Significance
The general perception that mRNA transcription of a gene is regulated by the functional requirements of the cell or tissue has never been systematically tested using genome-wide data. To assess the functional implications of mRNA transcription in mice directly, we analyzed gene expression data and phenotype data of mouse mutants as a proxy of gene function at the tissue level. Our results confirmed the important role transcriptional regulation has in maintaining the gene’s proper spatial and temporal functions. In addition, we found that mutations resulting in phenotypic defects in expressed tissues are more likely to occur in highly transcribed genes, tissue-specific genes, genes expressed during early embryonic stages, or genes with evolutionarily conserved mRNA expression.
Keywords: mRNA abundance, tissue specificity, developmental stages, ectopic expression, molecular evolution
Abstract
High-throughput gene expression profiling has revealed substantial leaky and extraneous transcription of eukaryotic genes, challenging the perceptions that transcription is strictly regulated and that changes in transcription have phenotypic consequences. To assess the functional implications of mRNA transcription directly, we analyzed mRNA expression data derived from microarrays, RNA-sequencing, and in situ hybridization, together with phenotype data of mouse mutants as a proxy of gene function at the tissue level. The results indicated that despite the presence of widespread ectopic transcription, mRNA expression and mutant phenotypes of mammalian genes or tissues remain associated. The expression-phenotype association at the gene level was particularly strong for tissue-specific genes, and the association could be underestimated due to data insufficiency and incomprehensive phenotyping of mouse mutants; the strength of expression-phenotype association at the tissue level depended on tissue functions. Mutations on genes expressed at higher levels or expressed at earlier embryonic stages more often result in abnormal phenotypes in the tissues where they are expressed. The mRNA expression profiles that have stronger associations with their phenotype profiles tend to be more evolutionarily conserved, indicating that the evolution of transcriptome and the evolution of phenome are coupled. Therefore, mutations resulting in phenotypic aberrations in expressed tissues are more likely to occur in highly transcribed genes, tissue-specific genes, genes expressed during early embryonic stages, or genes with evolutionarily conserved mRNA expression profiles.
It is widely assumed that transcription is under tight and sophisticated regulation (1) and that changes in transcription have phenotypic consequences. For example, spatial and temporal regulatory changes in transcription can impart major changes in the development of multicellular organisms (2, 3). Aberrations in transcription have been linked to the onset or progression of human diseases, including autism (4), schizophrenia (5), congenital heart defects (6), and cancers (7). The evolution of transcription regulation was proposed to have a more profound role than protein structural evolution in generating adaptive changes that lead to phenotypic diversity among species (8, 9). However, recent studies exploiting high-throughput gene expression profiling methods suggested widespread ectopic (or nonfunctional) mRNA expression (10) in eukaryotic genomes, according to the discoveries of pervasive transcriptional activities from nongenic regions (11, 12), coexpression in gene clusters without linkage conservation (13) or relatedness in annotated functions (14) of genes, neutral evolution of mRNA expression patterns among orthologs (15), and less correlation in mRNA abundance than in protein abundance among orthologous genes (16). Because the association between mRNA expression and gene function had never been directly assessed systematically and on a genome-wide basis, the general implications in function of mRNA expression became elusive.
The most straightforward approach to discern gene function is mutagenesis. Among mammals, the house mouse (Mus musculus) has been subjected to extensive mutagenesis (17), and more than 40% of mouse genes have been mutated, with the resulting mutant strain phenotyped (18). Using these data, we investigate whether genes function in the tissues where they are transcribed. Gene function is defined by the presence of abnormal phenotypes when a gene is mutated. Gene expression data come from oligonucleotide microarray, RNA-sequencing (RNA-seq), and RNA in situ hybridization. The combination of these phenotype data with spatial and temporal mRNA gene expression data (below) allows us to investigate how mRNA expression and phenotypes are connected in the presence of ectopic transcription. In addition, we investigate whether any association in mRNA expression and phenotypes affects the evolutionary conservation of gene expression. Our approach enables us to understand the underlying causes for and the biological features associated with the variations in expression-phenotype association among genes and tissues.
Results and Discussion
Measuring Expression-Phenotype Association.
Each gene has an mRNA expression profile, defined as the mRNA expression across the mouse tissues examined. Depending on the experiment (discussed in the following sections), mRNA expression indicates the presence or absence of detectable mRNA expression signals, mRNA expression within a certain range of abundance, or mRNA expression under a specific condition. Each gene also has a phenotypic profile, defined by the presence or absence of abnormal phenotypes in tissues when that gene is mutated. Similarly, each tissue has an mRNA expression profile defined by mRNA expression across all mouse genes surveyed. For each tissue, there is a phenotypic profile in which there is the presence or absence of abnormal phenotypes for each gene in that given tissue. Using these mRNA expression profiles and phenotypic profiles for mouse genes and tissues (Methods), we determined the extent to which mRNA expression profiles correspond to phenotypic profiles for each mouse gene or tissue.
The index for expression-phenotype connection (EPC) for each mouse gene or each mouse tissue was defined by the statistical deviation of the observed NEP/√(NE╳NP) from the expectation of randomness. When EPC is computed for a gene (EPCg), NE is the number of tissues where the gene is expressed, NP is the number of tissues with at least one abnormal phenotype when the gene is mutated, and NEP is the number of tissues that both have gene expression and abnormal phenotypes when the gene is mutated. When EPC is computed for a tissue (EPCt), NE is the number of genes that are expressed in the tissue; NP is the number of genes that, when mutated, result in abnormal phenotypes in the tissue; and NEP is the number of genes that both result in mutant phenotypes and exhibit mRNA expression. For each gene or tissue, the distributions of NEP/√(NE╳NP) under the null hypothesis of randomness were derived from 2,500 permutation experiments, each of which has a recomputed NEP/√(NE╳NP) by randomizing its phenotype profile while maintaining its mRNA expression profile. EPC was then defined by the NEP/√(NE╳NP) of the original data minus the averaged NEP/√(NE╳NP) of the 2,500 permutation experiments divided by the SD of NEP/√(NE╳NP) derived from 2,500 permutation experiments. We only calculated EPC when NE ≥ 1 and NP ≥ 1. EPC is equivalent to the Z-score in the Z-test methodologically. Fig. S1A shows the expression and phenotypic profiles of example genes, as well as their corresponding EPCg values.
EPC at the Gene Level.
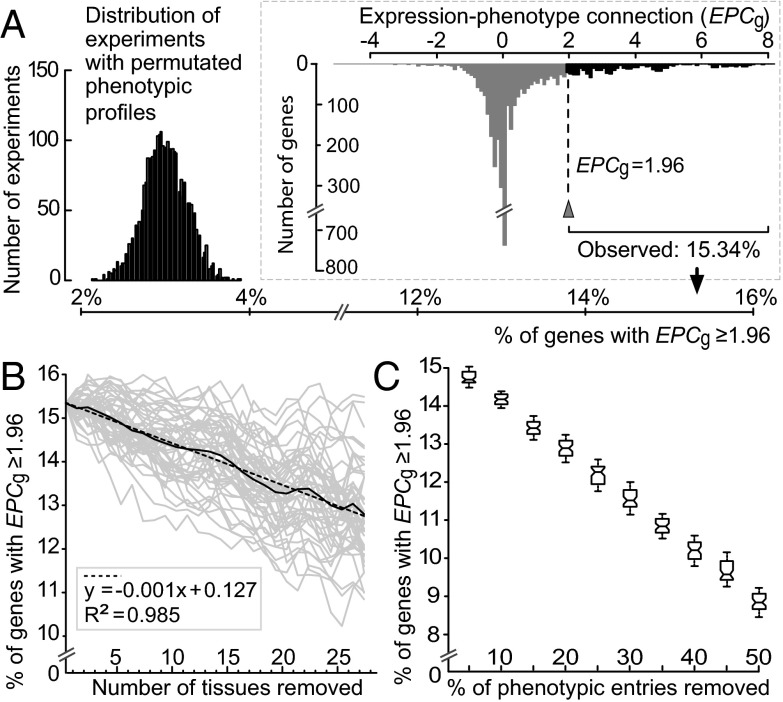
To determine whether mutations in a gene result in defects in tissues where the gene is expressed, we calculated mouse EPCg values using microarray-based mRNA expression data. Microarray expression signals were processed by the gcRMA (GeneChip robust multiarray averaging) method (19), and signals ≥200 indicated that a gene was expressed in a tissue (20) and were used to define NE and NEP. GeneAtlas v2 contains mRNA expression data from oligonucleotide array experiments on 60 mouse tissues (“spinal cord upper” and “spinal cord lower” expression data were merged into “spinal cord” for this study) (20). Of these 60 tissues, 47 have phenotype entries in Mouse Genome Informatics (MGI) from mutant strain phenotyping (Methods and Table S1). At present, GeneAtlas v2 contains the largest number of mouse tissues profiled for mRNA expression in a single study, allowing us to measure EPCg with minimal biases (below). In the distribution of EPCg from 3,859 mouse genes with NE ≥ 1 and NP ≥ 1 in 47 tissues (Fig. 1A), 15.34% (592 of 3,859) of the genes have EPCg ≥ 1.96 (P < 0.05, Z-test), which is significantly greater than the percentage of genes with EPCg ≥ 1.96 in the permutation experiments (3.00 ± 0.26% SD; P < 10−300, t test; Fig. 1A). For genes above this EPCg threshold, mRNA expression signals are directly tied to function in the tissues where they are expressed, and the loss of this function can result in abnormal phenotypes. A larger EPCg threshold resulted in a greater deviation of observed EPCg from the expectation under randomness (Fig. S2). Focusing on 1,216 tissue-specific genes (NE ≤ 5), the proportion of genes with EPCg ≥ 1.96 is 36.51% (444 of 1,216 genes). Therefore, the expression-phenotype association was observed despite intrinsic noise in mRNA expression (21) and measurement errors of microarrays (22) (below), and the association was particularly strong for tissue-specific genes.
Fig. 1.
EPC at the gene level, measured as EPCg. (A) Percentage of mouse genes with EPCg ≥ 1.96 was 15.34% (arrowhead from the distribution in Inset), which was significantly greater than the percentage of mouse genes with EPCg ≥ 1.96 estimated from permutation experiments. (B) Percentage of mouse genes with EPCg ≥ 1.96 decreased linearly with the number of mouse tissues removed in calculating EPCg. Each gray line is one of 50 experiments derived from the random removal of tissues one at a time. The solid black line is the average of the 50 gray lines. The linear regression line (dashed line with equation given in the gray box) for the average line is given. (C) Box plots for the percentage of mouse genes with EPCg ≥ 1.96 when a proportion of phenotypic entries were removed. Each box presents the distribution of estimations calculated based on 50 replicates. The values of the upper quartile, median, and lower quartile are indicated in each box, whereas the bars outside the box indicate semiquartile ranges.
Although significantly higher than random, the percentage of mouse genes with statistically significant EPC (15.34% of all genes) was small. For the remaining genes, there are two possible explanations for a lack of statistical support for EPC. First, a mutation in a gene might not cause discernable defects in the tissue where it is expressed. Second, there is a connection between mRNA expression and mutant phenotypes for a gene, but it cannot be detected due to data limitations, such as insufficient tissue sampling, incomplete phenotyping, and errors in mRNA quantification. A mouse has >150 cell types (23), which comprise even more tissues, but only 47 tissues were included in this analysis (Fig. 1A). To understand the influence of incomplete tissue sampling in estimating EPCg, we randomly removed the data on a tissue one at a time until only 20 tissues remained and recalculated EPCg after each random tissue removal. Based on 50 replicates of random tissue exclusion, we averaged the percentage of genes with EPCg ≥ 1.96 for each number of tissues removed (Fig. 1B). If the percentage was not affected by incomplete tissue sampling, the percentage should plateau to a value of ∼15.34% when the number of tissues removed is few; after a certain stage, the percentage decreases as the number of tissues removed increases. Alternatively, if the percentage was underestimated due to insufficient tissue sampling, the percentage of genes with EPCg ≥ 1.96 should decrease as the number of tissues removed increases from the beginning. Consistent with the insufficient tissue sampling, we found that the percentage of genes with EPCg ≥ 1.96 decreases linearly as the number of tissues removed increases (R2 = 0.985; P < 10−300, ANOVA for linear model fits; Fig. 1B).
Because phenotype screening of mutant mice can be biased by study design, the manifestation of a mutation in tissues unrelated to the study focus can be overlooked and remain undescribed. Thus, although 47 tissues are included in our analysis, for most genes, only a fraction of these tissues were examined for phenotypic abnormalities. To account for incomplete phenotyping, we randomly removed a proportion of phenotypic entries (5–50% in 5% increments) and recalculated the percentage of mouse genes with EPCg ≥ 1.96. Each of these 10 experiments had 50 replicates. The median percentage of genes with EPCg ≥ 1.96 decreased from 14.70% to 8.83% as the proportion of phenotypic entries removed increased (Fig. 1C), indicating that incomplete phenotyping leads to an underestimation of EPCg. When expression and phenotype data are available for more tissues (Fig. 1B) and when phenotyping of mutant strains is more complete (Fig. 1C), the percentage of genes with EPCg ≥ 1.96 will likely increase toward its true value.
Phenotyping mutant mouse strains can be biased by prior knowledge of a gene’s expression pattern. To determine if the significant EPCg in Fig. 1A is due to this bias, we limited the analysis to phenotype data published before the microarray dataset GeneAtlas v2 (20). If high EPCg values in the full dataset are due to phenotype inspection bias, EPCg of this subset of 2,084 genes should be lower. However, 16.21% (338 of 2,084) of phenotyped mouse genes had EPCg ≥ 1.96 (Fig. S3), which was slightly greater but not statistically different from the full dataset (P = 0.37, χ2 test). Therefore, the observed EPCg values were not due to biased phenotypic screening of tissues/organs with known gene expression signals.
Microarray signals harbor cross-hybridization noises (22). To account for experimental errors in the estimation of EPCg, we stochastically introduced noise within a range (±5% to ±50% of the experimental value) into the microarray signals and recalculated EPCg. Regardless of the magnitude of noise introduced, the median percentage of mouse genes with EPCg ≥ 1.96 stayed near 15.4% (Fig. S4), indicating that microarray noise had no effect on the estimate of EPCg.
Genes Without Overlapping Tissues of mRNA Expression and Mutant Phenotypes.
Of 3,859 genes in the full dataset, 996 genes resulted in abnormal phenotypes in tissues where they were not expressed (Fig. 1A; genes with NEP = 0, NE > 0, NP > 0). These genes had an average EPCg of −0.451 ± 0.338 SD, which was smaller than the average EPCg of the rest of the genes (1.064 ± 1.748 SD). Incomplete phenotyping and insufficient tissue sampling that underestimated EPCg (Fig. 1 B and C) can explain part of NEP = 0 genes. For example, although beta-1,4-N-acetyl-galactosaminyl transferase 1 gene (B4galnt1) is expressed in many adult tissues (Fig. S1B), only reproductive and neurological phenotypes have been examined for the viable and fertile knockout strain (24, 25). More complete phenotyping on B4galnt1 mutant strains across a wider range of tissues could reveal overlap between tissues with expression and tissues with abnormalities. All 47 tissues used to compute EPCg were from adult mice, but many abnormal phenotypes are only observed during fetal or neonatal stages due to premature death. For example, embryos from an NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase gene (Mthfd2) knockout strain have small pale livers and die before embryonic day 15.5, suggesting an important role of Mthfd2 in embryogenesis (26). Although the expression dataset indicates that Mthfd2 is highly expressed in the fertilized egg and embryonic stages (Fig. S1C), this information was not used in the calculation of EPCg. Another explanation for NEP = 0 genes is a bias in abnormal phenotypes that affect the whole organism rather than a specific tissue or cell type. Using Mammalian Phenotype Enrichment Analysis (MamPhEA) (18) for phenotypic enrichment analyses (Methods), compared with other genes (NEP > 0), NEP = 0 genes were enriched in phenotypes associated with the nervous system, behaviors, and cellular metabolism/homeostasis (the full results are shown in Fig. S5). These phenotypes have organism-level effects (e.g., behavioral changes, obesity) that cannot simply be coded as an abnormality in specific tissues. When these abnormalities are observed in tissues, they are often due to gross physiological changes in the mutant (e.g., increased/decreased fat amount due to changes in metabolic rate) rather than dysfunction of locally expressed genes. These examples indicated that the importance of mRNA transcription to gene function could be underestimated by the data and method used in this study.
EPC at the Tissue Level.
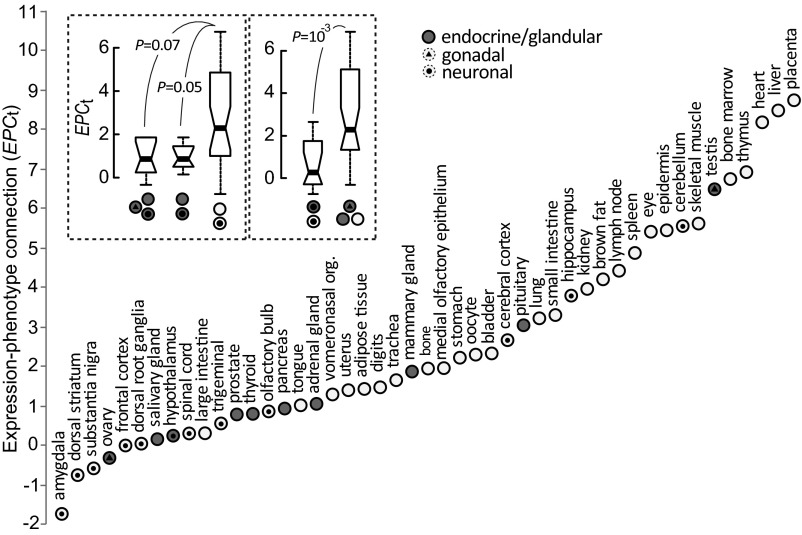
To assess the functional relevance of the transcriptome at the tissue level to that tissue’s morphology or physiology directly, we examined EPCt of 47 mouse tissues for 7,449 genes that have MGI phenotype entries and have mRNA expression profiles in GeneAtlas v2. Each mouse tissue has two gene profiles: one including genes with active (expression signals ≥200) transcription and another including genes that, when mutated, result in abnormal phenotypes in the tissue examined. To compute EPCt, the first expression profile was used to define NE and the second phenotypic profile was used to define NP. The two profiles were used together to define NEP. Of the 47 tissues, 22 (46.81%) have statistically significant EPC, indicated by EPCt ≥ 1.96 (Fig. 2A). By comparison, the average percentage of tissues that had EPCt ≥ 1.96 from permuted phenotypic profiles of each of the tissues was 2.52% (Fig. S6). A complementary approach exploiting hypergeometric tests (18) echoed the analysis by EPCt, showing that only gene sets expressed in tissues with high EPCt have significant statistical support for the enrichment of abnormal phenotypes in the same tissue (Table S2). These results suggest that genes transcribed in a tissue are often performing functions linked to the development and physiology of the tissue examined.
Fig. 2.
EPC at the tissue level, measured by EPCt, in 47 mouse tissues. Comparisons of EPCt between glandular/endocrine tissues (with and without gonadal tissues) vs. nonglandular/nonendocrine tissues or between neuronal tissues vs. nonneuronal tissues are shown (P values are from the Mann–Whitney U test). The values of the upper quartile, median, and lower quartile are indicated in each box, whereas the bars outside the box indicate semiquartile ranges. org., organ.
Tissue function likely underlies the observed variation in EPCt (Fig. 2). If colocalization of transcription and protein activity (either directly or indirectly) leads to high EPCt, tissues with primarily endocrine or glandular functions, which produce molecules that control the development or physiology of other tissues or interact with external environmental factors, should have smaller EPCt. Mutations in genes transcribed in these tissues manifest themselves as abnormal phenotypes in other tissues. The 47 mouse tissues include 10 endocrine/glandular tissues (Fig. 2). As predicted, these 10 endocrine/glandular tissues had lower EPCt compared with the remaining 37 tissues, although the difference was not statistically significant (P = 0.07, Mann–Whitney U test; left box in Fig. 2, Top Left). The 10 endocrine/glandular tissues include testis and ovary, which not only secrete hormones that function outside the tissue, but are involved in reproductive activities, such as the production of gametes. When testis and ovary were excluded from the 10 endocrine/glandular tissues, the difference in EPCt between the eight endocrine/glandular tissues and the 37 nonendocrine/glandular tissues was significant (P < 0.05, Mann–Whitney U test; Fig. 2). The higher EPCt of tissues producing proteins/molecules that function locally suggests that the location of transcription has functional implications.
Additionally, genes associated with neurological/behavioral traits [e.g., VGF nerve growth factor inducible (Vgf); Fig. S1A] can have global effects, which lead to lack of observable EPC (also Fig. S5). Consistent with this observation, neuronal tissues also tend to have lower EPCt compared with other tissues (P = 0.001, Mann–Whitney U test; right box in Fig. 2, Top Left).
mRNA Abundance and EPC.
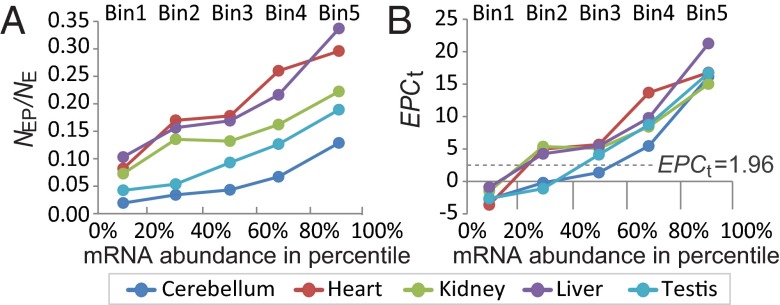
mRNA abundance can vary by several orders of magnitude in mammalian cells. Highly expressed genes are more easily identified experimentally (e.g., cDNA cloning) and tend to be well studied. To understand how mRNA abundance of a gene relates to its functional relevance, we used RNA-seq data from five mouse tissues (cerebellum, heart, kidney, liver, and testis) profiled in a study by Brawand et al. (27). These same investigators also profiled the tissue “brain,” but we omitted this organ because there was no corresponding “whole-brain” phenotypic code in MGI. RNA-seq data were used because RNA-seq quantifies mRNA abundance more accurately compared with microarrays and has low background noise to detect weakly expressed genes (28). RNA-seq expression signals were measured as reads per kilobase per million mapped reads (RPKM) (27). The number of transcribed genes (RPKM > 0) in cerebellum, heart, kidney, liver, and testis was 4,389, 4,326, 4,317, 4,022, and 4467, respectively. For each tissue, we divided all transcribed genes into five equal-sized bins based on mRNA abundance (1–5, lowest to highest). For each bin of each tissue, NE and NEP were defined by gene counts. For example, in bin 2 of the heart, NE was defined by the number of genes transcribed in the heart bin 2 (at 20–40% abundance rank), and the number of these genes also found to have abnormal phenotypes in the heart was NEP. We calculated the proportion of expressed genes showing abnormal phenotypes (NEP/NE) and EPCt for each bin of each tissue. When computing EPCt, 7,358 phenotyped genes with detectable RNA-seq expression signals (RPKM > 0) in at least one of the five tissues were used for the permutation analysis (2,500 permutations).
Because protein production can be regulated posttranscriptionally and proteins vary in their intrinsic range of functionally effective abundance levels (29), highly transcribed genes in the cell are not necessarily functionally more important than lowly expressed genes. However, the proportion of genes expressed resulting in abnormal phenotypes (NEP/NE) increased as the bin number increased for all of the tissues (Fig. 3A), indicating that genes with higher mRNA expression are more likely to be associated with mutant phenotypes. For bin 1 of all tissues (and also for bin 2 for testis and bins 2 and 3 for cerebellum), EPCt was <1.96 (Fig. 3B), suggesting lowly expressed genes are largely composed of genes functionally unimportant to the tissues where they are expressed. These results correspond to two major classes of transcribed genes with distinct mRNA abundances in metazoan cells: highly expressed, functionally important genes and lowly expressed genes with ectopic expression (30). NEP/NE and EPCt increased with mRNA abundance in all of the tissues, even among the highly expressed genes (i.e., bins 3–5), indicating that even among highly expressed genes, the absolute difference in mRNA abundance in tissues reflects a difference in the gene’s importance to tissue function.
Fig. 3.
Proportion of genes that causes phenotypic abnormalities in the tissue where expressed (NEP/NE) (A) and EPCt values (B) were higher for genes that have higher expression levels, defined by RNA-seq expression signals presented as RPKM. Genes were binned into five equal-sized bins based on mRNA abundance (1–5, lowest to highest) for five tissues.
Expression in Embryonic Stages and EPC.
Analysis of the spatial and temporal regulation of genes during embryogenesis is necessary to understand developmental genes. In situ hybridization is a classic technique used to visualize mRNA expression in embryos, although in situ hybridization expression signals are noisy and detection specificity is affected by probe design (31). Changes in gene regulation at earlier stages in embryogenesis tend to result in more severe phenotypes, indicated by a higher frequency of embryonic lethality (32), and enhancers responsible for earlier stages of organogenesis are proposed to be more evolutionarily conserved (33), suggesting that early regulatory activities are especially important. Therefore, temporal expression during embryogenesis could be a factor determining the influences of a gene’s expression to the physiology or morphology of the tissue.
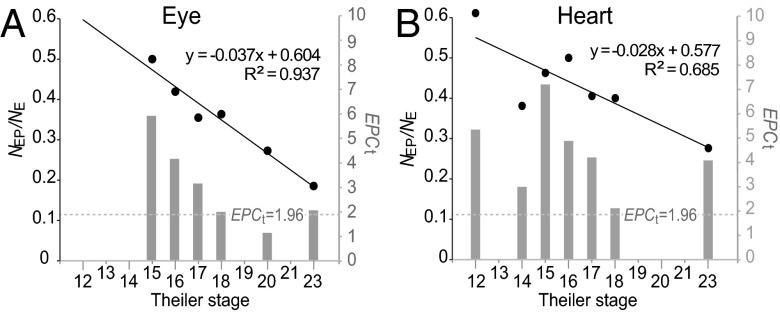
To test the hypothesis, we investigated two tissues: eye and heart. These tissues have discernable primordia with distinct developmental processes from their surrounding tissues, and their development is observable in early embryonic stages. mRNA in situ hybridization data from mouse embryos was retrieved from the e-Mouse Atlas of Gene Expression (EMAGE) (34) (Methods), which integrates in situ hybridization expression data from diverse sources. In the EMAGE data, Edinburgh Mouse Atlas Project (EMAP) IDs differ for embryonic tissues at different developmental Theiler stages. For eye and heart, expression data for Theiler stages 12–23 were included (Table S3). If a gene is expressed at multiple stages, the earliest stage was assigned for that gene. The number of genes with in situ hybridization signals in the developing eye (or heart) at each Theiler stage was defined as NE, and the number of genes that also showed mutant phenotypes in the adult eye (or heart) was defined as NEP. Stage-specific NEP/NE and EPCt were only computed for Theiler stages with a sufficient sample size (NE ≥ 10). When computing EPCt, 3,043 genes that have MGI phenotype data and EMAGE in situ hybridization data were used for phenotypic profile permutation (2,500 permutations). For both eye and heart, genes expressed at earlier Theiler stages in primordial embryonic tissues are more likely to result in phenotypic defects in the corresponding adult tissue (Theiler stages vs. NEP/NE: eye: Spearman’s correlation coefficient σ = −0.943, P < 10−3; heart: σ = −0.824, P = 0.02; Fig. 4A). This conclusion is also supported by EPCts, which are >1.96 for genes expressed at early embryonic stages (before Theiler stage 18) of both eye and heart (Fig. 4B).
Fig. 4.
Proportion of genes that causes phenotypic abnormalities in the tissue where expressed (NEP/NE) (black dots correspond to the left y axis) and EPCt values (gray bars correspond to the right y axis) for different embryonic stages of mouse developing eye (A) or heart (B). The linear regression line (and the equation) for NEP/NE and Theiler stages are shown.
EPC and the Evolution of Gene Regulation.
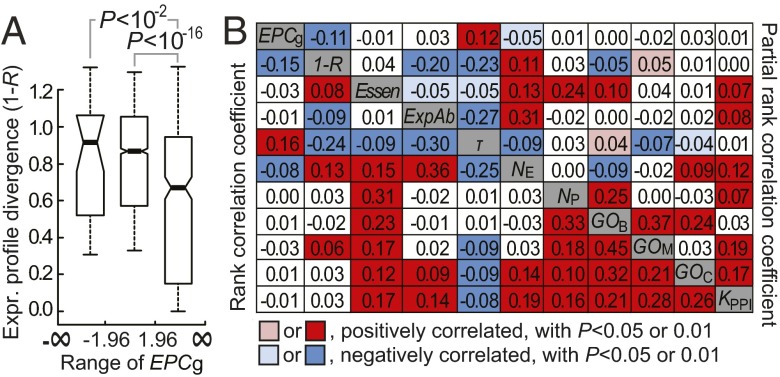
Natural selection acts on phenotypes that have an effect on an organism’s fitness. Gene properties tightly connected with phenotypes thus should more often be the subject of natural selection. To examine whether genes with a higher EPCg have more evolutionarily constrained mRNA expression profiles, we computed expression profile divergence between 1:1 human-mouse orthologs. Expression profile divergence was measured by 1 − R, where R is Pearson’s correlation coefficient between microarray expression signals across the 26 homologous tissues between human and mouse in GeneAtlas v2 (22) (Methods). Higher 1 − R indicates a greater expression profile divergence and more relaxed selective constraint in expression profile. Consistent with our expectation, 1 − R of genes with EPCg > 1.96 was significantly lower than 1 − R of other genes (Fig. 5A). This difference was not observed under the neutral model of transcriptome evolution approximated by randomizing expression profiles of human genes in calculating 1 − R (22, 35) (Fig. S7). Hence, mRNA expression profiles that are associated with the phenotypic profiles tend to be more evolutionarily conserved after the rodent-primate divergence.
Fig. 5.
Genes with higher EPCg have more conserved gene expression profiles following the rodent-primate divergence. (A) Box plots show the distributions of 1 − R for mouse genes with three different ranges of EPCg. The values of the upper quartile, median, and lower quartile are indicated in each box, and the bars outside the box indicate semiquartile ranges. P values are from the Mann–Whitney U test. (B) Rank correlation between EPCg and 1 − R before and after controlling for potential confounding factors. Rank correlation coefficients between the two examined properties are given in the bottom left corner, whereas the partial rank correlation coefficients after controlling for the all of the remaining factors are given in the upper right corner.
Evolutionarily conserved expression profiles, indicated by high 1 − R, were found in tissue-specific genes in mammals (36). We found that genes with high EPCg tend to be genes with high tissue specificity, indicated by lower NE or higher τ (Methods) (EPCg vs. NE: σ = −0.081, P < 10−4; EPCg vs. τ: σ = 0.163, P < 10−14), suggesting that tissue specificity has potentially confounded the relationship between EPCg and 1 − R. To measure the direct association between EPCg and 1 − R, we performed partial correlation analysis. In addition to τ, NE, and NP, we examined and controlled for other gene properties potentially governing regulatory evolution of genes, including gene essentiality (Essen), microarray-based mRNA abundance (ExpAb), number of associated Gene Ontology (GO) terms (GOM, GOB, or GOC represents the number of terms in “molecular function,” “biological processes,” or “cellular components,” respectively; Methods) and number of interacting partners in the protein–protein interaction network (KPPI). The partial rank correlation coefficient (σp) between 1 − R and EPCg after controlling for all of the other factors remained significantly negative (σp = −0.108, P < 10−6) (Fig. 5B). Thus, genes with evolutionarily constrained mRNA expression profiles tend to be associated with abnormal phenotypes in the expressed tissues. Consistent with the previous notion that tissue-specific genes tend to have greater EPCs, there was a positive correlation between EPCg vs. τ and a negative correlation between EPCg vs. NE after controlling for all of the remaining factors (Fig. 5B).
EPCg is an approximation of how well the mRNA expression pattern of a gene matches its physiological function, because ectopic expression in off-target tissues and a lack of detectable expression in tissue requiring a gene’s product decrease EPCg. The observed pattern that gene expression profiles with higher EPCg are more evolutionarily conserved supports the argument that gene expression profiles are shaped by purifying selection (22, 37). In addition to previously reported gene properties, such as tissue specificity and mRNA expression level (36), we found that EPCg is a factor correlated with the rate of regulatory evolution of mammalian genes.
Summary
Despite the presence of widespread ectopic transcription, mRNA expression and mutant phenotypes remain tightly associated at both the gene level and tissue level in mice. The expression-phenotype association at the gene level, indicated by EPCg, was particularly strong for tissue-specific genes, and could be underestimated in the present study due to incomplete tissue sampling and phenotyping. The variation in the association at tissue level, EPCt, can be explained by tissue functions. Mutations on genes expressed at higher levels or expressed at earlier embryonic stages more often result in abnormal phenotypes in the tissues where they are expressed. The mRNA expression profiles that have stronger connections with their phenotype profiles tend to be more evolutionarily conserved, indicating that the evolution of transcriptome and phenome are coupled. Our results suggest that changes in mRNA expression that cause more severe abnormal phenotypes or diseases in expressed tissues are more likely to occur in genes with abundant transcription levels, high tissue specificities, early expressed embryonic stages, or evolutionarily conserved expression profiles.
Methods
Phenotype Data of Mouse Genes.
Mouse genes and the associated mutant phenotypes were obtained from MGI (www.informatics.jax.org/), version 5.2. Ensembl IDs (v69) (38) of phenotyped mouse protein-coding genes were found at MRK_ENSEMBL.rpt, whereas the information on genotypes and phenotypes [presented as mammalian phenotype IDs (MP IDs)] (39) of the mutant generated was found at MGI_GenePheno.rpt. Phenotypes caused by mutations on multiple genes were discarded. We obtained a dataset of 7,449 mouse genes with one or more MP IDs when the gene is knocked out, knocked down, or mutated by transgenic insertions or point mutations. Organs or other anatomical parts with abnormal phenotypes are specified by MP IDs that are hierarchically structured. A parent MP ID represents a phenotype lineage that may include several child MP IDs to describe a more detailed abnormal phenotype. Genes with a child MP ID were also assigned to the parent MP IDs. MP ID terms used to define abnormal phenotypes in the 47 tissues are listed in Dataset S1. Publication dates for the literature describing abnormal phenotypes for MGI annotations were obtained from PubMed IDs (www.ncbi.nlm.nih.gov/pubmed).
mRNA Expression Data of Mouse Genes.
Expression signals of mouse genes measured by mRNA hybridization from 61 mouse tissues to the Affymetrix microarray chip (GNF1M) were obtained from the GeneAtlas v2 dataset (20). The processed mRNA expression signals, calculated by RPKM, for each mouse gene derived from 76-mer RNA-seq experiments in mouse cerebellum, heart, kidney, liver, and testis were obtained from supplementary material of a study by Brawand et al. (27). The mRNA in situ hybridization data from mouse embryonic tissues at various Theiler stages (40) were obtained from the BioMart interface of the EMAGE database (41). Genes annotated with “detected,” “strong,” “moderate,” or “weak” hybridization intensity were considered to be “expressed” in tissues/organs identified by EMAP IDs, according to anatomical ontology of developing mouse embryos (42). EMAP IDs are also hierarchically structured and are Theiler stage-specific. Accordingly, genes with a child EMAP ID were also assigned with the parent EMAP ID. To understand the influence of expression in embryonic stages to the EPC, we focused on EMAP IDs associated with eye and heart from Theiler stages 12–23 (Table S3).
Expression Profile Conservation of Mouse Genes.
Orthology relationships between human and mouse genes based on Ensembl annotation v69 were retrieved using BioMart (www.biomart.org/). Only 1:1 orthologs were used to compute expression profile divergence, as measured by 1 − R. The 26 homologous tissues between mouse and human used to compute 1 − R are from Liao and Zhang (22). ExpAb is defined as the average microarray signal across the 26 tissues. Tissue specificity of a mouse gene is calculated by (36), where n = 26 and Smax is the highest expression signal of the gene across the 26 tissues. Following Liao and Zhang (36), we arbitrarily let S(j) be 100 if it is lower than 100. The τ value ranges from 0 to 1, with higher values indicating higher tissue specificity. A gene is essential (Essen = 1) when mutations on it lead to premature death or infertility; otherwise, it is nonessential (Essen = 0) (43). GOM, GOB, or GOC was defined by the number of associated GO terms at level 5 annotated by Ensembl. The mouse protein–protein interaction network was obtained from the mammalian protein–protein interaction database at Munich Information Center for Protein Sequences (44). To identify factors contributing to 1 − R, partial correlation analysis was conducted using the “ppcor” package (45) for R (www.r-project.org/).
Supplementary Material
Acknowledgments
We thank anonymous reviewers for valuable comments and Wendy Grus for assistance in editing the manuscript. This work was supported by intramural funding from the National Health Research Institutes, Taiwan, and a research grant (MOST 101-2311-B-400-001-MY3) from the Ministry of Science and Technology, Taiwan (to B.-Y.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415046112/-/DCSupplemental.
References
- 1.Emerson BM. Specificity of gene regulation. Cell. 2002;109(3):267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Forrest ARR, et al. FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders AR, et al. MGS Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 2013;22(24):5001–5014. doi: 10.1093/hmg/ddt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierpont ME, et al. American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young Genetic basis for congenital heart defects: Current knowledge. Circulation. 2007;115(23):3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 7.Slamon DJ, deKernion JB, Verma IM, Cline MJ. Expression of cellular oncogenes in human malignancies. Science. 1984;224(4646):256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- 8.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188(4184):107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 9.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Trelles F, Tarrío R, Ayala FJ. Is ectopic expression caused by deregulatory mutations or due to gene-regulation leaks with evolutionary potential? BioEssays. 2005;27(6):592–601. doi: 10.1002/bies.20241. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21(2):93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Clark MB, et al. The reality of pervasive transcription. PLoS Biol. 2011;9(7):e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao B-Y, Zhang J. Coexpression of linked genes in Mammalian genomes is generally disadvantageous. Mol Biol Evol. 2008;25(8):1555–1565. doi: 10.1093/molbev/msn101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1(1):5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khaitovich P, et al. A neutral model of transcriptome evolution. PLoS Biol. 2004;2(5):E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrimpf SP, et al. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 2009;7(3):e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gondo Y. Trends in large-scale mouse mutagenesis: From genetics to functional genomics. Nat Rev Genet. 2008;9(10):803–810. doi: 10.1038/nrg2431. [DOI] [PubMed] [Google Scholar]
- 18.Weng M-P, Liao B-Y. MamPhEA: A web tool for mammalian phenotype enrichment analysis. Bioinformatics. 2010;26(17):2212–2213. doi: 10.1093/bioinformatics/btq359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99(468):909–917. [Google Scholar]
- 20.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 22.Liao B-Y, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol. 2006;23(3):530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- 23.Vogel C, Chothia C. Protein family expansions and biological complexity. PLOS Comput Biol. 2006;2(5):e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, et al. A genetic model of substrate deprivation therapy for a glycosphingolipid storage disorder. J Clin Invest. 1999;103(4):497–505. doi: 10.1172/JCI5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takamiya K, et al. Mice with disrupted GM2/GD2 synthase gene lack complex gangliosides but exhibit only subtle defects in their nervous system. Proc Natl Acad Sci USA. 1996;93(20):10662–10667. doi: 10.1073/pnas.93.20.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Pietro E, Sirois J, Tremblay ML, MacKenzie RE. Mitochondrial NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase is essential for embryonic development. Mol Cell Biol. 2002;22(12):4158–4166. doi: 10.1128/MCB.22.12.4158-4166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478(7369):343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heo M, Maslov S, Shakhnovich E. Topology of protein interaction network shapes protein abundances and strengths of their functional and nonspecific interactions. Proc Natl Acad Sci USA. 2011;108(10):4258–4263. doi: 10.1073/pnas.1009392108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebenstreit D, et al. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arvey A, et al. Minimizing off-target signals in RNA fluorescent in situ hybridization. Nucleic Acids Res. 2010;38(10):e115. doi: 10.1093/nar/gkq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damjanovski S, Sachs LM, Shi YB. Multiple stage-dependent roles for histone deacetylases during amphibian embryogenesis: Implications for the involvement of extracellular matrix remodeling. Int J Dev Biol. 2000;44(7):769–776. [PubMed] [Google Scholar]
- 33.Liao BY, Weng MP. Natural selection drives rapid evolution of mouse embryonic heart enhancers. BMC Syst Biol. 2012;6(Suppl 2):S1. doi: 10.1186/1752-0509-6-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christiansen JH, et al. EMAGE: A spatial database of gene expression patterns during mouse embryo development. Nucleic Acids Res. 2006;34(Database issue):D637–D641. doi: 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan IK, Mariño-Ramírez L, Koonin EV. Evolutionary significance of gene expression divergence. Gene. 2005;345(1):119–126. doi: 10.1016/j.gene.2004.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao B-Y, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23(6):1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- 37.Denver DR, et al. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37(5):544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- 38.Hubbard TJ, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(Database issue):D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CL, Eppig JT. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm Genome. 2012;23(9-10):653–668. doi: 10.1007/s00335-012-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theiler K. The House Mouse: Atlas of Embryonic Development. Springer; New York: 1972. [Google Scholar]
- 41.Stevenson P, Richardson L, Venkataraman S, Yang Y, Baldock R. The BioMart interface to the eMouseAtlas gene expression database EMAGE. Database (Oxford) 2011;2011:bar029. doi: 10.1093/database/bar029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayamizu TF, et al. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J Biomed Semantics. 2013;4(1):15. doi: 10.1186/2041-1480-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao B-Y, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23(8):378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21(6):832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 45.Kim SH, Yi SV. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica. 2007;131(2):151–156. doi: 10.1007/s10709-006-9125-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.