ABSTRACT
Methanococcus maripaludis has two surface appendages, archaella and type IV pili, which are composed of glycoprotein subunits. Archaellins are modified with an N-linked tetrasaccharide with the structure Sug-1,4-β-ManNAc3NAmA6Thr-1,4-β-GlcNAc3NAcA-1,3-β-GalNAc, where Sug is (5S)-2-acetamido-2,4-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose. The pilin glycan has an additional hexose attached to GalNAc. In this study, genes located in two adjacent, divergently transcribed operons (mmp0350-mmp0354 and mmp0359-mmp0355) were targeted for study based on annotations suggesting their involvement in biosynthesis of N-glycan sugars. Mutants carrying deletions in mmp0350, mmp0351, mmp0352, or mmp0353 were nonarchaellated and synthesized archaellins modified with a 1-sugar glycan, as estimated from Western blots. Mass spectroscopy analysis of pili purified from the Δmmp0352 strain confirmed a glycan with only GalNAc, suggesting mmp0350 to mmp0353 were all involved in biosynthesis of the second sugar (GlcNAc3NAcA). The Δmmp0357 mutant was archaellated and had archaellins with a 2-sugar glycan, as confirmed by mass spectroscopy of purified archaella, indicating a role for MMP0357 in biosynthesis of the third sugar (ManNAc3NAmA6Thr). M. maripaludis mmp0350, mmp0351, mmp0352, mmp0353, and mmp0357 are proposed to be functionally equivalent to Pseudomonas aeruginosa wbpABEDI, involved in converting UDP-N-acetylglucosamine to UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid, an O5-specific antigen sugar. Cross-domain complementation of the final step of the P. aeruginosa pathway with mmp0357 supports this hypothesis.
IMPORTANCE This work identifies a series of genes in adjacent operons that are shown to encode the enzymes that complete the entire pathway for generation of the second and third sugars of the N-linked tetrasaccharide that modifies archaellins of Methanococcus maripaludis. This posttranslational modification of archaellins is important, as it is necessary for archaellum assembly. Pilins are modified with a different N-glycan consisting of the archaellin tetrasaccharide but with an additional hexose attached to the linking sugar. Mass spectrometry analysis of the pili of one mutant strain provided insight into how this different glycan might ultimately be assembled. This study includes a rare example of an archaeal gene functionally replacing a bacterial gene in a complex sugar biosynthesis pathway.
INTRODUCTION
While N-linked glycosylation was initially identified as an exclusively eukaryotic process, it is now well established that this pathway is present in the prokaryotic domains of Archaea and Bacteria as well. The posttranslational modification of proteins with N-linked glycans is believed to be much more widespread in Archaea than in Bacteria. In N-linked glycosylation systems, an oligosaccharyltransferase is required for the transfer of assembled oligosaccharides from the lipid carrier onto target proteins; genes encoding this key signature protein of the pathway have been identified in 166 out of 168 sequenced archaeal genomes (1, 2). This is in contrast to Bacteria, where the N-glycosylation pathway seems restricted to members of the epsilon subdivision of Proteobacteria (Campylobacter, Wolinella, and Helicobacter) and a few Deltaproteobacteria, including Desulfovibrio species (3).
Though N-linked glycosylation is widespread in Archaea, the study of N-linked glycosylation pathways by both genetic and structural methodologies is concentrated on a few model species (4), including the thermoacidophile Sulfolobus acidocaldarius (5–9), the halophile Haloferax volcanii (10–14), and the methanogens Methanococcus voltae (15, 16) and Methanococcus maripaludis (17–23). In general, the archaeal N-linked glycosylation pathway contains aspects of both the eukaryotic and bacterial systems, with additional domain-specific traits that include unique glycan structures, a variety of linking sugars, and unusual dolichol carriers (4). Generally, archaeal N-linked glycans are initially assembled on the cytoplasmic face of the cytoplasmic membrane on dolichol lipid carriers through the activities of specific glycosyltransferases that catalyze the sequential addition of sugar monomers from nucleotide-charged sugar precursors. This lipid-linked oligosaccharide is then presumed to be flipped across the cytoplasmic membrane by a flippase prior to its en bloc transfer by the oligosaccharyltransferase AglB onto Asn residues within certain Asn-X-Ser/Thr sequons in the target protein, although additional sugars can still be added to the protein-bound glycan (4, 12).
N-linked glycans have been found to modify a variety of archaeal surface proteins, including S-layer proteins (5, 13), as well as the subunits that comprise appendages, including archaella (formerly archaeal flagella [24]) (14, 21) and pili (20). Typically, S-layer proteins and archaellins are used as reporters of archaeal N-linked glycosylation (25). The importance of N-linked glycosylation for archaellum assembly and function in S. acidocaldarius (7), H. volcanii (14), M. voltae (15), and M. maripaludis (21) is highlighted by archaellation and motility defects in mutants expressing archaellins that are either nonglycosylated or modified with truncated N-linked glycans.
The archaella of M. maripaludis S2 are composed of the major archaellins FlaB1 and FlaB2, located in the filament portion, and the minor archaellin FlaB3, found in the hook region (26). All three archaellins are modified by an N-linked tetrasaccharide at multiple sites (3 sites in FlaB1, 4 sites in FlaB2, and 2 sites in FlaB3) (17). The structure of the M. maripaludis S2 archaellin N-linked tetrasaccharide was determined to be Sug-1,4-β-ManNAc3NAmA6Thr-1,4-β-GlcNAc3NAcA-1,3-β-GalNAc, where Sug is (5S)-2-acetamido-2,4,-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose (17). An N-linked glycan also modifies the major pilin EpdE in M. maripaludis S2. However, the pilin glycan is a pentasaccharide that is identical to the archaellin tetrasaccharide, except for an additional hexose that branches from the first sugar (N-acetylgalactosamine [GalNAc]) (20).
Significant progress has been reported in the investigation of the N-linked glycosylation pathway in M. maripaludis. The oligosaccharyltransferase and the glycosyltransferases for the second, third, and fourth sugars of the archaellin tetrasaccharide have been identified as AglB (MMP1424), AglO (MMP1079), AglA (MMP1080), and AglL (MMP1088), respectively (21). Mutants carrying in-frame deletions of these genes exhibited archaellins bearing truncated N-linked glycans and defects in archaellum assembly or function. Archaellins from aglL and aglA mutants are modified by 3- and 2-sugar glycans, respectively. Though ΔaglL and ΔaglA cells are archaellated, they are less motile than wild-type cells; the motility defects correlate with the degree of glycan truncation. Mutants with archaellins modified by a 1-sugar glycan, such as those found in an aglO mutant, or nonglycosylated archaellins, as present in an aglB mutant, are nonarchaellated and thus nonmotile (21). Collectively, these results indicate that a minimum glycan of GlcNAc3NAcA-1,3-β-GalNAc is required for archaellum assembly in M. maripaludis S2; a similar requirement for archaellin modification by minimum N-linked glycans has been observed in M. voltae (15) and H. volcanii (14). In S. acidocaldarius, mutants producing a truncated N-glycan were shown to have severely reduced motility (7), although recent studies indicate that strains engineered to lack all six glycosylation sites in the archaellin FlaB were still motile (9). Recent studies have also shown that for FlaB2 of M. maripaludis, at least one of the four sequons used for N-glycan attachment must be occupied for archaellum assembly to occur (22), and for H. volcanii all sequons are necessary (14).
A number of gene products in M. maripaludis have been previously identified as involved in biosynthesis of the archaellin glycan. These include AglXYZ (MMP1081 to MMP1083), responsible for the acetamidino group modification at C-3 of the third sugar (18), AglU (MMP1084), a threonine transferase that modifies the third sugar at C-6, and AglV (MMP1085), a methyltransferase for the methyl group at C-5 in the fourth sugar (19). MMP0350 was also identified as a putative acetyltransferase involved in biosynthesis of the second sugar (27). Additionally, in vitro assays using heterologously expressed proteins have identified other M. maripaludis gene products that are involved in acetamido sugar biosynthesis (28, 29), although genetic analysis of these is still lacking. Furthermore, the lipid carrier for the N-glycan in M. maripaludis was identified as a dolichol monophosphate (23).
In this work, we identify several M. maripaludis genes whose products are predicted to account for the complete biosynthesis of the second sugar (2,3-diacetamido-2,3-dideoxyglucuronic acid [GlcNAc3NAcA]) of the archaellin glycan from UDP-GlcNAc as well as the biosynthesis of a direct precursor (2,3-diacetamido-2,3-dideoxymannuronic acid [ManNAc3NAcA]) of the third sugar that is likely further modified by the activities of AglXYZ and AglU to generate the final form of the third sugar (ManNAc3NAmA6Thr). We also present evidence that biosynthesis of UDP-ManNAc3NAcA occurs in a manner similar to the WbpABEDI pathway that generates UDP-ManNAc3NAcA as part of O-specific antigen (OSA) biosynthesis in Pseudomonas aeruginosa PAO1 lipopolysaccharide (30, 31).
MATERIALS AND METHODS
Strains and growth conditions.
Strains used in this study are listed in Table SA1 in the supplemental material. The Methanococcus maripaludis S2 Δhpt strain (Mm900) (32) and mutants derived from Mm900 were grown anaerobically under an atmosphere of CO2-H2 (20:80) in Balch medium III (33) at 37°C with shaking. During the in-frame deletion mutagenesis process, Mm900-derived transformants were grown in McCas medium supplemented with neomycin (1 mg/ml) or 8-azahypoxanthine (250 μg/ml; Acros Organics, NJ) as required (32). In complementation experiments, M. maripaludis mutants harboring their respective complementation plasmids were grown in nitrogen-free medium containing puromycin (2.5 μg/ml) for plasmid selection and supplemented with either l-alanine (10 mM) or NH4Cl (10 mM) as the lone nitrogen source (34). Escherichia coli TOP10 cells (Invitrogen, Burlington, ON, Canada) for plasmid cloning steps were grown in Luria-Bertani medium at 37°C supplemented with ampicillin (100 μg/ml) as needed. For cross-domain complementation experiments, Pseudomonas aeruginosa strains were grown in Luria-Bertani medium supplemented with gentamicin (50 μg/ml), carbenicillin (200 μg/ml), and tetracycline (50 μg/ml) when required. Antibiotics were obtained from Sigma-Aldrich (Oakville, ON, Canada).
Construction of plasmids for in-frame deletions and complementations.
In-frame deletions were constructed for mmp0351, mmp0352, mmp0353, mmp0355, mmp0357, and mmp0358 as previously described (32). In general, 1 kb of the up- and downstream flanking regions was amplified by PCR using the corresponding primer pairs (P1 with P2, P3 with P4) listed in Table SA2 in the supplemental material. Primers P2 and P3 introduced an AscI site so that ligation of the up- and downstream flanking regions yielded an in-frame deletion product for the target gene. BamHI or XbaI sites in P1 and P4 facilitated cloning of the in-frame deletion products into the vector pCRPrtNeo, resulting in pKJ1057 (mmp0351), pKJ1059 (mmp0352), pKJ980 (mmp0353), pKJ1148 (mmp0355), pKJ978 (mmp0357), and pKJ1182 (mmp0358) (see Table SA1 in the supplemental material).
For complementation of the mmp0351, mmp0352, mmp0353, and mmp0357 deletion mutants, each gene was amplified by PCR using the complementation primers listed in Table SA3 in the supplemental material. The resulting PCR products were digested with NsiI/MluI and cloned into NsiI/MluI-digested pHW40, a complementation vector in which transcription of the cloned gene is controlled by the inducible nif promoter (34). Internal NsiI sites were removed from mmp0352 and mmp0357 by a silent A-to-G and A-to-C change at nucleotides 891 and 663, respectively, using the site-directed mutagenesis (SDM) primers listed in Table SA3. The wild-type version of each respective gene was first cloned into the pCR2.1 TOPO vector, and these TOPO vector constructs were then used as the template for SDM.
The veracity of the in-frame deletion and complementation constructs was confirmed by DNA sequencing. Restriction enzymes were obtained from New England BioLabs, Fermentas, and Invitrogen. A PCR product purification kit from Qiagen (Toronto, ON, Canada) was used according to the manufacturer's protocols.
Generation of M. maripaludis mutants containing in-frame deletions of targeted genes.
Previously, mutants containing markerless in-frame deletions of mmp0350 and mmp0354 were reported (21, 27). Using the same methods established by Moore and Leigh (32), mutants containing markerless in-frame deletions of mmp0351, mmp0352, mmp0353, mmp0355, mmp0357, mmp0358, and mmp0359 were generated in M. maripaludis Mm900. Polyethylene glycol (PEG)-mediated transformation (35) was used to introduce pCRPrtNeo in-frame deletion constructs into M. maripaludis Mm900 cells. After subsequent plating of transformants on McCas-Noble agar containing 8-azahypoxanthine, single colonies were picked and then screened by PCR using primers designed to amplify across the target gene (listed in Table SA4 in the supplemental material) in order to identify deletion mutants. Mutants identified by PCR screening were restreaked and screened by PCR again to check for purity.
The complementation vectors were transformed into their corresponding deletion mutants, again using the PEG precipitation methodology (35). These complementation strains were subsequently grown under puromycin selection (2.5 μg/ml) in nitrogen-free medium supplemented with either l-alanine (nif promoter induced) or NH4Cl (nif promoter repressed) (34).
RT-PCR.
In order to determine if the genes mmp0350 to mmp0354 are cotranscribed, and if the genes mmp0359 to mmp0355 are cotranscribed, reverse transcriptase PCR (RT-PCR) was performed using primers (listed in Table SA5 in the supplemental material) designed to amplify across the intergenic regions between adjacent genes. The RNA template was isolated from M. maripaludis S2 wild-type cells using a High Pure RNA isolation kit (Roche, Mississauga, Ontario, Canada) followed by an additional DNase treatment (Turbo DNA-free kit; Ambion, Burlington, ON, Canada). A one-step RT-PCR kit (Qiagen) was used to amplify cDNA, using the supplied protocol. Two additional standard PCR amplifications were conducted, with each reaction using the same primer pairs but with different templates to verify amplicon sizes, using genomic DNA as the template, and to rule out genomic DNA contamination of the RNA sample, using purified RNA that did not undergo reverse transcription.
Western blot analysis of M. maripaludis archaellins and pilins.
Whole-cell lysates of M. maripaludis were separated by SDS-PAGE and transferred onto an Immobilon-P membrane (Millipore, MA). For detection of archaellin FlaB2, 12.5% acrylamide gels were used, while for detection of the EpdE pilin, 17.5% acrylamide gels were employed. FlaB2 was detected with anti-FlaB2-specific polyclonal chicken antibodies (18); horseradish peroxidase-conjugated rabbit anti-chicken IgY (Jackson Laboratories Inc., PA) was used as a secondary antibody. Since antibody specific for the major pilin EpdE is unavailable, deletion mutants were transformed with pWLG40 carrying epdE with an additional C-terminal FLAG tag to facilitate detection of EpdE-FLAG using monoclonal anti-FLAG (Sigma-Aldrich); horseradish peroxidase-conjugated goat anti-mouse IgG (Jackson Laboratories) was used as a secondary antibody. Blots were developed using a chemiluminescence kit (Roche) according to the manufacturer's protocols.
Isolation of archaella and pili.
Archaella from the mmp0357 deletion mutant were isolated as previously described by Bardy et al. (36). Pili were isolated from the nonarchaellated mmp0352 deletion mutant using the same method (20).
Mass spectrometry analysis.
Purified archaella (50 μg) from the mmp0357 deletion mutant was digested overnight with trypsin (Promega, Madison, WI) at a ratio of 30:1 (protein-enzyme [vol/vol]) in 50 mM ammonium bicarbonate at 37°C. The pilin isolated from the mmp0352 deletion mutant was first resolved by SDS-PAGE and then stained with Coomassie blue. The pilin band was excised and subjected to an overnight in-gel tryptic digestion. To improve peptide recovery, the digested pilin sample was then redigested with endoproteinase Asp-N (Roche, Indianapolis, IN). The digests were then analyzed by nano-liquid chromatography-tandem mass spectrometry (Nano-LC-MS/MS) by using a NanoAquity ultraperformance liquid chromatography (UPLC) system (Waters, Milford, MA) coupled to an Ultima hybrid quadrupole time of flight (QTOF) mass spectrometer (Waters). The digests were injected onto an Acclaim PepMax100 C18 μ-precolumn (5 mm by 300 μm [internal diameter]; Dionex/Thermo Scientific, Sunnyvale, CA) and resolved on a 1.7-μm BEH130 C18 column (100 μm by 100 mm [internal diameter]; Waters, Milford, CA) by using the following gradient conditions: 1 to 45% organic mobile phase (acetonitrile, 0.1% formic acid) over 36 min followed by a rapid increase to 95% acetonitrile and 0.1% formic acid in 2 min. The flow rate was 400 nl/min. MS/MS spectra were acquired on doubly, triply, and quadruply charged ions and searched against the NCBInr database using the Mascot search engine (Matrix Science, Ltd., London, United Kingdom). The spectral data sets were searched for glycopeptide MS/MS spectra, which were then interpreted by hand.
Cross-domain complementation studies.
Given the sequence similarity between mmp0353 and mmp0357 to the P. aeruginosa genes wbpA and wbpI, respectively, cross-domain complementation experiments were conducted to investigate these genes' potential functional equivalencies. mmp0353 and mmp0357 were each synthesized with a C-terminal His tag using P. aeruginosa codon preferences (GenScript, NJ); EcoRI/HindIII restriction sites were incorporated at the ends to facilitate cloning into the E. coli-P. aeruginosa shuttle vectors pUCP18/pUCP19 and pUCP26/pUCP27. The vector pairs each have their multiple cloning sites in opposite orientations; thus, cloned genes are in the correct orientation when in pUCP18 and pUCP26. The Shine-Dalgarno and spacer sequence AGGAGGACAAGCT (37) was included at the start of each gene to facilitate expression.
To transform P. aeruginosa cells with the pUCP18/pUCP19 and pUCP26/pUCP27 constructs (listed in Table SA1 in the supplemental material), electrocompetent cells were prepared according to Choi et al. (38), followed by electroporation using a MicroPulser (Bio-Rad). After a 2-h recovery, cells were plated on LB containing gentamicin and carbenicillin (for pUCP18/pUCP19) or gentamicin and tetracycline (for pUCP26/pUCP27). Transformants were confirmed by a Gmr Cbr (those harboring pUCP18/pUCP19 constructs) or Gmr Tcr (those harboring pUCP26/pUCP27 constructs) phenotype and recovery of the vector constructs using a QIAprep spin miniprep kit (Qiagen).
Bacteriophage D3 spot test and phage titration.
The restoration of wild-type O5-serotype OSA biosynthesis in complemented P. aeruginosa strains was assessed by a spot test using bacteriophage D3 (39), obtained from Andrew Kropinski. For these experiments, 10 μl of a 2 × 1010 PFU/ml stock of D3 was used. Phage titration was conducted using the double overlay method described by Adams (40).
LPS isolation and Western blot analysis.
Lipopolysaccharide (LPS) from overnight P. aeruginosa broth cultures was prepared as described by Hitchcock and Brown (41). LPS samples were separated on 15% PAGE gels (44:0.8 acryl-bis, 0.0824 M NaCl) and detected by silver staining (41). The LPS samples were also analyzed by Western blotting using murine monoclonal antibody MF15-4 (O5-serotype OSA specific; obtained from Joe Lam) (30).
Electron microscopy.
Electron microscopy of M. maripaludis cells was conducted using a Hitachi-7000 electron microscope operating at 75 kV. M. maripaludis cells were grown overnight; cells were pelleted and briefly washed with phosphate-buffered saline. Cells were loaded onto Formvar-coated copper grids and negatively stained with 2% phosphotungstic acid (pH 7.0).
RESULTS
We have previously reported on a number of genes involved in the biosynthesis and assembly of the tetrasaccharide N-linked to archaellins of M. maripaludis. The majority of these genes lay in a large cluster (mmp1079 to mmp1088) comprised of multiple operons (18, 19, 21). However, the deletion of one gene not located near this cluster, mmp0350, which encodes a putative acetyltransferase, was also previously shown to affect glycan structure and archaellum formation (27). In the immediate vicinity of mmp0350 are a number of other genes annotated as potentially involved in the biosynthesis of the glycan sugars or assembly of the glycan. Of these, one (mmp0354, annotated as an oligosaccharide transporter) was deleted previously in an unsuccessful attempt to identify the flippase responsible for flipping the lipid-linked oligosaccharide prior to its transfer to target proteins by AglB (21). The MMP0352 protein was shown after expression and purification in E. coli to efficiently catalyze UDP-GlcNAc oxidation (28), although deletion analysis has yet to be reported. This report focuses on the role of the remaining genes in this locus in the N-glycosylation pathway of M. maripaludis.
Construction of in-frame deletions.
In order to identify genes involved in sugar biosynthesis or glycan assembly in M. maripaludis, the genome was screened for clusters of genes whose annotations suggested roles in glycosylation. A cluster of 10 genes, which included the previously studied putative acetyltransferase mmp0350 (27), were identified and targeted for in-frame deletion to determine if any played a role in archaellin glycosylation. The annotations for these genes are listed in Table 1. In-frame deletion mutants were previously constructed for mmp0350 and mmp0354 (21, 27). Of the remaining eight genes, in-frame deletion mutants were successfully made in seven genes (mmp0351, mmp0352, mmp0353, mmp0355, mmp0357, mmp0358, and mmp0359). Screening of over 100 transformants in multiple experiments for a deletion of mmp0356 yielded only wild-type colonies. Successful deletion of each gene was determined initially by a PCR screen (see Fig. SA1 in the supplemental material), using the sequencing primers for each gene listed in Table SA4 in the supplemental material. In all cases, the sizes of amplicons obtained by PCR in the mutants and wild-type strains were as predicted. The in-frame nature of each deletion was subsequently confirmed by DNA sequencing.
TABLE 1.
M. maripaludis genes targeted for involvement in N-linked glycosylation in this study
| Gene | Gene product | BLAST result for P. aeruginosa Wbp proteins (% coverage [E value]) |
|---|---|---|
| mmp0350 (agl17) | Putative acetyltransferase | P. aeruginosa WbpD (82 [1e−59]) |
| mmp0351 (agl18) | Predicted pyridoxal phosphate-dependent aminotransferase | P. aeruginosa WbpE (94 [8e−66]) |
| mmp0352 (agl19) | Putative oxidoreductase | P. aeruginosa WbpB (37 [4e−6]) |
| mmp0353 (agl20) | UDP-glucose/GDP-mannose dehydrogenase-related protein | P. aeruginosa WbpA (93 [6e−52]) |
| mmp0354 | Putative oligosaccharide transporter (flippase) | |
| mmp0355 | Conserved hypothetical protein | |
| mmp0356 | Glycosyltransferase, group 1 | |
| mmp0357 (agl21) | UDP-N-acetylglucosamine 2-epimerase | P. aeruginosa WbpI (100 [5e−118]) |
| mmp0358 | Glycosyltransferase, family 1 | |
| mmp0359 | Glycosyltransferase, family 2 |
In-frame deletions result in a decrease in archaellin apparent molecular mass.
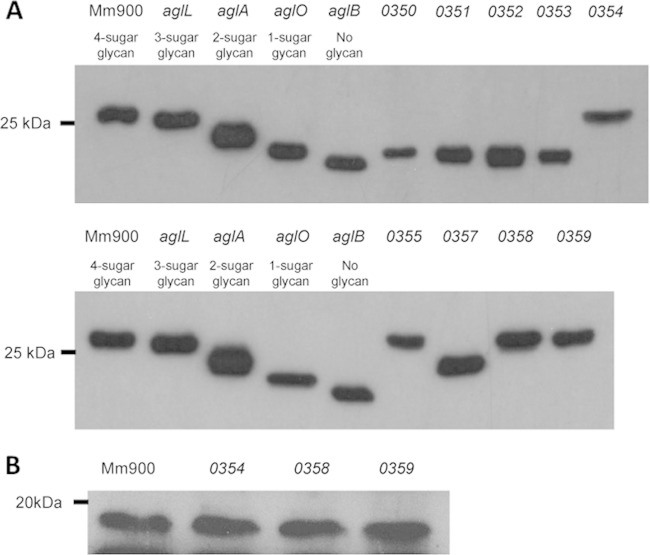
To determine if the in-frame deletion of mmp0351, mmp0352, mmp0353, mmp0355, mmp0357, mmp0358, or mmp0359 affected archaellin glycosylation, thereby resulting in archaellins of decreased apparent molecular mass, whole-cell lysates were initially analyzed by Western blots probed with antibody specific for FlaB2, a major archaellin filament glycoprotein (18). Even minor truncations to the M. maripaludis archaellin tetrasaccharide result in decreases of archaellin apparent molecular mass that are detectable by Western blotting as proteins of greater electrophoretic mobility than the wild type (18, 19, 21). As previously reported, archaellins from the ΔaglL, ΔaglA, and ΔaglO glycosyltransferase mutants have glycan lengths of 3 sugars, 2 sugars, and 1 sugar, respectively, while archaellins from the ΔaglB mutant are completely nonglycosylated (21). Whole-cell lysates from the ΔaglL, ΔaglA, ΔaglO, and ΔaglB strains collectively form a stepwise increase in FlaB2 electrophoretic mobility in Western blots which can be used effectively to initially estimate the degree of glycan truncation, if any, in novel deletion mutants. Archaellins from the Δmmp0351, Δmmp0352, and Δmmp0353 mutants displayed the same increase in electrophoretic mobility as those from the Δmmp0350 and ΔaglO mutants (i.e., with a predicted single-sugar glycan), while archaellins from the Δmmp0357 mutant displayed the same electrophoretic mobility as those from the ΔaglA mutant (i.e., with a predicted two-sugar glycan) (Fig. 1A).
FIG 1.
Western blot detection of archaellin FlaB2 and pilin EpdE-FLAG from in-frame deletion mutants. (A) Blots were developed with M. maripaludis anti-FlaB2. FlaB2 modified by known glycan lengths is as indicated for the wild type (Mm900), glycosyltransferase mutants (aglO, aglA, and aglL), and the oligosaccharyltransferase mutant (aglB). (B) Wild-type cells (Mm900) and the indicated deletion mutants were transformed with pKJ1107 and EpdE-FLAG detected with anti-FLAG antibodies.
Due to their annotations suggesting roles in sugar biosynthesis and archaellin apparent molecular masses equal to those of ΔaglO mutant archaellins, it is believed that mmp0351, mmp0352, and mmp0353 are all involved in the biosynthesis of the second sugar (GlcNAc3NAcA) of the archaellin tetrasaccharide, while the apparent molecular mass of archaellins from the Δmmp0357 mutant suggested that mmp0357 is involved in the biosynthesis of the third sugar (ManNAc3NAmA6Thr).
Archaellins from the Δmmp0354, Δmmp0355, Δmmp0358, and Δmmp0359 strains were all of apparent wild-type molecular mass, as estimated from Western blots, suggesting that deletion of these genes did not affect archaellin glycosylation. The results for the mmp0354 strain were consistent with the previous analysis of this deletion regarding effects on FlaB2 glycosylation (21). However, it is now known that EpdE, the major pilin of M. maripaludis, is also a glycoprotein, and it has an attached N-glycan that has the structure of the archaellin tetrasaccharide but with an extra hexose attached to the GalNAc linking sugar (20). Since mmp0358 and mmp0359 are annotated as glycosyltransferases, either could be the enzyme involved in the transfer of this hexose to the glycan. In addition, the annotation of mmp0354 suggests it could act as a flippase (a Wzx-like domain is present) that might be involved in the formation of the final pilin pentasaccharide. Thus, the possible involvement of all three of these genes was further examined for a role in pilin glycosylation. To investigate this, a FLAG-tagged version of EpdE was expressed in each of these mutants, as well as in wild-type cells, and the sizes of EpdE-FLAG proteins were determined by Western blots using anti-FLAG antibodies. If any of the deleted genes were involved in assembly of the pilin pentasaccharide, an increase in pilin electrophoretic mobility would be expected. However, in all cases, the electrophoretic mobility of EpdE-FLAG in the mutants was identical to that in the wild type (Fig. 1B). We conclude that none of these three genes encode proteins that are involved in assembly of either the archaellin or pilin glycans.
Gene complementation can restore archaellin molecular mass.
To confirm that the increase in electrophoretic mobility of archaellins in Western blots was due to the specific in-frame deletions, all deletion strains that showed an increase in the electrophoretic mobility of FlaB2 were complemented using a wild-type version of the deleted gene expressed in trans. mmp0350, mmp0351, mmp0352, mmp0353, and mmp0357 were each cloned into pHW40, a plasmid in which expression of the cloned gene is controlled by the inducible nif promoter (42). Complementation plasmids were transformed into their respective deletion mutants and maintained by using puromycin selection. The expression of cloned genes from pHW40 was induced by growing cells in nitrogen-free medium supplemented with l-alanine. Cells were also grown in nitrogen-free medium supplemented with NH4Cl. Though the nitrogen fixation pathway is not required in the presence of ammonia (43), a minor amount of transcription occurs from the nif promoter even in the presence of NH4Cl (21, 42).
When mmp0351 was expressed in deletion mutants grown in N-free medium supplemented with alanine, the archaellin apparent molecular mass was restored to that observed in wild-type cells. When the complemented strain was grown under NH4Cl-supplemented conditions, a ladder of FlaB2 molecular masses was detected, likely due to FlaB2 modified with partial glycans (Fig. 2B). This is believed to be a result of limited mmp0351 expression from pHW40 even when the nif promoter is repressed.
FIG 2.
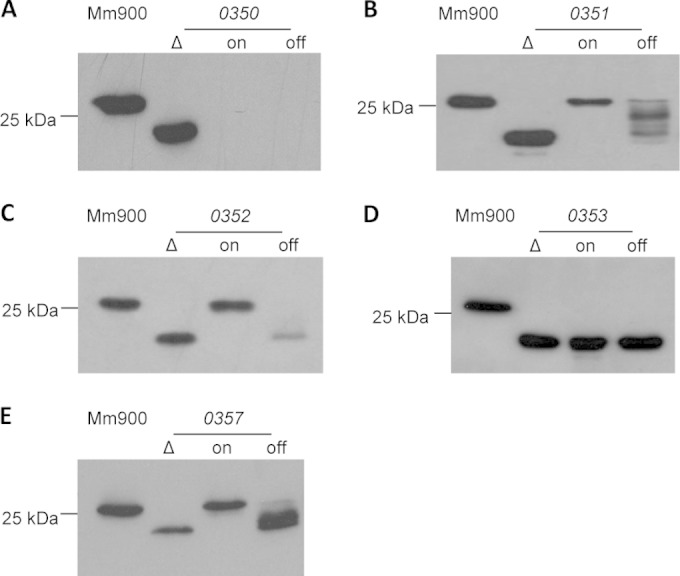
Western blot analysis of complementation experiments. Whole-cell lysates from mmp0350 (A), mmp0351 (B), mmp0352 (C), mmp0353 (D), and mmp0357 (E) deletion mutants as well as the same deletion mutants harboring their respective complementation vectors were separated by SDS-PAGE, transferred to an Immobilon membrane, and probed with anti-FlaB2. Complemented strains were grown in N-free medium with the addition of either alanine (transcription on) or NH4Cl (transcription off). Wild-type cell lysates (Mm900) were run as the control.
Complementation of the mmp0352 and mmp0357 deletion mutants also restored the archaellin apparent molecular mass to that of wild-type cells when the complemented cells were grown in N-free medium with the addition of alanine (Fig. 2C and E). Unlike the example of the mmp0351 complementation, though, in these cases, the archaellin apparent molecular mass was not restored to the wild-type size, even partially, in cells grown in the presence of NH4Cl.
Complementation of mmp0353 was also attempted multiple times. However, in each attempt, no change in the FlaB apparent molecular mass was observed upon complementation compared to the FlaB2 size observed in the mmp0353 mutant alone (Fig. 2D). An analysis of the complementation of mmp0350 by Western blotting detecting FlaB2 was attempted in the initial study of this gene (27) but thwarted as the mmp0350 deletion mutant stopped making FlaB2. In the current study, a new deletion of mmp0350 was generated and, again, complementation was attempted. Like before, the newly isolated mutant synthesized FlaB2 of increased electrophoretic mobility compared to that of the wild type when initially isolated. However, during the subculturing necessary in the course of the complementation experiment, the mutant again ceased making FlaB2, making the analysis of the complementation of archaellin glycosylation impossible (Fig. 2A).
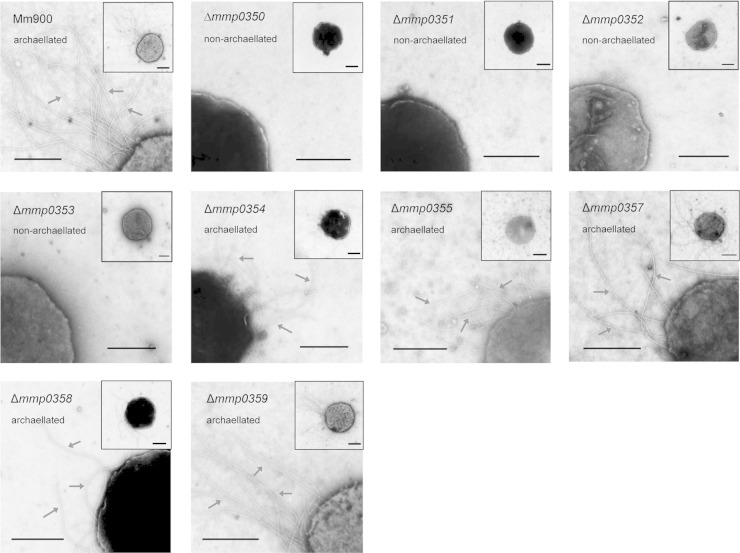
In-frame deletions affect archaellation.
The presence or absence of archaella on the surface of M. maripaludis cells can be predicted from Western blot results; the minimum glycan length required for archaellation in M. maripaludis is the disaccharide GlcNAc3NAcA-1,3-β-GalNAc (21). All deletion mutants were examined by electron microscopy to determine the effects of the deletions on archaellation (Fig. 3). The Δmmp0354, Δmmp0355, Δmmp0358, and Δmmp0359 mutants were all archaellated, as predicted based on their archaellins' wild-type apparent molecular mass in Western blots. Δmmp0357 mutant archaellins were thought to be modified by a disaccharide based on Western blot results, and these cells were predicted to be archaellated. This was confirmed by electron microscopy. Δmmp0351, Δmmp0352, and Δmmp0353 cells were all observed by electron microscopy to be nonarchaellated, again consistent with their archaellins having a glycan of a single sugar, as determined by Western blotting.
FIG 3.
Electron micrographs of the wild type and in-frame deletion mutants showing the archaellation state. Whole cells are shown in the inset with a portion of the whole cell enlarged to more readily observe archaella. All samples were negatively stained with 2% phosphotungstic acid (pH 7.0). Arrows indicate archaella. Scale bars represent 0.5 μm.
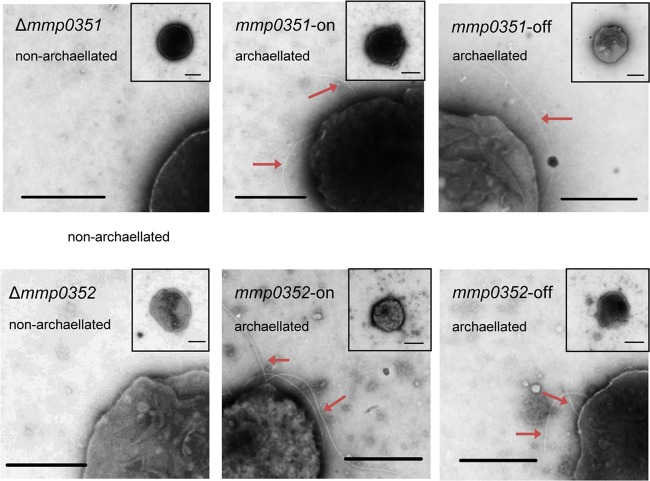
Δmmp0351 and Δmmp0352 complemented cells were examined by electron microscopy to determine if the restoration of archaellin electrophoretic mobility to that of wild-type archaellin was accompanied by a restoration of the wild-type archaellated phenotype. As expected, both Δmmp0351 and Δmmp0352 complemented cells were archaellated (Fig. 4). Complemented Δmmp0357 cells were not examined by electron microscopy because the deletion mutant was already archaellated.
FIG 4.
Electron micrographs of mmp0351 (A) and mmp0352 (B) deletion mutants complemented with the wild-type version of the respective gene. The deletions alone are shown in the left panel followed by the complement-on cells grown in N-free medium with the addition of alanine and complement-off cells grown in N-free medium with the addition of NH4Cl. All samples were negatively stained with 2% phosphotungstic acid (pH 7.0). Arrows indicate archaella. Scale bars represent 0.5 μm.
Mass spectrometry analysis of archaellins from the Δmmp0357 mutant identifies a truncated glycan structure.
Of all the mutant strains generated in this study, the Δmmp0357 mutant was the only strain that still produced archaella. To further characterize the glycan structure produced by this strain, archaella were purified, digested with trypsin, and analyzed by mass spectroscopy (MS). MS analysis of tryptic glycopeptides confirmed that the glycan produced was indeed truncated compared to that found in the wild type (Fig. 5a). Tryptic glycopeptides from Δmmp0357 mutant archaellin were shown to be modified with only a dimeric glycan species (258 Da to 203 Da; GlcNAc3NAcA-1,3-β-GalNAc). A representative spectrum of tryptic glycopeptide T53–81 is presented in Fig. 5b.
FIG 5.
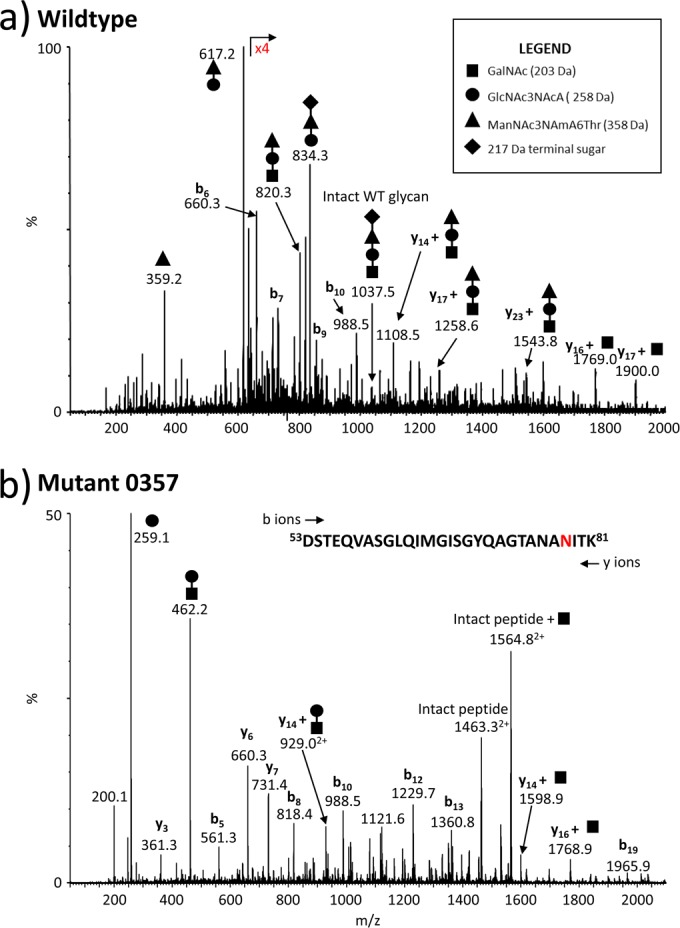
NanoLC-MS/MS analysis of the FlaB2 tryptic glycopeptide, T53–81, from the WT (a) and the Δmmp 0357 mutant strain (b). The FlaB2 tryptic peptide T53–81 contains one site of N-glycosylation (DSTEQVASGLQIMGISGYQAGTANANITK). The major carbohydrate oxonium ions are identified in the MS/MS spectra using symbols to indicate the sugar residues present: ◼, GalNAc; ●, GlcNAc3NAcA; ▲, ManNAc3NAmA6Thr; ◆, (5S)-2-acetamido-2,4-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose. The b and y ions arising from fragmentation of the peptide bonds are also shown. The structure of the tetrameric WT glycan has been described previously (17). The glycopeptide from the Δmmp0357 mutant strain (b) is modified with a disaccharide lacking the 3rd and 4th sugar residue of the WT glycan.
Mass spectrometry analysis of pilins from the Δmmp0352 mutant identifies a truncated glycan structure.
As the remaining Δmmp0350 to Δmmp0353 mutants, which all displayed an identical altered FlaB2 migration pattern in Western blots, are unable to produce archaella, it was necessary to examine pili from these strains to determine what specific changes to the glycan structure had occurred. As a representative of these mutant strains, pili were isolated and analyzed from the Δmmp0352 strain. Following trypsin/Asp-N double digestion of an excised gel band, a number of pilin glycopeptides were identified and characterized. In each case, only a single monosaccharide moiety was present at each site of glycosylation (203 Da; GalNAc). Representative spectra for pilin glycopeptide amino acids (aa) 52 to 62 and aa 34 to 47 are shown in Fig. 6a and b, respectively. The glycopeptide aa 34 to 47 contains two sequons that are adjacent to one another, and both sites (with the modified asparagines separated by only 2 amino acids) could be found modified with a single GalNAc. The MS data for FlaB2 is consistent with the Western blot results and in vitro studies showing that MMP0352 catalyzes UDP-GlcNAc oxidation at the 3′′ position (28), thus interfering with the synthesis of the second sugar of the N-linked glycan.
FIG 6.
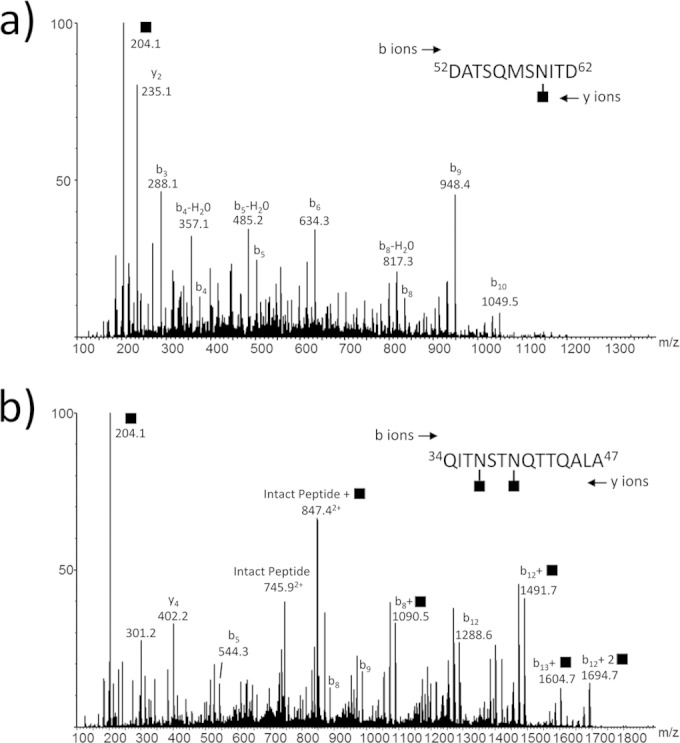
NanoLC-MS/MS analysis of Δmmp0352 mutant pilin tryptic/Asp-N-glycopeptides. (a) MS/MS spectrum of the doubly protonated glycopeptide ion at m/z 693.3 corresponding to 52DATSQMSNITD62 with a single GalNAc (◼). The same peptide was observed with no GalNAc residues attached (data not shown). (b) MS/MS spectrum of the doubly protonated glycopeptide ion at m/z 949.9 corresponding to 34QITNSTNQTTQALA47 modified with two GalNAc residues. The same peptide was also observed with 0 or 1 GalNAc modification (data not shown). Both MS/MS spectra are complicated by the strong loss of ammonia (−17 Da) and water (−18 Da) from the major fragment ions. Nevertheless, there is sufficient evidence in these spectra to confirm that these peptides are modified with one and two GalNAc residues, respectively.
RT-PCR analysis of the mmp0350 to mmp0359 genes.
Based on the orientations and short intergenic regions, it was thought that mmp0350 to mmp0354 were likely cotranscribed and that mmp0359 to mmp0355 also formed a single operon, transcribed from the complementary strand (see Fig. SA2A in the supplemental material). To test this hypothesis, primers were designed to amplify across the intergenic regions between adjacent genes within each possible operon. RT-PCR and standard PCR of RNA isolated from M. maripaludis Mm900 cells were performed as well as standard PCR with the same primers but using genomic DNA as the template to confirm amplicon size and primer specificity (see Fig. SA2B). In all cases, PCR products of the predicted size were amplified using DNA as the template or RNA subjected to reverse transcription. No products were obtained when RNA was not subjected to the reverse transcriptase step prior to standard PCR, thereby indicating that the RNA samples were not contaminated with DNA. The RT-PCR analysis shows that mmp0350 to mmp0354 are cotranscribed, and mmp0359 to mmp0355 are also cotranscribed.
Cross-domain complementation restores O-specific antigen biosynthesis in P. aeruginosa.
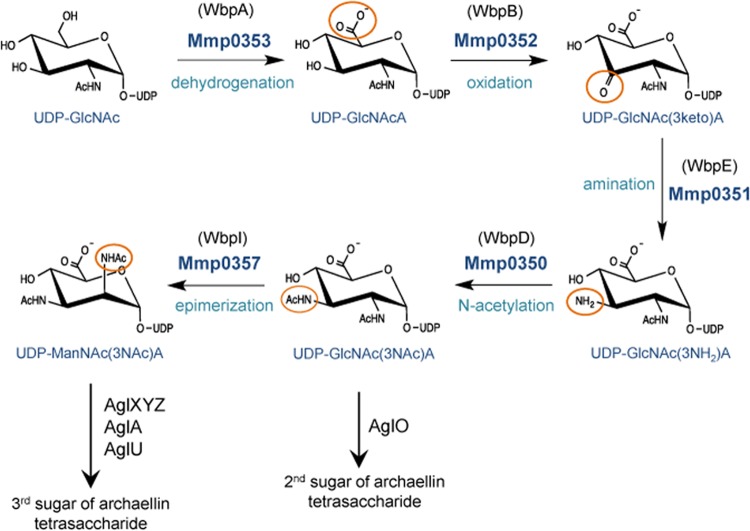
The gene annotations of mmp0350, mmp0351, mmp0352, mmp0353, and mmp0357 suggested various roles in sugar biosynthesis (Table 1). The amino acid sequences for each were used as queries for Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi). MMP0350, MMP0351, MMP0352, MMP0353, and MMP0357 were found to have high sequence similarity to the WbpD, WbpE, WbpB, WbpA, and WbpI Pseudomonas aeruginosa proteins, respectively. In P. aeruginosa PAO1, these five gene products are known to convert UDP-GlcNAc to UDP-ManNAc3NAcA in a stepwise fashion as part of O5-specific antigen biosynthesis (30, 31, 44).
Given the sequence similarities between the WbpD, WbpE, WbpB, WbpA, and WbpI P. aeruginosa PAO1 gene products and the MMP0350, MMP0351, MMP0352, MMP0353, and MMP0357 M. maripaludis gene products, it was hypothesized that the enzymatic activities of the M. maripaludis gene products were responsible for the biosynthesis of UDP-ManNAc3NAcA from UDP-GlcNAc in a pathway that is functionally equivalent to the WbpABEDI pathway in P. aeruginosa PAO1. ManNAc3NAcA is a component of OSA of O5-serotype P. aeruginosa PAO1; loss of UDP-ManNAc3NAcA biosynthesis results in loss of OSA expression (30, 31). ManNAc3NAcA is also likely a direct precursor of the third sugar of the M. maripaludis archaellin glycan and could then undergo further modifications by AglXYZ and AglU to generate ManNAc3NAmA6Thr.
Cross-domain complementation experiments were conducted for the first (WbpA) and last (WbpI) steps of the P. aeruginosa WbpABEDI pathway. The restoration of UDP-ManNAc3NAcA biosynthesis in P. aeruginosa was assessed by a spot test using bacteriophage D3, a lysogenic bacteriophage that uses O5 OSA as its receptor (39). Thus, successfully complemented P. aeruginosa deletion mutants were predicted to become sensitive to bacteriophage D3, as expression of functionally equivalent M. maripaludis gene products could restore UDP-ManNAc3NAcA biosynthesis and, in turn, the O5 OSA receptor for the phage.
MMP0353 and MMP0357 were expressed in trans in P. aeruginosa ΔwbpA and P. aeruginosa ΔwbpI mutants, respectively. For these experiments, mmp0353 and mmp0357 were synthesized using P. aeruginosa codon preferences, since the percentages of G+C content of the genome of each organism are dramatically different (P. aeruginosa PAO1, 66.6% [45]; M. maripaludis, 33.1% [46]). In addition, Shine-Dalgarno sequences were added to the sequence for expression purposes (37).
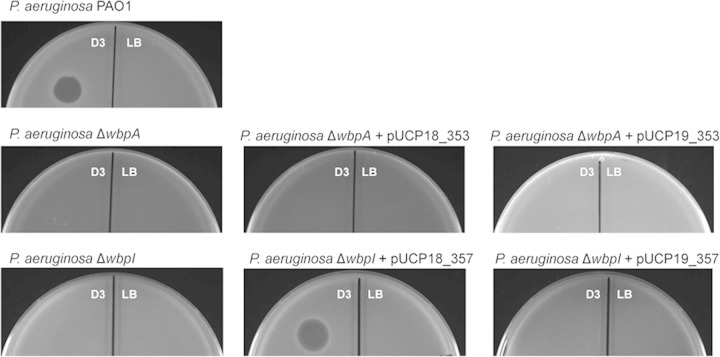
Spot testing of P. aeruginosa strains using bacteriophage D3 was used to screen for O5 OSA expression. The control P. aeruginosa PAO1 strain exhibited a zone of lysis when spotted with bacteriophage D3. As expected, neither the P. aeruginosa ΔwbpA mutant nor the P. aeruginosa ΔwbpI mutant were sensitive to D3 in spot tests, confirming the absence of O5 OSA expression in these deletion strains (Fig. 7). No lysis was observed when the P. aeruginosa ΔwbpA mutant was complemented with MMP0353 expressed from pUCP18. Removal of the C-terminal His tag from MMP0353 did not affect complementation results (data not shown); transcription of the non-His-tagged mmp0353 from pUCP18 was confirmed by RT-PCR (see Fig. SA3 in the supplemental material). Additional attempts to complement the P. aeruginosa ΔwbpA mutant using the non-His-tagged mmp0353 expressed from the alternate shuttle vector pUCP26 (47) did not result in restoration of O5 OSA expression when assessed by spot testing with bacteriophage D3 (data not shown).
FIG 7.
Spot test plates of P. aeruginosa strains using bacteriophage D3. Quadrants marked “D3” were spotted with 10 μl bacteriophage D3 (2 × 1010 PFU/ml stock). Quadrants marked “LB” were spotted with 10 μl of LB medium as a negative control. The complementation of the P. aeruginosa ΔwbpI mutant using MMP0357 expressed in trans restored D3 sensitivity.
In contrast to the complementation experiments with MMP0353, a zone of lysis was observed when the P. aeruginosa ΔwbpI mutant was complemented using MMP0357 expressed from pUCP18 but not when the complementation vector was pUCP19 (Fig. 7). Identical titers of D3 were obtained using P. aeruginosa PAO1 and the complemented P. aeruginosa ΔwbpI mutant as the host. Further confirmation of the restoration of O5 OSA expression in the P. aeruginosa ΔwbpI mutant when MMP0357 was expressed in trans was shown by Western blotting of isolated LPS using antibodies specific for O5 OSA (Fig. 8). As expected, no O5 OSA was detected in the P. aeruginosa ΔwbpI mutant, while the presence of O5 OSA was readily detected in the wild-type PAO1 strain. When the P. aeruginosa ΔwbpI mutant was complemented with pUCP18 carrying mmp0357, restoration of O5 OSA synthesis was observed, in agreement with the sensitivity to D3. As expected, no O5 OSA was detected using mmp0353 cloned into the pUCP19 complementation vector.
FIG 8.
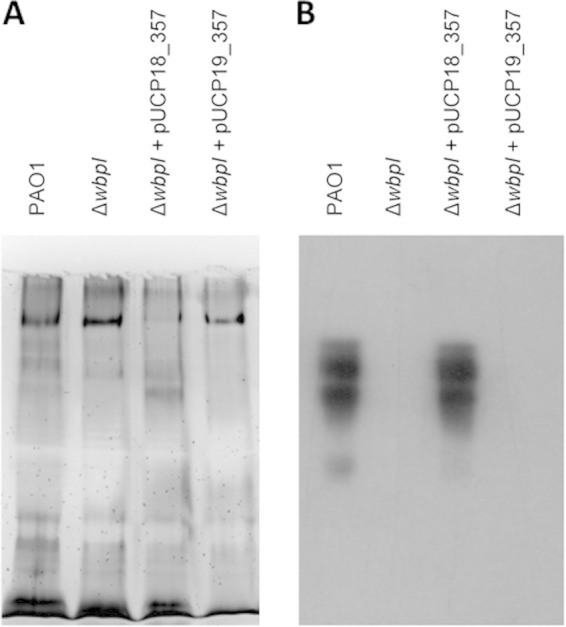
Restoration of O5 OSA synthesis in the complemented P. aeruginosa ΔwbpI strain. (A) Silver staining of LPS isolated from P. aeruginosa strains; (B) Western blot of LPS isolated from P. aeruginosa strains; the blot was developed using MF15-4, antibodies specific for O5 OSA. The complementation of the P. aeruginosa ΔwbpI mutant using MMP0357 expressed in trans restored O5 OSA biosynthesis.
DISCUSSION
In this study, five genes involved in the biosynthesis of the second and third sugars of the M. maripaludis archaellin N-linked tetrasaccharide glycan were identified: mmp0350, mmp0351, mmp0352, mmp0353, and mmp0357. The in-frame deletion of these genes resulted in archaellins bearing truncated glycans, as shown by increased electrophoretic mobility of archaellins in Western blots and by mass spectrometry analysis of archaellins and pilins. The genes collectively encode a five-step pathway for the biosynthesis of UDP-ManNAc3NAcA from UDP-GlcNAc. The final step of the pathway is the 2-epimerization of UDP-GlcNAc3NAcA to UDP-ManNAc3NAcA; the function of the M. maripaludis gene product responsible for this step was demonstrated by cross-domain complementation of an equivalent P. aeruginosa deletion strain.
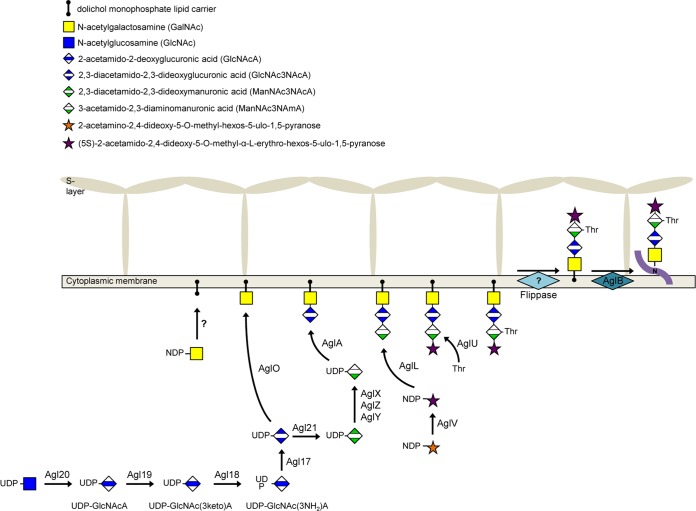
Deletions of mmp0351, mmp0352, and mmp0353 all demonstrated a nonarchaellated phenotype, and these genes, along with the Δmmp350 mutant (27), contribute to the biosynthesis of the second sugar (GlcNAc3NAcA). Western blotting showed that archaellins from the Δmmp0351, Δmmp0352, and Δmmp0353 mutants were all of decreased molecular mass, indicative of modification by a 1-sugar glycan (GalNAc). In M. maripaludis, the presence of a 1-sugar glycan on archaellins is known to be insufficient for archaellation, although the cells remain piliated (21). The major pilin, EpdE, is known to be N-glycosylated with a variant of the archaellin N-glycan (20). The pilin glycan is a pentasaccharide that is identical to the archaellin glycan but with a hexose attached as a branch to the GalNAc linking sugar. This knowledge allowed for a determination of the effect of gene deletions on N-glycosylation even in nonarchaellated cells by the MS analysis of isolated pili. We isolated pili from the mmp0352 deletion strain for analysis as a representative for the multiple deletion strains with archaellins bearing a 1-sugar glycan based on Western blots. MS analysis of EpdE pilins from the Δmmp0352 mutant confirmed our prediction that this group of genes was involved in the synthesis of the second sugar, as only tryptic glycopeptides bearing GalNAc were identified. Interestingly, the hexose branch found on GalNAc in pilin examined from wild-type cells was also absent. This result suggests that the attachment of the hexose to a glycan consisting of GalNAc alone cannot occur and perhaps occurs only after the tetrasaccharide is completely assembled on the lipid carrier. This may even occur after the tetrasaccharide has been transferred to the pilins. It has been previously shown in H. volcanii that the transfer, by AlgS, of the terminal mannose residue in the pentasaccharide N-linked to the S-layer occurs after the initial tetrasaccharide has been transferred to protein by AglB (48). In this scenario, the N-linked glycosylation system in M. maripaludis modifies both archaellins and pilins with the tetrasaccharide; however, pilins are further modified by the addition of a hexose group by a specific glycosyltransferase after the tetrasaccharide is already attached to pilins. Three potential glycosyltransferases are present in the mmp0350 to mmp0359 gene cluster, namely, mmp0356, mmp0358, and mmp0359. However, when EpdE-FLAG was expressed in the mmp0358 and mmp0359 deletion strains, no reduced molecular mass compared to its expression in wild-type cells was detected in Western blots, appearing to eliminate these two genes as encoding the glycosyltransferase responsible for this pilin-specific step. All attempts to delete mmp0356 were unsuccessful.
The mmp0357 gene was also targeted for its potential role in sugar biosynthesis. Deletion of mmp0357 resulted in archaellins of decreased apparent molecular mass, but these cells were archaellated when examined by electron microscopy. Western blotting with anti-FlaB2 initially suggested the presence of a 2-sugar glycan on Δmmp0357 mutant archaellins, and subsequent MS analysis of archaellins isolated from Δmmp0357 cells demonstrated a truncated glycan structure lacking ManNAc3NAmA6Thr and the terminal novel sugar [(5S)-2-acetamido-2,4,-dideoxy-5-O-methyl-α-l-erythro-hexos-5-ulo-1,5-pyranose]. Complementation of the Δmmp0357 mutant using the wild-type gene restored the archaellin molecular mass to that of the wild type. These data provide evidence that MMP0357 is involved in biosynthesis of the third sugar (ManNAc3NAmA) of the M. maripaludis archaellin glycan.
The five M. maripaludis genes (mmp0350, mmp0351, mmp0352, mmp0353, mmp0357) were assigned to specific steps of the UDP-ManNAc3NAcA biosynthesis pathway based on their high sequence similarity to gene products encoded by the wbp cluster in P. aeruginosa PAO1 (Fig. 9). In P. aeruginosa, the conversion of UDP-GlcNAc to UDP-ManNAc3NAcA requires the sequential enzymatic activities of WbpA, WbpB, WbpE, WbpD, and WbpI. In-frame deletions and subsequent complementation studies have shown that all steps are essential for the synthesis of UDP-ManNAc3NAcA and that without this pathway the O5 OSA is not made (30, 31). Biosynthesis of UDP-ManNAc3NAcA in Bordetella pertussis proceeds by the identical scheme, and successful cross-complementation of P. aeruginosa wbp deletion strains with the corresponding B. pertussis gene has been demonstrated (47). The evidence presented in this work demonstrates that this pathway also functions in Archaea. The evidence includes the extremely high sequence similarity of the five M. maripaludis genes to wbpABEDI and the MS analysis of N-linked glycans attached to either archaellins or pilins in the various M. maripaludis deletion strains, which are fully consistent with the proposed pathway and the cross-domain complementation of the final step of the pathway in the P. aeruginosa ΔwbpI mutant by the corresponding mmp0357 M. maripaludis gene, as evidenced by the restoration of O5 OSA synthesis. It is not clear why the cross-complementation of the first step of the pathway carried out by WbpA using MMP0353 was unsuccessful. Such complementation was possible using the corresponding B. pertussis gene, but the M. maripaludis gene represents a much greater evolutionary distance. In all cases, C-terminal His-tagged versions of the complemented proteins were used, and the same plasmid vectors were employed as well. Furthermore, it was not possible to show complementation even in the M. maripaludis mmp0353 deletion mutant. We do not attribute this inability to be due to polar effects, since the deletion of mmp0353 was made in frame and the only gene downstream and cotranscribed is mmp0354, which when deleted on its own had no detectable effect on the N-linked glycan. It may be that MMP0353 forms a multienzyme complex and needs to be translationally coupled to the other members of the complex. Alternatively, the stoichiometry of the MMP0353 subunits in comparison with the other subunits of the proposed complex may be significantly different when mmp0353 is expressed in trans from the nif promoter compared to its natural promoter. Unnatural variation in the stoichiometry of the subunits in a multisubunit complex can lead to formation of a nonfunctional complex (49, 50).
FIG 9.
Proposed biosynthetic pathway for UDP-d-ManNAc3NAcA in M. maripaludis, based on the WbpABEDI pathway determined for P. aeruginosa. P. aeruginosa proteins are shown in parentheses for each step. Additional steps required for generation of the final M. maripaludis sugars are indicated, as well.
Namboori and Graham (29) identified several enzymes involved in acetamido sugar biosynthesis in M. maripaludis. Particularly relevant to the current study are MMP0705 and MMP0706, which were shown in in vitro assays, using proteins purified after expression in E. coli, to convert UDP-GlcNAc to UDP-ManNAc (via the 2-epimerase activity of MMP0705) and then to UDP-ManNAcA (via the UDP-ManNAc 6-dehydrogenase activity of MMP0706). mmp0353 is a paralogue of mmp0706, and mmp0357 is a paralogue of mmp0705 (29). Our deletion analysis indicates that mmp0705 and mmp0706 cannot compensate for the loss of mmp0357 and mmp0353. Interestingly, Namboori and Graham (29) were able to demonstrate UDP-GlcNAc 2-epimerase activity for MMP0705 but not for MMP0357, despite the fact that MMP0357 is 53% identical to MMP0705. MMP0706 is homologous not only to UDP-ManNAc dehydrogenase but also to other enzymes, including UDP-Glc dehydrogenases, UDP-GalNAc dehydrogenase, and UDP-glucose/GDP-mannose dehydrogenase. In some organisms, such as B. pertussis, it has been shown that some 6-dehydrogenases are promiscuous and can use UDP-GlcNAc, UDP-GalNAc, and even UDP-Glc as substrates (47). While MMP0353 is 35% identical and 54% similar to MMP0706, its role in M. maripaludis is now believed to be as a UDP-GlcNAc dehydrogenase. Attempts to delete either mmp0705 or mmp0706 have been unsuccessful to date (unpublished data), indicating they might play a role in an essential pathway, such as in the synthesis of coenzyme M, a critical coenzyme for methanogenesis (51) that is reported to be modified with ManNAcA (52). The in vitro work of Namboori and Graham on MMP0352 enzymatic activity demonstrated its ability to catalyze the NAD-dependent oxidation of the 3′′ position of UDP-GlcNAc (28). The genetic and MS data presented in the current work and the role proposed for MMP0352 in the pathway for the generation of the second and ultimately the third sugars of the archaellin N-linked glycan are fully consistent with their findings.
The findings presented in this study significantly add to our knowledge of N-glycan biosynthesis in M. maripaludis, as the five gene products investigated in this contribution complete the entire biosynthetic pathways for the second and third sugars of the archaellin tetrasaccharide. The combined activities of MMP0353, MMP0352, MMP0351, and MMP0350 would convert UDP-GlcNAc to UDP-GlcNAc3NAcA, which is the second sugar in the archaellin tetrasaccharide. This sugar would be transferred to the dolichol monophosphate lipid carrier by AglO. In addition, in continuation of the pathway, UDP-GlcNAc3NAcA would be converted to UDP-ManNAc3NAcA through the 2-epimerase activity of MMP0357. This sugar could be seen as an immediate precursor to the third sugar of the N-linked glycan, ManNAc3NAmA6Thr, which could be generated by the activities of gene products previously identified. AglXYZ has been shown to be responsible for the acetamidino group at position 3 (18), and the AglA glycosyltransferase transfers this intermediate to the lipid carrier (21). The final, third sugar structure is obtained when AglU transfers a threonine to position 6. This threonine modification occurs only after the fourth sugar has been added to the lipid carrier (19). With the demonstrated involvement of mmp0350, mmp0351, mmp0352, mmp0353, and mmp0357 in the biosynthesis of the M. maripaludis N-linked glycan, these genes are now assigned agl gene designations (15) by following the protocol proposed by Eichler et al. (53). These designations are agl17 (mmp0350), agl18 (mmp0351), agl19 (mmp0352), agl20 (mmp0353), and agl21 (mmp0357). A model for the N-linked glycosylation pathway of M. maripaludis incorporating the proposed biosynthetic roles of Agl17, Agl18, Agl19, Agl20, and Agl21 is depicted in Fig. 10.
FIG 10.
Working model of the N-glycosylation pathway in M. maripaludis, highlighting the gene products analyzed in the current paper. The MMP nomenclature has been replaced with Agl designations: Agl17 (MMP0350), Agl18 (MMP0351), Agl19 (MMP0352), Agl20 (MMP0353), and Agl21 (MMP0357).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) (to K.F.J.).
We thank Andrew Kropinski, Joe Lam, and John Leigh for strains, bacteriophage, and antibodies.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00040-15.
REFERENCES
- 1.Magidovich H, Eichler J. 2009. Glycosyltransferases and oligosaccharyltransferases in Archaea: putative components of the N-glycosylation pathway in the third domain of life. FEMS Microbiol Lett 300:122–130. doi: 10.1111/j.1574-6968.2009.01775.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaminski L, Lurie-Weinberger MN, Allers T, Gophna U, Eichler J. 2013. Phylogenetic- and genome-derived insight into the evolution of N-glycosylation in Archaea. Mol Phylogenet Evol 68:327–339. doi: 10.1016/j.ympev.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Nothaft H, Szymanski CM. 2013. Bacterial protein N-glycosylation: new perspectives and applications. J Biol Chem 288:6912–6920. doi: 10.1074/jbc.R112.417857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarrell KF, Ding Y, Meyer BH, Albers SV, Kaminski L, Eichler J. 2014. N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol Rev 78:304–341. doi: 10.1128/MMBR.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peyfoon E, Meyer B, Hitchen PG, Panico M, Morris HR, Haslam SM, Albers SV, Dell A. 2010. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with the chitobiose-linked N-glycans. Archaea. doi: 10.1155/2010/754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer BH, Zolghadr B, Peyfoon E, Pabst M, Panico M, Morris HR, Haslam SM, Messner P, Schäffer C, Dell A, Albers SV. 2011. Sulfoquinovose synthase—an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius. Mol Microbiol 82:1150–1163. doi: 10.1111/j.1365-2958.2011.07875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer BH, Peyfoon E, Dietrich C, Hitchen P, Dell A, Albers SV. 2013. Agl16, a thermophilic glycosyltransferase, mediating the last step of the N-glycan biosynthesis in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J Bacteriol 195:2177–2186. doi: 10.1128/JB.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer BH, Albers SV. 2013. Hot and sweet: protein glycosylation in Crenarchaeota. Biochem Soc Trans 41:384–392. doi: 10.1042/BST20120296. [DOI] [PubMed] [Google Scholar]
- 9.Meyer BH, Birich A, Albers SV. 6 November 2014. N-glycosylation of the archaellum filament is not important for archaella assembly and motility, although N-glycosylation is essential for motility in Sulfolobus acidocaldarius. Biochimie. doi: 10.1016/j.biochi.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Guan Z, Naparstek S, Calo D, Eichler J. 2012. Protein glycosylation as an adaptive response in Archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ Microbiol 14:743–753. doi: 10.1111/j.1462-2920.2011.02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaminski L, Guan Z, Abu-Qarn M, Konrad Z, Eichler J. 2012. AglR is required for addition of the final mannose residue of the N-linked glycan decorating the Haloferax volcanii S-layer glycoprotein. Biochim Biophys Acta 1820:1664–1670. doi: 10.1016/j.bbagen.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichler J. 2013. Extreme sweetness: protein glycosylation in Archaea. Nat Rev Microbiol 11:151–156. doi: 10.1038/nrmicro2957. [DOI] [PubMed] [Google Scholar]
- 13.Kaminski L, Guan Z, Yurist-Doutsch S, Eichler J. 2013. Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. mBio 4(6):e00716-13. doi: 10.1128/mBio.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripepi M, You J, Temel S, Önder Ö, Brisson D, Pohlschröder M. 2012. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J Bacteriol 194:4876–4887. doi: 10.1128/JB.00731-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol 61:259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- 16.Voisin S, Houliston RS, Kelly J, Brisson JR, Watson D, Bardy SL, Jarrell KF, Logan SM. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J Biol Chem 280:16586–16593. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- 17.Kelly J, Logan SM, Jarrell KF, Vandyke DJ, Vinogradov E. 2009. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr Res 344:648–653. doi: 10.1016/j.carres.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Jones GM, Wu J, Ding Y, Uchida K, Aizawa S, Robotham A, Logan SM, Kelly J, Jarrell KF. 2012. Identification of genes involved in the acetamidino group modification of the flagellin N-linked glycan of Methanococcus maripaludis. J Bacteriol 194:2693–2702. doi: 10.1128/JB.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Y, Jones GM, Uchida K, Aizawa SI, Robotham A, Logan SM, Kelly J, Jarrell KF. 2013. Identification of genes involved in the biosynthesis of the third and fourth sugars of the Methanococcus maripaludis archaellin N-linked tetrasaccharide. J Bacteriol 195:4094–4104. doi: 10.1128/JB.00668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng SYM, Wu J, Nair DB, Logan SM, Robotham A, Tessier L, Kelly JF, Uchida K, Aizawa S, Jarrell KF. 2011. Genetic and mass spectrometry analysis of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J Bacteriol 193:804–814. doi: 10.1128/JB.00822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandyke DJ, Wu J, Logan SM, Kelly JF, Mizuno S, Aizawa SI, Jarrell KF. 2009. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol 72:633–644. doi: 10.1111/j.1365-2958.2009.06671.x. [DOI] [PubMed] [Google Scholar]
- 22.Ding Y, Uchida K, Aizawa SI, Murphy K, Khursigara CM, Chong JPJ, Jarrell KF. 20 February 2015. Effects of N-glycosylation site removal in archaellins on the assembly and function of archaella in Methanococcus maripaludis. PLoS One. doi: 10.1371/journal.pone.0116402._. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin A, Chang MM, Whitworth GE, Imperiali B. 2013. Biochemical evidence for an alternate pathway in N-linked glycoprotein biosynthesis. Nature Chem Biol 9:367–373. doi: 10.1038/nchembio.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarrell KF, Albers SV. 2012. The archaellum: an old motility structure with a new name. Trends Microbiol 20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Jarrell KF, Jones GM, Kandiba L, Nair DB, Eichler J. 2010. S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea. doi: 10.1155/2010/612948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa SI, Jarrell KF. 2007. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol 66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- 27.VanDyke DJ, Wu J, Ng SY, Kanbe M, Chaban B, Aizawa SI, Jarrell KF. 2008. Identification of putative acetyltransferase gene, MMP0350, which affects proper assembly of both flagella and pili in the archaeon Methanococcus maripaludis. J Bacteriol 190:5300–5307. doi: 10.1128/JB.00474-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namboori SC, Graham DE. 2008. Enzymatic analysis of uridine diphosphate N-acetyl-d-glucosamine. Anal Biochem 301:94–100. doi: 10.1016/j.ab.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Namboori SC, Graham DE. 2008. Acetamido sugar biosynthesis in the euryarchaea. J Bacteriol 190:2987–2996. doi: 10.1128/JB.01970-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burrows LL, Pigeon KE, Lam JS. 2000. Pseudomonas aeruginosa B-band lipopolysaccharide genes wbpA and wbpI and their Escherichia coli homologues wecC and wecB are not functionally interchangeable. FEMS Microbiol Lett 189:135–141. doi: 10.1111/j.1574-6968.2000.tb09219.x. [DOI] [PubMed] [Google Scholar]
- 31.Westman EL, McNally DJ, Charchoglyan A, Brewer D, Field RA, Lam JS. 2009. Characterization of WbpB, WbpE, and WbpD and reconstitution of a pathway for the biosynthesis of UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid in Pseudomonas aeruginosa. J Biol Chem 284:11854–11862. doi: 10.1074/jbc.M808583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore BC, Leigh JA. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol 187:972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lie TJ, Leigh JA. 2003. A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol Microbiol 47:235–246. doi: 10.1046/j.1365-2958.2003.03293.x. [DOI] [PubMed] [Google Scholar]
- 35.Tumbula DL, Makula RA, Whitman WB. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol Lett 121:309–314. doi: 10.1111/j.1574-6968.1994.tb07118.x. [DOI] [Google Scholar]
- 36.Bardy SL, Mori T, Komoriya K, Aizawa S, Jarrell KF. 2002. Identification and localization of flagellins FlaA and FlaB3 within flagella of Methanococcus voltae. J Bacteriol 184:5223–5233. doi: 10.1128/JB.184.19.5223-5233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang P, Habash M, Burrows LL. 2005. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J Bacteriol 187:829–839. doi: 10.1128/JB.187.3.829-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Taylor VL, Udaskin ML, Islam ST, Lam JS. 2013. The D3 bacteriophage α-polymerase inhibitor (Iap) peptide disrupts O-antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J Bacteriol 195:4735–4741. doi: 10.1128/JB.00903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams MH. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY. [Google Scholar]
- 41.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lie TJ, Wood GE, Leigh JA. 2005. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J Biol Chem 280:5236–5241. doi: 10.1074/jbc.M411778200. [DOI] [PubMed] [Google Scholar]
- 43.Kessler PS, Leigh JA. 1999. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics 152:1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wenzel CQ, Daniels C, Keates RAB, Brewer D, Lam JS. 2005. Evidence that WbpD is an N-acetyltransferase belonging to the hexapeptide acyltransferase superfamily and an important protein for O-antigen biosynthesis in Pseudomonas aeruginosa PAO1. Mol Microbiol 57:1288–1303. doi: 10.1111/j.1365-2958.2004.04767.x. [DOI] [PubMed] [Google Scholar]
- 45.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GKS, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 46.Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, Hackett M, Haydock AK, Kang A, Land ML, Levy R, Lie TJ, Major TA, Moore BC, Porat I, Palmeiri I, Rouse G, Saenphimmachak C, Soll D, Van Dien S, Wang T, Whitman WB, Xia Q, Zhang Y, Larimer FW, Olson MV, Leigh JA. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol 186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westman EL, Preston A, Field RA, Lam JS. 2008. Biosynthesis of a rare di-N-acetylated sugar in the lipopolysaccharides of both Pseudomonas aeruginosa and Bordetella pertussis occurs via an identical scheme despite different gene clusters. J Bacteriol 190:6060–6069. doi: 10.1128/JB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen-Rosenzweig C, Yurist-Doutsch S, Eichler J. 2012. AglS, a novel component of the Haloferax volcanii N-glycosylation pathway, is a dolichol phosphate-mannose mannosyltransferase. J Bacteriol 194:6909–6916. doi: 10.1128/JB.01716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cieslewicz M, Vimr E. 1997. Reduced polysialic acid capsule expression in Escherichia coli K1 mutants with chromosomal defects in kpsF. Mol Microbiol 26:237–249. doi: 10.1046/j.1365-2958.1997.5651942.x. [DOI] [PubMed] [Google Scholar]
- 50.Løvdok L, Bentele K, Vladimirov N, Müller A, Pop FS, Lebiedz D, Kollmann M, Sourjik V. 2009. Role of translational coupling in robustness of bacterial chemotaxis pathway. PLoS Biol 7:e1000171. doi: 10.1371/journal.pbio.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiMarco AA, Bobik TA, Wolfe RS. 1990. Unusual coenzymes of methanogenesis. Annu Rev Biochem 59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 52.Sauer FD, Blackwell BA, Kramer JK, Marsden BJ. 1990. Structure of a novel cofactor containing N-(7-mercaptoheptanoyl)-O-3-phosphothreonine. Biochemistry 29:7593–7600. doi: 10.1021/bi00485a008. [DOI] [PubMed] [Google Scholar]
- 53.Eichler J, Jarrell K, Albers S. 2013. A proposal for the naming of N-glycosylation pathway components in Archaea. Glycobiology 23:620–621. doi: 10.1093/glycob/cwt034. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.