Abstract
Background and Study Objectives:
Flies mutant for the canonical clock protein cycle (cyc01) exhibit a sleep rebound that is ∼10 times larger than wild-type flies and die after only 10 h of sleep deprivation. Surprisingly, when starved, cyc01 mutants can remain awake for 28 h without demonstrating negative outcomes. Thus, we hypothesized that identifying transcripts that are differentially regulated between waking induced by sleep deprivation and waking induced by starvation would identify genes that underlie the deleterious effects of sleep deprivation and/or protect flies from the negative consequences of waking.
Design:
We used partial complementary DNA microarrays to identify transcripts that are differentially expressed between cyc01 mutants that had been sleep deprived or starved for 7 h. We then used genetics to determine whether disrupting genes involved in lipid metabolism would exhibit alterations in their response to sleep deprivation.
Setting:
Laboratory.
Patients or Participants:
Drosophila melanogaster.
Interventions:
Sleep deprivation and starvation.
Measurements and Results:
We identified 84 genes with transcript levels that were differentially modulated by 7 h of sleep deprivation and starvation in cyc01 mutants and were confirmed in independent samples using quantitative polymerase chain reaction. Several of these genes were predicted to be lipid metabolism genes, including bubblegum, cueball, and CG4500, which based on our data we have renamed heimdall (hll). Using lipidomics we confirmed that knockdown of hll using RNA interference significantly decreased lipid stores. Importantly, genetically modifying bubblegum, cueball, or hll resulted in sleep rebound alterations following sleep deprivation compared to genetic background controls.
Conclusions:
We have identified a set of genes that may confer resilience/vulnerability to sleep deprivation and demonstrate that genes involved in lipid metabolism modulate sleep homeostasis.
Citation:
Thimgan MS, Seugnet L, Turk J, Shaw PJ. Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. SLEEP 2015;38(5):801–814.
Keywords: Drosophila melanogaster, lipid metabolism, lipid storage, microarray, sleep homeostasis, transcriptional changes
INTRODUCTION
Sleep is believed to support a restorative function, whereas extended sleep deprivation can lead to death.1–4 Between these two extremes, sleep deprivation results in graded cognitive and physiological decrements, suggesting that a lack of sleep exacts a graded toll as sleep debt accumulates.5–7 Symptoms that may hint at the performance deficits and cause of death have been noted,3,8–10 but the underlying causes have not yet been determined.
To identify genes that may be responsible for the accumulating deficits during sleep deprivation or that may protect an organism from the deleterious effects of extended waking, we evaluated flies with a mutation in the canonical clock protein cycle (cyc01). The cyc01 mutant is particularly sensitive to sleep deprivation as evidenced by an extremely large sleep rebound, cognitive deficits, and death after only 10 h of sleep deprivation, nearly eight times faster than wild-type flies.2,11,12 In contrast to sleep deprivation, when cyc01 mutants are starved they show an immediate and sustained increase in waking that is not compensated by a homeostatic response11,13 and does not induce cognitive impairment.11 Importantly, cyc01 mutants can sustain periods of waking 2.6 times longer when starved than when sleep deprived, suggesting that the graded effect of sleep loss may be attenuated during starvation.11 Interestingly, removing food also induces waking in humans14 and rats.15
Recent data suggest that lipid metabolism and sleep have a reciprocal interaction. Shorter sleep times are associated with increased weight,6 possibly due to changes in food preferences16,17 or in energy processing.18 Likewise, lipid metabolic enzymes have been associated with changes in theta oscillations during rapid eye movement (REM) sleep,19 and they play a critical role in the response to sleep deprivation.11 Indeed, mutations in the brummer (bmm) gene that reduces lipolysis result in a substantially increased sleep rebound following one night of sleep loss.11 In contrast, mutants in Lipid storage droplet-2 (Lsd2) that exhibit an increase fatty acid release, do not respond to sleep deprivation with either a sleep rebound or deficits in cognitive performance.11 Thus, lipid metabolism does not simply respond to sleep loss, but it plays an active role in sleep regulation and how an animal can cope with sleep deprivation. The mechanism for how the control of lipids modulates sleep and wake states remains unclear.
We therefore exploited the phenotypic difference in the cyc01 mutants' response to sleep deprivation and starvation to identify genes associated with performance and physiological decrements (those changed with sleep deprivation) or those that allow flies to remain awake without accruing negative consequences or cognitive impairment (those changed due to starvation-induced waking). Microarray analyses revealed transcriptional changes in multiple gene ontology categories, including channels and lipid metabolism genes. Given the limited exploration of lipid metabolism genes and the role that energy management is thought to play in sleep regulation,20 we tested the functional relevance of lipid metabolic enzymes in the response to sleep deprivation. We found that several putative lipid metabolism genes are involved in mediating the response to sleep deprivation. Moreover, using lipidomics, we demonstrate that one of the genes with the largest transcriptional changes, heimdall (hll, formerly CG4500) also alters lipid metabolism. Heimdall is the fictional character that for ages guarded the bridge to Asgard without sleep and without the consequences of sleep deprivation, similar characteristics to the hll knockdown. Together, these data highlight the role that the storage and catabolism of lipids play in sleep regulation after sleep deprivation. These results also confirm the utility of this array strategy in identifying genes that play a role in sleep homeostasis.
METHODS
Flies and Husbandry
Flies were reared in standard laboratory conditions, 12:12 light:dark (LD) schedule, standard food (yeast, sucrose, corn syrup, molasses, and agar), 25°C and 50% humidity. cyc01 and period01 (per01) mutant flies were obtained from Dr. Jeff Hall. UAS-hll RNAi was obtained from the Vienna Drosophila Resource Center.21 Wild-type Canton-S (CS), Actin-GAL4/CyO (Act-GAL4), bubblegumEY01376 (bgmEY03176), cueball2 (cue2), Df(3L)Ar14-8 and Df(3L)kto2/TMB,Tb1 were obtained from the Bloomington Stock Center (Bloomington, IN, USA). Tubulin GeneSwitch-Gal4 (TubGSw)22 was obtained from Dr Marc Tatar. Revertants for cue2 were generated using standard techniques that introduce the Δ2-3 version of the transposase to mobilize the P-element. Precise excision was confirmed by sequencing with primers designed using Primer3.23 The bubblegum1 (bgm1) deletion mutant and the precise excision (bgmrev) genetic background control were obtained from Dr Kyung-Tai Min. GeneSwitch GAL4 expression was induced by rearing adult flies immediately upon eclosion on either 100 μL/mL of RU486 (mifepristone, Sigma, Saint Louis, MO) or an equal volume of ethanol. A 50 mg/mL stock was diluted into normal food to a final concentration of 100 μL/mL.
Sleep Recording
Three-day-old flies were placed into 65-mm glass tubes containing standard laboratory food and monitored using the Drosophila activity monitoring system (Trikinetics, Waltham, MA, USA) as previously described.1,2 Briefly, activity was recorded in 1-min bins and episodes of quiescence ≥ 5 min were considered sleep. Total sleep time, sleep architecture, and sleep homeostasis were calculated using an in-house program according to criteria previously established.1,2,24
Sleep Deprivation
Flies were sleep deprived using the sleep-nullifying apparatus, which asymmetrically tilted −60° to +60° such that the sleeping flies were displaced during the downward movement six times/min.1,2 The clock mutants, cyc01 and per01, were maintained and sleep deprived under constant darkness, or dark:dark (DD); sleep deprivation occurred for 7 h during the day between CT0 and CT12. For transcriptional analysis, CS flies were maintained for 3 days under DD conditions and then deprived of sleep for 12 h during the primary sleep period. The primary sleep period was identified from the previous day's data based upon the average time that the CS flies initiated their longest sleep bout. All other flies were maintained on a 12:12 LD schedule and deprived of sleep for 12 h between ZT12 (lights out) to ZT0 (lights on). The standard sleep homeostasis protocol consisted of 2 days of baseline followed by sleep deprivation and then flies were released into recovery where they remained unperturbed for 48 h. Sleep homeostasis was calculated for each individual as a percentage of the minutes of sleep gained above baseline during the 48 h of recovery divided by the total min of sleep lost during 12 h of sleep deprivation (minutes gained/minutes lost).
Starvation
Starvation is defined as the desire for food without access to nutrients.25 Flies were starved according to the previously published protocol.11 Briefly, flies were moved from a tube containing standard Drosophila food to a tube with a 1% agar solution. For all genotypes, baseline and starvation were carried out in constant darkness for controls and starved conditions equivalent to sleep deprived animals. For flies with a functional clock, 3 days of baseline were taken before waking was induced for 12 h during the primary sleep period, which was determined using the previous day's average sleep time and longest sleep bout. cyc01 and per01 flies were starved or sleep deprived for 7 h so as not to induce lethality.
Microarray
Three-day-old female cyc01 mutants were monitored under baseline conditions for 2 days in DD. On the third day, they were either sleep deprived for 7 h according to standard procedures2 or starved for 7 h by replacing their food with 1% agar. The flies' behavior was evaluated during the treatment at which time, two thirds of the flies and their untreated controls were frozen so that RNA could be extracted from whole heads. The remaining one third of the flies were placed into tubes containing fresh food and their behavior was monitored for an additional 24 h to assay the size of the homeostatic response. Thus, we have behavioral data from siblings that were treated concurrently with flies that contributed to the micro-array. Each replicate was made up of 20 pooled heads and processed for microarray analysis. Eight independent experiments were conducted over 4 mo to ensure that the results would be reproducible. Partial complementary DNA (cDNA) microar-rays containing 6,240 elements that included > 4,500 unique cDNA expressed sequence tag (EST) clones representing approximately 30–40% of the total estimated number of genes in the Drosophila genome were evaluated.26,27 cDNA arrays were processed at the University of California, San Diego Biogem Biomedical Microarray Facility. Data from the 4,659 ESTs present were background subtracted, mean normalized, and subjected to loess transformation using Standardization and Normalization of MicroArray Data (http://pevsnerlab.kennedykrieger.org/snomadinput.html). The two most extreme values were identified from each condition (control, sleep deprived, and starved) and removed. For each gene, we calculated the median value of the eight replicates. Each value was then subtracted from the median to yield a difference score. The absolute values of the eight difference scores were ranked and the two highest values were excluded. Thus, statistical analysis was conducted for six replicates/group for each EST present. We were specifically interested in identifying differences between cyc01 flies that were kept awake by sleep deprivation versus those that were kept awake by starvation. Statistical differences between sleep deprived and starved flies were identified using the Cyber-T Bayesian statistical framework.28,29 A Bonferroni correction for multiple comparisons was performed resulting in significance threshold of P < 1.07 × 10−5. Untreated controls are presented to provide information on the direction of the change that occurred in the sleep deprived and starved conditions. An independent replicate of sleep deprived, starved, and untreated control was collected from cyc01 flies for confirmation using quantitative polymerase chain reaction (qPCR). Genes are annotated using FB2014_04. Microarray results have been deposited in the GEO database under identification GSE18550.
Quantitative Polymerase Chain Reaction
Total RNA was isolated from 20 fly heads with Trizol (Invitrogen, Carlsbad, CA) and DNAse I digested. In the case of whole flies, three to five flies were frozen and homogenized. cDNA synthesis was performed in quadruplicate using Superscript III (Invitrogen), according to manufacturer protocol. In order to evaluate the efficiency of each reverse transcription, equal amounts of cDNA were used as a starting material to amplify RP49 as previously described.2 cDNA from comparable reverse transcription reactions were pooled and used as a starting material to run four qPCR replicates. Expression values for RP49 were used to normalize results between groups. For flies maintained on an LD schedule, both experimental and untreated controls, were collected at the exact same circadian time ZT0-1 or CT0-1 for flies maintained in DD. For clock mutants, the control, sleep deprivation, and starvation experiments were run in parallel and the flies were collected at the same time.
Mass Spectrometric Analyses of Lipids
Samples for lipid spectra consisted of five flies homogenized in 1 mL of 0.63% LiCl. Aliquots (200 μL) of the homogenates were removed for protein measurements. Lipids were extracted from the remainder by the method of Bligh and Dyer. The extract was concentrated to dryness under nitrogen and reconstituted in chloroform/methanol (1:1) to which LiCl was added (final [Li], 2 mM), and lipids in the extract were infused (1 μl/min) with a Harvard syringe pump into the ESI source of a Finnigan (San Jose, CA) TSQ-7000 triple stage quadrupole mass spectrometer controlled by Finnigan ICIS software, as previously described.30–34 Glycerophosphocholine (GPC) lipids and triacylglycerols (TAG) were analyzed as Li+ adducts in positive ion mode. Free fatty acids (FFA) and glycerophosphoethanolamine (GPE), -glycerol (GPG), -serine (GPS), and -inositol (GPI) lipids were analyzed as [M-H]- ions in negative ion mode. For tandem MS, precursor ions selected in the first quadrupole were accelerated into a chamber containing argon to induce collisionally activated dissociation (CAD), and product ions were analyzed in the final quadrupole under described instrumental parameters.30–34 Identities of lipid species were determined from their tandem spectra, and their quantities were determined relative to an internal standard (e.g., 14:0/14:0-GPC for GPC and TAG species).
TAG Analyses and Identification of Molecular Species
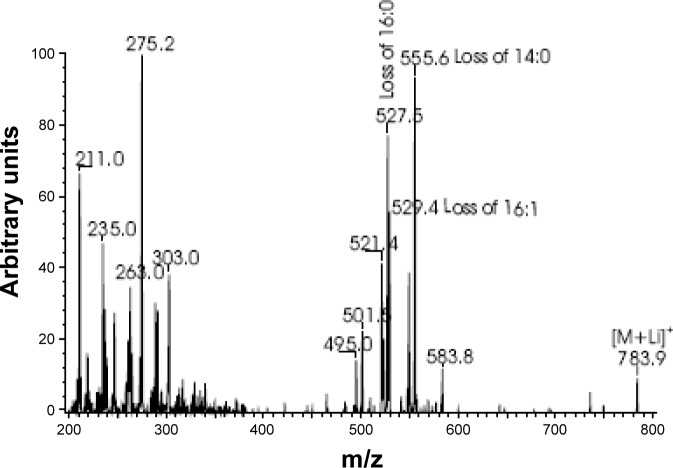
TAG were analyzed as Li+ adducts by positive ion electro-spray ionization mass spectrometry, or ESI/MSn (n = 1 or 2), as previously described.32 The identities of the molecular species represented by the parent ions in the full scan tracings (Figure 4) were determined from the MS/MS spectra obtained from ions with the m/z value of the peak in question upon CAD. For example, the ion of m/z 783 in the TIC tracing corresponds to the Li+ adduct of TAG species in which the total number of carbon atoms in the fatty acyl chains is 46 and the total number of double bonds is 1 (denoted 46:1-TAG). See the legend of Figure 4 for full explanation and rationale for assigning triglyceride species.
Figure 4.
Identification of the constituents that constitute the peak at m/z 783. Mass spectrometric MS/MS spectra obtained from ions with the m/z value of 783 in the total ion current tracing (Figure 4) corresponds to the Li+ adduct of triacylglycerol (TAG) species from the RU-treated flies in which the total number of carbon atoms in the fatty acyl chains is 46 and the total number of double bonds is 1 (denoted 46:1-TAG). The tandem spectrum obtained from collisionally activated dissociation (CAD) of m/z 783 is displayed in the figure, and it indicates that the predominant molecular species represented by that peak is 14:0/16:1/16:0-TAG. Features of the spectrum that establish that assignment are the presence of ions that represent neutral losses of each of the substituents as a free fatty acid at m/z 527 (loss of 16:0), 529 (loss of 16:1), and 555 (loss of 14:0), respectively. There are also ions reflecting loss of each substituent as a Li+ salt at m/z 521, 523, and 549, respectively. The ions representing loss of the sn-2 substituent of TAG-Li+ species are less abundant than the ions reflecting loss of the sn-1 or sn-3 substituent,34 and the sn-1 and sn-3 positions of TAG molecules are not distinguishable by mass spectrometry. Other ions consistent with this assignment are the acylium ion of 14:0 (m/z 211) and ions representing Li+ adducts of 14:0 (m/z 235) and 16:0 (m/z 263). There are also ions representing combined losses of 16:1 and 16:0 (m/z 275) or 14:0 (m/z 303) as an α,β-unsaturated fatty acid. Such combined losses always include the sn-2 substituent,85 indicating that 16:1 is the sn-2 substituent in the major TAG isomer contributing ion current to the m/z 783 peak. That there are less abundant isomers with the overall composition 46:1-TAG, e.g., 12:0/18:1/16:0-TAG, is reflected by relatively low abundance ions at m/z 501 and m/z 583 that represent loss of 18:1 and 12:0 as free fatty acids, respectively. MS and MS/MS analyses of lipid extracts of vehicle-treated flies revealed the same major TAG molecules observed for the RU-treated flies.
RESULTS
Transcriptional Profiling of Sleep Deprived and Starved Flies
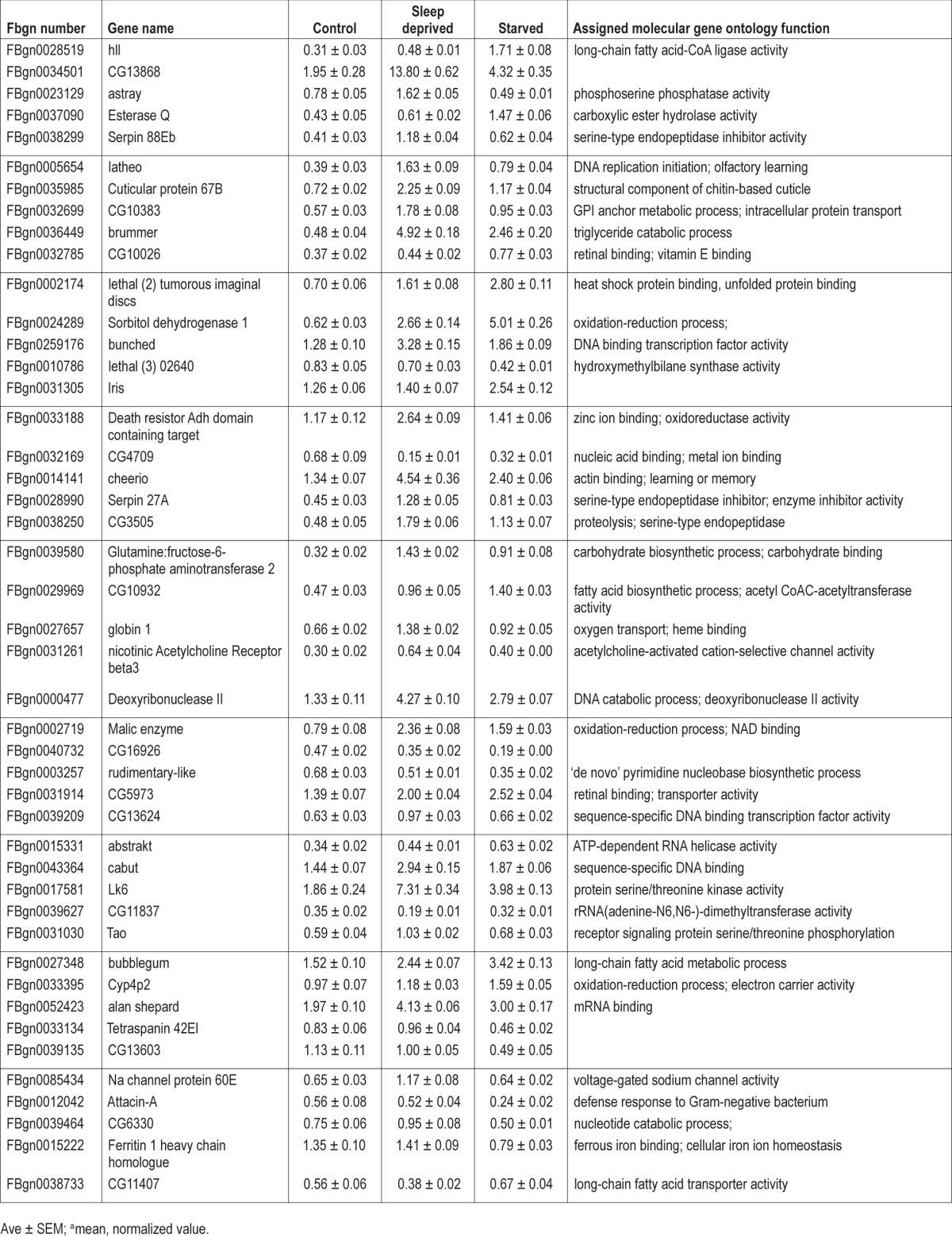
Striking phenotypic differences between flies that are sleep deprived and those that are starved have been previously observed.11 Thus, we hypothesized that transcripts that are differentially regulated between sleep deprived cyc01 mutants and their starved siblings should identify genes that confer either vulnerability or resilience to sleep loss, respectively. Sleep was evaluated in 3-day old cyc01 mutants maintained in constant darkness. Following 2 days of baseline in which baseline sleep was not different between the three groups (Figure S1), siblings were either sleep deprived or starved for 7 h. Six independent experiments were completed for each condition over the course of 3 mo. Two thirds of the flies from an experiment were frozen so that RNA could be extracted from whole heads. The remaining one third of the flies were monitored for an additional 24 h to assess the size of the homeostatic response; these latter behavioral results have been reported previously.11 Transcription profiling was conducted using partial cDNA microarrays processed at the University of California, San Diego Biogem Biomedical Microarray Facility. Because the cDNA microarray did not represent the entire genome, only 4,659 genes were identified as present in all replicates. Of the 4,659 genes that were present, 84 genes were statistically different between sleep deprived cyc01 and their starved siblings as assessed using the Cyber-T Bayesian statistical framework28 and confirmed with qPCR (Table 1). Raw array values are displayed in Table S1 and background subtracted, mean normalized data for all 4,659 present ESTs can be found in Table S2. Identified genes include chaperones, channels, proteolysis, transcription factors, kinases/phosphatases, carbohydrate metabolism, and lipid metabolism, suggesting multiple pathways are altered in the response to sleep deprivation.
Table 1.
Gene expression changes in sleep deprived and starved cyc01 flies identified by microarray analysis.

To further confirm the microarray results, we used qPCR to evaluate gene expression in an independent replicate consisting of sleep deprived and starved cyc01 flies. In addition, we extracted messenger RNA (mRNA) from heads of both per01 and CS flies under baseline and after either sleep deprivation or starvation. These latter experiments were designed to determine the extent to which the genes identified in the microarray on cyc01 mutants would be modified in a different clock mutant that responds differently to sleep loss2 and to a common background strain with an intact clock. cyc01, per01, and CS flies were maintained in DD to avoid differentially activating genes with light exposure. Because cyc01 mutants begin to die after only 10 h of sleep deprivation, both cyc01 and per01 flies were sleep deprived or starved for 7 h while CS flies were deprived of sleep or starved for 12 h during their primary sleep period. The primary sleep period was identified from the previous days' data based on the average time that the CS flies initiated their longest sleep bout.24 Changes in gene expression as assessed by qPCR can be seen in Figure 1A. To simplify the presentation, only those genes that show similar relative patterns in cyc01 flies and either CS and/or per01 flies are graphed in Figure 1A. Note that the cyc01, per01, and CS flies are in different genetic backgrounds such that it is not possible to directly assess quantitative changes in gene expression between the genotypes. However, within-genotype comparisons can be made by evaluating the relative direction of changes seen in siblings following sleep loss or starvation. After the relative changes in gene expression have been quantified within a genotype, it is informative to ask whether the relative changes are similar in the other genotypes, per01 and CS flies. Of the 84 transcripts identified on the microarray, numerous genes maintained the same relative relationship between sleep deprived and starved states seen in cyc01 flies in per01 or CS flies (Figure 1B), suggesting that changes in these transcripts withstand changes in background. Relative changes are highlighted for each genotype in gray. Together these data suggest that the identified genes may play a role in regulating the response to sleep loss.
Figure 1.
Gene expression profiles in sleep deprived and starved cyc01, per0, and Wild-type Canton-S (CS) flies. (A) Relative fold changes versus untreated genetic controls in representative genes derived from complementary DNA arrays (6 samples/condition) or quantitative polymerase chain reaction (qPCR, 1 sample/group, n = 20 flies). All flies were maintained in dark-dark cycle (DD). cyc01 and per01 were sleep deprived (SlDn) or starved (Stv) concurrently for 7 h while CS flies were deprived for 12 h during their primary sleep period. (B) Percent change versus untreated genetic controls expressed as mean ± standard error of the mean. Fold changes between sleep deprived and starved flies are highlighted in gray.
Many of the genes with the greatest transcriptional differences between the sleep deprived and starved flies were genes involved with lipid metabolism. The gene with the largest change was hll, but also near the top of our list was brummer (bmm). This latter observation is of interest in view of our previous finding that mutants of bmm have an increased sleep rebound after sleep deprivation.11 In addition, other lipid metabolism genes including CG11407, bubblegum (bgm), and cueball (cue) were present on our list. Thus, there appear to be broad alterations in transcription of lipid metabolic enzymes with sleep deprivation.
Behavioral Validation of Microarray
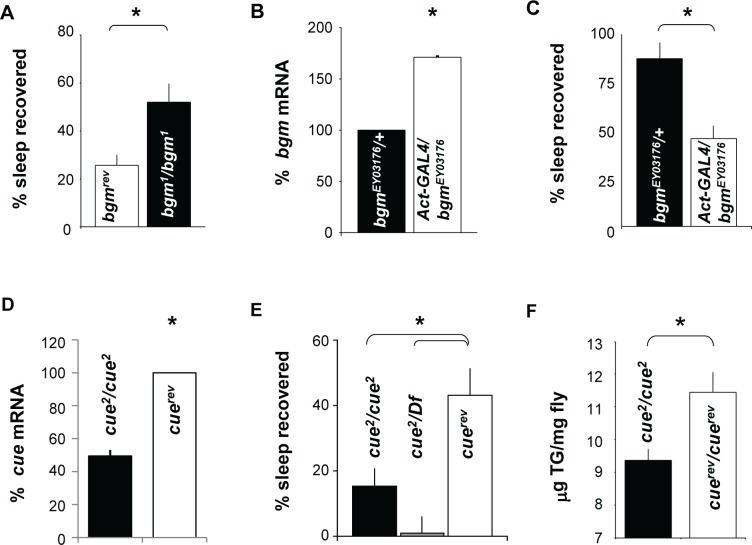
All gene profiling experiments are correlational in nature.35 Thus, transcriptional changes that result from extended waking induced by either sleep deprivation or starvation may represent changes that (1) regulate the homeostatic response, (2) induce physiologic impairments during sleep deprivation, (3) protect flies from the negative consequences of waking, or (4) have no direct involvement. Ideally, the precise role these genes play should be relevant for sleep regulation in general. That is, our goal is not to investigate the molecular mechanisms underlying cycle function, but rather it is to identify genes that influence sleep homeostasis. Given that many of the transcriptional changes we report between sleep deprived and starved flies are small and affect molecular pathways that are modified by post-transcriptional modifications, we have evaluated their role using genetics. If any of the genes we have identified play an active role in sleep homeostasis, their manipulation should alter the response to sleep deprivation. Thus, we used genetics to evaluate three genes, bubblegum, cueball, and hll, given their putative role in the processing of complex lipids. bubblegum (bgm) is an acyl-coenzyme A (CoA) synthetase differentially expressed during sleep deprivation and starvation. Several mutations for bgm are available, including bgm1 in which a PlacZ element is inserted in the first intron. This insertion results in large reductions in the expression of the bgm gene.36 Although young bgm1 flies show normal brain morphology, 15-day-old mutants show signs of degeneration in the optic lobe and retina. Precise excision of the P-element (bgmrev) restored the normal phenotype.36 The bgm mutants have a disruption in the processing of very long chain fatty acids (VLCFA) that results in their accumulation throughout the fly.36 Sleep parameters for bgm1 and their genetic controls bgmrev were consistent with the normal variation seen for wild-type flies and can be found in Table S3. bgm1 and bgmrev were sleep deprived for 12 h during the dark period when they were 5 days old so that the recovery period was completed by age 7 days. Sleep homeostasis is calculated for each individual as a ratio of the minutes of sleep gained above baseline during the 48 h of recovery divided by the total min of sleep lost during 12 h of sleep deprivation (min gained/min lost). As seen in Figure 2A, bgm1 mutants displayed a significantly larger sleep rebound than their genetic background control, bgmrev.
Figure 2.
Genetic validation of microarray. (A) Sleep homeostasis is increased in bgm1 mutants (n = 21) compared to its background genetic control, bgmrev (n = 24). * P = 0.0045, Student t-test. (B) bgm messenger RNA (mRNA) from flies in Actin-GAL4/bgmEY03176 compared to bgmEY01376/+. mRNA is expressed as a percentage of bgmEY01376/+ (n = 5 flies/group). *P < 0.05 by Student t-test. (C) bgmEY03176/+ flies (n = 45) also show an increased homeostatic response in comparison to Act-GAL4/bgmEY03176 (n = 27). *P = 0.00065 by Student t-test. (D) cue mRNA levels are decreased in cue2 homozygotes compared with the background control with the P-element precisely excised (cuerev). Levels are presented as a percentage of cuerev. *P < 0.05 by Student t-test. (E) Flies homozygous for cue2 (n = 129), or hemizygous cue2/Df(3L)Ar14-8 (n = 32) have a significantly reduced sleep rebound compared to genetic background controls in which the P-element has been excised, cuerev (n = 39). One-way analysis of variance F2,197 = 5.80; *P < 0.001 modified Bonferroni test. (F) Total triglyceride levels were significantly decreased in cue2/cue2 mutants compared to its background control cuerev/cuerev. *P = 0.017 using Student t-test with an n = 5 groups of 10 flies per genotype.
To confirm that decreased bgm resulted in an increased sleep rebound, we employed the Drosophila GAL4- Upstream Activating Sequence (UAS) system.37 In this system, a yeast transcription factor, GAL4, is used to drive tissue-specific expression of a given transcript by binding and transcribing genetic elements downstream of the yeast UAS. Because it has yet to be determined which are the critical tissues for modulating bgm expression, we used a driver that would express throughout the fly, Act-GAL4.38 To further evaluate bgm, we examined sleep homeostasis in a second allele of bgm, bgmEY03176. bgmEY03176 flies contain a P-element inserted into the 5' region of the bgm gene, just upstream of a putative transcript start site for one form of the bgm transcript. The P-element contains an integrated UAS element such that GAL4 drivers can be used to exogenously express bgm. Using Act-GAL4, we increased the expression of bgm throughout the fly compared to the background control, bgmEY03176/+. qPCR reveals that bgm mRNA expression is increased in Act-GAL4/+ > bgmEY03176/+ flies compared to bgmEY03176/+ controls (Figure 2B). Consistent with the findings from the loss-of-function in which bgm1 mutants have a large sleep rebound, gain-of-function Act-GAL4/+ > bgmEY03176/+ flies exhibited a reduced sleep rebound compared to bgmEY03176/+ controls (Figure 2C). The increased sleep rebound observed in bgmEY03176/+ is likely caused by the reduced levels of bgm compared to CS flies (Figure S2). Thus, the exogenous expression of bgm using the Act-GAL4 likely rescues a fly hypomorphic for bgm. Both the deletion mutant and the GAL4-UAS experiments support the conclusion that reduced levels of bgm result in an increased homeostatic response to sleep deprivation.
To further evaluate the possibility that genes associated with low-density lipoprotein receptor activity can influence sleep homeostasis, we identified a gene from our microarray that is predicted to have such activity, cueball (cue) (http://flybase.org/). The P-element mutant cue2 has been reported to be a hypomorphic allele resulting from a P-element insertion.39 As seen in Figure 2D, we confirmed that cue2 had decreased levels of cue mRNA compared to the precise excision genetic control (cuerev). Sleep data for cue2 and cuerev were within the normal variation found in wild-type flies and can be found in Table S3. Flies with a homozygous mutation in cue2 exhibit a low sleep rebound following 12 h of sleep deprivation compared to genetic controls (Figure 2E). To confirm that the low sleep rebound mapped to the cue locus, we crossed cue2 mutants to flies carrying a deficiency (Df) covering the cue gene. As seen in Figure 2E, cue2/Df flies also displayed a severely reduced homeostatic response following sleep deprivation. Interestingly, cue2 mutants show significantly reduced organismal triglyceride stores compared to their genetic background controls, cuerev confirming its role in lipid metabolism (Figure 2F).
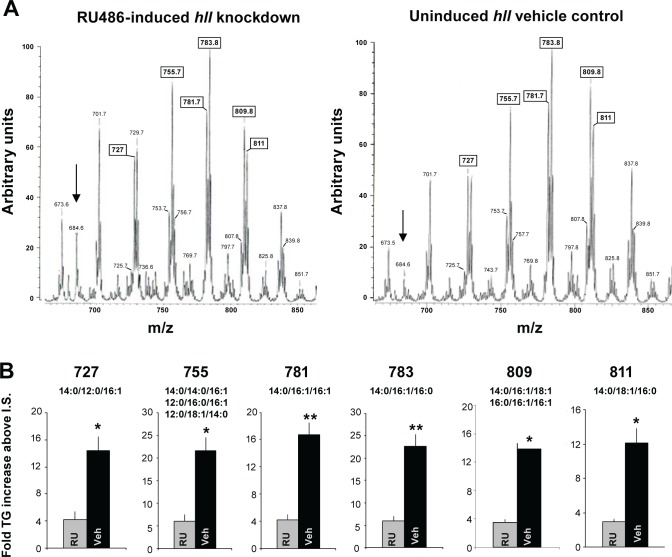
Finally, we evaluated the role of the gene with the greatest transcriptional change, hll, using RNA interference (RNAi). hll is a predicted acyl-CoA synthetase. We therefore determined whether knocking down hll using RNAi would alter lipid phenotypes. We used the inducible GeneSwitch GAL4 system to knockdown hll throughout the fly. In the GeneSwitch system, mifepristone is fed to the flies at the desired time to induce the expression of the RNAi transcript. Thus, adult sibling tubulin-GeneSwitch driving hll RNAi (tubGS > hllRNAi) are given food containing either mifepristone or the vehicle control. Thus, comparisons are made between the induced and uninduced siblings with the same background. Moreover, because induction of transcription occurs at the adult stage, flies avoid any developmental effects of RNA knockdown at earlier stages that may confound results. We analyzed lipid species in extracts from the flies by ESI/MS as previously described.30–34 Consistent with previous reports that changing acyl-CoA synthase expression results in altered triglyceride stores,40–42 we report that the global knockdown of hll in tubGS > hllRNAi flies fed RU486 (RU) results in an overall reduction of triglycerides compared to genetically identical siblings maintained on vehicle (Figures 3A and 3B). It should be noted that we and others have consistently reported that RU does not influence a variety of phenotypes including lifespan, sleep, sleep homeostasis, short-term memory, short-term memory following sleep deprivation, olfactory conditioning, phototaxis, geotaxis, locomotion, and the escape response.43–46 To determine whether the changes in the peaks involved changes in isobaric molecular constituents at a given m/z value, we conducted tandem mass spectrometry in which precursor ions selected in the first quadrupole were accelerated into a chamber containing argon to induce CAD and product ions were then analyzed in the final quadrupole. A representative tandem mass is shown in Figure 4. Although there is a decrease in overall triglycerides, we could not detect a shift in the distribution of triglyceride molecular species in tubGSw > hll flies compared to vehicle-fed genetically identical siblings. Together these data indicate that hll influences lipid metabolism.
Figure 3.
Triglyceride levels are decreased with ubiquitous hll knockdown. (A) Representative triglyceride profiles from flies with ubiquitous knockdown of hll (tubGS- > UAS-hllRNAi induced by RU486, left tracing) and the uninduced, genetically identical siblings on the vehicle control (veh, right tracing). We chose to use the GeneSwitch system to minimize the number of groups analyzed and to better control for genetic background. Tracings were generated using positive ion electrospray ionization (ESI) mass spectrometric (MS) analyses of lipid Li+ adducts from whole flies. Peaks are labeled by their mass to charge (m/z) ratios, and the boxed m/z values correspond to the peaks quantified in (B). The arrow denotes internal standard (m/z 684), and the most intense peak is normalized to 100. Although there is a decrease in overall triglyceride abundance, there is no change in the distribution of triglyceride molecular species. (B) Quantification of triglyceride peaks from the mass spectra of hll knockdown and the uninduced siblings (four samples per group; n = 5 flies/sample). Numbers above graph represent the m/z value from (A) and the smaller numbers are the lipid species that could correspond to the given m/z value. The intensity of the peak in question was divided by that of the internal standard (I.S.), and the resultant ratio was then normalized to protein content to determine the fold-increase of the triglyceride species represented by the peak. *P < 0.05) or **P < 0.01 as calculated by Student t-test with a Bonferroni correction.
We then confirmed that the lipid phenotype was associated with knockdown of hll in the RU486 induced flies compared to the vehicle-fed controls. qPCR results demonstrate that hll transcript levels are only 14% of the levels observed in controls (Figure 5A). To determine whether the ubiquitous knockdown of hll altered sleep homeostasis, we sleep deprived RU-fed tubGS > hllRNAi flies and their vehicle-fed siblings. As seen in Figure 5B, sleep rebound was significantly reduced compared to genetic background controls. To further rule out genetic background, we knocked down hll using the Act-GAL4 driver. As seen in Figure 5C, Act-GAL4/+ > UAS-hllRNAi/+flies showed a substantial reduction in hll mRNA compared to Act-GAL4/+ and UAS-hllRNAi/+ parental controls. Sleep data for parental lines (Act-GAL4/+ and UAS-hllRNAi/+) and the experimental line (Act-GAL4/+ > UAS-hllRNAi/+) can be found in Table S3. Sleep homeostasis was evaluated in Act-GAL4/+ > UAS-hll RNAi/+ flies and their background controls following 12 h of sleep deprivation. As seen in Figure 5D, flies in which hll was knocked down showed a significantly reduced sleep rebound compared to each of the parental lines, Act-GAL4/+ and UAS-hllRNAi/+. Together with the data presented for hll, bgm, and cue, these data indicate that a subset of the genes identified by transcriptional profiling can indeed influence sleep regulation as measured by sleep homeostasis.
Figure 5.
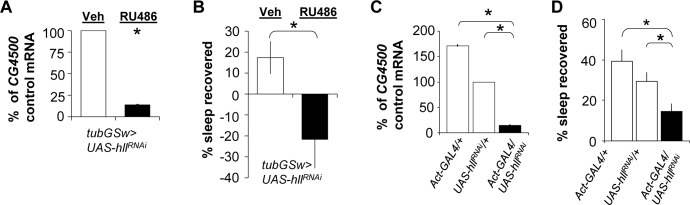
hll mutants exhibit reduced sleep homeostasis. (A) Levels of hll messenger RNA (mRNA) from tubGS > hllRNAi induced with RU486 are decreased compared to sibling flies uninduced using vehicle control. mRNA is expressed as a percentage of vehicle control (n = 5 flies/group). *P < 0.05 by Student t-test. (B) Sleep homeostasis is reduced when tubGS > hllRNAi are induced with the drug mifepristone (RU486, n = 28) compared to when the flies are treated with the vehicle control (veh, n = 28). (C) mRNA for hll was reduced in whole heads in Actin-Gal4/UAS-hllRNAi flies compared to parental lines (Actin-GAL4/+ and UAS-hllRNAi/+). mRNA levels were normalized to UAS-hllRNAi/+; n = 20 heads/group. *P < 0.05 by planned comparison Student t-test in indicated comparison. (D) Sleep homeostasis is reduced in Actin-Gal4/UAS-hllRNAi (n = 66) compared to Act-GAL4/+ (n = 39) and UAS-hllRNAi/+ (n = 66) parental lines. One-way analysis of variance F2,168 = 7.51; *P < 0.01 modified Bonferroni Test. *P < 0.01 by Student t-test.
DISCUSSION
We have described a novel strategy to identify genes that are likely to play a role in sleep homeostasis or that protect flies from the negative effects of sleep loss. This strategy takes advantage of the observation that waking induced by starvation leads to different functional outcomes than the same amount of waking induced by sleep deprivation.11 Our approach also takes advantage of the fact that cyc01 flies quickly accumulate the negative effects of sleep deprivation.2 Using this approach we have identified 84 transcripts that are differentially regulated between genetically identical sleep deprived and starved siblings. Importantly, genetic analysis confirms that three of these genes, bgm, cue, and hll modulate sleep homeostasis. Previous genetic studies have confirmed a role for other genes on this list including brummer, Delta, bunched, and positive regulators of Attacin.11,47,48 Interestingly, a tool recently developed to identify orthologs between flies and humans indicates that 68% of the genes identified from this array have human homologs, suggesting that these genes may inform the field about human sleep regulation and adaptation to sleep deprivation.49 In fact, transcripts for several genes identified on the microarray, including Amylase, malic enzyme, cheerio, and the human homologue of bunched (Tsc22) are also known to be modulated by sleep deprivation in humans.44,48,50,51
Because starvation is not an uncommon occurrence in nature, it is likely that the underlying molecular mechanisms mediating the effects of starvation on the response to sleep loss have been subjected to evolutionary forces. In fact, it has been suggested that animals able to remain alert and vigilant during periods of starvation may have a selective advantage over animals that accrue sleep debt at a normal rate.52,53 With this in mind, we expect that comparing waking induced by sleep deprivation to waking induced by starvation may expedite the identification of genes whose role in sleep regulation may be evolutionarily conserved and perhaps more related to mechanisms regulating sleep in humans. Our results have identified 84 genes, including channels, immune, signaling, peptide breakdown, and transcription factors, among many other types of functions. Similar types of transcriptional changes have been identified in other microarray studies.47,54–57 In each instance, many of the specific genes do not overlap but the broader categories are consistent between studies, implicating those pathways in these processes.
We chose to focus on lipid metabolism genes to validate the microarray results because lipids play a number of roles throughout the cell, including energy production, membrane maintenance, and signaling properties.58 Despite the fact that the first gene to be associated with sleep deprivation in flies was fatty acid synthase (Fas),1 and subsequent microarray studies in flies and mammals have identified lipid metabolism genes as being modulated by behavioral state,44,55,56 the relationship between sleep loss and lipid processing remains poorly understood. Given the diversity of roles that lipids play in the cell, the precise role of lipid metabolism in sleep regulation is likely to be complex. For example, modest increases in the expression of brain-type fatty acid binding protein (Fabp7) disrupt sleep whereas larger increases in Fabp7 expression result in both an increase in sleep and enhanced memory consolidation.59 In rodents, a deficiency in short-chain acyl-CoA dehydrogenase causes a slowing in theta frequency during paradoxical sleep without altering other sleep states or modulating the power of other oscillations.19 Quantitative genetic analysis of sleep in flies suggests a relationship between energy stores and total sleep time.60 However, as noted by the authors, the relationship between lipid stores and sleep time is likely to be dependent on how these stores are used rather than just their presence.
Interestingly, the lipid metabolism genes we have identified primarily influence sleep homeostasis while having little, if any, effect on baseline sleep time11 (Table S3). Thus, mutations in the previously characterized bmm mutant (which reduce lipolysis and block fatty acid release) result in a substantially increased sleep rebound following a night of sleep loss whereas mutations in Lsd2 (that exhibit an increase fatty acid release) do not respond to sleep deprivation with either a sleep rebound or deficits in cognitive performance.11 In this study, bgm and hll display an opposite response to sleep deprivation, though they both encode for acyl-CoA synthetases. Though male bgm mutants exhibit a prominent lipid phenotype, both male and female bgm mutants exhibit a neurodegenerative phenotype.36 These results, in combination with the sequence homology to proteins in humans and mice that show functional long chain fatty acid (LCFA) and VLCFA acyl-CoA synthase activity, indicate that bgm mutant animals suffer from a lipid defect.61–63 Hll currently does not have any published information on its specificity. Although the underlying mechanisms are not entirely clear, individual acyl-CoA synthetases are selective for FFA based on carbon chain length and they achieve additional specificity by cellular and subcellular localization.64,65 In fact, the expression pattern of these two genes is different according to Fly–FISH and BDGP expression pattern projects.66,67 Moreover, as a VLCFA transporter, bgm is likely active in the peroxisome whereas hll is likely active at the lipid droplet.36,65 These localization, developmental, and functional differences may account for the distinct responses to sleep deprivation. Unfortunately, we did not detect reductions in specific lipids upon RNAi knockdown of hll using lipidomics. Rather, the knockdown of hll resulted in the general reduction of all lipid species assayed. Therefore, future studies will be required to determine if the modulation of specific lipids are responsible for either enhancing or slowing the negative effects of sleep loss.
The interaction between sleep and metabolism has regained attention in recent years.68–71 For example, both insufficient sleep and sleep fragmentation correlate with increased body mass index (BMI).6,72 The increased BMI may be driven by a reversal of satiety hormones73 which could explain the observation that sleep deprived subjects eat more calorically dense snacks16,17,71,74 or potentially through increases in reactive oxygen species.75 Moreover, sleep deprivation alters molecular signaling in human adipose tissue, increases plasma nonesterified fatty acids and increases ketone bodies.76–77 Thus, inadequate sleep appears to alter the way that energy stores are used. Changes in metabolism are also known to alter sleep. That is, starvation induces waking in multiple species.11,13–15 In addition, mutations in lipid metabolism genes, such as Lsd2, have previously been shown to alter the sensitivity to sleep deprivation.11 Mutations in Lsd2 result in an increase in lipolysis which is predicted to increase energy availability.78,79 Interestingly, the increased presence of adenosine triphosphate and adenosine derived from neuronal activity and energy expenditure has been hypothesized to increase sleep drive.20,80,81 Thus, although the precise molecular mechanism translating metabolic signals into sleep need are not known, factors resulting from lipid metabolism are well situated to translate metabolic signals to sleep regulatory centers.
It is commonly recognized that sleep deprivation studies are correlational in nature: one applies a stimulus to keep animals awake and then measures a particular outcome.82,83 The outcome may be either due to the direct effects of sleep loss or the method used to keep the animal awake. Even if a confounding role of the stimulus used to keep the animal awake is excluded, it remains possible that the animal is forced to initiate adaptive mechanisms to defend against the sleep deprivation-induced impairment, and as a consequence, the adaptation rather than the direct effects of sleep loss per se may be primarily investigated.83 With this in mind, an advantage of evaluating the effects of sleep loss in the fly in general, and cyc01 mutants in particular, is the rapidity to which sleep deprivation results in negative outcomes in these animals.2,11,12,45,46,84 Given the speed with which deficits accrue, especially in cyc01 mutants, it is unlikely that they are able to mount an adaptive response of sufficient magnitude to mask the underlying pathology. If this proves to be the case, then the genes on our microarray that respond to sleep loss may be particularly interesting for elucidating the mechanisms associated with the negative effects of sleep loss. Similarly, flies also respond very rapidly to the absence of food and can sustain extended periods of waking without accruing adverse outcomes. Thus, the genes that are quickly modulated during waking induced by starvation may be particularly useful for understanding how to offset or slow the harmful effects sleep loss. Nonetheless, there is unlikely to be a magic bullet that can be used to protect animals from the effects of sleep deprivation. Indeed, we have previously shown that polymorphisms in a gene that confers resilience to sleep loss also increase the vulnerability of those animals to starvation.53 Thus, although these results indicate that lipids can be either positive or negative regulators of sleep homeostasis, it remains unclear how sleep regulatory centers process this information along with circadian and homeostatic signals, to adjust their outputs to match sleep need with environmental demands.
DISCLOSURE STATEMENT
This study was funded in part by 2 R01 NS051305-09 (P.J.S.), WM Keck Foundation Fellowship (M.S.T.), and CNRU grant NIH P30 DK56341. BIOGEM is supported by NIH/NIDDK - Award 1 P30 DK063491-03. Lipid MS analyses were performed in a facility supported by United States Public Health Service Grants P41-RR00954, P60-DK20579, P30-DK56341, and R37-DK34388. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank Laura Gottschalk, Alan Bohrer, and Trey Coleman for technical assistance. Also, we appreciate the critical reading and comment from Stephane Dissels and Krishna Melnattur.
SUPPLEMENTAL MATERIAL
Baseline sleep is similar in cyc01 flies prior to being sleep deprived, starved or serving as controls. One-way ANOVA for condition F2,766 = 1.3; P = 0.26. No significant differences were found between conditions using a modified Bonferroni Test.
Visit www.journalsleep.org to download the supplemental tables Tables S1–S3 (Microsoft Excel format).
bgm RNA levels in wildtype (Canton S) and the bgm mutant (bgmEY03176). *P < 0.05.
REFERENCES
- 1.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 2.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 3.Everson CA, Bergmann BM, Rechtschaffen A. Sleep deprivation in the rat: III. Total sleep deprivation. Sleep. 1989;12:13–21. doi: 10.1093/sleep/12.1.13. [DOI] [PubMed] [Google Scholar]
- 4.Kripke DF, Garfinkel L, Wingard DL, Klauber MR, Marler MR. Mortality associated with sleep duration and insomnia. Archives of general psychiatry. 2002;59:131–6. doi: 10.1001/archpsyc.59.2.131. [DOI] [PubMed] [Google Scholar]
- 5.Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–26. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- 6.Taheri S, Lin L, Austin D, Young T, Mignot E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLoS Med. 2004;1:e62. doi: 10.1371/journal.pmed.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandner MA, Sands-Lincoln MR, Pak VM, Garland SN. Sleep duration, cardiovascular disease, and proinflammatory biomarkers. Nat Sci Sleep. 2013;5:93–107. doi: 10.2147/NSS.S31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tononi G, Cirelli C. Sleep and synaptic homeostasis: a hypothesis. Brain Res Bull. 2003;62:143–50. doi: 10.1016/j.brainresbull.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Bergmann BM, Everson CA, Kushida CA, et al. Sleep deprivation in the rat: V. Energy use and mediation. Sleep. 1989;12:31–41. doi: 10.1093/sleep/12.1.31. [DOI] [PubMed] [Google Scholar]
- 10.Everson CA, Toth LA. Systemic bacterial invasion induced by sleep deprivation. Am J Physiol Regul Integr Comp Physiol. 2000;278:R905–16. doi: 10.1152/ajpregu.2000.278.4.R905. [DOI] [PubMed] [Google Scholar]
- 11.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- 13.Keene AC, Duboue ER, McDonald DM, et al. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20:1209–15. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danguir J, Nicolaidis S. Dependence of sleep on nutrients' availability. Physiol Behav. 1979;22:735–40. doi: 10.1016/0031-9384(79)90240-3. [DOI] [PubMed] [Google Scholar]
- 15.Borbely AA. Sleep in the rat during food deprivation and subsequent restitution of food. Brain Res. 1977;124:457–71. doi: 10.1016/0006-8993(77)90947-7. [DOI] [PubMed] [Google Scholar]
- 16.Nedeltcheva AV, Kilkus JM, Imperial J, Kasza K, Schoeller DA, Penev PD. Sleep curtailment is accompanied by increased intake of calories from snacks. Am J Clin Nutr. 2009;89:126–33. doi: 10.3945/ajcn.2008.26574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spiegel K, Tasali E, Penev P, Van Cauter E. Brief communication: sleep curtailment in healthy young men is associated with decreased leptin levels, elevated ghrelin levels, and increased hunger and appetite. Ann Intern Med. 2004;141:846–50. doi: 10.7326/0003-4819-141-11-200412070-00008. [DOI] [PubMed] [Google Scholar]
- 18.Spiegel K, Knutson K, Leproult R, Tasali E, Van Cauter E. Sleep loss: a novel risk factor for insulin resistance and Type 2 diabetes. J Appl Physiol. 2005;99:2008–19. doi: 10.1152/japplphysiol.00660.2005. [DOI] [PubMed] [Google Scholar]
- 19.Tafti M, Petit B, Chollet D, et al. Deficiency in short-chain fatty acid beta-oxidation affects theta oscillations during sleep. Nat Genet. 2003;34:320–5. doi: 10.1038/ng1174. [DOI] [PubMed] [Google Scholar]
- 20.Benington JH, Heller HC. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol. 1995;45:347–60. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- 21.Dietzl G, Chen D, Schnorrer F, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 22.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 24.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods Enzymol. 2005;393:759–72. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- 25.McCue MD. Starvation physiology: reviewing the different strategies animals use to survive a common challenge. Comp Biochem Physiol A Mol Integr Physiol. 2010;156:1–18. doi: 10.1016/j.cbpa.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 26.White KP, Rifkin SA, Hurban P, Hogness DS. Microarray analysis of Drosophila development during metamorphosis. Science. 1999;286:2179–84. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 27.Toma DP, White KP, Hirsch J, Greenspan RJ. Identification of genes involved in Drosophila melanogaster geotaxis, a complex behavioral trait. Nat Genet. 2002;31:349–53. doi: 10.1038/ng893. [DOI] [PubMed] [Google Scholar]
- 28.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 29.Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276:19937–44. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 30.Bao S, Li Y, Lei X, et al. Attenuated free cholesterol loading-induced apoptosis but preserved phospholipid composition of peritoneal macrophages from mice that do not express group VIA phospholipase A2. J Biol Chem. 2007;282:27100–14. doi: 10.1074/jbc.M701316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu FF, Bohrer A, Wohltmann M, et al. Electrospray ionization mass spectrometric analyses of changes in tissue phospholipid molecular species during the evolution of hyperlipidemia and hyperglycemia in Zucker diabetic fatty rats. Lipids. 2000;35:839–54. doi: 10.1007/s11745-000-0593-z. [DOI] [PubMed] [Google Scholar]
- 32.Hsu FF, Turk J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 1999;10:587–99. doi: 10.1016/S1044-0305(99)00035-5. [DOI] [PubMed] [Google Scholar]
- 33.Ramanadham S, Hsu FF, Bohrer A, Ma Z, Turk J. Studies of the role of group VI phospholipase A2 in fatty acid incorporation, phospholipid remodeling, lysophosphatidylcholine generation, and secretagogue-induced arachidonic acid release in pancreatic islets and insulinoma cells. J Biol Chem. 1999;274:13915–27. doi: 10.1074/jbc.274.20.13915. [DOI] [PubMed] [Google Scholar]
- 34.Ramanadham S, Hsu FF, Bohrer A, Nowatzke W, Ma Z, Turk J. Electrospray ionization mass spectrometric analyses of phospholipids from rat and human pancreatic islets and subcellular membranes: comparison to other tissues and implications for membrane fusion in insulin exocytosis. Biochemistry. 1998;37:4553–67. doi: 10.1021/bi9722507. [DOI] [PubMed] [Google Scholar]
- 35.Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: social life in molecular terms. Nat Rev Genet. 2005;6:257–70. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- 36.Min KT, Benzer S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science. 1999;284:1985–8. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- 37.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 38.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–71. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 39.Castrillon DH, Gonczy P, Alexander S, et al. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135:489–505. doi: 10.1093/genetics/135.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkes HA, Preston E, Wilks D, et al. Overexpression of acyl-CoA synthetase-1 increases lipid deposition in hepatic (HepG2) cells and rodent liver in vivo. Am J Physiol Endocrinol Metab. 2006;291:E737–44. doi: 10.1152/ajpendo.00112.2006. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, Naseem RH, Park BH, et al. Alpha-lipoic acid prevents lipotoxic cardiomyopathy in acyl CoA-synthase transgenic mice. Biochem Biophys Res Commun. 2006;344:446–52. doi: 10.1016/j.bbrc.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 42.Fujimoto Y, Onoduka J, Homma KJ, et al. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of Acyl-CoA synthetase. Biol Pharm Bull. 2006;29:2174–80. doi: 10.1248/bpb.29.2174. [DOI] [PubMed] [Google Scholar]
- 43.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci U S A. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seugnet L, Suzuki Y, Thimgan M, et al. Identifying sleep regulatory genes using a Drosophila model of insomnia. J Neurosci. 2009;29:7148–57. doi: 10.1523/JNEUROSCI.5629-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ. D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol. 2008;18:1110–7. doi: 10.1016/j.cub.2008.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanderheyden WM, Gerstner JR, Tanenhaus A, Yin JC, Shaw PJ. ERK phosphorylation regulates sleep and plasticity in Drosophila. PloS One. 2013;8:e81554. doi: 10.1371/journal.pone.0081554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams JA, Sathyanarayanan S, Hendricks JC, Sehgal A. Interaction between sleep and the immune response in Drosophila: a role for the NFkappaB relish. Sleep. 2007;30:389–400. doi: 10.1093/sleep/30.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ. Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol. 2011;21:835–40. doi: 10.1016/j.cub.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Y, Flockhart I, Vinayagam A, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seugnet L, Boero J, Gottschalk L, Duntley SP, Shaw PJ. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci U S A. 2006;103:19913–8. doi: 10.1073/pnas.0609463104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachmann V, Klaus F, Bodenmann S, et al. Functional ADA polymorphism increases sleep depth and reduces vigilant attention in humans. Cereb Cortex. 2012;22:962–70. doi: 10.1093/cercor/bhr173. [DOI] [PubMed] [Google Scholar]
- 52.Willie JT, Chemelli RM, Sinton CM, Yanagisawa M. To eat or to sleep? Orexin in the regulation of feeding and wakefulness. Annu Rev Neurosci. 2001;24:429–58. doi: 10.1146/annurev.neuro.24.1.429. [DOI] [PubMed] [Google Scholar]
- 53.Donlea J, Leahy A, Thimgan MS, et al. Foraging alters resilience/ vulnerability to sleep disruption and starvation in Drosophila. Proc Natl Acad Sci U S A. 2012;109:2613–8. doi: 10.1073/pnas.1112623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackiewicz M, Shockley KR, Romer MA, et al. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–57. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 55.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 56.Cirelli C, LaVaute TM, Tononi G. Sleep and wakefulness modulate gene expression in Drosophila. J Neurochem. 2005;94:1411–9. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 57.Jones S, Pfister-Genskow M, Cirelli C, Benca RM. Changes in brain gene expression during migration in the white-crowned sparrow. Brain Res Bull. 2008;76:536–44. doi: 10.1016/j.brainresbull.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitt F, Hussain G, Dupuis L, Loeffler JP, Henriques A. A plural role for lipids in motor neuron diseases: energy, signaling and structure. Front Cell Neurosci. 2014;8:25. doi: 10.3389/fncel.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerstner JR, Vanderheyden WM, Shaw PJ, Landry CF, Yin JC. Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PloS One. 2011;6:e15890. doi: 10.1371/journal.pone.0015890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harbison ST, Sehgal A. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics. 2008;178:2341–60. doi: 10.1534/genetics.107.081232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraisl P, Forss-Petter S, Zigman M, Berger J. Murine bubblegum orthologue is a microsomal very long-chain acyl-CoA synthetase. Biochem J. 2004;377:85–93. doi: 10.1042/BJ20031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pei Z, Oey NA, Zuidervaart MM, et al. The acyl-CoA synthetase “bubblegum” (lipidosin): further characterization and role in neuronal fatty acid beta-oxidation. J Biol Chem. 2003;278:47070–8. doi: 10.1074/jbc.M310075200. [DOI] [PubMed] [Google Scholar]
- 63.Steinberg SJ, Morgenthaler J, Heinzer AK, Smith KD, Watkins PA. Very long-chain acyl-CoA synthetases. Human “bubblegum” represents a new family of proteins capable of activating very long-chain fatty acids. J Biol Chem. 2000;275:35162–9. doi: 10.1074/jbc.M006403200. [DOI] [PubMed] [Google Scholar]
- 64.Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 2008;233:507–21. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grevengoed TJ, Klett EL, Coleman RA. Acyl-CoA metabolism and partitioning. Annu Rev Nutr. 2014;34:1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lecuyer E, Yoshida H, Parthasarathy N, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, Konikoff C, Van Emden B, et al. FlyExpress: visual mining of spatiotemporal patterns for genes and publications in Drosophila embryogenesis. Bioinformatics. 2011;27:3319–20. doi: 10.1093/bioinformatics/btr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jung CM, Melanson EL, Frydendall EJ, Perreault L, Eckel RH, Wright KP. Energy expenditure during sleep, sleep deprivation and sleep following sleep deprivation in adult humans. J Physiol. 2011;589:235–44. doi: 10.1113/jphysiol.2010.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–9. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanlon EC, Van Cauter E. Quantification of sleep behavior and of its impact on the cross-talk between the brain and peripheral metabolism. Proc Natl Acad Sci U S A. 2011;108(Suppl 3):15609–16. doi: 10.1073/pnas.1101338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markwald RR, Melanson EL, Smith MR, et al. Impact of insufficient sleep on total daily energy expenditure, food intake, and weight gain. Proc Natl Acad Sci U S A. 2013;110:5695–700. doi: 10.1073/pnas.1216951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Carreras A, Lee S, et al. Chronic sleep fragmentation promotes obesity in young adult mice. Obesity (Silver Spring) 2014;22:758–62. doi: 10.1002/oby.20616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spiegel K, Leproult R, Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354:1435–9. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- 74.St-Onge MP, Roberts AL, Chen J, et al. Short sleep duration increases energy intakes but does not change energy expenditure in normal-weight individuals. Am J Clin Nutr. 2011;94:410–6. doi: 10.3945/ajcn.111.013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khalyfa A, Wang Y, Zhang SX, Qiao Z, Abdelkarim A, Gozal D. Sleep fragmentation in mice induces nicotinamide adenine dinucleotide phosphate oxidase 2-dependent mobilization, proliferation, and differentiation of adipocyte progenitors in visceral white adipose tissue. Sleep. 2014;37:999–1009. doi: 10.5665/sleep.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Broussard JL, Ehrmann DA, Van Cauter E, Tasali E, Brady MJ. Impaired insulin signaling in human adipocytes after experimental sleep restriction: a randomized, crossover study. Ann Intern Med. 2012;157:549–57. doi: 10.7326/0003-4819-157-8-201210160-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chikahisa S, Shimizu N, Shiuchi T, Sei H. Ketone body metabolism and sleep homeostasis in mice. Neuropharmacology. 2013;79C:399–404. doi: 10.1016/j.neuropharm.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 78.Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13:603–6. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- 79.Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–81. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 80.Porkka-Heiskanen T, Strecker RE, Thakkar M, Bjorkum AA, Greene RW, McCarley RW. Adenosine: a mediator of the sleep-inducing effects of prolonged wakefulness. Science. 1997;276:1265–8. doi: 10.1126/science.276.5316.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krueger JM, Taishi P, De A, et al. ATP and the purine type 2 X7 receptor affect sleep. J Appl Physiol. 2010;109:1318–27. doi: 10.1152/japplphysiol.00586.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rechtschaffen A. Current perspectives on the function of sleep. Perspect Biol Med. 1998;41:359–90. doi: 10.1353/pbm.1998.0051. [DOI] [PubMed] [Google Scholar]
- 83.Rechtschaffen A, Bergmann BM, Everson CA, Kushida CA, Gilliland MA. Sleep deprivation in the rat: I. Conceptual issues. Sleep. 1989;12:1–4. doi: 10.1093/sleep/12.1.1. [DOI] [PubMed] [Google Scholar]
- 84.Huber R, Hill SL, Holladay C, Biesiadecki M, Tononi G, Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–39. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 85.Hsu FF, Turk J. Distinction among isomeric unsaturated fatty acids as lithiated adducts by electrospray ionization mass spectrometry using low energy collisionally activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 1999;10:600–12. doi: 10.1016/S1044-0305(99)00041-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baseline sleep is similar in cyc01 flies prior to being sleep deprived, starved or serving as controls. One-way ANOVA for condition F2,766 = 1.3; P = 0.26. No significant differences were found between conditions using a modified Bonferroni Test.
Visit www.journalsleep.org to download the supplemental tables Tables S1–S3 (Microsoft Excel format).
bgm RNA levels in wildtype (Canton S) and the bgm mutant (bgmEY03176). *P < 0.05.