Abstract
The precise splicing of genes confers an enormous transcriptional complexity to the human genome. The majority of gene splicing occurs cotranscriptionally, permitting epigenetic modifications to affect splicing outcomes. Here we show that select exonic regions are demarcated within the three-dimensional structure of the human genome. We identify a subset of exons that exhibit DNase I hypersensitivity and are accompanied by ‘phantom’ signals in chromatin immunoprecipitation and sequencing (ChIP-seq) that result from cross-linking with proximal promoter- or enhancer-bound factors. The capture of structural features by ChIP-seq is confirmed by chromatin interaction analysis that resolves local intragenic loops that fold exons close to cognate promoters while excluding intervening intronic sequences. These interactions of exons with promoters and enhancers are enriched for alternative splicing events, an effect reflected in cell type-specific periexonic DNase I hypersensitivity patterns. Collectively, our results connect local genome topography, chromatin structure and cis-regulatory landscapes with the generation of human transcriptional complexity by cotranscriptional splicing.
The human genome is prevalently expressed as complex, interleaved networks of transcription, with the majority of genes alternatively spliced to generate a range of distinct isoforms1,2. Splicing has been increasingly adopted for the expansion of transcriptional complexity during the evolution of eukaryotes. Introns have progressively lengthened, and splice sites have weakened, thereby broadening the range of available splicing choices and generating greater transcriptional diversity3,4. This transcriptional diversification poses an increasing challenge to the spliceosome to recognize correct consensus splice sites across often vast intronic distances that can overlap conflicting signals from other genes. Furthermore, these choices must often be made correctly in response to context-dependent cues for alternative splicing.
The conventional model of post-transcriptional splicing invokes a range of RNA-bin ding proteins that recognize and bind sequence-specific motifs within nascent RNA to regulate the inclusion or exclusion of an exon in a final mature transcript5. We now appreciate that splicing is also often a cotranscriptional process, occurring simultaneously with RNA polymerase transcription of the nascent RNA. Cotranscriptional splicing permits a range of additional epigenetic mechanisms to affect splicing regulation. Nucleosome positioning, histone modification, DNA methylation and CTCF occupancy can modulate the rate of RNA polymerase II (Pol II) elongation and splicing factor recruitment and thereby regulate splicing6–10. The combination and coordination of these multiple layers of post-and cotranscriptional splicing regulation presumably underlie the transcriptional complexity observed within eukaryotic organisms.
The impact of the genome's three-dimensional topography on the coordination of transcriptional processes has been increasingly appreciated in recent years11,12. The folding of the genome and movement of gene loci between subnuclear domains can mediate repression or activation of transcription initiation and termination13,14. Given the intimate association between transcription and splicing, we similarly considered the topography of the genome in relation to exon annotation and splicing. We show that the folding of chromatin loops can bring exons into close spatial proximity with cognate promoters or distal regulatory elements. Furthermore, this spatial localization of exons is enriched at alternatively spliced exons, suggesting a correlation between genome topography and cotranscriptional splicing.
Results
Exon subset shows DNase I hypersensitivity and ChIP-seq enrichments
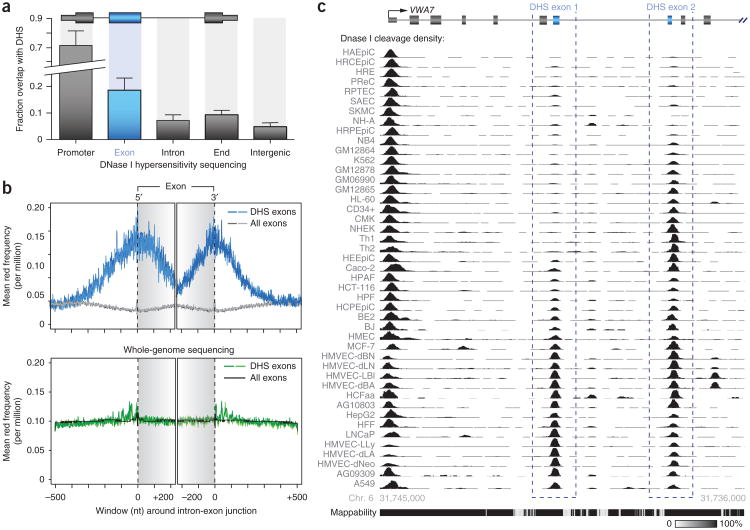
DNase I hypersensitive sites (DHSs) mark diverse classes of cis-regulatory regions, including promoters, enhancers, insulators and other sites of regulatory factor occupancy15. To capture the range of regulatory features associated with exons, we previously generated high-quality maps of DHSs from 86 diverse cell types in replicate16, identifying a total of 2.4 million DHSs, each of which is present in one or more cell type. We noted that a large number of DHSs closely approximated or overlapped annotated exons. In dientifying these DHSs, we first omitted those that overlapped confounding features, such as 5′ or 3′ UTRs, annotated promoters or sites containing evidence of transcription initiation from either strand (Supplementary Fig. 1a) that might otherwise explain DNase I sensitivity. This analysis subsequently resolved a subset of 10,734 exons (12.9% of total exons) that overlapped DHSs in one or more cell line (DHS exons; Fig. 1a).
Figure 1.
A subset of exons exhibits DNase I hypersensitivity. (a) Fractional overlap of various genomic regions with DHS peaks showing significant overlap with exons (P = 8.3 × 10−14, n = 10 cell types; error bars, s.d.). (b) Top, frequency distribution of DNase I–cleaved reads at DHS exons (two replicates shown in different shades) relative to all exons. Plots were created with data from K562 cells and aligned with reference to the 3′ or 5′ end of the exon. Bottom, matched whole-genome sequencing was performed to discern any bias in sequencing and alignment due to the repetitive and informational content of exons and introns. We observe little bias at either DHS exons or total exons. Gray background shading indicates exon boundaries. (c) DHS peaks (black; auto-scaled signal) overlapping exons (gray; DHS exons indicated by blue dashed boxes) of the VWA7 gene showing the cell type specificity of DNase I sensitivity across 45 cell types. The relative mappability of loci is indicated below52.
To determine whether the overlap between DHSs and exons was significant and to normalize for biases in the sequencing and alignment of reads that result from the varying copy number, repeat and informational content of introns and exons, we performed whole-genome shotgun sequencing of three matched cell lines (K562, HSMM and HUVEC; Fig. 1b and Supplementary Figs. 1b, 2 and 3). After normalization, we found that DHSs were significantly enriched at exons (3.29-fold; P = 8.3 × 10−14; Fig. 1b,c and Supplementary Fig. 1c), an unexpected finding given that exons are, on average, preferentially localized within nucleosomes17–19 and are thereby resistant to DNase I cleavage. Similarly, DHSs in exons were also sensitive to micrococcal nuclease, consistent with the absence of a residing nucleosome (Supplementary Fig. 1d–f).
To provide insight into regulatory features associated with DHS exons, we compared these annotations with results from Encyclopedia of DNA Elements (ENCODE) transcription factor and histone modification ChIP-seq experiments performed in seven cell types20 (Supplementary Table 1). This comparison showed an unexpectedly wide range of factors enriched at DHS exons relative to total exons or to control ‘matched’ exons (non–DNase I–sensitive exons incorporated in the same transcript as DHS exons; Supplementary Fig. 4a–c). Hierarchial clustering of DHS exons by ChIP-seq signal enrichment resolved three major subsets of exons with DHSs that could be broadly characterized as (i) promoter-like (enriched for general and gene-specific transcription factors), (ii) enhancer-like (enriched for enhancer-associated factors, such as P300) and (iii) cohesin-like (enriched for CTCF and/or cohesin components) (Supplementary Figs. 4d and 5a). The overlap of these three assignations with DHS exons was supported by comparison with combinatorial chromatin state segmentation maps that are annotated according to combinatorial histone modifications, which similarly parsed exons into three analogous promoter, enhancer and insulator categories21 (Supplementary Fig. 5b).
Genome loops fold exons to promoters and distal enhancers
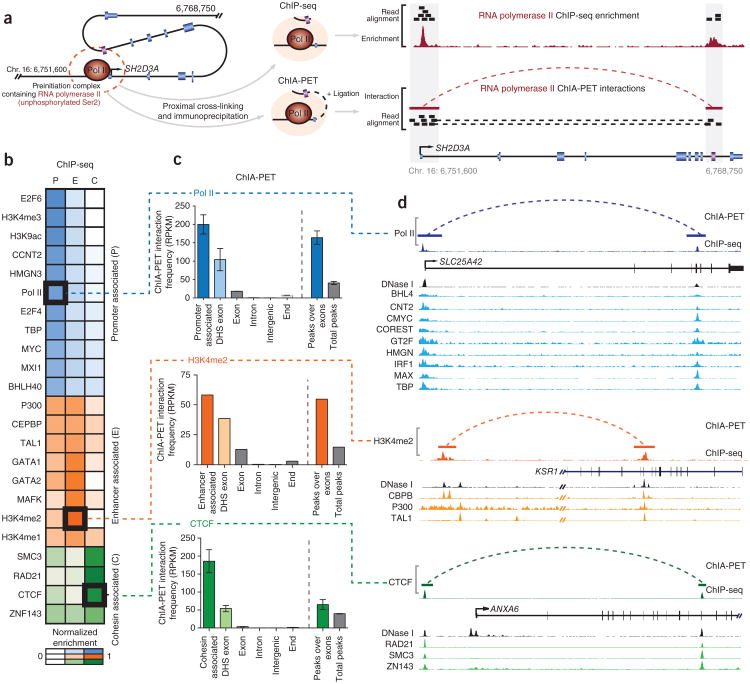
We first analyzed DHS exons with overlying ChIP-seq signals for features that are conventionally associated with promoters, such as Pol II and TBP binding and trimethylation of histone H3 at lyine 4 (H3K4me3). Although these ChIP-seq signals initially suggested that DHS exons might represent alternative or cryptic promoters, neither DHS exons nor their flanking regions showed any evidence of transcription initiation in either the sense or antisense direction, as measured by cap analysis of gene expression (CAGE) tags (Supplementary Fig. 1a). We therefore hypothesized that, rather than representing true promoters, the promoter-like ChIP-seq signals seen at DHS exons might be derived from the spatial localization of exons to promoters. Accordingly, the fixing of protein complexes during the initial steps of chromatin immunoprecipitation may also crosslink promoter-bound proteins to nearby exonic sequences, and the subsequent immunoprecipitation of targeted proteins would therefore return these collateral exonic sequences (Fig. 2a).
Figure 2.
Combined ChIP-seq and ChIA-PET analysis shows that DHS exons interact with promoters and distal regulatory elements. (a) Schematic showing how periexonic ChIP-seq enrichments indicate close spatial localization of exons and promoters. Initial formaldehyde treatment cross-links the preinitiation complex to occupied promoter and proximal exon sequences (purple) within the SH2D3A gene (left, dashed red circle). During ChIP-seq (top), immunoprecipitation of the initiating form of Pol II (hypophosphorylated at Ser2) yields sequenced reads that align to promoter and exonic sequences (right), resulting in periexonic ChIP-seq enrichment. ChIA-PET (bottom) employs an additional ligation step to join coprecipitating promoter and exon sequences in proximity. Alignment of reads derived from these ligated promoter-exon sequences spans interacting regions and confirms that the Pol II ChIP-seq signal observed at the SH2D3A exon results from close spatial proximity of the exon to the gene promoter upstream (right) within the genome's three-dimensional structure. (b) Heatmap indicating the relative enrichment of numerous transcription factors that distinguish DHS exons according to promoter-like (P), enhancer-like (E) and CTCF and/or cohesin (C) ChIP-seq signals. Marked boxes indicate proteins employed in downstream ChIA-PET validation. RPKM, reads per kilobase per million. (c) Histogram showing the frequency of various genomic regions undergoing ChIA-PET interactions with the promoter (top, blue), enhancer (middle, orange) or cohesin (bottom, green) sites as determined by coprecipitation with hypophosphorylated Pol II, H3K4me2 (ref. 27) and CTCF, respectively. DHS exons show enrichment of interactions with promoters (P = 0.0004, Mann-Whitney two-tailed test, n = 4; error bars, range), enhancer (n = 1) or CTCF and/or cohesin (P = 0.0005, Mann-Whitney two-tailed test, n = 3; error bars, range) sites. Similarly, DNase I peaks overlapping exons show enrichment of interactions with promoters (P = 0.0002, Mann-Whitney two-tailed test, n = 4; error bars, range), enhancers (n = 1) and CTCF and/or cohesin (P = 0.028, Mann-Whitney two-tailed test, n = 3; error bars, range) sites relative to total peaks. These findings validates the interactions between DHS exons and promoter or distal enhancer elements as anticipated by ChIP-seq enrichments. (d) Genome browser view showing selected examples of promoter (top), enhancer (middle) and CTCF and/or cohesin (bottom) interactions with exons as determined by matched ChIP-seq and ChIA-PET. DNase I sensitivity (black histogram) and additional supportive ChIP-seq enrichments (colored histograms) at genomic elements and the interacting exon are also indicated.
The conclusion that periexonic ChIP-seq signals result from arti-factual proximal ligation was supported by the lack of corresponding transcription factor binding motifs encompassed by ChIP-seq peaks (Supplementary Fig. 6a–c). Furthermore, we observed correlation between the spectrum of transcription factors enriched at exons and those at interacting promoters (mean Spearman's correlation of 0.642 between exon and promoter pairs, n = 289), albeit with a lower ChIP-seq signal intensity at exons relative to interacting promoters, which is consistent with lower efficiency in cross-linking due to an indirect association (Supplementary Fig. 6d,e). Lastly, we performed native ChIP-seq in K562 cells, targeting the active H3K4me3 modification that was enriched at DHS exons. Native ChIP-seq includes micro-coccal nuclease digestion that liberates individual nucleosomes before immunoprecipitation and should abolish chromatin interactions and reduce signal from proximal cross-ligation. We found that the enrichment of H3K4me3 at DHS exons was diminished (61.4% decrease; Supplementary Fig. 6f) in native ChIP-seq libraries relative to matching non-native ChIP-seq libraries. Collectively, these analyses support our assertion that periexonic ChIP-seq enrichments result, not from transcription factor occupancy, but rather from proximal cross-ligation to promoter-bound complexes.
This hypothesis can also be directly addressed by an orthogonal measure of promoter to distal site interaction, chromatin interaction analysis by paired-end tag sequencing (ChIA-PET)22. ChIA-PET combines ChIP-seq with a self-ligation step that is able to resolve whether coprecipitating genomic sequences are closely localized within the nucleus22. Therefore, by comparing matched ChIA-PET11 and ChIP-seq libraries, we were able delineate interactions between DHS exons and promoters (Fig. 2a).
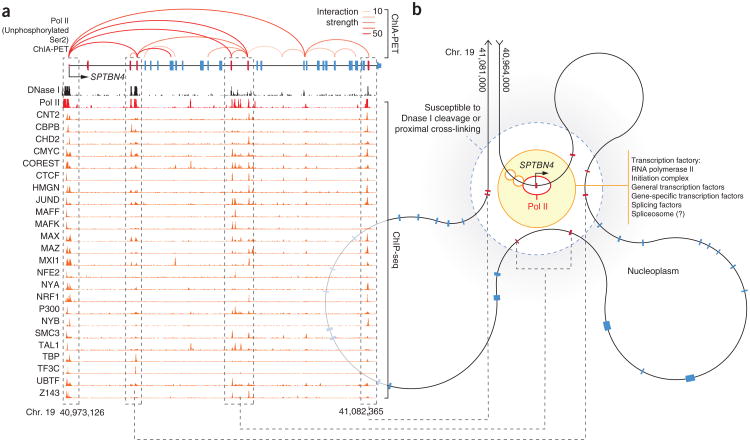
To assay the spatial proximity of DHS exons to their cognate promoters, we employed ChIA-PET libraries performed in two matched cell lines (MCF-7 and K562) using an antibody (8WG16) that targets the form of Pol II hypophosphorylated at Ser2 (refs. 11,23). The hypophosphorylated form of Pol II is found within the preinitiation complex and comprises a specific and characteristic mark at gene promoters that can be readily distinguished from the phosphorylated form of Pol II that elongates through downstream intragenic sequences24. Using this application of ChIA-PET, we could determine, across the genome, distal genomic sequences that are spatially localized to the preinitiation complexes assembled at gene promoters. We observed a marked enrichment of DHS exons residing within genomic regions interacting with promoters. DHS exons were significantly enriched for promoter interactions relative to local matched exons (12.65-fold; P = 2.1 × 10−4), an enrichment that was even more prominent at DHS exons exhibiting promoter-like features (22.28-fold; P = 2.9 × 10−4; Fig. 2b–d and Supplementary Fig. 7a,b). Reciprocally, DHS exons involved in Pol II ChIA-PET interactions also exhibited additional enrichment of ChIP-seq signals associated with promoters (Supplementary Fig. 7c). The complexity of local genome topography and its close convergence with exonic sequence structure are well illustrated by interactions related to the SPTBN4 gene (Fig. 3), in which local intragenic loops localize exons to the promoter while excluding intervening intronic sequences.
Figure 3.
Schematic of local genome folding of exons within the SPTBN4 gene. (a) Genome browser view showing ChIA-PET interactions (red; opacity indicates interaction frequency) and ChIP-seq signal (red histogram) for initiating form of Pol II (hypophosphorylated at Ser2) that correspond with the complex exon structure of the SPTBN4 gene, with the DHS (red) and matched (blue) exons indicated. DNase I hypersensitivity of loci is shown (black histogram), along with selected ChIP-seq libraries that are enriched at DHS exons (orange histograms; auto-scaled to view). Dashed boxes indicate corresponding exons. (b) Proposed model of local structure of the SPTBN4 gene interpreted from integrated ChIA-PET, ChIP-seq and DNase I annotations. SPTBN4 exons sensitive to DNase I cleavage and proximal cross-linking (red, within dashed circle) are located close to a transcription factory containing initiating Pol II (red eclipse) in association with the STPBN4 core promoter (black arrow). Additional protein features, anticipated by ChIP-seq enrichments, are also found within the transcription factory (orange circles).
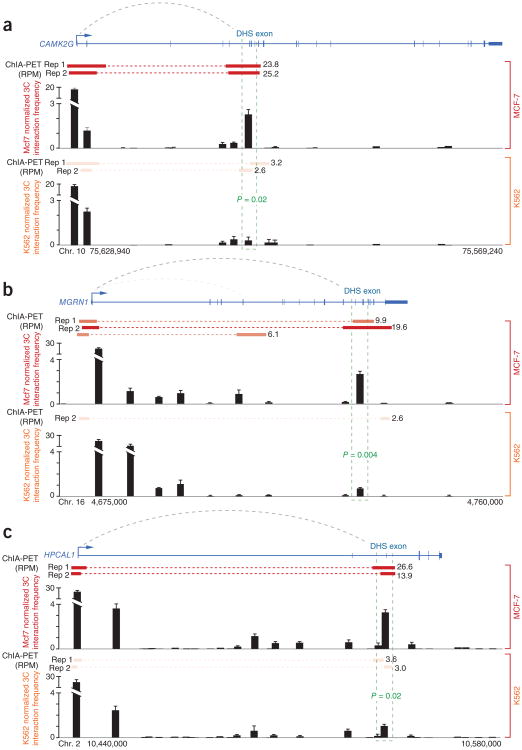
We independently performed chromatin conformation capture (3C) to validate cell type–specific interactions between promoters and exons for three genes as determined using ChIA-PET (Fig. 4). We were able to validate cell type–specific interactions between DHS exons and promoters, returning a significantly enriched 3C interaction frequency between DHS exons and cognate promoters in MCF-7 but not in K562 cells. This finding confirms not only the structure but also the cell type specificity of exon and promoter interactions as shown by ChIA-PET. More broadly, this finding also suggests that ChIP-seq enrichments can correlate with long-range interactions and illustrates the potential for ChIP-seq libraries to retain information on genome topography25.
Figure 4.
3C validates cell type–specific interactions between exons and promoters. (a–c) 3C interaction profiles for promoter and downstream exon-containing HindIII fragments in the CAMK2G (a), MGRN1 (b) and HPCAL1 (c) genes (n = 3 replicate libraries per cell type; error bars, s.d.). For each gene, ChIA-PET interactions (red bars with frequency) and 3C interaction frequency for both MCF-7 (top red histogram) and K562 (bottom orange histogram) cells are indicated. All genes show significant enrichments (P > 0.05, unpaired t test) for 3C interaction between DHS exons (green dashed boxes) and upstream promoters in MCF-7 cells relative to K562 cells. RPM, reads per million.
In addition to DHS exons with promoter-like features, we observed a substantial fraction of DHS exons (59.5%) enriched for ChIP-seq signals associated with distal regulatory sequences, such as the enhancer-associated factors P300, GATA1 and TAL1 (ref. 26) and the histone modifications of monomethylation (H3K4me1) and dimethylation (H3K4me2) at histone 3 lysine 4 (Supplementary Fig. 5)21. However, as for promoter-like DHS exons, these sites did not encompass corresponding sequence motifs, and we suggest that, similar to promoter-associated exons, these ChIP-seq patterns reflect a close spatial proximity of DHS exons to distal enhancers. To address this hypothesis, we applied publicly available ChIA-PET libraries targeting H3K4me2 (ref. 27), a broad marker of enhancers, to confirm that a subset of DHS exons indeed had proximal interactions with distal enhancer regulatory elements (Fig. 2b–d). Notably, these DHS exons exhibited a symmetrical peak profile for enhancer-associated ChIP-seq enrichments that differed from the asymmetric peak profile observed for promoter-like ChIP-seq enrichments (Supplementary Fig. 7d). This symmetrical profile may reflect the interaction of DHS exons with distal enhancers in both upstream and downstream directions. In contrast, promoter-like DHS exons are required to fold upstream to interact with promoters, a directional bias reflected in the slanted upstream signal of promoter-like ChIP-seq enrichments.
CTCF and cohesin have central roles in maintaining the spatial organization of the genome and in facilitating interactions between promoters and distal enhancers28,29. CTCF is known to be localized at or near certain exons9, and we similarly observed CTCF preferentially colocalized with components of cohesin (SMC3 and RAD21) at a substantial fraction (20.2%) of DHS exons (Supplementary Fig. 8a–d). To determine whether these factors might juxtapose DHS exons to distal genomic regions, including enhancer elements, we analyzed cell type-matched CTCF ChIA-PET libraries11,23. We found that DHS exons were significantly enriched (14.9-fold; P = 0.0087) in CTCF-centric interactions, an enrichment that was further amplified at sites with SMC3 and RAD21 co-occupancy (P = 1.6 × 10−4; Fig. 2c,d). Notably, these CTCF interactions encompassed exon 5 of the CD45 gene, which was recently shown to undergo exon inclusion in a CTCF-dependent manner9 (Supplementary Fig. 8g). As such, CTCF and cohesin likely fulfill a central role in arranging DHS exons within a complex higher-order structure that facilitates communication with distal genomic sequences (Supplementary Fig. 8e-f).
Promoter and enhancer interactions are associated with alternatively spliced exons
CTCF occupancy and several histone modifications, including H3K4me1, trimetylation of histone H3 at lysine 9 (H3K9me3) and trimethylation of histone H3 at lysine 36 (H3K36me3), have recently been shown to affect splicing outcomes6,9,30,31. Given that exon and intron sequences are demarcated within the genome's topology, we next considered whether the three-dimensional structure of the genome is associated with splicing outcomes.
We first investigated whether DHS exons are subject to cotran-scriptional splicing, a process in which epigenetic features can affect splicing8. To discern the degree to which DHS exons are subject to cotranscriptional splicing, we applied the completion of splicing index (CoSI) to DHS exons within matched RNA-seq libraries generated from fractionated chromatin-associated RNA populations that closely approximate nascent transcription32,33. We found that the majority of DHS exons were spliced within this subcellular RNA fraction (70.3% exhibited CoSI of >0.5), consistent with prevalent cotranscriptional splicing, with only a small minority of DHS exons remaining unspliced (<3% with CoSI of <0.10). Indeed, DHS exons and their host genes were enriched for cotranscriptional splicing relative to total exons34 (1.14-fold; P < 0.0001, Mann-Whitney test; Supplementary Fig. 9d). Additional analysis of ChIP-seq libraries also showed the accumulation of the elongating form (Ser2-phosphorylated) of Pol II at DHS exons (Supplementary Fig. 9e), suggesting that there is slower RNA polymerase–mediated elongation at DHS exons, which is consistent with current kinetic models of cotranscriptional splicing10.
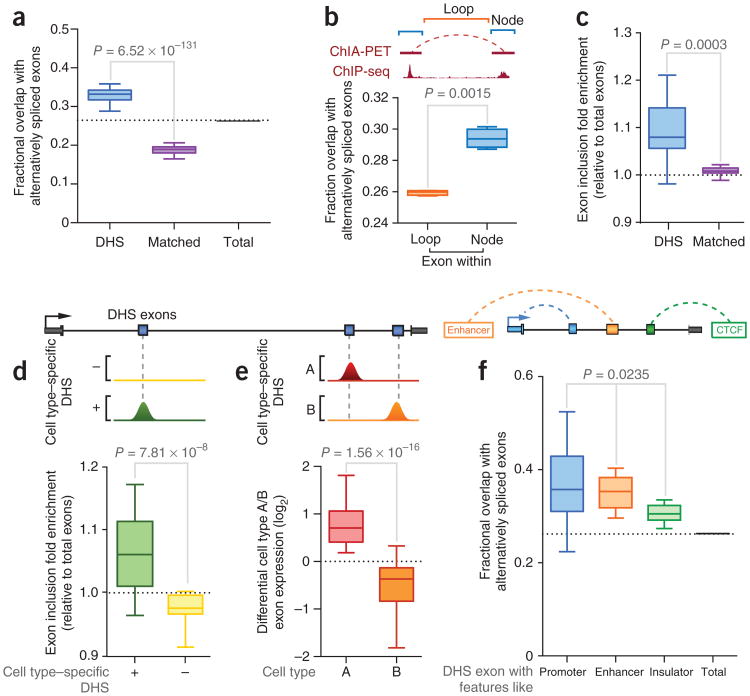
We next investigated any association between DNase I hypersensitivity and long-range interactions of exons and their alternative splicing status. First, we compared DHS exons annotated in 86 cell types to alternative splicing events catalogued within GENCODE annotations35. We found that DHS exons showed a large overlap with alternatively spliced cassette exons in all cell lines considered, with 34.5% of DHS exons being alternatively spliced, relative to the background levels of 26.2% seen for total exons (P = 8.7 × 10−102) and 18.8% seen for matched exons (Fig. 5a). Indeed, 48% of alternatively spliced exons overlapped DHSs in at least one of the ten cell types examined here (Online Methods). In support of this association, exons involved in Pol II–centric ChIA-PET interactions also exhibited a significant enrichment of alternative splicing events (P = 0.0015; Fig. 5b). The overlap between DHS exons and alternative splicing was markedly higher than previous attributed to histone modifications that affect exon exclusion6,30,36. DHS exons also showed depletion of H3K36me3 chromatin modifications, consistent with the reported depletion of this modification at alternatively spliced exons36 (Supplementary Fig. 9f).
Figure 5.
Association between DHS exons and cell type–specific alternative splicing. (a) Box-and-whisker plot (minimum-maximum range) indicating fractional overlap of DHS exons, matched exons and total exons with alternative splicing events (P = 6.52 × 10−131, two-tailed matched-pair t test, n = 86 cell types). (b) Top, definition of node and loop components for ChIA-PET interactions. Bottom, box-and-whisker plot indicating the fractional overlap of exons involved in chromatin interactions (within nodes) and exons within intervening regions (within loops) with alternative splicing events (P = 0.0015, two-tailed matched-pair t test, n = 4 library replicates). (c) Box-and-whisker plot showing fold enrichment for exon inclusion frequency for DHS exons and matched exons (P = 0.0003, Wilcoxon matched-pair signed-rank test, n = 12 cell types) relative to the background for all exons. (d) Box-and-whisker plot showing the fold enrichment of exon inclusion frequency for cell type–specific DHS exons relative to cell type–specific non-DHS exons (P = 7.81 × 10−6, Wilcoxon matched-pair signed-rank test, n = 12 cell types) relative to the background for all exons. (e) Box-and-whisker plot showing differential DHS exon usage between paired cell types (P = 1.56 × 10−6, Wilcoxon matched-pair signed-rank test, n = 5 randomly paired cell types). (f) Box-and-whisker plot showing fractional overlap of DHS exons with promoter-like, enhancer-like and insulator-like features with alternative splicing events (P = 0.0235, ANOVA, n = 8 cell types). In d–f, schematics (top) show the locations of DHS exons within genes.
The majority of alternative exons showed tissue-dependent variation in inclusion during splicing1,2, and we therefore next considered whether DNase I sensitivity correlates with alternative splicing outcomes. We first employed transcriptome annotations from RNA-seq libraries in ten matched cell types (with two replicates each; Supplementary Table 1) and employed DEXSeq37 to identify all exons with significant differential usage in different cell types (Online Methods). We found that DHS exons were subject to higher levels of exon inclusion relative to either total or matched exons (P = 0.0003, Wilcoxon matched-pair signed-rank test, n = 10; Fig. 5c and Supplementary Fig. 9g). Furthermore, this association was cell type specific, with exons exhibiting a higher rate of inclusion only in those cell types where overlapping DHSs were present (P = 7.81 × 10−6; Fig. 5d) but not in cell types where there was no DHS overlapping the exon, in which the rate of exon inclusion was not significantly different from the background rate (P = 0.1065, Wilcoxon matched-pair signed-rank test, n = 10). We also performed a complementary pairwise comparison of exon usage in cell lines, finding that exons exhibiting DNase I sensitivity in one cell line were enriched for inclusion relative to the other cell line in which no DNase I sensitivity was observed (P = 1.56 × 10−6; Fig. 5e and Supplementary Fig. 10).
We also observed distinctions between the associations of promoter-, enhancer- and cohesin-like DHS exons with alternative splicing. The fractional overlap with alternative splicing events was more prominent for exons undergoing interactions with promoters (37.5%) and enhancers (35.9%) than for cohesin-associated DHS exons (30.5% overlap; P = 0.0235; Fig. 5f). Similarly, promoter-like exons exhibited the highest rate of exon inclusion (P = 0.0485; Supplementary Fig. 9h), consistent with the long-standing finding that alternate promoter usage can result in changes to downstream alternative splic-ing38,39. Although further molecular studies are needed to explore the nature of this association, the correlation between genome topography and alternative splicing increases the potential that there is a structural influence on the regulation of tissue-specific cotranscriptional splicing40.
Discussion
Chromatin capture and ChIA-PET techniques have been able to reconstruct, with increasingly high resolution, the three-dimensional structure of the genome within the nucleus and have shown the importance of structural constraints imposed by the genome's topography on gene transcription11,12,41. Multiple genes spatially localize to transcriptional compartments—discrete subnuclear structures where RNA polymerase and transcriptional machinery is focused—in which genes subsequently undergo coregulated transcription42. Splicing factors and machinery similarly accumulate at discrete subnuclear foci closely associated with the transcriptional compartments, suggesting that genome topography may also impose similar structural constraints on processes of cotranscriptional splicing43,44.
Our study argues that current models of cotranscriptional splicing be extended into three-dimensional space in a manner analogous to the transcription factory model45 (Fig. 3b and Supplementary Fig. 11). The exonic sequences of these genes may be folded into transcriptional compartments harboring spliceosomal and transcriptional machinery, whose extensive connections have been well documented46. After transcription, these exons can be rapidly processed for exclusion or inclusion, according to the local regulatory context. This mechanism would permit a wide range of transcription factors and regulatory features, which otherwise regulate transcription initiation, to be localized to and have an effect on splicing, thereby leveraging an existing regulatory architecture for the organization of alternative splicing47.
The privileged demarcation of exons could readily generate tran-scriptional complexity from the modular architecture of the human genome, whereby a single exon can be spliced into a range of overlapping coding and noncoding transcripts48,49. The looping out of intervening regions could prevent confusion arising from conflicting splice signals from overlapping genes. Indeed, the folding of the genome into higher-order structures could compartmentalize transcription and splicing and thereby generate the complex, interleaved networks of transcripts that are a feature of the human genome50,51.
URLs
Sequence Read Archive, http://www.ncbi.nlm.nih.gov/sra/; Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/; in-house Perl scripts for computational analysis, http://matticklab.com/index.php?title=Marcel_Dinger#Genomic_and_Transcriptomic_Analysis_Tools; R, http://r-project.org/; GraphPad Prism, http://www.graphpad.com/scientific-software/prism/; Kent Source Utilities, http://genomewiki.ucsc.edu/index.php/Kent_source_utilities. Data were used from several sources: GENCODE gene assembly, comprehensive version 10 assembly (November 2011), http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwDnase/; DNase I hypersensitivity, release 4 (March 2012), http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwDnase/; Caltech RNA-seq, release 2 (January 2012), http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeCaltechRnaSeq/; CSHL long RNA-seq, release 1 (2010), http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeCshlLongRnaSeq/; histone ChIP-seq, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeBroadHistone/; Broad HMM chromatin state maps (release 1 June 2011), http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeBroadHmm/; ChIA-PET, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeGisChiaPet/; transcription factor binding sites, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeSydhTfbs/; nucleosome maps, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeSydhNsome/; Riken CAGE, release 2 (December 2011), http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRikenCage/.
Online Methods
Definitions
Genomic regions employed within this study are defined as follows.
Total exons. Exons were frst retrieved from GENCODE gene assemblies (version 10)29. All exons (i) overlapping 5′ or 3′ UTRs, (ii) overlapping transcripts annotated on the complementary strand, (iii) within 1 kb of an annotated transcript start site, (iv) comprising the frst or last exon of the transcript or (v) overlapping blacklisted regions (Kundaje; Release 3; October 2011) or excluded regions (Furey and Winter) due to mapping artifacts were omitted.
DHS exons. Total exons exhibiting overlap with matched DHSs. DHS exons overlapping CAGE-identifed RNA elements were omitted to occlude the possibility of cryptic promoters.
Matched exons. Total exons contained within the same mature transcripts as DHS exons (as determined by matching GENCODE prefx) but exhibiting no overlap with DHSs.
DHS exons (promoter-like). DHS exons overlapping promoter states, as determined by combinatorial chromatin state21 (Broad Chromatin HMM).
Promoter regions. Genome regions annotated as promoter state as determined by combinatorial chromatin state21 and overlapping annotated GENCODE transcriptional start sites.
Interacting promoter. Promoter regions that overlap, as determined from high-confdence ChIA-PET53, with DHS exons.
Gene 3′ end. Genome region encompassing the 1-kb window centered on the GENCODE 3′ terminus of the transcript and not overlapping any genomic region defned above.
Enhancers. Genome regions annotated as being in the enhancer state, as determined by combinatorial chromatin state21, with no overlap with any genomic region defned above.
Intronic. Introns were frst retrieved from GENCODE gene assemblies, with all nucleotides overlapping exons from alternative isoforms, or overlapping or antisense transcripts removed. All nucleotides overlapping the genomic regions defned above were removed.
Intergenic. Genome regions outside of the furthermost 5′ and 3′ boundaries of annotated GENCODE transcripts were retrieved. All nucleotides overlapping the genomic regions defned above were removed.
Alternative exons. Total exons that are fully overlapped by annotated introns from alternative isoforms (as determined by identical GENCODE prefx). Partial overlaps were omitted.
Constitutive exons. Total exons that have no overlap by introns retrieved from the GENCODE gene assembly. Partial overlaps were omitted.
Data usage
Data were employed in strict accordance with the ENCODE data release policy. The authors would like to acknowledge and thank the providers of these resources. The following data sets generated from human cell types as part of the ENCODE Consortium54 were employed within this study. Provided alignment (for example, *.bam) and annotation (for example, *.bed or *.narrowPeak) files were used wherever possible. Data were obtained from GENCODE gene assembly: comprehensive version 10 assembly (November 2011); DNase I hypersensitivity: release 4 (March 2012), Rep 1 and Rep 2 used, all cell types used; Caltech RNA-seq: release 2 (January 2012), Rep 1 and Rep 2 used, GM12878, HeLa S3, HSMM, K562, NHEK, HCT 116, HepG2, HUVEC, MCF-7 and NHLF cells used; CSHL long RNA-seq: release 1 (2010), Rep 1 and Rep 2 used, K562 nuclear PolyA+, nuclear PolyA- and total chromatin samples used; histone ChIP-seq: Rep 1 and Rep 2 used, GM12878, H1 hESC, HeLa S3, HepG2, HUVEC, K562, NHEK and NHLF cells used; Broad HMM chromatin state maps: (release 1 June 2011), GM12878, H1 hESC, HepG2, HMEC, HSMM, HUVEC, K562, NHEK and NHLF cells used; ChIA-PET: Pol II and CTCF Rep 1 and Rep 2 used, K562 and MCF-7 cells used; transcription factor binding sites: Rep 1 and Rep 2 used, GM12878, H1 hESC, HeLa S3, HepG2, HUVEC, K562 and MCF-7 cells used; nucleosome maps: Rep 1 and Rep 2 used, GM12878 and K562 cells used; Riken CAGE: release 2 (December 2011), whole-cell Rep 1 and Rep 2 used, GM12878, H1 hESC, HeLa S3, HepG2, HUVEC, K562 and NHEK cells used.
Computational analysis
Analysis was performed on Human Genome Build 37 (hg19). BEDTools30 was employed for data manipulation and analysis, as well as additional in-house Perl scripts. Statistical analysis was performed using R or GraphPad Prism. We also employed UCSC tools (Kent source utilities).
Normalization and analysis of DNase I sensitivity
DNase I hypersensitivity sensitivity sequencing was performed as previously described55. DNase I narrow peak elements were annotated with the HotSpot algorithm15. To normalize for alignment and sequencing artifacts between genomic regions that might result from differences in sequence composition, complexity and repeat content, we computed a mappability normalization constant. First, we performed whole-genome sequencing from purified genomic DNA in K562 cells using identical sequencing (36-mer Illumina Genome Analysis II sequencing) and alignment parameters as for DNase I hypersensitivity mapping. We then determined the normalized mean fold coverage provided by whole-genome sequence across defined genomic regions (Supplementary Fig. 1b). This provided a mappability normalization constant to correct for differential mapping of DNase I hypersensitivity reads to each genomic region. The enrichment for DNase I sites and reads (after normalization) within each genomic region was determined relative to genome background. To ascribe statistical significance to DHS enrichment, we first considered each independent replicate for each cell type (n = 86). A two-tailed Wilcoxon matched-pair signed-rank test was then performed to ascribe significance to the proportion of DHSs overlapping exons relative to genome background.
DHS exons were defined as follows. Exons were first retrieved from GENCODE gene assemblies (version 10, November 2011)13. All exons (i) overlapping 5′ or 3′ UTRs, (ii) overlapping transcripts annotated on complementary strand, (iii) within 1 kb of the annotated transcriptional start site or (iv) overlapping CAGE RNA Elements (ENCODE-RIKEN; release 2; December 2011) on either strand were omitted to exclude the possibility of cryptic promoters. All exons overlapping blacklisted (Kundaje; Release 3; October 2011) or excluded regions (Furey and Winter) were omitted.
ChIP-seq analysis
First, transcription factor peak sites (*.narrowPeak) were ascribed to overlapping DHS exons. Total ChIP-seq signal (as determined by the sum of individual reads encompassed within a peak site and normalized for peak size and library depth) was also determined to provide a quantitative measure of ChIP-seq signal enrichment. Hierarchial clustering according to quantitative ChIP-seq enrichment for selected transcription factors (showing enrichment at DHS exons relative to matched or total exons) was performed using Cluster3 (ref. 56). No normalization in either exon or ChIP-seq axis was performed, and distance measuring was determined by Pearson's correlation. We employed OverlapSelect (UCSC Genome Browser) to determine whether exons overlapped genomic features, such as peaks from ChIP-seq, using default parameters (including no minimum or maximum fractional or nucleotide overlap).
Presence of transcription factor motifs within corresponding ChIP-seq peaks was determined as follows. First, the entire human genome was scanned for transcription factor motifs represented within TRANSFAC57 using FIMO58, thereby providing a profile of predicted transcription factor motifs distributed across the entire genome. ChIP-seq peaks were then categorized according to their overlap with matching transcription factor motifs. Resulting peak groups (with or without the corresponding motif) were than overlapped with promoter or enhancer regions or with DHS exons. We performed a Friedman test to ascribe significance to the enrichment of peaks with motifs in both promoter and enhancer regions relative to DHS exons.
Correlation of transcription factor binding between interacting promoters and exons was determined as follows. First, promoters and DHS exons were paired according to specific and shared overlap with ChIA-PET interactions. The sum of all quantitative ChIP-seq signals was used to rank promoter and DHS exon pairs, with the top 200 being subjected to further analysis. Lastly, we compared the range of ChIP-seq enrichments between promoter and DHS exon pairs by Spearman's correlation.
Native ChIP-seq
Cells were resuspended in RSB buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5 mM spermidine), and cell membranes were lysed on ice for 10 min with addition of NP-40 to a final concentration of 0.02%. Nuclei were collected by centrifugation at 300g for 5 min at 4 °C and were washed once with RSB buffer. Nuceli were resuspended in 200 μl of MN buffer supplemented with protease inhibitor cocktail and digested with 17 U of micrococcal nuclease (Worthington) for 10 min at 37 °C. The digestion was stopped by adding 80 μl of MNase stop buffer. Supernatants (S1) were collected by centrifugation at 300g for 3 min at 4 °C. For chromatin immuno-precipitation, antibody (Cell Signaling Technology 9751 for tri-methyl-histone H3 Lys4) was conjugated to Dynabeads M-280 sheep anti-rabbit IgG (Life Technologies) in 1 ml of 1× PBS for at least 6 h at 4 °C and incubated with micrococcal nuclease–digested, diluted chromatin at 4 °C overnight. Complexes were washed four times with elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 5 mM sodium butyrate, 150 mM NaCl) and twice with TE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA) and were then washed briefly in incubation buffer (10 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 5 mM EDTA, pH 8.0, 0.5% SDS). Supernatants were recovered from the beads and treated with proteinase K at 55 °C for 4 h. DNA was purified by phenol-chloroform extraction and ethanol precipitation.
Chromatin segmentation state analysis
Periexonic enrichments at DHS exons were categorized according to overlap with the Broad HMM Chromatin State Segmentation map21. All promoter (1_Active_Promoter, 2_Weak_Promoter, 3_Poised_Promoter) and enhancer (4_Strong_Enhancer, 5_Strong_Enhancer, 6_Weak_Enhancer, 7_Weak_Enhancer) definitions were grouped for this analysis. In cases where exons overlapped with two different states, exons were ascribed both annotations and were included in both analyses.
ChIA-PET analysis
We employed two replicate ChIA-PET libraries for both CTCF (sc-15914, Santa Cruz Biotechnology) and Pol II (8WG16; Covance, MMS-126R) that had been prepared as previously described53. All ChIA-PET interactions reported and analyzed here were high-confidence interactions (with false discovery rate (FDR) of <0.05) that were filtered from background random ligation events. A detailed description of the technical and statistical protocols for linker filtering, short-read mapping, paired-end transcript classification, binding site identification, distinction between self- and interligation events and interaction cluster identification is reported in refs. 11,53. Briefly, this involved the following steps.
Embedded nucleotide barcode sequences were required to be matching and correctly orientated in the same direction. Tis flter permits the measure and omission of random ligation events, including events that follow immunoprecipitation, and thereby flters out technical noise.
Both PET ends were required to align uniquely with a maximum of one mismatch to the reference human genome.
PETs were classifed as self-ligating (representing protein binding sites) if both ends aligned closely or interligating (representing interactions) if ends ligated distally. For the libraries employed, fltered interactions were required to occur over a distance of 8 kb as determined from a comparison of distance lengths between correct to incorrect barcode orientations53.
An additional statistical analysis compared the rate of interligation to the rate expected at random to confrm the validity of each ChIA-PET library. Similarly, the frequency of each interaction between self-ligating clusters was required to exceed a random model determined from the density of each cluster.
Interactions were required to be represented by at least three reads that spanned the ligation event, with additional statistical analysis performed to assess data quality.
Those interactions that fulfilled the above criteria were deemed high confidence and were included in the analysis. Relative normalized overlap frequency was determined according to the number of ChIA-PET reads overlapping exons after normalization for library depth and exon size. A two-tailed Wilcoxon matched-pair signed-rank test was performed to ascribe significance to enrichments.
To determine enrichments at DHS exons, high-confidence interactions (represented by >3 sequenced reads) were divided into loop and node components. Node components were used to determine overlapping enrichment with total exons, DHS exons and promoter-like DHS exons. Relative normalized overlap frequency was determined according to the number of ChIA-PET reads overlapping exons after normalization for library depth and exon size. A two-tailed Wilcoxon matched-pair signed-rank test was performed to ascribe significance to enrichments.
3C
3C libraries were generated using HindIII as described previously59. 3C interactions were quantified by RT-PCR using primers designed within each HindIII restriction fragment (primer sequences available upon request). Quantitative RT-PCR was performed on a RotorGene 6000 platform using MyTaq HS DNA polymerase (Bioline) with the addition of 5 mM Syto9, an annealing temperature of 66 °C and extension of 30 s. 3C analysis of MCF-7 and K562 cells was performed in three independent experiments. BACs covering each gene were used to create an artificial library of ligation products to normalize for PCR efficiency. Data were normalized to the signal from the BAC library and, between cell lines, by reference to a region within the GAPDH gene. All quantitative RT-PCR products were electrophoresed on 2% agarose gels, gel purified and sequenced to verify the 3C product.
RNA-seq analysis
To determine the relative proportion of DHS exons subject to alternative or constitutive splicing, we performed the following analysis. We first categorized annotated exons within GENCODE as alternative or constitutive. Alternative cassette exons were required to be fully encompassed by an annotated intron from alternative isoforms (as determined by identical GENCODE prefix), and constitutive exons had no overlap by introns retrieved from the GENCODE gene assemblies. For each of the 86 cell types (retrieved from ENCODE; Supplementary Table 1), we then identified all DHS exons and, subsequently, the proportion of DHS exons that were either alternatively or constitutively spliced. To ascribe statistical significance to differences in these proportions, we performed a two-tailed Wilcoxon matched-pair signed-rank test, treating each cell line as an independent measure (n = 86).
To complement this analysis, we also determined the relative proportion of exons involved in chromatin interactions (as defined by ChIA-PET). First, we divided high-confidence Pol II interactions into loop and node components. Nodes correspond to the sites of interaction as determined from self-ligation PETs. Loops correspond to remaining intervening regions between two sites of interaction as determined by interligation PETs. Li et al. provide a detailed description of how PETs are filtered for technical noise and distinguished between self- and interligating PETs53. We determined the alternative or constitutive splicing status of all exons overlapped by nodes relative to the splicing status of exons within loop regions. Statistical significance was ascribed using a two-tailed Wilcoxon matched-pair signed-rank test, treating each ChIA-PET library as an independent measure (n = 4).
We next determined whether DHS exons had divergent rates of exon inclusion and exclusion relative to total exons. To determine relative exon usage, RNA-seq alignments were first retrieved from ENCODE (GM12878, HeLa S3, HSMM, K562, NHEK, HET116, HepG2, HUVEC, MCF-7 and NHLF cells were used). DEXSeq was employed to determine exon inclusion or exclusion, as it can process the multiple biological replicates provided by the ENCODE Consortium and ascribe significance to differences in exon usage between cell types37. Therefore, we employed DEXSeq to determine significant (P < 0.1, χ2 test) differential usage of each exon for each cell type (each cell type had between two and four replicate libraries) relative to a mixed reference (that consisted of all libraries combined in equal parts to generate an average reference, performed for each replicate from each cell line to generate two independent replicate reference libraries). Only exons exhibiting significant differential expression (adjusted P value of <0.1) were employed for further analysis. We then determined the fraction of DHS exons that had exon inclusion (increased exon usage in a particular cell type relative to the reference) relative to the fraction of all exons that had exon inclusion. Statistical significance was ascribed using a two-tailed Wilcoxon matched-pair signed-rank test, treating each cell type as an independent measure (n = 10).
We next identified exons undergoing cell type–specific DNase I sensitivity. We first catalogued all exons overlapping DHSs in the ten cell lines employed. From this list, we could then identify, for each cell line, those cell type–specific DHS exons and those remaining exons in the catalog that did not have DHSs. Exons with ubiquitous DNase I hypersensitivity were omitted from further analysis. The fold enrichment for exon inclusion relative to total significantly differentially expressed exons was then determined for each cell type. Statistical significance was ascribed using a two-tailed Wilcoxon matched-pair signed-rank test, treating each cell type as an independent measure (n = 10).
To complement this analysis of cell type–specific DNase I sensitivity, we also performed pairwise comparisons between cell types (denoted cell type A and B for each comparison), which permitted us to compare cell type–specific exon usage to cell type–specific DNase I sensitivity for each pair. Statistical significance was ascribed using a two-tailed Wilcoxon matched-pair signed-rank test, treating each cell type as an independent measure (n = 10).
Lastly, we considered splicing status and exon usage individually for DHS exons with promoter-like, enhancer-like and insulator-like enrichments. DHS exons were first categorized according to overlap with chromatin state segmentation map annotations21. We then determined the fractional overlap and exon inclusion rate relative to total exons for each category for each cell type. Statistical significance was ascribed by paired ANOVA.
Supplementary Material
Acknowledgments
The authors would like to thank the following funding sources: the Australian National Health and Medical Research Council (Australia Fellowships 631668 to J.S.M., T.R.M. and M.B.C. and 631381 and 1021731 to S.L.E.); the Queensland State Government (National and International Research Alliance Program to L.K.N.); the National Breast Cancer Foundation Australia (to S.L.E.); the National Human Genome Research Institute (NHGRI; ENCODE grant HG004456 to Y.R., G.L. and K.S.S.); and the US National Institutes of Health (NHGRI ENCODE grants U54HG004592 and U54HG007599 to J.A.S.).
Footnotes
Accession codes. Whole-genome sequencing libraries have been deposited in the Sequence Read Archive under accessions SRX285537, SRX285595 and SRX285596. ChIP-seq libraries have been deposited in the Gene Expression Omnibus under accession GSE46945.
Note: Supplementary information is available in the online version of the paper.
Author Contributions: T.R.M., G.L. and S.J.N. performed bioinformatic analysis. S.L.E. performed 3C analysis. G.L., K.S.S. and Y.R. performed ChIA-PET analysis. A.B.S., H.W., S.J. and R.S. performed native ChIP-seq and whole-genome sequencing. T.R.M., S.J.N., M.B.C., L.K.N., Y.R., J.S.M. and J.A.S. prepared the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 4.Pozzoli U, et al. Intron size in mammals: complexity comes to terms with economy. Trends Genet. 2007;23:20–24. doi: 10.1016/j.tig.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 6.Luco RF, et al. Regulation of alternative splicing by histone modifcations. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci USA. 2008;105:5160–5165. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, et al. ncRNA- and Pc2 methylation–dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabo PJ, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc Natl Acad Sci USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifcations. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 19.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 20.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fullwood MJ, et al. An oestrogen-receptor-α–bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheutin T, Cavalli G. Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion. PLoS Genet. 2012;8:e1002465. doi: 10.1371/journal.pgen.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassouf MT, et al. Genome-wide identifcation of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saint-André V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 31.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but ineffcient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatt DM, et al. Transcript dynamics of proinfammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(suppl. 1):S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz S, Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razin SV, et al. Transcription factories in the context of the nuclear and genome organization. Nucleic Acids Res. 2011;39:9085–9092. doi: 10.1093/nar/gkr683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melnik S, et al. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat Methods. 2011;8:963–968. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Allemand E, Batsche E, Muchardt C. Splicing, transcription, and chromatin: a menage a trois. Curr Opin Genet Dev. 2008;18:145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Moldón A, et al. Promoter-driven splicing regulation in fssion yeast. Nature. 2008;455:997–1000. doi: 10.1038/nature07325. [DOI] [PubMed] [Google Scholar]
- 48.Gerstein MB, et al. What is a gene, post-ENCODE? History and updated defnition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 49.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 50.Mercer TR, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 52.Derrien T, et al. Fast computation and applications of genome mappability. PLoS ONE. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.ENCODE Project Consortium. User's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 56.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 57.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.