In the thymus, high PU.1 expression persists through multiple cell divisions in early stages but then falls sharply during T-cell lineage commitment. Here, Champhekar et al. show that PU.1 is needed for full proliferation, restricting access to some non-T fates, and controlling the timing of T-cell developmental progression. Genome-wide transcriptome analysis identifies novel targets of PU.1-positive and PU.1-negative regulation affecting progenitor cell signaling and cell biology and indicating distinct regulatory effects on different subsets of progenitor cell transcription factors.
Keywords: conditional knockout, hematopoiesis, in vitro development, Notch, obligate repressor, RNA-seq
Abstract
The ETS family transcription factor PU.1 is essential for the development of several blood lineages, including T cells, but its function in intrathymic T-cell precursors has been poorly defined. In the thymus, high PU.1 expression persists through multiple cell divisions in early stages but then falls sharply during T-cell lineage commitment. PU.1 silencing is critical for T-cell commitment, but it has remained unknown how PU.1 activities could contribute positively to T-cell development. Here we employed conditional knockout and modified antagonist PU.1 constructs to perturb PU.1 function stage-specifically in early T cells. We show that PU.1 is needed for full proliferation, restricting access to some non-T fates, and controlling the timing of T-cell developmental progression such that removal or antagonism of endogenous PU.1 allows precocious access to T-cell differentiation. Dominant-negative effects reveal that this repression by PU.1 is mediated indirectly. Genome-wide transcriptome analysis identifies novel targets of PU.1 positive and negative regulation affecting progenitor cell signaling and cell biology and indicating distinct regulatory effects on different subsets of progenitor cell transcription factors. Thus, in addition to supporting early T-cell proliferation, PU.1 regulates the timing of activation of the core T-lineage developmental program.
The hematopoietic transcription factor PU.1 plays a central role in lymphoid and myeloid cell fates. PU.1-null animals die before birth due to hematopoietic failure, and conditional knockout PU.1 hematopoietic stem cells (HSCs) fail to contribute to lymphoid, myeloid, or dendritic cell lineages (Scott et al. 1994; McKercher et al. 1996; Dakic et al. 2005; Iwasaki et al. 2005; Nutt et al. 2005; Carotta et al. 2010). In the case of myeloid cells, dendritic cells, and B lymphocytes, PU.1 directly participates in the regulation of genes used for mature functions. However, it has other roles as well, since it is essential for the generation and survival of progenitors for multiple lineages, including those like T cells, where its role is restricted to early phases (Scott et al. 1994; Spain et al. 1999; Dakic et al. 2005). In fact, T-lineage cells must silence PU.1 completely before they are mature (Anderson et al. 2002). PU.1 binds to tens of thousands of genomic sites, which differ in erythroid cells, myeloid cells, B cells, and early T cells (Ghisletti et al. 2010; Heinz et al. 2010; Wontakal et al. 2011, 2012; Ridinger-Saison et al. 2012; Zhang et al. 2012; Ostuni et al. 2013), but in all of these cases, the majority of the binding sites detected are likely to be nonfunctional. To clarify why this regulatory protein is so important for hematopoiesis, we dissected its role in a developmental pathway where its function is restricted to the earliest uncommitted progenitor states: the T-cell pathway.
Multipotent precursors from the bone marrow home to the specialized microenvironment of the thymus, which induces their entry into the T-lineage developmental program. The earliest T-cell precursors in the thymus, called early thymic progenitors (ETPs) or Kit+ DN1 (DN is double negative, CD4− CD8−; Kit+ DN1 is Kit++ CD44+ CD25−), still harbor the potential to develop into at least five different blood lineages. These cells begin acquiring T-cell markers in the next stage, DN2a (Kit++ CD44+ CD25+), but commitment to the T lineage occurs only at the following stage, DN2b (Kit+ CD44+ CD25+) (Yui et al. 2010). T-cell receptor (TCR) expression begins in the following stage, DN3 (Kit− CD44− CD25+). Notch signals triggered by the Delta-like 4 (DL4) ligand expressed on thymic stromal cells drive this process by promoting the T-lineage fate and blocking access to alternate lineages (Ikawa et al. 2006; Rothenberg 2007; Hozumi et al. 2008). Development through these early DN stages is accompanied by the precisely coordinated expression of transcription factor-coding genes, including Lyl1, Scl (or Tal1), and Spi1 (encoding PU.1), in the early stages until commitment. Meanwhile Tcf7 (encoding TCF-1), Gata3, Tcf12 (encoding HEB), Runx1, Bcl11b, and Gfi1 are up-regulated, with sustained expression of Ikaros, Myb, and Tcfe2a (or Tcf3, encoding E2A), among others, which are indispensible for T-cell development (for review, see Yui and Rothenberg 2014).
Like the other factors expressed only in early stages, expression of PU.1 is inherited by early T cells from their prethymic precursors (Rothenberg et al. 2010). PU.1 expression in the thymus is highest in DN1 and DN2a cells, decreases with commitment at the DN2b stage, and is finally shut off in the DN3 stage (Anderson et al. 1999; Yui et al. 2010). The silencing of PU.1 expression is an important event, since continued PU.1 expression can block T-cell development at a DN2-like stage or divert early T cells to the myeloid lineage if Notch signaling from the environment is interrupted (Anderson et al. 2002; Lefebvre et al. 2005; Franco et al. 2006; Laiosa et al. 2006). Even inefficient silencing of PU.1 can cause T-cell leukemia (Rosenbauer et al. 2006). PU.1 may thus contribute to the initial developmental plasticity of the cells, especially their access to PU.1-dependent myeloid cell, B-cell, and dendritic cell fates. PU.1 expression in multipotent precursors is indispensable for the development of T-lineage cells, as T cells fail to develop in a germline PU.1 knockout model and PU.1 expression in adult HSCs is also required for their T-lineage potential (Scott et al. 1994; Dakic et al. 2005). However, the question has remained for PU.1 as to how it contributes to the development or maintenance of early intrathymic T-cell precursors in which it is expressed.
Understanding the function of PU.1 is challenging because it is critical for viability in prethymic lymphoid precursors. Loss of PU.1 function subjects lymphoid precursors to loss of competitiveness, whereas gain of function to levels like those in myeloid cells disrupts the T-cell program as well. To dissect stage-specific functions of PU.1 in the dynamic context of early T-cell development, we needed new approaches for synchronized, efficient removal of endogenous PU.1 activity. We therefore combined developmentally staged deletion of a conditional knockout allele with introduction of dominant-negative or hypomorphic competitor PU.1 constructs to perturb specific aspects of PU.1 activity. Our results show that PU.1 is important for early T-cell proliferation and may even help to restrict the developmental options available to these cells. Genome-wide analysis reveals multiple signaling and cell-biological functions of early pro-T cells that are directly regulated by PU.1. Importantly, we also found that PU.1 regulates the developmental timing of expression of several key T-lineage genes through an indirect network linkage revealed by the effects of an obligate repressor form of PU.1. PU.1 thus ultimately regulates the threshold for progression during early T-cell development.
Results
PU.1 is required for DN progression and survival of lymphoid precursors in T-cell differentiation culture
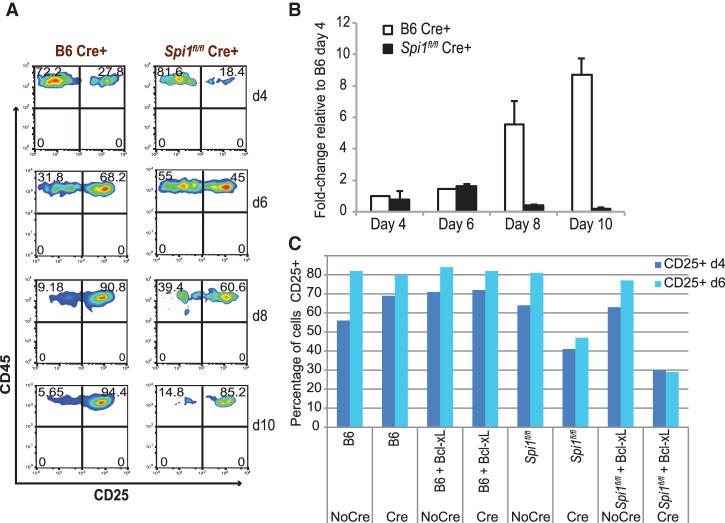
To investigate whether PU.1 contributes to T-cell development during the early intrathymic stages or is only important in prethymic precursors, we first used a conditional deletion strategy. We deleted PU.1 at specific stages in early T-cell precursors and tracked the effects on subsequent development in OP9-DL1 coculture, an in vitro system that mimics the thymic environment. First, retrovirally transduced Cre was used to delete Spi1 from c-Kit+ CD27+ Lin− multilineage hematopoietic precursors from embryonic day 14.5 (E14.5) fetal livers (fetal liver precursors [FLPs]). Cre+ cells from Spi1-targeted and control B6 mice were then sorted and tracked through culture by their coexpression of the retroviral NGFR marker. Deletion of Spi1 did not immediately reduce cell yield in PU.1-deleted cells as compared with Cre-treated wild-type controls. However, the PU.1-deleted FLPs were inefficient in their ability to differentiate into T-lineage cells, as seen by the delayed generation and reduced accumulation of CD25+ DN2 cells at days 4–8 (Fig. 1A,B). Over 8–10 d of T-cell differentiation culture, cells with deleted Spi1 generated considerably fewer progeny (Fig. 1B).
Figure 1.
Deletion of PU.1 in c-Kit+ CD27+ FLPs results in impaired DN progression and poor survival and recovery of early DN stage T cells. (A) E14.5 B6 and Spi1fl/fl FLPs were infected with Cre-expressing retroviral supernatant. One day after the infection, Cre+ Kit+ CD27+ cells were sorted and used to start OP9-DL1 cultures. These cultures were harvested on the indicated days and analyzed for DN progression as shown in the figure. FACS plots are representative of two independent experiments. (B) Plot summarizing Cre+ Spi1fl/fl cell counts expressed relative to the number of Cre+ B6 cells on day 4 of OP9-DL1 culture, from two independent experiments. Error bars indicate one standard deviation. (C) Failure of Bcl-xL to rescue progression to CD25+ (DN2/DN3) stages when PU.1 is deleted. Cells were prepared as in A, except that they were doubly transduced with Cre (NGFR+) and Bcl-xL (GFP+) retroviruses or controls. Results are from one experiment, representative of two independent experiments.
The reduced development of CD25+ DN2 cells from PU.1-deficient precursors could not be reversed simply by cotransducing Bcl-xL in a GFP+ retrovirus along with Cre to inhibit apoptosis (Fig. 1C; Supplemental Fig. S1A). Sustained expression of Cre itself was toxic to both wild-type and Spi1fl/fl cells (Supplemental Fig. S1B), but even at equal survival rates, the differentiation of the Spi1fl/fl Cre+ cells to CD25+ DN2 stage was specifically impaired in comparison with B6 Cre+ cells (Fig. 1C; Supplemental Fig. S1A, arrows). Bcl-xL improved the recovery of DN1 cells with complete Spi1 deletion, but, with or without Bcl-xL, production of CD25+ (DN2–DN3) cells was impaired in the absence of PU.1 (Fig. 1C; arrows in Supplemental Fig. S1A). Thus, PU.1 has an important role in the early T-cell developmental competence of Kit+ CD27+ FL precursors.
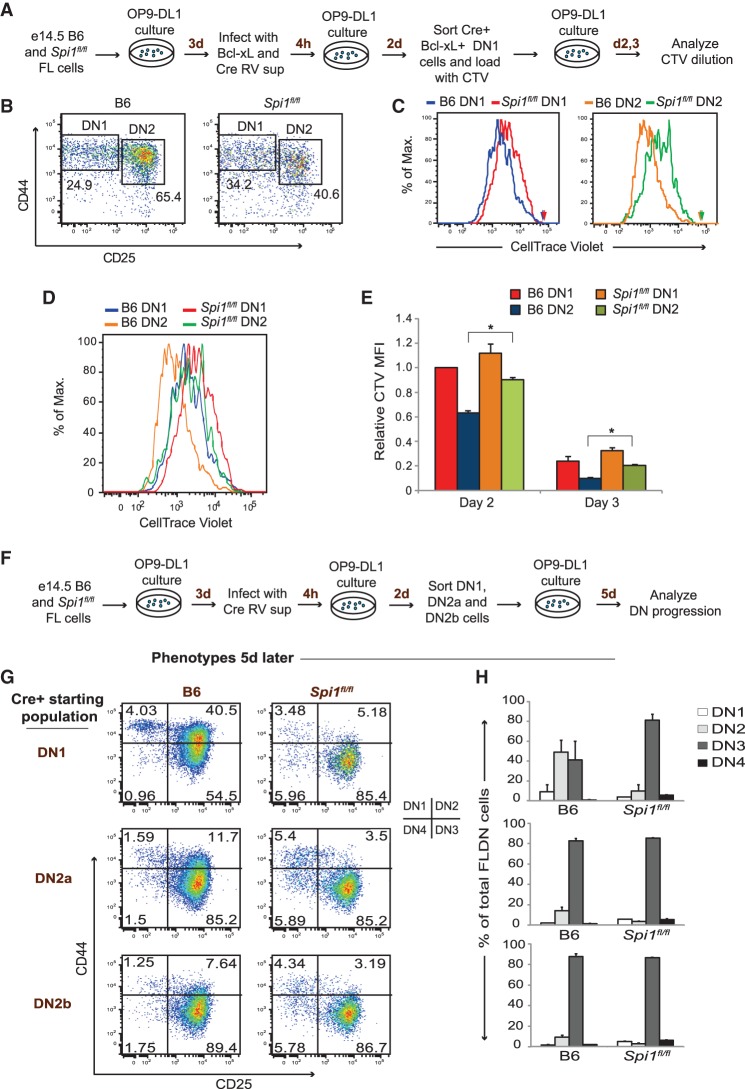
To test whether PU.1 still affected differentiation or proliferation once T-cell development was actually under way, we initiated T-cell development from wild-type and Spi1fl/fl precursors in OP9-DL1 coculture first, generating a pool of cells ranging from the ETP/DN1 to the DN2b stages, then transduced the cells with Cre and Bcl-xL for 48 h, and sorted the transduced DN1 cells for analysis and reculture. Once sorted, these cells could be tracked even if they later silenced retroviral expression (Anderson et al. 2002) to escape Cre toxicity. Although both wild-type and PU.1 knockout cells proliferated, PU.1 was indeed required for optimal proliferation. We stained the sorted DN1 cells with the cell cycle tracker CellTrace violet (CTV), returned them to culture, and then analyzed CTV dilution in their DN1 and DN2 progeny after two to three more days (Fig. 2A–E). Normally, proliferation accelerates between the ETP/DN1 and DN2 stages (Fig. 2B; Manesso et al. 2013), but this acceleration did not occur in PU.1-deficient cells. Instead, PU.1 knockout DN2 cells proliferated significantly less than their wild-type counterparts on both day 2 and day 3 of culture (Fig. 2D,E). Thus, PU.1 is needed for optimal proliferation at the DN2 stage.
Figure 2.
PU.1 retards DN progression and enhances proliferation of early T cells. (A) Scheme for cell cycle progression analysis of progeny of PU.1-deleted DN1 cells. B6 and Spi1fl/fl FLPs were cultured on OP9-DL1 cells for 3 d and infected with Cre-expressing and Bcl-xL-expressing retroviruses. Two days later, Cre+ Bcl-xL+ DN1 cells were sorted, loaded with CTV dye, and put back in fresh OP9-DL1 cultures. Two days and 3 d later, population phenotypes and CTV fluorescence were determined by FACS analysis. (B) Phenotypes of recultured cells at day 3. (C) Extents of CTV dilution for progeny of labeled DN1 cells, comparing PU.1-deleted DN1 and DN2 progeny with controls. Arrows indicate the day 0 CTV mean fluorescence intensity (MFI) for each population shown in the histogram. (D) All four populations from the histograms in C, superimposed to show the differences in proliferation of various subsets. (E) Relative proliferation of B6 and Spi1fl/fl DN cell populations measured by the differences in dilution of CTV at day 2 and day 3. CTV MFI for each population was normalized to the initial loading value and then plotted relative to the day 2 control B6 DN1 value. (F) Flowchart showing the design of DN stage progression experiments. E14.5 B6 and Spi1fl/fl FLPs were cultured on OP9-DL1 for 3 d and infected with a Cre-expressing retrovirus. Cre+ DN1, DN2a, and DN2b populations were sorted and used to seed the cultures analyzed 5 d later in G and H. (G) DN stage progression of cells descended from the indicated starting populations, based on CD44 and CD25 expression. The diagram at the right shows populations defined by the four quadrants. (H) Population distributions of progeny subsets descended from the indicated Cre+ starting populations, averaged from two independent DN progression experiments as in F and G. Error bars represent one standard deviation.
Over 3–5 d, the loss of PU.1 also had a striking effect on differentiation. Despite less overall proliferation, cells that had deleted PU.1 at the ETP or DN2a stages consistently differentiated faster than controls, as evaluated by the cell surface expression of classic DN subset markers CD44 and CD25 (Fig. 2F,G). When Cre was introduced in either the ETP or DN2a starting population, PU.1-deleted cells progressed more completely from the CD44+ CD25+ DN2 stage to the CD44− CD25+ DN3 stage than their B6 counterparts by the time of harvest. In contrast, there was no difference from the controls in the developmental profiles of cells that lost PU.1 at the DN2b stage, when endogenous PU.1 expression is already down-regulated and the cells have already committed to the T lineage. Thus, in early T cells, PU.1 normally supports population expansion while restraining differentiation.
PU.1 temporally restricts activation of the T-lineage developmental program
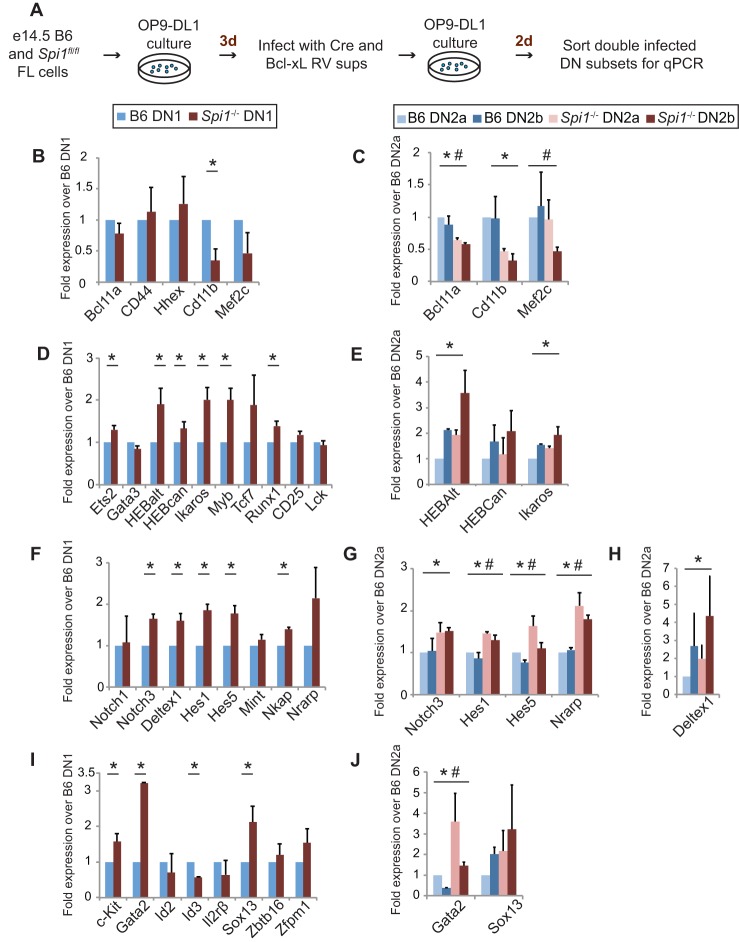
To evaluate the effect of PU.1 loss on T-lineage progression as a whole, we examined a well-characterized spectrum of genes under known developmental control (Zhang et al. 2012; Mingueneau et al. 2013; Yui and Rothenberg 2014). We transduced Cre and Bcl-xL into Spi1fl/fl or B6 control FL-derived DN (FLDN) cells after 3 d of differentiation as in Figure 2A, returned them to OP9-DL1 culture for two more days, and finally collected the Cre+, Bcl-xL+ cells sorted into DN1, DN2a, and DN2b fractions for RNA analysis (Fig. 3A; Supplemental Fig. S2A). Cre caused efficient stage-specific deletion of Spi1, as shown by the decrease in Spi1 (PU.1) RNA in subset populations that were still phenotypically matched to the controls (Supplemental Fig. S2B). As PU.1 protein is very stable (Kueh et al. 2013), it could take at least 2 d to clear pre-existing PU.1 protein from the cells, but early changes in gene expression emerged as PU.1 levels decreased.
Figure 3.
Deletion of Spi1 leads to derepression of T-lineage genes. (A) Flowchart for obtaining B6 and Spi1fl/fl DN1, DN2a, and DN2b cells to determine the effect of loss of PU.1 on the T-lineage developmental program. cDNA was prepared from DN subsets sorted in A, and gene expression changes were measured in the indicated populations using quantitative PCR (qPCR). (B–J) Actin-normalized expression values averaged from two independent experiments are expressed as the fold change relative to the B6 DN1 values (B,D,F,I) or the B6 DN2a values (C,E,G,H,J). The resulting data were plotted for phase 1 and alternate lineage genes (B,C), T-lineage genes (D,E), components of the Notch signaling pathway (F–H), and alternate lineage genes (I,J). Error bars represent one standard deviation. Significant differences at P < 0.05 are indicated with an asterisk (DN1 and DN2) and pound sign (DN2b).
Previous work had identified genes that can be up-regulated or down-regulated by forced high-level PU.1 expression in early T cells (Anderson et al. 2002; Dionne et al. 2005; Franco et al. 2006; Laiosa et al. 2006; Del Real and Rothenberg 2013), but it was not clear until now whether normal developmental expression levels of these genes depend on endogenous levels of PU.1. Although the effects were weak, acute deletion showed that several genes expressed specifically in the DN1 and early DN2 stages do depend on full expression of PU.1 in these early T-lineage cells. Cd11b (Itgam and Mac1), Mef2c, and Bcl11a were all detectably down-regulated in PU.1 knockout cells as compared with the corresponding B6 cells (Fig. 3B,C; Supplemental Fig. S3A,B). In contrast, T-lineage-associated genes—including two isoforms of Tcf12 (HEBalt and HEBCan), Ikaros, Runx1, Ets2, and Myb, most of which increase in expression between DN1 and DN2b/DN3—were significantly up-regulated in PU.1 knockout subsets as compared with their B6 counterparts, and increased Tcf7 expression was also seen in some DN1 samples (Fig. 3D,E; Supplemental Fig. S3A,C). Overexpressed PU.1 can antagonize Notch signaling in early T cells (Del Real and Rothenberg 2013), and, accordingly, multiple Notch-activated genes like Notch3, Deltex1, Hes1, Hes5, and HEBalt were up-regulated in the absence of PU.1 in all DN subsets (Fig. 3F–H; Supplemental Fig. S3A,D). This effect was not just an aspect of developmental acceleration. Notably, Nrarp, the lone Notch target gene that naturally decreases in expression from DN1 to DN2b, was also up-regulated in the PU.1-deficient DN2a and DN2b cells relative to controls. Thus, endogenous PU.1 normally restrains the expression of multiple Notch targets, with the strongest effects at stages when their expression is in transition.
Interestingly, loss of PU.1 also led to the up-regulation of additional genes that are expressed during the early T-cell stages but whose best-known function is to support the development of variant or alternate lineages. These included Sox13 (γδ T cells) and Gata2 (mast cells or progenitor cells), which were significantly up-regulated (Fig. 3I; Supplemental Fig. S3A,E), and, in some samples, Zbtb16 (PLZF and innate lymphocytes) as well (Supplemental Fig. S3). PU.1 loss also regularly resulted in higher Kit expression. Although these effects were weak, they were likely to be functionally significant. When PU.1-deleted cells were transferred to permissive conditions for natural killer (NK) cell development, progeny of PU.1-deficient FLPs regularly yielded more NK cells than controls (Supplemental Fig. S4). Taken together, these data raise the possibility that PU.1 not only limits the up-regulation of T-cell program genes but also may play a role in restricting the range of alternative lineage potentials of developing thymocytes during the stages when it is expressed.
PU.1-Engrailed (PU.1-Eng) fusion construct as a tool to investigate the role of Spi1 in early T cells
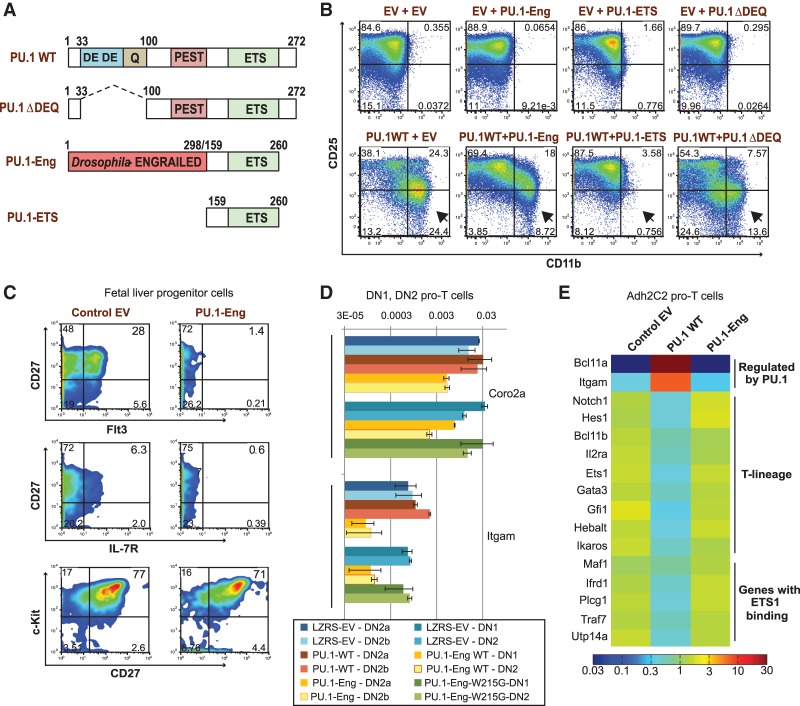
Conclusions from the knockout data alone were weakened by the modest size of the effects that we could measure in this dynamic developmental context. Although Cre recombinase could excise Spi1 DNA within 24 h after infection, the effects were weak until pre-existing PU.1 protein was cleared from the cells. Protein was clearly reduced by 48 h (data not shown), but the increased time could also increase the indirect effects from altered developmental progression. We therefore sought alternative strategies to neutralize PU.1 activity quickly and synchronously at a particular stage and time point. To competitively inhibit endogenous PU.1 protein, we designed modified PU.1 constructs that would incorporate the full PU.1-ETS DNA-binding domain but delete its transactivation and other protein interaction domains (Fig. 4A).
Figure 4.
PU.1 constructs retain target gene specificity and are able to block wild-type (WT) PU.1 function efficiently. (A) Cartoon showing the domain organization of wild-type PU.1 and the various PU.1 constructs used in this study. (B) PU.1-Eng, PU.1-ETS, and PU.1-ΔDEQ can block wild-type PU.1 activity. Adh-2C2 cells were infected with various constructs, as indicated at the top of each panel. After 48 h, diversion to the myeloid lineage was determined by analyzing the surface up-regulation of CD11b. The data in the FACS plots are representative of two independent experiments. (C) Repression of Il7r and Flt3 in progenitors by PU.1-Eng. E14.5 B6 FLPs were transduced with PU.1-Eng and cultured without OP9 stromal cells in medium supplemented with 1 ng/mL SCF and 5 ng/mL each Flt3L and IL-7. Cultures were harvested after 48 h and analyzed by flow cytometry for the indicated markers (representative of two independent experiments). (D) Repression of Itgam and Coro2a depends on the integrity of the PU.1-Eng DNA-binding domain. Plots show gene expression measured by qPCR in sorted FLDN cells 24 h after transduction with empty vector (EV; blue), PU.1 wild type (red), PU.1-Eng (gold), or PU.1-Eng W215G (green). The effects of PU.1-Eng were compared with PU.1 wild type in DN2a and DN2b cells (two experiments), and PU.1-Eng was compared with PU.1-Eng W215G in DN1 and total DN2 cells (two separate experiments). Values (average ± range) are shown relative to Actb on a log10 scale. (E) The lack of effect of PU.1-Eng in Adh.2C2 pro-T cells. Adh.2C2 cells were transduced with the constructs indicated and harvested at 48 h for gene expression analysis by qPCR. Average, Actin-normalized log10 expression values from two independent experiments were used to generate the heat map shown in the figure. All values are row-normalized, and the color scale at the right denotes 30-fold up-regulation (dark red) and 30-fold down-regulation (dark blue) of gene expression.
To enhance detection of competitive inhibitor effects, we created a dominant interfering form of PU.1 with known repressive activity. A PU.1-Eng fusion was constructed (Fig. 4A) in which the repression domain of the Drosophila Engrailed protein is fused to the DNA-binding ETS domain of PU.1, maintaining the DNA-binding specificity of PU.1. By recruitment of Groucho/TLE family corepressors to the Engrailed domain, this should consistently repress PU.1 target genes, antagonizing wild-type PU.1 function where wild-type PU.1 activates and paralleling wild-type PU.1 function where it represses (Han and Manley 1993; Jimenez et al. 1997; Tolkunova et al. 1998). Besides PU.1-Eng, we created PU.1-ETS, a construct expressing only the DNA-binding ETS domain of the PU.1-Eng fusion construct, to serve as a control for the passive target site-binding activity of PU.1-Eng. An additional construct, PU.1-ΔDEQ, deleted only the acidic and glutamine-rich transactivation domains.
Specificity of PU.1-Eng: antagonism of PU.1-dependent gene activation without effects on other ETS family targets
The target specificity and ability of these constructs to block wild-type PU.1 function were verified using lineage diversion of the DN3-like Adh.2C2 thymoma cell line by wild-type PU.1 (Dionne et al. 2005; Del Real and Rothenberg 2013). These cells do not express any PU.1, but ectopic expression of wild-type PU.1 in Adh.2C2 cells causes dose-dependent myeloid diversion marked by down-regulation of T-cell surface marker CD25 (Il2rα) and up-regulation of the myeloid marker CD11b (Itgam) (Fig. 4B). In contrast to wild-type PU.1 (Fig. 4B, WT), PU.1-Eng and PU.1-ETS could not transform the Adh.2C2 cells to CD25−/low CD11b+ cells (Fig. 4B, top) despite high-level expression and lack of toxicity. However, when coexpressed with wild-type PU.1, both PU.1-ETS and PU.1-Eng dominantly inhibited PU.1-driven myeloid diversion, verifying that they could competitively neutralize normal PU.1 (Fig. 4B, bottom row). While PU.1-ΔDEQ was able to reduce myeloid diversion, it was the weakest inhibitor of wild-type PU.1 in the group.
In primary multilineage precursor cells, Kit+ CD27+ Lin− FLPs, PU.1-Eng could also block expression of known positive regulatory targets of PU.1; i.e., growth factor receptor genes Flt3 and Il7r (DeKoter et al. 2002; Carotta et al. 2010). Transduction of PU.1-Eng caused sharp down-regulation of both surface Flt3 and IL-7 receptors within a day. The effect was gene-specific, since the Kit+ CD27+ cells were still present (Fig. 4C) at day 1. However, loss of these growth factor receptors was followed by severe loss of the precursors 1 d later, and subsequent lymphoid development was profoundly inhibited. Thus, PU.1-Eng acts faster but has effects on this population similar to those of PU.1 deletion (cf. Fig. 1; Carotta et al. 2010).
Transduction of PU.1-Eng into primary DN1 and DN2a/DN2b pro-T cells had dramatic effects on gene expression, as described in detail below. Here, PU.1-Eng down-regulated both Itgam and another gene, Coro2a, by more than fourfold within 24 h (Fig. 4D). These effects were abolished by the single amino acid mutation W215G, which disrupts DNA-binding activity (Fig. 4D). To assess whether these effects truly reflect competition with endogenous PU.1, we also measured PU.1-Eng “side effects” by testing whether it also affects gene expression in a later pro-T-cell context where endogenous PU.1 is already silent. For this we used the parental Adh.2C2 pro-T-cell line, which does not express any PU.1 naturally but can express other ETS family target genes through the action of other ETS factors; e.g., Ets1, Ets2, and Fli1. When we introduced PU.1-Eng into Adh.2C2 cells alone, we found that T-cell genes in general and even those previously linked to Ets1-binding peaks in human cells (Hollenhorst et al. 2009) were unchanged in expression (Fig. 4E) despite strong accumulation of PU.1-Eng protein (data not shown). The lack of PU.1-Eng effect contrasted with that of wild-type PU.1, which caused repression of many of these genes in Adh.2C2 cells, including Ets1 itself (Fig. 4E; Dionne et al. 2005; Del Real and Rothenberg 2013). Thus, the PU.1-Eng construct is selective and does not compete with ETS factors generally.
Negative regulation of T-cell development by PU.1 is indirect: PU.1-Eng causes derepression of the T-lineage developmental program
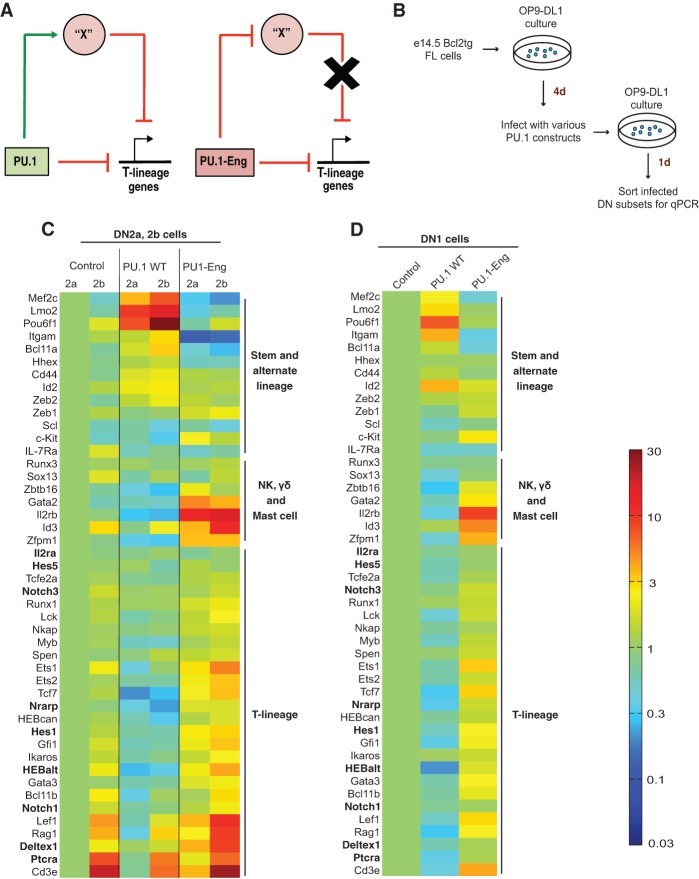
The fact that PU.1 deletion accelerates T-lineage developmental progression implies that the ability of overexpressed PU.1 to repress multiple T-lineage genes (Fig. 4E; Dionne et al. 2005; Franco et al. 2006; Del Real and Rothenberg 2013) is physiologically significant. On one hand, PU.1 could repress each of these genes individually through their cis-regulatory elements, either through dedicated repressive sites or by interfering with access by positively regulating ETS factors. Alternatively, it could positively regulate the expression of some intermediate repressors that in turn repress the T-lineage program. As shown in Figure 5A, if PU.1 directly repressed T-lineage genes, PU.1-Eng fusion protein should have the same effect owing to its obligate repressor activity. In contrast, if PU.1 inhibited them indirectly by positively regulating expression of a repressor, then these T-lineage identity genes would be among those up-regulated by PU.1-Eng.
Figure 5.
PU.1-Eng transduction up-regulates T-lineage-specific gene expression. (A) Cartoon showing alternative mechanisms by which wild-type PU.1 could repress T-cell genes and the use of PU.1-Eng to distinguish between these possibilities. (Left panel) PU.1 could directly repress T-lineage genes by recruiting repressors to silence each gene individually or up-regulate the expression of a repressor “X” that could in turn repress the T-cell program. The effects of obligate repressor PU.1-Eng under the two scenarios are shown in the right panel. (B) Schematic of experiments to generate the data shown in C and D and Supplemental Figure S6, A–F. Bcl2-tg FLDN cells were used to enhance viability under perturbation. They were processed as shown and sorted to purify DN1, DN2a, and DN2b cells for gene expression analysis by qPCR. (C,D) The retroviral supernatants used to express various constructs are indicated above each lane. Sorted DN stage cells from two to three independent experiments were pooled to obtain each set of qPCR data. Average, Actin-normalized log10 expression values from two such pooled data sets were used to generate the heat maps shown in the figure. All values were row-normalized to the control DN2a (C) or control DN1 (D) sample, and the common color scale at the right denotes 30-fold up-regulation (dark red) and 30-fold down-regulation (dark blue) of gene expression. Boldface gene names indicate Notch-activated genes.
In primary DN2a and DN2b cells, PU.1-Eng had a pronounced effect on T-lineage-specific genes. As previously reported, multiple T-lineage genes were down-regulated in primary pro-T cells overexpressing wild-type PU.1 protein, whereas the obligate repressor PU.1-Eng construct had the reverse effect, up-regulating T-lineage genes relative to empty vector (EV) controls (Fig. 5C,D; cf. Fig. 4E; Supplemental Figs. S5B, S6A–F [representative bar graphs]). Significantly, this set included almost all of the T-cell genes that were up-regulated in the PU.1 knockout cells described above (Fig. 3B–E), but the magnitudes of the effects here were much greater. Thus HEBAlt, HEBCan, Tcf7, Gata3, Ets1, Ets2, Lef1, Rag1, Cd3e, Ptcra, Deltex1, Gfi1, and Ikaros were all up-regulated by PU.1-Eng and down-regulated by PU.1 wild type. Strikingly, stage-specific T-lineage genes were also turned on precociously in PU.1-Eng-expressing cells. PU.1-Eng transduced ETP/DN1 cells turned on expression of genes that have strictly DN2- and DN3-specific timing of activation in normal cells (Fig. 5D; Supplemental Fig. S6A). These included important T-lineage regulators like Ets1, Bcl11b, and Lef1 as well as the recombinase gene Rag1 and the TCR complex gene Cd3e (Yui et al. 2010; Zhang et al. 2012). These results support the smaller effects in the same direction seen upon conditional deletion of the Spi1 gene (cf. Fig. 3), indicating that PU.1 normally limits the timing as well as magnitude of activation of T-cell genes.
These effects depended on the repression domain of PU.1-Eng rather than a simple competition for DNA occupancy because PU.1-ETS and also PU.1-ΔDEQ were virtually ineffective at activating any T-cell genes (Supplemental Fig. S5B). The effects were also not an artifact of forced Engrailed domain expression because expression of PU.1-Eng with a mutated DNA-binding domain (PU.1-Eng W215G) did not up-regulate these genes (Supplemental Fig. S5C). Thus, the up-regulation of T-cell genes by PU.1-Eng implies that endogenous PU.1 normally represses these genes indirectly to cause T-cell differentiation delay.
Activation of Notch target genes and alternative lineage genes by obligate repressor PU.1
PU.1 could indirectly repress these genes in normal cells by positively regulating the expression of a T-lineage-specific repressor or a Notch signaling antagonist. Indeed, complementing previous results with PU.1 gain of function (Del Real and Rothenberg 2013), expression of Notch1 itself was only slightly increased in PU.1-Eng-expressing cells, but many direct targets of Notch signaling—including Hes1, Deltex1, HEBalt, and Nrarp—were up-regulated (Fig. 5C,D, boldface gene names; Supplemental Fig. S5B). Since the cells were assayed just a day after PU.1-Eng transduction, these changes could reflect a rapid enhancement of signaling by existing Notch1 rather than longer-term pathway reinforcement due to network-level positive feedback. Thus, at least part of the activation of the T-cell program could reflect removal of a constraint on Notch signaling.
In addition to the effects on T-lineage genes, we also found that several alternate lineage genes were also down-regulated by wild-type PU.1 but up-regulated in response to PU.1-Eng expression in the DN1/ETP stage (Fig. 5D). These included NK or innate lymphoid cell (Zbtb16 and Il2rb) and mast cell (Cpa3 and Gata2) lineage-associated genes as well as the broadly used progenitor cell genes like Kit and Zfpm1. These stronger effects confirmed the results seen when many of these genes were also up-regulated in the PU.1 knockout FLDN cells described above (Fig. 3E). Thus, PU.1 may help to constrain access to innate lymphoid lineage fates even while it preserves access to myeloid and dendritic fates during the early phase of T-cell development.
Genome-wide analysis of genes regulated by PU.1-Eng
The interpretation of these PU.1-Eng effects relies on the assumption that this construct selectively represses physiological PU.1 target genes. To evaluate this critically, we carried out genome-wide analysis of the early impacts of PU.1-Eng and PU.1-ETS expression on the DN2a pro-T-cell transcriptome, comparing these with global patterns of endogenous PU.1 binding and global developmental regulation. To identify genes that undergo rapid changes in expression in response to PU.1-Eng, primary FLDN2a cells were sorted ∼20 h after transduction with EV, PU.1-Eng, or PU.1-ETS and were analyzed using RNA sequencing (RNA-seq). Such DN2a cells are unambiguously engaged in the T-cell pathway yet are still high in natural PU.1 expression (Yui et al. 2010; Scripture-Adams et al. 2014). Cells from Bcl2 transgenic (Bcl2-tg) mice were used as in Figure 5 to improve recovery of cells after regulatory perturbation (Franco et al. 2006; Taghon et al. 2007). The early time point minimized indirect effects and ensured that the cell surface phenotype was not yet altered (Supplemental Fig. S7). At this time point, PU.1-Eng had little if any impact on the major cell cycle regulators expressed (Supplemental Fig. S6G).
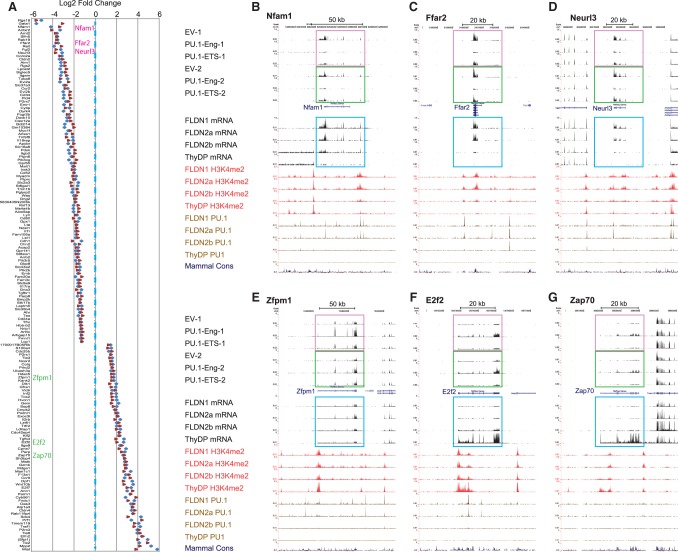
The six samples from two independent experiments were all still highly concordant with each other in gene expression levels at harvest (Pearson's r 0.84–0.91 for all pairwise comparisons), with EV and PU.1-ETS transduced samples being closest. DESeq comparisons showed few genes significantly affected in the PU.1-ETS transduced samples (Supplemental Table S1). However, in the PU.1-Eng transduced cells, there were selective, reproducible gene expression changes despite the short duration of the perturbation. Of the genes meeting the threshold statistical criteria (log2 fold change > |1|, false discovery rate [FDR] < 0.1 in at least one and FDR < 0.2 in both sets), 110 genes were reproducibly down-regulated, and 78 genes were up-regulated (Fig. 6A). Both down-regulated and up-regulated genes were comparable in their fold changes in expression, their reproducibility of response, and the geometric mean adjusted P-values for these effects in the two experiments (Fig. 6A; Supplemental Table S1). Examples of each group are shown in Figure 6, B–G, in which RNA-seq tracks for DN2 samples transduced with EV, PU.1-Eng, and PU.1-ETS in the two experiments (red and green boxes) are compared with RNA-seq tracks showing normal developmental expression change (blue boxes), with corresponding histone H3K4me2-marking tracks, and with PU.1-binding tracks from unperturbed DN1, DN2a, DN2b, and DP cell samples (Zhang et al. 2012). These results show that most of these regulated genes are associated with PU.1 occupancy sites in normal cells, often correlated with distinct H3K4me2 peaks, and that these genes may have highly pronounced changes in expression with development.
Figure 6.
Identification of positive and negative PU.1 target genes by genome-wide analysis. (A) Effects on expression of top-scoring PU.1-Eng-repressed and PU.1-Eng-activated genes in two independent RNA-seq experiments. Data from the two experiments are shown as fold change (log2 scale) from EV samples in the same experiment. Genes profiled in B–G are identified. (B–G) University of California at Santa Cruz (UCSC) genome browser portraits of three genes down-regulated by PU.1-Eng (B–D) and three genes up-regulated by PU.1-Eng (E–G). Boxed tracks show RNA-seq data from two different experiments (magenta and green) comparing EV, PU.1-Eng, and PU.1-ETS transduced cells with normal reference samples (blue) from DN1, DN2a, DN2b, and DP stages. Red tracks show positions of H3K4me2 histone marks, and brown tracks show binding of PU.1 based on chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) in the same reference cell types. Direction of transcription is right to left in B–D and left to right in E–G. Full scales for vertical axes in B–G are 2.5 reads per million (RPM) for all H3K4me2 ChIP-seq, 5 RPM for all PU.1 ChIP-seq, 1.5 RPM for all RNA-seq tracks in B–D, 5 RPM for all RNA-seq in E and F, and 3 RPM for all RNA-seq in G.
Selective regulation of genes by PU.1-Eng: correlation with endogenous PU.1 binding
If PU.1-Eng is really an obligate repressor and all up-regulation is indirect, then the repressed genes should be more enriched for direct targets of endogenous PU.1 than the up-regulated genes. We showed previously that endogenous PU.1-binding sites in DN1, DN2a, and DN2b pro-T cells are associated with many active promoters and enhancers genome-wide (Zhang et al. 2012). Among the genes rapidly affected by PU.1-Eng, many had multiple binding sites for endogenous PU.1 (Supplemental Table S2). These PU.1 peaks were not always found at promoters of the PU.1-Eng-affected genes: Many affected genes had only intronic or flanking intergenic PU.1 sites. Conversely, genes with PU.1 only at the promoters were often unaffected by PU.1-Eng entirely (e.g., Uggt1 and Arid5a, the genes flanking Neurl3 in Fig. 6D).
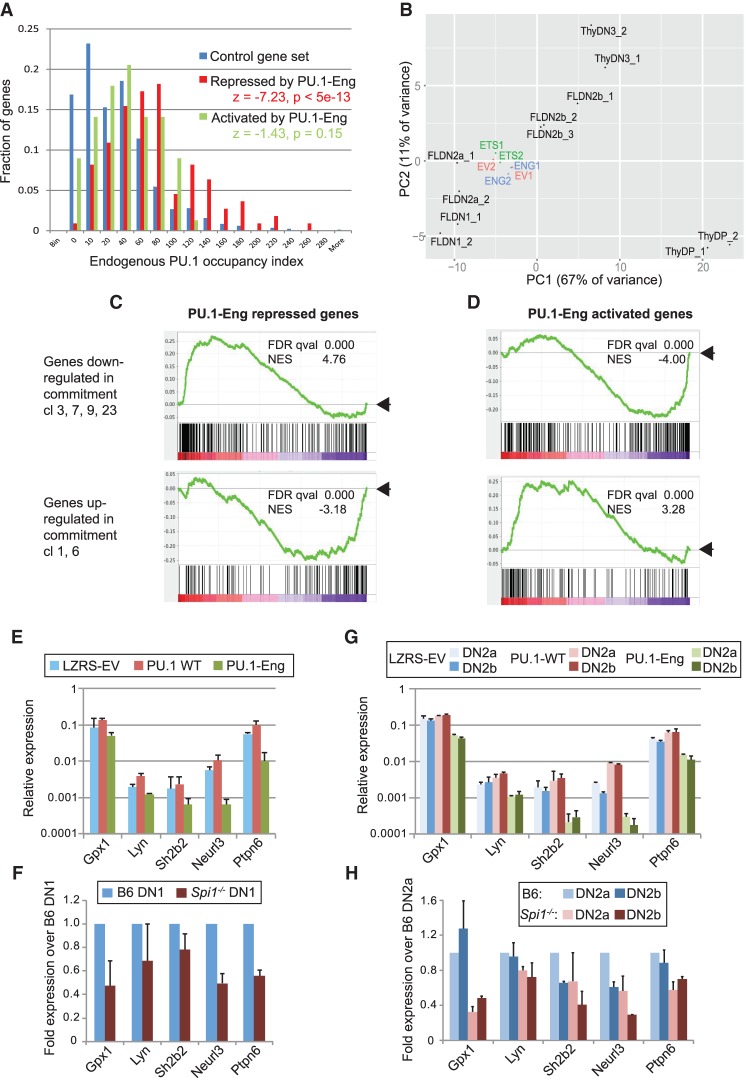
Notably, however, the densities and intensities of PU.1 occupancy in normal DN1 and DN2a cells (Zhang et al. 2012) were higher around the genes repressed by PU.1-Eng than around genes activated by PU.1-Eng or around genes unaffected by PU.1-Eng. For comparison, we calculated an in vivo PU.1 occupancy index for each gene by summing the intensities of chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) signals in DN1 and DN2a stages at the four strongest sites associated with that gene (Supplemental Table S2; see the Supplemental Material; data from Supplemental Table 5 in Zhang et al. 2012). We compared these occupancy index values for the highest-confidence lists of up-regulated and down-regulated genes (Fig. 6A) with each other and with a control set of 841 genes that are expressed stably throughout early T-cell development and are unaffected by PU.1-Eng (Fig. 7A, “control gene set”; Supplemental Table S2; Zhang et al. 2012). Figure 7A shows the distributions of PU.1 occupancy scores in the three groups as a histogram. PU.1 occupancy scores were not significantly different between the controls and the PU.1-Eng-activated genes but trended higher for the PU.1-Eng-repressed genes (Fig. 7A). The most strongly occupied genes were enriched among the genes most sensitive to PU.1-Eng repression, and these were also most likely to show weaker but detectable repression by PU.1-ETS (Supplemental Tables S1, S2). Thus, genes repressed by PU.1-Eng, but not genes activated by PU.1-Eng, were highly enriched for endogenous PU.1 occupancy at associated sites, as compared with the average occupancy for nonregulated genes.
Figure 7.
Characteristics of PU.1 target genes identified by genome-wide analysis. (A) Histogram comparing in vivo PU.1-binding distributions among genes repressed by PU.1-Eng (red), genes up-regulated by PU.1-Eng (light green), and control genes expressed stably and unaffected by PU.1-Eng (dark blue). The X-axis indicates histogram bins and summed ChIP-seq signals for each gene (at top four sites), and the Y-axis shows the fraction of genes in the set in the indicated bin. (B) Developmental indexing of the two independent PU.1-Eng (ENG), PU.1-ETS (ETS), and EV transduced samples as compared with normal reference cells (Zhang et al. 2012). Principal components were based on expression patterns of 173 regulatory genes, as described in the Supplemental Material (Supplemental Table S3). (C,D) Gene set enrichment analyses of the effects of PU.1-Eng on genome-wide transcription relative to normal developmental patterns of expression. The panels shown compare the enrichments of a set of developmentally down-regulated genes (cl. 3, 7, 9, and 23) and a set of developmentally up-regulated genes (cl. 1 and 6) against the full set of expressed loci (Supplemental Table S4J), ranked by fold change caused by PU.1-Eng. (NES) Normalized enrichment score; (positive NES) enriched; (negative NES) depleted. For full results, see Supplemental Table S4. (E–H) Effects of wild-type PU.1 addition or endogenous PU.1 deletion on PU.1-Eng-repressed target genes Gpx1, Lyn, Sh2b2, Neurl3, and Ptpn6 in FLDN1 cells (E,F) and FLDN2a and FLDN2b cells (G,H). Samples were prepared as described for PU.1 wild-type and PU.1-Eng transduced cells in Figure 5 (E,G) and for Cre+ control and Spi1fl/fl cells in Figure 3 (F,H). Gene expression levels determined by qPCR are shown on a log10 scale relative to Actb in E and G and on a linear scale relative to control levels in B6 DN1 or DN2a cells in F and H. Averages and the range of values from two independent experiments are shown.
Selective regulation of genes by PU.1-Eng: correlation with developmental regulation
All of the samples analyzed were still clearly DN2 cells, as shown by a developmental index based on expression of 173 regulatory genes (Fig. 7B; Scripture-Adams et al. 2014), which showed all six to cluster closely within the DN2a/DN2b interval (Supplemental Table S3; see the Supplemental Material). However, these cells express both genes that are down-regulated (including Spi1 itself) and genes that are up-regulated with development. To test whether PU.1-Eng preferentially affects genes that are expressed while endogenous PU.1 is present, we used gene set enrichment analysis (GSEA), exploiting our previous classification of all developmentally regulated genes into clusters according to expression patterns (Supplemental Table S4H,I; Zhang et al. 2012). We ranked 11,634 expressed genes according to their fold changes in response to PU.1-Eng or PU.1-ETS versus EV (see the Supplemental Material; Supplemental Table S4J) and then used GSEA to determine which expression pattern clusters were enriched at the top or bottom of the ranked list. PU.1-regulated gene sets previously defined in myeloid precursors by DeKoter and colleagues (Kamath et al. 2008) were also tested for enrichment. PU.1-ETS effects were insignificant, but PU.1-Eng had highly significant effects (Fig. 7C,D; Supplemental Table S4B–E). Genes with DN1–DN2a-biased expression patterns (clusters 3, 7, 9, and 23) were strongly overrepresented among genes repressed by PU.1-Eng (Fig. 7C, top), while genes that are up-regulated as endogenous PU.1 is shut off (clusters 1, 6, and 25) were depleted (Fig. 7C, bottom). Genes previously defined as PU.1 targets in myeloid cells (Kamath et al. 2008) were also enriched among PU.1-Eng-repressed genes (Supplemental Table S4B,I). In contrast, genes that increase in expression during T-cell commitment (clusters 1 and 6) and later (cluster 25) were more represented among genes up-regulated by PU.1-Eng (Fig. 7E,F; Supplemental Table S4D), confirming the acceleration effect inferred above (cf. Fig. 5). Thus, overall, genes repressed by PU.1-Eng, but not those activated by PU.1-Eng, were expressed at stages when endogenous PU.1 would naturally be present. In summary, although genes activated by PU.1-Eng include indirect targets, the PU.1-Eng-repressed genes are highly enriched for direct, functional targets of PU.1 regulation.
New targets for PU.1 activity in early T cells
The comparison of PU.1-Eng effects with those of forced expression of wild-type PU.1 and with PU.1 deletion revealed multiple target genes that had not been implicated previously in early T-cell development. Gene ontology (GO) analysis of PU.1-Eng-repressed genes (Supplemental Table S5) showed enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (P-value < 10−3), including cell adhesion, leukocyte transendothelial migration, chemokine signaling, B-cell receptor and TCR signaling, and cytokine–cytokine receptor signaling as well as developmental pathways such as osteoclast differentiation and hematopoietic cell lineage. Interestingly, several genes were associated with negative regulation of immune responses (P-value < 10−8). These genes should strongly affect the basic cell biology of environmental responsiveness and survival of the early pro-T cells, including not only signaling but also further aspects of chemotaxis, cytoskeletal activity, and resistance to oxidative stress.
Among the top PU.1-Eng-repressed genes were glutathione peroxidase Gpx1; Src family tyrosine kinase Lyn; E3 ubiquitin ligase Neurl3 (Lincr); signaling adaptor Sh2b2; NFAT-activating molecule Nfam1; actin organization factor Coro2a; and genes encoding two aquaporins and several regulator of G-protein signaling (Rgs) factors, GTPase-activating factors like Dock10, the G-protein γ2 subunit (Gng2), chemokine receptor Ccr2, and G-protein-coupled receptors, including fatty acid receptor Ffar2 (Gpr43), proton sensor Gpr65, and Gpr141. Two important protein tyrosine phosphatase genes—Ptprc (CD45) and Ptpn6 (SHP1 or “Motheaten”)—were also highly sensitive. Evidence from independent experiments confirmed PU.1 as a direct activator of Gpx1, Lyn, Neurl3, Sh2b2, and Ptpn6 in early T cells (Fig. 7E–H). For these genes, PU.1-Eng had effects similar to the deletion of endogenous PU.1 (Fig. 7F,H). Additional wild-type PU.1 further up-regulated many of these genes (Fig. 7E,G), suggesting that the endogenous PU.1 supply is not saturating for these (Figs. 4D, 7E,G). These target genes present myriad options for PU.1-dependent contributions to the cell biology and in vivo behavior of early T-lineage precursors.
Selective roles for PU.1 in the progenitor-associated regulatory gene network
A cohort of regulatory genes that we termed “phase 1” genes have their highest expression in the DN1/ETP stage, like PU.1, and are then silenced during commitment. PU.1 can positively regulate several of these genes in gain-of-function experiments (Del Real and Rothenberg 2013). Despite showing responses weaker than those of the high-confidence targets (Fig. 6A), a broader set of these regulatory genes was also scored as “enriched” among the PU.1-Eng-repressed genes in the GSEAs, supporting functional PU.1 input of some kind (Supplemental Table S4, gene scores). To determine whether these were positive or negative targets of wild-type PU.1, we compared their responses to overexpression of PU.1-Eng and wild-type PU.1 in primary pro-T cells.
As expected, Bcl11a, Lmo2, and Mef2c were up-regulated by added wild-type PU.1, in accord with effects of PU.1 deletion (Fig. 3) and PU.1-Eng, indicating that they are positive targets of PU.1 (Supplemental Fig. S8). However, another group of phase 1 regulatory genes—including Tal1, Mycn, and Erg—were repressed rather than activated by addition of wild-type PU.1, even more strongly than by PU.1-Eng (Supplemental Fig. S8). Interestingly, quantitative PCR (qPCR) signals from the exons not included in the PU.1-Eng or PU.1-ETS constructs indicated that Spi1 itself is under mild negative autoregulation in early T cells as well (data not shown). Thus, PU.1 works as a complex positive and negative modulator of multiple regulatory loci in early T-cell development and not simply by promoting a generally immature state.
Discussion
PU.1 plays an essential role for the development of myeloid and B-lineage cells (for which multiple lineage-specific PU.1 target genes have been identified), differentiation to the earliest myeloid and lymphoid precursors (Back et al. 2005; Iwasaki et al. 2005; Nutt et al. 2005), and HSC maintenance (Staber et al. 2013). While it has been clear that T-cell development is also severely affected in the absence of PU.1, these results came from experiments in which PU.1 was deleted at prethymic stages. Only limited roles of PU.1 have been revealed by deletion of PU.1 in T cells after the DN stages, when it is normally best expressed (Chang et al. 2010; Jabeen et al. 2011). Conversely, PU.1 overexpression can block T-cell development and repress the T-lineage program, sometimes diverting these cells to the myeloid lineage (Anderson et al. 2002; Lefebvre et al. 2005; Franco et al. 2006; Laiosa et al. 2006; Del Real and Rothenberg 2013), implying that its endogenous expression is mainly a regulatory hazard. Here, however, focusing on the earliest stages of T-cell development, we show that endogenous PU.1 in fact plays an important role in supporting the proliferation and distinctive cell-biological features of the earliest cells within the T-cell pathway itself and regulates the timing of access to T-lineage genes throughout the stages when it is expressed.
In this study, we used an acutely expressed obligate repressor form of PU.1 to search rapidly for genes with PU.1-sensitive expression on a genome-wide scale. This was necessary for two reasons. First, PU.1 binds to >30,000 genomic sites in developing T cells during the stages when it is expressed (Zhang et al. 2012), engaging a large fraction of all active genes throughout the genome of early T cells so that binding cannot be used to predict specific function. Second, deletion of the PU.1 gene does not affect PU.1 protein pools quickly enough to give developmentally synchronized effects in this dynamic context. Although forced expression of any dominant negative comes with caveats, many genes repressed by the obligate repressor met expected criteria for true PU.1 targets in that they were enriched for local PU.1 binding in occupancy strength and/or site numbers and were enriched for genes expressed preferentially during stages when PU.1 itself was present. Genes activated by PU.1-Eng treatment were not enriched for these criteria. Notably for future analysis, the results revealed numerous genes with moderate to strong local binding by endogenous PU.1 but no effect of the PU.1-Eng construct, even with PU.1 sites at the promoter or in the middle of the gene body. Conversely, many of the PU.1-Eng-repressed genes had no PU.1 binding normally at their promoters but only sites in intronic or flanking regions. Thus, the perturbation test was crucial to distinguish functional from nonfunctional binding. The results rule out trivial models for PU.1-Eng repression and suggest instead that the effect depends on recruitment to a particular subset of PU.1 sites that may be preferentially associated with function. Thus, distinctive features of these sites in future studies may help to determine the rules for the functional importance of a transcription factor-binding event.
The impact of endogenous PU.1 activity is to extend the proliferative capacity of the cells through DN2 stage while retarding onset of the T-cell differentiation program. Here, deletional and dominant-negative experiments confirm that PU.1 activity at normal as well as at supraphysiological levels restricts expression of many T-cell commitment genes even in the presence of environmental Notch ligands, which drive the T-cell program forward. In part, this may be because, despite an environment rich in Notch ligands, the intensity of Notch responses is blunted in the early T cells by PU.1 as long as it is present. However, PU.1 is likely to antagonize T-cell progression via additional targets as well. Moreover, PU.1 also blocks expression of genes associated with other variant cell fates, such as Gata2, Zbtb16, Prf1, and probably Il2rb, which could promote mast cell, innate lymphocyte, or NK cell pathways that are much less Notch-dependent. PU.1 is capable of acting as a direct repressor (Rekhtman et al. 2003; Stopka et al. 2005; Hu et al. 2011; Wontakal et al. 2012; de la Rica et al. 2013), and our results suggest that it down-regulates Sox13 and Il7r directly. However, its repressive effects on most T-cell genes work through an indirect mechanism. The T-cell differentiation genes in the T-lineage precursors are precociously activated by the obligate repressor form of PU.1 rather than repressed, indicating that the differentiation restraint function of PU.1 in these cells depends on the activation of intermediate genes. By implication, any binding sites around T-cell genes are likely to be sites where PU.1-Eng-binding impact is functionally ineffective or else is overwhelmed by the impact of positive T-cell activators when constraint is lifted. Importantly, the results also imply that the positive regulators for these genes are already available in DN1/ETP cells before the targets are normally expressed but are held in check as long as PU.1 levels are high.
The exact pathways through which this indirect regulation occurs still remain to be defined. Our results show PU.1-dependent targets enriched for a wide range of genes encoding effectors of signaling (particularly G-protein-related) and cell biology, many of which were unexpected in the early T-cell context. Some PU.1-regulated genes also reflect a continuum with its myeloid-promoting activity (Itgam, Itgax, Emr1, and Fcgr2b), reminiscent of the overlap of PU.1-regulated genes from myeloid and lymphoid programs reported before in myeloid cells (Kamath et al. 2008). Thus, it is important that our genome-wide analysis was of sorted DN2 cells that had already acquired positive markers of the T-cell program (rather than total early PU.1+ cells) to exclude any progenitors that might be initiating other programs. The genes affected are thus positive regulatory targets of PU.1 specifically within the early T-cell program. Although some targets like Ptpn6 may later be controlled by other factors once PU.1 is turned off, it is notable that many other new PU.1 targets are themselves restricted to the early PU.1+ stages. These results suggest that many of the signaling, morphological, and migratory behaviors of cells in the still-obscure early stages of T-cell development are actually PU.1-dependent.
PU.1 is coexpressed with multiple transcription factors that link early T-cell development with prethymic progenitors. Indeed, we found evidence for a positive role for PU.1 in maintaining the expression of at least three of these phase 1 regulatory genes: Bcl11a, Mef2c, and Lmo2. However, our results rule out a role for PU.1 as a global positive regulator of the phase 1 regulatory genes. In fact, at least two groups of these genes were affected by PU.1 in divergent ways: Whereas PU.1 enhanced expression of one set, it blunted expression of another, including Gata2, Mycn, Tal1, Erg, and Kit. Thus, the phase 1 regulatory state is complex, and the proliferative expansion and slow differentiation functions that it promotes could be mediated by distinct modules.
In summary, PU.1 operates in early uncommitted precursors, but its role is not limited to upper levels of the regulatory hierarchy in these cells. Direct PU.1 regulatory roles in early T-lineage cells include modulatory effects on other progenitor regulatory factors and strong impacts on genes involved in protection from oxidative stress, multiple signaling pathways, and cytoskeletal organization. Although PU.1 is not a “master regulator” of the precommitment regulatory state, it is thus a potent positive regulator of key elements of precursor cell biology as well as a highly selective regulator of specific lymphoid precursor transcription factors. Such targets can mediate its proliferation support and differentiation timing roles in early T-cell precursors.
Materials and methods
Mice
Spi1fl/fl mice were bred at the California Institute of Technology from stock originally derived at the Walter and Eliza Hall Institute (Dakic et al. 2005). Mice with the Spi1-targeted allele and a tamoxifen-inducible CreER allele were bred to B6 mice to separate these traits and then interbred to generate Spi1fl/fl homozygotes without CreER (referred to as Spi1fl/fl), which were used in this study. Bcl2-tg mice with a C57BL/6 genetic background [B6.Cg-Tg(BCL2)25Wehi/J] were used as previously described (Anderson et al. 2002). All strains were maintained as per institutional animal care and use committee guidelines.
Cell cultures
FL cells from E14.5 (day of plug, E0.5) from Spi1fl/fl × Spi1fl/fl, C57BL/6 × C57BL/6, or C57BL/6-Tg(Bcl2) × C57BL/6 crosses were used to initiate OP9-DL1 cultures using slight modifications of methods reported previously (Li et al. 2010; Del Real and Rothenberg 2013; Scripture-Adams et al. 2014). For details, see the Supplemental Material. The DN3-like Adh.2C2 cell line has been described previously (Dionne et al. 2005).
Retroviral constructs
Wild-type PU.1 and the W215G mutant form of PU.1 cloned in the LZRS-IRES-GFP vector were previously described (Anderson et al. 2002). PU.1-ΔDEQ was made by deleting the transcriptional activation domain (amino acids 33–100) of PU.1 while keeping the rest of the protein intact. The PU.1-Eng construct expresses a fusion protein, which contains the DNA-binding ETS domain of PU.1 (amino acids 159–260) fused to the obligate repressor domain (amino acids 1–298) of the Drosophila Engrailed protein (S. Acharya, M. Zarnegar, and E.V. Rothenberg, unpubl.). The PU.1-ETS construct only expresses the ETS domain of PU.1 (amino acids 159–260) and served as a control for the Engrailed construct. Versions of these constructs were also made using a C-terminal hemagglutinin antigen tag, and this was the form of PU.1-Eng W215G used in Figure 4. All the above constructs were cloned into the LZRS-IRES-GFP retroviral vector for use. Wild-type PU.1 was also cloned in the MSCV-IRES-NGFR vector and was used in Figure 4B. Tagged PU.1 wild type, PU.1-Eng, and PU.1-ETS and untagged wild-type PU.1 all bound to genomic DNA at PU.1 consensus sites in transduced cells (J. Ungerbäck, A. Champhekar, and E.V. Rothenberg, in prep.). Bcl-xL-MIG-R1 (Cheng et al. 2001) was obtained from Addgene. Constitutively active Cre in the LZRS-IRES-NGFR vector was described previously (Li et al. 2010).
Retroviral supernatants were packaged by transfecting Phoenix-Eco cells with retroviral constructs using Fugene 6 (Roche). Lymphoid cells were transduced with retrovirus particles adsorbed to RetroNectin-coated plates in 4-h incubations at 37°C.
RNA isolation and qPCR
Sorted FLDN populations were used to extract RNA with the RNeasy minikit (Qiagen).
Primers and conditions for qPCR are described in the Supplemental Material and Supplemental Table S6. Each reaction was run in triplicate, the data were normalized to Actin expression, and mean values from two to three experiments were expressed as a heat map (Matlab) or plotted as log10-scale graphs. For statistical analysis, actin-normalized gene expression values were log10-transformed, and two-tailed Student's t-tests (paired, equal variance) were performed to determine the significance of differences between expression values. Significant differences in expression with a P-value < 0.05 are indicated with an asterisk. Error bars in all plots represent one standard deviation (more than two biological replicates) or range (two biological replicates).
Transcriptome profiling and analysis of sequencing data: DESeq and HTSeq analyses
Total RNA was processed essentially as described (Zhang et al. 2012). cDNA libraries were sequenced with the Illumina HiSeq 2000 sequencer following the manufacturer's protocols (http://www.illumina.com) and produced between 11 million and 14 million mapped 50-base-pair (bp) single-end reads per sample (Gene Expression Omnibus accession no. GSE65344). Sequenced reads were mapped onto mouse genome build NCBI37/mm9 using TopHat, and read alignments were processed by HTSeq (Anders et al. 2015) as described in detail in the Supplemental Material. Count data were analyzed for differentially expressed genes using DESeq (version 1.16.0, http://bioconductor.org/packages/release/bioc/html/DESeq.html) (Anders and Huber 2010). Each replicate experiment was analyzed separately, and 168 genes with an adjusted P-value of ≤0.1 in both experiments were considered to be differentially expressed; this set was augmented by 36 genes with an adjusted P-value of <0.1 in one set and 0.1 < Padj < 0.2 in the other. Spi1 (Sfpi1) appears in these gene lists as an “up-regulated gene” only because of the exogenous PU.1-Eng or PU.1-ETS RNA.
Analysis of RNA-seq data: sample classification and natural PU.1 binding around repressed and activated genes
For details of genome-wide RNA expression analyses, see the Supplemental Material. Developmental indexing was carried out using a set of 173 developmentally regulated transcription factor genes to monitor the developmental state of our RNA-seq samples (Scripture-Adams et al. 2014). Briefly, samples were classified by principal component analysis of these transcript levels (Supplemental Table S3). Principal component loadings for the 173 genes from the normal samples were used to transform the values from the six experimental samples.
To determine whether PU.1-Eng-affected genes were endogenous PU.1 targets, genes that were up-regulated or down-regulated by PU.1-Eng were compared separately with a control set of nonregulated but well-expressed genes. The control set consisted of 841 genes from expression clusters 16 and 18 in Zhang et al. (2012), which are expressed stably from the DN1 to the DP stage, after the removal of four genes that were affected by PU.1-Eng. Briefly, supplemental Table 5 from Zhang et al. (2012) (with updated gene names) was used to identify all endogenous PU.1-binding peaks in the vicinity of each gene on the three lists in the DN1 and DN2a stages. The PU.1 occupancy index for each gene was then calculated as the sum of the occupancy levels (PU.1 ChIP-seq reads per million) from the DN1 and DN2a samples at the four strongest peaks assigned to that gene. See the Supplemental Material for details and statistics.
Analysis of RNA-seq data: GSEA and GO and pathway enrichment analyses
For a full description, see the Supplemental Material. For GSEA, we used the GSEAPreranked tool in order to fully characterize the effects of PU.1-Eng on developmentally regulated and lineage-restricted gene sets (http://www.broadinstitute.org/gsea/index.jsp). Briefly, genes were ranked in descending order of mean log2 ratios between all pairwise comparisons from both RNA-seq experiments. The ranked lists were then compared with distinct developmental expression pattern clusters from Zhang et al. (2012) and previously identified PU.1 target lists reported in myeloid precursors (Kamath et al. 2008). Gene sets with FDR < 0.01 were considered to be significantly enriched, but the sets discussed here had FDR < 0.001.
We performed GO term and KEGG pathway overrepresentation analysis on genes defined as differentially regulated using the package GOseq (version 1.16.2), which calculates enrichment after correcting for transcript length bias (Young et al. 2010). GO terms and KEGG pathways with an adjusted (Benjamini-Hochberg) P-value of ≤0.05 were considered to be enriched.
Supplementary Material
Acknowledgments
We thank Sanket Acharya and Mark Zarnegar for the PU.1-Eng construct; Arthur Skoultchi for related PU.1 constructs used in preliminary studies; Hao Yuan Kueh for discussion on PCA analysis; Jonas Ungerbäck for sharing unpublished data; Robert Butler and Maria Quiloan for technical assistance; Rochelle Diamond, Diana Perez, and Pat Koen at the Caltech Flow Cytometry and Cell Sorting Facility for cell sorting; Scott Washburn and Ingrid Soto for animal care; and members of E.V.R.’s laboratory for helpful discussions. This work was supported by National Institutes of Health grants (CA90233, AI95943, and HD076915), the Louis A. Garfinkle Memorial Laboratory Fund, the Al Sherman Foundation, and the Albert Billings Ruddock Professorship to E.V.R., and Victorian State Government Operational Infrastructure Support, National Health and Medical Research Council of Australia Independent Research Institute Infrastructure Support grant 361646, Program grant 105492, and Fellowship 1058238 to S.L.N.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.259879.115.
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126: 3131–3148. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. 2002. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16: 285–296. [DOI] [PubMed] [Google Scholar]
- Back J, Allman D, Chan S, Kastner P. 2005. Visualizing PU.1 activity during hematopoiesis. Exp Hematol 33: 395–402. [DOI] [PubMed] [Google Scholar]
- Carotta S, Dakic A, D'Amico A, Pang SH, Greig KT, Nutt SL, Wu L. 2010. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32: 628–641. [DOI] [PubMed] [Google Scholar]
- Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, et al. 2010. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol 11: 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH-Y, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. 2001. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell 8: 705–711. [DOI] [PubMed] [Google Scholar]
- Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. 2005. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med 201: 1487–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter RP, Lee HJ, Singh H. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity 16: 297–309. [DOI] [PubMed] [Google Scholar]
- de la Rica L, Rodriguez-Ubreva J, Garcia M, Islam AB, Urquiza JM, Hernando H, Christensen J, Helin K, Gomez-Vaquero C, Ballestar E. 2013. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol 14: R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Real MM, Rothenberg EV. 2013. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development 140: 1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. 2005. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol 280: 448–466. [DOI] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. 2006. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci 103: 11993–11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL. 1993. Functional domains of the Drosophila Engrailed protein. EMBO J 12: 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. 2009. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet 5: e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, Zuklys S, Hollander GA, Shima DT, Habu S. 2008. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med 205: 2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gu X, Baraoidan K, Ibanez V, Sharma A, Kadkol S, Munker R, Ackerman S, Nucifora G, Saunthararajah Y. 2011. RUNX1 regulates corepressor interactions of PU.1. Blood 117: 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Goldrath AW, Murre C. 2006. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med 203: 1329–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106: 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabeen R, Chang HC, Goswami R, Nutt SL, Kaplan MH. 2011. The transcription factor PU.1 regulates γδ T cell homeostasis. PLoS One 6: e22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. 1997. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev 11: 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath MB, Houston IB, Janovski AJ, Zhu X, Gowrisankar S, Jegga AG, DeKoter RP. 2008. Dose-dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B-cell identity. Leukemia 22: 1214–1225. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV. 2013. Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 341: 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. 2006. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25: 731–744. [DOI] [PubMed] [Google Scholar]
- Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. 2005. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol 174: 6184–6194. [DOI] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. 2010. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. 2013. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface 10: 20120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, et al. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15: 5647–5658. [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. 2013. The transcriptional landscape of αβ T cell differentiation. Nat Immunol 14: 619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. 2005. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med 201: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. 2003. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol 23: 7460–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridinger-Saison M, Boeva V, Rimmele P, Kulakovskiy I, Gallais I, Levavasseur B, Paccard C, Legoix-Ne P, Morle F, Nicolas A, et al. 2012. Spi-1/PU.1 activates transcription through clustered DNA occupancy in erythroleukemia. Nucleic Acids Res 40: 8927–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. 2006. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 38: 27–37. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. 2007. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity 26: 690–702. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Zhang J, Li L. 2010. Multilayered specification of the T-cell lineage fate. Immunol Rev 238: 150–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577. [DOI] [PubMed] [Google Scholar]
- Scripture-Adams DD, Damle SS, Li L, Elihu KJ, Qin S, Arias AM, Butler RR III, Champhekar A, Zhang JA, Rothenberg EV. 2014. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol 193: 3470–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain LM, Guerriero A, Kunjibettu S, Scott EW. 1999. T cell development in PU.1-deficient mice. J Immunol 163: 2681–2687. [PubMed] [Google Scholar]
- Staber PB, Zhang P, Ye M, Welner RS, Nombela-Arrieta C, Bach C, Kerenyi M, Bartholdy BA, Zhang H, Alberich-Jorda M, et al. 2013. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell 49: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. 2005. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J 24: 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. 2007. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol 8: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunova EN, Fujioka M, Kobayashi M, Deka D, Jaynes JB. 1998. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol Cell Biol 18: 2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wontakal SN, Guo X, Will B, Shi M, Raha D, Mahajan MC, Weissman S, Snyder M, Steidl U, Zheng D, et al. 2011. A large gene network in immature erythroid cells is controlled by the myeloid and B cell transcriptional regulator PU.1. PLoS Genet 7: e1001392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wontakal SN, Guo X, Smith C, MacCarthy T, Bresnick EH, Bergman A, Snyder MP, Weissman SM, Zheng D, Skoultchi AI. 2012. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci 109: 3832–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Rothenberg EV. 2014. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14: 529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV. 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol 185: 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell 149: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.