Abstract
Long noncoding RNAs (lncRNAs) are a novel class of RNA molecules defined as transcripts longer than 200 nucleotides that lack protein coding potential. They constitute a major, but still poorly characterized part of human transcriptome, however, evidence is growing that they are important regulatory molecules involved in various cellular processes. It is becoming increasingly clear that many lncRNAs are deregulated in cancer and some of them can be important drivers of malignant transformation. On the one hand, some lncRNAs can have highly specific expression in particular types of cancer making them a promising tool for diagnosis. The expression of other lncRNAs can correlate with different pathophysiological features of tumor growth and with patient survival, thus making them convenient biomarkers for prognosis. In this review we outline the current state of knowledge about the fast growing field of application of lncRNAs as tumor biomarkers.
Keywords: long noncoding RNA, biomarkers, cancer
Introduction
The development of high resolution microarray and genome wide sequencing technologies allowed comprehensive characterization of mammalian transcriptomes. The main conclusion from the pioneering in-depth transcriptome studies performed by FANTOM (Functional Annotation Of Mouse genome) and ENCODE (Encyclopedia of DNA Elements) consortia was discovery of the pervasive transcription of the mouse and human genomes with protein coding mRNAs constituting only a minor fraction of the transcribed sequences (Okazaki et al., 2002; Carninci et al., 2005; Birney et al., 2007). The most recent data from ENCODE consortium indicates that around 70% of human genome is transcribed, generating a vast range of noncoding RNAs (Djebali et al., 2012). Based on transcript size noncoding RNAs are classified into small noncoding RNAs (<200 nt) and long noncoding RNAs (lncRNAs; >200 nt). Small noncoding RNAs, particularly miRNAs are well characterized as post-transcriptional regulators of mRNAs and have well established roles in cancer (Garzon et al., 2009; Fabian and Sonenberg, 2012; Kong et al., 2012).
On the other hand, lncRNAs still remain poorly characterized, however, evidence for their importance and functionality is mounting. The number of known lncRNA genes is still rising as the process of their annotation is ongoing. The current version of GENCODE (encyclopedia of genes and gene variants) includes 15,877 human lncRNA genes encoding 26,414 transcripts based on manual curation, computational analysis, and experimental validation (Version 21, June 2014 freeze, GRCh38 – Ensembl 77; Dunham et al., 2012; www.gencodegenes.org).
Long noncoding RNAs share common features with mRNAs, such as many of them are transcribed by RNA polymerase II, undergo splicing, polyadenylation, and 5′-capping (Table 1). Similarly, to protein coding genes, lncRNA genes have histone marks of active promoters (H3K4me2/3, H2K9ac, H3K27ac) and actively transcribed gene bodies (H3K36me3; Guttman et al., 2009; Derrien et al., 2012). On the other hand, lncRNAs have several distinct features that distinguish them from protein coding mRNAs (Table 1). LncRNAs generally lack or have very little open reading frames (ORFs). Commonly, lncRNA transcripts are shorter and have fewer exons. Unlike mRNAs, which are mostly transported to the cytoplasm, the majority of lncRNAs are localized to the nucleus (Derrien et al., 2012). LncRNAs demonstrate relatively lower expression levels than protein coding genes, however, they exhibit more specific tissue expression patterns (Derrien et al., 2012).
Table 1.
Comparison of several important features of mRNAs and long noncoding RNAs (lncRNAs) based on GENCODE v7 catalog of human transcripts*.
| Feature | mRNAs | lncRNAs |
|---|---|---|
| Median transcript length | 2453 nt | 592 nt |
| Median number of exons | 3 | 8 |
| Expression levels | Higher | Lower |
| 5’ end supported by CAGE | 55% | 15% |
| 3’ end supported by polyA signal | 51% | 39% |
| Tissue specificity | Lower | Higher |
| Subcellular localization | Mostly cytoplasmic | Mostly nuclear |
*The data shown in Table 1 are taken from Derrien et al. (2012).
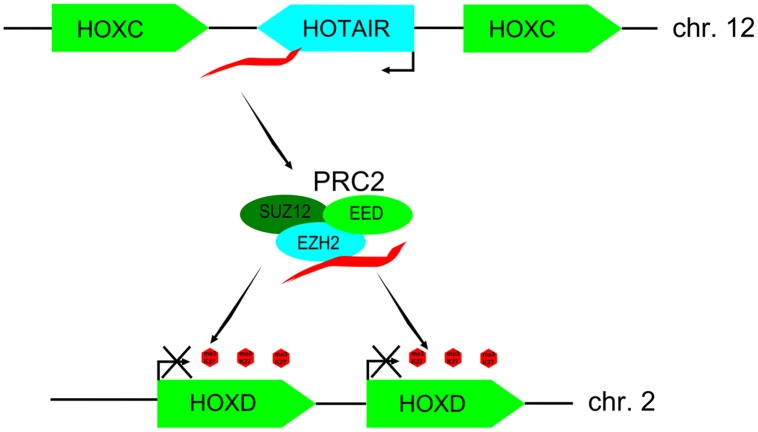
Contrary to the initial view that lncRNAs might be a mere transcriptional noise, several lines of evidence indicate to their functionality (Ponting and Belgard, 2010). Firstly, many lncRNAs demonstrate clear signs of evolutionary conservation and selection (Ponjavic et al., 2007; Guttman et al., 2009). Secondly, expression of many lncRNAs is developmentally and temporally regulated and restricted to certain tissues (Cawley et al., 2004; Ravasi et al., 2006; Mercer et al., 2008). It is becoming increasingly clear that lncRNAs are important regulatory molecules acting at different levels of gene expression, such as chromatin remodeling, transcription, stability, posttranscriptional modifications, and translation. For example, lncRNAs Xist and HOTAIR (HOX transcript antisense RNA) recruit chromatin remodeling complexes, such as Polycomb repressive complex 2 (PRC2) to induce heterochromatin state, thus repressing gene expression at target loci (Zhao et al., 2008; Gupta et al., 2010; Figure 1). Indeed, a considerable proportion of lncRNAs has been shown to associate with PRC2 complex, implying that it might be one of the prevalent mechanisms for their functionality (Khalil et al., 2009). LncRNAs directly regulate transcription by different mechanisms, such as via interaction with RNA binding proteins (Wang et al., 2008), repression of promoters (Martianov et al., 2007), acting as co-activators of transcription factors (Feng et al., 2006), or negatively regulating transcription factors by sequestration (Kino et al., 2010; Hung et al., 2011). At the level of pre-mRNA processing, lncRNA MALAT1 can regulate alternative splicing by interacting with several serine/arginine (SR) splicing factors (Tripathi et al., 2010).
FIGURE 1.
Model of long noncoding RNA (lncRNA) HOX transcript antisense RNA (HOTAIR) regulating expression of HOX genes in trans. LncRNA HOTAIR transcribed from the HOXC cluster of genes (chr. 2) binds PRC2 complex of polycomb-group of proteins and targets it to the HOXD cluster (chr. 12) leading to H3K27 methylation and silencing of neighboring HOXD genes. This figure is adapted from Rinn et al., 2007.
Long noncoding RNAs can interact with mRNAs and modulate their translation both positively and negatively. For example, lincRNA-p21 interacts with JUNB and CTNNB1 mRNAs and selectively lowers their translation through a mechanism that includes reduced polysome sizes or ribosome ‘drop-off’ (Yoon et al., 2012). In contrast, lncRNA antisense to Uchl1 gene increases translation of UCHL1 protein in a mechanism dependent on a 5′ overlapping sequence and an embedded inverted SINEB2 element in lncRNA (Carrieri et al., 2012). LncRNA can also regulate stability of target mRNAs through imperfect base pairing between an Alu element in the 3′ UTR of a Staufen 1 (STAU1)-mediated mRNA decay target and another Alu element in a cytoplasmic lncRNA (Gong and Maquat, 2011). This imperfect base-pairing creates a lncRNA–mRNA duplex that binds STAU1 resulting in degradation of mRNA.
With the growing evidence of lncRNA functionality it is of no surprise that they are implicated in diverse pathologic conditions, including cancer. Various lncRNAs have been found to be differentially expressed in cancer and their enforced expression or knock down could result in altered phenotypical responses related to malignant transformation, such as changes in proliferation, invasive potential, or apoptosis. Similarly, to protein coding genes, lncRNAs can be classified into oncogenes and tumor suppressors. This opens opportunities for application of lncRNAs as biomarkers for cancer diagnosis and prognosis. Indeed, the functions of lncRNAs are more likely to correlate with their abundance as they do not encode proteins, making them highly suitable as biomarkers. In this review, we summarize the current knowledge about lncRNAs as potential diagnostic and prognostic biomarkers of cancer.
Diagnostic Cancer Biomarkers
The tissue specific nature of expression of lncRNAs, which is generally higher than that of protein coding mRNAs (Derrien et al., 2012), makes them potentially advantageous for identification of highly specific diagnostic biomarkers. Whereas some well known lncRNAs, such as HOTAIR are known to be deregulated in a wide spectrum of cancers, several lncRNAs have been described to be highly specific for a particular cancer type.
For example, expression of lncRNA PCA3 (Prostate Cancer Antigen 3) is highly restricted to prostate tissue (Bussemakers et al., 1999). In the same study it was shown by differential display analysis that PCA3 is highly overexpressed in prostatic tumors in comparison with non-neoplastic prostatic tissue of the same patients. Moreover, its expression was not detected in other tumor types or cell lines (Bussemakers et al., 1999). PCGEM1 is another lncRNA gene with highly prostate tissue-specific expression. Interestingly, its elevated expression was associated with high risk groups, such as the African–American population, which is more susceptible to prostate cancer as compared to Caucasian–American or those individuals with family history of prostate cancer as compared to those without family history (Petrovics et al., 2004). Another lncRNA, encoded in 8q24 locus reported to be associated with prostate cancer susceptibility in European and African–American populations, termed PRNCR1 (prostate cancer noncoding RNA 1), was shown to be up-regulated in some of the prostate cancer cells as well as precursor lesion prostatic intraepithelial neoplasia (Chung et al., 2011).
HULC (highly up-regulated in liver cancer) has been identified as highly up-regulated in hepatocellular carcinoma (Panzitt et al., 2007) and colorectal carcinomas that produce liver metastases but not in the primary colon tumors or their non-liver metastases (Matouk et al., 2009). Interestingly, the specific genetic variant rs7763881 in HULC was found to be associated with decreased risk of HCC in persistent carriers of HBV (Liu et al., 2012).
Detection of cancer at the early stages significantly gnosis is the screening for biomarkers by non-invasive methods, such as sampling of extracellular fluids. A fraction of DNA and RNA molecules referred to as circulating nucleic acids (CNAs) are found in blood serum and other extracellular fluids. Changes in the levels of CNAs are associated with tumor burden and malignant progression, thus pointing to their potential as tumor biomarkers easily detected by PCR assays (Schwarzenbach et al., 2011). Several lncRNAs have been characterized as potential biomarkers in human body fluids. The most prominent example of such biomarkers is PCA3, a lncRNA highly expressed is prostate cancer (de Kok et al., 2002). The detection of PCA3 in the urine has been demonstrated to be a more specific marker to diagnose prostate cancer than the commonly used prostate-specific antigen (PSA) and already found wide application in clinics (Hessels et al., 2003; Tinzl et al., 2004; Lee et al., 2011). Similarly, UCA1 (urothelial carcinoma associated 1) transcript detected in urine was shown to be a highly sensitive and specific biomarker of bladder carcinoma (Wang et al., 2006). HULC was detected with high frequency in plasma of patients with hepatocellular carcinoma, making it a promising diagnostic biomarker for this type of cancer (Xie et al., 2013). Several other studies identify promising biomarkers for different types of cancer, such as MALAT1 derived fragment detected in plasma as a biomarker for prostate cancer (Ren et al., 2013), AA174084 found in gastric juice as indicator of gastric cancer (Shao et al., 2014), a set of salivary lncRNAs as potential markers for oral squamous cell carcinoma diagnosis (Tang et al., 2013).
Exosomes are nanovesicles secreted into the extracellular environment from the cells upon fusion of intracellular vesicles with the plasma membrane (Raposo and Stoorvogel, 2013). The molecular content of exosomes replicates that of the releasing cell and reflects its status. The abundance of exosomes in body fluids and possibility to detect tumor specific material of their parental cancer cells makes exosomes a promising tool for non-invasive diagnostics (Properzi et al., 2013). The content of exosomes comprises different kinds of RNA, including lncRNAs (Jenjaroenpun et al., 2013). Indeed, lncRNAs are highly enriched in exosomes compared to donor cells (Batagov et al., 2011; Gezer et al., 2014). For example, lncRNA TUC339 was found to be highly enriched in extracellular vesicles secreted from hepatocellular carcinoma cells where it modulated tumor cell growth and adhesion (Kogure et al., 2013). LincRNA–ROR, another lncRNA from hepatocellular carcinoma derived extracellular vesicles was shown to modulate chemotherapeutic response in this type of cancer (Takahashi et al., 2014). Given the fact that most transcriptome studies aimed to identify biomarkers were focused on miRNAs, shifting the focus to more unbiased characterization that would include other types of transcripts present in cancer derived exosomes would potentially lead to discovery of more lncRNA based biomarkers.
Prognostic Cancer Biomarkers
Different lncRNAs were shown to be aberrantly expressed in cancer and correlate with tumorigenesis, tumor progression, and metastatic properties in various cancer conditions. Such lncRNA can be involved in both oncogenic and tumor suppressor pathways and their expression can correlate with good or bad prognosis, making them promising prognostic biomarkers (Table 2).
Table 2.
lncRNAs with cancer biomarker potential.
| lnc-RNA | Cancer type | Reference | Oncogene Tumor suppressor | Biomarker usability potential |
|---|---|---|---|---|
| HOTAIR | Breast | Gupta et al. (2010), Bhan et al. (2013), Wang et al. (2013) | Oncogene | Predictor of metastasis and poor survival |
| Liver | Geng et al. (2011), Yang et al. (2011), Ishibashi et al. (2013) | Oncogene | Predictor of recurrence and poor survival | |
| Colorectal | Kogo et al. (2011) | Oncogene | Predictor of liver metastasis and pool survival | |
| Gastric | Hajjari et al. (2013) | Oncogene | Predictor of lymph node metastasis | |
| Pancreas | Kim et al. (2013) | Oncogene | Predictor of poor survival | |
| Lung | Liu et al. (2013a), Nakagawa et al. (2013) | Oncogene | Predictor of metastasis and poor survival | |
| Esophagus | Lv et al. (2013) | Oncogene | Predictor of metastasis and poor survival | |
| Cervical | Huang et al. (2014) | Oncogene | Predictor of metastasis and poor survival | |
| MALAT1 | Lung adenocarcinoma | Ji et al. (2003), Gutschner et al. (2013) | Oncogene | Predictor of metastasis and poor survival |
| Liver | Lai et al. (2012) | Oncogene | Predictor of recurrence after liver transplantation | |
| Colorectal | Zheng et al. (2014) | Oncogene | Predictor of poor postoperative prognosis | |
| Cervical | Guo et al. (2010) | Oncogene | ||
| Bladder | Ying et al. (2012), Han et al. (2013) | Oncogene | Predictor of metastasis | |
| Uterine | Yamada et al. (2006) | Oncogene | ||
| Osteosarcoma | Fellenberg et al. (2007) | Oncogene | Predictor of poor response to chemotherapy | |
| H19 | Esophagus | Hibi et al. (1996) | Oncogene | |
| Breast | Lottin et al. (2002) | Oncogene | ||
| Lung | Kondo et al. (1995) | Oncogene | ||
| Bladder | Ariel et al. (2000) | Oncogene | ||
| Ovarian | Kim et al. (1998) | Oncogene | ||
| Cervical | Kim et al. (2002) | Oncogene | ||
| Osteosarcoma | Ulaner et al. (2003) | Oncogene | ||
| Neck squamous carcinoma | el-Naggar et al. (1999) | Oncogene | ||
| Liver | Matouk et al. (2007) | Oncogene | ||
| ncRAN | Neuroblastoma | Bown et al. (1999), Yu et al. (2009) | Oncogene | Predictor of poor survival |
| Bladder | Zhu et al. (2011) | Oncogene | ||
| Colorectal | Qi et al. (2014) | Tumor suppressor | Low level predicts poor survival | |
| HULC | Liver | Panzitt et al. (2007), Matouk et al. (2009), Liu et al. (2012) | Oncogene | Diagnostics |
| GAS5 | Breast | Mourtada-Maarabouni et al. (2009) | Tumor suppressor | |
| Kidney | Qiao et al. (2013) | Tumor suppressor | ||
| Pancreas | Lu et al. (2013) | Tumor suppressor | ||
| Bladder | Liu et al. (2013b) | Tumor suppressor | ||
| Lung | Shi et al. (2013) | Tumor suppressor | ||
| Gastric | Sun et al. (2014) | Tumor suppressor | Low level predicts poor prognosis | |
| Pleural mesothelioma | Renganathan et al. (2014) | Tumor suppressor | Low level predicts poor prognosis | |
| Liver | Tu et al. (2014) | Tumor suppressor | Low level predicts poor prognosis | |
| PCA3 | Prostate | Bussemakers et al. (1999), Hessels et al. (2003) | Oncogene | Non-invasive diagnosis by sampling urine or blood |
| Tinzl et al. (2004), Lee et al. (2011) | ||||
| UCA1 | Bladder | Wang et al. (2006) | Oncogene | Non-invasive diagnosis by sampling urine |
| PCGEM1 | Prostate | Petrovics et al. (2004) | Oncogene | High risk predictor |
| PRNCR1 | Prostate | Chung et al. (2011) | Oncogene | High risk predictor |
| MEG3 | Leukemia | Benetatos et al. (2010) | Tumor suppressor | CpG methylation predicts poor survival |
| Pituitary adenoma | Gejman et al. (2008) | Tumor suppressor | ||
| Meningioma | Zhang et al. (2010) | Tumor suppressor | Loss associated with progression | |
| Glioma | Wang et al. (2012) | Tumor suppressor | ||
| NBAT-1 | Neuroblastoma | Pandey et al. (2014) | Tumor suppressor | Loss associated with progression |
| LincRNA–ROR | Liver | Takahashi et al. (2014) | Oncogene | Diagnostics by sampling cancer exosomes; indicator of chemoresistance |
| PCAT1 | Prostate | Prensner et al. (2011) | Oncogene | Associated with progression |
HOTAIR is one of the most well studied examples of lncRNA implicated in cancer (Wu et al., 2014a). Initially it was identified by Gupta et al. (2010) to be highly overexpressed in primary and metastatic breast cancer tissues as compared to normal breast epithelium. High HOTAIR expression level in primary tumors was shown to be a significant predictor of eventual metastasis and death (Gupta et al., 2010). Enforced expression of HOTAIR in breast carcinoma cells induced genome wide retargeting of PRC2 and, as a result, altered H3K27 methylation and gene expression patterns, increased invasiveness, and metastasis (Gupta et al., 2010). Interestingly, BRCA1, an important suppressor of breast cancer inhibits HOTAIR dependent PRC2 activity by competitive binding to its catalytic subunit EZH2 (Wang et al., 2013). In another breast cancer study it was shown that HOTAIR is regulated by oestradiol in estrogen receptor dependent manner (Bhan et al., 2013).
HOTAIR was also shown to be involved in hepatocellular carcinoma (Geng et al., 2011; Yang et al., 2011; Ishibashi et al., 2013). Overexpression of HOTAIR in tumor tissues was reported to be associated with increased risk of recurrence after hepatectomy, as well as positively correlate with lymph node metastasis (Geng et al., 2011). Similarly, increased HOTAIR was shown to be prognostic factor of tumor recurrence following liver transplantation (Yang et al., 2011). In the same study, knock down of HOTAIR in HCC cell line reduced cell viability and invasion, as well as increased sensitivity to cisplatin and doxorubicin (Yang et al., 2011). Patients with overexpressed HOTAIR had poorer prognoses and larger tumor sizes, more rapid proliferation of tumor cells (Ishibashi et al., 2013).
In colorectal carcinoma high expression of HOTAIR was also shown to correlate with poor prognosis and liver metastasis (Kogo et al., 2011). Positive correlation between expression levels of HOTAIR, the members of PRC2 complex, Suz12, and Ezh2, and H3K27me3 chromatin marks suggests the role of HOTAIR in PRC2 mediated chromatin reprogramming in metastasis (Kogo et al., 2011). Apart from the primary tumors, HOTAIR was also shown to be a negative prognostic factor in the blood of colorectal patients suggesting that measuring the HOTAIR blood levels may provide a minimally invasive test to identify patients with unfavorable prognosis (Svoboda et al., 2014). High expression of HOTAIR associated with advanced stage, lymphatic node metastasis and poor survival was also reported in gastric cancer (Endo et al., 2013; Hajjari et al., 2013; Xu et al., 2013). The increased expression level of HOTAIR positively correlates with Suz12, implying PRC2 dependent mechanism of epigenetic reprogramming in oncogenicity of gastric carcinoma (Hajjari et al., 2013). Also HOTAIR was shown to act as an endogenous sponge of miR-331-3p miRNA, thus abolishing repression of its target HER2, implicated in development of gastric cancer (Liu et al., 2014). In addition to the above described, HOTAIR was reported to be a negative prognosis biomarker for a number of other malignancies, such as pancreatic cancer (Kim et al., 2013), lung cancer (Liu et al., 2013a; Nakagawa et al., 2013), esophageal carcinoma (Lv et al., 2013), cervical cancer (Huang et al., 2014), and colon cancer (Wu et al., 2014b). A meta-analysis study of prognostic capability of HOTAIR in different types of cancer revealed that it is a more reliable predictor of overall survival in patients with digestive system malignancies (Zhang et al., 2014).
Another paradigmal case of lncRNA based cancer biomarkers is MALAT1 (Metastasis-associated Lung Adenocarcinoma Transcript 1). As its name implies, it was discovered as a predictive marker of metastasis development in early stage lung adenocarcinoma (Ji et al., 2003). In a MALAT1 knockout model of human lung cancer cells it was shown that MALAT1 regulates metastatic signature of genes (Gutschner et al., 2013). In such model metastatic capacities of lung adenocarcinoma mouse xenografts were significantly compromised (Gutschner et al., 2013). In the same study, targeting MALAT1 with antisense nucleotides significantly reduced metastasis making it an interesting target for therapy (Gutschner et al., 2013).
In hepatocellular carcinoma (HCC) MALAT1 overexpression is a predictive factor for tumor recurrence following liver transplantation (Lai et al., 2012). The depletion of MALAT1 by siRNA in HepG2 cell line resulted in reduction of cell viability, mobility and invasiveness, as well as increase of sensitivity to apoptosis (Lai et al., 2012). High expression of MALAT1 was shown to be a marker of poor postoperative prognosis in colorectal carcinoma (Zheng et al., 2014). Moreover, the obvious oncogenic role of MALAT1 was demonstrated in a number of other cancer models, including bladder cancer (Ying et al., 2012; Han et al., 2013), cervical cancer (Guo et al., 2010), uterine sarcoma (Yamada et al., 2006), and osteosarcoma (Fellenberg et al., 2007).
H19 is lncRNA expressed from the maternal allele that plays an important role in genomic imprinting during growth and development (Gabory et al., 2010). Loss of imprinting at the H19 locus in paternal allele results in biallelic expression and, therefore, elevated H19 levels in a wide range of cancers (Kondo et al., 1995; Hibi et al., 1996; Kim et al., 1998; el-Naggar et al., 1999; Ariel et al., 2000; Müller et al., 2000; Kim et al., 2002; Lottin et al., 2002; Ulaner et al., 2003). High expression of H19 was shown to be associated with a range of risk factors, such as smoking, exposure to carcinogens and hypoxia (Matouk et al., 2007). Indeed, hypoxia causes up-regulation of H19 in cell lines with mutated p53 (Matouk et al., 2010). H19 is also directly induced by MYC oncogene in different cell lines (Barsyte-Lovejoy et al., 2006). Further adding to the host of oncogenic pathways with the involvement of H19, it serves as a precursor of miR-675, miRNA that down-regulates RB1 tumor suppressor transcript in colorectal cancer (Tsang et al., 2010).
NcRAN (noncoding RNA expressed in aggressive neuroblastoma) is encoded by a gene mapped to chromosome arm 17q, whose amplification is one of the most common genetic abnormalities associated with poor prognosis in neuroblastoma (Bown et al., 1999; Yu et al., 2009). Indeed, the high expression levels of ncRAN were significantly associated with poor prognosis of the neuroblastoma patients (Yu et al., 2009). In addition, ncRAN was found to be upregulated in bladder cancer as compared to normal tissues and confer a set of oncogenic properties, such as increased cell proliferation, migration, and invasion (Zhu et al., 2011). In contrast, down-regulation of ncRAN was shown to be associated with metastatic properties and predict poor overall survival in colorectal carcinoma (Qi et al., 2014).
In addition to lncRNAs with pronounced oncogenic effects, which positively correlate with poor prognosis, a few lncRNAs have been characterized to act as tumor suppressors. One such example is GAS5 (growth arrest-specific 5), a tumor suppressor lncRNA which reduces growth, metabolism, and sensitizes cells to apoptosis by inhibiting glucocorticoid receptor (Kino et al., 2010). GAS5 is down-regulated in a number of cancers, such as breast cancer (Mourtada-Maarabouni et al., 2009), renal cell carcinoma (Qiao et al., 2013), pancreatic cancer (Lu et al., 2013), bladder cancer (Liu et al., 2013b), non-small-cell lung cancer (Shi et al., 2013), gastric cancer (Sun et al., 2014), malignant pleural mesothelioma (Renganathan et al., 2014), hepatocellular carcinoma (Tu et al., 2014). Low level of GAS5 indicates a poor prognosis in cancer patients (Sun et al., 2014; Tu et al., 2014).
Another example of a tumor suppressor lncRNA is lincRNA-p21. This lncRNA was shown to be directly induced by p53 and play a crucial role in p53 mediated transcriptional response (Huarte et al., 2010). Maternally expressed gene 3 (MEG3) is another prominent tumor suppressor lncRNA that acts by increasing p53 activity on specific transcription targets (Zhou et al., 2007). Besides, MEG3 is able to inhibit cell proliferation in the absence of p53 suggesting that this lncRNA may act also via p53-independent pathway (Zhou et al., 2007). Consistently, MEG3 is down-regulated in a number of cancers, including myeloid leukemia (Benetatos et al., 2010), pituitary adenoma (Gejman et al., 2008), meningioma (Zhang et al., 2010), and glioma (Wang et al., 2012).
In conclusion, lncRNAs may act both as oncogenes and tumor suppressors without obvious prevalence of one class over another. In this respect lncRNAs behave similarly, to protein-coding transcripts.
Prognostic Expression Signatures Based on lncRNAs
Characterizing tumor transcriptomes at the system’s level allows identification of molecular subtypes of cancer and development of predictive and prognostic gene expression signatures. Microarrays have been widely used for the past two decades in preclinical research to characterize tumor gene expression profiles on a genome-wide scale. Most of these signatures were based on protein coding genes as little was known about lncRNAs and their important roles in cancer. However, the widely used commercial microarray platforms in addition to protein coding mRNAs were designed to detect numerous ESTs and non-annotated RNAs. Many of these transcripts are now annotated as bona fide lncRNAs, therefore the vast collection of microarray datasets accumulated during the past two decades provides a valuable resource for identification of novel gene expression signatures and biomarkers based on lncRNAs. In our recent study we annotated probe sets detecting lncRNAs on widely used Affymetrix U133 series of microarray platforms by using two approaches: firstly by matching probe sets to Gencode database of lncRNAs and secondly by developing a stringent in silico protein coding potential prediction filter (Yarmishyn et al., 2014). Affymetrix U133 microarrays have been found to contain probe sets for the measurement of 1581 lncRNAs (Yarmishyn et al., 2014). Using this resource and previously deposited microarray-based data on expression profiling of neuroblastic tumors we identified 159 lncRNA signature that discriminates between localized and metastatic stages of neuroblastoma, as well as between relapsing and non-relapsing primary tumors (Yarmishyn et al., 2014). The data mining of Affymetrix U133 Plus datasets was also applied to identify lncRNA signatures associated with different histological subtypes and malignancy grades of glioma (Zhang et al., 2012). In the follow up study, the six-lncRNA prognostic signature of glioma significantly associated with overall survival was identified (Zhang et al., 2013). In another study the classification of glioma into three molecular subtypes based on lncRNA expression profiles was proposed (Li et al., 2014). Similarly, using lncRNA mining approach, Hu et al. (2014) identified six lncRNA prognostic signature significantly associated with disease free survival in colorectal carcinoma. LncRNA-based expression profiles were also utilized to classify colorectal cancer samples into five distinct molecular subtypes, each characterized by distinct biological pathways and clinical outcome (Chen et al., 2014).
As high throughput sequencing technology becomes cheaper and more accessible we might expect the influx of de novo data from transcriptome sequencing and identification of new biomarkers and diagnostic signatures. For example, Prensner et al. (2011) utilized RNA-Seq approach to identify 121 lncRNAs associated with prostate cancer progression in a cohort of 102 patients. One of these lncRNAs PCAT-1 was further characterized as a regulator of proliferation in prostate cancer cells (Prensner et al., 2011). In a similar study the sequencing of transcriptomes of high and low risk neuroblastoma identified a set of differentially expressed lncRNAs. One of them NBAT-1 was characterized as a tumor suppressor, whose loss contributes to aggressive neuroblastoma by increasing proliferation and impairing differentiation (Pandey et al., 2014). RNA-Seq analysis was also employed by White et al. (2014) to detect lncRNAs associated with lung cancer. In this study the transcriptome sequencing data from a cohort of 567 patients were used to identify 111 novel lncRNAs differentially regulated between tumor and adjacent normal tissue samples. Further meta-analysis revealed that a subset of these lncRNAs was deregulated in a broad range of tumors and another subset was highly deregulated specifically in lung cancers, making the latter a promising source of biomarkers (White et al., 2014).
Concluding Remarks
The last decade witnessed a vast expansion of knowledge regarding lncRNAs and their important roles in regulation of various biological processes and development of disease. A number of lncRNAs, such as HOTAIR, MALAT and H19 were found to be aberrantly expressed in a number of cancers and extensively characterized as important players affecting the hallmark events of carcinogenesis, such as proliferation, apoptosis, and metastasis. These examples demonstrate that lncRNAs, on par with protein coding genes and miRNAs, have a great potential to be used as cancer biomarkers. Indeed, since lncRNAs are implicated in cancer biology at the level of RNA unlike mRNAs, which represent an intermediate on the way to functioning protein, their levels of expression are more likely to correlate with cancer phenotypes. Also, since proteins and lncRNAs represent different regulatory realms, their combination might increase each other’s power as diagnostic and prognostic tools. It should be noted that the few examples described in this review are just a tip of the iceberg, as the field of lncRNAs is currently evolving and the work of annotation and characterization of lncRNAs is ongoing. With the deep sequencing technology becoming cheaper and more accessible one might expect an explosive growth of newly identified lncRNAs differentially expressed in cancers and associated with various clinical parameters.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Thomas Derrien and Rory Johnson for sharing the numerical data for the numbers of exons in lncRNAs and mRNAs, which was necessary for compiling Table 1.
References
- Ariel I., Sughayer M., Fellig Y., Pizov G., Ayesh S., Podeh D., et al. (2000). The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol. Pathol. 53 320–323 10.1136/mp.53.6.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsyte-Lovejoy D., Lau S. K., Boutros P. C., Khosravi F., Jurisica I., Andrulis I. L., et al. (2006). The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 66 5330–5337 10.1158/0008-5472.CAN-06-0037 [DOI] [PubMed] [Google Scholar]
- Batagov A. O., Kuznetsov V. A., Kurochkin I. V. (2011). Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics 12(Suppl. 3):S18 10.1186/1471-2164-12-S3-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L., Hatzimichael E., Dasoula A., Dranitsaris G., Tsiara S., Syrrou M., et al. (2010). CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 34 148–153 10.1016/j.leukres.2009.06.019 [DOI] [PubMed] [Google Scholar]
- Bhan A., Hussain I., Ansari K. I., Kasiri S., Bashyal A., Mandal S. S. (2013). Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 425 3707–3722 10.1016/j.jmb.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J. A., Dutta A., Guigó R., Gingeras T. R., Margulies E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 799–816 10.1038/nature05874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown N., Cotterill S., Lastowska M., O’Neill S., Pearson A. D., Plantaz D., et al. (1999). Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N. Engl. J. Med. 340 1954–1961 10.1056/NEJM199906243402504 [DOI] [PubMed] [Google Scholar]
- Bussemakers M. J., van Bokhoven A., Verhaegh G. W., Smit F. P., Karthaus H. F., Schalken J. A., et al. (1999). DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 59 5975–5979. [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309 1559–1563 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., et al. (2012). Long noncoding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491 454–457 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H. H., Kapranov P., Sekinger E. A., Kampa D., et al. (2004). Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116 499–509 10.1016/S0092-8674(04)00127-8 [DOI] [PubMed] [Google Scholar]
- Chen H., Xu J., Hong J., Tang R., Zhang X., Fang J. Y. (2014). Long noncoding RNA profiles identify five distinct molecular subtypes of colorectal cancer with clinical relevance. Mol. Oncol. 8 1393–403 10.1016/j.molonc.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Nakagawa H., Uemura M., Piao L., Ashikawa K., Hosono N., et al. (2011). Association of a novel long noncoding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 102 245–252 10.1111/j.1349-7006.2010.01737.x [DOI] [PubMed] [Google Scholar]
- de Kok J. B., Verhaegh G. W., Roelofs R. W., Hessels D., Kiemeney L. A., Aalders T. W., et al. (2002). DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 62 2695–2698. [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22 1775–1789 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., et al. (2012). Landscape of transcription in human cells. Nature 489 101–108 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I., Kundaje A., Aldred S. F., Collins P. J., Davis C. A., Doyle F., et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489 57–74 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Naggar A. K., Lai S., Tucker S. A., Clayman G. L., Goepfert H., Hong W. K., et al. (1999). Frequent loss of imprinting at the IGF2 and H19 genes in head and neck squamous carcinoma. Oncogene 18 7063–7069 10.1038/sj.onc.1203192 [DOI] [PubMed] [Google Scholar]
- Endo H., Shiroki T., Nakagawa T., Yokoyama M., Tamai K., Yamanami H., et al. (2013). Enhanced expression of long noncoding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE 8:e77070 10.1371/journal.pone.0077070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M. R., Sonenberg N. (2012). The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19 586–593 10.1038/nsmb.2296 [DOI] [PubMed] [Google Scholar]
- Fellenberg J., Bernd L., Delling G., Witte D., Zahlten-Hinguranage A. (2007). Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod. Pathol. 20 1085–1094 10.1038/modpathol.3800937 [DOI] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B. S., Mady R., Shah P., Kohtz J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20 1470–1484 10.1101/gad.1416106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A., Jammes H., Dandolo L. (2010). The H19 locus: role of an imprinted noncoding RNA in growth and development. Bioessays 32 473–480 10.1002/bies.200900170 [DOI] [PubMed] [Google Scholar]
- Garzon R., Calin G. A., Croce C. M. (2009). MicroRNAs in Cancer. Annu. Rev. Med. 60 167–179 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- Gejman R., Batista D. L., Zhong Y., Zhou Y., Zhang X., Swearingen B., et al. (2008). Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocrinol. Metab. 93 4119–4125 10.1210/jc.2007-2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y. J., Xie S. L., Li Q., Ma J., Wang G. Y. (2011). Large intervening noncoding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 39 2119–2128 10.1177/147323001103900608 [DOI] [PubMed] [Google Scholar]
- Gezer U., Özgür E., Cetinkaya M., Isin M., Dalay N. (2014). Long noncoding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 38 1076–1079 10.1002/cbin.10301 [DOI] [PubMed] [Google Scholar]
- Gong C., Maquat L. E. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470 284–288 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Li Y., Liu Y., Wang J., Li Y., Li G. (2010). Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim. Biophys. Sin. (Shanghai) 42 224–229 10.1093/abbs/gmq008 [DOI] [PubMed] [Google Scholar]
- Gupta R. A., Shah N., Wang K. C., Kim J., Horlings H. M., Wong D. J., et al. (2010). Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464 1071–1076 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., et al. (2013). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73 1180–1189 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Amit I., Garber M., French C., Lin M. F., Feldser D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature 458 223–227 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjari M., Behmanesh M., Sadeghizadeh M., Zeinoddini M. (2013). Up-regulation of HOTAIR long noncoding RNA in human gastric adenocarcinoma tissues. Med. Oncol. 30:670 10.1007/s12032-013-0670-0 [DOI] [PubMed] [Google Scholar]
- Han Y., Liu Y., Nie L., Gui Y., Cai Z. (2013). Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology 81 209e1–209e7. 10.1016/j.urology.2012.08.044 [DOI] [PubMed] [Google Scholar]
- Hessels D., Klein Gunnewiek J. M., van Oort I., Karthaus H. F., van Leenders G. J., van Balken B., et al. (2003). DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44 8–15 10.1016/S0302-2838(03)00201-X [DOI] [PubMed] [Google Scholar]
- Hibi K., Nakamura H., Hirai A., Fujikake Y., Kasai Y., Akiyama S., et al. (1996). Loss of H19 imprinting in esophageal cancer. Cancer Res. 56 480–482. [PubMed] [Google Scholar]
- Hu Y., Chen H. Y., Yu C. Y., Xu J., Wang J. L., Qian J., et al. (2014). A long noncoding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 5 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Liao L. M., Liu A. W., Wu J. B., Cheng X. L., Lin J. X., et al. (2014). Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch. Gynecol. Obstet. 290 717–723 10.1007/s00404-014-3236-2 [DOI] [PubMed] [Google Scholar]
- Huarte M., Guttman M., Feldser D., Garber M., Koziol M. J., Kenzelmann-Broz D., et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142 409–419 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., Grant G. D., et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43 621–629 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M., Kogo R., Shibata K., Sawada G., Takahashi Y., Kurashige J., et al. (2013). Clinical significance of the expression of long noncoding RNA HOTAIR in primary hepatocellular carcinoma. Oncol. Rep. 29 946–950 10.3892/or.2012.2219 [DOI] [PubMed] [Google Scholar]
- Jenjaroenpun P., Kremenska Y., Nair V. M., Kremenskoy M., Joseph B., Kurochkin I. V. (2013). Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. Peer J. 1:e201 10.7717/peerj.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P. M., et al. (2003). MALAT-1 a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22 8031–8041 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- Khalil A. M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106 11667–11672 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. T., Choi B. H., Niikawa N., Lee T. S., Chang S. I. (1998). Frequent loss of imprinting of the H19 and IGF-II genes in ovarian tumors. Am. J. Med. Genet. 80 391–395 [DOI] [PubMed] [Google Scholar]
- Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., et al. (2013). HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32 1616–1625 10.1038/onc.2012.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Park S. E., Lee C., Lee S. Y., Jo J. H., Kim J. M., et al. (2002). Alterations in promoter usage and expression levels of insulin-like growth factor-II and H19 genes in cervical carcinoma exhibiting biallelic expression of IGF-II. Biochim. Biophys. Acta 1586 307–315 10.1016/S0925-4439(01)00109-0 [DOI] [PubMed] [Google Scholar]
- Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3:ra8 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71 6320–6326 10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- Kogure T., Yan I. K., Lin W. L., Patel T. (2013). Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer 4 261–272 10.1177/1947601913499020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Suzuki H., Ueda R., Osada H., Takagi K., Takahashi T., et al. (1995). Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene 10 1193–1198. [PubMed] [Google Scholar]
- Kong Y. W., Ferland-McCollough D., Jackson T. J., Bushell M. (2012). microRNAs in cancer management. Lancet Oncol. 13:e249–e258 10.1016/S1470-2045(12)70073-6 [DOI] [PubMed] [Google Scholar]
- Lai M. C., Yang Z., Zhou L., Zhu Q. Q., Xie H. Y., Zhang F., et al. (2012). Long noncoding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 29 1810–1816 10.1007/s12032-011-0004-z [DOI] [PubMed] [Google Scholar]
- Lee G. L., Dobi A., Srivastava S. (2011). Prostate cancer: diagnostic performance of the PCA3 urine test. Nat. Rev. Urol. 8 123–124 10.1038/nrurol.2011.10 [DOI] [PubMed] [Google Scholar]
- Li R., Qian J., Wang Y. Y., Zhang J. X., You Y. P. (2014). Long noncoding RNA profiles reveal three molecular subtypes in glioma. CNS Neurosci. Ther. 20 339–343 10.1111/cns.12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. H., Liu Z. L., Sun M., Liu J., Wang Z. X., De W. (2013a). The long noncoding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 13:464 10.1186/1471-2407-13-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang W., Jiang J., Bao E., Xu D., Zeng Y., et al. (2013b). Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS ONE 8:e73991 10.1371/journal.pone.0073991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. H., Sun M., Nie F. Q., Ge Y. B., Zhang E. B., Yin D. D., et al. (2014). Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 13 92 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Pan S., Liu L., Zhai X., Liu J., Wen J., et al. (2012). A genetic variant in long noncoding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS ONE 7:e35145 10.1371/journal.pone.0035145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottin S., Adriaenssens E., Dupressoir T., Berteaux N., Montpellier C., Coll J., et al. (2002). Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23 1885–1895 10.1093/carcin/23.11.1885 [DOI] [PubMed] [Google Scholar]
- Lu X., Fang Y., Wang Z., Xie J., Zhan Q., Deng X., et al. (2013). Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 354 891–896 10.1007/s00441-013-1711-x [DOI] [PubMed] [Google Scholar]
- Lv X. B., Lian G. Y., Wang H. R., Song E., Yao H., Wang M. H. (2013). Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE 8:e63516 10.1371/journal.pone.0063516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. (2007). Repression of the human dihydrofolate reductase gene by a noncoding interfering transcript. Nature 445 666–670 10.1038/nature05519 [DOI] [PubMed] [Google Scholar]
- Matouk I. J., Abbasi I., Hochberg A., Galun E., Dweik H., Akkawi M. (2009). Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol. 21 688–692 10.1097/MEG.0b013e328306a3a2 [DOI] [PubMed] [Google Scholar]
- Matouk I. J., DeGroot N., Mezan S., Ayesh S., Abu-lail R., Hochberg A., et al. (2007). The H19 noncoding RNA is essential for human tumor growth. PLoS ONE 2:e845 10.1371/journal.pone.0000845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk I. J., Mezan S., Mizrahi A., Ohana P., Abu-Lail R., Fellig Y., et al. (2010). The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim. Biophys. Acta 1803 443–451 10.1016/j.bbamcr.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Sunkin S. M., Mehler M. F., Mattick J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105 716–721 10.1073/pnas.0706729105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M., Pickard M. R., Hedge V. L., Farzaneh F., Williams G. T. (2009). GAS5 a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28 195–208 10.1038/onc.2008.373 [DOI] [PubMed] [Google Scholar]
- Müller S., Zirkel D., Westphal M., Zumkeller W. (2000). Genomic imprinting of IGF2 and H19 in human meningiomas. Eur. J. Cancer 36 651–655 10.1016/S0959-8049(99)00328-7 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Endo H., Yokoyama M., Abe J., Tamai K., Tanaka N., et al. (2013). Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 436 319–324 10.1016/j.bbrc.2013.05.101 [DOI] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., et al. (2002). Analysis of the mouse transcriptome based on functional annotation of 60770 full-length cDNAs. Nature 420 563–573 10.1038/nature01266 [DOI] [PubMed] [Google Scholar]
- Pandey G. K., Mitra S., Subhash S., Hertwig F., Kanduri M., Mishra K., et al. (2014). The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26 722–737 10.1016/j.ccell.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Panzitt K., Tschernatsch M. M., Guelly C., Moustafa T., Stradner M., Strohmaier H. M., et al. (2007). Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132 330–342 10.1053/j.gastro.2006.08.026 [DOI] [PubMed] [Google Scholar]
- Petrovics G., Zhang W., Makarem M., Street J. P., Connelly R., Sun L., et al. (2004). Elevated expression of PCGEM1 a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene 23 605–611 10.1038/sj.onc.1207069 [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C. P., Lunter G. (2007). Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 17 556–565 10.1101/gr.6036807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Belgard T. G. (2010). Transcribed dark matter: meaning or myth? Hum. Mol. Genet. 19 R162–R168 10.1093/hmg/ddq362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Iyer M. K., Balbin O. A., Dhanasekaran S. M., Cao Q., Brenner J. C., et al. (2011). Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1 an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 29 742–749 10.1038/nbt.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Properzi F., Logozzi M., Fais S. (2013). Exosomes: the future of biomarkers in medicine. Biomark. Med. 7 769–778 10.2217/bmm.13.63 [DOI] [PubMed] [Google Scholar]
- Qi P., Xu M. D., Ni S. J., Shen X. H., Wei P., Huang D., et al. (2014). Down-regulation of ncRAN, a long noncoding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol. Carcinog. 9 1039–1045 10.1002/mc.22137 [DOI] [PubMed] [Google Scholar]
- Qiao H. P., Gao W. S., Huo J. X., Yang Z. S. (2013). Long noncoding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac. J. Cancer Prev. 14 1077–1082 10.7314/APJCP.2013.14.2.1077 [DOI] [PubMed] [Google Scholar]
- Raposo G., Stoorvogel W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200 373–383 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Pang K. C., Katayama S., Furuno M., Okunishi R., et al. (2006). Experimental validation of the regulated expression of large numbers of noncoding RNAs from the mouse genome. Genome Res. 16 11–19 10.1101/gr.4200206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Wang F., Shen J., Sun Y., Xu W., Lu J., et al. (2013). Long noncoding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer 49 2949–2959 10.1016/j.ejca.2013.04.026 [DOI] [PubMed] [Google Scholar]
- Renganathan A., Kresoja-Rakic J., Echeverry N., Ziltener G., Vrugt B., Opitz I., et al. (2014). GAS5 long noncoding RNA in malignant pleural mesothelioma. Mol. Cancer 13:119 10.1186/1476-4598-13-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129 1311–1323 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H., Hoon D. S., Pantel K. (2011). Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11 426–437 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- Shao Y., Ye M., Jiang X., Sun W., Ding X., Liu Z., et al. (2014). Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 120 3320–3328 10.1002/cncr.28882 [DOI] [PubMed] [Google Scholar]
- Shi X., Sun M., Liu H., Yao Y., Kong R., Chen F., et al. (2013). A critical role for the long noncoding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol. Carcinog. 10.1002/mc.22120 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Sun M., Jin F. Y., Xia R., Kong R., Li J. H., Xu T. P., et al. (2014). Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 14:319 10.1186/1471-2407-14-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda M., Slyskova J., Schneiderova M., Makovicky P., Bielik L., Levy M., et al. (2014). HOTAIR long noncoding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 35 1510–1515 10.1093/carcin/bgu055 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yan I. K., Kogure T., Haga H., Patel T. (2014). Extracellular vesicle-mediated transfer of long noncoding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 4 458–467 10.1016/j.fob.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wu Z., Zhang J., Su B. (2013). Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 7 761–766 10.3892/mmr.2012.1254 [DOI] [PubMed] [Google Scholar]
- Tinzl M., Marberger M., Horvath S., Chypre C. (2004). DD3PCA3 RNA analysis in urine—a new perspective for detecting prostate cancer. Eur. Urol. 46 182–186 10.1016/j.eururo.2004.06.004 [DOI] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39 925–938 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W. P., Ng E. K., Ng S. S., Jin H., Yu J., Sung J. J., et al. (2010). Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 31 350–358 10.1093/carcin/bgp181 [DOI] [PubMed] [Google Scholar]
- Tu Z. Q., Li R. J., Mei J. Z., Li X. H. (2014). Down-regulation of long noncoding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 7 4303–4309. [PMC free article] [PubMed] [Google Scholar]
- Ulaner G. A., Vu T. H., Li T., Hu J. F., Yao X. M., Yang Y., et al. (2003). Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum. Mol. Genet. 12 535–549 10.1093/hmg/ddg034 [DOI] [PubMed] [Google Scholar]
- Wang L., Zeng X., Chen S., Ding L., Zhong J., Zhao J. C., et al. (2013). BRCA1 is a negative modulator of the PRC2 complex. EMBO J. 32 1584–1597 10.1038/emboj.2013.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Ren Z., Sun P. (2012). Overexpression of the long noncoding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell Biochem. 113 1868–1874 10.1002/jcb.24055 [DOI] [PubMed] [Google Scholar]
- Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., et al. (2008). Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454 126–130 10.1038/nature06992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. S., Zhang Z., Wang H. C., Cai J. L., Xu Q. W., Li M. Q., et al. (2006). Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 12 4851–4858 10.1158/1078-0432.CCR-06-0134 [DOI] [PubMed] [Google Scholar]
- White N. M., Cabanski C. R., Silva-Fisher J. M., Dang H. X., Govindan R., Maher C. A. (2014). Transcriptome sequencing reveals altered long intergenic noncoding RNAs in lung cancer. Genome Biol. 15:429 10.1186/s13059-014-0429-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang L., Wang Y., Li H., Ren X., Wei F., et al. (2014a). Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 35 9531–9538 10.1007/s13277-014-2523-7 [DOI] [PubMed] [Google Scholar]
- Wu Z. H., Wang X. L., Tang H. M., Jiang T., Chen J., Lu S., et al. (2014b). Long noncoding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol. Rep. 32 395–402 10.3892/or.2014.3186 [DOI] [PubMed] [Google Scholar]
- Xie H., Ma H., Zhou D. (2013). Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed. Res. Int. 2013:136106 10.1155/2013/136106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. Y., Yu Q. M., Du Y. A., Yang L. T., Dong R. Z., Huang L., et al. (2013). Knockdown of long noncoding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 9 587–597 10.7150/ijbs.6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Kano J., Tsunoda H., Yoshikawa H., Okubo C., Ishiyama T., et al. (2006). Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 97 106–112 10.1111/j.1349-7006.2006.00147.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhou L., Wu L. M., Lai M. C., Xie H. Y., Zhang F., et al. (2011). Overexpression of long noncoding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18 1243–1250 10.1245/s10434-011-1581-y [DOI] [PubMed] [Google Scholar]
- Yarmishyn A. A., Batagov A. O., Tan J. Z., Sundaram G. M., Sampath P., Kuznetsov V. A., et al. (2014). HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics 15(Suppl. 9):S7 10.1186/1471-2164-15-S9-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L., Chen Q., Wang Y., Zhou Z., Huang Y., Qiu F. (2012). Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 8 2289–2294 10.1039/c2mb25070e [DOI] [PubMed] [Google Scholar]
- Yoon J. H., Abdelmohsen K., Srikantan S., Yang X., Martindale J. L., De S., et al. (2012). LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47 648–655 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Ohira M., Li Y., Niizuma H., Oo M. L., Zhu Y., et al. (2009). High expression of ncRAN, a novel noncoding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int. J. Oncol. 34 931–938. [DOI] [PubMed] [Google Scholar]
- Zhang S., Chen S., Yang G., Gu F., Li M., Zhong B., et al. (2014). Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS ONE 9:e105538 10.1371/journal.pone.0105538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gejman R., Mahta A., Zhong Y., Rice K. A., Zhou Y., et al. (2010). Maternally expressed gene 3 an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 70 2350–2358 10.1158/0008-5472.CAN-09-3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Q., Sun S., Lam K. F., Kiang K. M., Pu J. K., Ho A. S., et al. (2013). A long noncoding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 58 123–131 10.1016/j.nbd.2013.05.011 [DOI] [PubMed] [Google Scholar]
- Zhang X., Sun S., Pu J. K., Tsang A. C., Lee D., Man V. O., et al. (2012). Long noncoding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 48 1–8 10.1016/j.nbd.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322 750–756 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H. T., Shi D. B., Wang Y. W., Li X. X., Xu Y., Tripathi P., et al. (2014). High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 7 3174–3181. [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D. L., Gejman R., et al. (2007). Activation of p53 by MEG3 noncoding RNA. J. Biol. Chem. 282 24731–24742 10.1074/jbc.M702029200 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Yu M., Li Z., Kong C., Bi J., Li J., et al. (2011). ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology 77:e1–e5 10.1016/j.urology.2010.09.022 [DOI] [PubMed] [Google Scholar]