Abstract
Methionine can be reversibly oxidized to methionine sulfoxide (MetO) under physiological and pathophysiological conditions, but its use as a redox marker suffers from the lack of tools to detect and quantify MetO within cells. In this work, we created a pair of complementary stereospecific genetically-encoded mechanism-based ratiometric fluorescent sensors of MetO by inserting a circularly yellow fluorescent protein between yeast methionine sulfoxide reductases and thioredoxins. The two sensors, named MetSOx and MetROx for their ability to detect S and R-forms of MetO, respectively, were utilized for targeted analysis of protein oxidation, regulation and repair, as well as for monitoring MetO in bacterial and mammalian cells, analyzing compartment-specific changes in MetO, and examining responses to physiological stimuli.
Introduction
Reactive oxygen species (ROS) play critical roles in cell metabolism. Produced in a targeted or regulated manner, they can act as messengers in signal transduction, proliferation, differentiation, and apoptosis1. Particularly, hydrogen peroxide (H2O2) has emerged as a key signaling molecule which acts mainly by oxidizing critical thiol groups in various proteins2. However, ROS levels are tightly regulated to limit non-specific oxidation, which could damage proteins and metabolites and affect cell fitness. That is why oxidative processes are frequently found at the boundary between physiological and pathophysiological conditions3,4. An important advance in understanding the roles of ROS in cells was the development and utilization of genetically encoded fluorescent sensors. For example, using an approach developed for calcium quantification with engineered fluorescent proteins5, the H2O2 sensor HyPer was created by inserting a circularly permuted yellow fluorescent protein (cpYFP) into the bacterial H2O2 regulator OxyR6. This sensor, when expressed in living cells, is highly specific for H2O2 and can be targeted to any subcellular compartment6,7. It was used successfully in physiological and pathological models to dynamically monitor hydrogen peroxide8,9. Other genetically-encoded fluorescent sensors of the cellular redox state utilize redox-active Cys on YFP10 and GFP11, and their use dramatically improved our understanding of glutathione metabolism and redox homeostasis12.
Along with cysteine, which was thought to mediate most of the effects of ROS on proteins, methionine (Met) can be converted by biological oxidants to the R- and S-diastereomers of MetO. Met oxidation occurs under both physiological and pathophysiological conditions and, compared to the products of thiol oxidation, MetO is a more stable molecule, making it particularly attractive for the assessment of oxidative stress. However, it use as a redox biomarker and marker of oxidative damage to proteins and metabolites suffers from the lack of tools to detect and quantify MetO in vitro and in vivo. No chemical has been found to react specifically with MetO and the development of antibodies against MetO has also been unsuccessful13,14. In many organisms, the only molecules known to react specifically with MetO are the methionine sulfoxide reductases A (MSRA) and B (MSRB), which are the specific reductases of the S- and R-diastereomers of MetO, respectively15–17. Typical MSRs utilize several redox-active Cys to reduce their substrates in a three-step mechanism: i) Reduction of MetO leads to the formation of a sulfenic acid on the catalytic Cys. ii) A resolving Cys forms an intramolecular disulfide bond with the catalytic Cys, reducing the sulfenic acid. iii) The disulfide is reduced by a thioredoxin (Trx) in a process involving a transient intermolecular bond between catalytic Cys residues of the two partners18,19. The intermolecular bond between an MSR and a Trx could be stabilized by mutating a resolving Cys of the Trx20.
We took advantage of this mechanism to create genetically-encoded fluorescent sensors specific for each diastereomer of MetO. They were used to monitor MetO in vitro and in cells in response to physiological stimuli.
Results
Generation of diastereospecific ratiometric MetO sensors
We linked yeast MSRA or MSRB and their specific Trxs (Trx1 or Trx3, respectively; both Trxs were the mutants, in which resolving Cys were mutated in Ser) to two ends of a circularly permuted fluorescent protein (cpFP) to obtain three-protein fusions. Considering the mechanism of reduction of MSRs by Trx18,19, a stable disulfide bond is expected to be formed between an MSR and a Trx following reduction of a MetO-containing substrate (Fig. 1a). We utilized several cpFPs covering emission spectral range from blue to red6,21 and examined various arrangements of proteins and linkers in the fusion systems. All constructs yielded fluorescent probes, but only a circularly permuted yellow FP (cpYFP)6 fused at the N-terminus with an MSR and at the C-terminus with a Trx using relatively short linkers exhibited altered fluorescent spectra upon reaction with MetO. The sensors made based on MSRA and MSRB were named MetSOx and MetROx for their ability to sense S- and R-diastereomers of MetO, respectively (Fig. 1b, c, Supplementary Results, Supplementary Figs. 1 and 2).
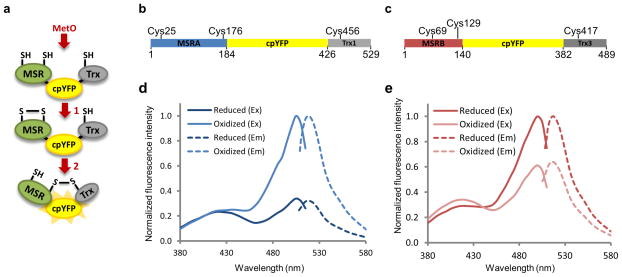
Figure 1. Design and spectra of MetSOx and MetROx sensors.
(a) Mechanism-based design of MetO sensors. 1) After reaction with MetO, disulfide is formed between two redox-active Cys of an MSR moiety. 2) Reduction of this bond by the catalytic Cys of the Trx moiety induces a conformational change in the sensor leading to changes in spectral properties of cpYFP. (b) Schematic representation of MetSOx, the sensor specific for the S-diastereomer of MetO. cpYFP (yellow) was fused in frame with yeast MSRA (blue) and yeast Trx1 (grey). C25, C176 and C456 represent redox-active Cys residues involved in catalysis. (c) Schematic representation of MetROx, the sensor specific for the R-diastereomer of MetO. cpYFP (yellow) was fused in frame with yeast MSRB (red) and yeast Trx3 (dark grey). For both MSRB and Trx3, mitochondrial targeting sequences were removed. C69, C129 and C417 represent redox-active Cys residues involved in catalysis. Excitation (Ex) (full line) and emission (Em) (dashed line) spectra of reduced (dark color) and oxidized (light color) MetSOx (d) and MetROx (e). Spectra were normalized to the oxidized form. The data presented are representative of 3 replicates. Sensors were reduced with 10 mM DTT and oxidized with 10 μM MRP4.
Reduced recombinant MetSOx showed two peaks of absorbance/excitation with maxima at 425 and 505 nm and a single peak of emission at 510–516 nm (Fig. 1d, Supplementary Fig. 3a, Supplementary Table 1). Reaction with a MetO-containing substrate led to a substantial increase in emission fluorescence intensity and the excitation peak at 505 nm without change at 425 nm, conferring ratiometric behavior, i.e., the ratio of fluorescence intensities at 505 nm and 425 nm increased with the addition of MetO. The fact that both MetSOx and MetROx use reactive Cys allowed us to create inactive fluorescent sensors, used as controls, in which single catalytic Cys residues within MSRs were mutated to Ser. As expected, the inactive MetSOx (C25S mutant) showed no change in fluorescence upon addition of MetO and behaved as a fully reduced MetSOx (Supplementary Fig. 3c).
Reduced MetROx showed two peaks of absorbance/excitation with maxima at 410 and 500 nm and a single peak of emission at 510–516 nm (Fig. 1e, Supplementary Fig. 3b, Supplementary Table 1). Reaction with MetO led to an increase in fluorescence intensity at 410 nm and a decrease at 500 nm with an isosbestic point at 447 nm. Thus, the 500/410 nm fluorescence intensity ratio decreased upon reaction with the substrate. Like the mutant MetSOx, the inactive MetROx (C129S mutant) did not react with MetO-containing substrates (Supplementary Fig. 3d).
Characterization of MetSOx and MetROx
When sensors were incubated with free Met, no change in the fluorescence ratio was observed, whereas incubation with free MetO, N-acetyl-MetO (a substrate mimicking MetO in proteins) and oxidized β-casein induced significant changes in both sensors (Fig. 2a, b, Supplementary Fig. 4). Kinetic measurements of sensor oxidation upon reaction with MetO-containing substrates showed fast reactivity, whereas no changes were observed for the inactive Cys-to-Ser forms. DTT treatment induced a decrease and increase in the fluorescence ratios of MetSOx and MetROx, respectively, indicating that this reducing agent could reduce both sensors (Fig. 2a, b).
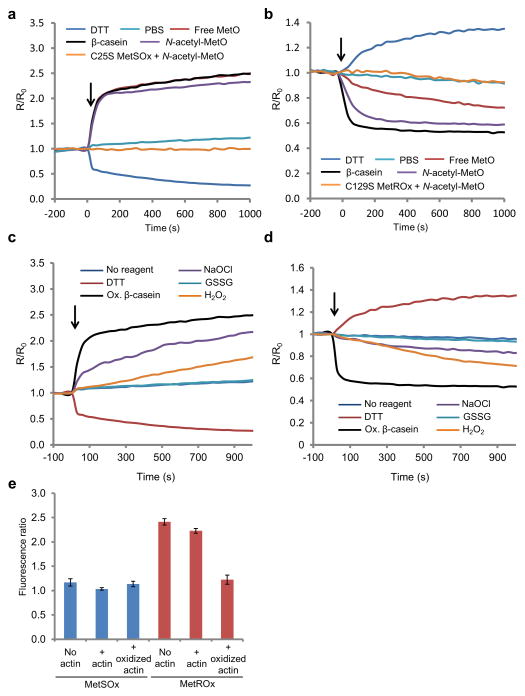
Figure 2. Response of recombinant MetSOx and MetROx to DTT and various oxidants.
Kinetics of reactivity of MetSOx (a) and MetROx (b) with MetO-containing substrates and DTT. Reduced sensors (1 μM) were incubated with free L-Met-R,S-O (100 μM, or 1,000 μM for MetSOx and MetROx, respectively), oxidized β-casein (10 μM), N-acetyl-L-Met-R,S-O (10 μM), PBS or DTT (1 mM), and the reduced inactive mutants (C25S MetSOx or C129S MetROx) were incubated with N-acetyl-L-Met-R,S-O (10 μM). F505 nm/F425 nm and F500 nm/F410 nm ratios (R) were normalized to the value at t = 0 (R0). Response of recombinant MetSOx (c) and MetROx (d) to DTT and various oxidants. Sensors (1 μM), partially reduced, were incubated with DTT (1 mM), oxidized β-casein (10 μM), NaOCl (10 μM), oxidized glutathione (GSSG, 100 μM) and H2O2 (100 μM). Variations in the ratio F505 nm/F425 nm or F500 nm/F410 nm (R) were normalized to the value at t = 0 (R0). In panels a, b, c and d, arrows indicate the addition of reagent. The data presented are representative of 3 replicates. (e) Comparison of MetSOx and MetROx reactivity towards MICAL1-oxidized actin. Sensors (0.12 μM) were incubated without actin (“No actin”), with 1.4 μM non-treated actin (“+ actin”) or with 1.4 μM MICAL1-oxidized actin (“+ oxidized actin”) for 15 min. Fluorescence excitation ratios (F505 nm/F425 nm for MetSOx and F500 nm/F410 nm for MetROx) were determined from spectra recorded at 530 nm emission wavelength. The data presented are the means (n = 3) ± SD. The data presented are representative of 3 replicates.
Recombinant MetO sensors were incubated with several oxidants (Fig. 2c, d). Treatment with oxidized glutathione did not affect fluorescence, indicating that this oxidant had no effect on MetSOx. In the case of sodium hypochlorite (NaOCl), the fluorescence ratio increased. This effect was likely due to oxidation of Met present in the sensor to MetO and its subsequent use as substrate, although we cannot exclude a potential direct oxidation of the reactive Cys of the sensor. For MetROx, neither oxidized glutathione nor NaOCl treatment led to oxidation of the sensor. Hydrogen peroxide had a marginal effect on both sensors compared to that of the MetO-containing protein substrate.
Cys alkylation/migration assay of the oxidized and reduced sensors confirmed that the catalytic Cys25 and Cys129 of MetSOx and MetROx, respectively, were necessary for the reactivity (Supplementary Fig. 5a). Indeed, whereas oxidized MetSOx and MetROx migrated faster than their reduced counterparts, C25S MetSOx and C129S MetROx showed no differences between oxidized and reduced states. The ratio of fluorescence determined for MetSOx and MetROx after incubation with mixtures of oxidized and reduced DTT of defined ambient redox potentials (Eh) allowed determining redox midpoint potential (Em) values: −276 ± 6 mV and −293 ± 4 mV for MetSOx and MetROx, respectively (Supplementary Fig. 5b). These potentials likely correspond to the formation of the mixed intermediate between an MSR and Trx.
As with other FPs, fluorescence of MetSOx and MetROx showed pH-dependence. We recorded fluorescence spectra of fully reduced and oxidized MetSOx and MetROx and their inactive forms and observed a maximum dynamic range at pH 7.5 for both sensors corresponding to a 6-fold change in the ratio of fluorescence intensity (Supplementary Fig. 6). Values of the excitation coefficient, quantum yield and molecular brightness for reduced and oxidized forms of MetSOx and MetROx at pH 7.5 were in the range of other sensors that utilized cpYFP5,22 (Supplementary Table 1).
Recently, MICAL proteins were identified as monooxygenases that stereospecifically oxidize two Met residues in actin to the R-diastereomer forms of MetO23,24. We employed a MICAL1-oxidized actin as a substrate and observed that MetROx, but not MetSOx, sensed Met oxidation, confirming stereospecificity of the sensors and their ability to monitor oxidation of individual Met residues (Fig. 2e).
To estimate the range of reactivity of the sensors, we performed a detailed characterization of their affinities towards free MetO and MetO-containing proteins. The data showed that, in vitro, MetSOx was similarly sensitive to free MetO and MetO in proteins, with K0.5~1 μM. MetROx was more sensitive to oxidized protein than to free MetO with K0.5~0.5 μM and 450 μM, respectively (Supplementary Fig. 7, Supplementary Table 2).
Use of MetSOx and MetROx in Escherichia coli
When expressed in E. coli, both MetSOx and MetROx showed fluorescence spectra similar to those of purified proteins (Supplementary Fig. 8a, b). Addition of free MetO induced an increase and decrease in the fluorescence ratio for MetSOx and MetROx, respectively, and thus caused their oxidation (Fig. 3a, b). After removing MetO, we observed opposite changes in the fluorescence ratio, indicating that the oxidation of both sensors was reversible in cells (Fig. 3a, b).
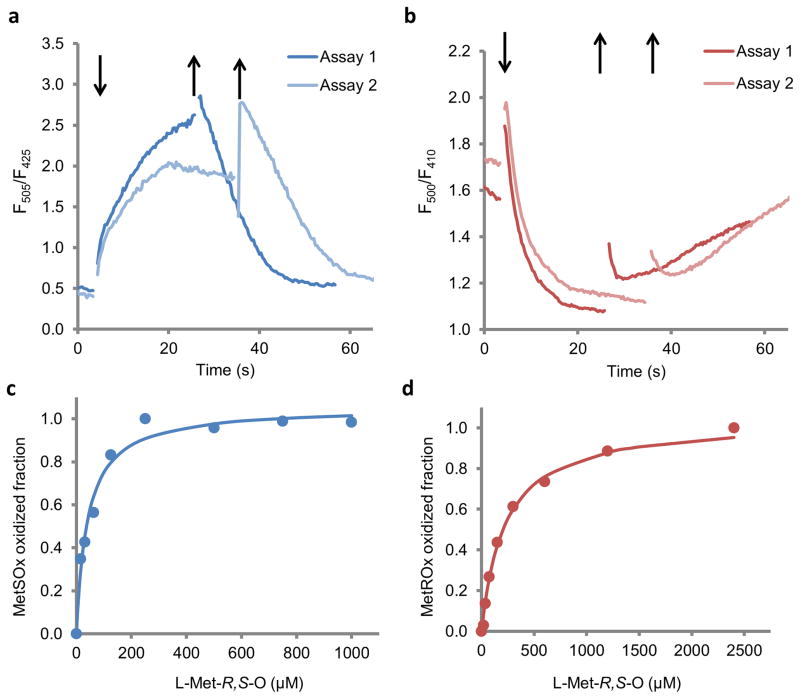
Figure 3. Characterization of MetSOx and MetROx sensors in E. coli.
E. coli cells expressing MetSOx (a) or MetROx (b) were incubated with MetO for ~30 min, and then rinsed to remove the oxidant. Arrows indicate the addition (↓) of MetO and rinsing (↑) the cells for two representative experiments (Assay 1 and Assay 2). MetSOx (c) and MetROx (d) oxidized fractions calculated from Supplementary Fig. 9 (n = 3), using equations 1 and 2, respectively, described in the “online methods” section.
The inactive C25S mutant of MetSOx and C129S mutant of MetROx expressed in E. coli had the fluorescence ratio similar to those of the reduced and oxidized forms of the sensors, respectively (Supplementary Fig. 8a, b). Incubation of E. coli expressing the active sensors with free MetO induced a rapid change in fluorescence for MetSOx and MetROx, but not for their mutant forms (Supplementary Fig. 8c, d). We systematically corrected MetSOx and MetROx signals by dividing the measured ratio of fluorescence by those of inactive sensors in subsequent experiments.
We analyzed reactivity of the sensors expressed in E. coli towards increasing concentrations of free MetO and observed changes in fluorescence starting at low micromolar concentrations (<20 μM) for both (Fig. 3c, d, Supplementary Fig. 9). In the case of MetSOx, the signals increased quickly, with the maximal values obtained around 200 sec. The signal was saturated at MetO concentrations above 250 μM, and the half saturation value was ~ 40 μM (Fig. 3c, Supplementary Fig. 9a, c). MetROx reacted more slowly than MetSOx and responded to higher concentrations of MetO (Fig. 3d). MetROx was saturated at concentrations above 2 mM MetO, and the half saturation value was ~ 200 μM (Fig. 3d, Supplementary Fig. 9b, d), similar to the purified MetROx (Supplementary Fig. 7b, Supplementary Table 2). Thus, both MetSOx and MetROx responded specifically to MetO in live cells and may be used to characterize reversible Met oxidation under physiological conditions.
We further prepared and characterized wild-type (Wt), single ΔmsrA and ΔmsrB mutants, and the double ΔmsrA/ΔmsrB mutant E. coli cells expressing MetO sensors. None of the mutants had a significant growth defect (Supplementary Fig. 10a), consistent with previous findings16,25. The MSR activity decreased to ~ 70% and ~ 40% in ΔmsrA and ΔmsrB cells, respectively, and was not detectable in the double mutant (Supplementary Fig. 10b). Following overnight growth (20 h), we measured the fluorescence ratio in cells expressing MetO sensors or their inactive forms. In Wt cells expressing MetSOx, the corrected F505 nm/F425 nm ratio was 1.0, indicating that the sensor was not oxidized. The ratio also did not change in ΔmsrA and ΔmsrB mutants, whereas the double ΔmsrA/ΔmsrB mutant showed a significant increase in the ratio (Fig. 4a). Thus, the S-diastereomer of MetO increased only in the double mutant after 20 h of growth. Wt and ΔmsrA cells expressing MetROx showed the corrected ratio of ~ 2.1, whereas these values were decreased to ~ 1.4 and ~ 1.5 in the single ΔmsrB mutant and the double ΔmsrA/ΔmsrB mutant, respectively (Fig. 4b). Thus, msrB deficiency led to an increase in the R-diastereomer of MetO following overnight growth.
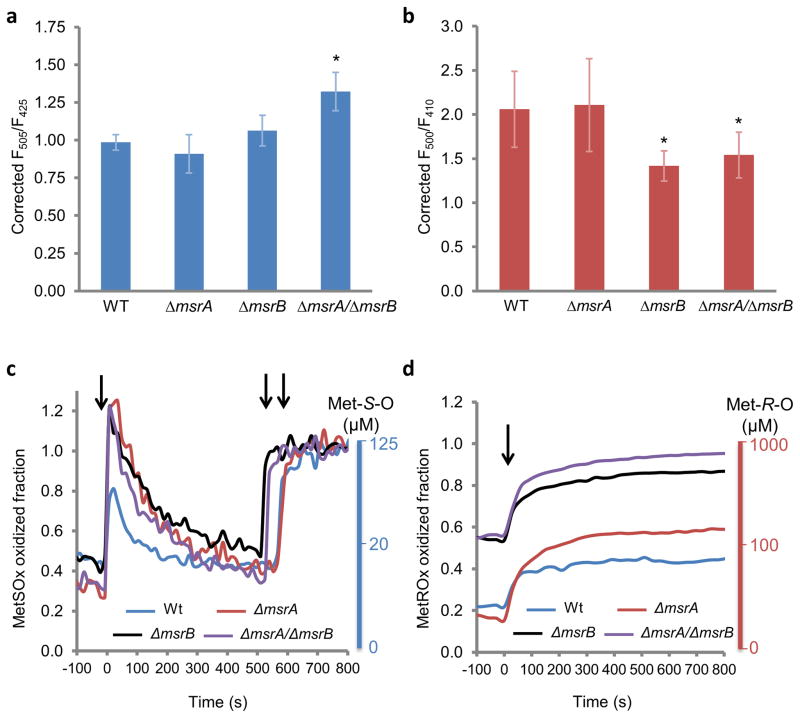
Figure 4. MetSOx and MetROx oxidation levels and the response to sodium hypochlorite in E. coli.
Static measurements of fluorescence in Wt and msr mutants expressing MetSOx (a) or MetROx (b). Strains were grown in LB for 20 h, rinsed and equilibrated in M9 medium and the ratio of fluorescence was recorded for cells expressing the sensor or its inactive form. The ratio was corrected by dividing the inactive sensor values. The values are presented as mean (n = 3) ± SD and are representative of 3 replicates. *p<0.05 (Student’s t-test) compared to Wt. Dynamics of the intracellular response of Wt and msr mutants expressing MetSOx (c) or MetROx (d) to NaOCl. Strains were grown in LB for 20 h, washed and equilibrated in M9 medium prior to the addition of NaOCl. The ratio of fluorescence was recorded for cells expressing the sensor or its inactive form, and was corrected by dividing the values of the inactive sensor. The fraction oxidized was calculated using equations 1 and 2 (online methods). MetS-O and Met-R-O scales were determined empirically from Fig. 3a, b. In panel c, the first arrow indicates the addition of NaOCl, and second and third arrows indicate the addition of free MetO. In panel d, the arrow indicates the addition of NaOCl. The data presented are representative of 3 replicates. Representative kinetics obtained for both MetO sensors and their inactive forms are shown in Supplementary Fig. 11.
Next, we examined the effect of the oxidant NaOCl in Wt and Δmsr mutant cells (Fig. 4c, d). In order to analyze MetO levels in cells using MetSOx and MetROx, we estimated the fluorescence ratio for the fully reduced and oxidized forms of sensors. In the case of MetSOx, the fluorescence ratio of the fully reduced sensor was obtained using C25S MetSOx as a reference, which behaved as the fully reduced sensor (Supplementary Fig. 8a), and the fully oxidized MetSOx ratio was obtained after the addition of saturating concentrations of free MetO in the cell suspension following reaction with NaOCl. This allowed us to determine the fraction of oxidized MetSOx upon treatment with NaOCl using equation 1 (online methods). The addition of 100 μM NaOCl to Wt E. coli expressing MetSOx induced rapid and transient oxidation of the sensor, the fluorescence ratio increased to ~ 0.8 in ~ 10 seconds, and then returned to the initial state in ~ 2 min (Fig. 4c). In single and double msr mutants, the increases were higher than in Wt cells, but all very similar, up to the maximal oxidation of MetSOx, and the decreases were slower than that observed in Wt. Using the saturation curves of the MetSOx signal in E. coli cells (Fig. 3e), we determined an empiric scale of changes in Met-S-O concentrations (Fig. 4c). The half and full oxidation of MetSOx was obtained for Met-S-O levels of 20 μM and higher than 125 μM, respectively. This allowed us to estimate that, under the conditions tested, Wt and msr mutant cells had ~ 15 μM Met-S-O, and the NaOCl treatment induced a 10-fold increase in the mutants, but only half of the increase in Wt cells.
We further empirically determined the fluorescence ratio of 2.5 of the fully reduced MetROx from maximal observed values (Fig. 4b), and the fluorescence ratio of 0.5 of the fully oxidized sensor was determined from MetO kinetics (Fig. 3d); these values were then used to calculate the fraction of oxidized MetROx in equation 2 (online method). MetROx was 3 times more oxidized in ΔmsrB and ΔmsrA/ΔmsrB than in Wt and ΔmsrA cells, with the oxidized fraction around 0.2 and 0.6 for Wt and ΔmsrA, and for ΔmsrB and ΔmsrA/ΔmsrB, respectively (Fig. 4d), consistent with static measurements (Fig. 4b). For Wt cells expressing MetROx, the oxidized fraction increased from 0.2 to 0.4 upon treatment with 40 μM NaOCl. The reaction was slower than that observed with MetSOx, and the value remained at 0.4 throughout the assay. ΔmsrA showed an increase from 0.2 to 0.6, slightly higher than that in Wt cells. Both ΔmsrB and ΔmsrA/ΔmsrB mutants showed an increase of the MetROx oxidized fraction from 0.6 to 0.9. Using an empiric scale for Met-R-O, similarly to MetSOx, we estimated that MetROx was half and fully oxidized with ~ 100 μM and ~ 1,000 μM Met-R-O, respectively (Fig. 4d). Thus, Wt and ΔmsrA showed initially ~ 50 μM MetO and the addition of NaOCl increased Met-R-O 2-fold. In the case of ΔmsrB and ΔmsrA/ΔmsrB mutants, the initial concentrations were ~ 200 μM and NaOCl induced a 5-fold increase in Met-R-O, up to ~ 1,000 μM. Although we cannot exclude a potential direct oxidation of MetSOx by NaOCl, this oxidant had no effect on the recombinant MetROx (Fig. 2d). Thus, the addition of low levels of NaOCl to the medium apparently led to a rapid formation of S- and R-diastereomers of MetO inside the cells, and msr mutants were more susceptible to oxidation than Wt cells.
MetROx response to physiological stimuli in HEK293 cells
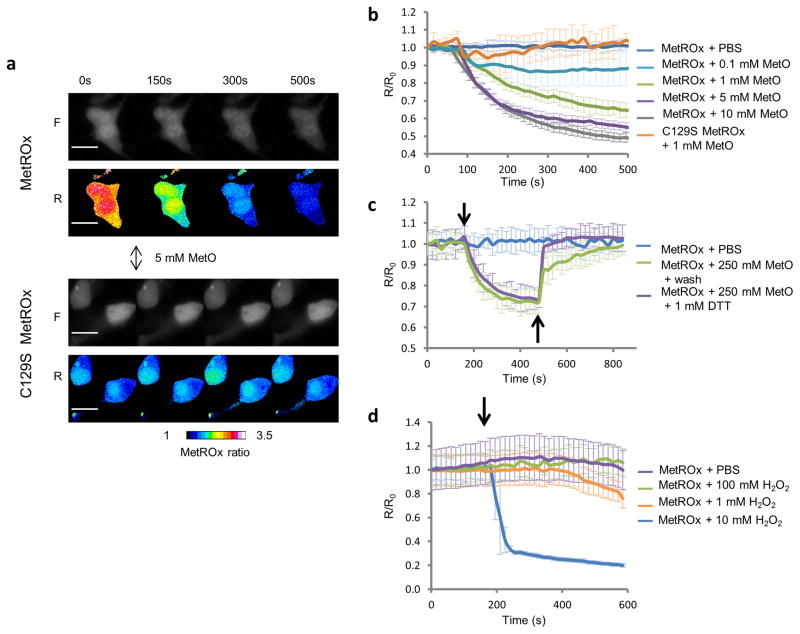
To characterize the use of the sensors in mammalian cells, we transfected HEK293 cells with MetSOx and MetROx expression vectors and followed the fluorescence changes by single cell fluorescence microscopy. Both sensors were expressed and detected by fluorescence. However, MetSOx showed very low fluorescence intensity and was not further characterized. As in bacteria, the addition of MetO induced a dose-dependent decrease in the fluorescence ratio of MetROx, but not of its inactive form (Fig. 5a, b). Subsequent treatment of cells with DTT or removal of MetO led to the reduction of the sensor showing that MetROx was fully functional in HEK293 cells (Fig. 5c). When MetROx expressing cells were treated with exogenous H2O2, changes in the fluorescence ratio were observed only at very high concentrations of the oxidant (Fig. 5d), arguing against the direct oxidation of the sensor by H2O2.
Figure 5. Characterization of MetROx in HEK293 cells.
(a) Time-series of raw fluorescent (F) and pseudocolored (R) ratio images of MetROx- and C129S MetROx-expressing cells subjected to 5 mM MetO. Scale bars represent 20 μm. (b) Kinetics of MetROx fluorescence in cells subjected to MetO (0.1 – 10 mM) detected by single cell live-microscopy. Background was subtracted and fluorescence ratios were normalized by the value at t = 0 s (n = 6–28). (c) Kinetics of fluorescence changes in MetROx-expressing cells treated with 250 μM MetO (↓), followed by washing or 1 mM DTT treatment (↑). (d) MetROx response to H2O2 in HEK293 cells. Time course analysis of HEK293 cells expressing MetROx and subjected to the indicated concentrations of H2O2. The fluorescence ratios were normalized by the value at t = 0 s. Data presented are the means (n = 6–8) ± SD and are representative of 3 replicates.
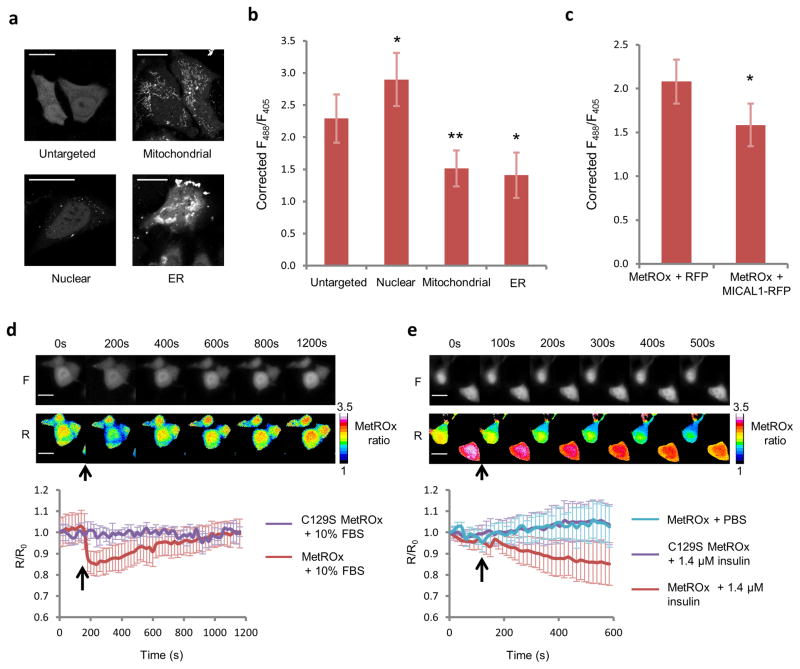
We further tested whether MetROx could detect protein-bound MetO in vivo and whether it was able to detect MetO under physiologically relevant conditions. In order to estimate Met-R-O variation detected by MetROx in HEK293 cells, we determined the value of the ratio for the fully reduced and oxidized sensor using saturating concentrations of DTT and MetO, and developed an empiric scale similarly to the E. coli assays (Supplementary Fig. 12). We further targeted MetROx and its inactive form to various cellular compartments (Fig. 6a). The corrected fluorescence ratios (Fig. 6b, Supplementary Fig. 13) allowed estimating MetROx oxidized fractions: ~ 0.3, ~0.1, ~0.7 and ~0.9 in the cytosol, nucleus, mitochondrial matrix and ER, respectively, suggesting that cytosol and nucleus had less than 100 μM Met-R-O whereas mitochondria and ER had 5–10 fold more Met-R-O. Next, to oxidize intracellular proteins, we overexpressed an RFP-fused constitutively active MICAL1 along with MetROx (Fig. 6c, Supplementary Fig. 14). A significant decrease in the fluorescence ratio was detected in MICAL1-overexpressing cells, but not in RFP-expressing control cells, indicating that MetROx detected Met-R-O generated by MICAL1 on actin and possibly other proteins. Since many growth factor and hormone signaling pathways and the regulation of the actin cytoskeleton dynamics involve redox mediators2, we tested whether serum stimulation of HEK293 cells could trigger Met oxidation. Following 6 h of serum starvation, 10% FBS induced a rapid and reversible oxidation of the sensor, showing that this treatment led to a transient increase in Met-R-O (Fig. 6d). We further tested whether a prototypical redox signaling process, the insulin pathway, could induce Met oxidation. Following serum starvation, 1.4 μM insulin induced a rapid oxidation of the sensor (Fig. 6e). Neither short-term (<12h) serum starvation nor cell confluency changed the MetROx ratio significantly (Supplementary Fig. 15).
Figure 6. MetROx response in subcellular compartments and physiological stimuli in HEK293 cells.
(a) Representative images of MetROx fluorescence in different cellular compartments. Scale bars represent 20 μm. (b) MetROx fluorescence ratio in different regions of interest was corrected by the ratio of the C129S MetROx targeted to the same compartment. Raw data are shown in Supplementary Fig. 13 (n = 13–27, *p < 0.05, **p < 0.005). (c) Corrected fluorescence ratio of MetROx in control (RFP-expressing) and MICAL1-expressing cells. Raw data are shown in Supplementary Fig. 14 (n = 17–26, p < 0.05). (d) Kinetics of MetROx response to serum stimulation. Raw fluorescent (F) and pseudocolored ratio (R) images of MetROx (upper panel) and kinetics of fluorescence (lower panel) of MetROx- and C129S MetROx-expressing cells subjected to 6 h of serum starvation followed by 10% serum treatment (↑) (n = 3). Scale bars represent 20 μm. (e) Kinetics of MetROx response to insulin. Raw fluorescent (F) and pseudocolored (R) ratio image-series of MetROx (upper panel), and kinetics of fluorescence (lower panel) of MetROx and C129S MetROx expressing cells subjected to 6 h serum starvation followed by 1.4 μM insulin treatment (↑) (n = 3). Scale bars represent 20 μm. Data presented are the means ± SD and are representative of 3 replicates.
Discussion
Prior to our study, there were no tools for examining MetO within live cells, and very limited tools have been available for in vitro analysis of Met oxidation in proteins or in the form of free amino acid. To develop the MetO sensors, we employed yeast S. cerevisiae MSRA and MSRB and their corresponding Trxs, as these enzymes are prototypical and have previously been well characterized26,27. We fused these proteins with cpYFP to create MetSOx and MetROx. Both sensors have two excitation maxima, which represent the two protonation states of the chromophore (neutral and anionic). We utilized this feature to carry out ratiometric fluorescence measurements in response to MetO (Fig 1d, e). This property, also observed in other sensors5,6,11,21,28, is ideal as it permits signal normalization regardless of variation in the concentration of the sensor itself. In vitro and in vivo characterization of reactivity toward MetO-containing substrates indicated that both sensors could detect changes in MetO levels from 1 to 1,000 μM (Fig. 3, Supplementary Table 2), which is in the range of the previous estimation of MetO levels in cells29. These characteristics make our sensors promising tools to monitor MetO in cells. However, several observations we made may need further clarification in order to make the best use of these unique tools.
We created our sensors based on the known mechanisms of MSR reduction by Trx18,19; thus, we expected that the change in fluorescence in response to MetO would involve the formation of a disulfide between the redox-active Cys (catalytic or resolving) of MSR and the Trx’s catalytic Cys (Fig. 1a). In the case of MetSOx, the inactive C25S form behaves as the fully reduced MetSOx and thus supports this mechanism. In the case of MetROx, however, the C129S mutant showed a spectrum similar to the oxidized MetROx raising a possibility of an alternative mechanism. Future work will be required to clarify the molecular mechanism behind oxidation and reduction of the sensors. For instance, mutations of the redox-active Cys in the Trx and MSR domains along with the characterization of fluorescence properties could provide further mechanistic insights into the operation of the sensors.
MetROx fluorescence was easily detected in vivo, but it was challenging to detect MetSOx fluorescence in mammalian cells as well as in bacteria. The low fluorescence might have been the reason why MetSOx was not able to distinguish between the MetO treated ΔmsrA and ΔmsrB mutant bacteria (Fig. 4c). This issue should be addressed in future studies by modifying the sensor to reach higher expression and/or greater sensitivity. It is also possible that MSR deficiency led to changes in redox homeostasis, precluding the expected response.
The oxidation of the sensors was reversible in cells, indicating that endogenous systems could reduce them (Fig. 3a, b, 5c). This reversibility indicates that the signal reflects the equilibrium between oxidation of the sensor by MetO and reduction capacity of the cells. To understand the sensor capabilities and correctly utilize the signals we measure, it would be of importance to identify which endogenous system (e.g. Trx and/or glutathione/glutaredoxin system) is responsible for the probe reduction. Characterization of these reduction systems is particularly important for the comparison of MetO content in different cellular compartments, because the detected differences in signals might be influenced by diverse reduction systems present in these compartments.
For both sensors, fluorescence intensity was pH-dependent, and at the optimal value of pH 7.5 recombinant MetSOx and MetROx showed a 6-fold increase and decrease of the fluorescence ratio after reaction with their substrates, respectively (Supplementary Fig. 6). Numerous genetically encoded fluorescent sensors suffer from pH-dependency, and variation of pH in cells can dramatically affect their response30. This is mainly due to the circular permutation of the fluorescent protein used to create cpYFP, which opens the β-barrel and exposes the chromophore phenoxy group to the environment31. This property requires precise control of pH in the experimental setting to validate the sensor’s response. Several methods were proposed to overcome this problem, such as mutating the sensor to remove the pH-dependency32 and monitoring pH changes in parallel experiments and correcting the signal of the sensor28. We chose a strategy to mutate the catalytic Cys of the MSR moieties of the sensors to Ser, and thus creating inactive versions, similarly to the inactive form SypHer of the H2O2 sensor HyPer33,34. Both in vitro and in vivo, the inactive versions of MetSOx (C25S MetSOx) and MetROx (C129S MetROx) did not respond to MetO (Fig. 2, Supplementary Fig. 3). By expressing these inactive forms and their active counterparts in parallel experiments, we could correct the signal observed (Fig. 3, 4). However, characterization of pH sensitivity indicated that their use may be limited to a pH range of ~6 to 8, as we observed significant changes between the spectra of the sensors and their inactive counterparts at higher pH values (Supplementary Fig. 6). This could be due to the fact that the mutant MSR moieties exhibit a slightly different structure compared to that of Wt forms, thereby modifying the overall structure of the sensor. MSRs are highly flexible enzymes and their structures are affected by mutations and redox states27,35.
Moreover, the inactive versions of MetSOx (C25S MetSOx) and MetROx (C129S MetROx) should not be considered as equally efficient negative controls. For the reduced MetSOx, the chromophore is preferentially protonated and the MetO-induced conformational change favors deprotonation to yield the anionic chromophore. Ambient increase in pH would cause the same effect as oxidation. Mutation of the catalytic Cys in the inactive C25S MetSOx does not change the chromophore protonation state, which remains protonated. As it is irresponsive to MetO, a signal increase would reflect pH changes, and thus C25S MetSOx can be considered as an appropriate pH control. One the other hand, for the reduced MetROx, the chromophore is preferentially deprotonated and the conformational change induced by MetO reduction favors protonation to yield the neutral chromophore. In this case, ambient acidification should cause a response similar to oxidation. Mutating the catalytic Cys to generate the inactive form of MetROx changes the chromophore state, which is preferentially protonated unlike its reduced active counterpart. This means that the use of C129S MetROx as a pH control should be considered very carefully. Acidification could mimic oxidation of MetROx, but as C129S MetROx is already preferentially protonated, it is not expected to respond to acidification as sensitively as MetROx. Future work will be needed to create the inactive and deprotonated version of MetROx, usable as optimal pH control.
Taken together, we developed two genetically encoded MetO sensors that could monitor relatively rapid changes in MetO levels in cells in a stereospecific manner. Being the first sensors of this type, they have from some limitations. Similarly to many other intracellular sensors, further work will be needed to improve them and address issues, such as optimization of fluorescence for MetSOx and developing additional negative controls. Nevertheless, the MetSOx and MetROx sensors developed in this study are highly promising tools which should find broad applications in studying free Met and protein oxidation and redox regulation under a variety of physiological and pathological conditions.
Online methods
Generation of MetSOx and MetROx fluorescent sensors and creation of expression vectors
Synthetic DNA oligonucleotides used for gene fusion, cloning and site-directed mutagenesis were purchased from Invitrogen™ Life Technologies (Carlsbad, CA, USA) (Supplementary Table 3). PCR was performed using Phusion® DNA Polymerase (New England Biolabs, Ipswich, MA, USA). FastDigest restriction enzymes were purchased from Thermo Fisher Scientific (Pittsburgh, PA, USA). Sequences coding for S. cerevisiae MSRA, MSRB, Trx1, and Trx3 were amplified from plasmids previously described26 using specific pairs of primers containing overlapping extensions to the cpYFP (Supplementary Table 3). The sequence coding for cpYFP was as in the hydrogen peroxide sensor HyPer6. MSR and Trx moieties were subsequently fused to cpYFP by PCR using equal amounts of purified PCR products. Final constructs were cloned into pGEM®T (Promega, Madison, WI, USA) and used as template for subsequent cloning. For expression in E. coli, constructs were amplified using specific pairs of primers (Supplementary Table 3), digested with BamHI and NotI restriction enzymes and cloned in pGEX-4T1 vector (GE Healthcare, Pittsburgh, PA, USA) in frame with an N-terminal glutathione-S-transferase tag. For cytosolic expression in mammalian cells, MetROx was cloned into pEGFP-C3 using NheI and BamHI restriction sites. For targeting to mitochondria, the mitochondrial targeting signal from subunit VIII of human cytochrome C oxidase was fused in the N-terminal position of MetROx and cloned into pEGFP-C3 using NheI and BamHI restriction sites7. Site-directed mutagenesis was made by whole plasmid amplification using primers containing mutated bases (Supplementary Table 3) as described26. All created plasmids are described in Supplementary Table 4.
Preparation and purification of recombinant MetSOx and MetROx fluorescent sensors
SoluBL21™ E. coli (Gelantis, San Diego, CA) cells were transformed with an indicated expression vector and grown in Luria-Bertani containing ampicillin (50 μg.ml−1) at 37°C. When the A600 reached ~ 0.6, synthesis of the recombinant protein was induced by the addition of 100 μM isopropyl β-D-1-thiogalactopyranoside. After 36 to 48 h incubation at 20°C, cells were harvested by centrifugation. Pellets were resuspended in PBS in the presence of Complete, EDTA-free, protease inhibitor mixture (Roche Applied Science, Indianapolis, IN, USA). Cells were disrupted by sonication. Proteins were purified using glutathione-Sepharose 4 Fast Flow (GE Healthcare) and eluted with 30 mM Tris-HCl, pH 8.0, supplemented with 20 mM glutathione. pH was neutralized with 5 M NaOH. Protein solutions were concentrated using 15 ml Amicon® Ultra concentrators with 30 kDa cutoffs (Millipore, Billerica, MA) and desalted in 30 mM Tris-HCl, pH 8.0, using 5 ml HiTrap™ Desalting columns (GE Healthcare). Initial protein concentrations were determined using the Pierce® bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). Protein purity was verified using SDS-PAGE gels stained with Imperial™ Protein Stain (Thermo Scientific). Recombinant sensors were aliquoted and stored at −20°C for use within a week or at −80°C for long-term storage.
Spectroscopic characterization of recombinant MetSOx and MetROx
For in vitro characterization, absorbance spectra were measured using Cary-60 spectrophotometer (Agilent Technologies, Santa Clara, CA, USA). Fluorescence was recorded in Costar™ black clear bottom 96-well microplates using a SpectraMax M5 fluorescence microplate reader (Molecular Devices, Sunnyvale, CA). Excitation spectra were recorded typically from 380 nm to 510 nm with 535 nm emission and a 530 nm cutoff. The device was typically set to 15 reads/wells with photomultiplier tube (PMT) set on “auto”, except for quantum yield determination, for which PMT was fixed to “medium”. Precise MetSOx and MetROx concentrations were measured in 1 M NaOH. Alkaline denaturation led to the appearance of a one-single peak of absorbance at 500 nm (Supplementary Fig. 2). Using three protein dilutions and the BCA assay, extinction coefficients of MetSOx and MetROx at 500 nm in 1 M NaOH were calculated at 39,800 and 32,700 M−1·cm−1, respectively. Prior to all assays, recombinant sensors were diluted to 5 to 10 μM and reduced with 10 mM DTT for 30 min at room temperature, then concentrated to ~ 100 μM with 4 ml/3,000 kDa cutoff Vivaspin™ concentrators (GE Healthcare) and desalted using Illustra NAP5™ columns (GE Healthcare) in 30 mM Tris-HCl, pH 8.0. For further spectroscopic characterization, purified and reduced sensors were diluted to 0.1 to 1 μM in PBS, pH 7.5.
Characterization of recombinant MetSOx and MetROx
For alkylation assays, MetSOx, MetROx, and their inactive versions C25S MetSOx and C129S MetROx (5 μM) were reduced with 10 mM DTT or oxidized with 10 μM oxidized Met-rich protein 4 (MRP4) for 30 min at room temperature. Oxidation and reduction states were verified by analyzing the fluorescence ratio. Reduced and oxidized sensors (2 μg) were incubated overnight at room temperature with 12.5 mM methoxypolyethylene glycol maleimide, 5,000 Da (Mal-PEG) in 50 mM Tris, and 0.5% SDS. Samples were reduced with 10 mM DTT and heated at 70°C for 10 min prior to loading onto 7% Tris-Acetate gels. Following SDS-PAGE, gels were stained with Imperial™ Protein Stain (Thermo Scientific). MetSOx and MetROx redox midpoints (Em) were determined similarly to described36. Briefly, MetO sensors (1 μM) were oxidized with 10 μM MRP4 for 1 h, then incubated with a mixture of oxidized and reduced DTT (10 mM) of defined Eh in 200 μl well microplates. Fluorescence spectra were recorded following 0.5, 1 and 3 h incubation. The percentage of oxidized sensor was determined using equations (1) and (2) and fitted to the Nernst equation by non-linear regression. The n value was set to 2 as the disulfide-dithiol exchange is a two-electron transfer process.
| equation (1) |
where R, measured F505 nm/F425 nm ratio; RReduced, F505 nm/F425 nm ratio of fully reduced MetSOx, and ROxidized, F505 nm/F425 nm ratio of fully oxidized MetSOx.
| equation (2) |
where R, measured F500 nm/F410 nm ratio; RReduced, F500 nm/F410 nm ratio of fully reduced MetROx and ROxidized, F500 nm/F410 nm ratio of fully oxidized MetROx.
Reactivity towards various MetO-containing substrates
Oxidized methionine-rich protein 4 (MRP4), oxidized folded and unfolded glutathione-S-transferase (GST) and oxidized β-casein were prepared as described26. N-Acetyl-MetO was prepared as described37. Sensors were diluted to 1 μM in 200 μl PBS in a microplate. Concentrations of reagents and oxidized proteins were adjusted to carry out assays using the same volume, typically 1 to 5 μl. For dose response assays, concentration ranges were as following: free L-Met-R,S-O, 12 nM to 500 mM; oxidized MRP4, 8 nM to 2 μM; oxidized GST, 8 nM to 8 μM; unfolded oxidized GST, 8 nM to 8 μM and oxidized β-casein, 6 nM to 7.6 μM. Oxidized fractions were calculated from recorded spectra according to the equations 1 and 2 for MetSOx and MetROx, respectively. Spectra recorded after 1, 2 and 3 h incubation gave similar results.
Stereospecific oxidation of actin and reaction with MetSOx and MetROx sensors
Recombinant mouse MICAL1 protein was prepared as described24. Purified rabbit skeletal muscle actin (Cytoskeleton, Inc) (1 mg) was solubilized in 100 μl distilled water and then diluted with fresh G-actin buffer (5 mM Tris-HCl, pH 8.0, 1 mM DTT, 0.2 mM ATP, 0.2 mM CaCl2) to 14 μM. Following 1 h incubation on ice, it was subjected to ultracentrifugation at 100,000 g at 4°C. The supernatant was collected and incubated with or without the recombinant mouse MICAL1 (5 μM) and NADPH (1 mM) at room temperature for 2 hours. Then, the reaction buffer was changed to fresh PBS buffer (pH 7.5) several times by using centrifugal filters (Amicon® Ultra-10K, EMD Millipore). Actin and MICAL1-oxidized actin were added to 1.4 μM in 100 μl PBS including 0.12 μM MetSOx or MetROx in a 96-well plate, and spectra were recorded.
pH calibration
For the analysis of pH dependence, buffers were as following: 100 mM Bis-Tris (Bis(2-hydroxyethyl)amino-tris(hydroxymethyl)methane), pH 5.8 and 6.4; 100 mM MOPS (4-Morpholinepropanesulfonic acid), 50 mM KCl, 150 mM NaCl, pH 7.0 and pH 7.5; Tris-HCl, pH 8.0, pH 8.3 and pH 9.0. All buffers were equilibrated at 25°C. Reduced sensors (1 μM) were incubated in a microplate (200 μl-final volume) for 1h at 25°C in the presence or absence of 1 mM free MetO or 10 μM oxidized Methionine-rich protein 4, which contains 31 MetO and has no significant absorbance or fluorescence because of the lack of Cys, Tyr and Trp residues13. Spectra recorded after 2 h and 3 h incubation showed no significant difference. Reproducibility was checked in PBS at pH 7.0, 7.5 and 8.0.
Determination of spectroscopic characteristics of reduced and oxidized MetSOx and MetROx sensors
Quantum yield (QY) was determined using five serial dilutions of absorbance ranging from ~ 0.01 to ~ 0.05; fluorescence vs. absorbance slopes were then determined. EGFP was used as a reference (QY = 0.6)38. MetSOx epsilon values at 425 nm and 505 nm, as well as MetROx epsilon values at 410 nm and 500 nm, were determined for reduced and oxidized forms in PBS, pH 7.5, using the BCA assay and calculated according to the Beer-Lambert equation.
Generation of ΔmsrA, ΔmsrB and ΔmsrA/ΔmsrB mutants of E. coli
SoluBL21 E. coli ΔmsrA, ΔmsrB and ΔmsrA/ΔmsrB mutants were created by homologous recombination with a kanamycin resistance cassette using Quick & Easy E. coli Gene Deletion Kit (Genes Bridges, Heidelberg, Germany) following manufacturer instructions and using specific primers (Supplementary Table 3). To create the double ΔmsrA/ΔmsrB mutant, the kanamycin resistance cassette was removed using the FLP recombinase (Genes Bridges) from the ΔmsrA, and then the msrB gene was deleted. All deletions were checked by PCR. Alternatively, total MSR activity using dabsylated-MetO was measured similarly to described39 (Supplementary Fig. 9b).
Transformation and expression of MetSOx and MetROx in E. coli
SoluBL21 E. coli (wild-type and ΔmsrA, ΔmsrB and ΔmsrA/ΔmsrB mutants) were rendered chimiocompetent according to a published method40 and transformed with pGEX-MetSOx, pGEX-C25S MetSOx, pGEX-MetROx or pGEX-C129S MetROx. After selection on LB plate containing ampicilin and/or kanamycin (50 μg.ml−1), one colony was used to prepare overnight preculture. One hundred ml of LB were inoculated to A600 ~ 0.05 and incubated at 37°C until A600 reached ~ 0.3. Induction was made with 100 μM isopropyl β-D-1-thiogalactopyranoside for 14 to 20 h at 20°C.
In vivo detection of MetO in E. coli
SoluBL21 E. coli (wild-type and ΔmsrA, ΔmsrB and ΔmsrA/ΔmsrB mutants) expressing the sensors in LB media were centrifuged at 5,000 g just prior to each assay, rinsed twice and resuspended in minimal M9 media. Cells expressing MetSOx and MetROx were diluted to A600 ~ 6 and ~ 3, respectively, and equilibrated at room temperature under moderate agitation for 30 min prior to the assays. Kinetics of the response to free MetO and sodium hypochlorite (NaOCl) assays with MetSOx expressing cells were carried out independently in a 1 ml quartz cuvette. Emission wavelength was 530 nm and excitation wavelengths were 425 and 505 nm. Similar assays with MetROx expressing cells were made simultaneously in microplates (200 μl reaction volume). Emission wavelength was 535 nm and excitation wavelengths were 410 and 500 nm.
In vivo microscopy
HEK293 cells were grown in high-glucose Dulbecco’s modified eagle medium (Invitrogen™ Life technologies) supplemented with 10 % fetal bovine serum, 100 U.ml−1 penicillin and 100 μg.ml−1 streptomycin. For in vivo live imaging experiments, the cells were plated onto glass bottom culture dishes (MatTek corporation, Ahsland MA, USA) and transfected the following day with the sensor-encoding vectors using FugeneHD transfection reagent (Promega). We let cells express the sensors for 24–48 h prior to treatments. MICAL1-RFP and pmRFP-N1 were expressed for 24 h prior to treatments. Constitutively active MICAL1 expression caused morphological changes and, in case of high expressors, toxicity in HEK293 cells. The cells with obvious moribund morphology (poor attachment, round shaped morphology, lost intracellular compartmentalization) were excluded from analysis. For imaging, cells were maintained in Live Cell Imaging Solution (Invitrogen™ Life technologies) supplemented with 4.5 g.l−1 glucose. Two microscopes were used to collect images. Confocal images (for Fig 6a, b, Supplementary Fig. 11–14) were captured on a Zeiss LSM710 confocal laser scanning microscope equipped with a 63×/1.4 oil immersion objective (Plan-ApoChromat, Zeiss). The excitation wavelengths were 405 nm and 488 nm and the emitted light was filtered below the wavelength of 505 nm. The optical slice was 4–6 μm. The rest of the live cell imaging experiments were carried out by using a fully motorized Olympus IX81 inverted microscope equipped with a Hamamatsu Orca ER cooled-CCD camera in conjunction with a zero-drift focus compensation system. MetROx fluorescence was excited with 420/40 and with 500/16 band-pass excitation filters. Emission was acquired every 15 to 30 s for 10 – 15 min using a 535/30 band-pass emission filter. Chemicals were added in 0.1 ml of buffer after the removal to the same volume from the incubation medium. We carried out the image acquisition by using MetaMorph software (Universal Imaging, Downingtown, PA). Following background fluorescence subtraction, the sensor ratio was calculated by dividing the image acquired at 488 (500) nm by the image at 405 (420) nm using the software ImageJ. If otherwise not indicated data are presented as means ± SD.
Supplementary Material
Acknowledgments
We thank Dr. Pascal Rey for the kind gift of dabsyl-MetO, and Drs. Vsevolod Belousov and Vladislav Verkhusha for discussion. This study was supported by the National Institutes of Health grants AG021518 and GM065204 to V.N.G and HL48743 to T.M.
Footnotes
Author contributions
L.T. designed, created and characterized the sensors, performed experiments with E. coli cells, analyzed the data and wrote the paper. Z.P. performed experiments with HEK293 cells, analyzed the data and wrote the paper. B.C.L. performed experiments with MICAL1-oxidized actin and analyzed the data. T.M. contributed reagents and tools and analyzed the data. V.N.G designed the sensors, supervised the research and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.D’Autréaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 2.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer AJ, Dick TP. Fluorescent Protein-Based Redox Probes. Antioxid Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 4.Maron BA, Michel T. Subcellular Localization of Oxidants and Redox Modulation of Endothelial Nitric Oxide Synthase. Circ J. 2012;76:2497–2512. doi: 10.1253/circj.cj-12-1207. [DOI] [PubMed] [Google Scholar]
- 5.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belousov VV, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 7.Malinouski M, Zhou Y, Belousov VV, Hatfield DL, Gladyshev VN. Hydrogen peroxide probes directed to different cellular compartments. PloS One. 2011;6:e14564. doi: 10.1371/journal.pone.0014564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin BY, Sartoretto JL, Gladyshev VN, Michel T. Endothelial nitric oxide synthase negatively regulates hydrogen peroxide-stimulated AMP-activated protein kinase in endothelial cells. Proc Natl Acad Sci. 2009;106:17343–17348. doi: 10.1073/pnas.0907409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Østergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson GT, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 12.Morgan B, et al. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat Chem Biol. 2013;9:119–125. doi: 10.1038/nchembio.1142. [DOI] [PubMed] [Google Scholar]
- 13.Le DT, et al. Analysis of methionine/selenomethionine oxidation and methionine sulfoxide reductase function using methionine-rich proteins and antibodies against their oxidized forms. Biochemistry. 2008;47:6685–6694. doi: 10.1021/bi800422s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wehr NB, Levine RL. Wanted and wanting: Antibody against methionine sulfoxide. Free Radic Biol Med. 2012;53:1222–1225. doi: 10.1016/j.freeradbiomed.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc Natl Acad Sci U S A. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimaud R, et al. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 17.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarrago L, Gladyshev VN. Recharging oxidative protein repair: catalysis by methionine sulfoxide reductases towards their amino acid, protein, and model substrates. Biochemistry (Mosc) 2012;77:1097–1107. doi: 10.1134/S0006297912100021. [DOI] [PubMed] [Google Scholar]
- 19.Boschi-Muller S, Olry A, Antoine M, Branlant G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim Biophys Acta. 2005;1703:231–238. doi: 10.1016/j.bbapap.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, et al. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilan DS, et al. HyPer-3: a genetically encoded H(2)O(2) probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem Biol. 2013;8:535–542. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 23.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol. 2013;15:1445–1454. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee BC, et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell. 2013;51:397–404. doi: 10.1016/j.molcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St John G, et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarrago L, Kaya A, Weerapana E, Marino SM, Gladyshev VN. Methionine sulfoxide reductases preferentially reduce unfolded oxidized proteins and protect cells from oxidative protein unfolding. J Biol Chem. 2012;287:24448–24459. doi: 10.1074/jbc.M112.374520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma XX, et al. Structural Plasticity of the Thioredoxin Recognition Site of Yeast Methionine S-Sulfoxide Reductase Mxr1. J Biol Chem. 2011;286:13430–13437. doi: 10.1074/jbc.M110.205161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J Off Publ Fed Am Soc Exp Biol. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tantama M, Hung YP, Yellen G. In: Progress in Brain Research. Knöpfel Thomas, Boyden Edward S., editors. Vol. 196. Elsevier; 2012. pp. 235–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarzländer M, et al. The ‘mitoflash’ probe cpYFP does not respond to superoxide. Nature. 2014;514:E12–14. doi: 10.1038/nature13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging Cytosolic NADH-NAD+ Redox State with a Genetically Encoded Fluorescent Biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishina NM, et al. In: Methods in Enzymology. Cadenas Enrique, Packer Lester., editors. Vol. 526. Academic Press; 2013. pp. 175–187. [Google Scholar]
- 34.Poburko D, Santo-Domingo J, Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranaivoson FM, et al. Methionine sulfoxide reductase B displays a high level of flexibility. J Mol Biol. 2009;394:83–93. doi: 10.1016/j.jmb.2009.08.073. [DOI] [PubMed] [Google Scholar]
- 36.Tarrago L, et al. Plant thioredoxin CDSP32 regenerates 1-cys methionine sulfoxide reductase B activity through the direct reduction of sulfenic acid. J Biol Chem. 2010;285:14964–14972. doi: 10.1074/jbc.M110.108373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira Dos Santos C, Cuiné S, Rouhier N, Rey P. The Arabidopsis plastidic methionine sulfoxide reductase B proteins. Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiol. 2005;138:909–922. doi: 10.1104/pp.105.062430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarrago L, et al. Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. J Biol Chem. 2009;284:18963–18971. doi: 10.1074/jbc.M109.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.