Abstract
During the industrial scale-up of bioprocesses it is important to establish that the biological system has not changed significantly when moving from small laboratory-scale shake flasks or culturing bottles to an industrially relevant production level. Therefore, during upscaling of biomass production for a range of metal transformations, including the production of biogenic magnetite nanoparticles by Geobacter sulfurreducens, from 100-ml bench-scale to 5-liter fermentors, we applied Fourier transform infrared (FTIR) spectroscopy as a metabolic fingerprinting approach followed by the analysis of bacterial cell extracts by gas chromatography-mass spectrometry (GC-MS) for metabolic profiling. FTIR results clearly differentiated between the phenotypic changes associated with different growth phases as well as the two culturing conditions. Furthermore, the clustering patterns displayed by multivariate analysis were in agreement with the turbidimetric measurements, which displayed an extended lag phase for cells grown in a 5-liter bioreactor (24 h) compared to those grown in 100-ml serum bottles (6 h). GC-MS analysis of the cell extracts demonstrated an overall accumulation of fumarate during the lag phase under both culturing conditions, coinciding with the detected concentrations of oxaloacetate, pyruvate, nicotinamide, and glycerol-3-phosphate being at their lowest levels compared to other growth phases. These metabolites were overlaid onto a metabolic network of G. sulfurreducens, and taking into account the levels of these metabolites throughout the fermentation process, the limited availability of oxaloacetate and nicotinamide would seem to be the main metabolic bottleneck resulting from this scale-up process. Additional metabolite-feeding experiments were carried out to validate the above hypothesis. Nicotinamide supplementation (1 mM) did not display any significant effects on the lag phase of G. sulfurreducens cells grown in the 100-ml serum bottles. However, it significantly improved the growth behavior of cells grown in the 5-liter bioreactor by reducing the lag phase from 24 h to 6 h, while providing higher yield than in the 100-ml serum bottles.
INTRODUCTION
Geobacter species are Gram-negative deltaproteobacteria, which have been recognized as an intrinsic component of a diverse range of natural subsurface environments, including soils and groundwater, as well as aquatic sediments (1). Geobacter species are considered to play a vital role in the global recycling of metals and carbon (2) in anaerobic environments, coupling the oxidation of acetate, and other key intermediates from fermentative metabolism of natural complex organic matter, to the reduction of Fe(III) and other metals (3, 4). G. metallireducens was the first organism with the ability of reducing insoluble Fe(III) oxides in tandem with oxidation of acetate to be isolated (5, 6).
G. sulfurreducens strain PCA is another member of the family Geobacteraceae, which also has the ability of coupling the oxidation of acetate to the reduction of sulfur, fumarate, Fe(III), and other metals (7). The availability of its complete genome sequence revealed not only unexpected insights into its complex metabolism (including evidence of aerobic metabolism [8] and one-carbon and complex carbon metabolism) but also multiple capabilities in terms of motility and chemotactic behavior (2). For a detailed introduction to both the physiology and ecology of Geobacter spp. (and other metal reducers), see reference 9.
It is perhaps not surprising that these multiple and complex metabolic and physiological capabilities make this fascinating species an ideal candidate for bioremediation applications (among others). Of particular interest is the treatment and removal of subsurface metal contaminants (10, 11), such as U(VI) (12, 13), Cr(VI) (14), and Tc(VII) (15, 16). For a comparative review of the microbial reduction of metals and metalloids, see reference 17, and for a recent review of systems-level and modeling approaches to bioremediation of uranium-contaminated groundwater, see reference 18.
Other related biotechnical applications linked to Geobacter species include the production of magnetite nanoparticles (19–21) and the potential role of this magnetic material in a wide range of applications such as cancer therapy (22), drug delivery (23), bioseparation (24, 25), catalysis (26), reductive bioremediation of contaminants (J. M. Byrne, H. Muhamadali, V. S. Coker, J. Cooper, and J. R. Lloyd, submitted for publication), and the production of nanomaterials for magnetic data storage devices (28). However, the ability to exploit these microorganisms to their full potential requires a deeper understanding of the basis of their biochemical composition, genetics, and metabolic behavior in a range of process environments (29), especially at scale.
There are a range of approaches available to optimize the biotechnological potential of bacteria, such as genetic modification and adjustment of medium components and/or incubation conditions, any of which could be applied to improve, for example, the growth kinetics and yields of these extremely useful microorganisms, particularly when there is a requirement to scale up for industrial/biotechnological applications. Here, we demonstrate a metabolomics approach for achieving this goal.
The metabolome is described as being the complement of all low-molecular-weight metabolites found within a biological sample, such as a single organism, which encompasses the end products of gene expression that are necessary for the maintenance, growth, and normal function of a cell (i.e., metabolic intermediates, signaling molecules, and secondary metabolites) (30). The field of metabolomics is accelerating at a considerable rate, which is perhaps not surprising considering improvements in the ability to measure multiple metabolites from complex biological systems with a high degree of accuracy and precision (31).
Since the metabolome is downstream from the genome, proteome, and transcriptome in biological systems and is affected via the changes and interactions of the above-described processes, it is said to reflect the activities of the cells at functional levels (32). Therefore, metabolomic approaches are considered complementary to genomics, transcriptomics, and proteomics (33) and have been used for optimizing microbial processes (34–36).
Metabolic fingerprinting is generally employed as a rapid, global, and high-throughput approach to provide sample classification and is also used as a screening tool to discriminate samples of different biological status (37) (such as the physiological state in this case) or origin, which may pertain to disease diagnostics (case-control) or longitudinal intervention, both aimed at biomarker discovery (38, 39).
Fourier transform infrared (FTIR) spectroscopy is a vibrational spectroscopy technique based on the principle of the absorption of infrared light by the sample of interest, causing intermolecular bond vibrations, which can be detected and directly correlated to (bio)chemical species. FTIR, in comparison to other metabolic fingerprinting techniques, offers the advantages of minimal sample preparation, high-throughput microbial analysis, extreme rapidity (spectral acquisition, 10 s to 1 min), high reproducibility, low cost, and portability. Since its potential for microbial analysis was first widely recognized by Naumann et al. (40), FTIR has been applied to a wide range of areas within microbiology (40), including clinical (41, 42), food (43–45), environmental (46–49), and biotechnological (50) applications.
While FTIR spectroscopy is a global, holistic approach, it does not of course have the sensitivity or specificity of mass spectrometry (MS) approaches. Therefore, in this study, we also employed gas chromatography-MS (GC-MS) as a metabolic profiling method which yields information on central carbon as well as amino acid metabolism (51) and has been used to analyze industrial bioprocesses and aid in metabolic engineering (34, 36).
This study was focused on the first part of a two-step biomagnetite nanoparticle production process which includes (i) biomass production and (ii) magnetite nanoparticle production (19, 52). The aim was to understand and extend our current knowledge regarding the complex metabolic potential of Geobacter species specifically at different stages of batch culture growth in contrasting serum bottle and bioreactor studies. If successful, this would support, aid, and potentially improve the industrial scale-up of biomass production for a range of Geobacter-specific biotechnological processes, most notably nanomagnetite production.
MATERIALS AND METHODS
Media and growth conditions.
G. sulfurreducens strain PCA was used in this study and cultivated under anoxic conditions at 30°C in a modified fresh water medium (NBAF) (53), which is described in Table S1 in the supplemental material.
For assessing potential differences in the metabolic state during growth at contrasting scales, two different batch volumes were used: 100 ml (in 120-ml serum bottles, in triplicate) and 5 liters (in a 7-liter Applikon dish-bottom reactor vessel; Applikon Biotechnology., Gloucestershire, United Kingdom) (see Fig. S1 in the supplemental material). Both volumes were inoculated with 10% (vol/vol) late-log-phase G. sulfurreducens, followed by incubation under anoxic conditions for 7 days at 30°C. NBAF was also used for the inoculum preparation step following the standard protocol. The same batch of medium and starting inoculum was used to start the experiment for both incubation conditions in order to reduce any potential variation.
At each time point, three 100-ml serum bottles were sacrificed to achieve sufficient biomass for GC-MS, FTIR, and optical density (OD) analysis. Aliquots (50 ml) were taken at 13 different time points: 0, 6, 12, 18, 24, 36, 48, 60, 72, 96, 120, 144, and 168 h. A fraction (15 ml) of the collected samples were quenched and used for GC-MS analysis, and the remaining (35 ml) were employed for FTIR analysis. The ODs of the samples were also recorded at 600 nm to monitor the growth of the organism during the incubation period.
FTIR. (i) Sample preparation.
Biomass was collected for FTIR analysis by centrifuging 35 ml of collected samples for 20 min at 5,240 × g at 4°C. The supernatant was removed, and the cell pellets were washed twice with 5 ml of sterile 0.9% (wt/vol) saline solution, followed by 10 min of centrifugation at 5,240 × g at 4°C. After the final washing step, the supernatant was removed and the pellets were flash frozen in liquid nitrogen for 1 min and stored at −80°C. Prior to analysis, all samples were thawed on ice and biomass loadings normalized according to OD at 600 nm (OD600).
A Bruker 96-well silicon sampling plate (Bruker Ltd., Coventry, United Kingdom) was washed three times with 5% sodium dodecyl sulfate (SDS) solution and twice with 70% ethanol and finally rinsed three times with deionized water and dried at 55°C. Samples were spotted randomly in triplicates as 20-μl aliquots onto an FTIR silicon plate and dried via heating for 30 min in an oven at 55°C in order to remove as much water as possible and fix the samples to the plate.
(ii) Instrument setup.
All FTIR bacterial spectra were collected in transmission mode on a Bruker Equinox 55 infrared spectrometer following previously published methods (54). All spectra were recorded in the mid-infrared range between 4,000 and 600 cm−1 wavenumbers, and these were acquired at a resolution of 4 cm−1, with 64 spectral coadds to improve the signal-to-noise ratio.
(iii) Data analysis.
FTIR spectra were analyzed using MATLAB version 9 (The MathWorks Inc., Natick, MA). In order to compensate for small unavoidable sample size differences, the spectra were scaled using the extended multiplicative signal correction (EMSC) method (55). CO2 vibrations (2,400 to 2,275 cm−1) were then removed from spectra and filled with a trend.
Principal component analysis (PCA) (56) and discriminant function analysis (DFA) were used for the statistical analysis of the preprocessed data. PCA is an unsupervised clustering method with the aim of reducing the dimensionality of a data set, without a priori knowledge of any groupings in the data, by decomposing it into new variables that are linear combinations of the original data. PC-DFA is a supervised method which discriminates the data into groups on the basis of PCs and a priori knowledge of the experimental class structure; PC-DFA functions by minimizing within-group variance while maximizing between-group variance (57). Comparison of intensities of different FTIR spectral regions (selected based on PC-DFA loadings) was carried out as follows: FTIR raw spectra of the appropriate samples were baseline corrected, followed by plotting the intensity (peak height to baseline) of each peak against time.
GC-MS analysis. (i) Quenching and extraction.
The 15-ml sample aliquots collected as described above were immediately quenched in 30 ml of 60% aqueous methanol (−48°C), followed immediately by centrifugation at −9°C for 10 min at 5,000 × g using a Sigma 6K15 centrifuge (Sigma-Aldrich, Dorset, United Kingdom). The supernatant was removed and cell pellets flash frozen in liquid nitrogen and stored at −80°C for further analysis (58).
Extraction was carried out by suspending the biomass in 0.5 ml of 80% methanol (−48°C), followed by snap-freezing in liquid nitrogen and thawing on ice. The freeze-thaw cycle was performed a total of three times to maximize the recovery of the intracellular metabolites. Extracts were then centrifuged at 15,871 × g for 5 min at −9°C. The supernatant was removed to a new sterile microcentrifuge tube and kept on dry ice while a second round of extraction using 0.5 ml of 80% methanol (−48°C) was carried out on the pellets following the steps described above. The second set of extracts (0.5 ml) was combined with the first set and normalized using 80% methanol according to the OD600 of each sample, followed by combining 50 μl from each of the samples in a new tube as the quality control (QC) sample (59). One hundred microliters of internal standard solution (0.2 mg ml−1 succinic-d4 acid, 0.2 mg ml−1 glycine-d5, 0.2 mg ml−1 benzoic-d5 acid, and 0.2 mg ml−1 lysine-d4) was added to all samples, and the samples were dried by speed vacuum concentration at 30°C for 12 h (concentrator 5301; Eppendorf, Cambridge, United Kingdom) prior to being stored at −80°C until further analysis.
(ii) Derivatization.
Derivatization followed a two-step procedure of methoxyamination followed by trimethylsilylation as described precisely by Wedge et al. (60).
(iii) GC-MS setup.
GC-MS analysis was carried out using an Agilent (Wokingham, United Kingdom) 6890N GC oven coupled to a Pegasus III mass spectrometer (Leco, St. Joseph, MI, USA), following previously published methods (60–62). All initial identifications adhered to minimum metabolite reporting standards (63) by providing quality assurance of data and removing mass spectral features within QC samples with high deviation. The final output from this procedure was a matrix of retention time versus mass data with related metabolite identities and peak areas linked to each sample injection. This data set was then used for statistical analysis.
(iv) Data analysis.
A total number of 104 unique GC-MS peaks were detected. Variables with more than 50% missing values were removed (64), resulting in a total of 67 peaks remaining. Peak areas of these peaks were first log10 scaled then subjected to PC-DFA. In addition, canonical correlation analysis (CCA) was also applied to gain a better view of the changes associated with time in the metabolic profiles measured by GC-MS. CCA is a commonly used statistical method to measure the correlation between two sets of random variables (65). In this study, we used CCA to measure the correlation between the GC-MS data and the corresponding time of incubation.
RESULTS AND DISCUSSION
There has been an increasing interest in the biotechnological exploitation of Geobacter species for a range of applications, including the formation of biomagnetite nanoparticles via the enzymatic reduction of Fe(III) oxyhydroxides. One particular challenge that needs to be addressed for successful commercial exploitation is the scalable production of biomass that is active against the Fe(III) precursor to magnetite. To date few studies have addressed the physiological state of Geobacter or other Fe(III)-reducing bacterial species in different bioreactor configurations, especially during scale-up. In this series of experiments, the goal was to assess the changes in growth behavior and metabolism of G. sulfurreducens cells associated with the scale-up of biomass production through monitoring growth (OD measurements), measuring metabolic fingerprints using FTIR, and generating metabolic profiles using GC-MS. In a parallel study (Byrne et al., submitted), which will be reported elsewhere, the surface reactivity, magnetic properties, and particle size distribution of the biomagnetite nanoparticles generated from 100-ml serum bottles and 5- and 50-liter bioreactors (under the same growth conditions) were compared and reported to be at comparable levels. However, during scaling up from serum bottles to fermentors, a prolonged lag phase occurred, extending the incubation period and potentially making the process less efficient and commercially attractive; hence, cellular metabolism is explored in detail here to identify the bottleneck and restore adequate cellular growth behavior.
Growth profiles.
Figure 1 shows the growth kinetics of G. sulfurreducens in NBAF at 30°C during a 7-day incubation period. A typical three-phase (lag, exponential, and stationary) growth pattern was observed, with similar final cell densities for both scales of batch cultures. However, increasing the vessel volume prolonged the lag phase significantly: cells grown in 100-ml serum bottles exhibited a 6-h lag phase, while the lag phase of the bioreactor-grown cells was ∼24 h.
FIG 1.
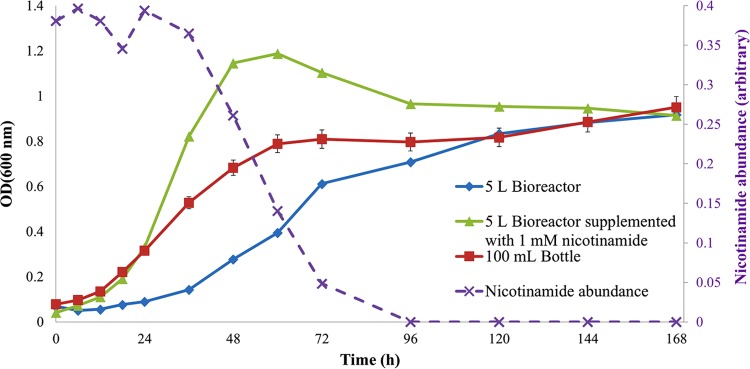
Growth curves of G. sulfurreducens grown on NBAF at 30°C for 168 h in 100-ml serum bottles (red line) and in a 5-liter bioreactor with (green line) and without (blue line) nicotinamide supplementation. Relative peak areas (normalized to an internal standard) of nicotinamide in the medium (purple line) detected by GC-MS in the 5-liter bioreactor supplementation experiment are plotted against the sampling time. The time point measurements for the serum bottles are means of three biological replicates with error bars indicating the standard deviation. Single measurements were recorded for the bioreactor samples due to volume constraint.
FTIR metabolic fingerprinting.
As microbial cells multiply and bacterial cultures proceed through their distinctive and well-documented growth phases, changes also occur in the distribution and composition of biochemical components within the cells (e.g., proteins, lipids, polysaccharides, and nucleic acids), which are not detectable by some conventional techniques such as OD measurement and bacterial plate counts. However, FTIR can reveal these changes by detecting different functional groups through molecular bond vibrations and recording them as spectral features, which can then be analyzed by a variety of multivariate statistical methods (66).
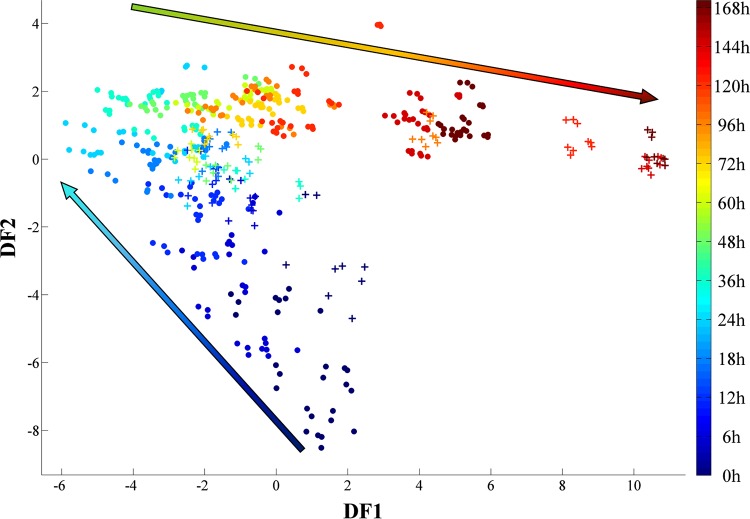
FTIR spectra collected in this study were preprocessed and analyzed using PC-DFA. The scores plot of all samples together (Fig. 2) display a gradual phenotypic change appearing in the G. sulfurreducens cells during the incubation period in both the 100-ml serum bottles and the larger 5-liter bioreactor samples, corresponding to different phases of bacterial growth. Furthermore, a delayed stationary phase associated with the scale-up process is clearly evident (Fig. 2, 120 to 168 h), which is in agreement with the turbidimetric findings (Fig. 1).
FIG 2.
PC-DFA score plot of the FTIR data collected for all the samples. Five PCs with a total explained variance of 97.39% were used for the DFA. The two sets of samples, i.e., the 100-ml serum bottles (circles) and the bioreactor (crosses), displayed similar growth patterns. However, compared to cells grown in the serum bottles, the bioreactor-grown cells displayed an extended lag phase (up to ∼24 h) and consequently entered the stationary phase at a later time point. The arrows illustrate the main trajectories with respect to time. The color bar on the right represents the duration of the incubation during which samples were collected.
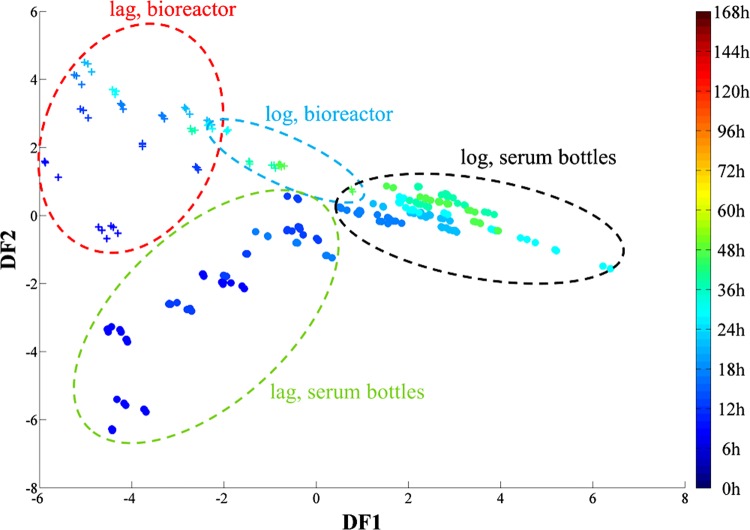
The PC-DFA scores of the samples grown from 0 to 48 h were plotted separately in order to investigate further the effects of the scale-up on the lag phase (Fig. 3). While data from both sets of samples displayed a gradual change, samples from the first 24 h of incubation showed a clear separation between those from the 100-ml serum bottles and those from the bioreactor (Fig. 3).
FIG 3.
PC-DFA score plot (using 3 PCs with a total explained variance of 96.18%) of G. sulfurreducens cells grown in 100-ml serum bottles (circles) and a 5-liter bioreactor (crosses) between 0 and 48 h of incubation at 30°C. There is a clear separation between the two different sets of samples on the basis of DF1 in the first 24 h of incubation. The color bar on the right represents the duration of the incubation during which samples were collected.
The main separation between the two sample sets seems to be on the DF1 axis. DF1 loading plots (see Fig. S2 in the supplemental material) were used to identify the most significant variables (wavenumbers) contributing toward this clustering pattern. Major variations within FTIR spectra for the lag and log phases of both culturing conditions were in the following main regions: (i) amide I (∼1,655 cm−1) and amide II (∼1,541 cm−1) bands for proteins, (ii) fatty acids (∼2,895 cm−1), and (iii) carboxylic acids (∼3,046, 1,709, 1,593, 1,402, and 1,223 cm−1).
Cells grown in the bioreactor started to proceed from lag phase and on to log phase after approximately 24 h of incubation, while those grown in the 100-ml serum bottles completed this transition within the first 6 h of incubation (Fig. 1). Interestingly, the PC-DFA score plot (Fig. 3) displayed the clustering of spectra from bioreactor-grown cells up to the first 24 h of incubation on the negative side of DF1 (red dashed cluster). The only spectra from the serum bottles clustering on the same side are from those samples taken within the first 6 h of incubation (green dashed cluster). Moreover, spectra from the bioreactor samples taken after 24 h of incubation display the occurrence of a phenotypic change by moving toward the serum bottle samples (Fig. 3, blue dashed cluster), which according to the growth curve (Fig. 1) coincides with the end of the lag phase and start of the log phase.
In summary, the FTIR results were consistent with the growth profiles recorded for both sets of samples, showing phenotypic changes that coincided with the occurrence of an extended lag phase during the scale-up process. To explore the accompanying phenotypic changes in more detail, we employed the complementary tool of the metabolic profiling approach using GC-MS.
GC-MS metabolic profiling.
In order to elucidate further the general metabolic changes occurring within and between clusters from the entire sample set, a PC-DFA score plot of the GC-MS data collected for all the samples was constructed (using the first 10 PCs with a total explained variance of 96.7%, with a priori groups being the individual time points) (Fig. 4a).
FIG 4.
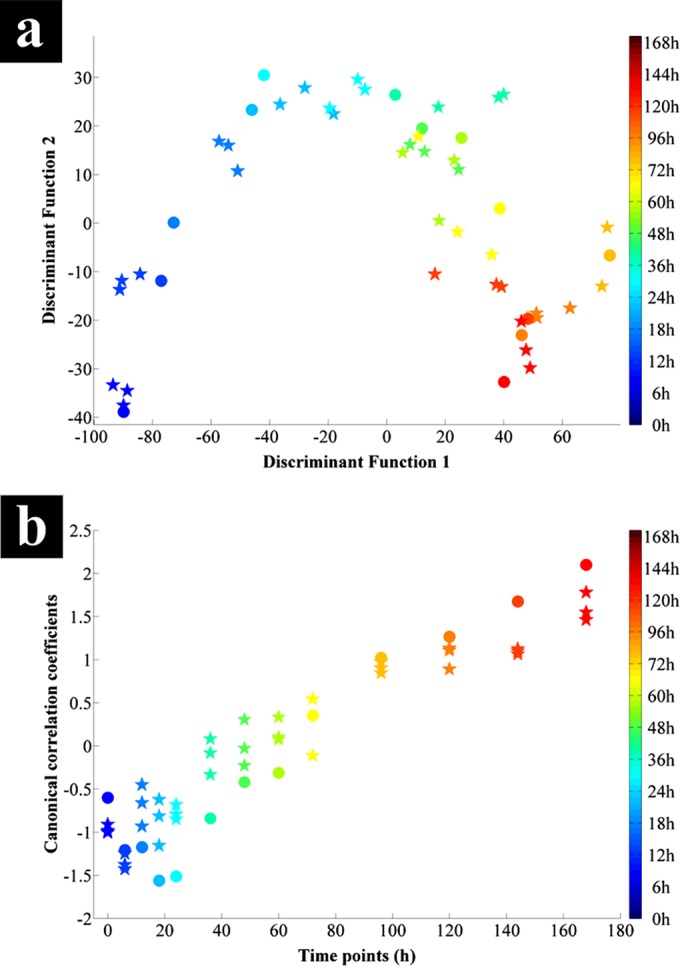
(a) PC-DFA score plot of the G. sulfurreducens cell extracts analyzed using GC-MS from 100-ml serum bottles (stars) and a 5-liter bioreactor (circles), collected from all time points. (b) CCA plot against time of the relative peak areas of detected metabolites by GC-MS for 100-ml serum bottle (stars) and bioreactor (circles) samples. Both samples displayed an overall linear time-dependent correlation. The color bars on the right represent the duration of the incubation during which samples were collected.
The PC-DFA score plot (Fig. 4a) of the cell extracts displayed a trajectory similar to that seen in the FTIR data (Fig. 3), showing the segregation of different growth phases and the transition of these distinct physiological states through the connecting time points. Furthermore, these results point to a gradual change occurring between the cells grown in the bioreactor and serum bottles after the first 6 h of incubation. This gradual change coincides with the transition from lag to log phase (Fig. 1) for the cells grown in the serum bottles, while the bioreactor-grown cells remained in the lag phase at these time points.
The bioreactor samples displayed a clear change of trajectory after the 24-h time point (Fig. 4a), clustering closer toward the serum bottle samples corresponding to the end of lag phase and start of log phase (Fig. 1). Since a significant metabolic difference was observed in the PC-DFA of the complete data set, these data were explored in more depth using CCA.
Similar to the PC-DFA, CCA was conducted on the PC scores of the first 10 PCs to investigate the correlation between the relative peak intensities of different metabolites with incubation time. Overall, these results (Fig. 4b) display a clear linear time-dependent effect with a canonical correlation coefficient of 0.9439, indicating a strong correlation between the GC-MS profiles and the incubation time. With the exception of the first time point (this is likely to be related to the lag phase), the 100-ml serum bottle samples displayed a positive correlation with incubation time. In contrast, the bioreactor samples displayed a negative correlation with time for samples collected in the first 24 h, followed by the same positive trend as for the 100-ml serum bottles. Furthermore, both sets of samples (bioreactor and serum bottles) seem to pass through an overlapping period after a 96-h of incubation period. The CCA findings from the GC-MS data were in agreement with the PC-DFA score plots of the FTIR (Fig. 2 and 3) and GC-MS data (Fig. 4a), as well as the growth curves (Fig. 1), coinciding with the occurrence of an extended lag phase for the bioreactor-grown cells associated with the scale-up process.
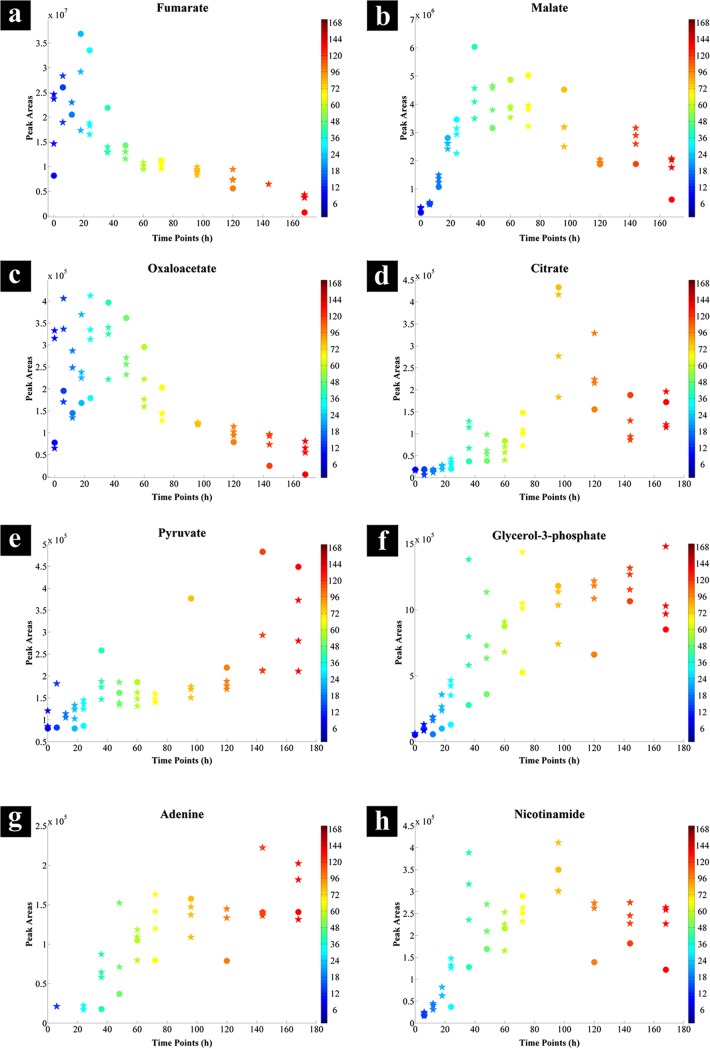
The CCA loading plot (Fig. 5) indicates the most significant variables (metabolites) that were positively or negatively correlated with incubation time. The plot illustrates that the most significant variables that were negatively correlated with time were fumarate, oxaloacetate, and an unknown metabolite (variable 59), while citrate shows a strong positive correlation (a full list of identified metabolites can be found in Table S2 in the supplemental material). Figure 6 shows the relative peak intensities of the significant metabolites selected by CCA and positively identified using an in-house library (discussed and interpreted below).
FIG 5.
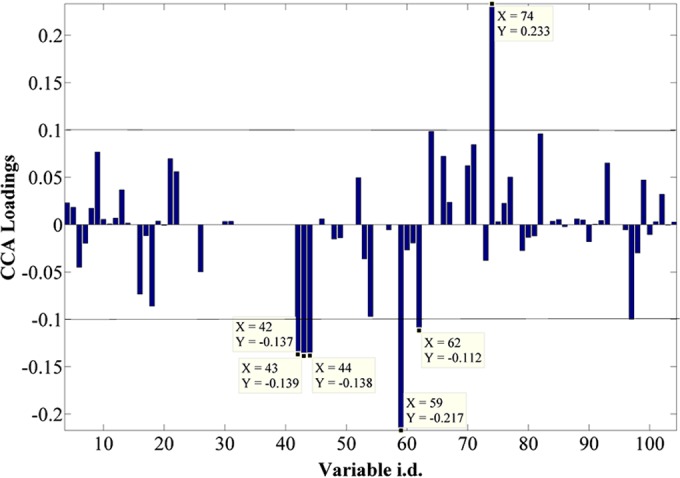
CCA loading plot of all the sample extracts analyzed by GC-MS, showing the most significant metabolites contributing toward the positive and negative correlation with time throughout the data set. The list of identified metabolites can be found in Table S2 in the supplemental material. Fumarate is seen as three peaks, as different derivatization products are seen in GC-MS.
FIG 6.
Relative GC-MS peak intensities of significant metabolites between 100-ml serum bottle (stars) and bioreactor (circles) cell extracts during the incubation period. The color bars on the right represent the duration of the incubation during which samples were collected.
Interpretation of GC-MS metabolic profiles.
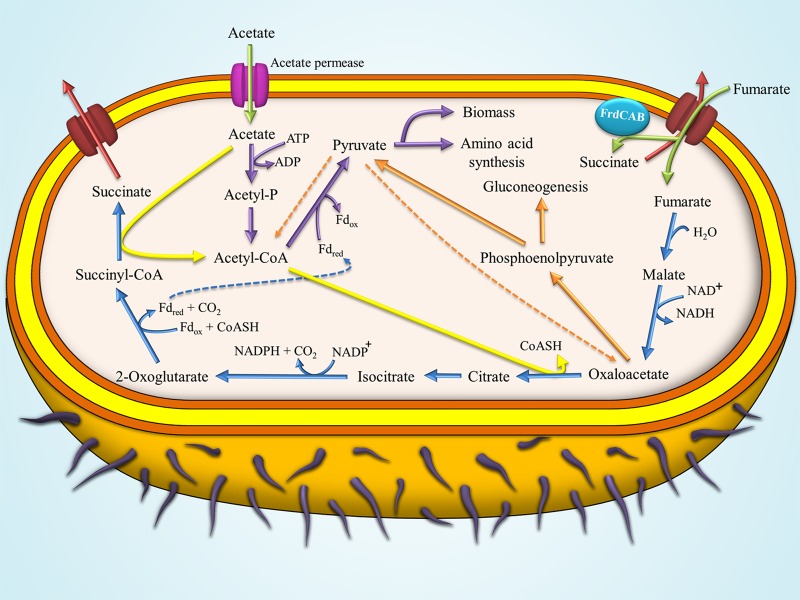
Since acetate is one of the key intermediate products in anaerobic metabolism of natural complex organic matter in the sedimentary environment (3, 4), the ability to take up and utilize this compound efficiently provides a competitive advantage (67). Studies have shown the presence of acetate permease-like proteins (68) (Fig. 7, magenta channel) in G. sulfurreducens, homologues to ActP in Escherichia coli which are known to be responsible for acetate uptake (69).
FIG 7.
Pathway of acetate metabolism in G. sulfurreducens during growth on NBAF medium with fumarate as the electron acceptor and acetate as the electron donor. Acetate is transported into the cells (green arrow) via acetate permease (magenta protein channel). Imported acetate can be activated via one of the following two pathways: (i) the acetate kinase (EC 2.7.2.1) followed by the activity of phosphotransacetylase enzyme (EC 2.3.1.8) (purple arrows) producing acetyl-CoA which is directed toward pyruvate synthesis and subsequently into biomass and amino acid synthesis pathways, or (ii) acetate being oxidized through conversion of succinyl-CoA to succinate via the activity of succinyl-CoA:acetate CoA-transferase enzyme (EC 2.8.3.18), resulting in the production of acetyl-CoA (yellow arrows) which is directed toward the TCA cycle. The fumarate provided in the medium is taken up (green arrow) via the fumarate transporter proteins (C4-dicarboxylic acid transporter). Imported fumarate can either be (i) directed toward the TCA cycle (blue arrow), which operates as an open loop and ends with the formation of succinate and its excretion into the medium (red arrow), or (ii) reduced to succinate via the activity of inner membrane-bound FrdCAB enzyme (fumarate reductase activity) followed by its excretion into the medium (red arrow) (66, 67, 76, 77).
The acetate imported by G. sulfurreducens can be activated via ligation with coenzyme A (CoA) through two main pathways (70). The first of these pathways involves the oxidation of acetate through the conversion of succinyl-CoA to succinate via the activity of the succinyl-CoA:acetate CoA-transferase enzyme (EC 2.8.3.18) (Fig. 7, yellow arrow). Acetyl-CoA produced via this pathway is directed toward the citrate synthase reaction in the tricarboxylic acid (TCA) cycle (70).
The second pathway involves the phosphorylation of acetate via acetate kinase (EC 2.7.2.1) followed by a second reaction involving phosphotransacetylase (EC 2.3.1.8) converting acetyl-phosphate to acetyl-CoA (Fig. 7, purple arrows) (70, 71). The acetyl-CoA produced via this pathway is converted to pyruvate via the activity of pyruvate oxidoreductase (EC 1.2.7.1) operating in the reverse direction (Fig. 7, purple arrow) (1). The reduced ferredoxin produced via the TCA cycle provides the reducing power to enable this reaction (Fig. 7, dashed blue arrow).
The fumarate provided in the medium as an electron acceptor is taken up by the cells via a C4-dicarboxylic acid transporter protein (Fig. 7, red transporter channel) (72), followed by its conversion to succinate either via a reducing reaction catalyzed by the membrane-bound fumarate reductase (EC 1.3.5.4) (Fig. 7, FrdCAB enzyme) or via the TCA cycle (Fig. 7, blue arrows) (71, 72).
The GC-MS results show an accumulation of fumarate (Fig. 6a) during the lag phase followed by a gradual depletion in intracellular levels throughout the following time points in both bioreactor- and 100-ml serum bottle-grown cells. This accumulation of fumarate reaches its peak point at around 24 h for the bioreactor samples and at around 6 h of incubation for the 100-ml serum bottle samples, followed by a decreasing trend corresponding to the consumption of fumarate by the cells and the transition from lag to log phase in both batch cultures (Fig. 1). A possible reason for accumulation of fumarate could be linked to its high concentration in the medium, resulting in cells adjusting to their new environment by importing fumarate.
Yang and coworkers (73) suggested the potential use of fumarate as a carbon source and its contribution to gluconeogenesis through its conversion to malate by fumarase and further to oxaloacetate via malate dehydrogenase (MDH) (EC 1.1.1.37). These authors also suggested that around 90 to 95% of the oxaloacetate produced via this pathway is directed toward gluconeogenesis and pyruvate production, via its conversion to phosphoenolpyruvate through the activity of phosphoenolpyruvate carboxykinase (EC 4.1.1.32) (Fig. 7, orange arrows).
The levels of oxaloacetate detected in our study supported these findings, as the 100-ml serum bottle samples showed a higher intensity of oxaloacetate (approximately double) than detected in the bioreactor samples during the first 24 h of incubation (Fig. 6c). This coincides with the consumption of fumarate (Fig. 6a) and an increase in pyruvate levels (Fig. 6e) for the 100-ml serum bottle samples, while there was an accumulation of fumarate (Fig. 6a) and no change in the pyruvate levels (Fig. 6e) in the bioreactor samples. These findings further highlight the potential role of fumarate as a carbon source, as stated by Yang et al. (73) and discussed above, through its conversion to oxaloacetate via the TCA cycle and the contribution of produced oxaloacetate toward gluconeogenesis (Fig. 7, orange arrows), resulting in higher pyruvate levels for biomass and amino acid biosynthesis.
It is known that conversion of malate to oxaloacetate via MDH activity is thermodynamically unfavorable in cells (74). This may constrain oxaloacetate homeostasis in cells grown with fumarate as the electron acceptor, while resulting in accumulation of malate, which could eventually be secreted into the medium (71).
The GC-MS data confirm these findings, as malate levels were not overly affected by the scaling-up process, since its intensities were similar in both bioreactor- and 100-ml serum bottle-grown cells throughout the experiments (Fig. 6b).
Nicotinamide was also identified by the CCA (Fig. 5; see Table S2 in the supplemental material). NAD is a cofactor involved in more than 300 oxidation-reduction reactions, of which the conversion of malate to oxaloacetate through the activity of MDH is one (75). The nicotinamide levels may reflect the levels of NAD availability within cells at different stages of growth. Interestingly, the levels of nicotinamide under both cultivation conditions displayed trends very similar to those of oxaloacetate and pyruvate, starting at low concentrations during the lag phase, increasing throughout the log phase, and finishing with a period of stability during the stationary phase (Fig. 6h).
The lower availability of NAD during the lag phase of cells grown in the bioreactor may affect the respiration rate by limiting the activity of MDH, an NAD-dependent enzyme, resulting in lower oxaloacetate formation. This may trigger a feedback loop which could be explained as follows: first, it limits the amount of oxaloacetate which could be fed into gluconeogenesis, pyruvate synthesis, and acetyl-CoA production (Fig. 7, orange arrows); second, it reduces the respiration rate, which is followed by subsequent limitation of the 2-oxoglutarate availability and its conversion to succinyl-CoA, which provides the reduced ferredoxin (Fig. 7, dashed blue arrow) required for catalyzing the conversion of acetyl-CoA to pyruvate (Fig. 7, purple arrow).
Furthermore, intracellular citrate levels in the 100-ml serum bottles started to increase after the first 6 h of incubation up to the 96-h time point, followed by a decreasing trend (Fig. 6d, stars). By contrast, citrate levels in the bioreactor samples were low and stable up to the first 24 h of incubation, after which they followed the same trend as in the serum bottle samples (Fig. 6d, circles). These findings further support our hypothesis that limited MDH activity restricts the pool of oxaloacetate, resulting in lower activity of the TCA cycle during the lag phase for G. sulfurreducens grown under both cultivation conditions.
To test the above hypothesis, NBAF was supplemented with nicotinamide (final concentration of 1 mM) to investigate the effects of nicotinamide on the growth of G. sulfurreducens cells in the 5-liter bioreactor. The growth curves displayed in Fig. 1 suggest that the nicotinamide-supplemented NBAF significantly improved the growth of the cells in the 5-liter bioreactor, while reducing the lag phase period from 24 h to 6 h, similar to that of the cells grown in the 100-ml serum bottles. Furthermore, levels of nicotinamide in the medium detected by GC-MS analysis (Fig. 1) show the consumption of nicotinamide. The NBAF medium prior to nicotinamide supplementation was also tested by GC-MS analysis; however, as expected, no peaks corresponding to nicotinamide were detected (data not shown). The nicotinamide supplementation effect on growth is illustrated in Fig. 1. As nicotinamide starts a depletion trend (60 h), the growth rate is declining and cells enter stationary phase. This is followed be the complete depletion of nicotinamide in the medium (96 h), which is concomitant with a steady period of growth.
We also examined the growth behavior of G. sulfurreducens on NBAF supplemented with nicotinamide (1 mM) and oxaloacetate (6 mM) in the 100-ml serum bottles; while oxaloacetate supplementation resulted in an extended lag phase, nicotinamide had no significant effects on the lag phase (see Fig. S4 in the supplemental material).
On the whole, during the first 24 h of incubation, the majority of the metabolites detected displayed lower intensities in the bioreactor-grown cells than in those grown in the 100-ml serum bottles, especially metabolites of TCA cycle, with the exception of malate and fumarate (Fig. 6). Comparisons of the significant FTIR vibrations (Fig. 8; see Fig. S3 in the supplemental material), identified via PC-DFA loadings (see Fig. S2), were in agreement with the GC-MS findings (Fig. 6). The intensity of the amide I band (Fig. 8a) for the serum bottle samples showed an increasing trend up to the first 24 h followed by a steady state, while the bioreactor samples displayed no significant change up to the 24-h time point followed by a slow increase thereafter. Furthermore, comparison of the intensity of carboxylic acid-specific FTIR vibrations (Fig. 8b; see Fig. S3) for the bioreactor samples also displayed a similar trend, with a steady state in the first 24 h followed by a slow increase, while the intensities detected for the serum bottle samples were almost 6 times higher than those for the bioreactor samples.
FIG 8.
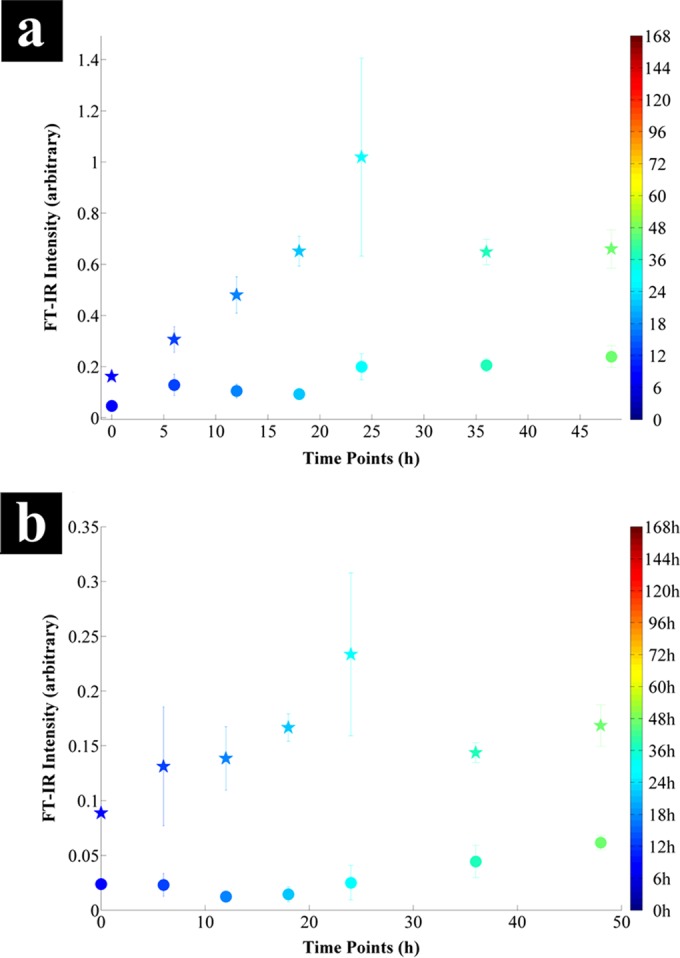
Comparison of the intensities of significant FTIR vibrations identified by the PC-DFA loading plot of serum bottle (stars) and bioreactor (circles) samples during the first 48 h of incubation (see Fig. S2 in the supplemental material). (a) The 1,655 cm−1 amide I region due to stretching of C=O bonds; (b) the 1,402 cm−1 region due to symmetric stretching of C=O bonds in carboxylic acids. Data points represent the mean of the three replicates, with bars indicating the relative standard deviation. The color bars on the right represent the duration of the incubation during which samples were collected.
Finally, the concentrations of glycerol-3-phosphate (Fig. 6f) did not change during the lag phase but increased sharply during the exponential phase, before finally reaching a period of stability during the stationary phase under both culturing conditions. This is of course perhaps not surprising, as glycerol-3-phosphate is one of the main precursors for the synthesis of triacylglycerides, which are major lipid components of bacterial cell membranes. Therefore, the concentration and necessity of this compound are directly related to the bacterial growth rate, which is in complete agreement with our GC-MS findings.
Concluding remarks.
This study clearly demonstrates the effects of scale-up on the growth and metabolism of G. sulfurreducens grown on NBAF with acetate as an electron donor and fumarate as an electron acceptor in batch culture. It also illustrates some of the challenges involved with scaling up bioprocesses, which is vital to underpin biotechnological exploitation of a wide range of microorganisms, including Geobacter species. The FTIR findings emphasize the advantages of this technique as a rapid, nondestructive, relatively inexpensive, and high-throughput screening tool and also reveal its potential for applications in a wide range of areas, including industrial scale-up. Our GC-MS metabolic profiling results suggested that the limited availability of oxaloacetate, potentially due to restricted nicotinamide levels and malate dehydrogenase activity, could be a significant metabolic bottleneck resulting from this scale-up process. Additional metabolomics experiments were performed to test this hypothesis. Guided by the hypothesis generation experiments from GC-MS (shown schematically in Fig. S5 in the supplemental material), which suggested that cells may be starved of oxaloacetate and/or nicotinamide, further cultures were grown that were supplemented with these metabolic intermediates to test these hypotheses (see Fig. S5). While supplementation with oxaloacetate had a negative effect on growth, when nicotinamide was added at the start of culturing in the bioreactor, the bacterial lag phase was significantly shorter and at a level comparable to that detected in the 100-ml serum bottles.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a United Kingdom BBSRC Ph.D. studentship grant (BB/H015868/1) for H.M. R.G. is also grateful to BBSRC for financial support. J.R.L. acknowledges support from the European Union 7th Framework Programme (FP7/2007-2013) under grant agreement no. 309517 (NANOREM). We also acknowledge support from the BBSRC/EPSRC Centre for Synthetic Biology of Fine and Speciality Chemicals (SYNBIOCHEM, grant BB/M017702/1).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00294-15.
REFERENCES
- 1.Mahadevan R, Palsson BO, Lovley DR. 2011. In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling. Nat Rev Microbiol 9:39–50. doi: 10.1038/nrmicro2456. [DOI] [PubMed] [Google Scholar]
- 2.Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 3.Lovley DR. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J Ind Microbiol 14:85–93. doi: 10.1007/BF01569889. [DOI] [PubMed] [Google Scholar]
- 4.Lovley DR. 2002. Dissimilatory metal reduction: from early life to bioremediation. ASM News 68:231–237. [Google Scholar]
- 5.Lovley DR, Stolz JF, Nord GL, Phillips EJP. 1987. Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature 330:252–254. doi: 10.1038/330252a0. [DOI] [Google Scholar]
- 6.Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caccavo F, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol 60:3752–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin WC, Coppi MV, Lovley DR. 2004. Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl Environ Microbiol 70:2525–2528. doi: 10.1128/AEM.70.4.2525-2528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol 49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- 10.Lovley DR, Anderson RT. 2000. Influence of dissimilatory metal reduction on fate of organic and metal contaminants in the subsurface. Hydrogeology J 8:77–88. doi: 10.1007/PL00010974. [DOI] [Google Scholar]
- 11.Lloyd JR, Lovley DR. 2001. Microbial detoxification of metals and radionuclides. Curr Opin Biotechnol 12:248–253. doi: 10.1016/S0958-1669(00)00207-X. [DOI] [PubMed] [Google Scholar]
- 12.Finneran KT, Anderson RT, Nevin KP, Lovley DR. 2002. Potential for bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sed Contam 11:339–357. doi: 10.1080/20025891106781. [DOI] [Google Scholar]
- 13.Lovley DR, Phillips EJP, Gorby YA, Landa ER. 1991. Microbial reduction of uranium. Nature 350:413–416. doi: 10.1038/350413a0. [DOI] [Google Scholar]
- 14.Telling ND, Coker VS, Cutting RS, van der Laan G, Pearce CI, Pattrick RAD, Arenholz E, Lloyd JR. 2009. Remediation of Cr(VI) by biogenic magnetic nanoparticles: an x-ray magnetic circular dichroism study. Applied Physics Lett 95:163701. doi: 10.1063/1.3249578. [DOI] [Google Scholar]
- 15.Lloyd JR, Sole VA, Van Praagh CVG, Lovley DR. 2000. Direct and Fe(II)-mediated reduction of technetium by Fe(III)-reducing bacteria. Appl Environ Microbiol 66:3743–3749. doi: 10.1128/AEM.66.9.3743-3749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui D, Eriksen TE. 1996. Reduction of pertechnetate in solution by heterogeneous electron transfer from Fe(II)-containing geological material. Environ Sci Technol 30:2263–2269. doi: 10.1021/es950627v. [DOI] [Google Scholar]
- 17.Lloyd JR. 2003. Microbial reduction of metals and radionuclides. FEMS Microbiology Rev 27:411–425. doi: 10.1016/S0168-6445(03)00044-5. [DOI] [PubMed] [Google Scholar]
- 18.Williams KH, Bargar JR, Lloyd JR, Lovley DR. 2013. Bioremediation of uranium-contaminated groundwater: a systems approach to subsurface biogeochemistry. Curr Opin Biotechnol 24:489–497. doi: 10.1016/j.copbio.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Byrne JM, Telling ND, Coker VS, Pattrick RAD, van der Laan G, Arenholz E, Tuna F, Lloyd JR. 2011. Control of nanoparticle size, reactivity and magnetic properties during the bioproduction of magnetite by Geobacter sulfurreducens. Nanotechnology 22:455709. doi: 10.1088/0957-4484/22/45/455709. [DOI] [PubMed] [Google Scholar]
- 20.Coker V, Pattrick R, van der Laan G, Lloyd J. 2007. Formation of magnetic minerals by non-magnetotactic prokaryotes. Microbiol Monogr 3:275–300. doi: 10.1007/7171_047. [DOI] [Google Scholar]
- 21.Cutting RS, Coker VS, Fellowes JW, Lloyd JR, Vaughan DJ. 2009. Mineralogical and morphological constraints on the reduction of Fe(III) minerals by Geobacter sulfurreducens. Geochim Cosmochim Acta 73:4004–4022. doi: 10.1016/j.gca.2009.04.009. [DOI] [Google Scholar]
- 22.Prijic S, Sersa G. 2011. Magnetic nanoparticles as targeted delivery systems in oncology. Radiol Oncol 45:1–16. doi: 10.2478/v10019-011-0001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majewskia P, Thierrya B. 2007. Functionalized magnetite nanoparticles—synthesis, properties, and bio-applications. Crit Rev Solid State Mat Sci 32:203–215. doi: 10.1080/10408430701776680. [DOI] [Google Scholar]
- 24.Franzreb M, Siemann-Herzberg M, Hobley T, Thomas O. 2006. Protein purification using magnetic adsorbent particles. Appl Microbiol Biotechnol 70:505–516. doi: 10.1007/s00253-006-0344-3. [DOI] [PubMed] [Google Scholar]
- 25.Berensmeier S. 2006. Magnetic particles for the separation and purification of nucleic acids. Appl Microbiol Biotechnol 73:495–504. doi: 10.1007/s00253-006-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coker VS, Bennett JA, Telling ND, Henkel T, Charnock JM, van der Laan G, Pattrick RA, Pearce CI, Cutting RS, Shannon IJ, Wood J, Arenholz E, Lyon IC, Lloyd JR. 2010. Microbial engineering of nanoheterostructures: biological synthesis of a magnetically recoverable palladium nanocatalyst. ACS Nano 4:2577–2584. doi: 10.1021/nn9017944. [DOI] [PubMed] [Google Scholar]
- 27.Reference deleted.
- 28.Zeng H, Black CT, Sandstrom RL, Rice PM, Murray CB, Sun S. 2006. Magnetotransport of magnetite nanoparticle arrays. Phys Rev B 73:020402. doi: 10.1103/PhysRevB.73.020402. [DOI] [Google Scholar]
- 29.Bundy J, Davey M, Viant M. 2009. Environmental metabolomics: a critical review and future perspectives. Metabolomics 5:3–21. doi: 10.1007/s11306-008-0152-0. [DOI] [Google Scholar]
- 30.Fiehn O. 2002. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol 48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- 31.Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R. 2007. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 8:1243–1266. doi: 10.2217/14622416.8.9.1243. [DOI] [PubMed] [Google Scholar]
- 32.Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, Oliver SG. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol 19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 33.Urbanczyk-Wochniak E, Luedemann A, Kopka J, Selbig J, Roessner-Tunali U, Willmitzer L, Fernie AR. 2003. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Rep 4:989–993. doi: 10.1038/sj.embor.embor944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellis DI, Goodacre R. 2012. Metabolomics-assisted synthetic biology. Curr Opin Biotechnol 23:22–28. doi: 10.1016/j.copbio.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 35.van der Werf MJ, Overkamp KM, Muilwijk B, Coulier L, Hankemeier T. 2007. Microbial metabolomics: toward a platform with full metabolome coverage. Anal Biochem 370:17–25. doi: 10.1016/j.ab.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 36.Winder CL, Dunn WB, Goodacre R. 2011. TARDIS-based microbial metabolomics: time and relative differences in systems. Trends Microbiol 19:315–322. doi: 10.1016/j.tim.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Al-Qadiri HM, Al-Alami NI, Lin M, Al-Holy M, Cavinato AG, Rasco BA. 2008. Studying of the bacterial growth phases using Fourier transform infrared spectroscopy and multivariate analysis. J Rapid Methods Automat Microbiol 16:73–89. doi: 10.1111/j.1745-4581.2008.00117.x. [DOI] [Google Scholar]
- 38.Mamas M, Dunn WB, Neyses L, Goodacre R. 2011. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch Toxicol 85:5–17. doi: 10.1007/s00204-010-0609-6. [DOI] [PubMed] [Google Scholar]
- 39.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. 2011. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 40:387–426. doi: 10.1039/B906712B. [DOI] [PubMed] [Google Scholar]
- 40.Naumann D, Helm D, Labischinski H. 1991. Microbial characterizations by FTIR spectroscopy. Nature 351:81–82. doi: 10.1038/351081a0. [DOI] [PubMed] [Google Scholar]
- 41.AlRabiah H, Correa E, Upton M, Goodacre R. 2013. High-throughput phenotyping of uropathogenic E. coli isolates with Fourier transform infrared spectroscopy. Analyst 138:1363–1369. doi: 10.1039/c3an36517d. [DOI] [PubMed] [Google Scholar]
- 42.Fischer G, Braun S, Thissen R, Dott W. 2006. FTIR spectroscopy as a tool for rapid identification and intra-species characterization of airborne filamentous fungi. J Microbiol Methods 64:63–77. doi: 10.1016/j.mimet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Ellis DI, Brewster VL, Dunn WB, Allwood JW, Golovanov AP, Goodacre R. 2012. Fingerprinting food: current technologies for the detection of food adulteration and contamination. Chem Soc Rev 41:5706–5727. doi: 10.1039/c2cs35138b. [DOI] [PubMed] [Google Scholar]
- 44.Ellis DI, Broadhurst D, Kell DB, Rowland JJ, Goodacre R. 2002. Rapid and quantitative detection of the microbial spoilage of meat by Fourier transform infrared spectroscopy and machine learning. Appl Environ Microbiol 68:2822–2828. doi: 10.1128/AEM.68.6.2822-2828.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roussel S, Bellon-Maurel W, Roger JM, Grenier P. 2003. Authenticating white grape must variety with classification models based on aroma sensors, FTIR and UV spectrometry. J Food Eng 60:407–419. doi: 10.1016/S0260-8774(03)00064-5. [DOI] [Google Scholar]
- 46.Wang H, Hollywood K, Jarvis RM, Lloyd JR, Goodacre R. 2010. Phenotypic characterization of Shewanella oneidensis MR-1 under aerobic and anaerobic growth conditions by using Fourier transform infrared spectroscopy and high-performance liquid chromatography analyses. Appl Environ Microbiol 76:6266–6276. doi: 10.1128/AEM.00912-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wharfe ES, Jarvis RM, Winder CL, Whiteley AS, Goodacre R. 2010. Fourier transform infrared spectroscopy as a metabolite fingerprinting tool for monitoring the phenotypic changes in complex bacterial communities capable of degrading phenol. Environ Microbiol 12:3253–3263. doi: 10.1111/j.1462-2920.2010.02300.x. [DOI] [PubMed] [Google Scholar]
- 48.Filip Z, Hermann S. 2001. An attempt to differentiate Pseudomonas spp. and other soil bacteria by FTIR spectroscopy. Eur J Soil Biol 37:137–143. doi: 10.1016/S1164-5563(01)01078-0. [DOI] [Google Scholar]
- 49.Pan J, Ge X, Liu R, Tang H. 2006. Characteristic features of Bacillus cereus cell surfaces with biosorption of Pb(II) ions by AFM and FTIR. Colloids Surf B Biointerfaces 52:89–95. doi: 10.1016/j.colsurfb.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Correa E, Sletta H, Ellis DI, Hoel S, Ertesvag H, Ellingsen TE, Valla S, Goodacre R. 2012. Rapid reagentless quantification of alginate biosynthesis in Pseudomonas fluorescens bacteria mutants using FTIR spectroscopy coupled to multivariate partial least squares regression. Anal Bioanal Chem 403:2591–2599. doi: 10.1007/s00216-012-6068-6. [DOI] [PubMed] [Google Scholar]
- 51.Fischer E, Sauer U. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur J Biochem 270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- 52.Cutting RS, Coker VS, Telling ND, Kimber RL, Pearce CI, Ellis BL, Lawson RS, van der Laan G, Pattrick RAD, Vaughan DJ, Arenholz E, Lloyd JR. 2010. Optimizing Cr(VI) and Tc(VII) remediation through nanoscale biomineral engineering. Environ Sci Technol 44:2577–2584. doi: 10.1021/es902119u. [DOI] [PubMed] [Google Scholar]
- 53.Lovley DR, Phillips EJ. 1986. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal Potomac river. Appl Environ Microbiol 52:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winder CL, Gordon SV, Dale J, Hewinson RG, Goodacre R. 2006. Metabolic fingerprints of Mycobacterium bovis cluster with molecular type: implications for genotype-phenotype links. Microbiology 152:2757–2765. doi: 10.1099/mic.0.28986-0. [DOI] [PubMed] [Google Scholar]
- 55.Martens H, Nielsen JP, Engelsen SB. 2003. Light scattering and light absorbance separated by extended multiplicative signal correction. Application to near-infrared transmission analysis of powder mixtures. Anal Chem 75:394–404. doi: 10.1021/ac020194w. [DOI] [PubMed] [Google Scholar]
- 56.Wold S, Esbensen K, Geladi P. 1987. Principal component analysis. Chemometrics Intelligent Lab Syst 2:37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]
- 57.Macfie HJH, Gutteridge CS, Norris JR. 1978. Use of canonical variates analysis in differentiation of bacteria by pyrolysis gas-liquid chromatography. J Gen Microbiol 104:67–74. doi: 10.1099/00221287-104-1-67. [DOI] [PubMed] [Google Scholar]
- 58.Winder CL, Dunn WB, Schuler S, Broadhurst D, Jarvis R, Stephens GM, Goodacre R. 2008. Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal Chem 80:2939–2948. doi: 10.1021/ac7023409. [DOI] [PubMed] [Google Scholar]
- 59.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, Moon S, Nikolau B. 2008. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 60.Wedge DC, Allwood JW, Dunn W, Vaughan AA, Simpson K, Brown M, Priest L, Blackhall FH, Whetton AD, Dive C, Goodacre R. 2011. Is serum or plasma more appropriate for intersubject comparisons in metabolomic studies? An assessment in patients with small-cell lung cancer. Anal Chem 83:6689–6697. doi: 10.1021/ac2012224. [DOI] [PubMed] [Google Scholar]
- 61.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R, The Human Serum Metabolome (HUSERMET) Consortium . 2011. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 62.Begley P, Francis-McIntyre S, Dunn WB, Broadhurst DI, Halsall A, Tseng A, Knowles J, HUSERMET Consortium, Goodacre R, Kell DB. 2009. Development and performance of a gas chromatography-time-of-flight mass spectrometry analysis for large-scale nontargeted metabolomic studies of human serum. Anal Chem 81:7038–7046. doi: 10.1021/ac9011599. [DOI] [PubMed] [Google Scholar]
- 63.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. 2007. Proposed minimum reporting standards for chemical analysis. Metabolomics 3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gromski PS, Xu Y, Kotze HL, Correa E, Ellis DI, Armitage EG, Turner ML, Goodacre R. 2014. Influence of missing values substitutes on multivariate analysis of metabolomics data. Metabolites 4:433–452. doi: 10.3390/metabo4020433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Härdle WK, Simar L. 2007. Applied multivariate statistical analysis. Springer, Berlin, Germany. [Google Scholar]
- 66.Brown M, Dunn WB, Ellis DI, Goodacre R, Handl J, Knowles JD, O'Hagan S, Spasić I, Kell DB. 2005. A metabolome pipeline: from concept to data to knowledge. Metabolomics 1:39–51. doi: 10.1007/s11306-005-1106-4. [DOI] [Google Scholar]
- 67.Mahadevan R, Bond DR, Butler JE, Esteve-Nuñez A, Coppi MV, Palsson BO, Schilling CH, Lovley DR. 2006. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol 72:1558–1568. doi: 10.1128/AEM.72.2.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Risso C, Van Dien SJ, Orloff A, Lovley DR, Coppi MV. 2008. Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. J Bacteriol 190:2266–2274. doi: 10.1128/JB.01841-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh M-K, Rohlin L, Kao KC, Liao JC. 2002. Global expression profiling of acetate-grown Escherichia coli. J Biol Chem 277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- 70.Segura D, Mahadevan R, Juarez K, Lovley DR. 2008. Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens. PLoS Comput Biol 4:e36. doi: 10.1371/journal.pcbi.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galushko AS, Schink B. 2000. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch Microbiol 174:314–321. doi: 10.1007/s002030000208. [DOI] [PubMed] [Google Scholar]
- 72.Butler JE, Glaven RH, Esteve-Núñez A, Núñez C, Shelobolina ES, Bond DR, Lovley DR. 2006. Genetic characterization of a single bifunctional enzyme for fumarate reduction and succinate oxidation in Geobacter sulfurreducens and engineering of fumarate reduction in Geobacter metallireducens. J Bacteriol 188:450–455. doi: 10.1128/JB.188.2.450-455.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang TH, Coppi MV, Lovley DR, Sun J. 2010. Metabolic response of Geobacter sulfurreducens towards electron donor/acceptor variation. Microb Cell Fact 9:90. doi: 10.1186/1475-2859-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lehninger AL, Nelson DL, Cox MM. 1993. Principles of biochemistry. Wiley, New York, NY. [Google Scholar]
- 75.Minarik P, Tomaskova N, Kollarova M, Antalik M. 2002. Malate dehydrogenases—structure and function. Gen Physiol Biophys 21:257–265. [PubMed] [Google Scholar]
- 76.Byrne JM, Coker VS, Moise S, Wincott PL, Vaughan DJ, Tuna F, Arenholz E, van der Laan G, Pattrick RAD, Lloyd JR, Telling ND. 2013. Controlled cobalt doping in biogenic magnetite nanoparticles. J R Soc Interface 10:20130134. doi: 10.1098/rsif.2013.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coker VS, Pearce CI, Pattrick RAD, Van der Laan G, Telling ND, Charnock JM, Arenholz E, Lloyd JR. 2008. Probing the site occupancies of Co-, Ni-, and Mn-substituted biogenic magnetite using XAS and XMCD. Am Mineralog 93:1119–1132. doi: 10.2138/am.2008.2681. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.