Abstract
Adult-onset autosomal dominant leukodystrophy (ADLD) is a slowly progressive neurological disorder characterized by autonomic dysfunction, followed by cerebellar and pyramidal features. ADLD is caused by duplication of the lamin B1 gene (LMNB1), which leads to its increased expression. The molecular pathways involved in the disease are still poorly understood. Hence, we analyzed global gene expression in fibroblasts and whole blood of LMNB1 duplication carriers and used Gene Set Enrichment Analysis to explore their gene signatures. We found that LMNB1 duplication is associated with dysregulation of genes involved in the immune system, neuronal and skeletal development. Genes with an altered transcriptional profile clustered in specific genomic regions. Among the dysregulated genes, we further studied the role of RAVER2, which we found to be overexpressed at mRNA and protein level. RAVER2 encodes a putative trans regulator of the splicing repressor polypyrimidine tract binding protein (PTB) and is likely implicated in alternative splicing regulation. Functional studies demonstrated an abnormal splicing pattern of several PTB-target genes and of the myelin protein gene PLP1, previously demonstrated to be involved in ADLD. Mutant mice with different lamin B1 expression levels confirmed that Raver2 expression is dependent on lamin B1 in neural tissue and determines an altered splicing pattern of PTB-target genes and Plp1. Overall our results demonstrate that deregulation of lamin B1 expression induces modified splicing of several genes, likely driven by raver-2 overexpression, and suggest that an alteration of mRNA processing could be a pathogenic mechanism in ADLD.
Introduction
Adult-onset autosomal dominant leukodystrophy (ADLD) is a slowly progressive demyelinating disease. Symptoms appear between the fourth and sixth decades of life and include autonomic dysfunction (bowel/bladder dysfunction, impotence, orthostatic hypotension and decreased sweating), followed by cerebellar (ataxia, dysmetria, nystagmus and action tremors) and pyramidal (spasticity and weakness in extremities) features. Mild cognitive, visual and auditory impairments have also been observed in some cases (1). ADLD is caused by the duplication of the lamin B1 (LMNB1) gene, resulting in increased expression of LMNB1 mRNA and protein (2,3). In one family with ADLD, there was no duplication of LMNB1, but its overexpression may be associated with a mutation in a regulatory region of the gene (4).
Lamin B1 is an essential protein during development in Drosophila melanogaster, Caenorhabditis elegans and mammalian cells (5–7). Lmnb1-deficient (Lmnb1Δ/Δ) mice display impaired embryonic development and die at birth due to lung and heart failure (7). Proper expression of Lmnb1 is crucial for brain development, and its deficiency results in a reduced brain size and impaired corticogenesis (7–9). Indeed, in the mouse brain, the level of lamin B1 protein varies during neurogenesis and neuronal differentiation (10) and peaks at birth (11). Lamin B1 levels are also important for maintaining normal cerebral function during adulthood. Overexpression of lamin B1 reduces myelination and induces neuronal axon degeneration in adult mice (12), consistent with the adult-onset phenotype of ADLD in humans.
Lamin B1 is a component of the nuclear lamina, a protein meshwork underlying the inner nuclear membrane, which contributes to the size, shape and stability of the nucleus (13). Its overexpression, for example, in ADLD cells, alters nuclear mechanics and ionic permeability (14). The nuclear lamina also regulates gene expression by acting as a platform that tethers chromatin at lamina-associated domains (LADs) and contributing to the spatial organization of chromosomes within the nucleus (15). In Lmnb1Δ/Δ mouse embryonic fibroblasts (MEFs), the lamin B1 deficiency induces differential expression of genes clustered in specific chromosomal locations unrelated to their function, due to a reorganization of chromosome positioning relative to the nuclear lamina (16). In oligodendrocytes purified from a transgenic mouse model overexpressing Lmnb1, the transcriptome is characterized by dysregulation of several genes, most of which are involved in myelin formation and maintenance (12). Among these genes, the proteolipid protein 1 gene (Plp1) shows downregulated expression, due to reduced occupancy of the transcription factor Yin Yang 1 (Yy1) at the promoter region (12). Nonetheless, how the pathological overexpression of lamin B1 influences gene expression in ADLD, and to what extent, remains unclear.
To address this fundamental question, we investigated how lamin B1 levels impact the whole-genome expression profile in tissues derived from ADLD patients. Overall, our results suggest that deregulation of lamin B1 induces modified splicing of several genes, which is at least partially driven by the enhanced expression of raver-2, a RNA-binding protein (RBP) that modulates the splicing repressor polypyrimidine tract binding protein (PTB) (17–20).
Results
Lamin B1 overexpression is associated with an altered transcriptional profile in whole blood and skin fibroblasts
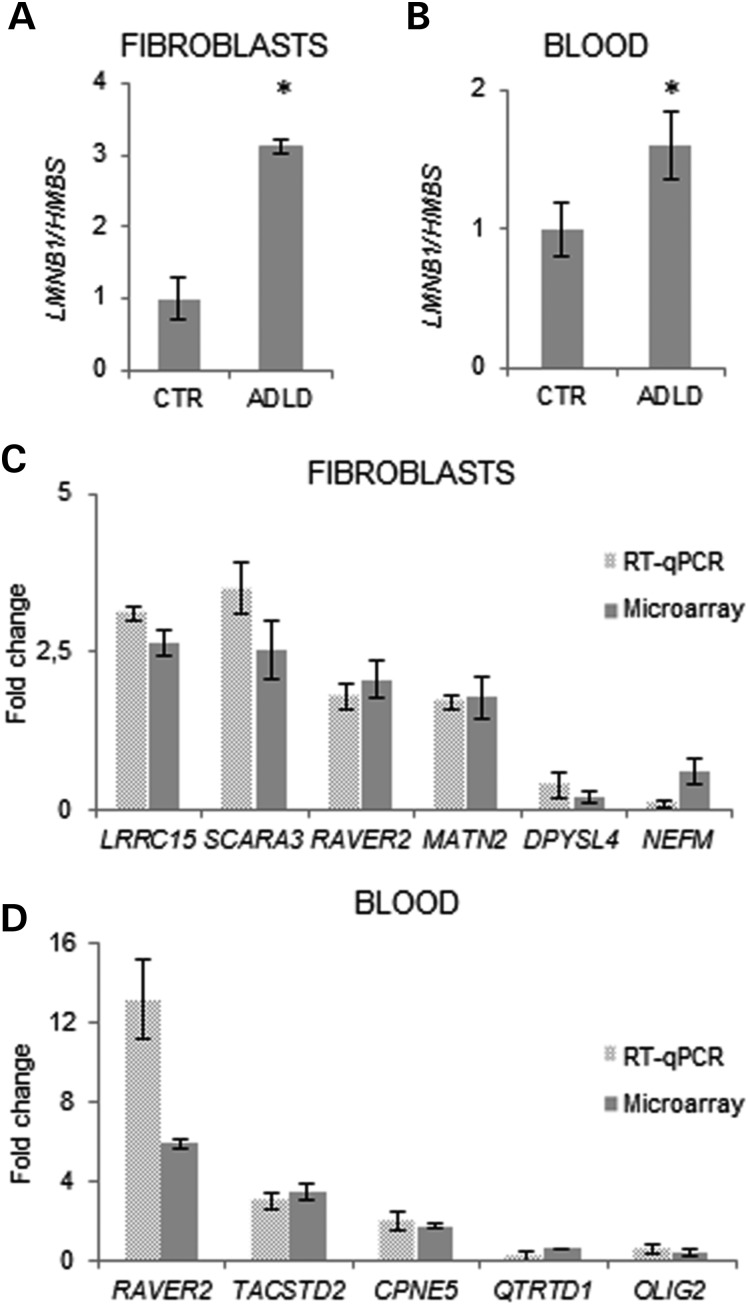
To investigate non-cell-type-specific, pathologically relevant pathways altered by LMNB1 overexpression, we performed whole-genome RNA profiling on human skin fibroblasts and whole blood of subjects with LMNB1 duplications versus controls. In mutated subjects, LMNB1 overexpression was previously verified by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR; Fig. 1A and B). We found 169 differentially expressed genes (DEGs) in ADLD fibroblasts (94 upregulated and 75 downregulated) and 485 DEGs in whole blood (256 upregulated and 229 downregulated; Supplementary Material, Tables S1 and S2). Microarray results were validated by RT-qPCR (Fig. 1C and D).
Figure 1.
Transcriptional signature of ADLD. (A and B) LMNB1 mRNA quantification in ADLD fibroblasts (A) and whole blood (B) relative to control samples (CTR). LMNB1 mRNA levels were normalized to HMBS mRNA (hydroxymethylbilane synthase gene) expression. Bars represent the mean ± SD of values obtained from fibroblasts (ADLD, n = 4; CTR, n = 6) or blood (ADLD, n = 4; CTR, n = 8); *P < 0.05, Student's t-test. (C and D) Validation of gene expression obtained from microarray data by RT-qPCR. Gene expression in ADLD (n = 4) fibroblasts (C) or blood (D). Results from microarray and RT-qPCR are shown. Bars represent the average fold change over CTR ± SD. Six/eight CTRs were used for analyses on fibroblasts and blood, respectively.
To identify common molecular changes in the two cell types, we compared fibroblasts and blood to identify shared DEGs. Three genes were deregulated in both tissues: the ribonucleoprotein PTB-binding 2 (RAVER2) and the solute carrier family 39, member 11 (SLC39A11) genes were upregulated in both tissues, while AF4/FMR2 family, member 3 (AFF3) was downregulated in whole blood and upregulated in fibroblasts (Supplementary Material, Tables S3).
Overall, these results indicate that lamin B1 overexpression is associated with dysregulation of several genes in whole blood or fibroblasts of ADLD patients, which may be relevant to pathology.
LMNB1 duplication alters expression of genes involved in different biological pathways
To gain insights into biological processes affected in ADLD, we performed an enrichment analysis of Gene Ontology categories using Gene Set Enrichment Analysis (GSEA) on pooled transcriptomes of ADLD versus CTR tissues (21). Consistent with previous findings, we identified the following lamin B1-dependent biological processes: cell cycle, chromosome segregation, response to oxidative stress and nervous system development (Table 1 and Supplementary Material, Tables S4–S6) (8,16,22,23). Moreover, we found that LMNB1 overexpression preferentially affects genes involved in the immune system response, skeletal development and cytoskeleton organization. Three gene sets enriched in ADLD tissues are related to RNA metabolism (tRNA metabolic process, rRNA processing and positive regulation of transcription). Moreover, RAVER2 and AFF3 are involved in the regulation of alternative splicing (19,20,24), suggesting that lamin B1 affects RNA processing.
Table 1.
Gene ontology categories altered in ADLD tissues
| GO | Name | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|
| Biological process | ||||
| GO:0006952 | Defense response | −2.34 | 0.000 | 0.000 |
| GO:0022402 | Cell cycle process | 2.12 | 0.000 | 0.001 |
| GO:0002376 | Immune system process | −2.30 | 0.000 | 0.001 |
| GO:0001501 | Skeletal development | 1.94 | 0.000 | 0.011 |
| GO:0000226 | Microtubule cytoskeleton organization and biogenesis | 1.94 | 0.000 | 0.011 |
| GO:0031589 | Cell substrate adhesion | 1.88 | 0.002 | 0.016 |
| GO:0006399 | tRNA metabolic process | 1.88 | 0.000 | 0.016 |
| GO:0008610 | Lipid biosynthetic process | 1.86 | 0.000 | 0.018 |
| GO:0006457 | Protein folding | 1.80 | 0.002 | 0.023 |
| GO:0006865 | Amino acid transport | 1.82 | 0.002 | 0.024 |
| Cellular component | ||||
| GO:0005819 | Spindle | 2.50 | 0.000 | 0.0 |
| GO:0005578 | Proteinaceous extracellular matrix | 2.33 | 0.000 | 0.0 |
| GO:0015630 | Microtubule cytoskeleton | 2.10 | 0.000 | 0.002 |
| GO:0005694 | Chromosome | 2.06 | 0.000 | 0.002 |
| GO:0005783 | Endoplasmatic reticulum | 2.04 | 0.000 | 0.002 |
| GO:0005605 | Basal lamina | 2.01 | 0.000 | 0.002 |
| GO:0000313 | Ribosome | 1.95 | 0.000 | 0.004 |
| GO:0005777 | Peroxisome | 1.86 | 0.002 | 0.006 |
| GO:0005739 | Mitochondrion | 1.83 | 0.000 | 0.007 |
| GO:0042175 | Nuclear outer membrane-endoplasmic reticulum membrane network | 1.81 | 0.000 | 0.009 |
| Molecular function | ||||
| GO:0008378 | Galactosyltransferase activity | 2.14 | 0.000 | 0.003 |
| GO:0004714 | Transmembrane receptor protein tyrosine kinase activity | 1.89 | 0.000 | 0.031 |
| GO:0003777 | Microtubule motor activity | 1.88 | 0.000 | 0.029 |
| GO:0016616 | Oxidoreductase activity | 1.83 | 0.000 | 0.027 |
| GO:0015171 | Amino acid transmembrane transporter activity | 1.80 | 0.000 | 0.039 |
| GO:0005201 | Extracellular matrix structural constituent | 1.77 | 0.006 | 0.039 |
| GO:0008168 | Methyltransferase activity | 1.75 | 0.002 | 0.050 |
| GO:0003682 | Chromatin binding | 1.74 | 0.002 | 0.049 |
| GO:0008484 | Sulfuric ester hydrolase activity | 1.71 | 0.014 | 0.056 |
| GO:0016741 | Transferase activity transferring one carbon groups | 1.70 | 0.004 | 0.058 |
NES, normalized enrichment score (analyzed using GO gene sets in the GSEA software); NOM p-val, nominal P-value; FDR q-val, false discovery rate, nominal P-value corrected for multiple hypotheses testing.
Top 10 GO gene sets altered in ADLD tissues are shown. No redundant gene ontology terms were obtained by using REVIGO bioinformatics tools.
LMNB1 duplication affects regulation of gene expression in specific genomic regions
Lamin B1 interacts with chromatin and determines chromosomal positioning in the mitotic spindle, regulating the repression of selected genes (15,16). Accordingly we found that chromosome, spindle and chromatin binding gene sets are enriched in ADLD tissues (Table 1). It was previously demonstrated that lamin B1 deficiency induces the upregulation of clusters of genes on specific chromosomes through their mislocalization (16). Therefore, to investigate if the overexpression of lamin B1 affects gene expression in specific genomic regions, we performed a positional gene set analysis using GSEA (21). We found that the deregulated genes statistically clustered in 19 cytobands (FDR <0.25 (21); Table 2). Three ADLD enriched gene sets were located on chromosome 1 (1p31, 1p32 and 1p34), suggesting that this chromosome may be relocated far from the nuclear lamina. In agreement with previous findings (16), these data support the view that changes in lamin B1 expression affect gene transcription by chromosome repositioning.
Table 2.
Chromosome enrichment identified regions that contain groups of genes that preferentially are increased or decreased in ADLD patients
| ADLD increased |
ADLD decreased |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CHR | Cytoband | NES | NOM p-val | FDR q-val | CHR | Cytoband | NES | NOM p-val | FDR q-val |
| 1 | 1p31 | 1.74 | 0.002 | 0.099 | 1 | 1q44 | −1.61 | 0.006 | 0.137 |
| 1p32 | 1.59 | 0.002 | 0.220 | 4 | 4p12 | −1.58 | 0.026 | 0.160 | |
| 1p34 | 1.60 | 0.000 | 0.230 | 4p14 | −1.51 | 0.035 | 0.245 | ||
| 2 | 2q13 | 1.81 | 0.002 | 0.223 | 4q13 | −1.65 | 0.000 | 0.109 | |
| 2q34 | 1.78 | 0.002 | 0.111 | 6 | 6p22 | −1.77 | 0.004 | 0.007 | |
| 2p23 | 1.53 | 0.015 | 0.247 | 6p23 | −1.80 | 0.004 | 0.152 | ||
| 6 | 6p25 | 1.75 | 0.002 | 0.118 | 7 | 7q36 | −1.67 | 0.000 | 0.109 |
| 9 | 9p21 | 1.80 | 0.000 | 0.136 | |||||
| 11 | 11p13 | 1.56 | 0.028 | 0.200 | |||||
| 11q13 | 1.59 | 0.000 | 0.200 | ||||||
| 15 | 15q22 | 1.59 | 0.008 | 0.190 | |||||
| 15q23 | 1.59 | 0.019 | 0.175 | ||||||
NES, normalized enrichment score (analyzed using GO gene sets in the GSEA software); NOM p-val, nominal P-value; FDR q-val, false discovery rate, nominal P-value corrected for multiple hypotheses testing. Only gene sets with FDR q-val <0.25 and NOM p-val <0.05 are shown.
Positional gene set analysis was performed by using GSEA.
RAVER2 expression varies as a function of lamin B1 levels in human tissues and mouse embryonic brain
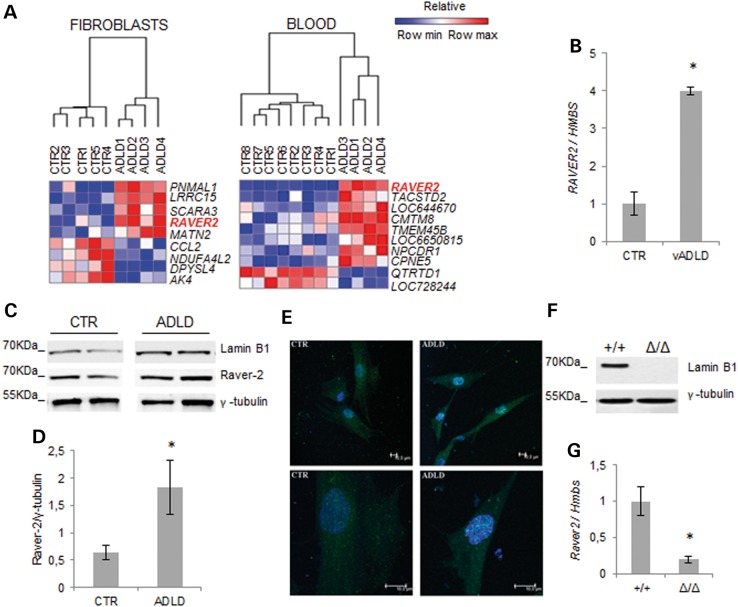
Using stringent selection criteria (correction for multiple analyses, FDR < 0.05), nine DEGs in ADLD fibroblasts and ten DEGs in ADLD blood were found deregulated. Among these, RAVER2 was the only gene deregulated in both tissues (Fig. 2A). To exclude interference of other genetic variants co-segregating with the LMNB1 duplication in our family, we confirmed RAVER2 mRNA overexpression also in fibroblasts from one patient with a variant form of ADLD (vADLD), characterized by lamin B1 overexpression in the absence of LMNB1 duplication (4) (Fig. 2B).
Figure 2.
RAVER2 expression varies as a function of lamin B1 levels. (A) HeatMap of DEGs with statistically significant changes (FDR-adjusted P-value smaller than 0.05) in fibroblasts (ADLD, n = 4; CTR, n = 5) and blood (ADLD, n = 4; CTR, n = 8). The GENE-E software (http://www.broadinstitute.org/cancer/software/GENE-E/) was used to generate the heatmaps. (B) Quantitative analysis of RAVER2 mRNA in vADLD and CTR fibroblast samples. The RAVER2 mRNA levels were normalized to HMBS mRNA expression. Bars represent the mean relative expression ± SD of one vADLD (two different batches of cells from the same donor) versus fibroblasts from six CTRs. *P < 0.05, Student's t-test. (C) Representative western blot of lamin B1 and raver-2 in fibroblasts from two ADLD patients and two CTRs. γ-Tubulin was used as loading control. (D) Quantitative analysis of raver-2 protein levels in fibroblasts from ADLD patients (n = 4) and age-matched CTR subjects (n = 4). The data are normalized to γ-tubulin and expressed as percentages of CTR levels. Bars represent average ± SEM. *P < 0.05, Mann–Whitney Rank Sum test. (E) Representative confocal images of primary fibroblasts from one ADLD patient and one age-matched CTR immunodecorated against raver-2 (green). Nuclei are counterstained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride. Scale bars: 10 µm. (F) Representative western blot of lamin B1 protein expression in brain of Lmnb1+/+ and Lmnb1Δ/Δ E18.5 embryos. (G) Raver2 mRNA expression in brain of E18.5 mouse embryos analyzed by RT-qPCR. *P < 0.05, Student's t-test.
To investigate whether raver-2 protein was also increased, we performed western blot analysis. Raver-2 protein was indeed upregulated in ADLD fibroblasts (Fig. 2C and D). Raver-2 immunoreactivity mainly localized in discrete nuclear clusters (Fig. 2E), as previously reported (19).
To directly test whether RAVER2 expression is regulated by lamin B1, we analyzed brains from E18 Lmnb1+/+ and Lmnb1Δ/Δ embryos (Fig. 2F) (7). Raver2 mRNA expression in Lmnb1Δ/Δ mice was reduced to 20% of Lmnb1+/+ (P = 0.002; Fig. 2G), demonstrating that lamin B1 positively regulates RAVER2.
The FANTOM5 promoter-based expression atlas (25) indicates that human RAVER2, although ubiquitous (18,26), is highly expressed in the nervous system (Table 3), consistent with its dysregulation playing a role in ADLD pathogenesis. Together, these results support the view that lamin B1 levels modulate RAVER2 expression and that this regulation occurs in the mammalian central nervous system (CNS), the target organ of ADLD in humans.
Table 3.
Top 10 ontology terms associated with cell types expressing RAVER2 promoter
| Ontology-based sample term enrichment analysis | P-value |
|---|---|
| Organ system subdivision | 5.13e−16 |
| Ectoderm-derived structure | 6.92e−14 |
| Presumptive ectoderm | 6.92e−14 |
| Neuroectoderm | 1.13e−11 |
| Adult organism | 1.36e−10 |
| Nervous system | 4.07e−10 |
| Presumptive neural plate | 7.14e−10 |
| CNS | 9.34e−10 |
| Structure with developmental contribution from neural crest | 7.30e−09 |
| Digestive system | 1.88e−08 |
HUMAN p1@RAVER2 promoter expression identified by FANTOM5 interrogation.
The splicing pattern of PTB-target genes is altered in blood and fibroblasts from ADLD patients and brain of Lmnb1 Δ/Δ mouse embryos
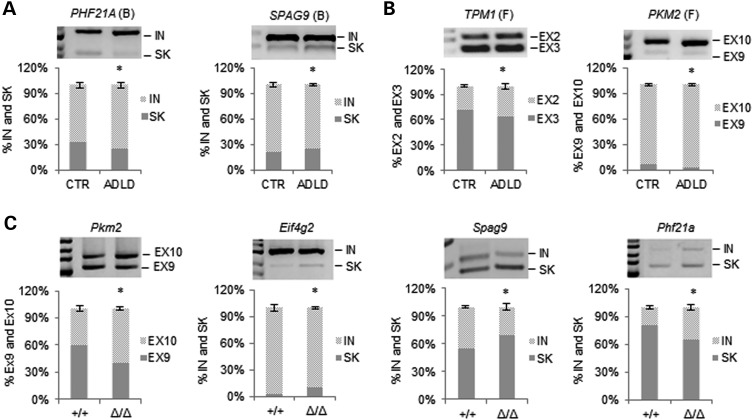
Raver-2 interacts with PTB (18–20), which is involved in all steps of mRNA metabolism and acts as repressor of alternatively spliced exons (27). To investigate potential changes in PTB-mediated splicing in ADLD, we examined a subset of previously characterized PTB-regulated exons (28–32) by semi-quantitative multiplex PCR and denaturant high performance liquid chromatography (dHPLC) (Table 4). In mRNA extracted from whole blood of LMNB1 duplication carriers, we found four abnormally spliced transcripts (PHF21A, SPAG9, PKM2 and GANAB) (Fig. 3A and Supplementary Material, Fig. S1). In ADLD fibroblasts, although RAVER2 expression was less increased than in ADLD whole blood (2- versus 13-fold change), we found three genes with altered splicing (TPM1, PKM2 and ACTN1) (Fig. 3B and Supplementary Material, Fig. S2).
Table 4.
PTB-regulated splicing events considered in this study
| Gene | Exon | Type of AS | Regulated eventa | Publication |
|---|---|---|---|---|
| TPM1 | 2/3 | Mutually exclusive | 2 | Gromak et al. (28) |
| PKM2 | 9/10 | Mutually exclusive | 10 | Xue et al. (29) |
| ACTN1 | NM/SM | Mutually exclusive | NM | Matlin et al. (30) |
| GANAB | 6 | Cassette | SK | Spellman et al. (31) |
| PHF21A | 14 | Cassette | IN | Xue et al. (32) |
| SPAG9 | 4 | Cassette | SK | Xue et al. (29) |
| EIF4G2 | 9 | Cassette | IN | Xue et al. (29) |
| FAM38A | 8 | Cassette | IN | Xue et al. (29) |
AS, alternative splicing.
aRegulated exon number (for mutually exclusive type) or name (NM/SM). Exon inclusion (IN) or exon skipping (SK).
Figure 3.
Abnormal lamin B1 expression is associated with altered splicing of PTB-target genes. Top significantly altered PTB-regulated alternative splicing identified in ADLD blood (A) and fibroblasts (B). Alternative splicing was analyzed by RT-PCR and dHPLC. For each gene, a representative RT-PCR is shown above the dHPLC data graph. Bars represent the average percentage of each splicing event ± SD. *P < 0.05, Student's t test, n = 4. EX, exon; IN, exon inclusion; SK, exon skipping; F, fibroblasts; B, blood. The splicing pattern of other PTB-regulated genes analyzed in blood and fibroblasts are shown, respectively, in Supplementary Material, Figures S1 and S2. (C) Significantly altered PTB-regulated alternative splicing events identified in brains of Lmnb1+/+ (n = 4) and Lmnb1Δ/Δ (n = 3) E18.5 embryos. Bars represent the average percentage of each splicing event ± SD. *P < 0.05, Student's t test. The splicing pattern of Fam38a, that was not significantly altered, is shown in Supplementary Material, Figure S3.
To determine whether changes in lamin B1 levels cause abnormal splicing, we examined the splicing pattern of PTB-target genes in the embryonic brains of Lmnb1Δ/Δ mice. Splicing was altered in four out of five target genes in Lmnb1Δ/Δ compared with Lmnb1+/+ mice (Pkm2, Eif4g2, Phf21a and Spag9; Fig. 3C and Supplementary Material, Fig. S3). In summary, in LMNB1 duplicated individuals, six of the eight PTB-regulated genes tested showed altered splicing in whole blood or fibroblasts, with one of those genes (PKM2) deregulated in both tissues. Mice lacking lamin B1 also show splicing defects, suggesting that this gene plays a causal role.
To verify whether the splicing reprogramming resulting from lamin B1 dysregulation is mediated by raver-2, we analyzed the splicing pattern of PTB-target genes in MEFs. Indeed, in mouse, raver-2 presents a specific spatio-temporal expression pattern during embryogenesis (18) and Raver2 is not expressed in fibroblasts (data obtained from FANTOM5). Accordingly, we found that the expression of the ribonucleoprotein was significantly reduced (97% decrease) compared with brain. (Supplementary Material, Fig. S4A and B). Consequently, in Lmnb1Δ/Δ MEFs we failed to detect any differences in the splicing pattern of PTB-target genes compared with Lmnb1+/+ MEFs (Supplementary Material, Fig. S4C).
Overall, these findings support the hypothesis that lamin B1 levels contribute to modulating PTB splicing activity, likely via raver-2.
PLP1 splicing is altered in ADLD fibroblasts and in the brain of Lmnb1Δ/Δ embryos
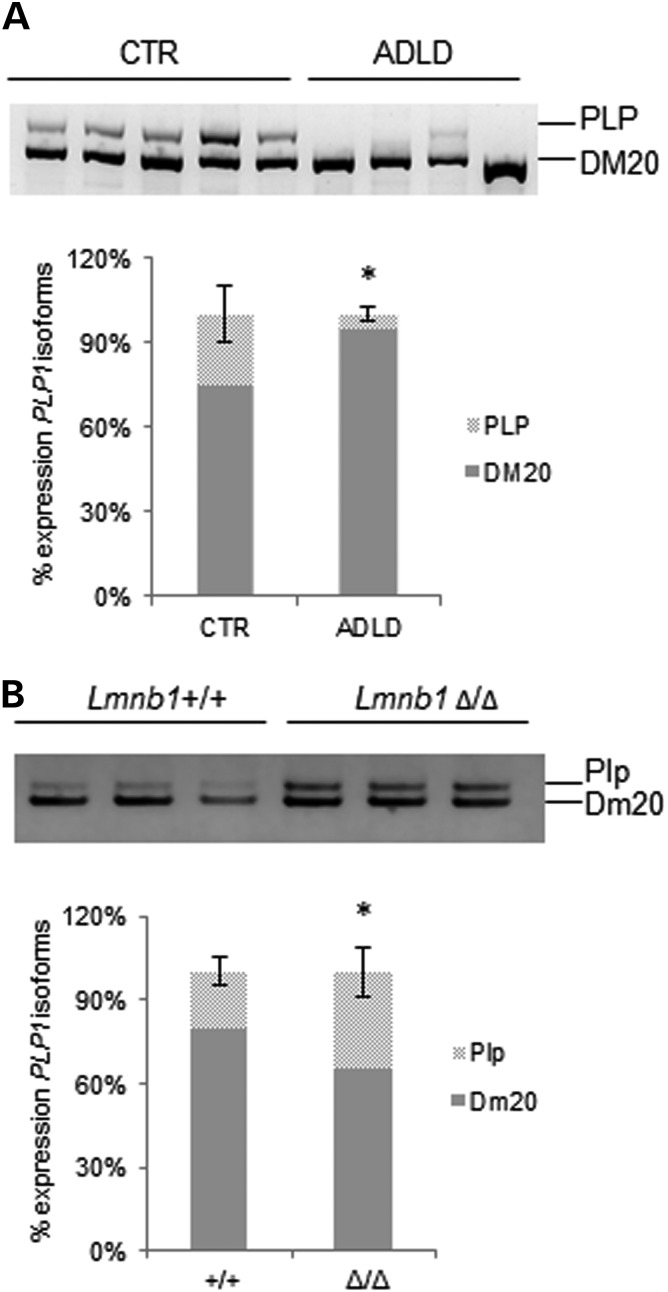
Lineage-specific overexpression of Lmnb1 in mouse oligodendrocytes leads to transcriptional repression of myelin-specific genes, including the proteolipid protein 1 gene (Plp1), which codes for the most abundant myelin protein (12). Altered expression of PLP1 characterizes another leukodystrophy, Pelizaeus–Merzbacher disease (PMD), in which duplication of the PLP1 gene leads to increased levels of the protein and an imbalance between its adult (PLP) and embryonic (DM20) isoforms (33). To investigate whether a PLP/DM20 isoform imbalance occurs in ADLD, we analyzed the alternative splicing of PLP1 in ADLD fibroblasts by reverse transcriptase-polymerase chain reaction (RT-PCR) and dHPLC. Fibroblasts of ADLD subjects showed a 20% increase in the embryonic PLP1 isoform (Fig. 4A). Conversely, in the brain of Lmnb1Δ/Δ embryos, the adult Plp1 isoform was increased relatively to the embryonic isoforms (Fig. 4B). Similarly to PTB-target genes, in Lmnb1Δ/Δ MEFs, we did not observe any differences of Plp1 splicing pattern (Supplementary Material, Fig. S5). These results indicate that lamin B1 plays a crucial role in the regulation of PLP1 expression, shifting the balance between adult and embryonic isoforms as a function of its levels.
Figure 4.
Abnormal levels of lamin B1 alter PLP1 splicing. (A) Analysis of embryonic (DM20) and adult (PLP) isoforms of PLP1 gene in ADLD (n = 4) and CTR (n = 5) fibroblasts analyzed through RT-PCR and dHPLC run. Bars represent the mean ± SD. *P < 0.05, Student's t-test, n = 5. (B) Analysis of Plp1 splicing pattern in brain of Lmnb1+/+ (n = 4) and Lmnb1Δ/Δ (n = 3) embryos. *P < 0.05, Student's t-test. Representative gels of mRNA splicing isoforms are shown above dHPLC data graphs.
Discussion
Changes in expression levels of lamin B1 are deleterious; duplication of the LMNB1 gene causes ADLD in humans (2), while Lmnb1 gene silencing has been demonstrated to be lethal in mice (7). However, the mechanisms whereby the pathological expression of lamin B1 causes its damaging effects are still unclear. To investigate this crucial issue we have demonstrated, by using different approaches, that lamin B1 dysregulation affects different steps of gene expression, namely transcription and alternative splicing.
First, to gain insights into pathologically relevant pathways that are altered in ADLD, we performed whole-genome expression profiling in two peripheral tissues. Because of the difficulty in obtaining human pathological tissues, other cell types, such as fibroblasts and lymphocytes, have been widely used (34–36). Whole blood samples have been demonstrated to have the most similar expression profile to different areas of the CNS (37), while skin fibroblasts have been used to model neurodegenerative diseases in vitro, given their availability and robustness (38). Therefore, to investigate global pathways altered by lamin B1 overexpression, we analyzed the transcriptome of whole blood and skin fibroblasts in ADLD patients and controls. ADLD cells show a significant number of deregulated genes that are involved in development of the nervous system. This is consistent with the notion that Lmnb1Δ/Δ embryos display reduced brain size and impaired corticogenesis (7–9). Moreover, since lamin B1 is broadly expressed (39), it is not surprising that enrichment analysis of gene ontology categories also indicates that lamin B1 overexpression affects other tissues/organs, such as the immune system. The fact that genes of the immune system are also affected is in line with increasing evidence that various immune system components are involved in neurodegenerative diseases, such as Alzheimer's (AD), Parkinson's (PD), Huntington's (HD) and Amyotrophic lateral sclerosis (ALS) (40,41). In addition, a cluster of genes involved in muscle contraction are deregulated in LMNB1 duplication carriers, consistent with the nuclear abnormalities and defects in actin turnover, which were described in one ADLD patient, where a myopathy is clinically evident (42).
Lamin B1 regulates gene expression by anchoring chromatin to the nuclear lamina, which acts as a scaffold for chromatin remodeling, and is thus critical for the spatial organization of chromosomes in the nucleus (15). Accordingly, in Lmnb1Δ/Δ MEFs, DEGs cluster in specific chromosomal locations due to changes in chromosome positions relative to the nuclear lamina (16). Similarly, in LMNB1 duplication carriers, we found that DEGs cluster in specific genomic regions, suggesting that changes in lamin B1 levels invariably induce the delocalization of chromosomes at the nuclear lamina. The reorganization of chromosome positioning was also found in skin fibroblasts from patients with cardiomyopathy and Hutchinson–Gilford progeria syndrome bearing mutations in the cognate lamin A/C gene (43,44). Together, these findings indicate that alterations to the levels of lamins are linked with changes in chromosome positioning.
Second, in order to identify the principal genes that result in the phenotype caused by LMNB1 overexpression, we compared the DEGs identified in whole blood and skin fibroblasts. During differentiation, changes in temporal and spatial expression of lamins cause distinctive chromatin patterns conferring unique transcriptomes in different cell lineages and developmental stages (45). Indeed, in a conditional transgenic mouse model overexpressing Lmnb1 in oligodendroglia, the majority of the deregulated genes are involved in typical oligodendrocyte functions, such as the formation and maintenance of myelin (12). Consistent with this cell-specificity of the transcriptome, we demonstrated that only three DEGs are common to both human fibroblasts and blood, the most significant being RAVER2. RAVER2 gene expression was up-regulated in both ADLD whole blood and fibroblasts, where we confirmed an increased protein level in the nucleus. In human fibroblasts, the RAVER2 locus located on cytoband 1p31 is physiologically bound by lamin B1 in a LAD (15,46), which typically represents a repressive location. Thus, RAVER2 overexpression in ADLD suggests that its locus is displaced as a result of chromosomal repositioning caused by increased lamin B1 levels. We were able to confirm these data by using positional gene set analysis, demonstrating that cytoband 1p31 contains a cluster of genes that are overexpressed in ADLD. A recent computational study has revealed a binding site for the transcription factor OCT-1 in the RAVER2 5′-flanking region (26). This evidence, along with the previous observation that OCT-1 recruitment to the nuclear periphery is increased in ADLD cells (42), further supports the view that lamin B1 levels modulate distinct pathways through repositioning of specific genomic loci.
To date, functions ascribed to raver-2, similarly to its homologous raver-1 (47), include the modulation of alternative splicing through the splicing repressor PTB (20).
Here, we demonstrated the increase of raver-2 expression in ADLD tissues associated with modified splicing of PTB-target genes. Likewise, the brains of Lmnb1Δ/Δ embryos are also characterized by an altered raver-2/PTB pathway, which could explain their abnormal development, with reduced brain size and impaired corticogenesis (7–9). Interestingly, our results are in line with recent findings demonstrating that the silencing of LMNB1 results in a high number of enlarged nuclear speckles and extensive changes in alternative splicing of multiple genes (48). Alternative splicing has an important role in defining tissue specificity, and this process is highly regulated during organogenesis, with different tissues (e.g. the placenta and embryo) expressing distinct isoforms during different developmental stages (49). Recent high-throughput studies have shown that more than 50% of alternative splicing isoforms are differentially expressed among tissues (50). Accordingly, we found that PTB-target genes altered by lamin B1/raver-2 overexpression are mainly tissue-specific. Raver-2 is probably not the only splicing regulator altered by lamin B1 overexpression, and possibly, other players are necessary to define the disease, but their number and identity is extremely difficult to be found given that the regulation of alternative splicing is highly tissue-specific and is likely that in each tissue one or more of them could regulate PTB.
The primary target tissue of ADLD is the CNS, specifically myelin. In ADLD patients, white matter defects are observed, particularly in the cerebellum, corticospinal tracts and corpus callosum, which lead to brain and spinal cord atrophy (2). Previous studies in ADLD patients have shown the decreased transcription of myelin proteins (mostly PLP1) in oligodendrocytes (12). Here, we have demonstrated an altered splicing pattern of PLP1 in ADLD fibroblasts and Lmnb1Δ/Δ embryos. It is worth noting that an abnormal balance of the two PLP1 isoforms was also observed in fibroblasts derived from patients with a different leukodystrophy, namely Pelizaeus–Merzbacher disease, although in the reverse direction compared with ADLD (33). Because a direct link between PTB and myelin genes has never been demonstrated, this finding suggests that other splicing factors could be affected by lamin B1/raver-2 dysregulation. Consistent with this hypothesis, in an ADLD transgenic mouse model, Hnrnpr protein is upregulated in the oligodendrocytes (12). Like raver-2, Hnrnpr protein belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins and interacts with the telomeric protein, survival of motor neuron 1 (SMN1), which is a regulator of RNA processing (51).
Recently, transcriptome profiling and computational predictions have indicated that an unexpectedly high proportion of diseases, including Fragile X-associated tremor/ataxia syndrome, spinal muscular atrophy and frontotemporal dementia, are characterized by aberrant alternative RNA splicing (52). Myotonic dystrophy 1, which is caused by dynamic and unstable expanded microsatellite sequences in DMPK gene, is the prototype of this group of diseases, also called spliceopathies. The nuclear export of the mutated RNAs is defective, and they aggregate to form nuclear foci, which then recruit and sequester RBP while stabilizing other splicing regulators (53). The unbalanced action of these regulatory proteins disrupts RNA metabolism, with alterations in the alternative splicing of numerous pre-messenger RNAs in several tissues (53). Other mechanisms causing aberrant splicing are disruption of cis-splicing sites or trans-acting factors; dysfunction of RBP required for the efficient assembly of small nuclear ribonucleoprotein (snRNP) complexes and spliceosomal snRNP biogenesis and mutations in proteins that regulate the splicing of several human transcripts or protect extra-long introns from incorrect splicing at cryptic sites (54). We suggest that also the deregulation of lamin B1 levels, possibly through modulation of RAVER2 expression, that was demonstrated to modify PTB pathway (20) could be an additional mechanism inducing disruption of RNA metabolism.
Based on the present findings, we confirm previous studies that report the role of lamin B1 in splicing regulation (48) and suggest that this biological function is mediated by raver-2 expression levels. Changes in lamin B1 levels induce through chromosome repositioning, modifications of the expression of multiple genes, including RAVER2, which is a modulator of PTB (20), and possibly other splicing regulators.
Consequently, we propose that ADLD could be partly regarded as a spliceopathy caused by increased levels of lamin B1/raver-2. Thus, adult-onset CNS demyelination could be the result of increased expression, during adulthood, of the embryonic isoform of the PLP1 protein, which has a crucial role in myelin maintenance. Accordingly, in ADLD, in addition to the already-demonstrated altered expression of myelin genes and increased nuclear rigidity (12,14), we propose that the dysregulation of alternative splicing could be one of the pathogenic mechanisms induced by LMNB1 overexpression. We believe that our results provide new insights into the molecular mechanisms underlying spliceopathies and laminopathies, and in doing so may reveal possible therapeutic targets for these diseases.
Materials and Methods
Patients and sample collection
The study has been approved by the AUSL Bologna Ethical Committee and informed consent was obtained from all participants. Peripheral whole blood of 12 individuals of an ADLD family (14,42) was collected in PAXgene™ Blood RNA Tubes (Qiagen, Mannheim, Germany). Primary cultures of fibroblasts from seven family members and two healthy controls, comparable for age, obtained by skin biopsies of the left deltoid region, and from one patient with the vADLD (4), were established and cultured as previously described (14). RNA was extracted from confluent passage-matched fibroblasts (5 × 106 cells, passage 3–5) cultured in serum-free medium for 24 h. LMNB1 duplication was identified (14) in four individuals, three symptomatic (42,55). No relevant pathologies were referred or found in the other participants of the study.
Microarray and bioinformatics analysis
RNA from fibroblasts and whole blood was extracted using the RNeasy Plus Kit and PAXgene™ Blood RNA System Kit (Qiagen). RNA quality was tested by Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and used when the integrity quality number (RIN) scored above 8. Four-hundred nanograms of RNA for each sample were amplified using the Illumina® TotalPrep™ RNA Amplification Kit (Ambion, Thermo Fisher Scientific, Waltham, MA, USA). Samples were analyzed with the Illumina whole-genome HumanHT-12 v4 (Illumina, San Diego, CA, USA) using the Illumina's Beadarray system 500 G Scanner. Image signal intensity was extracted; the background subtracted and normalized using Illumina Inc. BeadStudio software version 3.4.0 (Consorzio per il Centro di Biomedicina Molecolare, Trieste, Italy). All statistical analyzes were performed in the R environment using specific packages of Bioconductor (56). The raw signal intensities were processed with Lumi package applying variance-stabilizing transformation (VST) and quantile normalization (57). The Limma package was used to identify DEGs (58). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (59) and are accessible through GEO Series accession number GSE65368 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95368).
Genes with a P-value <0.01 and logarithm fold change (logFC) >|0.5| were considered DEGs. This selection method was demonstrated having an higher concordance degree of DEGs between different platforms when compared with DEGs selected only on P-value ranking (60). Significance levels (p-val) were corrected using Benjamini and Hochberg (BH) false discovery rate (FDR) method (p-adj) for multiple hypotheses testing (61).
GSEA was conducted on data from the full chip with no prior filtering (21). The GO gene sets databases (C5.bp.v4.0, C5.cc.v4.0, C5.mf.v4.0) and positional gene sets (C1.all.v4.0) from the Molecular Signatures Database (www.broadinstitute.org/gsea/msigdb/index.jsp) were used for enrichment analysis. A total of 1000 permutations were used to obtain the nominal P-value (NOM p-val). Normalized enrichment score (NES) was measured. Gene sets with nominal P-value <0.05 and false discovery rate (FDR q-val) <0.25 (21) were considered to be significantly deregulated. A summary of no redundant gene ontology terms was obtained by using REVIGO (62).
Validation of microarray results
Genes were selected on the basis of their significance, potential interest or the pathway in which they are involved. One microgram of RNA was retrotranscribed using the High Capacity RNA-to-cDNA kit (Applied Biosystems). RT-qPCR reactions were carried out in triplicate using TaqMan® Gene Expression Assays (Supplementary Material, Table S7) on an ABI-Prism7500 Fast Instrument (Applied Biosystems). Gene expression was normalized to hydroxymethylbilane synthase gene (HMBS) (3), while microarray to HMBS and X-prolyl aminopeptidase (aminopeptidase P) 1, soluble gene (XPNPEP1) (63). Gene expression fold changes were calculated using the ΔΔCt method (64).
Analysis of alternative splicing
Alternative splicing was analyzed by semi-quantitative multiplex PCR followed by non-denaturing dHPLC runs (Wave, Transgenomic, Omaha, NE, USA). Primers were designed on exons flanking the alternative cassette exon (Supplementary Material, Table S8). PCR were performed in a total volume of 25 μl, containing 1 μl of cDNA, 1 U FastStart High Fidelity PCR System (Roche), 120 μm dNTPs (Roche) and 20 pm of each primer (Sigma-Aldrich, St. Louis, MO, USA). Fragments obtained within the log-linear amplification range were analyzed without further purification on dHPLC. We calculated the ratio between the areas of the peaks representing the two isoforms, using WAVEMAKER Software (Transgenomic) in duplication carriers and control samples. Differences in relative expression of isoforms were statistically assessed by Student's t-test for three independent experiments.
Animals
Lmnb1-null (Lmnb1Δ/Δ) mice (7) were obtained from the Mutant Mouse Regional Resource Center (University of California, Davis, CA, USA). Animal health and comfort were veterinary-controlled. The mice were housed in filtered cages in a temperature-controlled room with a 12:12 h dark/light cycle with ad libitum access to water and food. All of the animal experiments were performed in accordance with the European Community Council Directive dated 24 November 1986 (86/609/EEC) and were approved by the Italian Ministry of Health and the IIT Ethical Committee.
Embryonic brains were collected from Lmnb1Δ/Δ embryos and wild-type (Lmnb1+/+) littermates at 18.5 days of gestation and processed using the RNeasy Lipid Tissue Mini Kit (Qiagen). MEFs were isolated from wild-type Lmnb1+/+ and Lmnb1Δ/Δ embryos at 13.5 days of gestation (14).
Western blot analysis
Fibroblasts were lysed by boiling for 5 min in 1% sodium dodecyl sulfate (SDS), 1 mm ethylenediaminetetraacetic acid, 5 mm 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (pH 7.4), mouse embryonic brains in 50 mm tris(hydroxymethyl)aminomethane, 0.5 M NaCl, 1% SDS and 1% Tx-100. Equal amounts of proteins were separated on NuPage 10% bis-tris polyacrylamide gels (Invitrogen, Thermo Scientific) and analyzed (14) using primary rabbit polyclonal antibodies against lamin B1 (Abcam, Cambridge, England, UK), γ-tubulin (Sigma), actin (Sigma) and raver-2 (Abcam). Densitometric analysis was performed using the NIH ImageJ program (65). The protein levels were normalized on γ-tubulin or actin content.
Immunofluorescence
Fibroblasts were plated onto poly-l-lysine-coated coverslips, serum-deprived for 24 h, fixed in 4% PFA and immuno-labeled as previously described (14), using the anti-raver-2 antibody (Abcam). The confocal optical sectioning was performed at room temperature using a Leica TCS SP5 AOBS TANDEM inverted confocal microscope that was equipped with a HCX PL APO 40× 1.25 Oil and a HCX PL APO blue 63× 1.4NA Oil objective lenses (Leica Microsystems, Wetzlar, Germany).
Statistical analysis
Student's t-test was used for data comparison; Mann–Whitney Rank Sum test was used for comparing raver-2 protein expression in fibroblasts. The differences were considered to be statistically significant when P < 0.05.
Supplementary Material
Funding
This work was supported by TELETHON (grant number GGP10184) and the Gino Galletti Foundation. Funding to pay the Open Access publication charges for this article was provided by Fondazione Telethon.
Supplementary Material
Acknowledgements
We are gratefully indebted with all family members who participated in the study. We thank Dr Silvia Piras for excellent technical assistance.
Conflict of Interest statement. None declared.
References
- 1.Cortelli P., Terlizzi R., Capellari S., Benarroch E. (2012) Nuclear lamins: functions and clinical implications. Neurology, 79, 1726–1731. [DOI] [PubMed] [Google Scholar]
- 2.Padiath Q.S., Saigoh K., Schiffmann R., Asahara H., Yamada T., Koeppen A., Hogan K., Ptácek L.J., Fu Y.H. (2006) Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat. Genet., 38, 1114–1123. [DOI] [PubMed] [Google Scholar]
- 3.Giorgio E., Rolyan H., Kropp L., Chakka A.B., Yatsenko S., Di Gregorio E., Lacerenza D., Vaula G., Talarico F., Mandich P., et al. (2013) Analysis of LMNB1 duplications in autosomal dominant leukodystrophy provides insights into duplication mechanisms and allele-specific expression. Hum. Mutat., 34, 1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brussino A., Vaula G., Cagnoli C., Panza E., Seri M., Di Gregorio E., Scappaticci S., Camanini S., Daniele D., Bradac G.B., et al. (2010) A family with autosomal dominant leukodystrophy linked to 5q23.2-q23.3 without lamin B1 mutations. Eur. J. Neurol., 17, 541–549. [DOI] [PubMed] [Google Scholar]
- 5.Lenz-Böhme B., Wismar J., Fuchs S., Reifegerste R., Buchner E., Betz H., Schmitt B. (1997) Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol., 137, 1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Rolef Ben-Shahar T., Riemer D., Treinin M., Spann P., Weber K., Fire A., Gruenbaum Y. (2000) Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell, 11, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vergnes L., Péterfy M., Bergo M.O., Young S.G., Reue K. (2004) Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA, 101, 10428–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffinier C., Jung H.J., Nobumori C., Chang S., Tu Y., Barnes R.H., 2nd, Yoshinaga Y., de Jong P.J., Vergnes L., Reue K., et al. (2011) Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell, 22, 4683–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y., Sharov A.A., McDole K., Cheng M., Hao H., Fan C.M., Gaiano N., Ko M.S., Zheng Y. (2011) Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science, 334, 1706–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takamori Y., Tamura Y., Kataoka Y., Cui Y., Seo S., Kanazawa T., Kurokawa K., Yamada H. (2007) Differential expression of nuclear lamin, the major component of nuclear lamina, during neurogenesis in two germinal regions of adult rat brain. Eur. J. Neurosci., 25, 1653–1662. [DOI] [PubMed] [Google Scholar]
- 11.Lin S.T., Fu Y.H. (2009) miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis. Model. Mech., 2, 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heng M.Y., Lin S.T., Verret L., Huang Y., Kamiya S., Padiath Q.S., Tong Y., Palop J.J., Huang E.J., Ptáček L.J., Fu Y.H. (2013) Lamin B1 mediates cell-autonomous neuropathology in a leukodystrophy mouse model. J. Clin. Invest., 123, 2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D. (2008) Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev., 22, 832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrera D., Canale C., Marotta R., Mazzaro N., Gritti M., Mazzanti M., Capellari S., Cortelli P., Gasparini L. (2014) Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. FASEB J., 28, 3906–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature, 453, 948–951. [DOI] [PubMed] [Google Scholar]
- 16.Malhas A., Lee C.F., Sanders R., Saunders N.J., Vaux D.J. (2007) Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol., 176, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., Krijgsveld J., Hentze M.W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–1406. [DOI] [PubMed] [Google Scholar]
- 18.Kleinhenz B., Fabienke M., Swiniarski S., Wittenmayer N., Kirsch J., Jockusch B.M., Arnold H.H., Illenberger S. (2005) Raver2, a new member of the hnRNP family. FEBS Lett., 579, 4254–4258. [DOI] [PubMed] [Google Scholar]
- 19.Henneberg B., Swiniarski S., Becke S., Illenberger S. (2010) A conserved peptide motif in Raver2 mediates its interaction with the polypyrimidine tract-binding protein. Exp. Cell Res., 316, 966–979. [DOI] [PubMed] [Google Scholar]
- 20.Das S.K., Holt D.G., Uehara H., Zhang X., Archer B., Ambati B.K. (2014) Raver2 preserves corneal avascularity by increasing sFlt1 production [ARVO E-abstract]. Invest. Ophthalmol. Vis. Sci., 55, 3249. [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. (2005) Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchison C.J. (2014) B-type lamins in health and disease. Semin. Cell Dev. Biol., 29, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhas A.N., Lee C.F., Vaux D.J. (2009) Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol., 184, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melko M., Douguet D., Bensaid M., Zongaro S., Verheggen C., Gecz J., Bardoni B. (2011) Functional characterization of the AFF (AF4/FMR2) family of RNA-binding proteins: insights into the molecular pathology of FRAXE intellectual disability. Hum. Mol. Genet., 20, 1873–1885. [DOI] [PubMed] [Google Scholar]
- 25.FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Lassmann T., Itoh M., Summers K.M., Suzuki H., Daub C.O., et al. (2014) A promoter-level mammalian expression atlas. Nature, 507, 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romanelli M.G., Lorenzi P., Diani E., Filippello A., Avesani F., Morandi C. (2012) Transcriptional regulation of the human Raver2 ribonucleoprotein gene. Gene, 493, 243–252. [DOI] [PubMed] [Google Scholar]
- 27.Spellman R., Rideau A., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W. (2005) Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans., 33, 457–460. [DOI] [PubMed] [Google Scholar]
- 28.Gromak N., Rideau A., Southby J., Scadden A.D., Gooding C., Hüttelmaier S., Singer R.H., Smith C.W. (2003) The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J., 22, 6356–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y.S., Zhang C., Yeo G., Black D.L., Sun H., et al. (2009) Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell, 36, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matlin A.J., Southby J., Gooding C., Smith C.W. (2007) Repression of alpha-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA, 13, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spellman R., Llorian M., Smith C.W. (2007) Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell, 27, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., Wang G., Wu Q., Wei C., Bi Y., et al. (2013) Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell, 152, 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regis S., Grossi S., Corsolini F., Biancheri R., Filocamo M. (2009) PLP1 gene duplication causes overexpression and alteration of the PLP/DM20 splicing balance in fibroblasts from Pelizaeus–Merzbacher disease patients. Biochim. Biophys. Acta, 1792, 548–554. [DOI] [PubMed] [Google Scholar]
- 34.Cooper-Knock J., Kirby J., Ferraiuolo L., Heath P.R., Rattray M., Shaw P.J. (2012) Gene expression profiling in human neurodegenerative disease. Nat. Rev. Neurol., 8, 518–530. [DOI] [PubMed] [Google Scholar]
- 35.Fogel B.L., Cho E., Wahnich A., Gao F., Becherel O.J., Wang X., Fike F., Chen L., Criscuolo C., De Michele G., et al. (2014) Mutation of senataxin alters disease-specific transcriptional networks in patients with ataxia with oculomotor apraxia type 2. Hum. Mol. Genet., 23, 4758–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernardini C., Lattanzi W., Bosco P., Franceschini C., Plazzi G., Michetti F., Ferri R. (2012) Genome-wide gene expression profiling of human narcolepsy. Gene Expr., 15, 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan P.F., Fan C., Perou C.M. (2006) Evaluating the comparability of gene expression in blood and brain. Am. J. Med. Genet. B. Neuropsychiatr. Genet., 141B, 261–268. [DOI] [PubMed] [Google Scholar]
- 38.Auburger G., Klinkenberg M., Drost J., Marcus K., Morales-Gordo B., Kunz W.S., Brandt U., Broccoli V., Reichmann H., Gispert S., Jendrach M. (2012) Primary skin fibroblasts as a model of Parkinson's disease. Mol. Neurobiol., 46, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber K.H., Kennedy B.K. (2013) When lamins go bad: nuclear structure and disease. Cell, 152, 1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Björkqvist M., Wild E.J., Tabrizi S.J. (2009) Harnessing immune alterations in neurodegenerative diseases. Neuron, 64, 21–24. [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues M.C., Sanberg P.R., Cruz L.E., Garbuzova-Davis S. (2014) The innate and adaptive immunological aspects in neurodegenerative diseases. J. Neuroimmunol., 269, 1–8. [DOI] [PubMed] [Google Scholar]
- 42.Columbaro M., Mattioli E., Maraldi N.M., Ortolani M., Gasparini L., D'Apice M.R., Postorivo D., Nardone A.M., Avnet S., Cortelli P., et al. (2013) Oct-1 recruitment to the nuclear envelope in adult-onset autosomal dominant leukodystrophy. Biochim. Biophys. Acta, 1832, 411–420. [DOI] [PubMed] [Google Scholar]
- 43.Mewborn S.K., Puckelwartz M.J., Abuisneineh F., Fahrenbach J.P., Zhang Y., MacLeod H., Dellefave L., Pytel P., Selig S., Labno C.M., et al. (2010) Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS ONE, 5, e14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCord R.P., Nazario-Toole A., Zhang H., Chines P.S., Zhan Y., Erdos M.R., Collins F.S., Dekker J., Cao K. (2013) Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson–Gilford progeria syndrome. Genome Res., 23, 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R.M., van Lohuizen M., et al. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell, 38, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadaie M., Salama R., Carroll T., Tomimatsu K., Chandra T., Young A.R., Narita M., Pérez-Mancera P.A., Bennett D.C., Chong H., et al. (2013) Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev., 27, 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romanelli M.G., Diani E., Lievens P.M. (2013) New insights into functional roles of the polypyrimidine tract-binding protein. Int. J. Mol. Sci., 14, 22906–22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camps J., Wangsa D., Falke M., Brown M., Case C.M., Erdos M.R., Ried T. (2014) Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J., 28, 3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Revil T., Gaffney D., Dias C., Majewski J., Jerome-Majewska L.A. (2010) Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics, 11, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M., Manley J.L. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol., 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mourelatos Z., Abel L., Yong J., Kataoka N., Dreyfuss G. (2001) SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J., 20, 5443–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R.K., Cooper T.A. (2012) Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med., 18, 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osborne R.J., Thornton C.A. (2006) RNA-dominant diseases. Hum. Mol. Genet., 15, R162–R169. [DOI] [PubMed] [Google Scholar]
- 54.Caillet-Boudin M.L., Fernandez-Gomez F.J., Tran H., Dhaenens C.M., Buee L., Sergeant N. (2014) Brain pathology in myotonic dystrophy: when tauopathy meets spliceopathy and RNAopathy. Front. Mol. Neurosci., 6, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guaraldi P., Donadio V., Capellari S., Contin M., Casadio M.C., Montagna P., Liguori R., Cortelli P. (2010) Isolated noradrenergic failure in adult-onset autosomal dominant leukodystrophy. Auton. Neurosci., 159, 123–126. [DOI] [PubMed] [Google Scholar]
- 56.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol., 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du P., Kibbe W.A., Lin S.M. (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics, 24, 1547–1548. [DOI] [PubMed] [Google Scholar]
- 58.Wettenhall J.M., Smyth G.K. (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics, 20, 3705–3706. [DOI] [PubMed] [Google Scholar]
- 59.Edgar R., Domrachev M., Lash A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi L., Jones W.D., Jensen R.V., Harris S.C., Perkins R.G., Goodsaid F.M., Guo L., Croner L.J., Boysen C., Fang H., et al. (2008) The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics, 9, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001) Controlling the false discovery rate in behavior genetics research. Behav. Brain Res., 125, 279–284. [DOI] [PubMed] [Google Scholar]
- 62.Supek F., Bošnjak M., Škunca N., Šmuc T. (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE, 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durrenberger P.F., Fernando F.S., Magliozzi R., Kashefi S.N., Bonnert T.P., Ferrer I., Seilhean D., Nait-Oumesmar B., Schmitt A., Gebicke-Haerter P.J., et al. (2012) Selection of novel reference genes for use in the human central nervous system: a BrainNet Europe Study. Acta Neuropathol., 124, 893–903. [DOI] [PubMed] [Google Scholar]
- 64.Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 65.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.