Abstract
The Mus81 endonuclease resolves recombination intermediates and mediates cellular responses to exogenous replicative stress. Here, we show that Mus81 also regulates the rate of DNA replication during normal growth by promoting replication fork progression while reducing the frequency of replication initiation events. In the absence of Mus81 endonuclease activity, DNA synthesis is slowed and replication initiation events are more frequent. In addition, Mus81 deficient cells fail to recover from exposure to low doses of replication inhibitors and cell viability is dependent on the XPF endonuclease. Despite an increase in replication initiation frequency, cells lacking Mus81 use the same pool of replication origins as Mus81-expressing cells. Therefore, decelerated DNA replication in Mus81 deficient cells does not initiate from cryptic or latent origins not used during normal growth. These results indicate that Mus81 plays a key role in determining the rate of DNA replication without activating a novel group of replication origins.
Introduction
The accurate transmission of genetic material requires the precise regulation of DNA replication. This involves controlling the rate of DNA replication and coordinating replication initiation events throughout the genome. Cellular regulatory safeguards ensure that each position within the genome replicates precisely once during each cell cycle. To guard against perturbations of DNA replication, cells have evolved several mechanisms to preserve genomic stability. For example, cell cycle checkpoints are activated when a minimum number of perturbed replication forks are detected. If DNA synthesis within early-replicating genomic regions cannot be completed, checkpoint-signaling cascades inhibit the initiation of DNA replication in late-replicating chromosomal regions1. When DNA synthesis rates slow down, or replication forks encounter a lesion that prevents DNA synthesis, cells can complete the replication process by increasing the frequency of initiation events, presumably in regions adjacent to stalled replication forks2.
The involvement of the S-phase checkpoint in initiating DNA replication emphasizes the importance of the initiation stage in ensuring genomic stability. Within eukaryotic cells, replication initiates at multiple, non-random sites called replication origins3, 4. Metazoa appear to possess potential replication origins in excess (i.e., more than are required for efficient replication), and for each cell division, initiation sites are chosen anew3, 5, 6. A critical part of the choice of replication initiation sites is the binding of the mini-chromosome maintenance (MCM) helicase complex to origins to license them for replication7, 8. Many licensed origins, however, do not initiate replication9. The apparent excess of potential origins may ensure coordination between replication, transcription, and other chromatin functions 10-12. The frequency of initiation can be affected by several factors including metabolic conditions (e.g., nucleotide availability 13), developmental stage14 and the presence of stalled replication forks. The fact that stalled replication forks can increase initiation frequency suggests that cells maintain a reservoir of unused origins that can be activated when replication is perturbed 3, 6. According to the terminology proposed by Mechali6, potential replication origins are defined as “flexible origins” if they are activated at various frequencies during normal cell proliferation, or as “dormant origins” (also defined as “cryptic origins” or “latent origins”) if they are not utilized during normal growth and are only activated under distinct metabolic conditions or following cell cycle perturbations. Extra replication initiation events that occur when cell cycle progression is temporarily perturbed may arise from pre-licensed back up origins to ensure that DNA within the affected region is eventually replicated 15-20. The stabilizing influence of flexible or dormant replication origins is underscored by observations in yeast that a reduced number of active origins elevates the rate of chromosome loss 21 and can trigger a cell-cycle checkpoint 22. In the absence of a cell-cycle checkpoint, reduced levels of initiation can lead to gross chromosomal abnormalities 23, 24 and DNA breakage 25, 26. Genomic regions with low initiation frequencies tend to be fragile 27 and DNA breaks are common in regions that replicate slowly (i.e., common mammalian fragile sites 28 and replication slow zones in yeast 4).
Although inhibition of DNA synthesis can activate the S-phase checkpoint, signaling thresholds associated with this process dictate that numerous forks must stall before the checkpoint is activated 22. As such, mild perturbations to DNA replication decelerate replication but do not activate the checkpoint. In this way, cells are protected from the potential deleterious effects associated with checkpoint activity (i.e., apoptosis and error-prone repair/recombination). We have previously shown29, 30 that exposures to low, non-toxic doses of replication inhibitors slow the pace of DNA synthesis and activate a specialized signaling pathway including the cancer-predisposing Bloom syndrome, RecQ helicase-like (BLM); Mus81 endonuclease homolog (S. cerevisiae) (Mus81); and the ataxia telangiectasia and Rad3 related (ATR) kinase. Activation of this pathway initially results in DNA breaks, which are rapidly repaired by the non-homologous end-joining pathway. This reaction involves DNA-PK and X-ray repair complementing defective repair in Chinese hamster cells 4 (XRCC4). Rapid repair of these DNA breaks prevents cell-cycle arrest. Following the initial deceleration of DNA synthesis, replication resumes at a reduced rate without further DNA breakage. These findings suggest that transient DNA breaks induced by the inhibition of DNA replication are actively generated as damage-repair intermediates.
Mus81 and BLM are involved in cellular responses to exogenously induced replicative stress. It is likely, however, that transient decelerations in DNA replication also occur under normal conditions. Here we ask whether Mus81 endonuclease plays a role in determining the rate of DNA replication during normal cellular growth. We also ask whether decreases in the rate of DNA replication are sufficient to alter replication initiation frequency and to trigger initiation from latent replication origins that are not normally utilized during unchallenged growth. In the absence of replication-perturbing agents, Mus81 promotes replication fork progression. An intact Mus81 endonuclease domain is required for a normal pace of DNA replication and for protecting cells from deleterious effects of replication inhibition (e.g., a low dose of aphidicolin or depletion of XPF endonuclease). In addition, loss of Mus81 increases initiation frequency from flexible replication origins that are utilized in both Mus81-proficient and deficient cells, but does not activate novel dormant replication origins.
Results
Mus81 Deficiency Slows DNA Synthesis
To determine whether Mus81 affects replication under normal conditions (i.e., in cells that are not exposed to DNA damaging agents), we measured replication fork progression and replication initiation frequency in HCT116 cells lacking Mus81 (Mus81-deficient cells or Mus81−/−) 30, 31. Control measurements were performed using an isogenic HCT116 cell line that carries a wild-type copy of Mus81 (Mus81-proficient cells or HCT116) 30, 31. Cell cycle profiles were similar in Mus81-deficient and proficient cells (Supplementary Fig. 1A and 1B).
We measured rates of replication fork progression by sequentially labeling cells with the thymidine analogs iodo-deoxyuridine (ldU) and chloro-deoxyuridine (CIdU). DNA fibers were then obtained from labeled cells and stretched onto microscope slides. Regions of nucleotide-analog incorporation (replication tracks) were visualized on these stretched DNA fibers using immunofluorescence, as described 30, 32 (Figures 1A and 1B). Using this technique, most replication tracks appeared as regions of green fluorescence (IdU incorporation) adjacent to regions of red fluorescence (CldU incorporation). This represents the fluorescent signature of replication fork progression. Lengths associated with ldU and CIdU incorporation were measured and rates of replication fork progression were calculated from these values. Fluorescent in-situ hybridization (FISH) was used to confirm these size ranges. The inclusion of a FISH signal serves an internal DNA size control. Despite the inclusion of a FISH marker, accurate comparisons of replication fork progression rates using dynamic molecular combing require that the studied samples be labeled and processed at the same experiment because minor variations in the duration of the thymidine analog pulses can cause variations in the length of replication tracks. Comparisons among measurements obtained from separate biological replicates necessitate a common sample that can serve as a standard, as reported below.
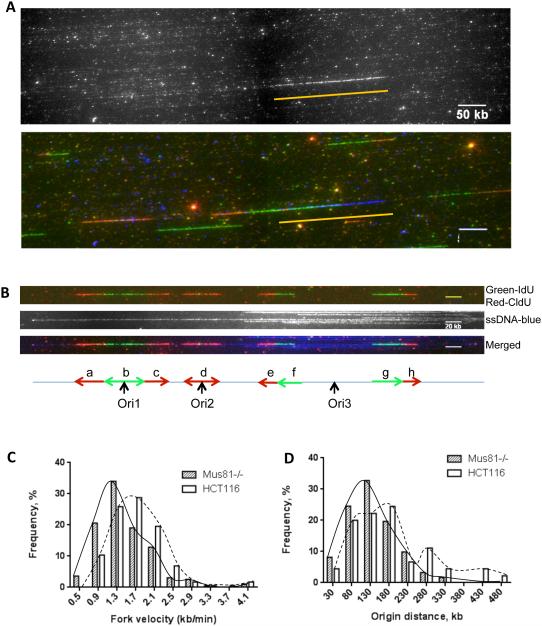
Figure 1. A high frequency of replication initiation and slower replication forks in Mus81 deficient cells.
Cells were sequentially labeled with ldU and CIdU. Extracted DNA was then stretched on silanized microscope coverslips. Regions of DNA that contained ldU and CIdU were fluorescently labeled via immunochemistry. DNA fibers were counterstained using an antibody directed against single-stranded DNA to exclude end-fiber replication tracks from progression-rate calculations, and to ensure that origins used to calculate an inter-origin distance were on the same fiber. (A) Raw images that include a FISH signal (top, traced by a yellow line) and a merged image of FISH (blue) and replication signals (green for IdU and red for CldU).A BAC of known size was used as the FISH probe for fiber size calibration. Scale bar, 50kb. (B) Typical replication signals (replicating DNA, top; anti-single strand DNA (ssDNA) antibody to trace DNA fiber, middle; Merged image, blue for ssDNA, bottom). The corresponding schematic diagram is shown underneath the fluorescence images (see Supplementary Fig. 2 for more DNA fiber examples). Replication initiation events occurred either during the initial IdU labeling (Ori1), during the secondary CldU labeling (Ori2), or before labeling was initiated (Ori3). Inter-origin distances were measured in fibers that contained at least two initiation events. Replication tracks that represented 20 min of DNA synthesis (a, c, e, f, g and h) were used to calculate replication fork velocity. Replication tracks that represented less than 20 min (b, d, initiated during a labeling), or that localized to the end of the DNA fiber (not showed) were not used. Scale bar, 20kb. (C, D). Data were binned and the percentage of replication signals that corresponded to a particular range of replication-rate (C) or inter-origin distance (D) was plotted for both Mus81-proficient (HCT116) and Mus81-deficient (Mus81−/−) cells. Mus81-deficient cells exhibited slower replication fork progression (C), and a higher frequency of replication initiation events (D). (C) and (D) are representative graphs from 3 experiments. Statistical analyses are shown in Supplementary Table 1.
Our analyses demonstrate that during unperturbed growth, cells deficient for Mus81 replicate more slowly than Mus81-proficient cells (Figure 1C and Supplementary Table 1). Cells lacking Mus81 had, on average, shorter replication tracks, which indicate a significantly slower DNA synthesis rate (the median rate of replication fork progression was 1.42 kb/min in Mus81-deficient cells, compared to 1.74 kb/min in Mus81-proficient cells (Supplementary Table 1). The slower average replication fork progression in Mus81-deficient cells could indicate either a mild but uniform overall reduction in DNA synthesis rates in all replication forks, or a substantial reduction in the pace of DNA synthesis in a group of replication forks.
Mus81 Deficiency Increases Replication Initiation frequency
Using this same sequential-labeling technique, distances between replication origins were also measured on stretched DNA fibers. As shown in Figure 1B, initiation events that occurred before the first label was introduced were detected as an unlabeled region flanked by IdU-positive regions (green), which were in turn flanked by CldU-positive regions (red). Replication initiation events that occurred during the initial labeling period (IdU) were detected as an IdU-positive region flanked by CldU-positive regions. Finally, replication initiation events that occurred during the second labeling period (CldU) were detected as a CIdU-positive region flanked by unlabeled DNA.
To determine whether the slow rate of DNA synthesis in Mus81-deficient cells was accompanied by a higher frequency of initiation events, we measured inter-origin distances in both Mus81-deficient and -proficient cells. As shown in the distribution of inter-origin distances plotted in Figure 1D, Mus81 proficient cells contained a group of fibers with longer inter-origin distances (hence the appearance of a bimodal distribution). The average inter-origin distance was significantly shorter in the Mus81-deficient cells compared to the Mus81-proficient controls (124.8 kb and 161.2 kb, respectively; Figure 1D and Supplementary Table 1). These observations suggested that cells that lacked Mus81 initiated replication more frequently than isogenic cells that carried a wild-type copy of Mus81. Fork density per Mb DNA fiber was also analyzed from four random selected images for HCT116 cells and Mus81−/− cells, respectively (Table 1) and the results confirmed the inter-origin data, i.e., Mus81-deficient cells initiated DNA replication more frequently than Mus81-proficient cells.
Table 1.
DNA replication fork density (forks/Mb DNA)
| HCT116 | Mus81−/− | |
|---|---|---|
| Image 1 | 0.63 | 0.83 |
| Image 2 | 0.69 | 0.76 |
| Image 3 | 0.73 | 0.94 |
| Image 4 | 0.64 | 0.78 |
|
| ||
| Average | 0.68 | 0.82* |
|
| ||
| Total forks | 235.00 | 317.00 |
| Total DNA length (Mb) | 345.60 | 384.80 |
p=0.0149 by student t test
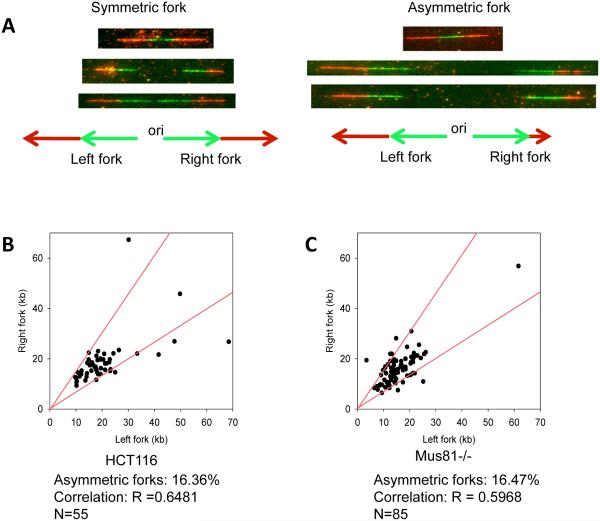
Shorter replication tracks reflect either a slower rate of DNA synthesis or replication fork stalling, preventing DNA synthesis from proceeding during a significant length of time relative to the labeling period. To distinguish between these two possibilities, replication fork asymmetry was analyzed in both Mus81-deficient and Mus81-proficient cells. Using the sequential-labeling technique, a stalled replication fork typically results in an asymmetric replication track (i.e., tracks that emanate from a common origin but are significantly different in length) (Figure 2A). A single-origin replication signal in which one side was 33% longer than the other side was defined as an asymmetric track. (For representative examples, see the rightmost fork in the fiber shown in Figure 1B and the leftmost fork in the fiber shown in Supplemental Figure 2C.) The frequency of asymmetric replication tracks was similar in both Mus81-deficient and proficient cells (Figure 2B and 2C) suggesting that Mus81 deficiency did not cause replication to stall. These observations suggest that the shorter replication tracks observed in Mus81-deficient cells result from slower DNA synthesis rates in those cells.
Figure 2. Mus81 deficiency does not trigger replication fork asymmetry.
HCT116 cells were sequentially labeled with IdU (green) and CldU (red) and then subjected to single fiber replication analyses as described in the legend to Figure 1. For each replication origin, the length of the right-fork signal was measured and plotted against the length of the left-fork signal. If the ratio between the left-fork length and the right-fork length deviated by more than 33% from 1 (i.e., outside the red lines in B and C), the fork was deemed asymmetric. (A) Examples of symmetric and asymmetric forks. (B,C) The percentage of asymmetric forks was calculated for both Mus81-proficient (B) and Mus81-deficient (C) cells. For Mus81-proficient and Mus81-deficient cells, 55 and 85 forks were counted, respectively.
Acute Mus81 Depletion Increases Replication Initiation rates
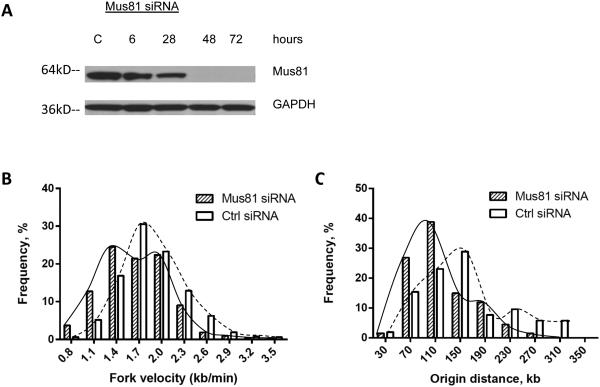
As the cell lines used for these analyses had been propagated in culture for several months, the Mus81 phenotype (i.e., the slow rate of DNA synthesis and the high rate of initiation frequency) may have resulted from long-term adaptation to the absence of Mus81. Alternatively, these phenotypes may indicate that Mus81 is constantly involved in determining the rates of replication fork progression and replication initiation. To distinguish between these possibilities, we asked what effect an acute knockdown of Mus81 would have on DNA replication. We used siRNA to deplete Mus8133 typically achieving complete depletion 2 days following the introduction of siRNA (Figure 3A). For cells exposed to siRNA directed against Mus81, we observed no significant cell-cycle changes (Supplementary Fig. 1C). When HCT116 cells were transfected with siRNA directed against Mus81, however, DNA synthesis was significantly slower compared to cells treated with control siRNA (Figure 3B and Supplementary Table 2). This suggests that both acute and chronic depletion of Mus81 results in slower replication fork progression. In addition, cells transfected with Mus81-specific siRNA exhibited shorter inter-origin distances, indicating an increased frequency of replication initiation events (Figure 3C and Supplementary Table 2). Similar results were obtained when the breast cancer cells MDA-MB-231 were used (Supplementary Fig. 3). These results indicate that the depletion of Mus81 rapidly slows DNA synthesis and increases the frequency of replication initiation events.
Figure 3. Acute Mus81 depletion triggers slower DNA synthesis and elevated initiation frequency.
HCT116 cells were transfected withsiRNA directed against Mus81 or with control siRNA. (A) Western-blot analysis showed that HCT116 cells transfected with siRNA against Mus81 exhibited a dramatic knockdown of Mus81 protein, compared to cells transfected with control siRNA 48 and 72 hours after transfection. Cells transfected with Mus81 siRNA or control siRNA for 48 hours were used for single fiber replication analyses: rates of replication fork progression (B) and inter-origin distances (C) in siRNA-treated cells. In comparisons between control- and Mus81-siRNA treated cells, the Mann Whitney test revealed significant differences in both replication fork velocity and inter-origin distance (Supplementary Table 2).
Mus81 Deficiency does not activate novel origins
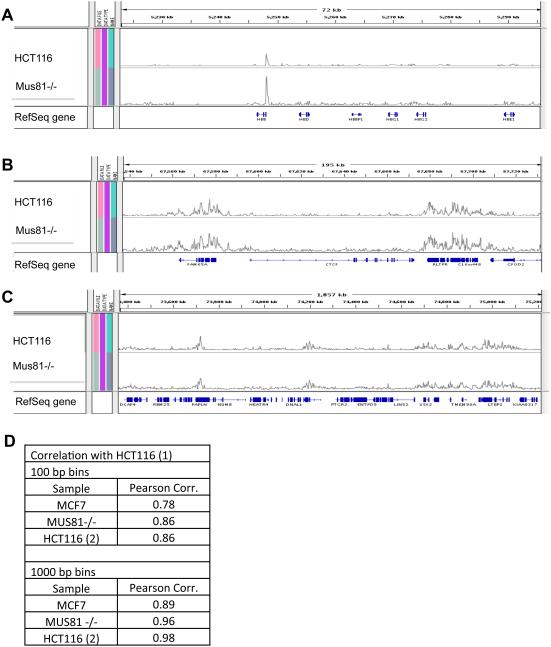
Slow replication can activate latent origins that do not initiate replication in rapidly replicating chromosomal loci 13. We asked, therefore, whether the increase in initiation frequency that was associated with Mus81 loss-of-function also reflected the activation of latent, cryptic, unused origins. Using both Mus81-proficient and Mus81-deficient cell lines we mapped all replication initiation events, as described 34. Massively parallel sequencing of newly replicated nascent DNA strands revealed strikingly similar replication-origin usage between these two cell lines (Figure 4). Similar patterns were evident in images of replication initiation patterns in individual loci (For examples, see Figure 4A-C), suggesting that similar genomic regions preferentially initiate DNA replication in both Mus81-proficient and deficient cells. When the overall reads per kilobase per million aligned reads (RPKM) values were compared between the Mus81-proficient and deficient HCT116 cells (bins that contained zero reads in both samples were excluded), Pearson correlation coefficients were 0.96 to 0.98 depending on bin size (Figure 4D). Control measurements comparing two technical replicates of sequence data obtained from HCT116 cells, resulted in a Pearson correlation coefficient of 0.98. Similar trends were observed when 100 bp bins were used, although Pearson correlation coefficients were generally lower (Figure 4D). Since the molecular combing data had demonstrated that Mus81-deficient cells exhibit shorter inter-origin distances than Mus81-proficient cells, the similar initiation patterns exhibited by the Mus81-proficient and deficient cells implies that replication initiation events in Mus81-deficient cells occur more frequently from the same sequences used by Mus81-proficient cells. Therefore, the higher percentage of origins that are activated in Mus81-deficient cells resulted from increased utilization of existing, pre-licensed flexible origins. Although the frequency of initiation was higher in Mus81-deficient cells, we have not observed activation of genomic regions that were not utilized as replication initiation sites in Mus81-proficient cells.
Figure 4. Mus81 depletion does not affect replication initiation patterns.
Locations of replication initiation events were determined by massively parallel sequencing of nascent strands from Mus81-proficient HCT116 cells and from HCT116 cells that lacked Mus81 (designated Mus81 (−/−). (A–C) Initiation profiles were visualized using the Broad Institute Integrated Genome Browser. Representative screen shots of replication-initiation profiles from three genomic loci are shown at different zoom levels. These loci include (A) 72 kb from the hemoglobin, beta (HBB) locus on Chromosome 11, (B) 195 kb from the CCCTC-binding factor (CTCF) locus on Chromosome 16, and (C) 1857 kb from Chromosome 14. It should be noted that the genome browser images are presented on a relative scale; therefore, the sizes of the peaks (calibrated per total numbers of aligned reads in each sample) indicate that initiation events preferentially occur at particular genomic locations but do not directly correspond to absolute initiation frequencies. (D) Locations of replication initiation events were determined by massively parallel sequencing of nascent strands from: 1) HCT116 cells (two separately sequenced samples designated HCT116 (1) and HCT116 (2)), 2) HCT116 cells that lacked Mus81 (designated Mus81 (−/−)), and 3) MCF7 cells. Locations of replication initiation sites were calculated using two bin sizes: 100 and 1000 bp. Shown are Pearson’s correlation coefficients concerning the location of replication initiation sites for comparisons between the HCT116 (1) sample and indicated samples.
The Endonuclease Activity of Mus81 Affects DNA Replication
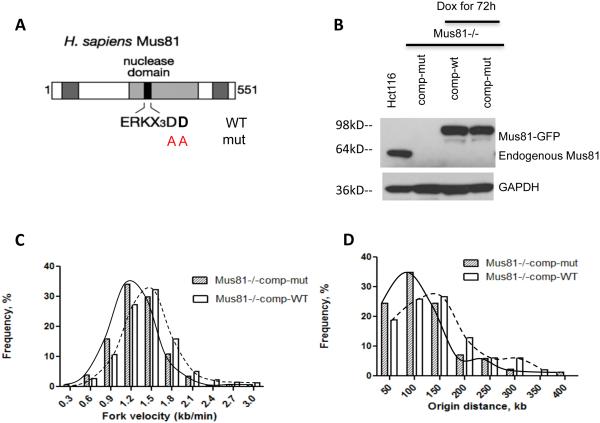
Mus81 is a structure-specific endonuclease, and its endonuclease activity is required for DNA-damage repair. Mus81 is part of a large protein complex that contains numerous enzymes involved in DNA repair 35. We asked, therefore, whether it is the enzymatic activity of Mus81 that affects the rate of DNA replication. Alternatively, Mus81 could play a structural role in this process by recruiting other proteins (such as the Mus81-containing “super complex”) to chromatin. The endonuclease activity of Mus81 requires two aspartic-acid residues at positions 338 and 339 within the conserved nuclease domain 36. Substituting these amino acids with alanine abolishes endonuclease activity. We used site-directed mutagenesis to reconstitute this mutation, generating an “endonuclease-dead” version of Mus81 (Figure 5A). We then transfected the Mus81-deficient HCT116 cells with constructs that encoded either a wild type or endonuclease-dead version of Mus81 (Figure 5B). Neither of these constructs affected cell cycle progression of Mus81-deficient cells (Supplementary Fig. 1D). Complementation with the wild-type Mus81 partially restored the slow replication observed in the Mus81 deficient cells. Expression of endonuclease-dead Mus81 resulted in slower replication fork progression and shorter inter-origin distances than expression of wild-type Mus81 (Figure 5C, 5D, and Figure Supplementary Table 3). This suggests that the endonuclease activity of Mus81 is involved in regulating the pace of DNA replication.
Figure 5. Mus81 endonuclease activity is required to maintain replication rates.
(A) Site-directed mutagenesis was used to replace two aspartic-acid residues within the conserved Mus81 nuclease domain (338 and 339) with alanine residues. (B–E) Wild type and mutant versions of Mus81 tagged with tGFP in a doxycyclin-inducible vector were introduced into HCT116 Mus81−/− cells and stable cell lines were established. (B) Western-blot analysis showed reproducible Mus81 expression in response to doxycycline for 72 hours in these stable cell lines. (C,D) Single fiber replication analyses were used to measure rates of replication fork progression (C) and inter-origin distances (D) in these cells after 72 hours of Mus81-tGFP induction by doxycycline. Cells complemented with the wild type Mus81 exhibited slower replication fork progression compared to the rates of DNA synthesis in the parental Mus81-proficient HCT116 cells shown in Figure 1, reflecting either experimental variation or incomplete complementation by Mus81. Cells expressing Mus81 with a mutant nuclease domain exhibited slower replication fork progression and more frequent initiation than cells expressing the wild-type Mus81. Statistical analyses are shown in Supplementary Table 3.
Mus81 Endonuclease Prevents Irreversible Fork Stalling
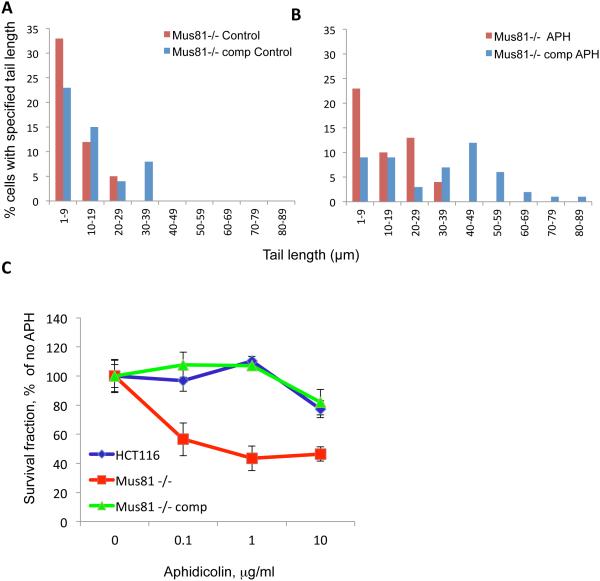
The Mus81 endonuclease induces DNA breakage during the repair of DNA damage (e.g. interstrand crosslinks and topoisomerase cleavage complexes)33, 37. Mus81 converts perturbed replication forks into DNA breaks, thereby facilitating the rapid resumption of DNA replication. This avoids irreversible inhibition of DNA synthesis, DNA damage, and activation of cell-cycle checkpoints 30, 38. Mus81 also cleaves stalled replication forks that are generated by topoisomerase I (TOP1) cleavage complexes 33 and is involved in DNA breakage at common fragile sites during early mitosis 39, 40. We measured the frequency of DNA breaks in Mus81-proficient and deficient cells using a comet assay. As shown in Figure 6A, Mus81-deficient cells exhibited similar, although somewhat lower levels of spontaneous DNA breaks than Mus81-proficient cells. To investigate whether Mus81 endonuclease activity played a role in the recovery from perturbed replication during S-phase, we exposed cells to low doses of aphidicolin (0.5 μg/ml) shown previously to slow down replication without triggering DNA damage in Mus81-proficient cells30, 38. When cells were exposed to 0.5 μg/ml aphidicolin for 10 minutes, Mus81-proficient cells exhibited high levels of double-strand breaks (Figure 6B) but the frequency of double-strand breaks did not notably change in Mus81-deficient cells (Figure 6B).
Figure 6. Mus81 deficiency decreases transient DNA breakage and increases sensitivity to low doses of aphidicolin.
Mus81 −/− (Mus81-deficient cells) and Mus81-comp (Mus81 −/− cells complemented with exogeneous Mus81 as described previously30, 31) were exposed to carrier (A) or to 0.5 μg/ml (about 1.5 μM) of aphidicolin (B) for 10 minutes and collected for neutral Comet assay. (C) HCT116 cells, Mus81−/− cells and Mus81−/−comp cells were treated with aphidicolin for 2 hours. The survival rate of cells was determined by clonogenic assay.
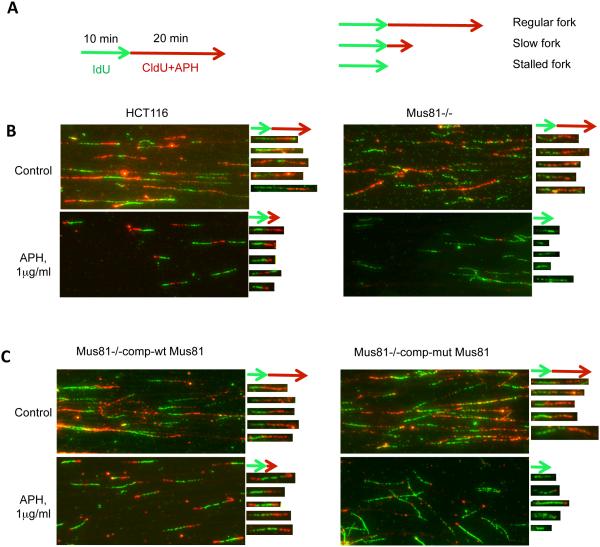
Although no significant DNA double-strand breaks were detected in Mus81-deficient cells, colony formation assays demonstrated that Mus81-deficient cells were markedly more sensitive to aphidicolin than Mus81-proficient cells (Figure 6C). These results suggested that the transient breaks generated by Mus81 in cells exposed to replication stress promoted cell survival by facilitating perturbed replication forks recovery. We asked, therefore, whether the endonuclease activity of Mus81 was involved in this process. Cells were labeled with IdU (green) for 10 min, then CldU (red) for 20 min. A low dose of APH was included during the CldU incubation period (Figure 7A). Cells were then harvested for single DNA fiber spread analysis 29, 30. During APH treatment, Mus81-proficient cells continued to replicate at a slow pace, whereas Mus81-deficient cells fully arrested replication (Figure 7B). This result is consistent with previous observations 30. Expression of wild-type Mus81 in Mus81-deficient cells rescued this phenotype, as a slow rate of DNA synthesis was observed during APH exposure (Figure 7C, left panels). When the endonuclease-dead version of Mus81 was expressed in Mus81-deficient cells, however, replication was not restored during APH exposure, as virtually no CldU incorporation was detected (Figure 7C, right panels). These results demonstrate that the endonuclease activity of Mus81, in addition to regulating the pace of DNA replication, facilitates the deceleration of DNA replication and prevents irreversible inhibition of DNA replication after exposure to low doses of APH.
Figure 7. Mus81 endonuclease activity prevents irreversible fork stalling in response to perturbed replication.
(A) Cells were labeled with IdU (green) for 10 min, then CldU (red) for 20 min. A low dose of APH was included during the CldU incubation period (left). Cells were harvested for single DNA fiber spread analysis. The right schematic graph showed 3 typical replication tracks: regular fork, slow fork and stalled fork based on the length of red replication tracks affected by APH treatment. (B) Images of DNA fibers from Mus81-proficient (HCT116) and Mus81–deficient (Mus81−/−) cells. Five representative fibers for each image were extracted and aligned to the right of their respective images. Effects of APH are shown in the bottom panels.Fibers that replicated with a similar pace during the IdU (green) and CldU (red) labeling periods exhibited a red signal that was longer than the green signal (regular fork), and most of replication forks in the control samples for both HCT116 and Mus81−/− cells belong to this category (2 top panels); fibers that slowed replication during the CldU labeling period exhibited a red signal that was clearly shorter than the green signal (slow fork), and most of replication forks in APH treated HCT116 cells belong to this category (bottom left panel); and fibers that could not recover from replication inhibition exhibited green signals and no red signals (stalled), and most of replication forks in APH treated Mus81−/− cells belong to this category (bottom right panel). (C) Images of DNA fibers from Mus81-deficient cells that express wild type Mus81 (Mus81−/− com-wt Mus81) or mutant Mus81 (Mus81−/−comp-mutt Mus81). Mutant Mus81 lacks endonuclease activity. Effects of APH are shown in the bottom panels. Five representative fibers from APH treated wild-type Mus81 and mutant Mus81 samples (figure 7C, 2 bottom panels) were selected and aligned under their original images, respectively. Most of wild-type Mus81 cells showed slow forks while most of mutant-type Mus81 cells showed stalled forks after low dose of APH treatment.
The inability to recover from inhibition of DNA replication in Mus81-deficient cells, combined with the reduced viability of Mus81-deficient cells in response to low doses of replication inhibitors, raise the possibility that Mus81 plays a role in preventing disruptions in DNA synthesis and activation of the ATR-Chk1-mediated checkpoint. To investigate whether the ATR-Chk1 pathway was activated, we measured Chk1 phosphorylation in Mus81-proficient and deficient cells during unchallenged growth and following exposure to APH. We observed low spontaneous levels of phosphorylation of the checkpoint kinase Chk1 on Serine 317 in Mus81-deficient cells or in Mus81-deficient cells complemented with endonuclease dead Mus81 but not in Mus81-proficient cells including Mus81-deficient cells that were complemented with a functional Mus81. Although we observed very low levels of phosphorylation of Chk1, these phosphorylation events were reproducible and indicated that small fraction of Chk1 was persistently phosphorylated during unperturbed growth in Mus81-deficient cells. By contrast, perturbation of DNA synthesis with aphidicolin produced a strong phosphorylation of Chk1 with enhanced Chk1 S317 phosphorylation in all cells; but the enhancement was more pronounced in Mus81-deficient cells (Supplementary Fig. 4).
Intact Okazaki Fragment Processing in Mus81-deficient Cells
Recent observations in yeast suggest that the Mus81-Eme1 (essential meiotic endonuclease 1 homolog 1 (S. pombe) complex plays a direct role in Okazaki fragment maturation in collaboration with the FEN1 endonuclease 41, 42. The efficient resolution of structures generated during Okazaki fragment processing may be required for rapid DNA replication. To test this possibility, we performed a BrdU comet assay, which can detect maturation of newly replicated DNA in individual cells43 in both Mus81-proficient and deficient HCT116 cells. Consistent with the molecular combing data, Mus81-deficient cells exhibited a longer tail than Mus81-proficient cells (Supplementary Fig. 4) suggesting that nascent DNA strands are on average shorter in those cells. However, the newly replicated DNA maturation rate (the rate for BrdU comet tail shortening) was similar in HCT116 cells with and without Mus81 (Supplementary Fig. 5). These observations suggest that Okazaki fragment processing rates are similar in both Mus81-proficient and deficient cells. Hence, slower Okazaki fragment maturation does not appear to be the main reason causing slower replication forks in Mus81-deficient cells.
Mus81 Prevents Cell Death in Response to XPF Deficiency
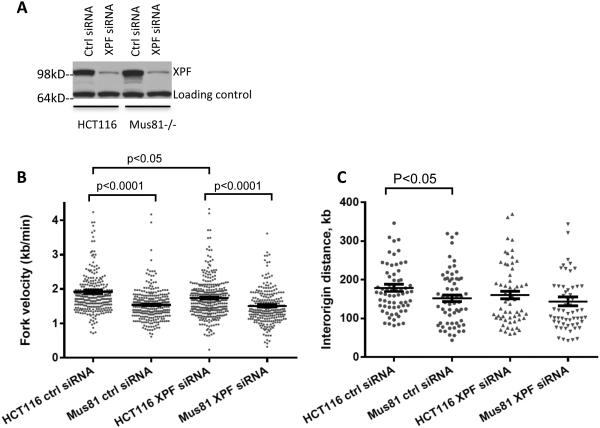
Our observations suggest that the endonuclease activity of Mus81 regulates both rates of DNA replication and recovery from mild DNA-replication perturbations. However, Mus81 did not affect cell survival during unperturbed growth. XPF is another member of the Mus81/XPF family, which plays important roles in the repair of DNA lesions beyond nucleotide excision repair 44. We asked, therefore, whether XPF could compensate for Mus81 deficiency during unperturbed growth. Mus81-proficient and Mus81-deficient HCT116 cells were transfected with siRNA directed against XPF (two times within 72 h). XPF expression was depleted 3 days after the second transfection (Figure 8A). As shown in Figure 8B, XPF depletion decreased replication fork speed in both Mus81 wild type and Mus81-deficient HCT116 cells. Notably, however, XPF depletion by itself affected DNA replication rates to a lesser extent than Mus81 depletion, and double depletion of Mus81 and XPF did not exhibit synergistic effects. Similar changes in inter-origin distances were shown in Figure 8C.
Figure 8. Effects of depletion of XPF on replication fork progression in Mus81−/− cells.
HCT116 and Mus81−/− cells were transfected with siRNA directed against XPF or control siRNA. (A) Western-blot analysis revealed an efficient knockdown of XPF protein via siRNA treatment. (B-C) Cells were sequentially pulsed with IdU and CldU for 20 minutes, respectively before harvesting for DNA combing. Scatter plot was used to show the distribution of replication fork speed (B) and interorigin distance (C). Median with interquartile range was shown for the scatter plot. Mann Whitney test was used for the statistical analysis. A few extreme numbers (>4.5 kb/min for fork speed, >400 kb for origin distance) were not shown for a better resolution of the scatter plot. Statistical analyses were shown in Supplementary Table 4.
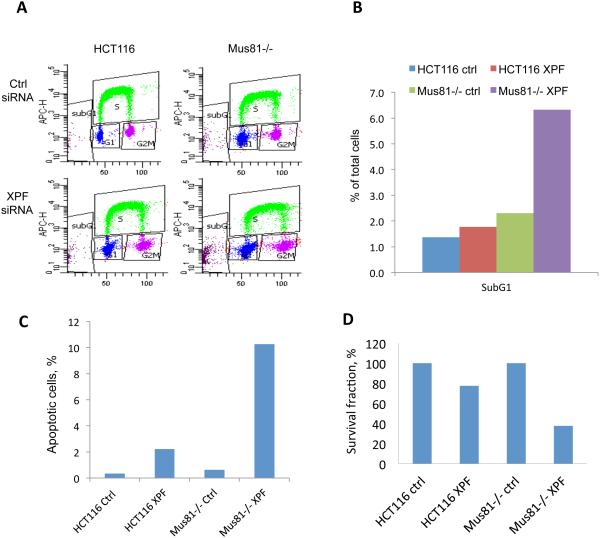
By contrast, when Mus81-deficient cells were treated with XPF siRNA, they exhibited a dramatic increase in sub-G1 or apoptotic cells as measured by flow cytometry analyses (Figure 9A, B and Supplementary Table 5). Annexin V and TUNEL apoptotic assay confirmed that there were more apoptotic cells in Mus81-deficient cells when XPF was depleted (Figure 9C, Supplementary Fig. 6). In agreement, clonogenic assays showed that depletion of XPF markedly decreased the survival of HCT116 cells. This effect was more pronounced in Mus81-deficient cells (Figure 9D). These results suggest that XPF may act in concert with Mus81 to prevent the formation of deleterious DNA lesions. These results also demonstrate that at least one of the two endonucleases is required for the normal progression of DNA synthesis during an unperturbed cell cycle, and that XPF may promote cell survival in the absence of Mus81 during unperturbed growth.
Figure 9. Effects of depletion of XPF on Mus81−/− Cell Survival.
HCT116 and Mus81−/− cells were transfected with control siRNA or XPF siRNA as described in Figure 8. (A) Cell cycle analysis by FACS. Cells were pulse-labeled with EdU for 45 min before harvesting. Cell cycle was tested with a Click-iT EdU kit by FACS. Examples of bivariate FACS were shown and percentage of cells in each cell cycle was shown in Supplementary Table 5. (B) Histogram of percentage of subG1 from Figure 9A. (C) The % of apoptotic cells was measured by TUNEL assay. Cells were transfected with control siRNA or XPF siRNA twice with 72 hours interval and collected 72 hours after the second transfection. TUNEL positive cells were analyzed by FACS (see Supplemental Figure 6B for the FACS dot plots). (D) Clonogenic assays. 24 hours after the second siRNA transfection, cells were seeded into 6-well plates at 50, 250, or 1000 cells per well. Two weeks later, cells were fixed and stained to count the colonies.
Discussion
Mus81 plays an important role in preserving genomic integrity by promoting homologous recombination-mediated repair, mitotic recombination, and the resolution of DNA lesions including interstrand crosslinks 31, 37, 45, 47. Importantly, Mus81 plays an active role in generating DNA breaks in cells exposed to campthotecin 33 and in the cellular response to replication inhibitors, facilitating breaks at common fragile sites during early mitosis 39, 40. When cells encounter conditions that slow DNA replication such as exposure to DNA polymerase inhibitors, Mus81 in coordination with BLM and ATR converts stalled replication forks into double-stranded DNA breaks. These breaks are subsequently repaired via non-homologous end joining to prevent the activation of DNA-damage checkpoints and allow cells to resume replication 29. While all the above studies suggest a role for Mus81 endonuclease in the resolution of lesions resulting from perturbation of DNA replication, here we show that unchallenged Mus81-deficient cancer cells exhibited slower rates of DNA replication and higher frequencies of replication initiation events. This suggests that Mus81 also plays a role during unperturbed S-phase to regulate DNA replication in the absence of DNA-damaging agents and stimuli that slow replication.
Mus81-deficient cells exhibit a higher frequency of initiation events and slow DNA replication. While these two phenomena are linked, the causal relationship between them is unclear: Loss of Mus81 activity may either result in slow replication, increasing initiation frequency as a secondary consequence, or loss of Mus81 can facilitate initiation, slowing replication indirectly. Our observations are consistent with both possibilities. Notably, the distribution of replication fork progression rates in Mus81-deficient cells could indicate either an overall mild but uniform reduction in the pace of DNA synthesis or a marked reduction in DNA synthesis rates that is limited to a subset of replication forks. If only a subset of replication forks is markedly affected, these data are consistent with a role for Mus81 in resolving lesions that occur in a small but significant fraction of replication forks during S-phase such as replication fork reversal (as shown in the working model illustrated in Supplementary Fig. 7). This hypothesis is consistent with the known substrate specificity of Mus81 endonuclease activity, since stalled replication forks (Supplementary Fig. 7b) and/or reversed forks (Supplementary Fig. 7g) can serve as potential substrates. Interestingly, our studies suggest that the endonuclease activity of Mus81 is required for setting the pace of DNA replication during unperturbed growth as well as for the response to replication inhibitors, eliminating the possibility that Mus81 primarily provide a scaffold to recruit other proteins that regulate replication fork progression.
What role can an endonuclease play in setting the pace of DNA replication? In yeast, the Mus81-Eme1 (essential meiotic endonuclease 1 homolog 1 (S. pombe) complex plays a direct role in Okazaki fragment maturation in collaboration with the FEN1 endonuclease 41, 42. In mammals, Mus81 binds late replicating loci during early mitosis and participates in the expression of fragility at common fragile sites.39, 40. We found that Okazaki fragment processing efficiency was similar in Mus81-deficient and proficient human cells, suggesting that the slower DNA replication forks in Mus81-deficient cells could not be primarily attributed to changes in Okazaki fragment processing. Since the endonuclease activity of Mus81 is required to maintain the normal rate of replication fork progression, our data suggest that Mus81-mediated DNA cleavage is required to resolve a lesion that occurs at a small but significant fraction of replication forks. Combined with the reported increase in replication fork reversal frequency in the absence of exogenous stress in Mus81-deficient cells 48, our data are in line with a role for Mus81 in resolving low-frequency reversed forks during unperturbed growth (see Supplementary Fig. 7 for a working model describing this hypothesis). Such a role might be critical for countering the deleterious effects of increased DNA replication forks reversal when DNA replication decelerates. An increased frequency of reversed replication forks (Supplementary Fig. 7f) might underlie the slow rate of DNA replication in Mus81−/− cells under normal cell growth condition.
A role for replication checkpoint proteins in regulating the pace of DNA replication was observed for ATR kinase, which suppresses over-activation of replication origins 49, 50 via the checkpoint kinase 1 (CHEK1 – Chk1) 51, 52. It is possible that the very limited phosphorylation of Chk1 on Serine 317 is involved in the slow replication phenotype observed in Mus81-deficient cells during unperturbed growth. However, cell cycle checkpoints mediated by Chk1 phosphorylation following perturbation of DNA synthesis can inhibit initiation of DNA replication51, 52, a phenomenon that was not observed in our study. In addition, the extent of phosphorylation observed during unperturbed replication was significantly lower than the phosphorylation typically exhibited in cells treated with APH, suggesting that the known S-phase checkpoint that involves massive Chk1 phosphorylation on Serine 317 and inhibition of initiation of DNA replication could not provide a sole explanation for the observed slow replication, frequent initiation phenotype of Mus81 depleted cells. Other complementary or parallel pathways might involve slowing replication while activating compensatory initiation. It is also important to note that since our study was performed in cancer cells (HCT116 and MDA-MB-213) we cannot rule out the possibility that the low levels of Ser317 phosphorylation and the observed response to Mus81 deficiency reflect the transformed nature of those cells.
The two structure-specific endonucleases, Mus81 and XPF, form dynamic heterodimers that exhibit partially overlapping but distinct substrate specificity53, 54. Recent studies have revealed a partial overlap in homologous recombination mediated repair functions of Mus81 and XPF following induction of double stranded DNA breakage 55. We report here that Mus81 protected cells from cell death in the absence of the XPF endonuclease suggesting that Mus81 could function as a backup for XPF activity (and vice versa) during unperturbed growth. The overlapping functions of the two endonucleases can underlie the relatively mild phenotype observed for Mus81 depletion. A complementary role of XPF could also account for the variability observed in the phenotype of Mus81 depletion in other cells. For example, Mus81-depleted Jeff, MRC5-SV and HeLa cells show no replication fork stalling and no changes in replication fork speed, Mus81 depleted U2OS cells exhibit replication fork stalling 39, 40, 56.
Conversely, cells require Mus81 to recover from replication inhibition following exposure to low doses of polymerase inhibitors30 and topoisomerase inhibitors 33. DNA lesions that form after mild perturbation of DNA replication may be impervious to XPF-mediated repair because they are too numerous, or because they are structurally incompatible. One speculative possibility is that replication lesions formed during unperturbed growth contain structures that that can be recognized by both Mus81 and XPF (for example, “splayed arms” that include an extended single-stranded DNA portion53), whereas structures formed following inhibition of DNA polymerases or DNA topoisomerases can predominantly be recognized by Mus81 (for example, replication forks with very short ssDNA regions53). Another possibility is that replication lesions that form during unperturbed growth could be repaired by single-strand annealing, which can be catalyzed by either Mus81 or XPF. In contrast, replication lesions that form after exposure to low doses of replication inhibitors require the resolution of replication forks or D-loops (via synthesis-dependent strand annealing), which can only be catalyzed by Mus8153.
During each cell cycle only a subset of replication origins are used to replicate the eukaryotic genome 57. Origins are selected anew each cycle 5 and slow replication can lead to the activation of a larger subset of origins, called “dormant origins”38, 58, 59. Single-fiber analyses per se cannot resolve whether activated dormant origins are selected from the pool of origins that are also activated during unperturbed growth, or if cells contain a class of latent, cryptic replication origins that are not used during normal growth but are only activated to rescue DNA replication when fork progression is perturbed (e.g., during replication stress)6. For example, at the Chinese hamster AMPD2 locus, origins that do not initiate replication during normal growth are activated when the concentration of nucleotide precursors is reduced13. The nascent strand sequencing results described here revealed that the same pool of DNA sequences initiated DNA replication in the slower-replicating Mus81-deficient cells and in the faster-replicating Mus81-proficient cells. These data support the notion that slow replication is accompanied by higher frequencies of flexible-origin activation (i.e., a greater utilization of pre-licensed origins). It is likely that the flexible origins activated at a higher frequency in Mus81 deficient cells represent origins that are normally activated later in the S-phase of the cell cycle in Mus81 proficient cells. This point could be investigated directly once data on replication timing in HCT116 cells become available. Our data show that Mus81 depletion does not lead to the use of novel DNA sequences, which do not initiate replication in Mus81 depleted cells, as replication origins. However, our observations do not preclude the possibility that such latent, cryptic origins exist, and that such origins can be activated under more extreme circumstances (e.g., DNA damage, or chromatin reprogramming during differentiation).
Methods
Cells and culture conditions
HCT116 and HCT116 Mus81−/− cells 31 were grown in Dulbecco Modified Eagle’s medium (Invitrogen), whereas MDA-MB-231 cells were grown in RPMI 1640 medium. Both media were supplemented with 10% heat-inactivated fetal calf serum. APH doses were calibrated based on a survival curve 30 to correspond to a 40 % survival.
DNA replication profiling by molecular combing
Asynchronous exponentially growing cells were labeled with 20 μmol/L IdU for 20 min. They were then washed in phosphate-buffered saline (PBS), and subsequently labeled with 50 μmol/L CldU for 20 min, then cells were chased with of 200 μmol/L of thymidine for 60-90 min. Cells were trypsinized and resuspended in PBS. Cells were then embedded in pulsed-field gel electrophoresis agarose plugs to prepare high-molecular-weight genomic DNA. After proteinase K digestion, agarose plugs were washed with TE buffer. Agarose plugs were then melted in 100 mmol/L MES (pH 6.5), and digested using 2 μL of β-agarase (Biolabs). The DNA solution was poured into a Teflon reservoir and DNA was combed onto silanized coverslips (Microsurfaces, Inc.) using a combing machine. Samples have included a BAC as a molecular marker, allowing us to convert labeled signal length (in microns) to fiber length (in kb) as molecular combing was previously shown to uniform stretching 32. Uniform stretching was confirmed by the uniform length of the stretched marker DNA. Coverslips were visually examined for DNA density and fiber length by a preliminary staining with YoYo1 (invitrogen). Sample preparations containing fibers that were mostly shorter than 200 kb or DNA concentration was too low or too high were discarded. The average fiber length in our studies is about 400 kb. For slide preparations that contained long fibers with appropriate density, coverslips with combed DNA were incubated at 60 °C for 2 h and denatured in 0.5 N NaOH for 20 min. IdU was detected using a mouse antibody directed against BrdU (IgG1, Becton Dickinson, cat.347580, 1:25). CldU was detected using a rat antibody directed against BrdU (Accuratechmecal, cat. OBT0030, 1:50). Single strand DNA (ssDNA) was detected using a mouse antibody directed against ssDNA (IgG 2a, Chemicon, cat. MAB3034, 1:100). Coverslips with DNA were incubated with primary antibodies for 1 h at room temperature. After washes, samples were incubated with secondary antibodies for 45 min. Secondary antibodies included Alexa Fluor 594 donkey antibody directed against rat (A21209, 1:100), Alexa Fluor 488 goat antibody directed against mouse IgG1 (γ1) (A-21121, Invitrogen, 1:100), and Alexa Fluor 647 goat antibody directed against mouse IgG2a (γ2a) (A21241, Invitrogen, 1:100). Slides were scanned with a BD pathway 855 controlled by AttoVision (Becton Dickinson). Fluorescent signals were measured using ImageJ (from the National Cancer Institute) with custom-made modifications. Only replication signals from high quality ssDNA (not from DNA bundles, not located at the end of a strand) were selected for analyses. Signals were marked for evaluation by “blind” measurers (not knowing which samples they were measuring); signal length was measured using the Image J software followed by automatic compilation of signal lengths into an Excel worksheet. Fork velocities and origin distances were calculated using Excel with a constant of 2kb/μm. Replication fork velocities were calculated using elongating fork signals only (initiating forks were eliminated; see a recent review62 for a discussion of good practices in dynamic molecular combing). Experiments were performed at least in duplicate using independent biological isolations of DNA fibers for each experimental condition. Statistical analysis was performed in Prism 5 or 6 (GraphPad software) using the non-parametric Mann-Whitney rank sum test. Normality of distributions was assessed using the Kolmogorov-Smirnov test.
Neutral comet assay
Neutral comet assay was performed using the CometAssay Kit (Trevigen) following the manufacturer's protocol. Briefly, cells were collected and suspended in low-melting-point agarose and applied to CometSlides™. Then cells were lyzed in lysis solution and were electrophoresed in TBE. The samples were then fixed in 70% (v/v) ethanol and dried overnight before staining with SyBr® Green (Molecular Probes, Eugene, OR) to visualize cellular DNA. Images of nuclei were captured using a CCD camera (Roter Scientific; Cool SNAP FX) with epifluorescence microscopy (Olympus; IX70) using a 20× objective lens. For each sample, 50 cells were scored for tail length. Tail length was manually measured using IPLab software.
Mus81 siRNA and XPF siRNA transfection and Western blotting
HCT116 cells were transfected with non-targeting control siRNA (Dharmacon, D-001810-1), Mus81 siRNA (Dharmacon, L-016143-01) or XPF siRNA (Dharmacon, L-019946-00) with DharmaFECT 2 siRNA Transfection Reagent (Dharmacon, T-2002) according to the manufacturer’s protocol. All siRNA transfection other than noted in the figure legends were done twice with a 72 hours interval, and cells were harvested for 3 days after the second transfection. Whole cell lysates by 1XSDS loading buffer were used for western-blot with anti-Mus81 antibody (Abcam, ab14387, 1:2000) and anti-XPF antibody (Lab vision Ab1, clone 219, 1:500) to verify the knockdown efficiency of Mus81 siRNA and XPF siRNA. Phospho-Chk1 (Ser317) and total Chk1 antibodies were purchased from Cell Signally (#2344, 1:1000 and #2345, 1:1000). GAPDH (Gene Tex, GTX627408, 1:5000) and PCNA (Santa Cruz Biotech, sc-7907, 1:2000) antibodies were used as loading controls.
Cell cycle analysis
Cells were pulse labeled with 10 µM EdU for 45 minutes before harvest. EdU staining using the Click-iT EdU kit (Invitrogen, C10424) were performed according to manufacturer’s protocol. DAPI was used for DNA staining. For cells without thymidine analogs incorporation, PI was used for DNA staining. BD LSRFortessa cell analyzer with FACSDiva software was used for cell cycle analysis.
Plasmid constructs and site-directed mutagenesis
Mus81 cDNA was purchased from Origene (RG203373). QuikChange II kit (Stratagene, 200523) was used to introduce the mutations into Mus81 (D338A and D339A) as described 63 with the following primers: sense primer was 5'-gcgcaagcgactggctgccctttgcagcagca-3', and the reverse primer was 5'-tgctgctgcaaagggcagccagtcgcttgcgc-3'. Then wild type and mutant Mus81tGFP cDNA were subcloned into Tet-On 3G Inducible Expression System (Clontech, 631168) and transfected into Mus81-deficient HCT116 to establish stable cell lines.
Clonogenic assay
After XPF siRNA treatment, cells were plated at a density of 50, 250 and 1000 per well in six-well plates and incubated for 12 days to allow formation of colonies. Cells were fixed with methanol, stained with 0.1% Commassie blue for 30min and washed with distilled water. Colonies were counted after air-drying. Plating efficiency (PE) was defined as the number of colonies counted/the number of cells seeded. The survival fraction (SF) was calculated as: PE treated/PE untreated×100.
Nascent strand abundance Assay
Genomic and nascent strand DNA samples were isolated from Mus81-proficient and Mus81-deficient cells as described 34, 64, 65 with some modifications. Nascent DNA samples were loaded onto a neutral 5–30% sucrose gradient in TNE. Gradients were centrifuged at 21,000 rpm for 20 h. Fractions that contained DNA fragments of 400–600 bp were pooled, dialyzed against TE, and precipitated. The sample was resuspended in DEPC water, and was digested overnight at 37°C with lambda exonuclease. Samples were heated to 95 °C for 10 min, ethanol precipitated, and resuspended in 50 µL of sterile water. Nascent strands were random-primed using the Klenow and DNA Prime Labeling System (Invitrogen). Double-stranded nascent DNA (1 µg) was sequenced using the Illumnia genome analyzer II (Solexa). Sheared genomic DNA was sequenced as a control.
Integrative genomics viewer tracks are reads per kilobase per million aligned reads (RPKM) values for nascent strand reads binned in 100 bp. Values were smoothed using a Gaussian smoothing algorithm with a window size of seven bins. RPKM values were normalized for copy-number variation using regional copy-variation values that were derived from our previous study of HCT116 cells using the Agilent aCGH 4x44 microarray (http://discover.nci.nih.gov). Pearson’s correlation coefficients between the binned and smoothed RPKM values across the entire genome were computed for HCT116 vs. MUS81-deficient cells. To avoid correlation skew, bins that were empty in both samples were excluded from the correlation calculation.
BrdU comet assay
The BrdU comet assay was performed as previously described 43. Briefly, HCT116 and Mus81−/− HCT116 cells were pulsed with 100 μM of BrdU for 30 minutes, then harvested immediately (0h) or chased for 1h or 6h in pre-warmed fresh medium without BrdU. Cells were collected and the cell pellet was resuspended in cold PBS. An alkaline assay was performed according to the manufacture’s instruction (Trevigen, 4250-050-K). Following electrophoresis, the gels were neutralized by 0.4 M Tris.HCl and washed with PBS prior to immunostaining. The BrdU labeled nascent DNA were detected by mouse anti-BrdU antibody (Becton Dickinson, cat.347580, 1:10), then goat anti-mouse IgG conjugated with Alexa 488 (Invitrogen, A-21121, 1:100). The gels were then counterstained with DAPI. The samples were then dried according to the manufacture’s instruction (Trevigen, 4250-050-K). Images were taken by a BD pathway 855 microscope controlled by AttoVision (Becton Dickinson) with 10X or 20X objective lens. Images were analyzed by CometScore software (TriTeK).
Apoptosis: Annexin V assay and TUNEL assay
Annexin V kit was from Becton Dickinson (#556547) and the assays were performed as described in the manufacturer ‘s instructions with minor modifications. Cells were kept on ice following trypsin-mediated detachment to avoid cell death.
TUNEL assay was performed as described in the manufacturer ‘s instructions (#11684795910, Roche).
Supplementary Material
Acknowledgements
We thank Drs. Paul Meltzer, Chiara Conti, Hong Fang Liu, Elisabeth Leo, and Kurt W. Kohn for helpful discussion. We are grateful to Dr. James Doroshow and William C. Reinhold for facilitating the bioinformatics pipeline data analysis. The intramural program of the CCR, National Cancer Institute, National Institutes of Health, funded this work.
Footnotes
Accession codes of NGS: GSE28911
Competing financial interests: The authors declare no competing financial interests
References
- 1.Shirahige K, et al. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 2.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nature reviews. Genetics. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 4.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 5.Wu JR, Gilbert DM. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- 6.Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 7.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 9.Woodward AM, et al. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggarwal BD, Calvi BR. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- 11.Danis E, et al. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- 12.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 13.Courbet S, et al. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455:557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- 14.Demczuk A, et al. Regulation of DNA replication within the immunoglobulin heavy-chain locus during B cell commitment. PLoS Biol. 2012;10:e1001360. doi: 10.1371/journal.pbio.1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert DM. Replication origin plasticity, Taylor-made: inhibition vs recruitment of origins under conditions of replication stress. Chromosoma. 2007;116:341–347. doi: 10.1007/s00412-007-0105-9. [DOI] [PubMed] [Google Scholar]
- 16.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 17.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santocanale C, Sharma K, Diffley JF. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 1999;13:2360–2364. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuduri S, Crabbe L, Tourriere H, Coquelle A, Pasero P. Does interference between replication and transcription contribute to genomic instability in cancer cells? Cell Cycle. 2010;9:1886–1892. doi: 10.4161/cc.9.10.11539. [DOI] [PubMed] [Google Scholar]
- 21.Dershowitz A, et al. Linear derivatives of Saccharomyces cerevisiae chromosome III can be maintained in the absence of autonomously replicating sequence elements. Mol Cell Biol. 2007;27:4652–4663. doi: 10.1128/MCB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang D, Koshland D. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 2003;17:1741–1754. doi: 10.1101/gad.1089203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- 25.Davidson IF, Li A, Blow JJ. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaziri C, et al. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 27.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 28.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 29.Shimura T, et al. DNA-PK is involved in repairing a transient surge of DNA breaks induced by deceleration of DNA replication. Journal of molecular biology. 2007;367:665–680. doi: 10.1016/j.jmb.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimura T, et al. Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. Journal of molecular biology. 2008;375:1152–1164. doi: 10.1016/j.jmb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiyama T, et al. Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells. Nucleic Acids Res. 2006;34:880–892. doi: 10.1093/nar/gkj495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conti C, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regairaz M, et al. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J Cell Biol. 2011;195:739–749. doi: 10.1083/jcb.201104003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin MM, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011 doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz IM, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Chen XB, et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 37.Hanada K, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006 doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao ZH, et al. Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 2007;67:8752–8761. doi: 10.1158/0008-5472.CAN-06-4554. [DOI] [PubMed] [Google Scholar]
- 39.Ying S, et al. MUS81 promotes common fragile site expression. Nat Cell Biol. 2013;15:1001–1007. doi: 10.1038/ncb2773. [DOI] [PubMed] [Google Scholar]
- 40.Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol. 2013;15:1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- 41.Ehmsen KT, Heyer WD. A junction branch point adjacent to a DNA backbone nick directs substrate cleavage by Saccharomyces cerevisiae Mus81-Mms4. Nucleic Acids Res. 2009;37:2026–2036. doi: 10.1093/nar/gkp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang MJ, et al. Genetic and functional interactions between Mus81-Mms4 and Rad27. Nucleic Acids Res. 2010;38:7611–7625. doi: 10.1093/nar/gkq651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGlynn AP, et al. The bromodeoxyuridine comet assay: detection of maturation of recently replicated DNA in individual cells. Cancer Res. 1999;59:5912–5916. [PubMed] [Google Scholar]
- 44.Zhang YW, et al. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–3620. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roseaulin L, et al. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matulova P, et al. Cooperativity of Mus81.Mms4 with Rad54 in the resolution of recombination and replication intermediates. J Biol Chem. 2009;284:7733–7745. doi: 10.1074/jbc.M806192200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marheineke K, Hyrien O. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J Biol Chem. 2004;279:28071–28081. doi: 10.1074/jbc.M401574200. [DOI] [PubMed] [Google Scholar]
- 50.Shechter D, Costanzo V, Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 2004;3:901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 51.Petermann E, et al. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 54.Pepe A, West SC. Substrate specificity of the MUS81-EME2 structure selective endonuclease. Nucleic Acids Res. 2014;42:3833–3845. doi: 10.1093/nar/gkt1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kikuchi K, et al. Structure-specific endonucleases xpf and mus81 play overlapping but essential roles in DNA repair by homologous recombination. Cancer Res. 2013;73:4362–4371. doi: 10.1158/0008-5472.CAN-12-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarbajna S, Davies D, West SC. Roles of SLX1-SLX4, MUS81-EME1, and GEN1 in avoiding genome instability and mitotic catastrophe. Genes Dev. 2014;28:1124–1136. doi: 10.1101/gad.238303.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DePamphilis ML. Origins of DNA replication that function in eukaryotic cells. Curr Opin Cell Biol. 1993;5:434–441. doi: 10.1016/0955-0674(93)90008-e. [DOI] [PubMed] [Google Scholar]
- 58.Abdurashidova G, et al. Functional interactions of DNA topoisomerases with a human replication origin. EMBO J. 2007;26:998–1009. doi: 10.1038/sj.emboj.7601578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conti C, et al. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herrick J, Michalet X, Conti C, Schurra C, Bensimon A. Quantifying single gene copy number by measuring fluorescent probe lengths on combed genomic DNA. Proc Natl Acad Sci U S A. 2000;97:222–227. doi: 10.1073/pnas.97.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Techer H, et al. Replication dynamics: biases and robustness of DNA fiber analysis. Journal of molecular biology. 2013;425:4845–4855. doi: 10.1016/j.jmb.2013.03.040. [DOI] [PubMed] [Google Scholar]
- 63.Huang L, et al. Prevention of transcriptional silencing by a replicator-binding complex consisting of SWI/SNF, MeCP1, and hnRNP C1/C2. Mol Cell Biol. 2011;31:3472–3484. doi: 10.1128/MCB.05587-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, et al. The human beta-globin replication initiation region consists of two modular independent replicators. Mol Cell Biol. 2004;24:3373–3386. doi: 10.1128/MCB.24.8.3373-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu H, et al. Mapping replication origin sequences in eukaryotic chromosomes. Current protocols in cell biology / editorial board, Juan S. Bonifacino … [et al.] 2014;65:22 20 21–22 20 17. doi: 10.1002/0471143030.cb2220s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.