Abstract
Sequence alignments of promoters in prokaryotes postulated that the frequency of occurrence of a base pair at a given position of promoter elements reflects its contribution to intrinsic promoter strength. We directly assessed the contribution of the four bp in each position in the intrinsic promoter strength by keeping the context constant in Escherichia coli cAMP-CRP regulated gal promoters by in vitro transcription assays. First, we show that bp frequency within known consensus elements correlates well with promoter strength. Second, we observe some substitutions upstream of the ex-10 TG-motif that are important for promoter function. Although the galP1 and P2 promoters overlap, only three positions were found where substitutions inactivated both promoters. We propose that RNA polymerase binds to the −12T bp as part of dsDNA while opening base pairs from −11A to +3 to form the single stranded transcription bubble DNA during isomerization. The cAMP-CRP complex rescued some deleterious substitutions in the promoter region. The base pair roles and their flexibilities reported here for E. coli gal promoters may help construction of synthetic promoters in gene circuitry experiments in which overlapping promoters with differential controls may be warranted.
Introduction
Initiation of transcription from a prokaryotic promoter occurs in several steps: i) binding of RNA polymerase (RNAP) to the promoter to form a closed complex (RPc); ii) isomerization of the closed (RPc) to an open complex (RPo); iii) conversion of the open (RPo) to an initiating complex (RPi); and iv) formation of an elongating complex (RPe) [1, 2]. Sequence alignments, mutational analysis, RNAP-DNA interaction studies, and in vitro transcription assays have shown that the amount of productive initiation of transcription from a promoter is guided by the presence of a combination of distinct DNA sequence elements in the promoter: the UP element (AT-rich), the −35 element (TTGACA), the ex-10 element (TG), the −10 element (TATAAT), the discriminator (dscr) element (G/C- or A/T-rich, −6 to −1), and the transcription start point (tsp) (+1) [3–11]. In the absence of any transcription factor, the intrinsic strength of a promoter depends on the presence of these elements—not all elements are present in every promoter—and on the closeness of DNA sequences of the elements to their consensus forms so that the frequency of occurrence of a base pair at a given position of the element reflects its relative importance in promoter function. Regulation of gene transcription may occur at any stage of transcription initiation. The function of specific base pairs (bp) in transcription initiation has been established for a few critical locations in several promoters by mutation and structural analysis [12–20]. The significance of the base pair frequency concept in promoter strength was developed without regard to the context sequence. It is probable that the contribution of a base pair to the promoter strength may depend upon the presence of a specific base pair at another seemingly unrelated position in the promoter. This would not be known by looking for consensus sequences among heterologous promoters and can only be assessed by analyzing each base pair at a given position and then changing base pairs at every other position within the context of the promoter under study, which would practically be an impossible task. We took a simpler, but nonetheless arduous, approach of assessing the contribution of a base pair at a given position in the promoter under the context sequence that was kept constant. Thus, we investigated the contribution of each base pair in the entire “promoter” DNA segment of the gal (galETKM) operon in Escherichia coli in determining transcription efficiency by mutational analysis. The analysis became more informative since the gal DNA sequence is embedded with overlapping promoters (Fig. 1). The operon is normally transcribed from two (P1 and P2) interspersed promoters [18, 19, 21, 22]. The cyclic AMP (cAMP) and cAMP receptor protein (CRP) complex (CCC) enhances P1 and represses P2 by binding to a DNA activating site (termed AS, activation site; Fig. 1a) [18, 19, 21, 23–26]. The DNA sequence of the entire region also contains two additional promoters (P3 and P4*), which are observed under specific conditions [16, 27]. [*Footnote: To avoid confusion in nomenclature, we referred to P3 described by Sur et al. [27] as P4 to distinguish it from another promoter previously described by Ponnabalam et al. as P3 [16]]. The promoter, P3, interspersed with the P1 and P2 is silent [16]. P3 can be activated by mutations (see below). The activated P3 is repressible by CCC binding to the AS. The fourth promoter, P4, with a functional −10 element (TATAAT) is independent of CCC [27]. Although our investigation of P1 and P2 touches on P3, P4 was not studied because the P4 promoter is located far upstream of the DNA segment that is not being considered here. In Fig. 1 the tsp for the promoter P1 is referred to as +1, and relative to it, P2 and P3 are referred to as −5 and +14, respectively [16] For easy comparison and interpretation of results described below, we subsequently noted each base pair position of P2 and P3 promoters counting from their respective tsp taken as +1. Both P1 and P2 are intrinsically fairly active and contain a perfect ex-10 and a reasonable −10 element, but not −35 and UP elements [17, 28–32]. P3 contains only a −10 element, which is not sufficient to make a promoter active [16].
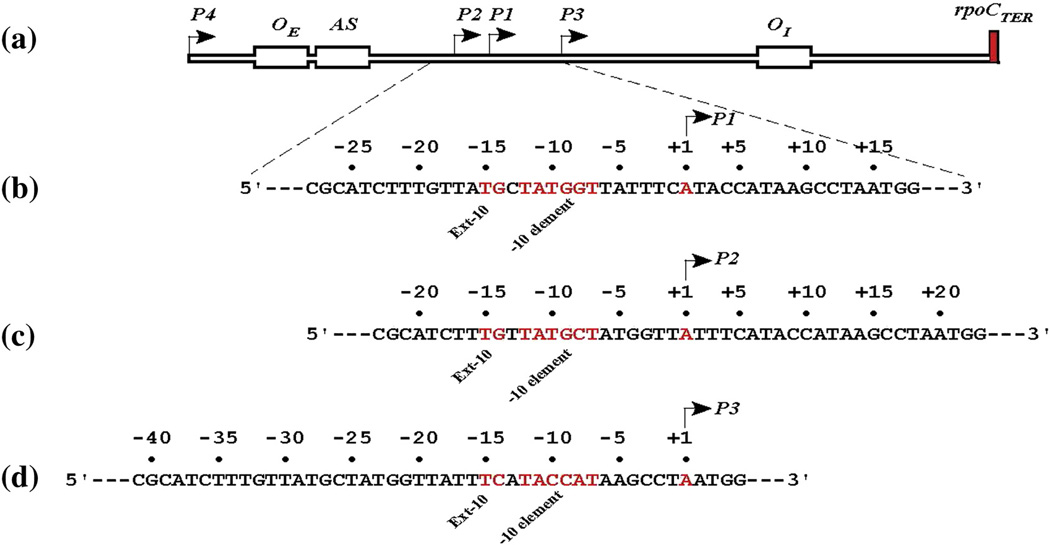
Fig. 1.
The sequence of gal promoters. (a) A schematic genetic map of the promoter region of the E. coli gal operon. P1, P2, P3, and P4 represent promoters with tsp at +1, −5, +14, and −96, respectively. In the following description negative (−) and positive (+) signs represent numbers upstream and downstream of corresponding transcription start point (+1). (b) DNA sequence from −28 to +18 showing the location of the −10 and extended −10 elements (red), and tsp (blue) of P1 (+1); operator: OE (−60.2) and OI (+53.5); cAMP-CRP binding site (AS, −40.5); rho independent terminator (rpoCter). (c) DNA sequence from −23 to +23 showing the location of the −10 and extended −10 elements, and tsp (+1) of P2. (d) DNA sequence from −41 to +5 showing the location of the −10 and extended −10 elements, and tsp (+1) of P3.
In this study, the contribution of each base pair in the segment −25 to +1 with reference to P1 coordinates was investigated by base pair substitutions. We systematically replaced each base pair in the 26 base pair segment by three other base pairs by site-directed mutagenesis and generated a total of 78 mutant DNAs, and then used them as supercoiled DNA templates in an in vitro transcription assay and the effects of the substitutions on P1, P2, and P3 were followed [33, 34]. Base pairs further upstream of the chosen 25 base pair segment were not studied because the promoters as mentioned do not contain any semblance −35 elements. Both intrinsic transcription of the promoters in each template and any regulatory effect of CCC on activities of the promoters were investigated. We successfully used such efforts previously to establish the role of individual base pairs in tsp selection [35], and in regulation of transcription elongation [36]. Here we report the finding of involvement of base pairs at new positions, besides UP and −10 elements, in efficient promoter function, and discuss the boundaries of some DNA elements.
Exhaustive kinetic studies only concluded that the P1 and P2 compete with each other for RNA polymerase binding at the level of closed complex formation; once the respective open complexes are formed, RNAP does not switch the promoter [37, 38]. The mechanisms by which CCC regulates the gal promoters are not totally understood. We argue that CCC helps the formation of both closed and open complexes at P1.
Results
The in vitro transcription results using 79 DNA templates (wild type and 78 mutants) in the absence and presence of CCC are presented in Figs. 2–3. For P1 and P2, the amount of full-length RNA from different mutant templates were expressed relative to the amount of full-length transcripts obtained for the respective promoter in the wild type DNA template taken as 1.0. For P3, the amount of RNA, if any, made relative to the control RNA (RNAI) made for each DNA template is presented because the wild type template did not show any P3 RNA. In CCC regulation, the ratio of P1 transcription in the presence of CCC to that in the absence of CCC represents fold of P1 activation. For P2 or P3, the ratio of transcription in the absence of CCC to that in the presence of CCC represents fold repression (Fig. 3; Tables S2 and S3). Note, although abortive initiation and read-through transcription beyond transcription termination signal can influence the strength of promoter in transcription [39, 40], we did not investigate such products in this study. Abortive products of P1 and P2 in the absence and presence of CCC have been studied previously on wild type gal DNA [33].
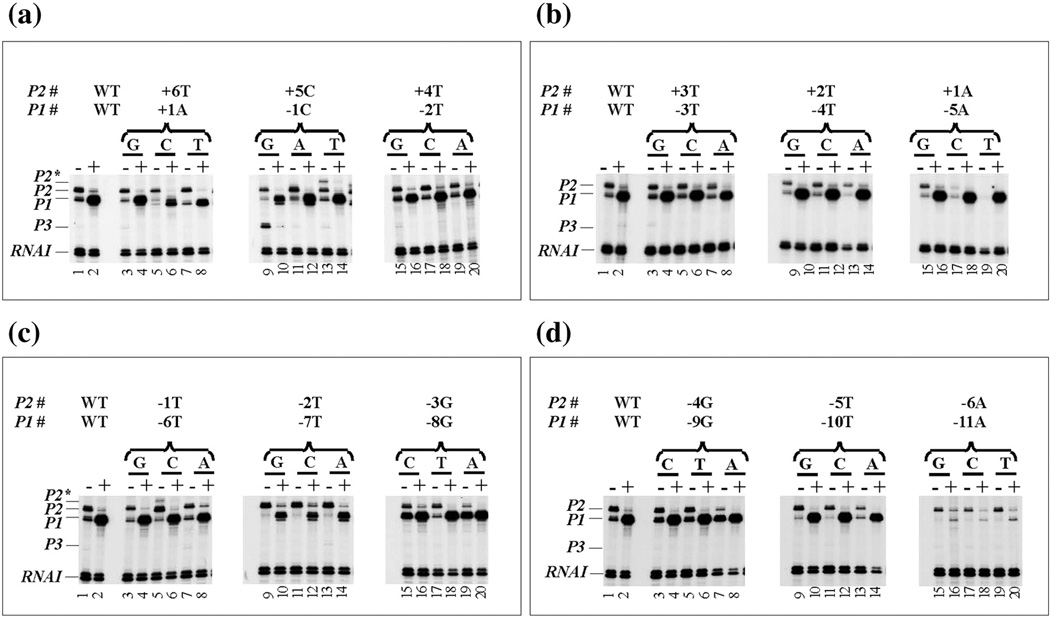
Fig. 2.
In vitro RNA synthesis from wild type and mutant gal DNA templates in the absence (−) and presence (+) of CCC. DNA templates with P1 numbering system from +1 to −25 and P2 from +6 to −20 are labeled as shown in the Fig. 1 The mutation above each lane is indicated: (a) +1 to −2, (b) −3 to −5, (c) −6 to −8 and (d) −9 to −11 represent P1, while (a) +6 to +4, (b) +3 to +1, (c) −1 to −3 and (d) −4 to −6 represent P2, P1, P2, P2*, and P3 represent gal promoters. RNAI transcript is used as an internal control.
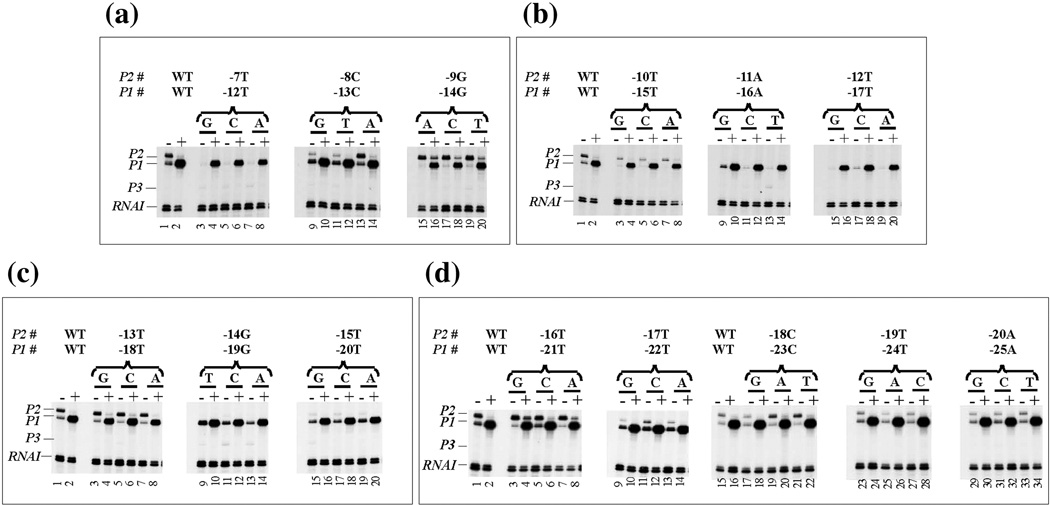
Fig. 3.
In vitro RNA synthesis from wild type and mutant gal DNA templates in the absence (−) and presence (+) of CCC. (see Fig. 2 legend for nomenclature). (a) −12 to −14, (b) −15 to −17, (c) −18 to −20, (d) −21 to −22 and (e) −23 to −25 represent P1, while (a) −7 to −9, (b) −10 to −12, (c) −13 to −15, (d) −17 to −18 and (e) −19 to −20 represent P2.
The P1 promoter
The dscr region (−6 to −1)
Intrinsic transcription
The dscr region −6 to −1, when rich in G/C base pairs, makes open complexes unstable and reduces intrinsic transcription, but when rich in A/T base pairs, makes open complexes more stable and increases intrinsic transcription levels [6, 11]. The −6 to −1 region of P1 is not G/C rich (Fig. 1b). We found no major changes in intrinsic transcription by any of the base pair changes in this region except for the −1C position, in which a change to −1G showed an approximately 2-fold decrease, and a change to −1A, which showed a 1.5-fold increase in P1 transcription (Figs. 2a lane 9 and 3a; black bars, -CCC). We did not put any special significance to either observation.
CCC effect
Although transcription from the wild type P1 promoter was stimulated 15fold by CCC, the stimulation of P1 varied from 5.5- to 15-fold with the substitutions in the −6 to −1 region (Figs. 2a-2c and 4a). The significance of the low level CCC stimulations with some of the mutants is not clear. It is noteworthy that in the presence of CCC, the P1 transcript of wild type, and some substitution templates, started both at the usual +1A (major transcript) as well as at +3A (minor transcript) because of the presence of a purine at +3, as expected from the axiom of tsp selection [35]. The optimal tsp is the 11th bp counting downstream from the −10 position [35].
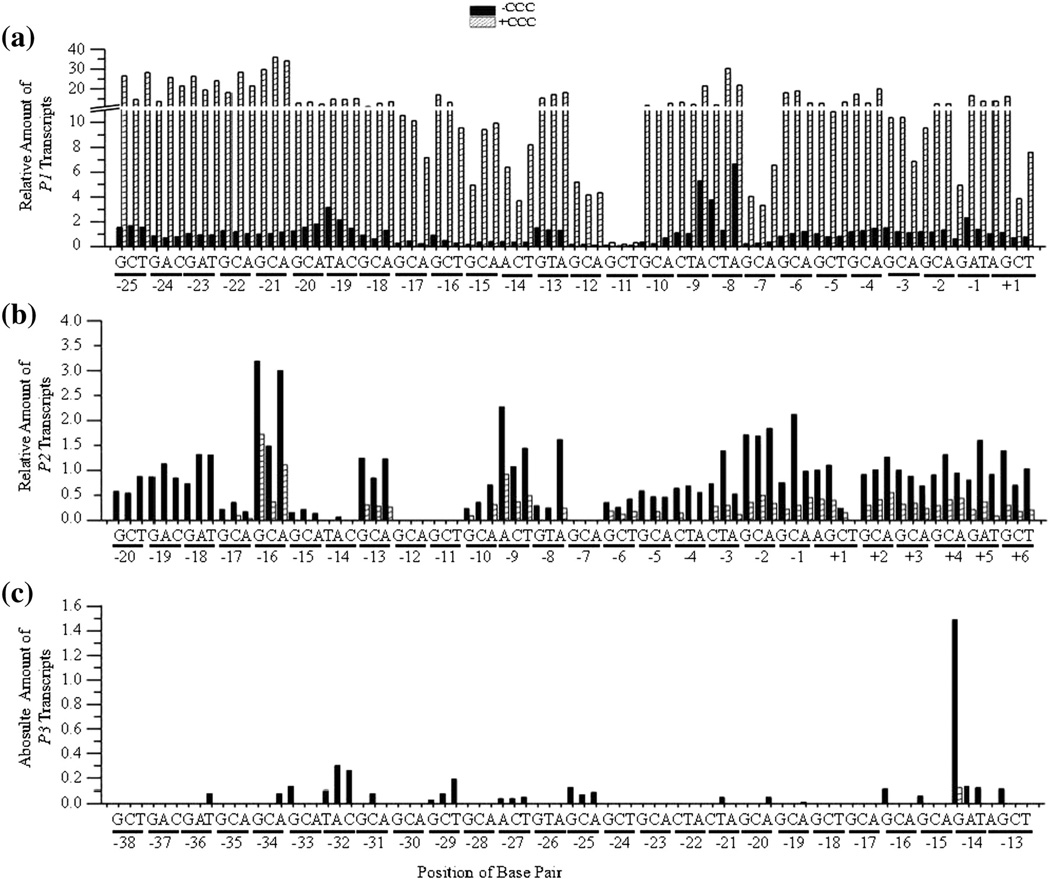
Fig. 4.
Bar graphs showing the quantification of the results shown in Fig. 2. (A) The relative amount of P1 transcripts from wild type and mutant DNA templates in the absence (shaded column) and presence (hatched column) of CCC. The y-axis contains a break from 11.0 – 11.1 with increment before and after the break of 2 and 10, respectively. The position of each base pair change from −25 to +1 is shown on the x-axis. (b) The relative amount of P2 transcripts from wild type and mutant DNA templates (positions +6 to −20). (c) The absolute amount of P3 transcripts from wild type and mutant DNA templates (positions −13 to −38).
The −10 element (−12 to −7)
Intrinsic transcription
The six base pairs-long −10 element is most critical for promoter function in bacteria (isomerization and transcription initiation) [14, 41] according to Harley and Reynolds [8], who analyzed 263 promoter sequences of E. coli and concluded a consensus sequence of −12TATAAT−7 with −7T being present 89%, −8A 49%, −9A 59%, −10T 52%, −11A 89%, and −12T 82% of the time implying their relative importance in terms of function. Mitchell et al. [30] improved the statistics further by analyzing 553 promoter sequences of E. coli, where −7T represent 90%, −8A 54%, −9A 50%, −10T 50%, −11A 87%, and −12T 79%. The −10 element of P1 contains 4/6 (−12TATGGT−7) conserved base pairs, including the critical −7T, −11A, and −12T for σ70 of RNAP used in our experiments. Heyduk and Heyduk [42] analyzed melting kinetics of 4096 variants of a bacterial promoter and also found the critical bases of −7T, −11A and −12T. Mekler and Severinov showed strong co-operative RNA interactions with individual non-template strand bases in the −10 region [43]. The transcription results are shown in Figs. 2c-2d, 3a and 4a. By changing −7T to the other 3 bases, we removed a highly conserved base. Clearly, the P1 promoter was largely (75% − 90%) inactivated in −7G, −7C, and −7A templates (Fig. 2c). Previous results also showed that −7T to −7A or −7C inactivated P1 [17, 44]. Based on crystal structures of RNAP sigma subunit and the −10 element, −7T has been shown to unstack and flip into a hydrophobic pocket of σ70 RNAP residues [45, 46].
Changes from −8G to −8C and −8A enhanced the intrinsic strength of P1 by 4-and 7-fold, respectively (Figs. 2c and 4a). The −8A improves the homology to consensus −10 element (5/6, TATGAT). The −8C (−12TATGCT−7) is present in 21% of 263 E. coli promoters [7, 8]. Our results show a hierarchy of the P1 promoter strength at position −8 as follows: A > C >> T > G (Fig. 5a). The frequency of base occurrence at the −8 position in E. coli follows the same order [7, 8].
Fig. 5.
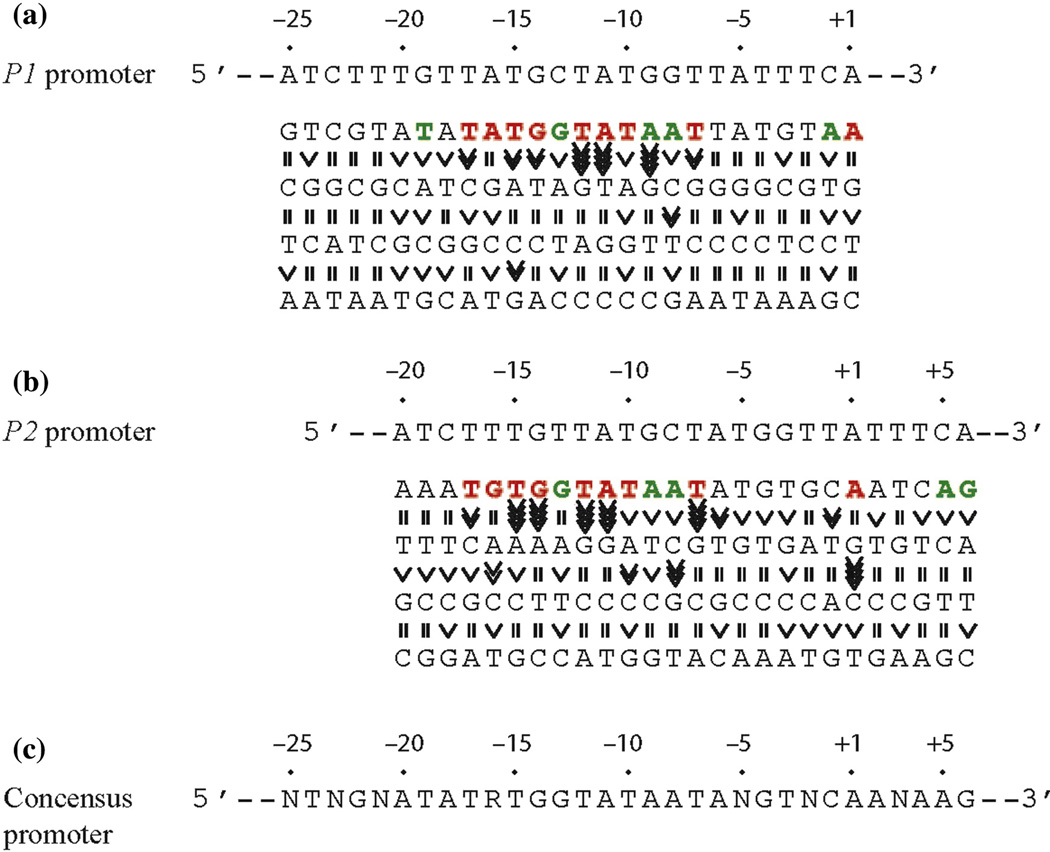
Base pair requirement in gal promoters. (a) The DNA sequence from −25 to +1 and a summary of the effect of base pair change from +1 to −25 on P1 transcription. (b) The DNA sequence from −20 to +6 and a summary of the effect of base pair change from −20 to +6 on P2 transcription. (c) Consensus promoter region of P1 and P2 derived from (a) and (B). R = A or G, N = any nucleotide. Base pair is in red if it is unique for promoter function, green if it improves promoter function, and black if it is degenerate. The symbol “>>>>” in vertical shapes represents 4.1-fold or more difference in promoter function from the wild type; “>>>”, 3.1 to 4-fold; “>>”, 2 to 3-fold; “>” less than 2-fold; “=“ indicates equal.
A change of −9G to −9A also improved (5/6, −12TATAGT−7) the −10 element homology to consensus and P1 activity was enhanced (5-fold) in the −9G to −9A change (Figs. 2d and 4a). P1 intrinsic level was unaffected in −9C and −9T templates.
The −10T is frequently conserved (52%) in −10 elements [7, 8]. Therefore, we expected that mutating it would reduce the intrinsic strength of P1. Precisely that was observed in the −10G, −10C, and −10A substituted templates (Figs. 2d and 4a); the substitutions reduced transcription significantly in these three cases. Note that with − 10G and −10C, a stretch of three G-C base pairs resulted in the −10 element (− 12TAGGGT−7 or −12TACGGT-7) of P1. Perhaps the GC-rich sequence makes it more difficult for RNAP to isomerize the −10 element explaining the relative defects. As mentioned, the strength of P1 decreased with changing base pairs at position −10 in the following order: T > A > G > C, which was also the order of frequency of occurrence of bases at the −10 position (Fig. 5a) [7, 8].
The −11 position of the −10 element of any promoter is the most critical base where the isomerization of the −10 region nucleates [42, 47–49], leading to a so-called “transcription bubble” and initiating transcription at +1. Substituting −11A by G, C, and T made P1 extremely defective (Figs. 2d and 4a). It was suggested that −11A has a “master” role for base pair unpairing and bubble formation [50, 51]. Consistently, the −11A has been shown recently to flip out of the stacked bases and insert into a hydrophobic pocket of the σ70 RNAP residues [45, 46]. Additionally, Y430 of σ70-subunit of RNAP was shown to quench 2-aminopurine (2AP) at position −11, suggesting interaction of Y430 with −11A that was flipped out during isomerization [52]. The transcription defect of −11 substituted templates has been previously reported; several independently isolated gal promoter mutants, p8-3, p211, and p11 are all −11A to −11T changes that inactivated P1 [18, 19, 21, 53].
The base T is conserved at position −12. The P1 promoter was also highly inactivated when −12T was changed to A, G, or C (Figs. 3a and 4a). Our results are in agreement with previous findings that a −12C change inactivated P1 [16, 54–56]. Overall, our results show the relative importance of the native base pairs within the −10 element of P1 as derived from bioinformatics analysis [8].
CCC effect
CCC stimulated the intrinsic P1 transcription in all of the base substitutions in the −10 region except the very defective substitutions at the −11 position. CCC restored the activation levels of P1 in the other two very defective substitution sets: −7G, −7C, and −7A, and −12G, −12C, and −12A, both in major (initiating at +1) and minor (initiating at +3A, whenever observed) transcripts. Among the substitutions that did not change the intrinsic level significantly, −8T, −9C, and −9T, CCC restored P1 transcription to normal levels. The −10G and −10C substitutions decreased the intrinsic transcription level by 3- and 5-fold, respectively. However, some of the base substitutions as −8C, −9A, and −8A templates showed 4-, 5-, and 7-fold higher intrinsic transcription than wild type. Interestingly, CCC further stimulated these three templates another 3 to 4 fold. The changes at −11 were the only examples in this study where CCC did not rescue any transcription (Fig. 2d, lanes 16, 18, and 20). In summary, CCC overcame the inactivity of −10 element substitutions in P1, except in the −11 mutants.
Ex −10 element (−15 to −14)
Intrinsic transcription
The ex-10 element (−15TG−14) is separated from its cognate −10 element by 1 bp (the position −13) (Fig. 1a). At the position −13, there is no consensus base pair in promoters; none of the substitutions of −13C had any significant effect on transcription in P1 (Fig. 3a). The ex-10 element provides contacts for RNAP to recognize a promoter with no −35 element as in P1 [29, 57]. Mutation of −14G to A, C, or T inactivated P1 (Figs. 3a and 4a). The −15T is the other part of the ex-10 element. Mutating −15T to −15G, −15C, or −15A also inactivated P1 drastically (Figs. 3b and 4a). These results are in agreement with previous studies with −15T and confirm that “TG” is the only sequence that is acceptable in the −15 and −14 positions for efficient P1 activity [30, 57–59].
CCC effect
Although compared to the wild type the intrinsic level of transcription of all six ex-10 substitutions are very low, the presence of CCC restored transcription to a reasonably high level, although not as high as in the wild type (Figs. 3a lanes 15–20 and 3b lanes 3–8).
The upstream “spacer” (−25 to −16)
Intrinsic transcription
In promoters with an active −35 element the DNA sequence of the region between −35 and −10 elements is called the “spacer”. The spacer is not supposed to contribute any sequence specificity [8]. But the length (number of base pairs) of the spacer is critical for optimal promoter activities among the class of promoters that have both −35 and −10 elements. The effective length varies from 15 to 19 base pairs with 17 being optimal. Since gal promoters do not contain a −35 element the spacer length should be a moot issue. But we did analyze part of the so-called spacer sequence—the region immediately upstream of the ex-10 element—by substitution analysis. We made DNA templates with changes in the −16 to −25 positions of P1. Mutation of −16A to −16G has a marginal effect on P1, but mutations to −16C and −16T were inactive for transcription (Fig. 3b). This implies that position − 16 prefers a purine instead of a pyrimidine for P1 activity. At position −17T, transcription in all three substitutions are low with a hierarchy for bases T >> C > G = A showing a relative preference of a pyrimidine over a purine in this position (Fig. 3b). Burr et al. (57) previously showed that the −17TG−16 sequence makes the gal P1 stronger and, relative to −15TG−14, called it a “second” TG motif, although it is more like a TR motif. These authors suggested that the two TG motifs function independently but we note that −17TG−16 motif action is dependent upon the presence of the −15TG−14 motif, whereas the reverse is not the case. We note that the importance of a −17TRTG−14 sequence in B. subtilis promoters has also been demonstrated [60–62]. The substitutions at position −18 showed no major effect in intrinsic P1 transcription (Fig. 3c). The intrinsic P1 activity was better with −19T (Fig. 3c), implying that a T at this position makes P1 better. In fact, the −19T change caused a substantial increase in intrinsic activity and has also been called a CCC-independent P1 mutation [63]. This result is in agreement with previous findings where a change from −19G to −19T or − 19A increases the intrinsic strength of P1 [16, 54, 55]. Mutations at positions from −20 to −25 did not show any noteworthy change in transcription level of P1 except at the position −20 where there was a hierarchy in base pairs in P1 transcription efficiency: A > C > G > T (Figs. 3c-3e and 5a). We further note that Busby et al. reported that a −23G to −23A change increased P1 activity by 40% in vivo [58]. We did not find any increase in the −23G to −23A case under in vitro conditions (Figs. 3e and 4a).
CCC effect
The intrinsic transcription levels of all substitutions in the −16 through −25 positions, including the defective −16C, −16T, −17G, −17C, and −17A, were all stimulated by CCC to the high levels, sometimes even to levels which are higher than that of the wild type (Fig. 4a).
The P2 promoter
The −6 to + 6 region
Intrinsic transcription
The 25-base pair substitutions experiments performed cover the segment −20 to +6 positions of the P2 promoter with its own tsp as +1 (Figs. 1c and 4b). No effect of base pair substitutions was expected in the +2 to +6 region of P2. The intrinsic transcription in all of the substitution templates in the segment was within −30% to +60% of the wild type and was not considered to be significant (Figs. 2a-2b and 4b). However, in the presence of +5T, a new transcript approximately three nucleotides longer was observed in a significant amount (labeled P2* in Fig. 2a lane 13). This change created a template with an AT-rich sequence of 10 bp (−2TTATTTTATA+8). Either the AT-richness makes a new start point at approximately −3G, which is very unlikely, or the RNA polymerase stutters three times at the 4Ts after initiating at +1A. UTP concentration-dependent stuttering of RNA polymerase at T-clusters has been demonstrated in several promoters including the P2 promoter [64, 65]. The precise origin of P2* could not be resolved by primer extension assays.
The +1 base pair substitution results are consistent with the axiom of start site selection established previously for P2 and, therefore, are not discussed in this paper [35]. In the dscr region (−6 to −1), −1T to C, −2T to G, C or A, and −3T substitutions increased intrinsic transcription by about 50% to 100% (Figs. 2c-2d and 4b). The rest of the substitutions in the region either did not affect or decreased intrinsic transcription by 25% to 75% as observed for corresponding positions at P1 (Figs. 2g-2d and 4b). The sequence of this region, like in P1, is not G/C rich and most likely does not play a dscr role. Like the +5C to +5T change, the −1T to −1C change also showed a P2*-like transcript.
CCC effect
CCC repressed P2 transcription in wild type [19, 21, 24–26]. In all of the substitutions from −6 to +6, CCC showed a more or less normal amount of repression (Figs. 2a-2d). The mechanism of P2 repression by CCC remains unknown at this stage. CCC’s normal repression activity in substitutions in the −6 to +6 region does not give any clue about CCC’s mechanism of action. CCC also repressed the synthesis of P2* RNA whenever observed (Figs. 2a lane 13 and 2c lane 5).
The −10 element (−12 to −7)
Intrinsic transcription
The −10 element of P2 has 4/6 (TATGCT) homology to the consensus sequence. The conserved −7T base pair when changed to A, G, or C inactive P2 as it did in P1 (Figs. 3a and 4b). Incidentally, the −7T position of P2 is the −12 position of the −10 element of P1, and the substitution also inactivated the latter promoter (Figs. 3a and 4b). The result is in agreement with previous findings where the gal promoter was inactivated when −7T was changed to −7C or −7A [17, 44, 55].
When −8C was mutated to −8G or −8T, the intrinsic strength of P2 was reduced 4-fold. However when −8C was changed to −8A, which increased the homology to the consensus −10 element (5/6, TATGAT), the level of P2 transcription increased by 60% (Figs. 3a and 4b).
A change of −9G to −9A improved the −10 element homology, too, (5/6, TATAGT) to the consensus [7, 8, 66–68]. Indeed, P2 activity was enhanced more than 2-fold (Figs. 3a and 4b). The −9G to T change also showed a 50% increase in intrinsic transcription (Figs. 3a and 4b). This is in agreement with several previous findings with mutations at −9A [17, 44, 55, 58, 59]. The −9C substitution did not make any difference in P2 transcription. All of the −9 substitutions inactivated P1 intrinsic transcription because −9 of P2 is the same as −14 (critical ex-10) of P1.
All three substitutions at the −10 position reduced P2 activity significantly as expected (Fig. 3b). The −10T to −10A substitution at P2 also reduced P1 transcription because −10 of P2 is −15T of the ex-10 region of P1 (Figs. 3b and 4b).
Mutation of −11A, which was the most critical of bases in the −10 element in P1, to G, C, or T practically destroyed P2 activity (Figs. 3b and 4b). Previous studies showed that P2 is indeed turned off when −11A was changed to −11C [44, 55, 58]. The results showed that the change of −12T (a highly conserved base) to G, C, or A also inactivated P2 transcription to undetectable levels (Figs. 3b and 4b).
CCC effect
For those substitutions at −7, −8, −10, −11, and −12, in which P2 transcription were extremely low; any more repression of P2 by CCC was beyond detection (Figs. 3a-3b). However, for other changes at position −8 (Fig. 3a) that showed detectable levels of P2 activity were normally repressible by CCC. Interestingly, CCC did not repress the substitutions at −9 of P2 as efficiently as in wild type template (Fig. 3a).
The ex-10 element (−15 to −14)
Intrinsic transcription
As mentioned for P1, the position −13, which separates an ex-10 from a −10 element does not have any consensus base. Substitution of −13T to the other three bases did not show any significant effect on P2 (Figs. 3c and 4b) similar to the significant effect on P1.
The −14G and −15T at P2 are conserved bases forming the “TG” motif of the ex-10 element. Without a −35 element as in P1, any change in this position should affect P2 activity. As expected, the results showed that P2 was inactivated by −14G to C, T, or A substitution (Figs. 3c and 4b). Our results are in total agreement with previous studies that showed P2 was turned off when −14G was mutated to −14T or −14A. [17, 29, 44, 54, 55, 58] According to Johnson et al., [17] −14T or −14A prevented RNAP from binding to P2. Moreover, very low levels of P2 were observed when −15T was changed to the three other bases (Figs. 3c and 4b). The −15T to G, C, or A change reduced P2 transcription dramatically.
CCC effect
Any repressive effect of CCC on base pair substitutions at the two ex-10 positions was not discernable because of very poor intrinsic transcription especially on −14 substitutions (Fig. 3c and 4b). CCC showed normal repression of transcription on substitutions at the −13 non-critical position.
The upstream spacer (−20 to −16)
Intrinsic transcription
Like P1, the P2 promoter does not contain a −35 element and thus naming the sequence upstream of ex-10 a spacer apparently is not relevant. We investigated the influence of any base pair substitutions from −16 to −20 in P2 to see whether the base sequence at this segment of DNA has any role in transcription. Substitution of −16T to G or A showed enhanced P2 activity (Figs. 3d and 4b); P2 increased substantially (3-fold) in −16G and −16A substitutions and only marginally (1.5-fold) in the −16C substitution indicating a preference for a purine at this location, similar to that in the −16 position of P1.
It was not known whether position −17T plays any role in P2. Our experiments showed that P2 activity was reduced substantially when −17T was substituted by the other base pairs (Figs. 3d and 4b). In summary, as in P1, the base pair at the −16 and −17 positions play a role in P2 transcription, the preferred sequence being −17TR−16. None of the base pair changes at positions further upstream, −18 to −20, showed much variation in P2 transcription (Figs. 3e and 4b).
CCC effect
CCC repressed the substitutions at positions −13, −16, −17 normally and at −18 to −20 very strongly (Figs. 3c-3e and 4b).
The P3 promoter
The P3 promoter in gal with a tsp at +1 (equivalent to +14 position of P1) is naturally defective, and Busby and colleagues discovered that P3 can be unmasked by a −25T to −25C change (−12 of P1), which at the same time inactivated P1 and P2 [16, 17, 63]. A strong signal of P3 was observed when −24A (−11 of P1) was replaced by adenine analogs 2-aminopurine or 2–6-diaminopurine [48, 50, 51]. Our gal DNA segment selected for analysis covered the region −13 to −38 of P3 (Fig. 1d). We did not observe P3 transcription in the −25C substitution both in supercoiled and relaxed DNA templates (results with relaxed template not shown) (Fig. 3a and 4c) [16, 17, 63].
The latent P3 promoter contains a reasonably good −10 element (4/6, −12TACCAT−7) but not any functional −35 or ex-10 sequences. A −10 sequence alone is not sufficient for promoter function [66]. P3 has a −15TC−14 sequence in its ex-10 region, which is 1 bp away from being an effective ex-10 element [16] (Fig. 1d). When −14C was changed to −14G, we observed a strong P3 transcript (Fig. 2a lane 9 and 4c). In this template, 43% of the total gal transcripts were from P3, 34% from P2, and 19% from P1. In the presence of −14G, P3 now contains an active ex-10 sequence (TG) and a reasonably good −10 element facilitating a high level of transcription. Consistently, the change of −15T to C or A was ineffective in carrying out any transcription (Figs. 2a and 4c). We are not sure why P3 (TGATACCAT) was stronger than P2 (TGTTATGCT) and P1 (TGCTATGGT) since each of them contains an ex-10 element. Low levels of P3 RNA were also observed with many other substitutions: −16G, −19A, −20A, −21T, −25G, −25C, −25A, −27A, −27T, −29G, −29C, −29T, −31C, −32T, −32A, −32C, −33G, −34A, and −36T (Figs. 2–3 and 4c).
CCC effect
Previously, it was reported that the resurrected P3 transcription was normally repressed by CCC [16]. In every case that we observed, CCC repressed transcription from P3, including the very high level obtained in the −14G substitution.
Discussion
Our analysis of the E. coli gal promoters shows that the base pair frequency in building consensus sequences correlates well with the promoter function, and emphasizes the contribution of base pairs outside the previously defined elements. The results of intrinsic transcription for P1 and P2 reported here are summarized in Fig. 5a and 5b. Based on this study, and observations previously reported in the literature, we classify base pairs and their positions that i) are essential for gal promoter activities, ii) affect more than one promoter, and iii) were not part of any previously defined consensus regions but enhance activities of the promoters over the native base pairs. We also discuss the boundaries of each sequence element. In Fig. 5, the base pair identity is in red if it is uniquely essential for the promoter function, green if presence at a given position enhances promoter function compared to the native base pair, and black if there is a degeneracy—when the nature of the base pair at a position does not show a significant defect in promoter function. The hierarchy of bases in the latter case with respect to transcription efficiency, or the lack of it, is also indicated (Fig. 5). Because of limited information obtained for the P3 promoter, we discuss here only P1 and P2.
Features of P1 and P2
The tsp for P1 and P2 is an A but can be a G [35]. This is a property of RNA polymerase containing sigma-70 and true for most promoters. The upstream dscr region in the two promoters is neither G/C- nor A/T-rich, does not determine the efficiency of the two promoters, and is not homologous between them. The exceptions are that a −1C to an A change in P1, a −1T to C, and any change in −2T in P2 show 2-fold better intrinsic activities. Both promoters make abortive transcripts [33]. It is possible that the nature of the base pair at the −1 position in the two promoters is linked to abortive transcription, and changes may reduce abortive transcription and increase productive transcription.
Base pair substitutions in the −10 and ex-10 elements demonstrated that these two elements in gal follow the conventional rule showing the value of the critical bases previously established.
However, in the region upstream of the ex-10, from positions −19 to −16 in P1 and in P2, a −19TNTR−16 sequence makes the promoters better than the wild-type versions (−19GTTA−16 in P1 and −19TCTT−16 in P2). As mentioned before, in a few promoters, the −16 and −17 bases have been grouped with the ex-10 element and counted as one larger element including the −12T as −17TRTGNT−12 [41]. However, we do not know whether the −19TNTR−16 motif in gal constitutes a separate functional DNA element or is part of the ex-10 −15TG−14 element. Two previous observations that base pair substitutions at positions from −22 to −13 in other promoters enhance promoter function [69, 70], and our current findings, warrant a re-evaluation of the commonly held view that the immediate upstream region (the spacer region in −35 promoters) of the ex-10 element does not contribute to promoter activities.
Promoter overlap
Given the considerable overlap between P1 and P2 base pair sequences, it is remarkable that most base pair changes that affected one promoter did not affect the other; only specific changes at three positions (out of 25 positions tested) inactivated both P1 and P2: substitutions: −12T of P1 (−7 of P2), −16A of P1 (−11 of P2), and −17T of P1 (−12 of P2) affected (different) critical elements of the two promoters and thus made the DNA doubly defective (P1−P2−).
If we consider the fact that base pair substitutions at positions −1 and at regions −16 to −20 influence both promoter efficiencies, one can build a consensus sequence that extends from −1 to further upstream of ex-10 at least to the −17 position at both P1 and P2. By comparing similar features of the two gal promoters we derived a consensus sequence (Fig. 5c). It includes the feature that specific base pairs (absent in the wild type promoters) upstream of the ex-10 elements enhance promoter activities.
Interactions of DNA elements and RNA polymerase
The DNA elements needed to form an active promoter interact with segments of RNA polymerase subunits in a specific manner; this interaction creates kinetic outcomes for productive transcription initiation at a promoter. Since the gal promoters investigated here do not contain UP and −35 elements, we can only discuss the ex-10, −10, and potential new DNA elements that were clearly identified here by transcription assays.
Since the transcription initiation step is a multi-step process, the process obviously would need multiple contacts between RNA polymerase in a temporal fashion. Previous genetic, biochemical, and structural studies established several contacts between specific bases in a promoter and amino acid residues in the RNA polymerase for the transcription initiation step [13–15]. For an ex-10 promoter, it has been suggested that the -15TG-14 sequence directly participates in RNA polymerase binding [28–30]. The two bases are involved in formation of closed complex by contact with residues (H455 and E458) in the region 3.0 of the σ70 subunit of RNA polymerase in double stranded form [12, 15, 29, 31, 71]. No structure is known for a closed complex. The -12TATAAT-7 sequence is involved in isomerization. We previously showed by 2-AP fluorescence assays that strand separation occurs from −12 to the +3 region [48]. The latter step initiates melting (base unpairing and flipping) of the DNA by starting at the “master base” A at the −11 position that propagates from −12 to at least the +3 position (not necessarily in a zipper like fashion), followed by initiation of phosphodiester bonds formation in the presence of NTPs [50]. Similar results were obtained by KMnO4 cleavage experiments in gal P2 [36]. Our current result shows that strand propagation through a segment −6 to −1 does not show base pair stringency.
RNA polymerase contacts double stranded −15TG−14 DNA during the binding step, and mostly with non-template single strand (−12 to +3 region) during isomerization. The current structure of open complex identifies the amino acid-base contacts during the isomerization step. Since −12T of the −10 element interacts with residues Q437 and T440 in region 2.4 of the sigma70 subunit in a double stranded form, unlike the rest of the five bases in the −10 element, it has been suggested that the closed complex formation includes −12T [49, 72–75]. In other words, instead of the ex-10 −15TG−14 element, the entire segment −15TGNT−12 is needed to form closed complex [41, 76]. The segment, termed −15 element, is involved in RNAP binding [41]. However, studies of the rate of base pair unpairing/unstacking by the use of 2-aminopurine fluorescence release clearly showed that the base −11A, opposite to −12T, un-pairs from T and flips out [50, 52] . This is inconsistent with the idea that −12T is exclusively involved in RNAP binding and remains in double stranded form. We propose that −12T is involved first in binding in a double stranded form and then opens up in the isomerization process. The latter step may also destabilize the closed complex.
The base pairs in the −10 element in the non-template strand bind to a large number of amino acid residues in the σ70 region 2.3 in single stranded form when generating an open complex [14]. In open complex formation the single-strandedness propagates at least to the +3 region allowing the template strand to land at the active site for transcription initiation. The strand separation begins with the master base −11A and extend to +3 with −11A and −7T flipping out of the DNA helix [42, 45, 48, 50, 52, 77, 78]. Zhang et al. also show that −11A, −7T, −6G and +2G flipped out into hydrophobic pockets during isomerization [45]. It appears that strand separation may not be a simple “unzipping” of the DNA from the −12 to +3 region because bases in this region unpair/unstack at different rates without synchrony, at least at the galP1 promoter [48]. Given these facts, the molecular mechanism of closed to open complex formation is intriguing. Whatever the mechanism, base pair substitution studies show that the contribution of each base pair in the −10 element proceeds in the following order of significance (from most to least): −11 = −7 = −12 > −10 > −9 = - 8 with a master role played by −11A.
We underscore that our base pair replacement studies do not reflect any aspect of the role of base pairs in kinetics of transcription initiation at a promoter. The structural studies have not hinted so far any interaction between this region and RNAP, although Arg-35 in the β’ subunit of RNA polymerase is favorably located to contact the minor groove of the −22 to −18 sequence [12, 69]. It was also shown that the β’-zipper of RNAP interacts with the spacer between −35 and −10 promoting open complex formation, and the deletion of the β’-zipper abolished transcription [69]. Additionally, it has also been reported that the RNAP interaction with the −12T in the −10 element is strongly stimulated by RNA polymerase interactions with base pair between −10 and −35 bp.
cAMP-CRP Complex
The galP1 promoter is a biochemically well-characterized CCC dependent Class II promoter in which the regulatory complex binds to a DNA site centrally at position −41.5 and activates P1 [19, 21–26]. Three domains of CCC, AR1, AR2, and AR3, make three sets of contacts with specific domains of RNA polymerase holoenzyme to activate transcription from P1 [79–85]. For all of the defects in intrinsic transcription created by base substitutions as described above, many of them (the −11 substitutions are the exceptions) are well rectified by CCC. These results suggest that CCC can overcome defects both closed and open complex formation. This is consistent with biochemical and biophysical observations that CCC stimulates both closed and open complex formation in the galP1 promoter [38]. Although the AR2-αNTDI and σ 4.0 contacts help in the isomerization step, the contacts are physically far away from the site of isomerization—the −10 DNA element. Thus the AR2- αNTDI and AR3- σ 4.0 contacts in turn must influence the σ2.3 region that directly participates in base unpairing/flipping steps by allosteric mechanisms within RNAP. Such allosteric changes may initiate within CRP, by cAMP binding, and may be transmitted to RNA polymerase [86–89]. Our results showed that transcription defects due to substitution in the −10 element are rescued by CCC, except in the case of substituted master bases at position −11 in which the derivatives are not discernable. These results imply that CCC may not participate in the initial master base opening but at later step(s) in the isomerization process. Consistently, making the −10 element in P1 a consensus sequence makes the promoter very active, which is regarded as CCC independent [44]. The base substitution analysis shows that the global activator cAMP-CRP complex (CCC) helps the galP1 promoter both at the level of closed and open complex formation.
How CCC represses P2 is not fully understood with respect to CCC binding at the −36.5 position of the P2 promoter to bring about repression. One model assumes that CCC binding at the −41.5 position sterically excludes RNA polymerase binding at P2 [90], while another model suggests that CCC represses P2 transcription by inhibiting RNA polymerase at a post-binding level [23, 38] (D. Jin, personal communication). CCC does not inhibit P2 transcription by preventing RNAP binding; CCC binding partially overlaps the −35 region of P2, which does not have a functional −35 element.
In summary, we analyzed the importance of each base pair in the gal promoter region from −25 to +1 of P1. This region contains three promoters, P1, P2, and P3. Therefore, any base pair change can affect all three promoters. We found that CCC restored inactivated P1 promoter with base changes in the −10 and ex-10 elements. Substitution of critical bases at positions −7, −10, −12, −14, −15, −16, and −17 were inactivated in the absence of CCC, but were restored in the presence of CCC. The only exception was −11 substitutions, which were not activated by CCC, suggesting that −11A is essential for the initiation of P1 in the absence and presence of CCC. We also found that base changes at positions −7T, −11, −12, −19, −20, and −22 inactivated P2 activity. There were only three positions (−12, −16 and −17) where substitutions inactivated both promoters.
Materials and Methods
Reagents
In this study, restriction endonucleases were purchased from New England Biolabs, Inc. (Beverly, MA). T4 DNA ligase was obtained from Invitrogen (Carlsbad, CA). E. coli RNA polymerase holoenzyme (specific activity: 2.5 × 103 U/mg) was supplied by USB/Affymetrix, Inc. (Cleveland, OH). Recombinant RNasin Ribonuclease Inhibitor (40 U/µl) was obtained from Promega (Madison, WI). Denaturing polyacrylamide gel solutions (Sequal Gel-6) was from National Diagnostics, Inc. (Atlanta, GA). Primers were purchased from BioServe Biotechnologies (Beltsville, MD) and Sigma-Aldrich Genosys Life Science (Woodland, TX). Adenosine 3’:5’-cyclic monophosphate (cAMP) was from Sigma-Aldrich (St. Louis, MO). XL PCR and DNA sequencing (dRhodamine terminator cycle sequencing ready reaction) kits were from Applied Biosystems (Rockville, MD). [α- 32 P]UTP (specific activity = 3000 Ci/mmol, 10 µCi/µl) was obtained from MP Biomedical, LLC (Aurora, OH).
Plasmids
The plasmids used in this study are listed in Table S1. They are derivatives of wild type plasmid, pSA850, which was generated by cloning a 166-bp fragment containing the galactose regulatory region from −75 to +91 into pSA508 [34, 91, 92]. The mutant DNA templates were constructed by PCR amplifications using the XL PCR protocol from Applied Biosystems. Briefly, primer XbaI-2 (5’ ATACGACTCACTATAGGGAATTTCTAGACCTTCCCGTTTCGC 3’), which mapped from −180 to −139, and the reverse primer (containing mutated nucleotides; H: A+T+C, B: G+T+C, D: G+A+T or V: G+A+C) were used to amplify the left PCR product. To construct base pair substitutions at −5A (−7TTATTTCA+1) in the left PCR product for example, the mutated reverse primer would contain a V at that position to generate G, T, or C at −5. The forward primer (containing complementary mutated nucleotides, e.g., B at position −5A) and reverse primer Hind3-6 (5’ GTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGG 3’), which mapped downstream from +631 to +608, were used to generate the right PCR product. Both PCR products were purified on a 1% agarose gel, which was electrophoresed in 1x TAE (10 mM Tris acetate, pH8.0, 1 mM EDTA) buffer. Vertical gel slices were excised and stained in 0.5 µg/µl Ethidium Bromide solution to use as a marker to localize the unstained PCR products. The stained gel slices were aligned to the unstained gels, and the unstained PCR products were sliced from the gels based on the alignment of the stain products. The DNAs were eluted from the gel slices according to the protocol outlined in the QIAquick gel extraction kit from Qiagen (Clarita, CA). The left and right purified PCR products were mixed and amplified by the two external primers (XbaI-2 and Hind3-6). The extended PCR products were gel purified as above and digested with EcoRI and HindIII. First, the digested fragments were purified from the enzymes and buffered by QIAquick PCR purification kit and then ligated to a pSA850 vector, which was also digested with EcoRI and HindIII, and dephosphorylated with shrimp alkaline phosphatase. The recombinant plasmids were transformed into maximum efficiency E. coli DH5α□ competent cells (Invitrogen). Purification of the plasmids was performed according to the protocol outlined in the Qiagen plasmid Midi kit. The concentration of the plasmid DNAs was determined spectrophotometrically at 260 and 280 nm. The plasmid DNAs were sequenced by using the dRhodamine terminator cycle sequencing kit and the reactions were applied on an ABI Prism 310 Genetic Analyzer to verify the correct base pair changes at the desired position of the gal promoter region.
In vitro transcription assays
To measure the effect of promoter mutations on the strength of P1 and P2 in the presence and absence of CCC, in vitro transcription reactions were performed according to the method described previously [34]. A ρ-independent transcription terminator, rpoC, was located downstream of OI to generate transcripts of 125- and 130-nt from P1 and P2, respectively [34, 93]. Supercoiled DNA template (2 nM) was preincubated with RNA polymerase (20 nM) to form open complexes at 37°C for 5 min in transcription buffer (20 mM Tris acetate, pH 8.0, 10 mM Magnesium acetate, 200 mM Potassium glutamate, 1 mM DTT, 1 mM ATP, 0.8 U/µl rRNasin, and 100 µM cAMP) with or without CRP (50 nM) in a total reaction volume of 45 µl. To start the elongation process, 5 µl of NTPs mixture (0.1 mM GTP, 0.1 mM CTP, 0.01 mM UTP, and 5 µCi [α-32P]UTP) was added to each reaction, which was centrifuged for a few seconds to mix the reaction. Incubation of the reactions was continued for an additional 10 min at 37°C before they were terminated by the addition of an equal volume (50 µl) of loading dye (90% formamide, 10 mM EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue). Samples were heated to 90°C for 2–3 min, chilled on ice, and loaded on a warm 6% sequencing gel, which was pre-electrophoresed for ~45 min. The gel was electrophoresed for 1 hr 30 min at a constant power of 60–65 W. The gel was transferred to a Whatman 3MM Chromatography paper, wrapped in plastic wrap, and dried on a gel dryer at 80°C for ~1 hr.
Quantification of RNA transcripts
To quantify the relative amount of gal RNA transcripts observed on each template, the dry gel was exposed to a PhosphorImager screen and scanned on a PhosphorImager (Molecular Dynamics, GE, Sunnyvale, CA). To normalize the amount of transcript per lane, RNAI transcripts (106–108 nts) were used as an internal control to quantify the relative amount of gal transcripts [94]. The RNAI transcipts, which are transcribed from the origin of the plasmid, are not affected by CCC. After RNA normalization, the wild type P1 and P2 levels in the absence of CCC were taken as 1.0 (Tables S2 and S3). The amount of activation or repression of P1 and P2 in the mutant templates were relative to the basal level of wild type P1 and P2. For P3, the amount of transcript was very low in many of the templates; therefore, we plotted the absolute amount of transcripts without normalizing it to the wild type template since P3 was not observed (Table S4). Previously, we showed that only 2–3 rounds of replication were obtained by our in vitro transcription assays [36]. The transcription results were repeated two to three times to check the reproduciblity of the mutations on P1 and P2.
Supplementary Material
Research Highlights.
Base pair frequency of known consensus elements correlates with promoter strength.
RNAP binds to −12T:A as part of dsDNA before the entire −10 element melts to ssDNA.
CCC helps RNAP bind to P1 at the level of both closed and open complex formation.
We found three substitutions in the overlapped area inactivated both P1 and P2.
The −11 substitutions are the only changes where CCC could not activate P1.
Acknowledgements
We thank Maxim Soukhodolets and Mofang Liu for preparing cAMP receptor protein (CRP), and Debbie Hinton, Susan Garges and Stephen Busby for critical reading of the manuscript. The authors would like to thank Cindy Clark, NIH Library Writing Center, for manuscript editing assistance. This work was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research. The authors have no conflict of interest to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McClure WR. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlin MJ. The selectivity of transcription. Annu Rev Biochem. 1974;43:721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- 3.Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975;72:784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 5.Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 6.Pemberton IK, Muskhelishvili G, Travers AA, Buckle M. The G+C-rich discriminator region of the tyrT promoter antagonises the formation of stable preinitiation complexes. Journal of molecular biology. 2000;299:859–864. doi: 10.1006/jmbi.2000.3780. [DOI] [PubMed] [Google Scholar]
- 7.Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic acids research. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill MC. Escherichia coli promoters I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J Biol Chem. 1989;264:5522–5530. [PubMed] [Google Scholar]
- 10.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, et al. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 11.Gummesson B, Lovmar M, Nystrom T. A proximal promoter element required for positive transcriptional control by guanosine tetraphosphate and DksA protein during the stringent response. The Journal of biological chemistry. 2013;288:21055–21064. doi: 10.1074/jbc.M113.479998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of ranscription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 13.Ebright RH. RNA polymerase-DNA interaction: structures of intermediate, open, and elongation complexes. Cold Spring Harbor symposia on quantitative biology. 1998;63:11–20. doi: 10.1101/sqb.1998.63.11. [DOI] [PubMed] [Google Scholar]
- 14.Decker KB, Hinton DM. Transcription Regulation at the Core: Similarities Among Bacterial, Archaeal, and Eukaryotic RNA Polymerases. Annu Rev Microbiol. 2013 doi: 10.1146/annurev-micro-092412-155756. [DOI] [PubMed] [Google Scholar]
- 15.Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. J Biol Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponnambalam S, Spassky A, Busby S. Studies with the Escherichia coli galactose operon regulatory region carrying a point mutation that simultaneously inactivates the two overlapping promoters. Interactions with RNA polymerase and the cyclic AMP receptor protein. FEBS Lett. 1987;219:189–196. doi: 10.1016/0014-5793(87)81214-0. [DOI] [PubMed] [Google Scholar]
- 17.Johnston F, Ponnambalam S, Busby S. Binding of Escherichia coli RNA polymerase to a promoter carrying mutations that stop transcription initiation. Journal of molecular biology. 1987;195:745–748. doi: 10.1016/0022-2836(87)90194-x. [DOI] [PubMed] [Google Scholar]
- 18.Busby S, Aiba H, de Crombrugghe B. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. Journal of molecular biology. 1982;154:211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- 19.Musso RE, Di Lauro R, Adhya S, de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977;12:847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- 20.Hopkins JD. A new class of promoter mutations in the lactose operon of Escherichia coli. J Mol Biol. 1974;87:715–724. doi: 10.1016/0022-2836(74)90080-1. [DOI] [PubMed] [Google Scholar]
- 21.Adhya S, Miller W. Modulation of the two promoters of the galactose operon of Escherichia coli. Nature. 1979;279:492–494. doi: 10.1038/279492a0. [DOI] [PubMed] [Google Scholar]
- 22.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. The Journal of biological chemistry. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 23.Spassky A, Busby S, Buc H. On the action of the cyclic AMP-cyclic AMP receptor protein complex at the Escherichia coli lactose and galactose promoter regions. The EMBO journal. 1984;3:43–50. doi: 10.1002/j.1460-2075.1984.tb01759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi T, O’Neill M, de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979;76:5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 26.DiLauro R, Taniguchi T, Musso R, de Crombrugghe B. Unusual location and function of the operator in the Escherichia coli galactose operon. Nature. 1979;279:494–500. doi: 10.1038/279494a0. [DOI] [PubMed] [Google Scholar]
- 27.Sur R, Debnath D, Mukhopadhyay J, Parrack P. A novel RNA polymerase binding site upstream of the galactose promoter in Escherichia coli exhibits promoterlike activity. Eur J Biochem. 2001;268:2344–2350. doi: 10.1046/j.1432-1327.2001.02114.x. [DOI] [PubMed] [Google Scholar]
- 28.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 29.Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. The EMBO journal. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell JE, Zheng D, Busby SJ, Minchin SD. Identification and analysis of ‘extended −10’ promoters in Escherichia coli. Nucleic acids research. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bown JA, Owens JT, Meares CF, Fujita N, Ishihama A, Busby SJ, et al. Organization of open complexes at Escherichia coli promoters. Location of promoter DNA sites close to region 2.5 of the sigma70 subunit of RNA polymerase. The Journal of biological chemistry. 1999;274:2263–2270. doi: 10.1074/jbc.274.4.2263. [DOI] [PubMed] [Google Scholar]
- 32.Minchin S, Busby S. Location of close contacts between Escherichia coli RNA polymerase and guanine residues at promoters either with or without consensus −35 region sequences. Biochem J. 1993;289(Pt 3):771–775. doi: 10.1042/bj2890771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choy HE, Adhya S. RNA polymerase idling and clearance in gal promoters: use of supercoiled minicircle DNA template made in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis DE. Identification of promoters of Escherichia coli and phage in transcription section plasmid pSA850. Methods in enzymology. 2003;370:618–645. doi: 10.1016/s0076-6879(03)70052-4. [DOI] [PubMed] [Google Scholar]
- 35.Lewis DE, Adhya S. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Molecular microbiology. 2004;54:692–701. doi: 10.1111/j.1365-2958.2004.04318.x. [DOI] [PubMed] [Google Scholar]
- 36.Lewis DE, Komissarova N, Le P, Kashlev M, Adhya S. DNA sequences in gal operon override transcription elongation blocks. J Mol Biol. 2008;382:843–858. doi: 10.1016/j.jmb.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodrich JA, McClure WR. Regulation of open complex formation at the Escherichia coli galactose operon promoters. Simultaneous interaction of RNA polymerase, gal repressor and CAP/cAMP. Journal of molecular biology. 1992;224:15–29. doi: 10.1016/0022-2836(92)90573-3. [DOI] [PubMed] [Google Scholar]
- 38.Herbert M, Kolb A, Buc H. Overlapping promoters and their control in Escherichia coli: the gal case. Proc Natl Acad Sci U S A. 1986;83:2807–2811. doi: 10.1073/pnas.83.9.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu LM, Cobb IM, Ozmore JR, Khoo M, Nahm G, Xia L, et al. Initial transcribed sequence mutations specifically affect promoter escape properties. Biochemistry. 2006;45:8841–8854. doi: 10.1021/bi060247u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- 41.Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Bio. 2007;1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 42.Heyduk E, Heyduk T. Next generation sequencing-based parallel analysis of melting kinetics of 4096 variants of a bacterial promoter. Biochemistry. 2014;53:282–292. doi: 10.1021/bi401277w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mekler V, Severinov K. Cooperativity and interaction energy threshold effects in recognition of the −10 promoter element by bacterial RNA polymerase. Nucleic acids research. 2013;41:7276–7285. doi: 10.1093/nar/gkt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhnke G, Krause A, Heibach C, Gieske U, Fritz HJ, Ehring R. The upstream operator of the Escherichia coli galactose operon is sufficient for repression of transcription initiated at the cyclic AMP-stimulated promoter. The EMBO journal. 1986;5:167–173. doi: 10.1002/j.1460-2075.1986.tb04192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, et al. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Molecular cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 47.deHaseth PL, Helmann JD. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Molecular microbiology. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 48.Roy S, Lim HM, Liu M, Adhya S. Asynchronous basepair openings in transcription initiation: CRP enhances the rate-limiting step. The EMBO journal. 2004;23:869–875. doi: 10.1038/sj.emboj.7600098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasse-Dwight S, Gralla JD. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. The Journal of biological chemistry. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 50.Lim HM, Lee HJ, Roy S, Adhya S. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HJ, Lim HM, Adhya S. An unsubstituted C2 hydrogen of adenine is critical and sufficient at the −11 position of a promoter to signal base pair deformation. The Journal of biological chemistry. 2004;279:16899–16902. doi: 10.1074/jbc.C400054200. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder LA, Gries TJ, Saecker RM, Record MT, Jr, Harris ME, DeHaseth PL. Evidence for a tyrosine-adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. Journal of molecular biology. 2009;385:339–349. doi: 10.1016/j.jmb.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Busby S, Irani M, Crombrugghe B. Isolation of mutant promoters in the Escherichia coli galactose operon using local mutagenesis on cloned DNA fragments. Journal of molecular biology. 1982;154:197–209. doi: 10.1016/0022-2836(82)90060-2. [DOI] [PubMed] [Google Scholar]
- 54.Busby S, Spassky A, Chan B. RNA polymerase makes important contacts upstream from base pair −49 at the Escherichia coli galactose operon P1 promoter. Gene. 1987;53:145–152. doi: 10.1016/0378-1119(87)90002-3. [DOI] [PubMed] [Google Scholar]
- 55.Bingham AH, Ponnambalam S, Chan B, Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41:67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- 56.Kolb A, Kotlarz D, Kusano S, Ishihama A. Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 1995;23:819–826. doi: 10.1093/nar/23.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burr T, Mitchell J, Kolb A, Minchin S, Busby S. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic acids research. 2000;28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Busby S, Truelle N, Spassky A, Dreyfus M, Buc H. The selection and characterisation of two novel mutations in the overlapping promoters of the Escherichia coli galactose operon. Gene. 1984;28:201–209. doi: 10.1016/0378-1119(84)90257-9. [DOI] [PubMed] [Google Scholar]
- 59.Chan B, Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989;84:227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- 60.Moran CP, Jr, Lang N, LeGrice SF, Lee G, Stephens M, Sonenshein AL, et al. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 61.Helmann JD. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voskuil MI, Chambliss GH. The TRTGn motif stabilizes the transcription initiation open complex. J Mol Biol. 2002;322:521–532. doi: 10.1016/s0022-2836(02)00802-1. [DOI] [PubMed] [Google Scholar]
- 63.Chan B, Minchin S, Busby S. Unwinding of duplex DNA during transcription initiation at the Escherichia coli galactose operon overlapping promoters. FEBS Lett. 1990;267:46–50. doi: 10.1016/0014-5793(90)80284-p. [DOI] [PubMed] [Google Scholar]
- 64.Turnbough CL., Jr Regulation of gene expression by reiterative transcription. Curr Opin Microbiol. 14:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin DJ. Slippage synthesis at the galP2 promoter of Escherichia coli and its regulation by UTP concentration and cAMP.cAMP receptor protein. J Biol Chem. 1994;269:17221–17227. [PubMed] [Google Scholar]
- 66.Miroslavova NS, Busby SJ. Investigations of the modular structure of bacterial promoters. Biochem Soc Symp. 2006:1–10. doi: 10.1042/bss0730001. [DOI] [PubMed] [Google Scholar]
- 67.Siebenlist U, Simpson RB, Gilbert WE. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 68.Csiszovszki Z, Lewis DE, Le P, Sneppen K, Semsey S. Specific contacts of the - 35 region of the galP1 promoter by RNA polymerase inhibit GalR-mediated DNA looping repression. Nucleic Acids Res. 2012;40:10064–10072. doi: 10.1093/nar/gks796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuzenkova Y, Tadigotla VR, Severinov K, Zenkin N. A new basal promoter element recognized by RNA polymerase core enzyme. The EMBO journal. 2011;30:3766–3775. doi: 10.1038/emboj.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh SS, Typas A, Hengge R, Grainger DC. Escherichia coli sigma(7)(0) senses sequence and conformation of the promoter spacer region. Nucleic Acids Res. 2011;39:5109–5118. doi: 10.1093/nar/gkr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanderson A, Mitchell JE, Minchin SD, Busby SJ. Substitutions in the Escherichia coli RNA polymerase sigma70 factor that affect recognition of extended - 10 elements at promoters. FEBS Lett. 2003;544:199–205. doi: 10.1016/s0014-5793(03)00500-3. [DOI] [PubMed] [Google Scholar]
- 72.Malhotra A, Severinova E, Darst SA. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 73.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waldburger C, Gardella T, Wong R, Susskind MM. Changes in conserved region 2 of Escherichia coli sigma 70 affecting promoter recognition. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- 75.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 76.Djordjevic M. Redefining Escherichia coli sigma(70) promoter elements: −15 motif as a complement of the −10 motif. J Bacteriol. 2011;193:6305–6314. doi: 10.1128/JB.05947-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qiu J, Helmann JD. Adenines at −11, −9 and −8 play a key role in the binding of Bacillus subtilis Esigma(A) RNA polymerase to −10 region single-stranded DNA. Nucleic Acids Res. 1999;27:4541–4546. doi: 10.1093/nar/27.23.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ebright RH. Transcription activation at Class I CAP-dependent promoters. Molecular microbiology. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 80.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) Journal of molecular biology. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 81.Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, et al. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. The EMBO journal. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou Y, Zhang X, Ebright RH. Identification of the activating region of catabolite gene activator protein (CAP): isolation and characterization of mutants of CAP specifically defective in transcription activation. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6081–6085. doi: 10.1073/pnas.90.13.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, et al. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 85.Busby S, Ebright RH. Transcription activation at class II CAP-dependent promoters. Molecular microbiology. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 86.Adhya S, Ryu S, Garges S. Role of allosteric changes in cyclic AMP receptor protein function. Subcell Biochem. 1995;24:303–321. doi: 10.1007/978-1-4899-1727-0_10. [DOI] [PubMed] [Google Scholar]
- 87.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 88.Garges S, Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985;41:745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- 89.Ryu S, Kim J, Adhya S, Garges S. Pivotal role of amino acid at position 138 in the allosteric hinge reorientation of cAMP receptor protein. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:75–79. doi: 10.1073/pnas.90.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shanblatt SH, Revzin A. Interactions of the catabolite activator protein (CAP) at the galactose lactose promoters of Escherichia coli probed by hydroxyl radical footprinting. The second CAP molecule which binds at gal and the one CAP at lac may act to stimulate transcription in the same way. J Biol Chem. 1987;262:11422–11427. [PubMed] [Google Scholar]
- 91.Choy HE, Adhya S. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lewis DE, Adhya S. In vitro repression of the gal promoters by GalR and HU depends on the proper helical phasing of the two operators. The Journal of biological chemistry. 2002;277:2498–2504. doi: 10.1074/jbc.M108456200. [DOI] [PubMed] [Google Scholar]
- 93.Squires C, Krainer A, Barry G, Shen WF, Squires CL. Nucleotide sequence at the end of the gene for the RNA polymerase beta’ subunit (rpoC) Nucleic acids research. 1981;9:6827–6840. doi: 10.1093/nar/9.24.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tomizawa J, Itoh T, Selzer G, Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.