooke M. C., Wasteneys G. O. (2001) MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613 [DOI] [PubMed] [Google Scholar]
- 21. Kronja I., Kruljac-Letunic A., Caudron-Herger M., Bieling P., Karsenti E. (2009) XMAP215-EB1 interaction is required for proper spindle assembly and chromosome segregation in Xenopus egg extract. Mol. Biol. Cell 20, 2684–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kosco K. A., Pearson C. G., Maddox P. S., Wang P. J., Adams I. R., Salmon E. D., Bloom K., Huffaker T. C. (2001) Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell 12, 2870–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tournebize R., Popov A., Kinoshita K., Ashford A. J., Rybina S., Pozniakovsky A., Mayer T. U., Walczak C. E., Karsenti E., Hyman A. A. (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13–19 [DOI] [PubMed] [Google Scholar]
- 24. Severin F., Habermann B., Huffaker T., Hyman T. (2001) Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hestermann A., Gräf R. (2004) The XMAP215-family protein DdCP224 is required for cortical interactions of microtubules. BMC Cell Biol. 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cassimeris L., Morabito J. (2004) TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell 15, 1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang P. J., Huffaker T. C. (1997) Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 139, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J., Hyman A. A. (2008) XMAP215 is a processive microtubule polymerase. Cell 132, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Widlund P. O., Stear J. H., Pozniakovsky A., Zanic M., Reber S., Brouhard G. J., Hyman A. A., Howard J. (2011) XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. U.S.A. 108, 2741–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spittle C., Charrasse S., Larroque C., Cassimeris L. (2000) The interaction of TOGp with microtubules and tubulin. J. Biol. Chem. 275, 20748–20753 [DOI] [PubMed] [Google Scholar]
- 31. Cassimeris L., Gard D., Tran P. T., Erickson H. P. (2001) XMAP215 is a long thin molecule that does not increase microtubule stiffness. J. Cell Sci. 114, 3025–3033 [DOI] [PubMed] [Google Scholar]
- 32. Lee M. J., Gergely F., Jeffers K., Peak-Chew S. Y., Raff J. W. (2001) Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule dynamics. Nat. Cell Biol. 3, 643–649 [DOI] [PubMed] [Google Scholar]
- 33. van der Vaart B., Manatschal C., Grigoriev I., Olieric V., Gouveia S. M., Bjelic S., Demmers J., Vorobjev I., Hoogenraad C. C., Steinmetz M. O., Akhmanova A. (2011) SLAIN2 links microtubule plus-end-tracking proteins and controls microtubule growth in interphase. J. Cell Biol. 193, 1083–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K., Goshima G. (2012) Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 199, 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Breugel M., Drechsel D., Hyman A. (2003) Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus-end-binding microtubule destabilizer. J. Cell Biol. 161, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Al-Bassam J., van Breugel M., Harrison S. C., Hyman A. (2006) Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 172, 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Slep K. C. (2009) The role of TOG domains in microtubule plus-end dynamics. Biochem. Soc. Trans. 37, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 38. Slep K. C., Vale R. D. (2007) Structural basis of microtubule plus-end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell 27, 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Al-Bassam J., Larsen N. A., Hyman A. A., Harrison S. C. (2007) Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure 15, 355–362 [DOI] [PubMed] [Google Scholar]
- 40. Ayaz P., Ye X., Huddleston P., Brautigam C. A., Rice L. M. (2012) A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337, 857–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ayaz P., Munyoki S., Geyer E. A., Piedra F.-A., Vu E. S., Bromberg R., Otwinowski Z., Grishin N. V., Brautigam C. A., Rice L. M. (2014) A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife 3, e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reber S. B., Baumgart J., Widlund P. O., Pozniakovsky A., Howard J., Hyman A. A., Jülicher F. (2013) XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol. 15, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 43. Lechner B., Rashbrooke M. C., Collings D. A., Eng R. C., Kawamura E., Whittington A. T., Wasteneys G. O. (2012) The N-terminal TOG domain of Arabidopsis MOR1 modulates affinity for microtubule polymers. J. Cell Sci. 125, 4812–4821 [DOI] [PubMed] [Google Scholar]
- 44. Al-Bassam J., Chang F. (2011) Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leahy D. J., Erickson H. P., Aukhil I., Joshi P., Hendrickson W. A. (1994) Crystallization of a fragment of human fibronectin: Introduction of methionine by site-directed mutagenesis to allow phasing via selenomethionine. Proteins 19, 48–54 [DOI] [PubMed] [Google Scholar]
- 46. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 47. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hasegawa H., Holm L. (2009) Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 19, 341–348 [DOI] [PubMed] [Google Scholar]
- 51. Leano J. B., Rogers S. L., Slep K. C. (2013) A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure 21, 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 53. Pecqueur L., Duellberg C., Dreier B., Jiang Q., Wang C., Plückthun A., Surrey T., Gigant B., Knossow M. (2012) A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus-end. Proc. Natl. Acad. Sci. U.S.A. 109, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rice L. M., Montabana E. A., Agard D. A. (2008) The lattice as allosteric effector: structural studies of αβ- and γ-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 5378–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brouhard G. J., Rice L. M. (2014) The contribution of αβ-tubulin curvature to microtubule dynamics. J. Cell Biol. 207, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nogales E., Wolf S. G., Downing K. H. (1998) Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 [DOI] [PubMed] [Google Scholar]
- 57. Löwe J., Li H., Downing K. H., Nogales E. (2001) Refined structure of αβ-tubulin at 3.5 Å resolution. J. Mol. Biol. 313, 1045–1057 [DOI] [PubMed] [Google Scholar]
- 58. Maurer S. P., Fourniol F. J., Bohner G., Moores C. A., Surrey T. (2012) EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maurer S. P., Bieling P., Cope J., Hoenger A., Surrey T. (2011) GTPγS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs). Proc. Natl. Acad. Sci. U.S.A. 108, 3988–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akhmanova A., Steinmetz M. O. (2011) Microtubule end binding: EBs sense the guanine nucleotide state. Curr. Biol. 21, R283–R285 [DOI] [PubMed] [Google Scholar]
- 61. Zanic M., Widlund P. O., Hyman A. A., Howard J. (2013) Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15, 688–693 [DOI] [PubMed] [Google Scholar]
- 62. Nakamura S., Grigoriev I., Nogi T., Hamaji T., Cassimeris L., Mimori-Kiyosue Y. (2012) Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS ONE 7, e51442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. (2006) Assembly dynamics of microtubules at molecular resolution. Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 64. Al-Bassam J., Kim H., Brouhard G., van Oijen A., Harrison S. C., Chang F. (2010) CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev. Cell 19, 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Akhmanova A., Hoogenraad C. C., Drabek K. (2001) CLASPs are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104, 923–935 [DOI] [PubMed] [Google Scholar]
- 66. Drabek K., van Ham M., Stepanova T., Draegestein K., van Horssen R., Sayas C. L., Akhmanova A., Ten Hagen T., Smits R., Fodde R., Grosveld F., Galjart N. (2006) Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr. Biol. 16, 2259–2264 [DOI] [PubMed] [Google Scholar]
- 67. Bratman S. V., Chang F. (2007) Stabilization of overlapping microtubules by fission yeast CLASP. Dev. Cell 13, 812–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mimori-Kiyosue Y., Grigoriev I. (2005) CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. De la Mora-Rey T., Guenther B. D., Finzel B. C. (2013) The structure of the TOG-like domain of Drosophila melanogaster Mast/Orbit. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]