ABSTRACT
Hepatitis C virus (HCV) entry involves binding to cell surface heparan sulfate (HS) structures. However, due to the lipoprotein-like structure of HCV, the exact contribution of virion components to this interaction remains controversial. Here, we investigated the relative contribution of HCV envelope proteins and apolipoprotein E in the HS-binding step. Deletion of hypervariable region 1, a region previously proposed to be involved in HS binding, did not alter HCV virion binding to HS, indicating that this region is not involved in this interaction in the context of a viral infection. Patient sera and monoclonal antibodies recognizing different regions of HCV envelope glycoproteins were also used in a pulldown assay with beads coated with heparin, a close HS structural homologue. Although isolated HCV envelope glycoproteins could interact with heparin, none of these antibodies was able to interfere with the virion-heparin interaction, strongly suggesting that at the virion surface, HCV envelope glycoproteins are not accessible for HS binding. In contrast, results from kinetic studies, heparin pulldown experiments, and inhibition experiments with anti-apolipoprotein E antibodies indicated that this apolipoprotein plays a major role in HCV-HS interaction. Finally, characterization of the HS structural determinants required for HCV infection by silencing of the enzymes involved in the HS biosynthesis pathway and by competition with modified heparin indicated that N- and 6-O-sulfation but not 2-O-sulfation is required for HCV infection and that the minimum HS oligosaccharide length required for HCV infection is a decasaccharide. Together, these data indicate that HCV hijacks apolipoprotein E to initiate its interaction with specific HS structures.
IMPORTANCE Hepatitis C is a global health problem. Hepatitis C virus (HCV) infects approximately 130 million individuals worldwide, with the majority of cases remaining undiagnosed and untreated. In most infected individuals, the virus evades the immune system and establishes a chronic infection. As a consequence, hepatitis C is the leading cause of cirrhosis, end-stage liver disease, hepatocellular carcinoma, and liver transplantation. Virus infection is initiated by entry of the virus into the host cell. In this study, we provide new insights into the viral and cellular determinants involved in the first step of HCV entry, the binding of the virus to host cells. We show that apolipoprotein E is likely responsible for virus binding to heparan sulfate and that N- and 6-O-sulfation of the heparan sulfate proteoglycans is required for HCV infection. In addition, the minimal HS length unit required for HCV infection is a decasaccharide.
INTRODUCTION
Hepatitis C virus (HCV) belongs to the genus Hepacivirus in the Flaviviridae family (1). It is a small enveloped virus with a positive single-stranded RNA genome of 9.6 kb. The genome is translated as a polyprotein of ∼3,000 amino acids, which is processed during translation by cellular and viral proteases to generate structural proteins (capsid, E1, and E2) and nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (2). The structural proteins are components of the viral particle. By being present at the surface of the viral particle, HCV envelope glycoproteins E1 and E2 play a major role in HCV entry (3, 4). These glycoproteins are type I transmembrane proteins which form a noncovalent heterodimer within infected cells, whereas they assemble as large covalent complexes stabilized by disulfide bonds on the viral particle (5). Within the E1E2 complex, E2 is currently the best-characterized subunit (5). Indeed, it is the major target of neutralizing antibodies (6), and it is also the receptor-binding protein, which has been shown to interact with tetraspanin CD81 (7) and scavenger receptor B1 (SRB1) (8).
A striking and unique feature of HCV biology is the unusually low buoyant density of the virion, which results from its physical association with lipoproteins, forming a hybrid particle called a lipoviroparticle (LVP) (9). Due to the association of the virion with lipoproteins, apolipoproteins, such as apolipoprotein E (apoE), apoB, apoA1, apoC1, apoC2, and apoC3, can also be found in association with HCV particles (10, 11). Furthermore, characterization of cell culture-produced particles indicates that their lipid composition resembles the lipid compositions of very-low-density lipoproteins (VLDLs) and low-density lipoproteins (LDLs) (12). Among HCV-associated apolipoproteins, there is a consensus about the involvement of apoE in HCV morphogenesis (13–15).
HCV entry is a complex process involving many cellular partners and viral components. Indeed, the initial attachment of the virus is followed by a series of sequential interactions with numerous host factors, internalization of the viral particle by clathrin-mediated endocytosis, and fusion of the viral envelope with endosomal membranes (16). It is now well established that heparan sulfate (HS) proteoglycans (HSPGs) serve as primary docking sites for many viruses. In the case of HCV, syndecan 1 and syndecan 4 have been reported to be involved in virion binding to hepatocytes (17, 18). This initial attachment of the virus to HSPGs and, potentially, the low-density lipoprotein receptor is followed by sequential interactions with at least four specific cellular entry factors: the scavenger receptor SRB1, the tetraspanin CD81, and two tight junction proteins, claudin-1 (CLDN1) and occludin (OCLN). In recent years, many cellular factors participating in or regulating different steps of the entry process have been identified. These factors include the epithelial growth factor receptor (EGFR) (19), the Niemann-Pick type C1-like 1 (NPC1L1) cholesterol uptake receptor (20), and transferrin receptor 1 (TSFR1) (21).
HSPGs are abundant in the matrix of the space of Disse and at the surface of hepatocytes. They are composed of a core protein and HS chains, which are linear polysaccharides consisting of a repeated disaccharide unit of an uronic acid and a derivative of glucosamine with various sulfation patterns (22). It was first reported that viral particles isolated from patients interact with glycosaminoglycans (GAGs) (23). Following this observation, it was shown that recombinant HCV envelope glycoprotein E2 and virion-associated glycoprotein complexes interact with HSPGs, suggesting a direct contact between the viral components of the virion and HSPGs (24, 25). Furthermore, E2 hypervariable region 1 (HVR1) has been proposed to contribute to this interaction (24, 26–28). However, other regions have also been proposed to be involved in interactions with HCV envelope glycoproteins (24, 26). These early studies were performed on genotype 1a (gt1a) or gt1b envelope proteins. To this day, no data concerning interactions of HS with HCV envelope proteins of other genotypes are available, except for data for HCV genotype 2a. On the other hand, apoE, which is found on the surface of LVPs, is also able to interact with HSPGs, and it has recently been reported that this apolipoprotein could be responsible for the binding of genotype 2a HCV virions to HSPGs (29, 30). Here, we investigated the relative contribution of HCV envelope proteins and apoE in the early event of HCV entry, specifically, in the HSPG-binding step, and we characterized the structural determinants of HS required for HCV infection. Our results support the hypothesis that apoE associated with the virus is responsible for HCV-HS interaction. We also found that N- and 6-O-sulfation but not 2-O-sulfation of HS is required for HCV infection. Finally, we show that the minimal HS length unit required for HCV infection is a decasaccharide.
MATERIALS AND METHODS
Cell culture.
The Huh-7 hepatoma cell line (31) was cultured in Dulbecco's modified Eagle medium (DMEM; Life Technologies) supplemented with 0.1 mM nonessential amino acids (NEAA; Life Technologies) and 10% fetal bovine serum (Life Technologies).
Antibodies and reagents.
Mouse anti-HCV monoclonal antibodies (MAbs) A4 (anti-E1) and A11 (anti-E2) (32), rat anti-HCV MAbs 9/27 and 3/11 (anti-E2; kindly provided by J. A. McKeating, University of Birmingham, Birmingham, United Kingdom) (4), and human anti-HCV antibodies AR3A and AR5A (anti-E2 and anti-E1E2 antibodies, respectively; kindly provided by M. Law, The Scripps Research Institute, La Jolla, CA, USA) (33) were used in this work. apoE-specific MAb 23 has been previously described (34). Control human MAb RO4 was kindly provided by S. Foung (Stanford University). Polyclonal anti-apoE antibody was from Millipore, and MAb E3 was from Progen. Residual laboratory sera were obtained from 5 patients according to the rules of the ethics commitee of the University Hospital of Lille. Soluble heparin extracted from bovine lungs was purchased from Sigma-Aldrich, and desulfated heparins were purchased from AMS Biotechnology (United Kingdom). An apoE-derived peptide (LRKLRKRLLLRKLRKRLL) was synthesized by ProteoGenix.
Preparation of heparin oligosaccharides.
Heparin-derived oligosaccharides were obtained as described previously (35). Briefly, 100 mg of heparin was incubated with 50 units of heparinase I (Iduron) at 30°C for 30 h. After desalting on a Sephadex G-10 column (Pharmacia Amersham Biotech), the digestion mixture was fractionated by filtration on Bio-Gel P-6 (Bio-Rad Laboratories). Pooled fractions corresponding to oligosaccharides with increasing degrees of polymerization (dp) were eluted by 0.2 M NH4Cl, pH 3.5, desalted, and freeze-dried.
Plasmid constructions.
The virus used in this study is based on the JFH1 strain (genotype 2a; GenBank accession number AB237837) (36) and was kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan). In addition, an intergenotypic H77/JFH1 chimera was also used in some experiments (37). Deletions were introduced in a modified version of the plasmid carrying the full-length JFH1 genome (pJFH1-CS-A4) engineered to reconstitute the A4 epitope in E1 (38) and titer-enhancing mutations (39). The deletion of hypervariable region 1 was introduced into the plasmid carrying the full-length genome by PCR. Two PCR products were generated by using the following primers: primers Sense1 (5′-CGTACGTGATGCGCGTCCCCG-3′) and Anti-Sense1 (5′-CTGCCGTTGGTGTTAATGAGCTGAATCGCGTCCACCCCAGCGGCCAG-3′) and primers Sense2 (5′-CTGGCCGCTGGGGTGGACGCGATTCAGCTCATTAACACCAACGGCAG-3′) and Anti-Sense2 (5′-GGTACCCACTCCTGAATCATGG-3′). The two fragments were assembled by a second PCR amplification by using the primers Sense1 and Anti-Sense2. The PCR product was ligated into pJFH1-CS-A4 after digestion with BsiWI and KpnI (New England BioLabs). The nucleotide sequence was verified by sequencing.
Virus production and purification.
Plasmids encoding the wild-type virus or the virus with the HVR1 deletion were digested with XbaI and treated with mung bean nuclease (New England BioLabs). In vitro transcriptions were performed using a MEGAscript kit (Ambion) according to the manufacturer's protocol. Viruses were rescued by electroporation of Huh-7 cells with 10 μg of in vitro-transcribed RNA as described in reference 39. To monitor virus production, viral supernatants were collected at 72 h and 96 h after electroporation. Viruses were amplified to obtain viral stocks. The infectivity of HCV in the different supernatants and the viral stocks was determined by counting the number of focus-forming units (FFU) as previously reported (40). RNAs were extracted by using a QIAamp viral RNA kit (Qiagen) to quantify the HCV genomes in the supernatant by real-time quantitative reverse transcription-PCR (RT-PCR) as previously described (41). For virus purification, highly infectious viral supernatants were collected and cleared by centrifugation. Viruses were concentrated by overnight precipitation with 8% polyethylene glycol (Fluka Chemie AG) and centrifugation at 13,000 × g for 20 min. Pellets were resuspended in 1 ml of cold phosphate-buffered saline (PBS) and layered on the top of a 10 to 50% continuous iodixanol gradient (Optiprep; Proteogenix). Gradients were ultracentrifuged for 16 h at 160,000 × g and 4°C in an SW41 rotor. Twelve fractions of 1 ml each were collected. The densities, titers, and HCV RNA contents of each fraction were determined. The two most infectious fractions were used for experiments involving purified virus.
Indirect immunofluorescence.
HCV-infected cells grown on glass coverslips were fixed in 100% cold (−20°C) methanol. After two washes with PBS, cells were blocked with 10% goat serum in PBS. Infected cells were detected by using anti-E1 MAb A4 and visualized with cyanine 3-conjugated goat anti-mouse IgG secondary antibodies (Jackson ImmunoResearch). Cell nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Coverslips were observed with a Zeiss Axiophot microscope equipped with ×10 magnification objective (Carl Zeiss AG, Oberkochen, Germany). Fluorescent signals were collected with a Coolsnap ES camera (Photometrix, Kew, Australia). For quantification, images of areas randomly picked from each coverslip were recorded.
Heparin pulldown assay of HCV envelope glycoproteins.
Infected cells or purified viruses were lysed with PBS–1% Triton X-100 in the presence of protease inhibitors (Complete; Roche Diagnostics). Different volumes of lysates (200, 100, or 50 μl) were incubated in the presence or absence of 500 μg/ml heparin for 1 h at 4°C and then overnight with heparin-Sepharose (CL-6B). Then, the beads were washed 5 times with 1% Triton X-100 in PBS. After the last wash, the beads were resuspended in Laemmli loading buffer and incubated for 10 min at 70°C. Protein samples were separated by SDS-PAGE and were then transferred onto nitrocellulose membranes (Hybond-ECL; Amersham). Proteins were probed with specific antibodies and the corresponding peroxidase-conjugated antispecies antibodies (anti-mouse; Jackson Immunoresearch). Peroxidase activity was detected by a chemoluminescent reaction (Pierce ECL substrate; Thermo Scientific).
Heparin pulldown assay of viral particles.
Purified virus was incubated in the presence or absence of different competitors (25 μg/ml heparin, 5 μg/ml MAb AR3A, 10 μg/ml MAb AR5A, 10 μg/ml MAb RO4, 20 μl MAb 9/27, 10 μg/ml MAb 6/16, 100 μg/ml purified IgG from 4 different HCV [gt1a]-positive patients and a noninfected control patient, 1/200-diluted anti-apoE polyclonal antibodies or anti-human IgG polyclonal antibodies, 20 μg/ml apoE-derived peptide, or 20 μg/ml control 3× Flag peptide) in PBS for 1 h at 37°C. Then, the mixture was cooled and incubated for 2 h at 4°C with heparin-Sepharose beads. The beads were washed 3 times with cold PBS, and RNAs were extracted by using the QIAamp viral RNA kit (Qiagen). HCV genomes were quantified by quantitative RT-PCR as described previously (41).
Immunoprecipitation of viral particles.
Protein G-Sepharose was incubated with the polyclonal anti-apoE antibody for 5 h at 4°C and then washed with PBS. Purified virus particles were preincubated for 1 h in the presence or absence of 25 μg/ml heparin at 37°C and then chilled before adding anti-apoE antibody-coated beads. The mixture was incubated overnight at 4°C. The beads were washed 3 times with cold PBS, and RNAs were extracted by using the QIAamp viral RNA kit (Qiagen). HCV genomes were quantified by quantitative RT-PCR.
Virus attachment and infection assay.
Purified virus was preincubated with or without heparin or antibodies in DMEM-HEPES (DMEM without bicarbonate containing 25 mM HEPES buffer) for 1 h at 37°C. Virus preparations were then cooled and incubated with target cells for 2 h at 4°C. The cells were rinsed with PBS. For measurement of binding, total RNA from cell lysates was extracted by using a NucleoSpin RNA kit (Macherey-Nagel) as recommended by the manufacturer. For infection assays, infected cells were further incubated for 30 h at 37°C, fixed, and processed for the detection of envelope proteins by immunofluorescence. As reinfection had not started at 30 h postinfection, we quantified the total number of infected cells. We used conditions that give about 30% infected cells under control conditions.
Real-time PCR of HS sulfotransferases.
Total RNA was isolated from 4 × 106 Huh-7 cells by using the NucleoSpin RNA kit (Macherey-Nagel) as recommended by the manufacturer. Reverse transcription from 1 μg of total RNA was performed by using a Maxima first-strand cDNA synthesis kit for quantitative RT-PCR (Thermo Scientific). Synthetic primers for N-deacetylase-N-sulfotransferases (NDSTs) 1 to 4 (NDST1, NDST2, NDST3, and NDST4, respectively) and 3-O-sulfotransferases (3-OSTs) 1, 2, 4, 5, and 6 (3-OST1, 3-OST2, 3-OST4, 3-OST5, and 3-OST6, respectively) are described in reference 42, and those for 2-O-sulfotransferase (2-OST), 3-OST3A, 3-OST3B, and 6-O-sulfotransferases 1 to 3 (6-OST1, 6-OST2, and 6-OST3, respectively) are described in reference 43. Real-time PCR amplifications were performed using an Mx3000P multiplex quantitative PCR system (Stratagene). The level of expression of the hypoxanthine phosphoribosyltransferase transcript was used as a control to normalize the expression of the genes of interest. Each PCR mixture consisted of 25 μl containing 2 μl of diluted cDNA sample (1:5), 12.5 μl of Maxima SYBR green quantitative PCR master mix (2×; Thermo Scientific), 1 μl of forward primer (7.5 μM for 2-OST, 22.5 μM for 3-OST1 and 3-OST2, 15 μM for all the other enzymes), 1 μl of reverse primer (7.5 μM for 2-OST, 22.5 μM for NDST4 and 3-OST6, 15 μM for all the other enzymes) (all primers were from Eurogentec), and 8.5 μl of water. It also included a nontemplate negative control to check for primer dimers. The conditions of PCR were as follows: 1 cycle of denaturation at 95°C for 10 min, followed by 40 cycles of 30 s at 95°C, 30 s at the specific annealing temperature (67°C for NDST4, 3-OST1, and 3-OST4; 68°C for 3-OST6; 60°C for all the other enzymes), and 30 s at 72°C. The fluorescence data were measured at the end of each cycle. A melting curve (55 to 95°C at 1°C intervals) was constructed for each primer pair to check for the presence of one gene-specific peak. The amplification efficiency of each primer pair was determined with serial dilutions of cDNA. Triplicate PCR mixtures were prepared for each sample. The point at which the PCR product was first detected above a fixed threshold, termed the cycle threshold (CT), was determined for each sample, and the average CT value for triplicate samples was used for further analysis. The relative quantification of the transcripts was calculated as described previously (44).
RNA interference.
Huh-7 cells were transfected with small interfering RNA (siRNA) pools (Dharmacon) targeting CD81 (CACGUCGCCUUCAACUGUA), NDST1 (CCUCCGACUUCUACUUUGA), 2-OST (On-Target plus Smart pool) or 6-OST1 (CCAGGAAGUUCUACUACA), and 6-OST2 (On-Target plus Smart pool) using the RNAiMAX Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. Briefly, 3 μl RNAiMAX Lipofectamine in 500 μl PBS was mixed with 50 pmol siRNA per well in 6-well plates. After 30 min of incubation, 2 ml of complete medium containing 2 × 105 cells was added to the siRNA-Lipofectamine mix for each well. The knockdown effects were determined at 96 h after transfection by Western blotting or quantitative RT-PCR analysis at the time of virus inoculation. For infection assays, the cells were incubated with virus for 1 h. The effects of silencing on virus infection were determined 30 h after inoculation by immunolabeling of the infected cells.
Graphs and statistics.
Prism (v5.0c) software (GraphPad Software Inc., La Jolla, CA) was used to prepare graphs and to determine the statistical significance of differences between data sets.
RESULTS
HVR1 is not the main determinant of the HCV interaction with HS.
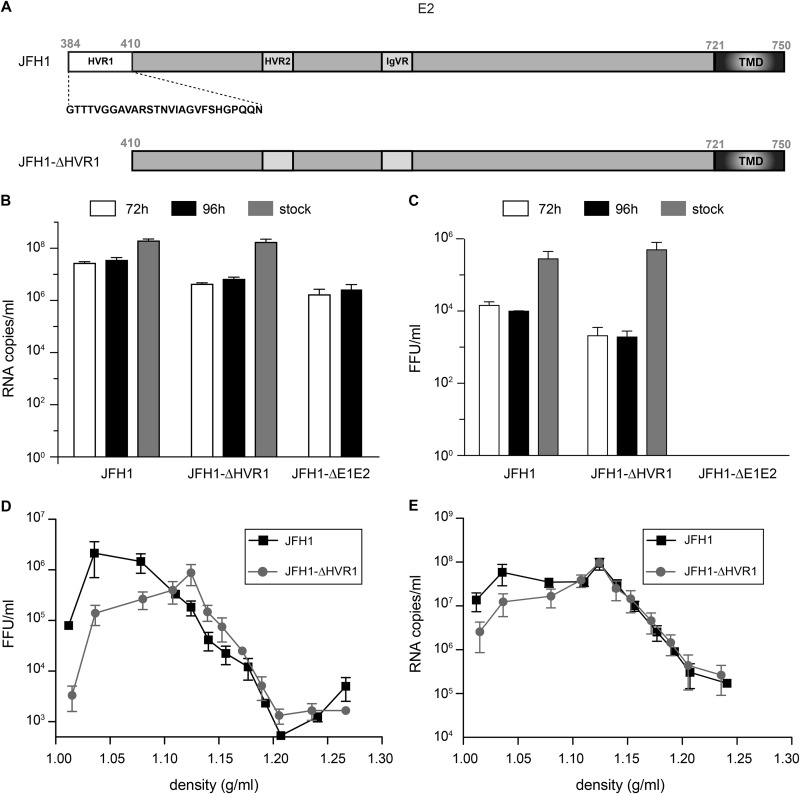
Due to a high level of N-linked glycosylation, the surface area of HCV envelope glycoproteins available to interact with HSPGs is reduced (45). However, in agreement with its high level of sequence variability, HVR1 is believed to be accessible at the surface of the viral particle, which fits with the hypothesis that this region might play a role in HCV interaction with HSPGs (24, 26–28, 45). We therefore reassessed the role of HVR1 in the HCV interaction with HSPGs. To this end, we first deleted hypervariable region 1, comprising the first 27 amino acids of E2, in a modified form of genotype 2a JFH1 virus (38, 39) (Fig. 1A, JFH1-ΔHVR1). This virus was amplified and purified to obtain infectious viral stocks with titers similar to those of the wild-type virus (Fig. 1B and C). The titers obtained for the virus with the HVR1 deletion were lower than those obtained for the wild type after electroporation. After several rounds of amplification, the titers of the JFH1-ΔHVR1 viral stocks were in the same range as those of the wild-type virus. The viral stock of JFH1-ΔHVR1 was sequenced to be sure that no adaptive mutation was present in the E1E2 region. As previously reported (46, 47), a slight difference in buoyant densities was observed (Fig. 1D and E).
FIG 1.
(A) Schematic representation of the HVR1 deletion in E2. TMD, transmembrane domain. (B to E) Characterization of the JFH1-ΔHVR1 mutant. Huh-7 cells were electroporated with in vitro-transcribed JFH1 or JFH1-ΔHVR1 RNA, and cell supernatants were collected either 72 h or 96 h later. Viral stocks were produced after further amplification. Amplified virus stocks were precipitated and loaded on a 10 to 50% iodixanol gradient. The HCV genomes were quantified by quantitative RT-PCR (B), and HCV titers were determined (C). Results are expressed as the means from three independent experiments, and error bars represent the standard deviations of the means. Amplified virus stocks were precipitated and loaded on a 10 to 50% iodixanol gradient. After ultracentrifugation, 12 fractions were collected and the HCV genome copies (D) and titers (E) were quantified for each fraction. Results are expressed as the means from three independent experiments, and error bars represent the standard deviations of the means.
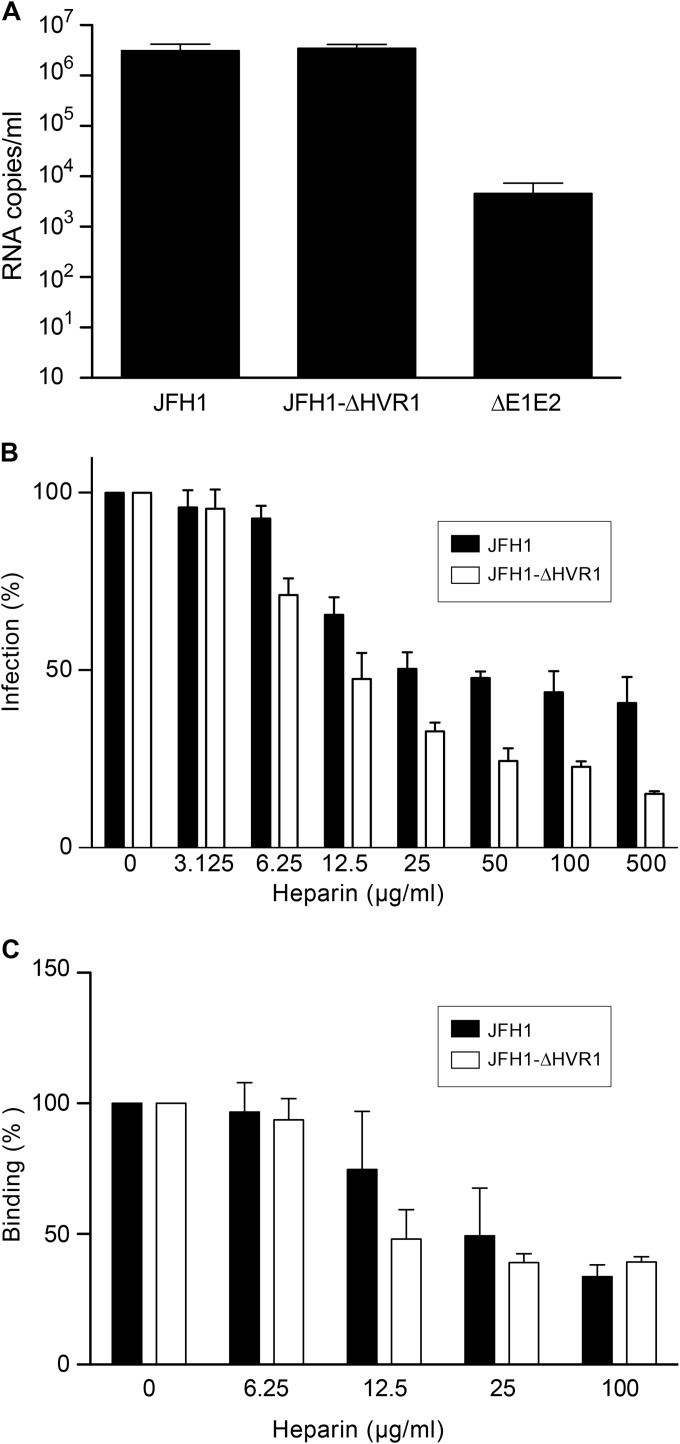
To investigate the binding properties of the JFH1-ΔHVR1 virus, we first determined its capacity to interact with hepatoma cells. Huh-7 cells were therefore incubated at 4°C with similar amounts of purified JFH1 and JFH1-ΔHVR1 viruses, and bound viral particles were determined by measuring the amount of HCV RNA associated with the cells after removing unbound virions. As shown in Fig. 2A, the JFH1-ΔHVR1 virus had a binding capacity similar to that of the wild-type virus, indicating that HVR1 is not essential for the initial binding of HCV particles to Huh-7 cells. Next we determined the capacity of heparin, a close structural homologue of highly sulfated HS, to inhibit viral attachment to Huh-7 cells in a competition assay. As shown in Fig. 2B, JFH1-ΔHVR1 virus infection was inhibited by heparin in a dose-dependent manner, indicating that this virus still relies on HS binding to initiate cell infection. We then confirmed that the cell attachment of both viruses is also inhibited by heparin in a dose-dependent manner (Fig. 2C). It is worth noting that JFH1-ΔHVR1 was even more sensitive than the wild-type virus to heparin competition. This difference could have been due to a global change in the HCV particle after HVR1 deletion (48) which could modulate virion binding to HS (see Discussion). It is worth noting that at high concentrations, inhibition of virus binding was similar for both JFH1 and JFH1-ΔHVR1 (Fig. 2C). This could potentially be due to the presence of noninfectious particles and/or exosomes which might not have the same properties as infectious virus for their interaction with HS. Similar observations have previously been reported when the results of an assay of neutralization by anti-apoE antibodies were compared with those of a binding assay (48). These data suggest that HVR1 of E2 is not involved in virus binding to HS.
FIG 2.
HCV with the HVR1 deletion relies on HS for attachment. (A) Huh-7 cells were inoculated with purified JFH1 or JFH1-ΔHVR1 viruses at an MOI of 5 at 4°C for 2 h. Then, the cells were washed with PBS to remove unbound viruses and lysed to extract RNAs. HCV genomes were measured by quantitative RT-PCR. Results are expressed as the means from three independent experiments, and error bars represent the standard deviations of the means. (B) Purified viruses were preincubated with increasing concentrations of heparin for 1 h at 37°C, and then the virus-heparin mixes were chilled and incubated with Huh-7 cells at 4°C for 2 h. The inoculum was removed, and the cells were washed and incubated for another 30 h at 37°C. Infected cells were quantified by indirect immunofluorescence with anti-E1 MAb A4. Results are expressed as the percentage of the value for the control infection without heparin and represent the means from three independent experiments. (C) Purified viruses were preincubated with increasing concentrations of heparin for 1 h at 37°C, and then the virus-heparin mixes were chilled and incubated with Huh-7 cells at 4°C for 2 h. The inoculum was removed, and the cells were washed and lysed to extract RNAs. HCV genomes were measured by quantitative RT-PCR. Results are expressed as the percentage of the control binding without heparin and represent the means from three independent experiments.
To determine whether isolated E2 with a deletion of HVR1 (E2-ΔHVR1) interacts with HS, we performed heparin-pulldown experiments with lysates of cells infected with JFH1 or JFH1-ΔHVR1 viruses. We used different volumes of cell lysates to avoid saturation of the beads. As shown in Fig. 3A, HCV envelope glycoproteins from these two viruses were pulled down, and the interaction was inhibited by preincubation of the lysates with 500 μg/ml of heparin. We quantified the E2 pulled down by heparin-coated beads for both viruses for the different volumes of lysates used (Fig. 3B). For the lowest volume of lysates, E2-ΔHVR1 was less efficiently precipitated than wild-type E2. This result suggests that E2-ΔHVR1 interacts with HS and that HVR1 might modulate this interaction. However, since a dramatic reorganization of HCV envelope glycoproteins occurs during virion budding and/or egress (25), we also performed heparin-pulldown experiments with lysates of purified JFH1 or JFH1-ΔHVR1 viruses. As shown in Fig. 3C, HCV envelope glycoproteins from JFH1-ΔHVR1 interacted with heparin-coated beads. To quantify the interaction of native JFH1 or JFH1-ΔHVR1 particles with HS, we performed a pulldown experiment with heparin-coated beads and we quantified the HCV genome by quantitative RT-PCR. As shown in Fig. 3D, JFH1 and JFH1-ΔHVR1 were equally precipitated. Taken together, these data indicate that HVR1 of E2 is not the main determinant for cell surface attachment and binding of HCV virions to HSPGs. However, in the context of isolated proteins, HVR1 with other regions of the viral glycoproteins can contribute to HS binding (24, 26).
FIG 3.
Interaction between heparin and HCV envelope proteins. Different volumes of lysates (200 μl, 100 μl, or 50 μl) obtained from cells infected with JFH1 or JFH1-ΔHVR1 were preincubated in the presence or absence of 500 μg/ml heparin for 1 h at 4°C and then precipitated with heparin-conjugated beads. Proteins were eluted with Laemmli buffer, and samples were separated on SDS-polyacrylamide gels. (A) HCV envelope proteins were detected by immunoblotting with A4 (anti-E1) and A11 (anti-E2) antibodies. (B) The quantities of E1 and E2 precipitated under each condition are presented. Results are expressed as the percentage of the total protein input. Data were analyzed by using Student's t test (*, P < 0.01). Mean values from three independent experiments are given. Error bars represent the standard deviations of the means. (C) HCV envelope proteins precipitated with heparin-conjugated beads from purified virus JFH1 or JFH1-ΔHVR1 lysates were detected by immunoblotting with A4 (anti-E1) and A11 (anti-E2) antibodies. (D) Purified JFH1 or JFH1-ΔHVR1 viruses were precipitated with heparin-coated beads, anti-apoE-coated protein G beads, or protein G beads. The amount of virus that bound to the beads was measured by quantitative RT-PCR. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means.
apoE is involved in HCV binding to the cell surface.
Due to the presence of envelope glycoproteins as well as apolipoproteins at the surface of the HCV particle, contradictory results concerning the virion determinant involved in the initial step of binding to host cells have been obtained (24, 26–30). Indeed, both HCV envelope glycoprotein E2 and apoE have been proposed to be the virion component involved in binding to target cells. First, to confirm the association of apoE with the viral particle, we performed a pulldown assay with anti-apoE-coated beads. As seen in Fig. 3D, both viruses were equally pulled down.
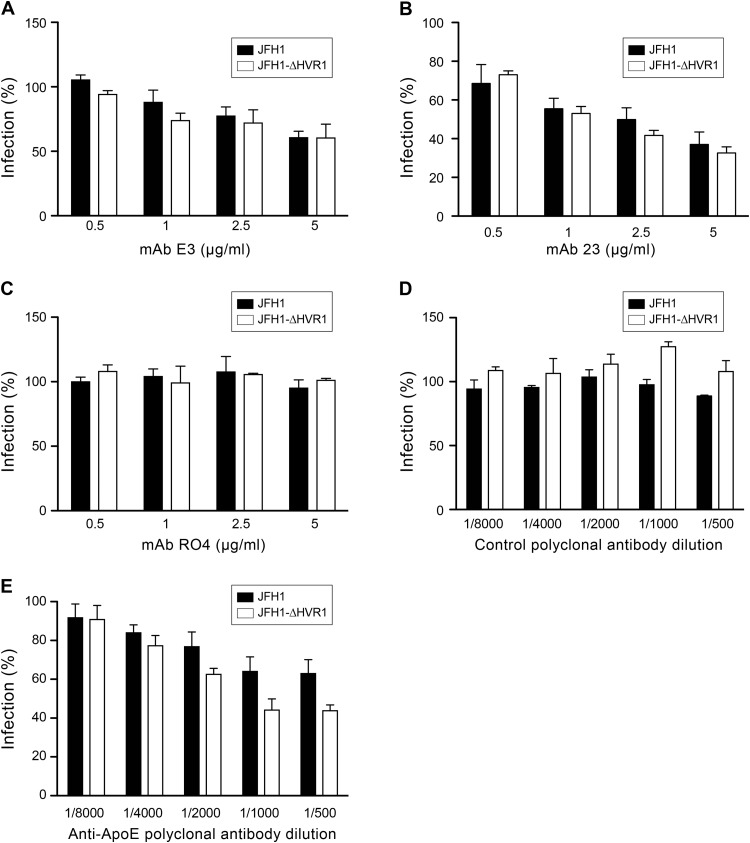
Therefore, to further characterize our JFH1-ΔHVR1 mutant, we determined its sensitivity to neutralization by anti-apoE antibodies. As shown in Fig. 4, both JFH1 and JFH1-ΔHVR1 viruses were equally inhibited by two different anti-apoE MAbs tested. However, JFH1-ΔHVR1 was more sensitive than the wild-type virus to neutralization by a polyclonal anti-apoE antibody. Indeed, a residual infectivity of 60% was observed for JFH1, whereas it went down to approximately 40% for the JFH1-ΔHVR1 virus (Fig. 4E).
FIG 4.
Anti-apoE antibody neutralization. Wild-type JFH1 and JFH1-ΔHVR1 viruses were preincubated with either control MAb RO4 (C), polyclonal antibody (E), or different anti-apoE antibodies, MAb E3 (A), MAb 23 (B), or polyclonal antibody 947 (D), at 37°C for 1 h and then added to the cells for 2 h. The inoculum was removed, and the cells were washed and incubated for another 30 h. Infected cells were detected by indirect immunofluorescence. Results are expressed as the percentage of HCVcc infectivity in the absence of antibodies. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means.
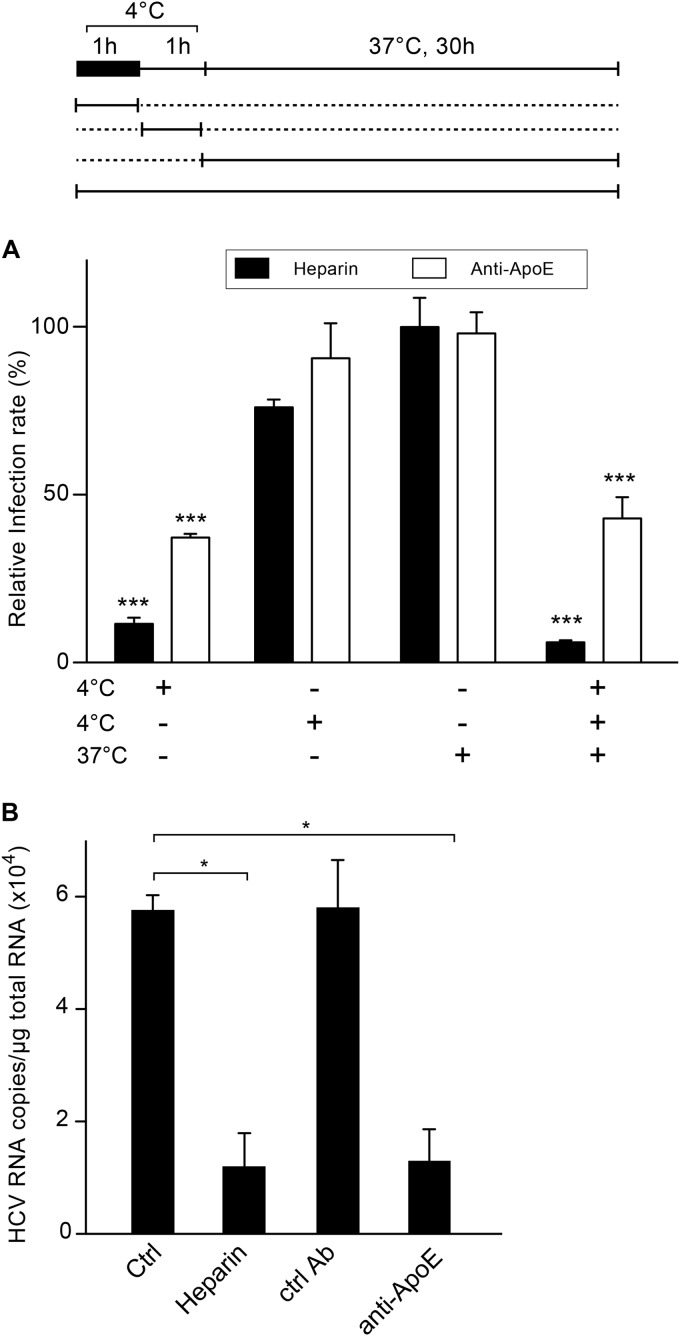
Due to the involvement of several cellular entry factors in HCV entry, kinetic studies of inhibition by specific antibodies have been used to determine the sequential implications of these different entry factors (49, 50). This can easily be done by incubating the virus with host cells at 4°C and then shifting the temperature to 37°C, with the specific antibodies or inhibitors being added at different time points (Fig. 5A). We therefore performed this type of experiment to determine whether apoE is involved in an early step of HCV entry. In parallel, we used heparin, which is known to inhibit the initial step of virion binding. As shown in Fig. 5A, the maximum inhibitory effect of anti-apoE antibody was observed when it was added together with the virus at 4°C. In contrast, no inhibition was observed if the virus was incubated with host cells at 4°C prior to treatment with the anti-apoE antibody. These results are very similar to the inhibitory effects observed with heparin, suggesting that apoE antibody inhibits HCV attachment at the cell surface. To confirm that the anti-apoE antibody inhibits virion attachment to host cells, Huh-7 cells were incubated at 4°C with purified JFH1 virus preincubated or not with a specific antibody or heparin, and the amounts of bound viral particles were determined by quantifying the HCV RNA associated with the cells after the removal of unbound virions. As shown in Fig. 5B, the anti-apoE antibody inhibited virion binding to host cells. Together, these data indicate that the apoE present at the surface of the viral particle is involved in virion attachment to host cells, which could likely be due to its binding to HSPGs.
FIG 5.
Anti-apoE antibodies inhibit HCV binding to the cell surface. (A) Kinetics of HCV entry inhibition by heparin or anti-apoE antibodies. Infection of the cells with purified JFH1 was divided into 3 steps. The schematic at the top shows that virus was inoculated into Huh-7 cells at 4°C for 1 h (black box). Then, the cells were rinsed and incubated at 4°C for an additional hour. Subsequently, the cells were washed and the temperature was shifted to 37°C for an additional hour. For each step, the presence of 500 μg/ml of heparin or 5 μg/ml of anti-apoE MAb 23 is depicted under the x axis. Thirty hours later, the cells were fixed and processed for immunofluorescence. Results are expressed as the percentage of the value for control infections without inhibitors. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means. Statistical analyses were performed by using a Bonferroni test (***, P < 0.001). (B) Cells were inoculated at 4°C for 2 h with purified virus with or without heparin (500 μg/ml) or with either anti-apoE MAb 23 or an irrelevant control (Ctrl) antibody (5 μg/ml). Cells were washed, and the amount of virus that bound to the cell surface was measured by quantitative RT-PCR. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means. Data were analyzed by using Student's t test (*, P < 0.05).
apoE is responsible for HCV interaction with HS.
Although our data do not support a role for HVR1 in HCV particle interaction with HS, we cannot definitively exclude the possibility that other regions in HCV envelope glycoproteins exposed on the virion are involved in binding to HSPGs. Furthermore, although our data indicate that apoE is involved in HCV virion binding to host cells, we still needed to confirm that virion-associated apoE directly binds to HS. Therefore, to clarify the relative contribution of apoE and envelope glycoproteins in the HCV interaction with HS, we performed pulldown experiments with heparin-coated beads in the presence of competitors that target the apoE-HS interaction or in the presence of different anti-HCV MAbs. Because MAbs 9/27 and 6/16 are directed against the E2 envelope protein of genotype 1a, we used an H77/JFH1 chimera for this experiment. As shown in Fig. 6A, none of the anti-HCV MAbs inhibited HCV binding to heparin. Interestingly, the epitopes that are recognized by the neutralizing antibody 9/27 and the 6/16 antibody are located in HVR1 (4); however, these antibodies were unable to inhibit the binding of the virus to heparin beads. Since these MAbs recognize two nonoverlapping epitopes, our data confirm that HVR1 is not crucial for virus binding to HS. To exclude the possibility that other regions of envelope glycoproteins can mediate the interaction with HS, we also used MAb AR5A, a conformational neutralizing antibody that recognizes a discontinuous epitope on the E1E2 heterodimer, and MAb AR3A, another conformational neutralizing antibody that disrupts E2 binding to CD81 (33). We also used patient-derived antibodies since such antibodies have previously been shown to be able to inhibit the HCV glycoprotein interaction with HS (26). However, neither the AR5A and AR3A antibodies nor the patient-derived sera were able to compete with HCV virion binding to heparin (Fig. 6A). In contrast, preincubation of the virions with anti-apoE antibody strongly inhibited HCV binding to heparin beads. Furthermore, incubation with an HS-binding peptide derived from apoE also markedly inhibited the interaction of HCV virions with heparin beads. Finally, heparin was able to compete with HCV binding to anti-apoE antibodies, as shown in a pulldown experiment with JFH1 and JFH1-ΔHVR1 virions (Fig. 6B). Together, these results are in favor of a major role of apoE in HCV binding to HS.
FIG 6.
apoE mediates HCV binding to HS. (A) Purified H77/JFH1 chimera was preincubated for 1 h at 37°C with 25 μg/ml heparin, antibodies (5 μg/ml MAb AR3A, 10 μg/ml MAb AR5A, 10 μg/ml MAb RO4, 10 μg/ml MAb 6/16, 20 μl MAb 9/27, 100 μg/ml purified IgG from a control patient [patient 5] or four different gt1a-infected patients [patients 1 to 4], 1/200-diluted anti-apoE polyclonal antibodies, or irrelevant [control] polyclonal antibodies [Ctrl pAb]), 20 μg/ml apoE-derived peptides, or 3× Flag-tagged control peptides before heparin-coated beads were added. The amount of virus that bound to the beads was measured by quantitative RT-PCR. The quantity of virus bound to the beads in the absence of any competitor was arbitrarily set equal to 100%. Mean values from three independent experiments are given. Data were analyzed by using a Dunnett's multiple-comparison test (***, P < 0.001). (B) Purified JFH1 or JFH1-ΔHVR1 viruses were incubated for 1 h at 37°C in the absence or presence of 25 μg/ml heparin and then precipitated with anti-apoE antibody-coated beads. Virus bound to the beads was measured by quantitative RT-PCR. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means.
HS structural determinants involved in HCV binding.
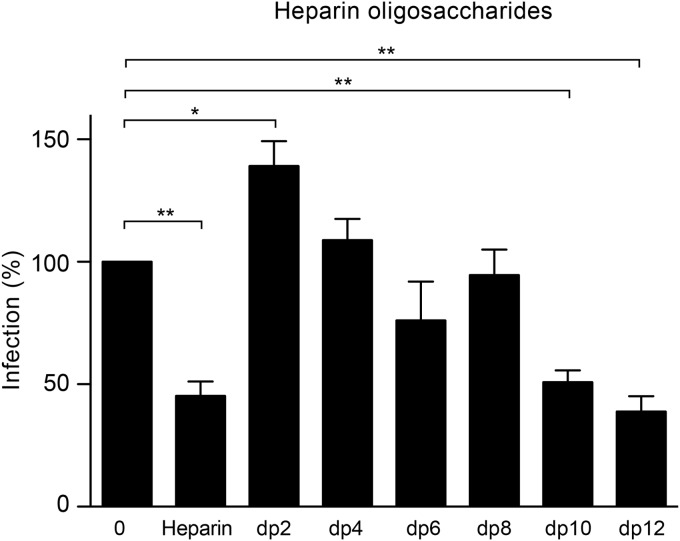
To address the HS structural determinants important for interaction with HCV, we first analyzed the minimal length of an HS unit required to inhibit HCV infection by using heparin-derived oligosaccharides of defined lengths. As shown in Fig. 7, we observed a 50% decrease in the level of infection in the presence of heparin, and preincubation of the virus with oligosaccharides with a degree of polymerization of 10 (dp10) saccharides or dp12 saccharides also inhibited HCV infection at levels similar to those for heparin. These data suggest that a minimum of 10 saccharides is necessary for HCV virion interaction with HS.
FIG 7.
HCV infection in the presence of heparin oligosaccharides of defined length. Purified viruses were preincubated with 1 mg/ml of heparin-derived oligosaccharides with different chain length (from dp2 to dp12) at 37°C for 1 h. Heparin was used as a positive control. Then, an equal volume of medium was added to the mixtures before they were chilled. Huh-7 cells were inoculated at 4°C for 2 h. The inoculum was removed, and the cells were washed and incubated for another 30 h at 37°C. Cells were processed for immunofluorescence to quantify the infection. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means. Data were analyzed by using Dunnett's multiple-comparison test (*, P < 0.05; **, P < 0.01).
To further characterize the HS determinants involved in HCV binding, we identified HS sulfotransferases expressed in Huh-7 cells. HS biosynthesis is divided into 3 main steps: chain initiation, polymerization, and modification. The initiation step is characterized by the linkage of a tetrasaccharide to the proteoglycan core protein. Then, the HS backbone is formed by the assembly of alternating glucuronate (GlcUA) and N-acetylglucosamine (GlcNAc) residues. As the chain assembles, it undergoes a series of modifications catalyzed by a C5 epimerase and multiple sulfotransferases. The first modification of the chain to occur is the removal of the N-acetyl group from subsets of GlcNAc and the addition of an N-sulfo group. This reaction is orchestrated by members of the NDST family. Further modifications of HS include epimerization of some glucuronate residues to iduronate residues and addition of sulfate groups at C-2 of uronic acid by 2-OST and at C-6 and/or at C-3 of GlcN residues by 6-OST and 3-OST. Although 2-OST is represented by a unique isoform, the 6-O-sulfation and 3-O-sulfation of HS can be catalyzed by three 6-OSTs and seven 3-OSTs. We therefore analyzed the expression profile of isoenzymes that are involved in HS sulfation. As shown in Fig. 8, we detected high levels of mRNA encoding NDST1 in Huh-7 cells. The level of expression of NDST2 was very low, while NDST3 and NDST4 transcripts were not detected. We also found a high level of 2-OST transcripts and a modest level of expression of 6-OST1 and 6-OST2, both of which are isoenzymes involved in the same 6-O-sulfation reaction. Even though seven enzymes are involved in the 3-O-sulfation reaction, we detected only low levels of 3-OST3A, 3-OST3B, and 3-OST5 in Huh-7 cells.
FIG 8.
HS biosynthetic enzyme expression profile. Total RNAs were extracted from Huh-7 cells, and the mRNA levels of enzymes involved in HS sulfation were quantified by quantitative RT-PCR. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means.
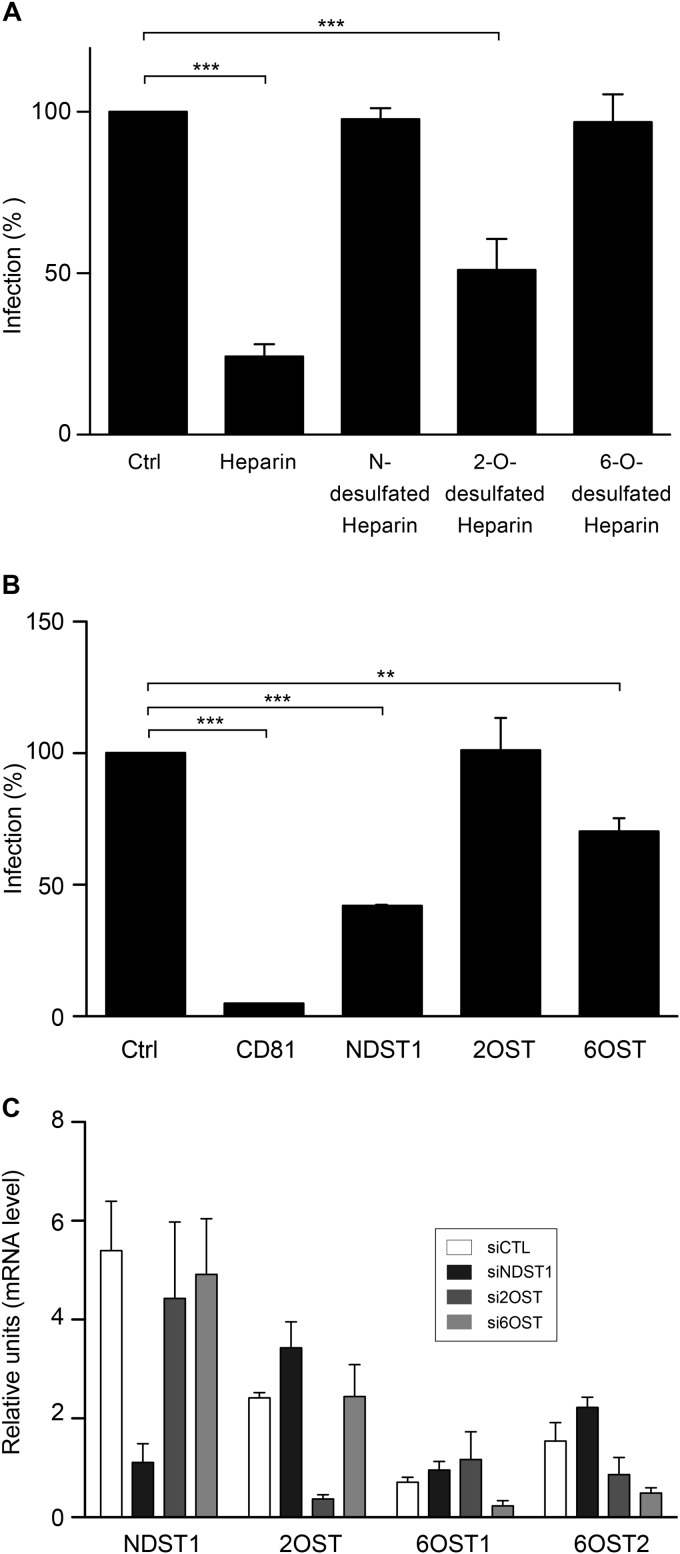
To determine if particular patterns of sulfation are important for HCV-HS interactions, we performed infection in the presence of chemically modified heparins. As shown in Fig. 9A, the 2-O-desulfated heparin was able to inhibit HCV infection by 50%, whereas unmodified heparin inhibited HCV infection by 80%. In contrast, N- or 6-O-desulfated heparins used at the same concentrations were unable to inhibit HCV infection. These results suggest that N- and 6-O-sulfo groups are important for HCV infection. To confirm these results, we used a silencing approach. We based our silencing strategy on the profile of expression of the enzymes expressed in Huh-7 cells. Consequently, small RNAs interfering with the expression of NDST1 and 2-OST and a combination of siRNAs targeting the two isoforms of 6-OST (6-OST1 and 6-OST2) were used. Furthermore, CD81 silencing was used as a positive control. The downregulation of the mRNA levels of the different enzymes was analyzed at 72 h posttransfection. The mRNA levels of NDST1, 2-OST, and 6-OST2 were efficiently decreased by their specific siRNAs (81% ± 7%, 85% ± 7%, and 74% ± 15%, respectively). Although we tested different siRNAs specific for 6-OST1, we could not achieve more than 60% silencing for this enzyme. It is worth noting that in cells silenced for the expression of NDST1, a slight upregulation of the other enzymes was observed (Fig. 9C). Then, we analyzed the consequence of the silencing of the different enzymes on HCV infection. As shown in Fig. 9B, infection was severely inhibited in cells in which CD81 was silenced. We observed a 60% decrease in the level of HCV infection in cells in which NDST1 was silenced, indicating that N-sulfation is necessary for HCV interaction with GAGs. HCV infection was unaffected by the silencing of 2-OST, whereas the silencing of both isoforms of 6-OST reduced HCV infection. We did not investigate the role of 3-O-sulfation in HCV entry because its requirement is very unlikely. Indeed, the 3-O-sulfotransferase that was the most expressed in Huh-7 cells was 3-OST3B, an enzyme that relies on prior 2-O-sulfation of the HS chain for its activity (51), and our results indicate that the 2-OST silencing has no effect on HCV infection. Together, our data are in agreement with the results obtained with the desulfated heparins and support the idea that N- and 6-O-sulfation but not 2-O-sulfation is required for HCV infection.
FIG 9.
Role of specific sulfation in HCV infection. (A) Purified virus was preincubated with heparin or its derivatives (100 μg/ml) for 1 h at 37°C, and then the mix was chilled to inoculate Huh-7 cells. After 2 h at 4°C, the cells were washed and incubated at 37°C for another 30 h. Infection was analyzed by immunofluorescence. Results are expressed as the percentage of the value for control infections without heparin. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means. Data were analyzed by using Dunnett's multiple-comparison test (***, P < 0.001). (B) Huh-7 cells were transfected with siRNA specific for CD81, NDST1, and 2-OST or a combination of siRNAs specific for 6-OST1 and 6-OST2. Five days later, the cells were inoculated with JFH1 virus for 1 h at 37°C. The cells were washed and further incubated for 30 h. HCV infection was quantified by immunofluorescence assay. Mean values from three independent experiments are given. Data were analyzed by using Dunnett's multiple-comparison test (**, P < 0.01; ***, P < 0.001). (C) Silencing of the different enzymes was measured by quantification of their mRNA levels 5 days after the transfection of the different siRNAs. Mean values from three independent experiments are given. Error bars represent the standard deviations of the means. siCTL, siNDST1, si2OST, and si6OST, siRNAs specific for the control, NDST1, 2-OST, and 6-OST, respectively.
DISCUSSION
As for many viruses, HCV interacts with HS, a complex group of cell surface-associated anionic polysaccharides. However, the exact contribution of HCV virion components to this interaction remains controversial. This is due to the unique nature of the HCV particle, which is associated with lipoproteins. Indeed, both apoE and HCV envelope glycoproteins have been proposed to play a role in HS binding (24, 26–30). However, after reinvestigating this question, our data support the suggestion that apoE plays a major role in HS binding, whereas the viral envelope glycoproteins present at the surface of the virion do not seem to be involved. We also characterized the structural determinants of HS that contribute to this interaction, and we found that N- and 6-O-sulfation but not 2-O-sulfation is required for HCV infection. Finally, we show that the minimum HS length unit required for HCV infection is a decasaccharide.
The HVR1 sequence of E2 glycoprotein is not involved in the binding of HCV virions to HS. Before the development of an HCV cell culture (HCVcc) system, the potential role of HS in HCV entry was first proposed on the basis of the findings of experiments performed with viral particles isolated from patients (23). However, in the absence of a culture system for HCV, recombinant viral envelope glycoproteins or surrogate models of viral particles have also been used to study this initial step of the HCV life cycle. Recombinant HCV envelope glycoprotein E2 was initially reported to interact with HS (24), and the capacity of HCV envelope glycoproteins to interact with HS has been confirmed with proteins isolated from purified virus (25). Furthermore, on the basis of the findings of studies with recombinant proteins or pseudoparticles containing HCV envelope glycoproteins, HVR1 of E2 has been proposed to contribute to this interaction (24, 26, 27). Here, we used a functional approach and a biochemical approach to reinvestigate the potential contribution of HVR1 to the HCV glycoprotein interaction with HS. In our biochemical approach, we found a partial decrease in the level of E2-ΔHVR1 binding to heparin-coated beads, suggesting that in the context of isolated proteins, HVR1 may contribute with other regions of E2 to mediate the interaction with HS, as suggested by previous studies (24, 26, 27). However, our functional studies indicate that in the context of HCVcc, it is unlikely that HVR1 mediates the HS interaction. Indeed, deletion of HVR1 does not reduce the binding capacity of the virion. JFH1-ΔHVR1 even showed an increased sensitivity to inhibition by heparin, but this difference could be due to a global change in the HCV particle after HVR1 deletion (48), which could modulate virion binding to HS. Concerning the functional studies, previous observations were first made with the help of pseudoparticles (26, 27). Nevertheless, these model systems are not the most appropriate tools to study such interactions, since pseudoparticles are devoid of the lipoprotein association that may affect the exposure of envelope proteins at the surface of the particles. Other experiments performed with the HCVcc system also suggested a role for HVR1 in HCV binding to HS (28). However, in this case only functional studies were performed without biochemical confirmation, and the phenotype observed could be due to indirect effects of the mutations, since HVR1 is involved in the interaction with SRB1 (8).
HCV envelope glycoproteins do not seem to play a role in the initial interaction of HCV particles with HS. The protein surface of HCV envelope glycoproteins is poorly exposed at the surface of viral particles; this is due in part to the association with lipoproteins (12) and to the presence of a large number of N-linked glycans (45, 52). These features dramatically reduce the possibilities for these glycoproteins to interact with HS when associated with the viral particle. They also explain the limited number of neutralizing epitopes identified on HCV glycoproteins. However, such neutralizing antibodies are interesting tools to probe the potential interaction of HCV envelope glycoproteins for their interaction with HS in the context of the virion. We therefore used several neutralizing antibodies targeting different regions of HCV envelope glycoproteins to determine their capacity to compete with HCV binding to heparin. As shown with MAb AR3A, the CD81-binding region is not involved in HCV-HS interaction, which is in agreement with a similar observation made with another MAb targeting this region (29). HVR1 is the most virion-exposed region of HCV envelope glycoproteins, and in agreement with the data obtained with our HVR1 deletion mutant, MAbs 9/27 and 6/16, recognizing epitopes in this region (4), did not inhibit HCV-HS interaction. We also used a neutralizing antibody recognizing an epitope shared by E1 and E2 and located outside the CD81-binding region (33), and again, no competition with the virus for binding to heparin was observed in the presence of this antibody. Finally, as it has been shown that anti-HCV antibodies isolated from patients are able to inhibit the interaction between recombinant E2 and HS, we also used antibodies isolated from several HCV-positive patients. However, we did not observe any effect of these antibodies on HCV-HS interaction. This lack of inhibition is likely due to the fact the heparin-binding site on HCV envelope glycoproteins is not exposed on native HCV particles.
Despite their potential lack of involvement in HCV virion attachment to HS, isolated HCV envelope glycoproteins are able to interact with heparin. Indeed, we confirmed that isolated HCV envelope glycoproteins expressed in the HCVcc system are able to interact with heparin. Although this might seem to be in contradiction with the lack of involvement in HCV virion attachment to HS, it is very likely that some HCV glycoprotein regions not exposed at the surface of the virion could have a heparin-binding motif. We can indeed expect that positively charged amino acids on the nonexposed face of the HCV glycoprotein complex interact with the phospholipid heads of the virion envelope and could have the capacity to interact with heparin after detergent dissociation. The structure of the core of the E2 envelope glycoprotein does not clearly show any extended area of positively charged residues that could correspond to a heparin-binding site (52, 53). However, one cannot exclude the possibility of the presence of such a region overlapping E1 and E2.
apoE is the virion component mediating the interaction with HS. This is in agreement with previously published data showing that viruses produced in cells expressing defective heparin-binding apoE mutants are not infectious (29). Indeed, our data on the inhibition of virion binding to heparin beads are in agreement with the lack of virus binding to heparin after ablation of the heparin-binding sequence of apoE. To discriminate the role of apoE and HVR1 in HCV entry, we performed neutralization experiments with different anti-apoE antibodies. Although JFH1-ΔHVR1 was more sensitive to neutralization by the anti-apoE polyclonal antibody, no difference was observed with two different MAbs. These results are in line with those recently reported by Bankwitz et al. (48). In addition, these authors also showed that HVR1 deletion does not affect the content of apoE associated with the virion (48). Furthermore, in line with the results of Jiang et al. (29), the findings of our kinetic analysis of HCV entry in the presence of anti-apoE antibody and the capacity of the anti-apoE antibody to inhibit virion binding to cells are other arguments supporting the role of apoE in binding to HS. The difference in the sensitivity of JFH1-ΔHVR1 to heparin and polyclonal anti-apoE antibody suggests that deletion of HVR1 affects the conformation of virion-associated apoE, as recently shown (48). A role for apoE in the interaction with HS is also supported by the abundance of this apolipoprotein on the virion, in contrast to the low number of HCV envelope proteins found on the viral particle (11, 12).
Binding of viral or cellular ligands to HS depends on defined patterns and orientations of the sulfo and carboxyl groups along the polysaccharide chain (54). In the liver, HS resembles heparin and has a high level of sulfation (1.34 sulfates/disaccharide, approximately twice the sulfation level observed in other tissues). More specifically, liver HS is rich in N-sulfated glucosamine and 2-O-sulfated iduronic acid and contains a high proportion of trisulfated disaccharides (55). These modifications are not spread equally along the glycosaminoglycan chains, but the highly sulfated motifs are clustered at the extremity of the chain distal from the core protein. The modifications of the HS chains require the action of specific sets of enzyme families. In this work, we analyzed the expression profiles of the enzymes involved in the different reactions of sulfation in the Huh-7 hepatoma cell line. It was reported that NDST1 and NDST2 are expressed in the liver; however, in Huh-7 cells, the level of NDST2 expression was very low and just above the detection limit. The specific functions of both enzymes are not completely understood. In mice, the knockout of NDST2 does not affect the structure of liver HS, whereas the knockout of NDST1 decreases the N-sulfation level by 50% (56, 57). The residual N-sulfation was attributed to NDST2, suggesting a compensatory effect of NDST2 in the absence of NDST1 expression. It is generally admitted that the reaction catalyzed by NDST is a prerequisite to further modifications. As expected, the knockout of NDST1 decreased the level of 2-O-sulfation; however, in hepatocytes, the level of 6-O-sulfated HS was barely affected. That can explain why the silencing of sulfotransferases was not as efficient as that of CD81 for the inhibition of HCV infection in Huh-7 cells. Indeed, we could expect that N-sulfation was not altered by the silencing of 6-OST1/2, while inhibition of the expression of NDST1 only partially reduced the level of 6-O-sulfate groups in HS chains. In line with these data, we observed an increase in the level of 6-OST1 expression when the expression of NDST1 was inhibited with a specific small interfering RNA. Interestingly, with the help of infectious viral particles produced in cell culture, we show for the first time that the reactions of HS sulfation catalyzed by NDST1 and 6-OST1/2, but not 2-OST, are required for HCV-HS interaction. Indeed, previous studies reported conflicting data concerning the role of specific sulfations in HCV entry. The binding of recombinant envelope proteins was shown to be dependent on N-sulfation (26), whereas by using HCV pseudoparticles, Basu et al. reported the role of O-sulfation in HCV entry (27). The different models used for these studies likely account for the discrepancies observed. These findings support the conclusion that specific sulfate groups on cellular HS rather than the total level of sulfation may be important for mediating HCV-host cell interaction. Besides the involvement of N- and 6-O-sulfate groups, the size of the saccharide chain appears to play an important role in efficient HCV-HS binding. Indeed, marked inhibition of HCV binding to target cells was observed for a dp10 oligosaccharide, in accordance with the findings of previous studies (26). These findings indicate that the interaction of hepatitis C virus with highly sulfated HS on target cells is not simply the result of charge interactions but requires a specific HS structure.
In conclusion, despite their capacity to interact with HS after dissociation from the virion with detergent, HCV envelope glycoproteins associated with the viral particle do not expose an HS-binding motif. In contrast, apoE associated with the HCV virion is responsible for HCV binding to specific HS involving N- and 6-O-sulfate groups and the minimal HS length unit required for HCV infection is a decasaccharide, indicating that HCV hijacks apoE to initiate its interaction with specific HS structures.
ACKNOWLEDGMENTS
This work was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS). Y.X. was supported by a fellowship from the China Scholarship Council (CSC; no. 2011610005), and S.B. was supported by a Marie Curie International Reintegration Grant (PIRG-GA-2009-256300).
We are grateful to Yves Rouillé and Czeslaw Wychowski for their scientific advice. We are grateful to Sophana Ung for his assistance with the illustrations. We also thank M. Law, J. McKeating, S. Foung, A. Goffard, and T. Wakita for providing us with reagents. The fluorescence microscopy data were generated with the help of the Bio-Imaging Center Lille Nord-de-France (BICeL).
REFERENCES
- 1.Ray SC, Bailey JR, Thomas DL. 2013. Hepatitis C virus. InKnipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed (electronic) Lippincott Wilkins & Williams, Philadelphia, PA. [Google Scholar]
- 2.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem 278:41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- 4.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A 100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vieyres G, Dubuisson J, Pietschmann T. 2014. Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion. Viruses 6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball JK, Tarr AW, McKeating JA. 2014. The past, present and future of neutralizing antibodies for hepatitis C virus. Antiviral Res 105C:100–111. doi: 10.1016/j.antiviral.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 8.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartenschlager R, Penin F, Lohmann V, Andre P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol 19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. 2013. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A 110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. 2012. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog 8:e1002466. doi: 10.1371/journal.ppat.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hueging K, Doepke M, Vieyres G, Bankwitz D, Frentzen A, Doerrbecker J, Gumz F, Haid S, Wolk B, Kaderali L, Pietschmann T. 2014. Apolipoprotein E codetermines tissue tropism of hepatitis C virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J Virol 88:1433–1446. doi: 10.1128/JVI.01815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol 83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeisel MB, Felmlee DJ, Baumert TF. 2013. Hepatitis C virus entry. Curr Top Microbiol Immunol 369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- 17.Shi Q, Jiang J, Luo G. 2013. Syndecan-1 serves as the major receptor for attachment of hepatitis C virus to the surfaces of hepatocytes. J Virol 87:6866–6875. doi: 10.1128/JVI.03475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefevre M, Felmlee DJ, Parnot M, Baumert TF, Schuster C. 2014. Syndecan 4 is involved in mediating HCV entry through interaction with lipoviral particle-associated apolipoprotein E. PLoS One 9:e95550. doi: 10.1371/journal.pone.0095550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin DN, Uprichard SL. 2013. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A 110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esko JD, Selleck SB. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 23.Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RW, Zarski JP, Drouet E. 2002. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J Med Virol 68:206–215. doi: 10.1002/jmv.10196. [DOI] [PubMed] [Google Scholar]
- 24.Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem 278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 25.Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. 2010. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol 84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barth H, Schnober EK, Zhang F, Linhardt RJ, Depla E, Boson B, Cosset FL, Patel AH, Blum HE, Baumert TF. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J Virol 80:10579–10590. doi: 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A, Kanda T, Beyene A, Saito K, Meyer K, Ray R. 2007. Sulfated homologues of heparin inhibit hepatitis C virus entry into mammalian cells. J Virol 81:3933–3941. doi: 10.1128/JVI.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koutsoudakis G, Dragun J, Perez-Del-Pulgar S, Coto-Llerena M, Mensa L, Crespo G, Gonzalez P, Navasa M, Forns X. 2012. Interplay between basic residues of hepatitis C virus glycoprotein E2 with viral receptors, neutralizing antibodies and lipoproteins. PLoS One 7:e52651. doi: 10.1371/journal.pone.0052651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. 2012. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol 86:7256–7267. doi: 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J, Wu X, Tang H, Luo G. 2013. Apolipoprotein E mediates attachment of clinical hepatitis C virus to hepatocytes by binding to cell surface heparan sulfate proteoglycan receptors. PLoS One 8:e67982. doi: 10.1371/journal.pone.0067982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res 42:3858–3863. [PubMed] [Google Scholar]
- 32.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. 2012. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A 109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J Virol 81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanpouille C, Denys A, Carpentier M, Pakula R, Mazurier J, Allain F. 2004. Octasaccharide is the minimal length unit required for efficient binding of cyclophilin B to heparin and cell surface heparan sulphate. Biochem J 382:733–740. doi: 10.1042/BJ20031453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurin G, Fresquet J, Granio O, Wychowski C, Cosset FL, Lavillette D. 2011. Identification of interactions in the E1E2 heterodimer of hepatitis C virus important for cell entry. J Biol Chem 286:23865–23876. doi: 10.1074/jbc.M110.213942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delgrange D, Pillez A, Castelain S, Cocquerel L, Rouille Y, Dubuisson J, Wakita T, Duverlie G, Wychowski C. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J Gen Virol 88:2495–2503. doi: 10.1099/vir.0.82872-0. [DOI] [PubMed] [Google Scholar]
- 40.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castelain S, Descamps V, Thibault V, Francois C, Bonte D, Morel V, Izopet J, Capron D, Zawadzki P, Duverlie G. 2004. TaqMan amplification system with an internal positive control for HCV RNA quantitation. J Clin Virol 31:227–234. doi: 10.1016/j.jcv.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Deligny A, Denys A, Marcant A, Melchior A, Mazurier J, van Kuppevelt TH, Allain F. 2010. Synthesis of heparan sulfate with cyclophilin B-binding properties is determined by cell type-specific expression of sulfotransferases. J Biol Chem 285:1701–1715. doi: 10.1074/jbc.M109.018184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotte M, Kersting C, Radke I, Kiesel L, Wulfing P. 2007. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast Cancer Res 9:R8. doi: 10.1186/bcr1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helle F, Vieyres G, Elkrief L, Popescu CI, Wychowski C, Descamps V, Castelain S, Roingeard P, Duverlie G, Dubuisson J. 2010. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol 84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prentoe J, Jensen TB, Meuleman P, Serre SB, Scheel TK, Leroux-Roels G, Gottwein JM, Bukh J. 2011. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol 85:2224–2234. doi: 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck ZY, Foung SK, Pecheur EI, Pietschmann T. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol 84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bankwitz D, Vieyres G, Hueging K, Bitzegeio J, Doepke M, Chhatwal P, Haid S, Catanese MT, Zeisel MB, Nicosia A, Baumert TF, Kaderali L, Pietschmann T. 2014. Role of hypervariable region 1 for the interplay of hepatitis C virus with entry factors and lipoproteins. J Virol 88:12644–12655. doi: 10.1128/JVI.01145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol 80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeisel MB, Cosset FL, Baumert TF. 2008. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology 48:299–307. doi: 10.1002/hep.22307. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Shworak NW, Sinay P, Schwartz JJ, Zhang L, Fritze LM, Rosenberg RD. 1999. Expression of heparan sulfate d-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem 274:5185–5192. doi: 10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- 52.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munoz EM, Linhardt RJ. 2004. Heparin-binding domains in vascular biology. Arterioscler Thromb Vasc Biol 24:1549–1557. doi: 10.1161/01.ATV.0000137189.22999.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyon M, Deakin JA, Gallagher JT. 1994. Liver heparan sulfate structure. A novel molecular design. J Biol Chem 269:11208–11215. [PubMed] [Google Scholar]
- 56.Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. 2004. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem 279:42732–42741. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 57.Ringvall M, Ledin J, Holmborn K, van Kuppevelt T, Ellin F, Eriksson I, Olofsson AM, Kjellen L, Forsberg E. 2000. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J Biol Chem 275:25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]