Abstract
Background
Chronic alcohol use causes widespread changes in the cellular biology of the amygdala's central nucleus (CeA), a GABAergic center that integrates autonomic physiology with the emotional aspects of motivation and learning. While alcohol-induced neurochemical changes play a role in dependence and drinking behavior, little is known about the CeA's dynamic changes during withdrawal, a period of emotional and physiologic disturbance.
Methods
We used a qRT-PCR platform to measure 139 transcripts in 92 rat CeA samples from control (N = 33), chronically alcohol exposed (N = 26), and withdrawn rats (t = 4, 8, 18, 32, and 48 hours; N = 5, 10, 7, 6, 5). This focused transcript set allowed us to identify significant dynamic expression patterns during the first 48 hours of withdrawal and propose potential regulatory mechanisms.
Results
Chronic alcohol exposure causes a limited number of small magnitude expression changes. In contrast, withdrawal results in a greater number of large changes within 4 hours of removal of the alcohol diet. Sixty-five of the 139 measured transcripts (47%) showed differential regulation during withdrawal. Over the 48-hour period, dynamic changes in the expression of γ-aminobutyric acid type A (GABAA), ionotropic glutamate and neuropeptide system–related G-protein-coupled receptor subunits, and the Ras/Raf signaling pathway were seen as well as downstream transcription factors (TFs) and epigenetic regulators. Four temporally correlated gene clusters were identified with shared functional roles including NMDA receptors, MAPKKK and chemokine signaling cascades, and mediators of long-term potentiation, among others. Cluster promoter regions shared overrepresented binding sites for multiple TFs including Cebp, Usf-1, Smad3, Ap-2, and c-Ets, suggesting a potential regulatory role.
Conclusions
During alcohol withdrawal, the CeA experiences rapid changes in mRNA expression of these functionally related transcripts that were not predicted by measurement during chronic exposure. This study provides new insight into dynamic expression changes during alcohol withdrawal and suggests novel regulatory relationships that potentially impact the aspects of emotional modulation.
Keywords: Alcohol Withdrawal, Gene Expression, Time Series, Central Nucleus of the Amygdala, Gene Regulatory Network
The central nucleus of the amygdala (CeA) is richly innervated with dopaminergic and adrenergic fibers from the ventral tegmental area and brainstem. Functionally, it integrates somatic and emotional information, processing autonomic and affective states, and plays a role in the motivation and stress of alcohol addiction (McBride, 2002). Subsequently, alcohol withdrawal affects all levels of amygdalar function, altering behavioral, emotional, physiological, and neuronal phenotype (Cruz et al., 2012; McBride, 2002; McBride et al., 2010). In part, these effects are thought to arise from changes in gene expression (Koob, in press). However, because alcohol alters CeA function broadly (Clapp et al., 2008), inhibiting glutamate excitation (Obara et al., 2009; Roberto et al., 2004), enhancing c-aminobutyric acid (GABA)-triggered chloride flux (Bajo et al., 2008), and altering intracellular signaling via multiple pathways (Hans-son et al., 2008), characterizing relevant gene expression changes during alcohol withdrawal is a complex undertaking. Improved insight into these expression changes may inform our understanding of the neurobiological substrates of alcohol dependence and compulsive use aimed at limiting the negative experiences of withdrawal and protracted abstinence (Koob, in press).
Despite the role of the CeA in mediating alcohol-related consumption and dependence behaviors, only modest gene expression changes have been reported in the region under various exposure paradigms. For instance, in a genome-wide study of alcohol-preferring rats, differential expression was shown in only 23 genes 24 hours after a last operant ethanol (EtOH) exposure period (Rodd et al., 2008). While these included a GABAA receptor subunit (Gabrr2) and a regulator of metabotropic glutamate receptors (Homer2), the limited changes observed were potentially attributable to random chance. In a follow-up study, McBride and colleagues (2010) measured CeA gene expression 1, 6, and 24 hours after an 8-week series of repeated 1-hour binge exposure sessions in alcohol-preferring rats. Here, the CeA showed a more robust response, with expression changes in 402 named genes from 34 Gene Ontology annotation categories including: GABA signaling, second messenger signaling systems, enzyme-linked receptor protein signaling, regulation of protein kinase, and the MAPKKK cascade. Together, these studies suggest that alcohol-induced expression changes in the CeA are likely to be both time- and context-dependent. Consequently, a more complete understanding requires frequent sampling under multiple experimental conditions. Toward this goal, this study examines the temporal patterns of gene expression in the CeA during the first 48 hours of alcohol withdrawal.
Recent work in our laboratory has focused on examining withdrawal-induced expression changes on a systems level, providing insight into regulatory dynamics in the dorsal vagal complex (DVC), a reciprocally connected portion of the medulla that supplies afferents to and receives efferents from the CeA (Freeman et al., 2012a,b). Here, we use a similar approach to study the CeA during dependence and withdrawal. By focusing on a limited gene set, representing GABAA, N-methyl-D-aspartic acid (NMDA), and functionally important G-protein-coupled receptor (GPCR) subunits that involve other neuromodulatory signals including corticotropin-releasing hormone, angiotensin-peptide system, metabotropic glutamate receptors, and other neuropeptide systems, as well as downstream signaling components, we gain a dynamic view of the transcriptional contributions to withdrawal following chronic alcohol exposure. With this approach, we discovered coexpressed gene relationships and potential underlying regulatory mechanisms. During withdrawal, 47% of measured transcripts experienced statistically significant expression changes. Often, these changes were withdrawal-specific—their expression levels were not significantly different from control values during chronic exposure. Over the course of withdrawal, we identified 4 significant temporal expression patterns that suggest roles for several transcription factors (TFs) previously implicated in alcoholic liver disease and inflammation in coordinating the amygdala's withdrawal-related expression change, including Cebp, Usf-1, Smad3, Ap-2, and c-Ets.
MATERIALS AND METHODS
Alcohol Exposure
The details of the method of alcohol exposure have been previously reported (Freeman et al., 2012a,b). Briefly, male Sprague-Dawley rats (>120 g; Harlan, Indianapolis, IN) were assigned to 3 treatment groups: control (N = 33), chronic exposure (N = 26), or withdrawal following chronic exposure (N = 33). Withdrawal animals were assigned to 1 of 5 time points: 4, 8, 18, 32, or 48 hours, as shown in Fig. 1 (N = 5, 10, 7, 6, and 5, respectively). Animals in the withdrawal and chronic exposure groups were fed the Lieber– DeCarli liquid alcohol diet (36% of calories as alcohol) ad libitum for at least 35 days (Lieber and DeCarli, 1994). Controls were fed a calorically matched liquid diet where alcohol was isocalorically replaced with carbohydrate and diet volume equaled the average consumption of alcohol-fed littermates on the previous day. No differences in weight gain were noted between alcohol-fed groups. Rats were individually housed in the Thomas Jefferson Alcohol Research Center Animal Core Facility and maintained at constant temperature and humidity with 12/12-hour light cycles (lights on at Zeitgeber time [ZT] 0). Protocols were approved by the TJU Institutional Animal Care and Use Committee.
Fig. 1.
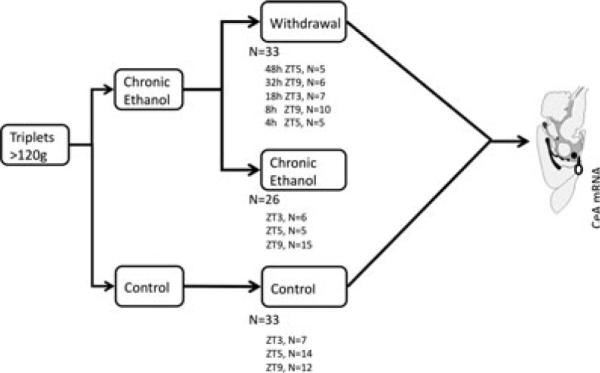
Experimental design. Animals were assigned to 1 of 3 treatment groups: control, chronic alcohol exposure, or withdrawal (with specific withdrawal time points covering the first 48 hours). Five time points were selected for study, ranging from 4 to 48 hours after withdrawal. These were selected to capture early transcription changes as well as later changes associated with adaptation to withdrawal and the typical behavioral response after alcohol removal in the Lieber–DeCarli rat model. Control and chronic exposure animals were killed at the same time as the withdrawal animal to account for differences in diurnal expression (Zeitgeber time [ZT] matched as indicated). CeA, central nucleus of the amygdala.
Average daily alcohol intake for rats on the alcohol diet was 15.2 g/kg and was not different among alcohol-fed rats (p > 0.25; Fig. S1). Studies using the Lieber–DeCarli liquid alcohol diet in this facility and elsewhere report 20 to 30 mM peak blood alcohol concentrations with 12 to 16 g/kg average daily alcohol intake in rats following long-term exposure (>3 weeks), comparable to this study (Freeman et al., 2012a,b; Lieber and DeCarli, 1994; Macey et al., 1996; Wilson et al., 1986).
At the withdrawal time, the alcohol diet was replaced with either water or control diet. Matched chronically exposed rats were given free access to alcohol until sacrifice. Previous studies and our experience show that following removal of the alcohol diet, rats exposed for more than 35 days experience symptomatic withdrawal within hours, with resolution over a 2- to 3-day period (Geisler et al., 1978; Macey et al., 1996; Walker et al., 1975). Similarly, liquid EtOH diet exposures longer than 10 days have been reported to induce physiologic dependence evidenced by autonomic and somatic dysfunction, susceptibility to audiogenic convulsions, and other behavioral signs within 4 hours of cessation (Hunter et al., 1975) that resolve over the first 48 to 72 hours (Geisler et al., 1978; Macey et al., 1996). Alcohol clearance rates following diet cessation are approximately linear and at 7 hours are <25% of original levels (Wilson et al., 1986). To examine gene expression changes during this period, we took CeA samples during chronic exposure and 4, 8, 18, 32, and 48 hours into withdrawal.
CeA Microdissection
Animals were killed by decapitation, and excised brains were placed into ice-cold artificial cerebral spinal fluid and secured for sectioning in agarose (4% UltraPureTM low melting point agarose [Invitrogen, Carlsbad, CA] in artificial cerebral spinal fluid; see Freeman et al., 2012a,b). A McIlwain Tissue Chopper (Gamshall, UK) was used to cut 625 lm coronal sections for CeA microdissection with size-matched micropunches ( ≤ 1.25 mm; Stoelting, Wood Dale, IL). Bilateral CeA samples from[notdef]a single animal were treated as 1 sample.
Gene Set Selection and System of Interest Identification
In this focused qRT-PCR study, gene selection was critical. We initially defined a system of interest based on previous CeA alcohol studies including GABAA (Bajo et al., 2008; Nie et al., 2004), NMDA (McCool et al., 2010; Obara et al., 2009; Roberto et al., 2004, 2006), GPCR subunits including Grm5, Crhr1, Oprm1, Cckbr, Gnb4, and genes related to GPCR function like Ace, Ace2, Agtrap, and Ren (Cruz et al., 2012; Kitanaka et al., 2008). This list was expanded to include associated regulatory elements including Rgs (Ho et al., 2010; Liu et al., 2006) and receptor trafficking proteins (Obara et al., 2009), downstream signaling components (Bajo et al., 2008; Sanna et al., 2002), TFs (Pandey, 2004; Radwanska et al., 2008; Vilpoux et al., 2009), and inducible targets (McBride et al., 2010) as shown in a schematic in Fig. 2 and in detail in Fig. 4A. The result is a focused and highly relevant gene set that can be examined over time during withdrawal.
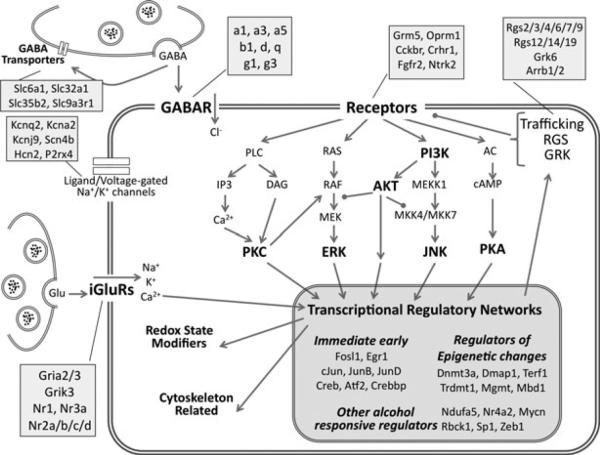
Fig. 2.
Neurobiological systems of interest assayed by qPCR. The schematic shows the biological systems targeted for study including excitatory and inhibitory neurotransmission, receptor-driven intracellular signaling, and transcriptional regulation. Major functional groups and signaling pathways are highlighted in bold. Compartments represented include the presynaptic terminal and membrane, and the postsynaptic membrane, cytoplasm, and nucleus. Receptor subunits, isoforms, and family members assigned to the functional categories are listed in the attached shaded boxes. This schematic's relative positioning is maintained for data mapping in Fig. 4.
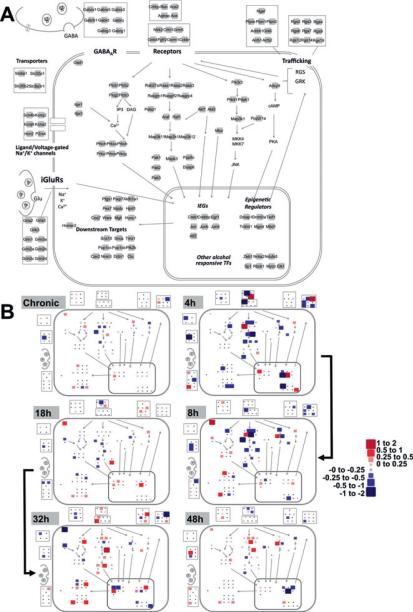
Fig. 4.
Cellular and signaling pathway view of gene expression changes during alcohol withdrawal. (A) Network layout of processes and molecules of interest. The 139 genes measured are represented as light gray squares and are grouped according to functional class. Relative positioning is maintained as in Fig. 2. (B) –ΔΔCT values were superimposed on the map presented in A with the color and size scale presented for all 65 genes found to have a statistically significant treatment effect by ANOVA regardless of whether the post hoc testing revealed significant changes at any individual time point. Small black points are included for positioning and represent the genes that did not show significant changes (NS). All –ΔΔCT values were calculated relative to the mean expression in sacrifice-time matched control animals. Values of 1 and –1 represent doubling and a halving of mRNA levels, respectively. The heavy black arrow indicates the progression of alcohol withdrawal from the chronic state through 48 hours of withdrawal at each of the 5 measured time points. Size changes are proportional to the absolute value of the –ΔΔCT changes. Positive values (red) indicate an increase in expression as compared to control animals, whereas negative values (blue) indicate a decrease.
RNA Handling and qRT-PCR
RNA was extracted with either the RNeasy or the AllPrep DNA/ RNA extraction kit (Qiagen, Valencia, CA) and then DNAase treated (DNA-Free RNA kit; Zymo Research, Orange, CA). Concentration and integrity were assessed with an ND-1000 (NanoDrop, Wilmington, DE) and RNA nano-6000 chips on an Agilent 2100 Bioanalyzer (Santa Clara, CA). Samples had approximately 800 ng of total RNA, a mean 260/280 ratio of 2.07 ± 0.2 (SEM) and were stored at −80°C until cDNA reverse transcription with SuperScript II (Invitrogen) per the manufacturer's protocol from 100 ng total RNA, which was determined to be an amount not requiring total RNA amplification yet sufficient for downstream analysis in this study. cDNA was stored at −20°C.
Roche's Universal Probe Library Assay Design Center (www.universalprobelibrary.com) was used to design intron-spanning PCR primers and probes. Expression assays target a single exon–exon junction and were designed without respect to isoform prevalence. One hundred thirty-nine assays passed quality control (Table S1). cDNA samples were selectively preamplified for 14 cycles with TaqMan® PreAmp Master Mix (Applied Biosystems, Foster City, CA) and a mix of all the selected primers, per the Bio-Mark™ manufacturer's protocol (Fluidigm, South San Francisco, CA). qPCR was performed on 96.96 BioMark™ Dynamic Arrays (Spurgeon et al., 2008) for 40 cycles (15 seconds at 95°C, 5 seconds at 70°C, 60 seconds at 60°C). Software-designated failed reactions were discarded. Threshold cycle (CT) values were calculated by the Real-Time PCR Analysis Software (Fluidigm, Table S2).
Data Normalization and Analysis
Data were quantile-normalized (Bolstad et al., 2003; Pradervand et al., 2009) (ΔCT) in R (http://www.r-project.org) to account for technical variability and batch effects in a manner analogous to the housekeeping gene-based normalization. This data-driven normalization approach is relatively resistant to the potential biases introduced by housekeeping gene normalization. Practically, quantile normalization has been shown to be superior to single housekeeping gene and rank-invariant normalization for reducing technical variation in large qRT-PCR studies (Mar et al., 2009). Following quantile normalization, diurnal effect-corrected –ΔΔCT values were determined for each gene on a per sample basis by subtracting the average value for a given treatment from the average ΔCT value of all controls killed at the same time of day, as previously reported (Freeman et al., 2012a,b). Prior to diurnal normalization, 21 (14%) of the genes showed significant differences between sacrifice times among controls. After sacrifice-specific normalization, none of the genes showed significant diurnal variation (ANOVA, p > 0.05). The –ΔΔCT values were used for further analysis.
Genes with significant treatment effects were identified via ANOVA with 7 possible levels: control, chronic, and the 5 withdrawal durations. Post hoc Tukey's HSD tests were used to identify differences between time points. Multiple testing corrections were performed using the q-value approach in R (Storey and Tibshirani, 2003). With false discovery rate thresholds of 10 to 20%, 70 to 80% of the genes show a statistically significant treatment effect (Table S3), a direct result of the very low π0 value (π0 = 0.25). This statistical outcome was expected from our experimental design where a high proportion (~50%) of genes change, which differs fundamentally from microarrays where ~5% of tens of thousands of genes change significantly. Further, in this enriched and limited 139-gene set, false positives are less likely (5% × 139 = 7, vs. 5% × tens of thousands of genes). The result is that an ANOVA p-value cutoff of 0.05 is more stringent and therefore was used for downstream analysis. All statistical tests were conducted at a 95% confidence level (p 0.05).
Time series clustering was performed to identify statistically significant expression patterns and gene groups among differentially expressed genes using short time series expression miner (STEM) (Ernst and Bar-Joseph, 2006). User-defined parameters were set to 35 profiles, maximizing the number of clustering genes, with a minimum correlation threshold of 0.7. Four statistically significant clusters were identified (p < 0.05). Functional enrichment analysis of these clusters was performed using the DAVID Bioinformatics Resource v6.7 against the user-defined 139-gene background (Huang da et al., 2007). DAVID is a freely available annotation tool that identifies statistically enriched functional terms from public databases including gene ontology (GO) and KEGG Pathways. Significance of enrichment was evaluated with Fisher's exact tests (p ≤ 0.05).
Shared TF binding sites among clustered genes were identified within the 1,000 bp upstream of the transcription start sites using the Promoter Analysis and Interaction Network Toolset (PAINT) (Vadigepalli et al., 2003, Table S4). This regulatory network analysis identifies TF binding sites that are statistically more common in the promoters of the gene subset than expected relative to their appearance in promoters randomly sampled from a reference (Fisher's exact test, p ≤ 0.05). Frequencies are compared to account for differences in binding site definitions, ensuring that issues of length, complexity, or degeneracy do not influence results. Each cluster was analyzed with PAINT twice, comparing clusters to both the rat gen ome and the 139-gene background. This enables a comprehensive view of the shared binding sites while identifying enrichment attributable to the initial 139-gene selection. Promoter sequences were retrieved using the TRANSFAC Professional 2009.4 algorithm (http://www.gene-regulation.com/pub/databases.html) and Entrez Gene IDs. The default settings in PAINT were used for binding site identification. Binding site enrichment suggests a role for the corresponding TF in the observed dynamics and can be interpreted as evidence for coregulation. Networks were visualized in Cytoscape (Cline et al., 2007).
RESULTS
Overall Transcription Dynamics
Using qRT-PCR to study CeA gene expression changes, we identified a marked contrast between withdrawn and chronically exposed animals. Overall, 65 of the 139 tested genes showed a significant treatment effect as identified by ANOVA. Of these, post hoc analysis identified a set of 61 that differed significantly between time points in at least 1 pairwise comparison (Tukey's HSD comparisons, p ≤ 0.05, Fig. 3, Table S3). Only 3 genes, Grin2c, Pebp1, and[notdef] Rgs6, were found to vary significantly between control and chronic exposure samples in post hoc testing. Additionally, 24 genes were found to have unique withdrawal expression states, where measures of at least 1 withdrawal time point differ statistically from both the control and chronically alcohol-exposed samples. Figure 4 shows the mean change in expression level at each time point of the 65 genes experiencing a statistically significant treatment effect. Figure 4B illustrates that the majority of the expression differences are seen during the withdrawal period, rather than following chronic exposure.
Fig. 3.
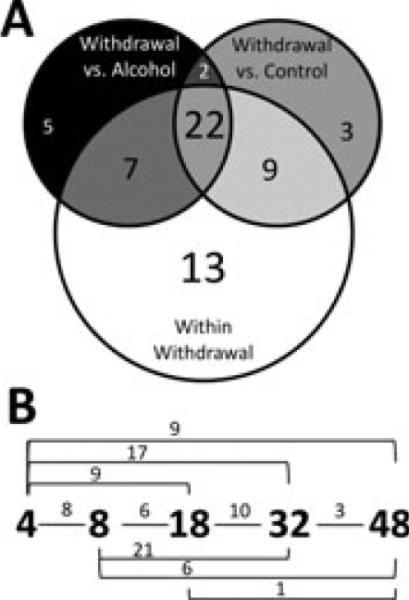
Statistical testing results. Of the 65 genes with a significant ANOVA alcohol treatment effect, post hoc analysis (Tukey's HSD comparisons, p ≤ 0.05) revealed that 61 genes showed statistically significant expression changes in 1 withdrawal time point when compared to at least 1 other treatment group (control, chronic ethanol, 4, 8, 18, 32, or 48 hours withdrawal). The Venn diagram in (A) shows the subsets of significant changes by Tukey's HSD comparison. The majority of these changes occurred during withdrawal (white circle) and did not always coincide with deviations from the control (gray circle) or alcohol-exposed (black circle) states. The numbers within each segment indicate the number of genes with statistically significant changes in gene expression under the indicated pairwise comparison. Genes with a change in any withdrawal time point comparison difference are included in the white group. (B) Significant gene expression differences between 2 time points during withdrawal by pairwise comparisons of the 61 genes with significant gene expression changes (white circle in panel A). The small numbers above the lines connecting time points represent the number of genes with significant differences in that comparison. These comparisons are not mutually exclusive; a single gene may be counted in multiple comparisons.
Expression Changes in Chronic Alcohol Exposure
Our initial hypothesis was that chronic alcohol exposure would result in adaptations in neurotransmitter and signaling systems activated by acute alcohol to counteract these effects, and that these changes would be reflected in CeA gene expression as is generally suggested (Koob, in press). This appeared to be the case for PKA/Creb signaling, a pathway known to be activated by acute alcohol exposure (Pandey, 2004; Pandey et al., 2005), which showed a mild increase in Creb mRNA levels, with balancing decreases in Adcy9 and Crebbp expression in chronic exposure, though only at nonsignificant levels (|fold change| = <0.1, p > 0.05). However, other measured transcripts did not behave according to this supposition. For ionotropic glutamate receptors, we observed mild decreases in the expression of NMDA subunits Grin2c and Grin3a and AMPA subunits Gria2 and Gria3, despite their known inhibition by alcohol (maximum fold change: Grin3a = –0.17). Similarly, Gad1, Gabra5, Gabrd, and Gabrg3 mRNA levels were mildly increased despite alcohol's activity as a central nervous system (CNS) depressant (maximum fold change: Gad1 = 0.35). Again, pairwise post hoc testing indicates that for the vast majority of genes, excluding Grin2c, Rgs6, and Pebp1, these changes were relatively minor and represent mean trends rather than significant differences between control and chronic exposure. However, these trends were opposite our initial expectations and suggest that complex regulatory mechanisms are involved in determining the dependent state, likely including other levels of transcription or translational regulation, not detectable by single measures during chronic exposure.
Withdrawal Expression Changes
In contrast, during withdrawal, more pervasive expression changes were seen (Fig. 4B), particularly early at 4 and 8 hours. These early expression changes included coordinated down-regulation in Ras/Raf signaling pathway components, with concomitant up-regulation of Dusp6, an inactivating phosphatase. We also observed down-regulation in transcripts associated with JNK and IP3/DAG signaling. Downstream of these pathways, transcriptional regulators Creb, Crebbp, Jun, and Elk1 underwent up-regulation. Concordantly, Zeb1 and Rbck1, transcriptional repressors, were strongly down-regulated in the early period. Beginning 4 hours into withdrawal, changes in mRNA levels of methylation state regulators were also noted, including down-regulation of Dmap1, Trdmt1, and Mbd1 and up-regulation of Mgmt expression, suggesting a potential role for epigenetic regulation in the CeA during alcohol withdrawal. There were also expression changes in transcripts encoding many membrane-bound proteins, although these were subunit-specific and not uniformly up or down within a functional group. Notable early down-regulation included the GABA transporter Slc6a1, GABAA receptor subunit Gabrd, and the GPCR modulators Rgs3, Rgs6, Rgs12, and Rgs19. Other early up-regulated transcripts included Ace, encoding angiotensin-converting enzyme, as well as Rgs4, Rasgrp1, and Grin2c.
Changes at 32 and 48 hours were distinct from early measures, most notably in the Ras/Raf signaling pathway whose associated kinases were largely up-regulated at these late points. Accordantly, the 18 hour measures appeared largely transitional between early and late expression. Overall, only 5 genes, Plcb1, Pebp1, Jun, Rbck1, and Rgs12, had average –ΔΔCT values that could be classified as consistently up- or down-regulated throughout the withdrawal period. However, while these genes showed statistically significant changes for overall treatment effect, post hoc testing revealed significant changes at only 1 or 2 time points relative to time-matched controls. All other measured genes had average –ΔΔCT values reflecting both up- and down-regulation over the 48 hours withdrawal period, which highlights the time and context-dependent nature of expression measures.
Correlated Expression and Regulatory Analysis
Clustering with the STEM algorithm identified transcripts with correlated expression over time (Ernst and Bar-Joseph, 2006). Four statistically significant patterns were identified and their temporal expression patterns are shown in Figs 5A, 6A, 7A, and 8A. Clusters 1 and 2, depicted in Figs 5 and 6, respectively, are both initially down-regulated with later induction, but differ in the magnitude of the initial decrease and the timing of peak induction. Cluster 3 (Fig. 7A) shows a more even increase throughout the withdrawal period, with only slight down-regulation at 4 hours. The fourth, as in Fig. 8A, shows oscillatory expression, varying appreciably over time, potentially reflecting transcriptional bursting (Golding et al., 2005; Raj and van Oudenaarden, 2008).
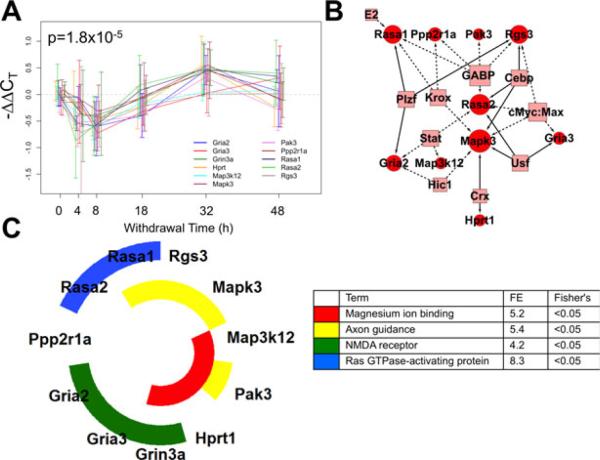
Fig. 5.
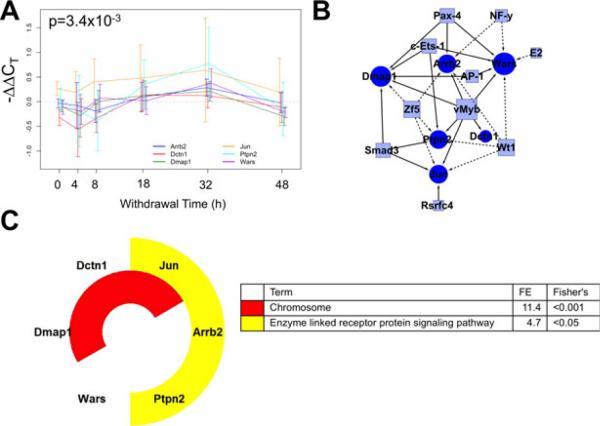
Correlated expression Cluster 1 discovered using the short time series expression miner (STEM) algorithm for time series profile matching. (A) The expression profile for this cluster including Rasa1, Rasa2, Rgs3, Mapk3, Map3k12, Pak3, Hprt1, Grin3a, Gria3, Gria2, and Ppp2r1a (p = 1.8 × 10–5). Values are the mean –ΔΔCT values as compared to sacrifice-time matched control animals. Error bars represent 95% confidence intervals around the mean. Individual series are offset to ease visualization, but all measurements were taken at the time indicated. (B) A schematic network of the proposed regulatory network illustrating transcription factor (TF) binding sites (boxes) shared among cluster members' promoters from the 1,000 bp upstream of the transcription start site, as annotated by PAINT. Circles represent cluster member genes, and connections between TFs and genes indicate a predicted regulatory relationship. Solid edges indicate that the binding site is overrepresented in the cluster in comparison with the 139-gene set, dashed lines indicate overrepresentation only in reference to the rat genome. (C) Nested pie chart of cluster members showing significantly enriched annotation terms from the DAVID Bioinformatics Resource. The 139-gene set was used as reference. Each ring is a single annotation term, indicated by the color noted in the legend along with fold enrichment score (FE) and the associated Fisher's exact test p-value. All genes in the cluster with that gene ontology annotation are colored in the nested pie chart.
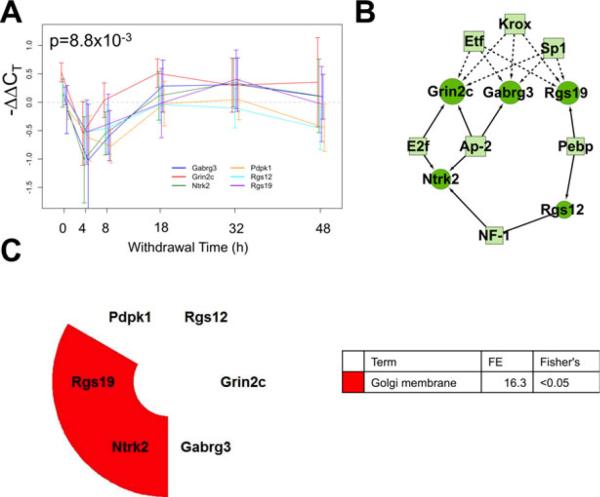
Fig. 6.
Correlated expression Cluster 2 discovered using the short time series expression miner (STEM) algorithm for time series profile matching. (A) The expression profile for this cluster including Pdpk1, Rgs12, Rgs19, Grin2c, Gabrg3, and Ntrk3 (p = 8.8 × 10–3). (B) A schematic of the proposed regulatory network. Circles represent cluster member genes and boxes are transcription factors (TFs). Connections between TFs and genes indicate a predicted regulatory relationship. Solid edges indicate that the binding site is overrepresented in the cluster in comparison with the 139-gene set, dashed lines indicate overrepresentation only in reference to the rat genome. (C) Results of the functional annotation analysis. FE, fold enrichment score.
Fig. 7.
Correlated expression Cluster 3 discovered using the short time series expression miner (STEM) algorithm for time series profile matching. (A) The expression profile for this cluster including Wars, Dmap1, Dctn1, Jun, Arrb2, and Ptpn2 (p = 3.4 × 10–3). (B) A schematic of the proposed regulatory network. Circles represent cluster member genes and boxes are transcription factors (TFs). Connections between TFs and genes indicate a predicted regulatory relationship. Solid edges indicate that the binding site is overrepresented in the cluster in comparison with the 139-gene set, dashed lines indicate overrepresentation only in reference to the rat genome. (C) Results of the functional annotation analysis. FE, fold enrichment score.
Fig. 8.
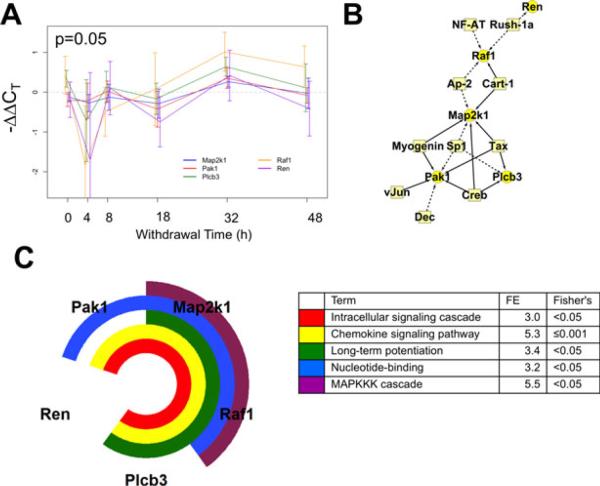
Correlated expression Cluster 4 discovered using the short time series expression miner (STEM) algorithm for time series profile matching. (A) The expression profile for this cluster including Pak1, Map2k1, Raf1, Plcb3, and Ren (p = 0.05). (B) A schematic of the proposed regulatory network. Circles represent cluster member genes and boxes are transcription factors (TFs). Connections between TFs and genes indicate a predicted regulatory relationship. Solid edges indicate that the binding site is overrepresented in the cluster in comparison with the 139-gene set, dashed lines indicate overrepresentation only in reference to the rat genome. (C) Results of the functional annotation analysis. FE, fold enrichment score.
Cluster 1—Initial Decrease Followed by Up-Regulation
As shown in Fig. 5A, the largest cluster has 11 members, including transcripts whose products function in the Ras/ Raf signaling pathway and as glutamate receptor subunits. Initially, the cluster showed a decrease in expression 4 and 8 hours into withdrawal. Then, at 18 hours, mRNA transcript levels began increasing, reaching a peak at 32 hours before nearing baseline levels at 48 hours. In addition, Cluster 1 transcripts share regulatory TF binding sites suggesting common regulatory relationships as shown in Fig. 5B. PAINT promoter analysis identified several binding sites more commonly found in the 1,000 bp promoter region of cluster members than would be expected based on their fre- quency in the rat genome including Gabp, cMyc:Max complex, and Krox. TFs with binding sites shown as solid arrows in Fig. 5B were overrepresented in comparison with the 139-gene set, including Cebp, Plzf, and Usf, offering additional statistical support of the regulatory relationship. Functional analysis (Fig. 5C) identified several statistically enriched annotation terms including NMDA receptor subunits (Gria2, Gria3, and Grin3a), Ras GTPase-activating (Rasa1 and Rasa2), Mg2+ ion binding (Hprt1, Pak3, and Map3k12), and axon guidance (Rasa1, Rgs3, Mapk3, and Pak3) in comparison with the 139-gene background.
Cluster 2—Initial Down-Regulation Followed by Return to Baseline
The second cluster has 6 transcripts including 2 Rgs family proteins, Rgs12 and 19, and GABAA and NMDA receptor subunits Gabrg3 and Grin2c. Depicted in Fig. 6A, this group underwent a rapid decrease in expression at 4 hours reaching a 30 to 50% reduction from control levels. mRNA levels began increasing at 8 hours and approached control levels without later increases in expression. Half the cluster's transcripts share binding sites for Sp1, Krox, and Etf family TFs, more commonly than would be expected based on the rat genome (Fig. 6B). Additionally, there is statistically significant enrichment for E2f, Ap-2, Pebp, and NF-1 binding sites in comparison with the 139 tested genes. Functionally significant enrichment for Golgi membrane localization was found, though based on annotations for only Rgs19 and Ntrk2, as shown in Fig. 6C.
Cluster 3—Up-Regulation With Dynamics
Cluster 3 has 6 members that undergo induction later in withdrawal, peaking around 32 hours before returning to baseline, as shown in Fig. 7A. While they also show a mild decrease in expression at 4 hours, this average decrease is smaller than Clusters 1 and 2. Wt1 and Zf5 binding sites were found in 4 of the 6 transcripts, more commonly than their frequency in the rat genome. vMyb, Smad3, c-Ets-1, Pax-4, and Ap-1 binding sites were overrepresented in the cluster in comparison with the tested background. Functional annotation analysis identified significant enrichment for chromosome and enzyme-linked receptor protein signaling-associated gene transcripts.
Cluster 4—Oscillatory Dynamics of Expression Changes
The final cluster is shown in Fig. 8. Five transcripts shared this oscillatory pattern, undergoing large changes in expression between measurements throughout the 48 hours withdrawal. Again, the most extreme changes were seen at 4 hours, where Raf1 and Ren underwent a 4-fold decrease from control expression levels. TF binding sites for Sp1 were shared by 3 members, more commonly than expected in the genome. Further Creb and related Tax binding sites were identified, occurring more frequently than expected in the 139-gene set. This gene group had multiple functional annotations: intracellular signaling cascades, chemokine signaling, the MAPKKK cascade, nucleotide binding, and long-term potentiation.
DISCUSSION
This study of gene expression dynamics during alcohol withdrawal in the CeA characterizes temporally coregulated changes during the first 48 hours following chronic exposure. Despite the limited expression changes during chronic exposure, CeA samples taken during withdrawal showed coordinated and widespread changes that involved transcripts associated with Rgs and Ras/Raf signaling, GABAA and glutamate receptor subunits, and associated downstream TFs. The largest magnitude changes were early, and both 4 and 8 hours were characterized by down-regulation in most measured transcripts. Time series clustering identified 4 significant patterns involving genes groups that share functional annotation for NMDA receptors, Ras GTPase-activating proteins, MAPKKK cascade, chemokine signaling, and enzyme-linked receptor protein signaling pathways, among others. Comparison of cluster promoters suggested roles for Cebp, Plzf, Usf, E2f, Ap-2, Ap-1 Pebp, NF-1, vMyb, Smad3, c-Ets-1, Pax-4, and Creb/Tax TFs in regulating the observed expression dynamics.
The use of a high sample capacity qRT-PCR platform to study gene expression changes during periods expected to require neuroadaptation because of an abrupt disruption in homeostasis is relatively novel. Similar work from this laboratory examining alcohol withdrawal in the DVC demonstrates that the approach is effective in quantifying dynamic expression changes as they unfold (Freeman et al., 2012a, b). However, the approach is not without limitations; as a focused gene expression study, gene set selection inevitably misses additional dynamic transcripts. Also, as both the DVC and CeA results suggest, levels of many transcripts early in alcohol withdrawal vary considerably among animals. In alcohol-treated animals, this may arise in part from combinations of variable patterns of intake and an unavoidable imprecise initiation of withdrawal in the ad libitum model. However, to a more limited degree, this variability is also observed in control animals killed at a single circadian time point, potentially representing true biologic heterogeneity. Consequently, a single isolated measure of an mRNA level may lead to false negatives. Therefore, it is remarkable that nearly half of the measured transcripts have significant expression changes despite this variability, suggesting a dramatic and consistent disruption in emotional and physiologic homeostasis. Further, while measurement of mRNA levels over time speaks to the biology of the system and its underlying regulatory networks, it also introduces potentially confounding diurnal variation. Although this study was designed to correct for diurnal gene expression differences by including sacrifice-time matched controls, it cannot account for unique withdrawal –diurnal expression states. Such compound interactions are likely and have been the focus of previously published withdrawal studies (Logan et al., 2012) and are the focus of additional work in our laboratory (Staehle et al., in preparation).
Alcohol-induced gene expression in the CeA has been the focus of previous studies. A limited number of changes were reported in a microarray study of CeA samples acquired from inbred alcohol-preferring (iP) rats 24 hours after the last operant conditioning session in a 10-week repeated exposure experiment; the study identified changes in only 23 genes, all of which were nonsignificant after false discovery rate correction (Rodd et al., 2008). This differs from our findings using outbred Sprague-Dawley rats, which show the CeA undergoing widespread expression changes within 4 hours of cessation of prolonged Lieber–DeCarli diet exposure. While this may reflect differences between strains, it may also suggest that expression is highly contextual, dependent upon alcohol exposure method and measurement timing. This context dependency is supported by an 8-week binge exposure study of iP rats where expression changes over the 24 hours following the last binge were identified in 402 genes associated with cellular functions similar to those identified here, including: GABA signaling, enzyme-linked receptor protein signaling, and regulation of protein kinase and MAPKKK cascade (McBride et al., 2010). The ad libitum rats in this study and the binge-exposed rats (McBride et al., 2010) consume larger cumulative alcohol doses than the rats in the operant exposure study (Rodd et al., 2008), which may also contribute to context dependency and the extent of gene expression changes.
The 4 clusters showing correlated expression throughout the 48-hour withdrawal encode mRNAs functionally associated with Ras GTPase activation and NMDA receptors (Cluster 1, Fig. 5), the Golgi membrane (Cluster 2, Fig. 6), chromosome structure and enzyme-linked receptor signaling pathways (Cluster 3, Fig. 7), and MAPKKK and chemokine signaling (Cluster 4, Fig. 8), among others. This suggests that changes in these cellular structures and functions occur as a result of alcohol exposure and withdrawal. Additionally, while unique in their specific temporal expression patterns, the clusters share a pattern of early down-regulation. This may seem counterintuitive as withdrawal is characterized by profound neural excitation; however, we argue that this is in direct concordance with published models of cellular adaptation during dependence (Koob, in press). Upon abrupt withdrawal from the adapted state, there are profound excitatory changes requiring a broad readjustment of cellular machinery. These changes are best understood in light of the preceding transition into chronic alcohol dependence and adaptation to the 35 days alcohol exposure. We believe that this prolonged exposure leads to broad cellular and molecular compensation to accommodate alcohol's effect as a CNS depressant, but that a single mRNA measure during dependence is unable to detect these because the CeA has reached a new steady state, no longer reliant on gene expression. However, secondary evidence of these adaptations can be observed in the transient mRNA expression changes following binge exposure (McBride et al., 2010) and in this study of withdrawal. Our expression data support this conceptualization, as only 3 of the 139 genes were found to be differentially expressed in chronically exposed compared to animals. In contrast, withdrawal was characterized by widespread down-regulation in signaling systems. We propose that the observed down-regulation occurs as a corrective response to the profound excitatory activation suggested by the concomitant increases in TFs Creb, Crebbp, Jun, and Elk1.
This interpretation agrees well with the expression response seen in the DVC (Freeman et al., 2012a,b), an interconnected autonomic brainstem nucleus that modulates vagal tone and that also experiences broad down-regulation in these signaling pathways early in withdrawal. Similar to these results, chronic exposure in the DVC resulted in few, small magnitude changes that became prominent after alcohol cessation. At the systemic level, the CeA and DVC display similar dynamics, with prominent down-regulation at 4 and 8 hours that transitions to up-regulation later, around 32 hours. This is particularly evident in the Ras/Raf signaling pathway. However, individual transcript changes appear to be region-specific. For example, DVC expression changes in the GABAA and NMDA receptor families involve more individual transcripts and are larger in magnitude than in the CeA. This was equally true for immediate early genes, which are generally thought of as ubiquitous and universally responsive; in the DVC, Creb1, Fosl1, Jund, and Egr1, all showed measurable up-regulation at 4 hours, while in the CeA only Creb1 showed a similar increase. These regional expression patterns may reflect functional differences between brain regions, as well as the timing of withdrawal's impact on the region's function. As the DVC is involved in vagal modulation of vital functions like circulation and respiration, its transcription response might reasonably be expected to occur over a relatively immediate timescale. In contrast, the CeA functions at a more integrative level combining information about autonomic and emotional state and may not require such immediate, large magnitude changes.
CeA-specific promoter sequence analysis identified binding sites for TFs that potentially contribute to the observed expression patterns. Of note, Cebp (Cluster 1) (Chen et al., 2009), Usf-1 (Cluster 1) (Potter et al., 2003), and Smad3 (Cluster 3) (Karaa et al., 2008) have been implicated in mediating alcoholic liver disease. Additionally, Usf-1 and Ap-2 (Clusters 2 and 4, respectively) are known to regulate the expression of monoaminergic neurotransmitter-associated genes including tyrosine hydroxylase and dopamine beta hydroxylase (Hong et al., 2008; Kim et al., 2001a,b; Schmidt et al., 2011). These regulatory relationships are particularly relevant in the CeA, which is richly innervated with adrenergic neurons. Plzf (Cluster 1) (Eidson et al., 2011; Spicuglia et al., 2011), Pebp (Cluster 2, polyoma enhancer-binding protein also known as CBF) (Giese et al., 1995; Puig-Kroger et al., 2000), Ap-1 (Cluster 3) (D’Aversa et al., 2008; Yang et al., 2012), E2f (Cluster 2) (Chen et al., 2011), vMyb (Cluster 3) (Jeon et al., 2006), and c-Ets-1 (Cluster 3) (Prosser et al., 1992) are known to regulate cellular oxidative stress and inflammation responses. These processes are implicated (Schmidt et al., 2011) by our CeA withdrawal gene expression data by the functional associations of MAPKKK and chemokine signaling in cluster 4 (Fig. 8) and additional CeA early withdrawal studies in this laboratory showing an up-regulation in TNF-α mRNA and other markers of innate immunity (Freeman et al., 2012a). A subset of the TFs implicated in the promoter analysis were measured experimentally, including Egr1 (the Krox family), Jun, Creb1, its related binding protein Crebbp, and Sp1. Of these, Jun, Creb1, and Crebbp show statistically significant expression changes over the experimental period. For Egr1 and Sp1, the measured changes were small in magnitude and highly variable. Rather than showing simple causal relationships, discernable as patterns in the mRNA expression of the predicted TFs, the data support a more complex interaction of TFs and their targets.
In summary, this quantitative expression study shows widespread changes in the expression of a set of functionally relevant genes in the CeA over the first 48 hours of alcohol withdrawal. Early changes measured in CeA samples taken only 4 hours into the withdrawal period suggest that the CeA responds rapidly to changes in the neurochemical environment as a result of cessation of alcohol intake. The changes included down-regulation in Ras/Raf and other signaling pathways, potentially consistent with a protective response to profound increases in excitatory signaling. Our results also suggest a role for TFs known to play roles in coordinating monoaminergic neurotransmission and cellular stress and inflammation responses, including Cebp, Usf-1, Smad3, Ap-2, and c-Ets. Moving forward, these insights can be examined in other brain regions, additional time points, and other functionally specific gene sets to increase our understanding of how gene expression changes contribute to the pathology of alcohol withdrawal as well as the neurobio-logical basis of emotional regulation during this potentially excitotoxic insult.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the grants from the NIH (R01 AA-015601, R01 GM-076495, and R33 HL-087361 to JSS, GM-083108 to JSS and RV, R33 HL088283 to RV, R24 AA-014986 to JBH, and T32 AA-007463 support of KF and MMS). The high-dimensional qPCR was performed with the generous help of Carmen N. Nichols, PhD, and Fluidigm, South San Francisco, CA. We also wish to thank Zeynep H. , Buğra zer, Monica Payne, and Peter Ucciferro for assistance with experiments and analysis, and the TJU Alcohol Research Center, especially Dr. Biddanda Ponnappa, John Mullen, and Permelia Mullen for their support with the animal model.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- Bajo M, Cruz MT, Siggins GR, Messing R, Roberto M. Protein kinase C epsilon mediation of CRF- and ethanol-induced GABA release in central amygdala. Proc Natl Acad Sci USA. 2008;105:8410–8415. doi: 10.1073/pnas.0802302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:2185–2193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Chen K, Ou XM, Wu JB, Shih JC. Transcription factor E2F-associated phosphoprotein (EAPP), RAM2/CDCA7L/JPO2 (R1), and simian virus 40 promoter factor 1 (Sp1) cooperatively regulate glucocorticoid activation of monoamine oxidase B. Mol Pharmacol. 2011;79:2308–2317. doi: 10.1124/mol.110.067439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Yang CM, Chang SP, Hu ML. C/EBP beta and C/EBP delta expression is elevated in the early phase of ethanol-induced hepatosteatosis in mice. Acta Pharmacol Sin. 2009;30:1138–1143. doi: 10.1038/aps.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp P, Bhave SV, Hoffman PL. How adaptation of the brain to alcohol leads to dependence: a pharmacological perspective. Alcohol Res Health. 2008;31:310–339. [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MT, Herman MA, Kallupi M, Roberto M. Nociceptin/orphanin FQ blockade of corticotropin-releasing factor-induced gamma-aminobutyric acid release in central amygdala is enhanced after chronic ethanol exposure. Biol Psychiatry. 2012;8:666–676. doi: 10.1016/j.biopsych.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aversa TG, Eugenin EA, Berman JW. CD40-CD40 ligand interactions in human microglia induce CXCL8 (interleukin-8) secretion by a mechanism dependent on activation of ERK1/2 and nuclear translocation of nuclear factor-kappaB (NFkappaB) and activator protein-1 (AP-1). J Neurosci Res. 2008;86:630–639. doi: 10.1002/jnr.21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidson M, Wahlstrom J, Beaulieu AM, Zaidi B, Carsons SE, Crow PK, Yuan J, Wolchok JD, Horsthemke B, Wieczorek D, Sant'Angelo DB. Altered development of NKT cells, cd T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS One. 2011;6:e24441. doi: 10.1371/journal.pone.0024441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman K, Brureau A, Vadigepalli R, Staehle MM, Brureau MM, Gonye GE, Hoek JB, Hooper DC, Schwaber JS. Temporal changes in innate immune signals in a rat model of alcohol withdrawal in emotional and cardiorespiratory homeostatic nuclei. J Neuroinflammation. 2012a;9:97. doi: 10.1186/1742-2094-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman K, Staehle MM, mu Vadigepalli R, Gonye GE, Nichols CN, Ogunnaike BA, Hoek JB, Schwaber JS. Rapid temporal changes in the expression of a set of neuromodulatory genes during alcohol withdrawal in the dorsal vagal complex: molecular evidence of homeo-static disturbance. Alcohol Clin Exp Res. 2012b doi: 10.1111/j.1530-0277.2012.01791.x. doi: 10.1111/j.1530-0277.2012. 01791.x [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler RF, Hunter BE, Walker DW. Ethanol dependence in the rat: temporal changes in neuroexcitability following withdrawal. Psychopharmacology. 1978;56:287–292. doi: 10.1007/BF00432851. [DOI] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein–protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Rimondini R, Neznanova O, Sommer WH, Heilig M. Neuroplasticity in brain reward circuitry following a history of ethanol dependence. Eur J Neurosci. 2008;27:1912–1922. doi: 10.1111/j.1460-9568.2008.06159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AM, MacKay RK, Dodd PR, Lewohl JM. Association of polymorphisms in RGS4 and expression of RGS transcripts in the brains of human alcoholics. Brain Res. 2010;1340:1–9. doi: 10.1016/j.brainres.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Lardaro T, Oh MS, Huh Y, Ding Y, Kang UJ, Kirfel J, Buettner R, Kim KS. Regulation of the noradrenaline neurotransmitter phenotype by the transcription factor AP-2beta. J Biol Chem. 2008;283:16860–16867. doi: 10.1074/jbc.M709106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter BE, Riley JN, Walker DW. Ethanol dependence in the rat: a parametric analysis. Pharmacol Biochem Behav. 1975;3:619–629. doi: 10.1016/0091-3057(75)90183-5. [DOI] [PubMed] [Google Scholar]
- Jeon GS, Byun HJ, Park SK, Park SW, Kim DW, Seo JH, Cha CI, Cho SS. Induction of transcription factor A-myb expression in reactive astrocytes following an excitotoxic lesion in the mouse hippocampus. Neurochem Res. 2006;31:1371–1374. doi: 10.1007/s11064-006-9184-x. [DOI] [PubMed] [Google Scholar]
- Karaa A, Thompson KJ, McKillop IH, Clemens MG, Schrum LW. S-adenosyl-L-methionine attenuates oxidative stress and hepatic stellate cell activation in an ethanol-LPS-induced fibrotic rat model. Shock. 2008;30:197–205. doi: 10.1097/shk.0b013e318160f417. [DOI] [PubMed] [Google Scholar]
- Kim CH, Ardayfio P, Kim KS. An E-box motif residing in the exon/ intron 1 junction regulates both transcriptional activation and splicing of the human norepinephrine transporter gene. J Biol Chem. 2001a;276:24797–24805. doi: 10.1074/jbc.M101279200. [DOI] [PubMed] [Google Scholar]
- Kim HS, Hong SJ, LeDoux MS, Kim KS. Regulation of the tyrosine hydroxylase and dopamine beta-hydroxylase genes by the transcription factor AP-2. J Neurochem. 2001b;76:280–294. doi: 10.1046/j.1471-4159.2001.00044.x. [DOI] [PubMed] [Google Scholar]
- Kitanaka N, Kitanaka J, Hall FS, Tatsuta T, Morita Y, Takemura M, Wang XB, Uhl GR. Alterations in the levels of heterotrimeric G protein subunits induced by psychostimulants, opiates, barbiturates, and ethanol: implications for drug dependence, tolerance, and withdrawal. Synapse. 2008;62:689–699. doi: 10.1002/syn.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Theoretical frameworks and mechanistic aspects of alcohol addiction: alcohol addiction as a reward deficit disorder. Curr Top Behav Neurosci. doi: 10.1007/7854_2011_129. in press doi: 10.1007/7854_2011_129 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber CS, DeCarli LM. Animal models of chronic ethanol toxicity. Methods Enzymol. 1994;233:585–594. doi: 10.1016/s0076-6879(94)33061-1. [DOI] [PubMed] [Google Scholar]
- Liu W, Yuen EY, Allen PB, Feng J, Greengard P, Yan Z. Adrenergic modulation of NMDA receptors in prefrontal cortex is differentially regulated by RGS proteins and spinophilin. Proc Natl Acad Sci USA. 2006;103:18338–18343. doi: 10.1073/pnas.0604560103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, McCulley WD, Seggio JA, Rosenwasser AM. Effects of withdrawal from chronic intermittent ethanol vapor on the level and circa-dian periodicity of running-wheel activity in C57BL/6J and C3H/HeJ mice. Alcohol Clin Exp Res. 2012;36:467–476. doi: 10.1111/j.1530-0277.2011.01634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey DJ, Schulteis G, Heinrichs SC, Koob GF. Time-dependent quantifiable withdrawal from ethanol in the rat: Effect of method of dependence induction. Alcohol. 1996;13:163–170. doi: 10.1016/0741-8329(95)02030-6. [DOI] [PubMed] [Google Scholar]
- Mar JC, Kimura Y, Schroder K, Irvine KM, Hayashizaki Y, Suzuki H, Hume D, Quackenbush J. Data-driven normalization strategies for high-throughput quantitative RT-PCR. BMC Bioinformatics. 2009;10:1–10. doi: 10.1186/1471-2105-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ. Central nucleus of the amygdala and the effects of alcohol and alcohol-drinking behavior in rodents. Pharmacol Biochem Behav. 2002;71:509–515. doi: 10.1016/s0091-3057(01)00680-3. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44:171–183. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool BA, Christian DT, Diaz MR, Lack AK. Glutamate plasticity in the drunken amygdala: the making of an anxious synapse. Int Rev Neurobiol. 2010;91:205–233. doi: 10.1016/S0074-7742(10)91007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Schweitzer P, Roberts AJ, Madamba SG, Moore SD, Siggins GR. Ethanol augments GABAergic transmission in the central amygdala via CRF1 receptors. Science. 2004;303:1512–1514. doi: 10.1126/science.1092550. [DOI] [PubMed] [Google Scholar]
- Obara I, Bell R, Goulding SP, Reyes CM, Larson L, Ary A, Truitt W, Szumlinski K. Differential effects of chronic ethanol consumption and withdrawal on homer/glutamate receptor expression in subregions of the accumbens and amygdala of P rats. Alcohol Clin Exp Res. 2009;33:1924–1934. doi: 10.1111/j.1530-0277.2009.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC. The gene transcription factor cyclic AMP-responsive element binding protein: role in positive and negative affective states of alcohol addiction. Pharmacol Ther. 2004;104:47–58. doi: 10.1016/j.pharmthera.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Chartoff EH, Carlezon WA, Jr, Zou J, Zhang H, Kreibich AS, Blendy JA, Crews FT. CREB gene transcription factors: role in molecular mechanisms of alcohol and drug addiction. Alcohol Clin Exp Res. 2005;29:176–184. doi: 10.1097/01.alc.0000153550.31168.1d. [DOI] [PubMed] [Google Scholar]
- Potter JJ, Rennie-Tankersley L, Mezey E. Endotoxin enhances liver alcohol dehydrogenase by action through upstream stimulatory factor but not by nuclear factor-kappa B. J Biol Chem. 2003;278:4353–4357. doi: 10.1074/jbc.M210097200. [DOI] [PubMed] [Google Scholar]
- Pradervand S, Weber J, Thomas J, Bueno M, Wirapati P, Lefort K, Dotto GP, Harshman K. Impact of normalization on miRNA microarray expression profiling. RNA. 2009;15:493–501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser HM, Wotton D, Gegonne A, Ghysdael J, Wang S, Speck NA, Owen MJ. A phorbol ester response element within the human T-cell receptor beta-chain enhancer. Proc Natl Acad Sci USA. 1992;89:9934–9938. doi: 10.1073/pnas.89.20.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Kroger A, Lopez-Rodriguez C, Relloso M, Sanchez-Elsner T, Nueda A, Munoz E, Bernabeu C, Corbi AL. Polyomavirus enhancer-binding protein 2/core binding factor/acute myeloid leukemia factors contribute to the cell type-specific activity of the CD11a integrin gene promoter. J Biol Chem. 2000;275:28507–28512. doi: 10.1074/jbc.M004323200. [DOI] [PubMed] [Google Scholar]
- Radwanska K, Wrobel E, Korkosz A, Rogowski A, Kostowski W, Bienkowski P, Kaczmarek L. Alcohol relapse induced by discrete cues activates components of AP-1 transcription factor and ERK pathway in the rat basolateral and central amygdala. Neuropsychopharmacology. 2008;33:1835–1846. doi: 10.1038/sj.npp.1301567. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto M, Bajo M, Crawford E, Madamba SG, Siggins GR. Chronic ethanol exposure and protracted abstinence alter NMDA receptors in central amygdala. Neuropsychopharmacology. 2006;31:988–996. doi: 10.1038/sj.npp.1300840. [DOI] [PubMed] [Google Scholar]
- Roberto M, Schweitzer P, Madamba SG, Stouffer DG, Parsons L, Siggins GR. Acute and chronic ethanol alter glutamatergic transmission in rat central amygdala: an in vitro and in vivo analysis. J Neurosci. 2004;24:1594–1603. doi: 10.1523/JNEUROSCI.5077-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna PP, Simpson C, Lutjens R, Koob G. ERK regulation in chronic ethanol exposure and withdrawal. Brain Res. 2002;948:186–191. doi: 10.1016/s0006-8993(02)03191-8. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Huber L, Majdazari A, Schutz G, Williams T, Rohrer H. The transcription factors AP-2beta and AP-2alpha are required for survival of sympathetic progenitors and differentiated sympathetic neurons. Dev Biol. 2011;355:89–100. doi: 10.1016/j.ydbio.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Spicuglia S, Vincent-Fabert C, Benoukraf T, Tiberi G, Saurin AJ, Zacarias-Cabeza J, Grimwade D, Mills K, Calmels B, Bertucci F, Sieweke M, Ferrier P, Duprez E. Characterisation of genome-wide PLZF/RARA target genes. PLoS One. 2011;6:e24176. doi: 10.1371/journal.pone.0024176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS. 2003;7:3235–3252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- Vilpoux C, Warnault V, Pierrefiche O, Daoust M, Naassila M. Ethanol-sensitive brain regions in rat and mouse: a cartographic review, using immediate early gene expression. Alcohol Clin Exp Res. 2009;33:945–969. doi: 10.1111/j.1530-0277.2009.00916.x. [DOI] [PubMed] [Google Scholar]
- Walker DW, Hunter BE, Riley J. A behavioral and electrophysiological analysis of ethanol dependence in the rat. Adv Exp Med Biol. 1975;59:353–372. doi: 10.1007/978-1-4757-0632-1_25. [DOI] [PubMed] [Google Scholar]
- Wilson JS, Korsten MA, Lieber CS. The combined effects of protein deficiency and chronic ethanol administration on rat ethanol metabolism. Hepatology. 1986;6:823–829. doi: 10.1002/hep.1840060504. [DOI] [PubMed] [Google Scholar]
- Yang CM, Lin CC, Lee IT, Lin YH, Yang CM, Chen WJ, Jou MJ, Hsiao LD. Japanese encephalitis virus induces matrix metalloproteinase-9 expression via a ROS/c-Src/PDGFR/PI3K/Akt/MAPKs-dependent AP-1 pathway in rat brain astrocytes. J Neuroinflammation. 2012;9:12. doi: 10.1186/1742-2094-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.