Abstract
The elegant architecture of the channel of bacteriophage phi29 DNA packaging motor has inspired the development of biomimetics for biophysical and nanobiomedical applications. The reengineered channel inserted into a lipid membrane exhibits robust electrophysiological properties ideal for precise sensing and fingerprinting of dsDNA at the single-molecule level. Herein, we used single channel conduction assays to quantitatively evaluate the translocation dynamics of dsDNA as a function of the length and conformation of dsDNA. We extracted the speed of dsDNA translocation from the dwell time distribution and estimated the various forces involved in the translocation process. A ~35-fold slower speed of translocation per base pair was observed for long dsDNA, a significant contrast to the speed of dsDNA crossing synthetic pores. It was found that the channel could translocate both dsDNA with ~32% of channel current blockage and ~64% for tetra-stranded DNA (two parallel dsDNA). The calculation of both cross-sectional areas of the dsDNA and tetra-stranded DNA suggested that the blockage was purely proportional to the physical space of the channel lumen and the size of the DNA substrate. Folded dsDNA configuration was clearly reflected in their characteristic current signatures. The finding of translocation of tetra-stranded DNA with 64% blockage is in consent with the recently elucidated mechanism of viral DNA packaging via a revolution mode that requires a channel larger than the dsDNA diameter of 2 nm to provide room for viral DNA revolving without rotation. The understanding of the dynamics of dsDNA translocation in the phi29 system will enable us to design more sophisticated single pore DNA translocation devices for future applications in nanotechnology and personal medicine.
Keywords: Bacteriophage phi29, DNA packaging, Nanomotor, Connector, Liposomes, Ion channel, Single channel conductance, Membrane channel, Nanostructure, Bionanotechnology, Nanobiotechnology, Nanomedicine, Nanopore
1. Introduction
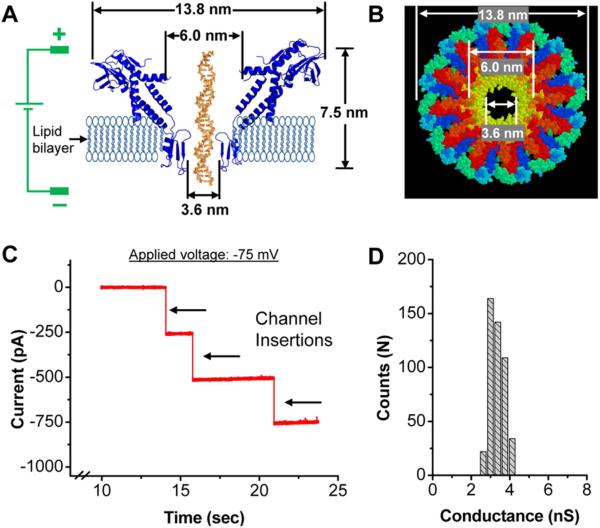
Translocation of macromolecules of protein, DNA and RNA across cellular compartments using nanomachines is a universal phenomenon in living systems [1-5]. The novelty and ingenious design of such machines have inspired the development of biomimetics. In the case of double-stranded DNA (dsDNA) viruses, the genome is packaged into a preformed protein shell called procapsid. This entropically and energetically unfavorable process is accomplished using an elegant nanomotor [6-10]. In bacteriophage phi29, the nanomotor consists of a protein channel called the connector, ATPase gp16, and a hexameric packaging RNA (pRNA) ring to gear the motor [11,12]. The connector consists of 12 protein subunits which encircle to form a dodecameric channel that enables dsDNA to translocate into the procapsid of the phage during maturation and exit during infection [11,13,14]. Based on the crystal structure [15,16], the ring is 13.8 nm at its wide end and 6.6 nm at its narrow end. The internal channel is 6 nm in diameter at the wide end and 3.6 nm in diameter at the narrowest constriction (Fig. 1A-B).
Figure 1. Structure and electrophysiological properties of membrane embedded phi29 connector nanopore.
(A) Illustration of membrane-embedded connector in the electrophysiological assay setup. A side view showing the dimensions of the connector. For simplicity, only 2 of the 12 subunits of the connector are shown. (B) Top view of the connector showing the dimensions of the dodecameric channel interior. (C) Insertion of connector channels in the lipid membrane, as demonstrated by step-wise increase in current at holding potential -75 mV. (D) Histogram showing uniform conductance (3.1 ± 0.3 nS) of the phi29 connector channels in presence of 1M NaCl, 5 mM Tris, pH 7.8, and at holding potential -75 mV (500 insertion events).
We have previously inserted the reengineered connector into a lipid bilayer to serve as a highly robust membrane-embedded nanopore [17,18] (Fig. 1A). The connectors were reconstituted into liposomes, followed by insertion into planar lipid membranes via vesicle fusion of liposome/connector complexes [17,18]. The insertion of the connector channels resulted in step-wise increase in conductance. The step size of the connector channels was homogenous and the channels exhibited equal conductance under both positive and negative trans-membrane potentials [17,18]. The connectors exhibit a perfectly linear Current-Voltage (I–V)relationship without displaying any voltage gating phenomenon at ±100 mV [17,19-21]. Furthermore, the nanopore system is stable under a wide range of solution conditions, including pH 2 - 12, and ionic strengths of 0.1 - 3 M NaCl or KCl [19].
Unlike other well studied protein nanopores, such as α-hemolysin [22] and MspA [23], the phi29 connector has a larger diameter and allows translocation of both ds- and ss- DNA or RNA under an electric field [17,20,21,24-26]. The connector channel exercises a one-way traffic property for dsDNA translocation from N-terminal entrance to C-terminal exit with a valve mechanism in DNA packaging [21]. In addition, the connector channel has shown three discrete step gating at higher trans-membrane voltages associated with conformational change in the channel subunits [25]. Since the crystal structure is available [15,16], explicit engineering of the connector channel can be made with atomic precision for added functionality. Recently, a reengineered connector with reduced channel size has shown the capability to discriminate ss-DNA and RNA [24]. By selectively introducing probes either at the terminal ends or within the channel lumen, single antibodies or single chemicals can be identified with high confidence at ultra-low concentrations based on their characteristic current fingerprints [27,28]. Procedures for large scale production and purification of the connector have been well developed [18,29-33]. These features make the connector nanochannel an ideal system for sensing and diagnostic applications. Nanopore based sensory techniques are currently an area of great interest for single molecule, high throughput, and label-free detection of a variety of biomacromolecules and chemicals based on modulations of the individual current blockage signatures [34-41].
Herein, we investigated dsDNA translocation dynamics based on the properties of dsDNA (conformation (linear vs. folded) and length); and the forces involved in DNA translocation including electrical driving, viscous drag, hydrodynamic drag and uncoiling/recoiling forces, and DNA-pore interactions. Two parameters were used to characterize the translocation events: current blockage percentage, and dwell time distribution, which follows an exponential decay. The dwell time was further used to extract the speed and estimate the forces associated with the translocation events. We also developed a simple automated MATLAB-based program for high throughput single molecule analysis of DNA translocation events. Taken together, the results will pave the way for studying more complex biological macromolecules at the single molecule level using protein nanopore-based detectors.
2. Experimental section
2.1 Materials
Organic solvents (n-Decane, hexane, chloroform, and DMSO) and standard chemical reagents were purchased from Sigma-Aldrich or Fisher. The phospholipids 1,2-diphytanoyl-sn glycerol-3-phosphocholine (DPhPC) and 1,2- Dioleoyl-sn-Glycero-3-Phosphocholine (DOPC) were purchased from Avanti Polar Lipids.
2.2 C-His wild-type connector expression and purification
The expression and purification of C-His gp10 has been described in detail previously [18]. Briefly, wild-type plasmids were constructed from vector pET-21a(+) (Novagen) with a two-step PCR. The plasmids were then transformed into the E. coli strain HMS174 (DE3) for protein expression. Purification of wild-type C-His tagged connector proteins was conducted with one-step immobilized metal affinity chromatography under native conditions.
2.3 Preparation of lipid vesicles containing the C-His connector channels
The preparation of connector reconstituted liposomes has been described previously [17]. Briefly, 1 ml of 1 mg/ml DOPC or DPhPC in chloroform was syringed in a round bottomed flask. The chloroform was removed under vacuum using the Rotary Evaporator (Buchi). The lipid film was then rehydrated with 1 ml of connector protein solution containing 200-300 mM sucrose to bud off vesicles into the solution. The lipid solution was then extruded through a polycarbonate membrane filter (100 nm or 400 nm) to generate unilamellar lipid vesicles. A final molar ratio of lipid vs. connector was established at 4000:1 to 16000:1.
2.4 Insertion of the connector into planar bilayer lipid membrane
The insertion of the connector reconstituted liposomes into a lipid bilayer has been described previously [17,18]. Briefly, a standard Bilayer Lipid Membrane (BLM) cell was utilized to form a free standing lipid bilayer. An aperture of 200 μm in diameter in a thin Teflon partition separated the compartment into cis and trans sides. The aperture was pre-painted with 0.5 μl 3% (wt/vol) DPhPC n-decane solution twice to ensure the complete coating of the entire edge of the aperture. The cis- and trans-compartments were then filled with conducting buffers (5 mM Tris/pH 7.9, 1 M NaCl). After confirming the formation of the lipid bilayer, the connector reconstituted lipid vesicles (0.5 - 2 μl) were directly incubated to fuse with the planar lipid membrane to form the membrane-embedded nanopore.
2.5 Measurements of current for each membrane-embedded channel
A pair of Ag/AgCl electrodes connected directly to both the compartments was used to measure the current traces across the BLM. The current trace was recorded using an Axopatch 200B patch clamp amplifier coupled with the Axon DigiData 1322A analog-digital converter (Axon Instruments) or the BLM workstation (Warner Instruments). All voltages reported were those of the trans-compartment. Data was low band-pass filtered at a frequency of 5 KHz or 1 KHz and acquired at a sampling frequency of 10 KHz. The PClamp 9.1 software (Axon Instruments) was used to collect the data, and the softwares Matlab and Origin Pro 8.0 was used for data analysis.
2.6 Preparation of DNA used in the experiment
Synthetic 12, 20, 35 and 141-nt DNA fragments and their complementary strands were custom ordered from IDT. To obtain dsDNA, we annealed two complementary ssDNA by heating them at 95°C for 5 min followed by slow coo ling to room temperature over a few hours. The dsDNA complex was then purified using 10-15% (wt/vol) native Polyacrylamide Gel Electrophoresis (PAGE) [18].
For 500, 2000 and 5000-bp dsDNA, we used restriction enzymes (such as EcoRV) to cut plasmid DNA (such as Cx43) into desired linear fragments with a blunt end, following manufacturer's guidelines and the buffers and reaction solutions provided with each enzyme. The DNA was then purified using 1% (wt/vol) agarose gel and eluted with a Qiaex II gel extraction kit [18].
For constructing double-crossover (DX-DNA) tetra-stranded DNA structures [42] the five strands were custom ordered from IDT. The strands were annealed and purified in 12% (wt/vol) native PAGE, following reported procedures [42].
2.7 Translocation experiments of DNA
Liposomes containing connector channels were added to the cis-compartment of the BLM cell. After definite insertion of connector channels, purified DNA was mixed with conducting buffer and added to the cis compartment in the BLM cell. Alternatively, DNA was premixed in the conducting buffer before the start of the experiment. For all the translocation experiments, DNA (1 pm-1 nM final concentration) was used. The current traces were then recorded over a period of 2-4 hours.
2.8 Development of computer program for automated analysis of dsDNA translocation events
To date, several groups have developed algorithms to analyze current blockade events [22,43-45]. Herein, we developed an automated single molecule analysis system using Matlab algorithm for detailed characterization of a large number of DNA translocation events using a set of parameters (Suppl. Fig. 1) (see Supplementary Information for mathematical details). Individual pore blockage events were first distinguished from noise events with high confidence. The range and center of background noise was determined to establish a threshold for possible translocation events. The current trace was then read sequentially until all possible translocation events were identified. Translocation events were then characterized by calculating the height and duration of each event waveform.
3. Results and discussion
The insertion of the C-His connector channels in the lipid membrane resulted in step-wise increase in conductance, as shown in the continuous current trace (Fig. 1C). The step size of the connector channels was homogenous (Fig. 1D) with a channel conductance determined to be 3.1 ± 0.3 nS, calculated using the ratio of the measured current jump (under 1 M NaCl, 5 mM Tris, pH 7.8) resulting from connector insertion to the applied voltage (75 mV). Furthermore, the C-His connectors exhibit a perfectly linear I–V relationship without displaying any voltage gating phenomenon under the reported conditions of ±100 mV, as established previously [17,19-21].
3.1 Parameters for characterizing translocation of dsDNA
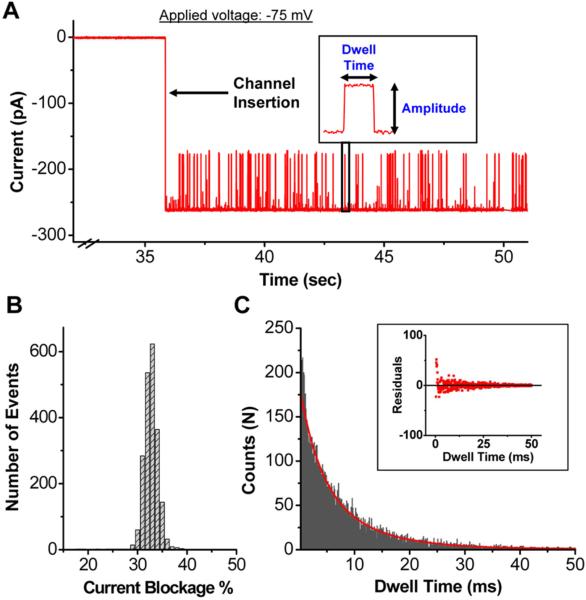
Following the addition of dsDNA, numerous current blockage events (Fig. 2A) were observed, indicating the translocation of dsDNA, which was previously verified unequivocally by quantitative PCR [17]. Since DNA is a non-conducting polymer, it physically blocks the flow of ions as it threads through the pore and gives rise to the transient blockage signals. We characterized each translocation event (insert in Fig. 2A) using two parameters: current blockage percentage, and dwell time.
Figure 2. Translocation of dsDNA through the connector channels.
(A) Current trace showing burst of transient current blockage events representing the translocation of 20 bp dsDNA upon channel insertion. DsDNA was premixed in the conducting buffer. Insert: magnified event of a single DNA translocation event with characteristic current amplitude and dwell time. (B) Histogram of current blockage percentage (31.7 ± 1.3 %) induced by 20 bp dsDNA (2100 translocation events). (C) Typical dwell time distribution of DNA translocation events following an exponential decay (11,500 events). Buffer: 1 M NaCl, 5 mM Tris, pH 7.8; Applied voltage: -75 mV; Red line: exponential fit; Insert: plot of the residuals.
Phi29 connector channel is not a uniform cylinder. Although the channel is 7.5 nm long with 3.6 nm to more than ten nm distribution in width, the 3.6 nm is the narrowest constriction and is the bottle-neck that determines the open pore current of the channel. The current blockage percentage was expressed as [(Ipore − IDNA)/Ipore] × 100, where Ipore is the current when the channel is open (i.e. the step size of one channel insertion, Fig. 2A), and IDNA is the blocked current level observed during DNA translocation. For dsDNA (irrespective of length), the current blockage percentage was deduced to be 31.7 ± 1.3 % (Fig. 2B), which is consistent with the cross-sectional areas of dsDNA (3.1 nm2) and the connector channel (10.2 nm2 at the narrowest constriction). The second parameter is the dwell time (τD) (insert, Fig. 2A), which is the time taken for dsDNA to traverse from the narrower N- to the wider C-terminal end of the connector [21]. Thousands of translocation dwell time events from several independent experiments were analyzed using custom MATLAB algorithm (see Methods and Suppl. Fig. 1), and plotted in a histogram (Fig. 2C, Suppl. Fig. 2). The data was fitted with an exponential model with density function: f(x; τD) = (1/τD) e(−x/τD). This is because the vast majority of events are spread over the tail of the distribution, and this model gives a more accurate value for τD than the peak τD. Timescales shorter than the peak time most likely represent partial entry of DNA without translocation or collisions. We note that our exponential dwell time distribution differs from the skewed Gaussians reported for ssDNA transport through α-hemolysin channels [22,46], presumably due to stronger DNA-pore interactions.
3.2 Dwell time distribution of linear dsDNA of varying lengths
We characterized different lengths of dsDNA (12, 20, 35, 141, 500, 2000 and 5000 bp) under identical experimental conditions, unless otherwise specified: -75 mV applied potential; buffer composed of 1 M NaCl, 5 mM Tris/pH 7.8; room temperature; and 1pM - 1nM dsDNA concentration. For a given length of dsDNA, the frequency of events depends upon the concentration of dsDNA in the solution, as described previously [21]. Typically, the number of analyzed translocation events ranged from 2000-20000 from at least 5 independent experiments to obtain statistically significant data (Suppl. Fig. 2).
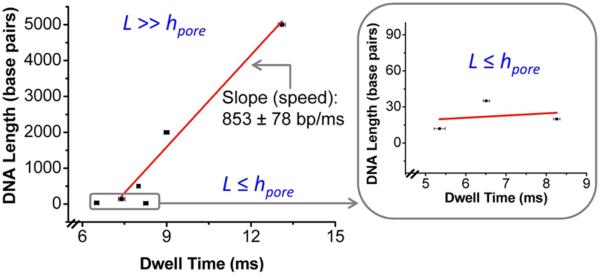
Two regimes of translocation events were observed: For shorter dsDNA (12, 20, and 35 bp dsDNA) that are less or around the length of the pore, hpore (7.5 nm), translocation dynamics is dominated by interactions of the DNA with the pore wall, and longer dwell times relative to their short length are observed (Fig. 3, boxed). For dsDNA longer than the length of the connector (141, 500, 2000 and 5000-bp), dwell time was observed to increase with length (Fig. 3). Similar trend was obtained at lower voltage of -40 mV.
Figure 3. Mean dwell time as a function of dsDNA length.
The mean dwell time for each dsDNA was obtained using an exponential model with density function. Data showing two classes of events for short dsDNA lengths (12, 20 and 35 bp) (boxed) and dsDNA much longer than the length of the connector (141, 500, 2000 and 5000 bp). Slower speed is observed for short length dsDNA (boxed), whereas a constant speed (slope) is observed for longer dsDNA. Applied voltage: -75 mV; error bars represent standard deviation; red line: linear fit.
From the dwell time, we calculated the average speed from the slope of the L vs τD plot, where is the length of dsDNA in base pairs. The speed for L > > hpore was determined to be 853 ± 78 bp/ms. This is a simplified calculation and does not account for factors such as hydrodynamic interactions, screening effects, electro-osmotic flow, and viscosity. Slower speed (~2 bp/ms) was observed for shorter length dsDNA (L ≤ hpore), presumably due to significant DNA-pore interactions. For L > > hpore, a constant speed was observed. Similar trend was observed at a lower voltage of -40 mV.
Although the physical properties of ss- and ds-DNA are different, we note that our observation is different from the finding in α-hemolysin pores, where the length dependency on translocation speed was observed for short ssDNA (L<hpore). For longer ssDNA (L > > hpore), the speed was independent of length [47], which is also observed in the phi29 connector. This phi29 connector acts like a one-way valve for dsDNA translocation [21], which can account for the difference.
Speed of DNA translocating through nanopores is of paramount importance for DNA sequencing applications. For the phi29 connector channel, the average speed is ~0.85 bp/μs. For comparison, in synthetic pores with dsDNA translocation capability, ~35× faster speed (~30 bp/μs) has been reported [48,49]. Although ssDNA translocation speed in α-hemolysin (1 base/μs [50]) and MspA (~5 base/μs [23]) pores cannot be compared directly, it is interesting to note that the phi29 system exhibits slower translocation dynamics. Nevertheless, even more slower speed is necessary to electrically read the sequence of an intact DNA strand given the limits of current instrumentations [35].
3.3 Forces influencing the translocation of dsDNA through the phi29 connector
There are several physical theories on how polymers reduce ion current in nanopores [51-53]. DNA translocation is a complex interplay between five dominant forces[46,47,54,55] acting on dsDNA as it threads through the connector pore: electrical driving force, viscous drag force, hydrodynamic drag force, uncoiling/recoiling forces and DNA-pore interactions, mathematically expressed as : −Fdriving ≈ FViscous drag + Fhydrodynamic drag + Funcoilling/recoilling + FDNA–pore. Here we provide a crude estimate of these forces.
(a) Electrical driving force
The dsDNA is subjected to a driving force [46,47], which is expressed as Fdriving = 2eV2/a, where e is the elementary charge (dsDNA has a charge of −2e per base pair; e = 1.6 × 10−19 C); V is the Voltage; and a is the spacing between nucleotides (0.34 nm). The Fdriving at 75 mV applied potential is calculated to be ~70 pN. However, due to electrostatic screening effects of the counter-ions in the solution that move with the DNA, the effective charge on the dsDNA and the resulting Fdriving is expected to be reduced by 15 - 50% [55-57]. This value is independent of the length of dsDNA (L > > hpore), as the driving force is only exerted on the DNA in the direct vicinity of the pore. We further investigated the minimum voltage required to translocate long 2 kbp dsDNA through the connector. Below 15 mV, translocation is rarely observed and those that occur randomly diffuse through the pore (data not shown). So application of 15 mV is probably sufficient to overcome the enthalpic and entropic costs of squeezing long dsDNA through the nanoscale volume of the connector.
In our crude analysis, we assumed that dsDNA molecules are exposed to an uniform electric field within the channel, which may not be the case. The equipotential lines in the bulk solution at the entrance and exit of the pore are not straight and parallel to the membrane plane. The access resistance may play an important role by 'lengthening' the pore. The resulting nonuniform electric field at the two openings may substantially contribute to the translocation pattern by imposing a dielectrophoretic force over the electrophoretic one, which may significantly increase the dwell time for short dsDNA molecules (L ≤ hpore), which is observed in Fig. 3.
(b) Viscous drag force of dsDNA in the connector
In the absence of specific DNA-pore interactions, FViscous drag = 2πrηhporevlinear/(R − r) [46,54], where η is the solution viscosity 1.0 × 10−3 Pa · s); r is the radius of dsDNA (1 nm); hpore is the height of the connector channel (7.5 nm); vlinear is the velocity of dsDNA; and R is the radius of the connector at the narrowest constriction (1.8 nm); For L ≤ hpore such as, for 12 bp dsDNA, vlinear = 2.24 bp/ms, and FViscous drag is estimated to be 0.04 pN; whereas for L > > hpore such as, 5 kbp dsDNA, the FViscous drag can be as high as 12 pN.
(c) Hydrodynamic drag on the untranslocated segment of dsDNA
The untranslocated part of dsDNA can be approximated as a sphere with a the radius of gyration, Rg (for 5 kbp dsDNA, Rg ≈ 0.18 μm [58]). The hydrodynamic drag force [54] on the untranslocated portion of dsDNA can be expressed as Fhydrodynamic drag = 6πηRgvsphere , where, vsphereis the velocity at which the center of mass progresses towards the connector, (expressed as dRg/dt) as the dsDNA translocates through the channel. For L > > hpore , such as, 5 kbp dsDNA, Fhydrodynamic drag is estimated to be ~30 pN. This force component is not present in the case of shorter dsDNA (L ≤ hpore).
(d) Uncoiling and recoiling forces acting on dsDNA
As dsDNA exists as random coil in solution, it is entropically unfavorable to pull a dsDNA from solution into a nanopore. Previous theoretical calculations [55] yielded an entropic uncoiling force of ~1 pN to thread dsDNA through a 5 nm cylindrical pore. After translocation, the recoiling force has been estimated to be ~0.1 fN. For both cases, only the segment of dsDNA entering the nanopore contributes to the forces. This force component again is applicable to dsDNA much longer than the persistence length; however, the sum total of uncoiling and recoiling forces only contributes a small component of the total forces.
(e) Forces involved in DNA-pore interactions
Significant interactions are known to occur during the translocation process between the phosphate backbone of dsDNA and lysine residues of the connector channel [4,59]. Furthermore, the connector channel acts as a oneway valve with respect to dsDNA translocation [21]. Taken together, the FDNA–pore values are expected to contribute to the translocation processes. This is apparent in the L vs τD plot (Fig. 3). Much slower speed (~2 bp/ms) is observed for short length dsDNA (12, 20, and 35-bp) (Fig. 3, boxed). The contribution of viscous drag, hydrodynamic drag and uncoiling/recoiling forces is expected to be minimal at these length scales. Hence the DNA-pore interactions is likely to be the dominant factor contributing to the slower translocation speed observed for short dsDNA and slower speed in general compared to solid state pores with dsDNA translocation capability [48,49].
3.4 Translocation of folded dsDNA
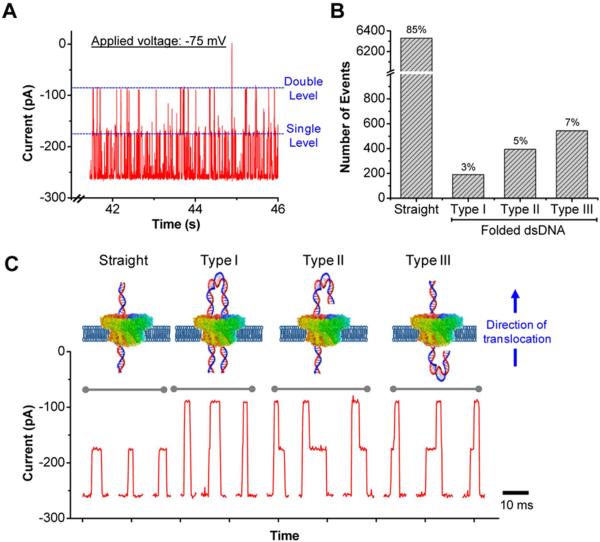
In solution DNA exists as a random coil. The persistence length of dsDNA, that is the length over which the DNA chain can fold back upon itself, is ~140 bp (~50 nm) in 1 M KCl buffer conditions [60]. Thus, it is conceivable that the DNA can translocate through the pore in a folded conformation, which has been observed in synthetic SiO2 nanopores of diameter 10 nm using λ-phage dsDNA [44,61-63] and graphene pores [64-66]. The phi29 channel is 3.6 nm at the narrowest constriction and only ~1/3 of the channel gets blocked by dsDNA (Fig. 2A-B). In presence of 5 kbp dsDNA, one or two plateaus at discrete current levels within a single translocation event are observed (Fig. 4), different from the single-level head-to-tail translocation of straight dsDNA (Fig. 2A). A MATLAB-based event sorting algorithm was used to classify the measured events into straight (head-to-tail) with characteristic ~32% current blockage, and double-level conformations with characteristic ~64% current blockage (Types I, II and III based on the random folding of leading and trailing edges). Quantitative analysis using a single channel in the membrane (to eliminate the possibility of simultaneous translocation events from multiple channels) revealed that ~15% of all the events accounted for the translocation of folded 5 kbp dsDNA whereby portions of the dsDNA folded upon itself and translocated in a doubled-up configurations. The capture of folded dsDNA is a stochastic process and the lower percentage of double-level events can be attributed to considerable energy barrier that is needed to overcome during the translocation. Similar events are observed, although less frequently with 500 bp dsDNA.
Figure 4. Translocation of folded 5 kbp dsDNA.
(A) Typical current trace showing the translocation of straight (single level) and folded dsDNA (double level). (B) Quantitative analysis of 7500 events showing the distribution of 5 kbp straight and folded dsDNA through a single channel in the membrane. (C) Illustration (top) and representative current signatures (bottom) of straight and folded (Types I, II and III) dsDNA translocating through a single connector channel in the membrane. For reference, the direction of translocation is from N- to C-terminal of the connector. Applied voltage: -75 mV.
The dwell time of folded dsDNA (i.e. double-level events only) at -75 mV was deduced to be 14.7 ± 2.2 ms, compared to 8.4 ± 0.1 ms for straight dsDNA. At -40 mV, even slower dynamics were observed: 22.5 ± 8.5 ms for folded dsDNA vs. 6.4 ± 0.1 ms for straight dsDNA. This is in contrast to the 10 nm SiO2 nanopores where near identical dwell times and corresponding velocities were observed for folded and straight dsDNA translocations [62]. While dsDNA can adopt a wide range of conformations in solution, the nature of the nanopore and more importantly, the dimensions of the pore can restrict a lot of these conformations of translocated dsDNA. In presence of two dsDNA strands, the strands repel each other to displace both strands from the pore axis. The electrical driving force and hydrodynamic drag forces are the same for linear and folded dsDNA; however, the viscous drag force and associated DNA-pore interactions may result in a smaller net force exerted on folded dsDNA conformations resulting in increased translocation time observed in our case.
3.5 Co-translocation of two dsDNA side by side (tetra-stranded DNA) through the connector
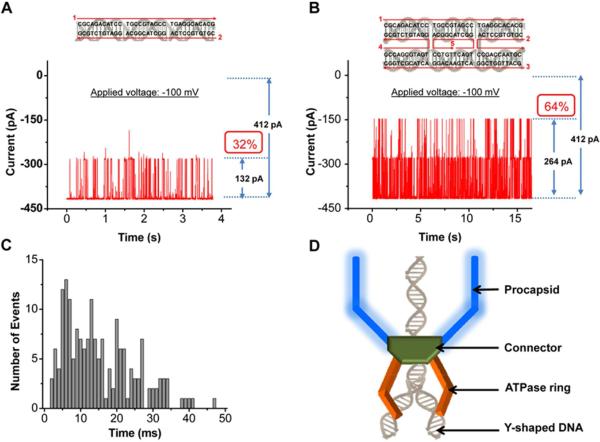
We have previously shown that circular DNA cannot pass through the connector channel, presumably due to supercoiling effects [17]. Moreover, the observed translocation of dsDNA is a random process and does not conclusively demonstrate whether the connector channel is indeed capable of translocating two dsDNA side by side simultaneously. To further validate this folded dsDNA translocation phenomena, we constructed double-crossover (DX-DNA) tetra-stranded DNA structures [42] (Fig. 5B). In presence of a single channel in the membrane, DXDNA was added; blockage events at the double-level (~64% pore current blockage) with longer and broader translocation events were observed, consistent with the cross-sectional areas of the DX-DNA and connector (Fig. 5B). Restricting the analysis to the subset of double-level DXDNA translocation events only, the dwell time was deduced to be 15.5 ± 1.5 ms (Fig. 5C), which is ~2.5× higher than one double strand 35 bp of DX-DNA. DsDNA is a helix with major and minor groove. Folded dsDNA as well tetra-stranded DNA (diameter of 3.4 - 4 nm) display certain degree of flexibility and there is possibility of stacking of the two dsDNA helices aligning with the major and minor groove, which results in a reduced effective cross-sectional diameter. Both folded dsDNA and tetra-stranded DNA physically only blocks 64% of the available channel area.
Figure 5. Translocation of DX-DNA structures.
(A) Schematic and 2D sequence of 35 bp dsDNA. Typical current trace showing the translocation of 35 bp dsDNA showing single-level events. (B) Schematic and 2D sequences of double crossover DNA (DX-DNA) [42]. Typical current trace showing the translocation of DX-DNA showing double-level events. (C) Dwell time distribution of DX-DNA translocation events (500 events). Applied voltage: -100 mV. (D) Model of DNA crunching and compression observed in T4 bacteriophage using Y-shaped branched DNA substrate [67].
From dsDNA packaging perspective, the results agree with the DNA crunching and compression phenomena observed in bacteriophage T4 [67] (Fig. 5D). The authors used Y-shaped branched DNA as substrate for DNA packaging and analyzed DNA entry into procapsid by FRET (Fluorescence Resonance Energy Transfer). With the distal dsDNA part composed of two dsDNA (tetra-stranded), FRET efficiency was observed to increase during DNA packaging. Although the authors could not confirm whether the tetra-stranded DNA passed through the connector channel, the observation of T4 crunching agrees with the phi29 “push-through a one way valve” mechanism reported previously [4,21,68]. At the very least, the tetra-stranded component of the Y-shaped DNA had entered the channel ring composed of T4 gp17 ATPase, otherwise, the FRET signal would not have increased and the crunching and compression of DNA substrate could not have occurred.
The finding of translocation of dsDNA with 32% blockage and tetra-stranded DNA with 64% blockage is in consent with the recently elucidated mechanism of viral DNA packaging via a revolution mechanism without rotation [68-71]. If rotation mechanism would have been applied, the channel size that is close to the size of the 2 nm dsDNA would have been observed. If revolution mechanism is applied, it requires a channel larger than the dsDNA diameter of 2 nm to provide additional room for dsDNA to revolve. It was demonstrated here that the phi29 connector DNA packaging channel is three times wider than the dsDNA that only blocked 32% of the channel, supporting the revolution mechanism.
4. Conclusions
In summary, we used a set of parameters to characterize the translocation dynamics of linear and folded dsDNA translocation through the membrane-embedded robust phi29 nanopore. Significantly slower translocation speed is observed for phi29 connector compared to synthetic nanopores with dsDNA translocation capability. We demonstrated for the first time the ability of the protein nanopores and in general nanopores smaller than 5 nm to distinguish folded dsDNA conformations. The results provided fundamental insights into the key factors governing the electrophoretic transport of dsDNA through the connector nanochannel and has provided a platform to explore complex biophysical processes involved in DNA packaging and transport, as well as structural studies of biological macromolecular complexes at the single molecule level in the future.
Supplementary Material
Acknowledgements
We thank Dr. Hui Zhang and Dr. Mario Vieweger for help with the data analysis. The work was supported by NIH grant EB012135 (P.G.) and, National Cancer Institute Cancer Center Support Grant P30CA177558. Funding to Peixuan Guo's Endowed Chair in Nanobiotechnology position is from the William Fairish Endowment Fund. Peixuan Guo is a co-founder of Kylin Therapeutics, Inc., and Biomotor and RNA Nanotechnology Development Corp. Ltd.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplementary data related to this article are available.
REFERENCES
- 1.Neher E, Stevens CF. Conductance fluctuations and ionic pores in membranes. Annu Rev Biophys Bioeng. 1977;6:345–81. doi: 10.1146/annurev.bb.06.060177.002021. [DOI] [PubMed] [Google Scholar]
- 2.Doyle DA, Morais CJ, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–29. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Z, Khisamutdinov E, Schwartz C, Guo P. Mechanism of one-way traffic of hexameric phi29 DNA packaging motor with four electropositive relaying layers facilitating anti-parallel revolution. ACS Nano. 2013;7:4082–92. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz C, De Donatis GM, Fang H, Guo P. The ATPase of the phi29 DNA-packaging motor is a member of the hexameric AAA+ superfamily. Virology. 2013;443:20–7. doi: 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi29. J Mol Biol. 1987;197:229–36. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 7.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–92. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Hwang Y, Catalano CE, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage lambda. Biochemistry. 1996;35:2796–803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 9.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93(4):L17–L19. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meifer WJJ, Horcajadas JA, Salas M. Phi29 family of phages. Microbiol Mol Biol Rev. 2001;65(2):261–87. doi: 10.1128/MMBR.65.2.261-287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi29 DNA. Science. 1987;236:690–4. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 12.Guo P, Zhang C, Chen C, Trottier M, Garver K. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–55. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 13.Guo P, Lee TJ. Viral nanomotors for packaging of dsDNA and dsRNA. Mol Microbiol. 2007;64:886–903. doi: 10.1111/j.1365-2958.2007.05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun S, Rao VB, Rossmann MG. Genome packaging in viruses. Curr Opin Struct Biol. 2010;20(1):114–20. doi: 10.1016/j.sbi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, et al. Structure determination of the head-tail connector of bacteriophage phi29. Acta Cryst. 2001;D57:1260–9. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- 16.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, et al. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J Mol Biol. 2002;315(4):663–76. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 17.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, et al. Translocation of double-stranded DNA through membrane-adapted phi29 motor protein nanopores. Nat Nanotechnol. 2009;4:765–72. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haque F, Geng J, Montemagno C, Guo P. Incorporation of Viral DNA Packaging Motor Channel in Lipid Bilayers for Real-Time, Single-Molecule Sensing of Chemicals and Double-Stranded DNA. Nat Protoc. 2013;8:373–92. doi: 10.1038/nprot.2013.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing P, Haque F, Vonderheide A, Montemagno C, Guo P. Robust Properties of Membrane-Embedded Connector Channel of Bacterial Virus Phi29 DNA Packaging Motor. Mol Biosyst. 2010;6:1844–52. doi: 10.1039/c003010d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang H, Jing P, Haque F, Guo P. Role of channel Lysines and “Push Through a Oneway Valve” Mechanism of Viral DNA packaging Motor. Biophysical Journal. 2012;102:127–35. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-Way Traffic of a Viral Motor Channel for Double-Stranded DNA Translocation. Nano Lett. 2010;10:3620–7. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93(24):13770–3. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci USA. 2008;105:20647–52. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng J, Wang S, Fang H, Guo P. Channel size conversion of Phi29 DNA-packaging nanomotor for discrimination of single- and double-stranded nucleic acids. ACS Nano. 2013;7(4):3315–23. doi: 10.1021/nn400020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng J, Fang H, Haque F, Zhang L, Guo P. Three reversible and controllable discrete steps of channel gating of a viral DNA packaging motor. Biomaterials. 2011;32:8234–42. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haque F, Guo P. Membrane-embedded Channel of Bacteriophage Phi29 DNA-Packaging Motor for Translocation and Sensing of Double-stranded DNA. In: Iqbal SM, Bashir R, editors. Nanopores, Sensing and Fundamental Biological Interactions. Springer; 2011. pp. 77–106. [Google Scholar]
- 27.Haque F, Lunn J, Fang H, Smithrud D, Guo P. Real-Time Sensing and Discrimination of Single Chemicals Using the Channel of Phi29 DNA Packaging Nanomotor. ACS Nano. 2012;6:3251–61. doi: 10.1021/nn3001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Haque F, Rychahou PG, Evers BM, Guo P. Engineered Nanopore of Phi29 DNA-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano. 2013;7:9814–22. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibanez C, Garcia JA, Carrascosa JL, Salas M. Overproduction and purification of the connector protein of Bacillus subtilis phage f29. Nucleic Acids Res. 1984;12:2351–65. doi: 10.1093/nar/12.5.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MA, Wood JP, Capaldi SA, Baron AJ, Gell C, Smith DA, et al. Affinity of molecular interactions in the bacteriophage phi29 DNA packaging motor. Nucleic Acids Res. 2006;34:2698–709. doi: 10.1093/nar/gkl318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Y, Blocker F, Xiao F, Guo P. Construction and 3-D computer modeling of connector arrays with tetragonal to decagonal transition induced by pRNA of phi29 DNA-packaging motor. J Nanosci Nanotechnol. 2005;5:856–63. doi: 10.1166/jnn.2005.143. [DOI] [PubMed] [Google Scholar]
- 32.Xiao F, Sun J, Coban O, Schoen P, Wang JC, Cheng RH, et al. Fabrication of Massive Sheets of Single Layer Patterned Arrays Using Lipid Directed Reengineered Phi29 Motor Dodecamer. ACS Nano. 2009;3:100–7. doi: 10.1021/nn800409a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, Xiao F, Guo P. The effect of N- or C-terminal alterations of the connector of bacteriophage phi29 DNA packaging motor on procapsid assembly, pRNA binding, and DNA packaging. Nanomedicine. 2008;4:8–18. doi: 10.1016/j.nano.2007.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haque F, Li J, Wu H-C, Liang X-J, Guo P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nature Nanotechnology. 2011;6:615–24. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 37.Healy K. Nanopore-based single-molecule DNA analysis. Nanomedicine. 2007;2:459–81. doi: 10.2217/17435889.2.4.459. [DOI] [PubMed] [Google Scholar]
- 38.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Current Opinion in Biotechnology. 2010;21(4):439–76. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasianowicz JJ, Robertson JW, Chan ER, Reiner JE, Stanford VM. Nanoscopic porous sensors. Annu Rev Anal Chem (Palo Alto Calif ) 2008;1:737–66. doi: 10.1146/annurev.anchem.1.031207.112818. [DOI] [PubMed] [Google Scholar]
- 40.Howorka S, Siwy Z. Nanopore analytics: sensing of single molecules. Chem Soc Rev. 2009;38(8):2360–84. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 41.Reiner JE, Balijepalli A, Robertson JW, Campbell J, Suehle J, Kasianowicz JJ. Disease detection and management via single nanopore-based sensors. Chem Rev. 2012;112(12):6431–51. doi: 10.1021/cr300381m. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Yang X, Qi J, Seeman N. Antiparallel DNA Double Crossover Molecules As Components for Nanoconstruction. J Am Chem Soc. 1996;118:6131–40. [Google Scholar]
- 43.Henrickson SE, DiMarzio EA, Wang Q, Stanford VM, Kasianowicz JJ. Probing single nanometer-scale pores with polymeric molecular rulers. J Chem Phys. 2010;132(13):135101. doi: 10.1063/1.3328875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nature Mater. 2003;2:611–5. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 45.Balijepalli A, Ettedgui J, Cornio AT, Robertson JW, Cheung KP, Kasianowicz JJ, et al. Quantifying short-lived events in multistate ionic current measurements. ACS Nano. 2014;8(2):1547–53. doi: 10.1021/nn405761y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lubensky DK, Nelson DR. Driven polymer translocation through a narrow pore. Biophys J. 1999;77(4):1824–38. doi: 10.1016/S0006-3495(99)77027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meller A, Nivon L, Branton D. Voltage-driven DNA translocations through a nanopore. Phys Rev Lett. 2001;86(15):3435–8. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 48.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5(9):1734–7. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wanunu M, Sutin J, McNally B, Chow A, Meller A. DNA translocation governed by interactions with solid state nanopores. Biophys J. 2008;95:4716–25. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97(3):1079–84. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson JWF, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc Natl Acad Sci USA. 2007;104(20):8207–11. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JW. Theory for polymer analysis using nanopore-based single-molecule mass spectrometry. Proc Natl Acad Sci USA. 2010;107(27):12080–5. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balijepalli A, Robertson JW, Reiner JE, Kasianowicz JJ, Pastor RW. Theory of polymer-nanopore interactions refined using molecular dynamics simulations. J Am Chem Soc. 2013;135(18):7064–72. doi: 10.1021/ja4026193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storm AJ. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–7. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 55.Chen L, Conlisk AT. Forces affecting double-stranded DNA translocation through synthetic nanopores. Biomed Microdevices. 2011;13(2):403–14. doi: 10.1007/s10544-011-9509-7. [DOI] [PubMed] [Google Scholar]
- 56.Padmanabhan S, Richey B, Anderson CF, Record MT., Jr. Interaction of an N-methylated polyamine analogue, hexamethonium(2+), with NaDNA: quantitative 14N and 23Na NMR relaxation rate studies of the cation-exchange process. Biochemistry. 1988;27(12):4367–76. doi: 10.1021/bi00412a025. [DOI] [PubMed] [Google Scholar]
- 57.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Driven DNA transport into an asymmetric nanometer scale pore. Phys Rev Lett. 2000;85:3057–60. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 58.Smith DE, Perkins TT, Chu S. Dynamical scaling of DNA diffusion coefficients. Macromolecules. 1996;29(4):1372–3. [Google Scholar]
- 59.Guo P, Schwartz C, Haak J, Zhao Z. Discovery of a new motion mechanism of biomotors similar to the earth revolving around the sun without rotation. Virology. 2013;446:133–43. doi: 10.1016/j.virol.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc Natl Acad Sci USA. 1997;94(12):6185–90. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen P, Gu JJ, Brandin E, Kim YR, Wang Q, Branton D. Probing single DNA molecule transport using fabricated nanopores. Nano Letters. 2004;4:2293–8. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E. 2005;71(5 Pt 1):051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 63.Fologea D, Gershow M, Ledden B, McNabb DS, Golovchenko JA, Li J. Detecting single stranded DNA with a solid state nanopore. Nano Lett. 2005;5(10):1905–9. doi: 10.1021/nl051199m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467(7312):190–3. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10(8):2915–21. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 66.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LM, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–7. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 67.Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398(2):224–32. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwartz C, De Donatis GM, Zhang H, Fang H, Guo P. Revolution rather than rotation of AAA+ hexameric phi29 nanomotor for viral dsDNA packaging without coiling. Virology. 2013;443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo P, Grainge I, Zhao Z, Vieweger M. Two classes of nucleic acid translocation motors: rotation and revolution without rotation. Cell Biosci. 2014;4(1):54. doi: 10.1186/2045-3701-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo P. Biophysical Studies Reveal New Evidence for One-Way Revolution Mechanism of Bacteriophage phi29 DNA Packaging Motor. Biophysical Journal. 2014;106(9):1837–8. doi: 10.1016/j.bpj.2014.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.De-Donatis G, Zhao Z, Wang S, Huang PL, Schwartz C, Tsodikov VO, et al. Finding of widespread viral and bacterial revolution dsDNA translocation motors distinct from rotation motors by channel chirality and size. Cell Biosci. 2014;4:30. doi: 10.1186/2045-3701-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.