Abstract
We developed a user-friendly program, Genome Profiler (GeP), to refine whole-genome multilocus sequence typing analysis by addressing gene paralogy with conserved gene neighborhoods. In comparison to similar programs, GeP produced overall the best results in terms of accuracy and is thus a useful alternative to resolve relationships of bacterial isolates.
TEXT
Whole-genome multilocus sequence typing (wgMLST) is a powerful tool to resolve the relationship of especially closely related bacterial isolates by indexing allele differences in the shared loci in the whole-genome sequencing (WGS) data (1–3). However, if gene paralogy is not adequately addressed, multicopy genes that are commonly observed in the bacterial genome might mislead the inferences of the genetic relationships (4). In closely related bacterial isolates, orthologs have a tendency to retain the same genome context, and synteny has been successfully used for automated clustering of orthologous genes in the pangenomic analysis of bacterial isolates (5, 6). Therefore, we hypothesized that conserved gene neighborhoods (CGN) can be used in the wgMLST to differentiate orthologs from recently duplicated paralogs. We implemented this idea into a novel program called Genome Profiler (GeP).
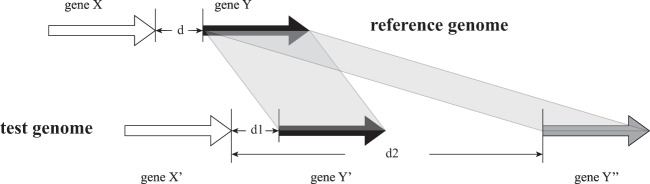
A detailed overview of GeP is presented in the supplemental material, including a flow chart of the logic of the program (see Fig. S1 in the supplemental material). Briefly, GeP starts by gathering information from the reference genomic sequence to build an ad hoc wgMLST scheme. The information of the new allele definitions and sequences will automatically accumulate by first using BLASTN or, in case it fails, BLASTX to locate the ortholog of the allele in the query genomes. For genes having multiple copies, CGN information in the reference genome is used to separate orthologs from paralogs. GeP assumes that the contiguity and the distance of any given two neighboring genes should be conserved between the reference genome and the tested genomes of the closely related isolates (Fig. 1). Therefore, GeP defines a value for each loci, namely, the “expected distance to the previous locus” (expected d), which is based on the CGN information gathered from the reference. If multiple valid BLAST hits for a given locus are found, GeP treats the hits as potential orthologs only when they are located inside the range of expected d, and it automatically selects the one with the smallest d value.
FIG 1.
Selection in the multiple BLAST hits in the GeP pipeline when the hits satisfy the initial screening (by default, percent coverage of >50% and percent identity of >80%). The CGN is used to select the ortholog of the gene Y from the hits. In the tested genome, BLAST hit gene Y′ is selected because the distance to the neighboring gene X is within the range of expected d (d + 10 bp). BLAST hit 2 gene Y′′ is excluded because the distance to the neighbor gene X is outside the expected range.
After locating all of the loci in the query genomes and assigning the corresponding allele number, GeP will summarize the genetic differences of all shared loci and write the results to several output files, allowing the user to easily visualize allelic differences of the isolates, as well as to perform downstream phylogenetic and population structure analyses. All the allele definitions and sequences are saved in files, allowing future analyses to use them and a standardized wgMLST scheme to be built upon.
We tested the GeP program on a WGS data set of 19 related Campylobacter jejuni isolates (see Table S1 in the supplemental material). Ten isolates were obtained from three independent waterborne outbreaks that occurred in 2000 to 2001 in Finland, and the others were from three Finnish chicken farms. The same data set was also analyzed using existing wgMLST programs, BIGSdb Genome Comparator (7) and SeqSphere+ version 1.0 (Ridom GmbH, Münster, Germany) (8), and compared to GeP.
An overview of the wgMLST results of the 19 C. jejuni genomes produced by the three programs using the genome of C. jejuni 4031 as a reference is presented in Table 1. The allele number of each locus in each genome, a summary of the pairwise allele differences, and the output.txt file from GeP can be found in Data Sets S1, S2, and S3, respectively, in the supplemental material. The topologies of the split graph generated by GeP and SeqSphere+ are identical and similar to the one produced by BIGSdb GC, with the exception of a visible netlike structure in the center of the graph (see Fig. S2 in the supplemental material). These results revealed that the core genomes of C. jejuni belonging to the same outbreak or isolated within the same farm were highly similar and separated from each other, confirming the results of our previous studies (2, 3). Despite the general similarity in the split graphs, the numbers of identical and polymorphic shared loci found by the three programs were different (Table 1), which affected pairwise allelic differences of the isolates (see Data Set S2). We manually inspected the loci differences between GeP and BIGSdb GC or SeqSphere+, and we classified the reasons for the observed dissimilarities into six categories, for simplicity here referred to as error types (Table 2).
TABLE 1.
Overview of the wgMLST results of 19 C. jejuni genomes produced by GeP, BIGSdb GC, and SeqSphere+a
| Loci | No. of loci |
||
|---|---|---|---|
| GeP | BIGSdb GC | SeqSphere+ | |
| Shared loci | 1,516 | 1,653 | 1,471 |
| Identical shared loci | 855 | 859 | 855 |
| Polymorphic shared loci | 661 | 793 | 616 |
| Excluded loci | 182 | 46 | 227 |
| Excluded loci solely because of gene duplication | 6 | 1 | 10 |
Using the genome of C. jejuni 4031 as a reference (total of 1,698 loci).
TABLE 2.
Summary of the error types affecting the results of BIGSdb GC and SeqSphere+ in comparison with GePa
| Type of error | Error present in: |
Locus tag | ||
|---|---|---|---|---|
| GeP | BIGSdb GC | SeqSphere+ | ||
| I | No | Yes | Yes | BN867_06930 |
| II | No | Yes | No | BN867_14050 |
| III | No | Yes | Yes | BN867_03400 |
| IV | No | Yes | No | BN867_09770 |
| V | No | Yes | No | BN867_02990 |
| VI | No | No | Yes | BN867_00440 |
An example of a locus affected by the error type is shown. Error type I, failing to choose the orthologous locus from the paralogous locus; error type II, treating a missing gene as a special allele; error type III, failing to identify an orthologous locus due to high nucleotide sequence divergence; error type IV, failing to exclude a sequence containing nucleotide ambiguity; error type V, allele sequence overlapping the neighboring locus; error type VI, failing to recognize a sequence containing simple sequence repeat variation as a new allele.
Error type I consists of cases failing to choose the orthologous gene from the paralogous gene. GeP found, in 306 cases, 34 loci containing possible paralogous genes in the tested genomes (see Table S2 in the supplemental material). GeP was able to use CGN to differentiate orthologs from the paralogs in 222 of these cases (see Data Set S3 in the supplemental material). SeqSphere+ failed in the identification of the duplication in several cases, which resulted in the omission of one locus (see Table S2). BIGSdb GC was more prone to error type I by including in the analysis 15 loci excluded by GeP.
The second error type consists of assigning the missing locus as an allele number. This strategy is implemented in BIGSdb GC and explains 83 out of 132 extra shared loci detected by the program than by GeP. It also explains the visible netlike structure in the BIGSdb GC split graph (see Fig. S2 in the supplemental material). The use of draft genome sequences limits the ability to recognize if a missing locus is a consequence of gene loss or simply a misassembly and might result in an incorrect estimation of the pairwise allele differences.
Error type III consists of the inability of both BIGSdb GC and SeqSphere+ to correctly assign allele numbers to the loci due to high sequence divergence at the nucleotide level. An example is the BN867_03400 locus (cmeB), the second unit of the RND efflux system (9), which was undetected by both programs in the genomes of three C. jejuni isolates. However, the complete cmeABC operon was annotated by RAST (10) in all C. jejuni genomes and correctly identified by GeP, indicating that the implementation of BLASTX in the GeP pipeline allowed a more accurate wgMLST analysis.
In addition, BIGSdb GC frequently failed in filtering out loci with nucleotide ambiguity (error type IV), and in some cases it forced the alignment, resulting in erroneous alleles composed by overlapping loci (error type V). Moreover, SeqSphere+ wrongly excluded from the analysis all loci containing homopolymeric tracts, which is commonly observed in C. jejuni genomes (3, 11), if the lengths of the tracts differ from that of the reference genome (error type VI). GeP takes homopolymeric tracts of different lengths into account by assigning different allele numbers. The user can later easily inspect the sequence alignment in the GeP output files and make a decision whether to include these loci.
In conclusion, we showed that GeP, by using CGN information, performed better overall in terms of accuracy in determining pairwise allelic differences in the wgMLST of genomes than BIGSdb GC and SeqSphere+. GeP is an open source program. It is easy to use even for unsophisticated users. It can also be easily integrated into bioinformatics workflow management systems, such as Galaxy (12). Here, we show only the test results of C. jejuni isolates; however, in principle, GeP could be applied to any other bacterial species. The accuracy and flexibility of GeP are thus a useful alternative for public health and clinical microbiologists and researchers who want to apply wgMLST in the investigation of bacterial infectious disease outbreaks.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Finnish Cultural Foundation (Suomen Kulttuurirahasto) (J.Z.) and by the Academy of Finland on behalf of CoE-MiFoSa (no. 11411405).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00051-15.
REFERENCES
- 1.Cody AJ, Bennett JS, Maiden MCJ. 2014. Multi-locus sequence typing and the gene-by-gene approach to bacterial classification and analysis of population variation, p 201–219. In Goodfellow M, Sutcliffe I, Jongsik C (ed), Methods in microbiology, vol 41 Academic Press, Waltham, MA. [Google Scholar]
- 2.Revez J, Llarena AK, Schott T, Kuusi M, Hakkinen M, Kivisto R, Hänninen ML, Rossi M. 2014. Genome analysis of Campylobacter jejuni strains isolated from a waterborne outbreak. BMC Genomics 15:768. doi: 10.1186/1471-2164-15-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Revez J, Zhang J, Schott T, Kivisto R, Rossi M, Hänninen ML. 2014. Genomic variation between Campylobacter jejuni isolates associated with milk-borne-disease outbreaks. J Clin Microbiol 52:2782–2786. doi: 10.1128/JCM.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 5.Fouts DE, Brinkac L, Beck E, Inman J, Sutton G. 2012. PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res 40:e172. doi: 10.1093/nar/gks757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oberto J. 2013. SyntTax: a Web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics 14:4. doi: 10.1186/1471-2105-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohl TA, Diel R, Harmsen D, Rothganger J, Walter KM, Merker M, Weniger T, Niemann S. 2014. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Revez J, Schott T, Llarena AK, Rossi M, Hänninen ML. 2013. Genetic heterogeneity of Campylobacter jejuni NCTC 11168 upon human infection. Infect Genet Evol 16:305–309. doi: 10.1016/j.meegid.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.