Highlight text
Overexpression of AtPP2CF1 led to increased plant biomass production through cell proliferation and expansion in stems. AtPP2CF1 is a candidate target gene for engineering plant growth production.
Key words: ABA (abscisic acid), biomass, cell proliferation and expansion, inflorescence stem, plant, PP2C.
Abstract
In contrast to mammals, higher plants have evolved to express diverse protein phosphatase 2Cs (PP2Cs). Of all Arabidopsis thaliana PP2Cs, members of PP2C subfamily A, including ABI1, have been shown to be key negative regulators of abscisic acid (ABA) signalling pathways, which regulate plant growth and development as well as tolerance to adverse environmental conditions. However, little is known about the enzymatic and signalling roles of other PP2C subfamilies. Here, we report a novel Arabidopsis subfamily E PP2C gene, At3g05640, designated AtPP2CF1. AtPP2CF1 was dramatically expressed in response to exogenous ABA and was expressed in vascular tissues and guard cells, similar to most subfamily A PP2C genes. In vitro enzymatic activity assays showed that AtPP2CF1 possessed functional PP2C activity. However, yeast two-hybrid analysis revealed that AtPP2CF1 did not interact with PYR/PYL/RCAR receptors or three SnRK2 kinases, which are ABI1-interacting proteins. This was supported by homology-based structural modelling demonstrating that the putative active- and substrate-binding site of AtPP2CF1 differed from that of ABI1. Furthermore, while overexpression of ABI1 in plants induced an ABA-insensitive phenotype, Arabidopsis plants overexpressing AtPP2CF1 (AtPP2CF1oe) were weakly hypersensitive to ABA during seed germination and drought stress. Unexpectedly, AtPP2CF1oe plants also exhibited increased biomass yield, mainly due to accelerated growth of inflorescence stems through the activation of cell proliferation and expansion. Our results provide new insights into the physiological significance of AtPP2CF1 as a candidate gene for plant growth production and for potential application in the sustainable supply of plant biomass.
Introduction
Protein phosphatase 2Cs (PP2Cs) are monomeric enzymes in which catalytic and regulatory domains are present on the same polypeptide, and which require divalent metal ions (typically Mn2+ or Mg2+) for their activity. PP2Cs play important roles in cell differentiation, growth, and metabolism, and in environmental stress signalling pathways, and are found in eukaryotes and some prokaryotes (Cohen, 1994; Schweighofer et al., 2004; Lu and Wang, 2008; Shi, 2009). While at least 18 PP2Cs have been identified in mammalian cells, plant PP2Cs are part of a larger and more diverse family of manganese/magnesium-dependent protein phosphatases (PPMs), with 80 isoforms identified to date in Arabidopsis and 90 in rice (Oryza sativa), implying that plant PP2C isoforms possess unique patterns of tissue and subcellular localization associated with diverse functions (Xue et al., 2008; Singh et al., 2010).
The plant hormone abscisic acid (ABA) regulates various aspects of plant growth and development, including seed maturation, dormancy, and tolerance to adverse environmental conditions—particularly drought, cold, and salinity (Verslues and Zhu, 2007; Cutler et al., 2010). Among the 13 subfamilies of Arabidopsis PP2C isoforms (A–L) classified based on phylogenetic analysis (Xue et al., 2008), members of subfamily A have been shown to be key negative regulators of ABA signalling pathways (Cutler et al., 2010; Hubbard et al., 2010; Hauser et al., 2011; Joshi-Saha et al., 2011). Considerable progress in biochemical and structural investigations of subfamily A PP2Cs has advanced the understanding of their molecular function in ABA signalling pathways. In the absence of ABA, members of this subfamily including ABI1, ABI2, and HAB1 dephosphorylate three members of the Snf1-related protein kinase 2 (SnRK2) family (SnRK2.2, -2.3, and -2.6), maintaining them in an inactive state. Under stress conditions, the presence of increased ABA is recognized by intracellular ABA receptors, namely the PYR/PYL/RCAR family of START proteins. These ABA-bound receptors bind to the catalytic site of subfamily A PP2Cs and inhibit their phosphatase activity, permitting the activation of SnRK2 kinases. In turn, SnRK2 kinases phosphorylate downstream factors such as the AREB/ABF transcription factors required for activation of ABA-responsive genes and SLAC1 anion channels involved in stomatal closure, thus relaying ABA signals (Cutler et al., 2010; Hubbard et al., 2010; Hauser et al., 2011; Joshi-Saha et al., 2011).
Plant subfamily E PP2Cs constitute the largest of the subfamilies (Supplementary Fig. S1) (Xue et al., 2008). A unique characteristic of subfamily E PP2Cs is the insertion of two amino acids in the β3-α1 loop of consensus motif 4, corresponding to residues 171–185 of ABI1 and comprising an important part of the active and substrate-binding sites (Supplementary Fig. S2) (Xue et al., 2008; Ma et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009, 2010; Park et al., 2009; Santiago et al., 2009; Yin et al., 2009). Dominant mutations in subfamily A PP2Cs, such as abi1-1 and abi2-1, which inhibit binding to PYR/PYL/RCAR receptors and induce the ABA-insensitive phenotype, reside within the β3-α1 loop, highlighting not only the general importance of this loop for substrate recognition and specificity (Miyazono et al., 2009; Umezawa et al., 2009), but also the functional differences between subfamily E PP2Cs and other PP2Cs. However, despite the potential physiological importance of subfamily E PP2Cs, much less is known about their enzymatic and signalling roles.
In this study, we characterized a novel PP2C family protein (designated AtPP2CF1) that can be classified as a subfamily E PP2C. Of the subfamily E members, AtPP2CF1 expression was strongly upregulated by ABA. An in vitro enzymatic assay revealed that AtPP2CF1 had serine/threonine phosphatase (PSP) activity typical of a classical PP2C, indicating that its activity was dependent on Mg2+. AtPP2CF1 exhibited an expression pattern similar to that of subfamily A PP2Cs such as ABI1, with transcripts detected in vascular tissues and stomatal guard cells. However, using a yeast two-hybrid (Y2H) system and homology-based structural modelling of AtPP2CF1, we found that AtPP2CF1 was unable to bind PYR/PYL/RCAR receptors or three SnRK2 kinases, both of which are ABI1-interacting proteins. Furthermore, overexpression of AtPP2CF1 caused Arabidopsis plants to exhibit weak hypersensitivity to ABA, which differed from the effect observed with overexpression of ABI1 and unexpectedly resulted in increased biomass production.
Materials and methods
Plant materials and growth conditions
All Arabidopsis thaliana plants used in this study, including wild-type, phyB-9 single mutants (phyB), cry2-1 phyB-9 double mutants (cry2 phyB), and transgenic lines, were of the Columbia ecotype (Mockler et al., 1999). Seeds were sown aseptically on Murashige and Skoog (MS) solid medium [0.5% (w/v) gellan gum or 0.8% (w/v) agar] with or without 1% sucrose or on soil Supermix A (Sakata, Kanagawa, Japan). After stratification for 3 d at 4°C, plants were grown in a growth chamber [22°C and approximately 60% relative humidity (RH)] under long-day conditions (16-h light/8-h dark cycle, approximately 50 μmol m–2 s–1 white fluorescent light). Seedlings of plants grown on solid medium were transferred to soil at 21 d after germination and fertilized weekly with Hyponex (1:1000 dilution; Hyponex Japan, Osaka, Japan).
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), and on-column DNase treatment was performed using an RNase-Free DNase Set (Qiagen, Hilden, Germany). cDNA was generated using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA) and subjected to RT-qPCR amplification using the Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). Quantification of RT-qPCR reactions was conducted using an ABI Prism 7000 sequence detection system and ABI Prism 7000 SDS software (Applied Biosystems) with gene-specific primers (Supplementary Table S1).
Construction of transgenic Arabidopsis plants
Protocols for the construction of plasmids are provided in Supplementary Methods S1 and S2. Arabidopsis transformation was carried out by the floral dip method (Clough and Bent, 1998).
Histochemical analysis of GUS staining
Arabidopsis seedlings were harvested into ice-cold 90% (v/v) acetone, incubated at room temperature for 20min to permeabilize the tissue, and then washed three times with 50mM sodium phosphate buffer, pH 7.0. β-glucuronidase (GUS) reactions were performed in GUS staining buffer [1.9mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide-cyclohexylammonium salt, 0.5mM K3Fe(CN)6, 0.5mM K4[Fe(CN)6]·3H2O, 0.3% Triton X-100, and 50mM sodium phosphate buffer, pH 7.0]. Samples were infiltrated under a vacuum for 5min and then incubated overnight at 37°C. The GUS-staining buffer was removed and the samples were washed and cleared in 70% ethanol. Observations were made with a stereomicroscope (SZX12; Olympus, Tokyo, Japan) and a fluorescence microscope (Biorevo BZ-9000; Keyence, Osaka, Japan).
Yeast ptc1 complementation test
The diploid Saccharomyces cerevisiae strain BY4743 (MATa/α his3∆1/his3∆1 leu2∆0/leu2∆0 lys2∆0/LYS2 MET15/met15∆0 ura3∆0/ura3∆0) was used in this study. The ptc1 single-knockout mutant strains were generated by the Saccharomyces Genome Deletion Project (Winzeler et al., 1999) and obtained from Open Biosystems (Huntsville, AL, USA). Protocols describing the construction of plasmids for the yeast complementation test are provided in Supplementary Methods S1 and S3. Yeast cells were grown and maintained in minimal synthetic medium containing either glucose (SD) or galactose (SG), or in complete medium containing either glucose (YPD) or galactose (YPG).
Purification of AtPP2CF1 and PP2C activity assays
Protocols detailing the construction of pGEX GST-AtPP2CF1 and pGEX GST-ABI1 are provided in Supplementary Methods S1 and S4. These constructs were introduced into Escherichia coli BL21 (Novagen, Madison, WI, USA). Recombinant fusion proteins were purified using a GSTrap 4B column (GE Healthcare, Uppsala, Sweden), concentrated with a Vivaspin 20 centrifugal concentrator (10 000 molecular weight cutoff; Sartorius, Goettingen, Germany), and passed through a PD-10 desalting column (GE Healthcare) to remove free glutathione, according to the manufacturer’s instructions. Protein concentrations were determined using the Pierce 660nm Protein Assay Kit (Thermo Scientific, Rockford, IL, USA).
PP2C activities were measured using separate Ser/Thr and Tyr phosphatase assay systems (Promega, Madison, WI), according to the manufacturer’s instructions. Briefly, the reactions were performed in 100 μl of assay buffer [50mM imidazole (pH 7.2), 0.2mM EGTA, 5mM MgCl2, 0.02% β-mercaptoethanol, and 0.1mg ml–1 BSA] by using 100 μM chemically synthesized phosphopeptide substrate [PSP substrate RRA(pT)VA, where pT represents phosphothreonine; or the tyrosine phosphatase (PTP) substrates END(pY)INASL or DADE(pY)LIPQQG, where pY represents phosphotyrosine] and 50–5250ng of recombinant protein (GST-AtPP2CF1 or GST-ABI1). The inhibitors used in the study were NaF (50mM), NaVO3 (1mM), EDTA (10mM), and okadaic acid (10 μM). For EDTA, the indicated concentrations of the chelator were added to the reaction mixtures without MgCl2. Absorbance was measured at 650nm in an Infinite F5000 microplate reader (Tecan Austria, Grödlg, Austria).
Homology-based structural modelling of AtPP2CF1
A homology-based model of AtPP2CF1 was built using the Build Homology Models protocol in Discovery Studio 3.1 (Accelrys, San Diego, CA, USA). The crystal coordinates of ABI1 in the PYL1-(+)-ABA-ABI1 complex (Protein Data Bank code 3kdj) were used as a template. Sequence alignment of AtPP2CF1 and ABI1 was made based on their structural superimposition and was adjusted by hand. Model quality was evaluated using the Profiles 3D protocol in Discovery Studio 3.1; the model structure with the highest Verify score (93.94, expected high score = 132.384) was selected as the most reliable model of AtPP2CF1.
Germination assays and leaf water loss measurements
For germination assays, ~60 sterilized seeds were plated on MS agar medium with increasing concentrations of (±)-ABA (Sigma-Aldrich, St Louis, MO, USA). After stratification for 4 d at 4°C, seedlings were grown in a growth chamber under long-day conditions. To determine seed germination rates, seeds that had germinated and developed green and expanded cotyledons were scored in six independent experiments. Leaf water loss was measured using the detached aerial parts of 3-week-old seedlings. Five seedlings per line were cut above the hypocotyl and fresh weight (FW) was determined under growth chamber conditions at the indicated times in three independent experiments.
Histological observation of inflorescence stems
For transverse sections, basal parts of 10- to 15-cm-long primary inflorescence stems or those of 9-week-old plants (10 or 20mm from the rosette base, respectively) were embedded in agarose blocks and sectioned into 100-μm-thick slices using a microslicer (DTK-1000; Dosaka, Kyoto, Japan). The sections were incubated with 0.05% (w/v) toluidine blue at room temperature for 1min, washed, and then observed using a fluorescence microscope (Biorevo BZ-9000; Keyence, Osaka, Japan). The images were processed and assembled with Adobe Photoshop (Adobe Systems, CA, USA). For each stem, the diameter and area imaged were measured using Paint.NET (http://www.getpaint.net/).
For observation of stem epidermal cells, the stem epidermis was peeled from the basal parts (20mm from the rosette base) of primary inflorescence stems in 9-week-old plants, incubated with 0.01% (w/v) safranin at room temperature for 1min, washed, and then observed using a fluorescence microscope (Keyence).
Dry biomass measurement
Arabidopsis plants were grown on soil in a growth chamber under long-day conditions for 12 weeks and fertilized weekly with Hyponex (1:500 dilution). At 12 weeks, the plants were maintained without watering for two weeks and were then cut at their hypocotyls. Dry seeds and plant tissues (excluding roots) were weighed separately on an XS205 Dual Range analytical balance (Mettler Toledo, Greifensee, Switzerland).
Measurement of flowering time
Flowering time was determined as both ‘days to flowering’ and ‘leaf number’. Days to flowering was determined as the number of days from sowing until the first flower bud became visible. Leaf number was determined by counting the number of rosette leaves at the time when the first flower bud became visible.
Accession numbers
The Arabidopsis Genome Initiative (AGI) numbers from The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/) are as follows: At3g05640 (AtPP2CF1), At4g26080 (ABI1), At4g17870 (PYR1), At5g46790 (PYL1), At2g26040 (PYL2), At1g73000 (PYL3), At2g38310 (PYL4), At5g05440 (PYL5), At2g40330 (PYL6), At4g01026 (PYL7), At5g53160 (PYL8), At1g01360 (PYL9), At4g27920 (PYL10), At5g45860 (PYL11), At5g45870 (PYL12), At4g18620 (PYL13), At3g50500 (SnRK2.2), At5g66880 (SnRK2.3), At4g33950 (SnRK2.6)
Results
AtPP2CF1 gene expression increased dramatically in response to exogenous ABA
In Arabidopsis, most subfamily A PP2C genes and several subfamily B and G PP2C genes are transcriptionally activated by ABA (Leung et al., 1997; Merlot et al., 2001; Saez et al., 2004; Kuhn et al., 2006; Nishimura et al., 2007; Brock et al., 2010; Liu et al., 2012; Zhang et al., 2012). Previous microarray analyses have suggested that the expression of a few subfamily E PP2Cs is also induced by exogenous ABA (Goda et al., 2008). Thus, we first investigated whether expression of subfamily E PP2C genes was regulated by exogenous ABA (Fig. 1A). RT-qPCR experiments using total RNA isolated from aerial parts of Arabidopsis plants treated with ABA demonstrated that transcripts of four subfamily E genes (At1g79630, At3g05640, At5g26010, and At5g27930) accumulated in response to ABA. Interestingly, peak expression of these genes was delayed by ~2–4h compared with that of ABI1, and accumulation of transcripts was downregulated by exogenous ABA only for one subfamily E PP2C gene, At4g32950 (Fig. 1A). Of the four ABA-inducible subfamily E PP2C genes, At3g05640 showed the strongest response to ABA, with an induction of at least 3.5-fold (Fig. 1A), indicating its physiological significance in the ABA signal transduction pathway. This gene was designated AtPP2CF1 and subsequent experiments were focussed on this target.
Fig. 1.
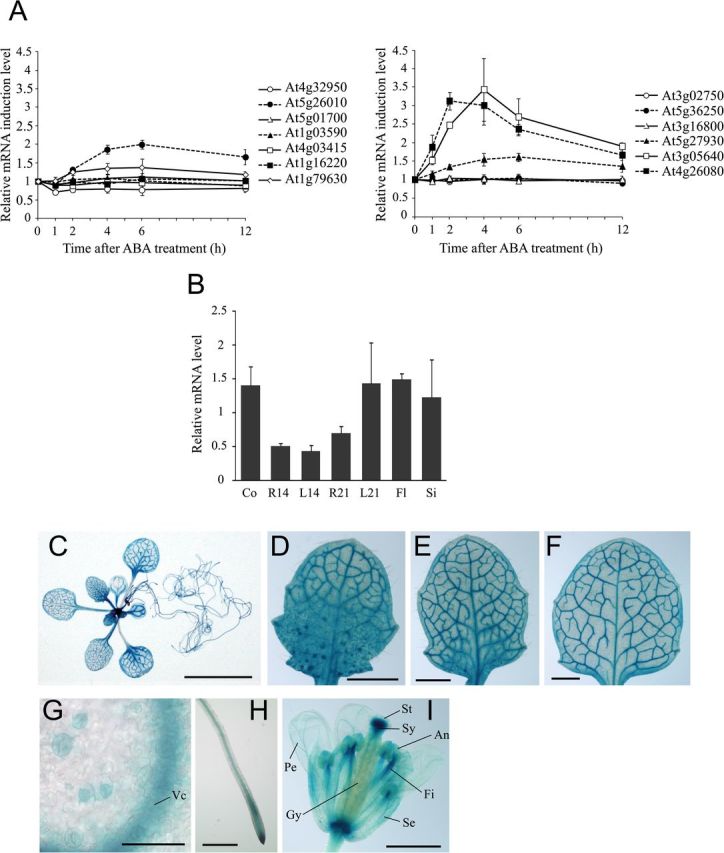
Spatial, temporal, and ABA-mediated regulation of AtPP2CF1 expression. Arabidopsis plants were germinated aseptically on MS medium with gellan gum. At 21 d after germination, seedlings were transferred to soil. (A) Time course of the induction of subfamily E PP2C mRNA expression in aerial parts of wild-type Arabidopsis plants in response to exogenous ABA. Two-week-old seedlings were sprayed with 10 μM ABA or 0.1% DMSO (mock control). Total RNA from aerial parts was isolated at the indicated time points after ABA or mock treatment and subjected to RT-qPCR. The expression ratios of individual subfamily E PP2C genes to UBC9 were calculated for each time point and treatment (raw data are shown in Supplementary Fig. S3), and induction of each PP2C mRNA after ABA treatment was calculated by dividing the expression after ABA treatment by the expression after mock treatment at each time point. Values represent the mean ± SD of three independent experiments. (B) Expression of AtPP2CF1 transcripts. Total RNA from different tissues of wild-type Arabidopsis plants was isolated and subjected to RT-qPCR. The expression ratios of AtPP2CF1 to UBC9 were calculated for each sample. Values represent the mean ± SD of three independent experiments. Co, 7-d-old cotyledons; R14, 14-d-old roots; L14, 14-d-old first and second rosette leaves; R21, 21-d-old roots; L21, 21-d-old first to fourth rosette leaves; Fl, flowers; Si, siliques. (C–I) Histochemical analysis of GUS activity in Arabidopsis pAtPP2CF1:GUS transgenic plants. (C) A 21-d-old seedling. (D–F) A developmental series of rosette leaves ranging from young (D) to old (F) within a 21-d-old transgenic seedling. (G) Guard cells of young leaves. (H) Roots. (I) Flowers. Vc, vascular cylinder; An, anther; Fi, filament, Gy, gynoecium; Pe, petal; Se, sepal; St, stigma; Sy, style. Scale bars: 10mm (C); 1mm (D–F, I); 500 μm (H); 50 μm (G).
AtPP2CF1 expression is subject to developmental and cell-type-specific regulation
Genomes of higher plants, including Arabidopsis and rice, contain a greater number of PP2C genes than mammalian genomes (Schweighofer et al., 2004; Lu and Wang, 2008; Xue et al., 2008; Singh et al., 2010). Therefore, it was reasonable to predict that each plant PP2C gene might exhibit a unique developmental expression pattern based on its physiological function. RT-qPCR analysis revealed that AtPP2CF1 was ubiquitously expressed in all plant tissues, with higher levels of expression in the cotyledons, mature leaves, flowers, and siliques (Fig. 1B).
In order to examine the tissue- or cell-type-specific expression pattern of AtPP2CF1 in greater detail, we prepared a construct expressing the GUS reporter gene under the transcriptional control of the AtPP2CF1 promoter and introduced this into Arabidopsis plants (Fig. 1C–I). Vascular-specific GUS staining was detected in most tissues, including the cotyledons, rosette leaves, roots, hypocotyl, sepals, and petals (Fig. 1C–I and Supplementary Fig. S4). AtPP2CF1 expression was also detectable at low levels in guard cells (Fig. 1G). These tissues and cells corresponded to the sites of ABA synthesis, consistent with the ability of ABA to induce AtPP2CF1 expression (Fig. 1A) (Koiwai et al., 2004). Furthermore, GUS staining revealed that AtPP2CF1 was expressed predominantly in root tips but weakly in the basal regions of young developing leaves, which have high rates of cell division, suggesting that AtPP2CF1 may play a role in proliferative growth (Fig. 1D–F, H).
AtPP2CF1 encoded an enzymatically active PP2C-type protein phosphatase
AtPP2CF1 showed high similarity to yeast PTC1 and Arabidopsis ABI1 (33.1% and 36.5%, respectively) (Supplementary Fig. S5). To examine whether AtPP2CF1 had PP2C activity, we carried out a yeast complementation test. In yeast (S. cerevisiae), loss of functional PTC1 leads to a temperature-sensitive growth defect in which mutant cells grow more slowly at the restrictive temperature (37°C) than at the permissive temperature (30°C) (Maeda et al., 1993). Therefore, only the yeast ptc1 knockout strain producing an active PP2C is able to grow normally at the restrictive temperature. In our study, transformation of ptc1 cells with the PTC1 expression vector restored normal growth at the restrictive temperature (Fig. 2A, construct 2), while transformation with an empty vector did not (Fig. 2A, construct 1). Some reports have demonstrated that expression of some subfamily A PP2C members, including ABI1, could complement the growth defect in this strain (Fig. 2A, construct 4) (Bertauche et al., 1996; Leung et al., 1997; Zhang et al., 2012). However, in the present study, expression of AtPP2CF1 did not restore the temperature-sensitive growth defect of this strain (Fig. 2A, construct 3), suggesting that AtPP2CF1 may not have functional PP2C activity or could not efficiently recognize the original substrates of yeast PTC1.
Fig. 2.
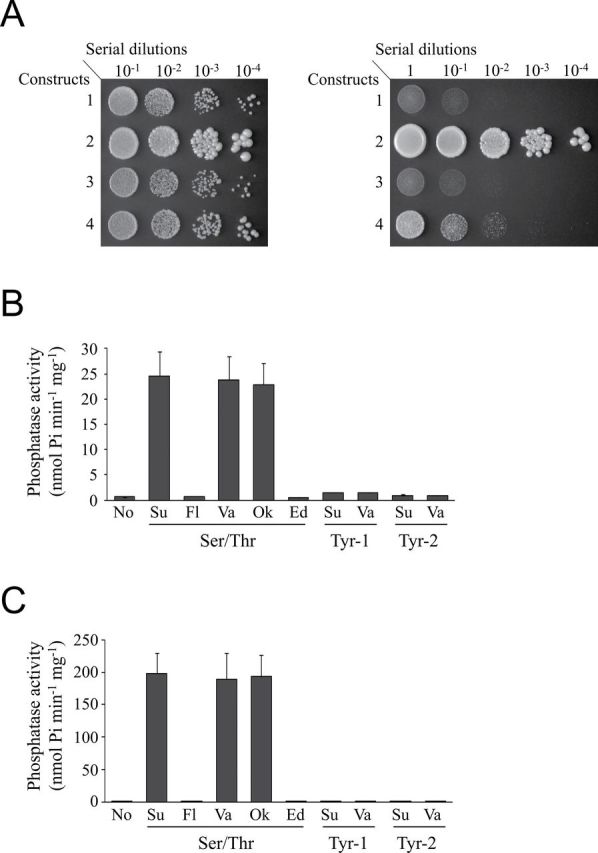
AtPP2CF1 had functional PP2C activity. (A) Complementation test of the yeast ptc1 mutation by AtPP2CF1. The yeast ptc1 knockout strain was transformed with constructs 1 (pYC2/CT, empty vector), 2 (pYC2/CT PTC1), 3 (pYC2/CT AtPP2CF1), or 4 (pYC2/CT ABI1) under the control of the galactose-inducible GAL1 promoter. Serial dilutions of exponentially growing yeast cell cultures were spotted on an SG plate lacking uracil, and growth was observed after 4 d at the permissive temperature (30°C, left) or the restrictive temperature (37°C, right). (B, C) Phosphatase activities of GST-AtPP2CF1 (B) and GST-ABI1 (C). Phosphatase assays were performed as described in the Materials and methods. Purified GST-AtPP2CF1 or GST-ABI1 was incubated with no substrate (No), 100 μM substrate (Su), 100 μM substrate plus 50mM NaF (Fl), 1mM NaVO3 (Va), 10 μM okadaic acid (Ok), or 10mM EDTA (Ed). Values represent the mean ± SD of three independent experiments.
To verify these possibilities, we assessed the in vitro enzymatic activity of AtPP2CF1 using one PSP and two PTP phosphopeptides. An AtPP2CF1 construct with GST fused to the N-terminal region (GST-AtPP2CF1) exhibited high substrate specificity for the PSP phosphopeptide, although this specific activity was approximately 8-fold lower than that of purified GST-tagged ABI1 (GST-ABI1), which was used as a positive control (Fig. 2B, C). Similar to GST-ABI1, the phosphatase activity of GST-AtPP2CF1 was inhibited by NaF, a nonspecific inhibitor of PSPs, but not by okadaic acid or NaVO3, inhibitors of protein phosphatases 1 and 2A (PP1 and PP2A) and PTPs, respectively. Moreover, Mg2+ ions were critical for the phosphatase activity of AtPP2CF1 (Fig. 2B). Together with the yeast ptc1 complementation test, these results suggested that AtPP2CF1 possessed functional PP2C activity but exhibited different substrate specificity from subfamily A PP2Cs.
AtPP2CF1 is unable to recognize and bind to the interacting proteins of some subfamily A PP2Cs
Accumulating evidence from Y2H assays indicates that subfamily A PP2Cs can interact with PYR/PYL/RCAR receptors and three SnRK2 kinases (SnRK2.2, -2.3, and -2.6), upstream and downstream regulators of subfamily A PP2Cs, respectively, albeit with different intensities (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Bhaskara et al., 2012; Soon et al., 2012). However, our Y2H assays revealed that AtPP2CF1 did not bind to any of the PYR/PYL/RCAR receptors or SnRK2 kinases tested in this experiment, either in the presence or absence of exogenous ABA (Supplementary Fig. S6), consistent with the different substrate specificities observed for AtPP2CF1 and ABI1 (Fig. 2A).
Next, to gain structural insight into the putative active and substrate-binding sites of AtPP2CF1, homology-based structural modelling of AtPP2CF1 was performed using the previously solved crystal structure of ABI1 in complex with PYL1 and (+)-ABA (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Yin et al., 2009). Two perpendicular views of the structure of AtPP2CF1 revealed that its catalytic domain adopted an overall conformation similar to that of ABI1 in the PYL1-(+)-ABA-ABI1 complex (Supplementary Figure S7). AtPP2CF1 comprised a central β-sandwich containing two five-stranded antiparallel β-sheets flanked by a pair of α-helices on each side. Notably, at least one metal ion was coordinated by three residues (Asp98, Asp294, and Asp340) in the active site of AtPP2CF1 (corresponding to residues Asp177, Asp347, and Asp413 of ABI1), consistent with the requirement of Mg2+ for its PP2C activity (Figs 2B and 3A; Supplementary Fig. S7). However, AtPP2CF1 exhibited differences within both the active site and catalytic domain residues (Fig. 3A). For example, although the Glu residue (Glu142) in the active site of ABI1 is key to the interaction with PYL1 (Miyazono et al., 2009; Yin et al., 2009), the corresponding residue in AtPP2CF1 was Gln76. Moreover, AtPP2CF1 contained the subfamily E PP2C characteristic insertion of two amino acids in the β3-α1 loop of consensus motif 4, which forms an essential part of the active- and substrate-binding site, causing this loop to protrude over the structure and form a steric hindrance against PYL1 (Fig. 3B; Supplementary Figures S2 and S7). It has been reported that Trp300 of ABI1 is an important residue for both direct interaction with PYR/PYL/RCAR and for recognition of (+)-ABA (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009). Although AtPP2CF1 appeared to have the corresponding Trp residue (Trp246) in a suitable position, the indole ring of Trp246 was significantly rotated compared to the corresponding residue in ABI1 (Trp300), causing steric hindrance that would prevent direct access to (+)-ABA and PYL1 (Fig. 3B; Supplementary Fig. S7). These observations suggested that AtPP2CF1 was no longer able to interact efficiently with R-(+)-ABA-bound PYR/PYL/RCAR receptors, reinforcing the idea that the substrate specificity of AtPP2CF1 differs from that of subfamily A PP2Cs.
Fig. 3.
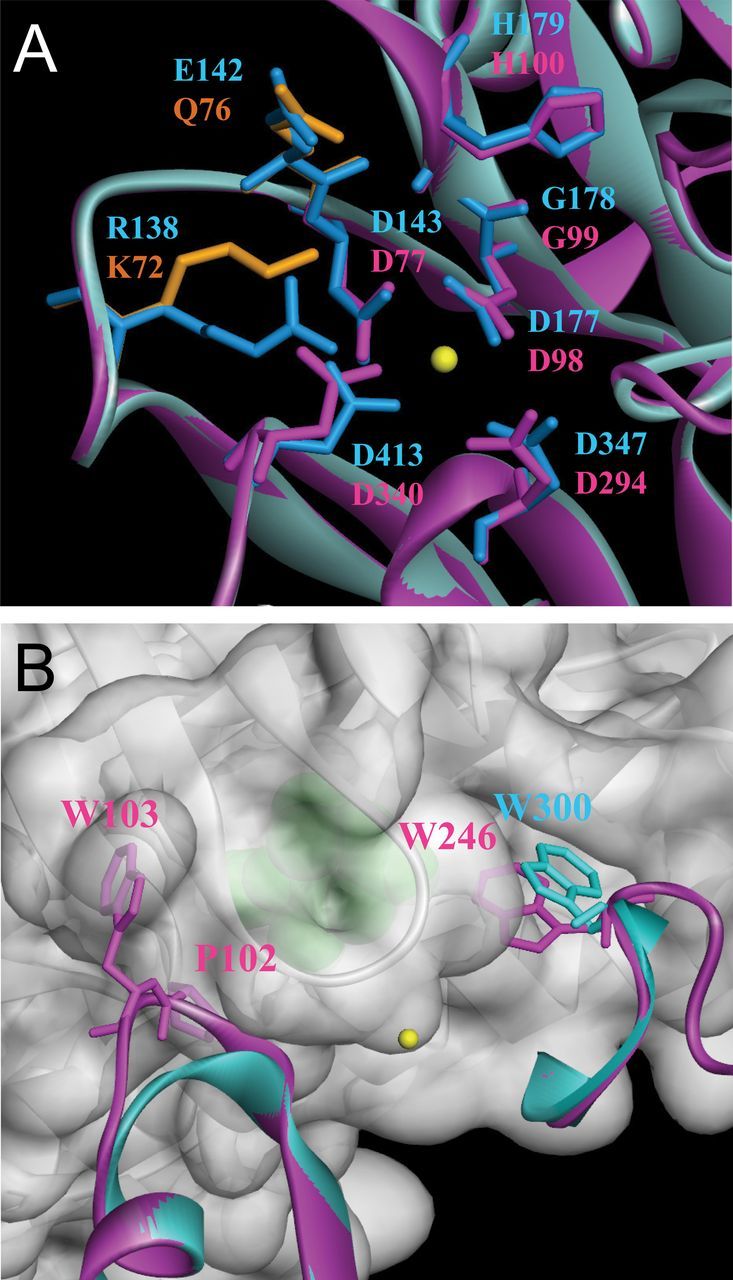
Homology-based structural modelling of AtPP2CF1. Ribbon representation of AtPP2CF1. (A) Close-up view of the putative active site of AtPP2CF1. The key residues comprising the active site of ABI1 are shown as blue-stick representations. Among the active site residues of AtPP2CF1, residues identical to ABI1 are shown as magenta-stick representations, and those specific for AtPP2CF1 are depicted as orange-stick representations. Note that at least one metal ion (yellow sphere) was bound to the active site of AtPP2CF1. (B) Close-up view of the AtPP2CF1 regions corresponding to the primary-binding interface of ABI1 with PYL1. PYL1 (grey) is shown in a semi-transparent surface representation. (+)-ABA is shown as a green sphere. For simplicity, only two regions of AtPP2CF1 (magenta), which correspond to the β3-α1 loop and the β9-β10 loop of ABI1 (blue), are shown. Trp246 of AtPP2CF1, Trp300 of ABI1, and Pro102 and Trp103 of AtPP2CF1, which correspond to an insertion of two amino acids in the β3-α1 loop of consensus motif 4, are highlighted in the stick representation.
Overexpression of AtPP2CF1 led to an ABA-hypersensitive phenotype in Arabidopsis
Next, we investigated whether AtPP2CF1 functioned in ABA signal transduction pathways by generating transgenic Arabidopsis plants overexpressing AtPP2CF1 or ABI1 under the transcriptional control of their 35SΩ promoters, AtPP2CF1oe and ABI1oe, respectively (Fig. 4A) and by examining the ABA sensitivities of these plants during seed germination and seedling development. RT-qPCR experiments showed that AtPP2CF1 transcript accumulation was markedly higher (3.5–7.1-fold) in two independent AtPP2CF1oe-line plants (#1 and #2) than in wild-type plants (Fig. 4B). Expression of ABI1 was also higher (3.4-fold) in ABI1oe plants than in wild-type plants (Fig. 4B).
Fig. 4.
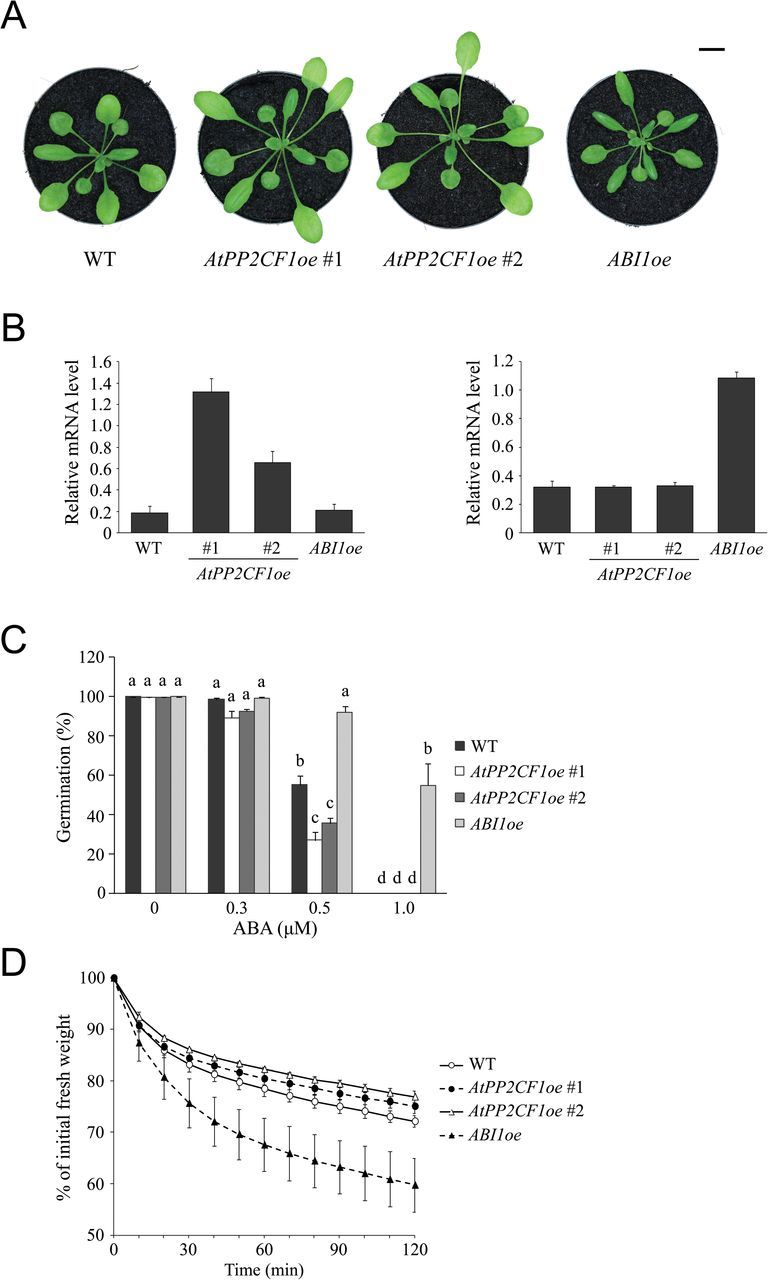
Phenotypic features of Arabidopsis AtPP2CF1oe and ABI1oe transgenic plants. (A) 32-d-old wild-type (WT), AtPP2CF1oe (#1 and #2), and ABI1oe plants. Scale bar: 10mm. (B) Expression of AtPP2CF1 (left) and ABI1 (right) transcripts in AtPP2CF1oe or ABI1oe plants. Total RNA from aerial parts of 2-week-old seedlings grown aseptically on MS medium with gellan gum was isolated and subjected to RT-qPCR. The expression ratios of AtPP2CF1 and ABI1 to UBC9 were calculated for each sample. Values represent the mean ± SD of three independent experiments. (C) Seed germination rate of WT, AtPP2CF1oe, and ABI1oe plants in the presence of the indicated ABA concentrations. Seed germination rate is expressed as the percentage of developed green and expanded cotyledons at 5 d after stratification. Values represent the mean ± SE of six independent experiments. Different letters above the bars indicate statistically significant differences between the lines in the presence of the indicated ABA concentrations, assessed by one-way ANOVA with Tukey-Kramer multiple comparison tests at P < 0.05. (D) Water loss from detached aerial parts of 3-week-old WT, AtPP2CF1oe, and ABI1oe plants. Water loss is expressed as the percentage of initial fresh weight. Values represent the mean ± SD of three independent experiments.
As expected from previous studies (Nishimura et al., 2010), ABI1oe plants clearly showed ABA insensitivity in seed germination tests (Fig. 4C). However, AtPP2CF1oe plants displayed the opposite phenotype; that is, germination efficiency in two independent AtPP2CF1oe plants was reduced to a lesser degree in the presence of exogenous ABA compared with that of wild-type plants, suggesting that AtPP2CF1oe plants were weakly hypersensitive to ABA during seed germination (Fig. 4C).
Under drought conditions, regulation of water loss in plants is a crucial ABA-mediated process involved in the stomatal response. We next investigated whether AtPP2CF1 may be involved in regulating water homeostasis by measuring FW loss in detached aerial parts of plants. Although ABI1oe plants showed a significant increase in the rate of water loss compared with the wild-type, two independent AtPP2CF1oe plants exhibited weaker reduction in water loss, suggesting the occurrence of ABA-hypersensitive stomatal closure consistent with AtPP2CF1 gene expression in guard cells (Figs 1G and 4D).
AtPP2CF1oe plants displayed increased inflorescence stem growth
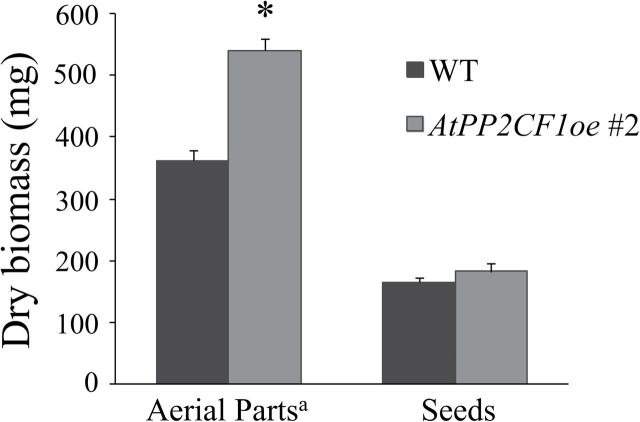
Arabidopsis plants that overexpress subfamily A PP2Cs such as ABI1 and HAB1 have been reported to exhibit significantly reduced size under moderate RH due to impaired stomatal closure and resulting deficiencies in water retention (Robert et al., 2006; Nishimura et al., 2010). Even under high RH, transgenic plants overexpressing HAB1 are smaller than corresponding wild-type plants (Robert et al., 2006). Here, under 60% RH, ABI1oe plants were smaller than wild-type plants (as expected) and exhibited leaf epinasty (Fig. 4A). In contrast, AtPP2CF1oe plants had similar morphology to wild-type plants, with the exception of long petioles (Fig. 4A). Furthermore, ABI1oe plants had short inflorescence stems but displayed shoot branching patterns (numbers of rosette and cauline branches) similar to those of wild-type plants (Fig. 5A–G and Supplementary Fig. S8). However, two independent AtPP2CF1oe plants developed longer and thicker inflorescence stems (Fig. 5A, B, F, G; Supplementary Fig. S8). There were also significant differences in numbers of shoot branches between wild-type and AtPP2CF1oe plants. In AtPP2CF1oe plants, a small but significant decrease in numbers of rosette branches was observed (one to two), while cauline branch numbers on the primary inflorescence were increased by one to two (Fig. 5C–E). Thus, overexpression of AtPP2CF1 resulted in a more erect, robust-stature phenotype and led to an increase in dry weight of the overexpressing plants (aerial parts excluding seeds) by a factor of 1.49 compared to wild-type plants (Figs 5 and 6).
Fig. 5.
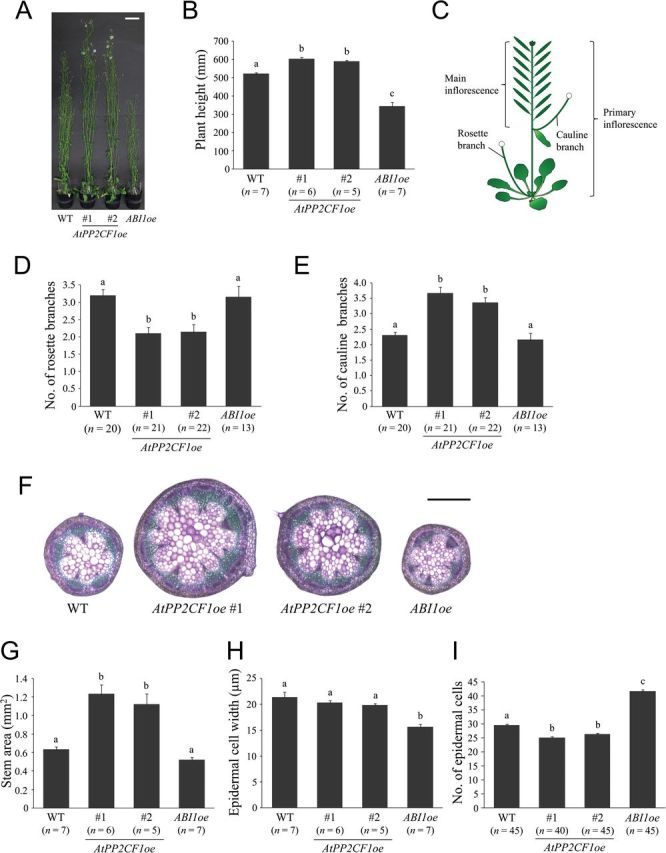
AtPP2CF1oe plants exhibited increased inflorescence stem growth. (A) Nine-week-old wild-type (WT), AtPP2CF1oe (#1 and #2), and ABI1oe plants. (B) Height of 9-week-old plants. (C) Schematic representation of Arabidopsis inflorescence stems. (D, E) Number of rosette branches (D) and cauline branches on primary inflorescence (E). Arabidopsis plants grown until global proliferative arrest stage (Hensel et al., 1994) were used for measurements. Short, thin rosette branches <10cm in length, which occurred infrequently, were excluded from the counts. (F–I) Basal parts (20mm from the rosette base) of primary inflorescence stems in 9-week-old plants were used for observations and measurements. (F) Transverse primary inflorescence stem sections from WT, AtPP2CF1oe, and ABI1oe plants. Sections were 100-μm thick and stained with toluidine blue. (G–I) Stem area (G) and epidermal cell width (H) and number (I). Stem epidermal cell width was determined as the stem circumference divided by the epidermal cell number in transverse stem sections. The number of epidermal cells (not including guard cells) per unit stem epidermal area (1mm long and 100 μm wide) was counted (see also Supplementary Fig. S9). Scale bars: 50mm (A); 500 μm (F). All values represent the mean ± SE. Different letters above bars indicate statistically significant differences between the lines, assessed by one-way ANOVA with Tukey-Kramer multiple comparison tests at P < 0.05 (B, D) and P < 0.01 (E, G–I).
Fig. 6.
Overexpression of AtPP2CF1 enhanced biomass production in plants. Dry biomass determination was performed as described in the Materials and methods. Values represent the mean ± SD for n = 4. a Aerial plant parts did not include root tissues or seeds. * Significantly different at P < 0.01 (Student’s t-test).
Microscopic observations of cross-sections of primary inflorescence stems revealed that the stem area in ABI1oe plants was the same as that in wild-type plants (Fig. 5F, G). However, the width of epidermal cells in ABI1oe inflorescence stems was reduced by 27% relative to the wild type, indicating increased cell division in the radial and circumferential directions in these stems (Fig. 5F–H). In contrast, stem area in AtPP2CF1oe was much larger than that in wild-type plants (Fig. 5F, G). AtPP2CF1oe inflorescence stems showed normal organization with no obvious differences in the size, shape, or degree of compactness of cells, reflecting increased cell division in the radial and circumferential directions in these stems (Fig. 5F, H).
Next, we examined the number of epidermal cells per unit stem surface (Fig. 5I and Supplementary Fig. S9). In ABI1oe inflorescence stems, the number of epidermal cells was increased by ~1.41-fold compared to wild-type plants (Fig. 5I). This increase was almost equal to the increase in cell numbers in the circumferential direction (37%) (Fig. 5H, I), suggesting that epidermal cells in ABI1oe inflorescence stems did not exhibit significant changes in a longitudinal direction. These observations suggested that the shorter stem phenotype of ABI1oe might be primarily a result of decreased levels of longitudinal cell-division activity. In contrast, in AtPP2CF1oe plants, epidermal cell numbers were substantially decreased (by 11–15%) compared with wild-type plants (Fig. 5I). These results, together with the finding of no differences in width of stem epidermal cells between wild-type and AtPP2CF1oe plants, indicated that stem epidermal cells in AtPP2CF1oe plants elongated in a longitudinal direction. The increased longitudinal length of AtPP2CF1oe stem epidermal cells corresponded to increased plant height (Fig. 5B, I), revealing that the promotion of AtPP2CF1oe stem height may result primarily from cell elongation rather than cell division.
AtPP2CF1 caused a slightly late-flowering phenotype in the overexpressing lines
Functional loss of flowering-time genes SOC1 and FUL has been reported to cause increased inflorescence stem thickness as a result of enhanced secondary growth (Melzer et al., 2008); we investigated flowering time in AtPP2CF1oe and ABI1oe lines in this context. The phyB single and cry2 phyB double mutants were also included as controls for early- and late-flowering phenotypes, respectively (Mockler et al., 1999). Under our growth conditions, AtPP2CF1oe and ABI1oe plants had the same flowering time and flowered significantly earlier than cry2 phyB plants (about 10 d earlier with two to three fewer leaves) (Fig. 7). However, both transgenic lines flowered slightly later than wild-type plants (about 3 d later with three to four more leaves), indicating that overexpression of AtPP2CF1 or ABI1 conferred slightly late flowering (Fig. 7).
Fig. 7.
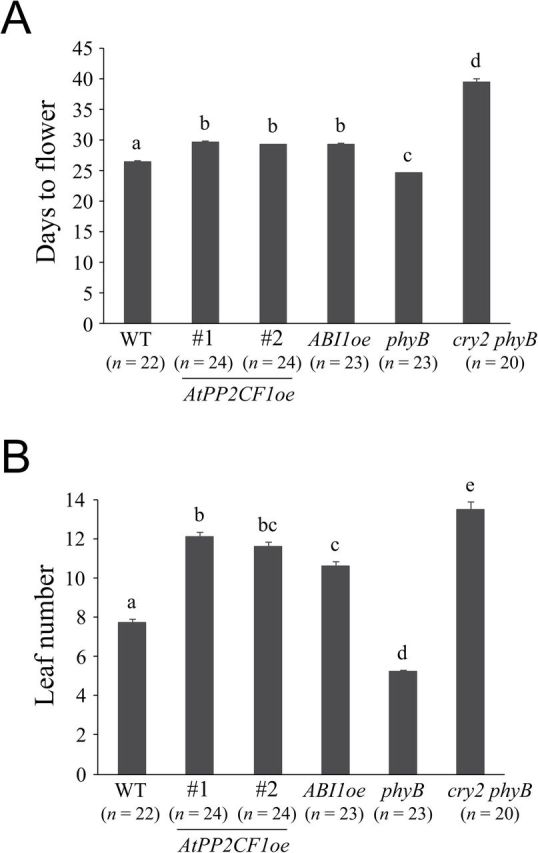
Flowering time of AtPP2CF1oe, ABI1oe, phyB, and cry2 phyB plants. Flowering time was determined by counting the days to flower (A) and leaf number (B). Values represent the mean ± SE. Different letters above bars indicate statistically significant differences between the lines, assessed by one-way ANOVA with Tukey-Kramer multiple comparison tests at P < 0.01.
These observations led us to wonder whether there is a relationship between flowering time and stem enlargement in these overexpressing lines. We next observed cross-sections of young primary inflorescence stems of AtPP2CF1oe, ABI1oe, and flowering mutant plants of the same developmental size rather than age (see Materials and methods for details). Stem area in the late-flowering cry2 phyB mutant was significantly larger than that in wild-type plants, while that of the early-flowering phyB mutant was substantially smaller (Fig. 8). Relative to the wild-type, phenotypes of young AtPP2CF1oe and ABI1oe inflorescence stems followed qualitatively similar trends to those seen in corresponding 9-week-old plants (Figs 5 and 8). Remarkably, despite the weaker late flowering of AtPP2CF1oe compared to cry2 phyB, AtPP2CF1oe plants had significantly thicker inflorescence stems than cry2 phyB plants (Fig. 8). Taken together, our data suggest that the enhanced stem growth and development of AtPP2CF1oe plants cannot be explained solely by the slight delay in flowering time, highlighting the functional significance of AtPP2CF1 in transverse stem cell division and enlargement at the primary growth stage.
Fig. 8.
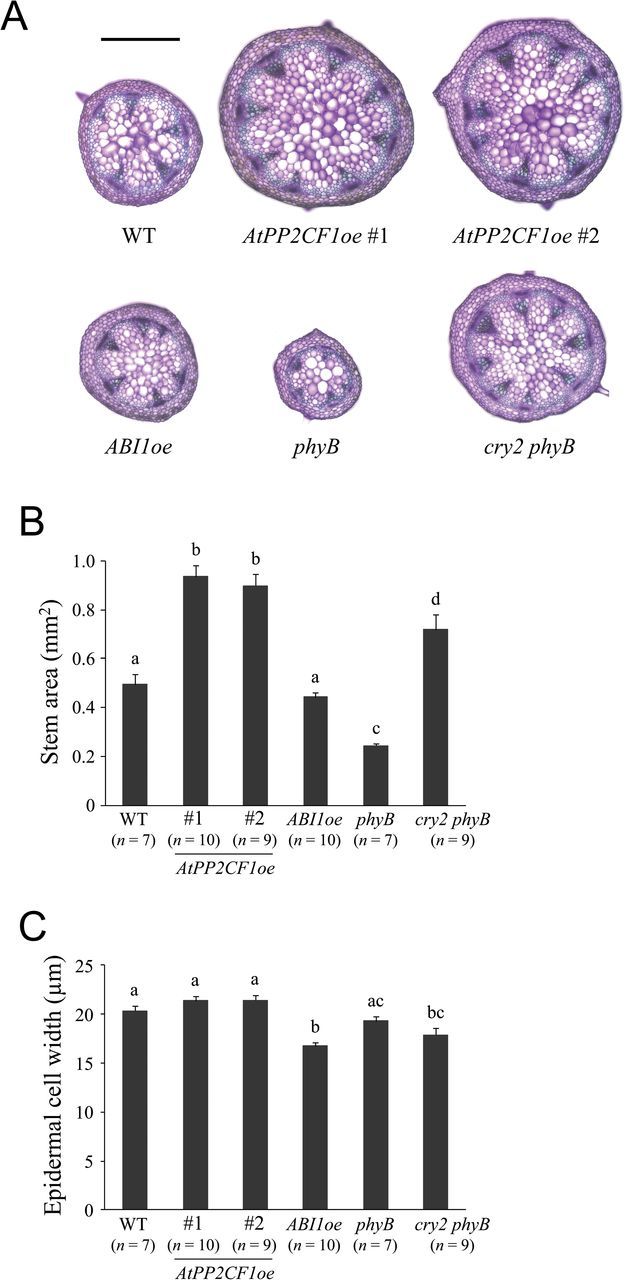
Comparison of young inflorescence stem of AtPP2CF1oe, ABI1oe, phyB, and cry2 phyB plants. Basal parts (10mm from the rosette base) of primary inflorescence stems that reached a length of 10–15cm were used for the observations and measurements. (A) Transverse primary inflorescence stem sections from wild-type (WT), AtPP2CF1oe (#1 and #2), ABI1oe, phyB, and cry2 phyB plants. Sections were 100-μm thick and stained with toluidine blue. Scale bar: 500 μm. (B, C) Stem area (B) and epidermal cell width (C). Stem epidermal cell width was determined as the stem circumference divided by the epidermal cell number in transverse stem sections. Values represent the mean ± SE. Different letters above bars indicate statistically significant differences between the lines, assessed by one-way ANOVA with Tukey-Kramer multiple comparison tests at P < 0.05.
AtPP2CF1oe plants exhibited normal fertility and seed set
Reduced fertility in plants has been reported to cause cessation of global proliferative arrest of the inflorescence meristem, thereby increasing inflorescence stem growth (Hensel et al., 1994). To test whether this was the case for AtPP2CF1oe plants, we analysed the effect of overexpression of AtPP2CF1 on fertility, expressed as the average number of seeds per silique on the main inflorescence. In two independent AtPP2CF1oe-line plants, average seed numbers per silique and seed weight were highly similar to the wild type, indicating that the fertility of AtPP2CF1oe plants was normal (Table 1). In contrast, ABI1oe plants showed significantly reduced seed production compared with the wild type (Table 1). Thus, at least for AtPP2CF1oe and ABI1oe plants, there was no association between inflorescence stem growth and fertility phenotype.
Table 1.
Fertility and seed set of AtPP2CF1oe and ABI1oe plantsa
| Line | Number of plants | Average seed number per siliqueb | Average seed weight (μg)c |
|---|---|---|---|
| WT | 20 | 34.6±1.8* | 18.99±0.46*,† |
| AtPP2CF1oe #1 | 21 | 36.5±1.4* | 17.60±0.48* |
| AtPP2CF1oe #2 | 22 | 34.5±1.7* | 17.63±0.43* |
| ABI1oe | 13 | 25.9±2.4† | 20.19±0.27† |
a The primary inflorescence stems of Arabidopsis plants grown until global proliferative arrest stage (Hensel et al., 1994) were used to determine, on the main inflorescence, the number of siliques (containing at least one seed), infertile ovaries, and arrested buds; and seeds in each silique (see also Fig. 5C). b Average seed number per silique was determined as the total number of seeds on the main inflorescence divided by the number of siliques. c Average seed weight was calculated using 50 seeds from each plant. Values are means ± SE. Different superscript symbols (*, †) indicate statistically significant differences between lines, assessed by one-way ANOVA with Tukey-Kramer multiple comparison tests at P < 0.05.
AtPP2CF1 exhibited different expression patterns than its most closely related paralogues
AtPP2CF1, At5g27930, and At3g16800 form a small clade within subfamily E PP2Cs (Supplementary Fig. S1). A previous report (Xue et al., 2008) and a Plant Genome Duplication Database (PGDD) (Lee et al., 2012) search revealed that this small clade developed through segmental chromosome duplication events (Supplementary Fig. S10). To confirm whether these genes underwent functional diversification after these events, their developmental expression profiles were compared. RT-qPCR analysis revealed that At5g27930 shared a similar expression pattern with AtPP2CF1, while At3g16800 was ubiquitously expressed in all tissues, with higher levels of expression detected in the roots (Figs 1B and 9A, B).
Fig. 9.
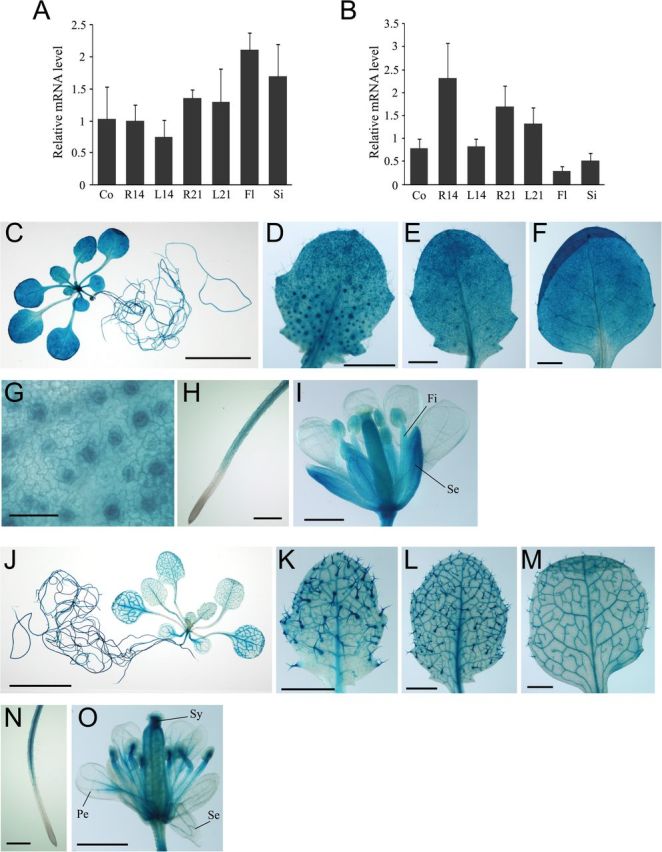
Spatial and temporal regulation of At5g27930 and At3g16800 expression. Arabidopsis plants were germinated aseptically on MS medium with gellan gum. At 21 d after germination, seedlings were transferred to soil. (A, B) Expression of At5g27930 (A) and At3g16800 (B) transcripts. Total RNA from different tissues of wild-type Arabidopsis plants was isolated and subjected to RT-qPCR. The expression ratios of At5g27930 and At3g16800 to UBC9 were calculated for each sample. Values represent the mean ± SD of three independent experiments. Co, 7-d-old cotyledons; R14, 14-d-old roots; L14, 14-d-old first and second rosette leaves; R21, 21-d-old roots; L21, 21-d-old first to fourth rosette leaves; Fl, flowers; Si, siliques. (C–O) Histochemical analysis of GUS activity in Arabidopsis pAt5g27930:GUS (C–I) and pAt3g16800:GUS (J–O) transgenic plants. (C, J) A 21-d-old transgenic seedling. (D–F, K–M) A developmental series of rosette leaves ranging from young (D, K) to old (F, M) within a 21-d-old transgenic seedling. (G) Guard cells of young leaves. (H, N) Roots. (I, O) Flowers. Fi, filament; Pe, petal; Se, sepal; Sy, style. Scale bars: 10mm (C, J); 1mm (D–F, I, K–M, O); 500 μm (H, N); 50 μm (G).
Next, to investigate the tissue- or cell-type-specific expression patterns of At5g27930 and At3g16800, we generated transgenic Arabidopsis plants expressing the GUS reporter gene under the transcriptional control of each gene’s promoter (Fig. 9C–O and Supplementary Fig. S4). Although RT-qPCR analysis revealed similar expression patterns for AtPP2CF1 and At5g27930, GUS staining revealed that the expression profiles of these AtPP2CF1 paralogues were different from those of AtPP2CF1 (Figs 1B–I and 9). As shown in Fig. 9C–I, GUS activity derived from the At5g27930 promoter was detected in mesophyll cells in the leaves, sepals, ovaries, guard cells, trichomes, hypocotyls, stamen filaments, and all root tissues except for root tip regions. Moreover, At5g27930 expression was detectable at low levels in vascular tissues (Supplementary Fig. S4). In contrast, strong GUS activity derived from the At3g16800 promoter was observed in all root tissues except for root tip regions (especially the root vasculature), vascular tissues of the leaves and hypocotyls, and trichomes; the style and vascular tissues of sepals and petals exhibited only weak GUS activity (Fig. 9J–O and Supplementary Fig. S4). Taken together, these results suggest that AtPP2CF1, At5g27930, and At3g16800 have completely divergent expression profiles during seedling development, indicating their sub- or neofunctionalization.
Discussion
In this study, we report a new Arabidopsis PP2C isoform, AtPP2CF1, as a member of PP2C subfamily E. AtPP2F1 encoded an ABA-inducible, functional PP2C. However, our evidence suggests that the physiological roles of AtPP2CF1 in plant cells differ from those of subfamily A PP2Cs, whose gene expression is also ABA-inducible (Goda et al., 2008).
Higher plants possess a larger number of PP2C genes than mammals as a result of chromosomal segment and tandem duplication events (Schweighofer et al., 2004; Xue et al., 2008; Singh et al., 2010), allowing for great functional diversification and variation in expression patterns, subcellular localization, and substrate specificity. Indeed, increasing evidence has indicated that subfamily A PP2Cs are involved in the ABA signal transduction pathway as key negative regulators with different expression and subcellular localization profiles (Saez et al., 2004; Nishimura et al., 2007; Umezawa et al., 2009; Zhang et al., 2012). Through similar genomic rearrangement events, AtPP2CF1, At5g27930, and At3g16800 represent a small clade within subfamily E (Xue et al., 2008). Furthermore, these three genes were expressed with partial overlap but with distinct tissue- and cell-specific and ABA-responsive patterns, indicating the sub- or neofunctionalization of each gene.
Here, we provide evidence that the substrate specificity of AtPP2CF1 may differ from that of ABI1. First, in contrast to ABI1, AtPP2CF1 could not rescue the yeast ptc1. Second, although heterogeneous, AtPP2CF1 could not interact with molecules that signal up- or downstream of subfamily A PP2Cs (i.e. PYR/PYL/RCAR receptors and SnRK2s). Structural modelling analyses provided a plausible explanation for this property of AtPP2CF1. For example, it was observed that a protrusion caused by a unique two-amino acid insertion in the β3-α1 loop of consensus motif 4 resulted in an important structural perturbation of the interaction with PYR/PYL/RCAR receptors. Furthermore, it has been reported that Glu142 in the active site of ABI1 directly interacts with PYL1 (Miyazono et al., 2009; Yin et al., 2009). However, AtPP2CF1 has a Gln residue (Gln76) in the corresponding position, a common characteristic of subfamily E PP2Cs. Although AtPP2CF1 has a conserved Trp246 residue at the position corresponding to Trp300 in ABI1, the function of which is to recognize PYR/PYL/RCAR receptors and (+)-ABA, the significant distortion of the AtPP2CF1-Trp246 indole ring compared to its counterpart residue in ABI1 causes steric hindrance and prevents these receptors from binding. These data suggested that AtPP2CF1 may function differently from subfamily A PP2Cs.
Arabidopsis plants overexpressing subfamily A PP2Cs have been reported to exhibit an ABA-insensitive phenotype (Saez et al., 2004; Kuhn et al., 2006; Robert et al., 2006; Nishimura et al., 2010; Zhang et al., 2012). In contrast, overexpression of AtPP2CF1 in Arabidopsis caused the opposite effect in seed germination and leaf evaporation assays. Furthermore, AtPP2CF1oe plants showed increased biomass production, in direct contrast with transgenic plants overexpressing subfamily A PP2Cs, including ABI1oe plants (Robert et al., 2006; Nishimura et al., 2010). These phenotypic differences are caused by differences in the biochemical and physiological functions of the various PP2Cs in plant cells.
The plant hormone ABA has been defined as a growth promoter under non-stress conditions, but is well known to act as a growth inhibitor under stress conditions such as drought and salinity, through the precise regulation of seed dormancy, seedling development, and stomatal behaviour (Sharp and LeNoble, 2002; Verslues and Zhu, 2007; Cutler et al., 2010). For example, ABA-deficient mutants such as aba1 and aba2/gin1 display significant reductions in size, which is especially pronounced in leaves, inflorescence stems, and flowers, as a result of reduced cell numbers and expansion; this size reduction is observed even under non-stress conditions (i.e. humidity near saturation) (Cheng et al., 2002; Barrero et al., 2005). Furthermore, ABA-insensitive plants, including srk2.2/2.3/2.6 triple mutants and transgenic lines overexpressing subfamily A PP2Cs, show a similar stunted phenotype, even under high RH (Robert et al., 2006; Fujii and Zhu, 2009; Nishimura et al., 2010). Thus, ABA plays an essential role in the promotion of leaf and stem development. Recent studies have demonstrated that PYR/PYL/RCAR receptors can be divided into two distinct subclasses, one comprising dimeric receptors with a lower affinity for ABA and the other comprising monomeric receptors with a significantly higher affinity for ABA (Dupeux et al., 2011; Hao et al., 2011). It has also been proposed that monomeric receptors may be essential for regulating basal activation of the ABA signal transduction pathway under non-stress conditions, while dimeric receptors may play a major role in the stress response under adverse environmental conditions, reinforcing the hypothesis that ABA has two distinct, opposing actions (Dupeux et al., 2011; Hao et al., 2011).
In the current study, we demonstrated that AtPP2CF1 expression was ABA-inducible and could be observed in young tissues such as root tips and the basal regions of young developing leaves that exhibit rapid cell division. Intriguingly, accumulation of AtPP2CF1 transcripts was shown to increase significantly in gra-D mutants, which develop large leaves with drastically increased cell numbers (Horiguchi et al., 2009). Also, AtPP2CF1oe inflorescence stems had a markedly different phenotype from those seen in ABI1oe plants. ABI1 overexpression reduced transverse stem cell enlargement and longitudinal stem cell division in inflorescence stems. In contrast, overexpression of AtPP2CF1 led to increased transverse stem cell division and longitudinal stem cell elongation, in turn leading to increased stem height and width, and greater biomass production. Taken together, our findings suggest that AtPP2CF1 may function as a growth-promoting factor in ABA signal transduction pathways under non-stress conditions, and may be modulated through interactions with factors downstream of monomeric PYR/PYL/RCAR receptors or with other or still-unidentified ABA receptors. It has recently been reported that ABA regulates branching by restricting the outgrowth of buds to form rosette branches (Reddy et al., 2013). We also found that AtPP2CF1oe-line plants showed reduced rosette branches compared with wild-type and ABI1oe plants, supporting the idea that AtPP2CF1 might function as a positive signal transducer in ABA signalling under non-stress conditions.
Plant growth is influenced by both intrinsic genetic and extrinsic environmental cues. Despite its importance, the complex genetic and molecular networks underlying plant growth processes are poorly understood. Although we propose a potentially important functional role of AtPP2CF1 in plant biomass production, definitive proof of this role is still lacking. Nevertheless, AtPP2CF1 may represent a useful tool for gene manipulation to engineer increased biomass in crop plants.
Supplementary Material
Supplementary data are available at JXB online.
Supplementary Method S1. PCR templates for plasmid construction.
Supplementary Method S2. Construction of plasmids for the establishment of transgenic Arabidopsis plants.
Supplementary Method S3. Construction of plasmids for the yeast ptc1 complementation test.
Supplementary Method S4. Construction of plasmids for purification of AtPP2CF1 and ABI1.
Supplementary Method S5. Y2H assays.
Supplemental Table S1. Gene-specific primers used in RT-qPCR.
Supplementary Fig. S1. Phylogenetic analysis of Arabidopsis PP2Cs belonging to subfamilies E and L.
Supplementary Fig. S2. Sequence alignment of conserved consensus motif 4 in all Arabidopsis PP2Cs.
Supplementary Fig. S3. Time course of subfamily E PP2C mRNA expression in aerial parts of wild-type Arabidopsis plants treated with ABA.
Supplementary Fig. S4. Histochemical analysis of GUS activity in transgenic Arabidopsis plants pAtPP2CF1:GUS, pAt3g16800:GUS, and pAt5g27930:GUS.
Supplementary Fig. S5. Multiple sequence alignments of the predicted phosphatase catalytic domain in AtPP2CF1 with those in PTC1 and ABI1.
Supplementary Fig. S6. AtPP2CF1 did not interact with PYR/PYL/RCAR receptors or with SnRK2 kinases in a Y2H assay.
Supplementary Fig. S7. Homology-based structural modelling of AtPP2CF1.
Supplementary Fig. S8. Kinematic analysis of the height of wild-type (WT), AtPP2CF1oe (#1 and #2), and ABI1oe plants.
Supplementary Fig. S9. Safranin-stained epidermises peeled from primary inflorescence stems of wild-type (WT), AtPP2CF1oe (#1 and #2), and ABI1oe plants.
Supplementary Fig. S10. Segmental chromosomal duplication regions of AtPP2CF1.
Acknowledgements
We would like to thank Arisa Tamura, Chie Sazuka, Masayo Suzuki, Yuriko Nakano, Miyu Miyachi, Megumi Sakai, and Ritsuko Kitagawa for technical assistance and Dr Minoru Hirano and Dr Hidenori Tanaka (Toyota Central R&D Labs. Inc.) for helpful discussions. We also thank Dr Toshinori Kinoshita (Nagoya University) for providing the phyB-9 mutant, Dr Tomonao Matsushita (Kyushu University) for providing the cry2-1 phyB-9 double mutant, and the RIKEN BRC (through the National Bio-Resource Project of the MEXT, Japan) for providing RIKEN Arabidopsis full-length (RAFL) cDNA clones developed by the plant genome project of RIKEN Genomic Sciences Center.
Glossary
Abbreviations:
- ABA
abscisic acid
- GUS
β-glucuronidase
- PP2C
protein phosphatase 2C
- PSP
Ser/Thr phosphatase
- PTP
Try phosphatase.
References
- Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL. 2005. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. Journal of Experimental Botany 56, 2071–2083. [DOI] [PubMed] [Google Scholar]
- Bertauche N, Leung J, Giraudat J. 1996. Protein phosphatase activity of abscisic acid insensitive 1 (ABI1) protein from Arabidopsis thaliana . European Journal of Biochemistry 241, 193–200. [DOI] [PubMed] [Google Scholar]
- Bhaskara GB, Nguyen TT, Verslues PE. 2012. Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiology 160, 379–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nürnberger T, Gust AA. 2010. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiology 153, 1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Endo A, Zhou L, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. The Plant Cell 14, 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen PTW. 1994. Nomenclature and chromosomal localization of human protein serine/threonine phosphatase genes. Advances in Protein Phosphatases 8, 371–376. [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Dupeux F, Santiago J, Betz K, et al. 2011. A thermodynamic switch modulates abscisic acid receptor sensitivity. The EMBO Journal 30, 4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proceedings of the National Academy of Sciences, USA 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, et al. 2008. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. The Plant Journal 55, 526–542. [DOI] [PubMed] [Google Scholar]
- Hao Q, Yin P, Li W, Wang L, Yan C, Lin Z, Wu JZ, Wang J, Yan SF, Yan N. 2011. The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Molecular Cell 42, 662–672. [DOI] [PubMed] [Google Scholar]
- Hauser F, Waadt R, Schroeder JI. 2011. Evolution of abscisic acid synthesis and signaling mechanisms. Current Biology 21, R346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB. 1994. The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis . Plant Physiology 106, 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Gonzalez N, Beemster GTS, Inzé D, Tsukaya H. 2009. Impact of segmental chromosomal duplications on leaf size in the grandifolia-D mutants of Arabidopsis thaliana . The Plant Journal 60, 122–133. [DOI] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. 2010. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes and Development 24, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C, Leung J. 2011. Abscisic acid signal off the STARting block. Molecular Plant 4, 562–580. [DOI] [PubMed] [Google Scholar]
- Koiwai H, Nakaminami K, Seo M, Mitsuhashi W, Toyomasu T, Koshiba T. 2004. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiology 134, 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. 2006. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiology 140, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T-H, Tang H, Wang X, Paterson AH. 2012. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Research 41, D1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. 1997. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. The Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhu Y, Zhai H, Cai H, Ji W, Luo X, Li J, Bai X. 2012. AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochemical and Biophysical Research Communications 422, 710–715. [DOI] [PubMed] [Google Scholar]
- Lu G, Wang Y. 2008. Functional diversity of mammalian type 2C protein phosphatase isoforms: new tales from an old family. Clinical and Experimental Pharmacology and Physiology 35, 107–112. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Maeda T, Tsai AYM, Saito H. 1993. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae . Molecular and Cellular Bioliogy 13, 5408–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng L-M, Zhou XE, et al. 2009. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. 2008. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana . Nature Genetics 40, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. 2001. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. The Plant Journal 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, et al. 2009. Structural basis of abscisic acid signalling. Nature 462, 609–614. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. 1999. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. 2009. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326, 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, et al. 2010. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. The Plant Journal 61, 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. 2007. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. The Plant Journal 50, 935–949. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA. 2013. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiology 163, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, Merlot S, N’Guyen V, Boisson-Dernier A, Schroeder JI. 2006. A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis . FEBS Letters 580, 4691–4696. [DOI] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. 2004. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. The Plant Journal 37, 354–369. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park S-Y, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. 2009. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science 9, 236–243. [DOI] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME. 2002. ABA, ethylene and the control of shoot and root growth under water stress. Journal of Experimental Botany 53, 33–37. [PubMed] [Google Scholar]
- Shi Y. 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. [DOI] [PubMed] [Google Scholar]
- Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. 2010. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon F-F, Ng L-M, Zhou XE, et al. 2012. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. 2007. New developments in abscisic acid perception and metabolism. Current Opinion in Plant Biology 10, 447–452. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y. 2008. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. 2009. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nature Structural and Molecular Biology 16, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan S-S. 2012. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. The Plant Journal 69, 667–678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.