Abstract
The control of cell migration by chemokines involves interactions with two types of receptors: seven transmembrane chemokine-type G protein-coupled receptors and cell surface or extracellular matrix associated glycosaminoglycans. Coordinated interaction of chemokines with both types of receptors is required for directional migration of cells in numerous physiological and pathological processes. Accumulated structural information, culminating most recently in the structure of a chemokine receptor in complex with a chemokine, has led to a view where chemokine oligomers bind to glycosaminoglycans through epitopes formed when chemokine subunits come together, while chemokine monomers bind to receptors in a pseudo two-step mechanism of receptor activation. Exploitation of this structural knowledge has and will continue to provide important information for therapeutic strategies, as described in this review.
Introduction
Host defense depends on the ability of chemokines to control cell migration during immune surveillance and inflammation. Similarly, chemokines regulate lymphocyte development, maturation and homing, as well as development of lymphoid and other organs1, 2. However, chemokines also have a dark side and contribute to numerous pathologies including inflammatory diseases and cancer3. In all of these scenarios, chemokines interact with both chemokine receptors (CKRs) and with glycosaminoglycans (GAGs) to promote migration and impart directionality to cell movement, respectively4.
CKRs are members of the seven transmembrane G Protein-Coupled Receptor (GPCR) family; they are expressed on migrating cells and couple chemokine binding on the outside of the cell, to activation of signaling pathways inside the cell that lead to cell motility. By contrast, GAGs are carbohydrate structures that are either attached to protein cores of proteoglycans on cells or shed into the extracellular matrix (ECM); among other functions, they facilitate the immobilization of chemokines and the formation of “haptotactic” chemokine gradients that effectively direct the migration of CKR bearing cells (Figure 1A). The architecture of chemokines is such that they must be able to accommodate both types of very different interactions, while maintaining receptor (and possibly GAG) specificity. Moreover the affinities of chemokines for GAGs and CKRs must be tuned in such a way that enables transfer of chemokine from GAG to receptor at the appropriate time. Structural, biophysical and functional studies are beginning to provide detailed insight into how these remarkable proteins work. In this review we describe the development and current state of structural knowledge of the chemokine system including structures of chemokines, their interactions with GAGs and interactions with CKRs. We also describe how investigations of chemokine interactions with both receptors and GAGs has led to diverse therapeutic strategies. Particularly exciting are recent breakthroughs in the structure determination of CKRs in complex with both synthetic antagonists and a natural chemokine ligand; these structures should significantly contribute to small molecule drug discovery efforts, have provided the first view of a CKR:chemokine complex, and have provided insight into the interaction of the HIV glycoprotein, gp120, with chemokine co-receptors CXCR4 and CCR5. Much remains to be understood, and some of the outstanding issues are highlighted at the end of the review.
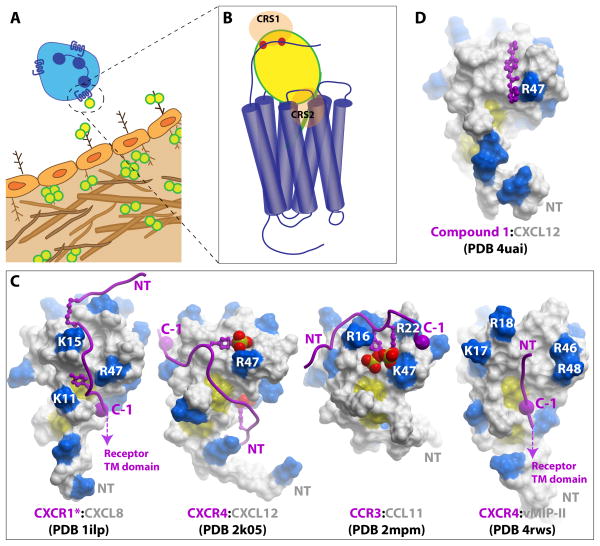
Figure 1. Chemokine interactions with receptors and GAGs.
(A) Central to the role of chemokines is their ability to form immobilized gradients to guide the migration of receptor bearing cells. The cartoon illustrates chemokines (yellow circles) immobilized on the luminal and basolateral endothelium. Interactions with chemokines, predominantly as oligomers, and GAGs (brown branched figures) contribute to the formation of the gradient. However, only chemokine monomers are required for activation of receptors; thus chemokine oligomers must dissociate in order to bind CKRs. (B) The general two-site model of CKR:chemokine binding involves the interaction of the receptor N-terminal domain with the core domain of the chemokine, chemokine recognition site 1 (CRS1), and the N-terminal signaling domain of the chemokine with the TM binding pocket of the receptor (CRS2). (C) CRS1 interactions in CXCR1*:CXCL8 (PDB ID 1ILP), CXCR4:CXCL12 (PDB ID 2K05), CCR3:CCL11 (PDB ID 2MPM) and CXCR4:vMIP-II (PDB ID 4RWS). The chemokines are shown as surface representations with basic residues highlighted in blue. The receptor N-termini are depicted as purple ribbons with sulfated tyrosine sidechains shown as sticks and spheres. The large purple spheres labeled “C-1” indicate the residue immediately N-terminal to the conserved Cys in the CKR N-terminus; this Cys is structurally constrained in the receptor at the mouth of the TM binding pocket and allows orientation of the “CRS1 structures” relative to the receptor TM domain. Thus the orientation of the CKR peptide in the CXCR1*:CXCL8 and CXCR4:vMIP-II structures allows the chemokine N-terminal signaling domain to be oriented in a manner compatible with the CRS2 interaction. The designation CXCR1* indicates that the CXCR1 peptoid is modified with a hexanoic acid moiety, shown as small purple sticks. (D) Structure of CXCL12 with a small molecule selected from a screen to block the CRS1 interaction by targeting the sTyr21 binding site on the chemokine.
Chemokine structure
In humans alone, there are approximately 45 chemokines and 22 CKRs, not including splice variants or isoforms5. The ligands are 7–12 kDa proteins that have been classified into four subfamilies based on a characteristic pattern of cysteine residues (CC, CXC, C and CX3C, where X is any amino acid) in proximity to the amino terminus of the mature proteins6. In general, chemokines bind receptors of their own class. However, within a given subfamily, many chemokines bind multiple receptors and multiple receptors bind many chemokines. A few exceptions to these generalizations exist including viral chemokines and receptors, and “atypical receptors” that in some cases have broad specificities across subfamilies5.
The first chemokine structure to be determined was that of CXCL8/IL-8 in 19907. It revealed the basic tertiary architecture characteristic of all chemokines -- a disordered N-terminal “signaling domain”, followed by a structured “core domain” consisting of an “N-loop”, a three-stranded β-sheet and a C-terminal helix (Figure 2A). As is the case for many CXC chemokines, CXCL8 forms dimers where the first strand of the three-stranded β-sheet of one subunit interacts with the same region of a second subunit, giving rise to an overall six-stranded β-sheet platform topped by two α-helices (Figure 2B). Subsequent to the structures of CXCL8 and other CXC chemokines, the structure of the CC chemokine, CCL4/MIP-1β, was solved8, followed by CCL5/RANTES9 and CCL2/MCP-110 shortly thereafter. These CC chemokine structures revealed a distinctly different dimer motif compared to CXC dimers. Formation of CC dimers occurs through interactions between residues near the N-terminus and encompassing the signature di-cysteine CC sequence, which leads to an overall elongated architecture (Figure 2C). The singular CX3C chemokine, CX3CL1/fractalkine is largely monomeric11 but can form dimers that resemble CC chemokines12, while XCL1/lymphotactin forms the canonical chemokine fold in equilibrium with a four-stranded β-sheet that associates in a head-to-tail dimer (Figure 2D)13.
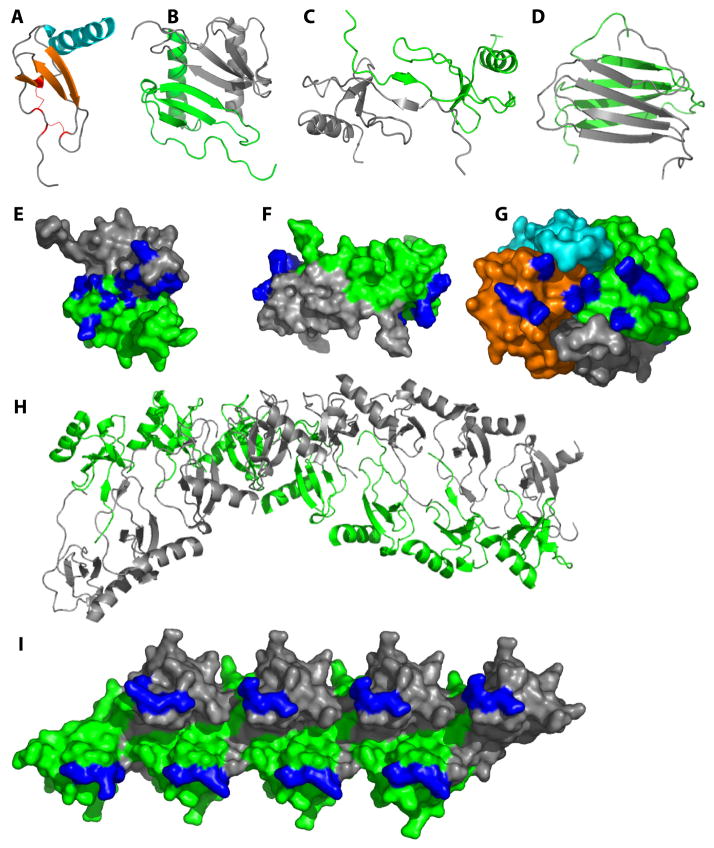
Figure 2. Chemokines form diverse oligomeric structures.
(A) The highly conserved tertiary structure typical of all chemokines is illustrated with a monomer subunit of CXCL8 (PDB ID 1IL8). (B) A CXC-type dimer, illustrated with CXCL8 (PDB ID 1IL8). (C) A CC-type dimer illustrated by CCL2 (PDB ID 1DOM). (D) The non-canonical XCL1 dimer (PDB ID 1J90). (E-G) GAG-binding epitopes (highlighted in blue) in the context of the CXCL12 dimer (E; PDB ID 2J7Z), the CCL2 dimer (F; PDB ID 1DOM) and the CCL2 tetramer (G; PDB ID 1DOL). In the case of CCL2, the oligomeric form favored appears to be dependent on the type of GAG present where HS stabilizes a dimer (F) and heparin stabilizes a tetramer (G). (H) Some chemokines form higher order oligomers, as illustrated by CCL4 at neutral pH. Note that CCL4 forms dimers as in (C) at low pH; thus the polymer is an assembly based on the dimer as a fundamental substructure (I) GAG-binding epitopes highlighted in the context of the CCL5 octamer.
Initially the different dimer motifs were thought to determine the specificity of CC and CXC chemokines for their respective CC vs. CXC receptors8. However, subsequent studies showed that oligomerization-deficient chemokines were as potent and efficacious as WT chemokines in functional assays, suggesting that monomers are the form that bind and activate receptors14, 15. Instead, chemokine oligomers are important for binding to GAGs, as described below, and they do so by adopting a wide range of oligomerization states including monomers (e.g. CCL7/MCP-3), dimers (e.g. CXCL8, CXCL12/SDF-1), tetramers (e.g. CXCL4/PF-4) and even polymers (e.g. CCL3/MIP-1α, CCL4, CCL5) (Figure 2)16, 17, 18. Thus the reversible equilibrium of chemokines between monomer and oligomeric states is critical for chemokine function, allowing them to interact with both CKRs and with GAGs, respectively (Figure 1A).
Chemokine interactions with GAGs and the role of oligomerization
Although it had been known for some time that chemokines bind GAGs, the relevance of GAG binding to chemokine function was not firmly established until several studies demonstrated its importance in vivo. These studies utilized GAG-binding deficient chemokines that were competent to bind and activate receptors in vitro, and demonstrated that the mutants were impaired in their ability to promote cell recruitment in vivo compared to the WT counterparts19, 20. The interaction of chemokines with GAGs is now thought to be critical for haptotactic migration of cells by establishing cell surface chemokine gradients that provide directional signals21, 22, 23. Cell surface immobilization of chemokines enables them to act locally rather than as paracrine molecules, and likely prevents inappropriate activation and desensitization of receptors on cells outside the region of interest for a given physiological situation24, 25. Chemokine:GAG interactions have also been shown to facilitate secretion of chemokines from tumor cells26 and T cells26, transcytosis across cells27, 28, chemokine stability29 and even signaling30.
In parallel with studies aimed at determining the relevance of GAG interactions, in vivo experiments using oligomerization-deficient variants demonstrated the importance of chemokine oligomerization to cell migration for a number of chemokines19. Moreover, further studies showing that many chemokines oligomerize on GAGs31,32, and that the affinity of chemokine:GAG interactions increases with GAG size up to ~dp20 (where dp=degree of polymerization)33, led to the hypothesis that oligomerization and GAG binding are coupled, and that oligomerization increases the affinity of chemokines for GAGs through an avidity effect. Biophysical studies of several chemokines confirmed this hypothesis and that interaction of chemokines with GAGs stabilizes chemokine oligomers32, 34. Moreover, a recent study of several chemokines that span a wide range of oligomerization states demonstrated that oligomerization can have a dramatic effect on GAG affinity, and can also be important for GAG specificity. For example, dimer variants of CXCL4 and CCL5 show almost negligible binding to cell surface GAGs compared to their WT counterparts that form tetramers and polymers respectively35. Furthermore, among the eight chemokines investigated in this study, WT CXCL4 and CCL5 were unique in their ability to bind with high affinity to chondroitin sulfate-A, which was lost with dimer variants of either chemokine. Perhaps one of the most unusual examples of the requirement for chemokine oligomerization and GAG binding relates to XCL1/lymphotactin. This chemokine rapidly interconverts between the canonical chemokine tertiary structure that binds XCR1 receptors, and a β-sandwich dimer that binds GAGs (Figure 2D)13.
The importance of both chemokine:GAG interactions and chemokine oligomerization has been underscored by the effectiveness of GAG mutants in disease models in vivo. Mutation of a “BBXB” motif (where B is a basic and X is any amino acid) in CCL5 resulted in a variant, [44-AANA-47]-CCL5, which has a dramatically reduced ability to bind GAGs, as well as an impaired ability to form oligomers larger than dimers36. It showed inhibitory effects in mouse models of inflammatory cell recruitment into the peritoneal cavity, lung and the CNS, apparently through formation of non-functional (e.g. GAG-binding deficient) heterodimers with WT RANTES. This same mutant was also effective in reducing plaque formation and myocardial reperfusion injury in mouse models of atherosclerosis37, 38 and injury and fibrosis in a model of liver damage39. GAG mutants of CXCL12 and CCL7 showed similar inhibitory effects on chemokine mediated inflammation in an air pouch model24, 40. Interestingly, the CCL7 GAG mutant antagonized cell migration not only towards WT CCL7, but also towards CXCL12, a chemokine that has no overlapping receptor specificity. In both cases, the data suggest that part of the mechanism is due to receptor desensitization caused by systemic distribution of the GAG-deficient chemokine. Finally, [P8A]-CCL2, a purely non-oligomerizing variant of CCL2, inhibited leukocyte recruitment in mouse models of lung inflammation, arthritis and experimental autoimmune encephalomyelitis (EAE)41, 42. Again, the reduced ability of [P8A]-CCL2 to bind GAGs likely causes it to have a more delocalized distribution than WT CCL2, leading to receptor desensitization at the wrong place and time.
While numerous structures of chemokines have been reported43, there are no structures of chemokines in complex with GAGs apart from two complexes containing heparin disaccharides 44, 45. Complexes with larger, more relevant GAGs are challenging because GAGs are heterogeneous in length and composition, which thwarts both crystallography and NMR studies. Moreover, commercially available homogeneous GAGs are limited to small heparin fragments. However, epitope mapping studies by NMR and mutagenesis have provided insight into the manner in which chemokines bind GAGs in a few cases. Some chemokines like CCL7 bind GAGs as monomers46. More frequently, however, complete GAG binding sites are assembled only when chemokines oligomerize4, 16, 32. For example, heparin stabilizes CXCL12 as a dimer through an interaction surface that runs approximately perpendicular to the dimer interface47 (Figure 2E), while the distribution of epitopes in CCL516, 18 imply that GAGs stabilize dimers of dimers within the polymer (Figure 2I). Different types of GAGs may also preferentially bind different oligomeric states of the same chemokine. For example, heparan sulfate (HS), a GAG with highly sulfated regions separated by unsulfated domains, binds CCL2 and CCL3 dimers46, 48, likely because its structure complements the discontinuous distribution of GAG-binding epitopes on the chemokine dimers (Figure 2F). By contrast, heparin, which is uniformly sulfated, stabilizes CCL2 as a tetramer in a manner that produces a continuous basic interaction surface32 (Figure 2G). While an important goal for future studies will be to determine high-resolution structures of oligomeric chemokines with GAGs larger than disaccharides, similar epitope mapping endeavors, combined with docking of GAGs to chemokine surfaces may go a long way towards understanding these complex interactions.
Interactions of chemokines with chemokine receptors: the two-site model
Similar to the challenges associated with structurally characterizing chemokine:GAG interactions, determining structures of CKR:chemokine complexes is also challenging, in this case because the receptors are conformationally flexible transmembrane (TM) proteins. Thus mutagenesis coupled with binding and functional assays have dominated efforts to determine molecular details of CKR:chemokine binding and activation. Early work on CXCL8 by Clark-Lewis revealed the critical role of the chemokine N-terminus in receptor activation and that receptor binding and activation could be uncoupled49. Specifically, N-terminal modifications (deletions or mutations) were identified that converted CXCL8 from an agonist into an antagonist50. Subsequent studies of many other chemokines also supported the generality of this phenomenon51. Moreover, proteolytic modification of chemokine N-termini was discovered as a natural mechanism for regulating chemokine function52. On the receptor side, mutagenesis studies revealed the general trend that the N-termini of CKRs are important for binding to the structured chemokine “core domain” 53, 54. Together, these findings gave rise to a paradigm referred to as the two-site model of receptor activation53, 55 in which the CKR N-terminus interacts with the chemokine core domain (chemokine recognition site 1, CRS15), while the N-terminus of the chemokine interacts with the receptor ligand-binding pocket (chemokine recognition site 2, CRS2) (Figure 1B). This paradigm has guided the field for many years, even in the absence of high-resolution structural information. Consistent with this model, an NMR study of CXCL12 in the presence of detergent solubilized CXCR4 demonstrated the ability of the small molecule compound, AMD3100, to specifically dislodge the CXCL12 N-terminus from its binding site on CXCR4 without displacing the bound chemokine core domain56; since AMD3100 binds to the TM binding pocket57, the logical conclusion was that the CXCL12 N-terminus binds in the pocket as well, and that the CRS1 core domain and CRS2 N-terminal interactions can be at least partially decoupled.
Exploiting the CRS2 interaction
Antagonist chemokine variants for exploring the role of specific receptors in disease
The ability to manipulate ligand pharmacology while still retaining high affinity binding with mutations, extensions or deletions of chemokine N-termini, has been exploited since the early 90s. Modified chemokines have proved useful for elucidating the roles of specific receptors in disease, particularly when small molecule receptor antagonists are unavailable. For example, [1+9-76]-CCL2, an antagonist variant of CCL2 lacking residues 2–8, showed efficacy in a number of diseases including arthritis58, cardiovascular disease59 and hepatic fibrosis60, implicating the receptor, CCR2. “Met-RANTES” (aka Met-CCL5) an antagonist variant of CCL5 extended with an N-terminal methionine, also showed efficacy in models of arthritis61, atherosclerosis62 and EAE63, among other disease models. One of the most interesting studies utilized Met-RANTES in a murine model of breast cancer, where it was shown to significantly slow tumor growth by inhibiting the macrophage infiltrate, as only the macrophages, but not the tumor cells, expressed relevant CKRs64. This study demonstrated for the first time that macrophages possess tumor-promoting activity, and also that CKR antagonists could be beneficial in cancer treatment by interfering with the tumor microenvironment. Finally, Kungl and coworkers developed a technology called CellJammer® which combines chemokine N-terminal modifications with enhanced GAG binding potential as a general design principle for generating anti-inflammatory protein therapeutics; the idea behind the technology is that the modified chemokines displace WT chemokines from requisite GAG interactions without activating CKRs65.
CCL5/RANTES variants as microbicides for the prevention of HIV
Many other examples of modified chemokines and their efficacy in disease models have been reported. However, a number of CCL5 variants deserve particular attention as they hold promise as future biologics and have also contributed significantly to understanding how HIV avoids inhibition by endogenous chemokines. The discovery of Met-RANTES was followed by AOP-RANTES, a synthetic analog containing an aminooxypentane group appended to the N-terminus. This variant proved to be more potent than Met-RANTES for inhibiting HIV infectivity, largely due to its ability to internalize CCR5 and delay its recycling back to the cell surface 66. With an aim to exploit this pharmacology, additional synthetically modified variants of CCL5 were subsequently discovered, the most potent being PSC-RANTES. In contrast to native CCR5 ligands, which are weak inhibitors of HIV infection, PSC-RANTES was fully protective in preventing vaginal transmission of SHIV in rhesus macaques67. Moreover, comparative studies of PSC-RANTES and related synthetic variants confirmed that their effectiveness in inhibiting HIV infectivity correlated more with their ability to internalize and sequester CCR5 inside the cell, than with their binding affinity for CCR568.
These successful proof-of-concept experiments motivated phage display efforts to identify fully recombinant CCL5 variants with similar properties, in order to avoid the high cost of chemical synthesis69. Three types of recombinant variants were identified: (1) those that signal and sequester CCR5 by internalization and prolonged inhibition of CCR5 recycling, (2) those that are high affinity antagonists that neither activate G proteins nor cause internalization and (3) those that show no signaling but still promote some level of internalization/sequestration. Two of these variants, the 6P4-RANTES agonist and the 5P12-RANTES antagonist, were subsequently tested in the rhesus macaques SHIV vaginal challenge model and shown to be fully protective, like PSC-RANTES70. Interestingly, however, the potency of 5P12-, 6P4- and PSC-RANTES turned out to involve different mechanisms. CCR5 exists in at least two conformations (e.g. G protein-coupled and uncoupled), and it appears that gp120 (the HIV envelope glycoprotein involved in the entry of HIV into immune cells by interactions with either CCR5 or CXCR4) binds indiscriminately with high affinity to both CCR5 populations, while native chemokines only bind with high affinity to the subpopulation of receptors in the nucleotide free G protein-coupled state71. By contrast, antagonist 5P12 is effective in HIV inhibition due to its ability to bind both populations of CCR5 with high affinity, and thereby block the gp120 interaction. The 6P4 agonist also has the ability to bind both receptor populations with high affinity but additionally can induce CCR5 internalization/downregulation, while PSC-RANTES, has low affinity for uncoupled CCR5, but still has significant antiviral activity due to its efficiency in internalizing/downregulating the receptor71. At this point it is unclear which type of modulator may prove to be the best microbicide, but there is some thought that non-signaling internalizers may be desirable for avoiding potential inflammation69.
In addition to the possibility that microbicides may emerge from continued exploration of CCL5 analogs, these studies demonstrated that it is possible to develop high affinity “biased ligands” with specifically defined pharmacological profiles by targeting the CRS2 interaction. Hopefully they will inspire efforts to develop small molecule biased ligands that can mimic the properties of these modified chemokines, as recently reported for the other HIV co-receptor, CXCR472. Another important goal will be to understand the molecular basis for the ability of 5P12-RANTES, 6P4-RANTES and gp120 to bind both CCR5 populations (or recruit all CCR5 receptors into a single population), when native chemokines cannot, as well as the conformational states of CCR5 dictated by these ligands that lead to different receptor signaling and trafficking responses.
These CCL5 variants will require stability in vivo if they are to become successful therapeutics. However, a follow-up study demonstrated that they retain antiviral activity under conditions related to their potential use as vaginal microbicides. Moreover, capitalizing on the affinity of CCL5 for GAGs, Wang and coworkers investigated the successful use of hydrogels containing GAGs for long-term delivery of 5P12-, 6P4- and PSC-RANTES with promising results73. If successful, GAG-based delivery vehicles may translate to other chemokine variants that have utility as anti-HIV biologics.
CCL5/RANTES variant chimeras targeting two steps in the HIV fusion reaction
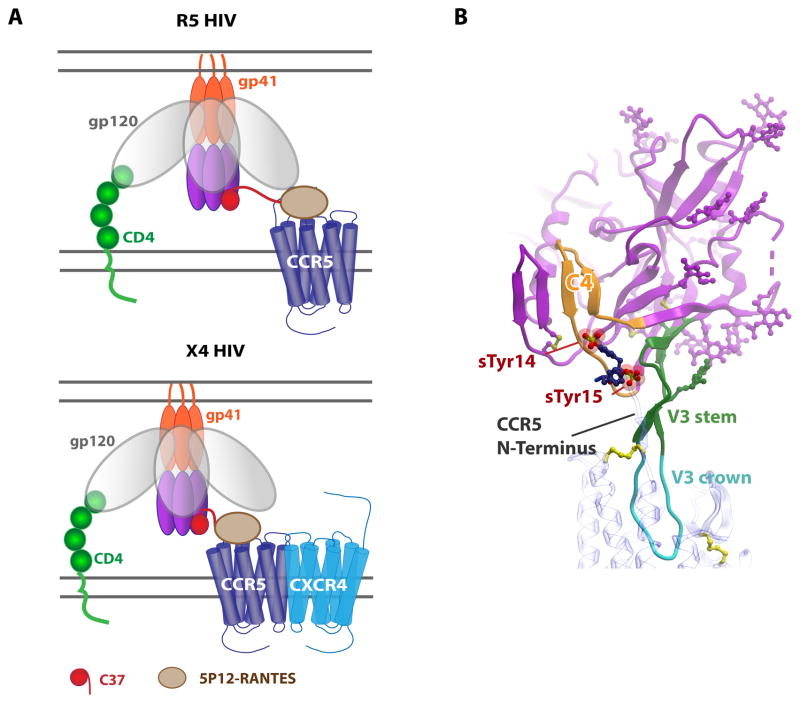
Another promising anti-HIV strategy has capitalized on the above N-terminally modified CCL5 variants. HIV entry into host cells initially involves the interaction of HIV gp120 with CD4 on host cell membranes, which causes conformational changes in gp120 that expose the third variable “V3-loop”. The V3-loop is then able to bind to either CCR5 or CXCR4, which in turn triggers conformational changes in HIV gp41 and penetration of its N-terminus into the host membrane. This so-called gp41 “prehairpin” intermediate state consists of a C-terminal coiled coil and an N-terminal coiled coil that ultimately collapse on each other to form a six-stranded helical bundle as the fusion reaction proceeds. However in the prehairpin state, gp41 is vulnerable to inhibition by a variety of molecules including peptides derived from its C-terminus74. Zhao and coworkers had the clever idea to engineer a chimera containing one of these gp41 inhibitors, C37, fused to the C-terminus of 5P12-RANTES or a related variant75. Once optimized with the appropriate linker length, the chimera showed IC50s in replication-competent viral assays as low as 4 pM and up to 100-fold more potent than either the RANTES variant or C37 alone. The authors proposed a model in which 5P12-RANTES and the C37 peptide simultaneously, or nearly simultaneously bind CCR5 and gp41, respectively, and that by binding CCR5, the chimera increases the local concentration of C37, enhancing its potency for trapping the prehairpin state (Figure 3A). Remarkably, the fusion also inhibited X4 (CXCR4-tropic) strains of HIV in addition to R5 (CCR5-tropic) strains (Figure 3A). It appears that the mechanism for inhibiting X4 virus also relates to increased local concentration of C37 by virtue of the CCR5 tether; however, the apparent ability of CXCR4 and CCR5 to heterodimerize may also be a contributing factor76, 77. The enhanced potencies of the chimera over related molecules that individually target CCR5 or gp41 are impressive and demonstrate the value of simultaneously targeting two different steps in the HIV fusion reaction.
Figure 3. Implications of structural information for HIV interactions and inhibition.
(A) A fusion of 5P12-RANTES with a C37 peptide inhibits HIV at the prefusion stage. Top: inhibition of R5 tropic viruses by binding of 5P12-RANTES to CCR5 and the C37 peptide to gp41. Bottom: inhibition of X4 tropic viruses requires the presence of CCR5 to increase the local concentration of C37. (B) Model of a gp120:CCR5 complex. Monomeric gp120 is shown in magenta (PDB 1D 2QAD) with the C4 “bridging sheet” highlighted in orange, the V3 stem highlighted in green and the V3 crown in cyan. CCR5 (PDB ID 4MBS) is shown with a blue transparent ribbon interacting with the V3 crown; sulfated N-terminal tyrosine residues (sTyr14 and sTyr15) are shown as sticks with sulfates shown as spheres.
Defining and exploiting CRS1 interactions
As described above, the first interaction thought to occur between chemokine and receptor is binding of the receptor N-terminus to the chemokine core domain (CRS1). CRS1 docking of the chemokine is then thought to orient the chemokine N-terminal signaling domain in a manner that enables it to bind to the CRS2 TM pocket (Figure 1B). Ideally, one would have structures of chemokines in complex with intact receptors to understand this recognition process. However, the challenge of working with membrane receptors has historically led to a divide and conquer approach, with focus on the more tractable CRS1 CKR:chemokine interactions that can be recapitulated as soluble complexes. These studies have provided important insights into the structural role of tyrosine sulfation (sTyr), a frequent post-translational modification observed in the N-termini of many CKRs.
Specifically many groups have utilized NMR methods to investigate CRS1 interactions of chemokines with peptides corresponding to CKR N-termini78, 79, 80. In all studies, the CKR peptides bind on the same face of the chemokine, and show interactions with the N-loop as expected from mutagenesis studies. Skelton and coworkers were able to determine a structure of CXCL8 in complex with a peptide from the N-terminus of CXCR1, which was modified with hexanoic acid to increase its affinity80 (Figure 1C). Two subsequent studies utilized sulfated receptor peptides; sulfation generally increases the affinity of chemokines for their receptors and thereby permitted structure determination78, 79. Veldkamp and coworkers reported the structure of an N-terminal sulfopeptide derived from CXCR4 bound to a “disulfide-locked” dimer of CXCL12 in a 2:2 complex, while Millard and coworkers revealed the structure of a peptide from CCR3 bound to a chemokine monomer, CCL11/eotaxin, in a 1:1 complex (Figure 1C).
In all three complexes, some common interactions were observed; specifically, the receptor peptides were found at a chemokine interface formed by the N-loop and β2-β3 strands and where present, the sulfotyrosines formed salt bridge interactions with homologous basic residues in the β2-β3 hairpin of the chemokine (e.g. R47/K47). However the receptor peptides differ quite dramatically in their orientation on the chemokine surface [76]. The CXCR1:CXCL8 structure is closest to what one might expect in order to accommodate CRS2 interactions in intact CKR:chemokine complexes since the receptor C-terminus points in the direction of the chemokine N-terminus (Figure 1C, far left). CXCR1:CXCL8 is also most similar to the orientation of the CXCR4 N-terminus on the surface of chemokine vMIP-II in the structure of an intact CXCR4:vMIP-II complex (described in the next section) (Figure 1C, far right). Quite the opposite, the CXCR4:CXCL12 complex cannot be reconciled with the expected CRS2 interaction as it suggests that the chemokine N-terminal signaling domain points away from the receptor binding pocket (Figure 1C, second from left)81. One possible explanation for these differences is that structural rearrangements occur after binding CRS1 in order to engage CRS2, and that these rearrangements differ from complex to complex. Another possibility for the incompatible orientation of the receptor peptide in the CXCR4:CXCL12 structure is that it is derived from a disulfide-locked dimer structure. As dimers of CXC chemokines have been shown to bind their respective receptors with high affinity and to function as partial agonists79, 82, the CXCR4:CXCL12 structure may therefore better reflect the chemokine dimer complex. In support of an alternative structure for the monomer bound form of CXCL12, a separate study using disulfide crosslinking to determine distance restraints between N-terminal residues in intact CXCR4 with monomeric CXCL12 led to a model in which the receptor peptide is oriented in the opposite direction from the dimeric CXCR4:CXCL12 NMR structure, and more compatible with the expected interaction of the chemokine N-terminus with the transmembrane CRS2 domain of the intact receptor81.
High-resolution structures of complexes with intact receptors will obviously be needed to understand how chemokine monomers and dimers bind their receptors, and as described in the next section, the first such structure has recently been determined83. Nevertheless, the described NMR studies provide insight into the structural role of sulfotyrosines. Furthermore, the disulfide-locked dimer of CXCL12 is under investigation as an antimetastatic biotherapeutic, due to its oligomerization-enhanced serum stability over WT CXCL1284. Finally, the CXCR4:CXCL12 complex motivated investigations of the druggability of the CRS1 interaction; compounds with micromolar affinity have been identified and a structure with a compound bound in the cleft formed by the CXCL12 N-loop and β2-β3 strands has been determined (Figure 1D)85. However, it remains to be seen whether CRS1-targeted compounds can be identified that have sufficient potency for overcoming the CRS2 interaction and sufficient specificity (given the sequence and structural homology of chemokines).
Structures of intact chemokine receptors in complex with small molecule, peptide and chemokine antagonists
CXCR4:IT1t and CXCR4:CVX15
Structures of membrane proteins are notoriously challenging and GPCRs are no exception, as they are difficult to express in sufficient quantities, difficult to purify, they are unstable and prone to aggregation in detergent, and they have little polar surface area for crystallization. Nevertheless, after years where bovine rhodopsin was the only GPCR to have been crystallized, additional GPCR structures began to appear in 2007, starting with the β2-adrenergic receptor86, 87. With few exceptions, the structures require appropriate ligands that provide sufficient stability for purification and stabilization into well-defined crystallizable constructs, making the best targets those for which drug discovery campaigns have identified high affinity ligands. Given the clear role of CXCR4 in HIV and in cancer metastasis, it is not surprising that this receptor was the first CKR to yield to structure determination in 201088.
CXCR4 was crystallized with two different synthetic ligands, a small molecule antagonist, IT1t, and a 16-residue cyclic peptide antagonist, CVX15 (Figure 4A and B)88. While the overall geometry of the receptor resembled structures of other GPCRs with the typical seven-helix topology, CXCR4 revealed for the first time the large acidic binding pocket of the receptor relative to other solved GPCR structures, which is consistent with the fact that the natural ligands are basic proteins. IT1t occupies a small part of the cavity (430 Å3) in the minor subpocket defined by TM helices I, II, III and VII and is stabilized by contacts with a number of polar side chains implicated in binding CXCL12. CVX15 fills a much larger fraction of the binding pocket (2200 Å3), particularly the major subpocket (TM helices III–VII). It is stabilized by interactions with some similar but also many different receptor side chains as IT1t, including residues identified as important for binding CXCL12. Along with pharmacological data, these structures suggest that IT1t and CVX15 act as orthosteric antagonists by interfering with the CRS2 CXCR4:CXCL12 interaction, while occupying very different parts and fractional volumes of the binding pocket. As described below, the CXCR4:CVX15 complex provided insight for the modeling of the interactions of CXCR4 with HIV gp12088.
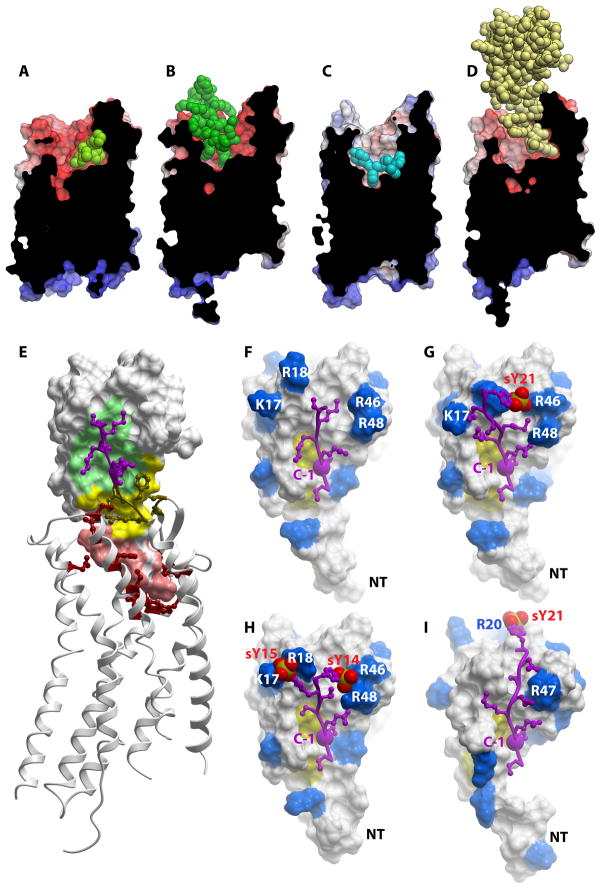
Figure 4. Chemokine receptor structures and models.
Surface representations of (A) CXCR4:IT1t (PDB ID 3ODU) (B) CXCR4:CVX15 (PDB ID 3OE0) (C) CCR5:Maraviroc (PDB ID 4MBS) and (D) CXCR4:vMIP-II (PDB ID 4RWS). Receptors are shown as cut-open surfaces, colored by electrostatic potential; the bound ligands are shown as spheres. (E) Structure of CXCR4:vMIP-II. vMIP-II is shown as a surface representation with regions corresponding to CRS1, CRS1.5 and CRS2 colored green, yellow and salmon, respectively. The receptor is shown as a ribbon, with residues making substantial contacts with chemokine shown as sticks. The purple region of the receptor (CRS1) N-terminus corresponds to the same region as shown in F-H. (F) Structure of the CRS1 interaction of CXCR4:vMIP-II. The chemokine is shown as a surface representation with basic residues highlighted in blue. The receptor N-terminus visible in the electron density is shown in purple. The yellow patch indicates the conserved Cys residues. The large purple spheres labeled “C-1” indicate the residue immediately N-terminal to the conserved Cys in the CKR N-terminus; this Cys is structurally constrained in the receptor at the mouth of the TM binding pocket and allows orientation of the “CRS1 structures” relative to the receptor TM domain. (G) Model of the CRS1 interaction of CXCR4:vMIP-II in which the CXCR4 N-terminus is extended by two residues to include sTyr21, shown as red spheres. (H) Model of the CRS1 interaction of CCR5:vMIP-II, represented as in G with sTyr14 and sTyr15. (I) Model of the CRS1 interaction of CXCR4:CXCL12, represented as in G with sTyr21.
In total, five structures of IT1t and CVX15 complexes were solved88, and all demonstrated that CXCR4 forms dimers with a roughly consistent dimer interface involving TM helices V and VI. This finding corroborated a wealth of cell-based studies suggesting that CXCR4 forms homo- and heterodimers76. It also raised the question of whether the stoichiometry of CKR:chemokine complexes is 1:1 as historically envisioned, or 2:1, which also seemed feasible and consistent with the two-site model81. However, a subsequent study demonstrated conclusively that CXCL12 binds a single subunit of CXCR481; thus the role of CXCR4 and other CKR dimers remains an open question. It may be that dimers coordinate intracellular signaling molecules like G proteins and β-arrestins. However, biochemical and structural studies have shown that other GPCRs can couple to G proteins89, be phosphorylated by kinases and interact with β-arrestins as monomers90. Another possibility is that dimers coordinate different intracellular signaling molecules in multi-protein complexes. Finally, it is also possible that chemokine homodimers represent a “reference” functional state and that heterodimerization provides a mechanism for modulating signaling, trafficking and even ligand binding behaviors76. This controversial topic will require much more investigation with functional approaches that avoid the hazards of overexpression systems to provide convincing evidence of the relevance of receptor dimers. However, if relevant, homo- versus heterodimerization of CKRs and other GPCRs may provide exciting opportunities for drug discovery by providing more specific therapeutic targets.
CCR5:Maraviroc
A structure of CCR5 in complex with Maraviroc was subsequently solved in 201391 (Figure 4C). Maraviroc is an FDA-approved “entry inhibitor” of HIV that functions as an inverse agonist of CCR5 by reducing its constitutive activity92. It has also been characterized as an allosteric modulator that binds to a site distinct from the orthosteric binding site of chemokines and gp12092, 93, 94. For example, it has been shown to act as an insurmountable antagonist of internalization by the CCR5 ligand, CCL3L1, with little effect on its receptor binding affinity93. Moreover, kinetic data suggest that it can bind to CCL3 or gp120-occupied receptor, and then change the conformation of the receptor making it incompatible for these interactions. Consistent with its allosteric nature, it also shows probe dependence and differential effects on the affinity and efficacy of different ligands, which are hallmarks of such inhibitors93. In the crystal structure of the CCR5 complex, Maraviroc binds deep within the binding pocket and positions its triazole, tropane and cyclohexane groups into subpockets, while making no contacts with the extracellular loops; thus it may have limited overlap with chemokine or gp120 within the binding pocket. One of the most intriguing characteristics of Maraviroc is that it is more than 100-fold more potent in inhibiting HIV entry than suggested by its IC50 for displacing gp120 when compared to another compound (TAK779) that is nearly equipotent in gp120 binding inhibition92. This finding led to the hypothesis that Maraviroc may affect other steps in the HIV entry process, such as negatively modulating the interaction of CD4 with CCR5 prior to engagement of gp120, or interfering with CCR5:gp120 conformations necessary to trigger gp41-mediated viral fusion. Structures of CCR5 with additional small molecules and with gp120-CD4 will be needed for comparison to the Maraviroc complex in order to fully understand the mechanisms underlying its antiviral effects, but it clearly engages a sweet spot in the CCR5 binding pocket that makes it a potent non-competitive inhibitor.
Structure of CXCR4 bound to the viral chemokine antagonist, vMIP-II
In 2015, the first structure of a CKR, CXCR4, in complex with a chemokine (vMIP-II) was solved, and provided detailed insight into the recognition of CKRs by their natural ligands (Figure 4D and E)83. vMIP-II is a virally encoded high affinity antagonist for CXCR4 and was chosen over the human agonist ligand, CXCL12, because CXCL12 requires G protein coupling for high affinity binding95, but antagonists generally do not. Additionally, vMIP-II binds promiscuously to both CC and CXC receptors, and is a CC chemokine; thus along with the solved structure of CCR5, the CXCR4:vMIP-II structure was expected to provide insight into the specificity of chemokines for CC and CXC CKRs.
In the vMIP-II-bound state, CXCR4 formed dimers that were spatially similar to those in the IT1t- and CVX15-bound structures; however, the structure of the complex confirmed the 1:1 stoichiometry anticipated from prior studies81, with ligand occupancy of both receptor subunits83. The structure generally conforms to the concept of the two-site model in that the chemokine core domain interacts with the receptor N-terminus (CRS1) while the chemokine N-terminus binds in the CRS2 TM binding pocket of the receptor (Figure 4D and E). Surprisingly, however, in contrast to the two-site model, where the expectation was that these two sites would be decoupled, the interaction between CXCR4 and vMIP-II involves an extensive contiguous interface, necessitating the introduction of an intermediate region termed CRS1.5 (Figure 4E). In fact, every residue of the chemokine N-terminal domain and N-loop (residue 1–16) as well as residues in the third β-strand, interact with the receptor83.
Part of the CXCR4 N-terminus (residues 1–22 involving the key sulfated tyrosine, sTyr21), is missing from the electron density. Nevertheless, the visible CRS1 region (CXCR4 residues 23–27) show interactions with the chemokine N-loop, as expected from numerous mutagenesis studies, and with the β3-strand in the chemokine core domain. Moreover, as the electron density stops just shy of sTyr21, it was fairly straightforward to generate a model containing sTyr21, which suggested a compelling interaction with nearby R46 of the chemokine (Figure 4F and 4G), similar to the basic residue interactions reported for sulfotyrosines in (CXCR4:CXCL12)2 and CCR3:CCL11 (Figure 1C). In CRS1.5, CXCR4 forms an anti-parallel β-sheet with the di-cysteine motif of vMIP-II. Finally, in CRS2 the chemokine makes numerous interactions within the binding pocket including many residues known as determinants of vMIP-II binding or CXCR4:CXCL12 binding and activation. The N-terminus of vMIP-II overlaps with the binding site of IT1t in the CXCR4:IT1t complex, while it only partially overlaps with CVX15 (Figure 4A, B, D), reflecting the structural plasticity of CXCR4 and its ability to accommodate diverse ligands via overlapping but distinct interfaces.
vMIP-II is also a high affinity antagonist of CCR5, and the availability of the CCR5:Miraviroc structure along with CXCR4:vMIP-II facilitated modeling of the complex83. Figure 4H and G illustrate the predicted CRS1 interaction of CCR5:vMIP-II, compared to the CXCR4:vMIP-II interaction, respectively. An important sequence difference relates to the presence of two adjacent sulfotyrosine residues (sTyr14 and sTyr15)96 proximal to the conserved N-terminal Cys residue in the receptors. These CCR5 sulfotyrosines along with E18 are predicted to interact with the basic residues in the vMIP-II N-loop (K17 and R18) and β2-β3 loop (R46 and R48). By comparison, CXCR4 has only a single proximal sulfotyrosine (sTyr21) but in concert with two acidic groups (D22 and E26), makes similar interactions with the basic residues on the vMIP-II surface. These CRS1 models thus provide a plausible explanation for the unusual ability of vMIP-II to interact with both a CC and CXC CKR. By contrast, the CRS1 interaction between CXCR4 and the CXCL12 monomer is predicted to be quite different due to the absence of basic residues in the CXCL12 N- and β2-β3 loops. Instead, the backbone of CXCR4 N-terminal residues S23 and M24 lie in a groove formed by the N- and β2-β3 loops, and position sTyr21 at the top of the core domain where it interacts with backbone amides of CXCL12 residues R20 and A21 (Figure 4I). Notably, these interactions mimic the placement of a small molecule CXCR4:CXCL12 inhibitor85 (Figure 1D) as well as sulfate groups and ions observed in several CXCL12 X-ray structures 45, which adds support for the CRS1 predictions. However, additional structures will be required to confirm the models and also to better understand the structural basis for the generally strict recognition of CC chemokines for CC receptors and CXC chemokines for CXC receptors.
Modeling based insights into gp120 interactions with co-receptors from the CXCR4 and CCR5 complex structures
After gp120 binds CD4 on the surface of host cells and induces a conformational change that exposes the previously buried V3 loop, the V3 loop and a region called C4 of the “bridging sheet” are thought to bind CCR5 or CXCR4. However, while there is a vast amount of structural information about gp120 and its complexes with CD4, antibodies and gp4197, 98, 99, CKRs are missing from these complexes with the exception of a NMR study involving a CCR5 N-terminal peptide combined with an X-ray structure of gp120100. This work, in conjunction with mutagenesis studies101, 102, 103, 104 led to a model in which the sulfated CCR5 N-terminus interacts at an interface near the gp120 C4 domain and the stem of the V3 loop100; however, structural information involving the TM domains of CCR5 (and CXCR4) were lacking.
With the solution of the CXCR4:CVX15 peptide complex, it was noted that the antagonist bears a great deal of structure and sequence similarity to the V3 loop “crown” of gp12088, 91, 105. Thus the CXCR4:CVX15 structure was used to model the interaction of the V3 loop bound in the CXCR4 TM binding pocket, and CCR5:V3 was subsequently modeled91, 105, 106. Considering these models, together with the N-terminal interactions of CCR5 described above, leads to a more inclusive model which suggests that the crown of the V3 loop binds in the CKR TM pocket (CRS2) while the sulfated N-terminus binds to the gp120 V3 stem and C4 domain (CRS1), in a manner that has some analogies to CRS1/CRS2 interactions of CKR:chemokine complexes (Figure 3B). As the V3 loop is a major determinant of viral tropism107, 108, the models provide starting points for understanding co-receptor usage. The structures of CXCR4 and CCR5 may also pave the way for structures of V3 loop complexes with co-receptors, and possibly intact gp120 in the future.
Conclusions and future directions
The structural biology of the chemokine system is rich with complexity at every level, which has led to the development of many therapeutic strategies. Although chemokines are small and homologous in tertiary structure, they have a broad range of oligomerization states, which are important for their ability to bind to GAGs. These interactions have been exploited in several ways including the use of oligomerization and GAG-binding deficient chemokines, as well as GAG-binding efficient and oligomerization-efficient chemokines, all of which have shown efficacy in disease models while also revealing fundamental mechanisms of chemokine function. By taking advantage of the ability to manipulate chemokine affinity and pharmacology (signaling and trafficking) through modification of their N-termini, it has been possible to develop chemokine-based antagonists as well as potential HIV entry inhibitors. CKR:chemokine interactions don’t just rely on interactions with the CRS2 transmembrane binding pocket; CRS1 interactions are also important which has encouraged discovery of CRS1 inhibitors. All of these translationally-motivated investigations preceded structures of intact complexes of CKRs with small molecule and peptide antagonist as well as the most recent structure involving a CKR complex with chemokine. These latest structures will no doubt accelerate efforts to develop therapeutics that target the chemokine system in many disease indications, but many more structures are needed. Some of the major areas in need of investigation are as follows:
In the area of chemokine:GAG interactions, there is a need for structures (or models) of oligomerized chemokines in complex with GAGs larger than disaccharides. However, identifying and synthesizing GAGs larger than tetrasaccharides with high affinity for specific chemokines is challenging. Additionally, chemokines complexed with GAGs may not be homogeneous in terms of oligomerization state. Thus traditional methods using NMR and crystallography may not be adequate and computational methods that integrate available biochemical and biophysical information will be required. Understanding the extent of the specificity in chemokine:GAG interactions will also be important for considerations of whether these interactions are viable therapeutic targets; techniques to understand the specificity of these interactions in vivo will therefore be required since interactions observed in vitro may not be relevant to the in vivo situation. The role of oligomerization in chemokine function beyond GAG binding is also not fully understood and requires further investigation. Finally chemokines can form heterodimers109 and the relevance of such interactions in vivo needs to be determined.
In the area of CKR complex structures, multiple structures of even the same CKR are needed with different ligands to understand active and inactive state conformations or conformations that determine different signaling responses. Complexes of CCR5 with 5P12-RANTES, 6P4-RANTES and PSC-RANTES are examples discussed herein; such complexes could facilitate the design of small molecule modulators that mimic the modified chemokines. Structures of different CKR:chemokine complexes will also be required to understand the structural basis for the specificity of chemokines for their receptors. Moreover, the issue of receptor homo- and heterodimerization also needs to be resolved and understood. Although CKR structures have now been reported, including a complex with chemokine, CKR structures remain extremely challenging goals. And of course, the next level of complexity is understanding how they interact with intracellular signaling partners.
While much has been learned since the first chemokine structure was determined, the state of the field is still at the proverbial “tip of the iceberg”. But it is clear what the important questions are and where the field needs to go.
Acknowledgments
The authors would like to thank Xu Wang for the coordinates of the CCL5 octamer. This work was funded by National Institutes of Health grants U01 GM094612 and R01 AI37113 to TMH. IK was partially supported by R01 GM071872 and U54 GM094618. The authors apologize to many researchers whose work could not be acknowledged appropriately due to space limitations.
Abbreviations
- GAG
glycosaminoglycan
- CCR
CC-type chemokine receptor
- CXCR
CXC-type chemokine receptor
- CCL
CC-type chemokine ligand
- CXCL
CXC-type chemokine ligand
- CKR
chemokine receptor
- HS
heparan sulfate
- NMR
nuclear magnetic resonance
- TM
transmembrane
- CRS
chemokine recognition site
References
- 1.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 2.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392(6676):565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 3.Gerard C, Rollins BJ. Chemokines and disease. Nat Immunol. 2001;2(2):108–115. doi: 10.1038/84209. [DOI] [PubMed] [Google Scholar]
- 4.Salanga CL, Handel TM. Chemokine oligomerization and interactions with receptors and glycosaminoglycans: the role of structural dynamics in function. Exp Cell Res. 2011;317(5):590–601. doi: 10.1016/j.yexcr.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholten DJ, Canals M, Maussang D, Roumen L, Smit MJ, Wijtmans M, et al. Pharmacological modulation of chemokine receptor function. Br J Pharmacol. 2011;165(6):1617–1643. doi: 10.1111/j.1476-5381.2011.01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadiere C, Farber JM, Graham GJ, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66(1):1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Three-dimensional structure of interleukin 8 in solution. Biochemistry. 1990;29(7):1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- 8.Lodi PJ, Garrett DS, Kuszewski J, Tsang ML, Weatherbee JA, Leonard WJ, et al. High-resolution solution structure of the beta chemokine hMIP-1 beta by multidimensional NMR. Science. 1994;263(5154):1762–1767. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
- 9.Skelton NJ, Aspiras F, Ogez J, Schall TJ. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry. 1995;34(16):5329–5342. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]
- 10.Handel TM, Domaille PJ. Heteronuclear (1H, 13C, 15N) NMR assignments and solution structure of the monocyte chemoattractant protein-1 (MCP-1) dimer. Biochemistry. 1996;35(21):6569–6584. doi: 10.1021/bi9602270. [DOI] [PubMed] [Google Scholar]
- 11.Mizoue LS, Bazan JF, Johnson EC, Handel TM. Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry. 1999;38(5):1402–1414. doi: 10.1021/bi9820614. [DOI] [PubMed] [Google Scholar]
- 12.Hoover DM, Mizoue LS, Handel TM, Lubkowski J. The crystal structure of the chemokine domain of fractalkine shows a novel quaternary arrangement. J Biol Chem. 2000;275(30):23187–23193. doi: 10.1074/jbc.M002584200. [DOI] [PubMed] [Google Scholar]
- 13.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105(13):5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajarathnam K, Sykes B, Kay C, Dewald B, Geiser T, Baggiolini M, et al. Neutrophil activation by monomeric interleukin-8. Science. 1994;264(5155):90–92. doi: 10.1126/science.8140420. [DOI] [PubMed] [Google Scholar]
- 15.Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, et al. Monomeric Monocyte Chemoattractant Protein-1 (MCP-1) Binds and Activates the MCP-1 Receptor CCR2B. J Biol Chem. 1998;273(50):33157–33165. doi: 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Sharp JS, Handel TM, Prestegard JH. Chemokine oligomerization in cell signaling and migration. Prog Mol Biol Transl Sci. 2013;117:531–578. doi: 10.1016/B978-0-12-386931-9.00020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren M, Guo Q, Guo L, Lenz M, Qian F, Koenen RR, et al. Polymerization of MIP-1 chemokine (CCL3 and CCL4) and clearance of MIP-1 by insulin-degrading enzyme. EMBO J. 2010;29(23):3952–3966. doi: 10.1038/emboj.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Watson C, Sharp JS, Handel TM, Prestegard JH. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure. 2011;19(8):1138–1148. doi: 10.1016/j.str.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U S A. 2003;100(4):1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campanella GS, Grimm J, Manice LA, Colvin RA, Medoff BD, Wojtkiewicz GR, et al. Oligomerization of CXCL10 is necessary for endothelial cell presentation and in vivo activity. J Immunol. 2006;177(10):6991–6998. doi: 10.4049/jimmunol.177.10.6991. [DOI] [PubMed] [Google Scholar]
- 21.Rot A. Neutrophil attractant/activation protein-1 (interleukin-8) induces in vitro neutrophil migration by haptotactic mechanism. Eur J Immunol. 1993;23(1):303–306. doi: 10.1002/eji.1830230150. [DOI] [PubMed] [Google Scholar]
- 22.Haessler U, Pisano M, Wu M, Swartz MA. Dendritic cell chemotaxis in 3D under defined chemokine gradients reveals differential response to ligands CCL21 and CCL19. Proc Natl Acad Sci U S A. 2011;108(14):5614–5619. doi: 10.1073/pnas.1014920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber M, Hauschild R, Schwarz J, Moussion C, de Vries I, Legler DF, et al. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science. 2013;339(6117):328–332. doi: 10.1126/science.1228456. [DOI] [PubMed] [Google Scholar]
- 24.Ali S, Robertson H, Wain JH, Isaacs JD, Malik G, Kirby JA. A non-glycosaminoglycan-binding variant of CC chemokine ligand 7 (monocyte chemoattractant protein-3) antagonizes chemokine-mediated inflammation. J Immunol. 2005;175(2):1257–1266. doi: 10.4049/jimmunol.175.2.1257. [DOI] [PubMed] [Google Scholar]
- 25.Dubrac A, Quemener C, Lacazette E, Lopez F, Zanibellato C, Wu WG, et al. Functional divergence between 2 chemokines is conferred by single amino acid change. Blood. 2010;116(22):4703–4711. doi: 10.1182/blood-2010-03-274852. [DOI] [PubMed] [Google Scholar]
- 26.Lebel-Haziv Y, Meshel T, Soria G, Yeheskel A, Mamon E, Ben-Baruch A. Breast cancer: coordinated regulation of CCL2 secretion by intracellular glycosaminoglycans and chemokine motifs. Neoplasia. 2014;16(9):723–740. doi: 10.1016/j.neo.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleton J, Neil S, Wintle J, Clark-Lewis I, Moore H, Lam C, et al. Transcytosis and surface presentation of IL-8 by venular endothelial cells. Cell. 1997;91(3):385–395. doi: 10.1016/s0092-8674(00)80422-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 2005;6(9):902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 29.Ellyard JI, Simson L, Bezos A, Johnston K, Freeman C, Parish CR. Eotaxin selectively binds heparin. An interaction that protects eotaxin from proteolysis and potentiates chemotactic activity in vivo. J Biol Chem. 2007;282(20):15238–15247. doi: 10.1074/jbc.M608046200. [DOI] [PubMed] [Google Scholar]
- 30.Roscic-Mrkic B, Fischer M, Leemann C, Manrique A, Gordon CJ, Moore JP, et al. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood. 2003;102(4):1169–1177. doi: 10.1182/blood-2003-02-0488. [DOI] [PubMed] [Google Scholar]
- 31.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, et al. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36(44):13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 32.Lau EK, Paavola CD, Johnson Z, Gaudry JP, Geretti E, Borlat F, et al. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1: implications for structure and function in vivo. J Biol Chem. 2004;279(21):22294–22305. doi: 10.1074/jbc.M311224200. [DOI] [PubMed] [Google Scholar]
- 33.Kuschert GS, Coulin F, Power CA, Proudfoot AE, Hubbard RE, Hoogewerf AJ, et al. Glycosaminoglycans interact selectively with chemokines and modulate receptor binding and cellular responses. Biochemistry. 1999;38(39):12959–12968. doi: 10.1021/bi990711d. [DOI] [PubMed] [Google Scholar]
- 34.Crown SE, Yu Y, Sweeney MD, Leary JA, Handel TM. Heterodimerization of CCR2 chemokines and regulation by glycosaminoglycan binding. J Biol Chem. 2006;281(35):25438–25446. doi: 10.1074/jbc.M601518200. [DOI] [PubMed] [Google Scholar]
- 35.Dyer DP, Salanga CS, Volkman BF, Kawamura T, Handel TM. The dependence of chemokine -glycosaminoglycan interactions on chemokine oligomerization. J Biol Chem. 2015 doi: 10.1093/glycob/cwv100. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson Z, Kosco-Vilbois MH, Herren S, Cirillo R, Muzio V, Zaratin P, et al. Interference with heparin binding and oligomerization creates a novel anti-inflammatory strategy targeting the chemokine system. J Immunol. 2004;173(9):5776–5785. doi: 10.4049/jimmunol.173.9.5776. [DOI] [PubMed] [Google Scholar]
- 37.Braunersreuther V, Steffens S, Arnaud C, Pelli G, Burger F, Proudfoot A, et al. A novel RANTES antagonist prevents progression of established atherosclerotic lesions in mice. Arterioscler Thromb Vasc Biol. 2008;28(6):1090–1096. doi: 10.1161/ATVBAHA.108.165423. [DOI] [PubMed] [Google Scholar]
- 38.Braunersreuther V, Pellieux C, Pelli G, Burger F, Steffens S, Montessuit C, et al. Chemokine CCL5/RANTES inhibition reduces myocardial reperfusion injury in atherosclerotic mice. J Mol Cell Cardiol. 2010;48(4):789–798. doi: 10.1016/j.yjmcc.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 39.Berres ML, Koenen RR, Rueland A, Zaldivar MM, Heinrichs D, Sahin H, et al. Antagonism of the chemokine Ccl5 ameliorates experimental liver fibrosis in mice. J Clin Invest. 2010;120(11):4129–4140. doi: 10.1172/JCI41732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Boyle G, Mellor P, Kirby JA, Ali S. Anti-inflammatory therapy by intravenous delivery of non-heparan sulfate-binding CXCL12. FASEB J. 2009;23(11):3906–3916. doi: 10.1096/fj.09-134643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handel TM, Johnson Z, Rodrigues DH, Dos Santos AC, Cirillo R, Muzio V, et al. An engineered monomer of CCL2 has anti-inflammatory properties emphasizing the importance of oligomerization for chemokine activity in vivo. J Leukoc Biol. 2008;84(4):1101–1108. doi: 10.1189/jlb.0108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahrara S, Proudfoot AE, Park CC, Volin MV, Haines GK, Woods JM, et al. Inhibition of monocyte chemoattractant protein-1 ameliorates rat adjuvant-induced arthritis. J Immunol. 2008;180(5):3447–3456. doi: 10.4049/jimmunol.180.5.3447. [DOI] [PubMed] [Google Scholar]
- 43.Thiele S, Rosenkilde MM. Interaction of chemokines with their receptors--from initial chemokine binding to receptor activating steps. Curr Med Chem. 2014;21(31):3594–3614. doi: 10.2174/0929867321666140716093155. [DOI] [PubMed] [Google Scholar]
- 44.Shaw JP, Johnson Z, Borlat F, Zwahlen C, Kungl A, Roulin K, et al. The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure. 2004;12(11):2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Murphy JW, Cho Y, Sachpatzidis A, Fan C, Hodsdon ME, Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1 alpha) binding to heparin. J Biol Chem. 2007;282(13):10018–10027. doi: 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salanga CL, Dyer DP, Kiselar JG, Gupta S, Chance MR, Handel TM. Multiple glycosaminoglycan-binding epitopes of monocyte chemoattractant protein-3/CCL7 enable it to function as a non-oligomerizing chemokine. J Biol Chem. 2014;289(21):14896–14912. doi: 10.1074/jbc.M114.547737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziarek JJ, Veldkamp CT, Zhang F, Murray NJ, Kartz GA, Liang X, et al. Heparin Oligosaccharides Inhibit Chemokine (CXC Motif) Ligand 12 (CXCL12) Cardioprotection by Binding Orthogonal to the Dimerization Interface, Promoting Oligomerization, and Competing with the Chemokine (CXC Motif) Receptor 4 (CXCR4) N Terminus. J Biol Chem. 2013;288(1):737–746. doi: 10.1074/jbc.M112.394064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stringer SE, Forster MJ, Mulloy B, Bishop CR, Graham GJ, Gallagher JT. Characterization of the binding site on heparan sulfate for macrophage inflammatory protein 1alpha. Blood. 2002;100(5):1543–1550. [PubMed] [Google Scholar]
- 49.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J Biol Chem. 1991;266(34):23128–23134. [PubMed] [Google Scholar]
- 50.Moser B, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. Interleukin-8 antagonists generated by N-terminal modification. J Biol Chem. 1993;268(10):7125–7128. [PubMed] [Google Scholar]
- 51.Clark-Lewis I, Kim KS, Rajarathnam K, Gong JH, Dewald B, Moser B, et al. Structure-activity relationships of chemokines. J Leukoc Biol. 1995;57(5):703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 52.Moelants EA, Mortier A, Van Damme J, Proost P. In vivo regulation of chemokine activity by post-translational modification. Immunol Cell Biol. 2013;91(6):402–407. doi: 10.1038/icb.2013.16. [DOI] [PubMed] [Google Scholar]
- 53.Pease JE, Wang J, Ponath PD, Murphy PM. The N-terminal extracellular segments of the chemokine receptors CCR1 and CCR3 are determinants for MIP-1alpha and eotaxin binding, respectively, but a second domain is essential for efficient receptor activation. J Biol Chem. 1998;273(32):19972–19976. doi: 10.1074/jbc.273.32.19972. [DOI] [PubMed] [Google Scholar]
- 54.Monteclaro FS, Charo IF. The Amino-terminal Domain of CCR2 Is Both Necessary and Sufficient for High Affinity Binding of Monocyte Chemoattractant Protein 1: Receptor Activation By a Pseudo-Tethered Ligand. J Biol Chem. 1997;272(37):23186–23190. doi: 10.1074/jbc.272.37.23186. [DOI] [PubMed] [Google Scholar]
- 55.Monteclaro FS, Charo IF. The Amino-terminal Extracellular Domain of the MCP-1 Receptor, but Not the RANTES/MIP-1a Receptor, Confers Chemokine Selectivity: Evidence for a Two-Step Mechanism fir MCP-1 Receptor Activation. J Biol Chem. 1996;271(32):19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 56.Kofuku Y, Yoshiura C, Ueda T, Terasawa H, Hirai T, Tominaga S, et al. Structural Basis of the Interaction between Chemokine Stromal Cell-derived Factor-1/CXCL12 and Its G-protein-coupled Receptor CXCR4. J Biol Chem. 2009;284(50):35240–35250. doi: 10.1074/jbc.M109.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerlach LO, Skerlj RT, Bridger GJ, Schwartz TW. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J Biol Chem. 2001;276(17):14153–14160. doi: 10.1074/jbc.M010429200. [DOI] [PubMed] [Google Scholar]
- 58.Gong JH, Ratkay LG, Waterfield JD, Clark-Lewis I. An antagonist of monocyte chemoattractant protein 1 (MCP-1) inhibits arthritis in the MRL-lpr mouse model. J Exp Med. 1997;186(1):131–137. doi: 10.1084/jem.186.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matoba T, Egashira K. Anti-inflammatory gene therapy for cardiovascular disease. Curr Gene Ther. 2011;11(6):442–446. doi: 10.2174/156652311798192888. [DOI] [PubMed] [Google Scholar]
- 60.Tsuruta S, Nakamuta M, Enjoji M, Kotoh K, Hiasa K, Egashira K, et al. Anti-monocyte chemoattractant protein-1 gene therapy prevents dimethylnitrosamine-induced hepatic fibrosis in rats. Int J Mol Med. 2004;14(5):837–842. [PubMed] [Google Scholar]
- 61.Plater-Zyberk C, Hoogewerf AJ, Proudfoot AE, Power CA, Wells TN. Effect of a CC chemokine receptor antagonist on collagen induced arthritis in DBA/1 mice. Immunol Lett. 1997;57(1–3):117–120. doi: 10.1016/s0165-2478(97)00075-8. [DOI] [PubMed] [Google Scholar]
- 62.Veillard NR, Kwak B, Pelli G, Mulhaupt F, James RW, Proudfoot AE, et al. Antagonism of RANTES receptors reduces atherosclerotic plaque formation in mice. Circ Res. 2004;94(2):253–261. doi: 10.1161/01.RES.0000109793.17591.4E. [DOI] [PubMed] [Google Scholar]
- 63.Matsui M, Weaver J, Proudfoot AE, Wujek JR, Wei T, Richer E, et al. Treatment of experimental autoimmune encephalomyelitis with the chemokine receptor antagonist Met-RANTES. J Neuroimmunol. 2002;128(1–2):16–22. doi: 10.1016/s0165-5728(02)00121-2. [DOI] [PubMed] [Google Scholar]
- 64.Robinson SC, Scott KA, Wilson JL, Thompson RG, Proudfoot AE, Balkwill FR. A chemokine receptor antagonist inhibits experimental breast tumor growth. Cancer Res. 2003;63(23):8360–8365. [PubMed] [Google Scholar]
- 65.Adage T, Piccinini AM, Falsone A, Trinker M, Robinson J, Gesslbauer B, et al. Structure-based design of decoy chemokines as a way to explore the pharmacological potential of glycosaminoglycans. Br J Pharmacol. 2012;167(6):1195–1205. doi: 10.1111/j.1476-5381.2012.02089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simmons G, Clapham PR, Picard L, Offord RE, Rosenkilde MM, Schwartz TW, et al. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276(5310):276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 67.Lederman MM, Veazey RS, Offord R, Mosier DE, Dufour J, Mefford M, et al. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science. 2004;306(5695):485–487. doi: 10.1126/science.1099288. [DOI] [PubMed] [Google Scholar]
- 68.Pastore C, Picchio GR, Galimi F, Fish R, Hartley O, Offord RE, et al. Two mechanisms for human immunodeficiency virus type 1 inhibition by N-terminal modifications of RANTES. Antimicrob Agents Chemother. 2003;47(2):509–517. doi: 10.1128/AAC.47.2.509-517.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaertner H, Cerini F, Escola J-M, Kuenzi G, Melotti A, Offord R, et al. Highly potent, fully recombinant anti-HIV chemokines: Reengineering a low-cost microbicide. Proc Natl Acad Sci USA. 2008;105(46):17706–17711. doi: 10.1073/pnas.0805098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veazey RS, Ling B, Green LC, Ribka EP, Lifson JD, Piatak M, Jr, et al. Topically applied recombinant chemokine analogues fully protect macaques from vaginal simian-human immunodeficiency virus challenge. J Infect Dis. 2009;199(10):1525–1527. doi: 10.1086/598685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colin P, Benureau Y, Staropoli I, Wang Y, Gonzalez N, Alcami J, et al. HIV-1 exploits CCR5 conformational heterogeneity to escape inhibition by chemokines. Proc Natl Acad Sci U S A. 2013;110(23):9475–9480. doi: 10.1073/pnas.1222205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castaldo C, Benicchi T, Otrocka M, Mori E, Pilli E, Ferruzzi P, et al. CXCR4 Antagonists: A Screening Strategy for Identification of Functionally Selective Ligands. J Biomol Screen. 2014;19(6):859–869. doi: 10.1177/1087057114526283. [DOI] [PubMed] [Google Scholar]
- 73.Wang NX, Sieg SF, Lederman MM, Offord RE, Hartley O, von Recum HA. Using glycosaminoglycan/chemokine interactions for the long-term delivery of 5P12-RANTES in HIV prevention. Mol Pharm. 2013;10(10):3564–3573. doi: 10.1021/mp3007242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 75.Zhao B, Mankowski MK, Snyder BA, Ptak RG, Liwang PJ. Highly potent chimeric inhibitors targeting two steps of HIV cell entry. J Biol Chem. 2011;286(32):28370–28381. doi: 10.1074/jbc.M111.234799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stephens B, Handel TM. Chemokine Receptor Oligomerization and Allostery. In: Kenakin T, editor. Prog Mol Biol Transl Sci. Vol. 115. Academic Press; 2013. pp. 375–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-Munoz L, Barroso R, Dyrhaug SY, Navarro G, Lucas P, Soriano SF, et al. CCR5/CD4/CXCR4 oligomerization prevents HIV-1 gp120IIIB binding to the cell surface. Proc Natl Acad Sci U S A. 2014;111(19):E1960–1969. doi: 10.1073/pnas.1322887111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Millard CJ, Ludeman JP, Canals M, Bridgford JL, Hinds MG, Clayton DJ, et al. Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1. Structure. 2014;22(11):1571–1581. doi: 10.1016/j.str.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 79.Veldkamp CT, Seibert C, Peterson FC, De la Cruz NB, Haugner JC, III, Basnet H, et al. Structural Basis of CXCR4 Sulfotyrosine Recognition by the Chemokine SDF-1/CXCL12. Sci Signal. 2008;1(37):ra4. doi: 10.1126/scisignal.1160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skelton NJ, Quan C, Reilly D, Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure. 1999;7(2):157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 81.Kufareva I, Stephens BS, Holden LG, Qin L, Zhao C, Kawamura T, et al. Stoichiometry and geometry of the CXC chemokine receptor 4 complex with CXC ligand 12: Molecular modeling and experimental validation. Proc Natl Acad Sci U S A. 2014;111(50):E5363–72. doi: 10.1073/pnas.1417037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nasser MW, Raghuwanshi SK, Grant DJ, Jala VR, Rajarathnam K, Richardson RM. Differential Activation and Regulation of CXCR1 and CXCR2 by CXCL8 Monomer and Dimer. J Immunol. 2009;183(5):3425–3432. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, et al. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015 doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takekoshi T, Ziarek JJ, Volkman BF, Hwang ST. A locked, dimeric CXCL12 variant effectively inhibits pulmonary metastasis of CXCR4-expressing melanoma cells due to enhanced serum stability. Mol Cancer Ther. 2012;11(11):2516–2525. doi: 10.1158/1535-7163.MCT-12-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith EW, Liu Y, Getschman AE, Peterson FC, Ziarek JJ, Li R, et al. Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine. J Med Chem. 2014;57:9693–9699. doi: 10.1021/jm501194p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, et al. GPCR Engineering Yields High-Resolution Structural Insights into 2-Adrenergic Receptor Function. Science. 2007;318(5854):1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 87.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, et al. High-Resolution Crystal Structure of an Engineered Human 2-Adrenergic G Protein Coupled Receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whorton MR, Jastrzebska B, Park PS-H, Fotiadis D, Engel A, Palczewski K, et al. Efficient Coupling of Transducin to Monomeric Rhodopsin in a Phospholipid Bilayer. J Biol Chem. 2008;283(7):4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang C-c, Tesmer JJG, et al. Monomeric Rhodopsin Is Sufficient for Normal Rhodopsin Kinase (GRK1) Phosphorylation and Arrestin-1 Binding. J Biol Chem. 2011;286(2):1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, et al. Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science. 2013;341(6152):1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Garcia-Perez J, Rueda P, Alcami J, Rognan D, Arenzana-Seisdedos F, Lagane B, et al. Allosteric Model of Maraviroc Binding to CC Chemokine Receptor 5 (CCR5) J Biol Chem. 2011;286(38):33409–33421. doi: 10.1074/jbc.M111.279596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muniz-Medina VM, Jones S, Maglich JM, Galardi C, Hollingsworth RE, Kazmierski WM, et al. The Relative Activity of “Function Sparing” HIV-1 Entry Inhibitors on Viral Entry and CCR5 Internalization: Is Allosteric Functional Selectivity a Valuable Therapeutic Property? Mol Pharmacol. 2009;75(3):490–501. doi: 10.1124/mol.108.052555. [DOI] [PubMed] [Google Scholar]
- 94.Lagane B, Garcia-Perez J, Kellenberger E. Modeling the allosteric modulation of CCR5 function by Maraviroc. Drug Discov Today Technol. 2013;10(2):e297–305. doi: 10.1016/j.ddtec.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 95.Nijmeijer S, Leurs R, Smit MJ, Vischer HF. The Epstein-Barr virus-encoded G protein-coupled receptor BILF1 hetero-oligomerizes with human CXCR4, scavenges Gαi proteins, and constitutively impairs CXCR4 functioning. J Biol Chem. 2010;285(38):29632–29641. doi: 10.1074/jbc.M110.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 97.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci USA. 2012;109(15):5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342(6165):1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pancera M, Majeed S, Ban Y-EA, Chen L, Huang C-c, Kong L, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107(3):1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang C-c, Lam SN, Acharya P, Tang M, Xiang S-H, Hussan SS-u, et al. Structures of the CCR5 N Terminus and of a Tyrosine-Sulfated Antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846):1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cormier EG, Tran DNH, Yukhayeva L, Olson WC, Dragic T. Mapping the Determinants of the CCR5 Amino-Terminal Sulfopeptide Interaction with Soluble Human Immunodeficiency Virus Type 1 gp120-CD4 Complexes. J Virol. 2001;75(12):5541–5549. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rizzuto CD, Wyatt R, Hernández-Ramos N, Sun Y, Kwong PD, Hendrickson WA, et al. A Conserved HIV gp120 Glycoprotein Structure Involved in Chemokine Receptor Binding. Science. 1998;280(5371):1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 103.Rizzuto C, Sodroski J. Short Communication: Fine Definition of a Conserved CCR5-Binding Region on the Human Immunodeficiency Virus Type 1 Glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16(8):741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 104.Dragic T. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J Gen Virol. 2001;82(8):1807–1814. doi: 10.1099/0022-1317-82-8-1807. [DOI] [PubMed] [Google Scholar]
- 105.Tamamis P, Floudas Christodoulos A. Molecular Recognition of CXCR4 by a Dual Tropic HIV-1 gp120 V3 Loop. Biophys J. 2013;105(6):1502–1514. doi: 10.1016/j.bpj.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tamamis P, Floudas CA. Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One. 2014;9(4):e95767. doi: 10.1371/journal.pone.0095767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Verrier F, Borman AM, Brand D, Girard M. Role of the HIV Type 1 Glycoprotein 120 V3 Loop in Determining Coreceptor Usage. AIDS Res Hum Retroviruses. 1999;15(8):731–743. doi: 10.1089/088922299310827. [DOI] [PubMed] [Google Scholar]
- 108.Jensen MA, van ‘t Wout AB. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev. 2003;5(2):104–112. [PubMed] [Google Scholar]
- 109.Carlson J, Baxter SA, Dreau D, Nesmelova IV. The heterodimerization of platelet-derived chemokines. Biochim Biophys Acta. 2013;1834(1):158–168. doi: 10.1016/j.bbapap.2012.09.010. [DOI] [PubMed] [Google Scholar]