Abstract
Thrombin contains electropositive patches at opposite poles of the molecule which represent potential exosites for the binding of macromolecular ligands. The function of anion-binding exosite I, the fibrin(ogen) recognition site, has been well described. Anion-binding exosite II, located near the carboxyl terminus of the molecule, has been proposed to bind heparin on the basis of chemical modification studies. To define the functional heparin-binding site on thrombin, purified recombinant alpha-thrombins were prepared with glutamic acid substitution for selected basic amino acid residues in exosite II or exosite I. Heparin affinity was assessed by NaCl gradient elution from heparin-agarose, and second-order rate constants for inhibition by antithrombin III were determined in the absence and presence of heparin. Affinity for heparin-agarose was reduced markedly by selected mutations in exosite II (R89E, R245E, K248E, and K252E, numbered from the amino terminus of the B chain) but not by other mutations in exosite II (K174E, K247E) or by mutations in exosite I (R68E, K154E). All recombinant thrombins had similar rate constants for inhibition by antithrombin III without heparin. However, affinity for heparin-agarose correlated directly with the rate of inhibition by antithrombin III with heparin. These results demonstrate that selected mutations in anion-binding exosite II define a functional heparin-binding site and support the template mechanism of heparin action.
Full text
PDF

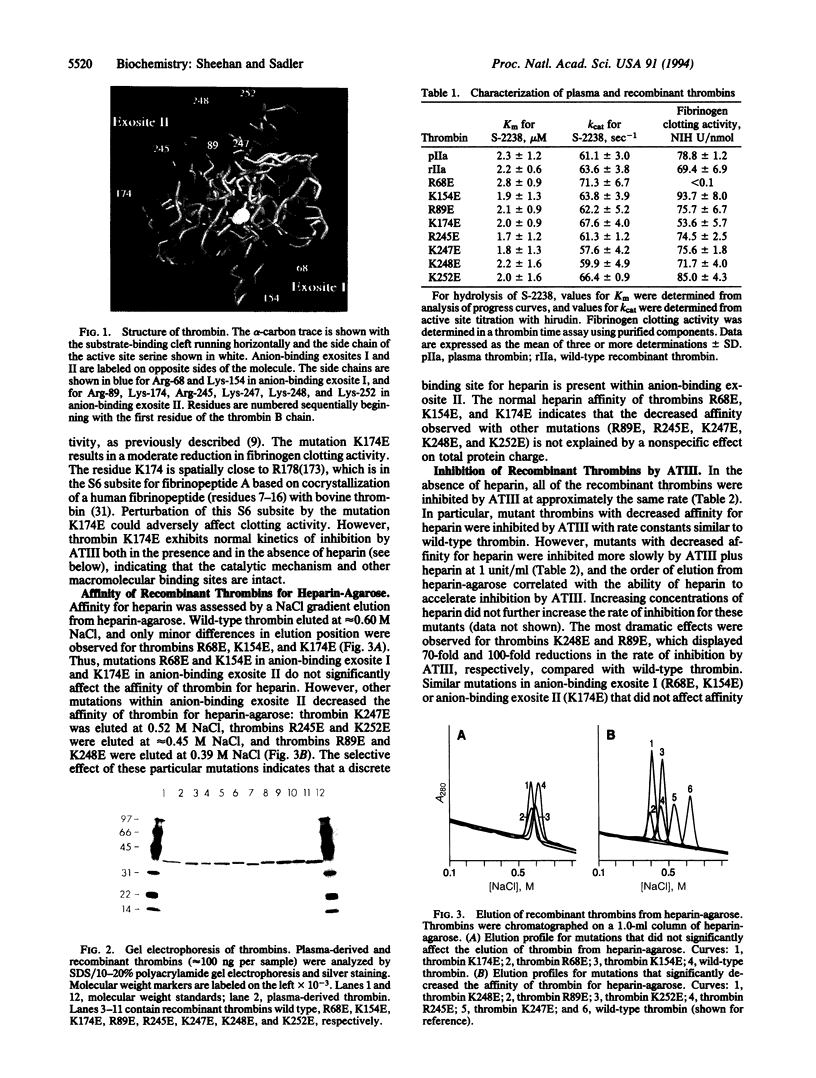
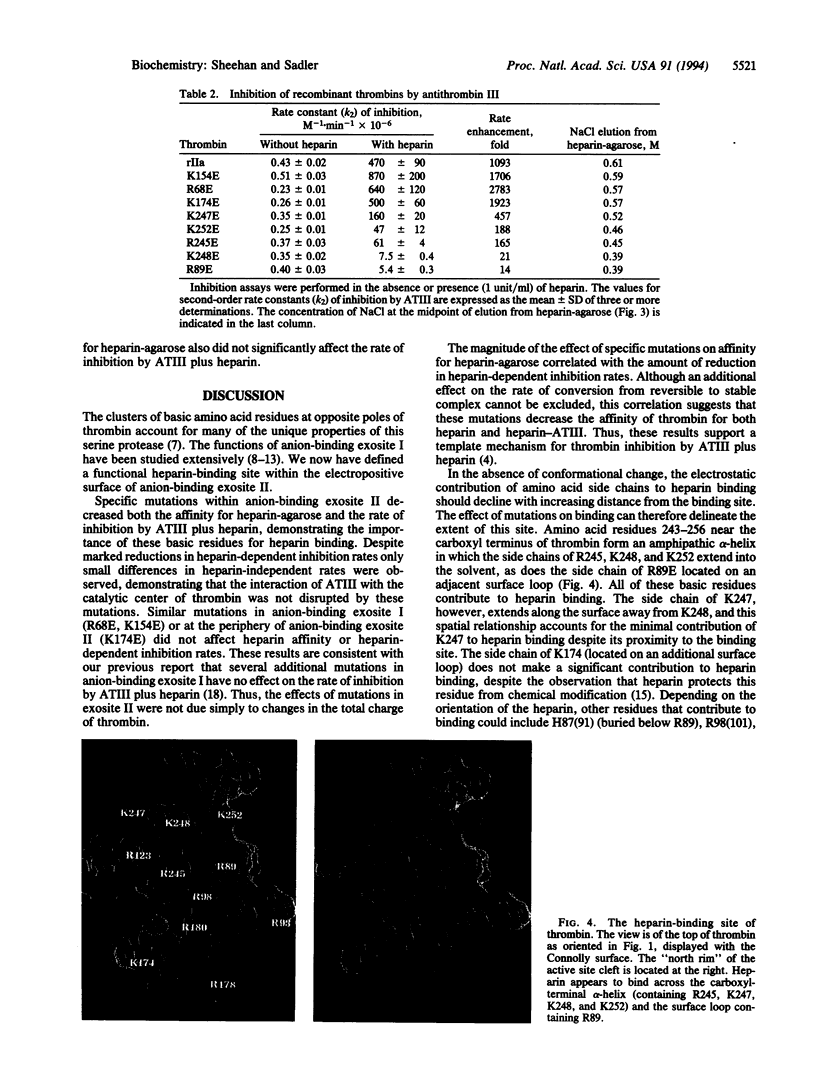

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arni R. K., Padmanabhan K., Padmanabhan K. P., Wu T. P., Tulinsky A. Structures of the noncovalent complexes of human and bovine prothrombin fragment 2 with human PPACK-thrombin. Biochemistry. 1993 May 11;32(18):4727–4737. doi: 10.1021/bi00069a006. [DOI] [PubMed] [Google Scholar]
- Bajaj S. P., Sabharwal A. K., Gorka J., Birktoft J. J. Antibody-probed conformational transitions in the protease domain of human factor IX upon calcium binding and zymogen activation: putative high-affinity Ca(2+)-binding site in the protease domain. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):152–156. doi: 10.1073/pnas.89.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Shavit R., Eldor A., Vlodavsky I. Binding of thrombin to subendothelial extracellular matrix. Protection and expression of functional properties. J Clin Invest. 1989 Oct;84(4):1096–1104. doi: 10.1172/JCI114272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshop B. A., Wrenn R. F., Frieden C. Analysis of numerical methods for computer simulation of kinetic processes: development of KINSIM--a flexible, portable system. Anal Biochem. 1983 Apr 1;130(1):134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- Bode W., Turk D., Karshikov A. The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992 Apr;1(4):426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin A. D., Weintraub H. J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989 Jan-Feb;9(1):21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- Chase T., Jr, Shaw E. p-Nitrophenyl-p'-guanidinobenzoate HCl: a new active site titrant for trypsin. Biochem Biophys Res Commun. 1967 Nov 30;29(4):508–514. doi: 10.1016/0006-291x(67)90513-x. [DOI] [PubMed] [Google Scholar]
- Church F. C., Pratt C. W., Noyes C. M., Kalayanamit T., Sherrill G. B., Tobin R. B., Meade J. B. Structural and functional properties of human alpha-thrombin, phosphopyridoxylated alpha-thrombin, and gamma T-thrombin. Identification of lysyl residues in alpha-thrombin that are critical for heparin and fibrin(ogen) interactions. J Biol Chem. 1989 Nov 5;264(31):18419–18425. [PubMed] [Google Scholar]
- Degen S. J., MacGillivray R. T., Davie E. W. Characterization of the complementary deoxyribonucleic acid and gene coding for human prothrombin. Biochemistry. 1983 Apr 26;22(9):2087–2097. doi: 10.1021/bi00278a008. [DOI] [PubMed] [Google Scholar]
- Eric D. Determination of the specific activity of recombinant hirudin. Thromb Res. 1990 Dec 15;60(6):433–443. doi: 10.1016/0049-3848(90)90228-5. [DOI] [PubMed] [Google Scholar]
- Fenton J. W., 2nd, Fasco M. J., Stackrow A. B. Human thrombins. Production, evaluation, and properties of alpha-thrombin. J Biol Chem. 1977 Jun 10;252(11):3587–3598. [PubMed] [Google Scholar]
- Fenton J. W., 2nd, Olson T. A., Zabinski M. P., Wilner G. D. Anion-binding exosite of human alpha-thrombin and fibrin(ogen) recognition. Biochemistry. 1988 Sep 6;27(18):7106–7112. doi: 10.1021/bi00418a066. [DOI] [PubMed] [Google Scholar]
- Gan Z. R., Li Y., Chen Z., Lewis S. D., Shafer J. A. Identification of basic amino acid residues in thrombin essential for heparin-catalyzed inactivation by antithrombin III. J Biol Chem. 1994 Jan 14;269(2):1301–1305. [PubMed] [Google Scholar]
- Hirsh J., Dalen J. E., Deykin D., Poller L. Heparin: mechanism of action, pharmacokinetics, dosing considerations, monitoring, efficacy, and safety. Chest. 1992 Oct;102(4 Suppl):337S–351S. doi: 10.1378/chest.102.4_supplement.337s. [DOI] [PubMed] [Google Scholar]
- Kisiel W., Hanahan D. J. Purification and characterization of human Factor II. Biochim Biophys Acta. 1973 Mar 30;304(1):103–113. doi: 10.1016/0304-4165(73)90119-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu L. W., Vu T. K., Esmon C. T., Coughlin S. R. The region of the thrombin receptor resembling hirudin binds to thrombin and alters enzyme specificity. J Biol Chem. 1991 Sep 15;266(26):16977–16980. [PubMed] [Google Scholar]
- MacGillivray R. T., Degen S. J., Chandra T., Woo S. L., Davie E. W. Cloning and analysis of a cDNA coding for bovine prothrombin. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5153–5157. doi: 10.1073/pnas.77.9.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcum J. A., McKenney J. B., Rosenberg R. D. Acceleration of thrombin-antithrombin complex formation in rat hindquarters via heparinlike molecules bound to the endothelium. J Clin Invest. 1984 Aug;74(2):341–350. doi: 10.1172/JCI111429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P. D., Robertson W., Turk D., Huber R., Bode W., Edwards B. F. The structure of residues 7-16 of the A alpha-chain of human fibrinogen bound to bovine thrombin at 2.3-A resolution. J Biol Chem. 1992 Apr 15;267(11):7911–7920. [PubMed] [Google Scholar]
- Nelken N. A., Soifer S. J., O'Keefe J., Vu T. K., Charo I. F., Coughlin S. R. Thrombin receptor expression in normal and atherosclerotic human arteries. J Clin Invest. 1992 Oct;90(4):1614–1621. doi: 10.1172/JCI116031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S. T., Björk I., Sheffer R., Craig P. A., Shore J. D., Choay J. Role of the antithrombin-binding pentasaccharide in heparin acceleration of antithrombin-proteinase reactions. Resolution of the antithrombin conformational change contribution to heparin rate enhancement. J Biol Chem. 1992 Jun 25;267(18):12528–12538. [PubMed] [Google Scholar]
- Olson S. T., Halvorson H. R., Björk I. Quantitative characterization of the thrombin-heparin interaction. Discrimination between specific and nonspecific binding models. J Biol Chem. 1991 Apr 5;266(10):6342–6352. [PubMed] [Google Scholar]
- Olson S. T., Shore J. D. Demonstration of a two-step reaction mechanism for inhibition of alpha-thrombin by antithrombin III and identification of the step affected by heparin. J Biol Chem. 1982 Dec 25;257(24):14891–14895. [PubMed] [Google Scholar]
- Olson S. T. Transient kinetics of heparin-catalyzed protease inactivation by antithrombin III. Linkage of protease-inhibitor-heparin interactions in the reaction with thrombin. J Biol Chem. 1988 Feb 5;263(4):1698–1708. [PubMed] [Google Scholar]
- Rovelli G., Stone S. R., Guidolin A., Sommer J., Monard D. Characterization of the heparin-binding site of glia-derived nexin/protease nexin-1. Biochemistry. 1992 Apr 7;31(13):3542–3549. doi: 10.1021/bi00128a031. [DOI] [PubMed] [Google Scholar]
- Rydel T. J., Ravichandran K. G., Tulinsky A., Bode W., Huber R., Roitsch C., Fenton J. W., 2nd The structure of a complex of recombinant hirudin and human alpha-thrombin. Science. 1990 Jul 20;249(4966):277–280. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- Sheehan J. P., Wu Q., Tollefsen D. M., Sadler J. E. Mutagenesis of thrombin selectively modulates inhibition by serpins heparin cofactor II and antithrombin III. Interaction with the anion-binding exosite determines heparin cofactor II specificity. J Biol Chem. 1993 Feb 15;268(5):3639–3645. [PubMed] [Google Scholar]
- Shimada K., Ozawa T. Evidence that cell surface heparan sulfate is involved in the high affinity thrombin binding to cultured porcine aortic endothelial cells. J Clin Invest. 1985 Apr;75(4):1308–1316. doi: 10.1172/JCI111831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman M. A., Majerus P. W. The measurement of thrombin in clotting blood by radioimmunoassay. J Clin Invest. 1976 Nov;58(5):1249–1258. doi: 10.1172/JCI108579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Speijer H., Govers-Riemslag J. W., Zwaal R. F., Rosing J. Prothrombin activation by an activator from the venom of Oxyuranus scutellatus (Taipan snake). J Biol Chem. 1986 Oct 5;261(28):13258–13267. [PubMed] [Google Scholar]
- Ternisien C., Jandrot-Perrus M., Huisse M. G., Guillin M. C. Effect of phosphopyridoxylation on thrombin interaction with platelet glycoprotein Ib. Blood Coagul Fibrinolysis. 1991 Aug;2(4):521–528. doi: 10.1097/00001721-199108000-00005. [DOI] [PubMed] [Google Scholar]
- Tollefsen D. M., Pestka C. A., Monafo W. J. Activation of heparin cofactor II by dermatan sulfate. J Biol Chem. 1983 Jun 10;258(11):6713–6716. [PubMed] [Google Scholar]
- Wu Q. Y., Sheehan J. P., Tsiang M., Lentz S. R., Birktoft J. J., Sadler J. E. Single amino acid substitutions dissociate fibrinogen-clotting and thrombomodulin-binding activities of human thrombin. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6775–6779. doi: 10.1073/pnas.88.15.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Picard V., Aiach M., Sadler J. E. Activation-induced exposure of the thrombin anion-binding exosite. Interactions of recombinant mutant prothrombins with thrombomodulin and a thrombin exosite-specific antibody. J Biol Chem. 1994 Feb 4;269(5):3725–3730. [PubMed] [Google Scholar]
- Wu Q., Tsiang M., Sadler J. E. Localization of the single-stranded DNA binding site in the thrombin anion-binding exosite. J Biol Chem. 1992 Dec 5;267(34):24408–24412. [PubMed] [Google Scholar]
- Zimmerle C. T., Frieden C. Analysis of progress curves by simulations generated by numerical integration. Biochem J. 1989 Mar 1;258(2):381–387. doi: 10.1042/bj2580381. [DOI] [PMC free article] [PubMed] [Google Scholar]