Abstract
Derived from any somatic cell type and possessing unlimited self-renewal and differentiation potential, induced pluripotent stem cells (iPSCs) are poised to revolutionize stem cell biology and regenerative medicine research, bringing unprecedented opportunities for treating debilitating human diseases. To overcome the limitations associated with safety, efficiency, and scalability of traditional iPSC derivation, expansion, and differentiation protocols, biomaterials have recently been considered. Beyond addressing these limitations, the integration of biomaterials with existing iPSC culture platforms could offer additional opportunities to better probe the biology and control the behavior of iPSCs or their progeny in vitro and in vivo. Herein, we discuss the impact of biomaterials on the iPSC field, from derivation to tissue regeneration and modeling. Although still exploratory, we envision the emerging combination of biomaterials and iPSCs will be critical in the successful application of iPSCs and their progeny for research and clinical translation.
Keywords: biomaterials, disease modeling, expansion, induced pluripotent stem cells, reprogramming
Introduction
Induced pluripotent stem cells (iPSCs) possess a regenerative therapeutic potential comparable to embryonic stem cells (ESCs) without many of the associated ethical concerns (Bilic & Izpisua Belmonte, 2012). Nevertheless, in vitro research applications and clinical translation of iPSCs have multiple challenges:
Costly and highly inefficient iPSC derivation and expansion protocols.
Incomplete reprogramming of somatic cells; or genetic instability occurring during in vitro expansion and differentiation, which may result in genetic abnormalities or potential immunogenicity following iPSC transplantation (Saha & Jaenisch, 2009; Araki et al, 2013).
Safety concerns, primarily attributed to the potential risk of iPSC progeny to form teratomas (due to the presence of residual undifferentiated iPSCs) or malignant transformation post-transplantation (Hong et al, 2014).
Overcoming these limitations depends on designing well-defined cell culture strategies for iPSC derivation, expansion, and differentiation. Limited reprogramming efficiency may be improved by modulating the presentation and kinetics of reprogramming factors, which is challenging with traditional reprogramming methods (Hu, 2014). Once reprogrammed, the efficiency of iPSC expansion and the control over their differentiation can be improved by carefully mimicking the native microenvironment of stem cells—also known as the stem cell niche (Dellatore et al, 2008). The stem cell niche orchestrates stem cell phenotype, proliferation, and differentiation through key elements such as specific extracellular matrix (ECM) composition, 3D architecture, chemical and mechanical signals, and cell–cell interactions among resident cells (Scadden, 2006; Dingal & Discher, 2014). Thus, approaches for closely mimicking stem cell niches may substantially increase the quality and efficiency of iPSC expansion and directed lineage specification. Such advancement will further enrich our understanding of iPSC biology and facilitate the development of new therapeutics (e.g. via establishment of patient-specific disease models), as well as advance iPSC-based cell replacement therapies (Robinton & Daley, 2012).
Biomaterials—materials selected or designed to interact with biological systems (Williams, 2009)—offer a unique and appealing strategy to advance iPSC research. For instance, biomaterials can be used to control the kinetics of reprogramming factors via nanoparticle- and microparticle-based systems. Biomaterials can also be used to overcome the issues associated with traditional iPSC expansion and differentiation protocols by creating stem-cell-like niches that incorporate key niche elements to enable precise regulation of stem cell fate and function (Lutolf et al, 2009; Murphy et al, 2014) (Fig1). Beyond guiding iPSC in vitro expansion and differentiation, biomaterials may also be used to facilitate iPSC transplantation (Higuchi et al, 2011; de Peppo et al, 2013; Villa-Diaz et al, 2013; Teng et al, 2014). In this review, we discuss in detail how emerging biomaterial-based strategies may solve key iPSC safety and efficiency challenges, which may soon allow iPSCs to realize their immense potential in the study and treatment of diseases.
Figure 1.
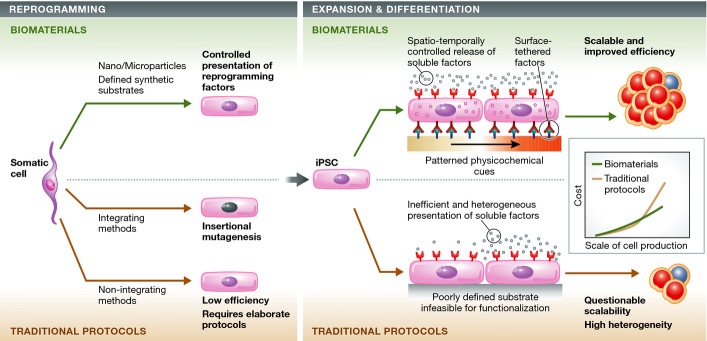
Overview of biomaterial-based strategies for enhancing the safety and efficiency of existing iPSC technologies and to better probe iPSC biology and control cell fate in vitro and in vivo
Biomaterials may be employed to facilitate all steps of iPSC production and consequently may help address pressing limitations of current derivation, expansion, and differentiation protocols. Traditional viral iPSC reprogramming methods, though efficient, are concerning due to insertional mutagenesis. Conversely, non-integrating iPSC reprogramming methods have lower efficiencies and require elaborate experimental procedures. To overcome the concerns associated with traditional iPSC reprogramming methods, biomaterial-based nano-/microparticles can be used to control the release kinetics of reprogramming factors to potentially avoid viral insertion, increase efficiency, and introduce simpler and less labor-intensive approaches for reprogramming. Furthermore, these nano-/microparticles can also be used to deliver soluble factors and small molecules for the expansion and differentiation of iPSCs (Corradetti et al, 2012). In parallel, biocompatible synthetic substrates can be engineered with patterned physicochemical cues and functionalized with surface-tethered factors to emulate native components of stem cell niches (Watt & Huck, 2013). Making use of biomaterial-based nano-/microparticles and biocompatible synthetic substrates can improve the scalability of traditional expansion and differentiation protocols because they can be reproducibly synthesized on large scales (as they are chemically defined) and at relatively low costs. The reduction in cost and labor will be key for the large-scale production of iPSCs and their progeny. Given the costs associated with the initial development and manufacturing of biomaterials, the cost of biomaterial-based iPSC production could be higher than that of traditional protocols at the early stage. However, in the long run, the application of biomaterials could render the iPSC production and differentiation processes more efficient (e.g. reducing the required concentrations of reprogramming factors via controlling their spatiotemporal presentation). We envision that the overall cost of biomaterial-based protocols will be significantly lower than that of traditional protocols.
Advanced biomaterials for the reprogramming of iPSCs
Limitations of traditional iPSC reprogramming systems
Current iPSC reprogramming protocols are often associated with safety and efficiency concerns. These concerns are especially critical for therapeutic applications where a large quantity of homogenous iPSCs with clinical grade quality is required. Two general approaches exist for iPSC derivation: integrating and non-integrating approaches. The integrating approach typically employs viral vectors to deliver and integrate pluripotency genes. Unfortunately, this approach has the potential for insertional mutagenesis, or malignant transformation (Ma et al, 2013b). Hence, clinical translation of integrating methods is severely limited due to safety and efficiency concerns. Alternatively, iPSCs may be derived with safer, non-integrating approaches, in which the pluripotency factors are transiently expressed or activated without genomic integration. Emerging non-integrating methods include episomal vectors and adenovirus-/plasmid-mediated transfection as well as pluripotency induction via chemically defined molecules [proteins (Kim et al, 2009; Zhou et al, 2009), mRNAs (Warren et al, 2010), and small molecules (Hou et al, 2013)]. Nevertheless, these methods remain labor-intensive and may still induce occasional genomic integration, leading to genomic instability (Hu, 2014). Even the seemingly straightforward DNA-free non-integrating methods [e.g. small molecule cocktails (Hou et al, 2013) or proteins (Zhou et al, 2009)] remain highly inefficient, requiring intricate protocols with multiple rounds of treatment, large doses, and uncontrolled presence/kinetics of reprogramming factors. Thus, new strategies are required to overcome the limitations associated with these traditional reprogramming methods.
Approaches employing biomaterials to surmount these barriers have shown promising results. Engineered biomaterials can potentially aid in iPSC derivation through controlling the kinetics of reprogramming factor delivery. Furthermore, well-defined biomaterial substrates can regulate the epigenetic state of iPSCs (Downing et al, 2013). This may work synergistically with traditional reprogramming approaches to improve reprogramming efficiency. In the following section, we highlight how these biomaterial-based strategies have been employed thus far and how these methods may impact iPSC derivation.
Biomaterial-based delivery systems for iPSC reprogramming
Biomaterial-based derivation of iPSCs—whether alone or in combination with existing non-integrating approaches—may improve reprogramming efficiency, safety, scalability, and reproducibility. Numerous compelling strategies involving biomaterials have emerged to deliver multiple reprogramming molecules with greater efficiency and more controlled kinetics than traditional methods (Fig2), which we discuss herein. We recognize that while biomaterial-based strategies have great implications for iPSC reprogramming, most approaches developed are too complicated to be commercialized into research tools. Concerted efforts from biologists, materials scientists, and engineers are merited to further simplify these strategies, and in particular, efforts are needed to limit costs as well as scaled batch-to-batch variability.
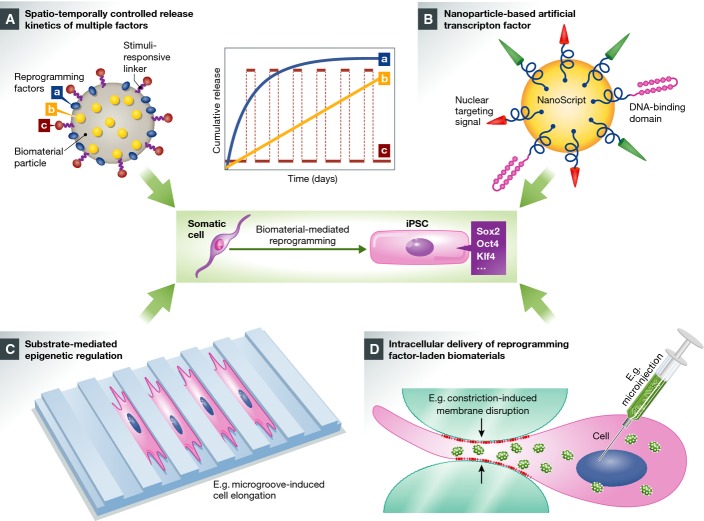
Figure 2.
Biomaterial-based approaches for improved iPSC reprogramming
(A) Well-defined, biomaterial-based micro-/nanoparticles can be formulated and engineered to load multiple reprogramming factors (e.g. Sox2, Oct4, and Klf4). The controlled distribution of different factors—on the surface of the particle (factor a) and entrapped in the particle (factor b)—can be readily achieved during the particle formulation process, and an additional factor (c), if required, can be loaded onto the particle via a stimuli-responsive linker. Given the biodegradability of chosen biomaterials, the varied distribution of multiple reprogramming factors (surface adsorption versus homogeneous encapsulation) and the degradation characteristics of the carrier particle dictate the spatiotemporally controlled release profiles of different factors such as sustained, zero-order release (curve b) and initial burst release followed by slower release or maintenance dose (curve a). Meanwhile, the surface-immobilized stimuli-responsive linker can be cleaved in response to environmental triggers, for example, pH and temperature, to release factor c on demand (e.g. pulsed release every other day, curve c). (B) Nanoparticle-based, smart, artificial transcription factor that is surface-functionalized with nuclear-targeting sequence, DNA-binding domains, and activators for the relevant transcription factors, enabling efficient nuclear localization and effective gene regulation. (C) The biomaterial substrate can be engineered with specific surface anisotropy or microgroove features, which in turn controls cell morphology and, as a result, mediates the epigenetic regulation and cellular reprogramming. (D) Reprogramming factor-laden biomaterials can be delivered into different intracellular loci via multiple engineering approaches, therefore efficiently modulating cell phenotype from inside-out. For example, high-throughput microfluidic technology can be employed to rapidly generate transient membrane disruption on the cells, enabling efficient intracellular localization of phenotype-altering agents without significantly impairing the target cells. Alternatively, this can be achieved via engineering methods such as microinjection or electroporation. Figure partially adapted with permission from Xu et al (2013b) and Patel et al (2014).
1. Biomaterials for potential spatial–temporal control of reprogramming factors
Maximizing efficiency of iPSC derivation depends on controlled spatial–temporal delivery of reprogramming factors since the timing and duration of cell exposure to extracellular stimuli significantly influence cell fate (Gaeta et al, 2013; Hou et al, 2013; Liu et al, 2013a). While these kinetics are poorly controlled with traditional reprogramming approaches (e.g. via media changes), biomaterial-based nano- and microparticles (MPs) can deliver and release payloads with spatial–temporal control [e.g. via sustained or pulsatile release profiles (Ge et al, 2012)], essential for maximizing the desired biological effects (Mohamed & van der Walle, 2008). These versatile particles may effectively deliver a spectrum of small molecules and biological cargos, including, but not limited to, growth factors, nucleic acids, and plasmids, making them a customizable platform for cellular reprogramming (Panyam & Labhasetwar, 2003).
Specifically, engineered degradable biomaterials can deliver factors to targeted subcellular locations with precise timing by leveraging the correlation between particle size and delivery kinetics. In general, smaller particles (typically < 1 μm, depending on target cell type and uptake mechanism) can rapidly enter cells and release encapsulated agents upon enzymolysis or hydrolysis (Woodward et al, 1985; Kou et al, 2013), whereas larger particles (typically > 1–2 μm) are not as efficiently internalized and are less accessible to degradation, permitting slower, sustained release of factors (Xu et al, 2009). This difference in release kinetics has been largely attributed to the surface area-to-volume ratios of particles with different sizes (Varde & Pack, 2007; Carpenedo et al, 2010). In addition to controlling particle size, manipulating other properties of biomaterials, including material composition, degradation rate, and inner architecture (e.g. porosity), enables tuning of delivery kinetics to produce controlled release systems (Varde & Pack, 2004; Klose et al, 2006; Giteau et al, 2008). Such tunability can be further realized by harnessing the potential responsiveness of biomaterials to environmental stimuli (Kost & Langer, 2012; Nakao, 2014; Patil & Shahiwala, 2014). Such delivery systems are capable of adjusting drug release in response to particular stimuli. In addition to microenvironmental stimuli, external stimuli can also be used to activate on-demand release and can include magnetism, electrical fields, ultrasound, and temperature changes (Mura et al, 2013). A broad set of biomaterials have been explored and designed to achieve desired temporal profiles such as daily pulsatile release and peak/plateaued release within a specific short interval. This technology could be particularly useful for controlling the temporal presentation of reprogramming factors, whether they are needed, for example, continuously for the first 5 days and then pulsatile thereafter each day every other day, or only on days 6 and 12 of reprogramming.
Given the versatility of the previously mentioned release platforms, the efficiency of their intracellular delivery, essential for transcriptional regulation of pluripotency induction, relies largely on the successful transmembrane trafficking of biomaterials. This process is primarily driven by endocytosis-mediated cellular uptake; however, it is generally associated with endosomal escape or loss of delivered materials due to vesicular degradation/recycling, leading to limited delivery efficiency (Kou et al, 2013; Sahay et al, 2013). To potentially obviate this issue and maximize the reprogramming efficiency, reprogramming factor-laden biomaterials may be forced into the cytosolic space via rapid and cytocompatible mechanical deformation of somatic cells (via a microfluidic device). This deformation induces transient membrane disruption, resulting in passive diffusion of biomaterials into the cells (Sharei et al, 2013). In combination with biomaterial-based delivery platforms, this microfluidic technology could further improve the spatial presentation of reprogramming factors and maximize the reprogramming efficiency in a safe and high-throughput manner [e.g. operated at a throughput rate of 20,000 cells/second per device (Sharei et al, 2013)]. It is conceivable that such a combination could be used to localize multiple factors that are difficult to deliver, such as macromolecules with diverse and sensitive structures (Yan et al, 2010) for a broad range of iPSC applications in addition to iPSC derivation. Alternative intracellular delivery methods include microinjection, electroporation, and sonoporation which have also been explored to facilitate cellular internalization of biomolecules or nanoparticles (Miller et al, 2002; Geng & Lu, 2013). They have demonstrated excellent spatiotemporal and dose control for delivery without significantly impairing the manipulated cells (Xie et al, 2013; Boukany et al, 2014).
The broad utility of the biomaterial-based controlled release strategies has been extensively demonstrated by a spectrum of materials including biodegradable polyester-based materials (Richards Grayson et al, 2003; Mohamed & van der Walle, 2008; Makadia & Siegel, 2011). For example, nano-/microparticles formulated by poly(lactic-co-glycolic acid) (PLGA) offer high stability, potential for narrow size distribution, tunability, and an excellent safety profile (FDA-approved) (Danhier et al, 2012; Ankrum et al, 2014b). It has been adopted as a gene delivery vehicle for multiple transfection applications (Seo et al, 2013; Tian et al, 2013). PLGA degrades into lactic acid and glycolic acid (natural metabolites found in the body). Its degradation rate can be tailored by balancing the ratio of lactic acid and glycolic acid and altering the molecular weight, thereby enabling controlled release of encapsulated genetic materials (Lu et al, 2000; Danhier et al, 2012). With fine-tuning of the surface chemistry, size, and drug loading, PLGA particles encapsulating phenotype-altering agents enable efficient, sustained stabilization/release of the encapsulated factors into individual cells or spheroids for prolonged control of cellular functions (Ankrum et al, 2014a,b) or morphogenesis (Carpenedo et al, 2009, 2010; Bratt-Leal et al, 2011), respectively. For instance, we have recently demonstrated loading of PLGA MPs with several agents into multiple cell types to control cell phenotype from the intercellular milieu [Fig3A (1–3)] (Sarkar et al, 2011; Ankrum et al, 2014a,b). By selecting agents that can easily cross cell membranes (e.g. certain positively charged agents and small molecules), it is also possible to use this strategy to control the microenvironment surrounding the modified cells. Specifically, we have shown that human mesenchymal stem cells (MSCs) loaded with dexamethasone (Dex)-doped PLGA MPs release Dex to regulate the differentiation of both the modified cells and cells in the adjacent microenvironment. We have also shown that budesonide-loaded PLGA MPs enhance the MSC immunomodulatory phenotype and that iron oxide nanoparticle-loaded PLGA MPs can be used to track MSCs in vivo (Xu et al, 2012; Ankrum et al, 2014a). This platform may be used to improve reprogramming efficiency and control factor delivery because phenotype-altering agents are presented from within cells. A similar approach may also be utilized within 3D cell aggregates rather than adding soluble factors in the media (in which factors must diffuse through cellular barriers, potentially reducing efficiency and kinetic control) (Carpenedo et al, 2009; Bratt-Leal et al, 2013). For example, it has been shown that compared to conventional soluble delivery methods, bone morphogenetic protein 4 (BMP4) locally delivered via gelatin MPs inside 3D ESC spheroids led to efficient mesoderm induction, despite nearly 12-fold less total growth factors being used (Bratt-Leal et al, 2013). We envision that such biomaterial particle platforms could be adopted as stable intracellular depots of reprogramming factors, yielding direct, efficient, and prolonged pluripotency induction without frequent introduction of soluble reprogramming factors.
Figure 3.
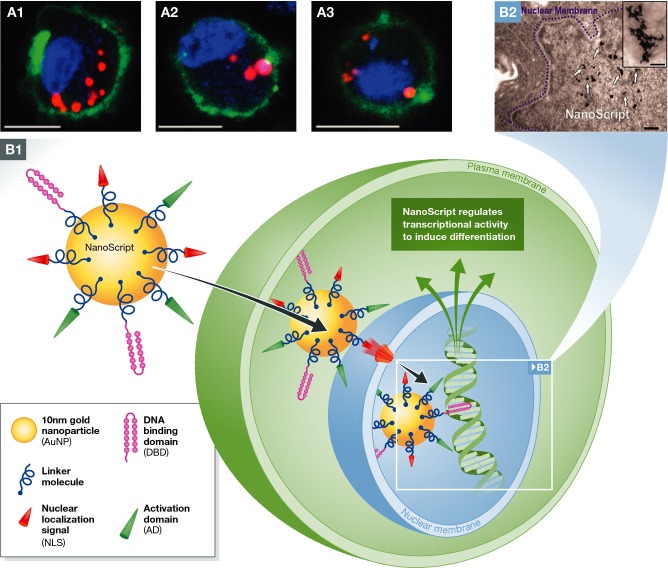
Emerging applications of biomaterial-based targeted modulation of cell phenotype and gene regulation with potential application for reprogramming somatic cells
(A) Confocal microscopy shows the intracellular localization of phenotype-altering agent-doped PLGA MPs in mesenchymal stem cells that can release agents for several weeks following internalization (A1) a mesenchymal stem cell, (A2) a MIN6 beta cell, and (A3) a RAW 264.7 macrophage. This robust particle platform could potentially serve as an intracellular depot for sustained presentation of reprogramming factors to achieve efficient iPSC derivation from multiple somatic cell types. Scale bars, 10 μm. Green (DiO stain), membrane; red (rhodamine 6 g), particles; blue (Hoechst), nuclei; Adapted with permission from Ankrum et al (2014b). (B) One potential biomaterial strategy for controlled regulation of gene expression is nanoparticle-based artificial transcription factors (NanoScript). This platform could be potentially adopted for the activation or expression of pluripotency-associated genes for improved iPSC derivation. B1: NanoScript is devised to emulate the structure and function of TFs by assembling the principle components, DBD, AD, and NLS, onto a single 10-nm gold nanoparticle via molecular linkers. This design enables the penetration through plasma membrane and entrance into the nuclear membrane through NLS–nuclear receptor coupling. NanoScript interacts with DNA and triggers transcriptional activity leading to desired gene regulation. B2: transmission electron microscopy (TEM) micrograph demonstrates the localization of NanoScript clusters within the nucleus (scale bar = 200 nm), with the inset showing individual nanoparticles (scale bar, 100 nm). Adapted with permission from Patel et al (2014).
2. Biomaterials for potential modulation of delivery kinetics of multiple reprogramming factors
Small-molecule- or protein-based iPSC derivation protocols employ multiple cocktails to reprogram cells (Kim et al, 2009; Zhou et al, 2009; Hou et al, 2013). The sequential introduction of individual time-sensitive reprogramming factors with varied timing and duration appears to be indispensable for the activation of early biological events [e.g. epithelial-to-mesenchymal transition (EMT)], essential for the following distinct reprogramming phases (David & Polo, 2014) that are important for improved pluripotency induction (Liu et al, 2013a, 2014a). Although these state-of-the-art regimens have significantly advanced the iPSC field, further improvements for tightening control over these systems is required. Biomaterial-based methods are poised to refine, simplify, and improve the efficiency and safety of these protocols. Biomaterials may be used to control timing of factor delivery in ways soluble small molecule cocktails cannot (e.g. biomaterials can achieve targeted release with pulsatile or sustained dosing) (Martinez et al, 2013). Furthermore, biomaterials may simplify derivation protocols by reducing the number/dosage of interventions necessary for stable pluripotency induction (e.g. responsive biomaterials that deliver multiple components at the desired times) (Bratt-Leal et al, 2013; Cheng et al, 2013). Biomaterials may also improve derivation safety by eliminating cytotoxic solvents used in the traditional small-molecule-based reprogramming methods.
Herein we outline how biomaterial-mediated iPSC derivation can regulate reprogramming events in somatic cells with superior precision and efficiency through controlled delivery of multiple reprogramming factors, further improving the current reprogramming protocols (Robinton & Daley, 2012; Hu, 2014). This regulatory potential has not yet been achieved experimentally. Although time is required to design and implement biomaterial strategies to deliver these intricate combinations of small molecules or proteins, we can envision how these platforms may advance the iPSC field by taking inspiration from successful delivery of drugs in other settings.
Scaffolds may also be doped with factors (including growth factors) prior to cell seeding to control cellular responses (Ma et al, 2013a). For example, Richardson et al (2001) developed a simple, single PLGA-based scaffold for the sequential release of dual angiogenic factors [vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF)]. The growth factors were loaded into PLGA scaffolds by either mixing with the polymer prior to scaffold formation (VEGF) or pre-encapsulated into PLGA MPs (PDGF) used for scaffold fabrication. The resultant dual factor-containing scaffold produced rapid release of VEGF, which was primarily associated with the surface of the scaffolds, and much slower release of PDGF, which was more evenly distributed throughout the scaffold, primarily released through the degradation of PLGA. Therefore, the balance of these two release profiles can be further tailored, if needed, by tuning the degradation rate of PLGA as discussed earlier. While this platform was designed for tissue regeneration use, such versatile, single polymer-based scaffolds may be used to codeliver combinations of reprogramming factors with distinct kinetics to achieve improved reprogramming efficiency. In addition, the robustness of a PLGA-based release platform can be used to deliver drugs with diverse physicochemical properties including simultaneous release of hydrophobic and hydrophilic agents (Zhang et al, 2007). Biomaterials can be used to regulate multiple and individual factor release with a single delivery system and this should be useful for next-generation iPSC derivation protocols.
3. Potential of artificial transcription factors for reprogramming
Biomaterials may also be harnessed to create nanoparticle-based artificial transcription factors (TFs) for efficiently controlling gene regulation and may be used for cellular reprogramming. Recently, Patel et al (2014) designed a platform to mimic TF domains (“NanoScript”) by conjugating cell-penetrating peptides and synthetic TFs onto gold nanoparticles. The synthetic TFs recapitulated their native gene regulation activity by mimicking the three principle TF components—nuclear localization signal (NLS), DNA-binding domain (DBD), and activation domain (AD)—which were tethered in close proximity on the gold nanoparticles (Fig3B). Furthermore, the gold nanoparticle not only served as the delivery vehicle, but also functioned as the linker domain (LD) of the synthetic TF. NanoScript effectively transcribed desired genes on endogenous DNA by localizing to the nucleus and initiating transcription of a reporter plasmid with a 15-fold increased efficiency compared to control groups (individually added TF components). This system may find utility in reprogramming somatic cells to iPSCs. Such biomaterial-based platforms may not only reduce safety concerns associated with viral vectors, but also enhance reprogramming efficiency with superior tunability.
4. Biomaterial-induced epigenetic regulation of iPSCs
In addition to direct delivery of reprogramming factors to improve reprogramming efficiency, existing iPSC derivation methods can be complimented through modulating the epigenetic state of somatic cells via engineering the cellular microenvironment. The physical properties of substrates on which iPSCs grow serve a vital role in regulating the cellular epigenetic state, and hence, reprogramming. A recent study by Li et al demonstrates that induction of iPSCs by exogenous transcription factors could be markedly enhanced by seeding murine or human fibroblasts onto polymer substrates with specialized surface topography or onto nanofibrous scaffolds with anisotropy (Downing et al, 2013). Specifically, micro- and nanopatterned polydimethylsiloxane (PDMS) substrates, especially those with microgrooves 10 μm wide/spaced, significantly promoted the expression of epithelial and pluripotency markers by triggering a more elongated cell morphology, which led to the induction of histone modifications (compared to cells with a circular shape) essential for epigenetic regulation and reprogramming. This process is thought to be mediated by mechanotransduction signaling through the cytoskeleton that interferes with acetylation and methylation of DNA-packing histones (Xu et al, 2013b). The microgrooved PDMS surface or aligned poly(L-lactide-co-caprolactone) nanofibers accelerated cellular reprogramming, in part by altering the expression of key epigenetic regulators. Modulating epigenetic regulators through biophysical features of the cellular substrate and cellular mechanosensing could serve as an important method for reprogramming somatic cells. Interestingly, substrate-induced mechanosensing may effectively bypass the effects of soluble biochemical factors for stem cell fate conversion (Li et al, 2011; Xu et al, 2013b; Dalby et al, 2014). Elucidating and harnessing the potential interplay between physical and biochemical cues, along with engineering the cellular microenvironment using well-defined biomaterials, may uncover new avenues for controlling the fate and function of stem cells with improved efficiency.
Engineered biomaterials for the expansion of iPSCs
Limitations of traditional iPSC expansion systems
Traditional PSC expansion systems are generally associated with safety and scalability issues. Many expansion protocols for PSC and iPSC require mouse embryonic fibroblasts (MEFs) as a feeder layer. Feeder cells produce essential cytokines, growth factors, and ECM molecules to support the self-renewal of PSCs (Hongisto et al, 2012). However, it is difficult to verify and ensure the reproducibility of their secretome contents, which are chemically undefined (Villa-Diaz et al, 2009). Additionally, human and murine PSCs show different dependence on MEF feeder layers, limiting the applicability of MEF feeder-based culture systems. Furthermore, potential animal-derived pathogens or xenogenic contaminants [e.g. N-glycolylneuraminic acid (Neu5Gc) (Heiskanen et al, 2007)] associated with MEF feeder layers and xeno-derived culture additives could compromise the quality and stability of iPSCs. Overall, these limitations may negatively impact scale-up and banking of iPSCs as a clinically compliant therapeutic product (Ahrlund-Richter et al, 2009).
To overcome the shortcomings of feeder layers, extensive effort has been devoted to the development of feeder-/xeno-free systems to expand iPSCs (Celiz et al, 2014; Lee et al, 2014). Matrigel™ has been widely employed as an alternative iPSC expansion substrate to bridge the gap between feeder-based and defined feeder-free iPSC expansion (Villa-Diaz et al, 2013). Matrigel™ is derived from mouse sarcomas and is enriched with ECM components such as laminin, fibronectin, collagen IV, heparin sulfate proteoglycans, and other growth factors (Kleinman et al, 1982). However, its undefined chemical compositions, in addition to being animal-derived, result in considerable lot-to-lot variability and potential xenogenic contamination, thus challenging its use in robust in vitro models and in large-scale production. Despite these challenges, Matrigel™ remains one of the most commonly used substrates for iPSC culture and serves as an important starting point to identify the required conditions for iPSC growth and to develop defined substrates for expanding iPSCs in an efficient and clinically compliant manner.
Alternative biomaterial platforms for high-efficiency iPSC expansion
To address the safety and efficiency issues associated with the aforementioned conventional expansion approaches, biomaterials have been explored in the development of chemically defined, xeno-/feeder-free culture platforms for (large-scale) efficient iPSC expansion. These biomaterial-based substrates or matrices primarily aim to harness or emulate the cell–matrix interactions occurring within the native stem cell microenvironment, which are crucial for the adhesion, growth, maintenance, and fate regulation of PSCs (Watt & Huck, 2013). Such matrix-mediated cellular responses are largely attributed to the specific physicochemical properties of the substrates/matrices such as matrix rigidity, surface chemistry, relative hydrophilicity (wettability), and topography. Rational engineering of these physicochemical features could involve, for example, the binding/pre-adsorption of essential ECM proteins found in the niche (e.g. vitronectin) with active conformations. Additionally, substrates can be engineered to sequester growth factors (supplemented or endogenous), creating an instructive, cell-interactive milieu to support the long-term self-renewal of hPSCs or their progeny (Chang et al, 2013; Belair et al, 2014). Rationally engineered biomaterials have been explored for iPSC expansion, ranging from complex combination of ECM proteins [analogous to Matrigel™, but with defined composition (Evseenko et al, 2009)] to simple UV-/ozone-treated plastic surfaces (Saha et al, 2011), which are becoming progressively simpler, more cost-effective, and scalable (Celiz et al, 2014) (Fig4). Most of these expansion substrates are applicable to chemically defined, xeno-free culture medium (e.g. mTeSR™), with a few of them exhibiting the support for iPSC expansion at levels similar or perhaps even greater to that of Matrigel™ (currently considered the gold-standard expansion substrate) (Celiz et al, 2014). Defined synthetic substrates, which comprise “off-the-shelf” components, are becoming easier to synthesize with low cost and display significant advantages for industrial scale-up of iPSCs. In this section, we will highlight several representative iPSC expansion substrates and provide our perspective on the promise of defined, synthetic iPSC expansion substrates for improved control of culture conditions and improved expansion efficiency and scalability.
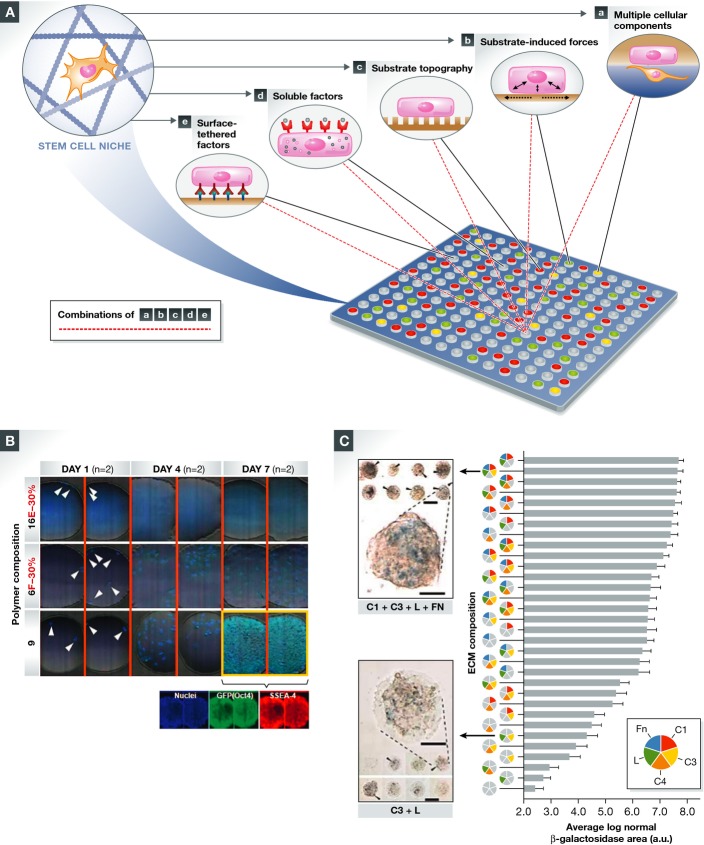
Figure 4.
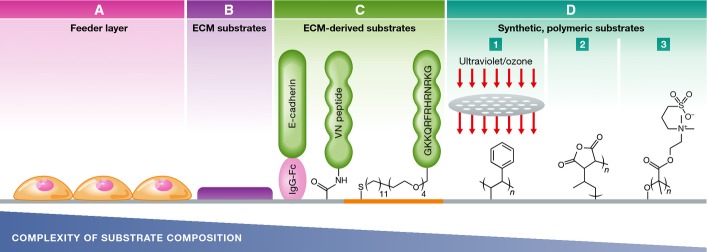
The development of PSC/iPSC expansion substrates: advancing from a complex, chemically undefined, feeder layer-based system to simple, synthetic polymeric substrates with improved efficiency, scalability, and reproducibility
(A) MEF feeder layers support PSC adhesion and self-renewal via their specific secretome contents. (B) ECM-coated substrates composed of an undefined mixture of ECM proteins such as Matrigel™. (C) Surface-tethered functional epitopes derived from ECM components such as E-cadherin and vitronectin (VN)-derived peptides [e.g. heparin-binding peptide, GKKQRFRHRNRKG (Klim et al, 2010)]. (D) Synthetic, polymeric substrates support the attachment and self-renewal of iPSCs via interface-mediated adsorption of essential adhesive ECM components from the culture medium. Examples for such substrates include the following: (1) ultraviolet-/ozone-modified TCPS; (2) poly(methyl vinyl ether-alt-maleic anhydride); (3) poly[2-(methacryloyloxy)ethyl dimethyl-(3-sulfopropyl)ammonium hydroxide] (PMEDSAH). Figure adapted and modified with permission from Celiz et al (2014).
1. ECM protein-based 2D substrates
To replace Matrigel™ for the growth of ESCs and iPSCs, several cell adhesion ECM proteins including, but not limited to, laminin (Lu et al, 2014), fibronectin (Amit et al, 2004; Kalaskar et al, 2013), and vitronectin (Braam et al, 2008; Chen et al, 2011) are actively being studied. For example, one laminin isoform, LN521, has been identified as a promising substrate for iPSC expansion (Rodin et al, 2014a). LN521 has been used as a sole substrate for iPSC derivation, expansion, and differentiation in a xeno-free medium (Rodin et al, 2014b). This laminin isoform supported the integration-free reprogramming (> 0.18% efficiency, comparable to Matrigel™ coating) and the expansion of human iPSCs, facilitating prolonged passaging (> 10 passages). Similarly, Kim et al (2013) established a xeno-free, vitronectin-based substrate for long-term production (> 100 passages) of human iPSCs. The iPSCs were transferred 5 days after reprogramming onto a vitronectin-coated culture plate in MesenGRO medium and cultured for additional 2 days, followed by switching the culture medium to a chemically defined medium (HF12SM) supplemented with a cocktail of signaling molecules (e.g. protein kinase C inhibitors). This chemically defined medium functioned synergistically with the vitronectin coating to induce and maintain iPSCs with high colony-forming efficiency and pluripotency. A similar vitronectin substrate has been utilized for generating and expanding functional iPSCs from human urine-derived cells as well (Lee et al, 2014). These findings reinforce that vitronectin is a robust substrate that can assist the self-renewal of iPSCs derived from different somatic cell types. Typical cell–ECM linkers such as integrin αVβ5 or α6β1 have been shown to be critically involved in the ECM-induced high-efficiency expansion of iPSCs (Sams & Powers, 2013). As our understanding of multiple cell adhesion receptor–ligand interactions improves, the list of ECM protein-based iPSC expansion substrates accrues (Miyazaki et al, 2012; Celiz et al, 2014). However, an important priority is to perform systematic studies that compare the impact of individual ECM proteins or their combinations on iPSC growth and expansion.
2. Synthetic peptide- and polymer-based 2D substrates
Despite their improved efficiency, native ECM protein substrates such as laminin and vitronectin are not ideal due to their large, temperature-/pH-sensitive conformations, which make them unstable for long-term use (e.g. > 1 month) (Wong et al, 2014). To overcome this limitation, stable synthetic peptide, polymer (or conjugates), and proprietary macromolecule substrates have been developed to facilitate long-term iPSC culture (Derda et al, 2007; Klim et al, 2010; Liu et al, 2011; Jin et al, 2012; Deng et al, 2013; Qian et al, 2014). Many of these systems often exploit certain biological motifs [e.g. adhesive peptides or heparin-mimicking moieties (Chang et al, 2013)] to attach cells on different substrates [e.g. tissue culture polystyrene (TCPS), hydrogels] and have been used in conjunction with chemically defined maintenance medium for high-efficiency iPSC propagation (Villa-Diaz et al, 2013).
For example, a commercially available material, Synthemax™, which is a biologically active peptide-functionalized acrylate polymer, has been shown to support the attachment, spreading, and proliferation of human iPSCs. The active, relatively short peptide sequences are synthetic and hence more stable and scalable compared to their complex, full-length ECM counterparts (Melkoumian et al, 2010). The Synthemax™ substrate is based on an Arg-Gly-Asp (RGD)-containing short peptide, which is derived from vitronectin and conjugated to a 2D acrylate surface via a poly(ethylene oxide) spacer. The presence of the spacer is thought to reduce steric hindrance, thus promoting peptide-induced iPSC adhesion and proliferation via the integrin machinery described above for vitronectin (Jin et al, 2012). The iPSCs maintained their pluripotency and normal karyotype on Synthemax™ for more than ten passages (Jin et al, 2012). Moreover, this system is amenable to standard sterilization techniques, such as γ-radiation, and can be scaled to large culture-vessel formats, suggesting its potential for large-scale manufacturing of iPSCs for in vitro applications (e.g. high-throughput drug screening) or clinical products (Melkoumian et al, 2010). This synthetic substrate further exemplifies the potential of harnessing cell–matrix interactions to enhance the efficiency and safety of iPSC production. Alternatively, synthetic substrates and peptides can help to minimize anoikis-induced cell death (caused when cells detach from their ECM) and facilitate iPSC expansion. For instance, QHREDGS, a conserved peptide fragment identified in angiopoietin-1 [a glycoprotein responsible for endothelial cell adhesion and vascularization (Miklas et al, 2013)], was utilized as a defined culture surface for iPSC maintenance (Dang et al, 2014). The QHREDGS peptides, which were chemically tethered to a non-fouling polyethylene glycol (PEG) hydrogel surface, markedly improved the adhesion of iPSCs, a process that appeared to be mediated by β1-integrin receptors. While this peptide promotes survival and proliferation of multiple other cell types (Rask et al, 2010), long-term survival of undifferentiated iPSCs on this peptide-immobilized substrate is currently unclear and needs to be assessed.
When considering the potential of bringing exogenous iPSC progeny to the clinic, scaling up with batch-to-batch consistency becomes a primary concern (Brindley et al, 2013). For that reason, synthetic peptide-functionalized substrates may be challenging owing to their high manufacturing costs and extra conjugation steps (Place et al, 2009). To lower the cost of iPSC expansion, a synthetic polymer-based substrate, poly[2-(methacryloyloxy) ethyl dimethyl-(3-sulfopropyl) ammonium hydroxide] (PMEDSAH, a zwitterionic hydrogel), could be useful (Villa-Diaz et al, 2010). Unlike the natural and recombinant ECM substrates discussed above, PMEDSAH is chemically defined and therefore offers the advantages of long-term stability and storage. Its reproducible synthesis is possible at large scales with relatively low costs (Villa-Diaz et al, 2010). PMEDSAH has supported prolonged iPSC colony formation and proliferation in an undifferentiated state for greater than 20 passages and consequently seems to be superior to Synthemax™ for scaling up iPSC production (Villa-Diaz et al, 2012). Alternatively, Chang et al (2013) reported a hydrogel-based substrate containing synthetic heparin-mimicking moieties that enables long-term self-renewal of hPSCs (> 20 passages). The hydrogel substrates bearing different functional groups, hydrophilicity, and matrix rigidity were synthesized via copolymerization of acrylamide (PAm) with synthetic heparin mimics [poly(sodium 4-styrenesulfonate) (PSS)] or with monomers enriched with carboxylate groups using varied molar ratios. The PSS moieties are capable of sequestering basic fibroblast growth factor (bFGF) that mediates FGF signaling (Sangaj et al, 2010) and self-renewal of hPSCs (Levenstein et al, 2006). Specifically, the hydrogel with PAm and PSS at a molar ratio of 6:2 (PAm6-co-PSS2) was most supportive for hPSC expansion. It also exhibited the ability to form compact colonies as observed with Matrigel™, albeit with slightly lower population doubling rate. Interestingly, the PAm6-co-PSS2-induced effect is highly dependent on its surface hydrophilicity and functional groups. The PAm hydrogel substrates copolymerized (equal 6:2 molar ratio) with the aforementioned PMEDSAH monomer (PAm6-co-PMEDSAH2, similar rigidity and functional groups but nearly threefold lower hydrophilicity) or with carboxyl-enriched monomer (similar rigidity and hydrophilicity but more carboxylate groups) exhibited minimal to no cell adhesion. These observations may be attributed to the significant alterations in the extent of BSA, vitronectin, or bFGF adsorbed/bound to surfaces of varied physicochemical properties. Moreover, compared to the carboxyl-enriched surface, the cells on PAm6-co-PSS2 surface showed higher endogenous expression levels of multiple integrin receptors, ECM proteins, and remodeling enzymes that are known to be essential for the self-renewal of hPSCs (Chang et al, 2013). These findings collectively highlight the significance of the cell–matrix interface for the adhesion and long-term self-renewal of hPSCs. We envision that the findings discussed herein for hPSCs can be readily applied to iPSCs. Chemically defined, synthetic substrates could offer significant advantages for the long-term expansion of iPSCs through rationally engineered cell–material interfaces.
3. Scalable, synthetic 3D expansion platforms
Despite their promise, the aforementioned 2D substrates lack important physiologically relevant interactions that 3D culture systems provide including native cell–cell and cell–matrix interactions (Kraehenbuehl et al, 2011). Compared to 2D monolayers, 3D culture is thought to be more efficient for providing spatial–temporal signals [e.g. 3D cell–ECM interactions and growth factor gradients (Sant et al, 2010)] that are essential for cell proliferation and function (Han et al, 2014). Furthermore, shifting from a 2D monolayer to 3D culture platform significantly increases the space for cells to propagate without causing unfavorable stacking/agglomeration in the third dimension, which is advantageous for the scale-up of iPSC expansion (McDevitt, 2013). Lei and Schaffer (2013) recently demonstrated the promise of using 3D scalable, synthetic platforms for high-efficiency iPSC expansion. Numerous synthetic or natural biomaterials including alginate, agarose, hyaluronic acid (HA), and poly(N-isopropylacrylamide)-co-poly(ethylene glycol) (PNIPAAm-PEG) hydrogels were compared for their ability to maintain the expansion and differentiation of iPSCs. Their multifactorial optimization studies revealed that under a specific predetermined cell encapsulation density, the thermoreversible PNIPAAm-PEG gel exhibited significant efficacy of promoting long-term iPSC expansion, with high growth rate, purity, and fidelity of pluripotency. Specially, this system achieved a ∼20-fold expansion of iPSCs per passage within only 4–5 days of cultivation and further attained long-term culture with high expansion rate (1072-fold over 280 days). This 3D iPSC expansion system can generate significantly higher cell densities and larger spheroids than 2D adherent systems, despite utilizing comparable amounts of culture media and exchange intervals (McDevitt, 2013). Additionally, the differentiation into all three germ layers and teratoma formation following long-term expansion in the PNIPAAm-PEG gel were confirmed in vitro and in vivo, respectively. The robustness of this 3D hydrogel expansion system was also validated in parallel with multiple human ESC cell lines. Chemically defined and scalable, this type of synthetic 3D matrix is a promising platform for the biological investigation and clinical development of iPSCs-based approaches.
4. Advantages of defined, synthetic polymer-based expansion platforms
Regardless of 2D or 3D culturing approaches, the utility and translation of ECM- or ECM-derived peptide-based substrates are often limited by their scalability, reproducibility, and cost (especially when it is critical to ensure high purity and pathogen-free conditions). In contrast, synthetic polymer-based substrates can be engineered to achieve minimal batch-to-batch variations and can offer reasonable scalability, which is a challenge with traditional substrates. Moreover, the physicochemical and mechanical properties of synthetic polymers are in general highly tunable. The ability to modulate the stiffness of the culture substrate can augment the synergy between integrin and soluble growth factor signaling, leading to enhanced proliferation and inhibited differentiation of ESCs (Saha et al, 2006; Xu et al, 2010; Dingal & Discher, 2014). Moreover, synthetic materials are typically amenable to nanoscale synthesis and automation technologies, thus can be easily customized into high-throughput platforms. Such platforms enable efficient manipulation of the physicochemical and mechanical properties of the substrates, along with the addition of soluble factors, to study their interplay with stem cell proliferation and differentiation with high levels of cellular readout. This principle has been recently demonstrated by several elegant studies involving high-throughput biomaterial arrays and ESCs/iPSCs (Anderson et al, 2004; Mei et al, 2010; Zonca et al, 2013).
Functionalizing synthetic substrates with biomolecules (e.g. growth factors) may promote iPSC expansion through enhancing the material interface-mediated signaling essential for stem cell self-renewal. For instance, the leukemia inhibitory factor (LIF) mitogen when covalently tethered onto polyester fiber substrates (Cetinkaya et al, 2007) or onto surfaces pre-engineered with a growth factor linker [poly(octadecene-alt-maleic anhydride) (Alberti et al, 2008)] showed significant support for the expansion of ESCs using a small quantity of growth factors (less than used to supplement media). Through rational selection of conjugation chemistry, the surface-tethered growth factors preserved their bioactivity and active conformation. In contrast to soluble factors that often require high concentrations in media, and whose delivery kinetics are burdened by media changes, surface-functionalized biomaterials can more efficiently and cost effectively display signals [e.g. activation of the downstream mitogenic signaling pathways required for stem cell self-renewal (Orford & Scadden, 2008; Cabanas-Danés et al, 2014)]. However, there are some caveats. Not all immobilized cues may function the same, some may require liberation from the surface to exhibit an effect (this can be achieved with enzyme cleavable linkers) (Lee et al, 2011). Also, combinations of soluble and immobilized cues may be required to achieve desired effects.
An ideal expansion platform should enable large-scale iPSC expansion with batch-to-batch consistency at low cost, demonstrate excellent stability for long-term storage and use, and be compatible with common biomedical sterilization techniques. Reusability would be an added benefit for prolonged cell propagation. Chemically defined, synthetic substrates may address all of these criteria. Alternatively, iPSC self-renewal can be enhanced on standard TCPS substrates through harnessing substrate-induced protein adsorption/cell adhesion without additional chemical modifications. For example, patterning TCPS with UV/ozone radiation can produce a favorable surface for iPSC growth and reprogramming, and gene-targeting protocols can be readily performed on the attached cells (Saha et al, 2011). To achieve the full potential for large-scale iPSC expansion (billions of cells), the aforementioned synthetic substrates could be readily engineered and incorporated into microcarrier/suspension bioreactor systems. Compared to a 2D static culture platform, microcarrier-/bioreactor-based expansion systems generally offer higher cell yield per volume, superior process controllability, and simpler handling, thus enabling cost-effective scaling-up of stem cell production (Liu et al, 2014b). Specifically, microcarriers (e.g. commercially available CultiSpher-S and Cytodex-3 systems) with well-defined microstructure and surface chemistry, when employed in conjunction with spinner flask-based dynamic culture, have been shown as a viable and economical alternative to conventional static adherent culture platforms for the large-scale expansion of multiple stem cell types (Alfred et al, 2011; Jung et al, 2012).
In addition, computational modeling (Epa et al, 2012) and high-throughput biomaterials/ECM screening technologies (Underhill & Bhatia, 2007; Mei, 2012) that enable systematic investigation of material property–cellular function relationships can be used to accelerate material discovery toward improved large-scale iPSC production. While available synthetic substrates used for iPSC expansion are far from ideal (e.g. mostly require the pre-adsorption of adhesive proteins), biocompatible, chemically defined synthetic controllable substrates could serve as the next generation of substrates for large-scale iPSC production with good manufacturing practices (GMP)-compliant quality (Chen et al, 2014).
Advanced biomaterials for the directed differentiation of iPSCs: regenerative medicine applications
While ESCs and iPSCs both represent a useful resource for tissue regeneration due to their unlimited self-renewal and capacity to differentiate into all three germ layers, unlike ESCs, iPSCs can be used to generate patient-specific cells and tissues. This makes iPSC more attractive for clinical translation as they can bypass the barriers related to immune rejection (Okano et al, 2013). Unfortunately, despite rapid advances in iPSC research, iPSC-based cell therapies suffer from limited quantities of transplantable cells, low engraftment efficiency, unstable lineage specification, and uncontrolled host tissue response (Dimmeler et al, 2014; Tabar & Studer, 2014). These problems must be addressed for iPSC-based technologies to realize their potential in regenerative medicine.
Progress in the development of biomaterials for iPSC research and therapy may help address these safety and efficiency limitations. Significant efforts are underway to engineer viable, tissue-mimetic microenvironments for directed cell proliferation, differentiation (i.e. highly efficient and lineage specific), and generation of functional tissue substitutes in vitro. For example, the aforementioned “particle-in-cell” platforms have been incorporated into ESC embryoid bodies (EBs) to present requisite extracellular cues or morphogenic factors in a biologically relevant and spatiotemporally controlled manner. This has resulted in organized differentiation of ESCs within the 3D EBs with efficiency superior to that achieved by direct presentation of soluble factors (Carpenedo et al, 2009; Bratt-Leal et al, 2011, 2013). A similar strategy uses 3D scaffold-guided ESC culture where controlled delivery of non-viral siRNA attained high levels of ESC transfection (up to 90% compared to < 40% on 2D culture substrates). Tight control over cell differentiation was achieved by specifically blocking genes representative of one germ layer (Zoldan et al, 2011). These strategies should be applicable for modulating the fate and functions of iPSCs or their progeny for efficient and safe regenerative applications. Moreover, the engineering approaches discussed herein may also provide supportive interfaces for facilitating graft acceptance, cell engraftment, and functional integration with host tissues in vivo. In this section, we discuss initial regenerative biomaterial-based iPSC applications and their promise for the field.
Neural tissue regeneration
Directed and efficient differentiation of stem cells to lineage-specific cells depends on the physicochemical properties of the ECM, such as the elastic moduli and topographical features, in addition to soluble factors (Lutolf et al, 2009). Biomaterials for neuronal differentiation tend to have low elastic moduli (< 1 kPa) to mimic the relative pliability of neural ECM (Leipzig & Shoichet, 2009). Recently, Musah et al (2014) robustly differentiated hESCs into neurons—in the absence of neurogenic factors—by solely tuning the mechanical properties of the underlying matrix. To assess the selective differentiation of hESCs on substrates having varying elasticity, they developed polyacrylamide (PA) hydrogels (0.7, 3.0, 10 kPa) and functionalized them with glycosaminoglycan (GAG)-binding peptides, which facilitated the attachment of hESCs (primed using embryoid body medium for 5 days). They showed that while stiffer gels (3.0 and 10 kPa) promoted hESC self-renewal, the compliant hydrogels (0.7 kPa) resulted in highly efficient and reproducible neuronal differentiation of hESCs (more than 80% of differentiated cells expressed neuronal marker Tuj1). Interestingly, the cells that attached to compliant hydrogels exhibited altered cytoskeleton primarily via decreased F-actin polymerization. This led to a decrease in the transcriptional regulatory activity of coactivator YAP—involved in signaling pathways regulated by chemical and mechanical stimuli—which was sequestered in the cytoplasm. On stiffer gels, YAP was present in the nucleus, while on compliant gels, YAP was excluded from the nucleus. Furthermore, to confirm that YAP depletion augmented neuronal differentiation, they knocked down YAP using RNAi. The YAP-depleted hESCs underwent neuronal differentiation when cultured on highly rigid polystyrene surfaces (106 kPa). Such studies underscore the importance of mechanical cues to control the differentiation of pluripotent stem cells.
Additionally, natural and synthetic surfaces engineered with topographical features enhance iPSC adhesion, promote neuronal differentiation, and guide the growth of axons (Wang et al, 2011; Kuo & Lin, 2013; Pan et al, 2013). In a study assessing the role of topographical features, Pan et al (2013) used PDMS substrates to investigate how differing widths of nanoscale channels for cell growth impacted neural differentiation of iPSCs. Surfaces with a narrower pitch (350 nm width) significantly enhanced the contact guidance and alignment of the seeded iPSCs. The nanostructured surfaces with either topographical cues alone or in conjunction with pre-neuronal induction of iPSCs markedly elevated the expression of neuronal markers. Although the specific mechanisms triggering the expression of neuronal markers in this study remain unexplored, cell–cell interactions (e.g. cell–cell distance controlled by pattern dimension) and cell–ECM interactions (laminin versus collagen) have previously been shown to affect neuronal differentiation of neural stem cells (Solanki et al, 2010) and may be involved.
To further improve the differentiation efficiency compared with using soluble inductive moieties alone, biomaterials can be chemically modified to display neural-inductive moieties. For example, the addition of synthetic neurotransmitter analogs has led to enhanced neuronal differentiation of iPSCs (Terashima et al, 2014; Zhang et al, 2014a). One can envision that immobilizing these moieties and enhancing their local availability may trigger relevant signaling cascades and promote the neuronal differentiation of iPSCs in a more robust manner (Kuo & Chung, 2012; Kuo & Wang, 2012; Kuo & Chang, 2013; Lei & Schaffer, 2013). In one example, a composite scaffold of alginate and poly(γ-glutamic acid) (γ-PGA) was engineered to control the neuronal differentiation of iPSCs through defined porosity and surface chemistries (Kuo & Chung, 2012). These porous scaffolds were surface-functionalized with a TATVHL peptide (transactivator of transcription (TAT)-VHL, known for stimulating neurite outgrowth), and exhibited a distinct stimulation of neurogenesis of iPSCs, as evidenced by the noticeable expression of neuronal marker Tuj1 and diminished expression of pluripotency markers over 7 days of culture (Kuo & Chung, 2012). Although utilizing tethered inductive moieties is a suitable approach to effect neuronal differentiation, potent soluble induction factors remain indispensable for stable and efficient iPSC neurogenesis. Numerous biomaterial-based approaches have been designed to control the presentation of neuron-inductive growth factors in a sustained fashion. Among these, functionalization of the synthetic substrates with growth factor-binding polydopamine (Yang et al, 2012) or heparin (Kuo & Wang, 2012) and utilization of nanoparticle-based delivery of plasmids expressing neurotrophin-3 (NT-3) (Chung et al, 2013) appear to be two viable approaches. These studies clearly suggest the potential of using biomaterials for inducing efficient neurogenesis of iPSCs.
Cardiovascular tissue regeneration
Many of the conventional methods for controlling the differentiation of PSCs/iPSCs into cardiac or vascular cell types require elaborate chemical induction regimens, but lack relevant physical contextual signals, for example, cyclic/mechanical loading or hypoxic conditions essential for the survival and maintenance of cardiac progenitor cells (Burridge et al, 2012; Liu et al, 2013b; Lane et al, 2014). Biomaterials offer integrative platforms for combining biochemical and physical signals. Ideally, biomaterials used for cardiac tissue repair should be mechanically robust to provide sufficient mechanical compliance matching with the host tissue. Multiple biopolymers including fibrin (Godier-Furnemont et al, 2011), alginate (Rosellini et al, 2009), collagen (Zhang et al, 2012), silk (Chi et al, 2012), poly(L-lactic acid) (PLLA) (Wang et al, 2014b), and poly(glycerol sebacate) (PGS) or its derivatives (Kharaziha et al, 2013; Lang et al, 2014) can be utilized for this purpose, and several have been studied in combination with iPSCs. For example, iPSC-derived smooth muscle cells (SMCs), when seeded in a porous PLLA scaffold, were able to maintain their mature SMC phenotype in vitro and promote the formation of vascular structures in vivo. Within 2 weeks of implantation in nude mice, the SMC-laden 3D PLLA constructs deposited substantial amounts of vascular tissue-specific matrix, and the restorative effect was dependent on the pore size and interconnectivity of the PLLA constructs (Xie et al, 2011; Wang et al, 2014b). These findings demonstrate the utility of biocompatible, synthetic scaffolds for achieving iPSC-derived vascular cell growth and function.
Under defined chemical induction cues, 3D cultures of iPSCs could elicit superior efficiency and control of fate decisions toward an endothelial lineage versus traditional 2D monolayer culture. For example, Zhang et al developed a 3D fibrin-based hydrogel system for generating functional endothelial cells (ECs) from iPSCs. This soft hydrogel network was chosen to potentially match the intrinsic softness of ESCs and embryos (400–600 Pa) and to facilitate EB formation (Chowdhury et al, 2010; Dingal & Discher, 2014) for subsequent endothelial induction. Additionally, the biocompatibility and cross-linkability of fibrin permits in situ cell encapsulation and implantation. Also, the fibrin network is amenable to easy digestion under cytocompatible conditions, enabling the release of EBs for downstream induction culture or cellular analysis. Fibrin gels allow 3D iPSC culture, which, beyond the geometric advantages already discussed, improves endothelial differentiation compared to 2D culture (Zhang et al, 2014b). Specifically, single human iPSCs were seeded either as a 2D monolayer (as control) or encapsulated into a fibrin gel and cultured in a defined endothelial induction medium for a total of 2 weeks. Immunohistological analysis revealed that the efficiency of endothelial induction from iPSCs was markedly improved (up to 45%) within the 3D fibrin scaffold. The resulting endothelial cells stably retained their functional phenotypes (uptake of acetylated low-density lipoprotein and formation of tubular structures on Matrigel™) for more than a month in vitro. Compared with 2D culture, the 3D environment appears to distribute precise stress and strain on the surrounding iPSCs and regulates their endothelial differentiation and morphogenesis (Rozario & DeSimone, 2010), leading to enhanced control of iPSC fate decision. Systematic studies elucidating the interactions of iPSCs and fibrin scaffolds engineered with a range of physicochemical properties (e.g. concentration, cross-linking density, porosity, density of ligands) are warranted to maximize the utility of the 3D environment for controlled iPSC differentiation.
In addition to architectural properties of biomaterial-based substrates, mechanical manipulation has also shown to be useful in promoting the cardiac differentiation and function of iPSC-derived cells. For example, human iPSC-derived cardiomyocytes encapsulated in a 3D collagen matrix and exposed to cyclic mechanical loading (Tulloch et al, 2011) significantly enhanced matrix fiber alignment and myofibrillogenesis of the cellular constructs, suggestive of cardiac tissue remodeling. While these architectural and mechanical signals show potential to increase differentiation efficiency, likely the combination of biomaterial scaffolds with the appropriate cellular niche elements will yield cardiac tissues possessing superior function. Coculture of stromal supporting cells and human iPSC-derived cardiomyocytes in these 3D collagen constructs showed a tenfold improvement in the proliferation of cardiomyocytes and the formation of vascular structures (Tulloch et al, 2011). When such constructs were transplanted in the hearts of athymic nude rats, the human cardiomyocytes survived, and the grafts closely integrated into host myocardium one week post-transplantation, as evidenced by the perfusion of neosynthesized human microvessels by the host coronary circulation. These results collectively highlight the potential synergism of 3D biomaterial matrices with physiologically relevant regulatory cues (stromal niche and mechanical stress) as an effective approach for functional cardiac repair.
Musculoskeletal tissue regeneration
Biomaterials with robust mechanical properties, high strength/stiffness, and amenability to processing into porous or fibrous structures are promising alternatives to traditional TCPS substrates for the controlled differentiation of iPSCs or their progeny toward musculoskeletal lineages and their eventual implantation. Examples include PLLA, hydroxyapatite (D'Angelo et al, 2012), PCL (Jin et al, 2013; Tong et al, 2014), and silk (Ye et al, 2011). These materials have been explored for the regeneration of musculoskeletal tissues given: (i) Stiffer substrates tend to foster the early fate commitment of stem cells toward musculoskeletal phenotypes (Mullen et al, 2013); (ii) mechanically competent scaffolds provide some physical support and protection of entrapped cells upon implantation, especially for tissues that are constantly load-bearing (Tong & Jia, 2012; Xiao et al, 2013; James & Laurencin, 2014); (iii) a porous/fibrous interface or 3D inner architecture can promote neovascularization and host tissue integration that can mimic properties of musculoskeletal tissues that exhibit inherent anisotropy (Place et al, 2009).
In addition, biomaterials may replace soluble osteogenic induction factors partially or entirely while maintaining high differentiation efficiency. For example, human iPSCs cultured with mineralized gelatin methacrylate-based matrices readily differentiated into osteogenic cells (within 2 weeks of culture) in the absence of typical osteoinductive factors both on 2D surfaces and in 3D scaffolds (Kang et al, 2014). This may also be achieved via naturally derived scaffolds (de Peppo et al, 2013). Specifically, iPSCs-derived mesenchymal progenitor cells interfaced with mechanically compliant biomaterials (a decellularized bovine trabecular bone scaffold) for 5 weeks exhibited distinct expression of bone lineage-specific genes and significant bone-like tissue formation. Such scaffold-induced effects were also significantly enhanced by the dynamic cultivation conditions created by a perfusion bioreactor. cDNA microarray analysis for the progenitor cells before and after 3D dynamic cultivation suggested the occurrence of a proliferation/differentiation switch during the 5 weeks of culture, indicative of committed bone lineage maturation and reduction in teratoma-forming potential. Furthermore, these in vitro engineered bone substitutes were implanted subcutaneously in nude mice for 12 weeks. The explants displayed a mature, dense bone-like tissue formation with the absence of undesired cell types. More importantly, distinct recruitment of osteoclasts and microvasculature ingrowth were observed along/across the entire explant construct, demonstrating successful bone tissue remodeling. These findings collectively suggest that efficient tissue-specific commitment of iPSC-derived progenitor cells down the osteogenic lineage can be achieved through the rational combination use of scaffolds and bioreactors.
Liver tissue regeneration
To address the huge need for transplantable liver tissue, approaches that harness liver tissue engineering principles are being investigated. Compared to conventional 2D monolayer cultures, 3D encapsulation of hepatocytes in alginate (Ozawa et al, 2013), collagen (Yip & Cho, 2013), or other synthetic hydrogels (Li et al, 2014; Malinen et al, 2014) accelerates the formation of hepatic spheroids with improved function. Hepatocytes and endothelial cells are two of the major cell types found in liver tissues (Kmiec, 2001). Cocultures of hepatocytes and neighboring non-parenchymal cells, for example, endothelial cells, show improved liver-specific drug metabolism function of hepatocytes and prolonged survival. This is primarily attributed to enhanced reciprocal signaling interactions. The impact of such interactions tightly relies on the spatial patterning of the cocultured cell types (Bhatia et al, 1999; Tukov et al, 2006; Li et al, 2014). For example, a composite polyelectrolyte fiber gel was formed by multi-interfacial polyelectrolyte complexation (MIPC), where fibers are drawn from two or more interfaces between two oppositely charged polyelectrolytes, resulting in isolated domains within single fibers. This gel was developed for liver tissue engineering using a modular approach involving multiple cell types derived from human iPSCs (Du et al, 2014). In this study, hepatocytes and endothelial cells were initially differentiated from human iPSCs via chemically defined factors and a multi-step protocol. These two cell types were separately encapsulated in different compartments of MICP fibers and subsequently assembled into a stratified liver-mimetic architecture. The spacing of endothelial domains in the construct was 200–250 μm such that hepatocyte domains resided within the diffusion limit of oxygen and nutrients (Jain et al, 2005). Coculture of hepatocytes and endothelial cells in their specific niche locations markedly improved hepatocyte function (e.g. albumin secretion) in vitro and facilitated neovascularization when implanted into a murine partial hepatectomy model. When the engineered constructs were implanted in mice for 2 weeks, human albumin was detected in the serum, indicating functional integration of vascularized implants with host blood vessels. The alternating gel fibers (chitin–collagen versus chitin–galactose) assembled via interfacial alginate represent a permissive matrix for the desired phenotypes of both cell types, yet the underlying molecular mechanism remains to be elucidated.
Unlike the aforementioned fiber system that is introduced after the derivation of hepatic cells from iPSCs, a continuous alginate hydrogel system employed for both iPSC colony/EB formation and hepatic differentiation was recently established for improved differentiation efficiency (Lau et al, 2013). This system relies on a unique, interconnective porous structure [microcavitary hydrogel (MCG)] designed to both enhance nutrient exchange and increase living space for rapid expansion of iPSCs and subsequent EB formation. To evaluate the system, single murine iPSCs were encapsulated in the alginate MCG scaffolds and cultured for 10 days until significant formation of EBs. Defined inductive factors were then applied stepwise to coax their differentiation into endodermal (100 ng/ml activin A) and subsequently hepatic lineages (20 ng/ml HGF, 10 ng/ml oncostatin M). The resulting cells encapsulated in the 3D MCG gel generated a markedly higher level of urea and albumin production compared to that of the 2D monolayer culture. Importantly, the cells maintained in 3D MCG gel minimally differentiated toward unfavorable lineages in the presence of leukemia inhibitory factor (LIF). Therefore, a single 3D biomaterial culture system permitted the entire process of iPSC expansion, EB formation, endoderm differentiation, and hepatic maturation within a month by simply switching soluble inductive factors. This versatile hydrogel system could serve as a potent platform for efficient, functional hepatocyte differentiation, as well as a practical cell delivery vehicle for liver regeneration. Compared to traditional 2D culture, the 3D porous scaffold utilized herein for liver tissue offers an additional dimension for efficient colony expansion (less constrained) and potentially elicits a synergistic role with soluble inductive cues for directed iPSC fate control.
Emerging applications of biomaterials with iPSCs
The combination of biomaterials and iPSCs for disease modeling
An important emerging application of iPSC technology includes creating patient-specific disease models (Rosenzweig, 2010). iPSCs may be derived from diseased and healthy tissue alike, and thus, patient-specific “disease-in-a-dish” models could be created to recapitulate stage-specific pathology (Park et al, 2008; Belmonte et al, 2009; Miller Justine et al, 2013; Badger et al, 2014; Woodard et al, 2014). These models could help us identify new disease targets, devise effective screening platforms (Takayama et al, 2013), and develop new therapeutic modalities. These tools are especially useful for investigating rare genetic disorders, where relevant animal models and patient samples are limited [e.g. ataxia telangiectasia (Lee et al, 2013) and Williams–Beuren syndrome (WBS) (Kinnear et al, 2013)]. iPSC-derived disease models may offer a significant advantage over animal models because they may more accurately reflect human disease pathology and may respond to therapies in a more physiologically relevant manner than traditional animal disease models. Furthermore, “disease-in-a-dish” strategies may significantly reduce costs and investigation time compared to animal studies. Although these models are limited by their lack of systemic context and disease-associated environmental cues [e.g. disorganized ECM that represents the hallmark of certain diseased tissues in situ (Bateman et al, 2009)], they would represent an important new method of disease investigation complementary to existing animal models.
The integration of biomaterials with existing iPSC-based disease models could better recapitulate disease pathology and may represent superior scalability and flexibility for creating large numbers of personalized models to meet diverse and urgent patient needs. This principle has recently been substantiated by multiple studies primarily involving cardiovascular or neurodegenerative diseases (Saha & Jaenisch, 2009; Wang et al, 2014a; Zhang et al, 2014a). For instance, to better understand cardiac arrhythmias and related cardiovascular diseases, a cardiac disease model was created using iPSCs-derived cardiomyocytes (CMs) seeded in a synthetic filamentous scaffold (Ma et al, 2014). The CMs were derived from healthy wild-type (WT) iPSCs and long QT syndrome type 3 (LQT3) iPSCs. Two-photon initiated polymerization (TPIP) with a UV-curable organic–inorganic hybrid polymer was used to create the 3D filamentous scaffolds with precisely controlled structural alignment, spatial resolution, and mechanical properties. Tailoring these parameters modulated the contractility of residing CMs and, more importantly, recapitulated the abnormal contractility of long QT syndrome in the LQT3 iPSC-CMs-seeded scaffold, which was not seen in WT iPSC-CMs-seeded counterparts. After identifying the most relevant cardiac model with LQT3 iPSC-CMs, the model was further tested by exposure to a spectrum of cardiotoxic compounds. The LQT3 iPSCs-CMs were found to be more sensitive to the pharmacological interference when grown in a 3D scaffold with lower fiber stiffness, compared to those cultured in 3D with stiffer fibers or on 2D surfaces. These findings collectively suggest that 3D tissue engineered models with defined cellular microenvironments hold great promise and may be useful for high-throughput drug screening and toxicity testing. Biomaterial-guided, personalized disease models may significantly reduce drug discovery costs by reducing the number of required animal studies and may offer broad flexibility for modeling diverse disease settings, thus aiding in the understanding of disease mechanisms and propelling the development of personalized medicine (van de Stolpe & den Toonder, 2013).
3D bioprinting: stacking iPSCs and biomaterials with improved physiological resemblance
Another biomaterial-based method for efficiently creating and studying iPSC-derived organ systems is 3D bioprinting. Three-dimensional printing, otherwise known as additive manufacturing, has attracted extensive attention over the past decade and is fostering major innovations in many fields. Three-dimensional printing of biocompatible materials, living cells, and essential supporting elements can be used to generate 3D functional tissues/organs suitable for transplantation or tissue modeling applications (e.g. “organ-on-a-chip”) (Chang et al, 2008; Levato et al, 2014; Murphy & Atala, 2014; Neufurth et al, 2014). Three-dimensional bioprinting enables layer-by-layer arrangement of biomaterials, biochemicals, and living cells with precise spatial control (Guillotin et al, 2010; Moon et al, 2010). With appropriately matched biomaterials and cell sources, the fabricated 3D constructs could offer desired biological and mechanical properties matching the needs for functional tissue/organ restoration.
Three-dimensional bioprinting holds great promise for closely mimicking the systemic complexities of physiological or pathological conditions (e.g. cross talk between immune and neurological systems) with sufficient micro-/nanoscale resolution (Xu et al, 2013a; Poldervaart et al, 2014). This could potentially address the challenges encountered by current 3D disease modeling, such as the lack of desired vascularization and innervation, hence unleashing the potential of iPSCs as research and drug discovery tools. To replicate biological tissues on a desired scale, thorough understanding of cellular microenvironments, including specific arrangement of different types of supporting cell, ECM compositions, and gradients of physical and soluble/insoluble biochemical cues, is required. Biomaterials are poised to emulate such microenvironmental elements while serving as the backbone for the printed constructs. In addition to providing essential biocompatibility, degradability, material biomimicry, and structural/mechanical support, biomaterials employed for 3D bioprinting should be amenable to existing 3D printing techniques and be accurately deposited with desired spatial and temporal control while displaying required post-printing properties. Moreover, the ability of biomaterials chosen to protect cells during and after the printing process is another critical consideration for the success of 3D bioprinting (Park et al, 2014).
These advantages of 3D bioprinting have been recently demonstrated in a proof-of-principle study by Kolesky et al (2014). They created an intricate, heterogeneous tissue construct replete with vasculature, multiple cell types, and ECM using a custom-designed 3D bioprinting method (Kolesky et al, 2014). They explored fugitive/sacrificial and cell-laden hydrogel ink materials that were designed to meet the following key criteria: (i) The inks are compatible with one another under the printing conditions; (ii) the patterned cells and surrounding ECM are not compromised during the process of removing sacrificial ink for creating vascular channels; that is, the fugitive ink should be biocompatible and should be removable under mild conditions; (iii) the resulting vascular channels are perfusable; and (iv) the cells introduced during printing (e.g. fibroblasts) and post-printing by perfusion (e.g. endothelial cells) should remain viable. Specifically, a 4-layered tissue construct composed of semi-woven features was generated by a layer-by-layer deposition of four ink materials through a 200-μm nozzle: PDMS, fugitive Pluronic F127 that undergoes thermally reversible gelation, and two different cell-laden GelMA inks (gelatin methacrylate and photopolymerizable by UV after printing). Afterward, pure GelMA ink was deposited onto the construct and UV-cross-linked to form the ECM matrix and encapsulate the printed features. The 3D printed construct, after the rapid removal of fugitive ink, effectively supported endothelialization under perfusion conditions within 2 days of in vitro culture. Moreover, the two printed fibroblast cell types exhibited similar levels of cell viability after 7 days of culture compared with the control cells without printing, suggesting the 3D bioprinting approach was non-destructive. This robust 3D bioprinting platform is a promising tool for producing functional tissue constructs that may be useful for screening drugs and studying tissue morphogenesis and stem cell niches.
High-throughput screening of maximally supportive substrates for iPSC expansion and differentiation
Recent advances in microscale technologies and their automation have begun to “upgrade” the traditional paradigm of cell culture (Levy et al, 2015). Miniaturizing an individual cell culture plate into small micron-sized patterns permits high-throughput parameter identification and optimization (Mei et al, 2007; Cimetta et al, 2013). Specifically for iPSCs, discovering biomaterial-based substrates that can overcome the limitations of traditional iPSC expansion/differentiation platforms in an efficient, high-throughput manner is of great significance (Fig5A). To this end, Mei et al (2010) have devised a high-throughput screening platform for identifying new substrates to culture ESCs/iPSCs to achieve accelerated colony expansion in a chemically defined, xeno-free condition. This platform, a biomaterial microarray, was developed to achieve rapid synthesis and analysis of synthetic substrates with diverse surface chemistries. Combinatorial synthesis of polymers with a broad range of acrylate monomers was employed for the polymer microarray fabrication. The substrates were evaluated by plating human iPSCs or ESCs on the microarray and measuring their colony-forming efficiencies. Material properties including surface wettability, topography, surface chemistry, and stiffness were assessed via high-throughput characterization methods aimed at establishing the correlation between material properties and cell behavior. The screening results revealed that monomers with relatively high acrylate content and moderate surface wettability led to efficient iPSC growth (Fig5B). Such correlation is mediated through the interactions between the surface-induced vitronectin adsorption and their corresponding integrin receptors on the iPSCs. This high-throughput biomaterial-screening platform enables efficient investigation of cell–matrix interactions and optimization of iPSC expansion substrates with minimal amounts of materials and cells required. Using a similar high-throughput polymerization and screening platform, Xie et al established a comprehensive library of 66 monomer-grafted surfaces and identified one “hit” substrate that maintained self-renewal of mouse ESCs for up to seven passages, primarily through integrin β1-mediated cell–matrix interactions (Zonca et al, 2013).
Figure 5.
High-throughput biomaterial arrays enable simultaneous evaluation of multiple cell–matrix and cell–cell interactions to identify conditions for efficient expansion and controlled differentiation of iPSCs or their progeny
(A) Multiple cellular microenvironmental cues and their combinations can be engineered and their interactions with iPSCs-/iPSC-derived progeny can be evaluated in a combinatorial manner on a single arrayed substrate. Microenvironmental cues can be presented to cells via (a) a coculture of multiple cellular components mimicking native cell–cell interactions; (b) substrate stiffness; (c) substrate-induced topographic cues generated by micro-/nanopatterning; (d) spatiotemporally controlled presentation of soluble factors; (e) tethered bioactive factors via substrate surface functionalization. (B) In a relevant example, single Oct4-GFP-positive human ESCs were plated onto the polymer arrays with 496 polymer combinations. Individual cells were initially seeded and attached at a near-clonal density within each spot (white arrowheads in the day 1 panel). Cell response was varied from days 1 to 7 on spots coated with three different polymers (two repeats of each), as shown by the staining of cell nuclei (Hoechst blue) and the level of GFP expression. The 16E-30% polymer was not supportive of cell attachment or survival, whereas the 6F-30% polymer exhibited moderate support for cell growth but did not maintain pluripotency. A hit polymer, the “9” homopolymer, supported robust proliferation of ESCs without differentiation, which was further confirmed by the immunostaining of pluripotency markers Oct4 (green) and SSEA4 (red). (C) In another relevant example, murine ESCs were placed on a combinatorial ECM array and stained with X-gal (blue area indicating the activity of a β-galactosidase reporter fused into a fetal liver-specific gene Ankrd17) after 3 days of culture. The combination of collagen I (C1), collagen III (C3), laminin (L), and fibronectin (Fn) (top red box) induced elevated reporter activity compared to cells on C3 + L (bottom gray box). Scale bar, 250 μm; inset scale bar, 50 μm. The right bar graph depicts the quantification of “blue” image area on each spot of the ECM mixtures, with specific compositions illustrated by the color legend (C4: collagen IV). The cells cultured on matrix mixture C1 + C3 + L + FN elicited ∼27-fold higher Ankrd17 reporter activity than that of the cultures on C3 + L. Error bars, s.e.m. (n = duplicate spots). (B) and (C) were adapted with permission from Flaim et al (2005) and Mei et al (2010), respectively.
In addition to searching for supportive substrates for high-efficiency iPSC expansion, one must consider stem cell niche interactions including cell–ECM (Flaim et al, 2005) and cell–cell interactions (Fig5A) that can support directed differentiation of iPSCs or their progeny. In a pilot study by Anderson et al (2004), high-throughput biomaterial arrays were developed for evaluating cell–material interactions to identify materials that support the controlled differentiation of human ESCs. The combinatorial polymer array was fabricated by mixing 24 acrylate co-monomers with different polarities, chain lengths, and branching in a multi-well format, followed by radical polymerization to form a library of nearly 600 materials. Over 1,700 ESC–material interactions were simultaneously characterized and a set of “hit” materials were found to effectively induce ESC differentiation toward epithelial-like cells, as shown by the cytokeratin-positive staining after 6 days of culture under proper solution conditions. The majority of these identified polymers also appeared to support cell attachment and spreading to display distinct cellular morphology, implying matrix-induced early fate decisions of ESCs (e.g. epithelial versus mesenchymal phenotype). Once the “hit” conditions are identified, this simple, one-step cell differentiation platform could be easily scaled up and allow for the production of large quantities of pure iPSC progeny, essential for the establishment of iPSC-derived research tools and cell replacement therapies. Furthermore, the array fabrication process (e.g. acrylate polymerization) is highly flexible and can be easily extended to the third dimension (e.g. scaffolds) or incorporated with bioactive motifs such as receptor-specific ligands or adhesive peptides.
In addition to the utility of polymer arrays for identifying materials that can control stem cell phenotype, alternatively, Bhatia et al developed an ECM microarray to investigate the combinatorial effect of ECM mixtures on the differentiation of ESCs (Flaim et al, 2005). This platform enables the investigation of multiple natural microenvironmental cues. For the fabrication process, the system requires only standard DNA spotter, “off-the-shelf” chemicals, and a significantly less (1,000 times) amount of ECM proteins than traditional approaches for studying cell–ECM interactions. And this platform is applicable for the deposition of nearly any kind of soluble or insoluble factors, which implies the potential of revealing a more global picture of the roles of diverse microenvironmental cues on stem cell fate and function. Specifically, 32 different combinations of five ECM proteins (collagens I, III, and IV, laminin, and fibronectin) were spotted with nanoliter volume onto the array and their effects on ESC differentiation were systematically studied. Particular combinations of ECMs were found to synergistically foster the early hepatic commitment of ESCs within 3 days of culture in the presence of retinoic acid (RA). For example, the combination of collagen I, collagen III, laminin, and fibronectin induced significantly higher hepatic reporter (β-galactosidase) expression than that of the cells cultured on collagen III + laminin (Fig5C). We envision that this versatile platform can be easily adapted to elucidate the cell–matrix interactions of iPSCs or their progeny and hence optimizing the manipulation of the microenvironment to achieve directed lineage specification.
Perspective and conclusion
Given the relatively short history and exploratory nature of iPSC research, it is remarkable that clinical trials have already begun. Namely, the world's first iPSC clinical trial for treating age-related macular degeneration (AMD) was approved in Japan, and the first patient was treated in September 2014 (David, 2014). Although promising, the major challenges discussed herein, that is, safety, efficiency, and scalability, will impact effective translation of current iPSC preclinical findings into clinical therapies.
Biomaterial science and its application to iPSCs or iPSC-derived cells holds far-reaching implications for advancing the utility of iPSCs as research tools and expediting their clinical translation. Biomaterials offer superior control over the spatiotemporal presentation and kinetics of soluble factors that are typically employed in traditional iPSC derivation, expansion, and differentiation protocols, leading to improved safety, efficiency, and scalability. Through rationally designed “outside-in” (e.g. engineering the cellular microenvironment) and “inside-out” (e.g. “particle-in-cell” platform) strategies, biomaterials may present opportunities to better probe iPSC biology and regulate their fate and function in vitro and in vivo. For example, libraries of materials can be systematically created with a varying stiffness, substrate topography, and degradation kinetics. Such approaches have recently been used to probe time-sensitive events at the cell–material interface (Burdick & Murphy, 2012; Tibbitt & Anseth, 2012).
The ability to achieve controlled presentation of soluble and substrate-based cues, particularly in the context of iPSC self-renewal and differentiation, motivates the need for future systematic studies to identify and establish relevant material structure–function relationships. Emerging technologies such as predictive computational modeling, high-throughput biomaterial arrays, and 3D bioprinting will help advance our comprehensive understanding of cell–cell/cell–matrix interactions. Until such time as research tool companies offer off-the-shelf biomaterials products to address these much needed areas of research, basic stem cell biologists will need to forge alliances with bioengineers who can engineer the required materials.
We recognize that, while promising and exciting, the application of biomaterials for iPSC research and therapeutic applications is challenging. There are additional complexities that must be considered. For example, the identification of relevant biomaterials can be laborious, and properties of biomaterials including surface chemistry, surface topography, stiffness, size, and shape can all impact the cellular response. One must also consider the intricate and often undefined cross talk between biomaterials and cells (e.g. the cell secretome can include enzymes that accelerate the degradation of materials and the degradation products can impact cell phenotype and function including the secretion of degradative enzymes). Synergistic and systematic efforts from the fields of engineering, biomaterial science, stem cell biology, and medicine will be required to tackle these challenges and, with the help of biomaterials, rapidly bring the potential of iPSCs to fruition.
Conflict of interest
JMK consults for Sanofi, Mesoblast, and Stempeutics in the field of regenerative medicine. The other authors declare that they have no conflict of interest.
References
- Ahrlund-Richter L, De Luca M, Marshak DR, Munsie M, Veiga A, Rao M. Isolation and production of cells suitable for human therapy: challenges ahead. Cell Stem Cell. 2009;4:20–26. doi: 10.1016/j.stem.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Alberti K, Davey RE, Onishi K, George S, Salchert K, Seib FP, Bornhauser M, Pompe T, Nagy A, Werner C, Zandstra PW. Functional immobilization of signaling proteins enables control of stem cell fate. Nat Methods. 2008;5:645–650. doi: 10.1038/nmeth.1222. [DOI] [PubMed] [Google Scholar]
- Alfred R, Radford J, Fan J, Boon K, Krawetz R, Rancourt D, Kallos MS. Efficient suspension bioreactor expansion of murine embryonic stem cells on microcarriers in serum-free medium. Biotechnol Prog. 2011;27:811–823. doi: 10.1002/btpr.591. [DOI] [PubMed] [Google Scholar]
- Amit M, Shariki C, Margulets V, Itskovitz-Eldor J. Feeder layer- and serum-free culture of human embryonic stem cells. Biol Reprod. 2004;70:837–845. doi: 10.1095/biolreprod.103.021147. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotech. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- Ankrum JA, Dastidar RG, Ong JF, Levy O, Karp JM. Performance-enhanced mesenchymal stem cells via intracellular delivery of steroids. Sci Rep. 2014a;4:4645. doi: 10.1038/srep04645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankrum JA, Miranda OR, Ng KS, Sarkar D, Xu C, Karp JM. Engineering cells with intracellular agent-loaded microparticles to control cell phenotype. Nat Protoc. 2014b;9:233–245. doi: 10.1038/nprot.2014.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R, Uda M, Hoki Y, Sunayama M, Nakamura M, Ando S, Sugiura M, Ideno H, Shimada A, Nifuji A, Abe M. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Badger JL, Cordero-Llana O, Hartfield EM, Wade-Martins R. Parkinson's disease in a dish – Using stem cells as a molecular tool. Neuropharmacology. 2014;76:88–96. doi: 10.1016/j.neuropharm.2013.08.035. (Pt A): [DOI] [PubMed] [Google Scholar]
- Bateman JF, Boot-Handford RP, Lamande SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- Belair DG, Le NN, Murphy WL. Design of growth factor sequestering biomaterials. Chem Commun. 2014;50:15651–15668. doi: 10.1039/c4cc04317k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte JCI, Ellis J, Hochedlinger K, Yamanaka S. Induced pluripotent stem cells and reprogramming: seeing the science through the hype. Nat Rev Genet. 2009;10:878–883. doi: 10.1038/nrg2700. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- Bilic J, Izpisua Belmonte JC. Concise review: induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- Boukany PE, Wu Y, Zhao X, Kwak KJ, Glazer PJ, Leong K, Lee LJ. Nonendocytic delivery of lipoplex nanoparticles into living cells using nanochannel electroporation. Adv Healthc Mater. 2014;3:682–689. doi: 10.1002/adhm.201300213. [DOI] [PubMed] [Google Scholar]
- Braam SR, Zeinstra L, Litjens S, Ward-van Oostwaard D, van den Brink S, van Laake L, Lebrin F, Kats P, Hochstenbach R, Passier R, Sonnenberg A, Mummery CL. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008;26:2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- Bratt-Leal AM, Carpenedo RL, Ungrin MD, Zandstra PW, McDevitt TC. Incorporation of biomaterials in multicellular aggregates modulates pluripotent stem cell differentiation. Biomaterials. 2011;32:48–56. doi: 10.1016/j.biomaterials.2010.08.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt-Leal AM, Nguyen AH, Hammersmith KA, Singh A, McDevitt TC. A microparticle approach to morphogen delivery within pluripotent stem cell aggregates. Biomaterials. 2013;34:7227–7235. doi: 10.1016/j.biomaterials.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindley DA, French A, Suh J, Roberts M, Davies B, Pinedo-Villanueva R, Wartolowska K, Rooke K, Kramm A, Judge A, Morrey M, Chandra A, Hurley H, Grover L, Bingham I, Siegel B, Rattley MS, Buckler RL, McKeon D, Krumholz K, et al. The implementation of novel collaborative structures for the identification and resolution of barriers to pluripotent stem cell translation. Stem Cells Dev. 2013;22(Suppl. 1):63–72. doi: 10.1089/scd.2013.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick JA, Murphy WL. Moving from static to dynamic complexity in hydrogel design. Nat Commun. 2012;3:1269. doi: 10.1038/ncomms2271. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanas-Danés J, Huskens J, Jonkheijm P. Chemical strategies for the presentation and delivery of growth factors. J Mater Chem B. 2014;2:2381–2394. doi: 10.1039/c3tb20853b. [DOI] [PubMed] [Google Scholar]
- Carpenedo RL, Bratt-Leal AM, Marklein RA, Seaman SA, Bowen NJ, McDonald JF, McDevitt TC. Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules. Biomaterials. 2009;30:2507–2515. doi: 10.1016/j.biomaterials.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenedo RL, Seaman SA, McDevitt TC. Microsphere size effects on embryoid body incorporation and embryonic stem cell differentiation. J Biomed Mater Res A. 2010;94A:466–475. doi: 10.1002/jbm.a.32710. [DOI] [PubMed] [Google Scholar]
- Celiz AD, Smith JG, Langer R, Anderson DG, Winkler DA, Barrett DA, Davies MC, Young LE, Denning C, Alexander MR. Materials for stem cell factories of the future. Nat Mater. 2014;13:570–579. doi: 10.1038/nmat3972. [DOI] [PubMed] [Google Scholar]
- Cetinkaya G, Turkoglu H, Arat S, Odaman H, Onur MA, Gumusderelioglu M, Tumer A. LIF-immobilized nonwoven polyester fabrics for cultivation of murine embryonic stem cells. J Biomed Mater Res A. 2007;81:911–919. doi: 10.1002/jbm.a.31107. [DOI] [PubMed] [Google Scholar]
- Chang R, Nam J, Sun W. Direct cell writing of 3D microorgan for in vitro pharmacokinetic model. Tissue Eng Part C Methods. 2008;14:157–166. doi: 10.1089/ten.tec.2007.0392. [DOI] [PubMed] [Google Scholar]
- Chang CW, Hwang Y, Brafman D, Hagan T, Phung C, Varghese S. Engineering cell-material interfaces for long-term expansion of human pluripotent stem cells. Biomaterials. 2013;34:912–921. doi: 10.1016/j.biomaterials.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KG, Mallon BS, McKay RD, Robey PG. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14:13–26. doi: 10.1016/j.stem.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Meng F, Deng C, Klok HA, Zhong Z. Dual and multi-stimuli responsive polymeric nanoparticles for programmed site-specific drug delivery. Biomaterials. 2013;34:3647–3657. doi: 10.1016/j.biomaterials.2013.01.084. [DOI] [PubMed] [Google Scholar]
- Chi NH, Yang MC, Chung TW, Chen JY, Chou NK, Wang SS. Cardiac repair achieved by bone marrow mesenchymal stem cells/silk fibroin/hyaluronic acid patches in a rat of myocardial infarction model. Biomaterials. 2012;33:5541–5551. doi: 10.1016/j.biomaterials.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Chowdhury F, Li Y, Poh YC, Yokohama-Tamaki T, Wang N, Tanaka TS. Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS ONE. 2010;5:e15655. doi: 10.1371/journal.pone.0015655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C-Y, Yang J-T, Kuo Y-C. Polybutylcyanoacrylate nanoparticle-mediated neurotrophin-3 gene delivery for differentiating iPS cells into neurons. Biomaterials. 2013;34:5562–5570. doi: 10.1016/j.biomaterials.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Cimetta E, Godier-Furnemont A, Vunjak-Novakovic G. Bioengineering heart tissue for in vitro testing. Curr Opin Biotechnol. 2013;24:926–932. doi: 10.1016/j.copbio.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti B, Freile P, Pells S, Bagnaninchi P, Park J, Fahmy TM, de Sousa PA. Paracrine signalling events in embryonic stem cell renewal mediated by affinity targeted nanoparticles. Biomaterials. 2012;33:6634–6643. doi: 10.1016/j.biomaterials.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Oreffo ROC. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- Dang LTH, Feric NT, Laschinger C, Chang WY, Zhang B, Wood GA, Stanford WL, Radisic M. Inhibition of apoptosis in human induced pluripotent stem cells during expansion in a defined culture using angiopoietin-1 derived peptide QHREDGS. Biomaterials. 2014;35:7786–7799. doi: 10.1016/j.biomaterials.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo F, Armentano I, Cacciotti I, Tiribuzi R, Quattrocelli M, Del Gaudio C, Fortunati E, Saino E, Caraffa A, Cerulli GG, Visai L, Kenny JM, Sampaolesi M, Bianco A, Martino S, Orlacchio A. Tuning multi/pluri-potent stem cell fate by electrospun poly(L-lactic acid)-calcium-deficient hydroxyapatite nanocomposite mats. Biomacromolecules. 2012;13:1350–1360. doi: 10.1021/bm3000716. [DOI] [PubMed] [Google Scholar]
- Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- David C. Next-generation stem cells cleared for human trial. Nature. 2014 doi: 10.1038/nature.2014.15897. [Google Scholar]
- David L, Polo JM. Phases of reprogramming. Stem Cell Res. 2014;12:754–761. doi: 10.1016/j.scr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Dellatore SM, Garcia AS, Miller WM. Mimicking stem cell niches to increase stem cell expansion. Curr Opin Biotechnol. 2008;19:534–540. doi: 10.1016/j.copbio.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zhang X, Zhao X, Li Q, Ye Z, Li Z, Liu Y, Zhou Y, Ma H, Pan G, Pei D, Fang J, Wei S. Long-term self-renewal of human pluripotent stem cells on peptide-decorated poly(OEGMA-co-HEMA) brushes under fully defined conditions. Acta Biomater. 2013;9:8840–8850. doi: 10.1016/j.actbio.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Derda R, Li L, Orner B, Lewis R, Thomson J, Kiessling L. Defined substrates for human embryonic stem cell growth identified from surface arrays. ACS Chem Biol. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- Dimmeler S, Ding S, Rando TA, Trounson A. Translational strategies and challenges in regenerative medicine. Nat Med. 2014;20:814–821. doi: 10.1038/nm.3627. [DOI] [PubMed] [Google Scholar]
- Dingal PC, Discher DE. Combining insoluble and soluble factors to steer stem cell fate. Nat Mater. 2014;13:532–537. doi: 10.1038/nmat3997. [DOI] [PubMed] [Google Scholar]
- Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Narayanan K, Leong MF, Wan ACA. Induced pluripotent stem cell-derived hepatocytes and endothelial cells in multi-component hydrogel fibers for liver tissue engineering. Biomaterials. 2014;35:6006–6014. doi: 10.1016/j.biomaterials.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Epa VC, Yang J, Mei Y, Hook AL, Langer R, Anderson DG, Davies MC, Alexander MR, Winkler DA. Modelling human embryoid body cell adhesion to a combinatorial library of polymer surfaces. J Mater Chem. 2012;22:20902–20906. doi: 10.1039/C2JM34782B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseenko D, Schenke-Layland K, Dravid G, Zhu Y, Hao Q-L, Scholes J, Wang XC, MacLellan WR, Crooks GM. Identification of the critical extracellular matrix proteins that promote human embryonic stem cell assembly. Stem Cells Dev. 2009;18:919–928. doi: 10.1089/scd.2008.0293. [DOI] [PubMed] [Google Scholar]
- Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- Gaeta X, Xie Y, Lowry WE. Sequential addition of reprogramming factors improves efficiency. Nat Cell Biol. 2013;15:725–727. doi: 10.1038/ncb2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Neofytou E, Cahill TJ, Beygui RE, Zare RN. Drug release from electric-field-responsive nanoparticles. ACS Nano. 2012;6:227–233. doi: 10.1021/nn203430m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng T, Lu C. Microfluidic electroporation for cellular analysis and delivery. Lab Chip. 2013;13:3803–3821. doi: 10.1039/c3lc50566a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giteau A, Venier-Julienne MC, Aubert-Pouessel A, Benoit JP. How to achieve sustained and complete protein release from PLGA-based microparticles? Int J Pharm. 2008;350:14–26. doi: 10.1016/j.ijpharm.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Godier-Furnemont AF, Martens TP, Koeckert MS, Wan L, Parks J, Arai K, Zhang G, Hudson B, Homma S, Vunjak-Novakovic G. Composite scaffold provides a cell delivery platform for cardiovascular repair. Proc Natl Acad Sci USA. 2011;108:7974–7979. doi: 10.1073/pnas.1104619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillotin B, Souquet A, Catros S, Duocastella M, Pippenger B, Bellance S, Bareille R, Remy M, Bordenave L, Amedee J, Guillemot F. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials. 2010;31:7250–7256. doi: 10.1016/j.biomaterials.2010.05.055. [DOI] [PubMed] [Google Scholar]
- Han YL, Wang S, Zhang X, Li Y, Huang G, Qi H, Pingguan-Murphy B, Li Y, Lu TJ, Xu F. Engineering physical microenvironment for stem cell based regenerative medicine. Drug Discov Today. 2014;19:763–773. doi: 10.1016/j.drudis.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Heiskanen A, Satomaa T, Tiitinen S, Laitinen A, Mannelin S, Impola U, Mikkola M, Olsson C, Miller-Podraza H, Blomqvist M, Olonen A, Salo H, Lehenkari P, Tuuri T, Otonkoski T, Natunen J, Saarinen J, Laine J. N-glycolylneuraminic acid xenoantigen contamination of human embryonic and mesenchymal stem cells is substantially reversible. Stem Cells. 2007;25:197–202. doi: 10.1634/stemcells.2006-0444. [DOI] [PubMed] [Google Scholar]
- Higuchi A, Ling QD, Ko YA, Chang Y, Umezawa A. Biomaterials for the feeder-free culture of human embryonic stem cells and induced pluripotent stem cells. Chem Rev. 2011;111:3021–3035. doi: 10.1021/cr1003612. [DOI] [PubMed] [Google Scholar]
- Hong SG, Winkler T, Wu C, Guo V, Pittaluga S, Nicolae A, Donahue RE, Metzger ME, Price SD, Uchida N, Kuznetsov SA, Kilts T, Li L, Robey PG, Dunbar CE. Path to the clinic: assessment of iPSC-based cell therapies in vivo in a nonhuman primate model. Cell Rep. 2014;7:1298–1309. doi: 10.1016/j.celrep.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongisto H, Vuoristo S, Mikhailova A, Suuronen R, Virtanen I, Otonkoski T, Skottman H. Laminin-511 expression is associated with the functionality of feeder cells in human embryonic stem cell culture. Stem Cell Res. 2012;8:97–108. doi: 10.1016/j.scr.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Hu K. All roads lead to induced pluripotent stem cells: the technologies of iPSC generation. Stem Cells Dev. 2014;23:1285–1300. doi: 10.1089/scd.2013.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Au P, Tam J, Duda DG, Fukumura D. Engineering vascularized tissue. Nat Biotechnol. 2005;23:821–823. doi: 10.1038/nbt0705-821. [DOI] [PubMed] [Google Scholar]
- James R, Laurencin CT. Musculoskeletal regenerative engineering: biomaterials, structures, and small molecules. Adv Biomater. 2014;2014:12. [Google Scholar]
- Jin S, Yao H, Weber JL, Melkoumian ZK, Ye K. A synthetic, xeno-free peptide surface for expansion and directed differentiation of human induced pluripotent stem cells. PLoS ONE. 2012;7:e50880. doi: 10.1371/journal.pone.0050880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G-Z, Kim T-H, Kim J-H, Won J-E, Yoo S-Y, Choi S-J, Hyun JK, Kim H-W. Bone tissue engineering of induced pluripotent stem cells cultured with macrochanneled polymer scaffold. J Biomed Mater Res A. 2013;101A:1283–1291. doi: 10.1002/jbm.a.34425. [DOI] [PubMed] [Google Scholar]
- Jung S, Panchalingam KM, Wuerth RD, Rosenberg L, Behie LA. Large-scale production of human mesenchymal stem cells for clinical applications. Biotechnol Appl Biochem. 2012;59:106–120. doi: 10.1002/bab.1006. [DOI] [PubMed] [Google Scholar]
- Kalaskar DM, Downes JE, Murray P, Edgar DH, Williams RL. Characterization of the interface between adsorbed fibronectin and human embryonic stem cells. J R Soc Interface. 2013;10:20130139. doi: 10.1098/rsif.2013.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Shih YR, Hwang Y, Wen C, Rao V, Seo T, Varghese S. Mineralized gelatin methacrylate-based matrices induce osteogenic differentiation of human induced pluripotent stem cells. Acta Biomater. 2014;10:4961–4970. doi: 10.1016/j.actbio.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharaziha M, Nikkhah M, Shin SR, Annabi N, Masoumi N, Gaharwar AK, Camci-Unal G, Khademhosseini A. PGS:Gelatin nanofibrous scaffolds with tunable mechanical and structural properties for engineering cardiac tissues. Biomaterials. 2013;34:6355–6366. doi: 10.1016/j.biomaterials.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Lee KI, Kim DW, Hwang DY. An ECM-based culture system for the generation and maintenance of xeno-free human iPS cells. Biomaterials. 2013;34:1041–1050. doi: 10.1016/j.biomaterials.2012.10.064. [DOI] [PubMed] [Google Scholar]
- Kinnear C, Chang WY, Khattak S, Hinek A, Thompson T, de Carvalho Rodrigues D, Kennedy K, Mahmut N, Pasceri P, Stanford WL, Ellis J, Mital S. Modeling and rescue of the vascular phenotype of Williams-Beuren syndrome in patient induced pluripotent stem cells. Stem Cells Transl Med. 2013;2:2–15. doi: 10.5966/sctm.2012-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman HK, McGarvey ML, Liotta LA, Robey PG, Tryggvason K, Martin GR. Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochemistry. 1982;21:6188–6193. doi: 10.1021/bi00267a025. [DOI] [PubMed] [Google Scholar]
- Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose D, Siepmann F, Elkharraz K, Krenzlin S, Siepmann J. How porosity and size affect the drug release mechanisms from PLGA-based microparticles. Int J Pharm. 2006;314:198–206. doi: 10.1016/j.ijpharm.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Kmiec Z. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol. 2001;161:III–XIII. doi: 10.1007/978-3-642-56553-3. , 1-151. [DOI] [PubMed] [Google Scholar]
- Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv Mater. 2014;26:3124–3130. doi: 10.1002/adma.201305506. [DOI] [PubMed] [Google Scholar]
- Kost J, Langer R. Responsive polymeric delivery systems. Adv Drug Deliv Rev. 2012;64(Suppl):327–341. doi: 10.1016/s0169-409x(00)00136-8. [DOI] [PubMed] [Google Scholar]
- Kou L, Sun J, Zhai Y, He Z. The endocytosis and intracellular fate of nanomedicines: implication for rational design. Asian J Pharm Sci. 2013;8:1–10. [Google Scholar]
- Kraehenbuehl TP, Langer R, Ferreira LS. Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat Methods. 2011;8:731–736. doi: 10.1038/nmeth.1671. [DOI] [PubMed] [Google Scholar]
- Kuo Y-C, Chung C-Y. TATVHL peptide-grafted alginate/poly(γ-glutamic acid) scaffolds with inverted colloidal crystal topology for neuronal differentiation of iPS cells. Biomaterials. 2012;33:8955–8966. doi: 10.1016/j.biomaterials.2012.08.073. [DOI] [PubMed] [Google Scholar]
- Kuo YC, Wang CT. Neuronal differentiation of induced pluripotent stem cells in hybrid polyester scaffolds with heparinized surface. Colloids Surf B Biointerfaces. 2012;100:9–15. doi: 10.1016/j.colsurfb.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Kuo Y-C, Lin C-C. Accelerated nerve regeneration using induced pluripotent stem cells in chitin–chitosan–gelatin scaffolds with inverted colloidal crystal geometry. Colloids Surf B. 2013;103:595–600. doi: 10.1016/j.colsurfb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Kuo YC, Chang YH. Differentiation of induced pluripotent stem cells toward neurons in hydrogel biomaterials. Colloids Surf B Biointerfaces. 2013;102:405–411. doi: 10.1016/j.colsurfb.2012.08.061. [DOI] [PubMed] [Google Scholar]
- Lane SW, Williams DA, Watt FM. Modulating the stem cell niche for tissue regeneration. Nat Biotechnol. 2014;32:795–803. doi: 10.1038/nbt.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang N, Pereira MJ, Lee Y, Friehs I, Vasilyev NV, Feins EN, Ablasser K, O'Cearbhaill ED, Xu C, Fabozzo A, Padera R, Wasserman S, Freudenthal F, Ferreira LS, Langer R, Karp JM, del Nido PJ. A blood-resistant surgical glue for minimally invasive repair of vessels and heart defects. Sci Transl Med. 2014;6:218ra216. doi: 10.1126/scitranslmed.3006557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau TT, Ho LW, Wang DA. Hepatogenesis of murine induced pluripotent stem cells in 3D micro-cavitary hydrogel system for liver regeneration. Biomaterials. 2013;34:6659–6669. doi: 10.1016/j.biomaterials.2013.05.034. [DOI] [PubMed] [Google Scholar]
- Lee K, Silva EA, Mooney DJ. Growth factor delivery-based tissue engineering: general approaches and a review of recent developments. J R Soc Interface. 2011;8:153–170. doi: 10.1098/rsif.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Martin NT, Nakamura K, Azghadi S, Amiri M, Ben-David U, Perlman S, Gatti RA, Hu H, Lowry WE. SMRT compounds abrogate cellular phenotypes of ataxia telangiectasia in neural derivatives of patient-specific hiPSCs. Nat Commun. 2013;4:1824. doi: 10.1038/ncomms2824. [DOI] [PubMed] [Google Scholar]
- Lee K-I, Kim H-T, Hwang D-Y. Footprint- and xeno-free human iPSCs derived from urine cells using extracellular matrix-based culture conditions. Biomaterials. 2014;35:8330–8338. doi: 10.1016/j.biomaterials.2014.05.059. [DOI] [PubMed] [Google Scholar]
- Lei Y, Schaffer DV. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc Natl Acad Sci USA. 2013;110:E5039–E5048. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipzig ND, Shoichet MS. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials. 2009;30:6867–6878. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Levato R, Visser J, Planell JA, Engel E, Malda J, Mateos-Timoneda MA. Biofabrication of tissue constructs by 3D bioprinting of cell-laden microcarriers. Biofabrication. 2014;6:035020. doi: 10.1088/1758-5082/6/3/035020. [DOI] [PubMed] [Google Scholar]
- Levenstein ME, Ludwig TE, Xu RH, Llanas RA, VanDenHeuvel Kramer K, Manning D, Thomson JA. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24:568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Mortensen LJ, Boquet G, Tong Z, Perrault C, Benhamou B, Zhang J, Stratton T, Han E, Safaee H, Musabeyezu J, Yang Z, Multon M-C, Rothblatt J, Deleuze J-F, Lin Charles P, Karp JM. A Small-Molecule Screen for Enhanced Homing of Systemically Infused Cells. Cell Rep. 2015;10:1261–1268. doi: 10.1016/j.celrep.2015.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhou J, Chowdhury F, Cheng J, Wang N, Wang F. Role of mechanical factors in fate decisions of stem cells. Regen Med. 2011;6:229–240. doi: 10.2217/rme.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Stevens KR, Schwartz RE, Alejandro BS, Huang JH, Bhatia SN. Micropatterned cell–cell interactions enable functional encapsulation of primary hepatocytes in hydrogel microtissues. Tissue Eng Part A. 2014;20:2200–2212. doi: 10.1089/ten.tea.2013.0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang X, Kaufman DS, Shen W. A synthetic substrate to support early mesodermal differentiation of human embryonic stem cells. Biomaterials. 2011;32:8058–8066. doi: 10.1016/j.biomaterials.2011.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, Guo Y, Qin D, Pan G, Chen J, Pei D, Zheng H. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat Cell Biol. 2013a;15:829–838. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou J, Wang H, Zhao M, Wang C. Current status of induced pluripotent stem cells in cardiac tissue regeneration and engineering. Regen Med Res. 2013b;1:6. doi: 10.1186/2050-490X-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Song Y, Yu H, Zhao T. Understanding the roadmaps to induced pluripotency. Cell Death Dis. 2014a;5:e1232. doi: 10.1038/cddis.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zang R, Yang S-T, Li Y. Stem cell engineering in bioreactors for large-scale bioprocessing. Eng Life Sci. 2014b;14:4–15. [Google Scholar]
- Lu L, Peter SJ, Lyman MD, Lai HL, Leite SM, Tamada JA, Uyama S, Vacanti JP, Langer R, Mikos AG. In vitro and in vivo degradation of porous poly(DL-lactic-co-glycolic acid) foams. Biomaterials. 2000;21:1837–1845. doi: 10.1016/s0142-9612(00)00047-8. [DOI] [PubMed] [Google Scholar]
- Lu HF, Chai C, Lim TC, Leong MF, Lim JK, Gao S, Lim KL, Wan AC. A defined xeno-free and feeder-free culture system for the derivation, expansion and direct differentiation of transgene-free patient-specific induced pluripotent stem cells. Biomaterials. 2014;35:2816–2826. doi: 10.1016/j.biomaterials.2013.12.050. [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Kohli M, Smith A. Nanoparticles for combination drug therapy. ACS Nano. 2013a;7:9518–9525. doi: 10.1021/nn405674m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Xie M, Laurent T, Ding S. Progress in the reprogramming of somatic cells. Circ Res. 2013b;112:562–574. doi: 10.1161/CIRCRESAHA.111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Koo S, Finnegan MA, Loskill P, Huebsch N, Marks NC, Conklin BR, Grigoropoulos CP, Healy KE. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials. 2014;35:1367–1377. doi: 10.1016/j.biomaterials.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makadia HK, Siegel SJ. Poly lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers. 2011;3:1377–1397. doi: 10.3390/polym3031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinen MM, Kanninen LK, Corlu A, Isoniemi HM, Lou Y-R, Yliperttula ML, Urtti AO. Differentiation of liver progenitor cell line to functional organotypic cultures in 3D nanofibrillar cellulose and hyaluronan-gelatin hydrogels. Biomaterials. 2014;35:5110–5121. doi: 10.1016/j.biomaterials.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Martinez JO, Chiappini C, Ziemys A, Faust AM, Kojic M, Liu X, Ferrari M, Tasciotti E. Engineering multi-stage nanovectors for controlled degradation and tunable release kinetics. Biomaterials. 2013;34:8469–8477. doi: 10.1016/j.biomaterials.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt TC. Scalable culture of human pluripotent stem cells in 3D. Proc Natl Acad Sci USA. 2013;110:20852–20853. doi: 10.1073/pnas.1320575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Goldberg M, Anderson D. The development of high-throughput screening approaches for stem cell engineering. Curr Opin Chem Biol. 2007;11:388–393. doi: 10.1016/j.cbpa.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Saha K, Bogatyrev SR, Yang J, Hook AL, Kalcioglu ZI, Cho SW, Mitalipova M, Pyzocha N, Rojas F, Van Vliet KJ, Davies MC, Alexander MR, Langer R, Jaenisch R, Anderson DG. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y. Microarrayed materials for stem cells. Mater Today. 2012;15:444–452. doi: 10.1016/S1369-7021(12)70196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, Martin AW, Nelson J, West P, Beltzer JP, Pal S, Brandenberger R. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- Miklas JW, Dallabrida SM, Reis LA, Ismail N, Rupnick M, Radisic M. QHREDGS enhances tube formation, metabolism and survival of endothelial cells in collagen-chitosan hydrogels. PLoS ONE. 2013;8:e72956. doi: 10.1371/journal.pone.0072956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Pislaru SV, Greenleaf JE. Sonoporation: mechanical DNA delivery by ultrasonic cavitation. Somat Cell Mol Genet. 2002;27:115–134. doi: 10.1023/a:1022983907223. [DOI] [PubMed] [Google Scholar]
- Miller Justine D, Ganat Yosif M, Kishinevsky S, Bowman Robert L, Liu B, Tu Edmund Y, Mandal PK, Vera E, Shim J-W, Kriks S, Taldone T, Fusaki N, Tomishima Mark J, Krainc D, Milner Teresa A, Rossi Derrick J, Studer L. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Futaki S, Suemori H, Taniguchi Y, Yamada M, Kawasaki M, Hayashi M, Kumagai H, Nakatsuji N, Sekiguchi K, Kawase E. Laminin E8 fragments support efficient adhesion and expansion of dissociated human pluripotent stem cells. Nat Commun. 2012;3:1236. doi: 10.1038/ncomms2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed F, van der Walle CF. Engineering biodegradable polyester particles with specific drug targeting and drug release properties. J Pharm Sci. 2008;97:71–87. doi: 10.1002/jps.21082. [DOI] [PubMed] [Google Scholar]
- Moon S, Hasan SK, Song YS, Xu F, Keles HO, Manzur F, Mikkilineni S, Hong JW, Nagatomi J, Haeggstrom E, Khademhosseini A, Demirci U. Layer by layer three-dimensional tissue epitaxy by cell-laden hydrogel droplets. Tissue Eng Part C Methods. 2010;16:157–166. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen C, Haugh M, Schaffler M, Majeska R, McNamara L. Osteocyte differentiation is regulated by extracellular matrix stiffness and intercellular separation. J Mech Behav Biomed Mater. 2013;28:183–194. doi: 10.1016/j.jmbbm.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura S, Nicolas J, Couvreur P. Stimuli-responsive nanocarriers for drug delivery. Nat Mater. 2013;12:991–1003. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotech. 2014;32:773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musah S, Wrighton PJ, Zaltsman Y, Zhong X, Zorn S, Parlato MB, Hsiao C, Palecek SP, Chang Q, Murphy WL. Substratum-induced differentiation of human pluripotent stem cells reveals the coactivator YAP is a potent regulator of neuronal specification. Proc Natl Acad Sci USA. 2014;111:13805–13810. doi: 10.1073/pnas.1415330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A. Temporal regulation of cytokines by the circadian clock. J Immunol Res. 2014;2014:614529. doi: 10.1155/2014/614529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufurth M, Wang X, Schroder HC, Feng Q, Diehl-Seifert B, Ziebart T, Steffen R, Wang S, Muller WE. Engineering a morphogenetically active hydrogel for bioprinting of bioartificial tissue derived from human osteoblast-like SaOS-2 cells. Biomaterials. 2014;35:8810–8819. doi: 10.1016/j.biomaterials.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Okano H, Nakamura M, Yoshida K, Okada Y, Tsuji O, Nori S, Ikeda E, Yamanaka S, Miura K. Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res. 2013;112:523–533. doi: 10.1161/CIRCRESAHA.111.256149. [DOI] [PubMed] [Google Scholar]
- Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- Ozawa F, Ino K, Arai T, Ramón-Azcón J, Takahashi Y, Shiku H, Matsue T. Alginate gel microwell arrays using electrodeposition for three-dimensional cell culture. Lab Chip. 2013;13:3128–3135. doi: 10.1039/c3lc50455g. [DOI] [PubMed] [Google Scholar]
- Pan F, Zhang M, Wu G, Lai Y, Greber B, Schöler HR, Chi L. Topographic effect on human induced pluripotent stem cells differentiation towards neuronal lineage. Biomaterials. 2013;34:8131–8139. doi: 10.1016/j.biomaterials.2013.07.025. [DOI] [PubMed] [Google Scholar]
- Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Choi JC, Shim JH, Lee JS, Park H, Kim SW, Doh J, Cho DW. A comparative study on collagen type I and hyaluronic acid dependent cell behavior for osteochondral tissue bioprinting. Biofabrication. 2014;6:035004. doi: 10.1088/1758-5082/6/3/035004. [DOI] [PubMed] [Google Scholar]
- Patel S, Jung D, Yin PT, Carlton P, Yamamoto M, Bando T, Sugiyama H, Lee KB. NanoScript: a nanoparticle-based artificial transcription factor for effective gene regulation. ACS Nano. 2014;8:8959–8967. doi: 10.1021/nn501589f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil SS, Shahiwala A. Patented pulsatile drug delivery technologies for chronotherapy. Expert Opin Ther Pat. 2014;24:845–856. doi: 10.1517/13543776.2014.916281. [DOI] [PubMed] [Google Scholar]
- de Peppo GM, Marcos-Campos I, Kahler DJ, Alsalman D, Shang L, Vunjak-Novakovic G, Marolt D. Engineering bone tissue substitutes from human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2013;110:8680–8685. doi: 10.1073/pnas.1301190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place ES, Evans ND, Stevens MM. Complexity in biomaterials for tissue engineering. Nat Mater. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- Poldervaart MT, Gremmels H, van Deventer K, Fledderus JO, Oner FC, Verhaar MC, Dhert WJ, Alblas J. Prolonged presence of VEGF promotes vascularization in 3D bioprinted scaffolds with defined architecture. J Control Release. 2014;184:58–66. doi: 10.1016/j.jconrel.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Qian X, Villa-Diaz LG, Kumar R, Lahann J, Krebsbach PH. Enhancement of the propagation of human embryonic stem cells by modifications in the gel architecture of PMEDSAH polymer coatings. Biomaterials. 2014;35:9581–9590. doi: 10.1016/j.biomaterials.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask F, Mihic A, Reis L, Dallabrida SM, Ismail NS, Sider K, Simmons CA, Rupnick MA, Weisel RD, Li R-K, Radisic M. Hydrogels modified with QHREDGS peptide support cardiomyocyte survival in vitro and after sub-cutaneous implantation. Soft Matter. 2010;6:5089–5099. [Google Scholar]
- Richards Grayson AC, Choi IS, Tyler BM, Wang PP, Brem H, Cima MJ, Langer R. Multi-pulse drug delivery from a resorbable polymeric microchip device. Nat Mater. 2003;2:767–772. doi: 10.1038/nmat998. [DOI] [PubMed] [Google Scholar]
- Richardson TP, Peters MC, Ennett AB, Mooney DJ. Polymeric system for dual growth factor delivery. Nat Biotechnol. 2001;19:1029–1034. doi: 10.1038/nbt1101-1029. [DOI] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Antonsson L, Hovatta O, Tryggvason K. Monolayer culturing and cloning of human pluripotent stem cells on laminin-521-based matrices under xeno-free and chemically defined conditions. Nat Protoc. 2014a;9:2354–2368. doi: 10.1038/nprot.2014.159. [DOI] [PubMed] [Google Scholar]
- Rodin S, Antonsson L, Niaudet C, Simonson OE, Salmela E, Hansson EM, Domogatskaya A, Xiao Z, Damdimopoulou P, Sheikhi M, Inzunza J, Nilsson AS, Baker D, Kuiper R, Sun Y, Blennow E, Nordenskjold M, Grinnemo KH, Kere J, Betsholtz C, et al. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat Commun. 2014b;5:3195. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
- Rosellini E, Cristallini C, Barbani N, Vozzi G, Giusti P. Preparation and characterization of alginate/gelatin blend films for cardiac tissue engineering. J Biomed Mater Res A. 2009;91:447–453. doi: 10.1002/jbm.a.32216. [DOI] [PubMed] [Google Scholar]
- Rosenzweig A. Illuminating the potential of pluripotent stem cells. N Engl J Med. 2010;363:1471–1472. doi: 10.1056/NEJMe1007902. [DOI] [PubMed] [Google Scholar]
- Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Ji L, de Pablo JJ, Palecek SP. Inhibition of human embryonic stem cell differentiation by mechanical strain. J Cell Physiol. 2006;206:126–137. doi: 10.1002/jcp.20441. [DOI] [PubMed] [Google Scholar]
- Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Mei Y, Reisterer CM, Pyzocha NK, Yang J, Muffat J, Davies MC, Alexander MR, Langer R, Anderson DG, Jaenisch R. Surface-engineered substrates for improved human pluripotent stem cell culture under fully defined conditions. Proc Natl Acad Sci USA. 2011;108:18714–18719. doi: 10.1073/pnas.1114854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotech. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams A, Powers M. Feeder-free substrates for pluripotent stem cell culture. In: Lakshmipathy U, Vemuri MC, editors. Pluripotent Stem Cells. 997, 7. New York: Humana Press; 2013. pp. 73–89. (eds), Vol. [DOI] [PubMed] [Google Scholar]
- Sangaj N, Kyriakakis P, Yang D, Chang C-W, Arya G, Varghese S. Heparin mimicking polymer promotes myogenic differentiation of muscle progenitor cells. Biomacromolecules. 2010;11:3294–3300. doi: 10.1021/bm101041f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant S, Hancock MJ, Donnelly JP, Iyer D, Khademhosseini A. Biomimetic gradient hydrogels for tissue engineering. Can J Chem Eng. 2010;88:899–911. doi: 10.1002/cjce.20411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Ankrum JA, Teo GS, Carman CV, Karp JM. Cellular and extracellular programming of cell fate through engineered intracrine-, paracrine-, and endocrine-like mechanisms. Biomaterials. 2011;32:3053–3061. doi: 10.1016/j.biomaterials.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Seo EJ, Jang IH, Do EK, Cheon HC, Heo SC, Kwon YW, Jeong GO, Kim BR, Kim JH. Efficient production of retroviruses using PLGA/bPEI-DNA nanoparticles and application for reprogramming somatic cells. PLoS ONE. 2013;8:e76875. doi: 10.1371/journal.pone.0076875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han MJ, Lytton-Jean A, Basto PA, Jhunjhunwala S, Lee J, Heller DA, Kang JW, Hartoularos GC, Kim KS, Anderson DG, Langer R, Jensen KF. A vector-free microfluidic platform for intracellular delivery. Proc Natl Acad Sci USA. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanki A, Shah S, Memoli KA, Park SY, Hong S, Lee K-B. Controlling differentiation of neural stem cells using extracellular matrix protein patterns. Small. 2010;6:2509–2513. doi: 10.1002/smll.201001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Stolpe A, den Toonder J. Workshop meeting report Organs-on-Chips: human disease models. Lab Chip. 2013;13:3449–3470. doi: 10.1039/c3lc50248a. [DOI] [PubMed] [Google Scholar]
- Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat Rev Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, Mizuguchi H. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- Teng S, Liu C, Krettek C, Jagodzinski M. The application of induced pluripotent stem cells for bone regeneration: current progress and prospects. Tissue Eng Part B Rev. 2014;20:328–339. doi: 10.1089/ten.TEB.2013.0301. [DOI] [PubMed] [Google Scholar]
- Terashima M, Amano M, Onodera T, Nishimura S-I, Iwasaki N. Quantitative glycomics monitoring of induced pluripotent-and embryonic stem cells during neuronal differentiation. Stem Cell Res. 2014;13:454–464. doi: 10.1016/j.scr.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Tian H, Chen J, Chen X. Nanoparticles for gene delivery. Small. 2013;9:2034–2044. doi: 10.1002/smll.201202485. [DOI] [PubMed] [Google Scholar]
- Tibbitt MW, Anseth KS. Dynamic microenvironments: the fourth dimension. Sci Transl Med. 2012;4:160ps24. doi: 10.1126/scitranslmed.3004804. [DOI] [PubMed] [Google Scholar]
- Tong Z, Jia X. Biomaterial-based strategies for the engineering of mechanically active soft tissues. MRS Commun. 2012;2:31–39. doi: 10.1557/mrc.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Zerdoum AB, Duncan RL, Jia X. Dynamic vibration cooperates with connective tissue growth factor to modulate stem cell behaviors. Tissue Eng Part A. 2014;20:1922–1934. doi: 10.1089/ten.tea.2013.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukov FF, Maddox JF, Amacher DE, Bobrowski WF, Roth RA, Ganey PE. Modeling inflammation–drug interactions in vitro: a rat Kupffer cell-hepatocyte coculture system. Toxicol In Vitro. 2006;20:1488–1499. doi: 10.1016/j.tiv.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill GH, Bhatia SN. High-throughput analysis of signals regulating stem cell fate and function. Curr Opin Chem Biol. 2007;11:357–366. doi: 10.1016/j.cbpa.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varde NK, Pack DW. Microspheres for controlled release drug delivery. Exp Opin Biol Ther. 2004;4:35–51. doi: 10.1517/14712598.4.1.35. [DOI] [PubMed] [Google Scholar]
- Varde NK, Pack DW. Influence of particle size and antacid on release and stability of plasmid DNA from uniform PLGA microspheres. J Control Release. 2007;124:172–180. doi: 10.1016/j.jconrel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz LG, Pacut C, Slawny NA, Ding J, O'Shea KS, Smith GD. Analysis of the factors that limit the ability of feeder cells to maintain the undifferentiated state of human embryonic stem cells. Stem Cells Dev. 2009;18:641–651. doi: 10.1089/scd.2008.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O'Shea KS, Lahann J, Smith GD. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat Biotechnol. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz LG, Brown SE, Liu Y, Ross AM, Lahann J, Parent JM, Krebsbach PH. Derivation of mesenchymal stem cells from human induced pluripotent stem cells cultured on synthetic substrates. Stem Cells. 2012;30:1174–1181. doi: 10.1002/stem.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa-Diaz LG, Ross AM, Lahann J, Krebsbach PH. Concise review: the evolution of human pluripotent stem cell culture: from feeder cells to synthetic coatings. Stem Cells. 2013;31:1–7. doi: 10.1002/stem.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Tang Z, Park IH, Zhu Y, Patel S, Daley GQ, Li S. Induced pluripotent stem cells for neural tissue engineering. Biomaterials. 2011;32:5023–5032. doi: 10.1016/j.biomaterials.2011.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang D-Z, Li K, Wang J, Wanders RJA, Kulik W, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014a;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu J, Jiao J, Liu Z, Zhou Z, Zhao C, Chang L-J, Chen YE, Ma PX, Yang B. Engineering vascular tissue with functional smooth muscle cells derived from human iPS cells and nanofibrous scaffolds. Biomaterials. 2014b;35:8960–8969. doi: 10.1016/j.biomaterials.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Huck WT. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol. 2013;14:467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- Williams DF. On the nature of biomaterials. Biomaterials. 2009;30:5897–5909. doi: 10.1016/j.biomaterials.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Wong S, Shim MS, Kwon YJ. Synthetically designed peptide-based biomaterials with stimuli-responsive and membrane-active properties for biomedical applications. J Mater Chem B. 2014;2:595–615. doi: 10.1039/c3tb21344g. [DOI] [PubMed] [Google Scholar]
- Woodard CM, Campos BA, Kuo S-H, Nirenberg MJ, Nestor MW, Zimmer M, Mosharov EV, Sulzer D, Zhou H, Paull D. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson's disease. Cell Rep. 2014;9:1173–1182. doi: 10.1016/j.celrep.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward SC, Brewer PS, Moatamed F, Schindler A, Pitt CG. The intracellular degradation of poly(epsilon-caprolactone) J Biomed Mater Res. 1985;19:437–444. doi: 10.1002/jbm.820190408. [DOI] [PubMed] [Google Scholar]
- Xiao L, Tong Z, Chen Y, Pochan DJ, Sabanayagam CR, Jia X. Hyaluronic acid-based hydrogels containing covalently integrated drug depots: implication for controlling inflammation in mechanically stressed tissues. Biomacromolecules. 2013;14:3808–3819. doi: 10.1021/bm4011276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Hu J, Ma H, Zhang J, Chang LJ, Chen YE, Ma PX. Three-dimensional growth of iPS cell-derived smooth muscle cells on nanofibrous scaffolds. Biomaterials. 2011;32:4369–4375. doi: 10.1016/j.biomaterials.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Xu AM, Leal-Ortiz S, Cao Y, Garner CC, Melosh NA. Nanostraw-electroporation system for highly efficient intracellular delivery and transfection. ACS Nano. 2013;7:4351–4358. doi: 10.1021/nn400874a. [DOI] [PubMed] [Google Scholar]
- Xu Q, Hashimoto M, Dang TT, Hoare T, Kohane DS, Whitesides GM, Langer R, Anderson DG. Preparation of monodisperse biodegradable polymer microparticles using a microfluidic flow-focusing device for controlled drug delivery. Small. 2009;5:1575–1581. doi: 10.1002/smll.200801855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Miranda-Nieves D, Ankrum JA, Matthiesen ME, Phillips JA, Roes I, Wojtkiewicz GR, Juneja V, Kultima JR, Zhao W, Vemula PK, Lin CP, Nahrendorf M, Karp JM. Tracking mesenchymal stem cells with iron oxide nanoparticle loaded poly(lactide-co-glycolide) microparticles. Nano Lett. 2012;12:4131–4139. doi: 10.1021/nl301658q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Zhao W, Zhu JM, Albanna MZ, Yoo JJ, Atala A. Complex heterogeneous tissue constructs containing multiple cell types prepared by inkjet printing technology. Biomaterials. 2013a;34:130–139. doi: 10.1016/j.biomaterials.2012.09.035. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu L, Laslett AL, Esteban MA. Cell reprogramming: into the groove. Nat Mater. 2013b;12:1082–1084. doi: 10.1038/nmat3821. [DOI] [PubMed] [Google Scholar]
- Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W, Priceman S, Wu L, Zhou ZH, Liu Z, Segura T, Tang Y, Lu Y. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nano. 2010;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- Yang K, Lee JS, Kim J, Lee YB, Shin H, Um SH, Kim JB, Park KI, Lee H, Cho S-W. Polydopamine-mediated surface modification of scaffold materials for human neural stem cell engineering. Biomaterials. 2012;33:6952–6964. doi: 10.1016/j.biomaterials.2012.06.067. [DOI] [PubMed] [Google Scholar]
- Ye J-H, Xu Y-J, Gao J, Yan S-G, Zhao J, Tu Q, Zhang J, Duan X-J, Sommer CA, Mostoslavsky G, Kaplan DL, Wu Y-N, Zhang C-P, Wang L, Chen J. Critical-size calvarial bone defects healing in a mouse model with silk scaffolds and SATB2-modified iPSCs. Biomaterials. 2011;32:5065–5076. doi: 10.1016/j.biomaterials.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip D, Cho CH. A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem Biophys Res Commun. 2013;433:327–332. doi: 10.1016/j.bbrc.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Zhang L, Radovic-Moreno AF, Alexis F, Gu FX, Basto PA, Bagalkot V, Jon S, Langer RS, Farokhzad OC. Co-delivery of hydrophobic and hydrophilic drugs from nanoparticle-aptamer bioconjugates. ChemMedChem. 2007;2:1268–1271. doi: 10.1002/cmdc.200700121. [DOI] [PubMed] [Google Scholar]
- Zhang T, Wan LQ, Xiong Z, Marsano A, Maidhof R, Park M, Yan Y, Vunjak-Novakovic G. Channelled scaffolds for engineering myocardium with mechanical stimulation. J Tissue Eng Regen Med. 2012;6:748–756. doi: 10.1002/term.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A. A 3D Alzheimer's disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials. 2014a;35:1420–1428. doi: 10.1016/j.biomaterials.2013.11.028. [DOI] [PubMed] [Google Scholar]
- Zhang S, Dutton JR, Su L, Zhang J, Ye L. The influence of a spatiotemporal 3D environment on endothelial cell differentiation of human induced pluripotent stem cells. Biomaterials. 2014b;35:3786–3793. doi: 10.1016/j.biomaterials.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoldan J, Lytton-Jean AK, Karagiannis ED, Deiorio-Haggar K, Bellan LM, Langer R, Anderson DG. Directing human embryonic stem cell differentiation by non-viral delivery of siRNA in 3D culture. Biomaterials. 2011;32:7793–7800. doi: 10.1016/j.biomaterials.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonca MR, Yune PS, Heldt CL, Belfort G, Xie Y. High-throughput screening of substrate chemistry for embryonic stem cell attachment, expansion, and maintaining pluripotency. Macromol Biosci. 2013;13:177–190. doi: 10.1002/mabi.201200315. [DOI] [PubMed] [Google Scholar]