Abstract
Adverse prenatal environments can promote metabolic disease in offspring and subsequent generations. Animal models and epidemiological data implicate epigenetic inheritance but mechanisms remain unknown. In an intergenerational developmental programming model affecting F2 metabolism, we demonstrate that the in utero nutritional environment of F1 embryos alters the germline DNA methylome of F1 adult males in a locus-specific manner. Differentially methylated regions are hypomethylated and enriched in nucleosome-retaining regions. A substantial fraction is resistant to early embryo methylation reprogramming, potentially impacting F2 development. Importantly, differential methylation is not maintained in F2 tissues, yet locus-specific expression is perturbed. Thus, in utero nutritional exposures during critical windows of germ cell development can impact the male germline methylome, associated with metabolic disease in offspring.
Introduction
The rapid global rise in the incidence of diabetes, obesity and cardiovascular disease suggests that non-genetic environmental factors are major contributors to disease risk. Epidemiological data and animal models have demonstrated that early life represents a window of phenotypic plasticity critically important for later adult metabolic health (1). The impact of the early life environment has been observed to extend over multiple generations in both human populations and animal models (2-8). There are at least two potential mechanisms mediating such non-Mendelian phenotypic inheritance: alterations in the parental metabolic milieu which induce fetal developmental exposures in the second generation; and epigenetic inheritance. The latter is strongly implicated when paternal transmission of environmentally-induced phenotypes is observed because rodent males, present solely at breeding, contribute to the future generation only through the sperm. Although a role for histone modifications and/or RNA has been proposed (4), the epigenetic mechanism(s) responsible for intergenerational inheritance of environmentally-induced phenotypes remains unknown.
Paternal transgenerational epigenetic inheritance of altered DNA methylation has been demonstrated previously: for example, in rodents exposed to the endocrine disruptor vinclozolin (9) and in mice with variable methylation at the Agouti viable yellow, Avy, and Axin-Fused, AxinFu, alleles formed by insertion of IAP elements into or nearby endogenous genes (10). In addition to repeat-mediated cis-acting effects, other endogenous loci which have an inherent epigenetic vulnerability to environmental conditions may contribute to intergenerational phenotypes and play an important role in the developmental origins of health and disease. Furthermore, recent studies have suggested that resistance to zygotic DNA methylation reprogramming extends beyond imprinted domains (11-13), raising the possibility that gametic methylation may play a larger role than previously recognised in early development. A key unanswered question is whether an altered in utero environment or nutritional insult might affect the DNA methylation profile of adult germ cells.
Our aim was to investigate the role of DNA methylation in epigenetic inheritance in an established in utero murine model of intergenerational developmental programming (3). To produce the most robust phenotype, the maximum caloric restriction which does not cause significant fetal loss was chosen (Fig.1A). This regime is largely incompatible with successful pregnancy in inbred mouse strains. Consequently we used the outbred ICR strain, also allowing us to better model the human population. In this model, F1 offspring of undernourished dams have low birth weight associated with early-life adiposity, reduced muscle stem cell number and function, impaired pancreatic function and progressive glucose intolerance (14-16). Importantly, inheritance of significantly reduced birth weight and glucose intolerance to the F2 generation is observed through the paternal line in the absence of any further environmental perturbation (Fig.S1D-H) (3). The period of experimentally-induced nutritional restriction in this model (day 12.5 to 18.5 of pregnancy) coincides with the re-acquisition of methylation in male primordial germ cells as they are epigenetically reprogrammed (17). The dynamics of such methylation changes have been best studied at imprinting control regions (ICRs). However, we have already excluded a substantial perturbation of methylation at ICRs in this model (18). Thus we now assess the whole-genome distribution of methylation in F1 sperm using immunoprecipitation of methylated DNA, combined with high-throughput sequencing (MeDIP-seq) (19-21), followed by independent validation by bisulphite sequencing.
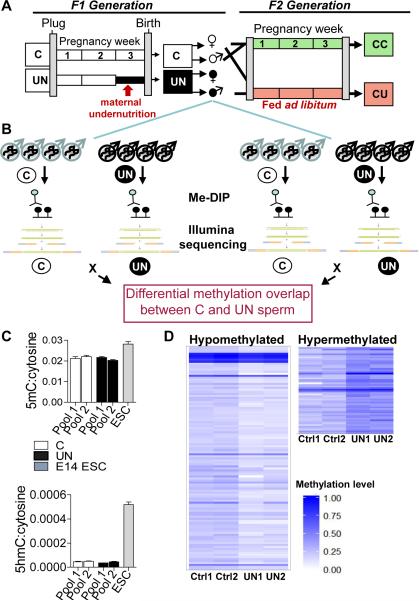
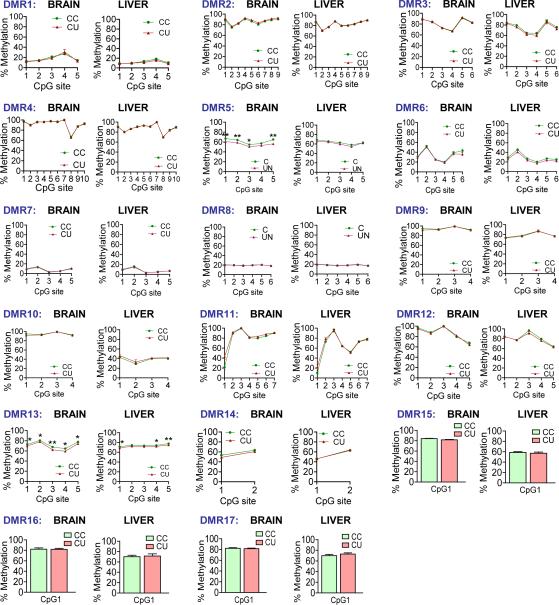
Figure 1. Total methylation is stable in UN sperm, with significant locus specific changes.
(A) Experimental design: F1 generation: Dams were randomised on pregnancy day 12.5 to control (C) or undernutrition (UN) groups and UN food intake restricted to 50%. Postnatal litters were equalised to eight pups and animals fed ad libitum. F2 generation: control F1 females mated at age 2 months with non-sibling control or UN males and fed ad libitum to produce: CC - both parents controls; CU - control dam, UN sire.
(B) Independent sperm DNA samples were quantified and pooled in equimolar ratios to generate two pools per condition. Control pools: n=8, 5 litters. UN pools: n=8, 4 litters. Following MeDIP-seq two independent C vs UN comparisons identified DMRs where methylation FC >1.5x and binomial p-value <0.0001 in both independent biological replicates.
(C) Mass spectrometry quantification of control and UN sperm 5-methyl-cytosine (above) and 5-hydroxymethyl-cytosine (below). E14 ESCs are shown for comparison.
(D) Heatmap of 111 hypomethylated DMRs (left) and 55 hypermethylated DMRs (right). Hypermethylated DMRs did not validate.
Results
Experimental design and metabolic phenotype
Mature sperm was isolated from F1 male mice fed standard chow, ad libitum, at 3 months of age, prior to the onset of glucose intolerance or any discernible metabolic phenotypes (14). These F1 males, previously exposed to experimental undernutrition in utero (UN), were smaller at birth (UN 1.34 ± 0.025g, controls 1.65 ± 0.028g, P<0.0001) and at 3 months of age (UN 41.4 ± 0.82g, controls 44.2 ± 0.94g, P=0.04) (Fig.S1B, C). Blood glucose and white adipose tissue mass at the time of sperm collection was not different between UN and control mice (Fig.S1C). We bred F1 control and UN males with control females prior to sperm isolation; offspring of these pregnancies were designated as CC (F2 offspring of control males) and CU (F2 offspring of UN males) (Fig.1A). The F2 offspring of F1 sperm donors were harvested at E16.5. A contemporaneous adult cohort of F2 CU mice demonstrates at 8 months of age similar metabolic phenotypes as previously observed (3), including reduced muscle mass and increased adiposity, with no difference in overall body or brain weight (Fig.S1D). Furthermore, this CU cohort also shows glucose intolerance, particularly in the first phase response to a glucose challenge (Fig.S1E), as was previously observed. Pyruvate tolerance tests suggest that increased gluconeogenesis may contribute to this glucose intolerance (Fig.S1F).
To assess whether a metabolic phenotype is discernable at E16.5 in the F2 generation, we examined lipid metabolism. There is an overall trend towards increased lipid abundance, particularly for saturated fatty acid-conjugated triglycerides (Fig.S1G). This is associated with a significant increase in expression of genes involved in lipid oxidation in E16.5 CU liver, such as PPARα, Pgc1α and Pgc1β and a trend towards down-regulation of genes involved in lipid synthesis, including Scd1, Srebp1 and Dgat1 (Fig.S1H), likely secondary to the increased hepatic lipid abundance at E16.5. Together, these data suggest that CU individuals have altered metabolism even in utero.
Hypomethylation of discrete loci in F1 adult sperm of males undernourished in utero
To confirm the purity of F1 sperm samples, bisulphite sequencing of imprinting control regions was carried out (Fig.S2A). Independent sperm DNA samples were pooled in equimolar ratios to make two pools for each condition, each pool comprising four individuals from four independent litters (Fig.1B), hence minimising outcomes that might be associated with inter-individual genetic differences. Mass spectrometry analysis of F1 sperm DNA demonstrates that in utero nutrition does not affect the total level of DNA methylation or hydroxymethylation (Fig.1C). It is also notable that the level of hydroxymethylation in sperm is only 2.1% of that observed in embryonic stem cells (Fig.1C). Consequently only the genomic distribution of DNA methylation was analysed further.
We assessed the genome-wide distribution of sperm methylation by MeDIP-seq (Fig.1B). This approach is most suited to the detection of robust regional changes in DNA methylation, offering near-unbiased genome-wide coverage with under-representation of low density mC/mCG (22), thus minimizing the possible influence of single nucleotide variants and allowing identification of clusters of differentially methylated cytosines. Optimisation of antibody specificity was carried out to ensure no cross sampling of hydroxymethylated or unmethylated cytosine (Fig.S2B, Materials and Methods). Sequencing of antibody-enriched samples generated a total of 322.6 million mappable reads for control and 301.8 million for UN libraries. Two independent comparisons between the control and UN pools were conducted using the MEDIPS package (23) (see Materials and Methods for more details). Loci with a methylation change >1.5 fold and a binomial P< 0.0001 in both of the independent comparisons were selected for further study and clustered into 166 differentially methylated regions (DMRs), of which 111 were hypomethylated and 55 hypermethylated in UN relative to control sperm (Fig.1D).
Bisulphite pyrosequencing validation of MeDIP-seq DMRs
To independently validate regions of altered methylation using a different technology, we employed bisulphite pyrosequencing assays on 32 regions using an expanded panel of sperm samples - 12 control males from 5 litters, and 11 UN males from 4 litters. Twenty four hypomethylated regions and eight hypermethylated regions were randomly chosen for validation, distributed throughout the range of fold change and P-values. No significant difference in methylation was found at any hypermethylated DMR, suggesting that these regions may be false positives (Table S1). In contrast, significant loss of methylation was confirmed at 17 of the hypomethylated regions in the expanded panel of F1 UN sperm samples (Fig.2, Table 1). The validation rate of the non-repetitive, unique hypomethylated regions was 90%. Strikingly, differences in methylation at these loci span multiple CpGs, with robust absolute changes of 10-30%, a relative reduction of up to 50% (Fig.2). Moreover, these differences are remarkably consistent among individual animals from multiple independent litters, indicating that they are unlikely to be caused by genetic variation (Fig.S3). The bisulphite sequencing data show identical absolute levels of methylation in the two replicate pools assessed by MeDIP-seq (Fig.S3). Furthermore, the absolute methylation level (generally under 50% in both groups) is consistent with these DMRs being “low methylated regions”, previously shown to be enriched in regulatory elements (24). Together, these data demonstrate that discrete loci in the adult male germline are susceptible to changes in methylation as a result of nutritional stress in utero.
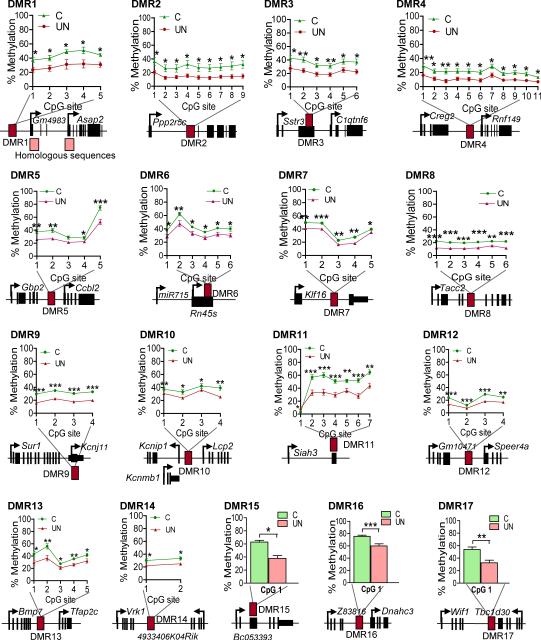
Figure 2. Bisulphite mutagenesis validation of hypomethylated DMRs in an expanded panel of F1 males’ sperm.
17 genomic regions validated (Table 1). Data plotted: mean +/− SEM. (C: n=12, 5 litters; UN: n=11, 4 litters) * P<0.05 ** P<0.01, *** P<0.001 unpaired two-tailed t-test.
Table 1.
Validation of hypomethylated DMRs by bisulphite pyrosequencing
| DMR coordinates | Sperm methylation | Blastocyst methylation | F2 E16.5 liver methylation | F2 E16.5 brain methylation |
|---|---|---|---|---|
| DMR1: MMU12:19181482-19182200 | Ctrl=50% UN=24% P<0.0001 |
4% (12) 8% (26) |
CC=13% CU=14% |
CC=10% CU=10% |
| DMR2: MMU12:111666100-111666400 | Ctrl=29% UN=14% P<0.0001 |
0% (12) 7% (26) |
CC=85% CU=88% |
CC=82% CU=83% |
| DMR3*: MMU15:78370350-78370800 | Ctrl=37% UN= 23% P<0.0001 |
0% (12) | CC=81% CU=81% |
CC=76% CU=73% |
| DMR4*: MMU1:39654450-39655100 | Ctrl=20% UN=12% P<0.0001 |
5% (12) | CC=88% CU=88% |
CC=88% CU=87% |
| DMR5: MMU3:142351001-142351500 | Ctrl=41% UN=29% P=0.0004 |
13% (12) 25% (26) |
CC=64% CU=61% |
CC=62% CU=56% |
| DMR6*: MMU17:39984601-39985700 | Ctrl=43% UN=33% P<0.0001 |
7% (26) 26% (12) |
CC=30% CU=26% |
CC=34% CU=33% |
| DMR7*: MMU10:80033801-80034300 | Ctrl=38% UN=30% P=0.001 |
22% (26) 28% (12) |
CC=8% CU=8% |
CC=8% CU=9% |
| DMR8*: MMU7:137835001-137836500 | Ctrl=21% UN=12% P<0.0001 |
22% (26) 25% (12) |
CC=18% CU=19% |
CC=20% CU=20% |
| DMR9*: MMU7:53354201-53354900 | Ctrl=18% UN=11% P=0.0003 |
0% (26) 9% (12) |
CC=78% CU=80% |
CC=95% CU=94% |
| DMR10: MMU11:33922001-33922500 | Ctrl=39% UN=29% P<0.0001 |
16% (26) | CC=38% CU=38% |
CC=28% CU=32% |
| DMR11*: MMU14:75925601-75926100 | Ctrl=48% UN=30% P<0.0001 |
0% (26) 3% (26) |
CC=64% CU=66% |
CC=78% CU=83% |
| DMR12*: MMU5:26397201-26397700 | Ctrl=23% UN=14% P<0.0001 |
24% (12, 26) | CC=80% CU=79% |
CC=85% CU=85% |
| DMR13*: MMU2:172688001-172688500 | Ctrl=41% UN=29% P<0.0001 |
6% (26) | CC=74% CU=72% |
CC=73% CU=69% |
| DMR14:MMU12:107752401-107752500 | Ctrl=32% UN=24% P=0.001 |
27.6% (26) | CC=54% CU=55% |
CC=58% CU=55% |
| DMR15*: MMU11:46390601-46391100 | Ctrl=63% UN=38% P<0.0001 |
4.2% (26) | CC=59% CU=57% |
CC=84% CU=82% |
| DMR16:MMU7:10836601-10837100 | Ctrl=75% UN=54% P<0.0001 |
56% (26) | CC=70% CU=71% |
CC=82% CU=82% |
| DMR17:MMU10:120699201-120699700 | Ctrl=54% UN=33% P=0.001 |
12.3% (26) | CC=70% CU=73% |
CC=82% CU=82% |
Absolute methylation level calculated by bisulphite mutagenesis combined with pyrosequencing in C and UN F1 sperm (n≥11, ≥ 4 litters), F2 E16.5 brain and liver (n≥12, ≥ 3 litters) at hypomethylated DMRs. DMRs at non-repetitive, unique loci are indicated by an asterisk. Blastocyst methylation level extracted from (12) and (26).
DMRs are not distributed randomly through the genome
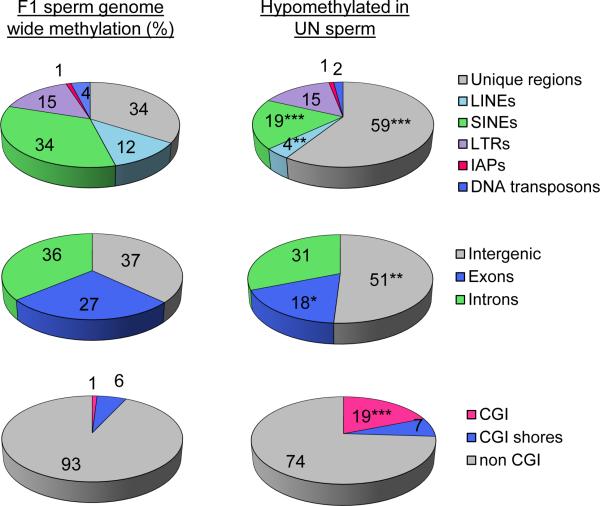
We examined the distribution of unique and repetitive elements among DMRs. Hypomethylated DMRs are significantly depleted from coding regions, but enriched in intergenic regions and CpG islands (Fig.3). Repetitive elements are significantly depleted from hypomethylated DMRs (χ2 P<0.0001) with under-representation of LINEs (χ2 P=0.001) and SINEs (χ2 P<0.0001) and no significant enrichment of IAPs (Fig.3).
Figure 3. DMRs are enriched in intergenic non-repetitive regions and CpG islands.
Top: Relative distribution (%) of F1 sperm methylated regions among unique sequence and repetitive elements genome wide (left) and among the F1 UN sperm hypomethylated DMRs (right). Unique regions are significantly enriched (χ2 P<0.0001) while LINEs and SINEs are significantly depleted from hypomethylated DMRs (χ2 P=0.001; χ2 P<0.0001 respectively), relative to all methylated regions detected in F1 sperm.
Middle: Relative distribution (%) of methylated regions among coding and non-coding sequence. Exons are significantly depleted (χ2 P=0.036), and intergenic regions significantly enriched (χ2 P=0.0012) among hypomethylated DMRs.
Bottom: Relative distribution (%) of methylated regions detected by MeDIP-seq among CpG islands (CGI) and CGI shores. CGIs are significantly enriched among hypomethylated DMRs (χ2 P<0.0001).
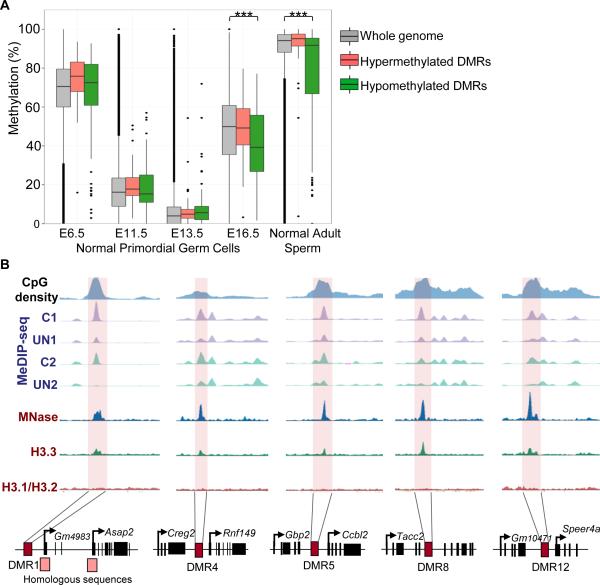
The predominance of hypomethylated DMRs is striking. This is consistent with in utero undernutrition during the final third of gestation impairing the re-acquisition of methylation in developing F1 male PGCs. The nutritional insult experienced by the foetus worsens with increasing gestation as maternal energy reserves are depleted. Therefore, we hypothesise that the likelihood of remethylation being disrupted by in utero undernutrition increases towards term. Analysis of the temporal dynamics of methylation reprogramming in normal PGCs (25) suggests that this is indeed the case. In normal male PGCs, whole-genome methylation is progressively reduced from E6.5 to 13.5, with evidence of remethylation by E16.5 (Fig.4A, grey bars). In contrast, those DMRs found to be hypomethylated in adult UN sperm (green bars) exhibit a distinct temporal pattern of reprogramming. These DMRs have significantly lower methylation levels at E16.5 in normal male PGCs (χ2 P<0.0001; Fig.4A), suggesting that these regions are late to re-methylate and may be susceptible to environmental perturbations which delay or impair remethylation at this stage. In normal adult sperm, methylation has largely been regained, but a minority of regions retain low methylation levels (26). UN-associated hypomethylated DMRs are enriched in these low methylated regions (χ2 P<0.0001; Fig.4A)
Figure 4. DMRs regain methylation late during PGC reprogramming and retain nucleosomes in mature sperm.
A) Methylation level of hypomethylated (green) and hypermethylated (red) DMRs in our data set versus the whole genome (grey) in normal PGCs (25) and mature sperm from adult males (26). Hypermethylated DMRs act as an additional negative control since they did not validate. E13.5 and E16.5 are male PGCs. E6.5 and E11.5: mixed sex PGCs (25).
B) Nucleosome enrichment (27) at 5 representative hypomethylated DMRs.
During spermiogenesis 99% of histones are exchanged for protamines, but nucleosomes are particularly retained in regions of high CpG density and low DNA methylation (27). Given the low methylation level of our DMRs we assessed whether these regions are also enriched in nucleosomes. 23/111 (21%) hypomethylated DMRs retain nucleosomes in mature sperm (Fig.4B). Bootstrap re-sampling of randomly selected regions from the background methylome demonstrates that this is a significant enrichment, P<0.0001, and a feature of low methylated regions (see Figure S4 for details). This suggests that at some loci, paternal germline hypomethylation induced by in utero undernutrition is transmitted in a chromatin context.
The developmental legacy of germline DMRs in late gestation of the F2 generation
With the exception of imprints, it has been thought that gene-associated methylation in the male germline is largely reprogrammed in the zygote by active DNA demethylation (17). However, recent studies suggest that resistance to DNA methylation reprogramming extends beyond imprinted domains (11-13). Indeed, 43% of our hypomethylated DMRs are resistant to zygotic reprogramming (26), suggesting that differential methylation in the paternal germline may persist into the early embryo and affect the development of the next generation (Table 2).
Table 2.
43% of hypomethylated DMRs are resistant to zygotic demethylation
| Number (%) of hypomethylated DMRs | |
|---|---|
| Blastocyst methylation < 20% (26) | 63 (57%) |
| Blastocyst methylation > 20% (26) | 48 (43%) |
Hypomethylated DMRs susceptible (<20% methylation) or partially resistant (>20% methylation) to blastocyst reprogramming (28).
To determine whether altered F1 sperm methylation persisted as a ‘memory’ of sperm compromise in F2 offspring, we bred F1 control and UN males with control females. Offspring were designated as CC (F2 offspring of control males) and CU (F2 offspring of UN males) as noted above (Fig.1A). Using liver and brain samples from late-gestation (E16.5) CC and CU embryos, we analysed DNA methylation at validated germline DMRs. Strikingly, differential methylation has been lost in F2 E16.5 brain and liver (Table 1, Fig.5). These data indicate that any functional consequences of germline DMRs are likely to be established early in development and/or linked to associated but currently unknown regulatory effects which may persist despite DNA remethylation in later development.
Figure 5. Analysis of methylation at F1 sperm DMRs in F2 brain and liver at E16.5.
F2 E16.5 CC and CU brain and liver methylation of F1 sperm previously validated hypomethylated DMRs, measured by bisulphite pyrosequencing. Data presented as mean +/− SEM. Brain per condition n = 16, ≥ 3 litters; Liver per condition n = 12, 3 litters.
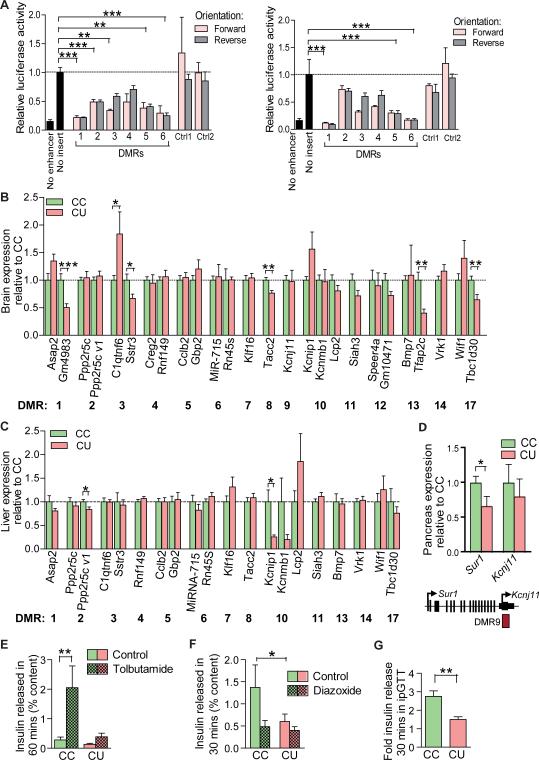
Analysis of validated DMRs in publicly available datasets (28) indicates that these loci have cell-type specific enrichment of histone modifications and transcription factor binding, characteristic of a role in cis regulation of transcription (Table S2). To assess the function of a randomly selected subset of six DMRs, we conducted luciferase reporter assays in neural stem cells (29) and NIH3T3 cells in culture, using methylation stable regions (non-DMRs) validated by pyrosequencing as additional negative controls. No significant enhancer function could be attributed to any of the regions tested in either cell type. In contrast, in vectors designed to assess a negative influence on transcription such as an enhancer blocking or silencer function, 5/6 regions significantly suppressed reporter activity when inserted in both the forward and reverse orientation in neural stem cells, and 3/6 regions in NIH3T3 cells (Fig.6A). Taken together, the data suggest that these germline DMRs may play cell-specific regulatory roles in the modulation of transcription.
Figure 6. Developmental legacy of altered UN sperm methylation in the F2 generation.
(A) Luciferase assay for a negative effect on transcription in 46C neural stem cells (29) (left) and NIH3T3 cells (right). Sequences were inserted between the promoter and enhancer of the Control pGL3 vector. The pGL3 Promoter vector (lacking an enhancer) was used as a positive control. Two regions validated by pyrosequencing as having unaffected F1 sperm methylation were used as negative controls. Control 1: MMU2:77723600–77723900, Control 2: MMU17:87639700-87640000. Data are plotted as mean +/− SEM, normalised to activity of the Control pGL3 vector with no insert. One way ANOVA, Dunnett's post-test **P<0.001, ***P<0.0001.
(B) F2 E16.5 brain expression of genes neighbouring F1 sperm DMRs. Data plotted as mean +/− SEM. MiR-715 expression normalised to SnoRNA 202, all other expression normalised to Hprt. Hprt and SnoRNA202 were unaffected. Unpaired two-tailed t-test: Gmf4983 P=0.0004, C1qtnf6 P=0.049, Sstr3 P=0.02, Tacc2 P=0.0018, Tfap2c P=0.015, Tbc1d30 P=0.006. Per condition n=16, ≥3 litters.
(C) F2 E16.5 liver expression of genes neighbouring F1 sperm DMRs. Data plotted as mean +/− SEM. Normalised as for (B). Unpaired two-tailed t-test: Ppp2r5c variant1 P=0.03, Kcnip1 P=0.011. Per condition n=12, 3 litters.
(D) F2 Pancreatic expression at 4 months. Per condition n ≥ 5 * P<0.05, unpaired two-tailed t-test (3).
(E) Tolbutamide (200μM) stimulated insulin secretion, freshly isolated 4 month old islets; n≥4, ≥ 2 isolations. **P<0.01, unpaired two-tailed t-test. (3)
(F) Diazoxide (250μM) inhibition of insulin secretion, freshly isolated 4 month old islets; n=4 per group, ≥ 2 isolations. *P<0.05, unpaired two tailed t-test. (3).
(G) Fold change in serum insulin 30 minutes following intraperitoneal glucose bolus (1mg/kg). **P<0.01, n≥8, unpaired two-tailed t-test. (3).
To assess this possibility, we examined expression of genes neighbouring the seventeen germ-line DMRs, using qPCR in liver and brain of F2 CC and CU fetuses at E16.5. Genes associated with DMRs 15 and 16 were not expressed in these tissues. Eight DMRs showed significant tissue-specific differences in expression of neighbouring genes (Fig.6B,C). In contrast, no change in expression was found at twelve genes not associated with DMRs (Fig.S5). Importantly, because methylation differences are not observed in E16.5 tissues of these same F2 offspring (Fig.5), it is unlikely that these expression changes are directly mediated by alterations in methylation. Rather, the cumulative effects of dysregulated epigenetic patterns earlier in development may yield sustained alterations in chromatin architecture, transcriptional regulatory networks, cell type or tissue structure.
Several affected genes, including Sstr3, C1qntf6, Tbc1d30, Kcnj11 and Sur1 are candidate contributors to the F2 phenotypes given their known roles in glucose tolerance and metabolism (30-36). For example, the DMR9 lies within the Kcnj11 gene, immediately downstream of Sur1. These genes encode the two subunits of the pancreatic β-cell ATP-dependent K+ channel which are necessary for the physiological control of insulin secretion (34, 36). Furthermore, polymorphisms at these loci are associated with Type 2 Diabetes (35, 37). In pancreatic islets isolated from 4 month old CU mice (F2 generation), expression of Sur1 is reduced by 33% (P<0.05) (Fig. 6D) (3). The function of β-cell ATP-dependent K+ channels in controlling insulin secretion can be assessed through measuring the response to treatment with agents which inhibit and activate these channels, such as sulphonylureas and diazoxide respectively; or through the insulin secretory response to a glucose challenge. Freshly isolated 4 month old CU pancreatic islets demonstrate impaired insulin secretion in response to the sulphonylurea tolbutamide (Fig.6E) and absence of suppression of insulin secretion to diazoxide (Fig.6F) (3). Furthermore, basal insulin secretion prior to diazoxide challenge was significantly reduced (Fig.6F) (3). Consistent with this, CU individuals secrete significantly less insulin during glucose tolerance testing (Fig.6G) (3). These data strongly suggest impaired function of ATP-dependent K+ channels in the adult CU pancreas, and implicate this in the altered glucose tolerance observed in CU individuals (Fig.S1E). Further work will be required to delineate the precise relationship between compromised F1 germline reprogramming at these loci and F2 phenotypes.
Discussion
Our data indicate that nutritional perturbations during in utero development can alter male germline methylation at discrete loci. In turn, some of these DMRs are associated with differential transcript expression during offspring embryonic life. Our findings contrast with the largely negative data of Carone et al. in which no significant changes were observed in sperm DNA methylation following dietary protein restriction in adult males (4). Disparities may be due to the use of protein rather than caloric restriction, strain differences, or the greater number of individuals assessed in our analysis. Alternatively it may be due to differences in the timing of the nutritional insult as Carone and colleagues’ imposed protein restriction during adult life. By contrast, the nutritional perturbation in our model occurs exclusively during late prenatal life, precisely when male primordial germ cells (PGCs) in the developing embryo are undergoing re-establishment of their epigenetic profile. At this time, PGCs may be particularly vulnerable to epigenetic perturbation. It is notable that intergenerational phenotypic inheritance caused by endocrine disruptors associated with altered sperm DNA methylation also involves prenatal exposure (9, 38). However, recent data has suggested that a high fat diet during adult life might alter sperm DNA methylation, indicating that the adult germline methylome may be more susceptible to environmental conditions than previously thought (8).
Our experiment was designed to minimise detection of single CpG methylation differences which we a priori hypothesise to be more likely to be due to genetic differences. Our results indicate that robust germ cell methylation changes do occur following in utero undernourishment at regions resistant to zygotic reprogramming. However, persistence of altered DNA methylation into late gestation somatic tissues of the subsequent generation was not observed. Nonetheless gene expression is altered in these F2 offspring at regions of F1 germline differential methylation. Such differences in gene expression could reflect the impact of altered methylation during early development, with subsequent transcriptional patterns which persist despite DNA remethylation in later gestation. Alternatively, altered F2 expression may be the cumulative result of multiple locus-specific defects in germline chromatin state. Further work will be required to explore these possibilities.
Recent work in cultured cells demonstrates that regional methylation levels can be a secondary consequence of changes in DNA-binding factors (24). Thus it is possible that the germline DMRs identified in our study are secondary to other chromatin perturbations. Consistent with this, we observed enrichment of nucleosome occupancy at DMRs. Further studies are required to examine whether these represent regions of vulnerability in the sperm genome. Histone modifications and small RNA molecules are known to be required for multigenerational gene silencing effects in C. Elegans (39, 40), an animal which lacks DNA methylation, and such mechanistic processes may also be involved in mammals. Indeed, there is evidence that sperm borne miRNAs play an important role in early mammalian development (41) and the early life environment may have the potential to alter the abundance of some sperm miRNAs (42).
Conclusion
Data presented here serve as a proof of principle that undernutrition during prenatal life, even when followed by normal postnatal nutrition, can compromise male germline development and epigenetic reprogramming, permanently altering DNA methylation in the germline of the adult offspring. Alterations in adult gamete methylation may serve as a legacy of earlier developmental exposures which may contribute to the intergenerational transmission of environmentally-induced disease.
Materials and Methods
Animal protocols
Maternal undernutrition
ICR mice were obtained from Taconic, Inc. (Hudson, NY). Outbred female ICR mice (Taconic, NY) aged 6-8 weeks were bred with ICR males. Pregnancies were timed by the presence of a vaginal plug (day 0.5). On day 12.5 of pregnancy, pregnant females were randomly assigned to “control” or “undernutrition” (UN) groups as described (14). Control dams had ad libitum access to standard chow (Purina 9F; Purina Mills, St. Louis, MO), with 21% of calories from protein, 21% from fat, and 56% from carbohydrate (wheat/corn), whereas undernutrition group dams were 50% food restricted from days 12.5 to 18.5 (calculated from intake in gestational day–matched controls). At birth, litter size was equalized to eight, and all dams resumed eating regular chow ad libitum. Offspring mice were designated as control or UN F1 offspring. To generate F2 offspring, control virgin females 8 weeks of age were bred with either control males or UN F1 males to generate CC and CU offspring, respectively (Figure 1A, right). Pregnancies were timed by vaginal plug. F2 fetal tissues were collected at E16.5 after pentobarbital anaesthesia.
Mice were housed in an OLAW-approved facility, with controlled temperature, humidity, and light-dark cycle (07:00-19:00). Protocols were approved by the Joslin Diabetes Centre Institutional Animal Care and Use Committee. ‘‘Principles of Laboratory Animal Care’’ (http://grants1.nih.gov/grants/olaw/references/phspol.htm) were followed.
Tissue dissection
For RNA and DNA extraction following dissection, tissues were snap frozen in liquid nitrogen. Prior to extraction, and to facilitate multiple extractions from the same tissue, samples were pulverised in liquid nitrogen and were never allowed to thaw.
RNA extraction
RNA was extracted using Trizol (Invitrogen) according to the manufacturer's instructions, with an overnight precipitation step at −20°C. Newly isolated RNA was quantified by spectrophotometric analysis and 28S and 18S ribosomal RNA bands quality assessed by electrophoresis with a 1% agarose gel. All samples were treated to remove DNA contamination with DNase (using the RNase-free DNase kit, Qiagen) according to the manufacturer's instructions, followed by re-precipitation.
Quantification of gene expression
cDNA was generated from 1μg total RNA per sample using the RevertAid H Minus cDNA synthesis kit (Fermentas) with random primers following the supplied protocol. cDNA samples were diluted 1/10 and a six-point standard curve of two-fold dilutions was prepared from pooled cDNA. The samples and standard curve were aliquoted to prevent freeze-thawing and stored at −80°C prior to use.
Real-time quantitative PCR with SYBR Green was performed with SensiMix (Quantace) according to the manufacturer's instructions using the primers in Table S3. Primers were designed to assay all annotated splice-variants of a gene. Quantification was performed and target gene expression normalised to Hprt, the expression of which did not differ between the groups. All primers amplified with efficiency estimated between 105% and 80%. Reactions were carried out on a DNA engine Opticon 2 thermocycler (MJ Research).
DNA extraction
gDNA isolation from somatic tissues was carried out by standard organic extraction as previously described (18).
Sperm DNA extraction
Two month old male offspring of control and UN pregnancies were sacrificed and sperm collected from the cauda epididymes and vas deferens as described previously (43, 44). Extruded sperm and sliced epididymes were suspended in 50ml of Solution A (0.75 mL 5M NaCl pH 8; 2.5 ml 0.5 M EDTA; H2O to 50 ml) and rocked on a platform for 10 min to release sperm. Non-sperm tissue was removed by 10 minutes of settling, followed by serial centrifugation at 500 × g for 15 min, and 700× g for 10 min. Sperm was harvested by centrifugation at 1100 × g for 5 min. 200μl Solution B (0.1 mL Tris-HCl pH 8; 0.2 ml 0.5 M EDTA; 2 ml 10% SDS; 8 ml 100mM DTT; H2O to 10 ml) was added, followed by a standard RNAseA and overnight proteinase K treatment at 55°C. DNA was extracted using DNEasy columns (Qiagen) according to the manufacturer's instructions. To accurately quantify sperm DNA concentration prior to MeDIP, Quant-iT™ PicoGreen® was used according to the manufacturer's instructions.
Mass spectrometry
DNA was digested into nucleosides using the DNA degradase Plus (Zymo research) kit according to manufacturer's instructions, and analysed for 5mC and 5hmC by LC-MS/MS as previously described (45).
Bisulphite treatment and pyrosequencing
Sodium bisulphite mutagenesis was carried out on 1μg of gDNA per sample using the Sigma Imprint® DNA Modification Kit according to the manufacturer's instructions, using the 2-step conversion protocol. Two samples with no template were run in parallel to confirm contamination had not occurred during bisulphite treatment.
Quantification of methylation was carried out by pyrosequencing as previously described (46). Pyrosequencing primers were designed using Qiagen PyroMark™ Assay Design software 2.0. Primers sequences are given in Table S4. Following PCR, 2.5μl of each product was run on a gel to ensure specificity of amplification and suitable concentration of product. The biotinylated strand was purified using strepdavidin sepharose high performance beads (GE healthcare) and PyroMark™ reagents (Qiagen) according to the manufacturer's instructions. Pyrosequencing was conducted on a PyroMark™ MD pyrosequencer (Biotage) using PyroMark™ Gold Qp6 SQA reagents (Roche) with quantification of methylated and unmethylated alleles using Pyro Q-CpG 1.0.9 software (Biotage).
MeDIP-seq
Optimisation of antibody specificity using artificially methylated and hydroxymethylated Arabidopsis Thaliana fragments (Diagenode AF-101-0002) was carried out to ensure no cross sampling of hydroxymethylated or unmethylated cytosine (Fig. S1). Sequences used are given below; primers used for qPCR amplification are in bold, underlined type:
Unmethylated control fragment
A. thaliana chromosome 3 TAIR9 (7 June 2009) bases 20074379 – 20074678 TGAAAGGTTGATGCGGGATGATGTCTACAACTATCCACCTGGACATAATACTCTGGGTGACTTGCCTCAATCTAGAAAGCGAAACTATGAAGAAAATTATCCCAGTGAACTTCGCAGGCAAGAAAAGCCTTACATCGATTCAAATTATGCTGCTGACTATTATCATGATAGTGAAGCTGGGAGTCGTAATGGACATTACCGTGATCATGAACACGAAAGGTCGTCCAGATATGATGGCTGCGATGACTATTCTTGTAATGATAACAACTATAGATCGAAAAATTACCATCATAGCAGAGA
Methylated control fragment
A. thaliana chromosome 1 TAIR9 (7 June 2009) bases 30084050 – 30084349 ATCCACTGCGCTATGCGGTCATTCTTGTTACTTATATCATGAACAACATGGTAAGCCAAAACTTCTTCTTTCCTCTTTCTTCTTTCTGTTTTCTGGTCGCTCGTTACCACCAAGACCTCGTCGAGGTTTCTCTCTCTCTGAAAAAGTGCTTTTGCTGCGTTTCCATGCTCTCGGAAGGAACTTGGCTGAATCGTTGGAGAATTATCGATCTGGCGTCGTTGGTGCATCTCTAGCTGCGCCAATTCATCGGATCCGGTTTGATTTCTCCCTTTCGCGAATCTAGCCTTCCGTGATCTTAGG
Hydroxymethylated control fragment
A.thaliana chromosome 1 TAIR9 (7 June 2009) bases 30003430 – 30003709 GGTGAGTAGGGGCGTCGGCTTCTTCTCCTCGGTGTGTCCCTTGGGCATCTGAGACTTGTGAGAATTTTTCATCTGCTGTCTATGTTTTTCGTCTTGCCTTTGTTTCTCTAACTCCCTCTTCTTCCTAAGCCTCCTCTCTTCTTCGGTCTCGATTTTCTTAGAATGAGGAAGATGAGACCTGTGGTGGTGCTTCGATGGATCCGGTTTAGCACGCTCCCGCCTTCCAACCTGCTTGGAGGCCCCTTTCTCGTGGTCATTGGGACCCTGGTGTCGAGGAGGA
Prior to MeDIP, DNA was sheared by sonication using the Diagenode Bioruptor UCD-200, re-quantified using a Nanodrop Spectrophotometer and fragment size was confirmed at 200-700bp. MeDIP-seq was carried out as described previously (19-21), taking into account modifications (47, 48), using 1μl of anti-5methyl-cytosine antibody diluted ¼ in PBS (Eurogentec BI-MECY-0100) and a modified immunoprecipitation buffer (10mM Na-Phosphate pH 7.0, 1M NaCl, 0.05% Triton X-100, 2.5% skimmed milk). Twelve cycles of whole-genome amplification were employed using Phusion™ High-Fidelity Taq and paired end Illumina primers. Libraries were electrophoresed in 2% 1x TAE agarose gels at 95V in 1x TAE buffer followed by excision of a 200bp range band either 200-400bp to 300-500bp according to the peak fragment size. Identical size ranges were obtained for control and UN libraries. Each sample was run in an independent gel tank pre-treated with 0.4N NaOH to prevent contamination. To minimise risk of contamination, separate reagents, pipettes and work spaces were used for the pre-amplification and post-amplification steps.
To control for quality of immunoprecipitation, an aliquot of each library was checked by qPCR for five regions of different CpG densities known to be methylated and unmethylated in sperm (49). The expected relationship between CpG density and IP efficiency at methylated regions was always observed. Fragment size was checked and library concentration was estimated using qPCR and by running a sample on an Agilent High Sensitivity DNA chip using the Agilent 2100 BioAnalyser.
Paired-end sequencing was carried out using Illumina GAIIx and HiSeq platforms. Pre-processing of the raw reads were performed using FastQC v0.10.1 (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and TrimGalore v0.2.7 (www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove poor quality bases and adapter sequences. All programmes were used with default setting unless otherwise specified. Reads post-QC were mapped to mouse reference genome NCBIM37/mm9 using BWA v0.6.2 (50). The mapped data were subject to further QC by SAMtools to remove unmapped, unpaired reads and reads with mapping quality score less than 10 (50). Mapped sequence yields are given in Table S5.
Differentially methylated regions (DMRs) were identified using MEDIPS (23) The mapped data were transformed into a MEDIPS compatible format using a custom Perl script. DNA methylation levels across the whole genome were assessed using sliding windows (window size 500bp and 200bp to shift the window). 500bp windows with average rpm (reads per million) score less than 0.5 were discarded. Overlapping 500bp windows with rpm ratios (between the UN and Control) greater than 1.5 or less than 0.666 and a binomial p-value less than 0.0001 were selected for further study and clustered together. Regions with rpm ratios greater than 1.5 or less than 0.666 and a binomial p-value less than 0.0001 in both independent Control vs UN comparisons were designated as differentially methylated regions, DMRs.
Luciferase assays
In vitro luciferase assays’ DNA fragments were generated by PCR from mouse gDNA using primers designed with either two SacI or two NheI restriction sites at each end in order to clone both in the sense and antisense orientation (Table S6). DMR1 was cloned from BAC RP23-451T11 which maps to MMU12:19,104,994-19,294,685. Each 25μl reaction contained 1X PCR Buffer (KOD Hot Start, Novagen), 300 μM dNTPs, 1 mM MgSO4, 0.5 U Hot Start KOD polymerase, 0.6 μM of each primer and 50 ng of template. The cycling parameters were as follows: 94°C for 2 minutes, 35 cycles at 94°C for 15 seconds, annealing temperature (specific for each primer) for 30 seconds, 72°C for 1min/kb of expected product, and a 5 minute extension cycle at 72°C. PCR products were digested with appropriate restriction enzyme and cloned into pGL3 Promoter (Promega) using standard molecular techniques.
The DNA luciferase constructs were co-transfected with pSV40PK Renilla (Promega, 1:200 dilution) as a transfection control into neural stem cells derived from a mouse embryonic stem cell line, 46C (29) and 3T3 fibroblasts. A positive pGL3 Control vector (Promega) containing both the SV40 promoter and enhancer was transfected to test the assay conditions, while the pSV40PK Renilla vector alone demonstrated no firefly luciferase activity acting as a negative control. The pGL3 Promoter vector (Promega) was used to assess sequences for enhancer activity. To test for a negative effect on transcription sequences were cloned into the XbaI site between the enhancer and promoter of the pGL3 Control vector (Promega). Each construct was tested in triplicate per plate.
The 46CNS cells were cultured in RHB A complete media (StemCells) supplemented with EGF and FGF. The NIH3T3 fibroblasts were cultured in DMEM (Sigma) with 10% FBS, 1% L-glutamine 1% pen/strep. For 46CNS on the day prior to transfection, cells were plated into 48 well plates at a concentration of 3 × 104 cells per well. After 24 hours 0.5 μg of DNA was added to 25 μl of diluent and 1.6 μl Nanofectin™ (PAA) to 25 μl of diluent then combined. This reaction was incubated at room temperature for 15 – 20 minutes. This reaction was then added drop-wise to the media in the well. Media was changed 3-4 hours after transfection. For NIH3T3 Fibroblasts 1.5 × 104 cells per well were seeded into 48 well plates. 0.5 μg of plasmid DNA was added to serum free media with 1 unit of transfection reagent TurboFect™ (Fermentas) in a 50ul total reaction. This reaction was then added drop-wise to the media in each well. The media was changed 3-4 hours after transfection.
The media was changed after a further 24 hours and after 48 hours cells were tested for their luciferase activity using the Dual-Luciferase® Reporter (DLR™) Assay System (Promega) according to the manufacturer's protocol. Each cell lysate was measured in duplicate on a single channel TD20/20 Luminometer (Turner Designs). The luminometer was programmed with a 2-second pre-read delay, followed by a 10-second measurement period. The firefly luciferase values were normalised to the renilla values and then each test construct was normalised to the pGL3-Control vector containing promoter and enhancer sequences. Statistics for the in vitro luciferase reporter assays were calculated using the one-way ANOVA test to calculate the overall statistical significance (P value), followed by Dunn's Multiple Comparison post-test to the empty vector to calculate the statistical significance between specific samples. P < 0.05 was deemed as statistically significant.
Supplementary Material
One sentence summary.
Nutritional DMRs are hypomethylated, retain nucleosomes and partly resist zygotic reprogramming but are not maintained in the next generation.
General summary for consideration for “This Week in Science”.
Prenatal environments can affect adult metabolic health and that of subsequent generations. Epigenetic inheritance has been implicated but mechanisms remain unknown. In a mouse intergenerational developmental programming model affecting two subsequent generations, in utero under-nutrition alters the sperm DNA methylome of male offspring. Affected regions lose methylation and are nucleosome-enriched. A substantial fraction is resistant to zygotic reprogramming with potential to affect development of the next (F2) generation. Importantly, methylation differences are not maintained in F2 tissues, but altered expression of neighbouring genes suggests additional epigenetic dysregulation. Thus, in utero nutrition can permanently alter male germline methylation and affect offspring metabolic health.
Acknowledgements
We are grateful to members of the Ferguson-Smith and Patti labs for useful discussions of this work and to Kristina Tabbada and David Oxley for expert technical assistance with Illumina sequencing and mass-spectrometry. Work was supported by grants from the MRC, Wellcome Trust and European Commission FP7, EpiGenesys and EpiHealth (to ACFS), from the Pediatric Endocrine Society and the NICHD (to EI), and from the American Diabetes Association, the Graetz Foundation and NIH P30DK036836 (to MEP).
References
- 1.Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake AJ, Walker BR. The intergenerational effects of fetal programming: non-genomic mechanisms for the inheritance of low birth weight and cardiovascular risk. J Endocrinol. 2004;180:1–16. doi: 10.1677/joe.0.1800001. [DOI] [PubMed] [Google Scholar]
- 3.Jimenez-Chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, Patti ME. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 6.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 7.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 12.Smallwood SA, Tomizawa SI, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011 doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez-Chillaron JC, Hernandez-Valencia M, Reamer C, Fisher S, Joszi A, Hirshman M, Oge A, Walrond S, Przybyla R, Boozer C, Goodyear LJ, Patti ME. Beta-cell secretory dysfunction in the pathogenesis of low birth weight-associated diabetes: a murine model. Diabetes. 2005;54:702–711. doi: 10.2337/diabetes.54.3.702. [DOI] [PubMed] [Google Scholar]
- 15.Isganaitis E, Jimenez-Chillaron J, Woo M, Chow A, DeCoste J, Vokes M, Liu M, Kasif S, Zavacki AM, Leshan RL, Myers MG, Patti ME. Accelerated postnatal growth increases lipogenic gene expression and adipocyte size in low-birth weight mice. Diabetes. 2009;58:1192–1200. doi: 10.2337/db08-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo M, Isganaitis E, Cerletti M, Fitzpatrick C, Wagers AJ, Jimenez-Chillaron J, Patti ME. Early life nutrition modulates muscle stem cell number: implications for muscle mass and repair. Stem Cells Dev. 2011;20:1763–1769. doi: 10.1089/scd.2010.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 18.Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, Didier N, Charalambous M, McEwen K, Marazzi G, Sassoon D, Patti ME, Ferguson-Smith AC. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 2012;8:e1002605. doi: 10.1371/journal.pgen.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 20.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 21.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, Beck S, Butcher LM. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc. 2012;7:617–636. doi: 10.1038/nprot.2012.012. [DOI] [PubMed] [Google Scholar]
- 23.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 25.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schubeler D, Vlag J, Stadler MB, Peters AH. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, Diekhans M, Fujita PA, Goldman M, Gravell RC, Harte RA, Hinrichs AS, Kirkup VM, Kuhn RM, Learned K, Maddren M, Meyer LR, Pohl A, Rhead B, Wong MC, Zweig AS, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 30.Iwanaga T, Miki T, Takahashi-Iwanaga H. Restricted expression of somatostatin receptor 3 to primary cilia in the pancreatic islets and adenohypophysis of mice. Biomed Res. 2011;32:73–81. doi: 10.2220/biomedres.32.73. [DOI] [PubMed] [Google Scholar]
- 31.Bruno JF, Xu Y, Song J, Berelowitz M. Pituitary and hypothalamic somatostatin receptor subtype messenger ribonucleic acid expression in the food-deprived and diabetic rat. Endocrinology. 1994;135:1787–1792. doi: 10.1210/endo.135.5.7956902. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi T, Adachi Y, Nagayama T. Expression of a secretory protein C1qTNF6, a C1qTNF family member, in hepatocellular carcinoma. Anal Cell Pathol (Amst) 2011;34:113–121. doi: 10.3233/ACP-2011-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, Stringham HM, Sim X, Yang L, Fuchsberger C, Cederberg H, Chines PS, Teslovich TM, Romm JM, Ling H, McMullen I, Ingersoll R, Pugh EW, Doheny KF, Neale BM, Daly MJ, Kuusisto J, Scott LJ, Kang HM, Collins FS, Abecasis GR, Watanabe RM, Boehnke M, Laakso M, Mohlke KL. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Babenko AP, Polak M, Cave H, Busiah K, Czernichow P, Scharfmann R, Bryan J, Aguilar-Bryan L, Vaxillaire M, Froguel P. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N Engl J Med. 2006;355:456–466. doi: 10.1056/NEJMoa055068. [DOI] [PubMed] [Google Scholar]
- 35.Laukkanen O, Pihlajamaki J, Lindstrom J, Eriksson J, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Tuomilehto J, Uusitupa M, Laakso M, G. Finnish Diabetes Prevention Study Polymorphisms of the SUR1 (ABCC8) and Kir6.2 (KCNJ11) genes predict the conversion from impaired glucose tolerance to type 2 diabetes. The Finnish Diabetes Prevention Study. J Clin Endocrinol Metab. 2004;89:6286–6290. doi: 10.1210/jc.2004-1204. [DOI] [PubMed] [Google Scholar]
- 36.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, Edghill EL, Frayling TM, Temple IK, Mackay D, Shield JP, Sumnik Z, van Rhijn A, Wales JK, Clark P, Gorman S, Aisenberg J, Ellard S, Njolstad PR, Ashcroft FM, Hattersley AT. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 37.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 38.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., Mello CC. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tash JS, Bracho GE. Identification of phosphoproteins coupled to initiation of motility in live epididymal mouse sperm. Biochem Biophys Res Commun. 1998;251:557–563. doi: 10.1006/bbrc.1998.9516. [DOI] [PubMed] [Google Scholar]
- 44.Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 46.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 47.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, Fujita PA, Learned K, Rhead B, Smith KE, Kuhn RM, Karolchik D, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.