Abstract
Follicular helper T (TFH) cells and follicular regulatory T (TFR) cells regulate the quantity and quality of humoral immunity. Although both cell types highly express the co-stimulatory receptor ICOS and require the transcription factor Bcl-6 for their differentiation, the ICOS-dependent pathways that coordinate their responses are not well understood. Here we report that ICOS activation in CD4+ T cells promotes the interaction of the p85α regulatory subunit of the signaling kinase PI3K and intracellular osteopontin (OPN-i), followed by nuclear translocation of OPN-i, interaction with Bcl-6 and protection of Bcl-6 from ubiquitin-dependent proteasome degradation. Post-translational protection of Bcl-6 expression by OPN-i is essential for sustained TFH and TFR cell responses and regulation of the germinal center B cell response to antigen. As such, the p85α–OPN-i axis represents a molecular bridge that couples ICOS activation to Bcl-6-dependent functional differentiation of TFH and TFR cells and suggests new therapeutic avenues to manipulate their responses.
The generation of long-lived high-affinity antibodies after microbial infection or vaccine induction requires precise control of the germinal center (GC) reaction. Follicular helper T (TFH) cells are specialized effector CD4+ T cells that provide help for GC formation and induce GC B cells to develop protective antibody responses to invading pathogens. Bcl-6, a proto-oncoprotein and transcriptional repressor belonging to the BTB-POZ family, has been identified as the central transcription factor that controls TFH differentiation and associated GC responses 1–3. Because Bcl-6 deficiency can result in increased susceptibility to chronic infection, while its excessive expression is associated with autoimmunity and lymphocytic transformation, precise control of Bcl-6 expression during T cell differentiation represents an essential component of the TFH cell response 4. Moreover, recently-defined Foxp3+ follicular regulatory T cells (TFR) that inhibit GC responses also require Bcl-6 expression for their differentiation and suppressive activity 5–7. However, in contrast to our insight into the molecular elements that regulate Bcl-6 expression in GC B cells 4, the mechanisms that govern Bcl-6 expression by both TFH and TFR cells are poorly understood.
The differentiation of TFH cells in vivo can be divided into several stages that include initiation, maintenance and full polarization 8. This process depends on early upregulation of Bcl-6 gene expression during T-cell activation and TFH commitment, followed by continued enhanced Bcl-6 expression during the maintenance and polarization phases of the TFH cell response 9. Although engagement of the ICOS receptor represents a key event in a process that culminates in Bcl-6 expression and acquisition of the TFH and TFR phenotypes, the essentials of this specialized inductive pathway have not been clarified.
ICOS binding its ligand (ICOSL) expressed by antigen-presenting cells (APC) results in recruitment of the phosphatidylinositol-3-OH kinase (PI3K) signaling complex that consists of a regulatory p85α subunit and a catalytic p110 component. Recruitment of PI3K to ICOS is an essential step in TFH cell differentiation, as mutations of the ICOS cytoplasmic tail that abrogate recruitment of PI3K impair TFH cell generation and GC responses 10. Although deficient expression of the p110δ component impairs follicular migration of TFH cells 11, 12, ICOS-dependent upregulation of Bcl-6 expression and development of CXCR5+ TFH-like cells proceed normally 11–13. In contrast, the contribution of the p85α component of PI3K to Bcl-6 expression and development of both TFH and TFR cells remains unclear. Because p85α regulates the activity and localization of intracellular proteins 14–16, we asked whether an interaction between p85α and downstream intracellular protein(s) in CD4+ T cells after ICOS stimulation might contribute to the Bcl-6-dependent TFH and TFR cell program.
The phosphoprotein osteopontin (OPN, encoded by Spp1) is expressed in activated T cells 17–23. Although dysregulated expression of OPN has been associated with a broad spectrum of TFH-associated autoimmune diseases and strongly correlates with autoantibody production 17–23, the precise contribution of OPN to TFH cell differentiation is uncertain. OPN is expressed as either a secreted (OPN-s) or intracellular (OPN-i) isoform that results from differential usage of Spp1 translational initiation sites 17. To clarify the contribution of each OPN isoform to the regulation of TFH responses, here we generated knock-in mice that expressed only OPN-i and compared them with wild-type mice that express both isoforms or OPN knockout (KO) mice that express neither OPN isoform. We find that OPN-i functions as a positive regulator of both TFH and TFR cell differentiation by enhancing Bcl-6 protein stability, and we identify the p85α–OPN-i complex as a critical molecular bridge that couples ICOS engagement to sustained TFH and TFR responses that combine to regulate the GC antibody response.
Results
Expression of OPN-i is essential for TFH and TFR cell differentiation
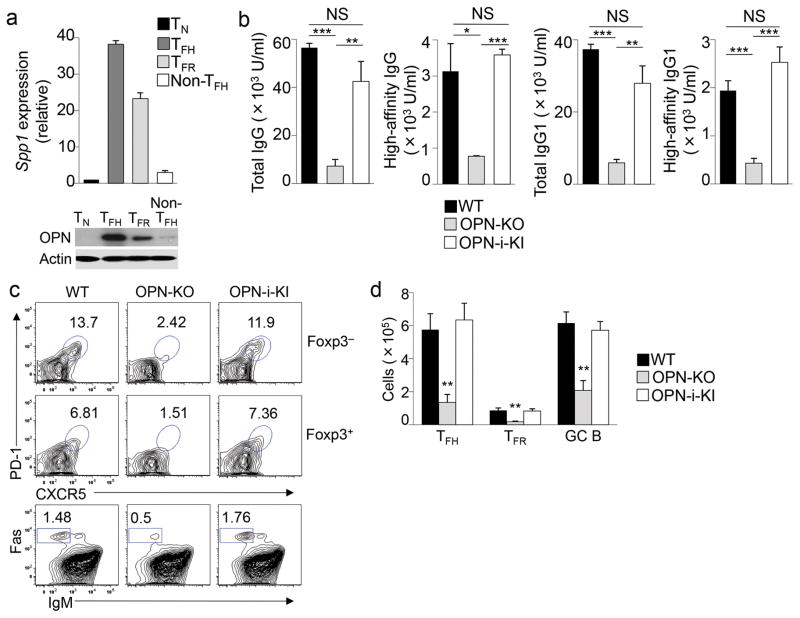
We first analyzed OPN mRNA and protein expression in different CD4+ T cell subsets after immunization with keyhole limpet hemocyanin (KLH) precipitated in complete Freund’s adjuvant (CFA). We noted that OPN was expressed most abundantly by the CD4+ TFH and TFR subsets compared with other CD4+ T cell subsets (Fig. 1a and Supplementary Fig. 1), suggesting a potential contribution of OPN to the development of these follicular effector and regulatory T cells.
Figure 1.
OPN regulates TFH and TFR cell differentiation.
a, Quantitative RT-PCR analysis of Spp1 mRNA (top) and immunoblot analysis of OPN and actin protein levels (bottom) expressed by CD4+ T cell subsets sorted (as shown in Supplementary Fig. 1) from pooled B6 mice (n = 16) 3 d after injection with KLH in CFA. Spp1 expression was normalized to the Rps18 control and results are presented relative to naïve T cells (TN), set as 1. b, Titers of total (NP23) and high-affinity (NP4) NP-specific IgG and IgG1 in serum of OT-II, OT-II OPN-KO and OT-II OPN-i-KI mice that were immunized i.p. with NP13-OVA in CFA, challenged with NP13-OVA in IFA 10 d later, and analyzed 7 d post-challenge. c, Flow cytometry of splenocytes from mice in b. Numbers indicate percent TFH (Foxp3−PD1+CXCR5+CD4+), TFR (Foxp3+PD1+CXCR5+CD4+) and GC B (Fas+IgM−B220+) cells. d, Numbers of TFH, TFR and GC B cells in c (n = 6 per group). *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired two-tailed Student’s t-test); NS, not significant. Data are representative of two (a) and at least three independent experiments (b–d) (error bars, mean ± s.e.m).
To define the contribution of OPN isoforms, we generated mice bearing a Spp1flstop allele knocked into the Spp1 locus. In this system, Cre/loxP-mediated recombination induces expression of the OPN-i isoform 17 after excision of a STOP cassette (Supplementary Fig. 2a). Crossing of Spp1flstop mice with EIIa-Cre mice, which express the Cre recombinase from the adenovirus EIIa promoter during embryonic development, generated Spp1flstop Cre+ (hereafter OPN-i KI), which express intracellular OPN), and the Spp1flstop Cre− mice (hereafter OPN KO), which do not express either OPN-i or OPN-s. Both genotypes were developmentally indistinguishable from wild-type littermates and PCR analysis confirmed expression of the Spp1flstop alleles at the Spp1 locus (Supplementary Fig. 2b). Secreted OPN was not detectable in supernatants of freshly isolated or activated T cells, DC, macrophages or NK cells from either OPN-i KI or OPN KO mice (Supplementary Fig. 2c). Moreover, immunoblot analysis showed equivalent intracellular expression of OPN protein in unfractionated splenocytes from OPN-i KI and OPN wild-type mice (Supplementary Fig. 2d). Expression of OPN-i, but not OPN-s, by plasmacytoid dendritic cells (pDC) is essential for efficient production of IFN-α after Toll-like receptor ligation 21. As an additional control, activated pDC from OPN-i KI and wild-type mice expressed similar levels of IFN-α, while pDC from OPN KO mice expressed no IFN-α (Supplementary Fig. 2e), confirming the functional phenotype of OPN-i KI mice.
We next investigated the contribution of OPN-i to TFH and TFR cell differentiation by comparing OPN-i KI mice with transgenic expression of the OT-II ovalbumin (OVA)-specific T cell antigen receptor (TCR) to OT-II and OT-II OPN KO mice. These mice were immunized with 4-hydroxy-3-nitrophenyl linked to OVA (NP13-OVA) to induce antigen-specific CD4+ T cell responses. Total and high-affinity antibody responses were reduced by 80–90% in OT-II OPN KO mice compared with OT-II mice (Fig. 1b), while antibody titers in OT-II OPN-i KI mice were similar to OT-II mice. Although T cell activation was not obviously impaired in OT-II OPN KO mice compared to OT-II and OT-II OPN-i KI mice as judged from CD44 expression (Supplementary Fig. 3a), OT-II OPN KO mice displayed defective formation of TFH cells, TFR cells (but not Foxp3+ Treg) and GC B cells (Fig. 1c, d and Supplementary Fig. 3a). These findings suggest that expression of OPN-i is essential for both TFH and TFR cell formation and for TFH-associated high-affinity antibody responses.
OPN-i specifically promotes TFH cell differentiation
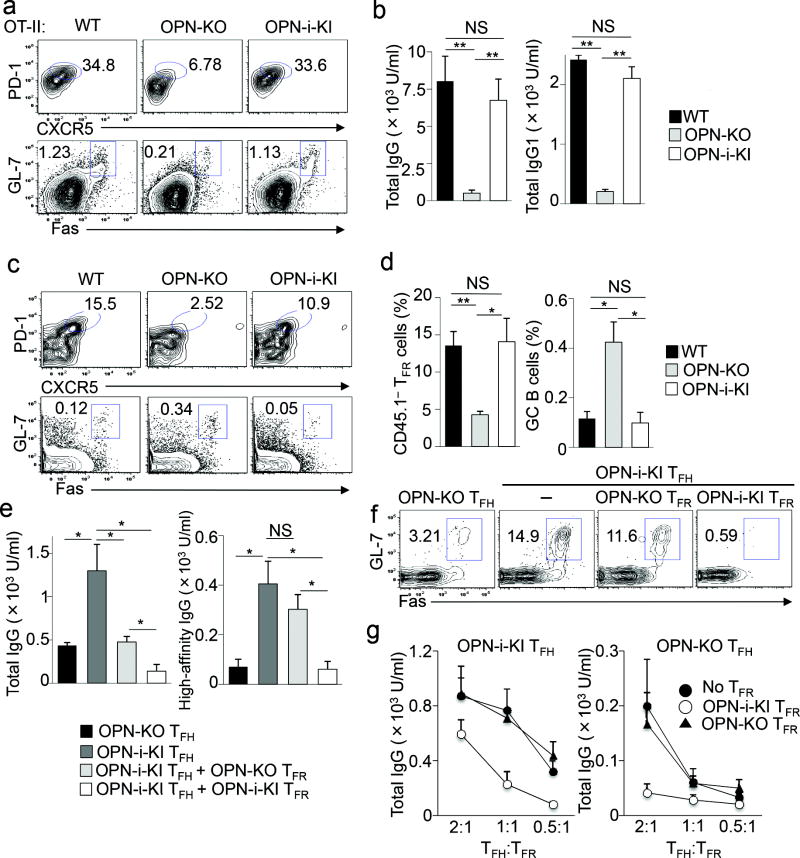
To address if the requirement for OPN expression is cell intrinsic to the TFH cells, we transferred OT-II OPN KO CD4+ T cells together with wild-type B cells into Rag2−/−Prf1−/− hosts, which are defective in T and B cells and lack perforin activity in NK cells, followed by immunization with NP13-OVA. We noted substantially reduced serum anti-NP total IgG and IgG1 and diminished splenic TFH and GC B cell formation in Rag2−/−Prf1−/− hosts reconstituted with OT-II OPN KO CD4+ T cells compared to the response of mice reconstituted with OT-II OPN-i KI or OT-II CD4+ T cells (Fig. 2a, b). When co-transferred with wild-type CD4+ T cells into Rag2−/−Prf1−/− hosts, OPN KO and wild-type B cells produced equivalent antibody responses suggesting OPN is not required in B cells for antibody production (Supplementary Fig. 3b). In addition, we analyzed OT-II OPN KO CD4+ T cells compared to OT-II OPN-i KI and OT-II CD4+ T cells after transfer into wild-type CD45.1 congenic mice that were immunized with OVA in CFA. Despite normal expansion and activation of OT-II OPN KO CD4+ T cells, according to numbers of Vβ5+ CD4 cells and expression of CD44 (Supplementary Fig. 3c), donor OT-II OPN KO CD4+ T cells displayed reduced CXCR5+ TFH cell frequency and expression of intracellular IL-21 compared to OT-II OPN-i KI and OT-II counterparts (Supplementary Fig. 3d). In contrast, the frequency of Vβ5+ CD4+ T cells expressing IFNγ, IL-4, and IL-17A, corresponding to TH1, TH2 and TH17 subsets respectively, was not reduced in OT-II OPN KO CD4 cells (Supplementary Fig. 3d). When differentiated in vitro in TH polarizing conditions, naïve OT-II OPN KO CD4+ T cells showed slightly increased differentiation into IL-17-producing cells compared to OT-II OPN-i KI and OT-II CD4+ T cells, while there was a modest, but not significant decrease in IFNγ expression by OT-II OPN KO CD4+ T cells compared to OT-II OPN-i KI and OT-II counterparts (Supplementary Fig. 3e). In addition, OPN KO CD4+ T cells differentiated into induced Treg (iTreg) cells with similar suppressive activity compared with wild-type and OPN-i KI CD4+ T cells (Supplementary Fig. 4a, b). These results suggest that OPN-i specifically promotes TFH cell differentiation and is not required for TH1, TH2, TH17 or iTreg cell differentiation.
Figure 2.
The OPN-i deficient TFH and TFR phenotype is cell-intrinsic.
a, Flow cytometry of donor OT-II CD4+ T cells from spleens of Rag2−/−Prf1−/− hosts transferred with naïve OT-II, OT-II OPN-KO and OT-II OPN-i-KI CD4+ T cells along with wild-type B cells, followed by immunization with NP13-OVA in CFA and analysis 10 d later. Numbers indicate percent TFH (Foxp3−PD1+CXCR5+CD4+) and GC B (Fas+GL7+B220+) cells. b, Serum titers of total (NP23) NP-specific IgG and IgG1 from recipient mice in a (n = 6 per group). c, Flow cytometry of donor Treg from spleens of Tcrα−/− hosts transferred with sorted CD45.2+ Treg (CD25hiCD44intCXCR5−CD4+) from the indicated mice and CD45.1+ wild-type naïve CD4+ T cells (CD25−GITR−CD44loCD62Lhi) at a ratio of 1:2, followed by immunization with NP26-KLH in CFA and analysis 10 d later. Numbers indicate percent TFR (CD45.1−CD44+Foxp3+PD1+CXCR5+CD4+) and GC B (Fas+GL7+B220+) cells. d, Frequency of TFR and GC B cells in c (n = 5 per group). e, Titers of total (NP23) and high-affinity (NP4) NP-specific IgG at d14 in immunized Rag2−/−Prf1−/− recipients of OPN-i-KI or OPN-KO TFH cells (5 × 104) and/or OPN-i-KI or OPN-KO TFR cells (2.5 × 104) (sorted as in Supplementary Fig. 1) and wild-type GL-7− B cells (1 × 105) from KLH-immunized mice. All recipients (n = 4 per group) were immunized with NP26-KLH in CFA. f, Donor GC B cells from spleens of recipients in e (in same order from left to right) at d22. g, Titers of NP-specific total IgG at d11 in immunized Rag2−/−Prf1−/− mice (n = 3 per transfer) given OPN-i-KI or OPN-KO TFH cells and/or OPN-i-KI or OPN KO TFR cells (2.5 × 104) at different ratios and wild-type GL-7− B cells (1 × 105) from KLH-immunized mice. Data are representative of three (a–b) and two (c–g) independent experiments. *P < 0.05 and **P < 0.01 (unpaired two-tailed Student’s t-test; error bars, mean ± s.e.m).
OPN-i expression in Treg cells is required for TFR cell differentiation
TFR cells originate from CXCR5−Foxp3+ natural Treg cells and are characterized as PD1+CXCR5+Foxp3+ T cells 6, 7. To determine whether OPN-i promotes TFR cell formation, we co-transferred CD45.1+ CD4+ naïve T cells (sorted as CD25−GITR−CD44loCD62Lhi) with CD45.2+CD4+ Treg cells (sorted as CD25hiCD44intCXCR5−) from either wild-type, OPN KO or OPN-i KI mice at a ratio of 2:1 into Tcrα−/− hosts, followed by immunization with NP26-KLH in CFA. We observed a significant reduction of OPN KO CD45.1−PD1+CXCR5+Foxp3+CD4+ TFR cells 10 d post-immunization (Fig. 2c, d), which was associated with increased GC B cell formation (Fig. 2c, d) as well as anti-NP (Supplementary Fig. 4c) and anti-KLH antibody titers (Supplementary Fig. 4d) in recipients of OPN KO CD4+ Treg, compared with the response after transfer of wild-type and OPN-i KI CD4+ Treg cells. These results suggest that OPN-i-deficiency impairs TFR cell differentiation from CXCR5− Treg precursors.
Transfer of differentiated OPN KO TFR cells along with wild-type B cells into Rag2−/−Prf1−/− hosts followed by NP26-KLH immunization resulted in greater expansion of GL7+Fas+ GC B cells and higher amounts of NP-specific total and high-affinity IgG compared to hosts transferred with the same numbers of OPN-i KI TFR cells (Fig. 2e–g and Supplementary Fig. 1), suggesting that OPN-i deficiency diminishes TFR suppressive activity on a per-cell basis in vivo. Transfer of mixtures of TFH cells and TFR cells at different ratios (2:1, 1:1 and 0.5:1) with wild-type B cells into Rag2−/−Prf1−/− hosts followed by NP26-KLH immunization showed increased serum anti-NP total IgG in hosts transferred with higher numbers of TFH cells, suggesting that the magnitude of TFR cell-mediated suppression is dependent on the extent of TFH-driven antibody responses (Fig. 2g). Taken together these results indicate that OPN-i expression is required for the generation of functional TFR cells.
OPN-i deficiency impairs Bcl-6 protein expression
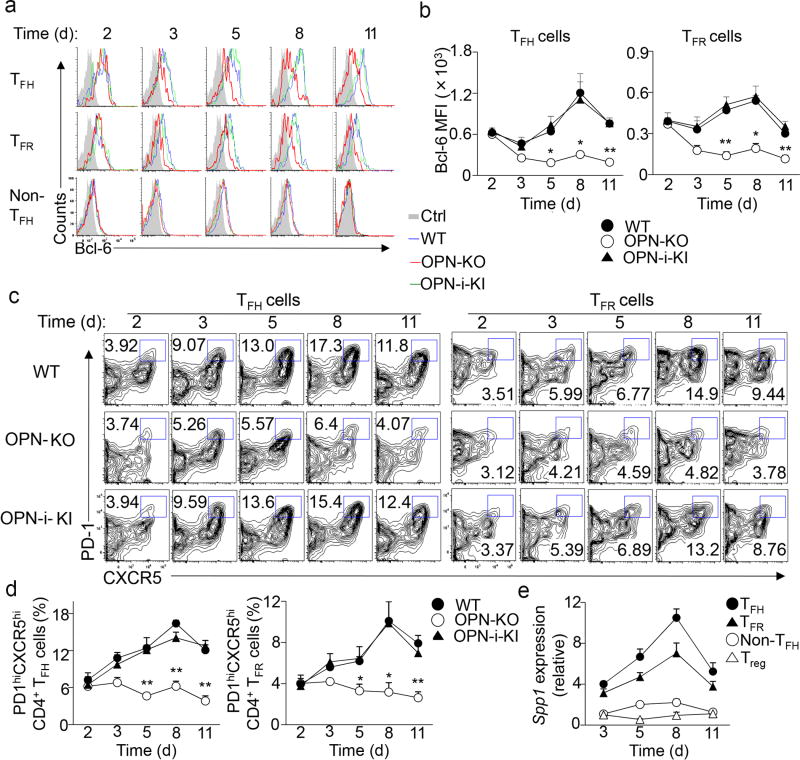
Because the transcription factor Bcl-6 is critical for TFH and TFR cell differentiation 6–8, we asked whether the impaired follicular T cell response of OPN KO CD4+ T cells was associated with reduced Bcl-6 expression. Flow cytometry and immunoblot analysis of Bcl-6 expression during TFH cell development in vivo in wild-type mice showed that Bcl-6 was detectable by day 1, peaked at day 7.5 and waned by day 10 after KLH immunization (Fig. 3a, b and Supplementary Fig. 1, 4e), consistent with a previous study 24. Expression of Bcl-6 in TFR cells followed a similar kinetic, albeit expression was lower than in TFH cells (Fig. 3a, b). OPN deficiency did not alter Bcl-6 and Prdm1 mRNA expression and did not impair early Bcl6 protein expression by OPN KO CD4+ T cells (within ~2 days), (Fig. 3a, b and Supplementary Fig. 4e, f). The OPN deficient response was marked by reduced generation of TFH and TFR cells by OPN KO CD4+ T cells and severely decreased expression of Bcl-6 protein in the few residual OPN KO TFH and TFR cells by day 3 and thereafter (Fig. 3a–d). These findings suggest that although OPN-i does not regulate Bcl-6 transcription, it may differentially affect expression of the Bcl-6 protein during differentiation into TFH and TFR cells.
Figure 3.
OPN-i deficiency impairs Bcl-6 protein expression.
a–b, Flow cytometry of splenocytes from the indicated mice at day 2–11 after immunization with KLH in CFA. Overlay of histograms of intracellular Bcl-6 expression in CD4+ TH subsets (as defined in Supplementary Fig. 1) (a) and quantitation of Bcl-6 MFI (b). c, TFH (CD4+CD44hiCXCR5+PD-1+Foxp3−) and TFR (CD4+CD44hiCXCR5+PD-1+Foxp3+) cell differentiation from mice in a. Numbers indicate percent PD1+CXCR5+ cells. d, Frequency of TFH and TFR cells in c (n = 3–4 per time point per group). e, Quantitative RT-PCR analysis of Spp1 mRNA in CD4+ T cell subsets sorted (as in Supplementary Fig. 1) from pooled OPN-i-KI mice (n = 3–7 per time point) at day 3–11 after immunization with KLH in CFA. Spp1 expression was normalized to the Rps18 control and results are presented relative to that of Treg at day 3, set as 1. Data represent two (a–e) independent experiments. *P < 0.05 and **P < 0.01 (unpaired two-tailed Student’s t-test; error bars, mean ± s.e.m or s.d. in e).
ICOS co-stimulation promotes an interaction between OPN-i and p85α
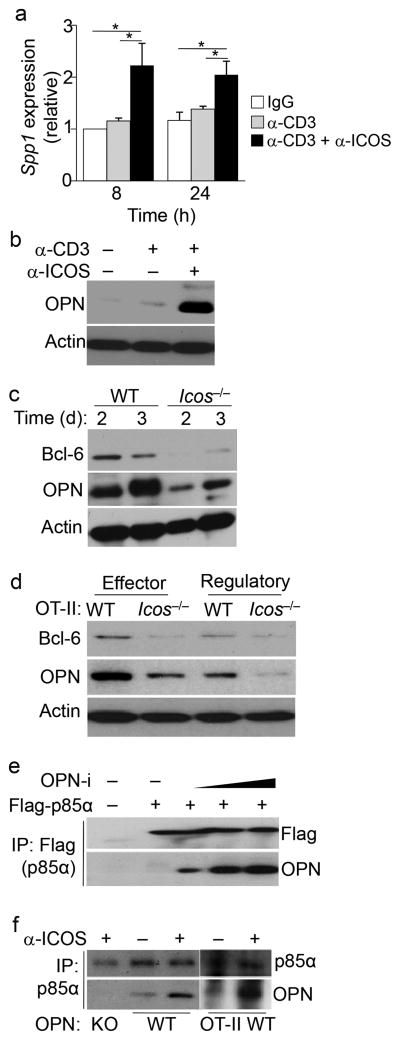
ICOS co-stimulation is essential for the induction and maintenance of Bcl-6 expression during TFH cell differentiation and is required for TFR cell formation 5, 25, 26. Because expression of OPN-i and Bcl-6 followed similar kinetics in TFH and TFR cells (Fig. 3e) and Bcl-6 protein expression was reduced in the few CXCR5+PD-1+ TFH cells generated in the OPN KO mice we tested the possibility that OPN-i mediates ICOS-induced Bcl-6 expression. Gene profile analysis (GSE61284) in CD4+ T cells 8 h after CD3 and ICOS ligation identified ~210 genes, including OPN (Spp1), that were significantly (P < 0.01) upregulated compared to CD3 ligation alone (Supplementary Fig. 5a). Pathway analysis (Ingenuity®) of these genes revealed that they were involved with many biological functions associated with T cell activation, antibody production and with systemic autoimmune disease (Supplementary Fig. 5b, c). We confirmed Spp1 mRNA and OPN protein upregulation by ICOS and CD3 co-stimulation compared to CD3 ligation alone in in vitro-activated CD4+ T cells (Fig. 4a, b). Icos−/− CD4+ T cells did not upregulate expression of OPN and Bcl-6 proteins compared with wild-type CD4+ T cells after KLH immunization (Fig. 4c). Impaired upregulation of OPN and Bcl-6 proteins was also observed in effector CD25−CD44hiGITR−CD4+ T cells and regulatory CD25+CD44hiGITR+CD4+ T cells from OT-II Icos−/− mice after OVA immunization (Fig. 4d). These findings suggest that ICOS ligation regulates OPN-i expression in in vivo-activated CD4+ T cells.
Figure 4.
ICOS co-stimulation promotes an interaction between OPN-i and p85α.
a, Quantitative RT-PCR analysis of Spp1 mRNA in naïve CD4+ T cells from B6 mice stimulated with anti-CD3 and anti-CD28 for 2 d, followed by resting overnight before 20 m incubation with the indicated Abs and cross-linking with goat anti-hamster Ab for 8 h or 24 h. Spp1 expression was normalized to the Rps18 control and results are presented relative to isotype-matched hamster IgG-treated cells at 8 h, set as 1. *P < 0.05 (unpaired two-tailed Student’s t-test; error bars, mean ± s.e.m of quadruplicates). b, Immunoblot analysis of OPN and actin of CD4+ T cells in a after 12 h cross-linking. c, Immunoassay of lysates of purified CD62L− CD4+ T cells from Icos−/− and wild-type mice 2 or 3 d after intraperitoneal injection with 100 μg KLH in CFA, probed with anti-Bcl-6, anti-OPN and anti-actin. d, Immunoblot analysis of lysates of sorted Vβ5+CD25−CD44hiGITR−CD4+ effector and CD25+CD44hiGITR+CD4+ regulatory T cells from pooled OT-II (n = 10) or OT-II Icos−/− mice (n = 15) 8 d post-immunization with OVA in CFA, probed as in c. e, Immunoassay of lysates of 293T cells transfected with plasmids encoding Flag-tagged p85α (Flag-p85α) and increasing concentrations of OPN-i, assessed by immunoprecipitation (IP) with anti-Flag and immunoblot analysis with anti-Flag and anti-OPN. f, Immunoassay of lysates of purified naïve CD4+ T cells from OPN-KO and wild-type mice stimulated with anti-CD3 and anti-CD28 for 2 d (left); or CD44+CD4+ T cells from OT-II mice 4 d post-immunization with OVA in CFA (right), followed by resting and re-stimulation with anti-CD3 and/or anti-ICOS (as in a) for 12 h, and assessed by IP with anti-p85α and immunoblot analysis with anti-p85α and anti-OPN. Data represent two (a, c, d) and three (b, e, f) independent experiments.
Because activation of the PI3K signaling pathway is induced by ICOS ligation 10, we determined whether OPN-i associates with the p110-p85 components of the PI3K complex. OPN-i did not interact with the p110α or p110δ subunits of PI3K in 293T cells transfected with plasmids encoding these proteins (Supplementary Fig. 6a, b). In addition, PI3K–Akt activation, as indicated by Akt phosphorylation, was not reduced in OPN KO CD4+ T cells stimulated with anti-CD3 and anti-ICOS compared to OPN-i KI CD4+ T cells (Supplementary Fig. 6c). Activation of IL-6 signals, as indicated by STAT1 and STAT3 phosphorylation, was also not impaired in OPN KO CD4+ T cells after incubation with IL-6 compared to OPN-i KI controls (Supplementary Fig. 6d). However, overexpressed OPN-i co-immunoprecipitated with Flag-p85α in transfected 293T cell lines (Fig. 4e). In CD4+ T cells stimulated with anti-CD3, endogenous OPN-i interacted with p85α in CD4+ T cells and co-ligation of TCR and ICOS substantially increased their association (Fig. 4f), suggesting that ICOS-dependent activation promotes the interaction between p85α and OPN-i.
p85α is required for TFH and TFR cell differentiation
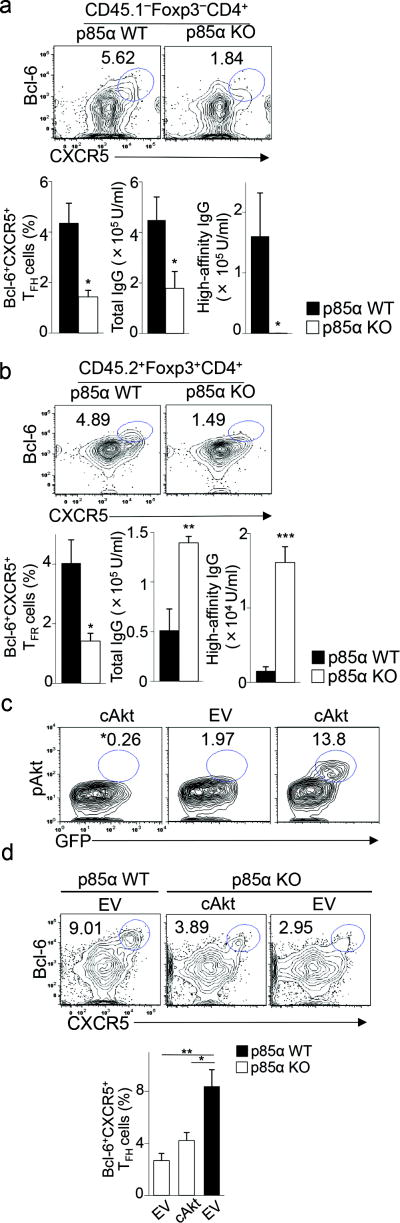
To determine whether p85α is required for Bcl-6 upregulation during TFH cell differentiation we used mice with loxP-flanked alleles encoding p85α (Pi3kr1fl) deleted by Cre recombinase expressed from the hematopoietic cell-specific Vav1 promoter (Pi3kr1flVav1-Cre+), termed p85α KO mice here. After immunization of p85α KO mice with KLH in CFA ×3d, development of TFH and TFR cells as well as Bcl-6 expression in these cells was impaired in p85α KO mice compared to p85α wild-type mice (Pi3kr1flVav1-Cre−) (Supplementary Fig. 6e). To specifically analyze the contribution of p85α to TFH cell differentiation, we transferred wild-type or p85α KO naïve CD4+ T cells together with wild-type B cells into Rag2−/−Prf1−/− hosts immunized with NP26-KLH in CFA. Despite the nearly normal expansion and activation of transferred p85α KO CD4+ T cells following immunization (Supplementary Fig. 6f), the frequency of p85α KO Bcl-6+CXCR5+Foxp3−CD4+ TFH cells was greatly reduced and anti-NP antibody responses were impaired in adoptive hosts compared to hosts transferred with p85α wild-type CD4+ T cells (Fig. 5a and Supplementary Fig. 6g). Loss of p85α in transferred CD4+ T cells did not significantly alter their differentiation into TH1, TH2 and TH17 cells in vivo (Supplementary Fig. 6h). However, p85α deficiency significantly impaired TFR cell formation after co-transfer of p85α KO (CD45.2+) CD4+ Treg cells with wild-type (CD45.1+) naïve CD4+ T cells into Tcrα−/− hosts immunized with NP26-KLH in CFA. Recipients of p85α KO CD4+ Treg produced increased amounts of total and high-affinity anti-NP antibodies compared with recipients of p85α wild-type CD4+ Treg cells (Fig. 5b and Supplementary Fig. 6g). Taken together these results indicate that p85α is required for TFH and TFR cell formation.
Figure 5.
p85α is required for TFH and TFR cell differentiation.
a, Flow cytometry of donor CD4+ T cells from spleens of Rag2−/−Prf1−/− hosts transferred with p85α WT or p85α KO naïve CD4+ T cells along with CD45.1+ wild-type B cells, followed by immunization with NP26-KLH in CFA and analysis 10 d later. Numbers indicate percent CD45.1−Foxp3−Bcl-6+CXCR5+ TFH cells. Bottom panel, frequency of TFH cells and titers of NP-specific total (NP23) and high-affinity (NP4) IgG in immunized recipients (n = 3–4 per group). b, Flow cytometry of donor CD4+ Treg from spleens of Tcrα−/− hosts transferred with CD45.2+ p85α WT or p85α KO CD4+ Treg along with CD45.1+ naïve CD4+ T cells, followed by immunization with NP26-KLH in CFA and analysis 10 d later. Numbers indicate percent CD45.2+Foxp3+Bcl-6+CXCR5+ TFR cells. Bottom panel, frequency of TFR cells and titers of NP-specific total (NP23) and high-affinity (NP4) IgG in immunized recipients (n = 3–6 per group). c, Flow cytometry of p85α KO CD4+ T cells transduced with retroviral vector expressing GFP alone (empty vector [EV]) or GFP and constitutively-active Akt (cAkt). Numbers indicate percent GFP+pAkt+ (phospho-Akt) cells. *, isotype control for pAkt staining. d, (top panel) Flow cytometry of donor CD4+ T cells from spleens of Rag2−/−Prf1−/− hosts transferred with p85α KO or p85α WT CD4+ T cells transduced with retroviral vectors (as described in c) along with wild-type B cells, followed by immunization with NP26-KLH in CFA and analysis 5 d post-immunization. Numbers indicate percent Bcl-6+CXCR5+ cells. Bottom panel, frequency of Bcl-6+CXCR5+CD4+ T cells (n = 4 per group). Data are representative of two (a–d) independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired two-tailed Student’s t-test; error bars, mean ± s.e.m).
To determine whether defective p85α expression diminished Bcl-6 expression, we transduced p85α KO CD4+ T cells with retroviral vector expressing green fluorescent protein (GFP) alone (empty vector) or GFP and a constitutively-active form of Akt 27, then sorted GFP+ cells and transferred them along with wild-type B cells into Rag2−/−Prf1−/− hosts, followed by immunization with NP26-KLH in CFA (Fig. 5c, d). Among the GFP+ p85α KO CD4+ T cells, more cells transduced with the constitutively active Akt (~14%) expressed phosphorylated Akt compared to cells transduced with the empty vector (2%; Fig. 5c). However, constitutively active Akt expression did not significantly increase expression of Bcl-6 and CXCR5 in p85α KO CD4+ Tcells compared to cells infected with a control virus (Fig. 5d and Supplementary Fig. 6g). These findings indicate that PI3K-p85α but not PI3K-p110 is required for Bcl-6-dependent TFH cell differentiation.
p85α chaperones nuclear translocation of OPN-i
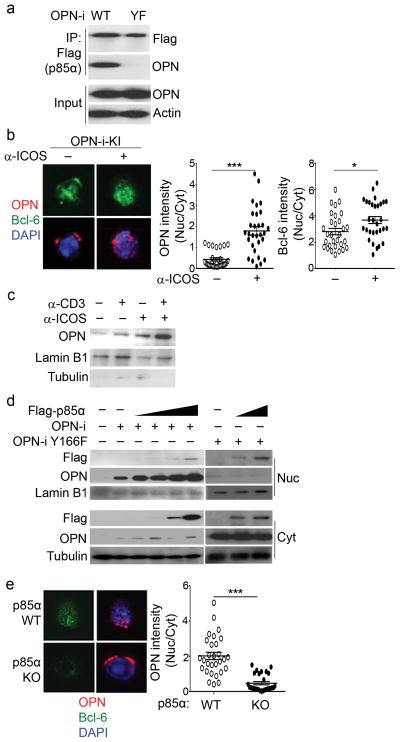
Because the above findings suggested that p85α and OPN-i might contribute to Bcl-6-dependent TFH cell differentiation, we sought to further define the interaction between p85α and OPN-i. In transfected 293T cells, the p85α-bound fraction of OPN underwent a shift in migration after treatment with protein phosphatases (Supplementary Fig. 6i), consistent with reports that p85α can recognize phosphorylated proteins. Using a web-based program (Scansite) 28, 29 we identified a sequence in the OPN protein near position Tyr 166 (UniProtKB: P10923) that might mediate an interaction with the SH2 domain of p85α (Supplementary Fig. 6j). Binding of a Y166F OPN-i mutant protein to p85α was substantially reduced compared to that of wild-type OPN-i protein (Fig. 6a), suggesting that interaction of p85α with OPN-i requires an intact OPN-i Y166 site.
Figure 6.
p85α chaperones nuclear translocation of OPN-i.
a, Immunoassay of lysates of 293T cells transfected with plasmids encoding Flag-p85α and OPN-i wild-type or OPN-i Y166F (YF) mutant, assessed by immunoprecipitation with anti-Flag and immunoblot analysis with anti-Flag and anti-OPN. Input, immunoblot analysis of an aliquot of cell lysates without IP. b, Confocal microscopy of OPN and Bcl-6 expression by CD44+CD4+ T cells from OPN-i-KI mice 3 d post-injection with KLH in CFA, followed by cross-linking with anti-ICOS Ab (as in Fig. 4a) in vitro. Cells were counterstained with DNA-intercalating dye DAPI to trace nuclear perimeters. About 25–30 cells stained with Bcl-6 were further analyzed for localization of OPN protein. Right, fluorescence intensity expressed as the mean ratio of nuclear (Nuc) to cytoplasmic (Cyt) fluorescence pixel intensity (n = 25–30 cells). c, Immunoblot analysis of nuclear fractions of OPN-i-KI CD62L−CD4+ T cells treated with anti-CD3 and/or anti-ICOS Abs (as in Fig. 4a), probed with anti-OPN, anti-Lamin B1 and anti-tubulin (to validate the integrity of nuclear separation). d, Immunoblot analysis of nuclear and cytosolic fractions of 293T cells transfected with plasmids encoding OPN-i wild-type or OPN-i Y166F mutant and increasing concentrations of Flag-p85α, probed with Abs, as indicated. e, Confocal microscopy of OPN and Bcl-6 expression by CD62L− CD4+ T cells from p85α WT or KO mice. Cell treatment and analysis as in a. Original magnification (b, e), 600×. *P < 0.05 and ***P < 0.001, Mann-Whitney test (error bars, mean ± s.e.m) (b, e). All results are representative of at least two independent experiments.
We next examined the cellular localization of OPN protein in CD44+CD4+ T cells enriched from OPN-i KI mice 3d post-immunization with KLH in CFA. Confocal microscopic analysis revealed that Bcl-6 protein stained with anti-Bcl-6 before labeling with a secondary fluorescence antibody was mainly nuclear in Bcl-6+ cells (representing ~10% of CD44+CD4+ T cells) and in vitro anti-ICOS restimulation further increased the abundance of intranuclear Bcl-6 (Fig. 6b and Supplementary Fig. 6k). While prior to ICOS stimulation OPN-i protein was located mainly in the cytosol of Bcl-6+CD4+ T cells, ICOS ligation caused most OPN-i protein to migrate within the nucleus (Fig. 6b, c). Because p85α can function as a chaperone to facilitate nuclear translocation of associated partner proteins 15, 16, we asked whether p85α assists in the nuclear translocation of OPN-i. In co-transfection experiments in 293T cells, nuclear accumulation of OPN-i increased in direct proportion to the amount of p85α (Fig. 6d), and OPN did not relocate to the nucleus following ligation of ICOS in p85α KO CD4+ T cells (Fig. 6e). In addition, the OPN-i Y166F mutant protein remained mainly in the cytosol despite co-transfection with increased concentrations of p85α (Fig. 6d and Supplementary Fig. 7a). These findings suggest that the nuclear translocation of OPN-i is facilitated by a specific interaction of p85α and OPN-i.
Intranuclear OPN-i interacts with and stabilizes Bcl-6 expression
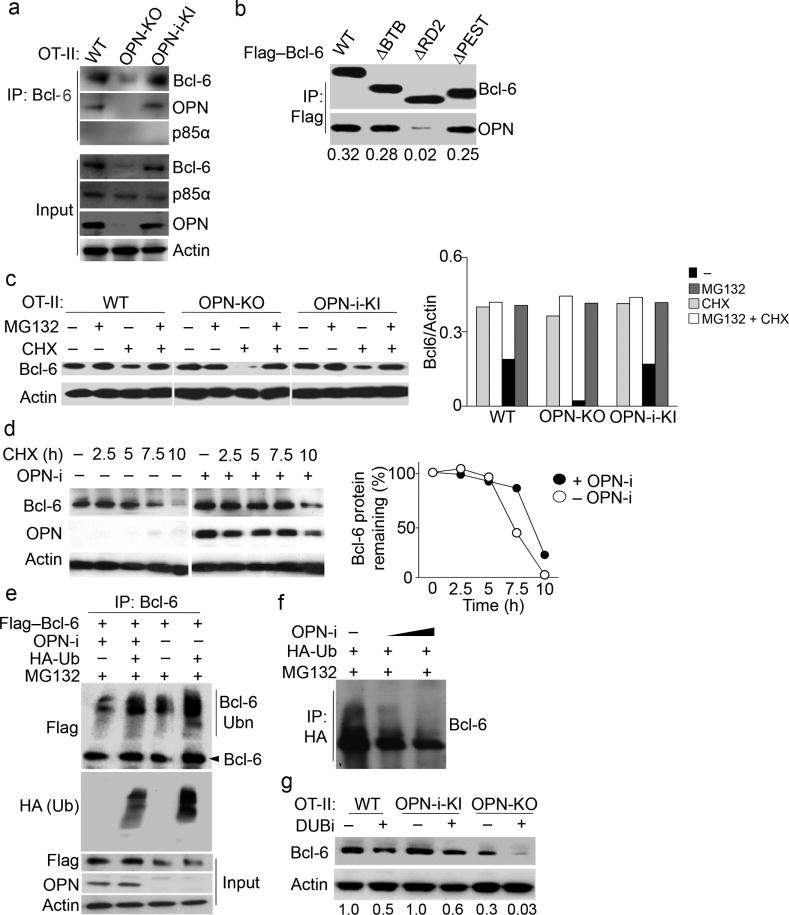
To characterize the potential interaction between OPN-i and Bcl-6, we immunoprecipitated Bcl-6 from Vβ5+ CD4+ T cells purified from OT-II and OT-II OPN-i KI mice post-immunization with OVA in CFA. Staining for endogenous OPN-i protein in the Bcl-6 immunoprecipitates indicated that the two proteins interacted (Fig. 7a). Cellular fractionation revealed that the majority of Bcl-6–OPN-i complexes were found in the nucleus (Supplementary Fig. 7b), consistent with the results of confocal microscopic analysis (Supplementary Fig. 7a). To specifically analyze the sequences of Bcl-6 required for interaction with OPN-i, we constructed Bcl-6 protein deletion mutants, including deletion of sequences in BTB, PEST or Zinc finger domains of Bcl-6 (Supplementary Fig. 7c). Co-immunoprecipitation analysis of overexpressed OPN-i and Flag–Bcl-6 wild-type or mutants in 293T cells indicated that sequences in RD2 of Bcl-6 (amino acids 120–300), but not the BTB, PEST or Zinc finger domains of Bcl-6 were required for interaction with OPN-i (Fig. 7b and Supplementary Fig. 7c, d). These findings indicate that intranuclear OPN-i may interact with Bcl-6.
Figure 7.
Intranuclear OPN-i interacts with and stabilizes Bcl-6 expression.
a, Immunoassay of lysates of purified CD44+CD4+ T cells from pooled OT-II (n = 8), OT-II OPN-KO (n = 10) and OT-II OPN-i-KI (n = 8) mice 7 d post-immunization with OVA in CFA, assessed by immunoprecipitation (IP) with anti-Bcl-6 and immunoblot analysis, as indicated. b, Immunoassay of lysates of 293T cells transfected with plasmids encoding OPN-i and Flag–Bcl-6 wild-type or Flag–Bcl-6 deletion mutants (lanes 1,4,7,10 in Supplementary Fig. 7d), assessed by IP with anti-Flag and immunoblot analysis, as indicated. Bottom, ratios of precipitated OPN to Bcl-6. c, Immunoblot analysis of lysates of purified CD62L−CD4+ T cells from the indicated OT-II mice 2.5 d post-immunization with OVA in CFA, followed by resting for 2 h, treatment with or without MG132 90 m after incubation with anti-CD3 and anti-ICOS, addition of cycloheximide (CHX) 30 m later, and analysis 3 h after treatment with or without CHX. Right, ratios of Bcl-6 to actin protein. d, Bcl-6 and OPN expression in 293T cells transfected with vectors expressing Flag–Bcl-6 and/or OPN-i, treated with CHX (100 μg/ml) for 10 h. Right, percent of residual Bcl-6 protein relative to that prior to addition of CHX. e–f, Immunoassay of lysates of 293T cells transfected with the indicated plasmids and pre-treated with MG132, assessed by denaturation of lysates, IP with anti-Bcl-6 (e) or anti-HA (f) and immunoblot analysis as indicated. Bcl-6(Ubn): polyubiquitinated Bcl-6. Increasing amounts of OPN-i plasmids in f. g, Immunoblot analysis of lysates of purified CD62L−CD4+ T cells from indicated OT-II mice 7 d post-immunization with OVA in CFA, treated with (+) or without (−) DUBi for 8 h, probed with anti-Bcl-6 and anti-actin. Bottom, ratios of Bcl-6 to actin. Data represent two (a–d, g) and three (e, f) independent experiments.
Because OPN KO CD4+ TFH and TFR cells expressed less Bcl-6 protein at day 3 to 10 post-KLH-immunization, we next tested if the absence of OPN-i rendered the Bcl-6 protein unstable. Addition of cycloheximide (CHX), which inhibits protein synthesis, into cultures of OT-II CD4+ T cells purified from OT-II OPN KO mice 2.5 d after OVA-immmunization followed by in vitro anti-CD3 and anti-ICOS stimulation, resulted in more reduction of Bcl-6 protein (90%) compared to OT-II and OT-II OPN-i KI CD4+ T cells (50%) (Fig. 7c). In addition, overexpression of OPN-i prolonged the stability of Bcl-6 protein in CHX-treated 293T cell lines (Fig. 7d). Reduced Bcl-6 amounts in all these CHX-treated CD4+ T cells were rescued by the addition of the proteasome inhibitor MG132 (Fig. 7c), suggesting loss of Bcl-6 was proteasome-dependent. Moreover, MG132 treatment resulted in the appearance of high molecular mass species of Bcl-6 in the denatured extracts of these cells (Fig. 7e, f). These high molecular forms of Bcl-6 were increased in the presence of overexpressed ubiquitin (Fig. 7e, f) suggesting they were ubiquitinated forms of Bcl-6. Addition of a pan deubiquitination inhibitor (DUbi) accelerated Bcl-6 degradation in wild-type, OPN KO and OPN-i KI CD4+ T cells as well as Flag–Bcl-6-transfected 293T cells. This effect was substantially prevented by endogenous expression of OPN-i by wild-type and OPN-i KI CD4+ T cells and overexpressed OPN-i in 293T cells (Fig. 7g and Supplementary Fig. 7e). These results suggest that OPN-i stabilizes Bcl-6 through interference with ubiquitin-mediated degradation of Bcl-6.
The p85α–OPN-i interaction regulates TFH and TFR responses in vivo
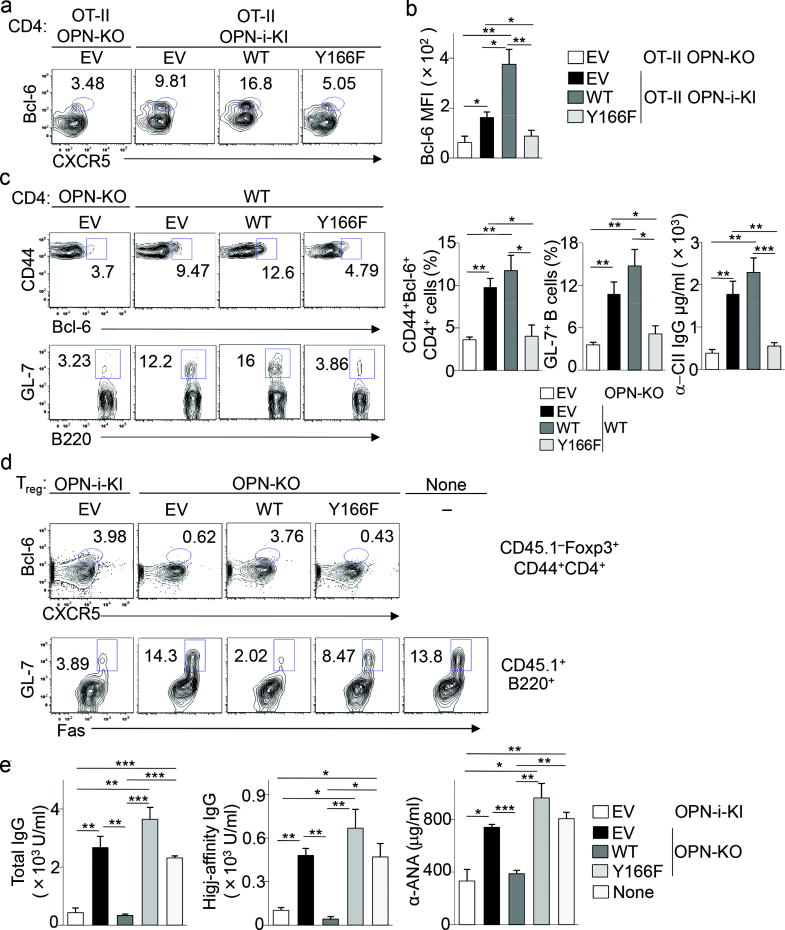
To test the physiological relevance of the p85α–OPN-i interaction in TFH differentiation, we transduced OT-II OPN-i KI CD4+ T cells with retroviral vector expressing GFP alone (empty vector) or GFP and either OPN-i wild-type or OPN-i Y166F mutant and co-transferred each with wild-type B cells into Rag2−/−Prf1−/− hosts followed by immunization with NP13-OVA in CFA. OT-II OPN KO CD4+ T cells transduced with empty vector were included as controls. TFH cell differentiation and intracellular expression of Bcl-6 in these TFH cells assessed by flow cytometry were substantially increased in OT-II OPN-i KI CD4+ T cells transduced with OPN-i wild-type compared to empty vector controls (Fig. 8a, b). Expression of the OPN-i Y166F in OT-II OPN-i KI CD4+ T cells markedly decreased Bcl-6 expression and TFH cell differentiation compared to these cells expressing the GFP alone (Fig. 8a, b) and to levels similar to those observed in OT-II OPN KO CD4+ T cells transduced with empty vector (Fig. 8a, b). Similar results were obtained in a model in which CD4+ T cells from wild-type or OPN KO mice immunized with type II collagen (CII) were transduced with retroviral vectors (as described above), transferred with wild-type B cells into Rag2−/−Prf1−/− hosts followed by immunization with CII in CFA. Expression of OPN-i Y166F in wild-type CII-immune CD4+ T cells decreased the differentiation of the collagen-specific T cells into Bcl-6+ TFH cells and reduced the frequency of GL7+ B cells and autoantibody responses to collagen to levels observed in OPN KO CD4+ T cells expressing empty vector (Fig. 8c). These results suggest that interaction between OPN-i and p85α is essential for Bcl-6-dependent functional differentiation of TFH cells.
Figure 8.
The p85α–OPN-i interaction regulates TFH and TFR responses in vivo.
a, Flow cytometry of donor CD4+ T cells from spleens of Rag2−/−Prf1−/− hosts given OT-II OPN-KO or OT-II OPN-i-KI CD4+ T cells, transduced with retroviral vectors encoding GFP alone (empty vector (EV)) or GFP and OPN-i wild-type or OPN-i Y166F mutant, sorted GFP+ cells and transferred (1 × 105) along with wild-type B cells (1 × 106), rested for 2 d in recipients before immunization with NP13-OVA in CFA and analysis 10 d post-immunization. Numbers indicate percent Foxp3−CXCR5+Bcl-6+ TFH cells. b, Bcl-6 protein expression (MFI) in a (n = 5 per group). c, Flow cytometry of spleens of Rag2−/−Prf1−/− hosts given type II collagen (CII)-immune CD4+ T cells from wild-type or OPN-KO mice, transduced with retroviral vectors (as described above), transferred (1 × 105) with wild-type B cells (2× 106) followed by immunization with CII and CFA at d0 and boosting with CII in IFA at d21. Right, frequency of Bcl-6+CD44+CD4+ T cells and GL7+B220+ cells, serum titers of anti-mouse CII Ab at d28 (n = 4–6 per group). d, Flow cytometry of spleens of Rag2−/−Prf1−/− hosts given CD45.2+ OPN-i-KI or OPN-KO CD25+CD4+ T cells (Treg), transduced with retroviral vectors (as in a), transferred sorted GFP+ T cells (4 × 104) together with 1 × 105 CD45.1+ CD25−CD4+ T cells and 2 × 106 wild-type B cells, followed by immunization with NP26-KLH in CFA at d0 and boosting with NP26-KLH in IFA at day 10. Numbers indicate percent CD45.1−Foxp3+Bcl-6+CXCR5+ TFR cells and CD45.1+Fas+GL-7+ GC B cells at d16. e, Serum titers of anti-NP and anti-ANA Ab in d (n = 3 per group). None: recipients given no Treg. Data represent three (c) and two (a–b, d–e) independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired two-tailed Student’s t-test; error bars, mean ± s.e.m).
Using a similar retroviral reconstitution system, we also evaluated the physiologic importance of the p85α–OPN-i interaction in the formation of TFR. CD45.2+ OPN KO CD25+CD4+ T cells were transduced with retroviral vectors expressing GFP alone (empty vector) or GFP and either OPN-i wild-type or OPN-i Y166F mutant and co-transferred with CD45.1+ CD25−CD4+ T cells and wild-type B cells into Rag2−/−Prf1−/− hosts, followed by immunization with NP26-KLH in CFA. CD45.2+ OPN-i KI CD25+CD4+ T cells transduced with empty vector were included as controls. Expression of wild-type OPN-i in OPN-KO CD25+CD4+ T cells markedly increased TFR differentiation to levels similar to that of OPN-i-KI CD25+CD4+ T cells expressing GFP alone (Fig. 8d). In contrast, expression of OPN-i Y166F in OPN-KO CD25+CD4+ T cells resulted in a substantially reduced frequency of Foxp3+Bcl-6+CXCR5+ TFR cells and an increase in GC B cells and the titers of anti-NP and antinuclear antibody (Fig. 8d,e). Decreased TFR cell formation was associated with an increase in GC B cell frequencies and markedly increased anti-NP and anti-ANA antibody titers in hosts transferred with OPN KO CD25+CD4+ T cells expressing OPN-i Y166F (Fig. 8d, e). Taken together, these results indicate that a specific interaction between OPN-i and p85α is essential for sustained expression of Bcl-6 and functional differentiation of both TFH and TFR cells (Supplementary Fig. 8).
Discussion
Signals from the ICOS receptor are essential for Bcl-6 expression and for the initiation and maintenance of TFH and TFR cell differentiation 5, 25, 26. Here we defined p85α–OPN-i as a molecular link that coupled ICOS engagement to sustained Bcl-6 expression and was essential for differentiation of both TFH and TFR cells. We found that ICOS ligation induced the interaction of the p85α component of PI3K with OPN-i, which promoted OPN-i translocation to the nucleus and protection of Bcl-6 from ubiquitination-dependent degradation. Although early steps of TFH cell differentiation resulting in enhanced Bcl-6 gene expression and follicular T cell migration depend on cytokines derived from activated DC 30, 31 and may be independent of OPN-i expression, sustained Bcl-6 expression and full TFH and TFR cell differentiation require ICOS-dependent translocation of OPN-i and interaction with Bcl-6. These findings indicate that TCR–ICOS signals known to be essential for sustained differentiation and expansion of TFH and TFR cells preserve Bcl-6 expression in these cells, in contrast to bystander CD4 cells that may undergo early cytokine-dependent activation by DC but fail to activate an ICOS–p85α–OPN-i pathway required for stable Bcl-6 expression.
ICOS activation induces two distinct but overlapping PI3K signaling pathways. The catalytic ICOS–PI3K(p110)–Akt pathway can promote the migration of TFH precursors into B cell follicles 11–13, 32, while we show here that the ICOS–p85α–OPN-i pathway contributed to stable Bcl-6 expression and was essential for sustained follicular T cell responses. The division of labor between the two ICOS-linked PI3K pathways may depend, in part, on distinct environmental cues. MHC-II-independent G-protein coupled signals may facilitate activation of the PI3K(p110)–Akt pathway 12, while engagement of the TCR and ICOS may promote the PI3K(p85α)–OPN-i pathway and sustained post-transcriptional expression of Bcl-6. TCR–ICOS signals may also indirectly regulate Bcl-6 expression by facilitating Itch-mediated degradation of Foxo1 33. Additional studies are needed to define how these pathways are integrated into a functional TFH differentiation program. p85α also functions as a chaperone for the nuclear translocation of the transcription factor XBP-1 after ligation of the insulin receptor 15, 16. The relatively low intranuclear expression of p85α and its absence from OPN-i–Bcl-6 complexes suggest that p85α may be released before OPN-i engages the intranuclear Bcl-6. The ability of the multi-functional OPN-i adaptor protein to interact with other intranuclear proteins to regulate gene expression by follicular T cells deserves further study 34.
Our findings also indicate that the appropriate balance between TFH and TFR cells is critical for optimal GC responses to infection and avoidance of excessive or autoimmune responses that may result in host tissue destruction. Although TFH and TFR cells share many surface receptors and both require Bcl-6 expression, the molecular elements responsible for their differentiation within the GC follicles have remained unclear. Here we show that the ICOS-dependent induction of OPN-i was required for sustained Bcl-6 expression in both follicular T cell subsets. The magnitude of the GC B cell selection and the associated antibody response depends on cognate TFH cell-antigen-specific B cell interactions. Although OPN deficiency resulted in reduced TFR activity, defective TFH cell responses in the OPN KO mice were not rescued by decreased inhibitory activity of TFR cells. In contrast, selective impairment of OPN-i expression in TFR cells led to substantially increased antibody responses, including the development of high affinity antibodies and autoantibodies, indicating that the OPN-dependent protection of Bcl-6 expression in both follicular CD4+ T cell subsets is essential for control of the germinal center response.
The OPN-i and OPN-s isoforms arise from differential translation of the same mRNA 17. Although increased expression of Spp1 gene has been associated with TFH-associated autoimmune disorders and malignancies, the finding that intranuclear OPN contributed to lineage-specific T cell differentiation is unexpected. Mice that overexpress OPN develop a systemic autoimmune disorder 18 that resembles sanroque mice with mutations of Roquin (Rc3h1), resulting in part from dysregulated ICOS expression 35–37. Intracellular OPN may promote excessive expression of IFN-α by plasmacytoid dendritic cells (pDC), and contribute to TH17 cell expansion in the context of SLE 21, 23. In support of the role of OPN in SLE pathogenesis, expression of OPN in humans with SLE and autoimmune-prone mice (MRL-lpr/lpr) correlates with disease activity 38, 39. Although high circulating levels of OPN-s may be a useful biomarker for SLE disease activity, separate analysis of OPN-i expression by CD4+ TFH cells and serum OPN may provide a more accurate assessment of SLE status. The finding that disruption of the ICOS–p85α–OPN-i pathway by overexpressing OPN-i mutant inhibited TFH responses and associated autoantibody production also suggests that targeting the p85α–OPN-i interaction may allow inhibition of TFH cell responses and amelioration of systemic autoimmune disease.
Our results also suggest a requirement for continued protection of the Bcl-6 protein from ubiquitination and proteosomal degradation. Bcl-6 represses a group of genes that control lymphocyte differentiation and cell division 4, 40. Studies of Bcl-6 expression by normal and neoplastic GC B cells have also suggested that Bcl-6 expression is highly sensitive to post-translational breakdown 4. Our findings indicate that overexpression of OPN-i leading to enhanced levels of OPN-i–Bcl-6 complexes may result in increased Bcl-6 expression and enhanced TFH cell responses. Indeed, overexpression and nuclear localization of OPN are associated with aggressive TFH-like lymphomas and poor prognosis 41, 42.
We found that OPN-i interacted with the RD2 domain of Bcl-6 to inhibit its ubiquitination-mediated degradation in follicular T cells. A similar interaction between Hsp90 and the Bcl-6 RD2 domain protects Bcl-6 from proteasomal degradation in neoplastic GC B cells 4, 43. Inhibition of Hsp90 expression or mutation of the BTB domain of Bcl-6 impairs normal and neoplastic GC B cell survival but spares TFH cell differentiation4, 43–45. We found that inhibition of OPN-i expression did not affect B cell activity. Excessive Bcl-6 expression may be inhibited by drugs that inhibit post-translational Bcl-6 metabolism 46, including those that target the p85α–OPN-i interaction defined here. Drugs that differentially target the OPN-i–Bcl-6 and Hsp90–Bcl-6 interaction may also allow separate control of follicular T cell and GC B cell responses.
Our findings also bear on efforts to define TFH plasticity and diversity through accurate analysis of Bcl-6 protein by CD4+ T cells. Although expression of Bcl-6 protein returns to basal levels by 2 weeks after immunization, Bcl-6 mRNA expression remains elevated in “TFH” cells. Precise definition of the TFH response, and its relationship to other TH subsets, may require coordinate measurements of both Bcl-6 protein and mRNA. In sum, the observations that intranuclear OPN-i protects Bcl-6 from proteasome-associated degradation and allows sustained Bcl-6 expression in TFH cells and TFR cells provides new insight into the ICOS-dependent differentiation of TFH and TFR cells and suggests new therapeutic avenues to manipulate the GC response.
Methods
Mice
C57BL/6J (B6), Pi3kr1fl, Tcrα−/−, Icos−/−, OT-II transgenic (Jackson Labs), Rag2−/−Prf1−/−, B6SJL (CD45.1) (Taconic Farms), B6.Foxp3GFP mice (kindly provided by Dr. H. von Boehmer), Spp1flstop Cre+ and Cre− littermates (Supplementary Fig. 2) were housed in pathogen-free conditions and used at 7–12 weeks of age. Deletion of loxP-flanked Pi3kr1 gene in hematopoietic cells was achieved by crossing Pi3kr1fl mice with Vav1-Cre (Jackson Labs) that express Cre under the Vav1 promoter. Experiments were performed in an unblinded fashion, with both sexes included for all experiments. For experiments with multiple time points, mice were assigned randomly to each group. All experiments were performed in compliance with federal laws and institutional guidelines as approved by DFCI’s Animal Care and Use Committee.
Antibodies and flow cytometry
Fluorescence dye labeled Abs specific for CD4 (L3T4), B220 (RA3-6B2), CD44 (IM7), Fas (15A7), IgM (II/41), T- and B-cell activation antigen (GL7), ICOS (C398.4A), PD-1 (J43), IFNγ (XMG1.2), IL-4 (11B11), IL-17A (eBio17B7), IL-21 (FFA21), Bcl-6 (K112-91) and FoxP3 (FJK-16s) were purchased from BD, eBioscience and Biolegend. Analysis of CXCR5 expression was performed using a biotinylated anti-CXCR5 (2G8, BD) antibody followed by incubation with APC- or APC.Cy7-labelled streptavidin, as described 25. Intracellular staining for Bcl-6, FoxP3 and cytokines was performed using the FoxP3 staining buffer set (eBioscience). Intracellular staining of phospho-S473-AKT (M89-61), pSTAT1 (14/P-STAT1), pSTAT3 (4/P-STAT3) was conducted according to manufacturer’s instructions (BD Bioscience). Cells were acquired on a FACSCantoII using FACSDIva software (BD Biosciences) and analyzed with FlowJo software (Tristar).
Adoptive transfer and immunization
FACS-sorted CD4+ T cell subsets and B cells purified using B lymphocyte enrichment set (BD Bioscience) were transferred into indicated hosts before immunization with protein antigens emulsified in CFA. In some experiments, reimmunization with protein antigens in IFA at the indicated times was performed. All immunizations were conducted by intraperitoneal injection. Serum was prepared at the indicated time for measurement of primary and secondary antibody titers.
Enzyme-linked immunosorbent assay (ELISA)
Detection of NP-specific antibodies was performed as described 47. Analysis of anti-mouse collagen antibody was performed as described 48. Determination of pAkt and Akt levels was conducted using InstantOne™ ELISA kit (eBioscience).
In vitro differentiation of TH subsets
OT-II CD25−CD4+ T cells (2 × 105 cells/ml) purified from the indicated mouse strains using CD4 T lymphocyte enrichment set (BD Bioscience) were stimulated with 1 μg/ml OT-II peptide (ISQAVHAAHAEINEAGR) in the presence of irradiated total splenocytes (2 × 106 cells/ml). The cytokine and antibody mixtures used for differentiation of CD4 cells to each TH cell subset were described previously 49. Cytokine production after incubation ×5 d was analyzed by ELISA, according to manufacturer’s instructions (RD systems).
Plasmids and generation of retrovirus
OPN-i expression vectors, pMLS5, OPN-i–Flag, and OPN-i–GFP were described previously 17. A tandem HA-Flag tag was introduced at the C-terminus of OPN-i cDNA by PCR using primers containing BamHI and EcoRI sites followed by cloning into pBABE-GFP vector. Bcl-6 cDNA was obtained from Open Biosystems (Bcl-6 Clone ID: 6309948), sequenced in full, before complete coding region sequences were cloned inframe with a Flag tag at the N-terminus into retroviral expression vector MSCV-IRES-GFP. The following plasmids were obtained from Addgene: p85α (plasmid 1399 and 1407), HA-p110 (plasmid 12522 and 15691) 50, 51 and HA-Ub (plasmid 17608) 52. Deletion constructs of Bcl-6, Flag–Bcl-6, and OPN-i Y166F mutants were generated by PCR-mediated mutagenesis with the QuickChange II XL Site-Directed Mutagenesis Kit (Agilent). The accuracy of all plasmids was confirmed by DNA sequencing. Retroviral stocks were generated by transfection of 293T cells with plasmids expressing pBABE-GFP control or pBABE-GFP plus OPN-i wild-type or OPN-i Y166F along with pCL-Eco packaging vector using TransIT-LT1 transfection reagent (Mirus). Viral supernatants were collected 72 h later before infection of CD4+ T cells, as described below.
Retroviral transduction
Purified naive CD4+ T cells from the indicated mouse strains were stimulated with plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (5 μg/ml) in the presence of 10 ng/ml human IL-2 (hIL-2). 24–36 h post-stimulation, CD4+ T cells were infected with retrovirus expressing GFP and the indicated protein in the presence of 8 μg/ml of polybrene before 1 h centrifugation at 2000 rpm followed by 6–8 h at 37°C and subsequent replacement of three quarters of hIL2-containing fresh medium. After 3 d of stimulation, CD4+ T cells were rested in the presence of hIL-2 for 1–2 d before sorting for GFP+CD4+ T cells followed by adoptive transfer into Rag2−/−Prf1−/− hosts and resting in vivo for 2 d prior to immunization as described above. In Fig. 8c, Rag2−/−Prf1−/− hosts were transferred with sorted GFP+ CII-immune CD4+ T cells and purified B cells followed by immunization with chicken CII in CFA and boosting with CII in IFA, as described previously 48. Retroviral infection of CD25+CD4+ T cells was adapted from Haxhinasto et al 27. Briefly, purified CD25+CD4+ T cells were stimulated with plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (5 μg/ml) in the presence of 1,000 U/ml hIL-2 and 20 ng/ml TGFβ. Three days post-stimulation, cells were infected with retrovirus expressing GFP and the indicated protein, as described above, for a total of 2 d before sorting GFP+ cells and transfer.
Immunoprecipitation and Immunoblot
The procedure was performed as described previously 21. The following antibodies were used: p85α (Z-8) and Bcl-6 (N-3 or C-19) (Santa Cruz); Flag (M2) and actin (AC-15) (Sigma); Lamin B1 (L-5) (Invitrogen); OPN (O-17) (IBL American); tubulin (AA2) (Millipore) and HA (C29F4) (Cellsignal). Band intensity was quantified using ImageJ software, version 1.45b (NIH).
Immunofluorescence staining
CD62L−CD4+ T cells (> 95%) from the indicated mouse strains injected with KLH in CFA were purified with MACS CD4+CD62L+ T-cell isolation kit (Miltenyi) and stimulated with anti-ICOS for the indicated times before fixation, permeabilization and immunostaining. Antibodies or dyes used include: rabbit Bcl-6 (N-3), anti-rabbit Alexa Fluor 568 (for Bcl-6); mouse OPN (AKm2A1, Santa Cruz), anti-mouse Alexa Fluor 647 (for OPN) and nuclear dye DAPI. Images were captured through a 63× objective lens with a Leica SP5X laser scanning confocal microscope and analyzed using ImageJ software, version 1.45b (NIH).
Gene expression profiling
Naïve CD4+ T cells (> 95%) were purified from single cell suspensions of B6 spleen using the MACS CD4+CD62L+ T-cell isolation kit (Miltenyi) and stimulated with anti-CD3 (5 μg/ml) and anti-CD28 (2 μg/ml) for 2 d followed by resting overnight before 20 min of incubation with anti-CD3 (0.2 μg/ml) and/or anti-ICOS (5 μg/ml) and cross-linking with goat anti-hamster Ab (20 μg/ml) for 8 h. RNA was prepared with the RNeasy plus micro kit according to the manufacturer’s instructions (Qiagen). RNA amplification, labeling and hybridization to Mouse Gene 1.0 ST Array (Affymetrix) were performed at the Microarray Core Facility (Dana Farber Cancer Institute), with quadruplicates for anti-CD3 ligation alone and duplicates for anti-CD3 and anti-ICOS co-stimulation. Data were deposited in the GEO database under accession number GSE61284.
Quantitative RT-PCR
RNA was extracted using RNeasy plus micro kit (Qiagen) before quantitative real time PCR was performed with TaqMan gene expression assays [Spp1 (Mm00436767_m1), Bcl-6 (Mm00477633_m1), Prdm1 (Mm00476128_m1), Rps18 (Mm02601777_g1)] and RNA-to-CT™ 1-Step Kit (Life Technologies). All results were first normalized to those of the Rps18 control and are presented as normalized expression for the sample relative to the appropriate comparison conditions, as indicated in the legends.
Statistical analyses
Statistical analyses were performed using two-tailed, unpaired Student’s t-test or Mann-Whitney test with the assumption of equal sample variance, with GraphPad Prism V6 software. Error bars indicate mean ± s.e.m. A P value <0.05 was considered to be statistically significant (* = <0.05, ** = <0.01, *** = <0.001). No exclusion of data points was used. Sample size was not specifically predetermined, but the number of mice used was consistent with previous experience with similar experiments.
Supplementary Material
Acknowledgments
We thank Kai Wucherpfennig for critical reading and Lewis Cantley, Ted Dawson, Shinya Yamanaka and Jean Zhao for provision of plasmids. We thank Dr. Lisa Cameron (DFCI Microscopy Core) for help with image preparation and analysis, Yue Shao (DFCI Microarray Facility) for microarray analysis, and A. Angel for manuscript and figure preparation. This work was supported in part by NIH Research Grant AI48125 and a gift from The LeRoy Schecter Research Foundation to HC; the Benacerraf Fellowship and NRSA Fellowship (T32 CA070083) to JWL; and a Belgian-American Educational Foundation Fellowship to BV.
Footnotes
Accession Codes. GEO: Microarray data have been deposited under accession number GSE61284.
Author Contributions
JWL and HC conceived and planned experiments; BV made the OPN-i KI genomic construct; JWL, JY and HH performed experiments; JWL and HC analyzed data and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Literature cited
- 1.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Bunting KL, Melnick AM. New effector functions and regulatory mechanisms of BCL6 in normal and malignant lymphocytes. Curr Opin Immunol. 2013;25:339–346. doi: 10.1016/j.coi.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14:152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linterman MA, et al. Foxp3(+) follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotty S. Follicular helper CD4 T cells (TFH) Annual Reviews of Immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 9.Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 10.Gigoux M, et al. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2009;106:20371–20376. doi: 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 12.Kang SG, et al. MicroRNAs of the miR-17 approximately 92 family are critical regulators of TFH differentiation. Nat Immunol. 2013;14:849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rolf J, Fairfax K, Turner M. Signaling pathways in T follicular helper cells. Journal of Immunology. 2010;184:6563–6568. doi: 10.4049/jimmunol.1000202. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, et al. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SW, et al. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winnay JN, Boucher J, Mori MA, Ueki K, Kahn CR. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinohara ML, Kim HJ, Kim JH, Garcia VA, Cantor H. Alternative translation of Osteopontin generates intracellular and secreted isoforms that mediate distinct biological activities in dendritic cells. Proc Natl Acad Sci USA. 2008;105:7235–7239. doi: 10.1073/pnas.0802301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantor H, Shinohara ML. Regulation of T-helper-cell lineage development by osteopontin: the inside story. Nat Rev Immunol. 2009;9:137–141. doi: 10.1038/nri2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashkar S, et al. Eta-1 (osteopontin): an early component of Type 1(cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 20.Shinohara ML, et al. T-bet-dependent expression of osteopontin contributes to T cell polarization. Proc Natl Acad Sci U S A. 2005;102:17101–17106. doi: 10.1073/pnas.0508666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinohara ML, et al. Osteopontin expression is essential for IFN-à production by plasmacytoid dendritic cells. Nat Immunol. 2006;7:498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patarca R, Wei FY, Iregui MV, Cantor H. Differential induction of interferon-gamma gene expression after activation of CD4+ T-cells by conventional antigen and Mls superantigen. Proceedings of the National Academy of Science USA. 1991;88:2736–2739. doi: 10.1073/pnas.88.7.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinohara ML, Kim JH, Garcia VA, Cantor H. Engagement of the Type-I interferon receptor on dendritic cells inhibits promotion of Th17 cells: Role of intracellular Osteopontin. Immunity. 2008;29:68–78. doi: 10.1016/j.immuni.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerfoot SM, et al. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi YS, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JH, et al. TRAF3 regulates the effector function of regulatory T cells and humoral immune responses. J Exp Med. 2014;211:137–151. doi: 10.1084/jem.20131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaffe MB, et al. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 30.Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol. 2013;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayamada S, et al. Type I IFN Induces Binding of STAT1 to Bcl6: Divergent Roles of STAT Family Transcription Factors in the T Follicular Helper Cell Genetic Program. J Immunol. 2014 doi: 10.4049/jimmunol.1300675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolf J, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 33.Xiao N, et al. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol. 2014;15:657–666. doi: 10.1038/ni.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue M, Shinohara ML. Intracellular osteopontin (iOPN) and immunity. Immunologic research. 2011;49:160–172. doi: 10.1007/s12026-010-8179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 36.Yu D, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 37.Glasmacher E, et al. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- 38.Patarca R, Wei FY, Singh P, Morasso MI, Cantor H. Dysregulated expression of the T-cell cytokine Eta-1 in CD4-8- lymphocytes during the development of murine autoimmune disease. Journal of Experimental Medicine. 1990;172:1177–1183. doi: 10.1084/jem.172.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong CK, Lit LC, Tam LS, Li EK, Lam CW. Elevation of plasma osteopontin concentration is correlated with disease activity in patients with systemic lupus erythematosus. Rheumatology (Oxford) 2005;44:602–606. doi: 10.1093/rheumatology/keh558. [DOI] [PubMed] [Google Scholar]
- 40.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell JA, et al. Expression profiling of a hemopoietic cell survival transcriptome implicates osteopontin as a functional prognostic factor in AML. Blood. 2009;114:4859–4870. doi: 10.1182/blood-2009-02-204818. [DOI] [PubMed] [Google Scholar]
- 42.Tun HW, et al. Pathway analysis of primary central nervous system lymphoma. Blood. 2008;111:3200–3210. doi: 10.1182/blood-2007-10-119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerchietti LC, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med. 2009;15:1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat Immunol. 2013;14:380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polo JM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 46.Kronke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HJ, Verbinnen B, Tang X, Lu L, Cantor H. Inhibition of follicular T helper cells by CD8 + Treg is essential for self tolerance. Nature. 2010;467:328–332. doi: 10.1038/nature09370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leavenworth JW, Tang X, Kim HJ, Wang X, Cantor H. Amelioration of arthritis through mobilization of peptide-specific CD8+ regulatory T cells. Journal of Clinical Investigation. 2013;123:1382–1389. doi: 10.1172/JCI66938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leavenworth JW, Wang X, Wenander CS, Spee P, Cantor H. Mobilization of natural killer cells inhibits development of collagen-induced arthritis. Proc Natl Acad Sci USA. 2011;108:14584–14589. doi: 10.1073/pnas.1112188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi K, Mitsui K, Yamanaka S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature. 2003;423:541–545. doi: 10.1038/nature01646. [DOI] [PubMed] [Google Scholar]
- 51.Zhao JJ, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim KL, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.