Abstract
The nonsense-mediated mRNA decay (NMD) pathway serves an important role in gene expression by targeting aberrant mRNAs that have acquired premature termination codons (PTCs) as well as a subset of normally processed endogenous mRNAs. One determinant for the targeting of mRNAs by NMD is the occurrence of translation termination distal to the poly(A) tail. Yet, a large subset of naturally occurring mRNAs contain long 3′ UTRs, many of which, according to global studies, are insensitive to NMD. This raises the possibility that such mRNAs have evolved mechanisms for NMD evasion. Here, we analyzed a set of human long 3′ UTR mRNAs and found that many are indeed resistant to NMD. By dissecting the 3′ UTR of one such mRNA, TRAM1 mRNA, we identified a cis element located within the first 200 nt that inhibits NMD when positioned in downstream proximity of the translation termination codon and is sufficient for repressing NMD of a heterologous reporter mRNA. Investigation of other NMD-evading long 3′ UTR mRNAs revealed a subset that, similar to TRAM1 mRNA, contains NMD-inhibiting cis elements in the first 200 nt. A smaller subset of long 3′ UTR mRNAs evades NMD by a different mechanism that appears to be independent of a termination-proximal cis element. Our study suggests that different mechanisms have evolved to ensure NMD evasion of human mRNAs with long 3′ UTRs.
Keywords: nonsense-mediated decay, long 3′ UTR, NMD inhibition, UPF1
INTRODUCTION
The fidelity of eukaryotic gene expression depends on a multitude of quality control pathways that eliminate faulty RNAs (Doma and Parker 2007; Schmid and Jensen 2010). One such pathway, the nonsense-mediated mRNA decay (NMD) pathway, detects and degrades mRNAs that have acquired premature termination codons (PTCs), for example, through mutation or faulty processing. NMD thereby prevents production of truncated proteins that could interfere with cell function via gain-of-function or dominant-negative effects (Frischmeyer and Dietz 1999; Kuzmiak and Maquat 2006; Schweingruber et al. 2013). In addition, evidence suggests that NMD participates in regulation of normal gene expression by targeting a subset of normally processed mRNAs (Mendell et al. 2004; Wittmann et al. 2006; Viegas et al. 2007; Huang et al. 2011; Yepiskoposyan et al. 2011; Tani et al. 2012; Hurt et al. 2013). Consistent with this, NMD contributes to important biological processes such as neuronal differentiation, myogenesis, T lymphocyte maturation, and inhibition of viral replication (Weischenfeldt et al. 2008; Gong et al. 2009; Bruno et al. 2011; Frischmeyer-Guerrerio et al. 2011; Balistreri et al. 2014; Lou et al. 2014).
The detection of NMD substrates is carried out by a set of UPF (UPF1, 2, and 3) and SMG (SMG1, 5, 6, and/or 7, depending on organism) proteins, which, as a consequence of translation termination at a PTC, assemble with NMD substrates and target them for degradation (Chang et al. 2007; Nicholson et al. 2010; Popp and Maquat 2013; Schweingruber et al. 2013). The mechanism by which PTCs are detected by the translation termination machinery and promote assembly of the UPF-SMG complex is still poorly understood but seems to be dictated in large part by the composition of the mRNA 3′ UTR. For example, the presence in the 3′ UTR of an exon junction complex (EJC), which is deposited on exon–exon junctions as a consequence of pre-mRNA splicing in many eukaryotes, stimulates NMD (Le Hir et al. 2000a,b, 2001; Kim et al. 2001; Lykke-Andersen et al. 2001; Gehring et al. 2003, 2005; Kashima et al. 2010). Moreover, the length of the 3′ UTR is an important characteristic that influences NMD. A number of studies have demonstrated that artificial 3′ UTRs of 800–900 nt in length promote NMD and increase UPF1 assembly with targeted mRNAs (Eberle et al. 2008; Singh et al. 2008; Rebbapragada and Lykke-Andersen 2009; Hogg and Goff 2010; Huang et al. 2011; Yepiskoposyan et al. 2011; Hurt et al. 2013). NMD can be prevented by placing cytoplasmic poly(A) binding protein (PABP) in proximity to the termination codon, suggesting that the increased distance between the translation termination event and cytoplasmic PABP that results from termination at a PTC contributes to NMD (Amrani et al. 2004; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008; Fatscher et al. 2014). The propensity of long 3′ UTRs to induce NMD is also supported by global studies showing enrichment of mRNAs containing long 3′ UTRs among endogenous NMD substrates (Mendell et al. 2004; Bruno et al. 2011; Yepiskoposyan et al. 2011; Hurt et al. 2013).
Surprisingly, more than one-third of human endogenous mRNAs have 3′ UTRs longer than 1000 nt (Pesole et al. 2000), despite artificial 3′ UTRs as short as ≈800 nt long triggering NMD (Eberle et al. 2008; Singh et al. 2008; Rebbapragada and Lykke-Andersen 2009; Hogg and Goff 2010; Yepiskoposyan et al. 2011). This raises the question of whether this large subset of mRNAs is targeted by NMD, or whether these mRNAs instead have acquired mechanisms to evade the NMD pathway. Global and mRNA-specific analyses have indicated that while a subset of endogenous long 3′ UTR mRNAs are indeed substrates of NMD, many are insensitive to depletion of NMD factors (Mendell et al. 2004; Wittmann et al. 2006; Viegas et al. 2007; Singh et al. 2008; Hogg and Goff 2010; Yepiskoposyan et al. 2011; Tani et al. 2012; Hurt et al. 2013). However, the mechanism by which a subset of long 3′ UTR mRNAs evades the NMD pathway is unknown. Here, we investigated the susceptibility of a set of human mRNAs with long 3′ UTRs (>1250 nt) to NMD and dissected their 3′ UTRs for elements promoting NMD evasion. Our findings revealed that a subset of NMD-evading long 3′ UTRs contain NMD-inhibiting cis elements located within the first 200 nt. These elements are A/U-rich, inhibit NMD in a position-dependent manner and can promote NMD evasion when inserted into a heterologous NMD-sensitive mRNA. Another subset of NMD-evading long 3′ UTRs appear to evade the NMD pathway by different mechanism(s) as their proximal 600 nt are not sufficient to promote NMD evasion. These observations suggest that at least two different mechanisms exist for long 3′ UTR mRNAs in humans to evade degradation by the NMD pathway.
RESULTS
Many human long 3′ UTR mRNAs are insensitive to UPF1 depletion
To investigate whether general mechanisms exist for human mRNAs with long 3′ UTRs to evade NMD, we selected several mRNAs containing 3′ UTRs of >1250 nt (Fig. 1A), significantly longer than artificial 3′ UTRs previously shown to promote NMD (Eberle et al. 2008; Singh et al. 2008; Hogg and Goff 2010). Using quantitative (q)RT-PCR assays, we tested the effect of UPF1 depletion on the accumulation of these mRNAs in human HeLa Tet-Off cells (Fig. 1B; Supplemental Fig. S1). As expected, UPF1 depletion resulted in increases in steady-state levels of SMG5 (1.6-fold; P < 0.01) and GABARAPL1 (2.6-fold; P < 0.05) mRNAs, both of which have been previously characterized as NMD targets (Mendell et al. 2004; Huang et al. 2011; Yepiskoposyan et al. 2011). The other tested mRNAs showed either moderate or no accumulation upon UPF1 depletion (Fig. 1B), which was consistent with findings in previous global studies (Mendell et al. 2004; Yepiskoposyan et al. 2011). These observations are consistent with the notion that many endogenous mRNAs with long 3′ UTRs are not targeted by the NMD pathway despite artificial long 3′ UTRs activating NMD (Singh and Lykke-Andersen 2003; Mendell et al. 2004; Tani et al. 2012; Hurt et al. 2013).
FIGURE 1.
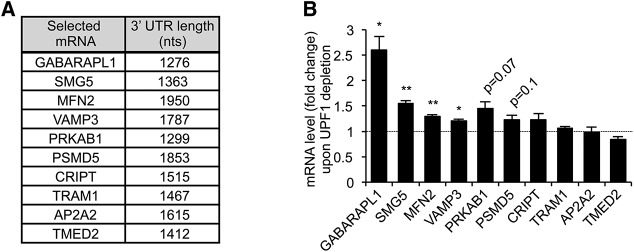
Levels of many human mRNAs containing a long 3′ UTR are not increased upon UPF1 depletion. (A) Table showing long 3′ UTR mRNAs used in this study as well as their 3′ UTR length. (B) Graph showing steady-state levels of tested mRNAs upon UPF1 depletion, normalized to mRNA levels upon Luciferase (LUC) depletion (fold change), as measured by quantitative RT-PCR. Error bars represent standard error of the means (SEM) from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01.
A subset of human long 3′ UTRs do not promote NMD when inserted into a heterologous mRNA
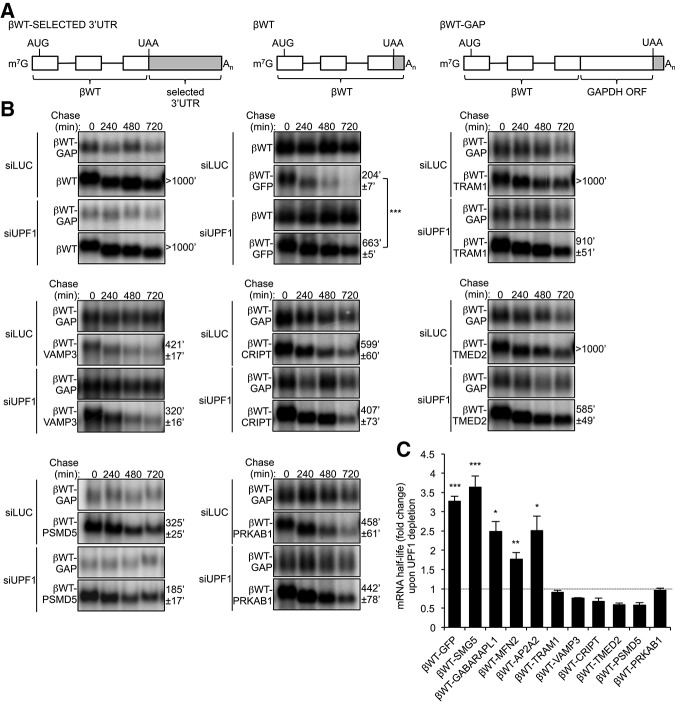
It is thought that the composition of the 3′ UTR is a major determinant for whether an mRNA is targeted for NMD (Le Hir et al. 2001; Lykke-Andersen et al. 2001; Gehring et al. 2003; Amrani et al. 2004; Maquat 2005; Singh et al. 2007, 2008; Eberle et al. 2008; Rebbapragada and Lykke-Andersen 2009; Hogg and Goff 2010; Yepiskoposyan et al. 2011). To test whether the long 3′ UTRs of our selected mRNAs confer NMD sensitivity in a heterologous context, we tested the effect of replacing the normal 3′ UTR of the wild-type β-globin (βWT) mRNA with the 3′ UTRs of the selected mRNAs (Fig. 2A). mRNA half-lives (t(1/2)) were compared between UPF1- and control-siRNA treated HeLa Tet-Off cells using tetracycline-regulated pulse-chase assays (Fig. 2B; Supplemental Fig. S2). Constitutively transcribed βWT or βWT-GAP mRNAs (shown in top panels for each condition) served as internal controls for half-life calculations; the use of βWT versus βWT-GAP mRNA in individual assays was dictated by the need for size differences with the tested reporter mRNAs. Consistent with previous observations (Singh et al. 2008), a βWT reporter containing an artificial long 3′ UTR (909 nt) consisting of the GFP open reading frame (βWT-GFP) is less stable (t(1/2) = 204′ ± 7′) than the βWT mRNA (t(1/2) > 1000′), and stabilized (t(1/2) = 663′ ± 5′; P < 0.001) upon UPF1 depletion (Fig. 2B; fold change over the control-siRNA condition graphed in Fig. 2C). Analyses of the individual long 3′ UTRs generally showed correlation with the behavior of the endogenous mRNAs from which they were derived. Insertion of the 3′ UTRs from the NMD target mRNAs SMG5 and GABARAPL1 into the βWT reporter mRNA resulted in destabilization as compared with βWT mRNA alone (Fig. 2B; Supplemental Fig. S2), and half-lives were increased 2.5 to 3.6 times upon UPF1 depletion (Fig. 2C; Supplemental Fig. S2) indicating that these mRNAs are targeted by NMD. In contrast, the 3′ UTRs of most (six of eight tested) of the long 3′ UTR mRNAs found to be resistant to NMD (Fig. 1B) failed to trigger NMD when inserted into the βWT reporter mRNA as judged by the lack of stabilization upon UPF1 depletion (Fig. 2B,C). A subset of these 3′ UTRs destabilized the βWT mRNA (Fig. 2B), but this destabilization was independent of NMD, as half-lives were not increased upon UPF1 depletion (Fig. 2B,C). For reasons that are unknown, some of the NMD-resistant reporter mRNAs exhibited greater instability upon UPF1 depletion, perhaps reflecting indirect effects of NMD repression. Two 3′ UTRs behaved differently with respect to NMD in the heterologous βWT context as compared with their corresponding endogenous mRNAs. The 3′ UTRs of MFN2 and AP2A2 mRNAs promoted NMD as judged by UPF1-dependent destabilization of the βWT mRNA (Fig. 2C; Supplemental Fig. S2), despite the corresponding endogenous mRNAs showing modest (MFN2) or no (AP2A2) increase in mRNA levels upon UPF1 depletion (Fig. 1B). This suggests that features outside of the 3′ UTR may contribute to NMD evasion by some long 3′ UTR mRNAs. Taken together, we conclude that a subset of long 3′ UTRs do not activate NMD, in contrast to both natural and artificial sequences of equal or shorter length. This suggests that these 3′ UTRs contain features that prevent activation of NMD.
FIGURE 2.
Several long 3′ UTRs do not promote NMD. (A) Schematics representing β-globin mRNA reporters used in B and C. 3′ UTRs are shown in gray. (B) Northern blots showing the decay of βWT, βWT-GFP, βWT-TRAM1, βWT-VAMP3, βWT-CRIPT, βWT-TMED2, βWT-PSMD5, and βWT-PRKAB1 mRNAs in HeLa Tet-Off cells treated with Luciferase (LUC) or UPF1 siRNAs. Numbers above panels refer to minutes after tetracycline-mediated transcriptional shutoff of reporters (chase). Values of t(1/2) were calculated after normalizing levels of βWT reporter mRNAs (lower panel) to levels of constitutively transcribed βWT or βWT-GAP internal control mRNAs (upper panel). t(1/2) are given as averages ± SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (***) P < 0.001. (C) Graph showing fold change in mRNA half-lives upon UPF1 depletion relative to LUC depletion, as calculated in B and Supplemental Figure S2. Error bars represent SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
TRAM1 and CRIPT 3′ UTRs do not inhibit mRNA polysome association
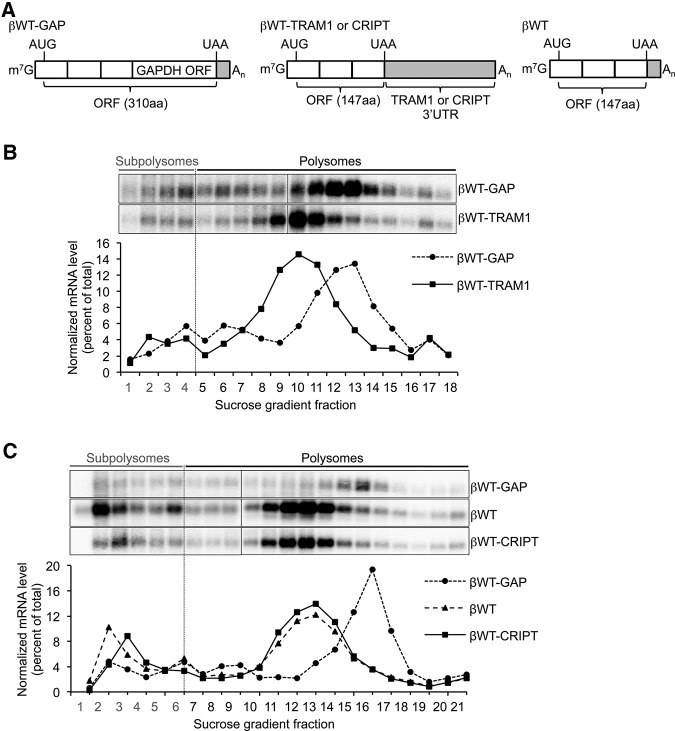
To investigate mechanisms by which natural long 3′ UTRs evade activation of NMD, we first focused on the TRAM1 3′ UTR. Given that NMD is a translation-dependent pathway, we first used polysome profiling to test whether the TRAM1 3′ UTR represses polysome association. As seen in Figure 3B (corresponding 260-nm absorbance traces are shown in Supplemental Fig. S3A), βWT-TRAM1 mRNA predominantly comigrated with polysomal fractions, consistent with the βWT-TRAM1 reporter mRNA being actively translated. An internal positive control mRNA (βWT-GAP), was similarly enriched in polysomal fractions, but was shifted toward heavier fractions as expected due to a longer open reading frame (ORF) (Fig. 3A). We also tested the CRIPT 3′ UTR in this assay. As seen in Figure 3C, βWT-CRIPT mRNA also concentrated in polysomal fractions and showed a similar profile to that of βWT mRNA (which shares the same ORF) (Fig. 3A), while βWT-GAP mRNA again migrated in heavier polysome fractions (260-nm absorbance traces are shown in Supplemental Fig. S3B). Taken together, these results suggest that the 3′ UTRs of TRAM1 and CRIPT do not repress polysome association of the βWT reporter, and thus that NMD evasion is not a result of translation repression.
FIGURE 3.
The long 3′ UTRs of TRAM1 and CRIPT do not inhibit mRNA polysome association. (A) Schematics representing β-globin mRNA reporters used in B and C. 3′ UTRs are shown in gray. (ORF) Open reading frame. ORF size is indicated in parentheses (aa, amino acid). “Subpolysome” designates fractions whose density is equal to or lower than the 80S ribosome. (B) Northern blots showing the levels of βWT-GAP and βWT-TRAM1 mRNAs in fractions collected after mRNP separation in sucrose gradients (polysome profiling). Graph showing Northern blot quantifications is shown below. The mRNA level of each individual fraction was normalized to the sum of mRNA levels in all fractions. (C) Same as in B for βWT-GAP, βWT, and βWT-CRIPT mRNAs.
The 5′ and 3′ halves of TRAM1 3′ UTR repress NMD in a distance-dependent manner
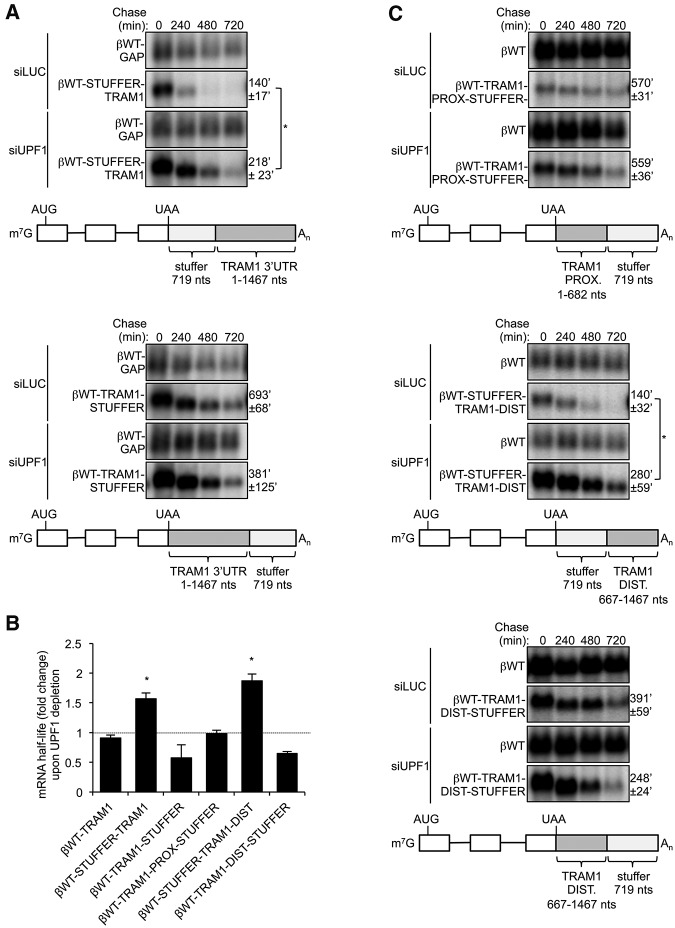
We next investigated the importance of the position of the TRAM1 3′ UTR relative to the termination codon. A stuffer sequence based on GFP ORF (719 nt) was inserted into the βWT-TRAM1 mRNA, upstream of (βWT-STUFFER-TRAM1) or downstream (βWT-TRAM1-STUFFER) from the TRAM1 3′ UTR, and the corresponding mRNAs were tested for their susceptibility to NMD. As seen in Figure 4A, insertion of the stuffer sequence upstream of the TRAM1 3′ UTR resulted in mRNA destabilization (t(1/2) = 140′ ± 17′; compare with βWT-TRAM1 mRNA in Fig. 2B). This destabilization is due at least in part to NMD as observed by the increased half-life (t(1/2) = 218′ ± 23′; P < 0.05) upon UPF1 depletion (Fig. 4A, top panels; fold difference graphed in Fig. 4B). In contrast, the βWT-TRAM1-STUFFER reporter containing the stuffer sequence downstream from TRAM1 3′ UTR was not subjected to NMD (Fig. 4A, bottom panels; Fig. 4B). Although the half-life of βWT-TRAM1-STUFFER mRNA was shorter than the half-life of βWT-TRAM1 mRNA (Fig. 2B), depletion of UPF1 did not result in its stabilization and actually caused a decrease in the mRNA half-life as was also observed for a subset of other mRNAs (Fig. 2B,C). Thus, the TRAM1 3′ UTR prevents NMD only when positioned in proximity of the termination codon.
FIGURE 4.
Sequences within the TRAM1 3′ UTR inhibit NMD when present in downstream proximity of the termination codon. (A) Northern blots showing the decay of βWT-STUFFER-TRAM1 (top panel) and βWT-TRAM1-STUFFER (bottom panel) mRNAs in HeLa Tet-Off cells treated with siRNAs targeting Luciferase (LUC) or UPF1. Numbers above panels refer to minutes after tetracycline-mediated transcriptional shutoff of reporter mRNAs (chase). Values of t(1/2) were calculated after normalizing levels of βWT-STUFFER-TRAM1 and βWT-TRAM1-STUFFER to levels of constitutively transcribed βWT-GAP mRNA. t(1/2) are given as averages ± SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (*) P < 0.05. Schematics representing βWT-STUFFER-TRAM1 or βWT-TRAM1-STUFFER mRNA reporters are shown below Northern blots. Numbers indicate nucleotides. 3′ UTRs are shown in light gray (GFP stuffer) and dark gray (TRAM1 3′ UTR). (B) Graph showing fold change in mRNA half-lives upon UPF1 depletion relative to LUC depletion, as calculated in A, C, and Figure 2B. Error bars represent SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (*) P < 0.05. (C) Same as in A for βWT-TRAM1-PROX-STUFFER (top panel), βWT-STUFFER-TRAM1-DIST (middle panel), and βWT-TRAM1-DIST-STUFFER (bottom panel) mRNAs.
To identify region(s) of TRAM1 3′ UTR required for NMD repression, the stuffer sequence was inserted into the TRAM1 3′ UTR to replace the distal (βWT-TRAM1-PROX-STUFFER) or the proximal (βWT-STUFFER-TRAM1-DIST) regions. As seen in Figure 4C (top panel), the reporter mRNA retaining the proximal region of the TRAM1 3′ UTR (βWT-TRAM1-PROX-STUFFER) remained stable (t(1/2) = 570′ ± 31′) and unaffected by UPF1 depletion (t(1/2) = 559′ ± 36′) (fold difference graphed in Fig. 4B). In contrast, the mRNA containing the distal half of the TRAM1 3′ UTR after the stuffer sequence is unstable (t(1/2) = 140′ ± 32′), and stabilized upon UPF1 depletion (t(1/2) = 280′ ± 59′; P < 0.05) (Fig. 4C, middle panels). To test whether the distal half of TRAM1 mRNA is capable of inhibiting NMD when placed in downstream proximity of the termination codon, we tested the effect of placing it in front of the stuffer sequence. As seen in Figure 4C bottom panel, this mRNA undergoes a moderate rate of mRNA decay but is not targeted by the NMD pathway as it is not stabilized upon UPF1 depletion (graphed in Fig. 4B). Taken together, the observations in Figure 4 suggest that 5′ and 3′ halves of TRAM1 3′ UTR can both repress NMD (Fig. 4B,C), and that the repression depends on being in downstream proximity to the termination codon.
An NMD repression element is located within the first 182 nt of the TRAM1 3′ UTR
To narrow down a minimal region of the 5′ half of TRAM1 3′ UTR capable of repressing NMD, we inserted the stuffer sequence at different positions within the proximal half of the TRAM1 3′ UTR (Fig. 5A). As expected, inserting the stuffer at position +1 relative to the termination codon (βWT-TRAM1-STUFFER-1) resulted in an unstable mRNA, which was stabilized upon UPF1 depletion (Fig. 5B, top left panel; fold stabilization graphed in Fig. 5C). Insertion of the stuffer at position +82 resulted in an mRNA that was also stabilized upon UPF1 depletion (middle left panel), but significantly less so (P < 0.001) than that observed with the stuffer at position +1 (Fig. 5B,C). Insertion of the stuffer sequence at, or downstream from, position +182 resulted in stable mRNAs that were not further stabilized upon UPF1 depletion (Fig. 5B,C). These results suggest the existence of a distance-dependent NMD-inhibiting cis element residing within the first 182 nt of the 3′ UTR of TRAM1. Moreover, the first 82 nt of TRAM1 3′ UTR appear to partially inhibit NMD when positioned in downstream proximity of the termination codon.
FIGURE 5.
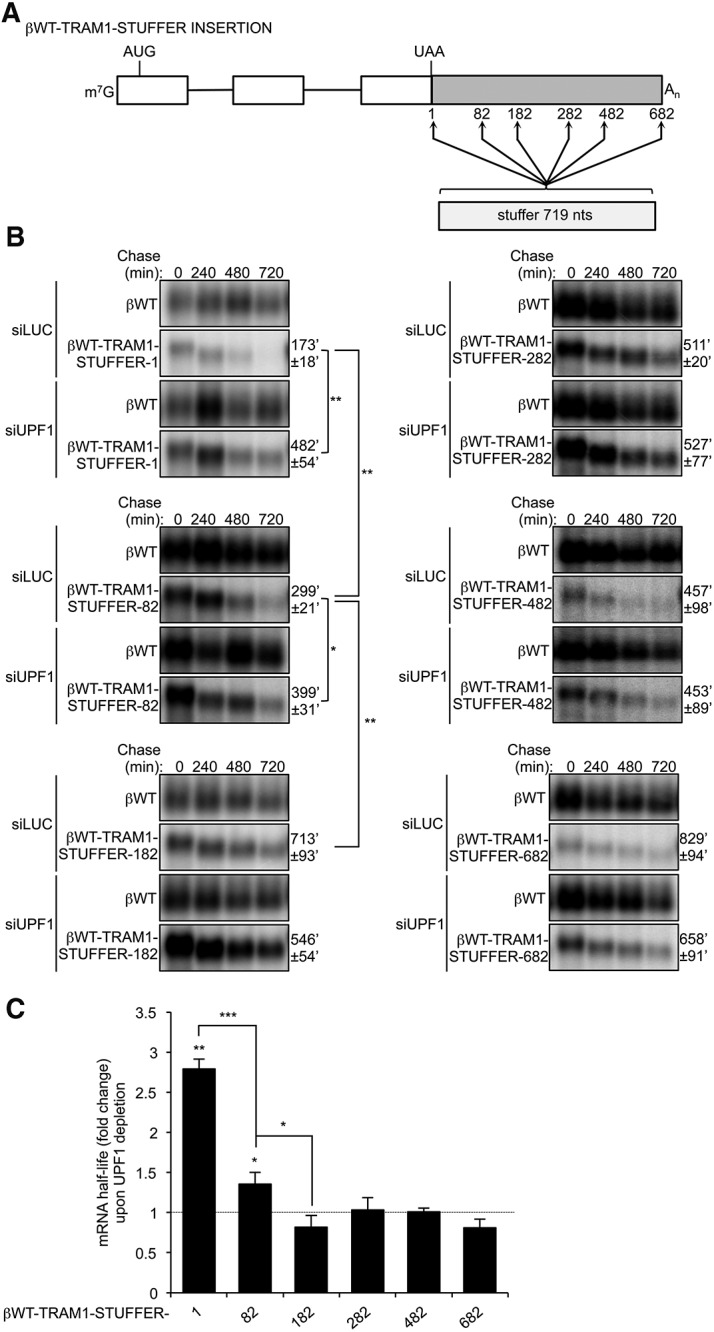
An NMD-inhibitory element is located within the first 182 nt of TRAM1 3′ UTR. (A) Schematic representing β-globin reporters containing GFP stuffer insertions at position 1, 82, 182, 282, 482, and 682 within the TRAM1 3′ UTR proximal region. Numbers indicate location of stuffer insertion relative to the position of the termination codon (UAA). (B) Northern blots showing the decay of βWT-TRAM1-STUFFER-1, −82, −182, −282, −482, and −682 mRNAs in HeLa Tet-Off cells treated with siRNAs targeting Luciferase (LUC) or UPF1. Numbers above panels refer to minutes after tetracycline-mediated transcriptional shutoff (chase). Values of t(1/2) were calculated after normalizing levels of βWT-TRAM1-STUFFER mRNAs to levels of constitutively transcribed βWT. t(1/2) are given as averages ± SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using either a paired or an independent unequal variance two-tailed t-test. (*) P < 0.05; (**) P < 0.01. (C) Graph showing fold change in mRNA half-lives upon UPF1 depletion relative to LUC depletion, as calculated in B. Error bars represent SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using either a paired or an independent unequal variance two-tailed t-test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
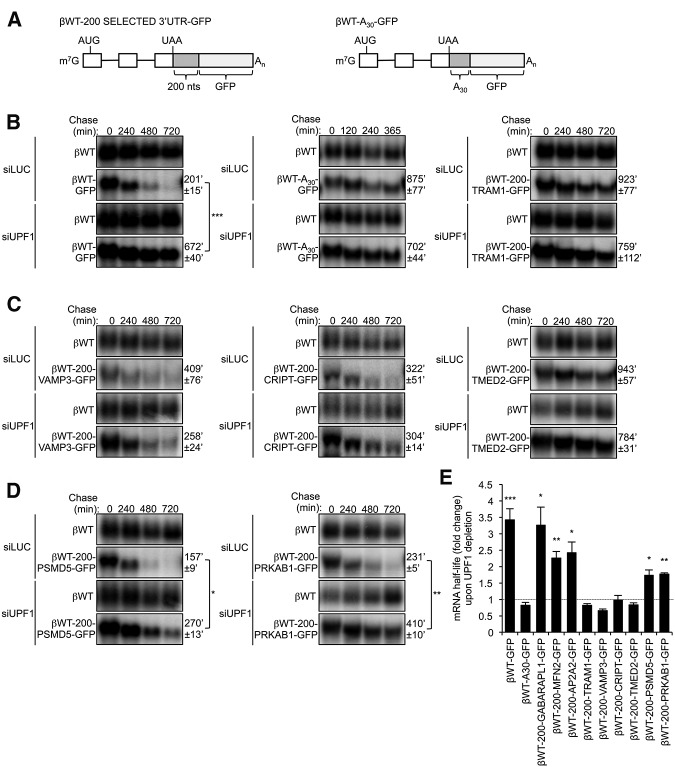
To test whether the termination-proximal fragment of the TRAM1 3′ UTR is sufficient to trigger NMD evasion in separation from the remaining TRAM1 3′ UTR, the first 200 nt of TRAM1 3′ UTR were inserted into the βWT-GFP NMD reporter in downstream proximity of the termination codon (βWT-200-TRAM1-GFP) (Fig. 6A). As shown in Figure 6B, this resulted in strong mRNA stabilization (t(1/2) = 923′ ± 77′, right panel, as compared wih 201′ ± 15′ observed for βWT-GFP, left panel). In addition, the βWT-200-TRAM1-GFP mRNA was not stabilized upon UPF1 depletion (Fig. 6B; quantified in Fig. 6E). Insertion of an oligo-A30 tract in downstream proximity of the termination codon of the βWT-GFP NMD reporter to mimic a termination-proximal poly(A) tail served as a positive control (Singh et al. 2008) and as expected also inhibited NMD (Fig. 6B, middle panel). We conclude that the first 200 nt of the TRAM1 3′ UTR contains an element sufficient to inhibit NMD. Insertion of the 200-nt element downstream from the stuffer sequence in the βWT-GFP mRNA confirmed that the element acts in a distance-dependent manner as the resulting mRNA was unstable and stabilized by UPF1 depletion (Supplemental Fig. S4).
FIGURE 6.
The first 200 nt of several long 3′ UTRs contain cis elements sufficient to inhibit NMD. (A) Schematics representing βWT-GFP reporters containing the first 200 nt of the 3′ UTRs of TRAM1, VAMP3, CRIPT, TMED2, PSMD5, and PRKAB1 mRNAs or a poly(A) track of 30 nt (A30). 3′ UTRs are shown in light gray (GFP) and dark gray (candidate 3′ UTR fragments, A30 track). (B) Northern blots showing the decay of βWT-GFP, βWT-A30-GFP, and βWT-200-TRAM1-GFP mRNAs in HeLa Tet-Off cells treated with siRNAs targeting Luciferase (LUC) or UPF1. Numbers above panels refer to minutes after tetracycline-mediated transcriptional shutoff (chase). Values of t(1/2) were calculated after normalizing levels of βWT-GFP, βWT-A30-GFP, or βWT-200-TRAM1-GFP mRNA to levels of constitutively transcribed βWT. t(1/2) are given as averages ± SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (***) P < 0.001. (C) Same as in B for βWT-200-VAMP3-GFP, βWT-200-CRIPT-GFP, and βWT-200-TMED2-GFP mRNAs. (D) Same as in B for βWT-200-PSMD5-GFP and βWT-200-PRKAB1-GFP mRNAs. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01. (E) Graph showing fold change in mRNA half-lives upon UPF1 depletion relative to LUC depletion, as calculated in B, C, D, and Supplemental Figure S5. Error bars represent SEM from at least three independent experiments. P-values compare UPF1 depletion to LUC depletion and were calculated using a paired two-tailed Student's t-test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
NMD-inhibiting cis elements are present within the first 200 nt of multiple long 3′ UTRs
We next investigated whether other NMD-evading long 3′ UTRs contain NMD-repressing elements within the first 200 nt. As seen in Figure 6C, the first 200 nt of the NMD-insensitive VAMP3, CRIPT, and TMED2 3′ UTRs, all resulted in mRNA stabilization (compared with βWT-GFP) (Fig. 6B) and insensitivity to UPF1 depletion when inserted immediately downstream from the termination codon in βWT-GFP mRNA (quantified in Fig. 6E). In contrast, the first 200 nt of GABARAPL1, MFN2, and AP2A2 3′ UTRs did not render βWT-GFP mRNA insensitive to NMD (Fig. 6E; Supplemental Fig. S5), which was expected given that the corresponding full-length 3′ UTRs promoted NMD (Fig. 2C; Supplemental Fig. S2). These results demonstrate that the first 200 nt of VAMP3, CRIPT, and TMED2 3′ UTRs, similar to TRAM1, contain NMD-inhibiting elements sufficient to inhibit NMD, suggesting this as a common mechanism of NMD evasion for long human 3′ UTR. However, not all NMD-insensitive long 3′ UTRs behave in this manner as the first 200, 400, or 600 nt of the NMD-insensitive PSMD5 and PRKAB1 3′ UTRs were not sufficient to inhibit NMD of the βWT-GFP reporter mRNA (Fig. 6D,E; Supplemental Fig. S6). This suggests that these 3′ UTRs are rendered insensitive to NMD by a different mechanism. Taken together, our observations reveal at least three classes of human long 3′ UTRs. Some (e.g., SMG5 and GABARAPL1 3′ UTRs) render their mRNAs sensitive to NMD, whereas others are NMD-resistant either due to the presence of sequence elements that inhibit NMD in a manner dependent on their proximity to the termination codon (TRAM1, VAMP3, CRIPT, and TMED2), or due to other, currently unknown, mechanisms (PSMD5 and PRKAB1).
DISCUSSION
The extension of the 3′ UTR that results from termination at a PTC has been proposed to be an important determinant for the targeting of mRNAs for NMD (Amrani et al. 2004; Mendell et al. 2004; Buhler et al. 2006; Singh et al. 2008; Hogg and Goff 2010; Bruno et al. 2011; Yepiskoposyan et al. 2011; Hurt et al. 2013). Yet a large subset of human mRNAs contain 3′ UTRs far longer than the length needed for artificial 3′ UTRs to trigger NMD, and many of these long 3′ UTR mRNAs are insensitive to depletion of NMD factors (Pesole et al. 2000; Mendell et al. 2004; Wittmann et al. 2006; Viegas et al. 2007; Singh et al. 2008; Hogg and Goff 2010; Yepiskoposyan et al. 2011; Tani et al. 2012; Hurt et al. 2013). This raises the question of how long 3′ UTR mRNAs evade the NMD pathway. Here, we investigated a set of long 3′ UTR mRNAs and confirmed that many are not targeted for NMD either in their endogenous context or when their 3′ UTRs were fused with the heterologous β-globin mRNA (Figs. 1, 2). We further dissected the 3′ UTR of TRAM1 mRNA, which we previously identified as being insensitive to NMD (Singh et al. 2008), and identified an NMD-inhibiting cis element located within the first 182 nt (Fig. 5). This cis element inhibits NMD when positioned in downstream proximity of the termination codon (Fig. 4) and is sufficient to block NMD of a heterologous mRNA (Fig. 6). The identified TRAM1 3′ UTR cis element likely acts redundantly with other regions of the TRAM1 3′ UTR to repress NMD given the capacity of the distal half of the TRAM1 3′ UTR to also inhibit NMD (Fig. 4). Dissection of other long 3′ UTR mRNAs resistant to NMD revealed the presence of NMD-inhibiting cis elements within the first 200 nt of many, but not all, of the mRNAs tested (Fig. 6). Thus, our observations suggest that a subset of long 3′ UTR mRNAs in humans contain termination codon-proximal cis elements that serve to promote evasion of the NMD pathway.
What is the mechanism by which the cis elements inhibit NMD? Cis elements were found in downstream proximity of the termination codon in TRAM1, VAMP3, CRIPT, and TMED2 3′ UTRs (Fig. 6) and, in the case of TRAM1, this location was critical for its ability to repress NMD (Figs. 4, 5; Supplemental Fig. S4). Previous studies indicate that translation termination at NMD-inducing termination codons is less efficient than translation termination occurring at normal termination codons (Amrani et al. 2004; Peixeiro et al. 2012). Thus, the cis elements might inhibit NMD by promoting efficient translation termination, as has been proposed for the poly(A) tail when in proximity of the termination codon (Amrani et al. 2004; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008; Fatscher et al. 2014). Alternatively, the cis elements could promote a level of termination codon readthrough, which has been previously associated with NMD evasion (Hogg and Goff 2010). Finally, the cis elements could inhibit NMD by directly preventing the assembly or function of NMD factors on the long 3′ UTR mRNAs. Consistent with either of these ideas, the central NMD factor UPF1 has been reported to associate less with TRAM1 and CRIPT 3′ UTRs, both of which contain NMD-inhibiting cis elements (Fig. 6), than with NMD-sensitive 3′ UTRs of similar length (Hogg and Goff 2010).
The NMD-inhibiting cis elements are likely to act through trans factors. Analysis of the nucleotide content of the cis elements uncovered a significant enrichment for A/U nucleotides (63%–71%) as compared with the same regions of the 3′ UTRs studied here that did not contain cis elements (35%–63%) (P = 0.001) (Fig. 7A). This observation raises the possibility that a trans factor with affinity for A/U-rich sequences may mediate the NMD-inhibitory effects of the cis elements. One candidate for this activity could be PABP, which has previously been found to show high affinity for A/U-rich sequences (Bollig et al. 2003; Sladic et al. 2004), and is a known antagonist of NMD (Amrani et al. 2004; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008; Fatscher et al. 2014). Alternatively, RNA binding proteins that either directly affect translation termination or NMD factor function, or that recruit other factors such as PABP, could be responsible (Fig. 7B). It is also a possibility that the A/U-rich cis elements base-pair with the poly(A) tail, thereby bringing the poly(A) tail, and hence PABP, in proximity of the termination event. In addition, a recent study found evidence that UPF1 associates most stably with G/C-rich sequences (Hurt et al. 2013). Thus, the A/U-rich composition of the cis elements could prevent stable UPF1 binding in downstream proximity of the termination event.
FIGURE 7.
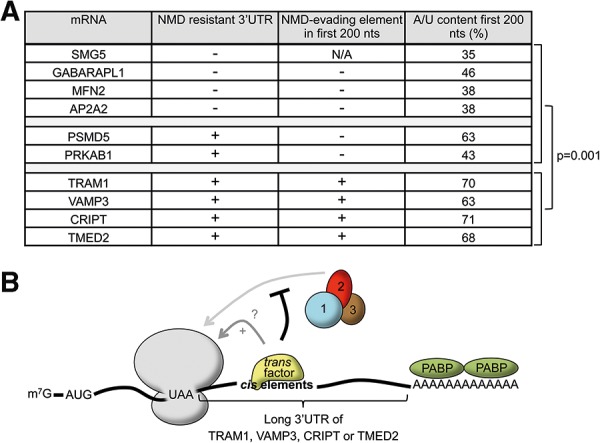
Model for NMD repression by termination-proximal cis elements. (A) Table showing A/U content within the first 200 nt of 3′ UTRs of mRNAs either containing (+) or not (−) NMD-inhibiting cis elements. P-values compare the two different groups and were calculated using an independent unequal variances t-test. (B) Schematic representing an mRNA with a long 3′ UTR that contains an NMD-inhibiting cis element. “+” indicates stimulation of translation termination. See Discussion for details. (PABP) Cytoplasmic poly(A) tail binding protein; 1, 2, 3: UPF1, UPF2, UPF3, respectively.
Interestingly, we found that the full-length 3′ UTRs of PSMD5 and PRKAB1 are insensitive to NMD but the first 600 nt of the 3′ UTRs do not contain a cis element sufficient to evade NMD. Thus, the 3′ UTRs of PSMD5 and PRKAB1 are likely to use a different mechanism to evade NMD. It is possible that the 3′ UTRs of PSMD5 and PRKAB1 do contain NMD-inhibiting cis elements, but that these fail to inhibit NMD in a heterologous context. Alternatively, a cis element could reside more distally within the 3′ UTRs. Mühlemann and coworkers reported evidence that structural arrangement of a long 3′ UTR that brings PABP in spatial proximity to the termination codon can inhibit NMD (Eberle et al. 2008). An interesting possibility to explore in future studies is that PSMD5 and PRKAB1 mRNAs may evade NMD by such a mechanism.
Our identification of termination-proximal NMD-inhibitory cis elements in a subset of long 3′ UTR mRNAs in human is reminiscent of elements previously described in yeast and retroviruses. In yeast, GCN4 and YAP1 mRNAs, which contain upstream (u)ORFs, contain elements immediately downstream from the uORFs that inhibit NMD (Ruiz-Echevarria et al. 1998; Ruiz-Echevarria and Peltz 2000). In addition, the Rous Sarcoma Virus (RSV) produces unspliced retroviral transcripts that contain multiple predicted NMD-activating features, including upstream ORFs as well as long 3′ UTRs. However, those viral transcripts are protected from NMD by the presence of a 400 nt cis-acting element located downstream from the gag ORF (Weil and Beemon 2006).
Our study describes several examples of NMD-inhibiting cis elements in human long 3′ UTRs. The lengthening of 3′ UTRs has been proposed to correlate with the evolution of morphological complexity (Chen et al. 2012). Hence, increased 3′ UTR size in higher organisms might have exposed genes to ancestral quality control mechanisms, such as NMD, therefore requiring compensatory evolution of gene features to evade those pathways. In addition to protecting the long 3′ UTR mRNAs from NMD, NMD-evading cis elements could also provide an additional level of post-transcriptional gene regulation by regulating mRNA stability according to cellular cues, such as, for example, if trans factors acting on the cis elements are regulated. Considering that one-third of human genes have 3′ UTRs longer than 1000 nt, NMD evasion may be a previously unappreciated widespread principle required for proper gene regulation.
MATERIALS AND METHODS
Plasmids
All plasmids and sequences are available upon request. For generation of pcTET2-βWT-3′ UTR reporter plasmids, amplification of candidate 3′ UTR cDNAs was performed by PCR using PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies). Forward and reverse primers contained NotI and, XbaI, SpeI or NheI restriction sites, respectively, for ligation into the pcTET2-βWT plasmid (Singh et al. 2008). pcTET2-βWT-STUFFER-TRAM1 and pcTET2-βWT-TRAM1-STUFFER were generated by ligating PCR-amplified GFP fragments into pcTET2-βWT-TRAM1 using NotI or XbaI restriction sites, respectively. pcTET2-βWT-TRAM1-PROX-STUFFER, pcTET2-βWT-STUFFER-TRAM1-DIST and pcTET2-βWT-TRAM1-DIST-STUFFER were made by ligating PCR-amplified GFP fragments into pcTET2-βWT-TRAM1-PROX (XbaI site) or pcTET2-βWT-TRAM1-DIST (NotI or XbaI sites), which were generated by inserting proximal (1–682 nt) or distal (667–1467 nt) fragments of TRAM1 using NotI and XbaI restriction sites. The resulting PCR products were inserted in the pcTET2-βWT-GFP plasmid using NotI or XbaI restriction sites (TRAM1 proximal and TRAM1 distal, respectively). For inserting GFP at different positions into the pcTET2-βWT-TRAM1, an NheI restriction site was first inserted at the desired location using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Then, GFP fragments were amplified using forward and reverse primers containing XbaI and NheI restriction sites respectively and ligated into pcTET2-βWT-TRAM1-PROX at different positions using the generated NheI restriction sites. For inserting 200, 400, or 600 bp fragments of candidate 3′ UTRs into pcTET2-βWT-GFP, the corresponding fragments were PCR-amplified using forward and reverse primers, both containing NotI restriction sites. Then, those fragments were inserted into the pcTET2-βWT-GFP plasmid using NotI. The pcTET2-βWT-GFP-200-TRAM1 plasmid was made by inserting PCR-amplified 200 bp of TRAM1 3′ UTR containing SpeI sites into pcTET2-βWT-GFP using XbaI restriction site.
Pulse-chase mRNA decay assays
HeLa Tet-Off cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 10% heat-inactivated Fetal Bovine Serum (FBS) (Life Technologies) and 50 ng/mL of tetracycline (Sigma-Aldrich). Each well of 12-well plates was transfected with 20 nM of siRNA targeting either Firefly Luciferase (LUC) or UPF1 (Singh et al. 2008) using SiLentFect (Bio-Rad) according to manufacturer's protocol. Twenty-four hours later, cells were transfected with 0.8 μg of indicated tetracycline-inducible plasmids, 40 ng of either pcβWT-GAP or pcβWT plasmids and 0.66 μg of pcDNA-Flag using TransIT HeLa Monster reagent (Mirus) according to manufacturer's protocol. Twenty-four hours later, cells were transfected again with siRNA as described above. Sixteen hours later, transcription from tetracycline-inducible plasmids was activated by washing cells twice with DMEM 10% FBS without tetracycline and incubating for 6 h in DMEM 10% FBS. Transcriptional shutoff was achieved by addition of 1 μg/mL tetracycline. Starting 30 min after tetracycline addition, cells were harvested in 600 μL TRIzol (Life Technologies) at times indicated for each experiment (chase). RNA isolation was performed as recommended by manufacturer's protocol. Northern blots were performed as previously described (Lykke-Andersen et al. 2000). Northern blots were scanned using a PhosphorImager (Typhoon Trio; Amersham Biosciences) and quantified using ImageQuant TL ID software. A paired two-tailed Student's t-test was used to calculate P-values.
qRT-PCR
Cells were transfected with 20 nM siRNA targeting either LUC or UPF1 as described above. Forty-eight hours later, cells were harvested in 600 μL of TRIzol and RNA was isolated according to the manufacturer's protocol. 0.5 μg of total RNA and 12.5 ng/μL of 8 nt random primers were used to generate cDNA using SuperScript III Reverse Transcriptase (Life Technologies) as recommended by the manufacturer's protocols. Polymerase chain reactions (PCRs) were performed on one one-hundredth of the reverse transcription (RT) reaction using Fast SYBER Green Master Mix (Life Technologies) according to manufacturer's protocol and the Applied Biosystems StepOnePlus Real-Time PCR System technology. Primers used for PCR are indicated in Supplemental Table 1 (GAPDH primers were generously provided by Dr. Shannon Lauberth). Each measurement was performed in duplicate to calculate the average Ct (Threshold Cycle) value. For each set of primers and each PCR assay, a 10-fold standard dilution was performed to calculate the PCR efficiency (E). When E value ranged between 1.8 and 2.1 mRNA levels were determined using average Ct value and E. RNA level fold changes represent ratios between mRNA levels upon UPF1 depletion and mRNA levels upon control (LUC depleted) condition. P-values were calculated using a two-tailed paired Student's t-test.
Western blots and antibodies
Cells were transfected as described in the Pulse-chase mRNA decay assay. At time zero after transcription shutoff, cells were harvested in 100 μL RIPA N Buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, 1% Nonidet P-40) and 100 μL of 2× SDS buffer (100 mM Tris–HCl pH 6.8, 4% SDS, 12% β-mercaptoethanol, 20% glycerol, and 0.1% bromophenol Blue). One-twentieth of each sample was analyzed by polyacrylamide gel electrophoresis followed by Western blotting. Blots were probed with rabbit polyclonal anti-UPF1 (1:1000) (Lykke-Andersen et al. 2000) or rabbit polyclonal anti-CBP80 (1:1000; antibody generously provided by Dr. Elisa Izaurralde) followed by horseradish peroxidase (HRP)-conjugated donkey anti-rabbit (1:20,000; Thermo Scientific) and visualization using HyGLO Chemiluminescent Antibody Detection (Denville Scientific Inc.).
Polysome profiling
Cells were grown in 10-cm plates with DMEM 10% heat-inactivated FBS and 50 ng/mL tetracycline. Cells were then transfected with 0.5 μg of pcβWT-GAP and 3.5 μg of pcTET2-βWT-CRIPT or pcTET2βWT-TRAM1 as well as 3.5 μg of pcβWT when indicated using TransIT HeLa Monster reagent (Mirus) according to manufacturer's protocol. Forty-eight hours after transfection, cells were washed with phosphate buffered saline solution (PBS, 8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, pH 7.4) and incubated with DMEM containing 10% heat-inactivated FBS without tetracycline. After 8 h, transcription was turned off by adding 1 μg/mL tetracycline and 0.1 μg/mL cycloheximide was added for 3 min to inhibit translation elongation. Polysome profiling was conducted as described previously (Johannes and Sarnow 1998). Fractions were analyzed by Northern blot as described previously (Lykke-Andersen et al. 2000). Northern blots were scanned using the PhosphorImager (Typhoon Trio; Amersham Biosciences) and quantified using ImageQuant TL 1D software. Normalized mRNA quantifications were determined by dividing individual fraction signals by the sum of all fraction signals.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Shannon Lauberth for providing GAPDH qRT-PCR primers and Dr. Elisa Izaurralde for anti-CBP80 antibody. We thank Drs. Rea Lardelli and Suzanne Lee for comments on the manuscript and members of the J.L.-A. laboratory for helpful discussions. This work was supported by grants from the US National Science Foundation (MCB-0946464) and National Institutes of Health (R01 GM099717) to J.L.-A. and a postdoctoral fellowship from the Fondation pour la Recherche Médicale en France to S.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048637.114.
REFERENCES
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. [DOI] [PubMed] [Google Scholar]
- Balistreri G, Horvath P, Schweingruber C, Zünd D, McInerney G, Merits A, Muhlemann O, Azzalin C, Helenius A 2014. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 16: 403–411. [DOI] [PubMed] [Google Scholar]
- Bollig F, Winzen R, Gaestel M, Kostka S, Resch K, Holtmann H 2003. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem Biophys Res Commun 301: 665–670. [DOI] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. 2011. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 42: 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Mühlemann O 2006. EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′ UTR length. Nat Struct Mol Biol 13: 462–464. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF 2007. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76: 51–74. [DOI] [PubMed] [Google Scholar]
- Chen CY, Chen ST, Juan HF, Huang HC 2012. Lengthening of 3′ UTR increases with morphological complexity in animal evolution. Bioinformatics 28: 3178–3181. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R 2007. RNA quality control in eukaryotes. Cell 131: 660–668. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatscher T, Boehm V, Weiche B, Gehring NH 2014. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA 20: 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC 1999. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8: 1893–1900. [DOI] [PubMed] [Google Scholar]
- Frischmeyer-Guerrerio PA, Montgomery RA, Warren DS, Cooke SK, Lutz J, Sonnenday CJ, Guerrerio AL, Dietz HC 2011. Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc Natl Acad Sci 108: 10638–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE 2003. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE 2005. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75. [DOI] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE 2009. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 23: 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 43: 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Sarnow P 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E 2010. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev 24: 2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Kuzmiak HA, Maquat LE 2006. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med 12: 306–316. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000a. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE 2000b. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev 14: 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF 2014. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 6: 748–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839. [DOI] [PubMed] [Google Scholar]
- Maquat LE 2005. Nonsense-mediated mRNA decay in mammals. J Cell Sci 118: 1773–1776. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O 2010. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 67: 677–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixeiro I, Inácio Â, Barbosa C, Silva AL, Liebhaber SA, Romão L 2012. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res 40: 1160–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Licciulli F, Larizza A, Makalowski W, Saccone C 2000. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 28: 193–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J 2009. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr Opin Cell Biol 21: 394–402. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW 2000. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101: 741–751. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, González CI, Peltz SW 1998. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J 17: 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH 2010. Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip Rev RNA 1: 474–485. [DOI] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zund D, Yamashita A, Mühlemann O 2013. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829: 612–623. [DOI] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inácio Â, Liebhaber SA, Romão L 2008. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Lykke-Andersen J 2003. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem Sci 28: 464–466. [DOI] [PubMed] [Google Scholar]
- Singh G, Jakob S, Kleedehn MG, Lykke-Andersen J 2007. Communication with the exon-junction complex and activation of nonsense-mediated decay by human Upf proteins occur in the cytoplasm. Mol Cell 27: 780–792. [DOI] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladic RT, Lagnado CA, Bagley CJ, Goodall GJ 2004. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur J Biochem 271: 450–457. [DOI] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N 2012. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE 2007. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res 35: 4542–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil JE, Beemon KL 2006. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 12: 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT 2008. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 22: 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jäck HM 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17: 2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.