SUMMARY
Research on archaeal extrachromosomal genetic elements (ECEs) has progressed rapidly in the past decade. To date, over 60 archaeal viruses and 60 plasmids have been isolated. These archaeal viruses exhibit an exceptional diversity in morphology, with a wide array of shapes, such as spindles, rods, filaments, spheres, head-tails, bottles, and droplets, and some of these new viruses have been classified into one order, 10 families, and 16 genera. Investigation of model archaeal viruses has yielded important insights into mechanisms underlining various steps in the viral life cycle, including infection, DNA replication and transcription, and virion egression. Many of these mechanisms are unprecedented for any known bacterial or eukaryal viruses. Studies of plasmids isolated from different archaeal hosts have also revealed a striking diversity in gene content and innovation in replication strategies. Highly divergent replication proteins are identified in both viral and plasmid genomes. Genomic studies of archaeal ECEs have revealed a modular sequence structure in which modules of DNA sequence are exchangeable within, as well as among, plasmid families and probably also between viruses and plasmids. In particular, it has been suggested that ECE-host interactions have shaped the coevolution of ECEs and their archaeal hosts. Furthermore, archaeal hosts have developed defense systems, including the innate restriction-modification (R-M) system and the adaptive CRISPR (clustered regularly interspaced short palindromic repeats) system, to restrict invasive plasmids and viruses. Together, these interactions permit a delicate balance between ECEs and their hosts, which is vitally important for maintaining an innovative gene reservoir carried by ECEs. In conclusion, while research on archaeal ECEs has just started to unravel the molecular biology of these genetic entities and their interactions with archaeal hosts, it is expected to accelerate in the next decade.
INTRODUCTION
The phylogenetic study by Carl Woese and his colleagues using sequences of the small-subunit rRNA genes revealed that life on Earth comprises three main lineages or domains, i.e., Bacteria, Archaea, and Eukarya (1, 2). This proposal has gained strong support from complete genome sequencing of several archaeal and bacterial organisms in the late 1990s as well as research on archaeal viruses and plasmids, which clearly have been shown to be equally distantly related to their bacterial and eukaryal counterparts as Archaea are related to Bacteria and Eukarya (3–5).
Viruses are considered to be the most abundant biological entities in the biosphere and thrive in every ecosystem on Earth (6–8), including extremely acidic, thermal, and hypersaline environments (3, 9). Because of their abundance, viruses may promote the turnover of a large sum of biomass every day (6) and play important roles in microbial population dynamics, genetics, and evolution (3, 9, 10).
Research on archaeal viruses started in the 1970s, when the concept of the archaeal domain was still in its infancy (11), but the early work essentially followed the logic of bacteriophage studies by looking for head-tail phage particles. A turning point occurred when Wolfram Zillig and colleagues started systematic investigations of archaeal viruses in the 1980s (4, 12). They discovered the first archaeal virus of a novel morphotype, known as Sulfolobus spindle-shaped virus 1 (SSV1 [formerly Sav-1]) (13). This was followed by tremendous efforts, led mainly by David Prangishvili's laboratory, to identify viruses infecting archaea, especially crenarchaea of the order Sulfolobales (14–27). These pioneering studies laid the groundwork for the rapid development of the new and exciting field of archaeal virology. By now, >60 archaeal viruses, isolated mainly from acidic hot springs and hypersaline lakes worldwide, have been described. They are morphologically diverse and genetically unique, representing the most fascinating group of the virosphere (3, 9). Notably, however, the host origins of currently known archaeal viruses represent only a very small proportion of identified archaeal species, and the number of known archaeal viruses accounts for ∼1% of all reported viruses (28). Clearly, we are only beginning to explore the vast archaeal virosphere.
Research on archaeal plasmids also started in the 1970s, when haloarchaeal plasmids were found to be related to gas vacuole formation in Halobacterium salinarium (29, 30). The recognition of Archaea as the third domain of life greatly stimulated archaeal plasmid research aiming to develop genetic tools for studying these unique organisms. This led to the immediate isolation of several plasmids from different archaeal hosts. Five of them were from methanogens, including pMP1, pME2001, and pME2200 from Methanobacterium thermoautotrophicum (31–34) and pFV1 and pFZ1 from Methanobacterium thermoformicicum (34). Two plasmids were isolated from hyperthermophilic archaea: pDL10 from Acidianus ambivalens, a thermophilic crenarchaeon isolated from a hot spring in Iceland (35), and pGT5 from Pyrococcus abyssi, a hyperthermophilic euryarchaeon isolated from a deep sea hydrothermal vent (36). Small plasmids were also isolated from haloarchaea. Among them, pHV2, pHSB1, and pHK2 were soon used as backbones for constructing cloning vectors (37–39).
Plasmids occur widely in archaea and are most common in haloarchaea. Among 15 haloarchaea for which complete genomes have been determined, only one lacks a plasmid (40). Systematic screening for plasmids has also been conducted for two families of thermophilic archaea, the Sulfolobaceae and Thermococcaceae. Whereas it was estimated that up to 3% of the isolates obtained from terrestrial hot springs in Iceland contained a conjugative plasmid (CP) (41), Daniel Prieur and colleagues found that >40% of >190 thermococcal isolates contained at least one plasmid with a genome size ranging from 2.8 to >35 kb (42).
To date, >60 viruses and >60 plasmids have been isolated from archaea (Table 1). Most viruses exhibit both morphological and genomic novelties, which have been extensively reviewed in several recent general reviews (3, 9, 43–45) and compared to those of bacterial viruses (46, 47). Many excellent reviews on different archaeal viruses have been published, and the most recent ones include those on structural genomics of archaeal viruses (48, 49); lytic viruses and their exceptional release mechanism (50, 51); the molecular biology of rudiviruses (52), fuselloviruses (53), and Sulfolobus turreted icosahedral virus (STIV) (51, 54, 55); as well as lipids of archaeal viruses (56). In comparison, archaeal plasmids are less frequently reviewed, with only a few reviews being found in the current literature (57–60). In this review, we focus on the molecular biology of archaeal viruses and plasmids, virus-plasmid interactions, as well as interactions of these genetic elements with their host organisms.
TABLE 1.
Overview of extrachromosomal genetic elements in archaea
| Archaeal phylum | Family | No. of ECEs identified |
|||
|---|---|---|---|---|---|
| Plasmids |
Viruses |
||||
| Cryptic | Conjugative | Families | Speciesc | ||
| Crenarchaeota | Sulfolobaceae | 9 | 12 | 6 | 24/4 |
| Desulfurococcaceae | 0 | 0 | 2 | 4/2 | |
| Thermoproteaceae | 1a | 0 | 2 | 2 | |
| Euryarchaeota | Haloarchaeaceae | >60/19b | 0 | 1 | 18/8 |
| Thermococcaceae | 14 | 0 | Unclassified | 2 | |
| Methanococcaceae | 10 | 0 | 0 | 0 | |
| Methanobacteriaceae | 8a | 0 | 1 | 3 | |
| Methanosarcinaceae | 2 | 0 | 0 | 0 | |
| Other euryarchaea | 5 | 0 | 0 | 0 | |
ECEs were identified from genome sequencing; thus, it is unclear if they are plasmids or viruses.
More than 60 are megaplasmids, many of which carry essential genes, whereas 19 are small haloarchaeal plasmids that appear to be cryptic.
The first number indicates the number of virus species classified, and the second indicates the number of viruses species not yet classified.
ARCHAEAL VIRUSES
The past decade has seen a rapid increase in the number of isolated and characterized archaeal viruses as well as a fast evolution of the taxonomy of these viruses. One order and 10 families of archaeal viruses have now been recognized (Table 2). However, there are still a number of archaeal viruses waiting to be classified (61, 62). Most of the currently recognized viruses with unusual morphotypes infect crenarchaea (Fig. 1). Crenarchaeal viruses with linear virions are grouped into the only archaeal virus order, Ligamenvirales, which comprises two families, i.e., Lipothrixviridae and Rudiviridae. The family Lipothrixviridae contains enveloped filamentous viruses, whereas the Rudiviridae include rod-shaped viruses without a lipid membrane (28). Among spindle-shaped or lemon-shaped viruses, some have been placed in the families Fuselloviridae (i.e., SSV1, SSV2, SSV4 to -7, SSV8 [also known as SSV K1], SSV9 [also known as SSV RH], and Acidianus spindle-shaped virus 1 [ASV1]) and Bicaudaviridae (Acidianus two-tailed virus 1 [ATV]). Unclassified spindle-shaped viruses (e.g., Sulfolobus tengchongenesis spindle-shaped virus 1/2 [STSV1/2] and Pyrococcus abyssi virus 1 [PAV1]) are believed to be associated with either of the above-mentioned two families on the basis of structural protein analysis (63). Of the spherical archaeal viruses, two (i.e., Pyrobaculum spherical virus [PSV] and Thermoproteus tenax spherical virus 1 [TTSV1]) have been assigned to the family Globuloviridae, and the rest are still unclassified (e.g., STIV/STIV2, spherical halovirus 1 [SH1], Pink Lake Haloarcula hispanica virus 1 [PH1], and Haloarcula hispanica icosahedral virus 2 [HHIV-2]). Bottle-shaped and droplet-like viruses are members of the families Ampullaviridae (i.e., Acidianus bottle-shaped virus [ABV]) and Guttaviridae (i.e., Sulfolobus neozealandicus droplet-shaped virus [SNDV] and Aeropyrum pernix ovoid virus 1 [APOV1]), respectively. Aeropyrum bacilliform virus 1 (APBV1) is the only member of the family Clavaviridae. Most of the isolated euryarchaeal viruses are of the head-tail type, some of which have been assigned to the families Myoviridae and Siphoviridae, although spindle-shaped and spherical icosahedral virus particles are often seen in samples taken from typical high-salt environments such as the Dead Sea (7) and Spanish solar salterns (64). The majority of isolated euryarchaeal viruses have yet to be classified (66, 67). The considerable variation in morphology and genomic properties found in known archaeal viruses has prompted a search for general principles applicable to the classification of some archaeal viruses in particular and all viruses in general. In this regard, structural proteins have been increasingly recognized as candidates for being a valuable marker in determining the relatedness of viruses, primarily because they are encoded by the true viral “self” genes (63, 68, 69). For example, structural studies have revealed that the structure of the coat protein of STIV is very similar to that of the coat protein of the bacterial virus PRD1 and those of the eukaryal viruses Paramecium bursaria Chlorella virus 1 (PBCV-1) and adenovirus (70). This observation offers clues to the evolutionary relatedness among viruses from the three domains of life (71). Conceivably, progress in this area will help delineate the evolutionary relationship among all viruses, including those infecting archaea.
TABLE 2.
Archaeal virusesa
| Virion morphology | Taxonomy |
Type species | Species name | Host | No. of species | Genome |
Reference(s) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Family | Genus | Size (kb) | Typeb | intc | ||||||
| Spindle | Fuselloviridae | Alphafusellovirus | SSV1 | Sulfolobus spindle-shaped virus 1 | Sulfolobus | 7 | 15.4 | ds, C | + | 13, 114, 277 |
| Betafusellovirus | SSV6 | Sulfolobus spindle-shaped virus 6 | Sulfolobus | 2 | 15.6 | ds, C | + | 276 | ||
| Bicaudaviridae | Bicaudavirus | ATV | Acidianus two-tailed virus | Acidianus | 1 | 62.7 | ds, C | + | 19, 20 | |
| Unclassified | Salterprovirus | His1 | His virus 1 | Haloarcula | 2 | 14.4 | ds, L | − | 86, 278 | |
| Unclassified | Unclassified | STSV1 | Sulfolobus tengchongenesis spindle-shaped virus 1 | Sulfolobus | 2 | 75.3 | ds, C | + | 95 | |
| PAV1 | Pyrococcus abyssi virus 1 | Pyrococcus | 1 | 18.1 | ds, C | − | 229, 230 | |||
| TPV1 | Thermococcus prieurii virus 1 | Thermococcus | 1 | 21.5 | ds, C | + | 98 | |||
| APSV1 | Aeropyrum pernix spindle-shaped virus 1 | Aeropyrum | 1 | 38.0 | ds, C | + | 22 | |||
| Bottle | Ampullaviridae | Ampullavirus | ABV | Acidianus bottle-shaped virus | Acidianus | 1 | 23.8 | ds, L | − | 15 |
| Bacilliform | Clavaviridae | Clavavirus | APBV1 | Aeropyrum pernix bacilliform virus 1 | Aeropyrum | 1 | 5.2 | ds, C | − | 16 |
| Unclassified | Unclassified | ACV | Aeropyrum coil-shaped virus | Aeropyrum | 1 | 24.9 | ss, L | − | 21 | |
| Droplet | Guttaviridae | Alphaguttavirus | SNDV | Sulfolobus neozealandicus droplet-shaped virus | Sulfolobus | 1 | 20 | ds, C | − | 78 |
| Betaguttavirus | APOV1 | Aeropyrum pernix ovoid virus 1 | Aeropyrum | 1 | 13.8 | ds, C | + | 22 | ||
| Lineard | Lipothrixviridae | Alphalipothrixvirus | TTV1 | Thermoproteus tenax virus 1 | Thermoproteus | 1 | 15.9 | ds, L | − | 279 |
| Betalipothrixvirus | SIFV | Sulfolobus islandicus filamentous virus | Sulfolobus | 6 | 40.8 | ds, L | − | 79 | ||
| Gammalipothrixvirus | AFV1 | Acidianus filamentous virus 1 | Acidianus | 1 | 21.9 | ds, L | − | 14 | ||
| Deltalipothrixvirus | AFV2 | Acidianus filamentous virus 2 | Acidianus | 1 | 31.7 | ds, L | − | 24 | ||
| Rudiviridae | Rudivirus | SIRV2 | Sulfolobus islandicus rod-shaped virus 2 | Sulfolobus | 3 | 35.4 | ds, L | − | 26 | |
| Unclassified | Unclassified | SRV | Stygiolobus rod-shaped virus | Stygiolobus | 1 | 28.1 | ds, L | − | 27 | |
| Spherical | Globuloviridae | Globulovirus | PSV | Pyrobaculum spherical virus | Pyrobaculum | 2 | 28.3 | ds, L | − | 25 |
| Unclassified | Unclassified | STIV | Sulfolobus turreted icosahedral virus | Sulfolobus | 2 | 16.6 | ds, C | − | 80 | |
| SH1 | Spherical halovirus 1 | Haloarcula/Haloferax | 1 | 30.9 | ds, L | − | 280 | |||
| HHIV-2 | Haloarcula hispanica icosahedral virus 2 | Haloarcula | 1 | 30.6 | ds, L | − | 281 | |||
| PH1 | Pink Lake Haloarcula hispanica virus 1 | Haloarcula | 1 | 28.1 | ds, L | − | 282 | |||
| Pleomorphic | Unclassified | Unclassified | HHPV1 | Haloarcula hispanica pleomorphic virus 1 | Haloarcula | 2 | 8.1 | ds, C | − | 96 |
| HRPV1 | Halorubrum pleomorphic virus 1 | Halorubrum | 3 | 7.0 | ss, C | − | 283 | |||
| HRPV3 | Halorubrum pleomorphic virus 3 | Halorubrum | 1 | 8.8 | ds, C | − | 66, 284 | |||
| HGPV1 | Halogeometricum pleomorphic virus 1 | Halogeometricum | 1 | 9.7 | ds, C | − | 66, 284 | |||
| Head-tail | Myoviridae | Phihlikevirus | phiH | Halobacterium phage phiH | Halobacterium | 1 | 59 | ds, L | − | 285, 286 |
| Siphoviridae | Psimunalikevirus | psiM1 | Methanobacterium phage psiM1 | Methanobacterium | 1 | 30.4 | ds, L | − | 287 | |
| Unclassified | Unclassified | phiCh1 | phiCh1 | Natrialba | 1 | 58.5 | ds, L | + | 288 | |
| HF1 | HF1 | Halorubrum | 2 | 75.9 | ds, L | − | 289 | |||
| BJ1 | BJ1 | Halorubrum | 1 | 42.3 | ds, C | + | 129 | |||
| SNJ1 | SNJ1 | Natrinema | 1 | 16.3 | ds, C | − | 290, 291 | |||
This table was prepared in accordance with the 2011 version of the International Committee on Taxonomy of Viruses catalog (99). Each family is represented by the type species. Unassigned species are also listed. More detailed information on archaeal viruses is shown in Fig. S1 in the supplemental material.
Genome type is indicated as follows: ds, double-stranded DNA genome; ss, single-stranded DNA genome; L, linear genome; C, circular genome.
Viruses known to encode a putative integrase are indicated with a plus symbol (+); those not known to encode a putative integrase are indicated with a minus symbol (−).
Linear viruses are assigned to the only order of archaeal viruses, Ligamenvirales.
FIG 1.
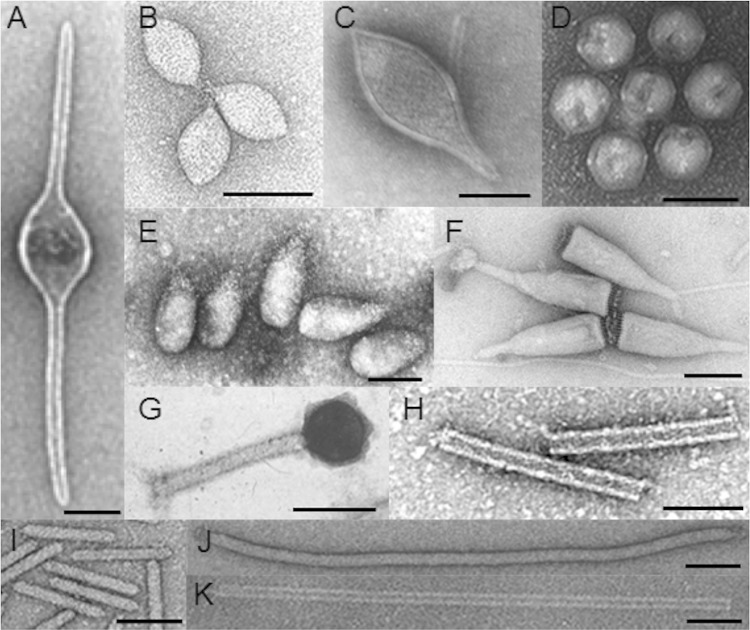
Electron micrographs of diverse morphotypes of archaeal viruses. (A) ATV. (Reprinted from reference 20 by permission from Macmillan Publishers Ltd. [copyright 2005].) (B) SSV1. (Reproduced from reference 147 [copyright 2003 Elsevier Masson SAS; all rights reserved].) (C) STSV1 (95). (D) STIV2 (47). (E) SNDV. (Reprinted from reference 3 by permission from Macmillan Publishers Ltd. [copyright 2006].) (F) ABV. (Reprinted from reference 15 with permission.) (G) phiH1. (Reprinted from reference 3 by permission from Macmillan Publishers Ltd. [copyright 2006].) (H) ACV. (Reproduced from reference 21 with permission.) (I) APBV1. (Reproduced from reference 16 [copyright Elsevier 2010].) (J) AFV2 (24). (K) SIRV2. (Reproduced from reference 52 with permission of the publisher [copyright the Biochemical Society].) Bar, 100 nm.
All known archaeal viruses, except for the crenarchaeal Aeropyrum coil-shaped virus (ACV) and the euryarchaeal Halorubrum pleomorphic virus 1/2/6 (HRPV1/2/6) and Haloarcula hispanica pleomorphic virus 1 (HHPV2), contain either a circular or a linear double-stranded DNA (dsDNA) genome of 5 to 144 kb (16). The linear dsDNA genomes are ended in different fashions: free, covalently closed, modified in an unknown manner, or linked to a specific protein. ACV, HRPV1/2/6, and HHPV2 have a circular single-stranded DNA (ssDNA) genome. Attempts to isolate an archaeal RNA virus have so far been unsuccessful. However, an RNA virus genome was assembled from a hot spring sample by using a metagenomic approach, suggesting that archaeal RNA viruses may exist in nature (72).
The genomes of nearly all isolated archaeal viruses have been sequenced. Only a small fraction of the annotated viral open reading frames (ORFs) encode a predictable function or share significant sequence similarity with genes of nonarchaeal origins in public databases. Therefore, approaches involving structural, biochemical, genetic, and transcriptomic analyses have been employed to gain insights into the functions of these unknown proteins in the life cycles of these viruses (48). The richness of unknown proteins reinforces the idea that archaeal viral genomes are a huge gene pool for the evolution of Archaea in particular and life in general.
There has been a tremendous expansion of our knowledge of archaeal viruses in the past 2 decades. Recently, more attention has been given to mechanisms underlining steps in the life cycle of these fascinating entities (52, 54, 55). Here we review some of the latest developments in molecular biology of the life cycle of archaeal viruses.
Adsorption and Entry
Entry into a host cell is the first step in the life cycle of a virus. It entails the recognition and binding of a receptor on the surface of the host cell by the virus and the subsequent delivery of the viral genome and, in some cases, proteins required for viral replication in the host cell (75–77). Very little is currently known about the mechanisms of host entry by archaeal viruses, but this important step has attracted increasing research attention in recent years.
As implied by their enormous diversity in morphology, archaeal viruses appear to be highly innovative in developing macromolecular protein appendages for specific interactions with and binding to their host cells, presumably in a habitat-adaptive fashion. Electron microscopic observations have revealed that a number of archaeal viruses are attached to the cell surface or debris of their host cells through a unique appendage at the tip or end of the virion (14, 15, 23, 78). These appendages include tail fibers with various shapes, lengths, thicknesses, rigidities, and stickinesses (e.g., SSV1 and SSV6); claws (Sulfolobus islandicus filamentous virus [SIFV] and Acidianus filamentous virus 1 [AFV1], etc.); and pointed ends without tail fibers (e.g., ATV, STSV1, and STSV2) (Fig. 2).
FIG 2.
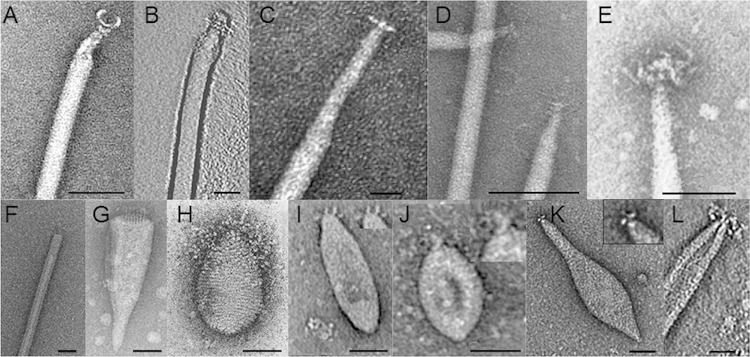
Appendages of selected archaeal viruses. (A) Claw-like terminal structure of AFV1. (Reproduced from reference 14 [copyright Elsevier 2003].) (B) The terminal structure of AFV2 (24). (C) The terminal structure of AFV3 (18). (D) T-bar or claw structure with a single thin filament at the end of AFV9. (Reproduced from reference 23 [copyright 2008 Elsevier Masson SAS; all rights reserved].) (E) SIFV tail fibers. (Reproduced from reference 79 [copyright Elsevier 2000].) (F) The plug-shaped end of SIRV2. (Reproduced from reference 52 with permission of the publisher [copyright the Biochemical Society].) (G) Thin sticks at the flat end of ABV. (Reprinted from reference 15 with permission.) (H) Beard-like tail fibers at the pointed end of SNDV. (Reproduced from reference 78 [copyright Elsevier 2000].) (I) Thick fibers at the tip of ASV1. (Reproduced from reference 276 with permission of John Wiley & Sons [copyright 2009 Society for Applied Microbiology and Blackwell Publishing Ltd.].) (J) Sticky ends of SSV7. (Reproduced from reference 276 with permission of John Wiley & Sons [copyright 2009 Society for Applied Microbiology and Blackwell Publishing Ltd.].) (K) Variable tails with appendages (insert) at the end of APSV1. (Reprinted from reference 22 with permission.) (L) Tail fibers of ATV. (Reproduced from reference 19 [copyright Elsevier 2006].) Bar, 50 nm.
Spindle-shaped viruses appear to have two types of tail structures. Virions of the family Fuselloviridae (e.g., SSV1, SSV2, and SSV4 to -9) have a bunch of short fibers at both ends, whereas those of other spindle-shaped viruses from hyperthermophilic crenarchaea (e.g., ATV, STSV1, STSV2, and Aeropyrum pernix spindle-shaped virus 1 [APSV1]) possess pointed ends. Interestingly, ATV was found to be able to undergo substantial morphological development outside host cells (20). Virions of ATV were initially protruded from host cells as lemon-shaped tail-less particles, which then developed tails at both ends at temperatures close to that of the natural habitat (i.e., 85°C) (19). It was speculated that this unusual property of ATV would allow the virus to increase the probability of cell contact and adsorption. On the other hand, no clearly visible end structures have been observed for spindle-shaped viruses infecting haloarchaea (e.g., His virus 1 [His1]) or hyperthermophilic euryarchaea (e.g., PAV1 and Thermococcus prieurii virus 1 [TPV1]).
The filamentous and rod-shaped viruses of the order Ligamenvirales exhibit a variety of end appendages (Fig. 2). Conceivably, viruses with different end appendages may adsorb to, and enter, host cells in different manners. Although studies on how these end structures function in viral entry are lacking, it has been shown that the mop-like tail fibers of SIFV existed in either an open or a closed form (79). The tail fibers of purified SIFV virions were in the closed form, whereas those of viral particles, which had been incubated with host membrane vesicles, were in the open form, suggesting that the conformational changes of the tail fibers likely occur during the adsorption of SIFV to its host cells. Notably, virions of the same family may possess appendages of different shapes. For example, although AFV1, AFV2, AFV3, AFV7, and AFV9 are all filamentous viruses of the family Lipothrixviridae, they differ considerably in their appendage structures (Fig. 2), raising the possibility that they may recognize different surface structures of host cells and/or employ different entry strategies.
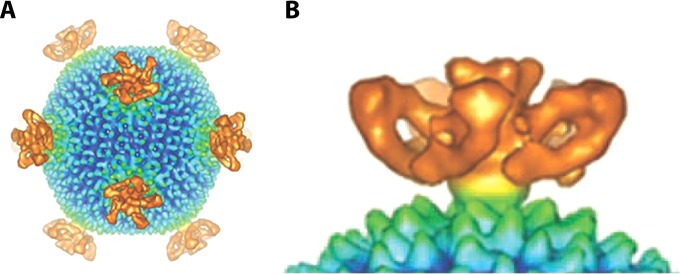
All sphere-shaped archaeal viruses (e.g., STIV, TTSV1, PH1, SH1, and HHIV-2) have an icosahedral protein capsid. Cryo-electron microscopy (cryo-EM) reconstruction has revealed detailed features of the surface structure of the STIV virion (Fig. 3) (80). The overall architecture of the virion resembles those found in bacterial and eukaryal icosahedral viruses. In addition, analysis of the crystal structure of the major capsid protein of STIV identified a common overall structure shared by viruses that infect bacteria (i.e., PRD1) and eukarya (i.e., adenovirus) (54, 55). A turret-like appendage projecting outwards from the virion surface was found at each of the 12 5-fold axes of symmetry. These structures are speculated to be involved in viral attachment to host receptors, viral genome entry into the host cell, and/or packaging of viral DNA into the capsid (54).
FIG 3.
Cryo-TEM reconstruction of STIV surface features. (A) Turret-like projections. (B) Side view of the turret-like projections. (Reproduced from reference 80 with permission of the publisher [copyright 2004 National Academy of Sciences, U.S.A.].)
How an archaeal virus employs its surface appendage to attach to and enter its host cell is poorly understood. However, a recent study of the entry process of Sulfolobus islandicus rod-shaped virus 2 (SIRV2) has offered some interesting clues (81). SIRV2 bound to the S. islandicus host rapidly, with the majority of the virions being irreversibly adsorbed to the host cell within 1 min. The calculated rate constant for adsorption by SIRV2 is 2 × 10−8 ml/min. This rate constant is substantially higher than those reported for host binding by a group of viruses from haloarchaea, i.e., Haloarcula hispanica head-tail virus 1 (HHTV-1) (2.9 × 10−13 ml/min), Haloarcula californiae head-tail virus 1 (HCTV-1) (5.1 × 10−11 ml/min), Halorubrum head-tail virus 1 (HRTV-1) (2.2 × 10−11 ml/min), HHPV1 (2.0 × 10−10 ml/min), and SH1 (1.9 × 10−12 ml/min) (82). The rapid adsorption as well as the relatively long intracellular phase of SIRV2 presumably allow the virus to minimize its exposure to hostile environments of high temperatures and low pH. Cryo-EM observations have established that the entry process of SIRV2 involves an initial binding to the tip of a filament on the surface of the host cell by the virus with its three terminal fibers at either end and subsequent movement of the virus along the filament toward the cell surface. Once it reaches the cell surface, the virion dissembles, presumably as the viral DNA is delivered into the cell interior (81). The entry process of SIRV2 bears superficial resemblance to that of pilus-binding bacterial viruses (e.g., Ff phages). However, these bacterial viruses are brought to the cell surface through pilus retraction (83). Therefore, the translocation of SIRV2 virions along the filament implies that the entry mechanism of SIRV2 differs from that employed by pilus-specific bacterial viruses.
Little is known about archaeal host receptors for viruses at the molecular level. As is the case for bacterial viruses, various components of the host surface structures may serve as receptors for archaeal viruses. In a preliminary study, a Sulfolobus solfataricus glycoprotein (the product of SSO1273), which specifically binds oligopeptides, was proposed to be involved in host receptor recognition by ATV through the predicted viral AAA ATPase p529 (84). Archaeal cell envelopes show great diversity and differ from their bacterial and eukaryal counterparts in structure and composition. It is possible, therefore, that archaeal viruses have evolved various novel mechanisms for viral entry in addition to those used by bacterial and eukaryal viruses. Also nearly uninvestigated are the detailed process and the mechanism of delivery of an archaeal viral genome into host cells. As probably the first experimental attempt in this area, DNA ejection from His1 was recently studied by using a single-molecule approach. The observed ejection appears to be unidirectional, paused, and incomplete, suggesting that cellular processes are required for the transfer of the viral genome into host cells (85).
Genome Replication
Much of the current knowledge about archaeal viral DNA replication is derived from bioinformatic analyses of viral genome sequences. Experimental verification of the properties and functions of putative replication proteins is available for only a few viruses. Studies on bacterial and eukaryal viruses show that viruses may replicate their genomes in different manners according to the nature (DNA or RNA) and structure (linear or circular double-stranded or single-stranded DNA/RNA) of their genome. The vast majority of reported archaeal viruses contain a dsDNA genome, and most of the dsDNA genomes are circular or linear, with covalently closed ends. Linear dsDNA genomes with ends covalently linked to a protein have also been found (23, 86, 87). Only five archaeal viruses (i.e., ACV, HRPV1/2/6, and HHPV2) are currently known to carry an ssDNA genome.
Linear dsDNA viruses.
Archaeal viruses containing a linear dsDNA genome are members of the families Rudiviridae, Lipothrixviridae, Ampullaviridae, Globuloviridae, Myoviridae, Siphoviridae, and Salterprovirus. The genomes of rudiviruses are covalently closed at both ends (26), whereas those of other linear dsDNA viruses are linked to a protein (23, 86, 87), modified in an unknown fashion at the ends (14), or yet to be determined (79). Genome replication among archaeal viruses is best understood for SIRV1 (88).
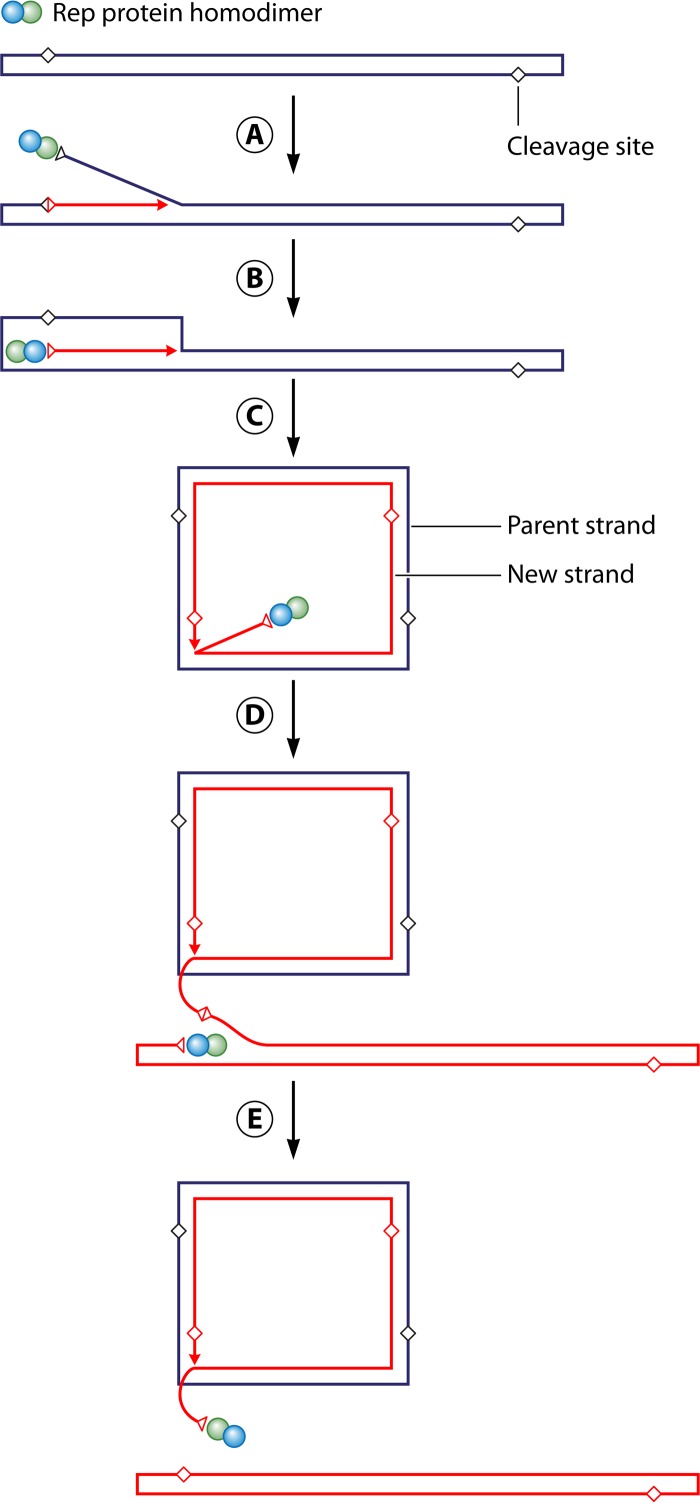
SIRV1 has a 32-kb linear dsDNA genome with 2,029-bp inverted terminal repeats (ITRs). The two DNA strands of the genome are covalently joined at both ends. Following infection of the viral genome in its host, head-to-head- and tail-to-tail-linked replication intermediates were identified in the cell extract, suggesting that the SIRV1 genome replicates in the same manner as the similarly structured linear genomes of eukaryal viruses, such as poxviruses, African swine fever virus, and Chlorella viruses (89). The viral DNA in ∼5% of the mature virions had a single-stranded nick 11 nucleotides (nt) from the terminus of the genome, as expected for the replication mechanism for a genome of a similar type, in which DNA replication is initiated through the introduction of a nick near either end of the linear genome. Structural analysis has shown that ORF119 of SIRV1 encodes a member of the replication initiator protein (Rep) superfamily, which includes proteins known to initiate rolling-circle replication (RCR) of a range of viruses and plasmids (88). The SIRV1 Rep protein existed as a dimer in solution and was capable of sequence-specific cleavage of ssDNA in vitro. Multiple nicks were generated by the protein on a DNA strand with a sequence matching that of the strand nicked in the viral DNA. However, the site of cleavage in vivo was preferentially nicked in vitro. No cleavage by Rep was detected when the cleavable strand was annealed to its complementary strand, suggesting that the terminal hairpin had somehow become at least partially single stranded when it was nicked by Rep in vivo. Once the viral genome was nicked, the newly formed 5′ end of the DNA was covalently attached to SIRV1 Rep, presumably via the active-site tyrosine residue in the protein, releasing a free 3′ end of the DNA to prime DNA synthesis. SIRV1 Rep was also able to catalyze the joining reaction, reforming the contiguous DNA strand and releasing the protein. Based on these results and previous models for rolling-circle DNA replication in bacterial and eukaryal viruses, SIRV1 DNA replication is proposed to proceed as follows (Fig. 4) (52, 88). The virus initiates DNA replication by introducing a single-stranded nick 11 nucleotides from either terminus, releasing a free 3′ end of the DNA. A new cleavage site is quickly regenerated with the extension of the 3′ end of the DNA. The newly formed cleavage site is then nicked by the second subunit of the Rep dimer. A joining reaction ensues, in which the second 3′ end of the DNA is ligated to the 5′ end of the DNA formed in the initial cleavage reaction, reforming a contiguous circular DNA strand. Strand displacement synthesis initiated from the first 3′ end continues to generate a head-to-head- or tail-to-tail-linked dsDNA circle. As replication goes around the circle, previously synthesized DNA is displaced and folded back into a linear SIRV1 dsDNA molecule. Folding of the DNA allows Rep to remain attached to the strand on the circular DNA after another round of nicking and joining, freeing a covalently closed SIRV1 genome. An alternative possibility is that the head-to-head and tail-to-tail intermediates adopt a cruciform structure at the borders between the two genome units by extrusion of the palindromic linkers formed by the ITRs. Resolution of these Holliday junction-like structures then yields new viral genomes. The latter scenario is supported by the finding that SIRV1 encodes a Holliday junction resolvase (ORF121 or Hjc), which was shown to recognize and cleave four-way DNA junctions in vitro (90). In agreement with this suggestion is the presence of a 23-nt sequence, which resembles the consensus sequence located near the hairpin termini of various poxviruses and is required for the resolution of poxvirus replication intermediates into unit genomes by a virally encoded Holliday junction-cleaving enzyme, near the termini of SIRV1 DNA (90, 91). Many Rep proteins are fused to a helicase, which serves to unwind dsDNA during strand displacement synthesis. However, a helicase function has not yet been assigned to any ORFs of SIRV1. Therefore, it would be interesting to determine if the virus encodes an unidentified DNA helicase or recruits a host helicase for DNA unwinding. Since homologs of SIRV1 Rep have been identified in the other four known members of the family Rudiviridae (i.e., SIRV2, Acidianus rod-shaped virus 1 [ARV1], Stygiolobus rod-shaped virus [SRV], and Sulfolobales Mexican rudivirus 1 [SMR1]), all rudiviruses may replicate their genomic DNA in the same fashion. Furthermore, SIRV1 Rep provides the first example in which a member of the Rep superfamily functions in a virus replicating its genome in neither an RCR nor a rolling-hairpin replication (RHR) mode. It appears, therefore, that initiator proteins of this Rep family are more widely employed by viruses, especially by linear dsDNA viruses of various morphotypes, than expected.
FIG 4.
Model for rudiviral DNA replication. (A) One subunit of the dimeric Rep protein introduces a single-stranded nick 11 nucleotides from a terminus in the viral genome, generating a free 3′ end for the initiation of viral DNA replication. A new cleavage site is quickly regenerated with the extension of the free 3′ end. (B) The newly formed cleavage site is nicked by the other subunit of the Rep dimer. The second 3′ end is rejoined with the 5′ end formed in the first cleavage reaction and bound by Rep, allowing the parental strand to be religated to form a continuous circular DNA strand. (C) Strand displacement synthesis initiated from the first 3′ end continues to generate a head-to-head- or tail-to-tail-linked dsDNA circle. (D) As replication goes around the circle, previously synthesized DNA is displaced and folded back into a linear SIRV1 dsDNA molecule. (E) Folding of the DNA allows Rep to remain attached to the strand on the circular DNA after another round of nicking and joining, freeing a covalently closed SIRV1 genome. (Reprinted from reference 88 with permission.)
Somewhat surprisingly, however, at least some members of the family Lipothrixviridae, which resembles the family Rudiviridae in morphology and genome content, appear to employ DNA replication mechanisms different from those employed by SIRV1 (14, 92). Lipothrixviruses are a large heterogeneous group of filamentous viruses that are divided into four genera (Alpha-, Beta-, Gamma-, and Deltalipothrixvirus) on the basis of genetic dissimilarity. Long ITRs (500 to 1,000 bp) resembling those present in rudiviral genomes are found only in betalipothrixviruses (18, 23). However, no Rep or Hjc proteins have been identified in any of the known filamentous viruses. Furthermore, the sequences at the extreme termini of all known lipothrixviruses have not been determined, presumably due to the presence of chemical modifications or covalent linkage to a protein. No data are available to suggest that the ends of the lipothrixviral genomes are covalently closed, as observed for rudiviruses. Intriguingly, lipothrixviruses, except for those of the genus Alphalipothrixvirus, share a conserved operon encoding a putative helicase and a putative nuclease (ORF593 and ORF203, respectively, from AFV3) (18). Taken together, these observations point to the possibility that lipothrixviruses employ genome replication strategies, or initiation mechanisms, different from those of rudiviruses. Consistent with the possible lack of a covalently closed terminal hairpin structure, the terminal sequences of the gammalipothrixvirus AFV1 contain smaller and less regular inverted terminal repeats than those in rudiviral genomes and multiple short direct repeats (TTGTT or its close variants) (14). Recently, it was found that AFV1 appears to exploit an unusual mechanism of DNA replication, which starts with the generation of a D loop at the 5′ end of the genome and progresses toward the right end via strand displacement synthesis. Recombination is speculated to play a key role in the termination of genome replication through the formation of terminal loops (92).
Linear dsDNA genomes are also found in archaeal viruses of other morphotypes. The Haloarcula hispanica spindle-shaped viruses His1 and His2 possess a dsDNA genome with terminal ITRs. The ends of both viral genomes are covalently attached to an unidentified protein. Both viruses encode a DNA polymerase similar to type B DNA polymerases from plants, fungal mitochondria, and some viruses (86). These type B polymerases are able to use proteins attached to the 5′ ends of linear dsDNA to prime DNA replication. Therefore, both His1 and His2 may replicate their genome by using a protein-priming mechanism. Protein-primed DNA replication is used by a range of viruses and plasmids, including some mammalian viruses (e.g., adenovirus), bacterial viruses (e.g., PRD1, Φ29, and CP-1), linear Streptomyces plasmids, and plant mitochondrial DNA (93). Acidianus bottle-shaped virus (ABV) (87) and the Haloarcula spherical virus SH1 (94) may also replicate their genome via a protein-primed mechanism.
For many other known linear dsDNA viruses (e.g., PAV1), attempts to identify the origin of replication and/or the homologs of replication proteins have been less than successful. It is therefore tempting to speculate that these viruses either rely heavily on the host replication machinery or have evolved replication proteins, and thus replication mechanisms, that remain to be elucidated. The former scenario would entail novel strategies for the initiation step of genome replication, which would be followed by the recruitment of host replication proteins.
Circular dsDNA viruses.
Circular dsDNA genomes are found in many archaeal viruses, including the majority of known crenarchaeal viruses. Surprisingly, however, how these genomes are replicated remains obscure. Origins of DNA replication and replication proteins have been predicted for a number of circular dsDNA viruses by using bioinformatic tools. However, none of these putative cis and trans replication elements have been experimentally verified. The θ and RCR modes of genome replication, among others, have been proposed for various circular dsDNA viruses. For example, S. tengchongenesis spindle-shaped virus 1 (STSV1) is speculated to replicate DNA in a θ mode since its 75-kb genome is highly asymmetric and divides into two halves with respect to gene orientation (95). The putative origin of DNA replication has been identified by the cumulative GC skew, and this prediction is supported by a high AT content as well as the presence of repeating sequences in the region. Presumably, replication initiated at the origin proceeds bidirectionally. However, STSV1 encodes no identifiable replication proteins. Haloarcula hispanica pleomorphic virus 1 (HHPV1) encodes a putative Rep protein, suggesting that the virus may replicate its 8-kb genome in an RCR mode (96). Homologs of replication proteins have been identified in the genomes of various archaeal circular dsDNA viruses. These include minichromosome maintenance (MCM) proteins (TPV1), highly putative DnaA (SSV1), and Cdc6 (APBV1) homologs (16, 97, 98). However, the biochemical properties of these proteins are unclear.
Circular ssDNA viruses.
Only five archaeal ssDNA viruses, i.e., Aeropyrum coil-shaped virus (ACV), Halorubrum pleomorphic virus (HRPV1/2/6), and Haloarcula hispanica pleomorphic virus 2 (HHPV2), have been isolated so far. All known ssDNA viruses replicate, or are believed to replicate, their genome in an RCR (for a circular genome) or RHR (for a linear genome) mode (297). In bacterial circular ssDNA viruses (e.g., ϕX174, M13, and fd), the single-stranded genome is first converted into a double-stranded replicative form (RF). A nick is then generated on one of the DNA strands by a Rep protein to release a free 3′ end to initiate genome replication. HRPV1 encodes a putative Rep protein (ORF1) containing all three key signature motifs, suggesting that the virus may replicate its genome by using an RCR mechanism (283). More recently, HRPV2 and HRPV6 were also found to encode a Rep protein similar to that of HRPV1 (284). Interestingly, HRPV1/2/6 shares remarkable similarity in genome organization and putative proteins with the circular dsDNA virus HHPV2, which infects H. hispanica. Since they all have a Rep protein of the RCR superfamily, these viruses appear to replicate their genomes in similar manners. The implications are that viruses are readily adapted to packaging a DNA genome in either its single-stranded or its double-stranded (RF) form. No putative Rep protein was identified in the genome of ACV, the largest known ssDNA virus, suggesting that the virus may have developed a novel replication mechanism (21). In this regard, ACV encodes a putative protein (ORF33) that appears to be a highly divergent member of the tyrosine recombinase family. Since both Rep and recombinase possess an active-site tyrosine involved in similar catalytic activities of strand nicking and joining, the ACV protein is suspected to function in viral genome replication.
Transcription
Transcription in Archaea has been extensively investigated. In their work representing a milestone in the understanding of Archaea ∼35 years ago, Wolfram Zillig and colleagues showed that DNA-dependent RNA polymerase (RNAP) from Sulfolobus acidocaldarius was far more complex than its bacterial counterpart and bore significant resemblance to eukaryal RNAP II (101). Archaeal RNAP consists of 13 subunits, which, as revealed by sequencing of their encoding genes, are homologous to the subunits of eukaryal RNAP II. Like RNAP II, archaeal RNAP requires additional protein factors for its recognition of promoter sequences. Archaeal TATA box-binding protein (TBP) and transcription factor B (TFB), homologs of eukaryal TBP and TFIIB, respectively, are basal factors minimally required to constitute transcription on archaeal promoters in vitro (102). Likewise, archaeal promoters are also of the eukaryal type and are often characterized by an 8-bp AT-rich TATA box ∼24 bp upstream of the transcriptional initiation site and a TFB recognition element (BRE) comprising two A's immediately upstream of the TATA box (103). Intriguingly, however, many of the archaeal transcriptional regulators are similar to members of the bacterial Lrp-like regulatory protein family (104, 105). The evolutionary implications of the use by Archaea of a eukaryal-type transcription apparatus regulated in a bacterial manner remain to be understood. Archaeal viruses appear to rely entirely on host RNAP and basal transcription proteins for transcription since they use the same promoter sequences as those of their host, and none of them are known to encode an RNAP, TBP, or TFB. Therefore, archaeal viruses must have developed strategies to coopt the host transcriptional apparatus for viral transcription. A diverse array of transcriptional regulatory proteins have been identified in archaeal viral genomes (48). These proteins are presumably involved in redirecting transcriptional activities in virus-infected cells such that viral genomes will be transcribed in a highly controlled fashion. In recent years, transcription profiles of the genomes of the rudiviruses SIRV1 and SIRV2 (106, 107), the fuselloviruses SSV1 and SSV2 (108, 110), and the icosahedral virus STIV (109) in infected host cells have been studied, providing useful clues to the mechanisms and control of gene expression in archaeal viruses.
SIRV2.
SIRV2 is one of the favored models for the study of archaeal virus-host interactions. Infection by SIRV2, long considered to be a temperate virus, resulted in the extensive degradation of host chromosomal DNA and the lysis of host cells (111). SIRV2, together with SIRV1, was the first crenarchaeal virus whose genome-wide transcription was systematically investigated. Following infection of S. islandicus LAL14/1 and S. islandicus REN2H1 with SIRV1 and SIRV2, respectively, patterns of transcription of the linear viral genomes were initially determined by Northern hybridization with specific DNA probes (106). Both rudiviruses display a relatively fast-replicating life cycle, with latent periods of 8 and 6 h for SIRV1 and SIRV2, respectively. Transcription of all but one of the viral genes (i.e., SIRV1 ORF55c/SIRV2 ORF55) was detected at 30 min postinfection (p.i.), the earliest sampling point, demonstrating the lack of apparent temporal control of genome transcription for the two viruses.
Recently, the infection cycle of SIRV2 was further investigated by using microarray and transcriptome sequencing (RNA-seq) approaches (107, 295). For microarray analysis, Okutan et al. (295) isolated an S. solfataricus strain, denoted S. solfataricus 5E6, from an S. solfataricus P2 stock. S. solfataricus 5E6 was highly susceptible to SIRV2 infection. The virus appeared to exhibit a one-step growth curve in S. solfataricus 5E6 similar to that in S. islandicus LAL14/1, with a latent period of 5 h. The infection was also lytic, with host chromosomal DNA being degraded in >80% of the infected cells at 8 h p.i. After SIRV2 infected S. solfataricus 5E6, a slight temporal control of viral genome transcription was observed. Transcription of the viral genome proceeded in three stages, i.e., early, middle, and late stages, as judged by the times when the transcription of specific regions peaked, and from both terminal regions toward the center of the linear genome. As found in the above-mentioned Northern hybridization studies, viral transcription was initiated rapidly upon infection. Some of the terminal genes were already weakly expressed at 15 min p.i. About 50% and 80% of the viral genes were significantly transcribed by 30 and 60 min p.i., respectively. All but one of the genes were expressed by 2 h p.i. Notably, among the early genes, ORF119C encodes the Rep protein presumably responsible for the initiation of viral genome replication. On the other hand, genes encoding the major coat protein (ORF134) and three minor structural proteins (ORF488, ORF1070, and ORF564) were all late genes. ORF98, which encodes the structural component of virus-associated pyramids (VAPs) (see “Assembly and Release,” below), was also expressed in the late stage. A total of 148 out of ∼3,000 detectable host genes were differentially expressed by >2-fold, with similar numbers of up- and downregulated genes, in response to SIRV2 infection, compared to those in the uninfected cells. Downregulated genes were primarily those responsible for the stress response and informational processing. Genes encoding proteins involved in protein folding (e.g., two thermosome subunits), protein degradation (e.g., a proteasome subunit and a protease), and the oxidative response (e.g., superoxide dismutase [SOD], rubrerythrin, and the peroxiredoxin homolog Bcp4) were downregulated. It was speculated that the stress response represents a host defense mechanism against viral infection. By downregulating the stress response genes, SIRV2 was able to circumvent the host defense and succeed in infecting the host. Among the most strongly downregulated genes was a gene encoding one of the four ESCRT-III-like proteins in S. solfataricus. Two of the three remaining genes were also downregulated. ESCRT-III proteins are believed to serve an important role in cell division. More than 15 downregulated genes are associated with transcription (e.g., two full-length TFB homologs), translation (e.g., four ribosomal proteins and two elongation factors), and chromosomal organization. Seventy-six host genes were upregulated by at least 2-fold upon SIRV2 infection. More than half of them are involved in transport and metabolism. Transcription of the gene encoding a ribonucleotide reductase, which converts ribonucleotides into deoxyribonucleotides and presumably contributes to viral DNA replication, was most strongly induced (∼6-fold). Several genes related to fatty acid degradation were also upregulated. Among them, genes encoding enoyl coenzyme A (enoyl-CoA) hydratase and a phenylacetic acid degradation-related protein were upregulated concomitantly with the viral structural genes and the pyramidal component gene, suggesting their possible role in host membrane modification for virus release.
Quax et al. (107) provided additional details about the virus-host interaction during SIRV2 infection of S. islandicus LAL14/1 by using the powerful RNA-seq technology. In their study, total RNA was isolated from infected host cells at different time points up to 9 h p.i. and from uninfected control cells. Transcripts were converted into cDNAs, which were subjected to deep sequencing. The number of viral sequencing reads increased steadily over the course of viral infection, leveling off at 5 h p.i., when viral reads accounted for ∼20% of the total mRNA reads. In agreement with the Northern hybridization and microarray results, viral transcription began very early during infection, i.e., immediately after viral infection (time [t] = 0 h p.i.), from both termini of the viral genome. Starting from the second sampling time (t = 1 h p.i.), essentially all viral genes were significantly expressed, again suggesting a weak temporal pattern in SIRV2 genome transcription. ORF83a and ORF83b, two identical genes located on the two distal ends of the genome, were most highly transcribed at the first sampling time. The genomic location of the two genes may represent an adaptation to the ability of SIRV2 to adsorb to and enter the host cell from either end. Gene products of ORF83a/ORF83b contain a helix-turn-helix (HTH) motif and are believed to be DNA-binding proteins. Intriguingly, a yeast two-hybrid screen, performed in that same study, identified an interaction between ORF83 and ORF121. ORF121 encodes the Holliday junction resolvase, which has been implicated in the resolution of viral replicative intermediates. Based on their transcriptional profiles and biochemical properties, ORF83 and ORF121 appear to function together in viral genome replication, possibly including the initiation of the process. Surprisingly, it was observed that the expression of ORF119c, which encodes the Rep protein, was very poor and peaked at the end of the infection cycle. Although this observation does not preclude the proposed function of the Rep protein in viral replication, as described above, it does raise a question concerning the respective roles of ORF83, ORF121, and the Rep protein in SIRV2 DNA replication. As shown by the microarray studies, the expression levels of genes coding for the viral structural proteins increased during the late stages of infection, with the transcripts for the major coat protein (ORF134) and the VAP component (ORF98) representing ∼35% and ∼13%, respectively, of the total viral transcripts at the end of the infection cycle. The host response to SIRV2 infection was surprisingly strong. More than one-third of all S. islandicus LAL14/1 genes were found to be significantly up- or downregulated in SIRV2-infected cells, compared to those in uninfected cells. This contrasts sharply with the finding that only a small fraction (∼5%) of the host genes were differentially expressed by >2-fold in S. solfataricus cells infected with SIRV2 in microarray assays. This discrepancy most likely reflects a difference in sensitivity between the two experimental approaches employed for transcriptomic analysis in those two studies, although it may also result from differences between the two host strains. The numbers of up- and downregulated genes were about the same. As found in the microarray assays, genes encoding components of the ESCRT-III sorting complex were downregulated. The complete cdv operon, which comprises these genes, was downregulated by 10-fold. The reduced expression levels of these genes probably resulted from DNA degradation caused by SIRV2 infection, since the Cdv proteins are known to be under the control of the checkpoint systems, which inhibit cell division in response to DNA damage (294). Among the upregulated host genes, those encoding CRISPR-Cas systems, which form the basis of prokaryotic adaptive immune systems against viruses and plasmids (see “CRISPR Systems,” below), were especially activated. The S. islandicus LAL14/1 genome possesses five complete and one incomplete CRISPR-cas loci. The complete CRISPR-cas loci consist of a CRISPR array and adjacently located cas genes, whereas the incomplete CRISPR-cas locus lacks the CRISPR array. Six cas operons, corresponding to the six CRISPR-cas arrays, encode two complexes each of three different types (subtypes I-A, I-D, and III-B). The transcription of all six operons, except for one encoding subtype III-B, which lacks the accompanying CRISPR array, was activated quickly in response to SIRV2 infection, and these cas genes were by far the most strongly upregulated host genes after viral infection. However, the responses of each of these operons to viral infection varied. Two operons of subtypes I-A and III-B were significantly expressed in uninfected cells, and their upregulation (∼10-fold) following SIRV2 infection was the most pronounced of all the cas operons. The subtype III-B operon without the associated CRISPR array was highly expressed in uninfected cells but was the only cas operon whose expression level decreased by 2-fold after viral infection. None of the spacers in the CRISPR arrays of S. islandicus LAL14/1 matches perfectly with the SIRV2 genome. This probably explains why the host cells were unable to survive SIRV2 infection. In comparison, genes encoding CRISPR-Cas systems were not among the genes considered to be upregulated in S. solfataricus 5E6 cells infected with SIRV2. Presumably, this strain was unable to activate CRISPR-based antiviral defense in response to SIRV2 infection because of the deletion or inactivation of part of its CRISPR-Cas systems. As a result of SIRV2 infection, many of the host toxin-antitoxin (TA) clusters were also abundantly upregulated. The TA clusters, two-gene elements ubiquitously present in prokaryotic genomes, are proposed to function in programmed cell death and in the stress response (293). Intriguingly, the general stress response was not found to be among the strongly suppressed processes in S. islandicus LAL14/1 infected with SIRV2, compared to that in S. solfataricus 5E6 upon infection by the virus. The basis for the differences between the different hosts in the physiological response to infection by the same virus awaits further investigation.
The rapid and nearly uniform initiation of genome transcription suggests that SIRV2, and presumably rudiviruses in general, may not use a delicate cascade control mechanism, as seen in viruses that exhibit clearly temporal patterns of viral transcription (see below). Efficient recruitment of the host transcription apparatus to viral promoters with or without the assistance of virus-encoded proteins may occur. Rudiviral promoters contain the canonical TATA box and BRE sequences as well as, in many promoters, the rudivirus-specific regulatory sequence element GTC. GTC-containing promoters may be recognized by specific host- and virus-encoded transcription factors. Several rudivirus-encoded transcription factors that may function in the regulation of viral transcription have been identified. For example, SvtR (P56b), a ribbon-helix-helix (RHH) DNA-binding protein, has been shown to repress the transcription of several viral genes, including ORF98 and ORF1070, which code for the VAP and tail fiber proteins, respectively, in vitro (112). Other putative DNA-binding proteins that may be involved in viral DNA replication and transcription include P59b (RHH motif), P55 (zinc-binding domain), and P114 (52). SIRV1 has also been shown to be able to coopt the host transcriptional activator Sta1, which possesses a winged helix-turn-helix (wHTH) fold, for transcription from viral promoters (113).
SSV1/SSV2.
SSV1, originally isolated from Sulfolobus shibatae, is a 15.5-kb circular dsDNA fusellovirus capable of infecting the foreign host S. solfataricus as a temperate virus, existing either as an integrated provirus in the host genome or in a plasmid form (114). Viral replication in host cells is induced upon UV irradiation, upon mitomycin C treatment, or in stationary phase, resulting in the production of a large number of SSV1 particles (up to 100 copies per cell) without apparent lysis of host cells (13, 113, 115). However, UV-induced viral replication is not seen for other members of the family Fuselloviridae.
When induced with UV irradiation, transcription of the SSV1 genome in S. solfataricus PH1 cells lysogenized with the virus followed a clearly temporal pattern, as revealed by microarray analysis (Fig. 5) (110). Early studies identified 10 transcripts of various lengths starting from seven promoters on the SSV1 genome (103). Among them, nine (transcripts T1 to T9) appear to be constitutively synthesized at low levels in uninduced host cells. T-ind, a short transcript whose encoding sequence lacks the canonical TATA box, is produced only upon UV irradiation of the host cell. Following UV induction, transcription of the viral genome proceeded through three discernible stages, i.e., immediately early, early, and late stages, in an ∼8.5-h transcription cycle ending with the release of mature progeny virus particles. T-ind, which appeared at 1 h and achieved a 16-fold increase in quantity by 2 h after UV induction, was the only viral transcript produced in the immediate early stage. This transcript or the protein that it encodes (B49) presumably activated the synthesis of transcripts T5 and T6, whose encoding genes flank that of T-ind and are oriented in opposite directions, in the early stage, as genes located at the 5′ end of the two transcripts were detected 1 h after the appearance of T-ind. Two inverted repeating sequences flanking the TATA box in the T5 and T6 promoters may represent sites of regulatory control by the T-ind product. T5 and T6 encode several known or putative DNA-binding proteins (i.e., HTH proteins E51, C80, and E73; zinc finger proteins A79, A45, and B129; wHTH proteins F63 and F112; and leucine zipper [LZ] protein D63), which probably play yet-to-be-defined regulatory roles in viral gene expression. While the sizes of the two transcripts appeared to increase over time, with their full-length products being observed 5 h after UV induction, the integrase mRNA located at the 3′ end of T5 was sporadically detected earlier, for unknown reasons. The early transcript T9 appeared 5 h after induction or shortly before the initiation of viral DNA replication (5 to 6 h). Six of the seven proteins encoded by this transcript are conserved among all known fuselloviruses. Among them, B251 was proposed to be a DnaA-like protein (97). It appears, therefore, that T9 serves a role in viral DNA replication. All remaining transcripts (T1/2, T3, and T4/7/8), products of the late genes, first appeared 6 h after UV induction. T2 and T7/8 encode the three structural proteins (VP1 to VP3). Other proteins encoded on these late transcripts may also be involved in viral assembly and release. Simultaneous transcriptomic analyses revealed only a small difference between the SSV1 lysogenic host and the uninfected control in response to UV irradiation. Among the few genes showing a significantly differential response are those encoding the two subunits of topoisomerase VI, which is probably involved in SSV1 DNA replication. In conclusion, the chronological transcription of the genome of SSV1 in the Sulfolobus host following UV induction is reminiscent of those of well-known bacterial and eukaryal viruses. However, the mechanistic basis for this regulation is unclear.
FIG 5.
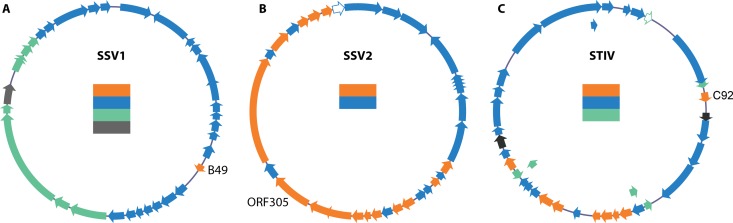
Temporal patterns of viral genome transcription in infected host cells. (A) Transcription of the SSV1 genome in the Sulfolobus solfataricus PH1 lysogen following UV irradiation. (Reproduced from reference 110 [copyright Elsevier 2007].) (B) Transcription of the SSV2 genome following viral infection of S. solfataricus P2. (Reproduced from reference 108 [copyright Elsevier 2013].) (C) Transcription of the STIV genome following viral infection of S. solfataricus strain 2-2-12. (Reprinted from reference 109 with permission.) The time sequence of viral genome transcription is shown by colored arrows in the order orange, blue, green, and gray. The ORF encoding B49 in SSV1 and ORF305 in SSV2 transcribed first. ORFs for which no probes were present or no transcription products were detected are shown by white or black arrows, respectively. The maps are prepared according to the latest gene annotation in GenBank.
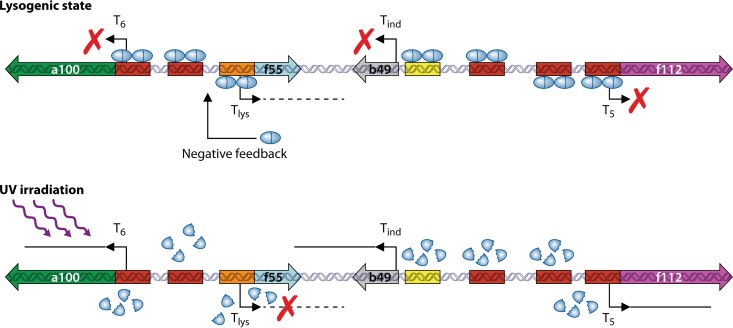
How is lysogeny established and maintained following SSV1 infection of its host? Recently, an ∼300-nt novel transcript, termed T-lys, was identified by studying SSV1 lysogeny in S. solfataricus InF1 (116). T-lys is located between T-ind and T6 and is transcribed in a direction opposite that of T-ind. In InF1(SSV1), an S. solfataricus InF1 lysogen carrying SSV1, T-ind was not produced, as expected, but T-lys was highly expressed. The amount of T-lys was reduced to 20% of the initial level in a late growth phase, consistent with the observation that SSV1 replication was induced in the stationary phase. T-lys was not detectable in the UV-irradiated InF1(SSV1) lysogen, suggesting that its role is restricted to the lysogenic state of the virus. T-lys encodes a 6.5-kDa protein termed F55. F55, existing as a dimer in solution, is a putative RHH transcription regulator and shares significant amino acid sequence similarity with proteins of the NikR and CopG families. DNA binding assays showed that F55 bound to an 11-bp specific site, located in the promoter sequences for T5, T6, T-ind, as well as T-lys. The binding site of F55 overlaps the transcription start site (TSS) and the BRE of these promoters. Therefore, binding by F55 to the target sequence would presumably interfere with the recruitment of RNAP to the promoter or the formation of the TBP-TFB-DNA ternary preinitiation complex, turning off transcription from the corresponding promoter (Fig. 6). In accordance with its differential binding affinities for different target sequences, F55 appeared to bind first to the promoters of T5 and T6, then to the promoter of T-ind, and finally to its own promoter, in a concentration-dependent manner. It was speculated that F55 would be degraded or inactivated following UV irradiation of the lysogenic host cell. These data suggest that SSV1 is, to a certain extent, analogous to bacteriophage λ in establishing and maintaining lysogeny. Like the CI repressor in a λ lysogen, F55 may play a key role in repressing SSV1 genome transcription by blocking transcription from the upstream promoters (T5 and T6 promoters) of a regulatory cascade. However, there are differences between the two systems, since CI is the only λ-encoded protein synthesized in a λ lysogen, whereas genes encoding several proteins, in addition to F55, were expressed in an SSV1 lysogen. This may be related to the fact that, unlike λ, which exists only as a prophage in the lysogen, both integrated and episomal forms of SSV1 are present in the lysogenic host.
FIG 6.
Model of the F55 interaction at its binding sites. In the lysogenic state, F55 (cyan ovals) binds as dimers to the target sequences in the promoters of T5, T6, and T-ind (red and yellow boxes) as well as to its own promoter (orange box) and represses transcription of T5, T6, and T-ind (red crosses). When the binding sites of F55 are saturated in the early promoters, F55 binds to its own promoter and downregulates the expression of its own gene following negative-feedback control. Upon UV irradiation, the F55 protein is degraded and/or inactivated by an as-yet-unknown mechanism, which releases F55 repression, allowing the subsequent transcription of the early T-ind, T5, and T6 transcripts to occur. The expression of early viral genes could be further activated by other viral transcription factors, which might repress F55 expression. (Reprinted from reference 116.)
A genome-wide transcriptional profile has also been determined for SSV2, a non-UV-inducible member of the family Fuselloviridae, during infection of S. solfataricus P2 by microarray analysis (108). The genome organization of SSV2 closely resembles that of SSV1, and the two viruses share a number of homologous proteins. Seven nonoverlapping transcripts were predicted according to, and were named after, their counterparts in the SSV1 genome. It took about the same length of time (∼9 h) for the infecting SSV2 to turn on all the viral genes in the host cell as that required for the transcription cycle of SSV1 in lysogenic host cells following UV irradiation. SSV2 also exhibited a temporal pattern of initiation of gene expression upon infection of its host (Fig. 5). However, the sequence of gene expression for SSV2 upon infection of the host differed distinctly from that for SSV1 following UV irradiation of an SSV1 lysogen. Transcription of early viral genes followed a distributive pattern, as genes activated successively were not adjacently located on the genome. Transcription was divided into an early stage and a late stage, separated at 4.5 h p.i. The first viral gene found to be expressed was ORF305. This gene, located on a monocistronic transcript equivalent to SSV1 T3, was detected at 1 h p.i. The ORF305 mRNA was the most abundant of all viral transcription products throughout the cycle of transcription. This contrasts sharply with SSV1 A291, a homolog of ORF305, which was upregulated 6 h after UV induction. SSV1 T3 was also one of the transcripts produced along with T-lys in the InF1(SSV1) lysogen. Homologs of ORF305 are encoded by all known fuselloviruses as well as S. islandicus, S. solfataricus, and Metallosphaera yellowstonensis. Primary structure prediction suggests that the product of ORF305 is a putative membrane protein with a signal peptide at its N terminus and a transmembrane segment in the middle, probably playing a role in virus docking and release. Expression of the genes encoding two structural proteins, VP1 and VP3, transcribed as single transcripts but located distal from ORF305, was detected at 2 h p.i. Transcription of ORF88a and ORF106, two genes oriented in opposite directions and linked with neither the ORF305 nor the VP1/VP3 genes, was detected at 3 p.i. By 4.5 h p.i., ∼15 genes residing exclusively on a ∼10-kb stretch of the genome were turned on, with only the T5 and T9 transcripts remaining undetected. SSV2 DNA replication started to accelerate after 4.5 h p.i., and mature virus particles were released into the culture fluid at the same time. Nine more genes, located mostly on T5 and T9 transcripts, were produced at 6 p.i. or at about the same time as when viral DNA replication accelerated. As observed for SSV1 genome transcription, the SSV2 integrase gene, located at the 3′ end of T5, was transcribed earlier than genes upstream of it, implying that transcription of the integrase gene alone occurred. At 7.5 h p.i., nearly all SSV2 genes were transcribed. ORF72 and ORF79a, located in a region corresponding to T-ind, were transcribed last, at 9 h p.i. These results show that following infection of its host cell, SSV2 appeared to be engaged first in preparation for virus assembly and release and subsequently in viral genome replication. In contrast, SSV1 replicates its genome prior to synthesizing the structural proteins and proteins likely involved in virus assembly and release, in the lysogenic host cell after UV irradiation. Integration of SSV2 DNA into the host genome apparently occurred concomitantly with the expression of the integrase gene and with the rapid accumulation of viral DNA in the host cell. Like SSV1, SSV2 appears to depend heavily on its host for viral genome replication and transcription. The expression of host genes encoding a number of replication and transcription proteins was upregulated in SSV2-infected cells. These proteins included Cdc6-1 (a replication initiator), MCM (a replicative helicase), PolB1 (a presumed replicative DNA polymerase), and PCNA2 (a sliding clamp subunit). Upregulated host genes also included those coding for reverse gyrase and topoisomerase VI, both of which are probably involved in DNA replication and virus packaging. Likewise, host genes for RNAP subunit F, TFBII, and a putative transcriptional regulator (SSO1255) were upregulated in SSV2-infected host cells, indicating a crucial role for the host in viral genome transcription. Increased expression of these host proteins may help prevent a decrease in host DNA replication and transcription, which would otherwise occur as a result of viral competition for the proteins. This also provides an explanation for the lack of inhibition of host growth by SSV2 infection during the infection cycle.
STIV.
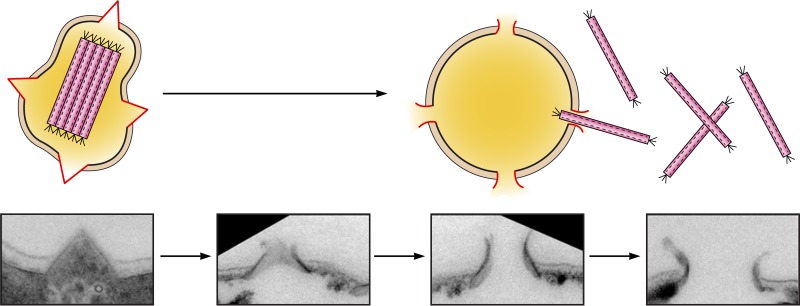
Genome transcription of the lytic icosahedral virus STIV following infection of S. solfataricus 2-2-12 has been studied by microarray assays (109). STIV showed a rather slow infection cycle of ∼32 h, with the viral genome first being detected in infected host cells at 16 p.i. and host cell death being observed toward the end of the infection cycle. Following STIV infection of its host, four viral transcripts with nine genes were first detected at 8 h p.i. (Fig. 5). Interestingly, among the early transcripts was a monocistronic transcript encoding C92, a 9.8-kDa membrane-bound protein. C92 has been shown to form pyramid structures for the release of mature virions (117). It is worth noting that the first viral gene (ORF305) expressed upon SSV2 infection of the host cell also encoded a membrane protein speculated to be involved in virus release. Transcription of most viral genes (31 in all) was detected at 16 h p.i. and peaked at 24 h p.i. Genes encoding eight out of nine virus-encoded proteins identified in STIV virions, located in three separate regions of the genome, were coordinately transcribed at 16 h p.i., and maximum levels of expression were achieved at 24 h p.i. Significant transcription of the last two genes and some intergenic regions was detected at 24 h p.i. Therefore, unlike the fuselloviruses SSV1 and SSV2, STIV does not show strong temporal regulation of genome transcription. In response to STIV infection, a small fraction (6%) of host genes was differentially expressed compared to uninfected cells. Of the 41 host genes upregulated by >4-fold, many were associated with functions in DNA replication and transcription. Of particular interest were genes encoding the replication initiation proteins Cdc6-1 and Cdc6-3. Other upregulated replication and transcription genes included those encoding reverse gyrase, a TFIIB paralog, the M subunit of RNAP, and a putative transcriptional regulator. The gene encoding Sso7d was also upregulated in STIV-infected cells, consistent with the proposed role for the chromatin protein in viral DNA packaging (118). STIV appears to share limited but potentially interesting similarity to SSV2 in eliciting a host response to infection. The expression level of the Cdc6 gene was significantly increased in both STIV- and SSV-infected host cells, suggesting a shared feature in the initiation of their genome replication. However, fewer host replication genes were upregulated in STIV-infected cells than in the SSV1-infected cells, presumably because STIV has a longer life cycle than does SSV1 and, thus, may not require the synthesis of additional host replication proteins for its genome replication. The two viruses also use similar strategies in recruiting host transcription machinery. A point of further interest is that among the 20 unannotated host ORFs upregulated in STIV-infected cells, 7 were also upregulated in SSV2-infected cells (108, 109). Conceivably, these host genes may encode functions required for steps shared by the two viruses in their infection cycles.
pSSVx.
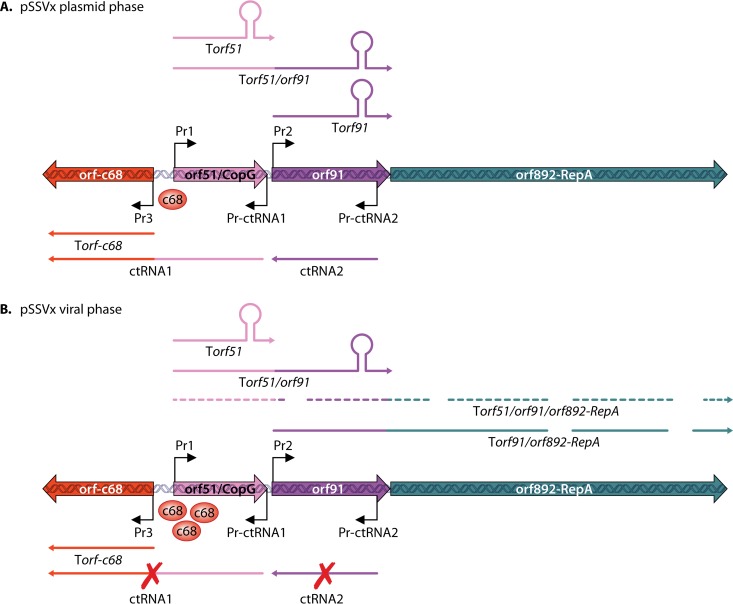
pSSVx is a plasmid-virus hybrid capable of coexisting with SSV2, the helper virus, in S. islandicus REY15/4 and spreading as virus particles (119). Like SSV2, it exhibits a pattern of inducible replication during host growth (120). Due largely to the genomic simplicity of pSSVx, gene expression in this genetic element has been investigated in detail (121, 122). There are 11 genes in pSSVx. Four plasmid genes, i.e., the putative copy number regulation genes copG (ORF51) and ORF91, the replication gene repA (ORF892), and the putative plasmid regulatory gene plrA (ORF76), are shared with the plasmids of the pRN1 family. Three viral ORFs are homologous to either fuselloviruses or other hybrid plasmid virus elements, whereas the remaining four are small RNA genes coding for CRISPR RNA1 (crRNA1) through crRNA4. In the early growth phase, pSSVx is present at a low copy number in the host, and only three genes, i.e., plrA and genes for ctRNA1 and ctRNA2, are expressed at high levels. Genes encoding ctRNA1 and ctRNA2, identified by Northern hybridization, are transcribed in a direction opposite that of copG (ORF51) and ORF91 (121). Therefore, the two RNAs are complementary to the copG and ORF91 mRNAs, respectively. Transcription from the rep operon of copG-ORF91-repA terminates prematurely at the 3′ end of copG or copG-ORF91, yielding two short transcripts lacking the repA gene. Upon induction, premature termination was attenuated, allowing the repA gene to be transcribed, yielding the long transcript of the rep operon covering the entire rep operon. During this stage, ctRNA1 and ctRNA2 are kept at undetectable levels, whereas two repA-containing transcripts (copG-ORF91-repA and ORF91-repA) are highly expressed. At the same time, induction of virus and plasmid replication occurs. These observations suggest that the two crRNAs might have inhibited the transcription of the repA operon by mediating premature termination. A model has been proposed for the predicted regulatory events in the induction of genome replication of the plasmid-virus hybrid (Fig. 7).
FIG 7.
Regulation of expression of genes at the rep locus in pSSVx, a virus satellite. Transcription of ORF892-RepA can start from either promoter Pr1 or Pr2. Solid lines, high relative levels of repA transcript; dashed lines, low relative levels of repA transcript. The thickness of the lines illustrating transcripts shows their relative abundance, and intrinsic transcription termination signals are indicated at the end of the transcripts. ORF892-RepA transcripts are sketched as interrupted lines to outline the nuclease susceptibility and consequently a fast turnover in the viral phase. (Reproduced from reference 53 [Fig. 6] with kind permission from Springer Science and Business Media [copyright Springer Japan 2014].)
Assembly and Release
Assembly.
Several archaeal viruses, such as SIRV2, STIV, HHIV-2, SH1, phiH, and phiCh1, have been shown to form progeny capsid particles in the host cytoplasm. Morphologically similar rudiviruses (SIRV1, SIRV2, ARV1, and SRV) all possess a tube-like superhelical structure formed by dsDNA and multiple copies of the major capsid protein (MCP), which is glycosylated and highly conserved (83 to 95% identity among the known rudiviruses) (52). The crystal structure of p134 (ORF134), the rudiviral MCP, reveals a unique four-helix-bundle topology (296). Recombinant ARV1 p134, overproduced in Escherichia coli, was able to self-assemble into filamentous superhelical structures of uniform widths and various lengths, and the optimal conditions for the self-assembly of p134 were close to those of the virus's habitat, indicating that the single MCP alone can generate the body of the virion in the host cytoplasm (27). The nucleocapsid and virion structures of lipothrixviruses are more complex than those of rudiviruses. AFV1, a gammalipothrixvirus, has two DNA-binding MCPs (AFV1-132 and AFV1-140) (123). Notably, both AFV1 MCPs possess a four-helix-bundle fold resembling that in the MCP of SIRV (SIRV-134), a finding not only in support of the recent grouping of the two viral families into the order Ligamenvirales but also suggestive of a possible similarity in the architectural roles of MCPs of the two viral families (69). Indeed, mixing of either of the two AFV1 MCPs with λ DNA fragments gave rise to long flexible filaments in vitro (123). Although these filaments are not directly relevant to virion structure, both proteins are likely involved in nucleocapsid assembly. Cryo-EM showed that native and SDS-stripped AFV1 displayed zipper-like structures characteristic of two-dimensional projections at different angles of a duplex DNA superhelix that would span the entire viral core. Intriguingly, the two MCPs differ drastically in physicochemical properties: the C terminus of AFV1-132 is highly positively charged (pI 9.5), like that of SIRV-134, whereas that of AFV1-140 shows a marked hydrophobic feature. Because both proteins bind DNA, it has been proposed that the DNA wraps around AFV1-132, and the AFV1-140 N terminus is attached to the DNA, while its globular lipophilic C-terminal domain is more exposed at the surface, such that it might become part of the envelope, forming a lipid-protein layer. How the AFV1 nucleocapsid is formed remains to be understood.
Aeropyrum coil-shaped virus (ACV), with a unique nonenveloped, hollow, cylindrical virion architecture, represents another mode of linear nucleocapsid organization. Based on transmission electron microscopy (TEM) images of dissembled ACV virions, the circular nucleoprotein formed on the ssDNA genome adopts two levels of organization by intertwining two halves of the circular molecule and supercoiling the resulting helix to produce the cylindrical helix of the virion (21).
STIV is a structurally well-studied Sulfolobus virus, which has an icosahedral protein shell dotted with turret-like structures pointing outwards at 12 5-fold vertices and an inner membrane enclosing a circular dsDNA genome (124). Mass spectrometry analyses have identified 11 proteins in the virion, 9 of which are virus encoded and 2 of which are host derived. In addition to the MCP, four viral proteins are believed to be involved in turret formation. The Sulfolobus chromatin protein Sso7d, one of the host proteins in the virion, is likely involved in the packaging of the viral genome (55). Observations of partially built virus particles in cellular electron tomograms of STIV-infected host cells suggest tight coupling in the assembly of the viral protein shell and the membrane. The viral membrane lipids are believed to be synthesized de novo. The insertion of the MCP into the lipid layer through its C terminus anchors the internal membrane to the protein shell. After the procapsid is formed, it is subsequently filled with the viral DNA in an unknown process.
Assembly of archaeal viruses of other morphotypes is less known. However, given the structural diversity of non-stick-shaped viruses, various strategies for virion assembly are conceivably adopted.
Release.
The release of mature virions from infected host cells completes the viral infection cycle. Recently, elegant analyses have begun to shed light on this step in several well-investigated archaeal virus-host systems. In their study of the SIRV2-host interaction, Prangishvili's laboratory discovered a novel virus release mechanism not found previously for other viruses (111). Following infection of S. islandicus by SIRV2, massive degradation of host chromosomal DNA started to occur in the early stage. This was followed by the assembly of virions in the cytoplasm in the form of several densely packed bundles of ∼50 virions, arranged side by side. Remarkably, heptagonal pyramidal structures, termed VAPs (virus-associated pyramids), appeared at the cell surface at the same time, rupturing the S layer and pointing outwards (Fig. 8). Mature virions and the host cytoplasm were released upon the opening of the VAPs, leaving behind the cell envelope as an empty shell. This mechanism of virus release differs distinctly from those used by lytic bacteriophages, which are released from the host cell either by degrading peptidoglycan or by inhibiting cell wall synthesis. VAPs were isolated from the membrane fraction of SIRV2-infected cells as stable, hollow, baseless pyramids with seven faces. These structures consisted of multiple copies of a single SIRV2 protein, P98, which was predicted to be a type II membrane protein and was self-sufficient to form the pyramid structure. However, questions remain concerning what drives the assembly of the virion bundles and how the well-orchestrated opening of VAPs and virion release are achieved. It has long been believed that most crenarchaeal viruses, including SIRV2, are nonlytic and exist in a stable carrier state on the basis of the absence of signs of cell lysis following viral infection. The finding that SIRV2 is a lytic virus, therefore, has necessitated a revisiting of these viruses.
FIG 8.
Schematic representation and micrographs of SIRV2 release from the host cell. Late during infection, progeny virions are assembled and aligned into clusters in the cytoplasm of the host cell and VAPs (red lines) at the cell surface, rupturing the S layer and pointing outwards. Mature virions and the cytoplasm were released upon the opening of the VAPs, leaving behind the cell envelope as a perforated and empty shell. (Reproduced from reference 111 with permission.)
Interestingly, this virion egress mechanism also appears to be used by STIV (54, 292). Young's laboratory showed that STIV virions were assembled in the host cell. While DNA-free precapsids were scattered around the cytoplasm, those filled with the viral genome were packed into well-organized quasicrystalline arrays in a way similar to that observed for the assembly of SIRV2 virion bundles before viral release. STIV-infected host cell developed pyramidal structures protruding from the cell surface. The pyramid was seven-sided, with sharply defined facets and apex. A 9.8-kDa membrane-bound viral protein, C92, was enriched in the membrane fraction of STIV-infected cells (117). This protein is homologous to P98 of SIRV2 (125, 126). Sulfolobus cells overproducing C92 developed pyramidal protrusions in the absence of viral infection, implying that the protein alone is sufficient to form pyramid structures. The use of similar egress mechanisms by morphologically and genomically distinct viruses is indicative of independently evolved morphogenesis and virion release pathways in archaeal viruses.
Recently, it was demonstrated that the S. solfataricus ESCRT-III machinery may be involved in the assembly and egress of STIV (127). The ESCRT gene cluster comprising vps4 (SSO0909), an ESCRT-III gene (SSO0910), and cdvA (SSO0911) was among the most highly upregulated gene clusters in S. solfataricus infected with STIV (109). Physical interactions between an ESCRT-III paralog (SSO0619) and the MCP of the virus and between another ESCRT-III paralog (SSO0910) and the pyramidal protein C92 were identified in a yeast two-hybrid screen. Immunolocalization by epifluorescence microscopy and transmission electron microscopy localized Vps4 to the pyramid lysis structure. When a mutant vps4 gene was overexpressed in the host cell, a productive STIV infection was abrogated. Therefore, it appears that STIV is able to hijack the host ESCRT machinery for its assembly and release.
Integration and Excision
Many archaeal viruses are capable of integrating their genome into the host genome, allowing their host to establish lysogeny. A growing list of these viruses now includes crenarchaeal viruses, e.g., SSV, ATV, STSV1, TPV1, APSV1, and APOV1, and euryarchaeal viruses, e.g., psiM2, phiCh1, and BJ1 (Table 2) (128). All of these viruses encode an integrase, which catalyzes both integration and excision of the viral DNA through site-specific recombination. However, integration activity has been experimentally demonstrated only for fuselloviruses and ATV (19, 114).
Integrases encoded by archaeal viruses and plasmids are all members of the tyrosine recombinase superfamily and fall into two types, the SSV1 type and the pNOB8 type. Both types of archaeal integrase target tRNA genes on the host chromosome. However, their integration mechanisms are distinguished by the location of the viral attachment sequences and the active-site residues of the enzymes. For SSV1-type integrases, the attachment site in the viral genome (attP) is located in the integrase gene proximal to the 5′ end, whereas for pNOB8-type enzymes, the attP sequence is upstream of the integrase gene (130, 131). Therefore, the SSV1-type integrase gene is split upon viral integration into two fragments, termed intN and intC, found at the boundaries of the integrated viral genome. Once the free form of the viral genome is lost, the provirus would become fixed in the host chromosomes. This may serve as a mechanism for horizontal gene transfer (HGT) (132). In addition, the conserved signature sequence in SSV1-type integrases (R…KXXR…Y) differs from that in the pNOB8-type integrases (R…YXXR…Y), both of which are different from the bacterial signature sequence (R…HXXR…Y) (131). Integrases encoded by all known archaeal viruses and some plasmids are of the SSV1 type, whereas those encoded by other archaeal plasmids and putative proviruses of euryarchaea belong to the pNOB8 type.
SSV1 integrase.
SSV1 integrase (IntSSV1) is the prototype of SSV1-type integrases. It is the only archaeal integrase that has been extensively studied so far. IntSSV1 catalyzes the integration of the SSV1 genome into a tRNAArg gene (CCG) in its natural host S. shibatae or in a foreign host. Integration entails site-specific recombination between viral and chromosomal attachment sites, attP and attA, respectively, to generate a right and a left proviral attachment site, attR and attL, respectively. A 44-bp invariant sequence is found at each att site. This 44-bp sequence comprises the downstream half of the tRNAArg gene and flanks the provirus as direct repeats (114, 133). Consequently, the integration of the SSV1 DNA into the host genome does not result in the disruption of the target tRNAArg gene, as shown for viral and plasmid integration in many other prokaryotes (133, 134). IntSSV1 is not an essential protein for SSV1. SSV1 lacking the IntSSV1 gene is capable of completing the infection cycle but is unable to integrate into the host genome (135).
The biochemical properties of IntSSV1 were first studied by Muskhelishvili et al. (136, 137). They showed that the enzyme catalyzed both integration and excision reactions. No additional accessory proteins were apparently required for the excision reaction, compared to those for the integration reaction. IntSSV1-mediated recombination occurred on linear and negatively and positively supercoiled DNAs but preferentially on linear templates. Those researchers found that the 44-bp invariant sequence was sufficient to support recombination, although the efficiency of recombination was affected by the flanking sequences. Furthermore, an 18-bp sequence within the att site was identified as the binding site for IntSSV1, as revealed by footprinting. For unknown reasons, however, their in vitro recombination assays of IntSSV1 have not been successfully established in other laboratories (138). Therefore, in their studies, Serre et al. employed assays targeting other activities of IntSSV1 (138, 139). They showed that IntSSV1 was able to cleave a 19-bp duplex DNA containing the above-mentioned 18-bp sequence, generating a nick on each of the two DNA strands in vitro. The two points of cleavage were offset by a stretch of 7 bp, referred to as the overlapping region, which corresponds perfectly to the anticodon loop of the target tRNA. IntSSV1 employed a cleavage mechanism depending on the active-site residue Tyr314 and formed a 3′-phosphoprotein intermediate during the cleavage reaction, as found for bacterial and eukaryal tyrosine recombinases. Like λ-Int, IntSSV1 also exhibited type IB topoisomerase activity, being able to relax both positively and negatively supercoiled DNA. Interestingly, in vitro complementation between the inactive IntSSV1 Y314F mutant and other inactive IntSSV1 mutants containing a substitution at each of the canonical residues forming the catalytic pocket of this family of recombinases restored the catalytic activity of the enzyme, suggesting that, as found with the yeast Flp recombinase (140), the active site of IntSSV1 is formed by Tyr314 from one monomer and the other residues from another monomer. Domain analysis revealed that IntSSV1 comprised an N-terminal domain and a C-terminal catalytic domain with similar sizes (residues 1 to 173 and residues 174 to 334, respectively), based on sensitivity to chymotrypsin treatment and amino acid sequence comparison with other tyrosine recombinases (142). Gel filtration assays showed that full-length IntSSV1 existed as a dimer in solution. The N-terminal domain was responsible for the dimerization of IntSSV1, whereas the C-terminal domain was capable of DNA cleavage and ligation although at efficiencies significantly lower than those of the full-length protein. In addition, neither domain alone showed a strong sequence preference for DNA binding. Presumably, recognition of the target sequence and efficient catalysis by IntSSV1 require covalent linkage and interdomain communication between the two domains. Recently, the crystal structure of the C-terminal domain of IntSSV1 was resolved (141, 142). The structure reveals a core fold similar to those of tyrosine recombinases of both bacterial and eukaryal origins, pointing to the conservation of these enzymes among the three domains of life. Five of the six catalytic residues cluster around a basic cleft on the surface of the structure, and the nucleophile Tyr314 is located on a flexible loop stretching away from the central cleft, supporting the suggestion from in vitro complementation experiments that the catalytic tyrosine is delivered to a neighboring subunit in trans.
Integrative genetic elements and their impact on genome evolution.
Interrogation of the rapidly accumulating genome sequencing data has made it increasingly clear that extrachromosomal genetic elements (ECEs) and their hosts exhibit more mutually beneficial interactions than previously appreciated. In many eukarya, integrated retroviruses and repetitive elements of a retroviral origin comprise a large fraction of their genomes. For example, ∼42% of the human genome sequence is derived from retroviruses, and these retroviral elements produce insertion mutations and facilitate DNA recombination, playing important roles in generating genetic novelty and driving genome evolution (143, 144). Archaea and Bacteria are single-celled organisms that can easily exchange genetic materials via three well-known mechanisms: direct uptake of naked DNA, plasmid transfer through conjugation, and virus transduction. After entering a host cell, the foreign DNA has two alternate fates: to become an integral part of the host genome or to be degraded. To ensure better survival, many ECEs have developed a strategy to integrate into their host chromosome via site-specific recombination, forming proviruses and integrated plasmids. Indeed, in the early 2000s, when only a few archaeal genome sequences were available, intact and degenerated integrated genetic elements were already identified in different archaeal genomes, including those from Sulfolobus, Aeropyrum, and Pyrococcus species (132). Some of these ECEs encode novel enzymes absent from their hosts, demonstrating that integrative genetic elements are an important vehicle of horizontal gene transfer (HGT) in Archaea. In addition, these integrative extrachromosomal genetic elements may interact with other mobile genetic elements such as insertion sequence (IS) elements and transposons and thus function as a vector in HGT. Once integrated into the host chromosome, these genetic elements may facilitate genomic rearrangements via homologous recombination, resulting in gene gain/loss and thereby providing an important mechanism for genome evolution in Archaea and Bacteria (130, 145, 146).
The integration of a mobile genetic element into its host genome is mediated by either an SSV1-type or a pNOB8-type integrase in Archaea. The former integrase, found only in Archaea, promotes the integration of genetic elements into their host chromosome, as discussed above for SSV1/2, whereas the latter enzyme catalyzes integration reactions in the same fashion as for the bacteriophage λ-type integrase but carrying a signature sequence distinct from that of the bacterial enzyme, as discussed above. All known archaeal conjugative plasmids encode a pNOB8-type integrase and are capable of integrating into their host chromosome at a tRNA gene. In archaeal genomes, some integrated or captured genetic elements are still recognizable as intact genetic entities. pXQ1 and XQ2, identified in the S. solfataricus P2 genome, were the first examples of these genetic elements. The two integrated elements are bordered by direct repeats (130, 131). The genomic regions harboring the two genetic elements are enriched in transposable insertion sequence elements, and one of the insertion sequences is inserted in the middle of the putative replication gene of pXQ1. In addition, a cluster of genes with sequences nearly identical in sequence to two regions of the Acidianus two-tailed virus (ATV) is present in the chromosome of S. solfataricus P2 (19). A close examination revealed that these gene fragments are located within the integrated genetic element XQ2 identified previously (132), and all of the genes encode unknown functions. Genes homologous to those of SSVs have also been identified in the S. solfataricus P2 genome, but the integration sites for the viruses are not recognizable (147). Similar studies on a few other archaeal genomes have led to the identification of a number of complete genetic elements or their remnants (130, 131).
While bioinformatic analyses revealed that the numbers of HGT genes were similar for all archaeal genomes, the numbers of identifiable integrated or captured genetic elements in a genome varied among archaeal species. There appears to be a positive correlation between the number of ECEs that have been isolated and the number of genetic elements integrated into the host genome in an archaeal species. To date, both integrated elements and plasmids/viruses have been identified most frequently from members of the Sulfolobaceae, Thermococcaceae, and Methanococcaceae. This may result from biased research efforts, which have focused mainly on genetic elements in these three archaeal families (see below). It is generally accepted that integrative genetic elements have played important roles in genome evolution in all archaea, as in bacteria. Indeed, when Cortez et al. employed an in silico Markov model-based strategy to analyze clusters of atypical genes (CAGs) in 119 archaeal and bacterial genomes, they found that only 7% of CAGs are likely transferred horizontally from distantly related cellular organisms, whereas 93% of CAGs are probably derived from ECEs that are either already known or not yet discovered (148).
ARCHAEAL PLASMIDS
All known archaeal plasmids are derived from the three physiological categories of extremophilic archaea, thermophiles, halophiles, and methanogens. These are exactly the three types of archaeal organisms that Carl Woese and colleagues employed to construct the phylogenetic tree of life. However, neither thermophiles nor methanogens represent a phylogenetic term since they are comprised of very diverse organisms. While some thermophiles are members of the Crenarchaeota, others belong to the Euryarchaeota together with methanogens and halophiles (1). Research on archaeal plasmids has been focused on a narrow range of archaea, including members of the Sulfolobaceae, Haloarchaeaceae, and Thermococcaceae, and various methanogens. Interestingly, archaeal plasmids isolated from more closely related organisms usually share more genes with one another. Because of this as well as the fact that replication mechanisms remain largely unknown for most archaeal plasmids at present, archaeal plasmids could be classified into four main groups, i.e., sulfolobus, haloarchaeal, thermococcal, and methanogenic plasmids. The first three groups include plasmids isolated from organisms of the Sulfolobaceae, Haloarchaeaceae, and Thermococcaceae, respectively, and the last group represents a collection of plasmids from various methanogens (see Table S2 in the supplemental material). This classification scheme is supported by the genetic dissimilarity shown by plasmids isolated from distantly related archaeal organisms using an operon similarity analysis (149).
Genomic Features
Sulfolobus plasmids.
A total of 24 crenarchaeal plasmids or plasmid-virus hybrids have been isolated from Sulfolobaceae. They are temporarily named sulfolobus plasmids because all but two of these plasmids were isolated from Sulfolobus species. The only two plasmids that were not derived from Sulfolobus were isolated from Acidianus species of the family Sulfolobaceae. Grouping of the two Acidianus plasmids with related sulfolobus plasmids is supported by sequence analysis (150, 151) and by genetic dissimilarity analysis (Fig. 9).
FIG 9.
Similarity dendrogram of sulfolobus plasmids. The dendrogram is based on the similarities of protein sequences encoded by the plasmid genomes, revealing their relationships as parts of plasmid families. The ruler at the upper left indicates a branch length corresponding to a 5% dissimilarity. Plasmids that do not show any common gene content are separated by branch lengths of 100%, while plasmids that do share sequences are separated by smaller branch lengths. The methodology employed for the dendrogram analysis was recently reported (149).
All sulfolobus plasmids are related, as they share a few DNA-binding proteins, although their putative replication proteins tend to differ. The most highly conserved ORF among these plasmids, represented by pRN1 ORF80, is a DNA-binding protein of unknown function. This protein was first identified in a few cryptic plasmids through genome comparisons and was designated plasmid regulatory protein A (PlrA) (150). Genes encoding homologs of PlrA are also present in some archaeal conjugative plasmids (152, 153) and in all known genomes of members of the Sulfolobaceae, including 10 Sulfolobus strains, Metallosphaera sedula, and Acidianus hospitalis. Since this ORF is absent from all other archaeal genomes and genetic elements, the genomic copies of plrA in the Sulfolobaceae are speculated to be derived from cryptic and conjugative plasmids via HGT.
The PlrA proteins of pRN1 and pSSVx have been characterized. The protein binds to two sites in its own promoter, and the two binding sites are located 60 bp apart (154, 155), suggesting that PlrA regulates its own expression. It was further shown that PlrA formed higher-order complexes upon DNA binding, presumably via protein-protein interactions (154, 155). Interestingly, PlrA has been implicated to play a role in plasmid segregation in in vivo experiments, since plasmid vectors derived from either pRN1 or pRN2 lacking the plrA gene are less stable than those constructed from the corresponding wild-type plasmids (65, 156). Currently, it remains unknown how the biochemical properties of the PlrA protein are related to its in vivo function.
Another conserved ORF in sulfolobus plasmids encodes a transcriptional repressor that shows a high level of sequence similarity to CopG, a ribbon-helix-helix (RHH) DNA-binding protein conserved in bacterial pMV158 family plasmids (157). For the bacterial systems, it was demonstrated that CopG employs two distinct mechanisms to regulate the expression of the rep operon: (i) hindering the binding of RNA polymerase to the promoter and (ii) displacing RNA polymerase once the enzyme has formed a stable complex with the promoter (158). Likewise, the archaeal CopG protein is expected to function as a transcriptional factor to repress the expression of replication proteins and thereby regulate the copy number of the plasmid (159). However, the function of these DNA-binding proteins has not yet been investigated in vivo for any archaeal genetic elements so far.
Cryptic plasmids.
Although all cryptic plasmids isolated from organisms of the Sulfolobaceae share several small DNA-binding proteins, including CopG and PlrA homologs, their putative replication proteins are highly diverse (60). Some of them clearly form a group, as they encode a homologous replication protein (RepA). These RepAs are very large proteins carrying a primase/DNA polymerase (Prim/Pol) domain at the N terminus and a helicase domain at the C terminus. The RepA gene often accounts for over one-third of the genome in these plasmids. pRN1 is the prototype of this group of plasmids, and therefore, these plasmids are referred to as the pRN1 family plasmids (Fig. 9). pRN1 coexists in S. islandicus REN1H1 with pRN2, another cryptic plasmid of the same type. However, the two plasmids replicate independently (12, 160). This group also includes pHEN7 from S. islandicus HEN7, pDL10 from the A. ambivalens, and pSSVx, a plasmid-virus hybrid, from S. islandicus REY15/4 (119, 150, 151).
The genomes of pRN1 family plasmids exhibit a modular organization of conserved and nonconserved sequences. The conserved modules consist of at least three genes, which encode RepA, its regulator(s), and PlrA, respectively, forming either one or two sequence modules separated by nonconserved DNA stretches at one or both junctions (151). A conserved sequence motif, 5′-TTAGAATGGGGATTC-3′, is present at the junctions between the conserved and nonconserved sequence regions in all of these plasmids, and this has led to the hypothesis that recombination at these conserved sequence motifs permits the formation of novel but related pRN1 plasmids (151).
The remaining Sulfolobus cryptic plasmids, including pSSVi, pXZ1, pIT3, pORA1, pTAU4, and pTIK4, encode putative replication proteins either distantly related or unrelated to those of the pRN1 family. Furthermore, they are distantly related among themselves, as shown by genetic dissimilarity analysis (Fig. 9). Two of these plasmids, i.e., pTAU4 and pSSVi, encode a helicase as their putative replication protein. pTAU4, a cryptic plasmid isolated from S. neozealandicus, encodes a homolog of MCM (161), a replicative helicase for chromosomal replication in Archaea and Eukarya (162), whereas pSSVi, a novel plasmid-virus hybrid identified in S. solfataricus P2, contains a gene for a superfamily 3 helicase (163). Putative replication proteins of pXZ1 and pORA1 (161, 164) show significant sequence similarity to that of the thermococcal plasmid pTN2 (PolpTN2), and these proteins carry a Prim/Pol domain and are very distantly related to the RepAs of the pRN1 family plasmids (165).
Bioinformatic analysis has furthermore shown that replication proteins containing the Prim/Pol domain are widespread in viruses and plasmids of both archaeal and bacterial origins. These proteins exhibit little conservation in sequence and domain organization, except for the Prim/Pol domain, among distantly related viruses and plasmids (166).
Conjugative plasmids.
A total of 12 conjugative plasmids (CPs) have been identified in Archaea, and they are all derived from members of the Sulfolobaceae. The first archaeal CP to be characterized was pNOB8 from Sulfolobus sp. strain NOB8H2, a strain isolated from a hot spring in Japan (167, 168). Subsequently, several CPs, including pING family plasmids (pKEF9, pHVE14, pARN3, and pARN4) and pSOG family plasmids, have been isolated from different S. islandicus strains derived from enrichment cultures of Icelandic hot spring samples (12, 41, 169). Recently, a conjugative plasmid, pAH1, was obtained from Acidianus hospitalis. A survey revealed that up to 3% of the Sulfolobus isolates contain a CP (41).
While some of the archaeal CPs are relatively stable, others exhibit striking genome instability. Interestingly, closely related CPs may differ significantly in stability. For example, pNOB8 shows a low copy number and is stably maintained in Sulfolobus NOB8H2, the native host. However, this plasmid readily forms a variant plasmid, pNOB8-33, following introduction into S. solfataricus through conjugation, and subsequent incubation of the transcipient leads to an increase in the copy number of pNOB8-33, which is followed by plasmid curing in recipient cells (41, 168). Plasmid pNOB8-33 carries a deletion of an ∼8-kb stretch of sequence, which encodes proteins homologous to the ParA ATPase and ParB DNA-binding protein, components of the bacterial plasmid partitioning and chromosomal segregation system (168). In bacteria, the Par proteins interact with the DNA element parC-parS and actively partition the replicated genomes of the plasmid or chromosome (170). Whether the pNOB8 ParA and ParB homologs serve a similar function in Sulfolobus and whether the loss of the parAB genes is responsible for the instability of pNOB8-33 remain to be investigated.
As a second example, pSOG1 and pSOG2, two CPs from S. islandicus SOG2/4, share ∼1/3 sequence homology (153). In the original host, pSOG1 exists at a very low copy number (153). After introduction into S. solfataricus through conjugation, pSOG1 was recovered as a high-copy-number plasmid. However, this plasmid was eventually cured from the foreign host during prolonged incubation of transcipients, presumably as a result of the activity of the host CRISPR-Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated) system. On the other hand, upon conjugation between S. islandicus SOG2/4 as the donor and S. islandicus HVE10/4 as the recipient, pSOG2 was obtained. Interestingly, this plasmid was stably maintained in both S. islandicus and S. solfataricus, suggesting that it did not activate the defense action of the host CRISPR systems. Since both CPs were derived from the same host, it was postulated that these CPs or their ancestors might have undergone genetic variation during conjugation (153).
The third example is the pING1 family CPs in S. islandicus HEN2P2, which exist as a mixture of unknown plasmids. Their conjugation into S. solfataricus resulted in various plasmid variants (41, 171). A few relatively stable pING1 family plasmids have been characterized by sequencing, whereas conjugative variants (pING4 and pING6) and nonconjugative but transferable variants (pING2 and pING3) have been identified (171). Strikingly, a large two-ORF IS element containing identical homologs of ORF213 and ORF408 in the S. solfataricus P2 genome is absent from pING1, the prototype of this plasmid family, but is present in all of the other stable variants. Conceivably, transposition of the IS element may have facilitated changes in pING1 plasmids in S. solfataricus, leading to the formation of relatively stable derivatives.
The nature of the instability of Sulfolobus conjugative plasmids is not understood so far, but some clues may be obtained from an analysis of available data. First, the arms race between CPs and host CRISPR-Cas systems may facilitate genetic changes in CPs. The host immune system is capable of specifically recognizing DNA sequence stretches (targeting sites) in CPs and degrading them by using a small-RNA-based mechanism (see “CRISPR Systems,” below). CPs lacking the targeting sites may be able to evade the host defense system. Furthermore, the CRISPR defense of CP conjugation may generate double-stranded DNA breaks, which will facilitate homologous recombination between CPs themselves and between CPs and host genomes, yielding CP variants that are subjected to CRISPR selection. Second, homologous recombination at specific DNA sequence motifs may lead to the generation of CP variants. The nonconserved genomic regions of conjugative plasmids are bordered by a sequence repeat capable of forming a hairpin structure (5′-TAAACTGGGGAGTTTA-3′) (152). This motif has been shown to be responsible for generating non-self-transferable pING2 and pING3 from the conjugative plasmids pING4 and pING6 of the pING family, respectively (41, 171). Interestingly, this motif shares a high level of sequence similarity with the putative recombination motif (5′-TTAGAATGGGGATTC-3′) identified in the cryptic plasmid family, suggesting that the recombination mechanism for plasmid diversification in Sulfolobaceae is conserved.
Comparative genomic analyses of six Sulfolobus conjugative plasmids revealed three conserved and functionally distinct modules in the genomes, i.e., sections A, B, and C (41, 152). Section A encodes probably all the components of the conjugative apparatus. All proteins encoded by section A ORFs are predicted to possess transmembrane helix motifs, a feature characteristic of proteins involved in DNA transfer. These include two ATPase proteins that are likely homologs of the bacterial conjugative proteins TraG and TraE (168). Section B contains a putative origin of replication with a highly conserved sequence, which features several perfect and imperfect direct and inverted repeats. Also encoded in this section are homologs of the CopG and PlrA proteins of pRN1 family plasmids as well as an integrase gene product of the tyrosine recombinase family. Further downstream, in section C, an operon encoding six to nine smaller proteins is implicated in the initiation and regulation of plasmid replication (152). So far, none of the above-described predictions has been verified experimentally. Since versatile genetic tools, including efficient host-vector systems, reporter gene assays, and highly efficient expression vectors (156, 172–174), have recently been developed for S. islandicus Rey15A, a host for all the above-mentioned conjugative plasmids except for pAH1 (175), it is now possible to conduct genetic analyses of predicted plasmid functions in this model crenarchaeon.
Haloarchaeal plasmids.
Haloarchaeal organisms form a monophyletic and coherent taxonomic group, the family Haloarchaeaceae, in 16S rRNA gene-based phylogenetic analyses of living organisms. Haloarchaea usually contain a main chromosome and several megaplasmids (176, 177). More than 15 haloarchaeal genomes are available in public databases, along with a large number of genomes of haloarchaeal plasmids, most of which are obtained from genome sequencing projects (40). Haloarchaeal plasmids often exhibit dynamic interactions with chromosomes due to the presence of a large number of insertion sequence elements. These plasmids either exist in an episomic form or integrate into the main chromosomes (178, 179), and host-plasmid interactions lead to frequent exchanges of genetic information. Therefore, some large plasmids (megaplasmids) carry genes essential for cell viability. For this reason, these plasmids are also termed the second chromosome in some haloarchaea in the literature (40, 180). Minimal replicons have been determined for two large haloarchaeal plasmids, pHH1 and pNRC100 (181, 182), both of which are involved in gas vacuole synthesis. Further analyses led to the identification of a putative replication protein, RepH (haloarchaeal replication protein homolog), a protein that does not show any sequence similarity to replication proteins of plasmids of bacterial origin or those obtained from nonhalophilic archaea. While it has been shown that some small haloarchaeal plasmids replicate in an RCR or a unidirectional θ mode, other haloarchaeal plasmids may replicate by using a mechanism involving a RepH or an ORC (origin recognition complex) homolog, although none of the putative replication proteins has been characterized so far (40).
The current GenBank database contains 24 full-length RepH homologs, most of which are from plasmids. Genes encoding a large number of truncated versions of these proteins are present on either the chromosomes or the plasmids of haloarchaea. A phylogenetic tree of RepH, constructed on the basis of sequences of the full-length RepH proteins, reveals at least two distinct types of RepH (see Fig. S1 in the supplemental material). At the amino acid sequence level, distantly related RepH proteins are so divergent that no meaningful consensus sequences can be derived, suggesting that these plasmid Rep proteins may function in distinct manners. Thus, to what extent these plasmids share the mechanisms of DNA replication remains to be investigated. The diversity of haloarchaeal plasmids has also been examined by constructing a plasmid gene content tree from all entries for haloarchaeal plasmids in GenBank (Fig. 10). This analysis shows a complex picture for the haloarchaeal plasmids. For example, there are two chromosomes and seven plasmids from Haloarcula marismortui, which fall into four distinct groups according to plasmid gene content analysis (183). The four smallest plasmids (pNG100 through pNG400, ranging from 33 to 50 kb in size) form one group, whereas three larger ones (pNG500 to pNG700, ranging from 132 to 410 kb in size) are different from one another (Fig. 10). Conceivably, there is a barrier to HGT between different plasmid groups in haloarchaea, contributing to the maintenance of genome stability of these organisms.
FIG 10.
Similarity dendrogram of haloarchaeal plasmids. The dendrogram is based on similarities of protein sequences encoded by the plasmid genomes, revealing their relationships as parts of plasmid families. The ruler at the upper left indicates a branch length corresponding to a 5% dissimilarity. Plasmids that do not show any common gene content are separated by branch lengths of 100%, while plasmids that do share sequences are separated by smaller branch lengths.
Thermococcal plasmids.
Members of the Thermococcaceae thrive in geothermal aquatic environments, mostly in deep-sea hydrothermal vents, where they play important roles in the ecology and metabolic activity of microbial consortia. This archaeal family has three genera at the present, Thermococcus, Pyrococcus, and Paleococcus, all of which are obligate heterotrophs growing anaerobically at temperatures of between 70°C and 105°C (42). A number of Thermococcus and Pyrococcus strains have been isolated from hydrothermal vents in the Atlantic, Indian, and Pacific Oceans, and a systematic screening for extrachromosomal genetic elements revealed that a large proportion of archaeal isolates carry plasmids. Fifteen plasmids have been isolated and characterized so far, and they fall into six different groups based on their putative replication proteins (184, 185).
The mode of replication is known only for the small plasmids pGT5 and pTN1, which were isolated from P. abyssi and Thermococcus nautilus, respectively. Both plasmids replicate via an RCR mechanism (186–188). Like pGT5 and pTN1, pRT1, another small plasmid isolated from Pyrococcus sp. strain JT1, codes for a protein containing two motifs similar to those of RCR proteins and was originally regarded as an RCR plasmid (189). However, its putative Rep protein does not share any sequence similarity to those of pGT5 and pTN1. Instead, the pRT1 protein resembles the putative replication protein of pAMT11 (190), a much larger plasmid (20.5 kb) from Thermococcus sp. strain AMT11. For this reason, it has been proposed that the two plasmids be classified into a new plasmid group (190). Consistent with this notion, plasmid tree analysis revealed that pGT5, pTN1, and pTP1, another thermococcal RCR plasmid (191), form one clade, which we propose to denote the pGT5 plasmid family, whereas pTA1 and pAMT11 form another clade (Fig. 11). This lends further support to the suggestion that pRT1 and pAMT11 most likely replicate differently from the pGT5 family plasmids.
FIG 11.
Similarity dendrogram of thermococcal plasmids. The dendrogram is based on similarities of protein sequences encoded by the plasmid genomes, revealing their relationships as parts of plasmid families. The ruler at the upper left indicates a branch length corresponding to a 5% dissimilarity. Plasmids that do not show any common gene content are separated by branch lengths of 100%, while plasmids that do share sequences are separated by smaller branch lengths.
Among all members of the pTN2 and pEXT9a plasmid families, only two ORFs are conserved. These ORFs encode a transcriptional factor containing a winged helix-turn-helix (wHTH) domain and a superfamily 1 (SF1) DNA helicase, respectively. The pTN2-encoded SF1 helicase (ORF1) enhances the synthesis of double-stranded DNA by PolpTN2 and T. nautilus PolB polymerase (192), presumably by catalyzing DNA unwinding in plasmid replication in both families. The other replication protein is family specific. In the pTN2 family, a Prim/Pol enzyme, represented by PolpTN2, was characterized recently (165, 184). The N-terminal domain of the protein showed both primase and DNA polymerase activities. Interestingly, the replication protein of the pEXT9a family carries a domain homologous to the C-terminal domain of PolpTN2, suggesting that the two classes of proteins are functionally related, although their shared function remains to be unraveled (185). Unlike the pTN2 or pEXA9a family plasmids, which devote two genes directly to plasmid replication, plasmids of the sulfolobus pRN1 family appear to have combined both the Prim/Pol activity and the helicase activity into a single protein, i.e., RepA. In other words, archaeal plasmids may have evolved their replication proteins by assembling functional modules of DNA replication in various assortments, leading to the diversification of replication mechanisms.
Thermococcal plasmids also exhibit a modular structure in gene organization: modules of conserved genes are separated by nonconserved DNA sequences. The conserved genes are clustered with apparent relevance to their functions (185). Both cis and trans elements responsible for plasmid replication, i.e., origins of replication and replication genes, are adjacently located, whereas genetic determinants for plasmid segregation as well as genes of unknown function are in a separate sequence stretch on the plasmids. The conserved regions are separated by hypothetical genes specific to each plasmid and, for relatively larger plasmids, also by genes with predicted functions. Each functional group of genes is often associated with a gene encoding a transcriptional factor, which may serve to regulate the expression of genes within the functional group. It is unclear whether there are conserved sequence motifs at the junctions between different modules. A detailed analysis of DNA sequences bordering the conserved modules may help determine if this large group of thermococcal plasmids also employs homologous recombination at conserved sequence motifs to assemble modular sequence regions, as for the crenarchaeal plasmids.
Plasmids of methanogens.
Methanogens include a diverse range of organisms that are distantly related phylogenetically. According to the current taxonomy of Archaea, methanogens comprise five orders of the Euryarchaeota: Methanococcales, Methanosarcinales, Methanobacteriales, Methanomicrobiales, and Methanopyrales (193). These organisms have been studied extensively, as they are the only organisms capable of producing methane, a unique ability that is exploited for the production of renewable energy on one hand and that contributes to global warming on the other hand. However, progress in the study of ECEs in methanogens has been slow, primarily due to the difficulty of culturing these strict anaerobes. Nevertheless, a few methanogenic plasmids have been obtained. These plasmids include pC2A from Methanosarcina acetivorans (194), pME2001 and pME2200 from Methanothermobacter marburgensis (195–197), pFV1 and pFZ1 from Methanobacterium formicicum (198), pURB500 from Methanococcus maripaludis (199), and three extrachromosomal genetic elements from Methanococcus jannaschii (200) and M. marburgensis through genome sequencing (201).
Very little is known about the mechanisms of DNA replication in this group of plasmids. Plasmids pFV1 and pFZ1 encode an Orc1/Cdc6 replication initiator showing a high level of sequence similarity to the replication initiator encoded in archaeal genomes (34). These Orc1/Cdc6 homologs are likely involved in the initiation of DNA replication in these methanogenic plasmids. In addition, both pME2001 and pME2200 encode a large protein containing a DNA-binding motif and an ATP/GTP-binding motif. Therefore, this protein is proposed to be the putative replication protein of these plasmids.
Horizontal gene transfer.
The Thermococcales and Methanococcales are phylogenetically distinct but coinhabit deep hydrothermal vents in oceans. Strikingly, some plasmids isolated from members of these two archaeal orders show a high level of sequence similarity. pT26-2 is the first integrative genetic element isolated from the Thermococcaceae, and this plasmid is closely related to several integrated genetic elements present in the genomes of Thermococcus kodakarensis; Pyrococcus horikoshii; Thermococcus gammatolerans; M. maripaludis S2, C2, and C7; and Methanococcus voltae; the former three belong to the Thermococcales, while the latter four are members of the Methanococcales (184). Since all of these genetic elements encode a putative integrase of the SSV1 type, it has been postulated that the occurrence of ECEs in different phyla of the Archaea may have resulted from events of cross-order HGT (130, 184).
HGT also occurred between pMETVU01 and pAMT7, as detected by genome comparisons. pMETVU01 was found in Thermococcus sp. strain AMT7, belonging to the Thermococcales, and pAMT7 was isolated from Methanococcus vulcanius M7, an organism belonging to the order Methanococcales. The two plasmids contain 10 conserved genes, which encode proteins sharing 53 to 97% identity at the amino acid sequence level. In fact, the two plasmids are the most closely related members of the pEXT9a family of plasmids (Fig. 11). Notably, the hosts of these plasmids are geographically close, as they were isolated from the same hydrothermal field at two sampling sites only ∼7 km apart (185, 202). Inhabitation of their hosts in the same environment apparently permits genetic exchanges between these plasmids.
Replication Mechanisms
Plasmids have served as models for investigations of the molecular mechanisms of DNA replication ever since their discovery. At present, three distinct mechanisms are known for plasmid replication in bacteria: θ replication, strand displacement, and rolling-circle replication (RCR). The detailed mechanisms of θ replication and RCR in bacterial plasmids have been extensively reviewed (203, 204). However, very little is known about the replication mechanisms of archaeal plasmids, largely due to the enormous diversity in the genomic content of archaeal plasmids and the lack of proteins homologous to the known replication (Rep) proteins of bacterial plasmids in most archaeal plasmids. This is consistent with the unique phylogeny of the domain Archaea and with the hypothesis that archaeal ECEs coevolve with their hosts. It appears likely that archaeal ECEs have developed novel mechanisms to replicate their genomes. While replication proteins in most archaeal viruses and quite a few archaeal plasmids remain to be identified, the majority of the putative replication proteins identified from ECEs have not been characterized experimentally. Among the few archaeal plasmids that have been studied for their biochemical properties are several very small archaeal plasmids encoding a Rep protein of the RCR type (205), two unidirectional θ plasmids, and pRN1, the prototype plasmid of the sulfolobus pRN1 family.
RCR.
Rolling-circle replication represents a common mode of plasmid and viral DNA replication in bacteria (204). The minimal replicon of an RCR plasmid includes a gene encoding a Rep protein; a DSO (double-stranded origin of replication), with which the Rep protein interacts, leading to the synthesis of the first strand of the plasmid DNA; and an SSO (single-stranded origin of replication), with which the host RNA polymerase interacts for primer synthesis. After priming, the host DNA replication machinery takes over and completes plasmid DNA synthesis.
All known RCR Rep proteins fall into two superfamilies (205). Rep proteins of superfamily I contain two tyrosine residues, and those of superfamily II have only one tyrosine residue in the active site. Both superfamilies have archaeal members. Whereas Rep proteins encoded in seven haloarchaeal plasmids are members of superfamily I, those of three thermococcal plasmids belong to superfamily II (see Table S2 in the supplemental material). Four of these archaeal plasmids have been investigated in detail.
Thermococcal RCR plasmids.
Three thermococcal RCR plasmids, i.e., pGT5, pTN1, and pTP1 from Pyrococcus abyssi, T. nautilus, and T. prieurii, respectively, carry two ORFs. Only the larger one is conserved among these plasmids. It carries three motifs characteristic of an RCR Rep protein (205). Two of the putative archaeal Rep proteins, i.e., Rep75 of pGT5 (187, 206, 207) and Rep74 of pTN1 (188), have been characterized in vitro. Both enzymes showed a high level of nicking-closing activity on single-stranded oligonucleotides containing their cognate putative DSO and remained covalently linked to the 5′ phosphate of the downstream fragment after nicking. As in the bacterial RCR plasmids, the thermococcal plasmid DSOs contain several direct and inverted DNA sequence repeats important for the initiation of plasmid DNA replication (186, 188). The DSO sequences show a high level of sequence similarity to those of the plasmids of the ϕX174/pC194 superfamily, and the cleavage site (5′-GTTGGGTTATCTTG↓ATA-3′ [identical nucleotides are underlined, and the site of cleavage is shown by an arrow]) is highly conserved. Cleavage of the DSO generated a 3′-OH on the DNA strand suitable for the host DNA polymerase to synthesize the first DNA strand. Rep75 remains attached to the 5′ end of the nick covalently. At the end of one replication round, Rep75 cleaves the DSO again and religates the newly synthesized strand, generating a circular single-stranded DNA that is converted to a double-stranded plasmid by host proteins (206).
Thermococcal RCR plasmids also exhibit features that are distinct from those of the known bacterial RCR plasmids. First, the DSO elements are located at the 5′ end of the coding sequence of Rep75/Rep74, whereas the SSO sequences are present in the noncoding region immediately upstream of the rep gene in the pGT5 and pTN1 genomes. Such an arrangement allows only the second round of DNA synthesis to occur on the ssDNA template after the completion of the first round of replication (206). Second, the archaeal Rep proteins are >2-fold larger than typical bacterial RCR Rep proteins and are homologous to the transposases of bacterial conjugative transposons, which replicate themselves via an RCR mechanism (208). Third, the pGT5 Rep75 protein has topoisomerase activity in vitro, an activity considered to be essential for pGT5 replication because this archaeal plasmid is relaxed in vivo, while Rep75 is unable to act on a relaxed plasmid (207). Together, these results indicate that thermococcal RCR plasmids replicate in an RCR mode with novel mechanistic features.
Haloarchaeal RCR plasmids.
Seven haloarchaeal plasmids, including pGRB1 from Halobacterium sp. strain GRB, pHSB1 and pHSB2 from Halobacterium sp. strain SB3, pHGN1 from Halobacterium sp. strain GN101, pHK2 from Haloferax sp. strain Aa2.2, pNB101 from Natronobacterium sp. strain AS-7901, and pZMX201 from Natrinema sp. strain CX2021, belong to superfamily I RCR plasmids (see Table S2 in the supplemental material) (38, 39, 209–213). Some of these plasmids were isolated decades ago have not been characterized further. Like typical bacterial RCR plasmids, all the haloarchaeal RCR plasmids except for pHK2 are small, ranging in size from 1,668 to 2,538 bp. pHK2 is exceptionally large (∼10.5 kb). The putative Rep proteins of these plasmids are conserved, sharing >30% amino acid sequence similarity and containing the three sequence motifs characteristic of an RCR Rep protein (205).
The Natrinema plasmids pZMX201 and pNB101 were the first archaeal RCR plasmids to be characterized in vivo (213). The double-stranded origins were identified as a stem-loop structure formed by an imperfect inverted repeat, and the sites of nicking were located within a heptanucleotide sequence (5′-TCTC↓GGC-3′) on the right side of the stem. The nicking site is conserved in all known haloarchaeal RCR plasmids. Furthermore, a hairpin structure that might function as an SSO was also identified in these plasmids, suggesting that they employ a conserved RCR mechanism. Zhou et al. employed a hybrid plasmid assay to study the specificity of in vivo nicking and closing activities by the Rep proteins of pZMX201 and pNB101 (213). Two test hybrid constructs containing a DSO and its cognate rep gene from one of the two plasmids and a DSO fragment from the other plasmid were prepared. The two DSOs on each test plasmid were located in such a way that the plasmid was divided into two unequal halves, i.e., a shorter half downstream of a DSO carrying a mevinolin selection marker and a longer half downstream of the other DSO containing an Escherichia coli plasmid sequence with an ampicillin selection marker. If the enzyme is capable of nicking and closing only at its own DSO, the complete test plasmid would be produced. If a Rep protein initiates and terminates plasmid DNA replication at both DSOs, replication products of two different sizes would also be generated, with the larger and the smaller ones containing the E. coli sequence and the mevinolin marker, respectively. This study demonstrated that pZMX201 and pNB101 are able to cross-recognize each other's DSOs, initiating and terminating RCR replication, because all three predicted RCR products were obtained for the two test plasmids (213). Furthermore, the roles of sequence elements in the pZMX201 DSO, including the conserved cleavage site TCTCGGC and its left and right arms, were investigated by using the same strategy. It appears that the five nucleotides in the middle of the conserved TCTCGGC sequence served important roles in both the initiation and termination phases of plasmid DNA replication, since mutation of a single nucleotide in this region abolished the replication of the test plasmids. In addition, DSO derivatives lacking the left arm of the DSO impaired the initiation, but not the termination, of plasmid replication (213).
θ replication.
θ replication involves the formation of a θ-like replication intermediate. The mechanism of θ replication has been studied most extensively for some plasmids from Gram-negative bacteria (203). The θ mode of replication is more widely used than the RCR mode of DNA replication. Replication of chromosomes in organisms of all three domains of life and a large group of bacterial plasmids proceeds in the θ mode. It is thus expected that most archaeal plasmids may have adopted this strategy to replicate their genome. A typical origin of θ replication consists of the following elements: DNA sequence motifs that serve as the binding sites for their cognate replication initiator proteins and an AT-rich sequence region that is unwound during the initiation of replication to form a replication bubble. Only two archaeal plasmids, i.e., pSCM201 from Haloarcula sp. (214) and pZMX101 from Halorubrum saccharovorum (215), are currently known to replicate via a θ mechanism.
pSCM201 encodes three ORFs, of which ORF1 contains a putative leucine zipper (LZ) motif, an HTH domain, and an ATPase domain. The latter two domains are common in replication initiation proteins (216), and the LZ motif is also found in the Rep proteins of many iteron-containing plasmids in bacteria (181, 182). Therefore, ORF1 was proposed to serve as the Rep protein of pSCM201. An AT-rich region flanked by perfect inverted repeats and seven 9-bp tandem repeats (or iterons) in the noncoding sequence region upstream of the rep gene were identified, and this region was suggested to be the origin of replication for this plasmid (214). Another haloarchaeal plasmid, pZMX101, was found to possess the same genetic organization and to code for a Rep homolog that shows 40% amino acid sequence similarity to Rep of pSCM201, suggesting that the two plasmids are of the same type. Indeed, it was furthermore demonstrated that the minimal replicon is comprised of the putative replication origin and the adjacent rep gene in both plasmids (214, 215).
The structure of the haloarchaeal θ origins of replication resembles that of haloarchaeal chromosomal origins of replication, such as those of Halobacterium sp. strain NRC-1 (217), H. marismortui (183), Haloferax volcanii (178), and Natronomonas pharaonis (218). The apparent difference is that the iteron element in the plasmid origins is replaced with the DNA-binding sites of the archaeal replication initiator Orc1/Cdc6. On the other hand, the AT-rich elements are widely conserved among many bacterial and eukaryal origins of replication. The replication origin of pSCM201 has been characterized in detail. The precise point of the initiation of pSCM201 replication was found to be located between the inverted repeats, and electron microscopic analysis showed that replication intermediates of the plasmid adopted a σ-like structure, with the origin located at one end of the structure, demonstrating that pSCM201 replication proceeded in a unidirectional θ mode (214).
Replication of the Sulfolobus pRN1 plasmid.
pRN1 has been used as a model for the study of the replication mechanism of the pRN1 family plasmids. Georg Lipps and colleagues characterized all three conserved replication proteins encoded in pRN1, including the copy number regulation protein CopG (encoded by ORF56) (219), a DNA-binding protein implicated in plasmid maintenance (PlrA) (encoded by ORF80) (154), and a multifunctional replication protein (RepA) (159, 220–222). They were the first to reveal that both primase and DNA polymerase (Prim/Pol) activities resided in the pRN1 RepA protein and localized the Prim/Pol activities to the N-terminal part of RepA (220, 221). They also found that the C-terminal helicase domain is structurally similar to a superfamily 3 helicase, displaying weak helicase activity in vitro (159). Structural analysis and site-directed mutagenesis of RepA demonstrated further that the Prim/Pol domain at the N terminus interacts with the helix bundle domain at the C terminus to synthesize a primer (222). Prim/Pol of pRN1 is the prototype of a novel DNA polymerase family whose members are encoded by various archaeal and bacterial plasmids as well as some bacterial viruses (166). Furthermore, the origin of replication of pRN1, located immediately downstream of the repA gene, has been experimentally verified (223).
Despite extensive studies on pRN1 plasmids for more than a decade, it is still unclear how these plasmids replicate. Early sequence analyses predicted putative SSOs and DSOs for several of these plasmids (150, 151). However, none of the RepAs encoded in the pRN1 family plasmids contains the three motifs characteristic of RCR Rep proteins, nor does the pRN1 RepA protein exhibit nicking-closing activity essential for RCR replication. Furthermore, shuttle vectors constructed with the putative minimal replicon of pRN1 or pRN2, which included the rep operon and the putative origin but lacked the predicted DSO element, were able to replicate in Sulfolobus cells (156), indicating that pRN1 plasmids do not replicate in an RCR mechanism.
Plasmid Functions
Only a few archaeal plasmids have a known function. Among these, two large haloarchaeal plasmids carry genes responsible for gas vacuole formation, and sulfolobus conjugative plasmids are able to spread from one host to another. Most archaeal plasmids, large or small, have no known functions. However, putative functions of some plasmid genes have been deduced from bioinformatic analysis. Small plasmids appear to be cryptic, since they encode only proteins with putative roles in their own replication, segregation, and gene regulation, but large plasmids have ORFs of other predicted functions. Several large thermococcal and methanococcal plasmids, e.g., pCIR10 from Thermococcus sp. strain CIR10, extrachromosomal element 1 (ECE1) and ECE2 from M. jannaschii (200), and pFS01 from Methanocaldococcus sp., encode Hfq-like domain proteins containing a C2H2 Zn finger (ZF) domain at the N terminus (185). Hfq-like proteins belong to a large family of Sm RNA-binding proteins that perform a range of important RNA-related functions in bacteria (224). In addition, a recent structural study identified a distant variant of an Hfq-like protein encoded by a Pyrobaculum spherical virus (48), raising the possibility that Hfq-like proteins may be widespread in archaeal plasmids and viruses.
Toxin-antitoxin (TA) systems, first identified in bacterial plasmids during studies of their segregational killing of plasmid-free cells, have been found in many archaeal and bacterial genomes. Recently, putative TA systems of the RelBE and VapBC families have been identified in several thermococcal and methanococcal plasmids. Plasmids pCIR10, pEXT9a, and pMETVU01 contain TA genes of the RelBE family, whereas pIRI33 from Thermococcus sp. strain IRI33 encodes a TA system of the VapBC family (185). In bacteria, VapC toxins are site-specific endonucleases that cleave tRNAfMet in the anticodon stem-loop, thereby inhibiting protein translation. VapB, the antitoxin, counteracts the toxic action of VapC through direct protein-protein interactions. In the bacterial RelBE systems, RelE, an RNase, inhibits translation by cleaving mRNAs at ribosomal A sites, whereas RelB antagonizes the action of RelE through both direct protein-protein interactions and transcriptional repression of the relBE operon. A general feature of these bacterial TA systems is that they encode a stable toxin and an unstable antitoxin. It has been shown that plasmid-borne TA systems mediate the postsegregational killing of plasmid-free cells, thereby contributing to the stable maintenance of the plasmids in their bacterial host cells. Archaeal plasmids encoding putative TA systems provide valuable models for the study of mechanisms and functions of the archaeal TA systems. However, none of these archaeal TA systems have functionally been characterized, except for a few Sulfolobus TA systems that were shown to be involved in the regulation of thermophily of the organism (225, 226).
Virus-Plasmid Interactions
The frequent coexistence of both viruses and plasmids in the same archaeal host has allowed these genetic elements to evolve unique and complex relationships. This is clearly demonstrated by the interactions between archaeal helper viruses and virus satellites. The first archaeal virus satellite was identified by analyses of virus-like particles in a S. islandicus REY15/4 culture infected by the fusellovirus SSV2. SSV2 was found to facilitate the spread of pSSVx, a plasmid-virus hybrid of the pRN1 plasmid family (119, 147). Like other members of the pRN1 family, pSSVx contains all three ORFs characteristic of this plasmid family. Interestingly, it also possesses homologs of two ORFs present in fuselloviral genomes (119). Later, a second plasmid-virus hybrid, pSSVi, was identified in S. solfataricus P2, and this virus satellite carries a replicon different from that of the pRN1 family plasmids as well as an SSV1-type integrase gene (164). Both pSSVx and pSSVi are capable of packaging into a spindle-like viral particle and spreading with the help of SSV1 or SSV2. More recently, a plasmid termed pXZ1 was found to have a genome organization similar to that of pSSVi, and pXZ1 coexists stably with the fusellovirus SSV4 in S. islandicus ARN3 (51). However, this plasmid lacks the ability to become packaged into a virion. Therefore, it appears that these cryptic plasmids have developed two distinct strategies to enhance their probabilities of survival. They may either exploit the packaging apparatus of fuselloviruses to form freely spreadable virus-like particles or employ a self-encoded integrase to facilitate gene exchanges with viruses and/or archaeal hosts.
Mechanisms for the packaging of the archaeal virus satellites into a virion are not clear. Three virus-like ORFs of pSSVx, i.e., ORF-c68, ORF154, and ORF288, were initially implicated in virion formation (119). However, no homologs of pSSVx ORF154 and ORF288 were found in the pSSVi genome, suggesting that these two ORFs are not essential for the formation of satellite virions (227). ORF-c68 has been shown to encode a transcriptional regulator (228) and, therefore, is unlikely to be involved in virion packaging. It is possible that the packaging machinery of a fusellovirus recognizes a sequence motif in the genome of a satellite virus to initiate virus packaging. Interestingly, the sequence motif 5′-AAGGGAAANAGNA-3′ is present in the genomes of pSSVx, pSSVi, and SSV2 (bp 989 to 1001, bp 2412 to 2424, and bp 1406 to 1418 on their linear map, respectively) and absent from all known pRN plasmids that replicate autonomously (45). Whether this sequence motif serves to initiate the packaging process awaits further investigation.
Transfer of interchangeable modules among ECEs has been postulated based on genomic analyses of Pyrococcus abyssi virus 1 (PAV1). Two thermococcal viruses have been obtained so far, PAV1 and Thermococcus prieurii virus 1 (TPV1) (98, 229). Interestingly, the two euryarchaeal viruses share only two ORFs that are implicated in viral infection. After several large plasmids were isolated from thermococci, it became clear that the PAV1 genome encodes plasmid protein homologs (185). In fact, the PAV1 genes homologous to those of archaeal plasmids and integrative genetic elements are clustered together and occupy roughly half of the PAV1 genome, while the viral genes that were shown (ORF121) or predicted (ORF676 and ORF678) to encode structural proteins are located in the other half of the genome (230). It is inferred from these observations that PAV1 or its ancestor virus may have resulted from recombination between a plasmid and a virus.
More complex interactions between archaeal viruses and plasmids have also been observed. For example, the conjugative plasmid pAH1 resides in both integrated and free forms in Acidianus hospitalis W1. Upon infection of Acidianus hospitalis W1 containing the plasmid with the lipothrixvirus AFV1, the level of the free circular form of pAH1 decreased, with a concurrent increase in the intracellular level of AFV1 DNA. It was inferred that AFV1 inhibited pAH1 replication because no plasmid degradation was detected (56, 57). Viral inhibition of plasmid propagation also provides a rationale for why some Sulfolobus conjugative plasmids have CRISPR clusters of the archaeal immune system (see below). Both pNOB8 and pKEF9 carry CRISPR clusters, and pKEF9 has a spacer very closely matching a sequence in the genome of the Sulfolobus rudivirus SIRV1 (48, 58). The processed spacer RNA is potentially capable of targeting and inactivating the virus intracellularly.
DEFENSE MECHANISMS OF ARCHAEAL HOSTS
Restriction-Modification Systems
Restriction-modification (R-M) systems were discovered along with the isolation of extrachromosomal genetic elements, since they provided an innate defense mechanism for bacterial hosts to restrict plasmid spread and virus infection (231). Four major types of R-M systems (types I to IV) are known, each of which is composed of two components: a methyltransferase (MTase) that modifies the host DNA in order to protect it and a restriction endonuclease (REase) that recognizes and cleaves nonmethylated invading DNA (232, 233). Based on bioinformatic prediction, at least one R-M system is encoded by nearly all prokaryotic genomes. While ∼95% of the sequenced bacterial genomes encode R-M systems (http://rebase.neb.com/rebase/rebase.html), only one of the 199 currently available archaeal genomes lacks the system, as of the time of writing of this review (234).
As in bacteria, restriction of foreign DNA by R-M systems was found soon after haloarchaeal viruses (halophages) were discovered. In 1985, Patterson and Pauling developed an assay to detect R-M systems present in six Halobacterium cutirubrum strains (235). In their experiments, a halophage was grown first in H. salinarium R1, a restriction-minus haloarchaeal strain, and then in each of the six strains for which R-M systems were to be analyzed. This would yield phage particles with the genomic DNA modified by host R-M systems, if they existed in a tested host. The modification was then analyzed by introducing the above-described phage particles individually into each of the H. cutirubrum strains; phages would be restricted in a new host if they lacked the modification of the host R-M system, exhibiting a 100- to 1,000-fold reduction in infection efficiency. Those researchers found that one of the host strains produced a halophage that was restricted in all of the other host strains, and the remaining five host strains released phages that were protected from restriction to different extents. Among the latter five strains, one yielded a phage infectious to all tested strains, whereas the rest of the strains produced phages infectious to only the corresponding phage-producing strains but restricted in the other two types of strains. Together, this led to the identification of two different R-M systems in H. cutirubrum (235).
In the current ReBase database, many archaea encode multiple R-M systems of types I, II, and IV (234). However, only a few archaeal R-M systems have been characterized so far. These include three functional R-M systems in hyperthermophilic archaea, i.e., the Pyrococcus PabI and PspGI R-M systems and the S. acidocaldarius SuaI system. These restriction endonucleases were isolated from P. abyssi (denoted R.PabI) (236), Pyrococcus sp. strain GI-H (R.PspGI) (237), and S. acidocaldarius (R.SuaI) (238). All three enzymes degraded E. coli DNA but showed no cleavage activity on DNA from their respective hosts. The three corresponding MTases (M.PabI, M.PspGI, and M.SuaI) were also isolated. As expected, modification by M.PabI protected DNA from cleavage by R.PabI (239). Two additional restriction enzymes were isolated from S. islandicus REN2H1 (SuiI) and Thermoplasma acidophilus (ThaI) (240, 241). Both enzymes are believed to be part of a functional R-M system, but their corresponding MTases have not yet been identified.
Host restriction poses a major challenge to genetic engineering since it greatly reduces the efficiency of plasmid transformation. For instance, the H. volcanii restriction system recognizes and cleaves adenine-methylated 5′-GATC-3′ (dam+) sites (242). To avoid host restriction, plasmid DNA to be used for transformation of H. volcanii needs to be passaged through an E. coli dam mutant. This would allow the treated DNA to be introduced into H. volcanii at a greatly improved efficiency (243). Since the S. acidocaldarius R-M system specifically cleaves unmethylated 5′-CCGG-3′, plasmid DNA may be introduced into the archaeon after treatment with the E. coli HaeIII methyltransferase to yield 5′-CCmGG-3′ on the plasmid. While this indicates that Archaea are capable of utilizing their R-M systems for defense against invading plasmid and viral DNAs, it also suggests that ECEs could produce a methylated genome to avoid host restriction.
Indeed, it has been shown that the genomes of several archaeal viruses are methylated, whereas their host chromosomal DNAs are not. These viruses include phiChi1 from Natrialba magadii, phi-N from Halobacterium salinarium (244, 245), and SNDV, STSV1, and STSV2 from Sulfolobus (78, 95, 246). In fact, the Sulfolobus viruses STSV1 and STSV2 encode three putative DNA MTases of >90% sequence identity (95, 246). All of these enzymes have homologs in many archaeal and bacterial genomes, as revealed by CCD analysis (247). The largest of them (>700 amino acid residues) is highly similar to adenine-specific methyltransferases containing a zinc ribbon, which are widely distributed in various archaeal and bacterial genomes. The two other methyltransferases are implicated in cytosine methylation, and their homologs are present in relatively few genomes. Importantly, the genes encoding closely related homologs of the two methyltransferases are found in the genomes of A. hospitalis, S. acidocaldarius DSM639, and S. islandicus YN 15.51 (248–250) but are absent from the nine sequenced genomes of other members of the family Sulfolobales. Methylation of a viral genome by multiple R-M systems may serve important roles for the virus to establish successful infection since the viral DNA methylated in multiple ways may better survive the innate defense system of the host.
Furthermore, methanogenic plasmids pFV1, pFZ1, and pFZ2 from M. thermoformicicum were found to carry different type II R-M systems (198), such as MthTI, a GGCC-recognizing R-M system (33), and MthZI and MthFI, both of which are CTAG-recognizing R-M systems (34). The genomes of these archaeal plasmids are thus methylated by the encoded MTases, which allow them to escape host restriction. Conceivably, an arms race between ECEs and their host will continue with respect to innate immune defense in Archaea. In this regard, the CRISPR adaptive immune defense may offer additional advantages in host defense against ECE invasion.
CRISPR Systems
The CRISPR-Cas system is a small-RNA-based adaptive defense system discovered recently in Archaea and Bacteria. This defense system is comprised of two parts: CRISPR loci consisting of an array of unique sequences (spacers interrupted by repeats) of similar lengths and clusters of genes coding for Cas proteins that are either DNA helicases or nucleases or repeat-associated mysterious proteins (RAMPs) that contain the classical RNA recognition motif (251). At each CRISPR locus, a leader sequence is located upstream of a repeat spacer array. The general mechanism underlining the action of CRISPR systems includes three steps: spacer acquisition, CRISPR RNA (crRNA) biogenesis, and invading nucleic acid destruction (Fig. 12) (100, 252–254). Upon the invasion of a newly encountered plasmid or virus, Cas1 and Cas2, which are widely conserved in different CRISPR systems, recognize specific sequence motifs on the invading element, and a small piece of the foreign DNA is cleaved and inserted into the CRISPR locus between the leader and the first repeat, generating the new first spacer. The entire CRISPR locus is expressed from the leader, which functions as a promoter, yielding a long precursor CRISPR RNA (pre-crRNA). Cas6 or its functional homologs bind to all repeat motifs in the pre-crRNA and cleave the RNA to generate mature crRNAs. RAMPs and crRNAs then form nucleoprotein complexes and identify the protospacer in the invading ECE, which is complementary to the crRNA generated from the spacer acquired during the first invasion event of the same genetic element. The complex recruits helicase and nuclease Cas proteins for the destruction of the invading nucleic acids. It has been demonstrated that Cas-crRNA nucleoprotein complexes obtained from both Archaea and Bacteria are capable of destroying invading DNA (255–257) and RNA (258–260), exhibiting DNA/RNA-targeting activities.
FIG 12.
Three functional stages of the CRISPR-Cas defense mechanism. Adaptation indicates the acquisition of a new spacer (in red) in a chromosomal CRISPR locus; Processing indicates that CRISPR clusters are transcribed, yielding precursor CRISPR RNAs (pre-crRNA), and processing of pre-crRNAs produces mature small guide crRNAs; and Interference indicates that crRNA forms a ribonucleoprotein complex with Cas proteins and nucleic acid targeting, resulting in the degradation of target DNA or/and RNA. (Reproduced from reference 253 [copyright Elsevier 2011].)
A number of Archaea and Bacteria harbor several CRISPR interference systems. These systems are categorized into three main classes, i.e., types I, II, and III, based on an elegant classification system proposed by Makarova et al. (261). Each type has its own conserved Cas protein(s) (i.e., Cas1 and Cas3 for type I, Cas9 for type II, and Cas10 for type III) (261). So far, type II CRISPR systems appear to be restricted to a few bacterial species, whereas type I and type III CRISPR systems are prevalent in Archaea. These archaeal CRISPR systems are further divided into several subtypes of types I and III, including subtypes I-A, I-B, I-D, III-A, III-B, and III-D (149, 261).
The interference activities mediated by CRISPR systems have been studied for several archaeal species. Type I systems have been shown to mediate DNA targeting in S. solfataricus, S. islandicus, H. volcanii, Haloferax mediterranei, Methanosarcina mazei, and T. kodakarensis (262–269). These archaeal type I systems identify invading DNA elements by recognizing protospacer-adjacent motifs (PAMs), as observed for the DNA-targeting mechanisms of bacterial type I and II CRISPR systems (254). So far, only type IIIB CRISPR systems (also known as CRISPR, mysterious RAMP, and Cmr modules), including Pyrococcus furiosus Cmr and S. solfataricus Cmr-β, have shown RNA-targeting activity. Cas-crRNA nucleoprotein complexes have been isolated for the two CRISPR systems, and they exhibited RNA cleavage activity in vitro (258, 259). Strikingly, a type IIIB system (Cmr-α) from S. islandicus REY15A was found to confer DNA interference in vivo, and its DNA-targeting activity did not depend on the presence of a PAM sequence. In addition, the dissimilarity between the protospacer and the residual repeat region in the crRNA facilitated the DNA-targeting activity, whereas the homology between them enhanced protection against DNA targeting (264). Recently, RNA targeting by the same CRISPR system was demonstrated, providing the first example of a CRISPR system with dual functionality in nucleic acid destruction (298).
CRISPR spacer acquisition in Archaea has been studied extensively by using S. solfataricus, S. islandicus, and H. hispanica as model systems. Unlike all other known CRISPR systems, infection of S. solfataricus or S. islandicus with a single virus did not lead to the insertion of new spacers in the CRISPR loci of these archaeal chromosomes. However, when both SMV1, a novel virus isolated from a hot spring in Yellowstone National Park, WY, USA, and the conjugative plasmid pMGB1 or STSV2 were introduced into S. solfataricus or S. islandicus, spacer acquisition was activated, and new spacers derived from pMGB1 or STSV2 were inserted into the CRISPR loci of each host genome (270, 271). Since none of the new spacers were derived from SMV1, this suggests that SMV1 might have induced spacer acquisition in these crenarchaea. In comparison, no induction was required for spacer acquisition in the euryarchaeon H. hispanica following infection by HHPV2. Intriguingly, the priming activity of Cas3 was found to be important for successful spacer acquisition in this euryarchaeon (272). Together, these results point to the diversity in the CRISPR-mediated defense against ECE invasion.
The importance of the CRISPR systems in host defense against viral infection has also been demonstrated using the SIRV2-Sulfolobus host system by transcriptomic analysis of host gene expression upon viral infection (107). It was found that most of the host defense genes, including cas genes and CRISPR arrays, were significantly upregulated in the host following SIRV2 infection. However, some of the Cas genes were already highly expressed prior to viral infection. The differential regulation of various cas genes/modules suggests that the interference complexes may have multiple functions. Further investigation is needed to learn more about the mechanistic details of the interference process and the function of the CRISPR systems.
CONCLUSIONS AND FUTURE DIRECTIONS
Archaea are believed to comprise ∼20% of the biomass on Earth (273). However, only a small fraction of these organisms have been isolated in pure cultures, and most of them are extremophiles. To date, >60 archaeal viruses have been isolated from six archaeal families, including three crenarchaeal families and three euryarchaeal families. They exhibit an exceptional diversity in morphotype and have been classified into one order, nine families, and 10 genera, with more awaiting classification. Approximately 60 plasmids have been isolated from Archaea. Many of these plasmids were obtained by systematic screening for integrated genetic elements in thermophilic archaea, including members of the Sulfolobaceae and Thermococcaceae, which thrived in terrestrial hot springs and hydrothermal vents in oceans, respectively, whereas others were identified through genome sequencing of archaea, including those of the Haloarchaeaceae and Methanococcaceae. Therefore, research on archaeal plasmids and viruses has touched only the tip of the iceberg, and many more efforts are needed to reveal novel features of archaeal extrachromosomal genetic elements and ultimately to understand their roles in the evolution of the biosphere.
However, even the small sampling of the tremendous diversity of archaeal viruses and plasmids has yielded many enlightening clues about these unique genetic elements and their roles in shaping the cellular form of life. The isolation of an increasing number of archaeal viruses with unusual morphologies and the finding that their genomes comprise mostly unknown genes suggest that these genetic entities are great inventors equipped with a vast array of uniquely designed genetic parts. As mobile genetic elements, viruses and plasmids are able to transfer these parts from and into the genome of their cellular hosts for possible use by HGT, driving the coevolution of these genetic elements with their hosts (274, 275). In addition, sequence modules are exchangeable between plasmids and probably also between viruses and plasmids. The apparent fluidity of the genetic elements is consistent with the observations that viral and plasmid genes constitute 15 to 20% of the prokaryotic genomes on average (274) and that sequences derived from mobile elements and endogenous viruses may account for at least 50% of mammalian genomes and up to 90% of plant genomes (274).
Highly divergent replication proteins have profound implications for the evolution of cellular DNA replication machineries. Compared to our current understanding of the shapes and genomes of archaeal viruses, our knowledge of the molecular mechanisms of viral and plasmid replication as well as virus-host interactions in Archaea is even more rudimentary. A number of basic questions await answers. (i) How many different modes of replication do archaeal viruses and plasmids employ, and what impact do these viral replication mechanisms have on the evolution of life? (ii) How does an archaeal virus recognize and attach to its host cell, and how does it transfer its genome into the host cell? (iii) How does a virus initiate the transcription and replication of its own genome, and how does it coopt the host machineries for the above-mentioned processes? (iv) How does a virus assemble and release its progeny virions in a lytic or nonlytic way? (v) How does a virus enter a lysogenic pathway and remain as a provirus, and how is a provirus induced? (vi) How does a virus overcome host defense? (vii) How do various genetic elements, e.g., viruses, virus satellites, and plasmids, interact in the same host cell? In-depth analyses of steps during viral infection were not performed until recently. With several viruses (e.g., SIRV2, STIV, and SSV1/2, etc.) chosen as model systems, several research groups have begun to dissect the complex process of archaeal viral infection and answer these questions. Currently, representatives of rod-shaped rudiviruses (SIRV2) and spherical icosahedral viruses (STIV) are among the better-understood archaeal viruses. For example, recent studies on SIRV2 have shed significant light on the molecular details of host binding, genome replication and transcription, and release of the virus during its infection cycle. However, even with these steps, many questions remain. Given the immense diversity of archaeal viruses, it would not be surprising to find that the molecular strategies that the viruses have evolved for various steps during infection are equally, if not more, diverse. Therefore, a challenging, and certainly rewarding, task lies ahead, to unravel the mystery of these least-known genetic entities on Earth.
Supplementary Material
ACKNOWLEDGMENTS
Research in Li Huang's laboratory was supported by the National Natural Science Foundation of China (grants 30921005 and 31130003) and by the Chinese Academy of Sciences (grant Y12A024BB1), and research in Qunxin She's laboratory was supported by Danish Councils of Independent Research (grants DFF-0602-02196B and DFF-1323-00330), by the National Natural Science Foundation of China (grant 31128011), and by the Scientific and Technological Self-Innovation Foundation of Huazhong Agricultural University (grant no. 2014RC011).
Biographies

Haina Wang is a Ph.D. student in Professor Li Huang's laboratory at the Institute of Microbiology, Chinese Academy of Sciences. She received a dual B.A. in Science and English Literature in Shandong Agricultural University, China, in 2005. During her Ph.D. studies (since 2009), she has been investigating novel archaeal viruses from acidic hot springs, focusing primarily on the life cycle of these viruses.

Nan Peng received his Ph.D. in Microbiology from Huazhong Agricultural University (China) in 2009. He worked in the laboratory of Dr. Qunxin She at the Archaea Centre, Department of Biology, University of Copenhagen, for two years as a joint Ph.D. student. He is now an associate professor at the College of Life Science and Technology of Huazhong Agricultural University. His main research interests include investigation of spacer acquisition by the CRISPR-Cas system of Sulfolobus and regulation of gene expression in Archaea.

Shiraz A. Shah has an M.Sc. in microbiology and received his Ph.D. in bioinformatics in 2012. He worked as a postdoctoral researcher in Professor Roger Garrett's laboratory at the Department of Biology, University of Copenhagen, where he characterized the CRISPR adaptive immune systems in archaeal genomes using bioinformatics. He now heads the recently established biocomputing core facility at the same university, where he helps the department's wet labs with incorporating bioinformatics analyses into their research.

Li Huang received his Ph.D. in microbiology at the University of Guelph (Canada) in 1988. From 1988 to 1993, he worked as a postdoctoral fellow at Johns Hopkins University, studying E. coli histone-like protein HU and regulatory control of the initiation of bacteriophage λ DNA replication. From 1993 to 1996, he joined the faculty at Pomona College, where he started to work on chromatin proteins from hyperthermophilic archaea of the genus Sulfolobus. Since 1996, he has been leading an archaeal research group at the State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences. He was promoted to the rank of full professorship in 1998. From 2008 to 2013, he served first as Executive Deputy Director General and later as Director General of the Institute of Microbiology. His current areas of research interest include chromosomal organization and DNA replication in Archaea and archaeal extrachromosomal genetic elements.

Qunxin She received his Ph.D. in Molecular Biology at the University of Aarhus (Denmark) in 1993. From 1993 to 1995, he worked as a postdoctoral fellow at the Technical University of Denmark, studying archaeal DNA replication. He joined the laboratory of Prof. Roger Garrett at the University of Copenhagen in 1996, studying archaeal genomics and genetic elements. He started his own laboratory in 2003, working on archaeal genetics, and obtained a tenured associate professorship in 2005. He was a founding member of the Archaea Centre at the university and has been the director of the center since 2011. He also holds the position of adjunct professor at the College of Life Science and Technology at Huazhong Agricultural University, China, and heads the Archaeal Molecular Biology Laboratory. His current research interests include genetic studies of archaeal CRISPR defense mechanisms, DNA replication and damage repair, and archaeal genetic elements.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00042-14.
REFERENCES
- 1.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A 87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, Zablen LB, Blakemore R, Gupta R, Bonen L, Lewis BJ, Stahl DA, Luehrsen KR, Chen KN, Woese CR. 1980. The phylogeny of prokaryotes. Science 209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 3.Prangishvili D, Forterre P, Garrett RA. 2006. Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 4.Albers SV, Forterre P, Prangishvili D, Schleper C. 2013. The legacy of Carl Woese and Wolfram Zillig: from phylogeny to landmark discoveries. Nat Rev Microbiol 11:713–719. doi: 10.1038/nrmicro3124. [DOI] [PubMed] [Google Scholar]
- 5.Garrett RA. 2014. A backward view from 16S rRNA to archaea to the universal tree of life to progenotes: reminiscences of Carl Woese. RNA Biol 11:232–235. doi: 10.4161/rna.28228. [DOI] [PubMed] [Google Scholar]
- 6.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 7.Oren A, Bratbak G, Heldal M. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143–149. doi: 10.1007/s007920050027. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F. 2004. Phage community dynamics in hot springs. Appl Environ Microbiol 70:1633–1640. doi: 10.1128/AEM.70.3.1633-1640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol Rev 35:1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 10.Rohwer F, Thurber RV. 2009. Viruses manipulate the marine environment. Nature 459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 11.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A 74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendorfer M, Kristjansson JK. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol 16:609–628. [Google Scholar]
- 13.Martin A, Yeats S, Janekovic D, Reiter WD, Aicher W, Zillig W. 1984. SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J 3:2165–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettstetter M, Peng X, Garrett RA, Prangishvili D. 2003. AFV1, a novel virus infecting hyperthermophilic archaea of the genus Acidianus. Virology 315:68–79. doi: 10.1016/S0042-6822(03)00481-1. [DOI] [PubMed] [Google Scholar]
- 15.Haring M, Rachel R, Peng X, Garrett RA, Prangishvili D. 2005. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol 79:9904–9911. doi: 10.1128/JVI.79.15.9904-9911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mochizuki T, Yoshida T, Tanaka R, Forterre P, Sako Y, Prangishvili D. 2010. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology 402:347–354. doi: 10.1016/j.virol.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 17.Vestergaard G, Haring M, Peng X, Rachel R, Garrett RA, Prangishvili D. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83–92. doi: 10.1016/j.virol.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Vestergaard G, Aramayo R, Basta T, Haring M, Peng X, Brugger K, Chen L, Rachel R, Boisset N, Garrett RA, Prangishvili D. 2008. Structure of the Acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J Virol 82:371–381. doi: 10.1128/JVI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prangishvili D, Vestergaard G, Haring M, Aramayo R, Basta T, Rachel R, Garrett RA. 2006. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol 359:1203–1216. doi: 10.1016/j.jmb.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 20.Haring M, Vestergaard G, Rachel R, Chen L, Garrett RA, Prangishvili D. 2005. Virology: independent virus development outside a host. Nature 436:1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- 21.Mochizuki T, Krupovic M, Pehau-Arnaudet G, Sako Y, Forterre P, Prangishvili D. 2012. Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome. Proc Natl Acad Sci U S A 109:13386–13391. doi: 10.1073/pnas.1203668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochizuki T, Sako Y, Prangishvili D. 2011. Provirus induction in hyperthermophilic archaea: characterization of Aeropyrum pernix spindle-shaped virus 1 and Aeropyrum pernix ovoid virus 1. J Bacteriol 193:5412–5419. doi: 10.1128/JB.05101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bize A, Peng X, Prokofeva M, MacLellan K, Lucas S, Forterre P, Garrett RA, Bonch-Osmolovskaya EA, Prangishvili D. 2008. Viruses in acidic geothermal environments of the Kamchatka Peninsula. Res Microbiol 159:358–366. doi: 10.1016/j.resmic.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Haring M, Vestergaard G, Brugger K, Rachel R, Garrett RA, Prangishvili D. 2005. Structure and genome organization of AFV2, a novel archaeal lipothrixvirus with unusual terminal and core structures. J Bacteriol 187:3855–3858. doi: 10.1128/JB.187.11.3855-3858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haring M, Peng X, Brugger K, Rachel R, Stetter KO, Garrett RA, Prangishvili D. 2004. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology 323:233–242. doi: 10.1016/j.virol.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Prangishvili D, Arnold HP, Gotz D, Ziese U, Holz I, Kristjansson JK, Zillig W. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the Sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vestergaard G, Shah SA, Bize A, Reitberger W, Reuter M, Phan H, Briegel A, Rachel R, Garrett RA, Prangishvili D. 2008. Stygiolobus rod-shaped virus and the interplay of crenarchaeal rudiviruses with the CRISPR antiviral system. J Bacteriol 190:6837–6845. doi: 10.1128/JB.00795-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackermann HW, Prangishvili D. 2012. Prokaryote viruses studied by electron microscopy. Arch Virol 157:1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- 29.Simon RD. 1978. Halobacterium strain-5 contains a plasmid which is correlated with presence of gas vacuoles. Nature 273:314–317. doi: 10.1038/273314a0. [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer F, Weidinger G, Goebel W. 1981. Characterization of plasmids in halobacteria. J Bacteriol 145:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomm M, Altenbuchner J, Stetter KO. 1983. Evidence for a plasmid in a methanogenic bacterium. J Bacteriol 153:1060–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meile L, Kiener A, Leisinger T. 1983. A plasmid in the archaebacterium Methanobacterium thermoautotrophicum. Mol Gen Genet 191:480–484. doi: 10.1007/BF00425766. [DOI] [PubMed] [Google Scholar]
- 33.Nolling J, de Vos WM. 1992. Characterization of the archaeal, plasmid-encoded type II restriction-modification system MthTI from Methanobacterium thermoformicicum THF: homology to the bacterial NgoPII system from Neisseria gonorrhoeae. J Bacteriol 174:5719–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolling J, de Vos WM. 1992. Identification of the CTAG-recognizing restriction-modification systems MthZI and MthFI from Methanobacterium thermoformicicum and characterization of the plasmid-encoded mthZIM gene. Nucleic Acids Res 20:5047–5052. doi: 10.1093/nar/20.19.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zillig W, Yeats S, Holz I, Bock A, Gropp F, Rettenberger M, Lutz S. 1985. Plasmid-related anaerobic autotrophy of the novel archaebacterium Sulfolobus ambivalens. Nature 313:789–791. doi: 10.1038/313789a0. [DOI] [PubMed] [Google Scholar]
- 36.Erauso G, Charbonnier F, Barbeyron T, Forterre P, Prieur D. 1992. Preliminary characterization of a hyperthermophilic archaebacterium with a plasmid isolated from a North Fiji Basin hydrothermal vent. C R Acad Sci III 314:387–393. [Google Scholar]
- 37.Charlebois RL, Lam WL, Cline SW, Doolittle WF. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci U S A 84:8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackett NR, DasSarma S. 1989. Characterization of the small endogenous plasmid of Halobacterium strain SB3 and its use in transformation of Halobacterium halobium. Can J Microbiol 35:86–91. doi: 10.1139/m89-013. [DOI] [PubMed] [Google Scholar]
- 39.Holmes ML, Dyall-Smith ML. 1990. A plasmid vector with a selectable marker for halophilic archaebacteria. J Bacteriol 172:756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z, Liu H, Liu J, Liu X, Xiang H. 2012. Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea. BMC Genomics 13:478. doi: 10.1186/1471-2164-13-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prangishvili D, Albers SV, Holz I, Arnold HP, Stedman K, Klein T, Singh H, Hiort J, Schweier A, Kristjansson JK, Zillig W. 1998. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid 40:190–202. doi: 10.1006/plas.1998.1363. [DOI] [PubMed] [Google Scholar]
- 42.Prieur D, Erauso G, Flament D, Gaillard M, Geslin C, Gonnet M, Le Romancer M, Lucas S, Forterrre P. 2006. Deep-sea Thermococcales and their genetic elements: plasmids and viruses. Methods Microbiol 35:253–278. doi: 10.1016/S0580-9517(08)70014-X. [DOI] [Google Scholar]
- 43.Ortmann AC, Wiedenheft B, Douglas T, Young M. 2006. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat Rev Microbiol 4:520–528. doi: 10.1038/nrmicro1444. [DOI] [PubMed] [Google Scholar]
- 44.Prangishvili D. 2013. The wonderful world of archaeal viruses. Annu Rev Microbiol 67:565–585. doi: 10.1146/annurev-micro-092412-155633. [DOI] [PubMed] [Google Scholar]
- 45.Peng X, Garrett RA, She Q. 2012. Archaeal viruses—novel, diverse and enigmatic. Sci China Life Sci 55:422–433. doi: 10.1007/s11427-012-4325-8. [DOI] [PubMed] [Google Scholar]
- 46.Pietila MK, Demina TA, Atanasova NS, Oksanen HM, Bamford DH. 2014. Archaeal viruses and bacteriophages: comparisons and contrasts. Trends Microbiol 22:334–344. doi: 10.1016/j.tim.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. 2011. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev 75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krupovic M, White MF, Forterre P, Prangishvili D. 2012. Postcards from the edge: structural genomics of archaeal viruses. Adv Virus Res 82:33–62. doi: 10.1016/B978-0-12-394621-8.00012-1. [DOI] [PubMed] [Google Scholar]
- 49.Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ. 2009. Structural and functional studies of archaeal viruses. J Biol Chem 284:12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prangishvili D, Quax TE. 2011. Exceptional virion release mechanism: one more surprise from archaeal viruses. Curr Opin Microbiol 14:315–320. doi: 10.1016/j.mib.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Snyder JC, Young MJ. 2013. Lytic viruses infecting organisms from the three domains of life. Biochem Soc Trans 41:309–313. doi: 10.1042/BST20120326. [DOI] [PubMed] [Google Scholar]
- 52.Prangishvili D, Koonin EV, Krupovic M. 2013. Genomics and biology of rudiviruses, a model for the study of virus-host interactions in Archaea. Biochem Soc Trans 41:443–450. doi: 10.1042/BST20120313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Contursi P, Fusco S, Cannio R, She Q. 2014. Molecular biology of fuselloviruses and their satellites. Extremophiles 18:473–489. doi: 10.1007/s00792-014-0634-0. [DOI] [PubMed] [Google Scholar]
- 54.Fu CY, Johnson JE. 2012. Structure and cell biology of archaeal virus STIV. Curr Opin Virol 2:122–127. doi: 10.1016/j.coviro.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fulton J, Bothner B, Lawrence M, Johnson JE, Douglas T, Young M. 2009. Genetics, biochemistry and structure of the archaeal virus STIV. Biochem Soc Trans 37:114–117. doi: 10.1042/BST0370114. [DOI] [PubMed] [Google Scholar]
- 56.Roine E, Bamford DH. 2012. Lipids of archaeal viruses. Archaea 2012:384919. doi: 10.1155/2012/384919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soler N, Gaudin M, Marguet E, Forterre P. 2011. Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem Soc Trans 39:36–44. doi: 10.1042/BST0390036. [DOI] [PubMed] [Google Scholar]
- 58.Lipps G. 2006. Plasmids and viruses of the thermoacidophilic crenarchaeote Sulfolobus. Extremophiles 10:17–28. doi: 10.1007/s00792-005-0492-x. [DOI] [PubMed] [Google Scholar]
- 59.Lipps G. 2008. Archaea plasmids, p 25–50. In Lipps G. (ed), Plasmids: current research and future trends. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 60.Garrett RA, Redder P, Greve B, Brügger K, Chen L, She Q. 2004. Archaeal plasmids, p 377–392. In Funnell BE, Phillips GJ (ed), Plasmid biology. ASM Press, Washington, DC. [Google Scholar]
- 61.Garrett RA, Prangishvili D, Shah SA, Reuter M, Stetter KO, Peng X. 2010. Metagenomic analyses of novel viruses and plasmids from a cultured environmental sample of hyperthermophilic neutrophiles. Environ Microbiol 12:2918–2930. doi: 10.1111/j.1462-2920.2010.02266.x. [DOI] [PubMed] [Google Scholar]
- 62.Rice G, Stedman K, Snyder J, Wiedenheft B, Willits D, Brumfield S, McDermott T, Young MJ. 2001. Viruses from extreme thermal environments. Proc Natl Acad Sci U S A 98:13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. 2014. Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol 88:2354–2358. doi: 10.1128/JVI.02941-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guixa-Boixareu N, Calderon-Paz JI, Heldal M, Bratbak G, Pedros-Alio C. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol 11:215–227. doi: 10.3354/ame011215. [DOI] [Google Scholar]
- 65.Berkner S, Grogan D, Albers SV, Lipps G. 2007. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res 35:e88. doi: 10.1093/nar/gkm449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. 2012. Global network of specific virus-host interactions in hypersaline environments. Environ Microbiol 14:426–440. doi: 10.1111/j.1462-2920.2011.02603.x. [DOI] [PubMed] [Google Scholar]
- 67.Dyall-Smith M, Tang SL, Bath C. 2003. Haloarchaeal viruses: how diverse are they? Res Microbiol 154:309–313. doi: 10.1016/S0923-2508(03)00076-7. [DOI] [PubMed] [Google Scholar]
- 68.Bamford DH. 2003. Do viruses form lineages across different domains of life? Res Microbiol 154:231–236. doi: 10.1016/S0923-2508(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 69.Prangishvili D, Krupovic M. 2012. A new proposed taxon for double-stranded DNA viruses, the order “Ligamenvirales.” Arch Virol 157:791–795. doi: 10.1007/s00705-012-1229-7. [DOI] [PubMed] [Google Scholar]
- 70.Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr Opin Struct Biol 15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 71.Krupovic M, Bamford DH. 2011. Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly. Curr Opin Virol 1:118–124. doi: 10.1016/j.coviro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 72.Bolduc B, Shaughnessy DP, Wolf YI, Koonin EV, Roberto FF, Young M. 2012. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol 86:5562–5573. doi: 10.1128/JVI.07196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reference deleted.
- 74.Reference deleted.
- 75.Haywood AM. 1994. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J Virol 68:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heller KJ. 1992. Molecular interaction between bacteriophage and the Gram-negative cell envelope. Arch Microbiol 158:235–248. doi: 10.1007/BF00245239. [DOI] [PubMed] [Google Scholar]
- 77.Poranen MM, Daugelavicius R, Bamford DH. 2002. Common principles in viral entry. Annu Rev Microbiol 56:521–538. doi: 10.1146/annurev.micro.56.012302.160643. [DOI] [PubMed] [Google Scholar]
- 78.Arnold HP, Ziese U, Zillig W. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409–416. doi: 10.1006/viro.2000.0375. [DOI] [PubMed] [Google Scholar]
- 79.Arnold HP, Zillig W, Ziese U, Holz I, Crosby M, Utterback T, Weidmann JF, Kristjanson JK, Klenk HP, Nelson KE, Fraser CM. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267:252–266. doi: 10.1006/viro.1999.0105. [DOI] [PubMed] [Google Scholar]
- 80.Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci U S A 101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quemin ERJ, Lucas S, Daum B, Quax TEF, Kuhlbrandt W, Forterre P, Albers SV, Prangishvili D, Krupovic M. 2013. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol 87:13379–13385. doi: 10.1128/JVI.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kukkaro P, Bamford DH. 2009. Virus-host interactions in environments with a wide range of ionic strengths. Environ Microbiol Rep 1:71–77. doi: 10.1111/j.1758-2229.2008.00007.x. [DOI] [PubMed] [Google Scholar]
- 83.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. 2011. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr Issues Mol Biol 13:51–76. [PubMed] [Google Scholar]
- 84.Erdmann S, Scheele U, Garrett RA. 2011. AAA ATPase p529 of Acidianus two-tailed virus ATV and host receptor recognition. Virology 421:61–66. doi: 10.1016/j.virol.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 85.Hanhijarvi KJ, Ziedaite G, Pietila MK, Haeggstrom E, Bamford DH. 2013. DNA ejection from an archaeal virus—a single-molecule approach. Biophys J 104:2264–2272. doi: 10.1016/j.bpj.2013.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bath C, Cukalac T, Porter K, Dyall-Smith ML. 2006. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology 350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 87.Peng X, Basta T, Haring M, Garrett RA, Prangishvili D. 2007. Genome of the Acidianus bottle-shaped virus and insights into the replication and packaging mechanisms. Virology 364:237–243. doi: 10.1016/j.virol.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 88.Oke M, Kerou M, Liu H, Peng X, Garrett RA, Prangishvili D, Naismith JH, White MF. 2011. A dimeric Rep protein initiates replication of a linear archaeal virus genome: implications for the Rep mechanism and viral replication. J Virol 85:925–931. doi: 10.1128/JVI.01467-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng X, Blum H, She Q, Mallok S, Brugger K, Garrett RA, Zillig W, Prangishvili D. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226–234. doi: 10.1006/viro.2001.1190. [DOI] [PubMed] [Google Scholar]
- 90.Birkenbihl RP, Neef K, Prangishvili D, Kemper B. 2001. Holliday junction resolving enzymes of archaeal viruses SIRV1 and SIRV2. J Mol Biol 309:1067–1076. doi: 10.1006/jmbi.2001.4761. [DOI] [PubMed] [Google Scholar]
- 91.Blum H, Zillig W, Mallok S, Domdey H, Prangishvili D. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281:6–9. doi: 10.1006/viro.2000.0776. [DOI] [PubMed] [Google Scholar]
- 92.Pina M, Basta T, Quax TE, Joubert A, Baconnais S, Cortez D, Lambert S, Le Cam E, Bell SD, Forterre P, Prangishvili D. 2014. Unique genome replication mechanism of the archaeal virus AFV1. Mol Microbiol 92:1313–1325. doi: 10.1111/mmi.12630. [DOI] [PubMed] [Google Scholar]
- 93.Salas M. 1991. Protein-priming of DNA replication. Annu Rev Biochem 60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 94.Bamford DH, Ravantti JJ, Ronnholm G, Laurinavicius S, Kukkaro P, Dyall-Smith M, Somerharju P, Kalkkinen N, Bamford JK. 2005. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J Virol 79:9097–9107. doi: 10.1128/JVI.79.14.9097-9107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xiang X, Chen L, Huang X, Luo Y, She Q, Huang L. 2005. Sulfolobus tengchongensis spindle-shaped virus STSV1: virus-host interactions and genomic features. J Virol 79:8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roine E, Kukkaro P, Paulin L, Laurinavicius S, Domanska A, Somerharju P, Bamford DH. 2010. New, closely related haloarchaeal viral elements with different nucleic acid types. J Virol 84:3682–3689. doi: 10.1128/JVI.01879-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koonin EV. 1992. Archaebacterial virus SSV1 encodes a putative DnaA-like protein. Nucleic Acids Res 20:1143. doi: 10.1093/nar/20.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. 2012. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol 14:503–516. doi: 10.1111/j.1462-2920.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2012. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom. [Google Scholar]
- 100.Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr Opin Microbiol 14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Langer D, Hain J, Thuriaux P, Zillig W. 1995. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci U S A 92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bell SD, Jackson SP. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol 6:222–228. doi: 10.1016/S0966-842X(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 103.Reiter WD, Palm P, Zillig W. 1988. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res 16:1–19. doi: 10.1093/nar/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aravind L, Koonin EV. 1999. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res 27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. 2003. The Lrp family of transcriptional regulators. Mol Microbiol 48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 106.Kessler A, Brinkman AB, van der Oost J, Prangishvili D. 2004. Transcription of the rod-shaped viruses SIRV1 and SIRV2 of the hyperthermophilic archaeon Sulfolobus. J Bacteriol 186:7745–7753. doi: 10.1128/JB.186.22.7745-7753.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quax TE, Voet M, Sismeiro O, Dillies MA, Jagla B, Coppee JY, Sezonov G, Forterre P, van der Oost J, Lavigne R, Prangishvili D. 2013. Massive activation of archaeal defense genes during viral infection. J Virol 87:8419–8428. doi: 10.1128/JVI.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ren Y, She Q, Huang L. 2013. Transcriptomic analysis of the SSV2 infection of Sulfolobus solfataricus with and without the integrative plasmid pSSVi. Virology 441:126–134. doi: 10.1016/j.virol.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 109.Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van de Oost J, Young MJ. 2008. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol 82:4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Frols S, Gordon PM, Panlilio MA, Schleper C, Sensen CW. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48–59. doi: 10.1016/j.virol.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 111.Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, Forterre P, Tenaillon O, Bernander R, Prangishvili D. 2009. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci U S A 106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guilliere F, Peixeiro N, Kessler A, Raynal B, Desnoues N, Keller J, Delepierre M, Prangishvili D, Sezonov G, Guijarro JI. 2009. Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1. J Biol Chem 284:22222–22237. doi: 10.1074/jbc.M109.029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kessler A, Sezonov G, Guijarro JI, Desnoues N, Rose T, Delepierre M, Bell SD, Prangishvili D. 2006. A novel archaeal regulatory protein, Sta1, activates transcription from viral promoters. Nucleic Acids Res 34:4837–4845. doi: 10.1093/nar/gkl502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci U S A 89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu DX, Huang L. 2002. Induction of the Sulfolobus shibatae virus SSV1 DNA replication by mitomycin C. Chin Sci Bull 47:923–927. doi: 10.1360/02tb9207. [DOI] [Google Scholar]
- 116.Fusco S, She Q, Bartolucci S, Contursi P. 2013. T(lys), a newly identified Sulfolobus spindle-shaped virus 1 transcript expressed in the lysogenic state, encodes a DNA-binding protein interacting at the promoters of the early genes. J Virol 87:5926–5936. doi: 10.1128/JVI.00458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. 2011. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol 85:6287–6292. doi: 10.1128/JVI.00379-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang Z, Guo L, Huang L. 2012. Archaeal chromatin proteins. Sci China Life Sci 55:377–385. doi: 10.1007/s11427-012-4322-y. [DOI] [PubMed] [Google Scholar]
- 119.Arnold HP, She Q, Phan H, Stedman K, Prangishvili D, Holz I, Kristjansson JK, Garrett R, Zillig W. 1999. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol Microbiol 34:217–226. doi: 10.1046/j.1365-2958.1999.01573.x. [DOI] [PubMed] [Google Scholar]
- 120.Contursi P, Jensen S, Aucelli T, Rossi M, Bartolucci S, She Q. 2006. Characterization of the Sulfolobus host-SSV2 virus interaction. Extremophiles 10:615–627. doi: 10.1007/s00792-006-0017-2. [DOI] [PubMed] [Google Scholar]
- 121.Contursi P, Cannio R, Prato S, She Q, Rossi M, Bartolucci S. 2007. Transcriptional analysis of the genetic element pSSVx: differential and temporal regulation of gene expression reveals correlation between transcription and replication. J Bacteriol 189:6339–6350. doi: 10.1128/JB.00638-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Contursi P, Cannio R, She Q. 2010. Transcription termination in the plasmid/virus hybrid pSSVx from Sulfolobus islandicus. Extremophiles 14:453–463. doi: 10.1007/s00792-010-0325-4. [DOI] [PubMed] [Google Scholar]
- 123.Goulet A, Blangy S, Redder P, Prangishvili D, Felisberto-Rodrigues C, Forterre P, Campanacci V, Cambillau C. 2009. Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage. Proc Natl Acad Sci U S A 106:21155–21160. doi: 10.1073/pnas.0909893106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu CY, Wang K, Gan L, Lanman J, Khayat R, Young MJ, Jensen GJ, Doerschuk PC, Johnson JE. 2010. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure 18:1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. 2010. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology 404:1–4. doi: 10.1016/j.virol.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 126.Snyder JC, Brumfield SK, Kerchner KM, Quax TE, Prangishvili D, Young MJ. 2013. Insights into a viral lytic pathway from an archaeal virus-host system. J Virol 87:2186–2192. doi: 10.1128/JVI.02956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Snyder JC, Samson RY, Brumfield SK, Bell SD, Young MJ. 2013. Functional interplay between a virus and the ESCRT machinery in Archaea. Proc Natl Acad Sci U S A 110:10783–10787. doi: 10.1073/pnas.1301605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pfister P, Wasserfallen A, Stettler R, Leisinger T. 1998. Molecular analysis of Methanobacterium phage psiM2. Mol Microbiol 30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- 129.Pagaling E, Haigh RD, Grant WD, Cowan DA, Jones BE, Ma Y, Ventosa A, Heaphy S. 2007. Sequence analysis of an archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics 8:410. doi: 10.1186/1471-2164-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.She Q, Brugger K, Chen L. 2002. Archaeal integrative genetic elements and their impact on genome evolution. Res Microbiol 153:325–332. doi: 10.1016/S0923-2508(02)01331-1. [DOI] [PubMed] [Google Scholar]
- 131.She Q, Shen B, Chen L. 2004. Archaeal integrases and mechanisms of gene capture. Biochem Soc Trans 32:222–226. doi: 10.1042/BST0320222. [DOI] [PubMed] [Google Scholar]
- 132.She Q, Peng X, Zillig W, Garrett RA. 2001. Genome evolution—gene capture in archaeal chromosomes. Nature 409:478–478. doi: 10.1038/35054138. [DOI] [PubMed] [Google Scholar]
- 133.Reiter WD, Palm P, Yeats S. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res 17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Williams KP. 2002. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res 30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clore AJ, Stedman KM. 2007. The SSV1 viral integrase is not essential. Virology 361:103–111. doi: 10.1016/j.virol.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 136.Muskhelishvili G, Palm P, Zillig W. 1993. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol Gen Genet 237:334–342. [DOI] [PubMed] [Google Scholar]
- 137.Muskhelishvili G. 1993. The archaeal SSV integrase promotes intermolecular excisive recombination in vitro. Syst Appl Microbiol 16:605–608. [Google Scholar]
- 138.Letzelter C, Duguet M, Serre MC. 2004. Mutational analysis of the archaeal tyrosine recombinase SSV1 integrase suggests a mechanism of DNA cleavage in trans. J Biol Chem 279:28936–28944. doi: 10.1074/jbc.M403971200. [DOI] [PubMed] [Google Scholar]
- 139.Serre MC, Letzelter C, Garel JR, Duguet M. 2002. Cleavage properties of an archaeal site-specific recombinase, the SSV1 integrase. J Biol Chem 277:16758–16767. doi: 10.1074/jbc.M200707200. [DOI] [PubMed] [Google Scholar]
- 140.Conway AB, Chen Y, Rice PA. 2003. Structural plasticity of the Flp-Holliday junction complex. J Mol Biol 326:425–434. doi: 10.1016/S0022-2836(02)01370-0. [DOI] [PubMed] [Google Scholar]
- 141.Eilers BJ, Young MJ, Lawrence CM. 2012. The structure of an archaeal viral integrase reveals an evolutionarily conserved catalytic core yet supports a mechanism of DNA cleavage in trans. J Virol 86:8309–8313. doi: 10.1128/JVI.00547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhan Z, Ouyang S, Liang W, Zhang Z, Liu ZJ, Huang L. 2012. Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase. Acta Crystallogr D Biol Crystallogr 68:659–670. doi: 10.1107/S0907444912007202. [DOI] [PubMed] [Google Scholar]
- 143.Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr. 2003. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 144.Brosius J. 2003. The contribution of RNAs and retroposition to evolutionary novelties. Genetica 118:99–116. doi: 10.1023/A:1024141306559. [DOI] [PubMed] [Google Scholar]
- 145.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 146.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 147.Stedman KM, She Q, Phan H, Arnold HP, Holz I, Garrett RA, Zillig W. 2003. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res Microbiol 154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 148.Cortez D, Forterre P, Gribaldo S. 2009. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol 10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vestergaard G, Garrett RA, Shah SA. 2014. CRISPR adaptive immune systems of Archaea. RNA Biol 11:156–167. doi: 10.4161/rna.27990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kletzin A, Lieke A, Urich T, Charlebois RL, Sensen CW. 1999. Molecular analysis of pDL10 from Acidianus ambivalens reveals a family of related plasmids from extremely thermophilic and acidophilic archaea. Genetics 152:1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Peng X, Holz I, Zillig W, Garrett RA, She Q. 2000. Evolution of the family of pRN plasmids and their integrase-mediated insertion into the chromosome of the crenarchaeon Sulfolobus solfataricus. J Mol Biol 303:449–454. doi: 10.1006/jmbi.2000.4160. [DOI] [PubMed] [Google Scholar]
- 152.Greve B, Jensen S, Brugger K, Zillig W, Garrett RA. 2004. Genomic comparison of archaeal conjugative plasmids from Sulfolobus. Archaea 1:231–239. doi: 10.1155/2004/151926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Erauso G, Stedman KM, van de Werken HJ, Zillig W, van der Oost J. 2006. Two novel conjugative plasmids from a single strain of Sulfolobus. Microbiology 152:1951–1968. doi: 10.1099/mic.0.28861-0. [DOI] [PubMed] [Google Scholar]
- 154.Lipps G, Ibanez P, Stroessenreuther T, Hekimian K, Krauss G. 2001. The protein ORF80 from the acidophilic and thermophilic archaeon Sulfolobus islandicus binds highly site-specifically to double-stranded DNA and represents a novel type of basic leucine zipper protein. Nucleic Acids Res 29:4973–4982. doi: 10.1093/nar/29.24.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Contursi P, Farina B, Pirone L, Fusco S, Russo L, Bartolucci S, Fattorusso R, Pedone E. 2014. Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx: a novel peculiar member of the winged helix-turn-helix transcription factor family. Nucleic Acids Res 42:5993–6011. doi: 10.1093/nar/gku215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Peng N, Xia Q, Chen Z, Liang YX, She Q. 2009. An upstream activation element exerting differential transcriptional activation on an archaeal promoter. Mol Microbiol 74:928–939. doi: 10.1111/j.1365-2958.2009.06908.x. [DOI] [PubMed] [Google Scholar]
- 157.Gomis-Ruth FX, Sola M, Acebo P, Parraga A, Guasch A, Eritja R, Gonzalez A, Espinosa M, del Solar G, Coll M. 1998. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J 17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hernandez-Arriaga AM, Rubio-Lepe TS, Espinosa M, del Solar G. 2009. Repressor CopG prevents access of RNA polymerase to promoter and actively dissociates open complexes. Nucleic Acids Res 37:4799–4811. doi: 10.1093/nar/gkp503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lipps G. 2009. Molecular biology of the pRN1 plasmid from Sulfolobus islandicus. Biochem Soc Trans 37:42–45. doi: 10.1042/BST0370042. [DOI] [PubMed] [Google Scholar]
- 160.Purschke WG, Schafer G. 2001. Independent replication of the plasmids pRN1 and pRN2 in the archaeon Sulfolobus islandicus. FEMS Microbiol Lett 200:97–102. doi: 10.1111/j.1574-6968.2001.tb10699.x. [DOI] [PubMed] [Google Scholar]
- 161.Greve B, Jensen S, Phan H, Brugger K, Zillig W, She Q, Garrett RA. 2005. Novel RepA-MCM proteins encoded in plasmids pTAU4, pORA1 and pTIK4 from Sulfolobus neozealandicus. Archaea 1:319–325. doi: 10.1155/2005/159218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bochman ML, Schwacha A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev 73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo X, Huang L. 2010. A superfamily 3 DNA helicase encoded by plasmid pSSVi from the hyperthermophilic archaeon Sulfolobus solfataricus unwinds DNA as a higher-order oligomer and interacts with host primase. J Bacteriol 192:1853–1864. doi: 10.1128/JB.01300-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Peng X. 2008. Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and the implications for plasmid survival. Microbiology 154:383–391. doi: 10.1099/mic.0.2007/012963-0. [DOI] [PubMed] [Google Scholar]
- 165.Gill S, Krupovic M, Desnoues N, Beguin P, Sezonov G, Forterre P. 2014. A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2. Nucleic Acids Res 42:3707–3719. doi: 10.1093/nar/gkt1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schleper C, Holz I, Janekovic D, Murphy J, Zillig W. 1995. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J Bacteriol 177:4417–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.She Q, Phan H, Garrett RA, Albers SV, Stedman KM, Zillig W. 1998. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles 2:417–425. doi: 10.1007/s007920050087. [DOI] [PubMed] [Google Scholar]
- 169.Zillig W, Arnold HP, Holz I, Prangishvili D, Schweier A, Stedman K, She Q, Phan H, Garrett R, Kristjansson JK. 1998. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2:131–140. doi: 10.1007/s007920050052. [DOI] [PubMed] [Google Scholar]
- 170.Mierzejewska J, Jagura-Burdzy G. 2012. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67:1–14. doi: 10.1016/j.plasmid.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 171.Stedman KM, She Q, Phan H, Holz I, Singh H, Prangishvili D, Garrett R, Zillig W. 2000. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J Bacteriol 182:7014–7020. doi: 10.1128/JB.182.24.7014-7020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Deng L, Zhu H, Chen Z, Liang YX, She Q. 2009. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13:735–746. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 173.She Q, Zhang C, Deng L, Peng N, Chen Z, Liang YX. 2009. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem Soc Trans 37:92–96. doi: 10.1042/BST0370092. [DOI] [PubMed] [Google Scholar]
- 174.Peng N, Deng L, Mei Y, Jiang D, Hu Y, Awayez M, Liang Y, She Q. 2012. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl Environ Microbiol 78:5630–5637. doi: 10.1128/AEM.00855-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guo L, Brugger K, Liu C, Shah SA, Zheng H, Zhu Y, Wang S, Lillestol RK, Chen L, Frank J, Prangishvili D, Paulin L, She Q, Huang L, Garrett RA. 2011. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J Bacteriol 193:1672–1680. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pfeiffer F, Broicher A, Gillich T, Klee K, Mejia J, Rampp M, Oesterhelt D. 2008. Genome information management and integrated data analysis with HaloLex. Arch Microbiol 190:281–299. doi: 10.1007/s00203-008-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Soppa J, Baumann A, Brenneis M, Dambeck M, Hering O, Lange C. 2008. Genomics and functional genomics with haloarchaea. Arch Microbiol 190:197–215. doi: 10.1007/s00203-008-0376-4. [DOI] [PubMed] [Google Scholar]
- 178.Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. 2007. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet 3:e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, Allers T. 2013. Accelerated growth in the absence of DNA replication origins. Nature 503:544–547. doi: 10.1038/nature12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ng WV, Ciufo SA, Smith TM, Bumgarner RE, Baskin D, Faust J, Hall B, Loretz C, Seto J, Slagel J, Hood L, DasSarma S. 1998. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res 8:1131–1141. [DOI] [PubMed] [Google Scholar]
- 181.Pfeifer F, Ghahraman P. 1993. Plasmid pHH1 of Halobacterium salinarium: characterization of the replicon region, the gas vesicle gene cluster and insertion elements. Mol Gen Genet 238:193–200. [DOI] [PubMed] [Google Scholar]
- 182.Ng WL, DasSarma S. 1993. Minimal replication origin of the 200-kilobase Halobacterium plasmid pNRC100. J Bacteriol 175:4584–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, Hung P, Date SV, Marcotte E, Hood L, Ng WV. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res 14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Soler N, Marguet E, Cortez D, Desnoues N, Keller J, van Tilbeurgh H, Sezonov G, Forterre P. 2010. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res 38:5088–5104. doi: 10.1093/nar/gkq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Krupovic M, Gonnet M, Ben Hania W, Forterre P, Erauso G. 2013. Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PLoS One 8:e49044. doi: 10.1371/journal.pone.0049044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Erauso G, Marsin S, Benbouzid-Rollet N, Baucher MF, Barbeyron T, Zivanovic Y, Prieur D, Forterre P. 1996. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication in a hyperthermophile. J Bacteriol 178:3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Marsin S, Forterre P. 1998. A rolling circle replication initiator protein with a nucleotidyl-transferase activity encoded by the plasmid pGT5 from the hyperthermophilic archaeon Pyrococcus abyssi. Mol Microbiol 27:1183–1192. doi: 10.1046/j.1365-2958.1998.00759.x. [DOI] [PubMed] [Google Scholar]
- 188.Soler N, Justome A, Quevillon-Cheruel S, Lorieux F, Le Cam E, Marguet E, Forterre P. 2007. The rolling-circle plasmid pTN1 from the hyperthermophilic archaeon Thermococcus nautilus. Mol Microbiol 66:357–370. doi: 10.1111/j.1365-2958.2007.05912.x. [DOI] [PubMed] [Google Scholar]
- 189.Ward DE, Revet IM, Nandakumar R, Tuttle JH, de Vos WM, van der Oost J, DiRuggiero J. 2002. Characterization of plasmid pRT1 from Pyrococcus sp. strain JT1. J Bacteriol 184:2561–2566. doi: 10.1128/JB.184.9.2561-2566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gonnet M, Erauso G, Prieur D, Le Romancer M. 2011. pAMT11, a novel plasmid isolated from a Thermococcus sp. strain closely related to the virus-like integrated element TKV1 of the Thermococcus kodakaraensis genome. Res Microbiol 162:132–143. doi: 10.1016/j.resmic.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 191.Gorlas A, Krupovic M, Forterre P, Geslin C. 2013. Living side by side with a virus: characterization of two novel plasmids from Thermococcus prieurii, a host for the spindle-shaped virus TPV1. Appl Environ Microbiol 79:3822–3828. doi: 10.1128/AEM.00525-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Beguin P, Baron B, Gill S, Charpin N, Forterre P. 2014. The SF1 helicase encoded by the archaeal plasmid pTN2 of Thermococcus nautili. Extremophiles 18:779–787. doi: 10.1007/s00792-014-0658-5. [DOI] [PubMed] [Google Scholar]
- 193.Liu Y, Whitman WB. 2008. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- 194.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A 94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bokranz M, Klein A, Meile L. 1990. Complete nucleotide sequence of plasmid pME2001 of Methanobacterium thermoautotrophicum (Marburg). Nucleic Acids Res 18:363. doi: 10.1093/nar/18.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stettler R, Pfister P, Leisinger T. 1995. Characterization of a plasmid carried by Methanobacterium thermoautotrophicum Zh3, a methanogen closely related to Methanobacterium thermoautotrophicum Marburg. Syst Appl Microbiol 17:484–491. doi: 10.1016/S0723-2020(11)80066-4. [DOI] [Google Scholar]
- 197.Luo Y, Leisinger T, Wasserfallen A. 2001. Comparative sequence analysis of plasmids pME2001 and pME2200 of Methanothermobacter marburgensis strains Marburg and ZH3. Plasmid 45:18–30. doi: 10.1006/plas.2000.1493. [DOI] [PubMed] [Google Scholar]
- 198.Nolling J, van Eeden FJ, Eggen RI, de Vos WM. 1992. Modular organization of related archaeal plasmids encoding different restriction-modification systems in Methanobacterium thermoformicicum. Nucleic Acids Res 20:6501–6507. doi: 10.1093/nar/20.24.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tumbula DL, Bowen TL, Whitman WB. 1997. Characterization of pURB500 from the archaeon Methanococcus maripaludis and construction of a shuttle vector. J Bacteriol 179:2976–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NS, Venter JC. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 201.Saunders E, Tindall BJ, Fahnrich R, Lapidus A, Copeland A, Del Rio TG, Lucas S, Chen F, Tice H, Cheng JF, Han C, Detter JC, Bruce D, Goodwin L, Chain P, Pitluck S, Pati A, Ivanova N, Mavromatis K, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Brettin T, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk HP, Kyrpides NC. 2010. Complete genome sequence of Haloterrigena turkmenica type strain (4k). Stand Genomic Sci 2:107–116. doi: 10.4056/sigs.681272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jeanthon C, L'Haridon S, Reysenbach AL, Corre E, Vernet M, Messner P, Sleytr UB, Prieur D. 1999. Methanococcus vulcanius sp. nov., a novel hyperthermophilic methanogen isolated from East Pacific Rise, and identification of Methanococcus sp. DSM 4213T as Methanococcus fervens sp. nov. Int J Syst Bacteriol 49(Part 2):583–589. doi: 10.1099/00207713-49-2-583. [DOI] [PubMed] [Google Scholar]
- 203.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62:434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Khan SA. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 205.Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marsin S, Forterre P. 1999. The active site of the rolling circle replication protein Rep75 is involved in site-specific nuclease, ligase and nucleotidyl transferase activities. Mol Microbiol 33:537–545. doi: 10.1046/j.1365-2958.1999.01498.x. [DOI] [PubMed] [Google Scholar]
- 207.Marsin S, Marguet E, Forterre P. 2000. Topoisomerase activity of the hyperthermophilic replication initiator protein Rep75. Nucleic Acids Res 28:2251–2255. doi: 10.1093/nar/28.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lee CA, Babic A, Grossman AD. 2010. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 75:268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kagramanova VK, Derckacheva NI, Mankin AS. 1988. The complete nucleotide sequence of the arcaebacterial plasmid pHSB from Halobacterium, strain SB3. Nucleic Acids Res 16:4158. doi: 10.1093/nar/16.9.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hall MJ, Hackett NR. 1989. DNA sequence of a small plasmid from Halobacterium strain GN101. Nucleic Acids Res 17:10501. doi: 10.1093/nar/17.24.10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Krebs MP, Rajbhandary UL, Khorana HG. 1990. Nucleotide sequence of ISH11, a New Halobacterium halobium insertion element isolated from the plasmid Pgrb1. Nucleic Acids Res 18:6699. doi: 10.1093/nar/18.22.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhou MX, Xiang H, Sun CM, Li Y, Liu JF, Tan HR. 2004. Complete sequence and molecular characterization of pNB101, a rolling-circle replicating plasmid from the haloalkaliphilic archaeon Natronobacterium sp strain AS7091. Extremophiles 8:91–98. doi: 10.1007/s00792-003-0366-z. [DOI] [PubMed] [Google Scholar]
- 213.Zhou L, Zhou MX, Sun CM, Han J, Lu QH, Zhou J, Xiang H. 2008. Precise determination, cross-recognition, and functional analysis of the double-strand origins of the rolling-circle replication plasmids in haloarchaea. J Bacteriol 190:5710–5719. doi: 10.1128/JB.00596-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sun CM, Zhou MX, Li Y, Xiang H. 2006. Molecular characterization of the minimal replicon and the unidirectional theta replication of pSCM201 in extremely halophilic archaea. J Bacteriol 188:8136–8144. doi: 10.1128/JB.00988-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhou LG, Zhou MX, Sun CM, Xiang H, Tan HR. 2007. Genetic analysis of a novel plasmid pZMX101 from Halorubrum saccharovorum: determination of the minimal replicon and comparison with the related haloarchaeal plasmid pSCM201. FEMS Microbiol Lett 270:104–108. doi: 10.1111/j.1574-6968.2007.00656.x. [DOI] [PubMed] [Google Scholar]
- 216.Lee DG, Bell SP. 2000. ATPase switches controlling DNA replication initiation. Curr Opin Cell Biol 12:280–285. doi: 10.1016/S0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- 217.Berquist BR, DasSarma S. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J Bacteriol 185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D. 2005. Living with two extremes: conclusions from the genome sequence of Natronomonas pharaonis. Genome Res 15:1336–1343. doi: 10.1101/gr.3952905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lipps G, Stegert M, Krauss G. 2001. Thermostable and site-specific DNA binding of the gene product ORF56 from the Sulfolobus islandicus plasmid pRN1, a putative archael plasmid copy control protein. Nucleic Acids Res 29:904–913. doi: 10.1093/nar/29.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lipps G, Rother S, Hart C, Krauss G. 2003. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J 22:2516–2525. doi: 10.1093/emboj/cdg246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Beck K, Lipps G. 2007. Properties of an unusual DNA primase from an archaeal plasmid. Nucleic Acids Res 35:5635–5645. doi: 10.1093/nar/gkm625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Beck K, Vannini A, Cramer P, Lipps G. 2010. The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis. Nucleic Acids Res 38:6707–6718. doi: 10.1093/nar/gkq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Joshua CJ, Perez LD, Keasling JD. 2013. Functional characterization of the origin of replication of pRN1 from Sulfolobus islandicus REN1H1. PLoS One 8:e84664. doi: 10.1371/journal.pone.0084664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hajnsdorf E, Boni IV. 2012. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie 94:1544–1553. doi: 10.1016/j.biochi.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 225.Cooper CR, Daugherty AJ, Tachdjian S, Blum PH, Kelly RM. 2009. Role of vapBC toxin-antitoxin loci in the thermal stress response of Sulfolobus solfataricus. Biochem Soc Trans 37:123–126. doi: 10.1042/BST0370123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Maezato Y, Daugherty A, Dana K, Soo E, Cooper C, Tachdjian S, Kelly RM, Blum P. 2011. VapC6, a ribonucleolytic toxin regulates thermophilicity in the crenarchaeote Sulfolobus solfataricus. RNA 17:1381–1392. doi: 10.1261/rna.2679911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang Y, Duan Z, Zhu H, Guo X, Wang Z, Zhou J, She Q, Huang L. 2007. A novel Sulfolobus non-conjugative extrachromosomal genetic element capable of integration into the host genome and spreading in the presence of a fusellovirus. Virology 363:124–133. doi: 10.1016/j.virol.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 228.Contursi P, D'Ambrosio K, Pirone L, Pedone E, Aucelli T, She Q, De Simone G, Bartolucci S. 2011. C68 from the Sulfolobus islandicus plasmid-virus pSSVx is a novel member of the AbrB-like transcription factor family. Biochem J 435:157–166. doi: 10.1042/BJ20101334. [DOI] [PubMed] [Google Scholar]
- 229.Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. 2003. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi.” J Bacteriol 185:3888–3894. doi: 10.1128/JB.185.13.3888-3894.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Geslin C, Gaillard M, Flament D, Rouault K, Le Romancer M, Prieur D, Erauso G. 2007. Analysis of the first genome of a hyperthermophilic marine virus-like particle, PAV1, isolated from Pyrococcus abyssi. J Bacteriol 189:4510–4519. doi: 10.1128/JB.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bickle TA, Kruger DH. 1993. Biology of DNA restriction. Microbiol Rev 57:434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tock MR, Dryden DT. 2005. The biology of restriction and anti-restriction. Curr Opin Microbiol 8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 233.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Patterson NH, Pauling C. 1985. Evidence for two restriction-modification systems in Halobacterium cutirubrum. J Bacteriol 163:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ishikawa K, Watanabe M, Kuroita T, Uchiyama I, Bujnicki JM, Kawakami B, Tanokura M, Kobayashi I. 2005. Discovery of a novel restriction endonuclease by genome comparison and application of a wheat-germ-based cell-free translation assay: PabI (5′-GTA/C) from the hyperthermophilic archaeon Pyrococcus abyssi. Nucleic Acids Res 33:e112. doi: 10.1093/nar/gni113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Morgan R, Xiao J, Xu S. 1998. Characterization of an extremely thermostable restriction enzyme, PspGI, from a Pyrococcus strain and cloning of the PspGI restriction-modification system in Escherichia coli. Appl Environ Microbiol 64:3669–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Prangishvili DA, Vashakidze RP, Chelidze MG, Gabriadze I. 1985. A restriction endonuclease SuaI from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett 192:57–60. doi: 10.1016/0014-5793(85)80042-9. [DOI] [PubMed] [Google Scholar]
- 239.Watanabe M, Yuzawa H, Handa N, Kobayashi I. 2006. Hyperthermophilic DNA methyltransferase M.PabI from the archaeon Pyrococcus abyssi. Appl Environ Microbiol 72:5367–5375. doi: 10.1128/AEM.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sollner S, Berkner S, Lipps G. 2006. Characterisation of the novel restriction endonuclease SuiI from Sulfolobus islandicus. Extremophiles 10:629–634. doi: 10.1007/s00792-006-0019-0. [DOI] [PubMed] [Google Scholar]
- 241.McConnell DJ, Searcy DG, Sutcliffe JG. 1978. A restriction enzyme Tha I from the thermophilic mycoplasma Thermoplasma acidophilum. Nucleic Acids Res 5:1729–1739. doi: 10.1093/nar/5.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Blaseio U, Pfeifer F. 1990. Transformation of Halobacterium halobium: development of vectors and investigation of gas vesicle synthesis. Proc Natl Acad Sci U S A 87:6772–6776. doi: 10.1073/pnas.87.17.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Holmes ML, Nuttall SD, Dyall-Smith ML. 1991. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J Bacteriol 173:3807–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Baranyi U, Klein R, Lubitz W, Kruger DH, Witte A. 2000. The archaeal halophilic virus-encoded Dam-like methyltransferase M.phiCh1-I methylates adenine residues and complements dam mutants in the low salt environment of Escherichia coli. Mol Microbiol 35:1168–1179. doi: 10.1046/j.1365-2958.2000.01786.x. [DOI] [PubMed] [Google Scholar]
- 245.Vogelsangwenke H, Oesterhelt D. 1988. Isolation of a halobacterial phage with a fully cytosine-methylated genome. Mol Gen Genet 211:407–414. doi: 10.1007/BF00425693. [DOI] [Google Scholar]
- 246.Erdmann S, Chen B, Huang X, Deng L, Liu C, Shah SA, Le Moine Bauer S, Sobrino CL, Wang H, Wei Y, She Q, Garrett RA, Huang L, Lin L. 2014. A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 18:51–60. doi: 10.1007/s00792-013-0591-z. [DOI] [PubMed] [Google Scholar]
- 247.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.You XY, Liu C, Wang SY, Jiang CY, Shah SA, Prangishvili D, She Q, Liu SJ, Garrett RA. 2011. Genomic analysis of Acidianus hospitalis W1 a host for studying crenarchaeal virus and plasmid life cycles. Extremophiles 15:487–497. doi: 10.1007/s00792-011-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chen L, Brugger K, Skovgaard M, Redder P, She Q, Torarinsson E, Greve B, Awayez M, Zibat A, Klenk HP, Garrett RA. 2005. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J Bacteriol 187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. 2009. Biogeography of the Sulfolobus islandicus pan-genome. Proc Natl Acad Sci U S A 106:8605–8610. doi: 10.1073/pnas.0808945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang R, Li H. 2012. The mysterious RAMP proteins and their roles in small RNA-based immunity. Protein Sci 21:463–470. doi: 10.1002/pro.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 253.Garrett RA, Vestergaard G, Shah SA. 2011. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol 19:549–556. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 254.Barrangou R, Marraffini LA. 2014. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, Young MJ, White MF, Lawrence CM. 2011. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem 286:21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Plagens A, Tripp V, Daume M, Sharma K, Klingl A, Hrle A, Conti E, Urlaub H, Randau L. 2014. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A cascade interference complex. Nucleic Acids Res 42:5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF. 2012. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, Masuda A, Dohmae N, Schaap PJ, Doudna JA, Heck AJ, Yonekura K, van der Oost J, Shinkai A. 2013. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, Garrett RA. 2011. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Manica A, Zebec Z, Teichmann D, Schleper C. 2011. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 264.Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 265.Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, Dyall-Smith M, Marchfelder A. 2012. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem 287:33351–33363. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Li M, Liu H, Han J, Liu J, Wang R, Zhao D, Zhou J, Xiang H. 2013. Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei. J Bacteriol 195:867–875. doi: 10.1128/JB.01688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nickel L, Weidenbach K, Jager D, Backofen R, Lange SJ, Heidrich N, Schmitz RA. 2013. Two CRISPR-Cas systems in Methanosarcina mazei strain Go1 display common processing features despite belonging to different types I and III. RNA Biol 10:779–791. doi: 10.4161/rna.23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Elmore JR, Yokooji Y, Sato T, Olson S, Glover CV III, Graveley BR, Atomi H, Terns RM, Terns MP. 2013. Programmable plasmid interference by the CRISPR-Cas system in Thermococcus kodakarensis. RNA Biol 10:828–840. doi: 10.4161/rna.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Peng W, Li H, Hallstrom S, Peng N, Liang YX, She Q. 2013. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol 10:738–748. doi: 10.4161/rna.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 85:1044–1056. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Erdmann S, Le Moine Bauer S, Garrett RA. 2014. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol Microbiol 91:900–917. doi: 10.1111/mmi.12503. [DOI] [PubMed] [Google Scholar]
- 272.Li M, Wang R, Zhao D, Xiang H. 2014. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res 42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.DeLong EF, Pace NR. 2001. Environmental diversity of bacteria and archaea. Syst Biol 50:470–478. doi: 10.1080/10635150118513. [DOI] [PubMed] [Google Scholar]
- 274.Forterre P, Prangishvili D. 2013. The major role of viruses in cellular evolution: facts and hypotheses. Curr Opin Virol 3:558–565. doi: 10.1016/j.coviro.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 275.Koonin EV, Dolja VV. 2013. A virocentric perspective on the evolution of life. Curr Opin Virol 3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Redder P, Peng X, Brugger K, Shah SA, Roesch F, Greve B, She Q, Schleper C, Forterre P, Garrett RA, Prangishvili D. 2009. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol 11:2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 277.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter WD, Zillig W. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 278.Bath C, Dyall-Smith ML. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J Virol 72:9392–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Janekovic D, Wunderl S, Holz I, Zillig W, Gierl A, Neumann H. 1983. TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic, anaerobic, sulfur reducing archaebacterium Thermoproteus tenax. Mol Gen Genet 192:39–45. doi: 10.1007/BF00327644. [DOI] [Google Scholar]
- 280.Porter K, Kukkaro P, Bamford JK, Bath C, Kivela HM, Dyall-Smith ML, Bamford DH. 2005. SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology 335:22–33. doi: 10.1016/j.virol.2005.01.043. [DOI] [PubMed] [Google Scholar]
- 281.Jaakkola ST, Penttinen RK, Vilen ST, Jalasvuori M, Ronnholm G, Bamford JKH, Bamford DH, Oksanen HM. 2012. Closely related archaeal Haloarcula hispanica icosahedral viruses HHIV-2 and SH1 have nonhomologous genes encoding host recognition functions. J Virol 86:4734–4742. doi: 10.1128/JVI.06666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Porter K, Tang SL, Chen CP, Chiang PW, Hong MJ, Dyall-Smith M. 2013. PH1: an archaeovirus of Haloarcula hispanica related to SH1 and HHIV-2. Archaea 2013:456318. doi: 10.1155/2013/456318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Pietila MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol Microbiol 72:307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- 284.Sencilo A, Paulin L, Kellner S, Helm M, Roine E. 2012. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res 40:5523–5534. doi: 10.1093/nar/gks215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schnabel H, Zillig W, Pfaffle M, Schnabel R, Michel H, Delius H. 1982. Halobacterium halobium phage Phi-H. EMBO J 1:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schnabel H, Schramm E, Schnabel R, Zillig W. 1982. Structural variability in the genome of phage phi-H of Halobacterium halobium. Mol Gen Genet 188:370–377. doi: 10.1007/BF00330036. [DOI] [Google Scholar]
- 287.Meile L, Jenal U, Studer D, Jordan M, Leisinger T. 1989. Characterization of Psi-M1, a virulent phage of Methanobacterium-Thermoautotrophicum Marburg. Arch Microbiol 152:105–110. doi: 10.1007/BF00456085. [DOI] [Google Scholar]
- 288.Witte A, Baranyi U, Klein R, Sulzner M, Luo C, Wanner G, Kruger DH, Lubitz W. 1997. Characterization of Natronobacterium magadii phage Phi Ch1, a unique archaeal phage containing DNA and RNA. Mol Microbiol 23:603–616. doi: 10.1046/j.1365-2958.1997.d01-1879.x. [DOI] [PubMed] [Google Scholar]
- 289.Nuttall SD, Dyallsmith ML. 1993. Hf1 and Hf2—novel bacteriophages of halophilic Archaea. Virology 197:678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- 290.Mei YJ, Chen D, Sun DC, Yang Y, Shen P, Chen XD. 2007. Induction and preliminary characterization of a novel halophage SNJ1 from lysogenic Natrinema sp F5. Can J Microbiol 53:1106–1110. doi: 10.1139/W07-072. [DOI] [PubMed] [Google Scholar]
- 291.Zhang ZQ, Liu Y, Wang S, Yang D, Cheng YC, Hu JN, Chen J, Mei YJ, Shen P, Bamford DH, Chen XD. 2012. Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology 434:233–241. doi: 10.1016/j.virol.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 292.Brumfield SK, Ortmann AC, Ruigrok V, Suci P, Douglas T, Young MJ. 2009. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus Sulfolobus turreted icosahedral virus. J Virol 83:5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Gerdes K, Christensen SK, Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 294.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. 2008. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A 105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Okutan E, Deng L, Mirlashari S, Uldahl K, Halim M, Liu C, Garrett RA, She Q, Peng X. 2013. Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells. RNA Biol 10:875–885. doi: 10.4161/rna.24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Szymczyna BR, Taurog RE, Young MY, Snyder JC, Johnson JE, Williamson JR. 2009. Synergy of NMR, computation, and X-ray crystallography for structural biology. Structure 17:499–507. doi: 10.1016/j.str.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Weigel C, Seitz H. 2006. Bacteriophage replication modules. FEMS Microbiol Rev 30:321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 298.Peng W, Feng M, Feng X, Liang YX, She Q. 2015. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res 43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.