Abstract
Immune checkpoint inhibitors1 result in impressive clinical responses2–5 but optimal results will require combination with each other6 and other therapies. This raises fundamental questions about mechanisms of non-redundancy and resistance. Here, we report major tumor regressions in a subset of patients with metastatic melanoma treated with an anti-CTLA4 antibody (anti-CTLA4) and radiation (RT) and reproduced this effect in mouse models. Although combined treatment improved responses in irradiated and unirradiated tumors, resistance was common. Unbiased analyses of mice revealed that resistance was due to upregulation of PD-L1 on melanoma cells and associated with T cell exhaustion. Accordingly, optimal response in melanoma and other cancer types requires RT, anti-CTLA4, and anti-PD-L1/PD-1. Anti-CTLA4 predominantly inhibits T regulatory cells (Tregs) to increase the CD8 T cell to Treg (CD8/Treg) ratio. RT enhances the diversity of the T cell receptor (TCR) repertoire of intratumoral T cells. Together, anti-CTLA4 promotes expansion of T cells, while RT shapes the TCR repertoire of the expanded peripheral clones. Addition of PD-L1 blockade reverses T cell exhaustion to mitigate depression in the CD8/Treg ratio and further encourages oligo-clonal T cell expansion. Similar to results from mice, patients on our clinical trial with melanoma showing high PD-L1 did not respond to RT + anti-CTLA4, demonstrated persistent T cell exhaustion, and rapidly progressed. Thus, PD-L1 on melanoma cells allows tumors to escape anti-CTLA4-based therapy, and the combination of RT, anti-CTLA4, and anti-PD-L1 promotes response and immunity through distinct mechanisms.
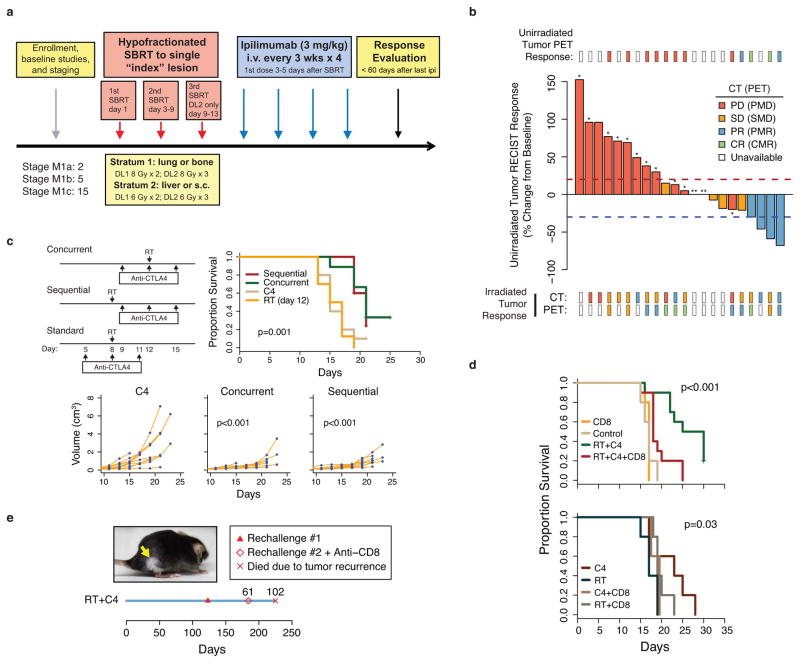
Anecdotal clinical reports suggest that RT may cooperate with anti-CTLA4 to systemically enhance melanoma response7; however, this combination has not been reported in a clinical trial. To examine the feasibility and efficacy of RT combined with immune checkpoint blockade, we initiated a phase I clinical trial of 22 patients with multiple melanoma metastases (Extended Data Table 1). A single index lesion was irradiated with hypofractionated RT, delivered over two or three fractions, followed by four cycles of the anti-CTLA4 antibody ipilimumab (Extended Data Fig. 1a). Accrual was completed in three out of four RT dose levels, and treatment was well tolerated (Extended Data Table 2). Evaluation of the unirradiated lesions by CT imaging using Response Evaluation Criteria in Solid Tumors (RECIST) demonstrated that 18% of patients had a partial response (PR) as best response, 18% had stable disease (SD), and 64% had progressive disease (PD) (Fig. 1a). For example, patient PT-402 showed a large reduction in sizes of unirradiated tumors and a partial metabolic response by positron emission tomography (PET) (Fig. 1b). None of the 12 patients evaluated by PET had progressive metabolic disease in the irradiated lesion (Extended Data Fig. 1b, Extended Data Table 3). The median progression-free survival (PFS) and overall survival (OS) was 3.8 and 10.7 months with median follow-up of 18.4 and 21.3 months (18.0 and 21.3 for patients without event), respectively (Fig. 1c).
Figure 1. RT + anti-CTLA4 promotes regression of irradiated and unirradiated tumors and is inhibited by PD-L1 on tumor cells.
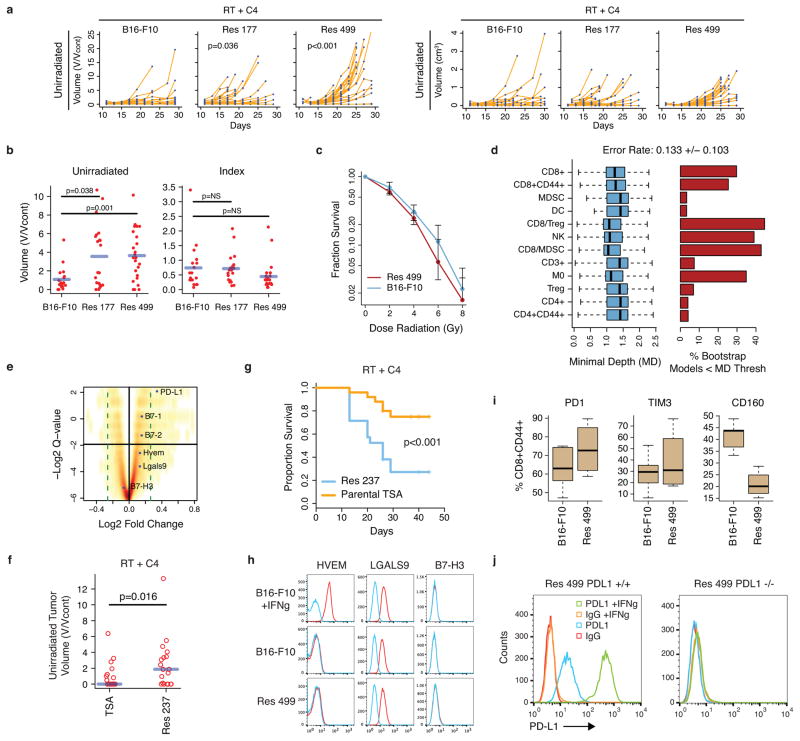
a) Waterfall plot of unirradiated tumors after RT to a single index lesion with anti-CTLA4. Dashed lines are thresholds for PD (red) and PR (blue). * Patients with new lesions. ** Clinical progression without imaging. b) PET/CT images of irradiated (white arrows) and unirradiated (yellow arrows) tumors from patient PT-402. c) PFS and OS for all patients (dashed lines: 95% CI). d) B16-F10 tumor growth after RT to the index tumor (n=8), anti-CTLA4 (C4) (n=9), anti-CTLA4 and RT to the index tumor (n=18), or no (control) treatment (n=9). The p-values are comparisons with control. Pie chart shows %CRs (yellow). See Fig. 2d for survival. e) Heat map showing relative abundance of immune cells or their ratios from tumors that are resistant (black hatch) or sensitive to RT + anti-CTLA4. Boxplot shows bootstrap importance scores for each variable. Higher values (red) are more predictive. f) Change in T cell subsets or their ratio after RT + anti-CTLA4 for sensitive parental (Sen) or resistant (Res) tumors. Values are subtracted from average of untreated controls. Red line is mean. g) Heat map of resistance gene signature and PD-L1 across human melanoma. p < 0.001 by gene set enrichment analysis. h) Expression of PD-L1 on Res 499 compared to B16-F10 melanoma cells and of Res 237 compared to TSA breast cancer cells. Isotype control (IgG). i) Total tumor volume from PD-L1 knockout (KO) or control (WT) Res 499 and corresponding survival.
Although responses were observed, the majority of patients in our trial did not respond. To understand the contribution of RT to immune checkpoint blockade and to discover mechanisms of resistance, we utilized the B16-F10 melanoma mouse model. Mice with bilateral flank tumors received anti-CTLA4, irradiation of one tumor (index) using a micro-irradiator, or both treatments delivered concurrently (Fig. 1d). The best responses in both tumors occurred with RT + anti-CTLA4. RT given before or concurrently with CTLA4 blockade yielded similar results (Extended Data Fig. 1c). Complete responses (CRs) were CD8 T cell-dependent, and mice with CRs also exhibited CD8 T cell-dependent immunity to tumor re-challenge (Extended Data Fig. 1d–e). However, similar to our clinical trial, only approximately 17% of mice responded. To better understand determinants of response, we derived cell lines from unirradiated tumors that relapsed after RT + anti-CTLA4 (Res 499 and Res 177). Resistance was confirmed in vivo and was not due to intrinsic RT resistance (Extended Data Fig. 2a–c). Random forest (RF) machine learning analysis8,9 of tumor infiltrating lymphocytes (TILs) demonstrated that the top predictor of resistance, as measured by variable importance scores and selection, was the CD8+CD44+ to Treg (CD8/Treg) ratio (Fig. 1e, Extended Data Fig. 2d). In resistant tumors, the CD8/Treg ratio failed to increase after RT + anti-CTLA4 as it did in sensitive tumors because CD8+CD44+ T cells did not significantly expand despite reduction in Tregs (Fig. 1f). Other immune variables associated with resistance were also related to the failure to accumulate CD8 TILs.
The prevalence of CD8 TILs can be blunted by mechanisms that interfere with T cell function. Transcriptomic profiling of Res 499/177 tumors revealed that PD-L1 was among the top 0.2% of upregulated genes that make up a RT + anti-CTLA4 “resistance gene signature” (Extended Data Fig. 2e, SI Table 1). Other genes include interferon stimulated genes, which may promote immune suppression through PD-L110,11. Similarly, PD-L1 was co-expressed with the resistance signature in tumors from a previously reported12 cohort of metastatic melanoma patients (Fig. 1g). This increase in PD-L1 was observed on melanoma cells devoid of contaminating stromal cells, and a comparable increase was similarly seen in the Res 237 murine breast cancer cells (Fig. 1h), which was selected from the TSA line for resistance to RT + anti-CTLA4 (Extended Data Fig. 2f–g). In contrast, expression of other inhibitory receptors and their ligands nominated by gene profiling did not suggest an obvious role in resistance (Extended Data Fig. 2h–i). Indeed, genetic elimination of PD-L1 on Res 499 cells by CRISPR (Extended Data Fig. 2j) restored response to RT + anti-CTLA4 by increasing survival from 0% to 60% (Fig. 1i). Thus, an increase in PD-L1 on tumor cells observed in multiple cancer types can be a dominant resistance mechanism to RT + anti-CTLA4.
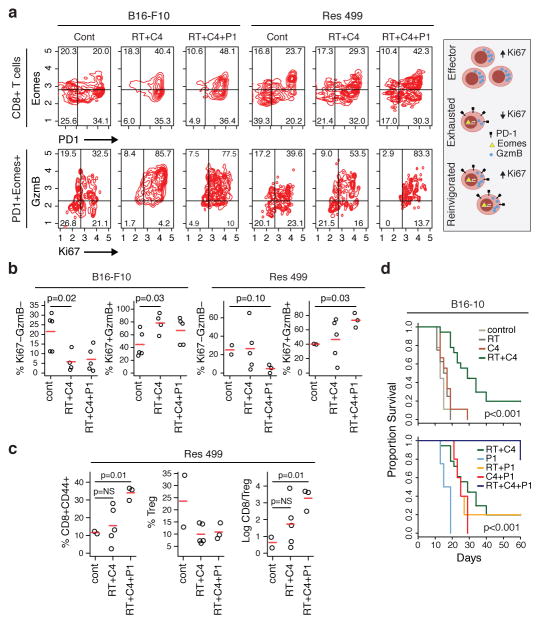
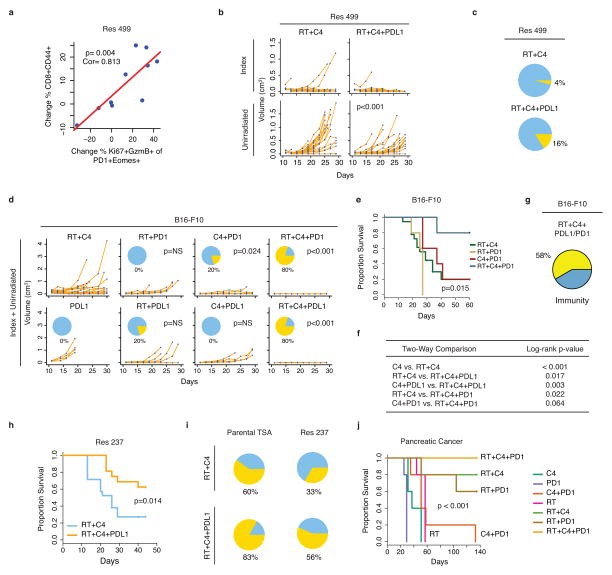
Elevated levels of PD-L1 can promote T cell exhaustion, a state characterized by dysfunction in T cell proliferation and effector function13. Exhausted T cells co-express the PD-L1 receptor PD-1 and the transcription factor Eomes14. Reversal of exhaustion, known as reinvigoration, is marked by an increase in the proliferation marker Ki67 and the cytotoxic protein GzmB within the exhausted T cell pool. In both untreated parental and resistant tumors, approximately 20% of CD8 TILs co-expressed PD-1 and Eomes, and only a minority of these cells were Ki67+GzmB+, indicating that a significant fraction was exhausted (Fig. 2a–b). In B16-F10 tumors, RT + anti-CTLA4 markedly increased both the proportion of PD-1+Eomes+ CD8 T cells and the proportion that were Ki67+GzmB+ within this subset. In contrast, in resistant tumors the average proportion of PD-1+Eomes+ T cells that were Ki67+GzmB+ only marginally increased after RT + anti-CTLA4; however, addition of anti-PD-L1 increased this to levels observed in parental tumors treated with only RT + anti-CTLA4. The frequency of CD8+CD44+ TILs and the CD8/Treg ratio also increased (Fig. 2c), and these were strongly correlated with the proportion of PD-1+Eomes+ CD8 TILs that were Ki67+GzmB+ (Extended Data Fig. 3a). Importantly, addition of anti-PD-L1 improved responses of resistant Res 499 tumors after RT + anti-CTLA4 (Extended Data Fig. 3b–c). For treatment naïve tumors, responses were even more dramatic as the addition of either anti-PD-L1 or anti-PD-1 to RT + anti-CTLA4 markedly improved survival and increased CRs to 80% (Fig. 2d, Extended Data Fig. 3d–f). On average, 58% of mice with CRs after adding anti-PD-L1 or anti-PD-1 were alive 90+ days after tumor rechallenge, and similar improvements were observed with Res 237 breast cancer tumors after addition of PD-L1 blockade (Extended Data Fig. 3g–i). Thus, elevated PD-L1 on tumor cells results in persistent T cell exhaustion that impairs the CD8/Treg ratio. Addition of PD-L1 blockade inhibits resistance and results in long-term immunity.
Figure 2. Addition of PD-L1 blockade reinvigorates exhausted T cells and improves response to RT + anti-CTLA4.
a) Representative contour plot of CD8 TILs from B16-F10 or Res 499 tumors after RT and anti-CTLA4 (C4) +/− anti-PD-L1 (P1) examined for PD-1 and Eomes (top row), followed by examination of the PD-1+Eomes+ subset for Ki67 and GzmB (bottom row). Schema shows exhaustion and reinvigoration markers. b) Proportion of PD-1+Eomes+ CD8 T cells that are either Ki67−GzmB− or Ki67+GzmB+. c) Changes in T cell subsets and their ratio from Res 499 tumors. d) Survival of mice with B16-F10 tumors (n=18 for RT+C4, n=5 for others). Shown are overall p-values.
Notably, RT is needed to achieve high CR rates as dual checkpoint blockade proved inferior to dual checkpoint blockade plus RT (Fig. 2d), a requirement additionally seen in a pancreatic cancer model (Extended Data Fig. 3j). The superiority of triple therapy in multiple cancer types suggests non-redundant mechanisms for each treatment. To examine this notion, we assessed treatment-related changes in TILs from unirradiated tumors. RF modeling of immune cell profiles confirmed that anti-CTLA4 predominantly caused a decrease in Tregs, anti-PD-L1 strongly increased CD8 TIL frequency, and the blockade of both increased the CD8/Treg ratio (Fig. 3a–b, Extended Data Fig. 4a). In contrast, RT caused only a modest increase in CD8 TILs; however, TCR sequencing revealed that this was accompanied by increased diversity of TCR clonotypes, which could be observed even in the presence of CTLA4 blockade (Fig. 3c–3d). Thus, within the tumor microenvironment, CTLA4 blockade primarily decreases Tregs, PD-L1 blockade predominantly reinvigorates exhausted CD8 TILs, and RT diversifies the TCR repertoire of TILs from unirradiated tumors.
Figure 3. RT, anti-CTLA4, and anti-PD-L1 have distinct effects on the TCR repertoire, Tregs, and T cell exhaustion.
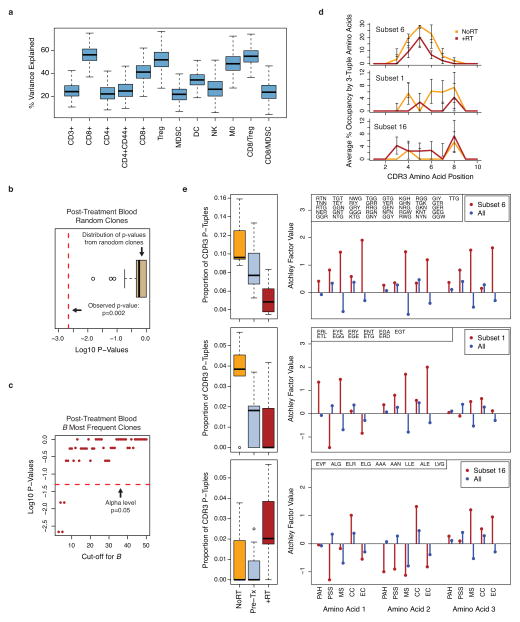
a) Heat map of changes in the frequency of immune cells or their ratios from B16-F10 tumors. Black hatches indicate treatment. Bar plots show bootstrap importance scores (mean +/− SE) that assess changes in immune parameters predicted by treatment type (read row-wise). Higher values (yellow) represent stronger association. b) T cell subsets and their ratios. c) Frequency distribution (dashed line is 0.5%) and d) boxplot of diversity index (0: clonal, 1: fully diverse) for most frequent TCR clonotypes found in TILs of unirradiated B16-F10 tumors after RT and/or anti-CTLA4. Boxplot summarizes data for mice treated with anti-CTLA4 (NoRT) or RT +/− anti-CTLA4 (+RT). e) Representative contour plots and f) ratios examining PD-1+Eomes+ splenic CD8 T cells from mice with B16-F10 tumors for Ki67+GzmB+ (reinvigorated) or Ki67−GzmB− (exhausted) subsets. g) TCR clonal frequency in post-treatment blood vs. TILs (top row) or vs. pre-treatment blood (bottom row). Quadrant boundaries are top 5% quantiles from the control. Clones below detection in pre-treatment blood are assigned upper bounds (blue). h) Maximum clonal frequency in post-treatment blood (dot) of the most frequent TCR clonotypes found in TILs. i) Distances to cluster centroids for the average CDR3 amino acid features of the five most frequent clones in pre- or post-treatment blood from mice treated with (red) or without (orange) RT. Membership into two clusters (circles and squares) determined by k-means.
To investigate if treatment effects on TILs were propagated to the peripheral T cell pool, we examined spleen and blood. As observed in TILs, RT + anti-CTLA4 reinvigorated exhausted PD-1+Eomes+ splenic CD8 T cells, and this reinvigoration was further enhanced by addition of anti-PD-L1 (Fig. 3e–f). Reinvigoration after addition of anti-PD-L1 was also accompanied by a large expansion of a small subset of the top 100 most frequent TCR clonotypes found in TILs (Fig. 3g). Remarkably, some clones reached a frequency in the post-treatment blood of over 20% after RT and dual checkpoint blockade (Fig. 3h). With anti-CTLA4 +/− RT, peripheral T cell clonal expansion was modest, which parallels the low CR rates following this treatment. RT alone was insufficient to drive peripheral T cell expansion, despite increasing TCR repertoire diversity of TILs, but did promote qualitative alterations in the TCR repertoire of the most expanded clonotypes. Unsupervised analysis using the average CDR3 amino acid features15,16 demonstrated that the TCRs of the most frequent clonotypes in the post-treatment blood formed two readily apparent clusters based on RT treatment (Fig. 3i). In contrast, the most frequent clonotypes from pre-treatment blood and randomly sampled clonotypes from post-treatment blood did not separate into clusters, consistent with differences in CDR3 amino acid properties being an effect of RT only observed in the most expanded clones (Extended Data Fig. 4b–c). The separation into two clusters was driven by differences in the CDR3 occupancy profile of short amino acid sequences belonging to distinct subsets differing in size, polarity, and electrostatic charge (Extended Data Fig. 4d–e). Together, these observations suggest that the favorable immune changes in TILs after immune checkpoint blockade promote their peripheral clonal expansion. When combined with increased TCR repertoire diversity afforded by RT, selection and oligo-clonal peripheral expansion of clones with distinct TCR traits are favored.
To determine if treatment and resistance-related changes in peripheral T cells can constitute a biomarker for tumor response, we modeled the effects of reinvigoration, exhaustion, and the CD8/Treg ratio. Specifically, we used 1) the percent PD-1+ splenic CD8 T cells that are Eomes+ to integrate the burden that exhausted T cells might exert, 2) the percent PD-1+ CD8 T cells that are Ki67+GzmB+ as a measure of potential reinvigoration, and 3) the CD8/Treg ratio as a barometer for the suppressive potential of Tregs. The overall prediction accuracy of the model was 84%, and variables for T cell reinvigoration and exhaustion were the most predictive, followed by the CD8/Treg ratio (Extended Data Fig. 5a–b). Moreover, the percentage of PD-1+ CD8 T cells that were Eomes+ was a striking modifier of the likelihood of CR as nearly all observed CRs occurred when the percent Ki67+GzmB+ in PD-1+ CD8 T cells was high but the relative size of the PD-1+Eomes+ exhausted population was not (Fig. 4a). Similar relationships existed with the CD8/Treg ratio, and prediction using T cells from peripheral blood yielded highly similar results (Extended Data Fig. 5c–e). In total, immune parameters from peripheral T cells that relate the size of the exhausted T cell population, reinvigoration, and the CD8/Treg ratio can predict response to RT combined with immune checkpoint blockade.
Figure 4. Tumor PD-L1 and T cell exhaustion and reinvigoration can predict response in mice and patients.
a) Percent PD-1+ CD8 T cells that are Eomes+ vs. Ki67+GzmB+ after RT combined with checkpoint blockade. Values are subtracted from average of untreated controls. Each circle represents a mouse. Probability of CR (proportional to circle size), prediction error rate, and quadrant boundaries are estimated from an RF model. b) Percent Eomes+PD-1+ CD8 T cells in post-treatment blood vs. change in % PD-1+Eomes+ CD8 T cells that are Ki67+GzmB+ after treatment. Each circle represents a patient. PFS is proportional to circle size and quadrant boundaries are average values for patients under the mean PFS. Concordance index of the RF model is 0.59. c) Contour plot of peripheral blood CD8 T cells from patients PT-102 and PT-402 examined for PD-1 and Eomes (top row), followed by examination of the PD-1+Eomes+ subset for Ki67 and GzmB (bottom row). d) PD-L1 staining from corresponding tumor biopsies. e) Change in % Ki67+GzmB+ in PD-1+Eomes+ CD8 T cells vs. PD-L1 status of melanoma cells from all patients with available pre- and post-treatment blood. f) RECIST response, g) PFS, and OS stratified by PD-L1 status of melanoma cells.
In order to assess whether immune predictors discovered in mice could be shared with patients, we examined peripheral T cells and tumor biopsies from patients on our clinical trial of RT + anti-CTLA4. For all 10 patients with available pre- and post-treatment blood, two had PRs in unirradiated tumors and PFS significantly longer than the median. For both of these patients, the percentages of Ki67+GzmB+ increased in PD-1+Eomes+ CD8 T cells after treatment while the proportion of PD-1+Eomes+ T cells remained at or below the mean (Fig. 4b). In contrast, patients with a high percentage of PD-1+Eomes+ T cells post-treatment did not have PRs and had a short PFS, regardless of reinvigoration. Comparison of patient PT-402, who had extended PFS/PR (Fig. 1a–b), with patient PT-102, who had short PFS/PD, demonstrates how reinvigoration is associated with response to RT + anti-CTLA4 as it is in mice (Fig. 4c vs. Extended Fig. 5f–g and Fig. 3e–f). Examination of pre-treatment tumor biopsies from patients PT-402 and PT-102 (Fig. 4d), and from all patients with available biopsy (Extended Data Table 4), revealed that PD-L1lo intensity on melanoma cells (Extended Data Fig. 6a) was associated with reinvigoration of PD-1+Eomes+ and of PD-1+ CD8 T cells after RT + anti-CTLA4, while PD-L1hi status was associated with persistent exhaustion (Fig. 4e, Extended Data Fig. 6b). None of the patients with PD-L1hi on melanoma cells had a CR/PR, and all rapidly progressed and died (Fig. 4f–g). PD-L1 status on macrophages was neither associated with reinvigoration nor independently predictive of PFS (Extended Data Fig. 6c–d). Thus, collective results from patients and mice suggest that elevated PD-L1 on melanoma cells inhibits T cell function and tumor response to RT + anti-CTLA4.
We investigated RT + anti-CTLA4 in mice and patients to understand mechanisms of both response and resistance (Extended Data Fig. 6e). Anti-CTLA4 predominantly inhibits Tregs, increasing the CD8/Treg ratio as previously described17, and results in modest peripheral expansion of TCR clonotypes in the tumor, also consistent with other reports18,19. RT diversifies the TCR repertoire of TILs and shapes the repertoire of expanded clones. Although the cause and consequence of these repertoire changes remain to be defined, RT can alter peptide presentation20, and CDR3 changes after M. tuberculosis infection have been hypothesized to be antigen-driven15. Resistance to RT + anti-CTLA4 can ensue due to elevated PD-L1 on cancer cells driving T cell exhaustion, a process that can be antagonized by PD-L1 blockade. However, severely exhausted T cells may regain only limited function after reinvigoration13,14, explaining why the correlation between reinvigoration and response declines when the exhausted T cell pool is large. Although tumors with genetic elimination of PD-L1 in melanoma cells can still relapse, suggesting resistance through other pathways and/or PD-L1 on non-tumor cells, the upregulation of PD-L1 by cancer cells is a dominant resistance mechanism in our models. Moreover, the shared findings between mice and patients predict that addition of PD-L1/PD-1 blockade to RT + anti-CTLA4 may show significant efficacy in clinical trials.
METHODS
Clinical trial patients and study design
The clinical protocol was registered on clinicaltrials.gov (NCT01497808). Eligible patients were at least 18 years of age with previously treated or untreated stage IV melanoma with multiple metastasis. Patients were required to have an Eastern Cooperative Oncology Group performance status of 0 or 1, adequate renal, hepatic, and hematological function, no current or history of CNS metastasis, no prior radiation that precludes use of stereotactic body radiation (SBRT), and at least one tumor between 1–5 cm that could be treated with SBRT. The primary objectives of this phase I study were to determine feasibility, dose-limiting toxicities (DLT) and maximum tolerated SBRT fraction when given in conjunction with ipilimumab. The secondary objectives were to determine late toxicity, immune-related clinical responses and changes. The study treated successive cohorts of patients with escalating doses of SBRT to a single tumor (index lesion), followed 3–5 days later by ipilimumab every three weeks for four doses. Moderate RT doses were used since higher RT dose has not been clearly correlated with better immune response but would likely increase toxicity. Patients were stratified into two stratum based on treatment site (lung or bone vs. liver or subcutaneous) and dose escalation of SBRT were determined as follows: For lung/bone lesion, dose level 1 (DL1) was 8 Gy x 2; dose level 2 (DL2) was 8 Gy x 3; and for liver/subcutaneous lesion, DL1 was 6 Gy x 2; DL2 was 6 Gy x 3. The study followed a “treat six” design with the goal of accruing 6 patients to each dose level, or 24 patients total. Enrollment to a dose level would stop if 2 or more patients had a DLT. If 0–1 patients out of the 6 had a DLT at DL1, escalation to DL2 would proceed. No DLTs were observed, defined by the protocol as any treatment-related grade 4 or higher immune-related toxicity (NCI CTC Version 4.0) or grade 3 or higher non-immune related toxicity experienced during study treatment or within 30 days after the last injection of ipilimumab. Pre- and post-treatment blood, CT, and PET/CT were obtained to follow tumor response and assess immune responses. Response evaluation by imaging was performed within 60 days of the last ipilimumab treatment using either RECIST v1.121 or PERCIST. The study protocol was approved by the University of Pennsylvania institutional review board. All participating patients provided written informed consent.
Cell lines and tissue culture
B16-F10 was purchased from ATCC. TSA was a gift from Sandra Demaria. PDA.4662 cell line was derived from single cell suspensions of PDA tissue from LSL-KrasLSL-G12D/+,LSL-p53LSL-R172H/+,Pdx1-Cre mice as previously described22. B16-F10 and PDA.4662 cell lines were cultured at 37° C in DMEM and TSA cells were cultured at 37° C in RPMI. Media was supplemented with 10% FBS, 100 U/ml penicillin and 100 ug/ml streptomycin, 2 mM L-glutamine. All cell lines were determined to be free of Mycoplasma (Lonza) and common mouse pathogens (IDEXX).
In vivo mouse studies
Five to seven week old female C57BL/6 and BALB/c mice were obtained from NCI Production (Frederick, MD) and Jackson Laboratory (Bar Harbor, ME) and maintained under pathogen free conditions. All animal experiments were performed according to protocols approved by the Institute of Animal Care and Use Committee of the University of Pennsylvania. For B16-F10 melanoma, 5×104 B16-F10 cells were mixed with an equal volume of Matrigel (BD Biosciences) and subcutaneously injected on the right flank of C57BL/6 mice on day 0 and the left flank on day 2. The right flank tumor site was irradiated with 20 Gy on day 8. Blocking antibodies were given on days 5, 8 and 11. For the concurrent vs. sequential RT experiment, the right flank was irradiated on either day 8 (sequential) or 12 (concurrent), while blocking antibodies were given on days 9, 12, and 15. For TSA breast cancer, 1×105 TSA cells were mixed with an equal volume of Matrigel (BD Biosciences) and subcutaneously injected on the right flank of BALB/c on day 0 and the left flank on day 2. The right flank of the mice was irradiated with 8 Gy on three consecutive days starting on day 10 or 11 post tumor implantation. Blocking antibodies were started 3 days prior to RT and given every 3 days for a total of 3 doses. For the pancreatic cancer model, 4×105 PDA.4662 cells were subcutaneously injected on the right flank. The right flank was irradiated with 20 Gy on day 8. Blocking antibodies were given on days 5, 8, and 11. For melanoma and breast cancer models, we used the optimal dose and fraction of radiation as previously reported23,24. All irradiation was performed using the Small Animal Radiation Research Platform (SARRP). Antibodies used for in vivo immune checkpoint blockade experiments were given intraperitoneally at a dose of 200 μg/mouse and include: CTLA4 (9H10), PD-1 (RMP1-14), PDL-1 (10F.9G2), CD8 (2.43), and rat IgG2B isotype (LTF-2) (BioXCell). Anti-CD8 was given 2 days prior to tumor implantations (day −2), day 0, then every 4 days for the duration of the experiment. Perpendicular tumor diameters were measured using calipers. Volume was calculated using the formula L × W2 × 0.52, where L is the longest dimension and W is the perpendicular dimension.
Survival and tumor response analysis
Differences in survival were determined for each group by the Kaplan-Meier method and the overall p-value was calculated by the log-rank test using the “survival” R package version 2.37+. For mouse studies, an event was defined as death or when tumor burden reached a protocol-specified size of 1.5 cm in maximum dimension to minimize morbidity. To help control for differences in treatment response due to experimental variation or intrinsic growth differences with sublines, tumor volume measurements were also analyzed after normalizing to the average volumes of untreated control mice. These average untreated tumor volumes were determined at day 11–12, a time when tumor dimensions could be accurately measured, and was considered a baseline tumor volume (Vcont). Normalized tumor response to treatment is the measured volume (V) relative to Vcont, or V/Vcont, a dimensionless value that is relative to a baseline volume. Measurements from different experiments separated by 1–2 days were binned. Differences in growth curves were determined by a mixed effect linear model with normalized data using the “lmerTest” R package version 2.0. Sample size estimations were based on preliminary pilot experiments. For control mice, we expected an average tumor volume of 0.4 cm3 at day 17–21. For most experiments, we assumed the treatment group would have an effect size resulting in a 50% reduction in average tumor volume. Sigma was estimated to be 1.5. For a 0.80 power at the 0.05 alpha level, this gave us a sample size of 5 mice. Mice were randomly assigned a treatment group. For experiments whereby the effect size was expected to be small and/or non-robust, two independent researchers with at least one researcher blinded to the treatment group assignments performed caliper measurements.
Flow cytometry
For flow cytometric analysis of in vivo experiments, blood, spleen, and tumor were harvested at either day 16 or 18 post tumor implantation. Single cell suspensions were prepared and red blood cells were lysed using ACK Lysis Buffer (Life Technologies). Live/dead cell discrimination was performed using Live/Dead Fixable Aqua Dead Cell Stain Kit (Life Technologies) or Sytox Red Dead Cell Stain (Life Technologies). Cell surface staining was done for 20–30 minutes. Intracellular staining was done using a fixation/permeabilization kit (00-5521-00, eBioscience.) T effector cells were phenotyped as CD8+CD44+, myeloid derived suppressor cells (MDSC) as CD11b+Gr-1+, and regulatory T cells (Tregs) as CD4+FOXP3+. All flow cytometric analysis was done using an LSR II (BD) or FACSCalibur (BD) and analyzed using FlowJo software (TreeStar) or the FlowCore package in the R language and environment for statistical computing. See Supplementary Methods for a list of antibodies used.
CRISPR gene targeting
Gene targeting by CRISPR/Cas9 was accomplished by co-transfection of a Cas9 plasmid (Addgene, 56503), the guide sequence (selected using ZiFit Targeter) cloned into the gBlock plasmid, and a plasmid with the puromycin selection marker. Successful targeting of PD-L1 was determined by flow cytometry screening of clones treated with and without 100 ng/mL of IFN-gamma (PeproTech). Confirmed clones were pooled. Clones without knockout were also pooled and used as controls. See Supplementary Methods for gRNA sequences.
Immunohistochemistry for PD-L1
Formalin-fixed, paraffin-embedded tumors were collected at the time of surgical resection or from biopsy. All patients with available recent biopsy, which was optional for trial enrollment, were used for analysis. After heat-induced antigen retrieval (Bond ER2, 20 min.), the tumor slides were stained with an anti-PD-L1 antibody (E1L3N, Cell Signaling) at 1:50 dilution. Intensity of staining on a 0–3+ scale, the percent tumor cells or macrophages with positive staining, and the cellular pattern (membrane vs. cytoplasm) were analyzed by two pathologists. Samples with membrane PD-L1 staining intensity score of 0–1 were classified as PD-L1lo, and samples with an intensity score of 2+ in at least 1% of the cells were classified as PD-L1hi. To confirm specificity, the anti-PD-L1 antibody was validated by staining Hodgkin’s lymphoma cells25 and placenta26.
Statistical methods and software
Computational analysis and predictions were performed using the R language and environment for statistical computing (version 3.0+) and Bioconductor (version 2.22+). The significance of all two-way comparisons was determined by two-sample, two-tailed t-test. An F-test was used to test for equal variance and a Shapiro-Wilks test was used to test for normality. For non-parametric data, a Wilcoxon test was used. A mixed effect linear model was used to determine significance of differences in tumor growth. Simple correlation between variables was done using a Pearson’s correlation. Unless noted, samples were independent biological replicates.
Microarray data processing and normalization
Total RNA was isolated and purified from cells using Isol-RNA Lysis Reagent (Fisher.) Total RNA from tumors was isolated and purified from frozen specimens using Isol-RNA Lysis Reagent and Qiagen RNAeasy extraction kit with DNAase I on column treatment. Labeled RNA was hybridized to the Affymetrix GeneChip Mouse Gene 1.0 and 2.0 ST Array. Affymetrix CEL files for all samples were processed using the RMA method as implemented in the “oligo” R package version 1.26.6. Probe annotations were provided by the “mogene10sttranscriptcluster.db” and “mogene20sttranscriptcluster.db” R package version 8.0.1 and 2.13.0, respectively. Since different array types and different batches were used, each expression set was z-score transformed27 and median centered. Multiple probes for the same gene were averaged and only genes common to the 1.0 and 2.0 ST arrays were kept. Batch effects were adjusted using the ComBat method as implemented in the “sva” R package version 3.8.0. The microarray data has been deposited at the GEO (GSE65503) and processed data provided as SI Table 2. Gene expression data for primary melanoma samples were downloaded from the GEO (GSE22155). For this data set, the post-processed data and provided annotations were used.
Determining differentially expressed genes and enriched gene sets
Non-specific filtering was used to remove genes with an interquartile range less than 0.05. To find differentially expressed genes (DEGs) between parental sensitive and resistant tumors, Significance Analysis of Microarray28 (“samr” R package version 2.0) was applied using a two class unpaired comparison, minimal fold change of 1.2, and median false discovery rate (FDR) of 0.05. Unannotated transcripts were not considered. To test whether gene sets were enriched in response to different conditions, we utilized Gene Set Analysis as implemented in the “GSA” R package version 1.0329. The “maxmean” test statistic was used to test enrichment using a two-class comparison. All p-values and false discovery rates were based on 500–1000 permutations. For restandardization, a method that combines randomization and permutation to correct permutation values of the test statistic and to take into account the overall distribution of individual test statistics, the entire data set was used rather than only the genes in the gene sets tested.
Flow cytometry data processing
Gating was performed using either FlowJo version 9.7.5 or the FlowCore R package version 1.28.24. For computational modeling, values were normalized by subtracting the average values of untreated controls. For the CD8/Treg ratio, the percent CD8+CD44+ cells were divided by the percent CD4+FOXP3+ cells. Because these data could be skewed with varying and wide distributions, these data were log transformed for downstream analysis.
Random forest for classification and survival analysis
Random forest (RF) for classification, regression, and survival analysis is a multivariable non-parametric ensemble partitioning tree method that can be used to model the effect of all interactions between genes on a response variable30,31. Each model was constructed using approximately two-thirds of randomly selected samples and cross-validated on the one-third of the samples left out of the model building process (out-of-bag samples). After many iterations, results of all models were averaged to provide unbiased estimates of predicted values, error rates, and measures of variable importance. Performance of an RF-RC model was measured by the misclassification error rate for classification, mean squared error for regression, and by a concordance index (one minus the error rate) for survival. For each variable, an importance score was determined, which measures the contribution of the variable to the error rate (higher scores are more predictive). When multiple response variables were modeled, as in the case of determining which treatment predicts changes in a set of immune parameters, treatment groups were converted to a design matrix and importance scores were determined for each response variable. We used the “randomForestSRCM” R package version 1.2 implementation32 and the following parameters: 1000 trees, node size of 1, mtry values equal to the number of variables in the model, and the Breiman-Cutler permutation method for importance score determination9. Gini index splitting rule was used for classification and a log-rank slitting rule was used for survival analysis. For classification, stratified sampling was used when the number of samples in each class was imbalanced. All predicted values, error rates, and importance scores were calculated using out-of-bag samples to provide unbiased estimates. To account for variance due to sample size and sampling error on the accuracy of these performance measures, bootstrapping was performed using 1000–5000 bootstrap iterations and the mean and standard deviation of the bootstrap distribution were determined. For presentation purposes, cut-off values for predictive variables were determined by using partial plots to estimate inflection points.
Minimal depth was used as a rigorous method to select predictive variables. Minimal depth (MD) is a dimensionless statistic that we have recently described8 that measures the predictiveness of a variable in tree-based models. Specifically, MD measures the shortest distance from the root node of a classification/regression tree to the parent node of a maximal subtree for a variable. The maximal subtree for a variable is the largest subtree whose root node splits on the variable. Thus, smaller values for MD indicate better predictiveness. A threshold value for MD that is calculated from the tree-averaged value determines whether a variable is strongly predictive. The entire MD-based variable selection is performed using two-third of the samples (in-bag samples). An unbiased prediction error rate for a model refit with the MD-selected variables is calculated using only out-of-bag samples. Using the “randomForestSRCM” package, we applied this MD-based variable selection with the same parameters used for RF as noted above. The tree-averaged MD threshold was used. Data were bootstrapped to provide robust estimates of MD values and prediction error rates. The frequency of bootstrap models whereby the MD values for a variable was less than the MD threshold determined how often a variable was selected as a top variable, which provides an estimate for the stability of variable selection.
TCR deep sequencing and clonotype diversity analysis
DNA from pre-treatment blood, post-treatment blood, and tumor was extracted on day 16 using the Qiagen DNA extraction protocol. Samples were sequenced by Adaptive Biotechnologies using “survey” sequencing depth for tumor and “deep” sequencing depth for blood samples. Processed data were downloaded and frequencies/counts for TCR clonotypes were examined by nucleotide sequences after non-productive reads were filtered out. The top 100 most frequent TCR clonotypes in the tumor were used to examine their frequencies in the pre- and post-treatment blood. The Shannon’s diversity index33 (DI) normalized to the number of reads (DI = −Σ (pi ln pi)/ln n, where n is the number of clones, pi is the clonal frequency of the ith clone, and sigma is summed from i=1 to i=n) was calculated for each sample. This gives a value between 0 and 1, where 0 is monoclonal and 1 is an even distribution of different clones.
Unsupervised and supervised analysis of CDR3 amino acid properties
Based on previously described methods15,16, Atchley factors were used to reduce a linear sequence of amino acids into analyzable numeric features of distinct amino acid properties. The five Atchley factors and the attributes they measure are: 1) PAH: accessibility, polarity, and hydrophobicity, 2) PSS: propensity for secondary structure, 3) MS: molecular size, 4) CC: codon composition, 5) EC: electrostatic charge. Each CDR3 was represented as a set of all possible contiguous amino acids of length p (p-tuple). We chose p=3 based on previous published reports but examined a range of p values, which gave comparable results (see below). For each p-tuple, the Atchley factors for the amino acids were then calculated to give a vector of length 5p, or 15 (3 amino acids x 5 Atchley factors). Thus, each CDR3 was represented by a set of these vectors. The average values for these vectors were calculated for the top B most frequent clones from the post-treatment blood. A cut-off of B=5 was chosen based on examination of the frequency distribution of the TCR clonotypes and an estimate of the number of clones with extreme values compared to the rest of the distribution. These averaged values were then clustered into two groups by k-means clustering with k=2. The association between cluster membership and treatment with or without RT was calculated by Fisher’s exact test. This entire process was repeated for the five clones in the pre-treatment blood, for randomly drawn clones from the post-treatment blood, for p-tuple lengths from p=2 to 10, and for cut-off values from B=3 to 50. In all cases, the distribution of p-values was compared to the p-value from the observed data.
Although averaging the Atchley factor values is a simple method to agglomerate CDR3 features for unsupervised classification, it does not provide insight into how treatment groups influence the amino acids that comprise the CDR3. To understand which sets of p-tuples were most strongly influenced by treatment groups with RT, without RT, and pre-treatment blood, we used previously described methods15 to assign p-tuples into n clusters based on their Atchley factor vector. Model based clustering with cluster number determination using the “mclust” R package was applied to all p-tuples from the top five clones in all treatment groups from pre- and post-treatment blood. This gave rise to 17 clusters, or subsets, of p-tuples. The proportion of p-tuples belonging to each of these 17 subsets, denoted Pi, was then calculated for each clonotype and used as features. The subsets that were most influenced by treatment group (treatment group with RT, without RT, or pre-treatment) were then analyzed by multivariable RF regression using a design matrix for treatment groups as the x-variable and Pi as the response variable. The variables Pi most affected by each treatment group were selected by comparing the observed importance scores to the importance scores generated by permutation. To determine the location and frequencies of amino acids belonging to the selected p-tuple subsets across the variable length CDR3 region, the CDR3 of each clone was divided into 10 bins of equal size. Then, the proportion of p-tuples in each of these 10 bins belonging to the selected subset was calculated and compared between treatment groups.
Extended Data
Extended Data Figure 1. Patients and mice treated with RT + anti-CTLA4 for melanoma.
a) Twenty-two stage IV melanoma patients (M stage indicated) were stratified by treatment site of a single index metastasis, which was the irradiated tumor. Two dosing levels (DL) for stereotactic body radiation (SBRT) were in each stratum. b) Waterfall plot of the RECIST % change from baseline of unirradiated tumors annotated to indicate metabolic responses by PET/CT (hatches above plot) and response of the irradiated index tumor as measured by CT and PET/CT (hatches below plot). RECIST criteria do not include irradiated tumors. Legend shows color-codes for response after CT or PET/CT (parenthesis). PMD: progressive metabolic disease; SMD: stable metabolic disease; PMR: partial metabolic response; CMR: complete metabolic response. White hatches indicate no imaging obtained. See Extended Data Table 2. c) Survival (right) and total tumor growth (bottom) after RT with either concurrent or sequential anti-CTLA4 compared to anti-CTLA4 (C4) or RT alone. The regimens and the standard regimen used for all other melanoma experiments are illustrated (left). The p-values for tumor growth are compared to anti-CTLA4. d) Survival after RT and/or anti-CTLA4 with or without T cell depletion (n=5–10) using anti-CD8 (CD8). Shown are overall p-values. The p-value for RT + anti-CTLA4 with and without anti-CD8 is p=0.005. Control is an isotype-matched antibody. e) Three mice with CRs were rechallenged with B16-F10 tumors. Shown is a representative mouse. Arrow indicates location of regressed tumor and vitiligo-like condition represented by non-pigmented fur (observed in approximately 50% of mice with CRs). Time line starts from original tumor implantation (day 0) and values above marks are days after first rechallenge. Recurrence occurred only after anti-CD8 treatment and second rechallenge.
Extended Data Figure 2. Tumor cells resistant to RT + anti-CTLA4 upregulate PD-L1 but not other candidate inhibitory receptor pathways.
a) Unirradiated tumor growth (left: normalized, right: raw values) for mice implanted with Res 177 (n=21), Res 499 (n=25), and B16-F10 (n=18) melanoma cells and treated with RT + anti-CTLA4. For normalization, volumes were divided by average of untreated controls (V/Vcont) to account for differences in growth between untreated tumor types. The p-values are for comparisons with B16-F10 tumors. b) Corresponding tumor volumes of unirradiated or irradiated index tumors at day 21 (blue line is mean). c) Clonogenic survival for Res 499 and B16-F10 cells (n=2). d) Selection of immune variables that robustly predict resistance to RT + anti-CTLA4 using minimal depth (MD). A variable was selected if its MD was less than a threshold value for significance. Shown are bootstrap distributions of MD values (left) and % bootstrap models for which the MD for the indicated variable was significant (right). Bootstrap mean +/− SD for the out-of-bag prediction error rate is listed on top. e) Volcano plot of differentially expressed genes from resistant tumors. Horizontal black line is 5% false-discovery rate and dotted green line is fold-change cut-off. Ligands for select inhibitory receptors are indicated. See SI Table 1. f) Unirradiated tumor volumes (day 26–29) and g) survival after RT + anti-CTLA4 for mice with bilateral tumors from TSA breast cancer cells (n=25) or from the Res 237 subline selected to be resistant (n=21). h) Expression of candidate T cell inhibitory receptor ligands on B16-F10 and Res 499. Interferon-gamma (IFNg) responsiveness was tested. i) Boxplots show distribution of % positive CD8+CD44+ T cells for the indicated inhibitory receptor compared to IgG control. j) PD-L1 surface expression for CRISPR PD-L1 homozygous knockout Res 499 and wild type control cells. IFNg was used to induce PD-L1 and confirm abrogated response.
Extended Data Figure 3. Addition of PD-L1/PD-1 blockade antagonizes resistance to RT + anti-CTLA4, and optimal response to checkpoint blockade requires RT.
a) Change in % CD8+CD44+ T cells after RT and checkpoint blockade vs. change in the degree of reinvigoration of exhausted T cells measured by % PD-1+Eomes+ T cells that are Ki67+GzmB+. Values are subtracted from average of untreated control. b) Growth of Res 499 tumors after RT + anti-CTLA4 (C4) with and without addition of anti-PD-L1 (PDL1). Shown are index and unirradiated tumors from n=25 mice in each group. The p-value is for comparison to RT + anti-CTLA4. c) Proportion of CRs (yellow) for mice with Res 499 tumors. d) Total tumor growth (index + unirradiated) for B16-F10 tumors after the indicated treatment that includes anti-PD-1 (PD1) or anti-PD-L1. The p-values are for comparisons to RT + anti-CTLA4 (n=18, n=5 for others). Pie charts show % CRs (yellow). e) Survival of mice after RT + anti-CTLA4 + anti-PD-1. Shown is the overall p-value, and f) the two-way comparisons that include those from Fig. 2d. g) Proportion of mice with CRs (yellow) after RT + anti-PD-L1 or anti-PD-1 that survived 90+ days after tumor rechallenge at day 60 (n=12). h) Survival of mice with bilateral Res 237 breast cancer tumors treated with RT + anti-CTLA4 with (n=16) or without (n=21) anti-PD-L1. i) Proportion of CRs (yellow) for mice with Res 237 or TSA breast cancer tumors. j) Survival of mice with pancreatic tumors from a cell line derived from KPC mice (KrasLSL-G12D/+;p53LSL-R172H/+;Pdx-1-Cre) (n=5 for each group). Select treatment groups are labeled on the plot for clarity. Overall p-value is shown.
Extended Data Figure 4. TCR clonotypes associated with RT are not observed in random clones from post-treatment blood and have distinct CDR3 features.
a) Boxplot of the bootstrap variance explained by multivariable RF regression model for effect of RT, anti-CTLA4, and/or anti-PD-L1 on immune variables from TILs. b) K-means clustering (k=2) was used on the average CDR3 amino acid features of randomly sampled clones from post-treatment blood after anti-CTLA4, anti-PD-L1, and/or RT. Membership into each cluster was determined and the p-value for separation into treatment groups with and without RT was calculated. Boxplot shows log10 p-values from 1000 random iterations. Comparison to the p-value from the observed data (red dotted line) gives a simulated p < 0.001. c) Log10 p-values for separation into treatment groups with and without RT vs. cut-off value used to select the most frequent clones. The 0.05 significance level is indicated (red dotted line). d) Average % occupancy in the CDR3 of the most frequent T cell clonotypes after RT +/− checkpoint blockade (+RT, red line) or checkpoint blockade alone (NoRT, orange line) by contiguous short amino acid sequences of length three (3-tuples) belonging to e) subsets with distinct treatment-related amino acid properties. These properties are characterized by Atchley factors, which measure 1) PAH: accessibility, polarity, and hydrophobicity, 2) PSS: propensity for secondary structure, 3) MS: molecular size, 4) CC: codon composition, and 5) EC: electrostatic charge. Shown (right) are the average values of each Atchley factor for amino acids that comprise the 3-tuples from the indicated subset (red) compared to all unselected 3-tuples (blue). Boxplots (left) show the proportion of 3-tuples from each of these subsets that are found in the CDR3s of the five most frequent clones after treatment. Compared to pre-treatment samples (Pre-tx), subset 6 is associated with RT +/− checkpoint blockade (+RT) or checkpoint blockade alone (NoRT). Subset 1 is primarily associated with checkpoint blockade alone, and subset 16 is primarily associated with RT +/− checkpoint blockade.
Extended Data Figure 5. Peripheral T cell exhaustion, reinvigoration, CD8/Treg ratio, and tumor PD-L1 predict response to RT + immune checkpoint blockade.
a) Heat map showing the relative proportions of PD-1+ CD8 T cells that are Ki67+GzmB+ or Eomes+ and the CD8/Treg ratio for each sample (columns) subtracted from the average values of untreated controls. Black hatches indicated CR and treatment with RT + anti-CTLA4 (C4) +/− anti-PD-L1 (P1). From these data, a multivariable RF predictor for CR was developed. Boxplot shows bootstrap distributions of variable importance scores (more predictive variables have higher values), and of b) minimal depth (MD), a statistic to measure predictiveness. Bar plot shows % bootstrap models for which the MD for the indicated variable was significant. Bootstrap mean +/− SD for the out-of-bag prediction error rate is listed on top. c) Probability of CR vs. change (treated vs. untreated control) in CD8/Treg ratio for mice with a high (blue dots) or low (red dots) change in % PD1+ splenic CD8 T cells that are Eomes+. d) Heat map similar to (a) except using T cells from peripheral blood. e) Percent peripheral blood PD-1+ CD8 T cells that are Eomes+ vs. Ki67+GzmB+ after RT + checkpoint blockade. Values are subtracted from average of untreated controls. Each circle represents a mouse. Probability of CR (proportional to circle size), prediction error rate, and quadrant boundaries are estimated from the RF model. f) Representative contour plots examining splenic CD8 T cells from B16-F10 or Res 499 tumors for PD-1 and Eomes (top), followed by examination of the PD-1+Eomes+ subset for Ki67 and GzmB (bottom). g) Ratios of PD-1+Eomes+ splenic CD8 T cells that are Ki67+GzmB+ (reinvigorated) compared to Ki67−GzmB− (exhausted) from mice with Res 499 tumors.
Extended Data Figure 6. Melanoma PD-L1 is associated with T cell exhaustion, response, and survival for patients treated on clinical trial of RT + anti-CTLA4.
a) Representative images (right) for patients with biopsies showing PD-L1 staining on tumor cells classified as PD-L1lo (top), 2+ (middle), or 3+ (bottom). Scores of 2+ and 3+ are classified as PD-L1hi. The arrow indicates PD-L1 staining on macrophages. An isotype antibody negative control and positive controls are shown (left). b) Changes in % Ki67+GzmB+ in PD-1+ CD8 T cells after RT + anti-CTLA4 vs. PD-L1 status on melanoma cells from all patients with available pre- and post-treatment blood. c) Changes in % Ki67+GzmB+ in PD1+Eomes+ CD8 T cells (left) or in PD1+ CD8 T cells (right) vs. macrophage PD-L1 status. d) Hazard ratio and 95% CI for PFS from a Cox regression model using PD-L1 status on tumor cells and macrophages. e) Model for non-redundant mechanisms and resistance to RT and immune checkpoint blockade.
Extended Data Table 1.
Demographics and baseline characteristics for patients on phase I clinical trial of RT + anti-CTLA4 for metastatic melanoma.
| Characteristic | n=22 |
|---|---|
| Age | |
| 18–44 | 0 |
| 45–64 | 8 |
| 65+ | 14 |
| Sex | |
| Male | 17 |
| Female | 5 |
| ECOG performance status | |
| 0 | 12 |
| 1 | 10 |
| Disease status | |
| M1a | 2 |
| M1b | 5 |
| M1c | 15 |
| Prior radiation therapy | 3 |
| Prior systemic therapy | |
| 0 | 11 |
| 1 | 6 |
| 2 | 4 |
| 3+ | 1 |
| Prior immunotherapy | 1 |
Extended Data Table 2.
Grade 3 and 4 toxicities from phase I clinical trial of RT + anti-CTLA4 for metastatic melanoma.
Two dose levels in two strata were tested. Stratum 1 (lung/bone) utilized 8 Gy x 2 or 8 Gy x 3. Stratum 2 (liver/subcutaneous) utilized 6 Gy x 2 or 6 Gy x 3. Six patients for each dose level were planned. All dose levels met accrual except 8 Gy x 3 prior to trial closure.
| Radiation Dose | |||||
|---|---|---|---|---|---|
|
| |||||
| Grade 3 Toxicities* | 6Gy x 2 | 8Gy x 2 | 6Gy x 3 | 8Gy x 3 | |
| n=6 | n=6 | n=6 | n=4 | Total | |
| Edema | 1 | 1 | |||
| Anaphylaxis | 1 | 1 | |||
| Hypotension | 1 | 1 | |||
| Fatigue | 1 | 1 | |||
| Anemia | 2 | 1 | 1 | 4 | |
| Gastric hemorrhage | 1 | 1 | |||
| Wound infection | 1 | 1 | |||
| Diarrhea | 1 | 1 | |||
| Cholecystitis | 1 | 1 | |||
| Weight loss | 1 | 1 | |||
| Colitis | 1 | 1 | |||
| Pneumothorax | 1 | 1 | |||
No Grade 4 toxicities were observed
Extended Data Table 3.
Stratum, irradiated sites, and response for patients on clinical trial.
Response of the local irradiated site (Local) and distant unirradiated sites (Dist) were determined by CT and PET/CT. Percent change from baseline for distant lesions measured by CT are indicated using RECIST and change measured by PET/CT are indicated by PERCIST. The irradiated tumor was not included in RECIST measurements per RECIST guidelines due to radiation-related effects precluding accurate CT measurements (e.g., patient ID 1 and 9). NA indicates the value was not measurable based on criteria. Progression of disease due to new lesion(s) prior to re-imaging (POD New) or due to clinical progression (POD Clin) is also indicated. PD: progression of disease, PR: partial response, SD: stable disease, CMR: complete metabolic response, PMR: partial metabolic response, PMD: progressive metabolic disease. Patient ID 3 is patient PT-102, and patient ID 4 is patient PT-402.
| ID | Strat | Irradiated Site | RECIST | PERCIST | POD New | POD Clin | Local CT | Local PET | Dist CT | Dist PET |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lung | Rt lower lung | 15% | NA | no | no | PD | CMR | SD | PMD |
| 2 | Sub-Q | Lt upper abdomen | no | yes | ||||||
| 3 | Lung | Lung | 13% | 6% | yes | no | PR | CMR | PD | PMD |
| 4 | Sub-Q | Rt gluteal region | −68% | −76% | no | no | PR | PMR | PR | PMR |
| 5 | Sub-Q | Lt axilla | 30% | 93% | yes | no | SD | PMR | PD | PMD |
| 6 | Lung | Rt middle lung | 96% | no | no | PD | PD | |||
| 7 | Lung | Lt middle lung | 96% | yes | no | PD | PD | |||
| 8 | Lung | Rt middle lung | 5% | −14% | yes | no | SD | CMR | PD | PMD |
| 9 | Lung | Rt middle lung | −20% | 0% | yes | no | PD | PMR | PD | PMD |
| 10 | Liver | Rt lateral liver | −50% | −100% | no | no | SD | CMR | PR | CMR |
| 11 | Liver | Posterior liver | 77% | 42% | yes | no | SD | SMD | PD | PMD |
| 12 | Sub-Q | Lt posterior abdomen | 49% | yes | no | PR | PD | |||
| 13 | Lung | Rt posterior lung | no | yes | ||||||
| 14 | Sub-Q | Lymph node | 69% | NA | yes | no | SD | SMD | PD | PMD |
| 15 | Sub-Q | Lt axilla | −59% | −100% | no | no | SD | SMD | PR | CMR |
| 16 | Lung | Lt lung hilum | −49% | no | no | PR | PR | |||
| 17 | Sub-Q | Lt axilla | 38% | 39% | yes | no | SD | PMR | PD | PMD |
| 18 | Sub-Q | Rt inguinal region | −21% | −25% | no | no | SD | PMR | SD | PMR |
| 19 | Liver | Middle liver | 71% | yes | no | SD | PD | |||
| 20 | Lung | Rt middle lung | −19% | no | no | CR | SD | |||
| 21 | Sub-Q | Lt SCV node | 153% | yes | no | PR | PD | |||
| 22 | Lung | Lt lower lobe | −7% | no | no | PR | SD |
Extended Data Table 4.
Melanoma biopsy sites and PD-L1(hi) status of melanoma cells from patients on clinical trial.
Recent biopsy was optional for enrollment on the clinical trial. Tumor tissue from all patients with recent biopsy was used. PD-L1 status was determined by examination of membrane staining on melanoma cells. PD-L1(hi) was classified as 2+ on at least 1% of cells. Patient ID 3 is patient PT-102, and patient ID 4 is patient PT-402.
| ID | Location | PD-L1(hi) |
|---|---|---|
| 2 | Skin (presumed) | yes |
| 3 | Skin | yes |
| 4 | Bowel | no |
| 7 | Skin (presumed) | yes |
| 8 | Stomach | no |
| 11 | Liver | no |
| 12 | Lymph node | yes |
| 13 | Lymph node | no |
| 15 | Skin | no |
| 17 | Skin | no |
| 18 | Skin (presumed) | no |
| 19 | Bowel | no |
Supplementary Material
Acknowledgments
C.T. was supported by an NIH training grant and career development award (T32DK007066, KL2TR000139). K.E.P. was supported by a Robertson Foundation/Cancer Research Institute Irvington Fellowship, T.C.G. and R.R. by the Melanoma Research Alliance, and X.X. and M.D.F by a grant from the NIH (P50CA174523). B.X. and A.J.M. were supported by the Basser Research Center for BRCA. A.J.M. is a Department of Defense Era of Hope Scholar (W81XWH-09-1-0339) and was supported by funding from the NIH/NCI (R01CA172651). H.I. and A.J.M. were supported by a grant from the NIH (R01CA163739). R.H.V. was supported by grants from the NIH (R01CA158186, P30CA016520) and by the Abramson Cancer Center Translational Center of Excellence in Pancreatic Cancer. E.J.W. was supported by funding from NIH (U19AI082630, R01AI105343, U01AI095608 and P01AI112521). The project was supported in part by the Institute for Translational Medicine and Therapeutics’ Transdisciplinary Program in Translational Medicine and Therapeutics, and the National Center for Research Resources (UL1RR024134).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions. C.T. and A.J.R. designed, performed, and/or analyzed mouse and in vitro experiments. H.D. assisted with mouse experiments. J.L.B. performed genetic ablation experiments for PD-L1. B.X. assisted with breast cancer mouse model. K.E.P. and P.M.O. designed and/or performed mouse immune profiling studies. E.S., R.S.H., and K.D.M. designed and/or performed human immune profiling studies. R.R. designed the clinical trial and was principle investigator along with S.M.H. and A.M., who oversaw its completion. D.P. assisted in coordinating the trial. A.M. evaluated the final clinical trial data. R.K.A., T.C.G., and L.M.S. were investigators on the trial, D.A.P. evaluated all imaging response for trial patients and assisted in study design, and M.D.F. and X.X. evaluated pathological biomarkers. R.M. was responsible for the statistical design of the clinical trial, and R.M. and A.J.M. performed statistical analysis. H.I. and A.J.M. performed and interpreted statistical analysis of pre-clinical data. A.J.M. wrote the manuscript, and E.J.W., R.H.V., A.M., C.T., and A.J.R. edited the manuscript. A.J.M., E.J.W., and R.H.V. together designed, interpreted, and oversaw the study.
The transcriptomic data are available at the GEO repository (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE65503.
The authors declare no competing financial interests. Code for computational analysis is available upon request.
References
- 1.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodi FS, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamid O, et al. Safety and Tumor Responses with Lambrolizumab (Anti–PD-1) in Melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer JR, et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolchok JD, et al. Nivolumab plus Ipilimumab in Advanced Melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postow MA, et al. Immunologic Correlates of the Abscopal Effect in a Patient with Melanoma. N Engl J Med. 2012;366:925–931. doi: 10.1056/NEJMoa1112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishwaran H, Kogalur UB, Gorodeski EZ, Minn AJ, Lauer MS. High-Dimensional Variable Selection for Survival Data. Journal of the American Statistical Association. 2010;105:205–217. [Google Scholar]
- 9.Ishwaran H, Kogalur UB, Chen X, Minn AJ. Random survival forests for high-dimensional data. Statistical Analy Data Mining. 2011;4:115–132. [Google Scholar]
- 10.Wilson EB, et al. Blockade of Chronic Type I Interferon Signaling to Control Persistent LCMV Infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teijaro JR, et al. Persistent LCMV Infection Is Controlled by Blockade of Type I Interferon Signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jönsson G, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16:3356–3367. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 13.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;131:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 14.Paley MA, et al. Progenitor and Terminal Subsets of CD8+ T Cells Cooperate to Contain Chronic Viral Infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas N, et al. Tracking global changes induced in the CD4 T-cell receptor repertoire by immunization with a complex antigen using short stretches of CDR3 protein sequence. Bioinformatics. 2014;30:3181–3188. doi: 10.1093/bioinformatics/btu523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atchley WR, Zhao J, Fernandes AD, Druke T. Solving the protein sequence metric problem. Proc Natl Acad Sci USA. 2005;102:6395–6400. doi: 10.1073/pnas.0408677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci USA. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cha E, et al. Improved Survival with T Cell Clonotype Stability After Anti-CTLA-4 Treatment in Cancer Patients. Sci Transl Med. 2014;6:238ra70–238ra70. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert L, et al. CTLA4 Blockade Broadens the Peripheral T-Cell Receptor Repertoire. Clin Cancer Res. 2014;20:2424–2432. doi: 10.1158/1078-0432.CCR-13-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reits EA. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. Journal of Experimental Medicine. 2006;203:1259–1271. doi: 10.1084/jem.20052494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) European Journal of Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Bayne LJ, et al. Tumor-Derived Granulocyte-Macrophage Colony-Stimulating Factor Regulates Myeloid Inflammation and T Cell Immunity in Pancreatic Cancer. Cancer Cell. 2012;21:822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, et al. Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: changing strategies for cancer treatment. Blood. 2009;114:589–595. doi: 10.1182/blood-2009-02-206870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dewan MZ, et al. Fractionated but Not Single-Dose Radiotherapy Induces an Immune-Mediated Abscopal Effect when Combined with Anti-CTLA-4 Antibody. Clin Cancer Res. 2009;15:5379–5388. doi: 10.1158/1078-0432.CCR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green MR, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holets LM. Trophoblast CD274 (B7-H1) Is Differentially Expressed Across Gestation: Influence of Oxygen Concentration. Biology of Reproduction. 2006;74:352–358. doi: 10.1095/biolreprod.105.046581. [DOI] [PubMed] [Google Scholar]
- 27.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of Microarray Data Using Z Score Transformation. The Journal of Molecular Diagnostics. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107–129. [Google Scholar]
- 30.Breiman L. Machine Learning. 2001;45:5–32. [Google Scholar]
- 31.Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–329. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishwaran H, Kogalur UB. Random Forests for Survival, Regression and Classification (RF-SRC) 2013. [Google Scholar]
- 33.Rempala GA, Seweryn M. Methods for diversity and overlap analysis in T-cell receptor populations. J Math Biol. 2012;67:1339–1368. doi: 10.1007/s00285-012-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.