Abstract
Most tropical mammal species are threatened or data-deficient. Data collection is impeded by the traditional monitoring approaches which can be laborious, expensive and struggle to detect cryptic diversity. Monitoring approaches using mammal DNA derived from invertebrates are emerging as cost- and time-effective alternatives. As a step towards development of blowfly-derived DNA as an effective method for mammal monitoring in the biodiversity hotspot of Peninsular Malaysia, our objectives were (i) to determine the persistence period of amplifiable mammal mtDNA in blowfly guts through a laboratory feeding experiment (ii) to design and test primers that can selectively amplify mammal COI DNA mini-barcodes in the presence of high concentrations of blowfly DNA. The persistence period of amplifiable mammal mtDNA in blowfly guts was 24 h to 96 h post-feeding indicating the need for collecting flies within 24 h of capture to detect mammal mtDNA of sufficient quantity and quality. We designed a new primer combination for a COI DNA mini-barcode that did not amplify blowfly DNA and showed 89% amplification success for a dataset of mammals from Peninsular Malaysia. The short (205 bp) DNA mini-barcode could distinguish most mammal species (including separating dark taxa) and is of suitable length for high-throughput sequencing. Our new DNA mini-barcode target and a standardized trapping protocol with retrieval of blowflies every 24 h could point the way forward in the development of blowfly-derived DNA as an effective method for mammal monitoring.
Introduction
According to the Global Mammal Assessment (2008), information on the distribution and abundance of most tropical mammal species remains data-deficient, and thus some species appear to be disproportionately threatened [1]. Lack of data on tropical mammal species can be associated with limitations of the existing monitoring approaches. Field-trapping techniques vary in efficiency [2] and are often a biased representation of diversity [3,4]. For example, identification of animal signs is laborious, requiring the input of specialists over an extended time period [5], and can be imprecise [6,7]. Likewise, expensive camera traps cannot identify individuals to species lacking easily observed diagnostic markings [8,9]. Impediments also include the challenge posed by cryptic species, which are not always morphologically distinct, and are more easily recognised by molecular techniques [10,11]. Considering that the current monitoring approaches are challenged by ethics [12], precision, and accuracy, a new approach is urgently needed.
Approaches deriving vertebrate DNA from associated invertebrate taxa are emerging as powerful tools for large-scale monitoring [13]. Such approaches have the potential to detect and identify rare and cryptic mammal species [14,15]. Two rare species—the Annamite striped rabbit (Nesolagus timminsi) and the Truong Son muntjac (Muntiacus truongsonensis)—as well as a “cryptic” species, the small-toothed ferret-badger (Melogale moschata), were detected from bloodmeals recovered from leeches collected in a forest in Vietnam [15].
Blowfly-derived DNA may present advantages over DNA recovered from other invertebrates, such as leeches, which are habitat restricted [15], or mosquitoes and tsetse flies, which have narrow host preferences [16,17]. Blowflies (Calliphoridae) are globally distributed in all habitats and exhibit broad host preferences both during larval and adult stages [18,19]. For example, Chrysomya megacephala is a saprophagous and coprophagous generalist and is among the first and most abundant species at mammal carrion in forests in Peninsular Malaysia [19]. Blowflies also potentially concentrate faecal DNA while feeding coprophagously [20,13].
One key factor affecting successful detection of ingested DNA is the digestion efficiency of the hematophagous, coprophagous or saprophagous feeder [13]. Knowledge of the persistence period of amplifiable mammal DNA in blowfly guts is essential for designing standardized trapping methods (e.g. [21]) for large-scale mammal monitoring via this approach. Typically, for ecological studies, blowflies are captured at baited traps, and may remain alive in the trap for hours [22] or days [23] before collection by the researcher. Forensic studies have determined the post-feeding detection period of human mtDNA in the guts of blowfly maggots (Calliphora vicina) to be between 24 h and 48 h [24]. However, the post-feeding persistence period of mammal DNA in the guts of adult blowflies still lacks any reliable data.
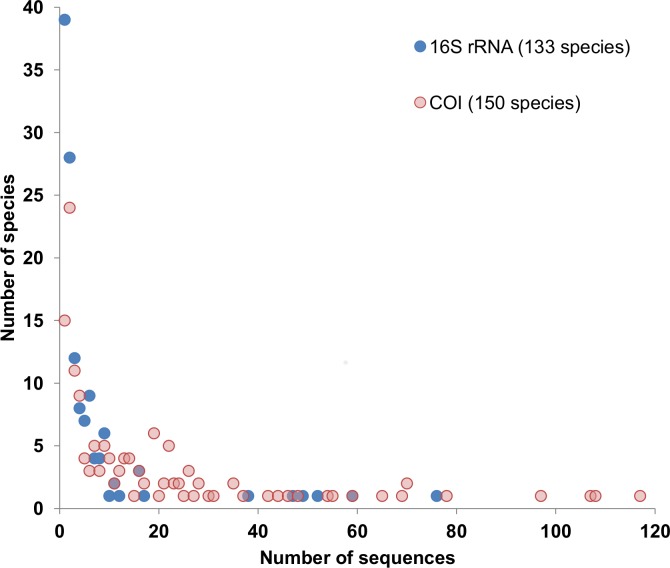
A second key factor for the success of invertebrate-derived DNA approaches is selection of the target DNA region. The region must be easy to PCR amplify from taxonomically unknown samples, must be variable among taxa to permit species identification, and reference sequences of known species must exist in order to match the amplified fragment. Previous studies have used 16S rRNA and 12S rRNA for detection of mammal mtDNA from blowflies [14], 16S rRNA from leeches [15], cytochrome b from mosquitoes and sandflies [25,26] and cytochrome c oxidase I (COI) from ticks and tsetse flies [17,27]. A fragment of COI is a preferred target for a number of reasons. Variation in COI has been used successfully to discriminate and identify mammals in Southeast Asia [28]. There are more COI sequences than 16S rRNA sequences on GenBank for mammal species (after excluding Homo sapiens sequences) (Fig 1) and this includes BARCODE standard records [29]. Therefore we suggest the chance of accurately assigning an unknown mammal to a species is higher for a fragment of COI than for other gene regions. Broad “universal” primers have been designed for amplification of mammal COI barcodes [30] but a short target is required for ingested DNA, which is likely to be partially degraded and difficult to amplify [13]. Considering that field-application will likely involve pooling a large number of blowflies for cost-effective high-throughput sequencing [31], a 100–300 bp fragment is preferred [32]. A “universal DNA mini-barcode for biodiversity analysis” has been published previously [33] but the primers have variable success amplifying mammal species (77% as reported by Meusnier et al. [33]; 0% as determined in silico by Ficetola et al. [34]; 80% as reported by Arif et al. [35]) and also will amplify blowfly DNA, which is likely to be present at higher concentration.
Fig 1. Number of 16S rRNA and COI sequences publicly available on GenBank for the 246 mammal species (excluding Homo sapiens) found in Peninsular Malaysia.
As a step towards development of blowfly-derived DNA as an effective method for mammal monitoring in a biodiversity hotspot, Peninsular Malaysia, our objectives were (i) to determine the persistence period of amplifiable mammal mtDNA in blowfly guts through a laboratory feeding experiment, and (ii) to design and test primers that can selectively amplify mammal DNA mini-barcodes in the presence of high concentrations of blowfly DNA.
Materials and Methods
Ethics statement
Our protocol for minimally-invasive collection of mammal DNA samples (hair, wing punches) has been approved by the University of Malaya Institutional Animal Care and Use Committee (UMIACUC) (Ref. ISB/02/1212013/JJW (R)) and the Department of Wildlife and National Parks, Peninsular Malaysia (Ref. JPHL&TN(IP): 80-4/2 Jld16(24)). No specific permits were required, however, approval was obtained from the land owners/managers for animal (mammal and blowfly) sampling and no protected species were sampled.
Persistence of mammal mtDNA in blowfly guts
To obtain a sample of blowflies of known age, physiological state, and feeding history a laboratory culture of Chrysomya megacephala was established. A rotting fish carcass was obtained from a local supermarket and placed outside the Museum of Zoology, University of Malaya, Kuala Lumpur, to encourage egg deposition by wild blowflies. The fish was then moved into a 39 x 25 x 33 cm sealed plastic container and the hatching larvae provided with more fish until pupation. Once all pupae had emerged C. megacephala adults were selected out (based on the morphological characters of the species) and sorted into three containers of approximately 100 blowflies each.
The adult blowflies were then starved for 24 h to allow digestion of any food taken and to adjust to similar hunger levels. After the starvation period pieces of market-supplied beef liver (Bos taurus) were placed into the containers for 4 h (06:00–10:00). We observed that all blowflies fed almost immediately upon provision of food. After the beef liver was removed sugar water was provided as the only food source. At 8, 16, 24, 48, 72, 96, 120, 144 h following removal of the beef liver, 9 individual, adult blowflies were selected arbitrarily (3 from each container) at each interval and were frozen at -20°C. The blowfly’s guts were then dissected out with sterile implements for DNA extraction using NucleoSpin Tissue kit (Macherey-Nagel, Germany), following the manufacturer’s instructions. To provide positive and negative controls respectively, DNA was also extracted directly from the market-supplied beef liver and wild-caught blowfly legs (Chrysomya megacephala).
PCR was performed using beef-specific primers targeting a 75 bp region of the Bos taurus cytochrome b mtDNA (BSP F: 5’-CCCGATTCTTCGCTTTCCAT-3’and BSP R: 5’-CTACGTCTGAGGAAATTCCTGTTG-3’) [36]. FastMix Frenche Hot Start PCR pre-mix (Intron Biotechnology, Korea) was used for all PCR reactions, adding 1 μL of each primer and 1 μL of DNA extract. The thermal cycling conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 1 min and a final extension at 72°C for 5 min. PCR products were visualised on a 1.5% agarose gel stained with 1 x GelRed (Biotium, USA). We also created beef/blowfly DNA mixtures from the control DNA extracts, and performed PCR to determine relative concentrations of beef DNA based on visual comparison of the electrophoresis images.
Design and testing of mammal primers
We assembled a test dataset of 41 DNA extracts from 41 mammal species (16% of the mammal species found in Peninsular Malaysia) collected during our previous field sampling in Peninsular Malaysia [37–39] and from collection at Ulu Gombak Forest Reserve Selangor, Gerik Perak, and nearby the Museum of Zoology, University of Malaya, Kuala Lumpur (see S1 Dataset). Based on our exploration of PCR amplification of COI with this dataset and other mammal samples, we have found that Uni-Mini-bar F [33] and RonM [30] have good success as a forward primers, but LepF1 [40], HCO2198 [41], and VF1d [30] have lower success. We have found that Uni-Mini-bar R [33] has low success as a reverse primer, but VR1d [30] has good success. Given the high success of Uni-Mini-bar F and RonM, and the 210 bp distance between these primers, we proceeded to design a reverse primer targeting the RonM binding site.
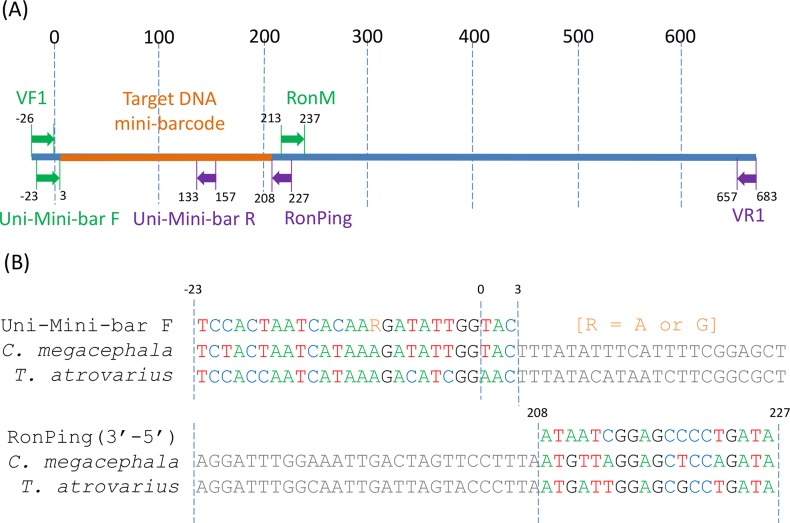
We aligned mammal COI sequences retrieved from GenBank and used the program Primer3 [42] to select a 19 bp region slightly upstream of RonM, which enabled design of a reverse primer with appropriate physical and structural properties. When used in combination with Uni-Mini-bar F, the new primer RonPing (5'-tatcaggggctccgattat-3') should amplify a 205 bp fragment at the 5’ end of the COI barcode region (Fig 2).
Fig 2. The binding sites of the primers were in relation to a Chrysomya megacephala (blowfly) and Thomomys atrovarius (Smooth-toothed pocket gopher) sequence.
A) Relative positions of mammal primers on the COI barcode region. B) The binding sites of the primers Uni-Mini-bar F and RonPing (reverse complement).
The success of Uni-Mini-bar F/ RonPing and Uni-Mini-bar F/ Uni-Mini-bar R across mammal species was then systematically compared using our 41 species dataset. PCR was performed using FastMix Frenche Hot Start PCR pre-mix (Intron Biotechnology, Korea) and COI Fast thermocycling program [43] for all reactions, with slight modification to the DNA volume (0.5–2 μL) depending on DNA extraction method.
Next we tested the ability of the Uni-Mini-bar F/ RonPing combination to amplify low concentrations of mammal DNA in the presence of high concentrations of blowfly DNA. We first confirmed (through testing on multiple DNA extractions with various thermocycling conditions) that the primer combination does not amplify Chrysomya megacephala COI. We then mixed DNA extracts from ten mammal species with DNA extracts from blowfly legs in a ratio of 1 part mammal (~0.43 ng) to 16 parts blowfly (~7.0 ng). PCR was performed for the mixed DNA samples as described above. Additionally, we used DNA extracts from the 48 h post-feeding samples from the feeding experiment above as templates for PCR with Uni-Mini-bar F/ RonPing.
A further test involved the detection of mammal DNA from wild-caught blowflies. Four baited traps (modified from [13]) were set at Rimba Ilmu, University of Malaya (S1 Fig). Rimba Ilmu is an 80 ha botanical garden and a habitat for small mammals such as bats, squirrels, tree shrews and rats. The traps were emptied every 24 h and blowflies were frozen at -20°C. The guts of 30 blowflies were then dissected out with sterile implements for DNA extraction and PCR with Uni-Mini-F/ RonPing as above. PCR products were sequenced in both directions by a local company (MYTACG-Kuala Lumpur, Malaysia). DNA sequences are available on BOLD in the dataset DS-RONPING (http://www.boldsystems.org/index.php/Public_SearchTerms?query=DS-RONPING).
To evaluate the success of the 205 bp DNA mini-barcode amplified by Uni-Mini-bar F/ RonPing for species assignment, COI sequences from mammal species found in Malaysia (based on [44]) were mined from GenBank. We retrieved COI sequences >600 bp and the sequences were trimmed to the 205 bp DNA mini-barcode target amplified by Uni-Mini-bar F/ RonPing. A neighbor-joining tree based on number of differences was produced from the aligned sequences using MEGA 6.0 [45].
Results
Persistence of mammal mtDNA in blowfly guts
Blowfly (Chrysomya megacephala) guts sampled at 24 h post-feeding, or earlier, all contained amplifiable beef mtDNA (S2 Fig). At 48 h post-feeding 78% of blowfly guts had amplifiable beef mtDNA, but this dropped sharply to 44% at 72 h and 22% at 96 h. At these later times, when amplification was successful, the bands were fainter indicating lower concentrations of DNA. There was no successful amplification from guts sampled at 120 h post-feeding and later.
Design and testing of mammal-specific primers
Our newly designed RonPing primer had a low number of mismatches (less than six) with most of the mammal COI sequences retrieved from GenBank (S3 Fig). RonPing had five mismatches with Chrysomya megacephala (Fig 2) and did not amplify this species. Canis lupus (Order: Carnivora) also had five mismatches located in approximately the same positions, with one near to 3’end of primer, but Canis lupus COI was amplified with RonPing. Other mammal species showing 3–4 mismatches were also amplified and sequenced successfully.
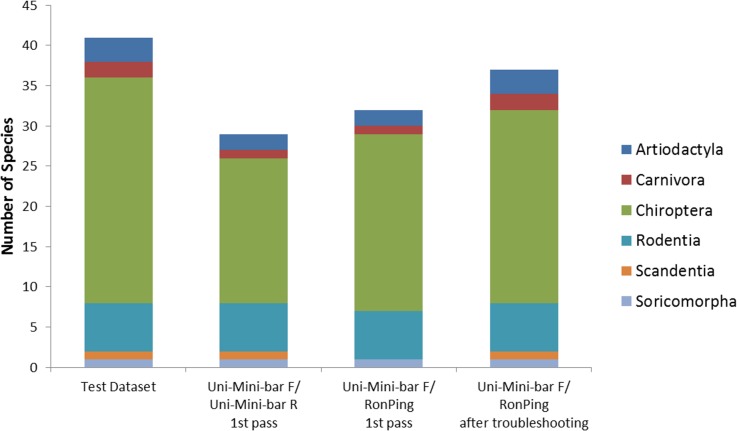
The success of primer pairs, Uni-Mini-bar F/ Uni-Mini-bar R [33] and Uni-Mini-bar F/ RonPing in amplifying COI from 41 mammal species extracts were 71% and 78% respectively (Fig 3). A further round of PCR using Uni-Mini-bar F/ RonPing yielded a higher proportion of species amplified (89%). Mammal sequences amplified from ten mammal/C. megacephala DNA mixtures using Uni-Mini-bar F/ RonPing had high quality peaks and were clear of contamination (S4 Fig). The Uni-Mini-bar F/ RonPing combination showed successful amplification from 27% of wild-caught C. megacephala with sequenced amplicons showing close matches (<95%) to Rhinolophus sp., Bos taurus and Gallus gallus.
Fig 3. Comparison of amplification success using primer pairs: Uni-Mini-bar F/ RonPing and Uni-Mini-bar F/ Uni-Mini-bar R for Artiodactyla (n = 3), Carnivora (n = 2), Chiroptera (n = 28), Soricomorpha (n = 1), Rodentia (n = 5) and Scandentia (n = 1).
Examining the target 205 bp region among mammals from Peninsular Malaysia mined from GenBank (113 species), all species, except for seven, possessed a unique COI sequence or unique sets of COI sequences for the 205 bp DNA mini-barcode (S5 Fig). The region was also able to separate 26 “dark” bat taxa recognized in previous DNA barcoding studies (e.g. Balionycteris maculate, Hipposideros armiger and Myotis muricola) [28].
Discussion
Calvignac-Spencer and colleagues [14] demonstrated the potential of sequencing invertebrate-derived DNA for mammal monitoring and suggested the advantage of blowflies over other invertebrates [13]. However, certain questions remained regarding the field-application in large-scale mammal monitoring. Blowflies have been trapped in a variety of ways depending on the purpose of sampling (e.g. for veterinary purposes [23, 46]). Calvignac-Spencer and colleagues [14] sampled flies opportunistically immediately upon arrival at bait. A probable field-application scenario for large-scale mammal monitoring would see a large number of baited traps, where the trapped blowfly is unable to make contact with the bait and is kept alive until collection (e.g. [47]). The traps would be set for a number of hours before the researcher is able to return to empty them. In field experiments Chrysomya megacephala usually arrived to fresh carrion within 24 h of exposure [19] meaning blowflies could potentially be in a trap for a number of hours digesting any mammal DNA already present in their guts. Our results indicate amplifiable mammal mtDNA persists in the guts of adult C. megacephala for 24 h—96 h post-feeding (89% at 48 h) which is consistent with that determined for blowfly maggots [24] and other dipterans (mosquitoes, tsetse flies) [48]. In contrast, mammal DNA can persist in ticks and leeches for at least several months [15, 48]. The longer persistence period may seem like an advantage to using these taxa but means it is more difficult to determine when the detected mammal species was present at the sampling site. Based on our findings we suggest that blowflies will need to be retrieved from traps and processed at least every 24 h to maximize the chance of amplifying mammal DNA from their previous meal. The interval between a blowfly feeding and looking for its next meal is unknown, but would also be a factor affecting the successful detection of ingested DNA. The high success in detecting mammal DNA from wild-caught blowflies (27% of blowflies contained detectable vertebrate DNA, with multiple species detected in some individuals) trapped over a period of 24 hours indicates the potential of a standardized trapping protocol with retrieval of blowflies every 24 h for effective mammal monitoring in the field.
Here, we provide the first experimental indication that successful detection of mammal DNA from blowflies is due to mammal DNA in their guts as opposed to mammal DNA being carried on their exoskeleton as a result of landing on mammal tissues or faeces. This was not addressed directly in previous studies [14]. The gradual decline in amplifiable mammal DNA could indicate a lack of severe enzymatic breakdown of ingested DNA in the fore-gut. For blowflies primary digestion is achieved by secretion of salivary enzymes onto food before it is ingested orally [24, 49]. The persistence period of amplifiable DNA may be different depending on the length of the target fragment chosen for amplification. It is worth to note that our experiment to assess mammal DNA persistence is likely to provide an upper estimate as the substrate the blowflies were fed with was of high quality (beef liver) and energy expenditure was limited (flies were kept in boxes). We found no difference between amplification success using the beef-specific primers targeting 75 bp and amplification success using Uni-Mini-bar F/ RonPing targeting 205 bp.
The use of 16S rRNA by Calvignac-Spencer and colleagues [14] was problematic. While the 16S rRNA fragment is easy to amplify using primers with broad taxonomic coverage across Mammalia, the short “barcode” produced has low species resolution i.e. multiple closely related species can share the same haplotype [34]. Our results suggest targeting COI, the animal DNA barcode [50], is a very practical option, providing the chance to exploit the identification capacity of BOLD [51] and the well-characterised patterns of species level divergence at this region (e.g. [28]). The new primer, RonPing, when used in combination with Uni-Mini-bar F amplifies a 205 bp fragment of COI. This appears to be an optimal length for a DNA mini-barcode allowing amplification from degraded samples, such as invertebrate-derived DNA, while not suffering a reduction in the ability to distinguish species [33]. This target also falls within the maximum read length for high-throughput sequencing (e.g. 300 bp for the Illumina MiSeq [32]), including spare length for MID tags to separate multiple samples [52].
The new primer combination was able to amplify a higher proportion of mammal species than previously proposed combinations [33]. Another significant advantage of the Uni-Mini-bar F/ RonPing combination is the fact that it does not amplify Chrysomya megacephala COI, even when the ratio of blowfly DNA to mammal DNA is high. This is likely due to RonPing having a double mismatch with C. megacephala within 5 bp of the 3’ end including a purine-purine mismatch (A-G) [53]. Sixteen mammal species also had an A-G mismatch in the same position, but they only had one mismatch within 5 bp from 3’end of primer not the C. megacephala double, which may be the main reason for successful amplification from these DNA templates. Lack of C. megacephala amplification negates the need for blocking primers [54] or, if no blocking probes are used in high-throughput sequencing, prevents significant wastage of sequencing effort due to amplification and sequencing of blowfly DNA [55]. The low number and even distribution of mainly pyrimidine-pyrimidine and purine-pyrimidine mismatches between the RonPing primer and GenBank mammal sequences suggest primer bias might not be too severe and consequently the primer may produce a less biased estimate of DNA templates present [53]. Although the ecoPCR program [34] predicted that the Uni-Mini-bar F/ RonPing combination could only amplify 19% of mammal species by allowing three mismatches to the whole mitochondrial genomes on GenBank (results not shown), results coherent with in vitro PCR can be obtained by allowing a higher number of mismatches.
The 205 bp COI fragment was successful in distinguishing nearly all examined mammal species from Peninsular Malaysia and separating “dark” bat taxa, previously recognised species that lack formal taxonomic status [39]. This suggested the potential of detecting cryptic taxa overlooked by traditional methods [28]. The species which shared haplotypes at the 205 bp region such as Rattus tiomanicus and R. rattus included sequences which have previously been identified as problematic and most likely represent misidentifications or contamination rather than shared haplotypes [56].
Conclusion
Our findings suggest our new DNA mini-barcode target and a standardized trapping protocol with retrieval of blowflies every 24 h could point the way forward in the development of blowfly-derived DNA as an effective method for mammal monitoring in Peninsular Malaysia. The next step would be a comprehensive comparison of diversity measures for mammals produced by the blowfly-derived DNA approach and by traditional monitoring approaches such as cage traps, mist nets, hair traps, camera traps, or scat samples.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Solid triangles represent clusters of multiple conspecifics.
(PDF)
Acknowledgments
Amanda Naaum provided advice and suggestions regarding the blowflies feeding experiment. Mr. Foo Yoke Wai provided sequencing support. Khairunnisa Syaripuddin and Noraishah Abdul-Aziz provided access to mammal DNA samples.
Data Availability
DNA sequences are available from the BOLD database at the following URL: http://www.boldsystems.org/index.php/Public_SearchTerms?query=DS-RONPING.
Funding Statement
This study was supported by grants from the Nagao Natural Environment Foundation Japan to LPS and the Ministry of Higher Education Fundamental Research Grant Scheme (FP042-2014A) to JJW and Yong Kien Thai (UM). The funding bodies had no role in the design, in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
References
- 1. Schipper J, Chanson JS, Chiozza F, Cox NA, Hoffmann M, Katariya V, et al. The status of the world’s land and marine mammals: diversity, threat, and knowledge. Science. 2008; 322: 225–230. 10.1126/science.1165115 [DOI] [PubMed] [Google Scholar]
- 2. Lambert TD, Malcolm JR, Zimmerman BL. Variation in small mammal species richness by trap height and trap type in southeastern Amazonia. J Mammal. 2005; 86: 982–990. [Google Scholar]
- 3. Fontúrbel FE. A methodological approach to assess the small mammal community diversity in the temperate rainforest of Patagonia. Mamm Biol. 2010; 75: 294–301. [Google Scholar]
- 4. Torre I, Guixe D, Sort F. Comparing three live trapping methods for small mammal sampling in cultivated areas of Ne Spain. Hystrix. 2010; 21(2): 147–155. [Google Scholar]
- 5. Campbell G, Kuehl H, Diarrassouba A, N’Goran PK, Boesch C. Long-term research sites as refugia for threatened and over-harvested species. Biol Lett. 2011; 7(5): 723–726. 10.1098/rsbl.2011.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davison A, Birks JDS, Brookes RC, Braithwaite TC, Messenger JE. On the origin of faeces: morphological versus molecular methods for surveying rare carnivores from their scats. J Zool. 2006; 257(2): 141–143. [Google Scholar]
- 7. Mumma MA, Soulliere CE, Mahoney SP, Waits LP. Enhanced understanding of predator–prey relationships using molecular methods to identify predator species, individual and sex. Mol Ecol Resour. 2014; 14(1): 100–108. 10.1111/1755-0998.12153 [DOI] [PubMed] [Google Scholar]
- 8. Brodie JF, Gibbs HK. Bushmeat hunting as climate threat. Science. 2009; 326: 364–365. 10.1126/science.326_364a [DOI] [PubMed] [Google Scholar]
- 9. Jansen PA, Muller-Landau HC, Wright SJ. Bushmeat hunting and climate: an indirect link. Science. 2010; 327(5961): 30 10.1126/science.327.5961.30-a [DOI] [PubMed] [Google Scholar]
- 10. Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, et al. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2006; 22(3): 148–155. [DOI] [PubMed] [Google Scholar]
- 11. Ceballos G, Ehrlich PR. Discoveries of new mammals and their implications for conservation and ecosystem services. Proc Natl Acad Sci U S A. 2008; 106(10): 3841–3846. 10.1073/pnas.0812419106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Powell RA, Proulx G. Trapping and marking terrestrial mammals for research: integrating ethics, performance criteria, techniques, and common sense. ILAR J. 2003; 44(4): 259–276. [DOI] [PubMed] [Google Scholar]
- 13. Calvignac-Spencer S, Leendertz FH, Gilbert MTP, Schubert G. An invertebrate stomach’s view on vertebrate ecology. Bioessays. 2013; 35: 1004–1013. 10.1002/bies.201300060 [DOI] [PubMed] [Google Scholar]
- 14. Calvignac-Spencer S, Merkel K, Kutzner N, Kühl H, Boesch C, Kappeler PM, et al. Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Mol Ecol. 2012; 22: 915–924. [DOI] [PubMed] [Google Scholar]
- 15. Schnell IB, Thomsen PF, Wilkinson N, Rasmussen M, Jensen LR, Willerslev E, et al. Screening mammal biodiversity using DNA from leeches. Curr Biol. 2012; 22(8): R262–R263. 10.1016/j.cub.2012.02.058 [DOI] [PubMed] [Google Scholar]
- 16. Lyimo IN, Ferguson HM. Ecological and evolutionary determinants of host species choice in mosquito vectors. Trends Parasitol. 2009; 25: 189–196. 10.1016/j.pt.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 17. Muturi CN, Ouma JO, Malele II, Ngure RM, Rutto JJ, Mithöfer KM, et al. Tracking the feeding pattern of tsetse flies (Glossina genus) by analysis of bloodmeals using mitochondrial cytochrome genes. PLoS ONE. 2011; 6: e17284 10.1371/journal.pone.0017284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Norris KR. The bionomics of blow flies. Annu Rev Entomol. 1965; 10: 47–48. [Google Scholar]
- 19. Azwandi A, Nina Keterina H, Owen LC, Nurizzati MD, Omar B. Adult carrion arthropod community in a tropical rainforest of Malaysia: analysis on three common forensic entomology animal models. Trop Biomed. 2013; 30(3): 481–494. [PubMed] [Google Scholar]
- 20. Stenglein JL, De Barba M, Ausband DE, Waits LP. Impacts of sampling location within a faeces on DNA quality in two carnivore species. Mol Ecol Resour. 2010; 10: 109–114. 10.1111/j.1755-0998.2009.02670.x [DOI] [PubMed] [Google Scholar]
- 21. Vogt WG, Havenstein DE. A standardized bait trap for blowfly studies. Aust J Entomol. 1974; 13(3): 249–253. [Google Scholar]
- 22. Amat E. Notes on necrophagous flies (Diptera: Calyptratae) associated to fish carrion in Colombian Amazon. Acta Amazon. 2010; 40(2): 397–400. [Google Scholar]
- 23. Akbarzadeh K, Rafinejad J, Nozari J, Rassi Y, Sedaghat MM, Hosseini M. A modified trap for adult sampling of medically important flies (Insecta: Diptera). J Arthropod Borne Dis. 2012; 6(2): 119–128. [PMC free article] [PubMed] [Google Scholar]
- 24. Campobasso CP, Linville JG, Wells JD, Introna F. Forensic genetic analysis of insect gut contents. Am J Forensic Med Pathol. 2005; 26(2): 161–165. [PubMed] [Google Scholar]
- 25. Abbasi I, Cunio R, Warburg A. Identification of blood meals imbibed by phlebotomine sand flies using cytochrome b PCR and reverse line blotting. Vector Borne Zoonotic Dis. 2009; 9: 79–86. 10.1089/vbz.2008.0064 [DOI] [PubMed] [Google Scholar]
- 26. Bataille A, Fournié G, Cruz M, Cedeño V, Parker PG, Cunningham AA, et al. Host selection and parasite infection in Aedes taeniorhynchus, endemic disease vector in the Galapagos Islands. Infect Genet Evol. 2012; 12: 1831–1841. 10.1016/j.meegid.2012.07.019 [DOI] [PubMed] [Google Scholar]
- 27. Gariepy TD, Lindsay R, Ogden N, Gregory TR. Identifying the last supper: utility of the DNA barcode library for bloodmeal identification in ticks. Mol Ecol Resour. 2012; 12: 646–652. 10.1111/j.1755-0998.2012.03140.x [DOI] [PubMed] [Google Scholar]
- 28. Francis CM, Borisenko AV, Ivanova NV, Eger JL, Kim BK, Guillen-Servent A, et al. The roles of DNA barcodes in understanding and conservation of mammal diversity in Southeast Asia. PLoS ONE. 2010; 5(9): e12575 10.1371/journal.pone.0012575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanner R. Data Standards for BARCODE Records in INSDC (BRIs). 2009. Available: http://barcoding.si.edu/pdf/dwg_data_standards-final.pdf. Accessed 2 July 2014.
- 30. Ivanova NV, Clare EL, Borisenko AV. DNA barcoding in mammals. Methods Mol Biol. 2012; 858: 153–182. 10.1007/978-1-61779-591-6_8 [DOI] [PubMed] [Google Scholar]
- 31. Stein ED, Martinez MC, Stiles S, Miller PE, Zakharov EV. Is DNA barcoding actually cheaper and faster than traditional morphological methods: results from a survey of freshwater bioassessment efforts in the United States? PLoS ONE. 2014; 9(4): e95525 10.1371/journal.pone.0095525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shokralla S, Gibson JF, Nikbakht H, Janzen DH, Hallwachs W, Hajibabaei MD. Next-generation DNA barcoding: using next generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol Ecol Resour. 2014; 14(5): 892–901. 10.1111/17550998122236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meusnier I, Singer GAC, Landry JL, Hickey DA, Hebert PDN, Hajibabaei M. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics. 2008; 9: 214 10.1186/1471-2164-9-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ficetola GF, Coissac E, Zundel S, Riaz T, Shehzad W, Bessiere J, et al. An In silico approach for the evaluation of DNA barcodes. BMC Genomics. 2010; 11: 434 10.1186/1471-2164-11-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arif IA, Khan HA, Al Sadoon M, Shobrak M. Limited efficiency of universal mini-barcode primers for DNA amplification from desert reptiles, birds and mammals. Genet Mol Res. 2011; 10(4): 3559–3564. 10.4238/2011.October.31.3 [DOI] [PubMed] [Google Scholar]
- 36. Tanabe S, Hase M, Yano T, Sato M, Fujimura T, Akiyama H. A real-time quantitative PCR detection method for pork, chicken, beef, mutton, and horseflesh in foods. Biosci Biotechnol Biochem. 2007; 71(12): 3131–3135. [DOI] [PubMed] [Google Scholar]
- 37. Sing KW, Syaripuddin K, Wilson JJ. Changing perspectives on the diversity of bats (Mammalia: Chiroptera) at Ulu Gombak since the establishment of the Field Study Centre in 1965. Raffles Bull Zool. 2013; S29: 211–217. [Google Scholar]
- 38.Syaripuddin K, Kumar A, Sing KW, Halim MRA, Nursyereen MN, Wilson JJ. Mercury accumulation in bats near hydroelectric reservoirs in Peninsular Malaysia. Ecotoxicology. 2014; 10.1007/s10646-014-1258-y [DOI] [PubMed]
- 39. Wilson JJ, Sing KW, Halim MRA, Ramli R, Hashim R, Sofian-Azirun M. Utility of DNA barcoding for rapid and accurate assessment of bat diversity in Malaysia in the absence of formally described species. Genet Mol Res. 2014; 13(1): 920–925. 10.4238/2014.February.19.2 [DOI] [PubMed] [Google Scholar]
- 40. Hebert PDN, Penton EH, Burns J, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly, Astraptes fulgerator . Proc Natl Acad Sci U S A. 2004; 101(41): 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxi–dase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994; 3: 294–299. [PubMed] [Google Scholar]
- 42. Rozen S, Skaletsky H. PRIMER 3 on the WWW for general users and for biologist programmers In: Krawetz SA, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. New Jersey: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 43. Wilson JJ. DNA barcoding in insects In: Kress WJ, Erickson DL, editors. DNA Barcodes: Methods and Protocols. Methods in Molecular Biology. New Jersey: Humana Press; 2012. pp. 17–46. 10.1007/978-1-61779-591-6_1 [DOI] [Google Scholar]
- 44. Davison GWH, Zubaid A. The status of mammalian biodiversity in Malaysia In: Chua LSL, Kirton LG, Saw LG, editors. Status of Biological Diversity in Malaysia and Threat Assessment of Plant Species in Malaysia. Kuala Lumpur: Forest Research Institute Malaysia; 2007. pp. 3–27. [Google Scholar]
- 45. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0 Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scholtz AJ, Cloete SWP, Laubscher JM, De Beer EF. A preliminary evaluation of a sheep blowfly trap in the Western Cape. J S Afr Vet Assoc. 2000; 71(3): 148–152. [DOI] [PubMed] [Google Scholar]
- 47. Whitworth TL. Keys to the genera and species of blow flies (Diptera: Calliphoridae) of the West Indies and description of a new species of Lucilia Robineau-Desvoidy. Zootaxa. 2010; 2663: 1–35. [Google Scholar]
- 48. Kent RJ. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour. 2009; 9: 4–18. 10.1111/j.1755-0998.2008.02469.x [DOI] [PubMed] [Google Scholar]
- 49. Hobson RP. Studies on the nutrition of blow-fly larvae. J Exp Biol. 1932; 9: 128–138. [Google Scholar]
- 50. Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci. 2003; 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ratnasingham S, Hebert PDN. Bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 2007; 7(3): 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carew EE, Pettigrove VJ, Metzeling L, Hoffmann AA. Environmental monitoring using next generation sequencing: rapid identification of macroinvertebrate bioindicator species. Front Zool. 2013; 10: 45 10.1186/1742-9994-10-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stadhouders R, Pas SD, Anber J, Voermans J, Mes THM, Schutten M. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5’ nuclease assay. J Mol Diagn. 2010; 12(1): 109–117. 10.2353/jmoldx.2010.090035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vestheim H, Jarman SN. Blocking primers to enhance PCR amplification of rare sequences in mixed samples—a case study on prey DNA in Antarctic krill stomachs. Front Zool. 2008; 5: 12 10.1186/1742-9994-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pinol J, Andres VS, Clare EL, Mir G, Symondson WOC. A pragmatic approach to the analysis of diets of generalist predators: the use of next-generation sequencing with no blocking probes. Mol Ecol Resour. 2014; 14: 18–26. 10.1111/1755-0998.12156 [DOI] [PubMed] [Google Scholar]
- 56. Shen YY, Chen X, Murphy RW. Assessing DNA barcoding as a tool for species identification and data quality control. PLoS ONE. 2013; 8(2): e57125 10.1371/journal.pone.0057125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Solid triangles represent clusters of multiple conspecifics.
(PDF)
Data Availability Statement
DNA sequences are available from the BOLD database at the following URL: http://www.boldsystems.org/index.php/Public_SearchTerms?query=DS-RONPING.