Summary
A proline-rich protein-like protein moves in vesicles on actin over the ER and influences root hair and etiolated hypocotyl cell elongation through subtle changes in cell wall composition.
Key words: Arabidopsis thaliana, cell expansion, hypocotyl, pollen Ole allergen, root hairs.
Abstract
The synthesis and composition of cell walls is dynamically adapted in response to many developmental and environmental signals. In this respect, cell wall proteins involved in controlling cell elongation are critical for cell development. Transcriptome analysis identified a gene in Arabidopsis thaliana, which was named proline-rich protein-like, AtPRPL1, based on sequence similarities from a phylogenetic analysis. The most resemblance was found to AtPRP1 and AtPRP3 from Arabidopsis, which are known to be involved in root hair growth and development. In A. thaliana four proline-rich cell wall protein genes, playing a role in building up the cross-connections between cell wall components, can be distinguished. AtPRPL1 is a small gene that in promoter::GUS (β-glucuronidase) analysis has high expression in trichoblast cells and in the collet. Chemical or mutational interference with root hair formation inhibited this expression. Altered expression levels in knock-out or overexpression lines interfered with normal root hair growth and etiolated hypocotyl development, but Fourier transform-infrared (FT-IR) analysis did not identify consistent changes in cell wall composition of root hairs and hypocotyl. Co-localization analysis of the AtPRPL1–green fluorescent protein (GFP) fusion protein and different red fluorescent protein (RFP)-labelled markers confirmed the presence of AtPRPL1–GFP in small vesicles moving over the endoplasmic reticulum. Together, these data indicate that the AtPRPL1 protein is involved in the cell’s elongation process. How exactly this is achieved remains unclear at present.
Introduction
The plant cell wall is a rigid extracellular matrix that is highly dynamic and involved in many developmental processes, such as cell shape, anisotropic growth, defence against pathogenic attacks, and other processes (Cosgrove, 1999, 2000; Huckelhoven, 2007). In dicotyledons, the cell wall is composed of cellulose microfibrils that are tethered by hemicelluloses. Xyloglucan, the major hemicellulose in plants such as Arabidopsis, is associated with cellulose by hydrogen bonds (Valent and Albersheim, 1974; Hayashi, 1989; Acebes et al., 1993; Hayashi et al., 1994) and this cellulose–hemicellulose network is embedded in a highly hydrated pectin matrix (Carpita and Gibeaut, 1993). The cell wall also contains structural proteins that comprise up to 10% of its dry weight (Varner and Lin, 1989). According to the amino acid composition and the glycosylation pattern, these wall proteins can be divided into different groups based on the high abundance of certain amino acids (Johnson et al., 2003). The two major groups are the hydroxyproline-rich glycoproteins (HRGPs) and the glycine-rich proteins (GRPs) (Sachetto-Martins et al., 2000). The former include extensins (Showalter, 1993; Cassab, 1998), arabinogalactan-proteins (AGPs) (Fincher et al., 1983; Nothnagel, 1997), and the proline-rich proteins (PRPs) (Showalter, 1993; Cassab, 1998).
These structural cell wall proteins are secreted as monomers into the wall where they can be cross-linked. How these cross-links are made still remains largely unknown. There is, however, evidence for a peroxidase-mediated reaction that forms intermolecular isodityrosine links (Bradley et al., 1992; Brisson et al., 1994; Schnabelrauch et al., 1996), which seems necessary to slow down or completely stop cell elongation in 1-aminocyclopropane-1-carboxylic acid (ACC)-treated Arabidopsis roots (De Cnodder et al., 2005). Recently Cannon et al. (2008) showed that in the root-, shoot-, hypocotyl-defective (rsh) mutant a mutation is found in the gene encoding an extensin (AtEXT3) which is located in the cell plate, the cross wall, and in mature cell walls. This extensin is a HRGP and contains a hydrophobic isodityrosine cross-link motif (YVY). In vitro, a peroxidase catalyses the formation of insoluble RSH gels with concomitant cross-linking and it is suggested that in planta this positively charged extensin scaffold binds with negatively charged pectins to create an extensin pectate coacervate (Valentin et al., 2010). MacDougall et al. (2001) indeed described that extensins have the potential to act as non-covalent cross-linking agents in pectin networks, forming elastic gels. Furthermore, extensin network formation can already occur without oxidative cross-linking, as shown by atomic force microscopy (Cannon et al., 2008).
PRPs are a group of structural cell wall proteins that were first identified as wound-induced gene products in carrot storage roots (Chen and Varner, 1985; Tierney et al., 1988), they are expressed in many plant species and their expression is spatially and temporally regulated during plant development. It has been shown that PRP genes are expressed during soybean leaf, root, stem, and seed coat development (Hong et al., 1990; Kleis-San Fransisco and Tierney, 1990; Lindstrom and Vodkin, 1991; Wyatt et al., 1992). PRP genes are also expressed in different stages of legume root nodule formation (Scheres et al., 1990; Van de Wiel et al., 1990; Wilson and Cooper, 1994), in the growth of bean seedlings (Sheng et al., 1991), in immature maize embryos (Jose-Estanyol et al., 1992), and in other tissues (Bernhardt and Tierney, 2000; reviewed in Johnson et al., 2003). Four PRP genes were characterized in Arabidopsis (AtPRP genes) and, based on their DNA sequence identity, repetitive motifs, and domain organization, divided into two different classes (Fowler et al., 1999). AtPRP1 and AtPRP3 are both exclusively expressed in roots, while AtPRP2 and AtPRP4 are expressed in leaves, stems, flowers, and siliques. Promoter analysis with β-glucuronidase (GUS) fusions showed that AtPRP3 is expressed in root hairs. Experiments with molecules that induce or inhibit root hair initiation coupled to gene expression analysis in mutant backgrounds proved that developmental pathways involved in root hair formation regulate AtPRP3 and that AtPRP3 is contributing to the cell wall structure in Arabidopsis root hairs (Bernhardt and Tierney, 2000; Hu and Tierney, 2001).
In this study, the analysis of At5g05500, a putative structural protein whose expression is mainly found in the trichoblasts of the Arabidopsis root, is described.
Materials and methods
Plant material and growth conditions
Plants were grown in a greenhouse with 16h light, 8h dark cycles at a temperature of 22 °C/18 °C during light and dark conditions, respectively, under a constant humidity of 70%. Dark-grown plants were grown on nutrient agar solidified medium (Estelle and Sommerville, 1987; Refrégier et al., 2004); light-grown plants were grown on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 1% sucrose and 0.7% agar (Duchefa, The Netherlands).
Mutant analysis
Atprpl1-1 was obtained from FagDB (Samson et al., 2002) in the Wassilevskija (Ws) background. Homozygous knock-out lines of Atprpl1-1 were selected by PCR screening. RNA was extracted from 7-day-old seedlings using the RNeasy RNA extraction kit (Qiagen) and used to generate cDNA (Superscript III reverse transcriptase H; Invitrogen). Semi-quantitative reverse transcription–PCR (RT–PCR) was performed to analyse the expression of AtPRPL1 in the T-DNA insertion lines with the primers OLERTfor 5ʹ ATGGA CTCA AGA GCCTTACCC 3ʹ (forward) and OLERTrev 5ʹ GTAAGTGGGTGGT GCAGTCG 3ʹ (reverse). ACT IN1 (At2g37620) was amplified as an internal control using the following primer set: RTactinfor 5ʹ GGCGATGAAGCTCAATCCAAA 3ʹ and reverse primer RTactinrev 5ʹ GGTC ACGACCAGCAAGATCAAG 3ʹ.
Generation of transgenic plants
DNA was isolated from wild-type (Col-0) Arabidopsis leaves with a DNA extraction kit (Plant DNA Mini kit, Omega Biotek, Inc.) following the manufacturer’s protocol. The promoter region, 2000bp upstream of the ATG start codon, was selected to study the expression pattern of PRPL1. This region was amplified using the forward primer pPRPLfor2kb 5ʹ GGGGAC AAGTTTGTACA AAAAAGCAG GCTGCCTCAGCT TTACCAGCTTT 3ʹ for the 2kb promoter and pPRPLfor446bp 5ʹ GGGACAAG TTTG TAC AAAAAAGCAGGCTCAAATTCCTCTGTTTGGTCCTT 3ʹ for the 446bp promoter, and reverse primer pPRPLrev 5ʹGGGGAC CACTTTGT ACAAGAAA GCT GGGT GTTACTTGTTATA CACTCTGCTTCTTAAAC 3ʹ for both constructs, including Gateway-compatible recombination sites. Amplified fragments were first cloned into pDONR207 and sequenced before cloning into the final binary vectors pGWB3 (GUS; 446bp promoter) and pGWB4 [green fluorescent protein (GFP) 2kb promoter]. This resulted in the final vectors pAtPRPL1::GUS and pAtPRPL1::GFP.
Overexpression of AtPRPL1 was achieved by amplifying the open reading frame (ORF) with primers PRP L1OEfor 5ʹ GGGG ACAAGTTT GTACAAA AAAGCAGG CTTC ATGGACTCAAGAGCCTTACCC 3ʹ (forward) and PR PL1OErev 5ʹ GGGGA CCA CTTTGTAC AAGAAAGCTGGGTTT CACTAAGTG GGTGGTGC AGTCG 3ʹ (reverse). After sequencing, the amplified ORF was cloned in the binary pH2GW7,0 (Karimi et al., 2002) that results in expression of the ORF under control of the Cauliflower mosaic virus (CaMV) 35S promoter.
To determine the subcellular localization of AtPRPL1, a C-terminal fusion gene with GFP under control of a 35S promoter was constructed. The ORF was amplified with the primers PR PL1OEfor 5ʹ GGGGAC AAGTTTGTA CAAAAAAGCAGGCT TC AT GG ACTC AAGAGCC TTACCC 3ʹ (forward) and PR PL1OEGFPr 5ʹ GGGGAC CACTTT GTA CA GAAAGC TGGGTTGTAAGT GGGTGGTGCAGTCG 3ʹ (reverse) and cloned into pGWB5 generating 35S::PRPL1-GFP. As a second construct, the fusion gene was placed under control of the AtPRPL1-own promoter. As the ORF does not contain any introns, the promoter region (446bp) and the ORF were amplified as one fragment with the primers PRPL1for446bp and PRP L1OEGFPr, and cloned into pDONR207. After sequencing, this fragment was recombined into pGWB4 generating pPRPL1::PRPL1-GFP.
All final vectors were transformed in Agrobacterium tumefaciens strain C58 that contained the pMP90 helper plasmid before Arabidopsis thaliana Col-0 was transformed by flower dip (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS medium (Murashige and Skoog, 1962) supplemented with 1% sucrose, 0.7% agar (Duchefa, The Netherlands), and the appropriate antibiotic. At least 30 independent transgenic lines were selected, and homozygous lines were identified after segregation analysis for further analysis.
Phylogenetic analysis
To elucidate the relationship of AtPRPL1 to other proteins, a phylogenetic tree was created from parsimony analysis. The protein sequence of AtPRPL1 was used to retrieve related sequences from the BLAST program (Altschul et al., 1990). These retrieved sequences were aligned in ClustalW 1.87 and a phylogenetic tree was made from Neighbor–Joining analysis using Mega 5.05 software (Tamura et al., 2011). The bootstrap values are shown in the tree, and 1000 replicates were used.
GUS staining
Seedlings were taken at different stages during plant development and fixed with 90% acetone for 20min. After rinsing with distilled water, whole seedlings were stained for 16h based on Jefferson et al. (1987) in GUS staining buffer [2mM 5-bromo-4-chloro-3-indolyl-β-d-glucoronide, 200mM sodium phosphate (pH 7.0), 10mM Na2EDTA, 1mM K4Fe(CN)6, and 1mM K3Fe(CN)6] at 37°C. After incubation, seedlings were fixed in ethanol/acetic acid (3/1) and cleared with 8M NaOH for 1h. Images were taken with a Zeiss Axioskop equipped with a Nikon DXM 1200 or Nikon Ds-Fi1 digital camera.
GFP visualization
GFP was localized in pAtPRPL1::GFP and protein–GFP fusion lines using a Nikon C1 confocal microscope or a Nikon Eclipse Ti-E inverted microscope attached to a microlens-enhanced dual spinning disk confocal system (UltraVIEW VoX; PerkinElmer). The seedlings were briefly dipped in 0.1mg ml–1 propidium iodide to counterstain the cell walls and to aid cell-specific localization of the GFP.
Transient expression to co-localize protein–GFP and mCherry-labelled markers for different organelles
A transient expression of mCherry-labelled markers for different organelles was performed in 35S::PRPL1-GFP and pPRPL1::PRPL1-GFP using the protocol described in Van Loock et al. (2010). The markers used were for the endoplasmic reticulum (ER), plasma membrane, peroxisomes, and the Golgi apparatus, and they are described in Nelson et al. (2007).
Transcript analysis
To quantify the expression levels of At5g05500 in the knock-out and the different generated transgenic lines, a quantitative RT–PCR (qRT–PCR) analysis was performed. RNA isolation was performed using TRIzol Reagent (Life Technologies) according to the guidelines of the manufacturer and its quality was checked with a nanodrop (ND1000). SuperScript™ II Reverse Transcriptase (Life Technologies) was used to perform cDNA synthesis according to the manufacturer’s instructions. All samples were diluted 1:8 with RNase-free water prior to the qPCR. Gene expression analysis was performed using TaqMan Universal Master Mix II, with uracil-N glycoslyase (UNG) and ROX (passive reference dye) as a technical control. The TaqMan probe At02181637_s1 was used to quantify the expression of At5g05500. To normalize the samples, the expression level of actin 8 (At1g49240) was monitored with the probe At02270958_gH. All probes span intron-separated exons and were ordered from Life Technologies. Results were analysed with the StepOnePlus Real-Time PCR System (Life Technologies) software at a confidence level of 95%.
FT-IR analysis of etiolated hypocotyls and root hairs
Fourier transform-infrared (FT-IR) analysis was performed as described by Mouille et al. (2003).
For hypocotyls, 4-day-old dark-grown seedlings were squashed between two BaF2 windows and rinsed thoroughly with distilled water before they were air-dried at 37 °C for at least 20min. Four biological repeats were grown and five spectra for each repeat were used to analyse changes in cell wall composition.
To study root hairs, 1-week-old light-grown seedlings were collected in absolute ethanol, and then samples were rehydrated for a few hours in distilled water and air-dried at 37 °C for at least 20min. Two biological repeats were grown, and spectra of 10 root hairs per seedling for each repeat were used to analyse changes in cell wall composition.
For both the hypocotyl and root hair study, the spectra were baseline corrected and area normalized before further analysis. A dendogram construction was performed as described before (Mouille et al., 2003; Robin et al., 2003). To analyse the spectral data with principal component analysis (PCA), WINDAS was used to normalize the data and to construct the data matrices. One-sample t-tests were executed to detect significant changes in the absorption at the different wavenumbers, which is indicative of compositional changes in the cell walls.
Results
AtPRPL1 is a proline-rich protein family member with an extensin-like domain
Several microarray data identified a large group of cell wall-related genes that are involved in A. thaliana root hair development (Ma and Bohnert, 2007; Velasquez et al., 2011; Bruex et al., 2012). One of these genes, At5g05500, is a cell wall-related gene that contains a pollen Ole allergen domain (PF01190, Pollen_Ole_e_I; Marchler-Bauer et al., 2007) and an extensin (IPR006041) domain. This gene contains no introns and has an ORF of 552bp that encodes a protein of 183 amino acids (Fig. 1a). At5g05500 has a putative signal peptide of 23 amino acids (Signal-3L; http://www.csbio.sjtu.edu.cn/cgi-bin/Signal3L.cgi) that with high probability targets the protein to the endomembrane system of the cell (Arabidopsis Cell eFP browser; based on Heazlewood et al., 2007). Using a BLASTP similarity search (Altschul et al., 1990, 1997) and the At5g05500 protein as a query sequence, phylogenetic analysis showed that the most closely related proteins are two unknown proteins of Vitis vinifera that share almost 70% identical amino acids with the At5g05500 protein. No closely related homologues were identified in Arabidopsis. A phylogenetic tree containing only Arabidopsis proteins was constructed with the Neighbor–Joining method using MEGA5.05 (Tamura et al., 2011). Figure 1b shows a part of the tree with only the 16 most related Arabidopsis proteins. The alignment of At5g05500 with five characterized proteins of the upper cluster is presented in Fig. 1c. All proteins more or less share a common Pollen_Ole_e_I and extensin family domain. This pollen Ole domain is named based on the high similarity with the major olive pollen allergen Ole 1 to which >70% olive allergic patients are sensitive (Lauzurica et al., 1988; Wheeler et al., 1990; Villalba et al., 1993). The most related of the characterized proteins in Arabidopsis are AtPRP1 and AtPRP3 (Fowler et al., 1999; Bernhardt and Tierney, 2000), two proteins that are 32% and 30%, respectively, identical to the At5g05500 protein, and both are 43% similar. The At5g05500 protein has 17 proline residues (or 9%; see grey shaded Ps in Fig. 1c) and a length of 183 amino acids, whereas AtPRP1 and AtPRP3 have 23% and 22% proline residues and a length of 313 and 310 amino acids, respectively. The percentage of proline residues in the whole protein is clearly lower than in the two AtPRPs; however, the C-terminal parts of the proteins share the greatest sequence identity (Fig. 1c). This suggests that At5g05500 could belong to the PRP class of genes (Showalter, 1993; Fowler et al., 1999). Given its close relationship to AtPRP1 and AtPRP3 and the lower percentage of proline, At5g05500 was named AtPRP-like1 (AtPRPL1). The hydrophobic isodityrosine cross-link motif (YVY), which is present in AtEXT3, a HRGP, is absent in AtPRPL1.
Fig. 1.
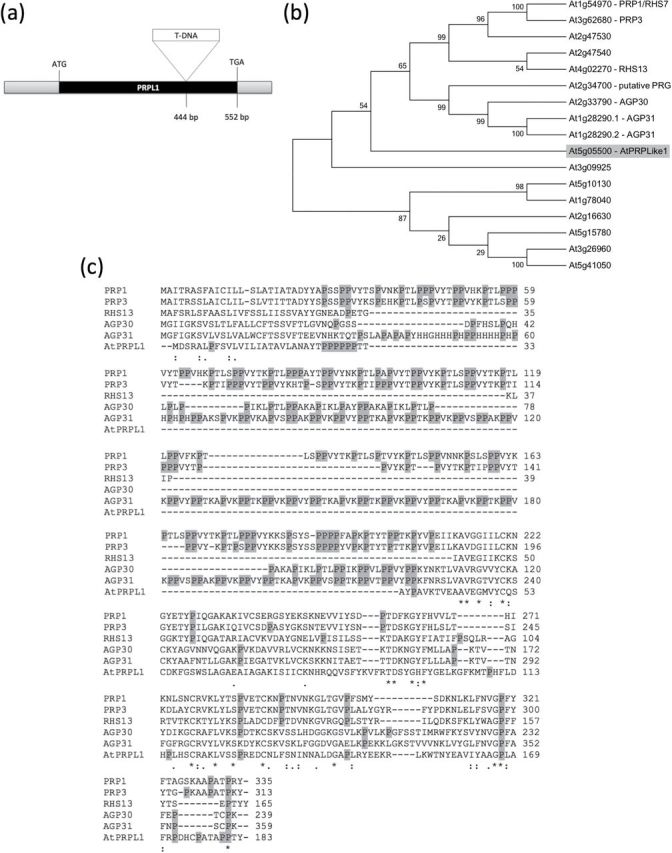
In silico analysis of At5g05500. (a) Schematic representation of At5g05500 gene structure, including the start and stop codon, one exon, and the T-DNA insertion position in the mutant. (b) Phylogenetic tree made by Neighbor–Joining analysis using Mega 5.05 software of the 15 most related proteins in Arabidopsis identified by BlastP. The bootstrap values are shown in the tree, and 1000 replicates were used. (c) Alignment of the DNA sequence of At5g05500 and the five most related genes. Prolines are shaded in grey.
Expression pattern of AtPRPL1 during different developmental stages
In silico promoter analysis revealed cytokinin- and ABA-responsive elements and pollen and root hair cis-elements (Supplementary Fig. S1 available at JXB online). The expression pattern of AtPRPL1 was investigated using a 446bp promoter fragment driving the GUS gene in homozygous T3 lines. In dark-grown hypocotyls, AtPRPL1 has a weak expression at the onset of the fast elongation, seen as faint blue staining after GUS assay (Fig. 2a). The expression level decreases once the fast elongation phase has started (Fig. 2b). No expression was detected after 4 d of growth, or after 8 d when the hypocotyl is fully grown (data not shown). These observations were confirmed in 2kb promoter::GFP lines (data not shown).
Fig. 2.
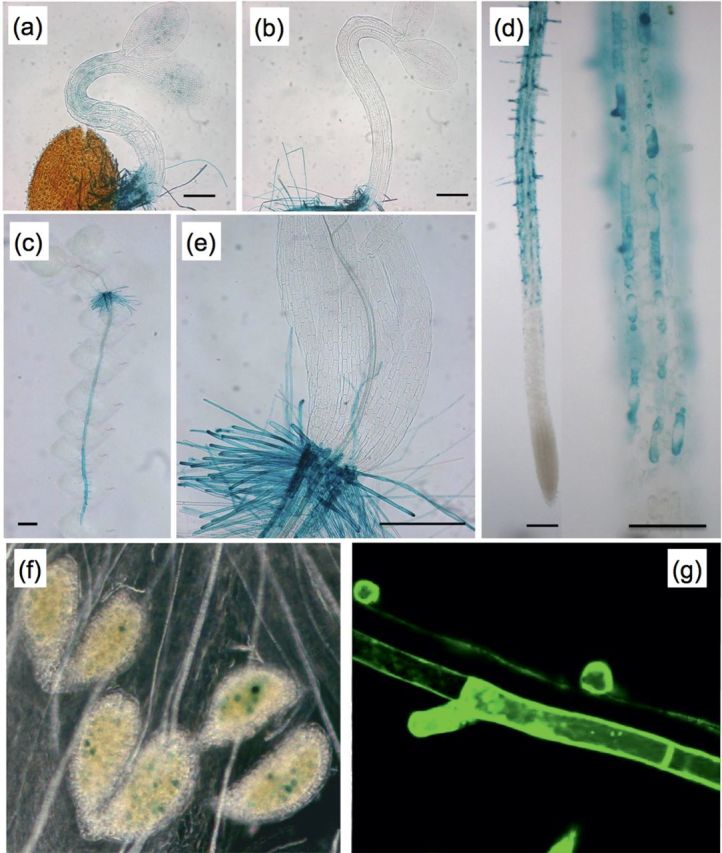
Expression analysis of At5g05500 using promoter–reporter lines. Bright-field images showing (a) faint GUS activity in a young etiolated hypocotyl, (b) absent GUS activity in a young etiolated hypocotyl where fast cell expansion has started, (c) enrichment in the root of a whole seedling, (d) trichoblast-specific expression in a root and in its close-up, (e) expression in root hairs at the collet, (f) expression in pollen. (g) represents a confocal image of promoter-driven GFP in the trichoblast cell files only. Scale bars are 200 μm in (a, b, and e), 100 μm in (d), and 1mm in (c).
AtPRPL1 is highly expressed in the root (Fig. 2a–d) and appears solely in the trichoblast cell file (overview of seedlings in Fig. 2c; close-ups in Fig. 2d). Also at the transition between hypocotyl and root (collet) AtPRPL1 is expressed in root hair cells only (Fig. 2e). No expression was detected in a light-grown hypocotyl or in cotyledons or leaves at any developmental stage (Fig. 2c, e, and data not shown). A transient expression was detected in developing pollen (Fig. 2f), while it was absent in other flower parts. The expression pattern of AtPRPL1 in roots was confirmed with GFP as reporter (Fig. 2g).
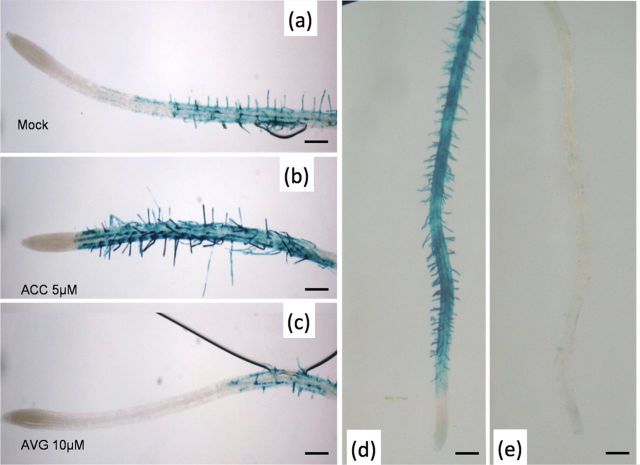
Since in the root, expression was found in trichoblasts only, the effect of ACC, which promotes root hair formation, and 1-α-(2-aminoethoxyvinyl)glycine (AVG), an inhibitor of ethylene biosynthesis, was studied on the expression pattern of AtPRPL1 (Masucci and Schiefelbein, 1996). Whereas staining was absent in the atrichoblasts of an untreated plant (Fig. 3a), after ACC treatment the cells showing ectopic root hairs also expressed AtPRPL1 (Fig. 3b). AVG treatment resulted in a strongly decreased or completely absent expression in correlation with decreased or absent root hair formation (Fig. 3c). These changes in AtPRPL1 expression were confirmed in both an eto2 and ein2-1 background that mimick the changes in ethylene levels after ACC and AVG treatment (Fig. 3d, e).
Fig. 3.
Effect of altered ethylene presence and mutated background on AtPRPL1 expression. (a) GUS activity is seen in the trichoblast cell files of a mock-treated root. (b) Treatment with the ethylene precursor ACC induces ectopic root hair formation and concomitant extra AtPRPL1 expression. (c) AVG-inhibited ethylene production interferes with root hair initiation/formation and reduces AtPRPL1 expression. (d, e) Both ethylene effects are mimicked in the eto2 and ein2-1 backgrounds. Scale bars are 100 μm.
Altered expression levels of AtPRPL1 interfere with cell elongation
Atprpl1-1, a T-DNA insertion line (A. thaliana, ecotype Ws) with an insertion in the exon (Fig. 1a), was obtained from the FlagDB collection (Samson et al., 2002). A semi-quantitative RT–PCR analysis of AtPRPL-1 expression in the wild type and Atprpl1-1 with actin and elongation factor as an internal control proved that it was a null mutant. Phenotypic analysis of root and etiolated hypocotyl length shows no significant difference between the knock-out line and the wild type (Fig. 4a, b). However, a clear reduction in root hair length was observed in the knock-out line (Fig. 4c). Wild-type root hairs were 536±12 μm long, whereas in the knock-out they were shorter: 393±12 μm. Supplementary Fig. S16 in Velasquez et al. (2011) mentions two other insertion lines in the same gene (in an exon and in the promoter), but at other positions, and these show a root hair length reduction as well, confirming that the absence of this gene causes the phenotype described here.
Fig. 4.
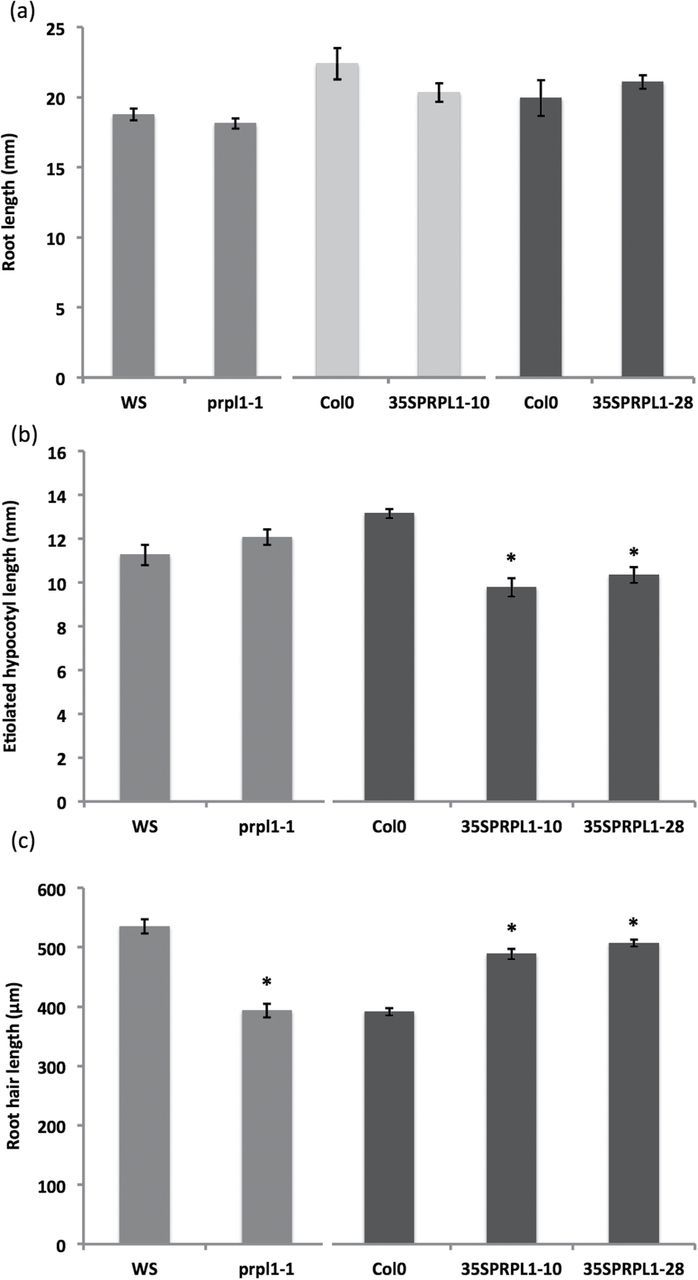
Root, etiolated hypocotyl, and root hair phenotyping in the wild type, knock-out, and overexpression lines of AtPRPL1. (a) Root length, (b) etiolated hypocotyl length, and (c) root hair length of wild type, knock-out, and overexpression plants. Asterisks refer to significant changes compared with the corresponding wild types (the wild type and lines that are statistically compared with it are presented in the same greyscale). Data shown are the average of three representative biological replicates having at least 15 seedlings; error bars represent the SE. Student’s t-test, P<0.05.
To analyse further the role of AtPRPL1 in cell elongation, transgenic lines overexpressing the ORF of AtPRPL1 under control of the constitutive CaMV 35S promoter (35S::AtPRPL1) were generated. Homozygous plants were identified after segregation analysis on hygromycin, and a semi-quantitative RT–PCR was performed to identify lines with a clear overexpression. Relative quantification with qPCR analysis revealed an AtPRPL1 expression increase of 19 and 16.5 times in 35S::AtPRPL1-10 and 35S::AtPRPL1-28 respectively.
Hypocotyls grown for 4 d in the dark were analysed and this revealed that the overexpression of AtPRPL1 reduced the hypocotyl length by 40% and 20% (Fig. 4b). Wild-type hypocotyl length was 13.16±0.21mm; 35S::AtPRPL1-10 and 35S::AtPRPL1-28 had hypocotyls of 9.79±0.42mm and 10.34±0.36mm, respectively. The overexpression also caused a significant increase in the root hair length when compared with the control plants. Root hair lengths of overexpression lines (489±9 μm for 35S::AtPRPL1-10 and 507±6 μm for 35S::AtPRPL1-28) were clearly longer, namely 24.74% and 27.80%, than those of the wild type (392±6 μm) (Fig. 4c).
FT-IR analysis of root hair cell walls reveals subtle compositional changes
As the PRPL1 protein is located in the root hairs, the composition of the cell wall of root hairs in wild-type plants, Atprpl1-1, and 35S::AtPRPL1-10 was investigated by FT-IR. Figure 5a shows the comparison of the average spectra where Ws serves as the wild type for Atprpl1-1 and Col-0 for 35S::AtPRPL1. These spectral data were analysed with one-sample t-test to reveal significant changes. The region between wave number 1384cm–1 and 1307cm–1 and between 1446cm–1 and 1415cm–1 showed that the null mutant Atprpl1-1 contains cell walls with significantly altered composition. However, those changes cannot be accurately assigned to specific bonds (Fig. 5b).
Fig. 5.
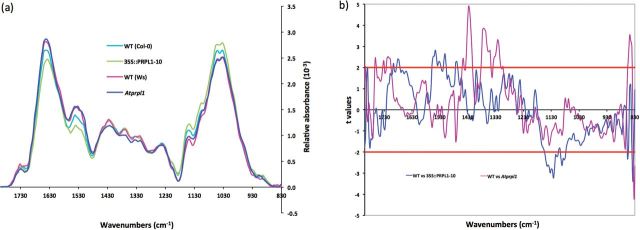
Cell wall compositional changes between root hairs of the wild type, knock-out, and overexpression lines of AtPRPL1 revealed by FT-IR. (a) Comparison of the average spectra of Atprpl1-1 (dark blue trace) and its corresponding WT (pink trace) and of 35S::AtPRPL1-10 (green trace) and its corresponding WT (light blue trace). (b) Student’s t-test (P=0.05) of a comparison of FT-IR spectra sampled from root hairs. Comparison between WT and Atprpl1-1 (violet trace) and WT and AtPRPL1-1 overexpression (blue trace). For t-values >2 the absorbance for the corresponding wavenumber is significantly higher in the wild type. Horizontal red lines mark the P=0.01 significance level.
The comparison of the spectra of the overexpressing line and wild type showed a significantly changed absorption in the region between 1550cm–1 and 1500cm–1. This region contains wave numbers that correspond to amide I (1546cm–1 and 1511cm–1) (Sene et al., 1994). Another region with a significantly changed absorption in the overexpression lines, from 1141cm–1 to 1070cm–1, contains mainly wave numbers that correspond to cellulose (1091cm–1) and xyloglucan (1120cm–1) (Kacuráková et al., 2000; Wilson et al., 2000; Carpita et al., 2001). Therefore, the data of AtPRPL1 overexpression strongly suggest that the presence of AtPRPL1 in root hairs is positively correlated with the presence of polysaccharides such as cellulose and xyloglucan.
FT-IR analysis of etiolated hypocotyl cell walls reveals no compositional changes and suggests that AtPRPL1 is not a non-conventional AGP
As shown in the phylogenetic analysis, AtPRPL1 is closely related to AtPRP genes and to AGP30 and AGP31. To test whether AtPRPL1 is an AGP or not and to identify potential cell wall compositional changes that could explain the hypocotyl phenotype in the overexpression lines, the composition of the cell wall of 4-day-old dark-grown hypocotyls in wild-type and overexpression plants was investigated by FT-IR microspectroscopy (Mouille et al., 2003; Pelletier et al., 2010). Spectra were taken next to the central cylinder in the middle of the hypocotyl. Student’s t-tests on the comparison of the average spectra sampled did not identify consistent significant changes in the cell wall composition of both overexpression lines when compared with the wild type (Supplementary Fig. S2 at JXB online), indicating that AtPRPL1 is not an AGP and that altering levels of AtPRPL1 affect wall composition in a complex fashion.
Subcellular localization of PRPL1
To investigate the subcellular localization of AtPRPL1, the ORF was fused to a GFP-coding sequence and used to generate transgenic lines with stable expression of 35S::AtPRPL1-GFP. Using confocal and spinning disc microscopy AtPRPL1–GFP was located in small vesicles that were actively moving around in the cell (see Supplementary Video S1 at JXB online). In plants expressing AtPRPL1-GFP driven by its own promoter, GFP was visible in similar moving vesicles, but only in the trichoblast cell files, confirming the expression analysis (data not shown). In comparison with the 35S promoter, the promoter of AtPRPL1 was, however, much weaker. To assess whether the movement was actin or microtubule based, 35S::AtPRPL1-GFP plants were grown for 2 d in the dark and treated with 5 μM latrunculin or 10 μM oryzalin to disrupt actin bundles and microtubules, respectively (see Supplementary Videos S2 and S3 at JXB online). The movies were created with the same settings of the confocal microscope, and bleaching of the GFP can be seen near the end of the movies. Only latrunculin (Supplementary Video S2 at JXB online) was capable of stopping the movement of the GFP-enriched vesicles, suggesting that they move on actin. To identify the nature of the AtPRPL1–GFP-enriched vesicles, different cell organelle markers (Nelson et al., 2007) were transiently expressed in 35S::AtPRPL1-GFP roots (Fig. 6a–d) according to Van Loock et al. (2010). From these co-localization experiments with plasma membrane-, Golgi apparatus-, peroxisome-, and ER-specific red fluorescent protein (RFP), it seems that AtPRPL1–GFP is localized on the ER, or in vesicles that move along or on the ER. To confirm these results, the mCherry-fused ER marker ER-rb CD3-960 (Nelson et al., 2007) was transformed in the 35S::AtPRPL1-GFP background. As in the transient expression, AtPRPL1–GFP is localized in small vesicles that move ‘on the ER’ in roots (Fig. 6e–g). From the pictures and the movies that were generated, it remains unclear whether or not the AtPRPL1–GFP is transferred from the ER to the cell wall.
Fig. 6.
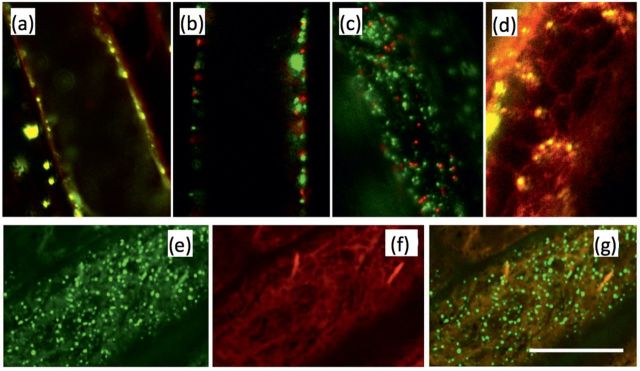
Protein–GFP localization. Transient co-localization analysis of AtPRPL1–GFP and RFP-tagged markers for the (a) plasma membrane, (b) Golgi apparatus, (c) peroxisomes, and (d) ER. (e) AtPRPL–GFP, (f) mCherry-fused ER marker ER-rb CD3-960, and (g) overlay of both in 35S::AtPRPL1-GFP lines stably transformed with the ER marker. Scale bar=10 μm.
Discussion
In this study, a novel PRP-like protein encoded by At5g05500, which is specifically expressed in root hair cells, was identified. The protein contains a pollen Ole allergen domain (pfam01190, Pollen_Ole_I; Marchler-Bauer et al., 2007) and an extensin (IPR006041) domain. Phylogenetic analysis showed that At5g05500 is closely related to AtPRP3 and AtPRP1, identified by Fowler and co-workers (1999). At5g05500 is a small gene that contains no introns and codes for a small protein of only 183 amino acids. The deduced protein contains an N-terminal putative signal peptide of 20 amino acids that probably directs this protein to the ER and possibly to the cell wall. This was confirmed in transgenic plants bearing protein–GFP constructs and using co-localization experiments. The protein is most related to PRPs, which indicates that AtPRPL1 could be a member of this PRP family (or subfamily) of genes. Also AGP30, reported to be more related to PRP genes than to other AGP genes (Showalter et al., 2010), and AGP31 were closely related to At5g05500. FT-IR results on transgenic lines overexpressing AtPRPL1 excluded that it is a NcAGP since an increase in a specific type II arabinogalactan side chain was not seen at the 1078cm–1 wavenumber (Kacuráková et al., 2000). Furthermore, AGP30 has been reported to be more related to PRPs than to other AGPs (Showalter et al., 2010), strengthening the present findings. As the closest relatives of At5g05500 are PRPs and as the percentage of proline residues in the protein is slightly lower, At5g05500 was named a proline-rich protein-like gene, AtPRPL1.
Expression analysis of AtPRPL1 using promoter::GUS and promoter::GFP lines indicated that its expression was positively correlated with the occurrence and outgrowth of root hairs. The expression after treatments (ACC and AVG) or in mutants (eto2 and ein2-1) that induced or inhibited the initiation of root hairs confirmed this. The two closest relatives in Arabidopsis, AtPRP1 and AtPRP3, were also shown to be involved in root hair growth (Bernhardt and Tierney, 2000; Hu and Tierney, 2001). AtPRP3 is expressed during root hair initiation and outgrowth of the root hairs. A null mutant of AtPRP3 shows an aberrant phenotype in root hair branching (Bernhardt and Tierney, 2000; Hu and Tierney, 2001). From the present expression data and the effect of the knock-out mutation and overexpression of AtPRPL1 on root hair length, it can be concluded that AtPRPL1 is positively involved in root hair formation, similar to AtPRP3.
The plant cell wall is a complex and diverse structure and consists of the polysaccharides cellulose, cross-linking glycans, pectins, and structural proteins. All components are organized into a complex extracellular matrix (Carpita and Gibeaut, 1993). Interactions between structural cell wall proteins and other cellular matrix components are critical for the integrity of this matrix and correct self-assembly of the cell wall (Cannon et al., 2008). PRPs belong to a superfamily of cell wall proteins called HRGPs. Besides the lightly glycosylated PRPs, moderately glycosylated extensins (EXTs) and highly glycosylated arabinogalactan-proteins (AGPs) also belong to the HRGPs (Showalter et al., 2010). HRGPs contribute to form an independent structure-determining network (Bernhardt and Tierney, 2000). Recently it was reported that post-translational modifications, such as proline hydroxylation and O-glycosylation, of HRGPs are essential in root hair growth (Velasquez et al., 2011, 2012a, b), whereby prolyl 4-hydroxylase (P4H) is the enzyme that converts proline to hydroxyproline. Velasquez et al. (2011) showed aberrancies in root hair elongation caused by inhibition of P4H activity. PRPs are thought to be secreted into the wall where, according to their ability, associate with and become covalently cross-linked to components within the cell wall (Bradley et al., 1992; Showalter, 1993). It was shown here that AtPRPL1 is a member of PRPs and suggest that it somehow participates in determining the root hair cell wall structure during plant development, being a member of the HRGP network. From the clear root hair phenotypes and PRPL1’s location in vesicles that move on/along the ER, it seems that AtPRPL1 is essential for creating a wall that grows normally. Differences in cell wall composition were seen in the FT-IR analysis of knock-outs and overexpression plants, but only in root hair walls, suggesting that AtPRPL1 affects cell wall composition in a rather complex manner. How AtPRPL1 interferes with cellulose deposition and xyloglucan sequestering into the root hair cell wall remains unknown. It could, however, be postulated that it somehow functions as a kind of chaperone in moving vesicles, but this has to be elucidated. How exactly it manages to play its role remains unclear. Furthermore, co-expression analysis revealed that At4g02270 (a closely related pollen Ole e 1 allergen and extensin family protein) is co-expressed with AtPRPL1 (www.genevestigator.ethz.ch; Zimmermann et al., 2004), pointing to possible redundancy. However, as seen here, the knock-out of At5g05500 already showed a clear root hair phenotype, suggesting that multiple knock-outs in extensin(-like) genes which were identified by Velasquez et al. (2011) could have even much more severe effects.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. In silico analysis of the putative promoter sequence of At5g05500.
Figure S2. Cell wall compositional changes between etiolated hypocotyls of the wild type and two overexpression lines of AtPRPL1 revealed by FT-IR.
Video S1. Movie showing green AtPRPL1–GFP proteins in 35S::AtPRPL1-GFP lines grown in control conditions.
Video S2. Movie showing green AtPRPL1–GFP proteins in 35S::AtPRPL1-GFP lines after treatment with 5 μM latrunculin.
Video S3. Movie showing green AtPRPL1–GFP proteins in 35S::AtPRPL1-GFP lines after treatment with 10 μM oryzalin.
Acknowledgements
The authors would like to thank the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT, Vlaanderen), the Hercules Foundation (AUAH-09-001 to DA), the University of Antwerp, the Research Foundation, Flanders (FWO, Vlaanderen), the Interuniversity Attraction Poles Programme – Belgian State – Belgian Science Policy [IUAP VI/33], and the French ‘Agence Nationale de Recherche’ (grant BLAN08-1_332764 to HH).
References
- Acebes JL, Lorences EP, Revilla G, Zarra I. 1993. Pine xyloglucan occurrence, localization and interaction with cellulose. Physiologia Plantarum 89, 417–422. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Tierney ML. 2000. Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiology 122, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. 1992. Elicitor- and wound-induced oxidative crosslinking of a proline-rich plant cell wall protein: a novel, rapid defence response. Cell 70, 21–30. [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C. 1994. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. The Plant Cell 6, 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, et al. 2012. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genetics 8, e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Cen Y, Kieliszewski MJ. 2008. Self-assembly of the plant cell wall requires an extension scaffold. Proceedings of the National Academy of Sciences, USA 105, 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal 3, 1–30. [DOI] [PubMed] [Google Scholar]
- Carpita N, Tierney M, Campbell M. 2001. Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Molecular Biology 47, 1–5. [PubMed] [Google Scholar]
- Cassab GI. 1998. Plant cell wall proteins. Annual Review of Plant Physiology and Plant Molecular Biology 49, 281–309. [DOI] [PubMed] [Google Scholar]
- Chen J, Varner JE. 1985. Isolation and characterization of cDNA clones for carrot extension and a proline-rich 33-Kda protein. Proceedings of the National Academy of Sciences, USA 82, 4399–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 1999. Enzymes and other agents that enhance cell wall extensibility. Annual Review of Cell and Developmental Biology 13, 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2000. Expansive growth of plant cell walls. Plant Physiology and Biochemistry 38, 109–124. [DOI] [PubMed] [Google Scholar]
- De Cnodder T, Vissenberg K, Van Der Straeten D, Verbelen J-P. 2005. Regulation of cell length in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid: a matter of apoplastic reactions. New Phytologist 168, 541–550. [DOI] [PubMed] [Google Scholar]
- Estelle MA, Somerville CR. 1987. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Molecular Genetics and Genomics 206, 200–206. [Google Scholar]
- Fincher GB, Stone BA, Clarke AE. 1983. Arabinogalactan-proteins: structure, biosynthesis and function. Annual Review of Plant Physiology 34, 47–70. [Google Scholar]
- Fowler TJ, Bernhardt C, Tierney ML. 1999. Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiology 121, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. 1989. Xyloglucans in the primary cell wall. Annual Review of Plant Physiology and Plant Molecular Biology 40, 139–168. [Google Scholar]
- Hayashi T, Baba K, Ogawa K. 1994. Macromolecular complexes of xyloglucan and cellulose obtained by annealing. Plant and Cell Physiology 35, 219–223. [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. 2007. SUBA: the Arabidopsis subcellular database. Nucleic Acids Research 35, D213–D218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JC, Nagao T, Key JL. 1990. Characterization of a proline-rich cell wall protein gene family of soybean. Journal of Biological Chemistry 265, 2470–2475. [PubMed] [Google Scholar]
- Hu J, Tierney ML. 2001. Arabidopsis lines lacking expression of a root hair-specific proline-rich cell wall protein (AtPRP3) are altered in root hair shape. Proceedings of the 9th International cell wall meeting, Toulouse, France, p. 65.
- Huckelhoven R. 2007. Cell-wall associated mechanism of disease resistance and susceptibility. Annual Review of Phytopathology 45, 101–127. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions, β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. 2003. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology 133, 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josè-Estanyol M, Ruiz-Avila J, Puigdomènech P. 1992. A maize embryo-specific gene encodes a proline-rich and hydrophobic protein. The Plant Cell 4, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacuráková M, Capek P, Sasinková V, Wellner N, Ebringerová A. 2000. FT-IR study of plant cell wall model compounds: pectic polysaccharides and hemicelluloses. Carbohydrate Polymers 43, 195–203. [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kleis-San Fransisco SM, Tierney ML. 1990. Isolation and characterization of a proline-rich cell wall protein from soybean seedlings. Plant Physiology 94, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzurica P, Maruri N, Galocha B, Gonzalez J, Diaz R, Palomino P, Hernandez D, Garcia R, Lahoz C. 1988. Olive (Olea europea) pollen allergens—II. Isolation and characterization of two major antigens. Molecular Immunology 25, 337–344. [DOI] [PubMed] [Google Scholar]
- Lindstrom JT, Vodkin LO. 1991. A soybean cell wall protein is affected by seed color genotype. The Plant Cell 3, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert H. 2007. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biology 8, R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall AJ, Brett GM, Morris VJ, Rigby NM, Ridout MJ, Ring SG. 2001. The effect of peptide–pectin interactions on the gelation behaviour of a plant cell wall pectin. Carbohydrate Research 335, 115–126. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, et al. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Research 35, D237–D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1996. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. The Plant Cell 8, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H. 2003. Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. The Plant Journal 35, 393–404. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nothnagel EA. 1997. Proteoglycans and related components in plant cells. International Review of Cytology 174, 195–291. [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, et al. 2010. A role for pectin de-methylesterification in a developmentally-regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytologist 188, 726–739. [DOI] [PubMed] [Google Scholar]
- Refrégier G, Pelletier S, Jaillard D, Höfte H. 2004. Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiology 135, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin S, Lecomte M, Höfte H, Mouille G. 2003. A procedure for the clustering of cell wall mutants in the model plant Arabidopsis based on Fourier-transform infrared (FT-IR) spectroscopy. Journal of Applied Statistics 30, 669–681. [Google Scholar]
- Sachetto-Martins G, Franco LO, de Oliveira DE. 2000. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochimica et Biophysica Acta 1492, 1–14. [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. 2002. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Research 30, 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, van de Wiel C, Zalensky A, et al. 1990. The ENOD12 gene product is involved in the infection process during the pea–Rhizobium interaction. Cell 60, 281–294. [DOI] [PubMed] [Google Scholar]
- Schnabelrauch LS, Kieliszewski MJ, Upham BL, Alizedeh H, Lamport DTA. 1996. Isolation of pI 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. The Plant Journal 9, 477–489. [DOI] [PubMed] [Google Scholar]
- Sene C, McCann MC, Wilson RH, Grinter R. 1994. Fourier-transform Raman and Fourier-transform infrared spectroscopy (an investigation of five higher plant cell walls and their components). Plant Physiology 106, 1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, D’Ovidio R, Mehdy MC. 1991. Negative and positive regulation of a novel proline-rich protein mRNA by fungal elicitor and wounding. The Plant Journal 1, 345–354. [DOI] [PubMed] [Google Scholar]
- Showalter AM. 1993. Structure and function of plant cell wall proteins. The Plant Cell 5, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. 2010. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiology 153, 485–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney ML, Wiechert J, Pluymers D. 1988. Analysis of the expression of extensin and p33-related cell wall proteins in carrot and soybean. Molecular and General Genetics 211, 393–399. [Google Scholar]
- Valent BS, Albersheim P. 1974. The structure of plant cell walls: V. On the binding of xyloglucan to cellulose fibers. Plant Physiology 54, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin R, Cerclier C, Geneix N, Aguie-Beghin V, Gaillard C, Ralet MC, Cathala B. 2010. Elaboration of extensin–pectin thin film model of primary plant cell wall. Langmuir 26, 9891–9898. [DOI] [PubMed] [Google Scholar]
- Van de Wiel C, Scheres B, Franssen H, van Lierop MJ, van Lammeren A, van Kammen A, Bisseling T. 1990. The early nodulin transcript ENOD2 is located in the nodule parenchyma (inner cortex) of pea and soybean root nodules. EMBO Journal 9, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loock B, Markakis MN, Verbelen J-P, Vissenberg K. 2010. High-throughput transient transformation of Arabidopsis roots enables systematic co-localization analysis of GFP-tagged proteins. Plant Signaling and Behavior 5, 261–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner JE, Lin LS. 1989. Plant cell wall architecture. Cell 56, 231–239. [DOI] [PubMed] [Google Scholar]
- Velasquez SM, Iusem ND, Estevez JM. 2011. Root hair sweet growth. Plant Signaling and Behavior 6, 1600–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, et al. 2012. a O-Glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403. [DOI] [PubMed] [Google Scholar]
- Velasquez M, Salter JS, Dorosz JG, Petersen BL, Estevez JM. 2012. b Recent advances on the posttranslational modifications of EXTs and their roles in plant cell walls. Frontiers in Plant Science 3, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba M, Batanero E, López-Otín C, Sánchez LM, Monsalve RI, González de la Peña MA, Lahoz C, Rodríguez R. 1993. The amino acid sequence of Ole e I, the major allergen from olive tree (Olea europaea) pollen. European Journal of Biochemistry 216, 863–869. [DOI] [PubMed] [Google Scholar]
- Wheeler AW, Hickman BE, Fox B. 1990. Heterogeneity of a major allergen from olive (Olea europea) pollen. Molecular Immunology 27, 631–636. [DOI] [PubMed] [Google Scholar]
- Wilson E, Cooper JB. 1994. Characterization of PRP1 and PRP2 from Medicago truncatula . Plant Physiology 105, 445–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RH, Smith AC, Kacuráková M, Saunders PK, Wellner N, Waldron KW. 2000. The mechanical properties and molecular dynamics of plant cell wall polysaccharides studied by Fourier-transform infrared spectroscopy. Plant Physiology 124, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt RE, Nagao RT, Key JL. 1992. Patterns of soybean proline-rich protein gene expression. The Plant Cell 4, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.