DNA sequencing and hence genomics have been transformed over the last decade by the commercialization of inexpensive, massively parallel, short-read, sequencing technology. Nonetheless, a new generation of single-molecule DNA sequencers, which uses nanopore technology, is initiating a further upheaval in genomics. These instruments are portable, capable of reads of over 100 kb, cheap and fast. Nanopore sequencing, a huge technical challenge, took over 25 years to develop. Today's availability of a commercial nanopore sequencing device can be traced to the 1990s when nucleic acid translocation through nanopores was first observed, stochastic sensing developed and the high-resolution structure of a protein nanopore solved. The nanopore platform that has been developed is also capable of the single-molecule detection of a wide variety of additional analytes of medical interest, ranging from small molecules to post-translationally modified proteins.
When my group began work on the α-hemolysin (αHL) pore in the 1980s (1), the possibility of nanopore sequencing was not on our agenda. Following the molecular characterization of the pore by pioneers including Sidney Harshman and Sucharit Bhakdi, we sought to investigate its mechanism of assembly. αHL is secreted by Staphylococcus aureus as a monomeric water-soluble 293-amino acid protein, which forms an oligomeric pore in lipid bilayers. To us, this appeared to be a relatively simple system from which basic principles in membrane protein assembly might be learned. Over the years, this has indeed proved to be the case. In particular, the prepore concept, in which an oligomer forms on a membrane surface before penetrating the bilayer, has proved to be generally applicable to pore-forming proteins (2).
Nonetheless, in the late 1980s, we began to think about applications of protein pores in biotechnology. Our ideas included the incorporation of nanopores into filters for rapid purifications and separations, the permeabilization of cells both to introduce reagents for applications in basic research and for drug delivery, and the use of reversible pore-forming proteins to transport molecules into cells for protection during preservation by freezing or desiccation. Of these early efforts, only our work on molecular sensing has been sustained. We were most fortunate to obtain funding for these speculative endeavors from the US Department of Energy and the Office of Naval Research.
Our general approach to sensing was founded on knowledge about the interactions of channel blockers with natural ion channels, which had been investigated for many years. Currents carried through channels by aqueous ions can be measured by electrophysiological techniques. In the presence of blockers, which generally bind within the lumen of a channel, the ionic current is reduced. The current is restored when the blocker is removed. In early work, we used mutagenesis to build a binding site into the lumen of the αHL pore based on educated guess-work (in the absence of a structure), and used the mutant pores to detect divalent metal ions by macroscopic (many pore) current recording. The extent of current block revealed the concentration of the blocker. Even so, it was clear then that electrical recordings from individual pores (single-channel recording) would reveal far more about the nature of a blocker. At the same time, the structure of the heptameric αHL pore was solved by Eric Gouaux and his colleagues (3), which allowed the placement of designed binding sites within the lumen of the pore opening up the possibility of ‘stochastic sensing’ (4), a single molecule detection technique that allows the identification of analytes.
Stochastic sensing
In stochastic sensing, a single protein nanopore is placed in a lipid bilayer and the ionic current driven through it by an applied potential is monitored. Analyte molecules that bind to engineered sites within the lumen of the pore are detected by transient changes in the current (usually a decrease) manifested as square-wave blockades (Fig. 1A). The concentration of an analyte is revealed by the frequency of occurrence of the blockades. Information about the identity of an analyte is contained within the mean duration of the current blockades, the amplitude of the blockades, and additional characteristics of the blockades, such as an increase in current noise while the analyte is bound. Because signals from related analytes (e.g. various divalent metal ions or structurally related organic molecules) differ, the engineered binding site does not have to be absolutely selective for an analyte, which is often very difficult to achieve. It is indeed an advantage of stochastic sensing that related analytes can be recognized and distinguished by a single detection element. This is essential for nanopore DNA sequencing. Current recording has been central to stochastic sensing; the current carried by a single αHL pore is typically 30 pA (1 pA = 6.25 × 106 charges per second) and readily measured. However, variations on the theme can be imagined, for example the detection of analyte binding to protein receptors by single-molecule fluorescence.
Fig. 1. Stochastic sensing with nanopores.
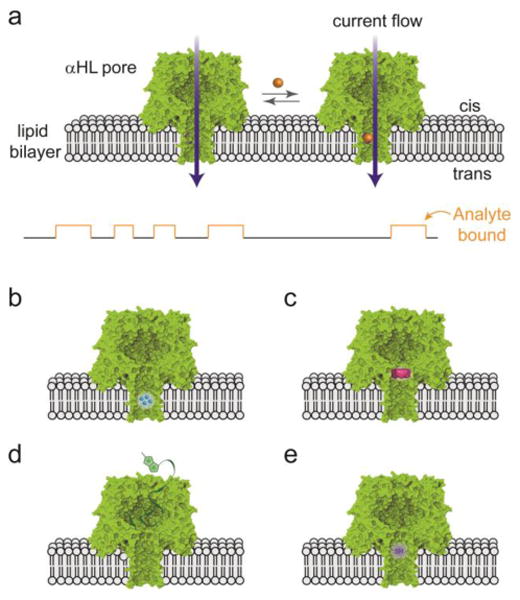
A), Sensing with the αHL pore. The current carried by aqueous ions through an individual pore is monitored. Analytes binding at a site within the pore lumen are detected through a transient change in the current. Binding sites can be formed by: B), genetic engineering, C), targeted chemical modification, e.g. with a molecule adapter (as shown). Large analytes can be detected by a ligand presented outside the pore lumen: D), ligand on a polymer chain. E), Covalent chemical reactions occurring within the pore can also be detected. Here a single reactive thiol group is shown. Adapted from (5).
The first experiments on stochastic sensing with a purposefully engineered protein nanopore demonstrated that divalent metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+) can be distinguished and quantified (4). This was achieved by using site-directed mutagenesis to introduce coordinating histidine residues with side chains that project into the transmembrane β barrel of the αHL pore (Fig. 1B). Knowledge of the structure of the pore was required to design the geometry of the locations of the residues. Subsequently, numerous analytes were detected by stochastic sensing (5), including a wide variety of small organic molecules, peptides and proteins (enzymes, lectins, antibodies), and nucleic acids. For stochastic sensing, analytes must be water-soluble, but that is not an issue for molecules relevant to biology and medicine. During these developments, several enabling technologies were acquired and exploited including the use of adapter molecules lodged in the pore lumen (Fig. 1C) for the detection of small organic molecules (6), and molecular fishing lines (Fig. 1D) to detect molecules too large to enter the lumen of the pore (7). Additional highlights included the detection of proteins by using peptide ligands genetically fused to the pore (8) or covalently-attached aptamers (9), the construction of metal centers as analyte binding sites (10), and the ability to distinguish enantiomers based on the chiral environment within the pore lumen (11). These findings suggested that it might be relatively simple to distinguish the four bases of DNA with the αHL pore, though not perhaps in the context of an intact DNA strand.
Covalent chemistry in a nanopore
If non-covalent chemistry (the association and dissociation of complexes) could be observed with the αHL nanopore, it occurred to us that covalent chemistry might also be monitored. We have used the nanopore approach to monitor a variety of aqueous chemistries on the wall of the pore with millisecond temporal resolution (12) (Fig. 1E), e.g., photodeprotection and photoisomerization, thiol-disulfide chemistry, polymer chain growth, organoarsenic chemistry, metal chelation, a kinetic isotope effect, and S-nitrosothiol chemistry. In recent work, a complex reaction network involving seven states was analyzed (13). Many analytes of physiological relevance are chemically reactive, ranging from chemical warfare agents to components of garlic and onion, and we have demonstrated detection based on covalent chemistry (14).
Analysis of nucleic acids with protein nanopores
Following the 1996 finding that short nucleic acids are translocated through the αHL pore under an applied potential (15), several groups examined the interactions of DNA and RNA with the pore, producing a variety of important results. It was immediately clear that single-stranded (ss) but not double-stranded (ds) nucleic acids are translocated (15). By determining the most probable translocation time and the residual current during translocation, the base compositions (e.g. of homopolymers) and lengths of nucleic acids can be distinguished (15, 16). Further, transitions between homopolymer segments can be detected as RNAs move through the αHL pore (16), as can bulky base modifications (17). The directionality of entry (5′ or 3′ first) can also be determined (18). In the case of dsDNA, duplex dissociation (19) and unzipping (20) can be examined. Notably, these studies indicated that translocation is quick at 1 to 10 μs per nucleotide for ssDNA, an observation of importance for nanopore sequencing. Nevertheless, there was no progress on sequencing itself for almost a decade. To a substantial extent, it was the NIH $1000 Genome Project, initiated in 2004, that stimulated progress in nanopore sequencing by melding the physicists' focus on translocation with the biological chemists' emphasis on molecular recognition, as manifested in the exploration of stochastic sensing.
Steps towards nanopore sequencing
The $1000 Genome Project led to a reassessment of what had to be done to achieve DNA (or RNA) sequencing. In nanopore strand sequencing, bases would be identified by a “reading head” as a single strand of DNA passes through the nanopore. Strand sequencing (Fig. 2A) would require: (i) efficient threading of ssDNA into protein nanopores; (ii) identification of individual DNA bases, which might be demonstrated with static strands; (iii) slow ratcheting of DNA through the pore to allow time-resolved base identification; and (iv) parallelization of the system to produce a competitive overall rate of sequencing. The first three of these criteria were demonstrated in academia and then developed in industry. The fourth has been developed in industry, and remains an area where substantial improvements must be made.
Fig. 2. Nanopore strand sequencing.
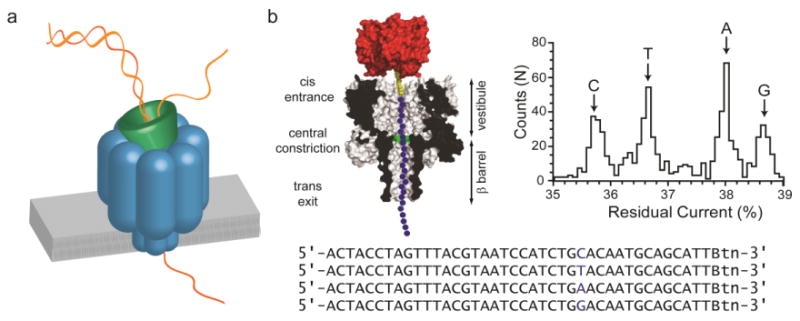
A), Basis of nanopore sequencing. ssDNA is fed through an individual protein pore by an enzyme that handles dsDNA. The sequence is determined by analysis of fluctuations in the ionic current. B), Early base identification experiments. ssDNAs were suspended in an αHL pore by attachment to streptavidin to mimic the ratcheting motion of the enzyme. The bases G, A, T and C in a DNA hetero-oligomer each gave a different residual ionic current. Adapted from (25).
The efficient capture of DNA was shown to require an internal positive charge within the protein nanopore (21, 22). While the precise mechanism of threading is unclear, this observation has proved useful. The discrimination of an individual base within an immobilized DNA strand was first demonstrated with the αHL pore (23). Soon afterwards, all four nucleoside monophosphates were identified with a modified αHL pore containing a cyclodextrin adapter (24). Isolated nucleotide identification would only be useful in exo rather than strand sequencing. (In exo sequencing, individual “bases” are cleaved sequentially from the end of a DNA strand by an enzyme held at the mouth of a pore and identified after they migrate to a binding site within the pore). Nonetheless, in 2006, the clear identification of all four bases, albeit out of the context of a DNA strand, provided a powerful impetus to proceed with the nanopore approach. Shortly afterwards, all four bases were distinguished in ssDNA, immobilized within the αHL pore by the attachment of streptavidin to one end of the strand (Fig. 2B) (25, 26). The stationary DNA, stretched in the electric field, is presumably in a similar conformation to that offered by the ratcheting motion of an enzyme (see below). Modified DNA bases, such as 5-methyl cytosine, were also identified by the same approach. Soon after, in a significant discovery, an alternative to the αHL pore, the MspA porin, was found to distinguish the four canonical bases with superior dispersion of the residual currents (27). It was also clear that the available pores would not produce a simple one-base/one-current output, but rather the reading of bases in ssDNA would be context dependent (28). Simply put, a small moving window (or windows) would identify words rather than letters. The current signal contains additional information beyond a straightforward letter code, but requires more sophisticated decipherment.
The third requirement was to ratchet the DNA through the pore to allow time-resolved base identification. The noise associated with the rapid data acquisition required for freely moving DNA (μs per base) does not allow separation of the current levels for sequence determination. Base-by-base ratcheting by a DNA polymerase was first demonstrated by the Ghadiri group (our $1000 Genome Project partners) (29), and by 2010 considerably refined by the Akeson (30) and Ghadiri (31) groups. The use of a processive enzyme (whether it be a polymerase, helicase, or nuclease) to control DNA movement allows long dsDNAs to be handled, and thereby obviates the need to deal with the difficulties of handling ssDNA. As the result of an applied potential, the ssDNA within the nanopore is stretched, limiting its conformational flexibility. Because the motion of the DNA is punctuated in the millisecond range by the action of the enzyme, base reading is facilitated and analogous to analyte identification in stochastic sensing.
At this point, it was clear that nanopore sequencing was achievable, although it would require a substantial effort, and it was pursued vigorously by Oxford Nanopore. Academic groups for the most part have moved on to other activities.
Commercialization of nanopore sequencing
Oxford Nanopore was founded in 2005 to exploit stochastic sensing. While the nanopore platform remains a versatile technology, by 2007 the company was largely turned over to nanopore sequencing. Oxford Nanopore worked initially on exo sequencing, for which clear-cut base identification had been demonstrated (24). As the viability of strand sequencing became recognized in work from academic laboratories, Oxford Nanopore transferred its emphasis to that area. In February 2012, the company announced success in DNA nanopore strand sequencing at the Advances in Genome Biology and Technology meeting in Marco Island, Florida. Notably, Oxford Nanopore scientists were able obtain reads of tens of kilobases. Details of the process were not released, but could be deduced in outline, at least, from presentations and patent filings. Two years since, a portable parallel sequencing device, the size of a cell phone, the MinION, has been released in a worldwide early access program (MAP). In the spirit of open development, sequencing data, software improvements and sample preparation protocols are being shared on the internet. The first data release (11th June 2014) was from Loman's group at the University of Birmingham who shared a 8476-base read from Pseudomonas aeruginosa strain 910 (Fig. 3) (32). More recently, the Loman group have shown that an entire Escherichia coli genome can be sequenced in a single MinION run [ REF].
Fig. 3. MinION data from the access program.

A), The first accessible sequence, from Pseudomonas aeruginosa 910: released on twitter on 11th June 2014 by N. Loman, University of Birmingham. B), Ionic current data from the MinION in the form of a “wiggle plot” (32).
Efforts in academia have continued, notably in the Akeson and Gundlach groups. In June of 2014, Gundlach's group documented and provided data and software for the sequencing of the bacteriophage ϕX174 genome using an approach closely similar to that of Oxford Nanopore (33). They used 256 quadromer (4 sequential bases) levels to analyse the current output from MspA through which ssDNA was threaded with ϕ29 DNA polymerase, reading sequences of up to 4500 bases. While short Illumina sequences could be aligned with the output, it was concluded that the accuracy was not yet good enough for de novo sequencing and that only 77% of single nucleotide polymorphisms (SNPs) would be identified during comparative sequencing.
Parallel sequencing
There is a pressing need for rapid sequencing, as well as cheap sequencing. One nanopore working at 10 ms/base would take about 20 years to sequence a human genome with 10X coverage, emphasizing the need for parallel sequencing, the fourth recognized requirement. Ten thousand pores would take less than a day for a genome, and a million pores just 10 minutes. This speed, combined with long read lengths and portability would elicit the full power of the nanopore approach.
The MinION uses a chip that contains ∼2000 robust polymer, rather than lipid bilayer, membranes over microwells that are individually electrically addressed. Assuming a Poisson distribution, a maximum of 37% of the membranes will contain a single nanopore when pores are allowed to insert randomly at the most favorable concentration. Therefore, at least several hundred nanopores are active simultaneously in the MinION, yielding an enormous advantage over sequencing with a single pore.
Attempts to improve the stability and throughput of sequencing chips continue. The ability to position individual protein nanopores in lipid-free apertures in solid-state arrays is a potential breakthrough in this regard, which beats the Poisson limit and provides considerable stability (34). Alternative detection methods might be further developed to increase throughput (35).
The future
The potential power and scope of nanopore sequencing is enormous, considering its low cost (including a minimal initial outlay), speed, portability, generally applicability (e.g. to RNA as well as DNA), ability to directly identify modified bases, etc. A critical improvement will be the minimization of the time required for sample preparation, to allow the “immediate” analysis of nucleic acids from human fluids, including circulating DNA and RNA (36).
Many improvements to nanopore sequencing will now be made in industry, while academic research turns towards other targets. For example, the analysis of proteins presents a tough challenge. While the human genome encodes just 20,000 or so proteins, they appear in countless modified forms. It was recently demonstrated that proteins can be unfolded as they are pulled through the αHL pore, either by using a DNA leader sequence (37) or with a motor protein (38). It was further shown that a post-translation modification (phosphorylation) can be detected (39).
Finally, it should not be forgotten that nanopore sensing is a platform technology, readily adapted for the detection of virtually any water-soluble analyte (5). The commercial hardware and software that has been developed for nanopore sequencing will support a wide variety of analytical applications. In the longer term, nanopore devices, including sequencers, based on the detection of analytes with transverse electronics (e.g. tunneling currents) might increase the rate of detection by two or three orders of magnitude (40). But the feasibility of such approaches has not yet been demonstrated.
Acknowledgments
The author thanks Mariam Ayub for her comments and help with the figures, and Nick Loman for permission to describe his work with the MinION.
Footnotes
Conflict of interest statement: Hagan Bayley is the Founder, a Director and a share-holder of Oxford Nanopore Technologies, a company engaged in the development of stochastic sensing and nanopore sequencing technology. Work in the Bayley laboratory at the University of Oxford is supported in part by Oxford Nanopore Technologies. Work on this review was not supported by Oxford Nanopore Technologies and the opinions expressed in it are the author's.
References
- 1.Tobkes N, Wallace BA, Bayley H. Secondary structure and assembly mechanism of an oligomeric channel protein. Biochemistry. 1985;24:1915–20. doi: 10.1021/bi00329a017. [DOI] [PubMed] [Google Scholar]
- 2.Bayley H. Membrane-protein structure: Piercing insights. Nature. 2009;459:651–2. doi: 10.1038/459651a. [DOI] [PubMed] [Google Scholar]
- 3.Song L, et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–65. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 4.Braha O, et al. Designed protein pores as components for biosensors. Chem Biol. 1997;4:497–505. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- 5.Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–30. doi: 10.1038/35093038. 2001. [DOI] [PubMed] [Google Scholar]
- 6.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature. 1999;398:686–90. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 7.Movileanu L, Howorka S, Braha O, Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nature Biotechnology. 2000;18:1091–5. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 8.Cheley S, Xie H, Bayley H. A genetically-encoded pore for the stochastic detection of a protein kinase. ChemBioChem. 2006;7:1923–7. doi: 10.1002/cbic.200600274. [DOI] [PubMed] [Google Scholar]
- 9.Rotem D, Jayasinghe L, Salichou M, Bayley H. Protein detection by nanopores equipped with aptamers. J Am Chem Soc. 2012;134:2781–7. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boersma AJ, Bayley H. Continuous stochastic detection of amino acid enantiomers with a protein nanopore. Angew Chem Int Ed Engl. 2012;51:9606–9. doi: 10.1002/anie.201205687. [DOI] [PubMed] [Google Scholar]
- 11.Kang XF, Cheley S, Guan X, Bayley H. Stochastic detection of enantiomers. J Am Chem Soc. 2006;128:10684–5. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 12.Bayley H, Luchian T, Shin S-H, Steffensen MB. Single-molecule covalent chemistry in a protein nanoreactor. In: Rigler R, Vogel H, editors. Single Molecules and Nanotechnology. Springer; Heidelberg: 2008. pp. 251–77. [Google Scholar]
- 13.Steffensen MB, Rotem D, Bayley H. Single-molecule analysis of chirality in a multicomponent reaction network. Nat Chem. 2014;6(7):603–7. doi: 10.1038/nchem.1949. [DOI] [PubMed] [Google Scholar]
- 14.Wu HC, Bayley H. Single-molecule detection of nitrogen mustards by covalent reaction within a protein nanopore. J Am Chem Soc. 2008;130(21):6813–9. doi: 10.1021/ja8004607. [DOI] [PubMed] [Google Scholar]
- 15.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–3. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–33. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell N, Howorka S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew Chem Int Ed Engl. 2008;47:5565–8. doi: 10.1002/anie.200800183. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Dunning JE, Huang APH, Nyamwanda JA, Branton D. DNA heterogeneity and phosphorylation unveiled by single-molecule electrophoresis. Proc Natl Acad Sci USA. 2004;101:13472–7. doi: 10.1073/pnas.0405568101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vercoutere W, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nature Biotechnology. 2001;19:248–52. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 20.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in nanopores. Phys Rev Lett. 2003;90:238101–238104. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 21.Maglia G, Rincon Restrepo M, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc Natl Acad Sci U S A. 2008;105:19720–5. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci U S A. 2008;105:20647–52. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashkenasy N, Sánchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: a step toward nanopore DNA sequencing. Angew Chem Int Ed Engl. 2005;44:1401–4. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–10. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 25.Stoddart D, Heron A, Mikhailova E, Maglia G, Bayley H. Single nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106:7702–7. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purnell RF, Schmidt JJ. Discrimination of single base substitutions in a DNA strand immobilized in a biological nanopore. ACS Nano. 2009;3:2533–8. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 27.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS One. 2011;6(10):e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoddart D, Maglia G, Mikhailova E, Heron A, Bayley H. Multiple base-recognition sites in a biological nanopore– two heads are better than one. Angew Chem Int Ed. 2010;49:556–9. doi: 10.1002/anie.200905483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–20. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieberman KR, et al. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J Am Chem Soc. 2010;132:17961–72. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu J, Gonzalez-Lopez M, Cockroft SL, Amorin M, Ghadiri MR. Real-time monitoring of DNA polymerase function and stepwise single-nucleotide DNA strand translocation through a protein nanopore. Angew Chem Int Ed. 2010;49:10106–9. doi: 10.1002/anie.201005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loman N, Quick J, Calus S. A P. aeruginosa serotype-defining single read from our first Oxford Nanopore run. 2014 figshare: http://figshare.com/articles/Wiggle_plot_showing_Oxford_Nanopore_signal_data_for_a_P_aeruginosa_read/1053026.
- 33.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014 doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall AR, et al. Hybrid pore formation by directed insertion of a-haemolysin into solid-state nanopores. Nature Nanotechnology. 2010;5:874–7. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heron AJ, Thompson JR, Cronin B, Bayley H, Wallace MI. Simultaneous measurement of ionic current and fluorescence from single protein pores. J Am Chem Soc. 2009;131:1652–3. doi: 10.1021/ja808128s. [DOI] [PubMed] [Google Scholar]
- 36.Lo YM. Noninvasive fetal whole-genome sequencing from maternal plasma: feasibility studies and future directions. Clin Chem. 2013;59:601–3. doi: 10.1373/clinchem.2012.191098. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Larrea D, Bayley H. Multistep protein unfolding during nanopore translocation. Nature Nanotechnology. 2013;8:288–95. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nivala J, Marks DB, Akeson M. Unfoldase-mediated protein translocation through an alpha-hemolysin nanopore. Nat Biotechnol. 2013;31:247–50. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen CB, Rodriguez-Larrea D, Bayley H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat Biotechnol. 2014;32:179–81. doi: 10.1038/nbt.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branton D, et al. The potential and challenges of nanopore sequencing. Nature Biotechnology. 2008;26:1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]


