Abstract
Background
Cryptosporidium hominis is a dominant species for human cryptosporidiosis. Within the species, IbA10G2 is the most virulent subtype responsible for all C. hominis–associated outbreaks in Europe and Australia, and is a dominant outbreak subtype in the United States. In recent yearsIaA28R4 is becoming a major new subtype in the United States. In this study, we sequenced the genomes of two field specimens from each of the two subtypes and conducted a comparative genomic analysis of the obtained sequences with those from the only fully sequenced Cryptosporidium parvum genome.
Results
Altogether, 8.59-9.05 Mb of Cryptosporidium sequences in 45–767 assembled contigs were obtained from the four specimens, representing 94.36-99.47% coverage of the expected genome. These genomes had complete synteny in gene organization and 96.86-97.0% and 99.72-99.83% nucleotide sequence similarities to the published genomes of C. parvum and C. hominis, respectively. Several major insertions and deletions were seen between C. hominis and C. parvum genomes, involving mostly members of multicopy gene families near telomeres. The four C. hominis genomes were highly similar to each other and divergent from the reference IaA25R3 genome in some highly polymorphic regions. Major sequence differences among the four specimens sequenced in this study were in the 5′ and 3′ ends of chromosome 6 and the gp60 region, largely the result of genetic recombination.
Conclusions
The sequence similarity among specimens of the two dominant outbreak subtypes and genetic recombination in chromosome 6, especially around the putative virulence determinant gp60 region, suggest that genetic recombination plays a potential role in the emergence of hyper-transmissible C. hominis subtypes. The high sequence conservation between C. parvum and C. hominis genomes and significant differences in copy numbers of MEDLE family secreted proteins and insulinase-like proteases indicate that telomeric gene duplications could potentially contribute to host expansion in C. parvum.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1517-1) contains supplementary material, which is available to authorized users.
Keywords: Cryptosporidium, Genomics, Whole genome sequencing, Genetic recombination, Virulence
Background
Cryptosporidium spp. inhabit the brush borders of the gastrointestinal and respiratory epithelium of various vertebrates, causing enterocolitis, diarrhea, and cholangiopathy in humans [1]. Immunocompetent children and adults with cryptosporidiosis usually have a short-term illness accompanied by watery diarrhea, nausea, vomiting, and weight loss. In immunocompromised persons, however, the infection can be protracted and life-threatening [2]. Cryptosporidiosis is one of the most important causes of moderate-to-severe diarrhea and diarrhea-associated deaths in children in developing countries [3] and a major cause for waterborne and foodborne outbreaks of human illness in industrialized nations [4,5]. In the United States the number of reported cases of cryptosporidiosis has increased more than twofold since 2005 [6-9]. Currently, it is estimated that there are approximately 750,000 annual cases of cryptosporidiosis in the United States [5].
Among the many established Cryptosporidium species and genotypes, C. hominis and C. parvum are the two responsible for greater than 90% of the human cryptosporidiosis cases in most countries. C. hominis is largely human-specific and responsible for anthroponotic transmission of cryptosporidiosis. C. parvum infects both humans and some farm animals, especially pre-weaned calves and lambs and thus can be transmitted both anthroponotically and zoonotically [10]. Within C. hominis, subtype IbA10G2 is the dominant strain for C. hominis-associated waterborne outbreaks of cryptosporidiosis in the United States, Europe, and Australia [10-16]. The dominant subtype associated with waterborne cryptosporidiosis outbreaks in the United States since 2005 is a new subtype, IaA28R4 [17-20].
Whole genome sequencing of Cryptosporidium spp. has greatly facilitated the development of genotyping, subtyping and multilocus sequence typing (MLST) tools for characterizing the transmission of C. hominis and C. parvum [21,22]. These tools have played a major role in improving our understanding of cryptosporidiosis epidemiology [10,23]. Nevertheless, genomic studies of Cryptosporidium spp. lag far behind those on other related apicomplexan parasites largely because of the lack of effective cultivation and animal models. Thus far, only the genomes of one laboratory isolate each of C. parvum, C. hominis, and C. muris have been sequenced using traditional Sanger sequencing technology [22,24,25]. More recently, the genome of an anthroponotic II subtype (IIcA5G3b) of C. parvum serially propagated in immunosuppressed mice has been sequenced using Illumina technology [26]. The lack of whole genome sequence data, especially from field specimens obtained from outbreaks, has hampered our understanding of genetic determinants for host specificity, virulence, and the biological fitness of various Cryptosporidium species and C. parvum and C. hominis subtypes.
In this study, we sequenced the genomes of two dominant outbreak subtypes (IbA10G2 and IaA28R4) of C. hominis by using 454 and Illumina technologies. Prior to sequencing, oocysts were isolated directly from field specimens without propagation in laboratory animals, and extracted DNA was amplified to generate enough material for sequencing. Results of this study have (1) filled some gaps in our understanding of Cryptosporidium genomics, (2) identified some major deletions and one large insertion in the C. hominis genome, and (3) showed the high genetic similarity of the two outbreak subtypes. We have also demonstrated the occurrence of genetic recombination in chromosome 6.
Results
Cryptosporidium hominis sequence data and de novo assemblies
After sequencing using 454 technology, 1,048,412 reads (382.5 Mb) were obtained from specimen 30974 (IbA10G2) and 1,157,140 reads (431.7 Mb) were obtained from specimen 33537 (IaA28R4). They produced an assembly of 8,841,752 bp in 443 contigs for specimen 30974 (N50 = 78,110 bp) and an assembly of 14,065,231 bp in 1,464 contigs for specimen 33537 (N50 = 27,749 bp). Using Illumina paired-end sequencing, 64,449,544 reads (5,780.0 Mb) were obtained from specimen 30976 (IaA28R4) and 30,886,077 reads (2,798.3 Mb) were obtained from specimen 37999 (IbA10G2). They produced an assembly of 22,133,082 bp in 6,140 contigs for specimen 30976 (N50 = 145,968 bp) and 9,054,010 bp in 78 contigs for specimen 37999 (N50 = 406,678 bp; Table 1).
Table 1.
Summary of sequence data from whole genome sequencing of four Cryptosporidium hominis specimens in comparison with data from the published C. hominis (TU502) and C. parvum (IOWA) genomes
| Specimen (gp60 subtype) | Technique | Total nucleotides | Total sequence reads | Assembly | Average coverage (fold) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| # of Contigs | Length (bp) | Mean (bp) | Minimum (bp) | Maximum (bp) | N50 (bp) | |||||
| 30976 (IaA28R4) | Illumina Genome Analyzer IIx 100 bp paired end | 5,780,028,818 | 64,449,544 | 6,140 | 22,133,082 | 3,605 | 502 | 1,279,890 | 145,968 | 257 |
| 37999 (IbA10G2) | Illumina Genome Analyzer IIx 100 bp paired end | 2,798,259,889 | 30,886,077 | 78 | 9,054,010 | 116,077 | 510 | 1,029,232 | 406,678 | 307 |
| 33537 (IaA28R4) | 454 GS-FLX Titanium | 431,742,212 | 1,157,140 | 1,464 | 14,065,231 | 9,607 | 501 | 154,507 | 27,749 | 31 |
| 30974 (IbA10G2) | 454 GS-FLX Titanium | 382,520,957 | 1,048,412 | 443 | 8,841,752 | 19,959 | 513 | 325,032 | 78,110 | 43 |
| C. hominis TU502 (IaA25R3) | Sanger | - | - | 1,422 | 8,743,570 | 6,149 | 251 | 90,444 | 14,504 | 12 |
| C. parvum IOWA (IIaA15G2R1) | Sanger | - | - | 18 | 9,102,324 | 504,874 | 17,388 | 1,278,458 | 1,014,526 | 13 |
Genome coverage and bacterial contamination
Among the two specimens sequenced by 454 technology, 424 of the 443 contigs generated from the IbA10G2 specimen 30974 and 767 of the 1,464 contigs from the IaA28R4 specimen 33537 mapped to the eight chromosomes of the C. parvum IOWA isolate, representing 8,816,174 and 8,590,919 bp, thus giving a 96.9% and 94.4% coverage of the genomes, respectively (Table 2, Additional file 1: Figure S1). The C. parvum genome was used as the reference because it is the most complete genome fully assembled into eight chromosomes with the aid of a physical HAPPY map generated prior to the sequencing effort [27], and has ~97% sequence similarity to the reference C. hominis TU502 (IaA25R3) genome. The latter has 1,422 scaffolds and contigs, but was estimated to have synteny to many contigs in the C. parvum IOWA (IIaA15G2R1) genome sequences [25]. For the two specimens sequenced by paired-end Illumina, 64 of the 78 contigs generated from the IbA10G2 specimen 37999 and 45 of the 6,140 contigs from the IaA28R4 specimen 30976 mapped to the eight chromosomes of the C. parvum reference genome. The mapped contigs represented 9,041,990 and 9,054,312 bp and thus had 99.34% and 99.47% coverages of the genomes, respectively. In contrast, 1,269 of 1,422 contigs from the published genome of TU502 (IaA25R3) mapped to the eight C. parvum chromosomes, representing a 95.5% coverage (Table 2, Figure 1A). No physical map is available for any of the C. hominis specimens sequenced to date to aid the assembly of genomic sequences.
Table 2.
Coverage of four Cryptosporidium hominis genomes sequenced in this study and sequence similarities to published C. parvum (IOWA) and C. hominis (TU502) genomes
| Chromosome | C. parvum length (bp) | 37999 (IbA10G2) | 30976 (IaA28R4) | 30974 (IbA10G2) | 33537 (IaA28R4) | TU502 (IaA25R3) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Contigs mapped | Length (bp) | Coverage (%) | Similarity to IOWA (%) | Similarity to TU502 (%) | Contigs mapped | Length (bp) | Coverage (%) | Similarity to IOWA (%) | Similarity to TU502 (%) | Contigs mapped | Length (bp) | Coverage (%) | Similarity to IOWA (%) | Similarity to TU502 (%) | Contigs mapped | Length (bp) | Coverage (%) | Similarity to IOWA (%) | Similarity to TU502 (%) | Contigs mapped | Length (bp) | Coverage (%) | Similarity to IOWA (%) | ||
| 1 | 875659 | 15 | 867575 | 99.08 | 96.8 | 99.79 | 1 | 873289 | 99.73 | 96.81 | 99.86 | 36 | 863586 | 98.62 | 96.84 | 99.82 | 54 | 840604 | 96 | 96.93 | 99.86 | 124 | 859754 | 98.18 | 96.88 |
| 2 | 985969 | 7 | 987017 | 100.11 | 96.79 | 99.58 | 8 | 983830 | 99.78 | 96.82 | 99.81 | 41 | 970191 | 98.4 | 96.81 | 99.77 | 69 | 944487 | 95.8 | 96.9 | 99.82 | 115 | 946071 | 95.95 | 96.73 |
| 3 | 1099352 | 13 | 1098355 | 99.91 | 96.89 | 99.62 | 13 | 1096430 | 99.73 | 96.86 | 99.79 | 40 | 1081251 | 98.35 | 96.86 | 99.75 | 87 | 1064015 | 96.78 | 96.98 | 99.81 | 158 | 1079381 | 98.18 | 96.78 |
| 4 | 1104417 | 3 | 1103687 | 99.93 | 96.76 | 99.72 | 4 | 1105075 | 100.06 | 96.76 | 99.8 | 86 | 1056974 | 95.7 | 96.81 | 99.74 | 123 | 1024384 | 92.75 | 96.93 | 99.82 | 195 | 1007110 | 91.19 | 96.77 |
| 5 | 1080900 | 13 | 1092751 | 101.1 | 96.78 | 99.74 | 11 | 1107822 | 102.49 | 96.78 | 99.84 | 71 | 1013283 | 93.74 | 96.93 | 99.77 | 118 | 940540 | 87.01 | 97.09 | 99.83 | 186 | 972978 | 90.02 | 96.79 |
| 6 | 1332857 | 5 | 1304591 | 97.88 | 96.91 | 99.76 | 2 | 1298888 | 97.45 | 96.93 | 99.86 | 66 | 1263267 | 94.78 | 97.01 | 99.76 | 124 | 1237394 | 92.84 | 97.09 | 99.83 | 192 | 1240122 | 93.04 | 96.82 |
| 7 | 1278458 | 5 | 1268482 | 99.22 | 97.19 | 99.79 | 4 | 1269257 | 99.28 | 97.2 | 99.87 | 27 | 1267106 | 99.11 | 97.18 | 99.82 | 79 | 1258429 | 98.43 | 97.25 | 99.88 | 124 | 1282777 | 100.34 | 97.18 |
| 8 | 1344712 | 3 | 1319172 | 98.1 | 96.78 | 99.76 | 2 | 1319721 | 98.14 | 96.8 | 99.83 | 57 | 1300516 | 96.71 | 96.81 | 99.78 | 113 | 1281066 | 95.26 | 96.9 | 99.81 | 175 | 1304075 | 96.98 | 96.74 |
| Total | 9102324 | 64 | 9041990 | 99.34 | 96.86 | 99.72 | 45 | 9054312 | 99.47 | 96.87 | 99.83 | 424 | 8816174 | 96.93 | 96.9 | 99.78 | 767 | 8590919 | 94.36 | 97 | 99.83 | 1269 | 8692268 | 95.49 | 96.84 |
Figure 1.
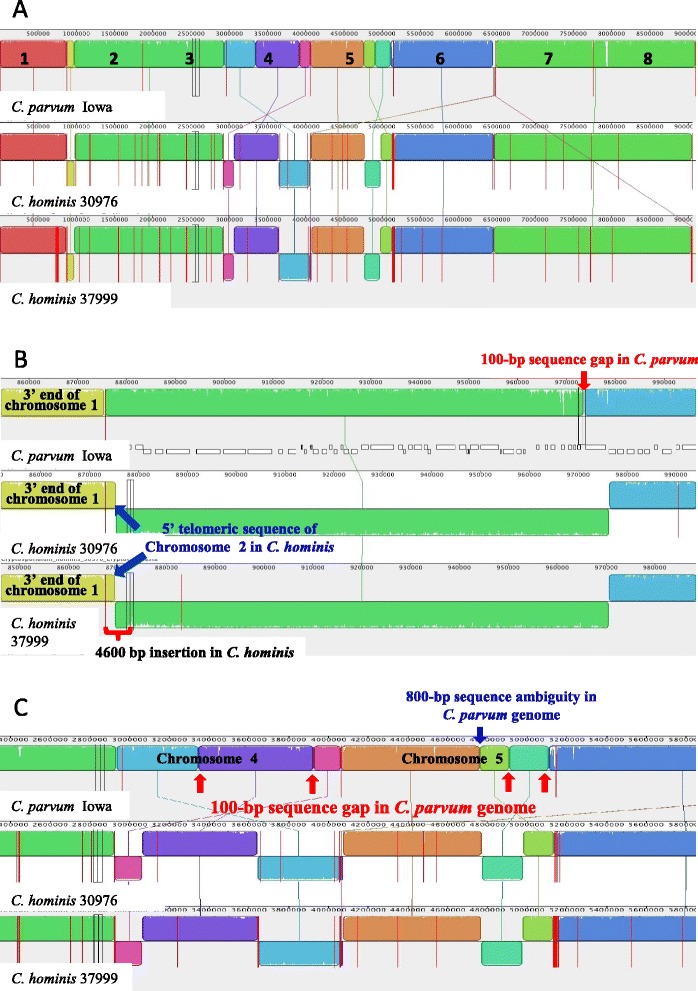
Structural organization of two Illumina-sequenced genomes of Cryptosporidium hominis comparing to eight chromosomes (numbered and separated by vertical red lines) of published Cryptosporidium parvum genome. The color blocks (known as Locally Collinear Blocks) are conserved segments of sequences internally free from genome rearrangements, whereas the inverted white peaks within each block are sequence divergence between the reference C. parvum (IOWA) genome and C. hominis genome under analysis. A. Coverage of two C. hominis genomes showing possible sequence rearrangements in chromosomes 2, 4, 5 and 6. Assembled contigs are bordered by vertical red lines. For specimens 30976, only Cryptosporidium contigs were used in mapping. B. Possible sequence rearrangements at the 5′ end of chromosome 2. C. Possible sequence rearrangements in chromosomes 4 and 5.
Most of the 14 unmapped contigs from specimen 37999 were small (≤1,824 bp) and were sequences of multicopy genes (ex. rRNA units) and genes with paralogs in the genome or large repetitive sequences (ex. fatty acid synthase and cgd5_1210 and cgd5_1220). However, sequences of four contigs (45, 66, 74, and 77) had no similarity to any published sequences, and one contig (#76) had 95% sequence similarity to a 500-bp region of the genome of Strentrophomonas maltophilia (CP002986). Similarly, most of the unmapped contigs from specimen 30974 were small (≤3,170 bp) and were sequences of multicopy genes (ex. rRNA units), genes with paralogs in the genome and large repetitive sequences (ex. fatty acid synthase and cgd5_2180), and telomeric sequences of Cryptosporidium. Sequences of 18 contigs (#303, 357, 392, 415, 416, 436, 492, 503, 521, 524, 529, 537, 542, 543, 551, 562, 563, and 564) had no similarity to any published Cryptosporidium sequences, and one contig (#392) had 98-100% sequence similarity to Bacteroides fragillis plasmids from humans (AB646744 and U25716). Similar observations were made for TU502. In addition, the 547 bp at the 5′ end of contig AAEL01000108 (19,113 bp) had 98% sequence similarity to cgd3_530 on chromosome 3, while the remaining part of the sequence mapped to chromosome 8. Similarly, the 5′ (15,709-bp) region of contig AAEL01000024 (36,266 bp in length) mapped to chromosome 7, the 3′ region (nucleotides 25,790-36,266) mapped to chromosome 2, while the middle region containing the rRNA unit mapped to chromosomes 1, 2, 7, and 8.
In contrast, most of the unmapped contigs from IaA28R4 specimens 30976 and 33537 had non-Cryptosporidium sequences. For example, the largest 100 unmapped contigs (16,411-138,945 bp) from specimen 33537 were 99-100% similar to the genome (CP006252) of the enterobacteria Serratia liquefaciens, with the exception of contig 0018 (94,132 bp), which was from its plasmid. As the genome of S. liquefaciens is about 5.2 Mb, the 1,464 contigs of 14,065,231 bp from specimen 33537 were from the combined C. hominis and S. liquefaciens genomes, with all S. liquefaciens contigs positioned behind the mapped Cryptosporidium sequences (Additional file 1: Figure S1). Evidence of contamination from several bacterial species was present in data from specimen 30976, as the 6,140 contigs totaled 22.13 Mb, which is larger than the combined genomes of C. hominis and one bacterial species. BLAST analysis of contigs indicated that ~28% of the total nucleotides were from members of Enterobacteriaceae and 8% from Bacteroidaceae. The 20 largest unmapped contigs (88,676-515,888 bp) had 75-85% sequence similarities to genomes of members (Serratia, Yersinia, Klebsiella, E. coli, Salmonella, etc.) of Enterobacteriaceae, except for one (contig #51), which had a 98% sequence similarity to a 21,307 bp region of an uncultured organism from the human gut (GQ873945).
Sequence similarity to published C. parvum genome and physical characteristic of C. hominis genomes
The genomes of specimens 30974, 30976, 33537, 37999 and TU502 had 96.90%, 96.87%, 97.0%, 96.86%, and 96.84% sequence similarities to the C. parvum IOWA genome in the mapped regions, respectively (Table 2). The alignment of whole genome sequences generated by Mauve showed near complete sequence synteny of the four C. hominis genomes to the published C. parvum reference genome, which is the only complete Cryptosporidium genome available for comparison. Some possible inversions and translocations of sequence fragments were seen in chromosomes 2, 4, 5 and 6 in the two almost fully sequenced C. hominis genomes from specimens 30976 and 37999 (Figure 1A). However, these inversions and translocations all occurred in sequence gap regions of the reference C. parvum genome. For example, specimens 30976 and 37999 generated sequences that cover the sequence gap in chromosome 2 of the C. parvum genome and have ~4,600-bp extra sequences at the 5′ end of the fragment, with telomeric repeats (TTTAGG) (Figure 1B). The inversions of sequences in chromosomes 4 and 5 also happened around sequence gaps in the C. parvum IOWA genome (Figure 1C). Near the 3′ end of chromosome 5, the large C. hominis contigs in 30976 and 37999 that are upstream of the two small scaffolds (coding for cgd5_4510 to cgd5_4610) in the C. parvum IOWA genome both end with telomeric repeats (data not shown). Similarly, the large C. hominis contigs in 30976 and 37999 that are upstream of the small scaffold (coding for cgd6_5460 to cgd6_5520) at the 3′ end of chromosome 6 in the C. parvum IOWA genome both end with telomeric repeats. In addition, one or two of the genes coded by the C. parvum scaffold (cgd6_5460 for specimen 37999 and cgd6_5460 and cgd6_5470 for specimen 30976) are located at the 5′ end of chromosome 5 in C. hominis, which has telomeric repeats at the 5′ end (Figure 2A). Most of the remaining genes are missing in C. hominis, except for cgd6_5500, whose ortholog is present in C. hominis in an unknown chromosome together with the ortholog of cgd5_4600. Although the genome sequences of specimens 30974 and 33537 were more fragmented, the same inversion of scaffolds was seen in chromosome 5 (Additional file 1: Figure S1).
Figure 2.
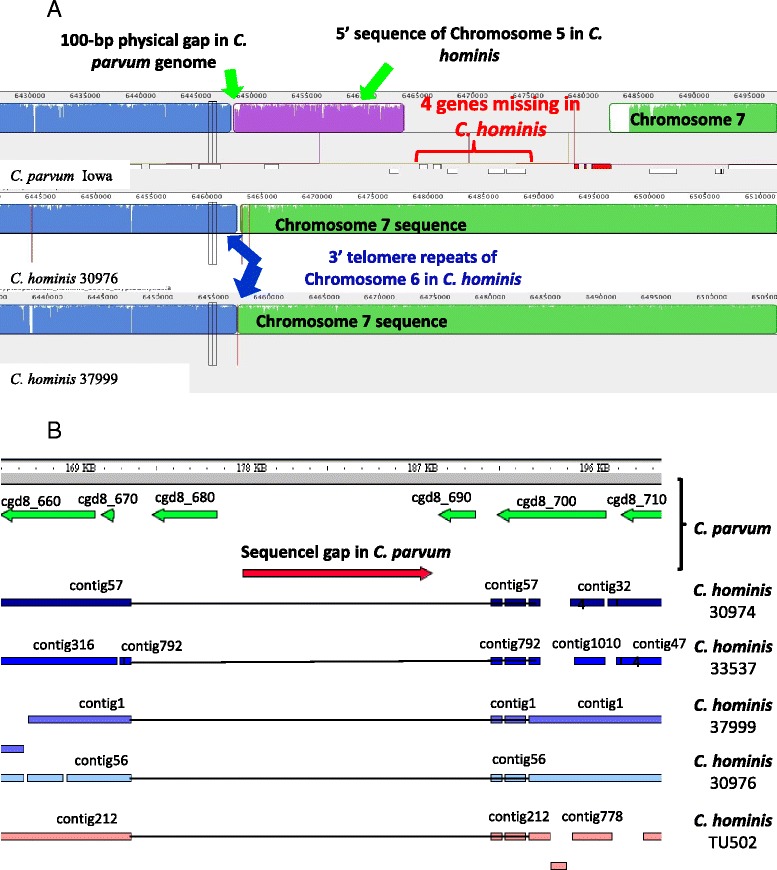
Deletion of genes in Cryptosporidium hominis genomes in comparison with Cryptosporidium parvum. A. Deletion of four genes (cgd6_5480, cgd6_5490, cgd6_5510, and cgd6_5520) at the 3′ end of chromosome 6 (probably should be the 5′ end of chromosome 5) in C. hominis. B. A major 19,048-bp deletion in C. hominis genome in chromosome 8, including the cgd8_680 and cgd8_690 genes. Note the ~10 kb sequence gap in C. parvum.
Most of the missing sequences in the sequenced genomes were in the telomeric regions or low sequence complexity areas of the eight chromosomes of C. parvum, and generally occurred in the two 454-sequenced and the published C. hominis genomes. Half of the ten sequence gaps (unsequenced regions present in clones) in the C. parvum genome were missing in the 454-sequenced and the published C. hominis genomes (data not shown). The two Illumina-sequenced genomes, however, fully covered most of the ten sequence gaps in the C. parvum IOWA genome (Table 3). Most of the sequences generated from C. hominis specimens 30976 and 37999 were longer than the estimated length of sequence gaps in the C. parvum IOWA genome, with the noticeable exception of the ~10,000-bp sequence gap in C. parvum, which is not present in all five C. hominis genomes (Table 3). This region was fully covered by contig 11 of 30976, contig 2 of 37999, contig 0057 of 30974, contig 0792 of 33537, and contig 212 (AAEL01000212) of TU502 (Figure 2B). In C. hominis, the ~100 bp sequence downstream of the deletion is almost identical to the beginning sequence of the insert and the immediate sequence downstream of the insert in C. parvum. This might have contributed to the large deletion in the C. hominis genome. The size of the deletion was 19,048 bp if the sequence gap in C. parvum was indeed 10,000 bp.
Table 3.
Coverage of two Illumina-sequenced Cryptosporidium hominis genomes in sequence gaps of the published C. parvum IOWA genome
| Chromosome | Gap in C. parvum IOWA (bp) | Sequence length in C. hominis specimen (bp) | |
|---|---|---|---|
| 37999 | 30976 | ||
| 2 | 100* | 1481 | 1481 |
| 3 | 500 | 5582 | 5576 |
| 4 | 100 (1st)* | >458 | 1450 |
| 4 | 100 (2nd)* | 2937 | 2714 |
| 5 | 100 (1st)* | 14,926 | 14,929 |
| 5 | 100 (2nd)* | 1788 | 1311 |
| 5 | 1,000 (3rd) | Not covered | 467 |
| 6 | 2,500 | 245 | 245 |
| 6 | 100* | >538 (ending with telomeric repeats) | >857 (ending with telomeric repeats) |
| 8 | 10,000 | 19,048 bp deletion spanning entire gap | 19,048 bp deletion spanning entire gap |
*Regions where inversions and translocations of sequences occurred in sequenced C. hominis genomes.
C. parvum- and C. hominis- specific sequences
In addition to the large 19,048-bp deletion, which contained the C. parvum-specific cgd8_680 and cgd8-690 genes and other potential genes in the ~10,000-bp sequence gap, the comparative genomic analysis identified several other deletions in the C. hominis genome (Table 4), including those mentioned above at the 3′ end of chromosomes 5 (containing orthologs of cgd5_4580, cgd5_4590, and cgd5_4610) and 6 (containing orthologs of cgd6_5480, cgd6_5490, cgd6_5510, and cgd6_5520). Most of the C. parvum-specific genes were members of multicopy gene families. Thus, cgd5_4580, cgd5_4590, cgd5_4600, cgd5_4610, cgd6_5480, and cgd6_ 5490 in C. parvum are genes of the Cryptosporidium telomeric MEDLE family of secreted proteins, all with signal peptides and similar sequences. As mentioned above, these genes are located in tandem in telomeric regions of these two chromosomes, but C. hominis has only one such gene, Chro.50507 or the ortholog of cgd5_4600. Likewise, cgd6_5510 and cgd6_5520 both code for telomeric insulinase-like proteases with signal peptides. Members of the insulinase-like proteases all have very different nucleotide sequences, and both C. parvum and C. hominis have 11 such genes in tandem near the 3′ end of chromosome 3. Thus, C. hominis lacks two of the subtelomeric genes at the 5′ end of chromosome 5 as well as five copies of the Cryptosporidium telomeric MEDLE family of secreted proteins in chromosomes 5 and 6. The C. parvum-specific nature of cgd6_5510 (also known as ZPT) was previously known [28].
Table 4.
Species-specific genes in genomes of Cryptosporidium parvum and C. hominis
| Chromosome | Length (bp) | Genes | Specificity |
|---|---|---|---|
| 8 | 19,048 | cgd8_680, cgd8_690 and other potential genes in 10,000 bp sequence gap | C. parvum |
| 6 | 15,314 | cgd6_5480, cgd6_5490, cgd6_5510, cgd6_5520 | C. parvum |
| 5 | 5,620 | cgd5_4580, cgd5_4590, cgd5_4610 | C. parvum |
| 3 | ~4800 | Chro.50011 | C. hominis |
Notes:
1. cgd5_4580, cgd5_4590, cgd5_4600, and cgd5_4610: four genes with similar sequences at the 3′ end of chromosome 5 in C. parvum, all called telomeric MEDLE family of secreted proteins. C. hominis has only one such gene here (Chro.50507, the ortholog of cgd5_4600).
2. cgd6_5480 and cgd6_ 5490: two genes of the telomeric MEDLE family of secreted proteins with similar sequences at 3′ end of chromosome 6 in C. parvum. C. hominis has no such gene here. The two genes have sequences similar to the four genes above. This fragment and cgd6_5510 (ZPT) and cgd6_5520 below are located at the 5′ end of chromosome 5 in the C. hominis genomes sequenced. C. hominis specimen 37999 does not appear to have the ortholog for cgd6_5470, although 30976 clearly has it. Ortholog of cgd6_5500 is apparently translocated to an unknown chromosome in C. hominis, downstream of the ortholog of cgd5_4580.
3. cgd6_5510 (ZPT) and cgd6_5520: telomeric insulinase-like protease with a signal peptide (the two genes have very different sequences). C. parvum has 11 such genes near 3′end of chromosome 3.
4. cgd8_680: a large low complexity protein with repeats. cgd8_690: a signal peptide containing protein with 2 Cryptosporidium-specific paralogs (cgd8_660 and its ortholog chro.80081).
The comparative genomic analysis further identified several large contigs not found in the published C. parvum IOWA genome, including contigs AAEL01000413, AAEL01000717, and AAEL01000728 in the published TU502 genome. Contig AAEL01000413 is 6,056 bp in length (29% GC) and has 260 copies of telomeric sequence TTTAGG at its 3′ end. It mapped to contigs 442 and 290 of specimen 30974 and contigs 1743, 1586, and 768 of specimen 33537, and was fully covered by contig 82 of specimen 30976 and contig 35 of specimen 37999. It codes at nucleotides 1,798-3,267 for the hypothetical protein Chro. 50011 with RS/HS repeats at the carboxyl end. The coding region was fully covered in all C. hominis genomes sequenced in this project. Sequence alignment indicates that the gene is located at the 3′ end of chromosome 3, with the entire insertion (4,795 bp in 30976 and 4,830 bp in 37999, excluding telomere repeats) all ending with copies of the telomere repeat sequence TTTAGG (Figure 3A). As the 3′ end of chromosome 3 of C. parvum also had the telomeric repeats, this insertion appears valid. The coding region is not present in C. parvum EST library data and five MS/MS peptide libraries deposited in CryptoDB (http://cryptodb.org/cryptodb/). PCR amplification of DNA from five C. parvum and C. hominis specimens each confirmed its presence in only C. hominis (Figure 3B).
Figure 3.
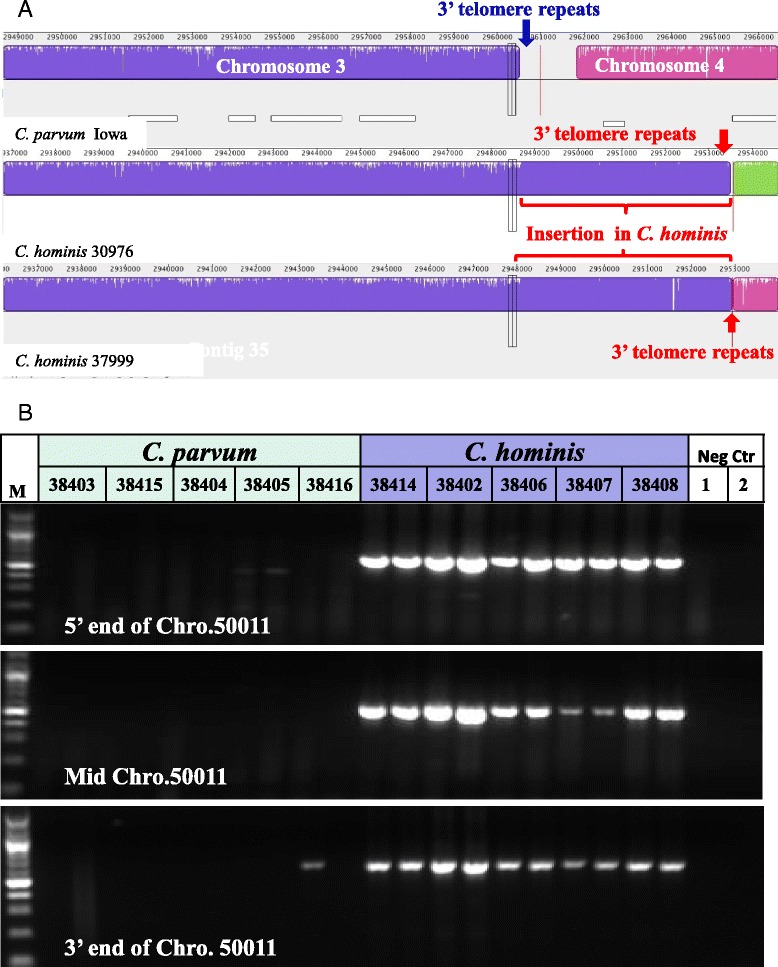
Cryptosporidium hominis-specific nature of Chro.50011. A. Insertion of ~4,860 bp containing the Chro.50011 gene at the 3′ end of chromosome 3 in C. hominis. B. Confirmation of the absence of the ortholog of Chro.50011 in four specimens of C. parvum by PCR analysis of three regions of the Chro.50011 gene. The faint band in PCR analysis of the 3′ end of the gene in C. parvum specimen 38416 produced a nucleotide sequence identical to Chro.50011 in C. hominis.
In contrast, contig AAEL01000728 is 2,277 bp in length (23% GC) and mapped to contig 257 of specimen 30974 and contigs 1238, 1367, and 1487 of specimen 33537. It is located in chromosome 5 of specimens 30976 (contig_6) and 37999 (contig_1), and within a sequence gap area in the C. parvum IOWA genome. PCR analysis using primers based on the AAEL01000728 sequence amplified DNA of C. hominis, C. parvum, and C. andersoni, with the sequences from C. parvum and C. hominis differing from each other by two nucleotides in the 413-bp region, and from C. andersoni having 97% sequence similarity to nucleotide 19,798-20,202 of XM_002142452 (coding for a large hypothetical protein CMU_010870) from C. muris (data not shown). Contig AAEL01000717, which contains the sensor histidine kinase gene (Chro.00003, nucleotide 673–2,319), was probably not of Cryptosporidium origin. It is 2,333 bp in length had a 66% GC content. It has no equivalents in the published C. parvum and C. muris genomes and C. hominis genomes sequenced in the present study, but has a 77% sequence similarity to the sensor histidine kinase gene of Rhizobium etli (nucleotides 1,334,700-1,334,429 of CP001074). PCR primers based on this sequence did not amplify DNA of C. parvum or C. hominis (data not shown).
Sequence similarity to published C. hominis genomic data
The genomes of specimens 30974 (IbA10G2), 30976 (IaA28R4), 33537 (IaA28R4), and 37999 (IbA10G2) had 99.78%, 99.83%, 99.83%, and 99.72% sequence similarities to the published C. hominis genome of TU502 (of the IaA25R3 subtype), respectively (Table 2). Mapping of Illumina reads from specimens 30976 and 37999 to the contigs of the published C. hominis TU502 genome indicated that single nucleotide polymorphisms (SNPs) were distributed across all eight chromosomes of the genome. A few loci on several chromosomes, however, had higher sequence polymorphisms (Figure 4A, Additional file 2: Figure S2). Most of the highly polymorphic loci occurred in genes coding for mucins (orthologs of cgd2_430, cgd2_440, cgd2_450, cgd3_720, and cgd6_1080 or gp60), other secreted proteins with signal peptides (orthologs of cgd1_150, cgd1_3810, cgd3_3430, cgd6_1030, and cgd6_5270), and proteases (orthologs of cgd3_4260 and cgd6_60) (Table 5). A CryptoDB search of transcriptome data generated from an in vitro culture of C. parvum previously by real-time PCR [29] indicated that most of the genes are differentially expressed at various developmental stages.
Figure 4.
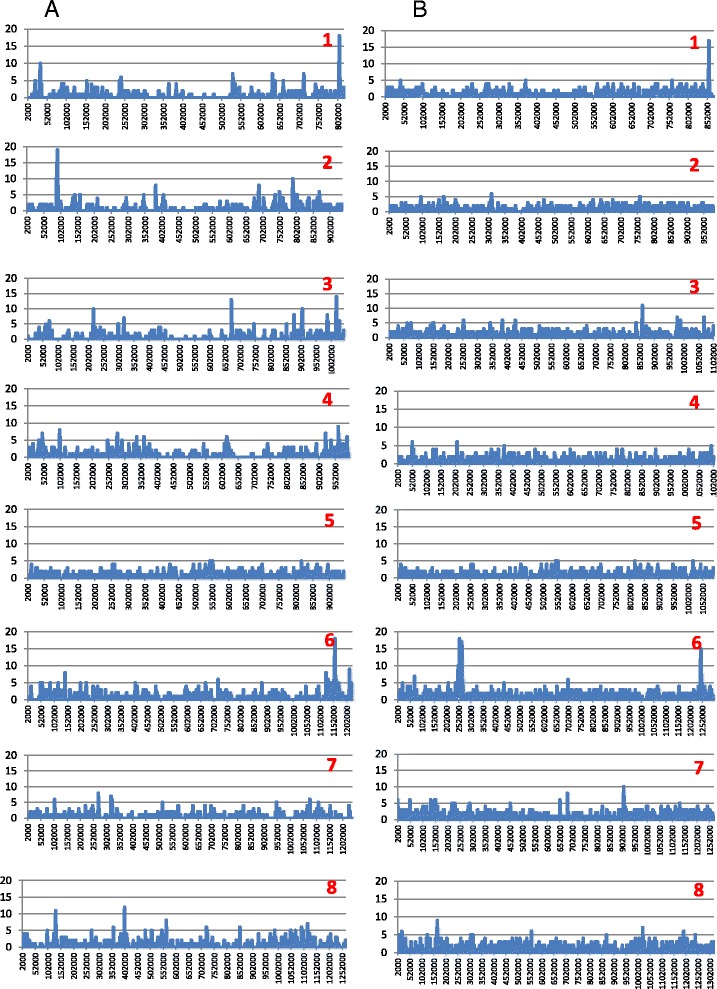
Distribution of SNPs in Cryptosporidium hominis genome by chromosome. The number of SNPs in a sliding window of 2,000 bp with 200 bp steps across each of the eight chromosomes is shown. A. Sequence divergence between specimen 30976 of the IaA28R4 subtype and the published isolate TU502 of the IaA25R3 subtype. B. Sequence divergence between specimen 37999 of the IbA10G2 subtype and specimen 30976 of the IaA10G2 subtype.
Table 5.
Highly polymorphic loci in Cryptosporidium hominis genomes
| Locus | Contig in 30976 | SNP/kb (30976 vs Tu502) | Gene in C. hominis* | Ortholog in C. parvum* | Annotation |
|---|---|---|---|---|---|
| chr1_var1 | contig_5 | 5.0 | Chro.10024 | cgd1_150 | Hypothetical protein with a signal peptide |
| chr1_var2 | contig_5 | 9.0 | Chro.10427 | cgd1_3810 | Conserved hypothetical protein with a signal peptide |
| chr2_var1 | contig_32 | 9.5 | Chro.20050-52 | cgd2_430-450 | Mucin glycoprotein with a signal peptide |
| chr2_var2 | contig_20 | 5.0 | Intergenic downstream of Chro.20394 | Intergenic downstream of cgd2_3690 | WD repeat protein (cgd2_3690) |
| chr3_var1 | contig_59 | 5.0 | Intergenic downstream of Chro.30096 | Within cgd3_720 | Very large mucin with a signal peptide |
| chr3_var2 | contig_31 | 6.5 | Chro.30315 | cgd3_2770 | Hypothetical conserved protein |
| chr3_var3 | contig_293 + contig_255 | 7.0 | Chro.30479 | cgd3_4260 | Insulinase-like protease |
| chr6_var1 | contig_3 | 9.0 | Chro.60606 | cgd6_5270 | Hypothetical protein with a signal peptide |
| chr8_var1 | contig_11 | 5.5 | Chro.80070 | cgd8_550 | Large uncharacterized protein |
| chr8_var2 | contig_2 | 6.0 | Chro.80189 | cgd8_1610 | Sacsin-like HSP90 chaperone domain |
*Additional polymorphic genes identified by comparative analysis of other isolates with C. hominis TU502: Chro.60016 (ortholog of cgd6_60), and Chro.60138 (ortholog of cgd6_1080).
Sequence similarity among sequenced C. hominis genomes and occurrence of genetic recombination
The genomes of the four C. hominis specimens sequenced in this study were similar to each other, except for a subtelomeric region at the 3′ end of chromosome 1 and three regions on chromosome 6. This was supported by SNP analyses through both mapping of Illumina reads to assembled contigs (Figure 4B) and direct comparison of sequence alignments of assembled contigs. At the 3′ end of chromosome 1 (within Chro.10427 or the ortholog of cgd1_3810), two types of sequences were seen among the specimens sequenced in this study: one from the two IbA10G2 specimens (30974 and 37999) and another from the two IaA28R4 specimens (30976 and 33537). Both were very divergent from the sequence in TU502 (Figure 4, Additional file 2: Figure S2). In contrast, at the three loci in chromosome 6, the sequence polymorphism was biallelic, with each genome showing one of the two types of nucleotide sequences, including the reference C. hominis genome (Table 6). Thus at the 5′ end of the chromosome (containing Chro.60016, the ortholog of cgd6_60, coding for a protease), all specimens except for specimen 30974 had sequence identical to the published sequence from TU502 (Additional file 3: Figure S3). At the gp60 locus (Chro.60138, the ortholog of cgd6_1080: a well-known subtyping locus for Cryptosporidium), specimens 30974 and 37999 had the Ib type sequence whereas others, including TU502 had the Ia type sequence. Similarly at the 3′ end of the chromosome downstream of Chro.60606 (the ortholog of cgd6_5270), specimens 30974 and 37999 had sequences similar to the published sequence from TU502, whereas specimens 30976 and 33537 had a different type of sequence (Figure 4, Additional file 2: Figure S2, Table 6). The breakpoints for the three genetic recombination areas occurred at intergenic regions upstream and downstream of cgd6_60 (ortholog: Chro.60016), upstream of cgd6_1000 (ortholog: Chro.60130) and downstream of cgd6_1100 (ortholog: Chro.60142), and upstream of cgd6_5240 (ortholog: Chro.60603) and downstream of cgd6_5320 (its ortholog in C. hominis is unnamed) for the three regions, respectively. Because of the occurrence of genetic recombination, the two IbA10G2 specimens (30974 and 37999) sequenced in the study had different types of sequence at the 5′ end of chromosome 6 (Table 6).
Table 6.
Genetic recombination in chromosome 6 of two virulent Cryptosporidium hominis subtypes
| Specimen (subtype) | Sequence characteristics | ||
|---|---|---|---|
| 5′ end (cgd6_60) | gp60 area (cgd6_1000-cgd6_1100) | 3′ end (cgd6_5240-cgd6_5320) | |
| 30974 (IbA10G2) | IbA10G2 | IbA10G2 | IaA25R3 |
| 37999 (IbA10G2) | IaA25R3* | IbA10G2 | IaA25R3 |
| 30976 (IaA28R4) | IaA25R3 | IaA28R4 | IaA28R4 |
| 33537 (IaA28R4) | IaA25R3 | IaA28R4 | IaA28R4 |
| TU502 (IaA25R3) | IaA25R3 | IaA25R3 | IaA25R3 |
*15/16 SNPs at the 3′ end of cgd6_60 are unique in 37999.
Intra-specimen sequence diversity at the trinucleotide repeat region of gp60
Because of a recent report on intra-specimen genetic heterogeneity seen in Illumina sequencing of a PCR-WGA product from a C. parvum specimen [30], we examined intra-specimen sequence diversity at the trinucleotide repeat region of the gp60 gene in the four C. hominis specimens sequenced in this study. In the specimens sequenced by using 454 technology, 205 and 310 sequence reads mapped to the gp60 gene for specimens 30974 and 33537, respectively. Among them, 78 and 59 reads had sequences fully covering the entire trinucleotide repeats for the IbA10G2 and IaA28R4 subtypes, respectively. No intra-specimen sequence diversity was seen (Additional file 4: Figure S4, Additional file 5: Figure S5). Similarly, 2,781 and 5,576 sequence reads mapped to the gp60 gene in specimens 37999 and 30976 sequenced by using Illumina, respectively. Among them, 73 and 30 reads had sequences fully covering the entire trinucleotide repeats for the IbA10G2 and IaA28R4 subtypes, respectively. No intra-specimen diversity was seen in specimen 37999, whereas in 30976 (of the IaA28R4 subtype), 28 reads had 28 copies of the TCA repeat, one had 27 copies of the TCA repeat, and one had 29 copies of the TCA repeat (data not shown).
Discussion
Genome similarity between C. hominis and C. parvum, gene deletions, and species-specific genes
Results of this study have confirmed the genetic similarity between the almost fully sequenced C. parvum and C. hominis genomes. The genomes of the two species are nearly 97% similar in nucleotide sequences, with complete synteny in gene organization. This is similar to the previous conclusion based on comparison of the fully assembled genome of the C. parvum IOWA isolate and the more fragmented genome from the C. hominis TU502 isolate [24,25]. Some potential genetic rearrangements in several chromosomes were observed in the current study, but they all occurred in the ten sequence gaps and several sequence ambiguity areas in the reference C. parvum genome. As there are no HAPPY maps and genomic libraries with large inserts for C. hominis, the observations on genome organization of C. hominis need to be supported by PacBio sequencing. Nevertheless, comparative genomic analysis in this study has identified several major deletions and one insertion in C. hominis, which were overlooked in previous studies probably because of the fragmented nature of the published C. hominis genome. The significance of these gene insertions and deletions (indels) is not clear. Because of the high sequence similarity in most genes between C. parvum and C. hominis, these major indels could potentially be responsible for some biological differences between C. parvum and C. hominis.
Gene duplication and interallelic recombination could contribute to the gene expansion and losses seen between C. parvum and C. hominis genomes. Most of the genes deleted in the C. hominis genome are members of multigene families and have paralogs nearby. Thus, of the six MEDLE family of secreted protein genes possibly present in tandem in C. parvum (cgd5_4580, cgd5_4590, cgd5_4600, cgd5_4610, cgd6_5480, and cgd6_ 5490), only one, the ortholog of cgd5_4600, is present in C. hominis. Similarly, two genes (cgd6_5510 and cgd6_5520) that code for insulinase-like are absent in C. hominis. The subtelomeric locations of these genes facilitate the expansion and deletions of multicopy genes by interallelic recombination. Sequence homology is probably also involved in the loss of cgd8_680 and cgd8_690 orthologs in chromosome 8 of C. hominis, as the ~100 bp region upstream of the fragment containing the two genes and the ~100 bp region downstream of the fragment have almost identical sequences. The sequence homology in two nearby regions could have resulted in the deletion of the two genes in C. hominis during species evolution. As cgd8_690 is a paralog of cgd8_660 and has some sequence similarity to the 5′ end of cgd8_670, this gene loss in chromosome 8 of C. hominis also involves a multigene family. In compact apicomplexan genomes with mostly single copy genes, members of multigene families usually play very important biological functions [31,32]. The function of the MEDLE family of secreted proteins in apicomplexan parasites has not been examined. However, insulinase-like proteases have been shown recently to be rhoptry or microneme-associated in Toxoplasma gondii and are probably involved in cell invasion [33,34]. Indeed, both cgd6_5510 and cgd6_5520 have peak expression during the invasion process. The expression of cgd6_5480 and cgd6_ 5490 in C. parvum may also be developmentally regulated, as they showed identical expression patterns in in vitro culture [29]. As sequence differences in non-coding regulatory elements can also affect the timing or expression levels of invasion-associated proteins, more studies are needed to determine whether the duplications of MEDLE and insulinase genes are indeed the cause of the host expansion of in C. parvum.
Compared to the deletion of at least nine genes, C. hominis appears to have only one unique gene that is absent in C. parvum. This gene, Chro.50011, is located at the 3′ end of chromosome 3 instead of the original annotation at the 5′ end of chromosome 5 (Figure 2A). It codes for a 489 aa hypothetical protein that contains RS and HS repeats at the carboxyl end, and has recently been identified as a C. hominis–specific gene, Chos-1, by Bouzid and colleagues [35]. Although the function of the protein is not clear, it has been suggested that this protein is a member of a new Cryptosporidium-specific protein family that are candidate mediators of host specificity and virulence [35]. It remains to be determined whether the C. hominis genome codes for additional species-specific genes in areas of the ten sequence gaps in the C. parvum IOWA genome.
Sequence similarity among C. hominis genomes and genetic recombination in virulent C. hominis subtypes
As expected, much higher genetic similarity is present among C. hominis genomes. The four C. hominis specimens sequenced in this study had whole genome sequences that are 99.72-99.83% similar to the published C. hominis genome from TU502. Genes coding for some secreted proteins (especially mucins) and proteases contribute more to the sequence differences than others, suggesting they are under selection and therefore may serve as good targets for the development of diagnostic tools and intervention measures. For example, some of the polymorphic mucin genes such as cgd2_430 (Mucin5) and cgd6_1080 (gp60) are well known targets of host immune responses [36,37] and have been used widely in subtyping C. parvum and C. hominis [10]. Proteases (especially cysteine proteases) and protein kinases have been recently shown to play important roles in cell invasion of Cryptosporidium and thus have been used as common targets in the development of therapeutic treatments [38-41].
In contrast to the relatively high nucleotide sequence differences between the genomes sequenced in this study and the published C. hominis TU502 genome, the genomes of four specimens from two virulent C. hominis subtypes (IbA10G2 and IaA28R4) in the United States are very similar to each other except for the 3′ end of chromosome 1 and three areas in chromosome 6. In particular, chimeric sequences were seen in chromosome 6 (Table 6), indicating the occurrence of genetic recombination in the two subtypes. One of the three areas with genetic recombination is where gp60 (cgd6_1080) is located, a locus widely known for its extremely high sequence diversity and occurrence of genetic recombination [42]. Recently, population genetic analyses of chromosome 6 sequences have shown the exclusive occurrence of genetic recombination in the virulent C. hominis subtypes IbA10G2 and IaA28R4, especially around gp60 [43,44]. It was postulated that the fitness of the two subtypes as a result of genetic recombination was likely responsible for the wide dissemination of IbA10G2 around the world and the emergence of IaA28R4 in the United States. The two IbA10G2 specimens sequenced in this study also differ from each other at the 5′ end of chromosome 6, especially in the ortholog of cgd6_60 (coding for a protease) as a result of genetic recombination. It was previously shown by MLST analysis of chromosome 6 that IbA10G2 specimens from different areas are genetically different [45]. Although the two IaA28R4 specimens sequenced in this study are mostly identical, data from a recent population genetic study of IaA28R4 specimend in the United States suggest that there are at least two origins of the subtype [44]. Therefore, multiple genetic recombination events are probably involved in the evolution of both IbA10G2 and IaA28R4 and are likely responsible for the observed emergence of the same virulent gp60 subtypes in different geographical locations in response to selection pressure [46]. The occurrence of genetic recombination in virulent C. hominis subtypes also suggests that the widely used gp60-based typing alone is insufficient in molecular epidemiologic characterizations of field specimens, as pointed out previously [46]. Therefore, the use of MLST and other multilocus subtyping tools can provide new insights into the transmission of Cryptosporidium spp. [44,47-49]. As expected, the three loci in chromosome 6 where genetic recombination occurs, cgd6_60, cgd6_1080, and cgd6_5270 (coding for a hypothetical protein with a signal peptide and paralogs) are all highly polymorphic in the present study. The biological functions of proteins coded by cgd6_60 and cgd6_5270 thus should be studied.
Conclusion
In conclusion, this comparative genomic analysis has revealed some unique genetic differences between C. parvum and C. hominis and identified some multigene families that can potentially contribute to differences in host specificity of the two closely related species. It has further supported the potential role of genetic recombination in the emergence and evolution of virulent C. hominis subtypes. Improvements in knowledge in these two areas are still hampered by the lack of genomic studies of other Cryptosporidium species of significant public health and economic importance, the incompleteness of the reference C. parvum and C. hominis genomes, and poor understanding of the functions of the thousands of hypothetical proteins in Cryptosporidium genomes and regulatory elements in non-coding areas. With the increased recognition of the importance of cryptosporidiosis in pediatric health in developing countries [3], common occurrence of large waterborne outbreaks in industrialized nations [15,16,50], and a major increase in cryptosporidiosis incidence in the United States in recent years [6,8,9], more effort should be directed toward studies on functional genomics and the basic biology of Cryptosporidium spp. [51].
Methods
Cryptosporidium specimens
Four C. hominis specimens were used in whole genome sequencing in the study: specimens 30974 and 37999 of the IbA10G2 subtype and 30976 and 33537 of the IaA28R4 subtype. Specimen 30974 was collected from a patient from a cryptosporidiosis outbreak in July 2010 in Columbia, South Carolina associated with a splash pad that had problems with filtration and chlorination. Testing of filter backflush and stools from six patients all identified the presence of the C. hominis IbA10G2 subtype. Specimen 30976 was collected from a patient in a cryptosporidiosis outbreak in July 2010 in the St. Louis area in Illinois and Missouri associated with swimming pools and a water park. Testing of nine patient specimens identified the occurrence of C. hominis IaA28R4 in seven patients, IaA24R4 in one patient, and IdA15G1 in another patient. Specimen 33537 was collected from a patient from a cryptosporidiosis outbreak in July 2011 in Walsenburg, Colorado associated with a waterpark that had problems with the chlorinator. Testing of filter backflush and stools from five patients identified IaA28R4 in all. Specimen 37999 was collected from a sporadic cryptosporidiosis patient in Twin Falls, Idaho in September 2012. All stool specimens were collected fresh from symptomatic patients and stored in 2.5% potassium dichromate at 4°C prior to being used in Cryptosporidium oocyst isolation for whole genome sequencing within 6 months. Cryptosporidium species and subtypes were determined by PCR-RFLP analysis of the small subunit rRNA and sequence analysis of the 60 kDa glycoprotein (gp60) genes, respectively [17].
Oocyst isolation and whole genome amplification
Cryptosporidium oocysts were isolated from stool specimens by discontinuous sucrose and cesium chloride gradients as previously described [52]. They were further purified by immunomagnetic separation using the Dynabeads Anti-Cryptosporidium kit (Invitrogen, Carlsbad, CA). After treating the purified oocysts with 10% commercial bleach on ice for 10 min and five cycles of freezing and thawing, DNA was extracted from them by using the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). Whole genome amplification (WGA) of the 25–100 ng of extracted DNA was conducted by using the REPLI-g Midi Kit (Qiagen). The quality of the WGA products was verified by sequencing BamHI-digested WGA products cloned into a pUC19 vector (Fermantas, Pittsburgh, PA). The sequencing was done by using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
454 and Illumina sequencing and de novo contig assembly
The WGA products from specimens 30974 and 33537 were sequenced with 454 technology on a GS-FLX Titanium System (Roche, Branford, CT) by using approximately 1 μg of DNA for library construction and following standard Roche library protocols, with an average insert size of 600 bp. One full PTP plate was used in the analysis of each specimen. The sequence reads from each run were assembled using Newbler in the GS De Novo Assembler (http://www.454.com/products/analysis-software/) with the default settings.
The WGA products from specimens 30976 and 37999 were used to generate Illumina TruSeq (v3) libraries (average insert size: 350 bp) and sequenced 100×100 bp paired-end on an Illumina Genome Analyzer IIx (Illumina, San Diego, CA). The sequence reads with a minimum quality of 20 were trimmed by using CLC Assembly Cell 4.1.0 (http://www.clcbio.com/products/clc-assembly-cell/). The data were then assembled with default parameters and a minimum contig length of 500 bp, with scaffolding using paired-end data.
Comparative genomic analyses
For comparisons of sequences at the genome level, contigs of each specimen were aligned with reference sequences of the near complete genome of the C. parvum IOWA isolate (version AAEE00000000.1) and the 1,422 contigs of the C. hominis TU5205 isolate (version NZ_AAEL00000000.1) using Nucmer, a tool in MUMmer 3.23 (http://mummer.sourceforge.net/) [53]. Multiple genome alignments were also constructed by using the progressive alginment algorithm of the Mauve 2.3.1 (http://asap.genetics.wisc.edu/software/mauve/) with default options [54]. In-house perl scripts were developed to calculate the average nucleotide identities. For the detection of SNPs, Fastqc 0.10.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for the QC analysis of Illumina sequence reads, and PRINSEQ 0.20.3 (http://prinseq.sourceforge.net/) [55] was used to remove low quality reads, with a min_qual_mean setting of 20 and min_len of 65. Reads were then aligned to reference sequences by using Bowtie 0.12.7 (http://bowtie-bio.sourceforge.net/index.shtml) [56]. The resulting SAM files were processed, sorted and duplicates were removed by using Picard 1.126 (http://broadinstitute.github.io/picard/). The mpileup in SAMtools (http://samtools.sourceforge.net/) was finally used to create the pileup file for SNP variant calls using the mpileup2snp in VarScan 2.3.7 (http://varscan.sourceforge.net/) [57]. Default parameters for VarScan were used except that min-avg-qual was set to 30.
PCR verification
As the comparative genomic analysis had identified some nucleotide sequences (AAEL01000413, AAEL01000728, and AAEL01000717) in the published C. hominis that had not been seen in the published C. parvum genome, primers were designed based on these sequences to verify the source of these sequences by PCR (Additional file 6: Table S1). Five specimens each of C. parvum and C. hominis were used in PCR analysis of each target. In addition, two C. andersoni specimens were used in confirmation of Cryptosporidium-origin of contig AAEL01000728. Each specimen was analyzed in duplicate nested PCR using 50 μl PCR mixture consisting of 1 μl (~100 ng) of extracted DNA or 2 μL of primary PCR products (in secondary PCR), 200 μM deoxynucleoside triphosphate, 1× PCR buffer (Applied Biosystems), 3.0 mM MgCl2, 5.0 U of Taq polymerase (Promega, Madison, WI), 100 nM primers, and 400 ng/μl of non-acetylated bovine serum albumin (Sigma-Adrich, St. Louis, MO). The primary and secondary PCR reactions were performed in a GeneAmp PCR 9700 thermocycler (Applied Biosystems) for 35 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s, with an initial denaturation (94°C for 5 min) and a final extension (72°C for 7 min). The secondary PCR products were sequenced in both directions using Sanger technology described above. Nucleotide sequences obtained were aligned with reference sequences downloaded from GenBank by using ClustalX (http://www.clustal.org/).
NCBI BioProject No.
Nucleotide sequences generated from the project, including all SRA data and assembled contigs, were submitted to the NCBI BioProject under the accession number PRJNA252787.
Ethics statement
The study was done on delinked residual diagnostic specimens. It was covered by Human Subjects Protocol No. 990115 “Use of residual human specimens for the determination of frequency of genotypes or sub-types of pathogenic parasites”, which was reviewed and approved by the Institutional Review Board of the Centers for Disease Control and Prevention (CDC). No personal identifiers were associated with the specimens at the time of submission for diagnostic service at CDC.
Additional files
Coverage of two Roche 454 sequenced genomes of Cryptosporidium hominis comparing to the published C. parvum (IOWA) and C. hominis (TU502) genomes. The eight chromosomes of C. parvum are numbered and assembled contigs are bordered by vertical red lines. The color blocks are conserved segments of sequences internally free from genome rearrangements, whereas the inverted white peaks within each block are sequence divergence between the reference C. parvum genome and C. hominis genome under analysis.
Sequence divergence between Cryptosporidium hominis IaA28R4 (specimen 30976) and IaA25R3 (isolate TU502) subtypes by chromosome. The number of the SNPs in a sliding window of 2,000 bp with 200 bp steps across each of the eight chromosomes is shown.
Polymorphic nucleotide sequences in mid part of the Chro.60016 (ortholog of cgd6_60, coding for a protease) gene in Cryptosporidium hominis specimens. Dots denote sequence identity to the reference TU502 sequence. Specimen 37999 also has different sequence at the 3′ end of the gene (not shown).
Lack of variation in sequence diversity in the trinucleotide repeat region in the gp60 gene of specimen 30974 of the Cryptosporidium hominis IbA10G2 subtype. Of 205 reads from 454 sequencing that mapped to gp60, 78 had complete sequence of the trinucleotide repeats, with no variation in repeat numbers. Dots denote sequence identity to the reference sequence, whereas dashes denote deletions of nucleotides.
Lack of variation in sequence diversity in the trinucleotide repeat region in the gp60 gene of specimen 33537 of the Cryptosporidium hominis IaA28R4 subtype. Of 310 reads from 454 sequencing that mapped to gp60, 59 had complete sequence of the trinucleotide repeats, with no variation in repeat numbers. Dots denote sequence identity to the reference sequence, whereas dashes denote deletions of nucleotides.
Primers used in the PCR verification of possible Cryptosporidium hominis-unique nucleotide sequences.
Footnotes
Yaqiong Guo and Kevin Tang contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Acknowledgements
We thank Scott Sammons of the Centers for Disease Control and Prevention for technical assistance. This work was supported by the National Natural Science Foundation of China (31229005, 31425025 and 31110103901), National Special Fund for State Key Laboratory of Bioreactor Engineering, China (No.2060204), and Centers for Disease Control and Prevention, USA. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Authors’ contributions
YF and LX conceived and designed the experiments; YG, LAR, NL, DMR, and MF performed the experiments; YG, KT, KK, CY, and LX analyzed the data; YG, KT, YF and LX wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Yaqiong Guo, Email: guoyaqiong1987@sina.com.
Kevin Tang, Email: kht7@cdc.gov.
Lori A Rowe, Email: ioy0@cdc.gov.
Na Li, Email: nli@ecust.edu.cn.
Dawn M Roellig, Email: iyd4@cdc.gov.
Kristine Knipe, Email: wgg9@cdc.gov.
Michael Frace, Email: mff3@cdc.gov.
Chunfu Yang, Email: cxy0@cdc.gov.
Yaoyu Feng, Email: yyfeng@ecust.edu.cn.
Lihua Xiao, Email: lxiao@cdc.gov.
References
- 1.Chalmers RM, Davies AP. Minireview: clinical cryptosporidiosis. Exp Parasitol. 2010;124:138–46. doi: 10.1016/j.exppara.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Mor SM, Tzipori S. Cryptosporidiosis in children in sub-saharan Africa: a lingering challenge. Clin Infect Dis. 2008;47(7):915–21. doi: 10.1086/591539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet. 2013;382(9888):209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2004–2010. Water Res. 2011;45(20):6603–14. doi: 10.1016/j.watres.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoder JS, Beach MJ. Cryptosporidiosis surveillance–United States, 2003–2005. MMWR Surveill Summ. 2007;56(7):1–10. [PubMed] [Google Scholar]
- 7.Hlavsa MC, Watson JC, Beach MJ. Cryptosporidiosis surveillance–United States 1999–2002. MMWR Surveill Summ. 2005;54(1):1–8. [PubMed] [Google Scholar]
- 8.Yoder JS, Harral C, Beach MJ. Cryptosporidiosis surveillance - United States, 2006–2008. MMWR Surveill Summ. 2010;59(6):1–14. [PubMed] [Google Scholar]
- 9.Yoder JS, Wallace RM, Collier SA, Beach MJ, Hlavsa MC. Cryptosporidiosis surveillance - United States, 2009–2010. MMWR Surveill Summ. 2012;61(5):1–12. [PubMed] [Google Scholar]
- 10.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–9. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Thomas E, Watkins J, et al. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north west Wales. J Water Health. 2010;8(2):311–25. doi: 10.2166/wh.2009.185. [DOI] [PubMed] [Google Scholar]
- 12.Mayne DJ, Ressler KA, Smith D, Hockey G, Botham SJ, Ferson MJ. A community outbreak of cryptosporidiosis in sydney associated with a public swimming facility: a case–control study. Interdis Perspect Infect Dis. 2011;2011:341065. doi: 10.1155/2011/341065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng JS, Pingault N, Gibbs R, Koehler A, Ryan U. Molecular characterisation of Cryptosporidium outbreaks in Western and South Australia. Exp Parasitol. 2010;125(4):325–8. doi: 10.1016/j.exppara.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Waldron LS, Ferrari BC, Cheung-Kwok-Sang C, Beggs PJ, Stephens N, Power ML. Molecular epidemiology and spatial distribution of a waterborne cryptosporidiosis outbreak in Australia. Appl Environ Microbiol. 2011;77(21):7766–71. doi: 10.1128/AEM.00616-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournet N, Deege MP, Urbanus AT, Nichols G, Rosner BM, Chalmers RM, et al. Simultaneous increase of Cryptosporidium infections in the Netherlands, the United Kingdom and Germany in late summer season, 2012. Euro Surveill. 2013;18(2):20348. [PubMed] [Google Scholar]
- 16.Widerstrom M, Schonning C, Lilja M, Lebbad M, Ljung T, Allestam G, et al. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply. Sweden Emerg Infect Dis. 2014;20(4):581–9. doi: 10.3201/eid2004.121415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao L, Hlavsa MC, Yoder J, Ewers C, Dearen T, Yang W, et al. Subtype analysis of Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa in 2007: widespread occurrence of one Cryptosporidium hominis subtype and case history of an infection with the Cryptosporidium horse genotype. J Clin Microbiol. 2009;47(9):3017–20. doi: 10.1128/JCM.00226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantey PT, Kurian AK, Jefferson D, Moerbe MM, Marshall K, Blankenship WR, et al. Outbreak of cryptosporidiosis associated with a man-made chlorinated lake–Tarrant County, Texas, 2008. J Environ Health. 2012;75(4):14–9. [PubMed] [Google Scholar]
- 19.Valderrama AL, Hlavsa MC, Cronquist A, Cosgrove S, Johnston SP, Roberts JM, et al. Multiple risk factors associated with a large statewide increase in cryptosporidiosis. Epidemiol Infect. 2009;137:1781–8. doi: 10.1017/S0950268809002842. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease CaP Outbreak of cryptosporidiosis associated with a splash park - Idaho, 2007. MMWR Morb Mortal Wkly Rep. 2009;58(22):615–8. [PubMed] [Google Scholar]
- 21.Zhu G, Xiao L. Cryptosporidium species. In: Fratamico P, Liu Y, Kathariou S, editors. Genomes of Foodborne and Waterborne Pathogens. Washington, DC: American Society for Microbiology; 2011. pp. 271–86. [Google Scholar]
- 22.Widmer G, Sullivan S. Genomics and population biology of Cryptosporidium species. Parasite Immunol. 2012;34(2–3):61–71. doi: 10.1111/j.1365-3024.2011.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalmers RM, Katzer F. Looking for Cryptosporidium: the application of advances in detection and diagnosis. Trends Parasitol. 2013;29(5):237–51. doi: 10.1016/j.pt.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, et al. Complete genome sequence of the apicomplexan. Cryptosporidium parvum Sci. 2004;304(5669):441–5. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 25.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, et al. The genome of Cryptosporidium hominis. Nature. 2004;431(7012):1107–12. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 26.Widmer G, Lee Y, Hunt P, Martinelli A, Tolkoff M, Bodi K. Comparative genome analysis of two Cryptosporidium parvum isolates with different host range. Infect Genet Evol. 2012;12(6):1213–21. doi: 10.1016/j.meegid.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper MB, Bankier AT, Dear PH. A HAPPY map of Cryptosporidium parvum. Genome Res. 1998;8(12):1299–307. doi: 10.1101/gr.8.12.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao L, Ryan UM. Molecular epidemiology. In: Fayer R, Xiao L, editors. Cryptosporidium and Cryptosporidiosis. 2. Boca Raton, FL: CRC Press and IWA Publishing; 2008. pp. 119–71. [Google Scholar]
- 29.Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS, Rutherford MS. The Cryptosporidium parvum transcriptome during in vitro development. PLoS One. 2012;7(3):e31715. doi: 10.1371/journal.pone.0031715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grinberg A, Biggs PJ, Dukkipati VS, George TT. Extensive intra-host genetic diversity uncovered in Cryptosporidium parvum using Next Generation Sequencing. Infect Genet Evol. 2013;15:18–24. doi: 10.1016/j.meegid.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 31.DeBarry JD, Kissinger JC. A Survey of innovation through duplication in the reduced genomes of twelve parasites. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0099213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anantharaman V, Iyer LM, Balaji S, Aravind L. Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: insights from comparative genomics. Int Rev Cytol. 2007;262:1–74. doi: 10.1016/S0074-7696(07)62001-4. [DOI] [PubMed] [Google Scholar]
- 33.Hajagos BE, Turetzky JM, Peng ED, Cheng SJ, Ryan CM, Souda P, et al. Molecular dissection of novel trafficking and processing of the Toxoplasma gondii rhoptry metalloprotease toxolysin-1. Traffic. 2012;13(2):292–304. doi: 10.1111/j.1600-0854.2011.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laliberte J, Carruthers VB. Toxoplasma gondii toxolysin 4 is an extensively processed putative metalloproteinase secreted from micronemes. Mol Biochem Parasitol. 2011;177(1):49–56. doi: 10.1016/j.molbiopara.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouzid M, Hunter PR, McDonald V, Elwin K, Chalmers RM, Tyler KM. A new heterogeneous family of telomerically encoded Cryptosporidium proteins. Evol Appl. 2013;6(2):207–17. doi: 10.1111/j.1752-4571.2012.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connor RM, Burns PB, Ha-Ngoc T, Scarpato K, Khan W, Kang G, et al. The polymorphic mucin antigens CpMuc4 and CpMuc5 are integral to Cryptosporidium parvum infection in vitro. Eukaryot Cell. 2009;8:461–9. doi: 10.1128/EC.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ajjampur SS, Sarkar R, Allison G, Banda K, Kane A, Muliyil J, et al. Serum IgG response to Cryptosporidium immunodominant antigen gp15 and polymorphic antigen gp40 in children with cryptosporidiosis in South India. Clin Vaccine Immunol. 2011;18(4):633–9. doi: 10.1128/CVI.00464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndao M, Nath-Chowdhury M, Sajid M, Marcus V, Mashiyama ST, Sakanari J, et al. A cysteine protease inhibitor rescues mice from a lethal Cryptosporidium parvum infection. Antimicrob Agents Chemother. 2013;57(12):6063–73. doi: 10.1128/AAC.00734-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang JM, Ju HL, Yu JR, Sohn WM, Na BK. Cryptostatin, a chagasin-family cysteine protease inhibitor of Cryptosporidium parvum. Parasitology. 2012;139(8):1029–37. doi: 10.1017/S0031182012000297. [DOI] [PubMed] [Google Scholar]
- 40.Keyloun KR, Reid MC, Choi R, Song Y, Fox AM, Hillesland HK, Zhang Z, Vidadala R, Merritt EA, Lau AO, et al. The gatekeeper residue and beyond: homologous calcium-dependent protein kinases as drug development targets for veterinarian Apicomplexa parasites. Parasitology. 2014;141(11):1499–1509. doi: 10.1017/S0031182014000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellanos-Gonzalez A, White AC, Jr, Ojo KK, Vidadala RS, Zhang Z, Reid MC, et al. A novel calcium-dependent protein kinase inhibitor as a lead compound for treating cryptosporidiosis. J Infect Dis. 2013;208(8):1342–8. doi: 10.1093/infdis/jit327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leav BA, Mackay MR, Anyanwu A, RM OC, Cevallos AM, Kindra G, et al. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect Immun. 2002;70(7):3881–90. doi: 10.1128/IAI.70.7.3881-3890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, et al. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis. 2013;19(10):1573–82. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Y, Tiao N, Li N, Hlavsa M, Xiao L. Multilocus sequence typing of an emerging Cryptosporidium hominis subtype in the United States. J Clin Microbiol. 2014;52(2):524–30. doi: 10.1128/JCM.02973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gatei W, Barrett D, Lindo JF, Eldemire-Shearer D, Cama V, Xiao L. Unique Cryptosporidium population in HIV-Infected persons. Jamaica Emerg Infect Dis. 2008;14(5):841–3. doi: 10.3201/eid1405.071277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widmer G. Meta-analysis of a polymorphic surface glycoprotein of the parasitic protozoa Cryptosporidium parvum and Cryptosporidium hominis. Epidemiol Infect. 2009;137:1800–8. doi: 10.1017/S0950268809990215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int J Parasitol. 2013;43(14):1141–7. doi: 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Drumo R, Widmer G, Morrison LJ, Tait A, Grelloni V, D'Avino N, et al. Evidence of host-associated populations of Cryptosporidium parvum in Italy. Appl Environ Microbiol. 2012;78(10):3523–9. doi: 10.1128/AEM.07686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herges GR, Widmer G, Clark ME, Khan E, Giddings CW, Brewer M, et al. Evidence that Cryptosporidium parvum populations are panmictic and unstructured in the Upper Midwest of the United States. Appl Environ Microbiol. 2012;78(22):8096–101. doi: 10.1128/AEM.02105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hlavsa MC, Roberts VA, Anderson AR, Hill VR, Kahler AM, Orr M, et al. Surveillance for waterborne disease outbreaks and other health events associated with recreational water –- United States, 2007–2008. MMWR Surveill Summ. 2011;60(12):1–32. [PubMed] [Google Scholar]
- 51.Striepen B. Parasitic infections: time to tackle cryptosporidiosis. Nature. 2013;503(7475):189–91. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- 52.Arrowood MJ, Donaldson K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol. 1996;43(5):89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- 53.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics (Oxford, England) 2011;27(6):863–4. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]


