Abstract
Constitutive androstane receptor (CAR) and pregnane X receptor (PXR) are xenobiotic sensors that enhance the detoxification and elimination of xenobiotics and endobiotics by modulating the expression of genes encoding drug-metabolizing enzymes and transporters. Elevated levels of drug-metabolizing enzymes and efflux transporters, resulting from CAR activation in various cancers, promote the elimination of chemotherapeutic agents, leading to reduced therapeutic effectiveness and acquired drug resistance. CAR inhibitors, in combination with existing chemotherapeutics, could therefore be used to attenuate multidrug resistance in cancers. Interestingly, all previously reported CAR inverse-agonists are also activators of PXR, rendering them mechanistically counterproductive in tissues where both these xenobiotic receptors are present and active. We used a directed high-throughput screening approach, followed by subsequent mechanistic studies, to identify novel, potent, and specific small-molecule CAR inhibitors that do not activate PXR. We describe here one such inhibitor, CINPA1 (CAR inhibitor not PXR activator 1), capable of reducing CAR-mediated transcription with an IC50 of ∼70 nM. CINPA1 1) is a specific xenobiotic receptor inhibitor and has no cytotoxic effects up to 30 µM; 2) inhibits CAR-mediated gene expression in primary human hepatocytes, where CAR is endogenously expressed; 3) does not alter the protein levels or subcellular localization of CAR; 4) increases corepressor and reduces coactivator interaction with the CAR ligand-binding domain in mammalian two-hybrid assays; and 5) disrupts CAR binding to the promoter regions of target genes in chromatin immunoprecipitation assays. CINPA1 could be used as a novel molecular tool for understanding CAR function.
Introduction
Constitutive androstane receptor (CAR) (MB67 and NR1I3) (Baes et al., 1994) is a transcription factor that acts as a xenobiotic sensor and is capable of regulating cellular function in response to xenobiotics and endobiotics (Timsit and Negishi, 2007; Ma et al., 2008; di Masi et al., 2009). Activated CAR in tissues heterodimerizes with retinoid X receptor (RXR) and translocates to the nucleus. CAR shares a large portion of its metabolic functions with pregnane X receptor (PXR; NR1I2), which has structural features in its ligand-binding domain (LBD) that allow it to bind a diverse set of chemical motifs. CAR’s LBD is more compact and yet capable of binding varied structural entities (Wu et al., 2013). CAR and PXR bind similar response elements on chromatin and hence regulate an overlapping set of genes (Wei et al., 2002). CAR remains a major player in xenobiotic metabolism by controlling the transactivation of many cytochrome P450 (P450) enzymes and transporters, particularly CYP2B6 and multidrug resistance 1 (MDR1). Overexpression of CAR in hepatic-induced human embryonic stem cells enhanced expression of hepatic phenotype markers and has been hypothesized as having a key functional role in directing human hepatogenesis (DeKeyser et al., 2011; Zamule et al., 2011; Chen et al., 2013). CAR is the molecular target of phenobarbital (PB)-induced hepatocellular carcinoma, and activation of this receptor is an essential requirement for liver tumor development (Yamamoto et al., 2004; Huang et al., 2005). CAR overexpression in neuroblastoma drives doxorubicin resistance by increasing MDR1 levels (Takwi et al., 2014) in ovarian cancer cells, whereas pharmacologic activation of CAR leads to upregulation of MDR1 and decreased effectiveness of anticancer drugs; downregulation of CAR by RNA interference enhances the cell growth inhibition and apoptosis mediated by these anticancer drugs (Wang et al., 2014). CAR activation also causes hepatic lipogenesis and insulin insensitivity by upregulating the thyroid hormone–responsive spot 14 gene, which might promote fatty liver diseases and insulin resistance (Breuker et al., 2010). The involvement of CAR in various diseases emphasizes its clinical and pharmacologic significance (Kachaylo et al., 2011).
Exogenously expressed human CAR1 in immortalized cell lines spontaneously accumulates in the cell nuclei and tends to be constitutively active in the absence of agonists (Baes et al., 1994). CAR also exhibits high basal but low agonist-induced activation in immortalized cells (Faucette et al., 2007). The splice variant hCAR3 is inducible by agonists when it is overexpressed in cells (Auerbach et al., 2003; Chen et al., 2010), but the agonist-inducible activity is not substantial without simultaneously overexpressing RXRα (Auerbach et al., 2005). In tissues expressing endogenous hCAR, the receptor is predominantly cytoplasmic unless activated by direct-binding ligands such as 6-(4-chlorophenyl)imidazo [2,1-b][1,3] thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl) oxime (CITCO) (Maglich et al., 2003) or indirect activators such as PB (Honkakoski et al., 1998; Mutoh et al., 2013). Among the reported inhibitors of hCAR, PK11195 [1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide] is the most potent small molecule inhibitor (Li et al., 2008). Clotrimazole (CLZ), meclizine, and androstanol are moderate inhibitors of CAR that function in both in vitro biochemical assays and cell-based transfection assays (Forman et al., 1998; Moore et al., 2000; Tzameli et al., 2000; Moore et al., 2002; Huang et al., 2004; Omiecinski et al., 2011). We and others, however, have observed that all these CAR inhibitors are also moderate to potent activators of PXR function (Jones et al., 2000; Moore et al., 2002; Li et al., 2008; Lau et al., 2011).
In addition to a smaller set of target genes unique for each receptor, CAR and PXR coregulate an overlapping set of metabolizing genes, depending on the activation status of these receptors (Wei et al., 2002). The CYP2B6 gene, although considered a predominantly CAR-regulated gene, is also induced equally well by activated PXR (Faucette et al., 2006; Faucette et al., 2007). In tissues such as liver or colon that express both PXR and CAR, treatment with CAR inhibitors that also activate PXR would result in a confused gene expression profile, and interpretation of receptor function is confounded by the opposing dual activity of such CAR inhibitors. Therefore, the goal of our directed screening approach was to discover CAR inhibitors that either antagonize PXR or at least do not activate PXR; such CAR inhibitors are expected to reduce the induction of target genes either uniquely regulated by CAR or coregulated by both PXR and CAR.
We identified CAR inhibitor not PXR activator 1 (CINPA1) as a potent inhibitor of hCAR that does not activate PXR. CINPA1 represents a novel class of chemicals that can be used as investigative tools to study CAR biology without concern about PXR activation.
Materials and Methods
Human embryonic kidney cell line HEK293T, liver hepatocellular cell line HepG2, human intestinal cell line LS174T, and human osteosarcoma U2OS cells were obtained from American Type Culture Collection (Manassas, VA). GeneBLAzer Validated Assays for Nuclear Receptors, Tb-anti–GST antibody, GST-hCAR-LBD, fluorescein-PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1-α) coactivator peptide, and time-resolved fluorescence resonance energy transfer (TR-FRET) coregulator buffer G were obtained from Life Technologies (Carlsbad, CA). Fetal bovine serum (FBS) and charcoal-dextran–treated FBS were obtained from HyClone (Logan, UT). Anti-mouse IRDye secondary antibody was purchased from LI-COR Biosciences (Lincoln, NE). Anti-CAR antibody (Clone N4111) was purchased from R&D Systems (Minneapolis, MN) and anti–RNA polymerase II from EMD Millipore (Darmstadt, Germany). PK11195, rifampicin, CLZ, anti-FLAG M2 antibody, and protease inhibitor cocktail were obtained from Sigma-Aldrich (St. Louis, MO). CITCO was obtained from Tocris Bioscience (Bristol, UK). Dimethylsulfoxide (DMSO) was purchased from Fisher Scientific (Pittsburgh, PA). CINPA1 was obtained from ChemDiv (San Diego, CA). The following chemicals were used in the GeneBLAzer assays: dexamethasone, GW9662 [2-chloro-5-nitro-N-phenylbenzamide], rosglitazone, (E)-guggulsterone, 9-cis retinoic acid, RU-486 (mifepristone), and 22(S)-hydroxycholesterol from Sigma-Aldrich; fenofibrate, TO901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]benzenesulfonamide], and vitamin D3 were from Cayman Chemicals (Ann Arbor, MI); and HX531 [4-(7,8,9,10-tetrahydro-5,7,7,10.10-pentamethyl-2-nitro-5H-benzo[b]naphtho[2,3-e][1,4]-diazepin-12-yl)-benzoic acid] was from Tocris Bioscience.
Cell Culture, Plasmids, and Transfection.
A humidified incubator at 37°C with 5% CO2 was used to maintain all cell lines. HEK293T, HepG2, HepG2-PXR clone 1 (Lin et al., 2008; Li et al., 2012), and LS174T cells were maintained in minimum Eagle’s medium supplemented with 10% FBS and PenStrep (100 μg/ml; Life Technologies). GeneBLAzer cell lines were maintained according to the manufacturer’s instructions. A clone of HepG2 cells stably overexpressing FLAG-hCAR1 (HepG2-hCAR1 clone 17) was generated by antibiotic selection with G418 (Life Technologies) using a limited dilution method. HepG2-hCAR1 and HepG2-PXR Clone 1 cells were maintained in media containing G418 (500 μg/ml). U2OS (human osteosarcoma–derived) cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% FBS, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin. Primary human hepatocytes were obtained through the Liver Tissue Cell Distribution System (donors 1–6 and 8, corresponding to case no. 13-001 [M], 13-003 [F], 13-005 [M], 13-006 [M], 14-005 [F], 14-008 [M], and 14-016 [F] respectively; Pittsburgh, PA) or from Triangle Research Laboratories [donor 7, case number HUM4043 (F); Research Triangle Park, NC] and maintained in Williams’ E medium containing Primary Hepatocyte Maintenance Supplement (Life Technologies).
The CAR expression vector (FLAG-hCAR1 in pcDNA3.1 vector) and CYP3A4-luciferase reporter (CYP3A4-luc; in pGL3 vector) have been previously described (Li et al., 2012). A luciferase reporter gene under the control of the CYP2B6 promoter region (PB enhancer module/xenobiotic responsive enhancer module, or PBREM/XREM), CYP2B6-luc, was kindly provided by Dr. Hongbing Wang (Wang et al., 2003). The pK-Renilla luciferase plasmid was purchased from Promega (Madison, WI). Plasmids used for mammalian two-hybrid assays, pG5-Luc (a GAL4-luciferase reporter construct) and pACT, were obtained from Promega (CheckMate). The pBIND-SRC-1 (621–765) plasmid was previously described (Wang et al., 2013). The pBIND-mNCoR (1958–2401) and pBIND-SMRTα (2004–2517) plasmids were constructed as previously described (Wang et al., 2013). The pACT-hCAR1 plasmid was prepared by polymerase chain reaction (PCR) amplification of hCAR1 by using oligonucleotides 5′-GTA CCG AGC TCG GAT CCA ACT AGT AA-3′ and 5′-CAG GAT CCG CGG CCG CTC AGC TGC AGA T-3′, digested using BamHI (Promega) and NotI (Promega) and ligating the resulting fragment into BamHI- and NotI-cleaved pACT vector plasmid at a 1:7 molar ratio. The pBIND-TIF2 plasmid was similarly prepared by PCR amplification of TIF2 using oligonucleotides 5′-ATT CGG ATC CAT ACC ATG GAG AGA GCT-3′ and 5′-ATA AGA TCT GGA TCC CTA GCT CTG TGA-3′, digested using BamHI (Promega), and ligating the resulting fragment into BamHI-cleaved pBIND vector plasmid at a 1:10 molar ratio. All transfections were performed by using FuGENE 6 or FuGENE HD (Promega) according to the manufacturer’s recommendations.
Gene Expression Assays and Luciferase Assays.
HepG2 cells grown in flasks were transfected with FLAG-hCAR1 and CYP2B6-luciferase reporter at a 1:3 ratio with FuGENE HD (Promega) and incubated for 24 hours. Cells were trypsinized and plated in 384-well plates (CulturPlate-384, PerkinElmer, Waltham, MA) at 5000 cells/well for the primary or secondary screening assays. Cells were treated with chemicals transferred by using a pin tool 24 hours before measuring reporter luciferase activity with SteadyLite firefly luciferase reagent and an EnVision plate reader (PerkinElmer). The percentage of CAR inhibition was calculated by setting 50 µM PK11195 (positive control) to 100% inhibition and DMSO (negative control) to 0%. Unless otherwise noted, total DMSO (vehicle) concentration in all assays was maintained at or below 0.56%. Compounds were screened in a dose-responsive format (56 µM to 2.8 nM, 1:3 dilutions for 10 concentrations). PXR activity was measured in HepG2 cells stably transfected with hPXR and CYP3A4-luciferase reporter (previously described as PXR Clone1 cells (Lin et al., 2008; Li et al., 2012). Briefly, cells were treated with CLZ, PK11195, or CINPA1 at the indicated concentrations with or without 5 µM of hPXR agonist rifampicin in phenol red–free Dulbecco’s modified Eagle’s medium supplemented with 5% charcoal-dextran–treated FBS and incubated for 24 hours before Steadylite luciferase assays. The luminescence signal was detected by using an Envision plate reader (PerkinElmer), and the percentage of PXR activation was calculated.
Gene Expression Assays and GeneBLAzer Assays.
GeneBLAzer cells individually expressing the ligand-binding domains of farnesoid X receptor (FXR), glucocorticoid receptor (GR), liver X receptor (LXR) α, LXRβ, peroxisome proliferator-activated receptor (PPAR) γ, RXRα, RXRβ, or vitamin D receptor (VDR) fused to the GAL4-DNA binding domain (GAL4-DBD) were obtained from Life Technologies (Li et al., 2012; Yu et al., 2013). On activation with the respective agonist, β-lactamase is expressed under the transcriptional control of an Upstream Activator Sequence, and a FRET-based substrate (CCF2-AM) is used to measure the enzyme activity of β-lactamase. The following agonists were used at an excitatory concentration that resulted in at least 75% receptor activation (EC75): 375 nM GW4064 for FXR; 3 nM dexamethasone for GR, 50 nM TO901317 for LXRα, 32 nM TO901317 for LXRβ, 52 nM rosglitazone for PPARγ; 267 nM 9-cis retinoic acid (9-cisRA) for RXRα and β; and 0.2 nM 1α, 25-dihydroxyvitamin D3 for VDR. Control antagonists for each receptor were used as suggested by the manufacturer: 25 µM eGuggulsterone for FXR, 100 nM RU-486 for GR, 50 µM fenofibrate for LXRα/β, 10 µM GW9662 for PPARγ, 10 µM HX531 for RXRα/β, and 10 µM 22(S)-hydroxycholesterol for VDR.
Gene Expression Assays and Quantitative Real-Time PCR.
Cell lines or human hepatocytes were treated with chemicals for 24–48 hours before RNA was extracted and purified by using the Maxwell 16LEV simplyRNA tissue kit (Promega). Then, cDNA was prepared from 2 μg of RNA, and diluted cDNA was used to perform quantitative RT-PCR assays by using TaqMan probes (ABI, Thermo Fisher Scientific, Waltham, MA; 7500 Thermocycler) with 18S as the internal standard.
Mammalian Two-Hybrid Assay.
The pACT-hCAR1, pBIND-coregulator peptide and pG5-luc were cotransfected into HEK293T cells. Renilla luciferase is constitutively expressed from the pBIND plasmids. The Dual-Glo luciferase assay (Promega) was used to measure pG5-luc luciferase activity. The relative luciferase activity was determined by normalizing firefly luciferase activity with Renilla luciferase activity.
Time-Resolved Fluorescence Resonance Energy Transfer Coactivator Recruitment Assay.
The effect of putative CAR ligands on the recruitment or repression of PGC-1α binding to hCAR was evaluated by using a LanthaScreen TR-FRET assay according to the manufacturer’s instructions. Briefly, GST-hCAR-LBD (5 nM) and a mixture of Tb-anti–GST antibody/Fl-PGC-1α peptide (5 nM/125 nM) were added to each well containing titrations of test compounds or DMSO solvent control. The final chemical concentrations were 70 µM to 3.5 nM (1:3 dilutions for 10 concentration levels). DMSO and CLZ (42 µM) were used as negative (0% inhibition) and positive (100% inhibition) controls, respectively. The final DMSO concentration was 0.7% in all assay wells. Assay plates were then briefly centrifuged and incubated at room temperature for 1 hour, and TR-FRET emissions at 490 and 520 nm were measured after a 340-nm excitation. Emission signals collected on a PHERAstar plate reader (BMG Labtech, Durham, NC) were used to calculate the 520:490 TR-FRET ratio and normalized to positive and negative controls to derive individual % inhibition values. The % inhibition values were then plotted for individual chemicals. When applicable, the graphic software GraphPad Prism 5.04 (GraphPad Software, La Jolla, CA) was used to fit the data into a one-site competitive-binding equation to derive IC50 values.
Immunofluorescence.
U2OS cells were transiently transfected to express FLAG-hCAR1. After 24 hours, cells were treated with DMSO (control), 1 µM CITCO, 5 µM CINPA1, or 5 µM PK11195. Cells were fixed by using a 4% paraformaldehyde solution (Sigma-Aldrich), permeabilized by using 0.5% Triton X-100 in PBS, and incubated with anti–FLAG-antibody overnight at 4°C. Cells were washed three times with PBS after each step. Secondary antibody labeled with Alexa Fluor 555 dye was used to visualize FLAG-tagged hCAR1 (red) using a Nikon C1Si microscope (Tokyo, Japan). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue).
Chromatin Immunoprecipitation Assays.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Cherian et al., 2012). Briefly, human hepatocytes in six-well plates were maintained for 3 to 4 days in Williams’ E medium containing Primary Hepatocyte Maintenance Supplement (Life Technologies), with daily media changes. Nine wells were used for each treatment group. Hepatocytes in Fig. 8 were treated overnight with DMSO, 1 μM CITCO, 5 μM CINPA1, or 5 μM PK11195. In Fig. 9, the hepatocytes from donor 7 were treated with DMSO, 0.1 μM CITCO, 1 μM CINPA1, or 0.1 μM CITCO + 1 μM CINPA1 for 45 minutes. Proteins were cross-linked with 1% formaldehyde for 10 minutes. Cell extracts were digested for 10 minutes with 50 U of micrococcal nuclease (New England Biolabs, Ipswich, MA) at 37°C and further sonicated to yield sheared DNA fragments with an average length of 200–1000 base pairs. The sonicated samples were pelleted by centrifugation, and the supernatant was diluted 3- to 5-fold with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, and protease inhibitor cocktail). Then, 100 μl of diluted supernatant was reserved as input (10%) for each treatment. The samples were precleared with protein G-Sepharose 4 Fast Flow (GE Healthcare, Buckinghamshire, UK)) in ChIP dilution buffer (1:1) preblocked with sheared herring sperm DNA (200 μg/ml) and bovine serum albumin (500 μg/ml; Roche, Basel, Switzerland). The samples were then divided, and the remaining proteins were incubated with either anti-CAR (Clone N4111, R&D Systems), anti–RNA polymerase II (clone CTD4H8, Millipore), or control mouse IgG overnight at 4°C. The antibody-protein-DNA complex was precipitated by incubating the samples with protein G-Sepharose beads for 2 hours at 4°C. The protein-DNA complex was washed and eluted from the beads with elution buffer (1% SDS, 0.1 M NaHCO3). Cross-links were reversed, and DNA was eluted from the protein-DNA complexes by adding 200 mM NaCl and incubating overnight at 65°C. Protein was digested by incubation with proteinase K at 45°C for 2 hours. DNA was recovered and purified. Quantitative real-time PCR assays were performed to determine the change in CAR occupancy at various sites of CAR binding. The double-negative controls were a nonspecific antibody (normal mouse IgG) and primers coding for intergenic regions that do not bind CAR. Thermal cycling conditions were 95°C for 10 minutes followed by 40 cycles of 25 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. The primers used were as follows: CYP2B6-PBREM forward 5′-AGG CCC TTG GTT CAG GAA AG-3′, CYP2B6-PBREM reverse 5′-CTG CCT GTC TCA TCC TAC GC-3′; CYP2B6-XREM (dNR3) forward 5′-ATT GCA CAA CAC AGC AGG AG-3′, CYP2B6-XREM (dNR3) reverse 5′-CAA CCC ACA CTT TCC TGA CC-3′; CAR-free region forward 5′-CAG CTG GAG GGG TCA TCA AA-3′, CAR-free region reverse primer 5′-GCT AGC CAG AGA CCC TTC AC-3′; CYP3A4-XREM forward – 5′-AAG GTC ATA AAG CCC AGT TTG T-3′, CYP3A4-XREM reverse-5′-CAC CTG GGG TCA ACA CAG GAC-3′, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter (ChIP positive control with RPol) forward 5′-TAC TAG CGG TTT TAC GGG CG-3′, reverse 5′-TCG AAC AGG AGG AGC AGA GAG CGA-3.
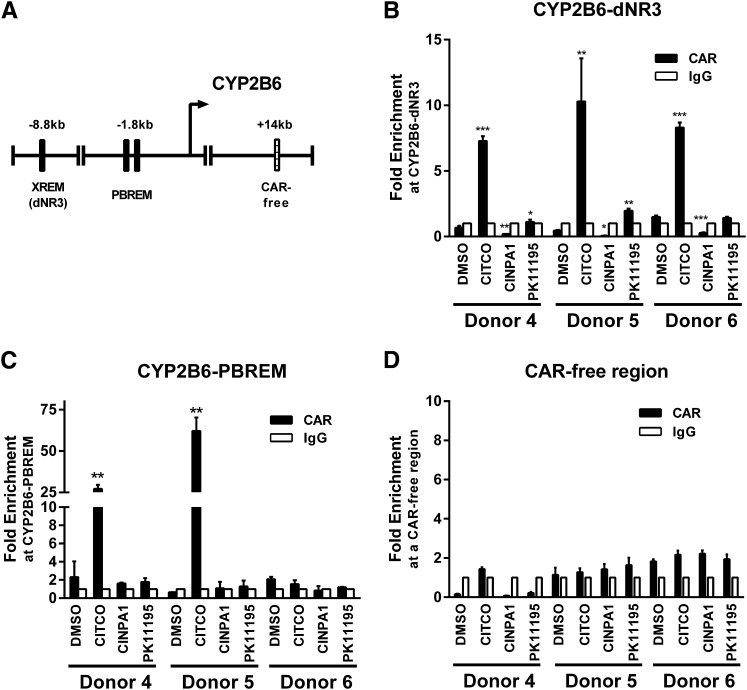
Fig. 8.
CINPA1 treatment does not allow CAR binding to DNA response elements at the CYP2B6 gene promoter in human hepatocytes. (A) PBREM and XREM (dNR3) in the CYP2B6 promoter region. (B–D) Human primary hepatocytes from three separate donors were treated overnight with DMSO, 1 µM CITCO, 5 µM CINPA1, or 10 µM PK11195. Protein complexes were cross-linked and chromatin was immunoprecipitated by using anti-CAR antibody or control IgG. CAR occupancy at two separate CYP2B6 promoter regions (dNR3 and PBREM) was determined by performing quantitative real-time PCR assays. CAR occupancy at a CAR-free intergenic region around +14 kb was also analyzed as a negative control for recruitment. Fold-enrichment normalized to IgG control was plotted. Data represent mean ± S.D. of three experiments. P values (denoted *, **, or ***) were determined by comparing compound treatments with DMSO-treated samples.
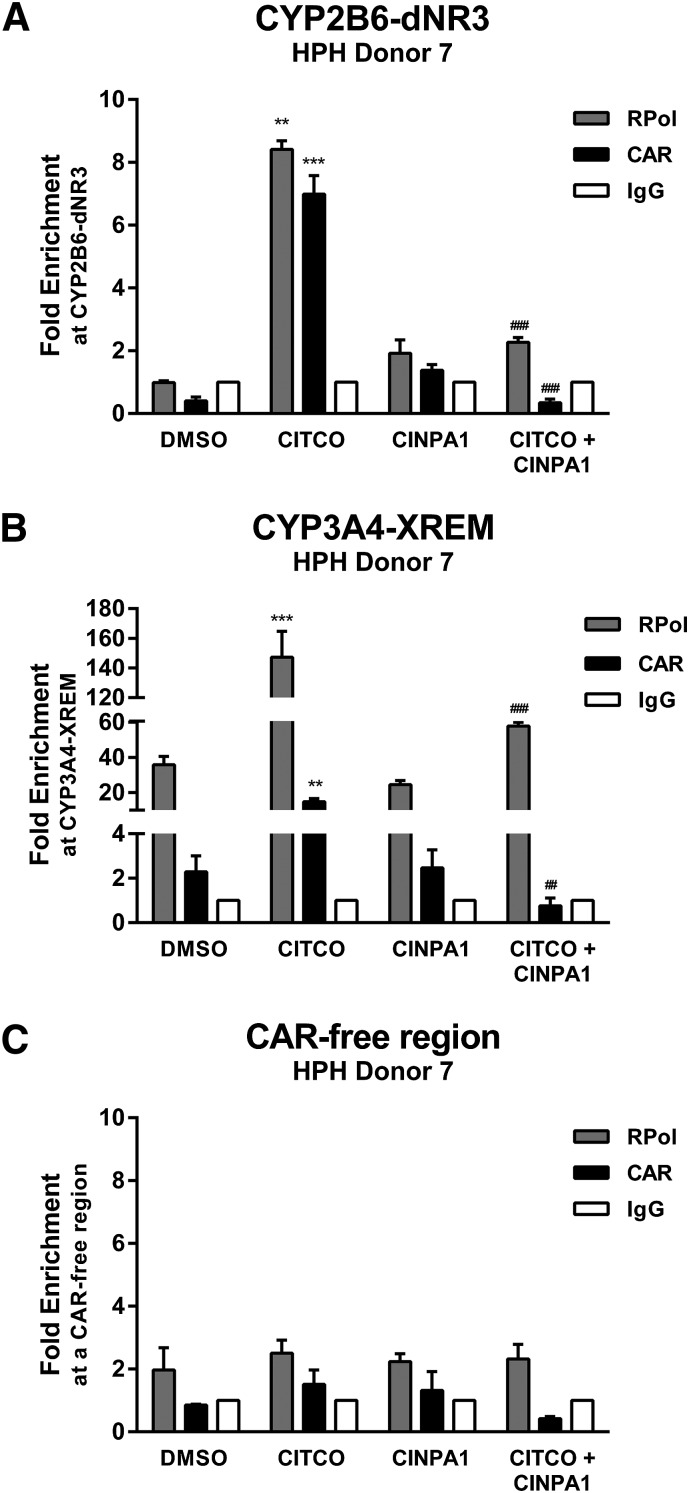
Fig. 9.
CINPA1 disrupts CITCO-activated CAR binding to DNA response elements at the CYP2B6 and CYP3A4 gene promoter regions. (A–C). Freshly plated human hepatocytes (HPH) were treated for 45 minute with DMSO, 0.1 µM CITCO, 1 µM CINPA1, or 0.1 µM CITCO + 1 µM CINPA1. Protein complexes were cross-linked, and chromatin was immunoprecipitated by using anti-CAR antibody, anti–RNA polymerase II (RPol) antibody, or control IgG. CAR or RPol occupancy at the CYP2B6-dNR3 region, CYP3A4-XREM region, and a CAR-free intergenic region (ChIP negative control) was determined by performing quantitative real-time PCR assays. Fold-enrichment was normalized to IgG control. Data represent the mean ± S.D. of three PCRs and are representative of other experiments. One-way analysis of variance with Fisher’s least significant difference test was used to compare multiple treatment groups. **P < 0.01; ***P < 0.001 compared with DMSO samples. ##P < 0.01; ###P < 0.001 compared with CITCO samples.
Statistical Analysis.
Results are expressed as the mean ± S.D. of at least three independent experiments. Significance was established when P < 0.05. Student's t test was used for comparison of the means between two groups as specified. One-way analysis of variance was used to analyze data sets with multiple comparisons between treatments, and experiment-wide significance level α was set to 0.05.
Results
Identification of CINPA1 as an Inhibitor of CAR-Mediated Transactivation.
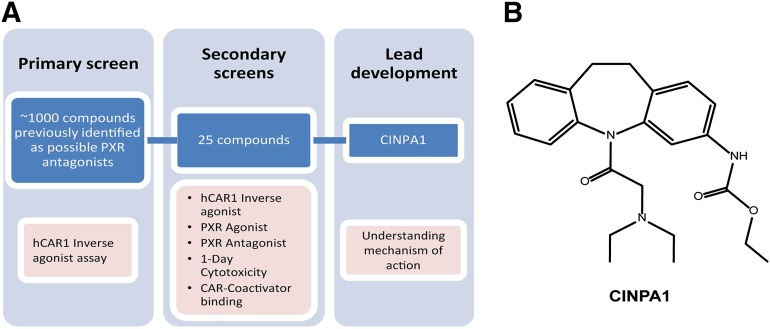
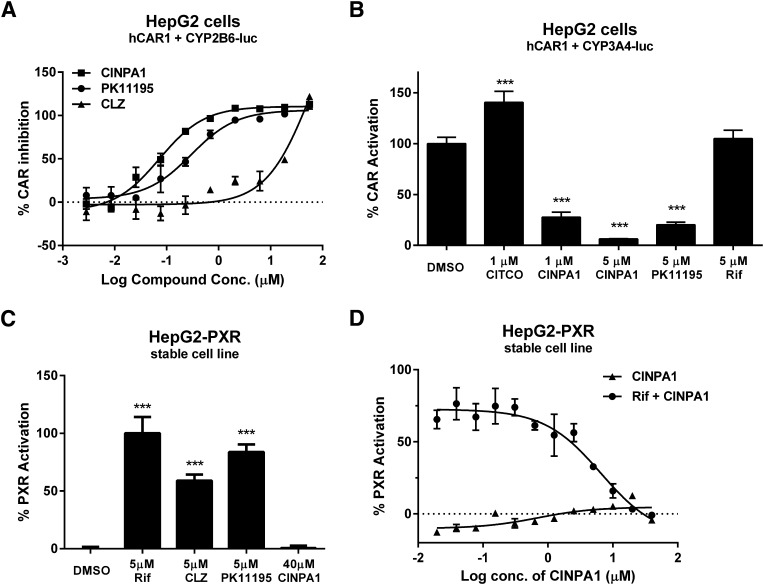
We focused our efforts on identifying CAR inhibitors that are not agonists of PXR. For this purpose, we performed our primary screen using a library of ∼1000 chemicals previously identified as possible antagonists of PXR (Wenwei Lin et al., unpublished work) in a dose-responsive format. A flowchart of our screening strategy is illustrated in Fig. 1A. HepG2 cells were transiently transfected with pcDNA-FLAG-hCAR1 and CYP2B6-luciferase plasmids and treated with varying concentrations of the test compounds in 384-well plates for 24 hours, and CAR-mediated luciferase activity was measured. Twenty-five hits, defined as those displaying dose-responsive inhibitory effect on the constitutively active hCAR1, were selected and tested in various secondary screening assays. CINPA1 (Fig. 1B) was selected based on its potent inhibitory effect on hCAR1 and lack of detectable agonistic activity against PXR. CINPA1 inhibited CAR-mediated CYP2B6-luciferase activity with an IC50 of 70 nM, which is slightly more potent than PK11195 (Fig. 2A). CINPA1 inhibition of CAR was not limited to the CYP2B6 gene but was also seen in CAR-mediated CYP3A4-luciferase activity (Fig. 2B). In contrast to other known CAR inhibitors, such as PK11195 and CLZ, CINPA1, even at 40 µM, did not exhibit any agonistic effect on PXR-regulated gene expression (Fig. 2, C and D). CINPA1 is a weak antagonist of PXR with an estimated IC50 value of 6.6 µM in a stable HepG2 cell line coexpressing PXR and a CYP3A4-luciferase plasmid (HepG2-PXR clone 1, as previously described (Lin et al., 2008; Li et al., 2012) (Fig. 2D). Interestingly, CINPA1 is also able to modestly inhibit the CITCO-induced activation of the hCAR3 isoform, with no partial agonistic activity (Supplemental Fig. 1).
Fig. 1.
CINPA1 is identified as an inhibitor of CAR activity through a directed small-molecule screening. (A) Flowchart of screening strategy. (B) Chemical structure of CINPA1.
Fig. 2.
CINPA1 inhibits CAR-mediated transactivation without activating PXR. (A) HepG2 cells were transfected with plasmids expressing hCAR1 and CYP2B6-luc reporter. After 24 hours’ incubation, cells were treated with DMSO (negative control) or indicated concentrations (Conc.) of CINPA1, CLZ, or PK11195 (positive control). Luciferase activity was measured 24 hours after treatment by using SteadyLite reagent. Activity in DMSO-treated samples was set as 0% inhibition; activity in samples treated with 50 µM PK11195 was set as 100% CAR inhibition. (B) HepG2 cells were transiently transfected with plasmids expressing hCAR1, CYP3A4-luc reporter and control plasmid pTK-RL. After 24 hours’ incubation, cells were treated with DMSO (control), 1 µM CITCO, 5 µM PK11195, 1 µM or 5 µM CINPA1, or 5 µM rifampicin (Rif) for 24 hours. Dual luciferase activity was measured 24 hours after treatment using Dual-Glo luciferase reagent. Firefly luciferase was normalized to Renilla luciferase, and % CAR activation was calculated by setting DMSO to 100%. ***P < 0.001 comparing treatments with DMSO. (C) HepG2 cells stably expressing hPXR and CYP3A4-luciferase reporter (PXR clone 1) were treated with indicated compounds for 24 hours. Luciferase activity was measured 24 hours after treatment by using SteadyLite reagent. ***P < 0.001 comparing treatments with DMSO. (D) HepG2-PXR Clone 1 cells were treated with CINPA1 in a dose-responsive format (40 µM to 20 nM; 1:2 dilutions for 12 concentrations) in the presence or absence of 5 µM rifampicin (Rif) for 24 hours before luciferase assay using Steadylite. % PXR activation was calculated by setting DMSO- (negative control) treated cells with 0% and 5 µM rifampicin-(positive control) treated cells with 100%. GraphPad Prism was used to fit the data into a dose-response stimulation equation to derive IC50 values.
CINPA1 is a Potent CAR-Specific Inhibitor.
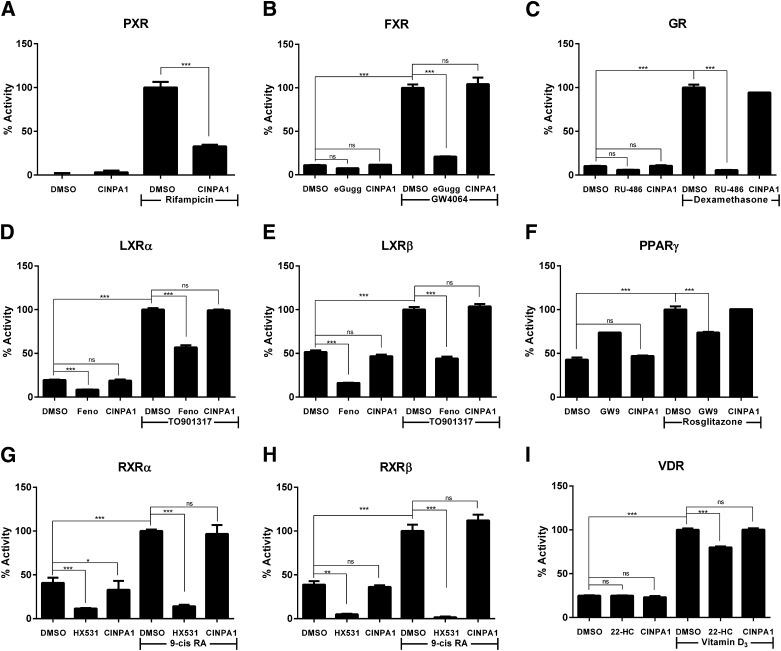
CAR and PXR have flexible ligand-binding pockets (Wu et al., 2013) that allow binding of many different chemical core structures. CAR inhibitors identified in the past had limitations because of their PXR agonistic effect, which made them less useful for dissecting CAR-specific gene manipulations, especially in the context of genes regulated by many nuclear receptors. We evaluated CINPA1 in a panel of GeneBLAzer cells individually expressing the ligand-binding domains of selected nuclear receptors closely related to PXR or CAR: FXR, GR, LXRα, LXRβ, PPARγ, RXRα, RXRβ, or VDR, fused to the GAL4-DNA binding domain (GAL4-DBD). CINPA1 did not activate any of the nuclear receptors tested (Fig. 3, A–I). Whereas CINPA1 weakly inhibited PXR-mediated gene expression (Fig. 3A), it did not attenuate the agonist-induced activation of any of the receptors tested in the GeneBLAzer assays (Fig. 3, B–I).
Fig. 3.
CINPA1 is a weak antagonist of PXR, but it does not modulate the activity of other nuclear receptors. (A) HepG2-PXR Clone 1 cells were treated with DMSO or 18 µM of CINPA1 in the presence or absence of 5 µM rifampicin for 24 hours before luciferase assay using Steadylite. (B–I) GeneBLAzer cells were plated according to the manufacturer’s instructions and treated with DMSO (control), a predetermined concentration of the indicated nuclear receptor (NR) antagonist (as detailed in the description of our experimental procedures), or 18 µM of CINPA1. NR-mediated β-lactamase activity was measured as a FRET ratio after 24-hour treatment. Data are presented as % Activity, determined by setting the maximum FRET signal obtained for each receptor (in the respective agonist + DMSO wells) to 100%. One-way analysis of variance with Bonferroni corrections for multiple comparisons was used to analyze data (as noted in brackets), and statistical significant differences are indicated by *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant, P > 0.05. eGugg, Guggulsterone; Feno, Fenofibrate; GW9, GW9662; 22HC, 22(S)-hydroxycholesterol; 9-cis RA, 9-cis retinoic acid.
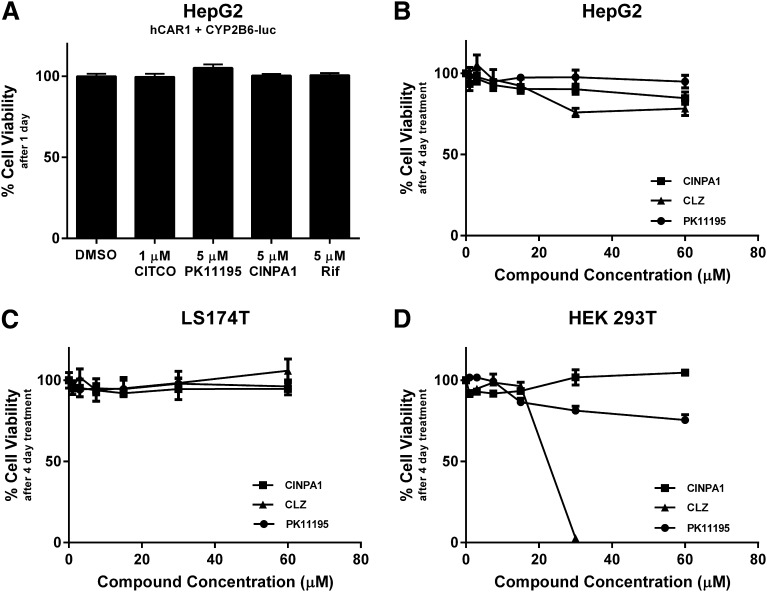
In a 1-day cell viability assay, 5 µM CINPA1 did not exert any toxic effects on HepG2 cells prepared similarly as the luciferase screening assays. Within the concentration range effective in inhibiting CAR and PXR, CINPA1 does not exhibit general cytotoxicity in any of the cells lines evaluated in an extended 4-day cell viability assay (Fig. 4, B–D). At higher concentrations (30 and 60 µM), all chemicals tested, CINPA1, CLZ, and PK11195 were cytotoxic to varying degrees in different cell lines.
Fig. 4.
CINPA1 is not cytotoxic within the concentration range effective for modulating CAR and PXR. (A) HepG2 cells were transfected with hCAR1 and CYP2B6-luc plasmid to mimic conditions used in the luciferase screening assays. Cells were treated with indicated compounds for 24 hours, and viability was measured by using CellTiter Glo reagent. Rif, rifampicin. No significant change was observed between treatments. (B) HepG2. (C) LS174T. (D) HEK293T cells were treated with DMSO (control), CINPA1, CLZ, or PK11195 at the indicated concentrations. Cell viability was measured 4 days posttreatment by using CellTiter Glo reagent. Viability of cells treated with DMSO (negative control) was set to 100% in all assays.
CINPA1 Inhibits Expression of CAR-Regulated Genes in Cell Lines and Human Hepatocytes.
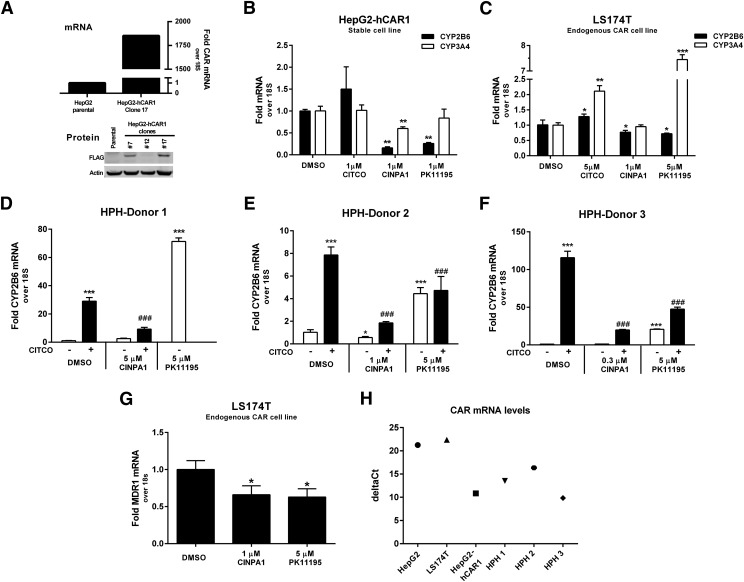
CAR is a transcription factor that regulates the expression of various gene families, including those involved in drug metabolism and lipogenesis, and CYP2B6 is considered a principal CAR-regulated gene. We established a stable clone of HepG2 cells that overexpress hCAR1 (HepG2-hCAR1, clone 17; Fig. 5A). We also examined various cell lines and identified LS174T as moderately expressing endogenous CAR and its downstream targets; LS174T has previously been reported to express endogenous PXR with inducible activity (Wang et al., 2013). In HepG2-hCAR1, where CAR but not PXR is overexpressed and known to be constitutively active, CINPA1 substantially and significantly attenuated the levels of both endogenous CYP2B6 and CYP3A4 genes although PK11195 attenuated the level of CYP2B6 without significantly affecting that of CYP3A4 (Fig. 5B); however, in LS174T cells that endogenously express both CAR and PXR, CINPA1 moderately reduced the levels of CYP2B6 without significantly affecting CYP3A4, whereas PK11195 reduced the levels of CYP2B6 but increased that of CYP3A4 (Fig. 5C), most likely working through PXR activation as previously reported (Li et al., 2008).
Fig. 5.
CINPA1 attenuates CAR-mediated gene expression in CAR-expressing cell lines and human primary hepatocytes. (A) FLAG-CAR1 expression in the HepG2-hCAR1 stable clone was evaluated. CAR mRNA expression in HepG2-hCAR1 Clone 17 cells was compared with parental HepG2 cells using quantitative RT-PCR. 18S was used as the internal control and showed no change in threshold cycle (Ct) between the two cell lines. FLAG-tagged CAR protein expression levels in a few selected clones were analyzed by Western blot using anti-FLAG antibody. Clone 17 had the highest expression levels of hCAR1 and was used in subsequent studies. (B) HepG2-hCAR1 cells were treated with DMSO (control), 1 µM CITCO, 1 µM CINPA1, or 1 µM PK11195 in triplicated wells for 24 hours. RNA was extracted, cDNA synthesized, and gene expression analyzed by performing real-time PCR using Taqman probes for each gene. (C) LS174T cells were maintained in 10% charcoal-dextran–treated, FBS media for 4 days before the experiment. Cells were treated with DMSO, 5 µM CITCO, 1 µM CINPA1, or 5 µM PK11195 for 24 hours before RNA extraction. (B) and (C), P values (*, **, or ***) were determined by comparing compound-treated samples to DMSO-treated samples in a Student’s t test. (D–F) Human primary hepatocytes (HPHs) were maintained in Williams’ E media with supplements for 4 days before treatment. Cells were treated in the presence or absence of 1 µM CITCO, and DMSO, 5 µM PK11195, or CINPA1 (5 µM treatment of donor 1, 1 µM for donor 2, and 0.3 µM for donor 3) for 48 hours. RNA extracted was used for cDNA synthesis and measured by performing quantitative real-time PCR assays with Taqman probes. (D) HPH-donor 1, PK11195 alone was tested, but CITCO + PK11195 could not be tested because of limited hepatocyte availability. Statistical significance was determined using one-way analysis of variance by comparing compound-treated samples to DMSO-treated samples (* or ***) or to CITCO-treated samples (###). (G) LS174T cells were maintained in 10% charcoal-dextran–treated FBS media for 4 days before the experiment. Cells were treated with DMSO, 1 µM CINPA1, or 5 µM PK11195 for 24 hours. MDR1 mRNA was normalized to the internal control 18S, and DMSO-treated samples were set to 1. *P < 0.05 compared with DMSO samples in a Student’s t test. (H) Comparison of CAR mRNA levels in the cells lines and human primary hepatocytes used for gene expression analysis. RNA from the DMSO-treated cells (B–F) was used for cDNA synthesis, and hCAR was measured by quantitative real-time PCR. deltaCt (Ct = threshold cycle) values were calculated by normalizing to the internal control 18S. Lower deltaCt values indicate more CAR mRNA detected in the respective cell line/donor.
Primary human hepatocytes express varying levels of endogenous PXR and CAR, resulting in varying levels of P450 enzyme. CAR in primary hepatocytes is predominantly cytoplasmic; therefore, the basal levels of CAR target genes such as CYP2B6 are likely not CAR-mediated (Honkakoski et al., 1998; Maglich et al., 2003) but can be induced by CAR activators such as CITCO. We tested the effect of CINPA1 on PXR and CAR in primary human hepatocytes from seven different donors and show the results of a representative set of three donors. Fresh or cryopreserved hepatocytes from each donor were obtained and treated with CINPA1 or PK11195 in the presence or absence of CAR activator CITCO. Because hepatocyte sample availability was limited, each donor shown was tested with decreasing concentrations of CINPA1 (5 µM treatment of donor 1, 1 µM for donor 2, and 0.3 µM for donor 3). In all three donors, CITCO induced CYP2B6 levels, indicating that CAR is functional. CINPA1 treatment effectively inhibited CITCO-induced CAR transactivation of the CYP2B6 gene in all three donors (Fig. 5, D–F). We noticed that in donor 2 (Fig. 5E), but not in donors 1 and 3 (Fig. 5, D and F), CINPA1 alone significantly reduced basal CAR transactivation, reflecting the donor variations typically observed among primary human hepatocytes. In contrast to CINPA1, PK11195 alone increased CYP2B6 transcription in all three donors (Fig. 5, D–F), consistent with a previous report demonstrating that PK11195 induces CYP2B6 through PXR activation (Li et al., 2008). Interestingly, both CINPA1 and PK11195 reduced MDR1 levels in LS174T cells (Fig. 5G). Taken together, these data show that unlike PK11195, CINPA1 specifically targets and reduces CAR-mediated gene expression without activating PXR. Figure 5H shows the CAR mRNA levels in the various cell models used to obtain these data.
CINPA1 Disrupts CAR Interaction with Coactivators and Enhances Recruitment of Corepressors but Does Not Reduce CAR Protein Levels or Nuclear Translocation.
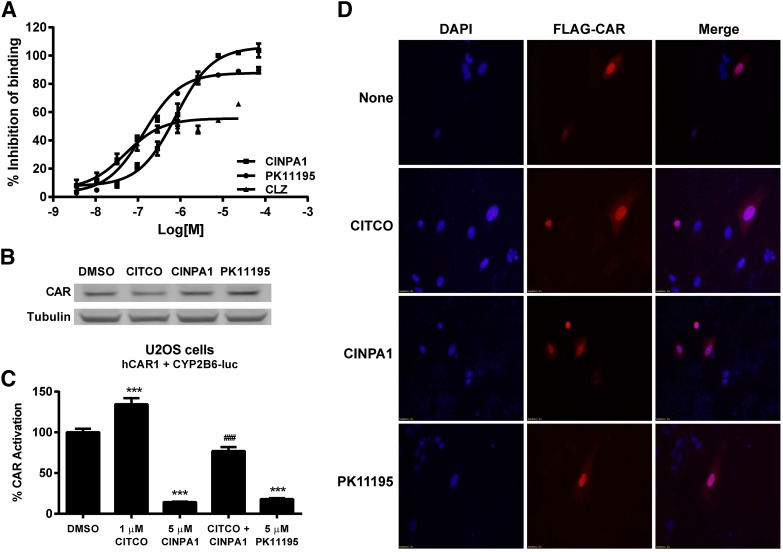
We first used a LanthaScreen TR-FRET assay to determine the effect of CINPA1 on the constitutive interaction between CAR-LBD and a coactivator peptide. In this assay, GST-tagged CAR-LBD constitutively interacts with a fluorescently labeled PGC-1α coactivator peptide and emits a strong FRET signal. CAR inverse agonist–binding to the CAR-LBD results in a reduced FRET signal. We compared CINPA1 with known CAR inverse agonists CLZ and PK11195 in this assay. As shown in Fig. 6A, CINPA1 efficiently inhibits CAR-LBD interaction with the coactivator peptide, indirectly suggesting that CINPA1 is a ligand of CAR. CINPA1 had no effect on the protein levels of CAR in human hepatocytes as illustrated by Western blotting in Fig. 6B, which is representative of similar results from two other hepatocyte donors. CINPA1 did not affect the mRNA levels of either PXR or CAR in human hepatocytes from the same donor used for protein quantification (donor 8) (Supplemental Fig. 2). Exogenous CAR is functional in U2OS cells (Fig. 6C), and these cells were chosen for immunofluorescence assays for their flat morphology and large cytoplasmic extensions (Fritz et al., 2009). In U2OS cells expressing FLAG-hCAR1 and immunostained with FLAG antibody, CAR localized to the nucleus irrespective of treatment conditions (DMSO, CITCO, CINPA1, or PK11195). This result indicates that CINPA1 treatment does not alter the nuclear localization of CAR (Fig. 6D).
Fig. 6.
CINPA1 does not reduce protein levels or alter nuclear localization of CAR. (A) CAR binding to PGC-1α coactivator peptide was determined by using the LanthaScreen fluorescent coregulator peptide assay. Fluorescein-labeled PGC-1α peptide (125 nM) was complexed with GST-hCAR-LBD (5 nM) and Tb-anti–GST antibody (5 nM) as described in experimental procedures. Compounds (CLZ, PK11195, or CINPA1) at concentrations ranging from 70 µM to 3.5 nM (1:3 dilutions for 10 concentration levels) were used along with DMSO as a negative control, and CLZ (42 µM) was used as a positive control. % Inhibition of binding was calculated for each treatment, and the data were normalized to the positive control (42 µM CLZ, 100% inhibition) and negative control (DMSO, 0% inhibition). (B) Human primary hepatocytes (donor 8) were treated with DMSO, 1 µM CITCO, 5 µM CINPA1, or 5 µM PK11195 for 48 hours. Lysates were prepared, and CAR expression was analyzed by Western blot using anti-CAR monoclonal antibody (clone N4111). Tubulin was used as a loading control. Data shown are representative of at least three hepatocyte donors tested. (C) U2OS cells were transiently transfected with plasmids expressing FLAG-hCAR1, CYP2B6-luc reporter, and control plasmid pTK-RL. After 24-hour incubation, cells were treated with DMSO (control), 1 µM CITCO, 5 µM CINPA1, 1 µM CITCO + 5 µM CINPA1, or 5 µM PK11195. Dual luciferase activity was measured 24 hours after treatment. Renilla luciferase was used to normalize firefly luciferase values, and % CAR activation was calculated by setting DMSO to 100%. Statistical significance was determined using one-way analysis of variance for comparisons between multiple treatment groups. ***P < 0.001 compared with DMSO treatment; ###P < 0.001 comparing CITCO + CINPA1 with CITCO-treated cells. (D) U2OS cells were transfected to express FLAG-hCAR1. Cells were treated with indicated chemicals for 2 hours before fixing and staining with anti–FLAG-M2 antibody (Sigma-Aldrich). Secondary antibody labeled with Alexa Fluor 555 dye was used to visualize FLAG-tagged hCAR1 (red) using a Nikon C1Si microscope. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue).
Remarkable species selectivity is exhibited by CAR in its ligand binding and activation profiles. Ligands that bind human CAR are largely unable to bind rodent CAR and vice versa. We analyzed CAR-mediated transcription at the Cyp2b10 gene in mouse primary hepatocytes and found that CINPA1 does not inhibit mouse CAR activity (Supplemental Fig. 3A). Furthermore, CINPA1 did not inhibit luciferase reporter activity driven by transiently expressed mouse CAR in HepG2 cells (Supplemental Fig. 3B) (Sueyoshi et al., 1999; Li et al., 2008).
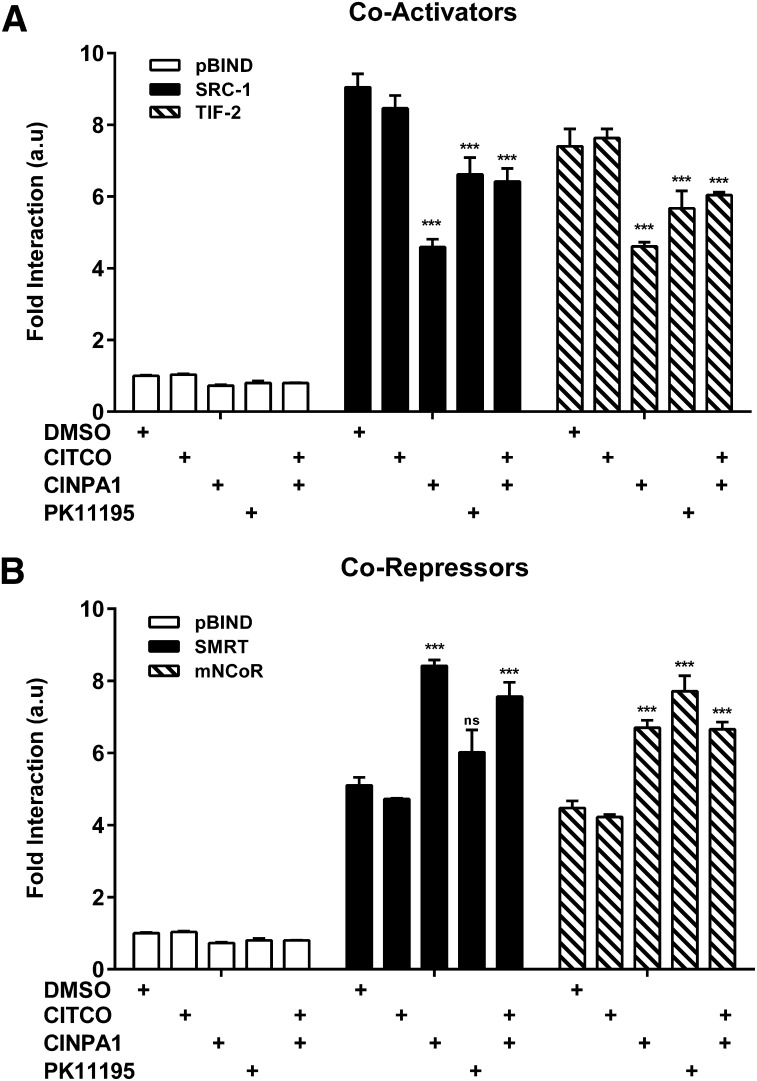
Activated CAR protein binds response elements on DNA and recruits coactivator proteins that are essential for transcriptional activation (Forman et al., 1998; Min et al., 2002), whereas inverse-agonist bound CAR binds corepressor proteins (Jyrkkarinne et al., 2003; Bae et al., 2004; Kublbeck et al., 2011). We further determined the effect of CINPA1 on the interaction between CAR-LBD and coregulators by using mammalian two-hybrid systems. As shown in Fig. 7A, CINPA1 treatment resulted in reduced SRC-1 and TIF-2 coactivator binding to CAR-LBD in the presence or absence of agonist CITCO. This finding is in congruence with that in earlier gene expression assays, in which CINPA1 reduced basal and ligand-initiated CAR activity (Fig. 5, B–F) and the TR-FRET assay (Fig. 6A). Conversely, CINPA1 treatment resulted in increased interaction of CAR-LBD with the corepressor proteins SMRTα and mNCoR. Thus, we show that CINPA1 binding to CAR-LBD results in reduced coactivator recruitment and increased corepressor interaction, which contributes to explaining the inhibitory mechanism of CINPA1.
Fig. 7.
CINPA1 disrupts interaction of CAR with coactivators and enhances corepressor binding. (A and B) Mammalian two-hybrid assays were set up in HEK293T cells transfected with expression plasmids encoding VP16AD-hCAR1 fusion protein, GAL4DBD-coregulator fusion proteins, and the reporter plasmid pG5luc . Cells were treated with DMSO (control), 5 µM CITCO, 5 µM CINPA1, 10 µM PK11195, or 1 µM CITCO + 5 µM CINPA1 for 24 hours before measurement of luciferase activities. Fold interaction represents pG5luc reporter activity normalized to the Renilla luciferase internal control. Data presented are the mean ± S.D. of at least three independent transfections. ***P < 0.001 compared with DMSO treatment within the same coregulator set. ns, not significant.
CINPA1 Inhibits CAR Recruitment to the CYP2B6 Gene Promoter.
Chromatin immunoprecipitation assays were used to evaluate the effect of CINPA1 on CAR recruitment to promoter regions of CAR-regulated genes in human hepatocytes. CYP2B6 is a well characterized CAR-regulated gene with a defined PBREM around −1.8kb upstream and an XREM (which contains a DR4 motif also called an NR3 binding site) around −8.8kb upstream of its transcriptional start site, respectively (Fig. 8A) (Wang et al., 2003). Both PXR and CAR are capable of binding to these response elements, depending on their differential activation states. As shown in Fig. 8, B and C, CITCO treatment enhances recruitment of CAR to the XREM (dNR3) regions of the CYP2B6 gene in all three donors (Fig. 8B) and to the PBREM in donors 4 and 5 but not donor 6 (Fig. 8C), consistent with the increased CYP2B6 mRNA levels in response to CITCO shown in Fig. 5, D–F. CINPA1 treatment prevents CAR recruitment to the dNR3 in all three donors (Fig. 8B), but the effect on the PBREM is less consistent among different donors (Fig. 8C). Similar donor variation on the basal level of CYP2B6 in response to CINPA1 was also observed in Fig. 5, D–F. CAR protein was below detection levels (when compared with normal mouse IgG) in ChIP experiments at the CYP2B6 promoter region in two other human hepatocyte donors (data not shown), further demonstrating the donor-to-donor variation in human hepatocytes. Figure 8D illustrates the relative absence of CAR compared with mouse IgG at a CAR-free intergenic region within the CYP2B6 gene, a negative control for ChIP.
In hepatocytes, CITCO treatment consistently enhanced recruitment of CAR to the XREM (dNR3) region of the CYP2B6 promoter (Fig. 8B) and induced the expression of CYP2B6, which is effectively inhibited by CINPA1 cotreatment (Fig. 5, D–F). Consistent with these observations, CINPA1 effectively reduced the CITCO-mediated recruitment of CAR and RNA polymerase II to the XREM (dNR3) region of the CYP2B6 promoter (Fig. 9A). As expected, CINPA1 also reduced the CITCO-mediated recruitment of CAR and RNA polymerase II to the XREM region of the CYP3A4 promoter (Fig. 9B). As a negative control, CAR and RNA polymerase II were not recruited to the CAR-free region (Fig. 9C). The parallel recruitment of CAR and RNA polymerase II to the promoter regions of CYP2B6 observed in the ChIP assays (Fig. 9) is consistent with the corresponding transcriptional activity shown in Fig. 5. Taken together, these data suggest that CINPA1 acts, in part, by reducing the association of CAR with its target promoter.
Discussion
CAR is an important xenobiotic sensor that controls the metabolism and elimination of many xenobiotics and endobiotics. Downstream target genes of CAR are involved in various important physiologic processes such as energy homeostasis, cell cycle regulation, and cell-cell communication (Maglich et al., 2004; Molnar et al., 2013). Dysregulation of these genes often results in pathologic conditions like insulin resistance, fatty liver disease, chemical carcinogenesis, and tumor promotion (Huang et al., 2005; Breuker et al., 2010; Takwi et al., 2014). There is a need to develop potent and specific chemical tools to uncover the molecular mechanisms governing CAR activation and its regulation of target gene expression.
CAR displays several distinctive characteristics in its activation. Endogenously expressed CAR, such as that in human hepatocyte cultures, is activated by both direct and indirect activators, resulting in nuclear accumulation of the receptor; but CAR exogenously expressed in immortalized cell lines, such as HepG2 cells, results in spontaneous nuclear accumulation and constitutive activation in the absence of an agonist (Baes et al., 1994; Honkakoski et al., 1998; Maglich et al., 2003). Many naturally occurring splice variants of CAR have also been identified, but their individual contributions to CAR physiology remain unclear.
A few inhibitors of hCAR with varying potencies have been previously reported. CLZ, meclizine, and androstanol moderately repress hCAR function in various assays (Moore et al., 2000, 2002; Tzameli et al., 2000; Huang et al., 2004). Interestingly, CLZ, in addition to being a good mouse CAR agonist, may act as an activator of human CAR depending upon cell-type–specific factors (Jyrkkarinne et al., 2003; Auerbach et al., 2005; Simonsson et al., 2006). Li and colleagues identified a well known peripheral benzodiazepine receptor ligand, PK11195, as a CAR inhibitor—the most potent inhibitor of CAR identified thus far (Li et al., 2008). Interestingly, all the known CAR inhibitors are also moderate-to-potent activators of PXR function, complicating expression levels of their shared target genes (Jones et al., 2000; Moore et al., 2002; Huang et al., 2007; Li et al., 2008; Lau et al., 2011).
With the aim of identifying specific CAR inhibitors that do not activate PXR function, we used a directed high-throughput screening strategy and identified CINPA1 as a potent inhibitor of hCAR activity. The flexible LBD of PXR and, to some extent, CAR allow them to bind a broad range of chemical structures, making it hard to design structure-based chemical libraries for screening. For our primary screen of CAR inhibition, we started with ∼1000 compounds shortlisted as putative PXR antagonists from a previous screen, which allowed us to search for CAR inhibitors from a set of chemicals that would not activate PXR. CINPA1 was identified as the most potent among these in its ability to inhibit hCAR1. As expected, since the screen was started with compounds that inhibit PXR, CINPA1 is a weak antagonist of PXR. Most importantly, CINPA1 does not activate PXR-mediated gene expression, distinguishing it from previously known CAR inhibitors. CINPA1 binding results in reduced coactivator interaction with CAR-LBD, suggesting that the site of CINPA1 binding is most likely the ligand-binding pocket of CAR.
Because CAR has distinctive subcellular localization and basal activity in primary hepatocytes and immortalized cell lines, we have used both primary hepatocytes and cell lines to evaluate the effect of CINPA1 on CAR. CYP2B6 is the best-characterized CAR-regulated P450 enzyme. CYP2B6 expression can be upregulated via activation of either CAR or PXR, but it is regulated primarily by activated CAR. The CYP2B6 gene promoter has multiple xenobiotic-response elements concentrated mainly at two loci. The PBREM region at ∼1.8 kb upstream, harboring two DR4 motifs, and the 400-bp distal XREM region, containing one DR4 motif (and a few half sites), are the main binding sites for CAR (Wang et al., 2003). Human hepatocytes express endogenous CAR, which is not constitutively active but inducible by activators such as CITCO. There is considerable variation between donors, especially the basal CAR activity and its response to CINPA1 treatment; however, the induction of CAR target genes and CAR binding to the target promoter by CAR activator CITCO and the inhibitory effect of CINPA1 on such inductions by CITCO are more consistent among donors. Importantly, although CAR target genes such as CYP2B6 are induced by the activation of CAR, the expression of these CAR target genes is also regulated by pathways independent of CAR and PXR. The activation states of other nuclear receptors, such as GR (Timsit and Negishi, 2007; Lim and Huang, 2008) and hepatocyte nuclear factor 4α (HNF4α) (Tirona et al., 2003; Ding et al., 2006), could also contribute to the regulation of PXR and CAR-mediated gene expression. Therefore, it is not surprising that the response of the basal CYB2B6 levels to CINPA1 varies among primary hepatocytes from different donors.
Treatment with CINPA1 does not alter the nuclear localization or reduce the levels of CAR in the cells. Consistent with previous reports (Baes et al., 1994), exogenously expressed CAR in immortalized cell lines accumulates in the nucleus, which was not altered by CINPA1 treatment as illustrated by the results of immunofluorescence assays; however, CAR interaction with coregulators is modulated by the presence of CINPA1. The results of mammalian two-hybrid assays revealed reduced CAR binding to the coactivators SRC-1 and TIF-2 in the presence of CINPA1. No substantial increases in CAR-coactivator interaction were seen with the CAR agonist CITCO, possibly due to the high constitutive activity of CAR. This finding is consistent with gene expression data from immortalized cells overexpressing hCAR1 (Fig. 5B) and has been previously reported by others (Chen et al., 2010). Conversely, increased interaction of CINPA1-associated CAR to the corepressors SMRTα and mNCoR was evident and reinstates the inhibitory effect of CINPA1. It is possible for small-molecule inhibitors that bind the LBD of a nuclear receptor to change receptor conformation such that coactivator binding and/or DNA-binding are compromised. For instance, MDV-3100 (enzalutamide) is an androgen receptor antagonist that binds in the ligand binding pocket of the receptor but effectively reduces nuclear translocation, inhibits coactivator binding, and prevents androgen receptor binding to DNA (Tran et al., 2009). Our data suggest that CINPA1 might have a similar mechanism of action for CAR inhibition wherein it binds the LBD of CAR but alters the coactivator recruitment (Fig. 6A) and DNA-binding properties of CAR.
CAR and PXR exhibit a high level of redundancy in their role as xenobiotic sensors. Their flexible ligand-binding domains make it hard to design chemical entities that bind either receptor specifically. Lack of specific chemical modulators makes dissecting the separate functions of these receptors a difficult task. All CAR inhibitors previously identified are also potent activators of PXR, rendering them ineffective in assay environments in which both receptors are present and active, such as hepatocytes or whole-animal models. Our studies identify and characterize CINPA1 as a potent inhibitor of CAR that does not activate PXR. CINPA1 interacts with CAR-LBD and reduces coactivator recruitment. The reduction in CAR-mediated target gene expression with CINPA1 treatment is consistent with reduced CAR and RNA polymerase II recruitment to the promoter regions of those genes. CAR is responsible for PB-induced hepatocellular carcinoma (Yamamoto et al., 2004; Huang et al., 2005) and decreased effectiveness of anticancer drugs in ovarian cancer (Wang et al., 2014). A unique chemical entity like CINPA1 could be further developed for therapeutic purposes.
Supplementary Material
Acknowledgments
The authors thank Dr. Martin Privalsky for providing the SMRTα construct, Dr. Hongbing Wang for providing the CYP2B6-luc and pCR3-mouse CAR constructs, Dr. David Moore for providing the hCAR1 construct, Asli Goktug for technical assistance, other members of the Chen research laboratory for valuable discussions, and Dr. Cherise Guess for editing the manuscript.
Abbreviations
- CAR
constitutive androstane receptor
- ChIP
chromatin immunoprecipitation
- CITCO
6-(4-chlorophenyl)imidazo [2,1-b][1,3] thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl) oxime
- CLZ
clotrimazole
- DMSO
dimethylsulfoxide
- FBS
fetal bovine serum
- FXR
farnesoid X receptor
- GR
glucocorticoid receptor
- GW9662
2-chloro-5-nitro-N-phenylbenzamide
- HX531
4-(7,8,9,10-tetrahydro-5,7,7,10.10-pentamethyl-2-nitro-5H-benzo[b]naphtho[2,3-e][1,4]-diazepin-12-yl)-benzoic acid
- LBD
ligand-binding domain
- LXR
liver X receptor
- P450
cytochrome P450
- PB
phenobarbital
- PBREM
phenobarbital-responsive enhancer module
- PCR
polymerase chain reaction
- PK11195
1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide
- PPAR
peroxisome proliferator-activated receptor
- PXR
pregnane X receptor
- RXR
retinoid X receptor
- TO901317
N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]benzenesulfonamide
- TR-FRET
time-resolved fluorescence resonance energy transfer
- VDR
vitamin D receptor
- XREM
xenobiotic responsive enhancer module
Authorship Contributions
Participated in research design: Cherian, Lin, Chen.
Conducted experiments: Cherian, Lin, Wu, Chen.
Performed data analysis: Cherian, Lin, Chen.
Wrote or contributed to the writing of the manuscript: Cherian, Chen.
Footnotes
This work was supported by the American Lebanese Syrian Associated Charities, St. Jude Children’s Research Hospital, and the National Institutes of Health (NIH) [Grants R01-GM086415, R01-GM110034, and P30-CA21765]. Human primary hepatocytes were obtained through the Liver Tissue and Cell Distribution System (Pittsburgh, PA), which is funded by NIH Contract N01-DK-7-0004/HHSN267200700004C.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. (2003) Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res 31:3194–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Stoner MA, Su S, Omiecinski CJ. (2005) Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3). Mol Pharmacol 68:1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y, Kemper JK, Kemper B. (2004) Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP). DNA Cell Biol 23:81–91. [DOI] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. (1994) A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol 14:1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuker C, Moreau A, Lakhal L, Tamasi V, Parmentier Y, Meyer U, Maurel P, Lumbroso S, Vilarem MJ, Pascussi JM. (2010) Hepatic expression of thyroid hormone-responsive spot 14 protein is regulated by constitutive androstane receptor (NR1I3). Endocrinology 151:1653–1661. [DOI] [PubMed] [Google Scholar]
- Chen F, Zamule SM, Coslo DM, Chen T, Omiecinski CJ. (2013) The human constitutive androstane receptor promotes the differentiation and maturation of hepatic-like cells. Dev Biol 384:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Tompkins LM, Li L, Li H, Kim G, Zheng Y, Wang H. (2010) A single amino acid controls the functional switch of human constitutive androstane receptor (CAR) 1 to the xenobiotic-sensitive splicing variant CAR3. J Pharmacol Exp Ther 332:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian MT, Wilson EM, Shapiro DJ. (2012) A competitive inhibitor that reduces recruitment of androgen receptor to androgen-responsive genes. J Biol Chem 287:23368–23380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKeyser JG, Laurenzana EM, Peterson EC, Chen T, Omiecinski CJ. (2011) Selective phthalate activation of naturally occurring human constitutive androstane receptor splice variants and the pregnane X receptor. Toxicol Sci 120:381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Masi A, De Marinis E, Ascenzi P, Marino M. (2009) Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol Aspects Med 30:297–343. [DOI] [PubMed] [Google Scholar]
- Ding X, Lichti K, Kim I, Gonzalez FJ, Staudinger JL. (2006) Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J Biol Chem 281:26540–26551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucette SR, Sueyoshi T, Smith CM, Negishi M, Lecluyse EL, Wang H. (2006) Differential regulation of hepatic CYP2B6 and CYP3A4 genes by constitutive androstane receptor but not pregnane X receptor. J Pharmacol Exp Ther 317:1200–1209. [DOI] [PubMed] [Google Scholar]
- Faucette SR, Zhang TC, Moore R, Sueyoshi T, Omiecinski CJ, LeCluyse EL, Negishi M, Wang H. (2007) Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther 320:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. (1998) Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature 395:612–615. [DOI] [PubMed] [Google Scholar]
- Fritz J, Strehblow A, Taschner A, Schopoff S, Pasierbek P, Jantsch MF. (2009) RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol 29:1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. (1998) The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol 18:5652–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. (2007) Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene 26:258–268. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. (2005) Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol 19:1646–1653. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Wei P, Schrader WT, Moore DD. (2004) Meclizine is an agonist ligand for mouse constitutive androstane receptor (CAR) and an inverse agonist for human CAR. Mol Endocrinol 18:2402–2408. [DOI] [PubMed] [Google Scholar]
- Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, Tomkinson NC, LeCluyse EL, Lambert MH, Willson TM, et al. (2000) The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol 14:27–39. [DOI] [PubMed] [Google Scholar]
- Jyrkkärinne J, Mäkinen J, Gynther J, Savolainen H, Poso A, Honkakoski P. (2003) Molecular determinants of steroid inhibition for the mouse constitutive androstane receptor. J Med Chem 46:4687–4695. [DOI] [PubMed] [Google Scholar]
- Kachaylo EM, Pustylnyak VO, Lyakhovich VV, Gulyaeva LF. (2011) Constitutive androstane receptor (CAR) is a xenosensor and target for therapy. Biochemistry (Mosc) 76:1087–1097. [DOI] [PubMed] [Google Scholar]
- Küblbeck J, Jyrkkärinne J, Molnár F, Kuningas T, Patel J, Windshügel B, Nevalainen T, Laitinen T, Sippl W, Poso A, et al. (2011) New in vitro tools to study human constitutive androstane receptor (CAR) biology: discovery and comparison of human CAR inverse agonists. Mol Pharm 8:2424–2433. [DOI] [PubMed] [Google Scholar]
- Lau AJ, Yang G, Rajaraman G, Baucom CC, Chang TK. (2011) Differential effect of meclizine on the activity of human pregnane X receptor and constitutive androstane receptor. J Pharmacol Exp Ther 336:816–826. [DOI] [PubMed] [Google Scholar]
- Li G, Lin W, Araya JJ, Chen T, Timmermann BN, Guo GL. (2012) A tea catechin, epigallocatechin-3-gallate, is a unique modulator of the farnesoid X receptor. Toxicol Appl Pharmacol 258:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. (2008) The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol 74:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YP, Huang JD. (2008) Interplay of pregnane X receptor with other nuclear receptors on gene regulation. Drug Metab Pharmacokinet 23:14–21. [DOI] [PubMed] [Google Scholar]
- Lin W, Wu J, Dong H, Bouck D, Zeng FY, Chen T. (2008) Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem 283:30650–30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Idle JR, Gonzalez FJ. (2008) The pregnane X receptor: from bench to bedside. Expert Opin Drug Metab Toxicol 4:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem 278:17277–17283. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Watson J, McMillen PJ, Goodwin B, Willson TM, Moore JT. (2004) The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J Biol Chem 279:19832–19838. [DOI] [PubMed] [Google Scholar]
- Min G, Kemper JK, Kemper B. (2002) Glucocorticoid receptor-interacting protein 1 mediates ligand-independent nuclear translocation and activation of constitutive androstane receptor in vivo. J Biol Chem 277:26356–26363. [DOI] [PubMed] [Google Scholar]
- Molnár F, Küblbeck J, Jyrkkärinne J, Prantner V, Honkakoski P. (2013) An update on the constitutive androstane receptor (CAR). Drug Metabol Drug Interact 28:79–93. [DOI] [PubMed] [Google Scholar]
- Moore LB, Maglich JM, McKee DD, Wisely B, Willson TM, Kliewer SA, Lambert MH, Moore JT. (2002) Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol 16:977–986. [DOI] [PubMed] [Google Scholar]
- Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel JB, Goodwin B, Liddle C, Blanchard SG, Willson TM, et al. (2000) Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J Biol Chem 275:15122–15127. [DOI] [PubMed] [Google Scholar]
- Mutoh S, Sobhany M, Moore R, Perera L, Pedersen L, Sueyoshi T, Negishi M. (2013) Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling (Abstract). Sci Signal 6:ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omiecinski CJ, Coslo DM, Chen T, Laurenzana EM, Peffer RC. (2011) Multi-species analyses of direct activators of the constitutive androstane receptor. Toxicol Sci 123:550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson US, Lindell M, Raffalli-Mathieu F, Lannerbro A, Honkakoski P, Lang MA. (2006) In vivo and mechanistic evidence of nuclear receptor CAR induction by artemisinin. Eur J Clin Invest 36:647–653. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. (1999) The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274:6043–6046. [DOI] [PubMed] [Google Scholar]
- Takwi AA, Wang YM, Wu J, Michaelis M, Cinatl J, Chen T. (2014) miR-137 regulates the constitutive androstane receptor and modulates doxorubicin sensitivity in parental and doxorubicin-resistant neuroblastoma cells. Oncogene 33:3717–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. (2007) CAR and PXR: the xenobiotic-sensing receptors. Steroids 72:231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. (2003) The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 9:220–224. [DOI] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. (2009) Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324:787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. (2000) The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol 20:2951–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. (2003) A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem 278:14146–14152. [DOI] [PubMed] [Google Scholar]
- Wang Y, Masuyama H, Nobumoto E, Zhang G, Hiramatsu Y. (2014) The inhibition of constitutive androstane receptor-mediated pathway enhances the effects of anticancer agents in ovarian cancer cells. Biochem Pharmacol 90:356–366. [DOI] [PubMed] [Google Scholar]
- Wang YM, Lin W, Chai SC, Wu J, Ong SS, Schuetz EG, Chen T. (2013) Piperine activates human pregnane X receptor to induce the expression of cytochrome P450 3A4 and multidrug resistance protein 1. Toxicol Appl Pharmacol 272:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zhang J, Dowhan DH, Han Y, Moore DD. (2002) Specific and overlapping functions of the nuclear hormone receptors CAR and PXR in xenobiotic response. Pharmacogenomics J 2:117–126. [DOI] [PubMed] [Google Scholar]
- Wu B, Li S, Dong D. (2013) 3D structures and ligand specificities of nuclear xenobiotic receptors CAR, PXR and VDR. Drug Discov Today 18:574–581. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Moore R, Goldsworthy TL, Negishi M, Maronpot RR. (2004) The orphan nuclear receptor constitutive active/androstane receptor is essential for liver tumor promotion by phenobarbital in mice. Cancer Res 64:7197–7200. [DOI] [PubMed] [Google Scholar]
- Yu DD, Lin W, Chen T, Forman BM. (2013) Development of time resolved fluorescence resonance energy transfer-based assay for FXR antagonist discovery. Bioorg Med Chem 21:4266–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamule SM, Coslo DM, Chen F, Omiecinski CJ. (2011) Differentiation of human embryonic stem cells along a hepatic lineage. Chem Biol Interact 190:62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.